Abstract
Considering malaria as a local and focal disease, epidemiological understanding of different ecotypes of malaria can help in devising novel control measures. One of the major hurdles in malaria control lies on the evolution and dispersal of the drug-resistant malaria parasite, Plasmodium falciparum. We herewith present data on genetic variation at the Single Nucleotide Polymorphism (SNP) level in four different genes of P. falciparum (Pfcrt, Pfmdr1, Pfdhfr, and Pfdhps) that confer resistance to different antimalarials in two different eco-epidemiological settings, i.e. Hilly-Forest (HF) and Riverine-Plain (RP), in a high malaria endemic district of Odisha state, India. Greater frequency of antimalarial resistance conferring SNPs and haplotypes was observed in all four genes in P. falciparum, and Pfdhps was the most variable gene among the four. No significant genetic differentiation could be observed in isolates from HF and RP ecotypes. Twelve novel, hitherto unreported nucleotide mutations could be observed in the Pfmdr1 and Pfdhps genes. While the Pfdhps gene presented highest haplotype diversity, the Pfcrt gene displayed the highest nucleotide diversity. When the data on all the four genes were complied, the isolates from HF ecotype were found to harbour higher average nucleotide diversity than those from RP ecotype. High and positive Tajima’s D values were obtained for the Pfcrt and Pfdhfr genes in isolates from both the HF and RP ecotypes, with statistically significant deviation from neutrality in the RP ecotype. Different pattern of Linkage Disequilibrium (LD) among SNPs located in different drug-resistant genes was found in the isolates collected from HF and RP ecotypes. Whereas in the HF ecotype, SNPs in the Pfmdr1 and Pfdhfr were significantly associated, in the RP ecotype, SNPs located in Pfcrt were associated with Pfmdr1, Pfdhfr and Pfdhps. These findings provide a baseline understanding on how different micro eco-epidemiological settings influence evolution and spread of different drug resistance alleles. Our findings further suggest that drug resistance to chloroquine and sulfadoxine-pyrimethamine is approaching fixation level, which requires urgent attention of malaria control program in India.
Keywords: malaria, Plasmodium falciparum, ecotypes, drug resistant genes, Odisha
1. Introduction
Epidemiological outcome of malaria infection differs in different ecotypes of the globe (Das et al., 2012; Kaewwaen and Bhumiratana, 2015; Kar et al., 2014; Okwa et al., 2009; Sharma, V.P. et al., 2015), and therefore, malaria is considered as a local and focal disease (Conn et al., 2015; Dash et al., 2008; Rath, 2004). Several studies involving different ecological and climatic settings have provided evidence that malaria epidemiology can be significantly variable across small eco-climatic scales (Jambulingam et al., 1991; Kaga and Ohta, 2012; Schapira and Boutsika, 2012). For example, malaria epidemiological outcomes including distributional prevalence of mosquito vectors and malaria transmission were correlated with different ecotypes in Nigeria (Okwa et al., 2009), Kenya (Ingasia et al., 2015), Brazil (Rosa-Freitas et al., 2007), Southeast Asia (Seng et al., 1999) and India (Jambulingam et al., 1991; Ramar et al., 2014; Shukla et al., 2007; Singh et al., 2015). Moreover, malaria outcome was found to be significantly higher in forested ecotype in comparison to no-forest ecotype (Kar et al., 2014) as observed in Belize (Hakre et al., 2004), Bangladesh (Haque et al., 2011), Nepal (Reisen et al., 1993), and India (Nath and Mwchahary, 2012; Sharma et al., 2006; Shukla et al., 2008). In India, studies conducted in the Sundargarh districts of Odisha state (high malaria endemic) showed that villages in forest and plain areas (separated by short geographical distances) have distinct malaria transmission pattern (Sharma et al., 2006), which could have been a consequence of prevalence of different species and vectorial behavior of a particular species of the mosquito vectors (Das, 2015; Manguin et al., 2008; Nanda et al., 2000; Singh et al., 1996). Considering the evolution and spread of malaria parasites resistant to different antimalarials [viz. Chloroquine (CQ), Sulfadoxine and Pyrimethamine (SP) etc.] that highly influence malaria epidemiological outcome and pose strong impediment to malaria control programs (Das and Dash, 2007; Hastings, 2003; Mallick et al., 2013b; Singh, V. et al., 2009), whether different local micro eco-climatic factors have influenced genetic changes at the genes conferring resistance to different antimalarials, needs to be evaluated (Sorosjinda-Nunthawarasilp and Bhumiratana, 2014). Needless to mention, such information will be of enormous benefit to the local malaria control program (Dash et al., 2008). This is because, in high malaria transmission areas resistance against CQ and SP in the malaria parasite Plasmodium falciparum spreads fast, whereas in low transmission areas, drug pressure plays a much crucial role (Hastings and Watkins, 2005; Malisa et al., 2016; Mallick et al., 2013a; Mallick et al., 2013b; Talisuna et al., 2002). It has further been proposed that the predominance of tribal groups along with unrestricted use of inappropriate antimalarials, population movements, resettlements, and presence of sylvatic mosquito vectors promote rapid evolution of antimalarial resistance and therefore high malaria transmission settings encompassing this type of ecotype were proposed to be centre of origin of drug resistance (Chareonviriyaphap et al., 2000; Kar et al., 2014; Keiser et al., 2005; Malakooti et al., 1998; Singh, N. et al., 2009).
India is endemic to malaria and accounts for about 52% of the total malaria morbidity in Southeast Asia (Pradhan et al., 2016). Interestingly, majority of the malaria morbidity (about 26.9%) and mortality (about 17.6%) is contributed by Odisha state alone, although it comprises about 3% of Indian population (including some aboriginal tribes) (Pradhan et al., 2016). Intense and stable malaria has been reported from tribal areas of Odisha and neighbouring states (http://www.malariasite.com/tag/orissa/)(Das et al., 2012; Kumar et al., 2012; Kumar et al., 2007; Nanda et al., 2000). The state of Odisha consists of two highly malarious clusters; the North-Western (comprising of five districts, viz. Deogarh surrounded by Keonjhar, Sundergarh, Anugul and Sambalpur) and the South-Western (comprising of seven districts, viz. Koraput, surrounded by Malkangiri, Nawarangpur, Kalahandi, Raygada, Nuapada, and Kandhamal (Mohanty et al., 2009; Pradhan et al., 2016; Rao et al., 2015; Sahu et al., 2013), although other districts too contribute to the total malaria cases. Interestingly, the districts in both the clusters are rich in hills and forests and home for aboriginal tribes (Pradhan et al., 2016; Ramar et al., 2014; Sahu et al., 2013). The Deogarh district is one of the epicentres of high malaria endemicity (Pradhan et al., 2016); comprising of two distinct ecotypes [Hilly-Forested (HF) and Riverine-Plain (RP)], and therefore can serve as a model to understand the influence of micro eco-typical habitats on malaria epidemiological outcome (in this case mutational pattern in different genes conferring drug resistance in the malaria parasite, P. falciparum). This is important, as treatment in case of failure to the antimalarial CQ and SP has now become very common in almost all malaria endemic regions of the globe including India (Cui et al., 2015; Klein, 2013). In Odisha, high level of resistance to both CQ and SP has been reported from many malarious districts including Keonjhar, Sundargarh, Anugul and Sambalpur districts (Mohanty et al., 2009; Peterson et al., 1988; Srivastava et al., 2013; Sutar et al., 2011), with no report from the Deogarh district.
Tracking the patterns of mutations, estimating genetic diversities at the Single Nucleotide Polymorphism (SNP) level and asserting linkage among the SNPs in populations are the most efficient ways to understand the evolution of that particular gene (Carlton et al., 2015; Malisa et al., 2016; Pelleau et al., 2015; Sutar et al., 2013). Several studies following these methodologies in genes conferring resistance to antimalarials in P. falciparum have indicated evolutionary potential of these genes both at the global scale and also in India (Awasthi et al., 2011; Brown et al., 2015; Das and Dash, 2007; Kumar et al., 2015; Li et al., 2015; Rouhani et al., 2015). Mutations in the gene encoding a P. falciparum CQ resistance transporter (Pfcrt) and resulting change in single amino acid (AA) locus 76 from K to T (K76T) were proven a strong marker for CQ (Fidock et al., 2000; Valderramos et al., 2010). Similarly, mutations in the P. falciparum multi-drug resistance gene (Pfmdr1) conferring single AA change at point 86 from N to Y (N86Y) and/or multi-copy number of Pfmdr1 was further reported to be linked with K76T and CQ resistance conferring synergistic increase in resistance when combined (Babiker et al., 2001; Chauhan et al., 2014; Foote et al., 1990; Mwai et al., 2009; Price et al., 1999). Furthermore, mutations (S436A and A437G) in the P. falciparum dihydrofolate reductase enzyme coding gene singly pose mild resistance to SP drugs, and when linked with mutations A581G and /or K540E and /or A613S/T, confer high resistance (McCollum et al., 2012; Peterson et al., 1988; Rouhani et al., 2015). Moreover, polymorphism in the Pfdhps gene encoding S108N is the core mutation, but this confers comparatively lower resistance when present singly (Brooks et al., 1994; McCollum et al., 2012; Reeder et al., 1996; Triglia et al., 1997). Of particular importance is the correlation on the number of different mutations a parasite possesses to the ability to resist an antimalarial. For example, double mutant of the Pfcrt-S72V73M74N75T76 and triple mutant C72V73I74E75T76 haplotypes were more prevalent along with wild type C72V73M74N75K76 and reflected more successful resistant haplotypes (Ghanchi et al., 2011; Nagesha et al., 2003). Moreover, accumulation of multiple mutations in Pfdhfr gene resulting in double mutants (C59R/S108N and N51I/S108N) shows moderate levels of resistance, the triple mutants N51I/C59R/S108N show a significant level, and the quadruple mutant parasite (N51I/C59R/ S108N/I164L) is considered to be completely resistant to pyrimethamine (McCollum et al., 2012; Sirawaraporn et al., 1997). Very similarly, co-accumulation of resistance-conferring mutations in both the Pfdhfr and Pfdhps genes synergistically diminishes the success of SP (Rouhani et al., 2015).
In the present study, we have performed DNA sequencing of four genes conferring drug-resistance in the malaria parasite P. falciparum in field isolates collected in the two different ecotypes in the Deogarh district of Odisha state, India. We identified SNPs in the four genes that are differentially segregating in these two populations and compared both the occurrence and distributional prevalence of different SNPs between the high endemic (HF) and moderately endemic (RP) areas. Pattern of genetic diversity between populations for each gene and linkage disequilibrium (LD) between different SNPs of a particular gene were also estimated. The results as a whole were interpreted in term of ongoing and past use of CQ and SP on evolution of drug resistance genotype of Indian P. falciparum in the two different malaria ecotypes in Deogarh district of Odisha, India.
2. Methodology
2.1. Study area, sample collection and malaria species diagnosis
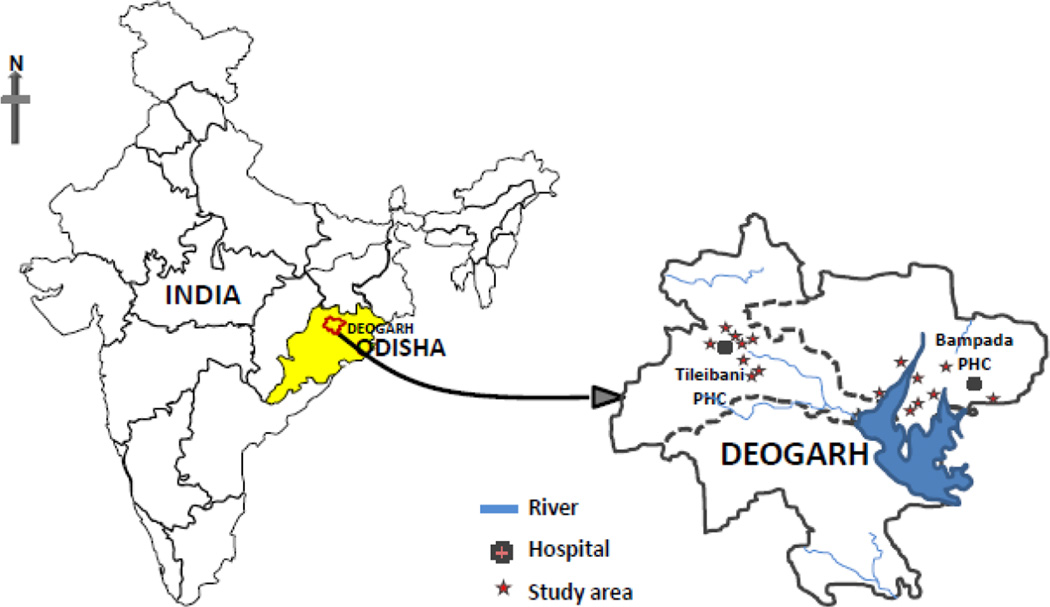
The Deogarh district (21° 31' N Latitude and 84° 43' E Longitude) is located in the western part of Odisha (Figure 1). It covers a 2781.66 Sq. kilometre area with a total population of 27,41,08 (http://www.ordistricts.nic.in/district_profile/aboutus.php) and is highly endemic to P. falciparum malaria (Annual Parasite Incidence > 25) (http://nrhmorissa.gov.in/mis/SearchDetail.aspx). About 22% of the total land area of the Deogarh district is covered by forest, and therefore, this district is comprised of two distinct ecological settings (ecotypes); Hilly-Forest (HF) and Riverine-Plain (RP) (http://www.odishasampad.in/). We have collected isolates of P. falciparum from villages falling under these two ecotypes (HF and RP) that are under the two primary health centres (PHCs) located in about 40-kilometre distance (PHC Tileibani - HF and PHC Bampada – RP) (Figure 1).
Figure 1.
Map of India highlighting Deogarh district (Odisha state). Sample collection sites in Hilly-Forest ecotype (PHC Tileibani) and Riverine-Plain ecotype (PHC Bampada) are demarcated.
Since malaria transmission in the Deogarh district occurs almost throughout the year, we have collected P. falciparum samples though active field collection during three different transmission periods [pre-monsoon (February-March), monsoon (July-August) and post-monsoon (September-November)] in the years 2011 and 2012 (Table 1). Malaria symptomatic individuals were finger-pricked and six drops of blood were collected from each patient. While three drops were used for diagnosis by (i) microscopic examination, preparation of thick and thin films and stained with Giemsa and (ii) rapid diagnostic test with bivalent kit, the rest three drops were put in a Whatman filter paper (for DNA isolation in the lab). The patients diagnosed with malaria with the rapid diagnosis test were provided appropriate treatment support. The blood samples in the slides and the filter paper (after dried) were brought to the laboratory in New Delhi for further analyses. In the lab, both the thick and thin films were stained with Giemsa and the results were matched with the observations on the rapid diagnostic test. Since diagnostic tests by PCR is considered to be highly sensitive (Gupta et al., 2010; Johnston et al., 2006), DNA was isolated from all the field collected samples using Qiagen kit. Considering high mixed parasite infections prevalent in India (Gupta et al., 2010), we used PCR diagnostic assay using published primers (Gupta et al., 2010) for determination of mono infection by P. falciparum in the collected samples. Monoclonality of P. falciparum infections were determined on the basis of determination of a single haplotype of each of the four genes conferring drug resistance (see below). In total, 1,000 samples have initially been collected from both the two ecological zones (354 from HF and 646 from RP), out of which only 229 were pure and single-clonal P. falciparum infections (118 from HF and 111 from RP) were used for DNA sequencing of the four genes conferring drug resistance in P. falciparum. The study was approved by the human ethics committee of the National Institute of Malaria Research, New Delhi, India and written informed consents have been obtained from each adult participant and from parents/guardians of patients below 18 years of age.
Table 1.
Sampling detail of P. falciparum isolates in two different ecotypes in Odisha, India.
| Seasonal collection of P. falciparum isolates |
|||||
|---|---|---|---|---|---|
| Ecotype | February and March 2011 |
September and November 2011 |
July and August 2012 |
Total | |
| Hilly-Forested | 20 | 69 | 29 | 118 | 229 |
| Riverine-Plain | 24 | 47 | 40 | 111 | |
2.2. PCR Amplification of the four drug-resistant genes
In this study, we have considered four genes (Pfcrt, Pfmdr1, Pfdhfr, and Pfdhps) that are reported to be associated with resistance to antimalarials, chloroquine (CQ) and sulfadoxine-pyrimethamine (SP) in P. falciparum. For all the four genes, the genetic regions containing SNPs that are associated with in vitro drug-resistance in P. falciparum were only sequenced (for details of the genetic regions, primers for each gene fragment, see supplementary Table 1). For this, nested PCR protocols have been followed (for details of PCR protocols for each individual gene, please refer to the supplementary Table 1). For example, two different reaction volumes (15µl and 30µl) were used in primary and nested PCR. All PCR amplifications were performed by using “AmpliTaq Gold™” polymerase (PE Applied Bio Systems, Foster City, CA) and for visual quantification, only 3µl PCR products were used in 1.5% Agarose with 100bp DNA ladder (Bangalore Genei, Bengaluru, India). Details of protocols for PCR amplification of each gene have been provided in supplementary Table 1.
2.3. DNA Sequencing and population genetic analyses
All PCR products were purified using shrimp alkaline phosphatase (SAP) and exonuclease I (Exo I) enzymes (Fermentas, USA) before processed for DNA sequencing. For each 25 µl PCR product, one unit of Exo I and one unit of SAP with 10X SAP buffer were used and final reaction volume was made up to 30.0 µl with autoclaved, nuclease-free water (Ambion, Life Technologies). The reaction mixtures were incubated in Eppendorf Master Cycler Pro gradient thermal cycler for 50 min at 37 °C (digestion) and then for 20 min at 85 °C (inactivation of enzymes). Purified PCR products were sequenced commercially (Macrogen Inc., Seoul, Korea, http://dna.macrogen.com/english).
Multiple DNA sequences from each gene were collectively imported to the DNADynamo computer program (Blue Tractor Software, North Wales, United Kingdom; (http://www.bluetractorsoftware.co.uk/) along with respective reference sequence of the wild type (Pf3D7) for viewing the sequence chromatogram, manual editing, and multiple sequence alignment. The edited sequences were deposited in GenBank with accessions XXXXXX– YYYYY. Single nucleotide polymorphisms (SNPs) were spotted by scanning mismatch highlights from the split window of DNADynamo base-call alignment window and reconfirmed by referring aligned chromatograms in lower split window. Since all the four sequenced DNA fragments are located in the coding regions (exon) of the genes, the aligned sequences were translated to amino acid sequences and synonymous and non-synonymous mutations, if any, were spotted. Any change in the amino acid was ascertained by looking at the responsible SNP in comparison to the standard reference sequence. Since specified SNPs in designated genetic regions have been implicated with drug-resistance phenotype of P. falciparum, SNPs located in those specific regions were specifically looked at. For example, SNPs causing amino acid changes in the Pfcrt-C72V73M74N75K76 (capital letters representing wild type amino acids (AA) with their positions in the gene subscripts), Pfmdr1-N86, Pfdhfr-A16N51C59S108I164, and Pfdhps-S436A437K540A581A613 were noted. Similarly, mutations in other regions of the sequenced DNA fragments of each gene and corresponding change in amino acid were also noted.
Differential arrangements of SNPs for each gene in every P. falciparum isolate form a particular haplotype. In order to know genetic differences between the HF and RP ecotypes for each independent gene, first the frequencies of different haplotypes were calculated using the Statistical Package for Social Sciences 16.0 (SPSS, Inc., Chicago, IL, USA) for each ecotype separately. Further, in order to know if differential distribution of different haplotype in a particular gene exists between isolates from HF and RP ecotypes, chi-square tests were performed independently for each gene. In addition, haplotype diversity (Nei, 1987) and two measures of nucleotide diversity (θw and π) were estimated for each of the HF and RP ecotypes. While the nucleotide diversity parameter π is estimated based on average number of pair wise nucleotide difference per site, (Tajima, 1989) the θ w estimate is dependent on the number of segregating sites (Watterson, 1975). All these parameters were estimated using the DnaSP 5.02 computer program (Librado and Rozas, 2009). In order to ascertain if the four genes conferring drug resistance in P. falciparum follow neutral equilibrium model of molecular evolution, the Tajima’s D test was performed for each gene for the isolates from each ecotypes separately using the DnaSP 5.02 computer program (Librado and Rozas, 2009). The Tajima’s D (Tajima, 1989) statistic calculates the normalized differences between the two measures of nucleotide diversity (θ w and π) for isolates from each ecotype. Whereas an excess of low frequency polymorphism generates negative value of TD indicating directional selection or population size expansion, low level of high frequency polymorphism generates positive value indicating balancing selection or population size reduction (Das et al., 2004). Moreover, in order to determine if differential association exists among SNPs segregating in all the four gene for isolates from a particular ecotype (either HF or RP), linkage disequilibrium (LD) tests were performed for each possible pair-wise SNP implicated as drug-resistant marker in the four genes by calculating the r2 values using the Haploview computer program (Barrett, 2005).
1. Results
Out of the total 1000 malaria positive blood samples collected (354 from HF and 646 from RP), only 229 isolates (118 from HF and 111 from RP) were found to be infected with P. falciparum employing three different types of malaria diagnosis, viz. microscopy, RDT and PCR. In addition, in order to ascertain if all the 229 isolates are monoclonal infections, sequence chromatograms of the four genes conferring drug resistance (Pfcrt, Pfmdr1, Pfdhfr, and Pfdhps) of all the 229 isolates were carefully checked for double peaks. For every gene, mono-clonality was established based on single peaks, resulting fractions of the 229 isolates independently in each of the genes (Pfcrt-64/229; Pfmdr1-73/229; Pfdhfr-29/229 and Pfdhps-84/229) to be monoclonal. The distribution of single clonal infections was however not found to be significantly different between the HF and RP ecotypes based on the data of single peak in the four genes (for HF; Pfcrt-34, Pfmdr1-36, Pfdhfr-13 and Pfdhps-41 and for RP; Pfcrt-30, Pfmdr1-37, Pfdhfr-16 and Pfdhps-43). In total, only 24 isolates (12 from HF and 12 from RP) were found to be monoclonal based on single peak of nucleotide chromatogram when all the four genes were considered.
All the mutations implicated in conferring resistance to different antimalarials in P. falciparum were sequenced in multiple isolates from both the HF and RP ecotypes. In addition, with multiple sequence alignment, additional mutations (some reported and some novel) were also detected. The distribution of different haplotypes (due to combinations of different mutations implicated in drug resistance in four genes) in both the HF and RP is depicted in Table 2. Whereas in three genes similar haplotypes (Pfcrt three haplotypes, in Pfmdr1 two and on Pfdhfr two haplotypes) were found between the HF and RP ecotypes, in case Pfdhps, six haplotypes in HF and four in RP were detected (Table 2). Although the frequency of different haplotypes of the four genes was different between isolates from two ecotypes, but no marked difference could be seen (Table 2). The distribution of the number of haplotypes were subjected to chi-square test for all the four genes between the isolates from HF and RP ecotypes, which indicate statistically significantly deviation from the expected distribution only in case of Pfcrt gene. Therefore, the data in general on the frequency distribution of different haplotypes that are associated with phenotype drug resistance in P. falciparum indicate no significant difference between the two ecotypes.
Table 2.
Frequency (in percent) of different drug resistant marker haplotypes in four genes implicated in drug resistance in two ecotypes of P. falciparum
| Ecotypes | Genes |
|||||||
|---|---|---|---|---|---|---|---|---|
| Pfcrt | Pftndr | Pfdhfr | Pfdhps | |||||
| C72V73I74E75T76 | 25(73.5%) | N86 | 8 (22.2%) | C59S108 | 4(30.8%) | A436G437E540A581 | 3 (7.3%) | |
| C72 V73M74N75K76 | 7 (20.6%) | Y86 | 28 (77.8%) | R59N108 | 9 (69.2%) | A436G437K540A581 | 1 (2.4%) | |
| S72 V73M74N75T76 | 2(5.9%) | S436A437K540A581 | 28 (68.3%) | |||||
| Hilly-Forested | S436A437K540G581 | 1 (2.4%) | ||||||
| S436G437K540A581 | 1 (2.4%) | |||||||
| S436G437K540G581 | 7(17.1%) | |||||||
| Total | 34 | 36 | 13 | 41 | ||||
| C72V73I74E75T76 | 15 (50.0%) | N86 | 10 (27.0%) | C59S108 | 7(43.8%) | A436G437E540A581 | 4 (9.3%) | |
| C72 V73M74N75K76 | 6 (20.0%) | Y86 | 27(73.0%) | R59N108 | 9(56.3%) | S436A437K540A581 | 33 (76.7%) | |
| Riverine-Plain | S72V73M74N75T76 | 9 (30.0%) | S436A437K540G581 | 1 (2.3%) | ||||
| S436G437K540G581 | 5(11.6%) | |||||||
| Total | 34 | 37 | 16 | 43 | ||||
For Pfcrt: χ2=6.808*, P=0.033; Pfmdr: χ2=0.227, P=0.634; Pfdhfr: χ2=0.513, P=0.474; Pfdhps: χ2=2.840, P=0.725
Statistically significant at 0.05 level.
Interestingly, many other mutations (other than the mutations implicated in drug resistance) were found to be segregated in the two (Pfmdr1 and Pfdhps) out of the four genes. Whereas the Pfmdr1 harbours four mutations (one in HF and three in RP), only one mutation (C228T) was common between the isolates of two ecotypes and the rest two were confined to RP ecotype (Table 3). While the A269G mutation (found in the RP ecotype) has been reported elsewhere, the other two mutations were novel (Table 3). On the other hand, the Pfdhps gene possessed as many as 11 mutations with one mutation common to the isolates from two ecotypes and the rest are unique to an ecotype (five in HF and five in RP) (Table 3). Surprisingly, the mutation that is common in both the ecotypes (T1632Z) was found in multiple isolates (seven in each ecotype) and identified to be a stop codon. The rest 10 mutations were found in individual P. falciparum isolates in two ecotypes of Deogarh district (Table 3).
Table 3.
List of novel mutations (other than the marker mutations) in the Pfmdr1 and Pfdhfr genes in the two different ecotypes of Indian P. falciparum. No such novel mutations could be detected in the Pfcrt and Pfdhfr genes in the present study.
| Genes | Nucleotide mutations and resulting changes in amino acids (in parentheses) |
|
|---|---|---|
| Hilly Forested | Riverine Plain | |
| C228T (S76S) | C228T (S76S) | |
| Pfmdr | A269G (D90G)*$ | |
| T317C (F106S)$ | ||
| T1632G (Y544Z) | T1632G (Y544Z) | |
| Cl305A(S435S)**# | Cl378T (Q460Z)$ | |
| Pfdhps | A1335G (K445K)# | A1454G (N485S)$ |
| T1373C (L458S)# | A1508G (D503G)$ | |
| A1375C (F459L)# | A1537G (I513V)$ | |
| A1426I (K476Z)# | T1685C (L562P)$ | |
Reported in earlier studies *(Okombo et al., 2014)
Unique to HF;
Unique to RP; ‘Z’ represents a stop codon.
In this study, Single Nucleotide Polymorphisms (SNPs) were identified by direct DNA sequencing of the DNA fragments in the four genes conferring drug resistance in P. falciparum. In order to ascertain if genetic differentiation exists between the isolates from two ecotypes, we segregated the DNA sequences of the four genes according to ecotypes and performed different population genetic analyses of DNA sequence variation. In general, the Pfdhps gene was found to be highly polymorphic among the four genes (Table 4). This pattern was more pronounced in the HF ecotype than in the RP. The TD values were found to be variable across the genes and between the ecotypes. Surprisingly, for Pfdhfr, high and positive values of TD were observed in isolates from both the ecotypes (Table 4). However, in no case statistically significant deviation from neutral expectation could be observed. However, in case of the Pfcrt gene, positive value of TD with statistically significant deviation from neutral model of molecular evolution could be observed in isolates from RP ecotype. The data therefore suggest that DNA sequence polymorphisms do exist among different genes conferring drug resistance in P. falciparum and also between the isolates from two different ecotypes.
Table 4.
Summary statistics of DNA sequence polymorphism in four different genes containing drug resistance in two ecotypes of P. falciparum.
| Ecotypes | Genes | No. of isolates |
Number of haplotypes |
Haplotype diversity |
Nucleotide diversity | Test of neutrality |
|
|---|---|---|---|---|---|---|---|
| π | θ | Tajima's D | |||||
| Pfcrt | 34 | 3 | 0.426 | 0.02297 | 0.01698 | 0.93263 | |
| Pfrndr | 36 | 3 | 0.367 | 0.00106 | 0.00124 | −0.28681 | |
| Hilly-Forested | Pfdhfr | 13 | 3 | 0.5 | 0.0029 | 0.00221 | 1.89943 |
| Pfdhps | 41 | 13 | 0.745 | 0.00307 | 0.00479 | −1.08329 | |
| Average | 0.5095 | 0.0075 | 0.006305 | ||||
| Pfcrt | 30 | 3 | 0.641 | 0.03218 | 0.01753 | 2.28081* | |
| Pfrndr | 37 | 5 | 0.495 | 0.00146 | 0.00247 | −0.9918 | |
| Riverine-Plain | Pfdhfr | 16 | 2 | 0.525 | 0.0036 | 0.00206 | 1.89943 |
| Pfdhps | 43 | 7 | 0.607 | 0.0026 | 0.00387 | −0.94829 | |
| Average | 0.567 | 0.00996 | 0.006483 | ||||
Statistically significant at 0.05 level.
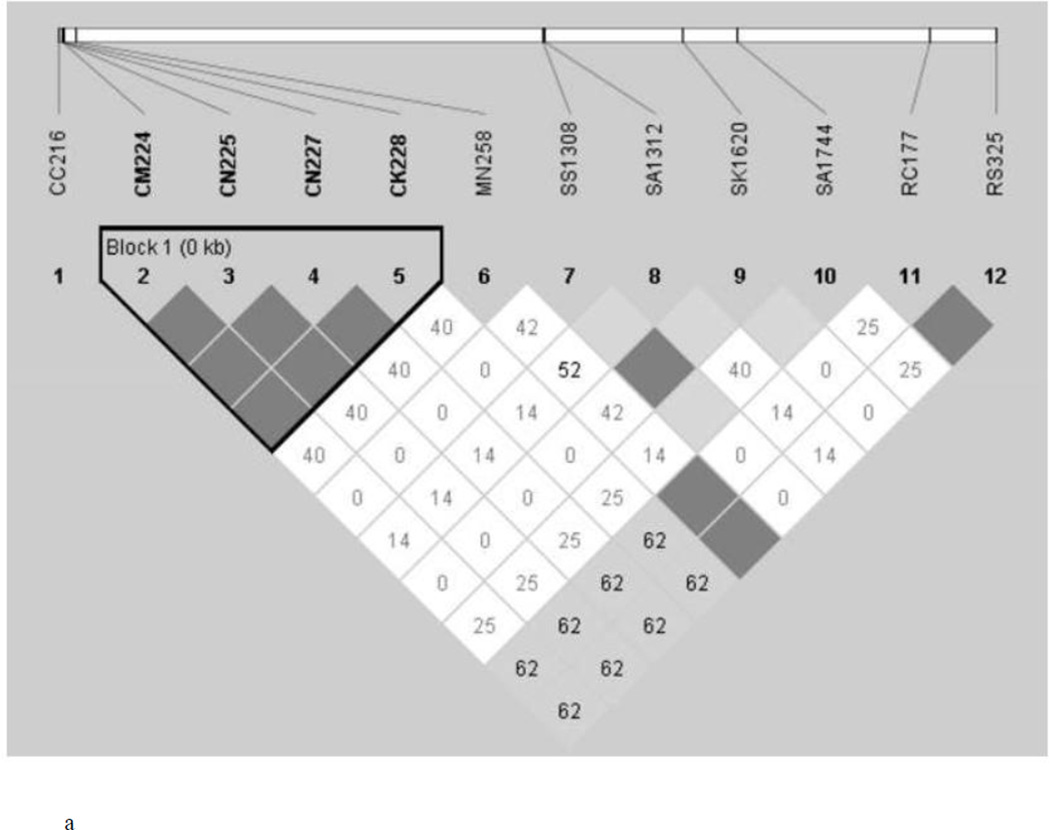
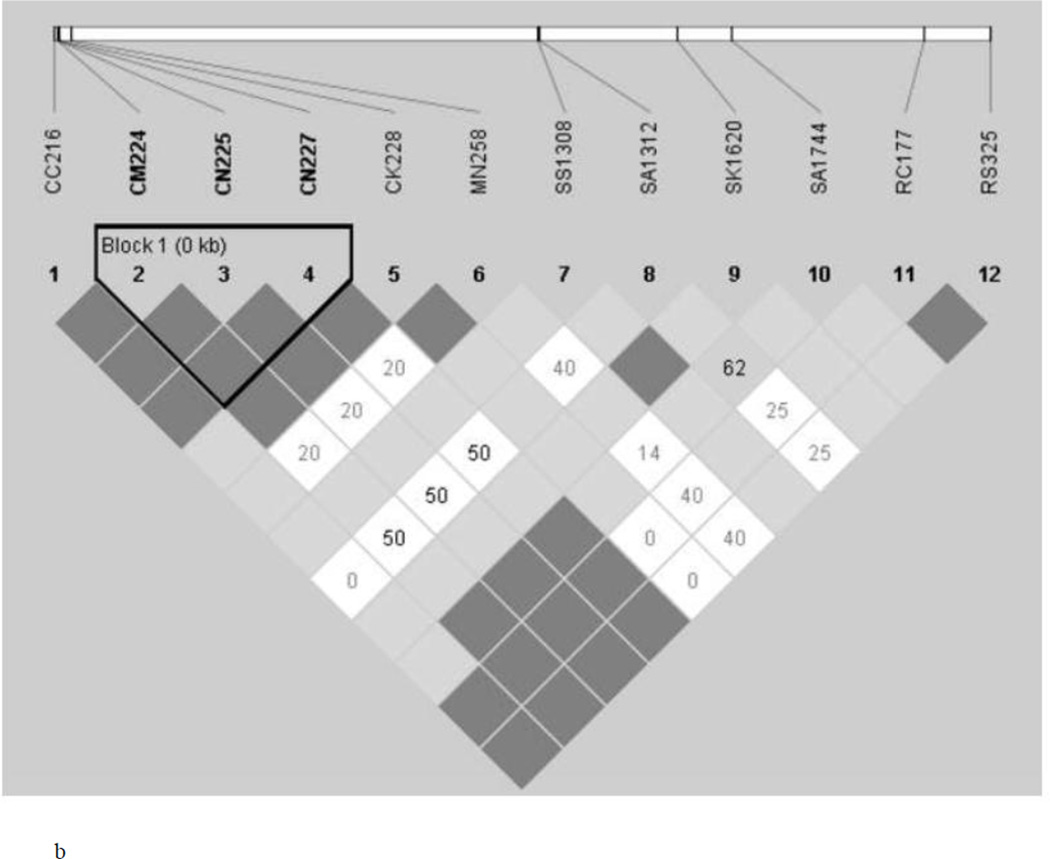
Finding of 12 isolates from each of the two ecotypes to be monoclonal in all the four genes and determination of 12 SNPs (variably present in four genes) to be segregating in the two ecotypes provides us an opportunity to determine if any two SNPs either present in a single gene or in two different genes are associated with each other. For this, we performed LD test and determined the R2 value of each SNP pair-wise association (Figure 2). In general, more number of statistically significant associations between SNP pairs was found in the RP in comparison to the HF ecotype. As expected, several statistically significant associations were found among SNPs located in the Pfcrt gene in both the ecotypes (Figure 2). Similarly, significant associations were found between SNPs located in both Pfdhfr and Pfdhps genes. Surprisingly, several cases of statistically significant associations were found between the SNPs present inside the Pfcrt gene and Pfdhfr gene and Pfdhps in RP ecotype. However, no such association could be detected in the HF ecotype. In contrast, in HF ecotype, the sole SNP of the Pfmdr1 gene was significantly associated with both the SNPs of the Pfdhfr gene; and no such association was found in the RP ecotype. The result on the whole thus indicate that while association of SNPs present in a particular gene is a common phenomenon in both the ecotypes, association of SNPs between different genes seems to be ecotype-specific.
Figure 2.
Linkage Disequilibrium (LD) between pairs of SNPs located in 4 different genes (Pfcrt, Pfmdr, Pfdhfr and Pfdhps) implicated in drug resistance in P. falciparum in Hilly-Forest (a) (HF) and (b) Riverine-Plain (RP)
2. Discussion
Considering malaria as a highly local and focal disease, local ecological conditions play vital role in malaria transmission (Conn et al., 2015; Das, 2015; Dash et al., 2008; Rath, 2004). To this extent, what amount of micro ecological settings influence malaria epidemiological outcome is poorly understood. We herewith have considered differential prevalence, frequency, and evolutionary pattern of mutations in the four genes that are known to confer drug resistance in the most dreadful malaria parasite, P. falciparum prevalent and predominant in two ecotypes (HF and RP). We have chosen these two ecotypes placed very closely (about 40 kilometres apart) to each other but with different ecological and topographical settings in a single district of Odisha state of India, which significantly contributes (about 26.9%) to malaria in India (Pradhan et al., 2016). Furthermore, it is reported that intense malaria transmission occurs in the HF, whereas the RP is moderately endemic for malaria (Kar et al., 2014; Pradhan et al., 2016). Since (i) resistance to different antimalarials is highly prevalent in India (and in Odisha), (ii) such conditions contribute considerably to malaria epidemiology, and (iii) genetic basis of drug resistance in malaria is widely established, we attempted to retrieve epidemiological information from population genetic studies of the genes conferring resistance and compare with ecological and other micro-variables between the two different ecotypes (HF and RP).
A general observation on the prevalence and distribution of different mutations associated with drug resistance in the four genes (Pfcrt, Pfmdr1, Pfdhfr, and Pfdhps) of P. falciparum is the presence of all the mutations that too in appreciable frequency in samples from both the HF and RP ecotypes. However, some deviations exist; for example, the Pfdhps gene harbours high diversity in both the type and number of mutations and therefore the corresponding haplotypes. All the four commonly reported amino acid mutations that confer resistance to sulfadoxine-pyrimethamine (S436A437K540A581) could be found in the present study, except one (A613S) that is rare in India (Biswas et al., 2000; Kumar et al., 2015). The present findings therefore substantiate similar outcomes from other Indian states including Odisha with respect to the mutational pattern at the Pfdhps gene (Sharma, D. et al., 2015). For the Pfcrt gene, three different haplotypes were found including the wild type (chloroquine sensitive) haplotype (C72V73M74N75K76), which reconfirm its prevalence in Odisha. However, in other Indian states this haplotype is very rarely found (Mishra et al., 2006; Mixson-Hayden et al., 2010). Similarly, the C72V73I74E75T76 haplotype is highly prevalent, and the frequency of the S72V73M74N75T76 haplotype is comparatively low (Table 2), reconfirming previous observations from Odisha (Okombo et al., 2014; Ramani et al., 2016; Sutar et al., 2013; Sutar et al., 2011). The skewed distributional prevalence of the C72V73I74E75T76 haplotype is reflected by the observation of statistically significant χ² value (Table 2). To be noted that the C72V73I74E75T76 type is known to confer higher resistance to CQ than the S72V73M74N75T76 type (Mittra et al., 2006), suggesting high level of CQ resistance in P. falciparum in Odisha (Ramani et al., 2016). The principal mutation conferring resistance to antimalarial at the Pfmdr1 gene (Y86) could be found in high frequency (73% in RP and 77.8% in HF; Table 2). Interestingly, not all the four amino acid substitutions (I51R59N108L164) associated with drug resistance could be found in the present study, whereas, these have been reported from India including Odisha (Sharma, D. et al., 2015). In the present study, we could find higher prevalence of the R59N108 combination (haplotype) in comparison to the C59S108 (drug sensitive type). These two mutations are considered to be highly dominate ones across many malaria endemic populations of the globe including India (Sharma, D. et al., 2015) and the other two mutations (I51L164) are reported to be surfacing in India relatively recently (Sharma, D. et al., 2015). Since it is argued that P. falciparum parasites with triple and quadruple mutations in the Pfdhfr gene are highly resistant to SP, and currently sulfadoxine-pyrimethamine is used in a combination therapy with Artemisinin, it can be noted that P. falciparum isolates in Deogarh district of Odisha is less resistant to SP. However, recent finding on the prevalence of triple mutations [(I51R59N108) or (R59N108L164)] in Odisha (Sharma, D. et al., 2015) indicate that resistance to SP is emerging in Odisha. The overall pattern of mutations associated with resistance to different antimalarials in P. falciparum in Deogarh district of Odisha indicates that (i) haplotypes associated with drug resistance in all the four genes are prevalent in isolates from both HF and RP ecotypes, (ii) the Pfdhps gene harbours a comparatively larger number of haplotypes than other three genes and (iii) no significant differences could be observed for the patterns of mutations associated with drug resistance (and their corresponding haplotypes) between the HF and RP ecotypes.
Interestingly, only two (Pfmdr1 and Pfdhps) out of the four genes harbour mutations other than the ones that are associated with drug resistance (Table 3). Based on the observed pattern of mutations in the Pfcrt gene in Indian P. falciparum, it has been previously demonstrated that this gene is under massive genetic reconstruction (Chauhan et al., 2013; Das and Dash, 2007). For the Pfdhfr gene, although we have sequenced a larger DNA fragment in comparison to the Pfcrt gene, occurrence of mutations other than the ones conferring to SP resistance is reported to be minimal (Sharma, D. et al., 2015) For the Pfmdr1 gene, three amino acid substitutions (other than the mutations associated with drug resistance) could be found; one common in isolates from both the HF and RP ecotypes, and two only confined to RP (Table 3). While the common one (C228T) and the T317C (confined to RP) are entirely novel, the A269G (confined to RP) has been reported earlier in Kenya (Okombo et al., 2014). For the Pfdhps gene, as many as 11 nucleotide substitutions (resulting in two synonymous substitutions and nine non-synonymous, including three stop codons) could be found. Out of these two synonymous substitutions one has been recently reported from Odisha (Kumar et al., 2015). On the whole, the HF contains two stop codons; two synonymous and three non-synonymous substitutions and the RP comprises two stop codons (one common with HF) and six non-synonymous substitutions. All the three non-synonymous in HF and five (out of six) non-synonymous substitutions and the three stop codons are novel and unique to that particular ecotype (Table 3); whereas with the T544Z non-synonymous substitution is common between the two ecotypes. Such an observation indicates that (i) pattern of mutations (other than the ones associated with drug resistance) in the four genes (Pfcrt, Pfmdr1, Pfdhfr and Pfdhps) are highly gene-specific, and (ii) no significant differentiation in the overall pattern could be observed between the isolates from HF and RP ecotypes.
In order to know if differential patterns of nucleotide diversity and signature of molecular evolution and association of commonly occurring SNPs in the four genes implicated in providing drug resistance in P. falciparum exist between the isolates from HF and RP ecotypes, we have conducted population genetic analyses of DNA sequence data. As found in case number and prevalence of different mutations (see above), the Pfdhps gene harbours the highest number of haplotypes (13 in HF and 7 in RP) as well as haplotype diversity among the other genes (Table 4). However, the Pfcrt gene displays the highest nucleotide diversity as measured by π (0.02297 in HF and 0.03218 in RP) among the four genes. High and positive TD values could be observed in case of the Pfcrt and Pfdhfr genes in both the ecotypes, with highest values of 2.28081 (Pfcrt-RP) (Table 4). This value is statistically significantly deviated from the expectation under neutral model of molecular evolution. Since positive TD values indicate evidence of balancing selection (Chauhan et al., 2014), it seems that alleles of both the Pfcrt and the Pfdhfr genes are maintained in stable frequencies. In case of Pfdhps, the TD values are high and negative in both the ecotypes, but not statistically significantly deviated from the neutral equilibrium model. This might indicate an initial genetic hitchhiking in the presence of large effective population size of P. falciparum (due to high malaria transmission in Odisha) as observed earlier in Pfmdr1 gene in Odisha (Chauhan et al., 2014) and in case of microsatellite polymorphisms in case of Pfcrt gene in India (Chauhan et al., 2013). To be noted that such evolutionary events (invoked by both natural selection and demography) might have created high haplotype diversity in the Pfdhps gene, as suggested by Chauhan et al. 2013.
The test of linkage disequilibrium between a pair of SNP yielded interesting results (Figure 2). As expected, all the four mutations in the Pfcrt gene are found to be strongly linked in P. falciparum isolates sampled from both the HF and RP ecotypes. However, in the RP ecotype, all the four mutations of the Pfcrt gene were statistically significantly associated with mutations of the Pfmdr1, Pfdhfr, and Pfdhps genes. In the HF ecotype, however, significant LD was observed between SNPs of the Pfmdr1 and Pfdhfr genes. The N86Y mutation is widely prevalent in almost all malaria endemic countries of the globe including India (Mita et al., 2009; Thomsen et al., 2013) and often found to be associated with mutations in the Pfcrt gene (Chauhan et al., 2014). Therefore, differential associations of mutations among the four genes implicated in drug resistance in the two ecotypes seem to be highly ecotype-specific. Different ecological and topographical conditions prevailing in the two different ecotypes might have contributed to the observed differential association of SNPs present in different genes. However, distributional prevalence of different species of Anopheles (vectors to malaria parasites) (An. fluviatilis in HF and An. culicifacies in RP) might also have contributed to the observation (Nanda et al., 2000; Sharma et al., 2006; Sorosjinda-Nunthawarasilp and Bhumiratana, 2014). It is known that individuals adjust their genomes through new genetic associations for adaptation to adverse eco-environmental conditions (Van Tyne et al., 2011). Whether the observed patterns of mutations and their evolution in the isolates from HF and RP ecotypes are propelled by adaptation by natural selection could not be established from the present study, but it could be ascertained that differential evolutionary forces (exerted by drug pressure in the field) are in operation at the four different genes implicated in drug resistance in the malaria parasite P. falciparum in the Deogarh district of Odisha.
In conclusion, the results of the present study, although limited to a single district of a state that contributes the highest number of cases and deaths due to malaria in India, provide many meaningful insights into the patterns of mutations in the four genes implicated in drug resistance in the malaria parasite, P. falciparum. Although in many aspects, there were no significant differences between the two ecotypes (HF and RP), but some underlying differences could be noted. Considering the amount of genetic diversity to be associated with the intensity of malaria transmission in India (Chauhan et al., 2014; Sharma, D. et al., 2015), the isolates from HF ecotype harbours more average genetic diversity (as estimated by nucleotide diversities, π and θ) than those from the RP. Further, the observed associations between pairs of SNPs present in four different genes could be due to different drug pressure applied in the field due to different treatment practices. The results from genetic studies indicate that CQ resistant genotypes are approaching fixation level and SP resistant genotypes are evolving very fast, and the existing drug policy needs to be reviewed, as SP is a partner drug administered with artemisinin. However, genetic studies of this kind in wide areas (e.g. the whole Odisha state) including different micro ecological settings with different malaria transmission patterns are needed before a conclusive decision on change in drug policy.
Supplementary Material
Highlights.
Patterns of genetic variations in genes conferring drug-resistance in micro eco-epidemiological settings in malaria are not known
We performed SNP analyses in four different drug-resistant genes in P. falciparum form two different eco-epidemiological locations of Odisha state of India.
Results revealed higher genetic variation in general in all the four genes in the Hilly-Forest ecosystem than the Riverine-Plain one
More cases of genetic linkage between different drug-resistant genes were evident in Riverine-Plain ecosystem.
High observed genetic variation and genetic linkage suggest strong resistance capabilities of P. falciparum in Odisha, calling for change in drug policy
Acknowledgments
We are obliged to the Director of the National Institute of Malaria Research for facilitating the study, the NIMR staff who helped in the field and laboratory. Special thanks to Dr. Prasant Mallick to help in PCR trouble shooting. We are very grateful to the Chief District Malaria Officer of Deogarh district and his staff for facilitating the fieldwork. This study was supported by NIH/National Institute of Allergy and Infectious Diseases award U19AI089676, and by an NIH/Fogarty International Centre Global Infectious Disease research-training grant D43TW007884. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the Fogarty International Centre or the National Institutes of Health. NPK is a Ph. D. student of the Goa University, Goa, India and extends his gracious thanks to Professor R. Roy, Head of the Department of Zoology, Professor S. N. Bole, Dean Faculty of Life Sciences and Environment and Rd. S. R. Shetty, the Vice Chancellor of Goa University. This manuscript bears the NIMR publication screening committee approval no. 66/2015.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
3. Competing interests
The authors declare that they have no competing interests.
4. Authors’ contributions
NPK, NN, JMC, and AK designed the study. NPK led the survey team, collected samples, and data and did experiments. NPK, AD, and KC analysed the data. NPK, NN, JMC, AD, AK, and KC wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Narayani Prasad Kar, Email: narayani_d1@yahoo.co.in.
Kshipra Chauhan, Email: kshipra.chauhan@gmail.com.
Nutan Nanda, Email: nutanmrc@yahoo.co.in.
Ashwani Kumar, Email: ashwani07@gmail.com.
Jane M. Carlton, Email: carltj01@nyu.edu.
Aparup Das, Email: aparup@mrcindia.org.
References
- Awasthi G, Prasad G, Das A. Population genetic analyses of Plasmodium falciparum chloroquine receptor transporter gene haplotypes reveal the evolutionary history of chloroquine-resistant malaria in India. Int. J. Parasitol. 2011;41:705–709. doi: 10.1016/j.ijpara.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Babiker HA, Pringle SJ, Abdel-Muhsin A, Mackinnon M, Hunt P, Walliker D. High-level chloroquine resistance in Sudanese isolates of Plasmodium falciparum is associated with mutations in the chloroquine resistance transporter gene pfcrt and the multidrug resistance Gene pfmdr1. J. Infect. Dis. 2001;183:1535–1538. doi: 10.1086/320195. [DOI] [PubMed] [Google Scholar]
- Barrett JC. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;2:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Biswas S, Escalante A, Chaiyaroj S, Angkasekwinai P, Lal AA. Prevalence of point mutations in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum isolates from India and Thailand: a molecular epidemiologic study. Trop Med Int Health. 2000;5:737–743. doi: 10.1046/j.1365-3156.2000.00632.x. [DOI] [PubMed] [Google Scholar]
- Brooks DR, Wang P, Read M, Watkins WM, Sims PF, Hyde JE. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase: dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur. J. Biochem. 1994;224:397–405. doi: 10.1111/j.1432-1033.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- Brown TS, Jacob CG, Silva JC, Takala-Harrison S, Djimdé A, Dondorp AM, Fukuda M, Noedl H, Nyunt MM, Kyaw MP. Plasmodium falciparum field isolates from areas of repeated emergence of drug resistant malaria show no evidence of hypermutator phenotype. Infect., Genet. Evol. 2015;30:318–322. doi: 10.1016/j.meegid.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Volkman SK, Uplekar S, Hupalo DN, Alves JMP, Cui L, Donnelly M, Roos DS, Harb OS, Acosta M. Population genetics, evolutionary genomics, and genome-wide studies of malaria: A view across the international centers of excellence for malaria research. Am. J. Trop. Med. Hyg. 2015;93:87–98. doi: 10.4269/ajtmh.15-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chareonviriyaphap T, Bangs MJ, Ratanatham S. Status of malaria in Thailand. Southeast Asian J Trop Med Public Health. 2000;31:225–237. [PubMed] [Google Scholar]
- Chauhan K, Pande V, Das A. Analyses of genetic variations at microsatellite loci present in-and-around the Pfcrt gene in Indian Plasmodium falciparum. Infect., Genet. Evol. 2013;20:476–487. doi: 10.1016/j.meegid.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Chauhan K, Pande V, Das A. DNA sequence polymorphisms of the pfmdr1 gene and association of mutations with the pfcrt gene in Indian Plasmodium falciparum isolates. Infect., Genet. Evol. 2014;26:213–222. doi: 10.1016/j.meegid.2014.05.033. [DOI] [PubMed] [Google Scholar]
- Conn JE, Norris DE, Donnelly MJ, Beebe NW, Burkot TR, Coulibaly MB, Chery L, Eapen A, Keven JB, Kilama M. Entomological monitoring and evaluation: diverse transmission settings of ICEMR projects will require local and regional malaria elimination strategies. Am. J. Trop. Med. Hyg. 2015;93:28–41. doi: 10.4269/ajtmh.15-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Mharakurwa S, Ndiaye D, Rathod PK, Rosenthal PJ. Antimalarial Drug Resistance: Literature Review and Activities and Findings of the ICEMR Network. Am. J. Trop. Med. Hyg. 2015;93(3 Suppl):57–68. doi: 10.4269/ajtmh.15-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. The distinctive features of Indian malaria parasites. Trends Parasitol. 2015;31:83–86. doi: 10.1016/j.pt.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Das A, Anvikar AR, Cator LJ, Dhiman RC, Eapen A, Mishra N, Nagpal BN, Nanda N, Raghavendra K, Read AF. Malaria in India: the center for the study of complex malaria in India. Acta Trop. 2012;121:267–273. doi: 10.1016/j.actatropica.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Dash AP. Evolutionary paradigm of chloroquine-resistant malaria in India. Trends Parasitol. 2007;23:132–135. doi: 10.1016/j.pt.2007.01.012. http://dx.doi.org/10.1016/j.pt.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Das A, Mohanty S, Stephan W. Inferring the population structure and demography of Drosophila ananassae from multilocus data. Genetics. 2004;168:1975–1985. doi: 10.1534/genetics.104.031567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash A, Valecha N, Anvikar A, Kumar A. Malaria in India: challenges and opportunities. J. Biosci. (Bangalore) 2008;33:583–592. doi: 10.1007/s12038-008-0076-x. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Naudé B, Deitsch KW, Su X-z. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote S, Kyle D, Martin R, Oduola A, Forsyth K, Kemp D, Cowman A. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature. 1990;345:255–258. doi: 10.1038/345255a0. [DOI] [PubMed] [Google Scholar]
- Ghanchi NK, Ursing J, Beg MA, Veiga MI, Jafri S, Martensson A. Prevalence of resistance associated polymorphisms in Plasmodium falciparum field isolates from southern Pakistan. Malar. J. 2011;10:18. doi: 10.1186/1475-2875-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta B, Gupta P, Sharma A, Singh V, Dash AP, Das A. High proportion of mixed-species Plasmodium infections in India revealed by PCR diagnostic assay. Trop Med Int Health. 2010;15:819–824. doi: 10.1111/j.1365-3156.2010.02549.x. [DOI] [PubMed] [Google Scholar]
- Hakre S, Masuoka P, Vanzie E, Roberts DR. Spatial correlations of mapped malaria rates with environmental factors in Belize, Central America. Int J Health Geogr. 2004;3:1. doi: 10.1186/1476-072X-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque U, Sunahara T, Hashizume M, Shields T, Yamamoto T, Haque R, Glass GE. Malaria prevalence, risk factors and spatial distribution in a hilly forest area of Bangladesh. PLoS One. 2011;6:e18908. doi: 10.1371/journal.pone.0018908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings IM. Malaria control and the evolution of drug resistance: an intriguing link. Trends Parasitol. 2003;19:4. doi: 10.1016/s1471-4922(02)00017-x. [DOI] [PubMed] [Google Scholar]
- Hastings IM, Watkins WM. Intensity of malaria transmission and the evolution of drug resistance. Acta Trop. 2005;94:218–229. doi: 10.1016/j.actatropica.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Ingasia LA, Cheruiyot J, Okoth SA, Andagalu B, Kamau E. Genetic variability and population structure of Plasmodium falciparum parasite populations from different malaria ecological regions of Kenya. Infect., Genet. Evol. 2015;39:372–380. doi: 10.1016/j.meegid.2015.10.013. [DOI] [PubMed] [Google Scholar]
- Jambulingam P, Mohapatra S, Govardhini P, Das LK, Manoharan A, Pani S, Das P. Microlevel epidemiological variations in malaria & its implications on control strategy. Indian J. Med. Res. 1991;93:371–378. [PubMed] [Google Scholar]
- Johnston SP, Pieniazek NJ, Xayavong MV, Slemenda SB, Wilkins PP, da Silva AJ. PCR as a confirmatory technique for laboratory diagnosis of malaria. J. Clin. Microbiol. 2006;44:1087–1089. doi: 10.1128/JCM.44.3.1087-1089.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewwaen W, Bhumiratana A. Landscape ecology and epidemiology of malaria associated with rubber plantations in Thailand: integrated approaches to malaria ecotoping. Interdiscip. Perspect. Infect. Dis. 2015;2015:1–7. doi: 10.1155/2015/909106. http://dx.doi.org/10.1155/2015/909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaga T, Ohta S. Ecophysiological and climatological effects on distribution of vector species and malaria incidence in India. Int. J. Env. Res. Public Health. 2012;9:4704–4714. doi: 10.3390/ijerph9124704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar N, Kumar A, Singh O, Carlton J, Nanda N. A review of malaria transmission dynamics in forest ecosystems. Parasit Vectors. 2014;7:12. doi: 10.1186/1756-3305-7-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser J, Singer BH, Utzinger J. Reducing the burden of malaria in different eco-epidemiological settings with environmental management: a systematic review. Lancet Infect Dis. 2005;5:695–708. doi: 10.1016/S1473-3099(05)70268-1. [DOI] [PubMed] [Google Scholar]
- Klein E. Antimalarial drug resistance: a review of the biology and strategies to delay emergence and spread. Int. J. Antimicrob. Agents. 2013;41:311–317. doi: 10.1016/j.ijantimicag.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Chery L, Biswas C, Dubhashi N, Dutta P, Dua VK, Kacchap M, Kakati S, Khandeparkar A, Kour D, Mahajan SN, Maji A, Majumder P, Mohanta J, Mohapatra PK, Narayanasamy K, Roy K, Shastri J, Valecha N, Vikash R, Wani R, White J, Rathod PK. Malaria in South Asia: Prevalence and control. Acta Trop. 2012;121:246–255. doi: 10.1016/j.actatropica.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Moirangthem R, Gahlawat SK, Chandra J, Gupta P, Valecha N, Anvikar A, Singh V. Emergence of sulfadoxine-pyrimethamine resistance in Indian isolates of Plasmodium falciparum in the last two decades. Infect., Genet. Evol. 2015;36:190–198. doi: 10.1016/j.meegid.2015.08.027. [DOI] [PubMed] [Google Scholar]
- Kumar A, Valecha N, Jain T, Dash AP. Burden of malaria in India: retrospective and prospective view. Am. J. Trop. Med. Hyg. 2007;77:69–78. [PubMed] [Google Scholar]
- Li J, Chen J, Xie D, Eyi UM, Matesa RA, Obono MMO, Ehapo CS, Yang L, Yang H, Lin M. Molecular mutation profile of Pfcrt and Pfmdr1 in Plasmodium falciparum isolates from Bioko Island, Equatorial Guinea. Infect., Genet. Evol. 2015;36:552–556. doi: 10.1016/j.meegid.2015.08.039. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Malakooti MA, Biomndo K, Shanks GD. Reemergence of epidemic malaria in the highlands of western Kenya. Emerging Infect. Dis. 1998;4:671–676. doi: 10.3201/eid0404.980422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malisa A, Pearce R, Mutayoba B, Abdullah S, Mshinda H, Kachur P, Bloland P, Roper C. Quantification of markers of antimalarial drug resistance from an area of high malaria transmission: Comparing frequency with prevalence. Afr. J. Biotechnol. 2016;11:13250–13260. [Google Scholar]
- Mallick PK, Singh R, Singh OP, Singh AK, Bhasin VK, Valecha N. Reduced heterozygosity at intragenic and flanking microsatellites of pfcrt gene establishes natural selection based molecular evolution of chloroquine-resistant Plasmodium falciparum in India. Infect., Genet. Evol. 2013a;20:407–412. doi: 10.1016/j.meegid.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Mallick PK, Sutton PL, Singh R, Singh OP, Dash AP, Singh AK, Carlton JM, Bhasin VK. Microsatellite analysis of chloroquine resistance associated alleles and neutral loci reveal genetic structure of Indian Plasmodium falciparum. Infect., Genet. Evol. 2013b;19:164–175. doi: 10.1016/j.meegid.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manguin S, Garros C, Dusfour I, Harbach R, Coosemans M. Bionomics, taxonomy, and distribution of the major malaria vector taxa of Anopheles subgenus Cellia in Southeast Asia: an updated review. Infect., Genet. Evol. 2008;8:489–503. doi: 10.1016/j.meegid.2007.11.004. [DOI] [PubMed] [Google Scholar]
- McCollum AM, Schneider KA, Griffing SM, Zhou Z, Kariuki S, Ter-Kuile F, Shi YP, Slutsker L, Lal AA, Udhayakumar V, Escalante AA. Differences in selective pressure on dhps and dhfr drug resistant mutations in western Kenya. Malar. J. 2012;11:77. doi: 10.1186/1475-2875-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Raj DK, Hazra R, Dash A, Supakar PC. An efficient PCR-SSCP-based method for detection of a chloroquine resistance marker in the PfCRT gene of Plasmodium falciparum. Trans. R. Soc. Trop. Med. Hyg. 2006;100:243–247. doi: 10.1016/j.trstmh.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Mita T, Tanabe K, Kita K. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol. Int. 2009;58:201–209. doi: 10.1016/j.parint.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Mittra P, Vinayak S, Chandawat H, Das MK, Singh N, Biswas S, Dev V, Kumar A, Ansari MA, Sharma YD. Progressive increase in point mutations associated with chloroquine resistance in Plasmodium falciparum isolates from India. J. Infect. Dis. 2006;193:1304–1312. doi: 10.1086/502979. [DOI] [PubMed] [Google Scholar]
- Mixson-Hayden T, Jain V, McCollum AM, Poe A, Nagpal AC, Dash AP, Stiles JK, Udhayakumar V, Singh N. Evidence of selective sweeps in genes conferring resistance to chloroquine and pyrimethamine in Plasmodium falciparum isolates in India. Antimicrob. Agents Chemother. 2010;54:997–1006. doi: 10.1128/AAC.00846-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty A, Swain S, Singh DV, Mahapatra N, Kar SK, Hazra RK. A unique methodology for detecting the spread of chloroquine-resistant strains of Plasmodium falciparum, in previously unreported areas, by analyzing anophelines of malaria endemic zones of Orissa, India. Infect., Genet. Evol. 2009;9:462–467. doi: 10.1016/j.meegid.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Mwai L, Kiara SM, Abdirahman A, Pole L, Rippert A, Diriye A, Bull P, Marsh K, Borrmann S, Nzila A. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob. Agents Chemother. 2009;53:5069–5073. doi: 10.1128/AAC.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagesha HS, Casey GJ, Rieckmann KH, Fryauff DJ, Laksana BS, Reeder JC, Maguire JD, Baird JK. New haplotypes of the Plasmodium falciparum chloroquine resistance transporter (pfcrt) gene among chloroquine-resistant parasite isolates. Am. J. Trop. Med. Hyg. 2003;68:398–402. [PubMed] [Google Scholar]
- Nanda N, Yadav R, Subbarao SK, Joshi H, Sharma V. Studies on Anopheles fluviatilis and Anopheles culicifacies sibling species in relation to malaria in forested hilly and deforested riverine ecosystems in northern Orissa, India. J. Am. Mosq. Control Assoc. 2000;16:199–205. [PubMed] [Google Scholar]
- Nath DC, Mwchahary DD. Malaria Prevalence in Forest and Nonforest Areas of Kokrajhar District of Assam. ISRN Public Health. 2012;2012:9. [Google Scholar]
- Nei M. Molecular evolutionary genetics, illustrated, reprint ed. Columbia: Columbia university press; 1987. [Google Scholar]
- Okombo J, Kamau AW, Marsh K, Sutherland CJ, Ochola-Oyier LI. Temporal trends in prevalence of Plasmodium falciparum drug resistance alleles over two decades of changing antimalarial policy in coastal Kenya. Int. J. Parasitol. 2014;4:152–163. doi: 10.1016/j.ijpddr.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okwa O, Akinmolayan F, Carter V, Hurd H. Transmission dynamics of malaria in Nigeria. Ann. Afr. Med. 2009;8:1–9. doi: 10.4103/1596-3519.55756. [DOI] [PubMed] [Google Scholar]
- Pelleau S, Moss EL, Dhingra SK, Volney B, Casteras J, Gabryszewski SJ, Volkman SK, Wirth DF, Legrand E, Fidock DA. Adaptive evolution of malaria parasites in French Guiana: Reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc. Natl. Acad. Sci. U. S. A. 2015;112:11672–11677. doi: 10.1073/pnas.1507142112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc. Natl. Acad. Sci. U. S. A. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan A, Anasuya A, Pradhan MM, Kavitha A, Kar P, Sahoo KC, Panigrahi P, Dutta A. Trends in Malaria in Odisha, India—An Analysis of the 2003–2013 Time-Series Data from the National Vector Borne Disease Control Program. PLoS One. 2016;11:e0149126. doi: 10.1371/journal.pone.0149126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price R, Cassar C, Brockman A, Duraisingh M, Van Vugt M, White N, Nosten F, Krishna S. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob. Agents Chemother. 1999;43:2943–2949. doi: 10.1128/aac.43.12.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani S, Parija SC, Mandal J, Hamide A, Bhat V. Detection of chloroquine and artemisinin resistance molecular markers in Plasmodium falciparum: A hospital based study. Tropical Parasitology. 2016;6:69. doi: 10.4103/2229-5070.175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramar M, Mohanty SC, Sahu SS, Gunasekaran K, Jambulingam P. Epidemiological Investigation for the Incidence of Plasmodium falciparum Malaria from Tribal Areas in Koraput District of Odisha State, India. Am. J. Epi. Infect. Dis. 2014;2:109–114. [Google Scholar]
- Rao MRK, Padhy RN, Das MK. Prevalence of dengue viral and malaria parasitic co-infections in an epidemic district, Angul of Odisha, India: An eco-epidemiological and cross-sectional study for the prospective aspects of public health. J Infect Public Health. 2015 doi: 10.1016/j.jiph.2015.10.019. http://dx.doi.org/10.1016/j.jiph.2015.10.019. [DOI] [PubMed]
- Rath SS. Indian tribes and their urgent and emerging health status: an overview. In: Kalla AK, Joshi PC, editors. National Workshop on Emerging Issues in Tribal, Health Medicines. Concept Pub. Co: New Delhi; 2004. pp. 46–70. [Google Scholar]
- Reeder JC, Rieckmann KH, Genton B, Lorry K, Wines B, Cowman AF. Point mutations in the dihydrofolate reductase and dihydropteroate synthetase genes and in vitro susceptibility to pyrimethamine and cycloguanil of Plasmodium falciparum isolates from Papua New Guinea. Am. J. Trop. Med. Hyg. 1996;55:209–213. doi: 10.4269/ajtmh.1996.55.209. [DOI] [PubMed] [Google Scholar]
- Reisen W, Pradhan S, Shrestha J, Shrestha S, Vaidya R, Shrestha J. Anopheline mosquito (Diptera: Culicidae) ecology in relation to malaria transmission in the inner and outer terai of Nepal, 1987–1989. J. Med. Entomol. 1993;30:664–682. doi: 10.1093/jmedent/30.4.664. [DOI] [PubMed] [Google Scholar]
- Rosa-Freitas MG, Tsouris P, Peterson AT, Honorio NA, de Barros FS, de Aguiar DB, Gurgel Hda C, de Arruda ME, Vasconcelos SD, Luitgards-Moura JF. An ecoregional classification for the state of Roraima, Brazil: the importance of landscape in malaria biology. Mem. Inst. Oswaldo Cruz. 2007;102:349–357. doi: 10.1590/s0074-02762007005000052. [DOI] [PubMed] [Google Scholar]
- Rouhani M, Zakeri S, Pirahmadi S, Raeisi A, Djadid ND. High prevalence of pfdhfr, pfdhps triple mutations associated with anti-malarial drugs resistance in Plasmodium falciparum isolates seven years after the adoption of sulfadoxine-pyrimethamine in combination with artesunate as first-line treatment in Iran. Infect., Genet. Evol. 2015;31:183–189. doi: 10.1016/j.meegid.2015.01.020. [DOI] [PubMed] [Google Scholar]
- Sahu SS, Gunasekaran K, Vanamail P, Jambulingam P. Persistent foci of falciparum malaria among tribes over two decades in Koraput district of Odisha State, India. Malar. J. 2013;12:72. doi: 10.1186/1475-2875-12-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira A, Boutsika K. Malaria ecotypes and stratification. Adv. Parasitol. 2012;78:97–167. doi: 10.1016/B978-0-12-394303-3.00001-3. [DOI] [PubMed] [Google Scholar]
- Seng CM, Matusop A, Sen FK. Differences in Anopheles composition and malaria transmission in the village settlements and cultivated farming zone in Sarawak, Malaysia. Southeast Asian J. Trop. Med. Public Health. 1999;30:454–459. [PubMed] [Google Scholar]
- Sharma D, Lather M, Mallick PK, Adak T, Dang AS, Valecha N, Singh OP. Polymorphism in drug resistance genes dihydrofolate reductase and dihydropteroate synthase in Plasmodium falciparum in some states of India. Parasit Vectors. 2015;8:1–9. doi: 10.1186/s13071-015-1080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SK, Tyagi PK, Padhan K, Upadhyay AK, Haque MA, Nanda N, Joshi H, Biswas S, Adak T, Das BS, Chauhan VS, Chitnis CE, Subbarao SK. Epidemiology of malaria transmission in forest and plain ecotype villages in Sundargarh District, Orissa, India. Trans. R. Soc. Trop. Med. Hyg. 2006;100:917–925. doi: 10.1016/j.trstmh.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Sharma VP, Dev V, Phookan S. Neglected Plasmodium vivax malaria in northeastern States of India. Indian J. Med. Res. 2015;141:546–55. doi: 10.4103/0971-5916.159511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla R, Sharma S, Dhiman R. Seasonal prevalence of malaria vectors and its relationship with malaria transmission in three physiographic zones in Uttaranchal state, India. J. Vector Borne Dis. 2007;44:75–77. [PubMed] [Google Scholar]
- Shukla RP, Sharma SN, Nanda N, Dhiman RC, Dash AP. Malaria Persistence in Kumaon Foothills of District Nainital, Uttarakhand, India. J. Am. Mosq. Control Assoc. 2008;24:214–218. doi: 10.2987/5567.1. [DOI] [PubMed] [Google Scholar]
- Singh N, Dash AP, Thimasarn K. Fighting malaria in Madhya Pradesh (Central India): Are we loosing the battle? Malar. J. 2009;8:1–8. doi: 10.1186/1475-2875-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Mishra AK, Chand SK, Bharti PK, Singh MP, Nanda N, Singh OP, Sodagiri K, Udhyakumar V. Relative Abundance and Plasmodium Infection Rates of Malaria Vectors in and around Jabalpur, a Malaria Endemic Region in Madhya Pradesh State, Central India. PLoS One. 2015;10:e0126932. doi: 10.1371/journal.pone.0126932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Singh O, Sharma V. Dynamics of malaria transmission in forested and deforested regions of Mandla District, central India (Madhya Pradesh) J. Am. Mosq. Control Assoc. -Mosquito News. 1996;12:225–234. [PubMed] [Google Scholar]
- Singh V, Mishra N, Awasthi G, Dash AP, Das A. Why is it important to study malaria epidemiology in India? Trends Parasitol. 2009;25:5. doi: 10.1016/j.pt.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Sirawaraporn W, Sathitkul T, Sirawaraporn R, Yuthavong Y, Santi DV. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1124–1129. doi: 10.1073/pnas.94.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorosjinda-Nunthawarasilp P, Bhumiratana A. Ecotope-based entomological surveillance and molecular xenomonitoring of multidrug resistant malaria parasites in Anopheles vectors. Interdiscip. Perspect. Infect. Dis. 2014 doi: 10.1155/2014/969531. http://dx.doi.org/10.1155/2014/969531. [DOI] [PMC free article] [PubMed]
- Srivastava P, Ratha J, Shah NK, Mishra N, Anvikar AR, Sharma SK, Das MK, Srivastava B, Valecha N. A clinical and molecular study of artesunate+ sulphadoxine-pyrimethamine in three districts of central and eastern India. Malar. J. 2013;12:247–252. doi: 10.1186/1475-2875-12-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutar SK, Dhangadamajhi G, Kar SK, Ranjit M. Molecular monitoring of antimalarial drug resistance among Plasmodium falciparum field isolates from Odisha, India. Acta Trop. 2013;126:84–87. doi: 10.1016/j.actatropica.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Sutar SK, Gupta B, Ranjit M, Kar SK, Das A. Sequence analysis of coding DNA fragments of pfcrt and pfmdr-1 genes in Plasmodium falciparum isolates from Odisha, India. Mem Inst Oswaldo Cruz. 2011;106:78–84. doi: 10.1590/s0074-02762011000100013. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talisuna AO, Langi P, Bakyaita N, Egwang T, Mutabingwa TK, Watkins W, Van Marck E, D'Alessandro U. Intensity of malaria transmission, antimalarial-drug use and resistance in Uganda: what is the relationship between these three factors? Trans R Soc Trop Med Hyg. 2002;96:310–317. doi: 10.1016/s0035-9203(02)90108-2. [DOI] [PubMed] [Google Scholar]
- Thomsen TT, Madsen LB, Hansson HH, Tomás EV, Charlwood D, Bygbjerg IC, Alifrangis M. Rapid selection of Plasmodium falciparum chloroquine resistance transporter gene and multidrug resistance gene-1 haplotypes associated with past chloroquine and present artemether-lumefantrine use in Inhambane District, southern Mozambique. Am. J. Trop. Med. Hyg. 2013;88:536–541. doi: 10.4269/ajtmh.12-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T, Menting JG, Wilson C, Cowman AF. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13944–13949. doi: 10.1073/pnas.94.25.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderramos SG, Valderramos JC, Musset L, Purcell LA, Mercereau-Puijalon O, Legrand E, Fidock DA. Identification of a mutant PfCRT-mediated chloroquine tolerance phenotype in Plasmodium falciparum. PLoS Path. 2010;6:e1000887. doi: 10.1371/journal.ppat.1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tyne D, Park DJ, Schaffner SF, Neafsey DE, Angelino E, Cortese JF, Barnes KG, Rosen DM, Lukens AK, Daniels RF. Identification and functional validation of the novel antimalarial resistance locus PF10_0355 in Plasmodium falciparum. PLoS Genet. 2011;7:e1001383. doi: 10.1371/journal.pgen.1001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. On the number of segregating sites in wbination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
Web references
- http://nrhmorissa.gov.in/mis/SearchDetail.aspx, 10. 03. 2016.
- http://www.malariasite.com/tag/orissa/, 10. 03. 2016.
- http://www.ordistricts.nic.in/district_profile/aboutus.php, 10. 03. 2016.
- http://www.odishasampad.in/, 10. 03. 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.