Abstract
Due to the genetic variability of the HIV-1 envelope glycoproteins (Env), the elicitation of neutralizing antibodies to conserved neutralization determinants including the primary receptor binding site, CD4 binding site (CD4bs), is a major focus of vaccine development. To gain insight into the evolution of Env-elicited antibody responses, we utilized single B cell analysis to interrogate the memory B cell Ig repertoires from two rhesus macaques following five serial immunizations with Env/adjuvant. We observed that the CD4bs-specific repertoire displayed unique features in the third complementarity determining region (CDR3) of Ig heavy chains with minor alterations along the immunization course. Progressive affinity maturation occurred as evidenced by elevated levels of somatic hypermutation (SHM) in antibody sequences isolated at late immunization time point compared to the early time point. Antibodies with higher SHM were associated with increased binding affinity and virus neutralization capacity. Moreover, a notable portion of the CD4bs-specific repertoire was maintained between early and late immunization time points, suggesting that persistent clonal lineages were induced by Env vaccination. Furthermore, we found that the predominant persistent CD4bs-specific clonal lineages had larger population sizes and higher affinities than that from the rest of the repertoires, underscoring the critical role of antigen affinity selection in antibody maturation and clonal expansion. Genetic and functional analyses revealed that the accumulation of SHM in both framework regions and CDRs contributed to the clonal affinity and antigenicity evolution. Our longitudinal study provides high resolution understanding of the dynamically evolving CD4bs-specific B cell response following Env immunization in primates.
Introduction
A successful HIV-1 vaccine is expected to elicit both T cell-mediated immunity and protective long-lasting antibody responses mediated by B cells (1). However, elicitation of potent and broadly neutralizing antibody (bNAb) response against circulating HIV-1 strains has been a major challenge. So far, all monoclonal antibodies that display broadly neutralizing activity are isolated from chronically HIV-1-infected individuals. These bNAbs bind to conserved epitopes on the HIV-1 envelope glycoprotein (Env), which consists of the exterior gp120 and the transmembrane gp41 envelope proteins. Various conserved epitopes on Env are targeted by bNAbs, such as the membrane proximal external region of gp41 targeted by 2F5, 4E10, and 10E8, glycan-containing epitopes associated with V2 by PG9/PG16 and V3 by PGT121/PGT135/PGT128, the CD4 binding site (CD4bs) by VRC01, b12 and others, and quaternary epitopes overlapping gp120 and gp41 by PGT151, 35O22, and 8ANC195 (reviewed by references (2–4)). bNAbs in general display unique features such as high degree of somatic hypermutations (SHM), long length of the 3rd complementarity determining region (CDR) of immunoglobulin (Ig) heavy chain, and restricted Ig germline usages (2–4). Among these epitopes, the CD4bs is an ideal target for a bNAb-eliciting vaccine as it is highly conserved functional element, engaging Env to bind its primary receptor, CD4.
To efficiently elicit CD4bs-specific bNAb response in a vaccine setting, current effort in the field is focused on designing immunogens that closely mimic the antigenicity of the native Env functional trimer spike (5–7), robustly activate the bNAb germline precursors, especially CD4bs-specific antibody germlines (8–10), using Env variants isolated from natural infection at time points associated with the broadening of serum neutralization activities (11), or Env trimer cocktails to elicit cross-reactive neutralizing antibody responses (12). In parallel with the development of the next generation of vaccine candidates with the improved potential to elicit CD4bs-specific neutralizing antibodies, it is crucial to understand how HIV-1 vaccine-elicited CD4bs-specific antibody responses dynamically evolve throughout immunization.
Several HIV-1 vaccine efficacy trials in humans have been performed, with only the RV144 trial displaying a modest protection efficacy (13). In one study, antibody response targeting HIV-1 gp120 major variable regions, V1/V2, was found to correlate with protection (14). Vaccine efficacy in this trial was not durable, peaking at ~60.5% and then waning to background in three years, thus resulting in an average efficacy of 31.2% (15). This transient protection may be due to the fact that the protective antibody responses elicited by the vaccine are not persistent (16). However, it is not clear if HIV-1 vaccines generally induce only transient B cell responses. Therefore, investigating the persistence of antibody responses is critical and a comprehensive longitudinal analysis of the genetics, functionality, and durability of antibody responses induced by HIV-1 Env immunization should be conducted, as done here.
Non-human primate (NHP) models offer unique opportunities for HIV-1 vaccine studies, due to the similarity in the genetics and physiology between macaques and humans, as well as their susceptibility to infection with simian immunodeficiency viruses and chimeric simian-human immunodeficiency virus. The technologies to isolate antigen/epitope-specific NHP B cells followed by single-cell RT-PCR amplification, recombinant monoclonal antibodies (MAbs) expression and Next-Generation Sequencing (NGS) technology can be successfully applied to analyze Env antibody responses in vivo at high resolution (17–19). However, it is not well known how the Env- or CD4bs-specific memory B cell response is initially elicited and how it evolves during the immunization course as well as how this process leads to the antibody affinity maturation and virus neutralization.
In the present study, we integrated these technologies to characterize and delineate the evolution of CD4bs-specific clonal lineages during serial immunizations of NHPs. We found that compared to the highly diversified Env-specific antibody response, the CD4bs-specific antibody response had lower diversity with unique genetic and functional properties. A notable fraction of the Env- and CD4bs-specific memory B cell lineages was maintained and persisted during the course of the immunization regimen. Primarily driven by antigen affinity selection, these progressively accumulated SHM led to elevated Env-binding affinity and expanded virus neutralization capacity. Investigation of persistent and predominant CD4bs-specific clonal lineages revealed that affinity maturation attributed to SHM occurred in both frameworks and CDR regions, which led to substantial change of the antibody molecule thermostability and partial footprint shifts on the gp120 surface. The high resolution longitudinal analysis of antibody clonal lineages targeting the HIV-1 primary receptor binding site epitope presented here provides insight into the evolution of B cell responses following HIV-1 Env immunization, expanding information important for vaccine development.
Materials and Methods
Samples for B cell repertoire data collection
Rhesus macaques were immunized five times on a monthly interval with recombinant YU2 gp140-F trimer/AbISCO and CpG adjuvant. Peripheral blood was collected 1–3 weeks after each immunization, followed by PBMC preparation as described previously (20). In this study, frozen PBMCs from Rhesus macaques F125 and F128 at 1–3 weeks after the 2nd and 5th immunizations were used.
Cell staining and single-cell flow cytometric sorting
PBMCs were stained as described previously (18). Briefly, frozen PBMCs were thawed and treated with 10,000 U/ml DNase I (Roche) in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) media, followed by Aqua Dead Cell Staining (Life Technologies). A cocktail of antibodies containing CD3 (APC-Cy7; SP34-2, BD Pharmingen), CD8 (Pacific blue; RPA-T8, BD Pharmingen), CD14 (Qdot 605; M5E2, VRC), CD20 (PE-Alexa Fluor 700; 2H7, VRC), CD27 (PE-Cy7; M-T271, BD Pharmingen), IgG (FITC; G18-145, BD Pharmingen), and IgM (PE-Cy5; G20-12, BD Pharmingen) was used to stain the PBMCs. To sort Env and CD4bs-specific B cells, gp140-F conjugated with streptavidin-allophycocyanin conjugate (SA-APC, Life Technologies) and gp140-F-D368R with streptavidin-phycoerythrin conjugate (SA-PE, Life Technologies) were incorporated in the above-stated antibody cocktail. Following staining, the cells were sorted at a single-cell density into 96-well plates with lysis buffer using a four-laser FACS Aria III cell sorter. Env-specific memory B cells were defined as CD3−CD8−Aqua Blue−CD14−CD20+IgG+CD27+IgM−gp140-Fhi, while CD4bs-specific memory B cells were defined as CD3−CD8−Aqua Blue−CD14−CD20+IgG+CD27+IgM−gp140-Fhigp140-F–D368Rlo.
Single-cell RT-PCR
The sorted cells were lysed by the lysis buffer followed by single cell reverse transcription and PCR reactions to amplify Ig sequences as described previously (21, 22). Briefly, in each well containing lysed cell, 150 ng random hexamers, 0.4 mM dNTPs, 100 U SuperScript III (Life Technologies), and 3.5 μl of water were added, followed by incubation at 42°C for 10 min, 25°C for 10 min, 50°C for 60 min, and 94°C for 5 min to generate cDNA. Nested PCR was then performed with 2 μl of cDNA in 25-μl reactions with the HotStar Taq Plus Kit (QIAGEN), using 5′ leader sequence–specific and 3′ IgG-specific primers (22). In the second round of nested PCR, 1.75 μl of the 1st PCR product was used as template. All nested PCRs were incubated at 94 °C for 5 min followed by 50 cycles of 94 °C for 30 s, 50 °C (heavy chain) or 52 °C (kappa or lambda chain) for 45 s, and 70 °C for 1 min with a final elongation at 70 °C for 10 min before cooling to 4 °C. Nested PCR products were evaluated on 2% 96-well E Gel (Life Technologies), and positive wells with a specific band of ∼450 bp were purified and sequenced.
Antibody cloning
A cloning PCR reaction was performed in a total volume of 50 μl with 1 μl of the 2nd nested PCR product as template, using Expand High-Fidelity PCR Kit according to the manufacturer’s instructions (Roche). Briefly, the PCR reaction consisted of 5 μl of 10× reaction buffer, 1 μl of 10 mM dNTPs, 1 μl each of 25 μM of 3′ and 5′ cloning primers, 1 μl of DNA polymerase and water to 50 μl. The cloning primers were described previously (22). The PCR reaction had an initial denaturation at 95°C for 3 min, followed by 20 cycles of 95°C for 30 s, 50°C for 30 s and 68°C for 2 min. There was a final elongation at 68°C for 8 min before cooling to 4°C followed by evaluation on 1% agarose gel. Positive bands [∼400 bp for heavy chain VDJ and ∼350 bp for light chain VJ] were then purified, digested with restriction enzymes, and ligated into eukaryotic expression vectors containing human Igγ1H, Igγ2, or Igκ1 L chain Ab expression cassettes (23, 24). After transformation of ligated products into competent cells, bacterial colonies were sequenced for insertion of correct sequences.
Antibody expression and specificity binding test
The antibody heavy and light chain expression vectors carrying the correct insertions were transfected at an equal ratio into 293F cells, cultured at an initial density of 1 million cells per milliliter, using 293fectin Transfection Reagent (Life Technologies) as described previously (22). Four days after the transfection, the cell culture supernatants were harvested and purified by Protein A Sepharose columns packed in-house (GE Healthcare). The purified antibodies were further tested for specificity by ELISA.
MaxiSorp plates (Nunc, Thermo Scientific) were coated with 0.2 μg of antigen gp140-F or mutant gp140-F-D368R (CD4bs binding knockout) in 100 μl of phosphate buffered saline (PBS) per well, overnight at 4°C. After the plates were washed with PBS/0.2% Tween 20 and blocked with in PBS/2% dry milk/5% FBS for 1h at 37°C, purified antibodies were added into wells in 5-fold dilution series, starting from 10 μg/ml, and incubated for 1h at 37°C. HRP-conjugated goat-human IgG (Jackson ImmunoResearch) was then used at a 1:10,000 dilution in PBS/0.2% Tween 20 and incubated for 1 h at room temperature. The bound antibody was detected with 100 μl/well of TMB substrate (Life Technologies) for 5 min before the reaction was stopped by addition of 100 μl of 3% H2SO4. The optical density (OD) was measured at 450 nm. Between each step of the ELISA, plates were washed four times with PBS supplemented with 0.2% Tween 20.
Single-cell sorted B cell Ig sequence genetic analysis
The single-cell RT-PCR-generated antibody V(D)J gene segments were analyzed by two approaches. For overall heavy chain VDJ family analysis, sequences were initially analyzed by IMGT/High V-Quest (25) (version 1.1.3, reference directory release 201338-1) (http://imgt.cines.fr) to define Ig subdomains. For a more exhaustive annotation of the individual heavy and light chain V(D)J gene segments, antibody sequences were processed through IgBLAST version 1.1.0. (26) using previously published rhesus macaque germline databases (18, 22) as well as IGD and IGJ databases from IMGT to deduce gene segment usage. Somatic hypermutation (SHM) was evaluated at the nucleotide level by alignment of single-cell sorted sequences against the corresponding assigned germline sequences using IgBLAST version 1.1.0 (26). When SHM at the level of amino acid was calculated, nucleotide sequences in rhesus macaque germline databases were translated into peptide sequences to form germline peptide sequence database. The query nucleotide sequences (single-cell sorted sequences) were also subsequently translated and aligned to its corresponding identified sequence in rhesus macaque germline peptide databases by blastx in blast-2.2.26+ package (27) to evaluate SHM. Clonal lineages are defined by the usage of V and J segments, CDR3 length and CDR3 homology (over 90%). CDR3 was defined using IMGT/High V-Quest, as stated below. The Ig molecule encoding sequences generated from single B cell sorting and RT-PCR amplification are available with accession numbers KU750814-KU751671 and KU761300 at GenBank (http://www.ncbi.nlm.nih.gov/genbank/). Sequences with GenBank accession numbers KF947536-KF948098 (Env-specific), KF948099-KF948514 (Total), and JQ885990-JQ886005 (CD4bs) generated from previous studies (17, 18) were also included in the analysis.
CDR3 analysis
CDR3 amino acid sequences, including the conserved 5′ cysteine (C) and 3′ tryptophan (W) residues were obtained by querying antibody nucleotide sequences with IMGT/High V-Quest function using the rhesus macaque and human germline databases. The contribution of each amino acid to the individual CDR3 was then determined. CDR3 hydrophobicity was investigated by calculating the GRAVY score, using the online calculator provided by Dr. Stephan Fuchs (http://www.gravy-calculator.de). The GRAVY score is defined by the sum of hydrophobicity of all amino acids divided by the peptide length, with a higher score indicating a higher hydrophobicity of the peptide.
Bioinformatics analysis of antibody NGS data
Single-cell RT-PCR-amplified sequences, as template sequences were querying against the NGS database obtained from animal F128 Imm 5 [17] to identify somatic variants and to generate identity/divergence plots. The NHP antibody NGS data was processed and analyzed using a bioinformatics pipeline developed based on the framework of a human antibodyomics pipeline (28). Given a data set of NGS-derived NHP antibody sequences, each sequence was (1) reformatted and labeled with a unique index number; (2) assigned to V, D (for heavy chain only), and J gene families using the rhesus macaque germline gene database (18, 22) and an in-house implementation of IgBLAST, with sequences possessing an E-value > 10−3 for V gene assignment removed from the data set; (3) subjected to a template-based, error-correction procedure, in which insertion and deletion (indel) errors of less than three nucleotides in the V gene segment were detected by alignment to their respective germline gene sequences and subsequently corrected; (4) compared to the template antibody sequences at both nucleotide level and amino acid level using a global alignment module in CLUSTALW2 (29), which provides the basis for identity/divergence analysis (e.g., Fig S2); (5) subjected to a multiple sequence alignment (MSA)-based procedure to determine CDR3, which was further compared to the template CDR3 sequences at the nucleotide level, and to determine the sequence boundary of the V(D)J coding region. After full-length variable region sequences were obtained, a bioinformatics filter was applied to detect and remove erroneous sequences that may contain swapped gene segments due to PCR errors. Specifically, a full-length read would be removed if the V-gene alignment was less than 250 bp.
Antibody binding affinity
Biolayer light interferometry (BLI) was performed using an Octet RED96 instrument (ForteBio, Pall Life Sciences). The purified antibodies were captured onto anti-human IgG Fc biosensors and monomeric YU2gp120, purified by affinity chromatograph followed by size-exclusive chromatography, was diluted in 2-fold series starting from 250 nM to 7.8 nM as the analyte in solution. The biosensors were hydrated for 10 min in PBS with 0.2% Tween 20, as the binding buffer. Briefly, the biosensors were first immersed in binding buffer for 60 s to establish a baseline and then in a solution containing 10 μg/ml of NHP or human Env-specific mAbs for 60 s to capture the mAb. The biosensors were then submerged in binding buffer for a wash of 60 s. The biosensors were then immersed in a solution containing various concentrations of gp120 for 120 s for analyte/ligand association, followed by 120 s in binding buffer to assess analyte/ligand dissociation. Binding affinity constants (dissociation constant, KD; on-rate, Kon; off-rate, Koff) were determined using Octet Analysis version 7 software.
HIV-1 neutralization assays
Antibody neutralization assays were performed with the format of single-round of infection using HIV-1 Env-pseudoviruses and TZM-bl target cells, as described previously (30–32). Neutralization curves were fit by nonlinear regression using a 5-parameter hill slope equation as previously described (31). The 50% inhibitory concentrations (IC50) of antibody were reported as the concentration of antibody required to inhibit infection by 50%. The IC50 geometric mean (Geomean) indicating mAb neutralization potency was derived from IC50 values against each individual tier 1 virus for each mAb. When IC50 value is above 50 μg/ml for certain viruses (no neutralization), a value of 50 µg/ml is designated for calculation. The number of viruses neutralized by mAb (IC50 < 50 μg/ml) out of the total number of tested tier 1 viruses was used to calculate the neutralization breadth.
GEAT361 lineage germline precursor inference
Germline V(D)J genes were assigned using previously published custom databases (18, 22) in IgBLAST (PMID 23671333). Assigned V(D)J germline genes were aligned to the query sequence and manually inspected to determine junctional diversity due to the trimming P- and N-nucleotides. In the germline reverted clones, the junctions were retained while the V(D)J genes were reverted to germline nucleotide sequence.
Differential scanning calorimetry (DSC)
DSC experiments were performed using N-DSC II differential scanning calorimeter (Calorimetry Sciences Corp). Antibody samples, purified by protein A column followed by size-exclusive chromatography to remove aggregates and dimers, at concentration of 0.57 mg/ml in PBS were scanned at rate of 1 K/min under 3.0 atm of pressure. DSC data were analyzed using NanoAnalyze software (TA Instruments).
Ala scanning and molecular modeling
A panel of 27 JRCSF gp120 Ala mutants containing alterations known to affect sCD4 or a set of known CD4bs-specific mAb binding (18, 22) was selected. The Ala mutations were generated previously (33) in the context of the full-length JRCSF gp160. The gp120 was released from the pseudoviruses by detergent lysis and captured by the sheep anti-gp120 C5 Ab, D7324 (Aalto Bio Reagents) previously coated on ELISA plates. After washing, antibody binding affinity to gp120 variants was assessed as below. The level of binding by the human mAb 2G12 and HIVIg was used to normalize the variant gp120 expression levels. The effect of a given Ala mutation on Ab binding was represented by apparent affinity (avidity) relative to the binding level to the wild type (WT) gp120, calculated with the formula of {(EC50_WT/EC50_mutant)/[EC50_WT for 2G12 (or HIVIg)/EC50_mutant for 2G12 (or HIVIg)]} × 100, where EC50 is the median effective concentration as previously described (33). Based on alanine mapping results, we highlighted the binding specificities of representative NHP Imm 2 and Imm 5 mAbs by using the gp120 core crystal structure PDB-2NY3 from Protein Data Bank (34).
Statistical analysis
The comparison of V(D)J-family distributions between the Env- and CD4bs-specific Ig repertoires was analyzed with the χ2 test. Comparisons of individual V-gene segment contributions to the Env- and CD4bs-specific Ig repertoires were carried out with two-way ANOVA. Evaluation of three or more groups was done with one-way ANOVA. Unless mentioned otherwise, statistical evaluation between two groups was performed with t test or Mann-Whitney test. The correlation was determined with the nonparametric Spearman correlation test, with a two-tailed p value calculated for significance. Statistical significance was determined as *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. All statistical analysis was performed with GraphPad Prism version 6.
Results
HIV vaccine-elicited Ig repertories from rhesus macaques immunized with trimeric YU2 gp140F
Chinese rhesus macaques, designated as F125 and F128, were immunized monthly with well-characterized first generation of trimeric HIV-1 envelope glycoprotein YU2 gp140-F in adjuvant as previously described (20). Vaccine-elicited Env- and CD4bs-specific memory B cells were sorted from peripheral blood mononuclear cells (PBMCs) 1–3 weeks after the 2nd (Imm 2) and 5th immunizations (Imm 5), respectively, using FACS-based multi-color epitope-specific single B cell sorting technology developed previously (Fig 1A) (17). Env- and CD4bs-specific memory B cells were gated as CD20+IgG+IgM-CD27+gp140-Fhi and CD20+IgG+IgM-CD27+gp140-Fhi/gp140-F–D368Rlo, respectively (Fig 1A). We observed that Env- and CD4bs-specific memory B cells accounted for approximately 5% and 0.2% of total memory B cells, respectively (Fig 1A), consistent with the results of our previous studies (18). After cell lysis, reverse transcription (RT) of the mRNA and nested polymerase chain reaction (PCR) were conducted to recover the variable regions of Ig heavy/light chains (HC/LC). Following sequence verification and phylogenetic analysis, we characterized the genetic composition of the Env- and CD4bs-specific Ig repertories.
Figure 1. Isolation of vaccine-induced NHP antigen-specific single cell and antigen-specific Ig heavy chain gene segment usage.
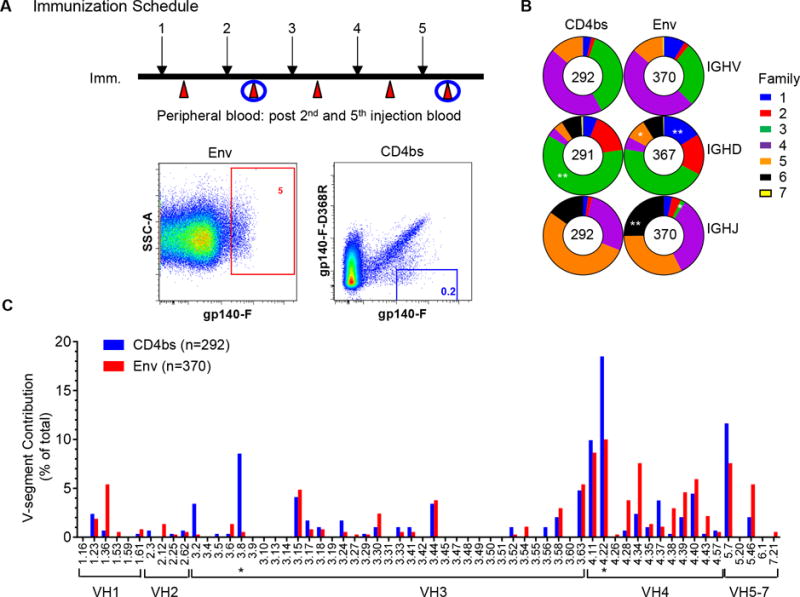
(A) Schematic presentation of the immunization schedule and antigen-specific B cell sorting. Upper panel, immunization/sampling schedule. Whole blood samples from two macaques inoculated with HIV Env YU2gp140-F trimer 5 times in a monthly interval (black arrow) were collected 1–3 weeks after each inoculation (red arrow) to prepare PBMCs (20). PBMCs from time points following the 2nd and 5th immunization (Imm 2 and Imm5) were subjected to single cell sorting, indicated with blue circles. Lower panel, single cell sorting for Env-and CD4bs-specific memory B cells. IgG memory B cells were defined as CD3−/CD8−/ Aqua Blue−/ CD14−/ CD20+/ IgG+/ CD27+/ IgM−. Env- and CD4bs-specific memory B cells were then gated by phenotype of gp140-Fhi and gp140-Fhi/gp140-F–D368Rlo, respectively. Gate frequency (percent) of Env- and CD4bs-specific memory B cells out of total memory B cells is depicted in red and blue, respectively. (B) IGHV-, D-, and J-family Ig-gene usage of sorted Env- and CD4bs-specific memory B cells, defined by IgBLAST and IMGT/High V-Quest. Heavy chain gene families are color coded, with size of the colored area corresponding to the frequency (percent) out of the total number of sequences indicated in the center of the graphs. Differences in the gene family usage between the Env- and CD4bs-specific Ig repertoires were evaluated using χ2 test with *p<0.05, **p<0.01. (C) Env- (red) and CD4bs (blue) -specific Ig repertoire V-gene segment contribution to the total number of sequences of each individual repertoire. VH3.8 and VH4.22, over-expressed in CD4bs-specific Ig repertoire, were marked with asterisk. Two-way ANOVA was used for statistical analysis with *p<0.05.
The sorting specificity was confirmed by cloning selected amplicons into Ig HC/LC expression vectors (22–24) to produce a panel of NHP mAbs for Env-binding and virus neutralization assays. The criteria for selecting amplicons for cloning and expression were that the sequences should represent the genetic diversity of the amplicons by phylogenetic analysis and have matched heavy and light chain amplicons available after PCR amplification. A total of 34 mAb clones, accounting for more than 10% of the total amplified 292 CD4bs-specific heavy chain Ig clones, were selected for expression and characterization. The binding specificity of the sorted CD4bs-specific mAbs was confirmed by binding affinity to trimeric YU2gp140-F and the CD4bs-knockout mutant, YU2gp140-F-D368R via ELISA assays. Among the 34 cloned putative CD4bs-specific mAbs, all 34 (100%) bound gp140-F, and 30 (88%) displayed the expected gp140-Fhi/gp140-F-D368Rlo binding phenotype (data not shown), which is consistent with the sorting precision in previous studies (18). Taken together, the antigen binding specificity of the single-cell sorted Ig repertoires is validated, enabling further genetic analysis of the Env- and CD4bs-specific antibody repertoire along the immunization course.
Genetic characterization of HIV vaccine-elicited Env- and CD4bs-specific Ig repertoires
To investigate the genetic composition of the HIV vaccine-elicited Env- and CD4bs-specific Ig repertoires, the single-cell sorted sequences were analyzed based on the rhesus monkey V(D)J database as described previously (18). Particularly, the germline gene segment usage was defined and the level of SHM, which represents the Ig sequence divergence from their germline ancestor and is critical for antibody affinity maturation, was evaluated. We found that both Env- and CD4bs-specific Ig repertoires had similar heavy chain gene family distributions, where most of the VH genes belong to germline VH3 and 4 families, DH genes to DH3 family, and JH genes to JH4 and 5 families (Fig1B), respectively. However, a significant overrepresentation of the DH3 family was noted in CD4bs-specific Ig repertoire compared with the Env-specific repertoire, while DH1, 5 and JH3, 6 families were significantly overrepresented in Env-specific Ig repertoire than CD4bs-specific repertoire (*p<0.05, **p<0.01, χ2 test) (Fig1B). Upon closer examination of individual V-gene segment use of the heavy chain, VH3.8 and 4.22 were found to be used with significantly greater frequency in the CD4bs-specific repertoire than their usages in the Env-specific repertoire (*p<0.05, two-way ANOVA) (Fig 1C).
In addition, the light (kappa [κ] and lambda [λ]) chains of both Env- and CD4bs-specific Ig repertoires also showed similar family distributions, where most of VK genes belonged to the germline VK1 family, the VL genes to the VL2 family, the JK genes to the JK4 family, and the JL genes to the JL2 family (data not shown). Of note, there was significant overrepresentation of VL2 family use in the CD4bs-specific repertoire and VL3 use in the Env-specific repertoire (*p<0.05, χ2 test) (data not shown). Upon closer examination of the individual V-gene segment use of light chains, VK2.51 was found to be more common in the Env-specific repertoire and VL2.7 more common in the CD4bs-specific repertoire (*p<0.05, two-way ANOVA) (data not shown), respectively.
In summary, although single-cell sorted Env- and CD4bs-specific Ig repertoires possess overall similar germline gene segment profiles, significant biases do exist which might contribute to distinct biologic functions and relative degrees of clonal expansion.
CDRH3 sequences of the CD4bs-specific repertoire display unique features regarding length, hydrophobicity and electrostatic properties
By sequence inspection, we observed several unique features of the CD4bs-specific Ig repertoire, which may be responsible for the distinct epitope-recognition of these antibodies, compared to the Env-specific and the total IgG repertoire. Examination of heavy chain CDR3 (CDRH3) regions of HIV-1 vaccine-elicited Env- and CD4bs-specific Ig repertoires revealed that the CD4bs-specific Ig repertoire displayed overall increased CDRH3 length (****p<0.0001, one-way ANOVA) (Fig 2A) with skewed length distribution than the Env-specific and total IgG repertoire. CD4bs-specific antibodies isolated from HIV-infected individuals also displayed longer CDRH3 length (35–37), which could facilitate accessing the epitope including the CD4 binding loop on gp120 situated in a recessed position (37, 38). The CDRH3 length of CD4bs-specific Ig repertoire also has a more confined distribution, ranging from 10 to 25 (mean=17) amino acid residues (Fig 2A), while that of the Env-specific repertoire was broader, ranging from 6 to 32 (mean=15.5) residues (Fig 2A). Compared to the total Ig repertoire recovered from memory B cells with an average of 14.4 residues of CDRH3 length (17), both the Env and CD4bs-specific Ig repertoires recovered here had an overall longer CDRH3 (****p<0.0001, one-way ANOVA) (Fig 2A). In contrast, there was no difference in light chain CDR3 lengths between the Env- and CD4bs-specific Ig repertoires (p>0.05, Mann-Whitney test) (Fig S1A). This observation is consistent with the notion that the heavy chain CDR3 is the primary determinant for antigen recognition. In addition, there was no significant difference in mean of CDRH3 lengths in sequences isolated at Imm 2 time point compared to those isolated at Imm 5 in either the Env- or CD4bs-specific repertoires (data not shown), suggesting that the CDRH3 length is relatively stable during the course of immunization.
Figure 2. The unique genetic and functional features of CD4bs-specific Ig repertoire heavy chain CDR3 (CDRH3) region.
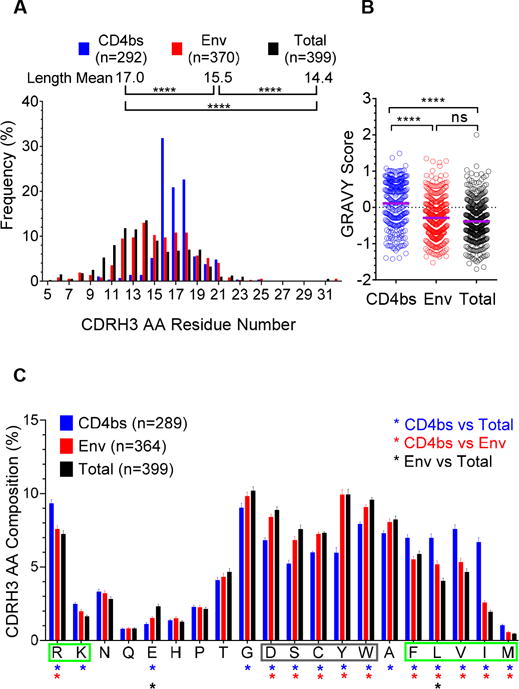
(A) The comparison of CDRH3 amino acid (AA) length of single-cell sorted CD4bs- (blue), Env-specific (red) and total (no antigen-specificity) B cell (black) Ig sequences. CDRH3 region is defined based on the IMGT CDR3 definition. The individual CDRH3 length frequency out of the total number of CDRH3 region length was calculated and depicted. Statistical differences of CDRH3 length among CD4bs-, Env-specific, and total Ig compartments were evaluated using one-way ANOVA with ****p<0.0001. The mean of AA length of each of Ig repertoires are indicated. (B) The CDRH3 of CD4bs-specific Ig repertoire displayed hydrophobicity higher than that of the Env-specific and the total Ig repertoires indicated by GRAVY score. Statistical differences of CDRH3 GRAVY scores among the CD4bs- (blue), Env- (red) specific, and total (black) Ig repertoires were evaluated using one-way ANOVA with ****p<0.0001, ns = nonsignificant. (C) Comparison of amino acid residue contribution to CD4bs-, Env-specific and total Ig CDRH3 regions. The contribution of each amino acid residue to individual CDRH3 was calculated and summarized for each Ig compartment. Statistical differences were evaluated using one-way ANOVA with *p<0.05. AA residues highlighted by a green box had an increased frequency in the CD4bs-specific compartment compared to the total compartment, while residues highlighted by a grey box had reduced frequency. AA residues are displayed on the X-axis according to polarity, with charged residues to the left and hydrophobic residues to the right.
We also observed another unique feature of CD4bs-specific repertoire, the increased degree of hydrophobicity (****p<0.0001, one-way ANOVA) (GRAVY score, Fig 2B) in the CD4bs-specific CDRH3 regions, which was attributed to a higher content of the hydrophobic amino acid residues F, L, V, I and M (*p<0.05, one-way ANOVA) compared to the Env-specific and the total Ig repertories (Fig 2C), partly explained by the prevalent usage of JH5 segment (Fig 1B). The higher CDRH3 hydrophobicity displayed by the vaccine-elicited CD4bs-specific Ig repertoire is consistent with CD4bs-specific mAbs isolated from natural infection, which interact with the receptor binding site and possess a high degree of hydrophobicity (39).
Furthermore, the positively charged AA residues, arginine (R) and lysine (K), were preferentially located in the CD4bs-specific CDRH3 (*p<0.05, Fig 2C), suggesting that they are responsible for the recognition of the negatively charged CD4 binding loop of gp120 (38), including the critical CD4bs residues on the gp120 target, D368 and E370.
Affinity maturation progressively occurs as evidenced by the elevated levels of somatic hypermutation during the immunization course
To investigate the antibody affinity maturation process, the amplified Ig sequences were queried against the macaque Ig germline database to assess the level of SHM (18). Both Env and CD4bs-specific Ig repertoires isolated following Imm 5 displayed ~2% more VH mutations at the nucleotide (Nt) level and ~4% at the amino acid (AA) level, compared to the amplicon sequences following Imm 2 (****p<0.0001, one-way ANOVA) (Fig 3A). Similar findings were observed for the CD4bs-specific repertoire light chain (V genes of both kappa and lambda chains, ****p<0.0001, one-way ANOVA) (Fig S1B). The higher level of SHM in the later stage of repeated immunization indicates that affinity maturation occurs progressively over the course of immunization. Upon closer examination of the regions of V-gene segment use for heavy chains, we found that the mutations within all of the subdomains of the VH region (FRs1-3 and CDRs1-2 increased from Imm 2 to Imm 5 (**p<0.01, ***p<0.001, ****p<0.0001, Mann-Whitney test) (Fig 3B), indicating that both frameworks and variable CDR regions evolve during affinity maturation. Similarly, the mutations in all of the subdomains of V-gene segment of light chains increased during immunization (data not shown). There were no significant differences in SHM for heavy and light chain V genes between the Env and CD4bs-specific Ig repertoires (data not shown), suggesting the same level of affinity maturation occurs for both repertories.
Figure 3. Somatic hypermutations (SHM) of the VH segments in CD4bs- and Env-specific Ig repertories indicate progressive affinity maturation during immunization.
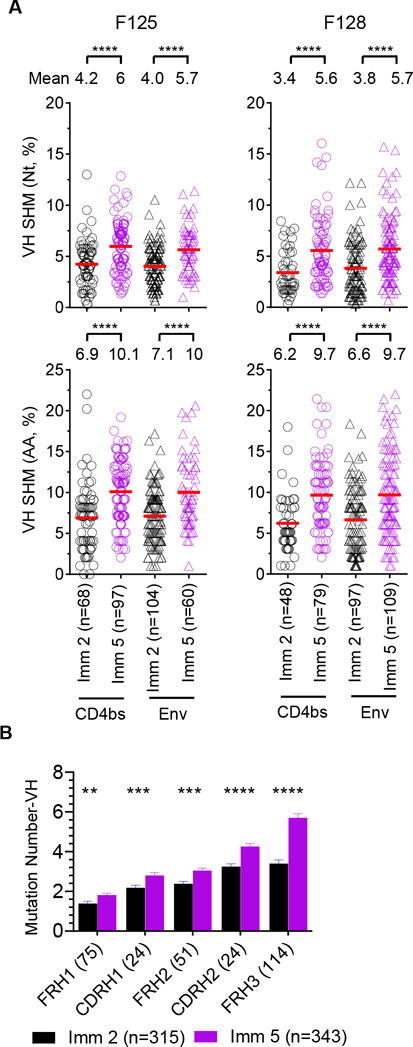
(A) VH SHM levels of single-cell sorted antigen-specific Ig repertoires increase during the course of immunization from animals F125 (left) and F128 (right). Heavy chain V-gene SHMs of single-cell sorted Env (triangles) and CD4bs-specific (circles) repertoires at Imm 2 (black) and 5 (purple) are shown at nucleotide (Nt, upper panel) or amino acid (AA, lower panel) level. The mean of SHM percentage in each compartment is indicated. Statistical differences in SHM between Imm 2 and 5 were evaluated using one-way ANOVA with ****p<0.0001. (B) Accumulated mutations within subdomains of VH region of all the VH amplicons from animals F125 and F128 during the course of immunization. Each antibody heavy chain VDJ sequence was divided into the subdomains of frameworks (FR) and complementarity determining regions (CDRs) based on IMGT definition, with the number in parenthesis indicating subdomain length. Each subdomain was further aligned to the corresponding germline to obtain nucleotide mutation number from Imm 2 (black) and 5 (purple) with VH sequences pooled from both animals. Statistical differences in mutation frequency were evaluated using the Mann-Whitney test with **p<0.01, ***p<0.001, ****p<0.0001.
HIV-1 vaccine-elicited CD4bs-specific antibody affinity maturation is associated with functionality progression
Given the affinity maturation of the Env- and CD4bs-specific Ig repertoire, indicated by the increasing level of SHM of the Ig heavy/light chain, we sought to investigate if the elevated level of SHM was associated with improvements in antibody antigen-binding affinity and virus neutralization capacity. We selected 40 additional clones from Imm 2 and Imm 5, along with the antibodies cloned earlier in the study to a total of 74 clones to characterize their epitope binding specificity, antigen-binding affinity, and virus neutralization breadth and potency. The selected clones demonstrated the typical CD4bs-specific gp140-Fhi/gp140-F-D368Rlo binding phenotype, consistent with that we observed previously. We then assessed their binding affinity for gp120 by biolayer light interferometry (BLI) as described previously (18). We observed that the binding affinities (KD) of mAbs isolated from Imm 5 were significantly higher than those from Imm 2 (**p<0.01, Mann-Whitney test) (Fig 4A, left panel). Interestingly, the off-rates significantly decreased from Imm 2 to Imm 5 (**p<0.01, Mann-Whitney test) (Fig 4A, right panel), while the on-rates remained relatively constant (Fig 4A, middle panel), suggesting that the increased antigen binding affinity was primarily achieved by enhanced antibody-antigen complex stability, possibly through accumulated SHM. This observation is consistent with the affinity maturation of other antigen-specific antibody responses such as the tetanus toxoid-specific antibodies isolated from human subjects vaccinated with tetanus toxoid vaccine (40).
Figure 4. The functionality evolution of representative mAbs from CD4bs-specific Ig repertoire during the course of immunization.
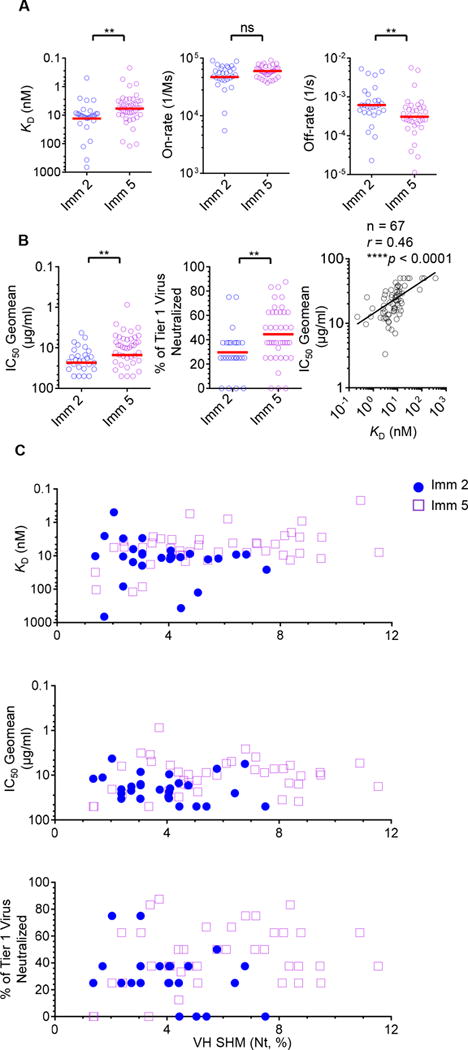
(A) Binding affinity of CD4bs-specific mAbs increased during the course of immunization. Representative mAbs were cloned from single-cell sorted CD4bs-specific Ig sequences from Imm 2 and Imm 5 immunization time points with binding affinity to gp120 analyzed by Biolayer light interferometry. The dissociation constants (KD), association rates (on-rate) and dissociation rates (off-rate) of mAbs isolated from Imm 2 (blue) and 5 (purple) were compared using the Mann-Whitney test with **p<0.01, ns = non-significant. (B) The tier 1 virus neutralization capacity of CD4bs-specific mAbs increased during the course of immunization and was correlated with binding affinity. Eight tier 1 pseudoviruses were tested with the results reported as the neutralization IC50 value, the concentration of the mAb at which the virus entry is inhibited by 50%. The neutralization potency (left panel) and breadth (middle panel) were compared between mAbs from Imm 2 (blue) and Imm 5 (purple) and evaluated using the Mann-Whitney test with **p<0.01. The correlation between binding affinity and neutralization potency was determined using the non-parametric Spearman test (right panel). Total mAb number (n), correlation coefficient (r) and the p value are shown. (C) CD4bs-specific mAb repertoire possesses distinctive functionality and maturation properties at two different immunization time points. CD4bs-specific mAb VH Nt SHMs were plotted vs. antigen binding affinity (KD, upper panel), tier 1 virus neutralization potency (IC50 Geomean, middle panel), and neutralization breadth (% of tier 1 virus neutralized, lower panel) of time points Imm 2 (blue circles) and Imm 5 (purple hollow square), respectively.
We further analyzed the neutralization capacity of the cloned CD4bs-specific mAbs against a panel of 8 neutralization-sensitive HIV-1 strains (tier 1) including clades A, B, and C pseudoviruses, along with 3 neutralization-resistant primary strains (tier 2, clade B) (Table S1). All cloned mAbs displayed various capacities for neutralizing tier 1 viruses, albeit no neutralization against tier 2 viruses was observed (Table S1). Consistently, the tier 1 virus HXBc2 was preferentially neutralized by most of the CD4bs-specific mAbs (~88% of total cloned mAbs). The allogeneic (non-clade B) tier 1 viruses, DJ263 and MW965, were neutralized by 34% and 22% of these mAbs, respectively (Table S1). Moreover, the neutralization potency and breadth against tier 1 viruses were found to increase during the course of immunization (**p<0.01, Mann-Whitney test) (Fig 4B, left and middle panels). Finally, we found that the antigen-binding affinity (KD) and virus neutralization potency (IC50 geomean) were highly correlated (****p<0.0001, nonparametric Spearman correlation) (Fig 4B, right panel), indicating that the improved functionality of CD4bs-specific antibodies evolved in parallel with the elevated levels of SHM. Interestingly, CD4bs-specific clones isolated early during the course of immunization (Imm 2) had moderate levels of SHM, antigen-binding affinity, and neutralization potency (Fig 4C). Somehow contrastingly, clones at late immunization stage (Imm 5) had broader ranges (from moderate to high) of SHM level, affinity and neutralization capacity distribution (Fig 4C), thus possess higher degree of heterogeneity.
Env- and CD4bs-specific Ig repertoires are diversified with moderate degree of clonal dominance
To gain further insight into how the Env-and CD4bs-specific B cell repertoires evolve during the immunization course at clonal level, we sought to dissect the clonal lineage composition of these repertoires. The clonal lineage was defined using the criteria initially deployed for T cell receptor analysis (41) and subsequently adopted for B cell receptor (BCR) studies (17, 42), briefly listed as: 1) same V and J gene usage, 2) same CDR3 length, and 3) CDR3 nucleotide sequence homology > 90%. Based on this definition, we classified the heavy chain Ig repertoire into different clonal lineages to first assess the diversity of the repertoire.
We found that the Env-specific Ig repertoire was highly diversified, with the majority of the repertoire consisting of a single clone per lineage, while a few lineages consisting of multiple clonal expansions (Fig 5A, left). We observed a similar trend for the CD4bs-specific repertoire (Fig 5B, left), although there were more lineages consisting of multiple clones compare to the Env-specific repertoire. Of note, occasionally there were predominant clonal lineages with expansion frequencies accounting for more than 5% of the total repertoire in both repertoires (marked as *) (Fig 5A & B, left). The heterogeneity of each respective repertoire was further assessed by the ratio of the number of clonal lineages (c) to the total number of expansions (n). In sum, the Env-specific repertoire displayed a higher degree of heterogeneity (>60%) (Fig 5A, right upper panel) than the CD4bs-specific repertoire (40–50%) (Fig 5B, right lower panel), which is consistent with that the Env-specific repertoires recognize more diversified epitopes than the relatively focused CD4bs-specific repertoires. There was a slight decrease of heterogeneity in both repertories from time point Imm 2 to Imm 5 (Fig 5A & B, right panel). However, given the limited sampling numbers, it is not clear whether this observed decrease in heterogeneity is significant.
Figure 5. The clonal lineages of Env- and CD4bs-specific Ig repertoires display various degree of heterogeneity at different immunization time points (Imm 2 vs. Imm 5).
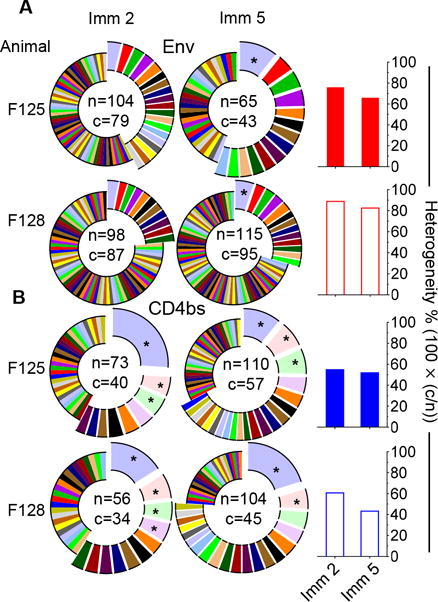
(A) The clonal lineage distribution and heterogeneity of single-cell sorted Env-specific Ig repertoire. Pie charts, each slice of the pie chart represents one clonal lineage (c), with the size of the colored area corresponding to the lineage frequency out of the total number of expansions (n). Clonal lineages with multiple expansions are displayed as exploded portions, lineages that have expanded to constitute > 5% of the total number of expansions are marked with asterisks. Bar graph, the heterogeneity {ratio of clonal lineage number (c) to expansion number (n)*100 as (c/n)*100} is summarized on the right panel for Env- specific repertoire (red). (B) The clonal lineage distribution and heterogeneity of single-cell sorted CD4bs-specific Ig repertoire, displayed as in (A). The heterogeneity is summarized on the right panel for CD4bs-specific repertoire (blue).
Similarly, the Ig light chain amplicons were also analyzed for clonal lineage compositions with over 50% and 30% heterogeneity detected for the Env- and CD4bs-specific repertoires, respectively (data not shown). Together, the Env- and CD4bs-specific Ig repertoires were highly diversified with moderate portion of predominant lineages.
Persistent clonal lineages exist in both of the Env- and CD4bs-specific Ig repertoires
The clonal lineage classification of the Env- and CD4bs-specific Ig repertoires allowed us to longitudinally interrogate their level of prevalence (defined as the frequency of all expansions from a given clonal lineage within the entire repertoire) at different immunization time points. We observed that a small but notable portion of lineages consist of expansions identified at both Imm 2 and Imm 5 repertoires (Fig 6, left, in yellow), while the majority of the clonal lineages were only identified at one of the time points (Fig 6, left, in blue or purple). Based on the frequency of each clonal lineage within the whole repertoire along the immunization course, we classified them into three types according to their different degree of clonal persistence: (1) high persistence (Psthi), with expansion present at both Imm 2 and Imm 5 time points (Fig 6, right, in yellow); (2) low persistence (Pstlo), with expansion present only at Imm 2 (Fig 6, right in blue); and (3) newly emerging, with expansions present only at Imm 5 (Fig 6, right, in purple). The latter type was most likely activated after the second immunization. The existence of the Psthi clonal lineages accounted for about 9–28% of the Env-specific and 23–35% of the CD4bs-specific repertories, respectively (Fig 6, right), of which are substantial portions.
Figure 6. Persistent clonal lineages of Env- and CD4bs-specific repertoires from animals F125 & F128 have established during the immunization.
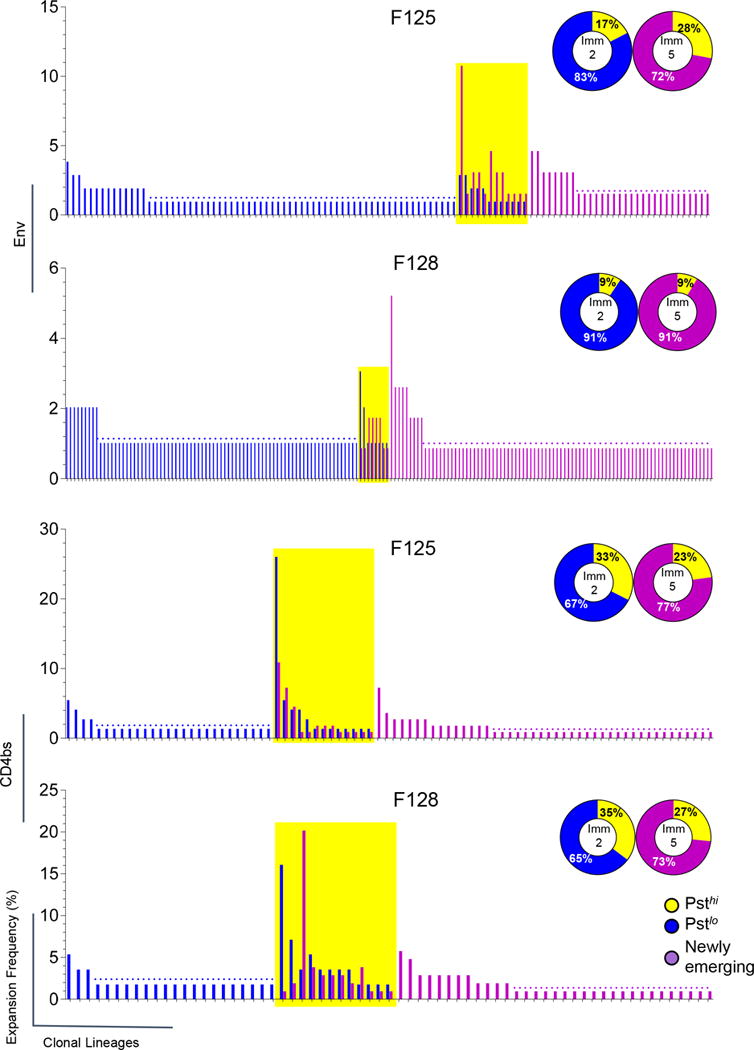
Bar graph, the frequency of expansions from each individual clonal lineage at different immunization time points. Clonal lineages isolated at Imm 2 are in blue and Imm 5 in purple. The clonal lineages, which have expansions at both time points, are highlighted with yellow boxes. Dot lines depict the frequency of clonal lineages with single expansion in the repertoire at each time point. Pie chart, the persistence of clonal lineages of the Env- and CD4bs-specific repertories. Clonal lineages that have expansions at both time points are referred as highly persistent and designated as Psthi compartment (yellow), that have expansions only at Imm 2 were classified as of low persistence (Pstlo, blue), while lineages have expansions only at Imm 5 were designated as Newly emerging (purple) compartment. The frequencies of these three categories of clonal lineages with different clonal persistence out of the total clonal lineages were indicated. Env-specific lineages are shown in the upper two panels and CD4bs lineages in the lower two panels.
Since it is possible that the prevalence of certain clonal lineages may be below the level of detection in the repertoires generated by single-cell sorting, due to their low frequencies in the total B cell repertoire or sample limitations, we interrogated the presence of the representative clonal lineages by querying their related sequences in a more comprehensive repertoire generated by B cell next generation sequencing (NGS) to overcome the depth limitation imposed by single-cell sorting (17). We found that, consistent with our clonal lineage classification based on single cells sorting method, the Psthi clonal lineages always present at a higher frequency (~3-fold) of expansions than the Pstlo clonal lineages for both Env- and CD4bs-specific repertoires (Fig S2). Furthermore, we constantly observed recurring isolation of most of (approximately 70%) CD4bs-specific memory B cells from the same clonal lineage in independent single-cell sorting experiments using the same PBMC sample. Therefore, the clonal lineage persistence classification based on the clonal prevalence sampled by single cell sorting properly illustrated the repertoire clonal durability.
Affinity for antigen plays a critical role in determining clonal lineage persistence
To investigate the genetic properties of the clonal lineages with high persistence, we asked if the SHM level of the clones from the CD4bs-specific lineages primarily determined their degree of persistence. We found that at the same immunization time point, there was no significant difference in SHM levels between the clones in the Psthi and the Pstlo or Newly emerging lineages at both the Nt and AA levels (data not shown).
To investigate the functional properties associated with Psthi clonal lineages, we selected representative Ig sequences from each cluster of the Psthi, Pstlo, and the Newly emerging lineages for further analysis. Interestingly, clones from the Psthi lineages displayed significantly higher gp120 binding affinity than the clones from Pstlo or Newly emerging lineages at the same immunization time point (Imm 2 or Imm 5) (*p<0.05, Mann-Whitney test) (Fig 7A, upper left panel), which indicates that CD4bs-specific memory B cell clones with high binding affinity to gp120 at the early immunization time point are more likely to persistently evolve and expand. Within the Psthi clonal lineages, increasing gp120 binding affinity was also associated with elevated neutralizing potency from Imm 2 to Imm 5 (**p<0.01, one-way ANOVA) (Fig 7A, upper right panel). Consistent with the observation of the total CD4bs-specific repertoire, the selected mAbs demonstrated no significant difference in SHM at either Nt or AA levels at the same immunization time point (Fig 7A, lower panels).
Figure 7. Affinity selection is strongly associated with clonal persistence of CD4bs-specific repertoire.
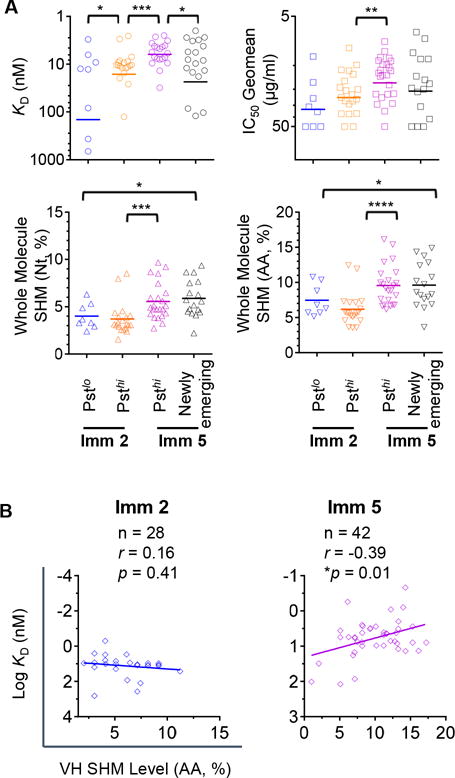
(A) Genetic and functional characterization of clones from the Psthi, Pstlo and Newly emerging lineages in the CD4bs-specific repertoire. The statistical differences of binding affinity KD (upper left panel), neutralization potency (upper right panel), SHM Nt level (lower left panel), and AA level (lower right panel) between each two of the four compartments were evaluated using one-way ANOVA or Mann-Whitney test with *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. (B) The correlation between binding affinity (transformed KD log) and VH AA SHM level was determined at Imm 2 (left panel) and Imm 5 (right panel), respectively, using the non-parametric Spearman test.
In light of the observation that it is the antigen affinity, not SHM, drives antibody persistence, we examined the correlation between the VH SHM level and affinity for antigen gp120 of each CD4bs-specific antibody clone. We found that such correlation varied between different immunization time points. At early immunization time point (Imm 2) when the overall SHM level of the repertoire was low, there was no correlation between the SHM level and the antibody affinity for gp120 (Fig 7B, left panel). In contrast, at later immunization time point (Imm 5) with overall increasing SHM level, there was a correlation between the SHM level and the antibody affinity for the antigen (*p<0.05, nonparametric Spearman correlation) (Fig 7B, right panel). Overall, our data suggest that 1) at early immunization time point, clonal lineage persistence is determined by antigen affinity selection, less dependent on SHM levels; and 2) once clonal lineage persistence is established, the accumulated SHM results in elevated antigen affinity. Our data reinforce the critical role of antigen affinity selection plays during the early immunization time point in determining the clonal persistence of desirable clonal lineages including that of the bNAbs.
The affinity maturation of a predominant CD4bs-specific clonal lineage of high persistence
In this study, we observed that CD4bs-specific clonal lineages displayed various degrees of clonal persistence and predominance. There was occasional occurrence of clonal lineages with high persistence and moderate predominance (Fig 5). To gain insight into the affinity maturation process of the highly persistent and predominant clonal lineages, we selected the most predominant CD4bs-specific persistent clonal lineage of the repertoire of animal F125, the GEAT361 lineage, to investigate its genetic and functionality evolution. GEAT361 lineage expansions accounted for 49% and 32% of the total Psthi expansions identified by single B cell sorting from animal F125 at Imm 2 and 5 time points, respectively (Fig 8A). This clonal lineage uses VH4.22_JH5 with 16 AA residues in CDRH3 and VL2.13_JL3 with 10 AA residues in CDRL3. Representative antibodies within this clonal lineage were cloned and their binding specificities to the CD4bs were verified (Fig S3A).
Figure 8. The affinity maturation pathway of a predominant persistent CD4bs-specific clonal lineage.
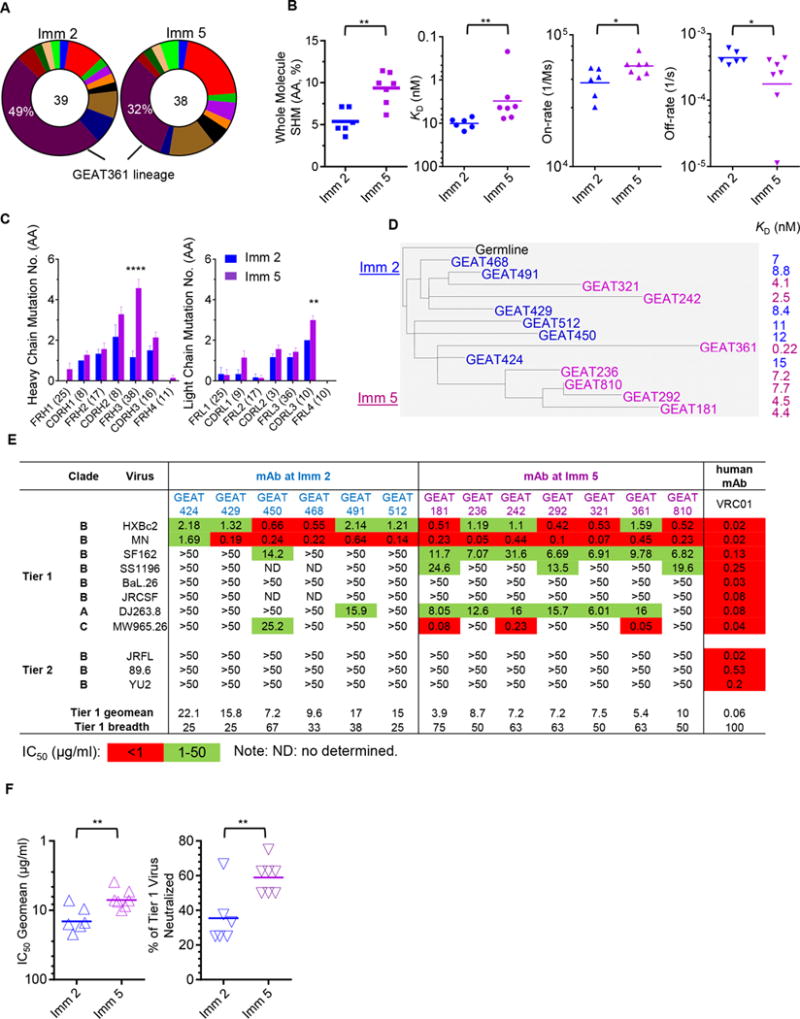
(A) The clonal composition of the highly persistent lineages (Psthi) in the CD4bs Ig repertoire from animal F125. The expansion frequency (percent) of each CD4bs-specific Psthi clonal lineage out of the total Psthi compartment is color coded and illustrated with the total expansion numbers of the Psthi compartment, indicated in the center of the pie chart. One of the most dominant clonal lineages, namely GEAT361 lineage, possesses the highest expansion frequencies of 49% and 32% at Imm 2 and 5 time points respectively, is marked and selected for further characterization. (B) SHM and binding affinity {dissociation constants (KD), association (on-rate) and dissociation (off-rate) rates} of clones from the GEAT361 lineage at Imm 2 and Imm 5 time points (t test with *p<0.05, **p<0.01). (C) Average accumulated mutations within the heavy and light chain subdomains of variant clones within the GEAT361 lineage at time point Imm 2 and Imm 5, respectively (two-way ANOVA with **p<0.01, ****p<0.0001). (D) Phylogenetic analysis of the Ig molecules of the selected variant clones in the GEAT361 lineage. Both the heavy chain VDJ sequence and the matched light chain VJ sequence were used to generate the phylogenetic tree of the whole antibody molecule from both Imm 2 (blue) and Imm 5 (purple) time points with the germline precursor molecule colored in black. The corresponding mAb binding affinity (KD) to gp120 is also displayed on the right side. (E) The neutralization capacity of the selected variant clones of GEAT361 lineage isolated at Imm 2 (blue) and Imm 5 (purple), respectively, were tested to neutralize a panel of HIV-1 pseudoviruses from different clades. Virus neutralizing potency was color-coded as indicated and tier 1 virus neutralization geomean and breadth was calculated with CD4bs bNAb VRC01 as reference antibody. (F) Comparison of the tier 1 virus neutralization potency (IC50 Geomean) and breadth (% of Tier 1 Viruses Neutralized) between the CD4bs mAbs isolated from Imm 2 and Imm 5 time points (t test with **p<0.01).
Consistent with our early observation, clones of GEAT361 lineage at Imm 5 displayed significantly higher SHM level of both VH/VL chains and binding affinity (both on-rates and off-rates) than those at Imm 2 (*p<0.05, **p<0.01, t test) (Fig 8B). Upon closer investigation of SHM mutations in VH and VL subdomains (Fig S3B), we found that the most prominently accumulated mutations occurred within framework region 3 (FR3) of the heavy chain (****p<0.0001, two-way ANOVA) and CDR3 of the lambda chain (**p<0.01, two-way ANOVA) from Imm 2 to Imm 5 (Fig 8C). Interestingly, we found that a loop region within the heavy chain FR3 subdomain, which connects the strands D and E of FR3 (IMGT definition), and is referred to as TCR HV4-like region (43–45), or sometimes as CDR4, contained high levels of SHM (Fig S3B). The HV4-like loop region of Ig heavy chain FR3 usually is in the same plane with the heavy chain CDRs (46), and could contribute to antigen recognition or interaction with super antigens (46–48). To examine the spatial distribution of the residues of SHM within the FR3 subdomain, we modeled the structure of the VH region of GEAT361 lineages using the CD4bs-specific MAb GE136 Fab as template to reveal the approximate 3D locations of these residues (Fig S3C). In this model, we found that most of the FR3 residues modified by SHM are distal from the CDRs, while some of the mutations within the HV4-like region are proximal to the CDRs (Fig S3C) and could directly contribute to antigen binding affinity. In a sum, our data suggest that the overall increasing affinity for antigen is likely caused by accumulated SHM in the antibody molecule, particularly enriched in both of the framework region of the VH and variable region of the VL gene.
Consistently, phylogenetic analysis of the whole molecular sequences revealed that clones at Imm 2 segregated into clusters closer to the germline precursor sequence (Fig 8D, in blue) with overall lower affinity (Fig 8D, greater KD value) than those at Imm 5 (Fig 8D, lower KD value, in purple), which form clusters in more distal positions from the inferred germline precursor (Fig 8D). These data demonstrated that for HIV-1 CD4bs-specific Psthi predominant clonal lineages, the increased genetic divergence from the assigned germline ancestor (level of SHM) from Imm 2 to Imm 5 was associated with elevated affinity for cognate antigen.
We also characterized the virus neutralization capacity of the mAbs from this lineage (Fig 8E) and found that its viral neutralization potency and breadth also elevated from Imm 2 to Imm 5 (**p<0.01, t test) (Fig 8F). Interestingly, while the neutralization breadth of mAb clones isolated from Imm 2 was overall restricted within Clade B tier 1 viruses, most of the mAb clones isolated from Imm 5 possessed the capacity for neutralizing clade A virus DJ263.8 and some of them neutralized clade C virus MW965.26 at high potency (Fig 8E). Three mAbs isolated from Imm 5, GEAT181, 242 and 361, which displayed higher antigen affinity and SHM (Fig 8D), demonstrated superior neutralization breadth and potency against these selected viruses (Fig 8E). These data indicate that the affinity maturation process leads to improved cross-clade virus neutralization.
Antibody affinity maturation pathway involves decreased thermostability and moderately shifted footprint on gp120 surface
The substantial effects of somatic mutations within antibody HC/LC framework subdomains on the antigen binding affinity and virus neutralization capacity of the antibody may result from the improved thermodynamic folding of antibody molecules. Thus, we assessed the thermostability of the fragment antigen-binding (Fab) portions of selected mAbs from this GEAT361 lineage by differential scanning calorimetry (DSC). Interestingly, we found the melting temperature (Tm) of Fab portions of the Imm 2 mAbs (Tm=80.7 °C) displayed ~2 °C higher than that of the Imm 5 mAbs with Tm = 78.9 °C (Fig 9A, left). Referring to the human bNAb, VRC01 with an average SHM of 25% as a whole molecule (28) and a fairly low Tm of 68.7 °C (Fig 9A, left), we speculated that antibodies with higher SHM may lose thermostability. As expected, we observed a negative correlation between the thermostability (Fab Tm) and SHM (*p<0.05, nonparametric Spearman correlation) (Fig 9A, right). Therefore, during antibody affinity maturation with elevated antigen recognition affinity, the thermostability of the free antibody molecule could decrease as the cost of antigen affinity selection.
Figure 9. GEAT361 lineage thermostability and footprints shift from Imm 2 to Imm 5.
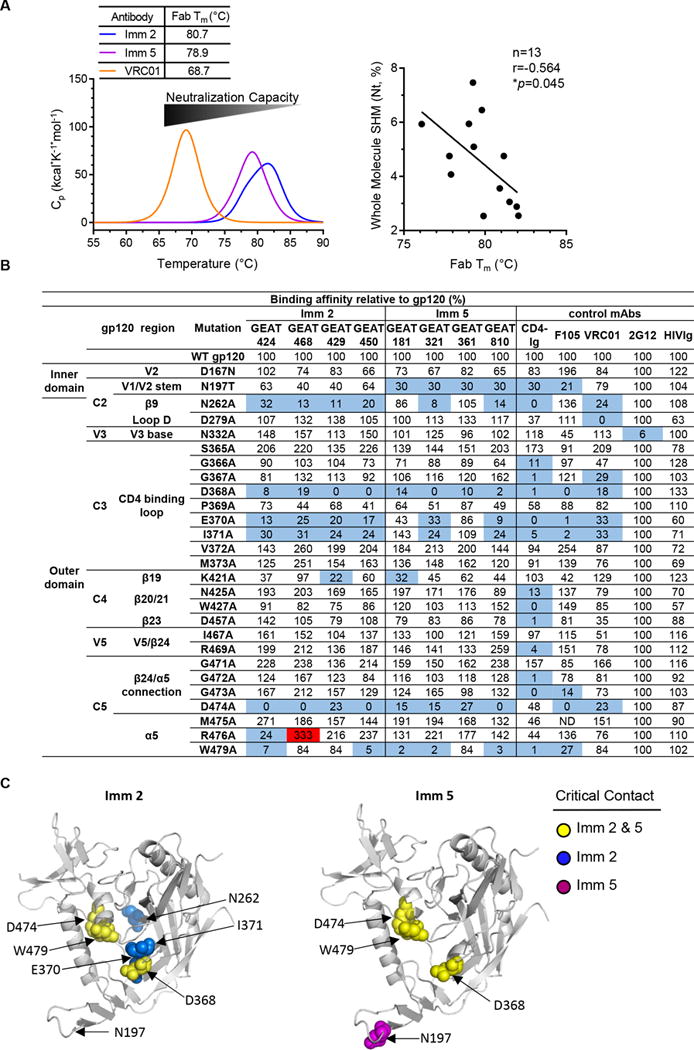
(A) The antibody affinity maturation is associated with the change of thermal stabilities of the free antibody molecule. Left, the average DSC melting profiles of the Fab portion of the mAbs (Fab Tm) from the Imm 2 (blue) and Imm 5 (purple) time points. Note that from Imm 2 to Imm 5 time point, the average Fab Tm decreases, conversely with the increasing trend of antibody affinity for gp120 and virus neutralization capacity. The bNAb VRC01 (orange) has an even lower Fab Tm. Right, correlation between the whole molecule SHM (Nt) and the Fab Tm of the cloned mAbs (non-parametric Spearman test). (B) Representative clones from the GEAT361 linage and selected human antibodies as references were tested to bind a panel of JRCSF Env mutants containing single Ala point mutations in gp120. Both 2G12 and HIVIg served as controls. The effect of Ala mutation on antibody binding is shown relative to wild-type (WT) gp120. A three-fold reduction by mutation was highlighted in blue and a three-fold increase highlighted in red. (C) gp120 core surface (PDB: 2NY3) used to highlight the recognition sites on gp120 by selected GEAT361 clonal variants inferred by Ala scanning. Left, footprints of Imm 2 mAb variants of lower affinity maturation level. Right, footprints of the variants with the highest degree of affinity maturation, GEAT181 and 361, isolated from Imm 5 time point. gp120 backbone is in grey and residues affecting antibody binding for both Imm 2 and Imm 5 variants highlighted in yellow, while mutations on residues only affecting binding at Imm 2 or Imm 5 are highlighted in blue and purple, respectively.
To understand how accumulated SHM affects the antibody affinity and virus neutralization capacity, we investigated if somatic variants from different immunization time points display different footprints on the surface of antigen gp120. We tested the apparent binding affinity of the representative clones from both time points with a panel of alanine scanning mutants of the gp120 of virus isolate JRCSF by ELISA assay. Shown in Fig 9B, with a panel of known CD4bs-specific mAbs as references, we were able to compare the footprints of these mAb variants. As typical CD4bs-specific antibodies, all clones had abolished binding affinity to gp120 variants with the mutations in the CD4 binding loop (D368A) and β24/α5 connection (D474A) (Fig 9B), suggesting that the interaction interface between the antibody and these two major recognition regions of gp120 were established by time point Imm 2.
In addition, we found that the mutations on the V1/V2 stem (N197T), β9 (N262A) and CD4 binding loop (E370A and I371A) of gp120 surface displayed different effects on the gp120 binding affinity for mAbs at these two different time points (Fig 9C). In general, from Imm 2 to Imm 5, the effect of V1/V2 stem mutation N197T on antibody binding affinity to gp120 greatly increased, suggesting that the Imm 5 antibodies have increased contact with V1/V2 stem (Fig 9C). For the clones GEAT181 and 361, isolated from Imm 5 with the best neutralization breadth (Fig 8E), there were profound differences in their footprints in CD4 binding loop (E370 and I371) and β9 (N262) compared to the other somatic variants (Fig 9C). Such footprint differences on gp120 may confer to the expanded tier 1 virus neutralization breadth and was established later during the immunization regimen through accumulated SHM. Consistent with the tier 1 virus-restricted neutralization capacity, the footprints of the selected clones in the clonal lineage evolved during the course of immunization were similar to the footprints of the non-bNAb, F105, distinct from the footprints of the bNAb, VRC01 (Fig 9B) (49).
Discussion
To efficiently elicit CD4bs bNAb response in a vaccine setting, it is important to understand the mechanistic evolution of CD4bs-specific antibody responses by vaccination. Questions remain unclear in the field about the degree of diversity, durability, and persistence of the response elicited by Env vaccine candidates. In this study, we focused on the developmental pathways of CD4bs-specific Ig repertoire during the immunization course of NHPs immunized with the first generation of soluble trimeric form of Env immunogen, gp140-F. We investigated the genetic composition and functional evolution of CD4bs-specific Ig repertoire responses, including gene segment usage preference, clonality delineation, as well as the role of SHM and affinity selection on clonal lineage evolution to fill the knowledge gaps. Although tier 2 neutralizing antibody responses targeting the CD4bs have not yet elicited following Env vaccination, here we establish the foundations of a high resolution analytical system that will be of high value as our capacity increases to efficiently elicit such responses in NHPs.
The CDRH3 regions confer the fine specificity of antibodies. Strikingly, about 80% of the CD4bs-specific Ig repertoire displays CDRH3 length from 15–19 (mean= 17) residues, whereas the CDRH3 lengths of most Env-specific antibodies possess a broader range from 12–19 (mean=15.5) residues, as well as do antibodies comprising the total IgG repertoire (mean=14.4). The CDRH3 sequences of the CD4bs-specific Ig repertoire are more hydrophobic and as well contain more positively charged residues than those of Env-specific repertoire. The unique CDRH3 features of CD4bs-specific Ig repertoire are under pressure of functional selection, which is consist with the result from a recent study which demonstrated that DH germline origins could profoundly influence antibody recognition (50).
During the immunization course, both CD4bs- and Env-specific Ig repertoires display substantial increasing levels of SHM, antigen binding affinity, and improved virus neutralization capacity limited to tier 1 viruses. We observed that mAb binding affinity was significantly associated with neutralization potency against tier 1 viruses (Fig 4B, right panel), suggesting mAbs with higher binding affinity possess more efficient neutralization capacity. We also found that the increasing antigen-binding affinity during the immunization course (Fig 4A) correlated with the decreases in antibody-antigen complex dissociation rates (off-rate), presumably due to the accumulation of the interactions within the antigen-antibody recognition interface possibly caused by SHM. The Imm 5 CD4bs repertoires display higher degree of heterogeneity in terms of the SHM level, affinity for antigen and tier 1 virus neutralization capacity than that of Imm 2 (Fig 4C). Therefore, it appears that the repeated immunization with the same immunogen continuously drives affinity maturation for persistent clonal lineages, while recruits newer clones and maintains the clonal diversities of the B cell repertoire.
Memory B cell clonal lineages of both Env- and CD4bs-specific Ig repertoires undergo dynamic evolution during the immunization course and display various degrees of persistence. A notable portion of the CD4bs-specific clonal lineages displayed a high degree of persistence and maintained substantial population sizes. Interestingly, we found that antigen affinity selection plays a primary role in determining clonal lineage persistence. The administered immunogen in this study is likely in favor of the clonal lineages with relatively high antigen cognate affinity leading to the efficient clonal expansion, which subsequently maintains high persistence. As demonstrated in our previous study, the imperfect antigenicity of the gp140-F immunogen resulted in the elicitation of CD4bs-specific B cell responses with limited neutralization capacity due to suboptimal binding angles and footprints (18, 51). However, the strong association of antigen affinity and lineage persistence shown here provides hope that recently developed well-ordered envelope glycoprotein trimers will likely lead to the elicitation of antibody responses with improved neutralizing capacity in the future (7, 52).
Characterization of a representative predominant persistent CD4bs-specific clonal lineage revealed that SHM in both frameworks and CDR regions of Ig molecule contributed to the functionality evolution, including the overall increased antibody binding affinity to cognate immunogens, decreased thermostability, and subtle shifts in antibody footprints on the cognate Env gp120 surface, as well as the virus neutralization breadth against heterologous viruses. It is possible that the accumulated SHM during affinity maturation leads to a higher degree of Ig molecular flexibility, which favors antigen-antibody induced fit and complex stability. This observation is consistent with the limitations of antigen affinity evolution previously reported in a tetanus toxoid vaccination study (40). Extensive SHM accumulated in the HIV-1 Env-specific antibodies frameworks that are critical for virus neutralization breadth and potency has been observed in antibody repertoires elicited during natural infection (53). The accumulated SHM associated with functionality within the heavy chain FR3 of antibodies, including the HV4-like region, highlights the potential of this subdomain to modulate antigen recognition. Compared to the observed considerable SHM within the heavy chain FRs and light chain CDRs, CD4bs-specific Ig repertoire CDRH3 were relatively consistent during the immunization course (Fig 8C & Fig S3B), with minimal changes, suggesting that CDRH3 predominantly conferring antibody specificity is mostly predetermined.
Despite the great genetic diversity of the CD4bs-specifc B cell repertoire, we observed a few predominant clonal lineages in the persistent compartment from these two immunized animals, which accounted for more than 5% of the sampled repertoire. The persistent CD4bs-specific B cell repertoire of animal F125 consisted of predominant heavy chain VH4.22_JH4/5_CDRH3 16 AA, which used light chains of either VL2.13_JL3_CDRL3 10 AA or VL2.7_JL2_CDRL3 10 AA (data not shown). We also observed that heavy chain VH4.11_JH5_CDRH3 18 AA usage is predominant in the persistent CD4bs-specific repertoire, with light chain VL2.7_JL2_CDRL3 10 AA usage in animal F128 (data not shown). Therefore, light chain clonal lineage, VL2.7_JL2_CDRL3 10 AA, commonly used by the repertoires from both animals, together with heavy chain clonal lineages of VH4.11_JH5_CDR3 18 AA and VH4.22_JH4/5_CDR3 16 AA might be common signatures for rhesus monkey CD4bs-specific B cell clones.
It is not surprising that footprints of the persistent predominant GEAT361 lineage isolated from animal F125 resembles the non-broadly neutralizing antibody F105 more than the bNAb, VRC01 on gp120 surface (Fig 9B), in light of the overall moderate virus neutralization breadth of the serum from animal F125 (22). It has been increasingly appreciated that the model immunogen, YU2gp140-F, exposed antigenic Env surfaces that are not exposed on the native functional Env spike, explaining the elicitation of non-broadly neutralizing antibodies (51). However, by using an immunization regimen based on purified glycoprotein in adjuvant in this study, we were able to robustly elicit total Env- as well as CD4bs-specific memory B cell responses, with ~5% memory B cells being Env-specific, and ~5% of Env-specific memory B cells being CD4bs-specific by flow cytometry. Furthermore, the strong correlation of SHM, affinity for cognate antigen, and virus neutralization capacity of the highly persistent predominant CD4bs-specific clonal lineage elicited by vaccination suggest that it is feasible to elicit persistent antibody responses to conserved determinants with the next generation of HIV-1 trimeric Env vaccine candidates that display improved antigenicity (5–7) at protective titers in the future.
In conclusion, our data demonstrate that affinity maturation of the CD4bs-specific Ig repertoire occurred progressively and clonal lineage persistence is achievable with Env vaccine immunization in NHPs. Among different individual immunized NHP animals, there are CD4bs-specific clonal lineages of high persistence with notable genetic convergence, which have important implication for vaccine development.
Supplementary Material
Acknowledgments
The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: anti-HIV-1 gp120 antibodies including 2G12 from Dr. Hermann Katinger, 39F from Dr. James Robinson (Tulane University), F105 from Dr. Marshall Posner (Dana Farber Cancer Institute), and HIV immune globulin (HIVIG). We are grateful to Dr. Joseph Sodroski (Dana Farber Cancer Institute) for providing the plasmid encoding CD4-Ig, and Dr. Dennis Burton (The Scripps Research Institute) for providing JRCSF gp120 alanine scanning mutant panel plasmids. We thank James Steinhardt for proofreading this manuscript.
Abbreviations
- mAbs
monoclonal antibodies
- bNAb
broadly neutralizing antibody
- Env
envelope glycoprotein
- CD4bs
CD4 binding site
- TCR
T cell receptor
- BCR
B cell receptor
- rMAbs
recombinant monoclonal antibodies
- PBMC
peripheral blood mononuclear cell
- GC
germinal center
- Ig
immunoglobulin
- CDR3
complementarity-determining region 3
- FR
framework region
- SHM
somatic hypermutation
- RT
reverse transcription
- PCR
polymerase chain reaction
- AA
amino acids
- Nt
nucleotide
- NGS
next generation sequencing
- BLI
biolayer light interferometry
- SEC
size-exclusion chromatography
- DSC
differential scanning calorimetry
- Pst
persistence
Footnotes
This work was supported by grants from National Institutes of Health/National Institute of Allergy and Infectious Diseases (R01 AI102766 to Y.L., P01 AI104722 to R.T.W., Y.L. and G.B.K.H., and UM1 AI100663 to R.T.W. and J.Z.), Karolinska Institutet funds and foundations (to C.S.), the Swedish Physicians against AIDS Research Foundation (to C.S.), Swedish Research Council International Postdoc Fellowship (to C.S.), the Swedish Research Council (to G.B.K.H.), and the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health (to J.R.M.). This work was also partially funded by the International AIDS Vaccine Initiative (IAVI) (to R.T.W.) with the generous support of USAID, Ministry of Foreign Affairs of the Netherlands, and the Bill & Melinda Gates Foundation; a full list of IAVI donors is available at www.iavi.org. The contents of this manuscript are the responsibility of the authors and do not necessarily reflect the views of USAID or the US Government.
References
- 1.Lewis GK, DeVico AL, Gallo RC. Antibody persistence and T-cell balance: two key factors confronting HIV vaccine development. Proc Natl Acad Sci U S A. 2014;111:15614–15621. doi: 10.1073/pnas.1413550111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 2015;16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwong PD, Mascola JR, Nabel GJ. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat Rev Immunol. 2013;13:693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- 4.Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, Ward AB, Wyatt RT. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog. 2015;11:e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015;11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, Pietzsch J, Gray MD, Cupo A, van Gils MJ, Yao KH, Liu C, Gazumyan A, Seaman MS, Bjorkman PJ, Sanders RW, Moore JP, Stamatatos L, Schief WR, Nussenzweig MC. Immunization for HIV-1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice. Cell. 2015;161:1505–1515. doi: 10.1016/j.cell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, Menis S, Jones M, Kubitz M, Spencer S, Adachi Y, Burton DR, Schief WR, Nemazee D. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGuire AT, Dreyer AM, Carbonetti S, Lippy A, Glenn J, Scheid JF, Mouquet H, Stamatatos L. HIV antibodies. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science. 2014;346:1380–1383. doi: 10.1126/science.1259206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malherbe DC, Pissani F, Sather DN, Guo B, Pandey S, Sutton WF, Stuart AB, Robins H, Park B, Krebs SJ, Schuman JT, Kalams S, Hessell AJ, Haigwood NL. Envelope variants circulating as initial neutralization breadth developed in two HIV-infected subjects stimulate multiclade neutralizing antibodies in rabbits. J Virol. 2014;88:12949–12967. doi: 10.1128/JVI.01812-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bricault CA, Kovacs JM, Nkolola JP, Yusim K, Giorgi EE, Shields JL, Perry J, Lavine CL, Cheung A, Ellingson-Strouss K, Rademeyer C, Gray GE, Williamson C, Stamatatos L, Seaman MS, Korber BT, Chen B, Barouch DH. A multivalent clade C HIV-1 Env trimer cocktail elicits a higher magnitude of neutralizing antibodies than any individual component. J Virol. 2015;89:2507–2519. doi: 10.1128/JVI.03331-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH, M.-T. Investigators Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 14.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robb ML, Rerks-Ngarm S, Nitayaphan S, Pitisuttithum P, Kaewkungwal J, Kunasol P, Khamboonruang C, Thongcharoen P, Morgan P, Benenson M, Paris RM, Chiu J, Adams E, Francis D, Gurunathan S, Tartaglia J, Gilbert P, Stablein D, Michael NL, Kim JH. Risk behaviour and time as covariates for efficacy of the HIV vaccine regimen ALVAC-HIV (vCP1521) and AIDSVAX B/E: a post-hoc analysis of the Thai phase 3 efficacy trial RV 144. Lancet Infect Dis. 2012;12:531–537. doi: 10.1016/S1473-3099(12)70088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yates NL, Liao HX, Fong Y, deCamp A, Vandergrift NA, Williams WT, Alam SM, Ferrari G, Yang ZY, Seaton KE, Berman PW, Alpert MD, Evans DT, O’Connell RJ, Francis D, Sinangil F, Lee C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Pinter A, Zolla-Pazner S, Gilbert PB, Nabel GJ, Michael NL, Kim JH, Montefiori DC, Haynes BF, Tomaras GD. Vaccine-induced Env V1-V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci Transl Med. 2014;6:228ra239. doi: 10.1126/scitranslmed.3007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundling C, Zhang Z, Phad GE, Sheng Z, Wang Y, Mascola JR, Li Y, Wyatt RT, Shapiro L, Karlsson Hedestam GB. Single-cell and deep sequencing of IgG-switched macaque B cells reveal a diverse Ig repertoire following immunization. J Immunol. 2014;192:3637–3644. doi: 10.4049/jimmunol.1303334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sundling C, Li Y, Huynh N, Poulsen C, Wilson R, O’Dell S, Feng Y, Mascola JR, Wyatt RT, Karlsson Hedestam GB. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Sci Transl Med. 2012;4:142ra196. doi: 10.1126/scitranslmed.3003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai K, He L, Khan SN, O’Dell S, McKee K, Tran K, Li Y, Sundling C, Morris CD, Mascola JR, Karlsson Hedestam GB, Wyatt RT, Zhu J. Rhesus Macaque B-Cell Responses to an HIV-1 Trimer Vaccine Revealed by Unbiased Longitudinal Repertoire Analysis. MBio. 2015;6:e01375–01315. doi: 10.1128/mBio.01375-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sundling C, Forsell MN, O’Dell S, Feng Y, Chakrabarti B, Rao SS, Lore K, Mascola JR, Wyatt RT, Douagi I, Karlsson Hedestam GB. Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. J Exp Med. 2010;207:2003–2017. doi: 10.1084/jem.20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sundling C, Phad G, Douagi I, Navis M, Karlsson Hedestam GB. Isolation of antibody V(D)J sequences from single cell sorted rhesus macaque B cells. J Immunol Methods. 2012;386:85–93. doi: 10.1016/j.jim.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 25.Alamyar E, Giudicelli V, Li S, Duroux P, Lefranc MP. IMGT/HighV-QUEST: the IMGT(R) web portal for immunoglobulin (IG) or antibody and T cell receptor (TR) analysis from NGS high throughput and deep sequencing. Immunome Res. 2012;8:26. [Google Scholar]
- 26.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O’Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 30.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu XL, Zhou TQ, O’Dell S, Wyatt RT, Kwong PD, Mascola JR. Mechanism of Human Immunodeficiency Virus Type 1 Resistance to Monoclonal Antibody b12 That Effectively Targets the Site of CD4 Attachment. Journal of virology. 2009;83:10892–10907. doi: 10.1128/JVI.01142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pantophlet R, Ollmann Saphire E, Poignard P, Parren PW, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J Virol. 2003;77:642–658. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 36.Breden F, Lepik C, Longo NS, Montero M, Lipsky PE, Scott JK. Comparison of antibody repertoires produced by HIV-1 infection, other chronic and acute infections, and systemic autoimmune disease. PloS one. 2011;6:e16857. doi: 10.1371/journal.pone.0016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, Ernandes MJ, Kong R, Longo NS, Louder MK, McKee K, O’Dell S, Schmidt SD, Tran L, Yang Z, Druz A, Luongo TS, Moquin S, Srivatsan S, Yang Y, Zhang B, Zheng A, Pancera M, Kirys T, Georgiev IS, Gindin T, Peng HP, Yang AS, Program NCS, Mullikin JC, Gray MD, Stamatatos L, Burton DR, Koff WC, Cohen MS, Haynes BF, Casazza JP, Connors M, Corti D, Lanzavecchia A, Sattentau QJ, Weiss RA, West AP, Jr, Bjorkman PJ, Scheid JF, Nussenzweig MC, Shapiro L, Mascola JR, Kwong PD. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L, Kwon YD, Zhou T, Wu X, O’Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poulsen TR, Jensen A, Haurum JS, Andersen PS. Limits for antibody affinity maturation and repertoire diversification in hypervaccinated humans. J Immunol. 2011;187:4229–4235. doi: 10.4049/jimmunol.1000928. [DOI] [PubMed] [Google Scholar]
- 41.Davis MM, Chien YH. T cell antigen receptors. Lippincott-Raven; Philidelphia: 1999. [Google Scholar]
- 42.Moody MA, Yates NL, Amos JD, Drinker MS, Eudailey JA, Gurley TC, Marshall DJ, Whitesides JF, Chen X, Foulger A, Yu JS, Zhang RJ, Meyerhoff RR, Parks R, Scull JC, Wang L, Vandergrift NA, Pickeral J, Pollara J, Kelsoe G, Alam SM, Ferrari G, Montefiori DC, Voss G, Liao HX, Tomaras GD, Haynes BF. HIV-1 gp120 Vaccine Induces Affinity Maturation in both New and Persistent Antibody Clonal Lineages. Journal of virology. 2012;86:7496–7507. doi: 10.1128/JVI.00426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi YW, Herman A, DiGiusto D, Wade T, Marrack P, Kappler J. Residues of the variable region of the T-cell-receptor beta-chain that interact with S. aureus toxin superantigens. Nature. 1990;346:471–473. doi: 10.1038/346471a0. [DOI] [PubMed] [Google Scholar]
- 44.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Llera A, Mariuzza RA. Structure-function studies of T-cell receptor-superantigen interactions. Immunological reviews. 1998;163:177–186. doi: 10.1111/j.1600-065x.1998.tb01196.x. [DOI] [PubMed] [Google Scholar]
- 46.Potter KN, Li Y, Capra JD. Staphylococcal protein A simultaneously interacts with framework region 1, complementarity-determining region 2, and framework region 3 on human VH3-encoded Igs. J Immunol. 1996;157:2982–2988. [PubMed] [Google Scholar]
- 47.Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, Goudsmit J. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PloS one. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, O’Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, Graham BS, Haynes BF, Burton DR, Wyatt RT, Mascola JR. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J Virol. 2011;85:8954–8967. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Kapoor P, Parks R, Silva-Sanchez A, Alam SM, Verkoczy L, Liao HX, Zhuang Y, Burrows P, Levinson M, Elgavish A, Cui X, Haynes BF, Schroeder H., Jr HIV-1 gp140 epitope recognition is influenced by immunoglobulin DH gene segment sequence. Immunogenetics. 2016;68:145–155. doi: 10.1007/s00251-015-0890-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tran K, Poulsen C, Guenaga J, de Val N, Wilson R, Sundling C, Li Y, Stanfield RL, Wilson IA, Ward AB, Karlsson Hedestam GB, Wyatt RT. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proc Natl Acad Sci U S A. 2014;111:E738–747. doi: 10.1073/pnas.1319512111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, Gnanapragasam PN, Oliveira TY, Seaman MS, Kwong PD, Bjorkman PJ, Nussenzweig MC. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


