Abstract
Systemic delivery of mRNA-based therapeutics remains a challenging issue for preclinical and clinical studies. Here, we describe new lipid-like nanoparticles (TT-LLNs) developed through an orthogonal array design, which demonstrates improved delivery efficiency of mRNA encoding luciferase in vitro by over 350-fold with significantly reduced experimental workload. One optimized TT3 LLN, termed O-TT3 LLNs, was able to restore the human factor IX (hFIX) level to normal physiological values in FIX-knockout mice. Consequently, these mRNA based nanomaterials merit further development for therapeutic applications.
Keywords: Lipid-like nanoparticles, mRNA delivery, orthogonal array design, hemophilia, genetic disorders
Graphical abstract
Messenger RNA (mRNA) based therapeutics have shown great promise for expressing functional antibodies and proteins.1–5 Clinical studies have explored mRNA for use as vaccines through local administration of naked mRNA or mRNA-transfected dendritic cells in order to induce antigen-specific immune responses.1,6–8 Recently, extensive efforts have been devoted to achieving the systemic delivery of mRNA using liposomes, polymeric nanoparticles, and mRNA-protein complexes.9–13 Although significant advances have been made, new mRNA carriers optimization issues remain in order to improve delivery efficiency and maximize therapeutic windows of mRNA therapeutics in different human conditions.
Previously, lipid-like nanoparticles (LLNs) have demonstrated efficient delivery of small interfering RNA (siRNA) in rodents and nonhuman primates.14–18 siRNA and mRNA possess common physicochemical properties, including components of nucleic acids and negative charges; therefore, LLNs may also serve well as mRNA delivery materials. However, LLNs-assisted mRNA delivery is relatively unexplored and understanding of this system is very limited. Herein, we report a new class of N1,N3,N5-tris(2-aminoethyl)benzene-1,3,5-tricarboxamide (TT) derived LLNs for mRNA delivery (Figure 1). Our previous studies offered a useful set of starting design criteria including a six-member ring core structure, as well as linker and lipid chain length.14,15 On the basis of these experiences, TTs are designed to consist of a phenyl ring, three amide linkers, and three amino lipid chains (Figure 1a). An orthogonal experimental design was utilized in order to optimize the formulation of lead material TT3 LLNs, which improved delivery efficiency over 350-fold with significantly reduced experimental workload. Moreover, correlation analysis of TT LLNs properties and mRNA translation identified key determinants of LLNs properties for mRNA delivery. In addition, PEGylation of TT3 LLNs showed dramatic effects on particle stability, particle size, and mRNA delivery efficiency. Consistent with in vitro observations, an optimized TT3 LLN (O-TT3 LLNs) efficiently delivered mRNA encoding human factor IX (hFIX) and produced hFIX at a therapeutically relevant level in both wild-type and FIX-knockout mice. This study offers new insights into the development of mRNA delivery materials.
Figure 1.
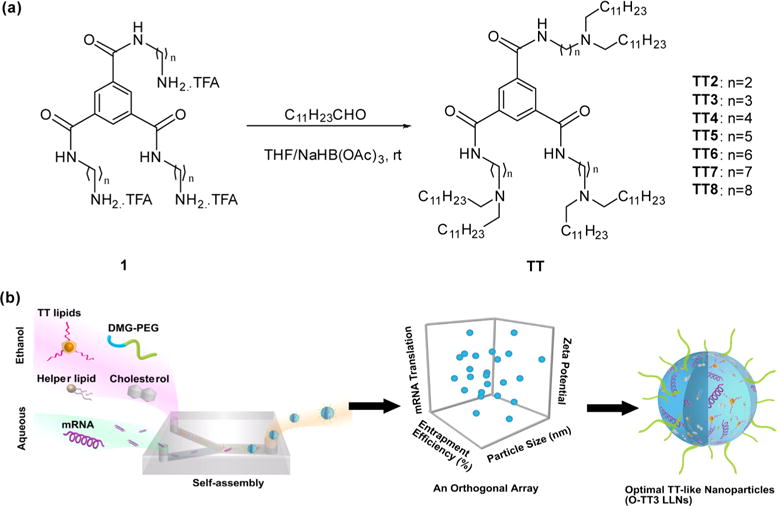
(a) A synthetic route to N1,N3,N5-tris(2-aminoethyl)benzene-1,3,5-tricarboxamide derivatives (TT2–TT8). Compound 1 underwent a reductive amination in order to afford desired products TT2–TT8. (b) Illustration of material development for mRNA delivery. TT2–TT8 were formulated with helper lipids, cholesterol (Chol), DMG-PEG2000, and mRNA to form TT2–TT8 LLNs via pipetting for in vitro studies or using Precision NanoSystems for in vivo studies. An orthogonal experimental design was utilized to optimize the formulation of a lead material TT3 LLNs.
First, we designed a synthetic route to N1,N3,N5-tris(2-aminoethyl)benzene-1,3,5-tricarboxamide (TT) derivatives (Supporting Information Figure S1 and Figure 1a). Benzene-1,3,5-tricarbonyl trichloride (2) was reacted with Boc-protected diamine (3) in order to produce the intermediates (4).19 Deprotection of (4) gave compound (1), which underwent reductive amination to afford the desired products TT2 through TT8.14 The structures of TT2–TT8 were confirmed by 1H NMR spectroscopy and mass spectrometry (Supporting Information). On the basis of our previous formulation experience for siRNA,14,20 newly synthesized TT2–TT8 were then formulated with 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol (Chol), 1,2-dimyristoyl-sn-glycerol, methoxypolyethylene glycol (DMG-PEG2000) (TT/DOPE/Chol/DMG-PEG2000 = 50/10/38.5/1.5, mole ratio) as well as mRNA encoding firefly luciferase (FLuc mRNA) in order to form TT2–TT8 LLNs (Figure 1b). Meanwhile, particle properties including size, zeta potential, and entrapment efficiency of TT2–TT8 LLNs were measured using a dynamic light-scattering instrument and a ribogreen assay (Supporting Information Figure S2).15,21 Particle size of TT2–TT8 LLNs ranged from 99 ± 2 to 178 ± 1 nm with PDI < 0.2. Most TT LLNs were positively charged and entrapment efficiency of mRNA was in the range of 15–82%.
Next, we evaluated delivery efficiency and cytotoxicity of TT2–TT8 LLNs-FLuc mRNA in Hep3B cells, a human hepatoma cell line. As shown in Figure 2a, TT3 LLNs showed significantly higher expression of the firefly luciferase compared to other TT LLNs at a dose of 1.2 μg/mL of FLuc mRNA. In addition, TT2–TT8 LLNs showed minimal to moderate inhibitory effects on Hep3B cells (Supporting Information Figure S2). A correlation analysis was then performed between transfection efficiency and particle size, surface charge, entrapment efficiency, and cell viability (Figure 2c–e). A significant positive correlation between transfection efficiency and entrapment efficiency was observed, while there was no significant correlation between transfection efficiency and particle size, surface charge, and cell viability.
Figure 2.
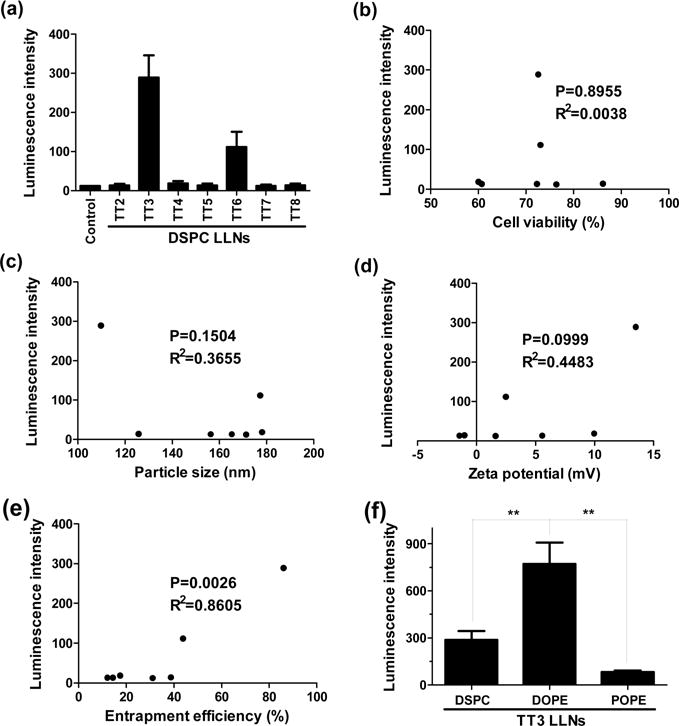
Delivery efficiency of TT2–TT8 LLNs to Hep3B cells (a human hepatoma cell line) and correlation analysis between transfection efficiency and cell viability, particle size, zeta potential, and entrapment efficiency. (a) TT3 LLNs showed significantly higher expression of the firefly luciferase compared to other TT LLNs at a dose of 1.2 μg/mL of luciferase mRNA. (b–e) Correlation analysis between transfection efficiency of TT LLNs and cell viability, particle size, zeta potential, or entrapment efficiency. A significant correlation between transfection efficiency and entrapment efficiency was observed, while no significant correlation between transfection efficiency and particle size, surface charge, and cell viability. (f) Effects of helper lipids on mRNA delivery efficiency. DOPE formulated TT3 LLNs were more potent than DSPC and POPE formulated TT3 LLNs. (triplicate; **, P < 0.01; t test, double-tailed).
To study the effects of helper lipids on delivery efficiency, we formulated the lead material, TT3 with 2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) or 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) in the presence of other formulation components. DOPE formulated TT3 LLNs were more potent than DSPC and POPE formulated TT3 LLNs (Figure 2f). On the basis of the above results, the formulation components of TT3 LLNs were determined as TT3, DOPE, cholesterol, and DMG-PEG2000.
In order to optimize the formulation of TT3 LLNs, we applied an orthogonal experimental design, which is a well-established approach for biomedical studies.22–27 This approach enabled us to fine-tune the formulation ratios with a minimum number of experiments. As shown in Figure 3a, we investigated four levels for each formulation component: TT3, DOPE, cholesterol, and DMG-PEG2000. Theoretically, four levels of four formulation components can yield 256 combinations. By utilizing the orthogonal experimental design, we were able to evaluate the effects of the 4 components with only 16 combinations (Supporting Information Table S1a and Figure S3). Because the impact of the four components was unknown, Figure 3a displayed the arbitrarily assigned four levels for each component based on the molar ratio (50/10/38.5/1.5) tested in Figure 2. From the first round of optimization, we ranked the effects of the four components (TT3 > DOPE > Chol = DMG-PEG2000) by comparing the ΔK values (Supporting Information Table S1b). Luciferase expression of TT3 LLNs was increased over 6-fold after the first round of optimization (TT3 LLNs 1–13 vs TT3-DSPC LLNs, Figure 3b). In order to further optimize the formulation ratio and improve mRNA delivery efficiency, we conducted a second round of orthogonal optimization. Figure 3c–f provided guidance for assigning four levels of each component (Figure 4a). The trend in Figure 3c showed that TT3 with reduced levels may facilitate mRNA delivery. Hence, we gradually decreased the ratio of TT3 from 40 to 10. TT3 exhibited the highest impact on delivery efficiency in the first orthogonal array (Supporting Information Table S1b); therefore, two levels of TT3 were overlapped with that in the first round of optimization. We then increased the ratio of DOPE from 10 to 40 and decreased the ratio of DMG-PEG2000 from 0.75 to 0 based on the trend in Figure 3d and 3f. One level of DOPE and DMG-PEG2000 was overlapped with that in the first round of optimization in order to avoid discontinuity. Lastly, because the peak of cholesterol was between 18.5 and 38.5 (Figure 3e) its level was reassigned with a reduced interval.
Figure 3.
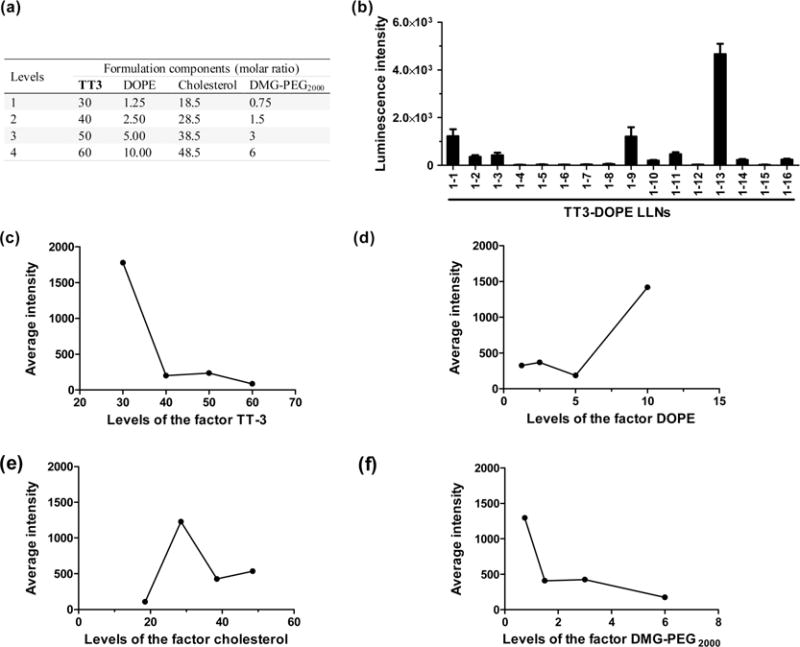
A first round of orthogonal experimental design and analysis. (a) Four levels for each formulation component: TT3, DOPE, cholesterol, and DMG-PEG2000. (b) Sixteen combinations (1–1 to 1–16) were evaluated with Hep3B cells for their relative luminescence intensity through the first orthogonal array. (c–f) The impact trend of TT3 (c), DOPE (d), cholesterol (e), and DMG-PEG2000 (f) on delivery efficiency. Increased TT3 and DMG-PEG2000 reduced mRNA delivery efficiency, while increased DOPE facilitated mRNA delivery efficiency. The optimal ratio for cholesterol ranged from 20 to 40.
Figure 4.
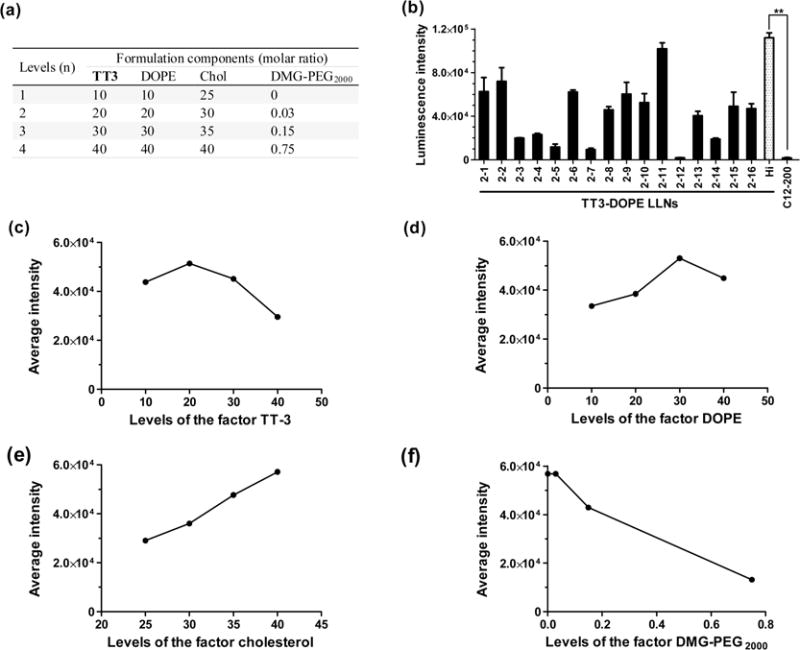
A second round of orthogonal experimental design and analysis. (a) Four levels for each formulation component: TT3, DOPE, cholesterol, and DMG-PEG2000. (b) Sixteen combinations (2–1 to 2–16) were evaluated with Hep3B cells or their relative luminescence intensity through the second orthogonal array. The most efficient formulation was validated with a designated code of Hi-TT3 LLNs (formulation ratio is TT3/DOPE/Chol/DMG-PEG2000 = 20/30/40/0). (triplicate; **, P < 0.01; t test, double-tailed). (c–f) The impact trend of TT3 (c), DOPE (d), cholesterol (e), and DMG-PEG2000 (f) on delivery efficiency.
In the second round of orthogonal optimization, 16 formulation combinations were evaluated as described above (Figure 4, Supporting Information Table S2a and Figure S5). Figure 4b–f displayed the relative intensity of luciferase expression and the impact trend of the four components. The predicted best formulation was identified by selecting the highest Kn values of these four components (Supporting Information Table S2b, highlighted in bold), which was TT3/DOPE/Chol/DMG-PEG2000 = 20/30/40/0 (named Hi-TT3 LLNs). Hi-TT3 LLNs were further evaluated by their luciferase expression level (Figure 4b), which showed comparable delivery efficiency to the formulation of TT3 LLNs 2–11 (TT3/DOPE/Chol =30/30/40) (Figure 4b and Supporting Information Table S2a). Cationic lipids have potential toxicity;28 therefore, Hi-TT3 LLNs with the lower percentage of TT3 were selected for further studies. Importantly, Hi-TT3 LLNs increased delivery efficiency over 20-fold compared to the best formulation (TT3 LLNs 1–13) identified in the first round of orthogonal optimization and over 350-fold compared to the original start-point TT3-DSPC LLNs (Figure 4b). More importantly, Hi-TT3 LLNs were over 65-fold more efficient than C12-200-DSPC LLNs, a previously reported material.15 These results indicate that an orthogonal experimental design represents a powerful approach to the goal of optimizing nanoparticle formulations. Consistent with previous findings (Supporting Information Figure S4), significant correlation was observed between transfection efficiency and entrapment efficiency, while there was no significant correlation with particle size and cell viability in the two rounds of orthogonal optimization (Supporting Information S6). Interestingly, zeta potential also showed significant correlation with transfection efficiency in the second round of orthogonal experiments (Supporting Information Figure S6).
Previous studies report that PEGlyation of polymer-based nanoparticles significantly affects the stability and cellular uptake.29,30 Interestingly, we noticed that Hi-TT3 LLNs are not stable without the incorporation of DMG-PEG2000 in the formulation. We then investigated the impact of DMG-PEG2000 on delivery efficiency, particle size, and stability. Consistent with reports in the literature,29,30 the results showed that the ratio of DMG-PEG2000 was negatively correlated with delivery efficiency and particle size; that is the higher ratio of DMG-PEG2000, the lower the luciferase expression and the smaller the particles will be (Figure 5a,b). The particle size of TT3 LLNs increased dramatically 5 h after formulation with a low ratio of DMG-PEG2000 (Figure 5b). When formulated with the molar ratio TT3/DOPE/Chol/DMG-PEG2000 = 20/30/40/0.75 (named O-TT3 LLNs), these nanoparticles were stable for a minimum of 2 weeks (Figure 5c). In order to balance delivery efficiency with particle stability, we chose the formulation O-TT3 LLNs for further studies. A Cryo-EM image showed the spherical morphology of O-TT3 LLNs (Figure 5d). To virtualize the cellular uptake of O-TT3 LLNs, we treated Hep3B cells using O-TT3 LLNs loaded with Alexa-Fluor 647-labeled RNA and FLuc mRNA (weight ratio: 1/1). Three hours after treatment, cells were fixed with formaldehyde. Cell membranes and nuclei were then stained by Alexa fluor 488 conjugate of wheat germ agglutinin (green) and NucBlue fixed cell ready probes reagent (blue), respectively. Compared to untreated and free RNA-treated cells, significant cellular uptake of TT3 LLNs was observed (Figure 6). Reflecting all of the above results, we selected O-TT3 LLNs formulation (TT3/DOPE/Chol/DMG-PEG2000 = 20/30/40/0.75) for in vivo studies.
Figure 5.
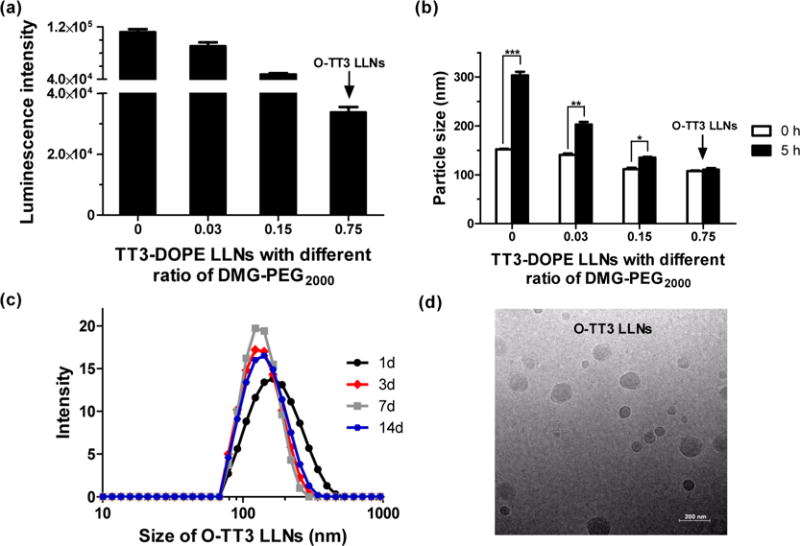
Impact of PEGlyation on TT3 LLNs. The ratio of DMG-PEG2000 was negatively correlated with delivery efficiency in Hep3B cells (a) and particle size (b). TT3 LLNs were increasingly stable with addition of DMG-PEG2000. (triplicate; *, P < 0.05; **, P < 0.01; ***, P < 0.001; t test, double-tailed). (c) O-TT3 LLNs were stable for at least 2 weeks. (d) A representative Cryo-TEM image of O-TT3 LLNs. Scale bar: 200 nm.
Figure 6.
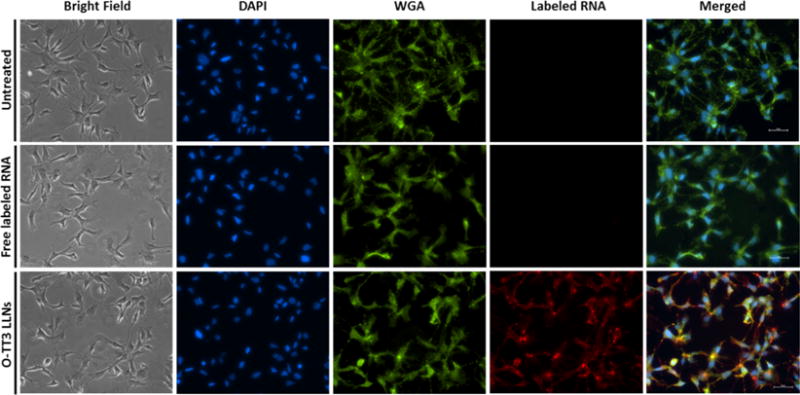
Cellular uptake of O-TT3 LLNs. Cell nuclei and membranes of Hep3B cells were stained with DAPI (blue) and WGA (green), respectively. Alexa-Fluor 647-labeled RNA (red). Scale bar: 50 μm.
To understand biodistribution of O-TT3 LLNs in vivo, we injected O-TT3 FLuc LLNs intravenously at an mRNA dose of 0.5 mg/kg with control groups of free FLuc mRNA, C12-200-DSPC LLNs, and the original TT3-DSPC LLNs. Six hours post administration, we measured bioluminescence intensity of dissected organs using the IVIS imaging system. O-TT3 LLNs-treated group showed significantly higher bioluminescence signal in the liver and spleen compared to C12-200-DSPC LLNs, and TT3-DSPC LLNs-treated groups. No signal was detected in the kidney, lung, and heart (Supporting Information Figure S7). These in vivo results further validated that in vitro optimizations of TT3 LLNs was an effective approach.
To further study the delivery efficiency of O-TT3 LLNs in vivo, we selected an mRNA-encoding human factor IX (hFIX), a blood clotting factor, and therapeutically relevant protein.31 Deficiency of hFIX protein leads to the inherited genetic disorder, hemophilia B, which impairs the process of hemostasis and results in serious complications, including joint and muscle hemorrhage.31–33 In this study, we formulated O-TT3 LLNs with hFIX mRNA and then injected the formulation intravenously in wild-type mice. Six hours after administration, the level of hFIX was measured by a well-established chromogenic ELISA assay.34 Wild-type mice produced 1020 ng/mL hFIX at a dose of 0.55 mg/kg and 2057 ng/mL at a dose of 1.1 mg/kg, respectively. No hFIX was detected in the plasma of mice injected with untreated, free hFIX mRNA-, or TT3 LLNs-treated groups (Figure 7a). Meanwhile, we observed no significant alterations in clinical appearance in the O-TT3 LLNs treated groups compared to the control groups. Histopathology analysis was consistent with these observations, which suggested that O-TT3 LLNs were well tolerated at the current dose (Supporting Information Figure S8). Next, we tested the delivery efficiency of O-TT3 LLNs in FIX-knockout mice.35 Compared to O-TT3 FLuc LLNs, O-TT3 hFIX LLNs induced significant production of hFIX in a dose-dependent manner (608 ng/mL at a dose of 0.55 mg/kg and 1740 ng/mL at a dose of 1.1 mg/kg) in the FIX-knockout mice (Figure 7b). Lastly, we quantified the hFIX activity using a well-described chromogenic assay in which the coagulation factor IX is activated by activated Factor eleven (FXIa) with simultaneous activation of Factor ten (FX) in the presence of Factor eight (FVIII), phospholipid and Ca2+ followed by the hydrolysis of a chromogenic FXa substrate.34 The normal plasma level of hFIX in humans ranges from 500 to 1500 mIU/mL.36,37 As shown in Figure 7c, the plasma level of hFIX was 304 mIU/mL at a dose of 0.55 mg/kg and 791 mIU/mL at a dose of 1.1 mg/kg in the O-TT3 hFIX LLNs treated group, while no detectable signal was found in O-TT3 FLuc LLNs treated FIX-knockout mice. Taken together, we concluded that O-TT3 hFIX LLNs were indeed capable of restoring the hFIX to normal physiological values in FIX-knockout mice, which demonstrates that this delivery system has the potential for therapeutic applications.
Figure 7.
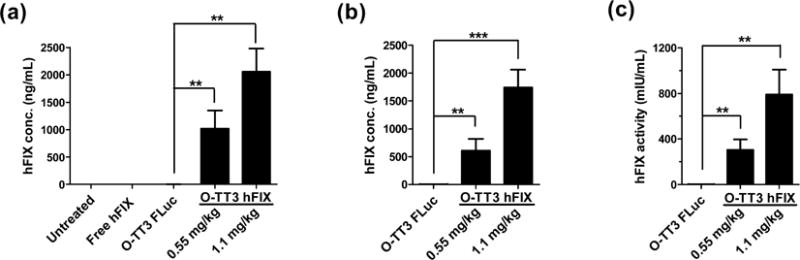
hFIX level in wild-type (a) and FIX knockout (b) mice 6h after intravenous administration. (O-TT3 FLuc, FLuc mRNA formulated O-TT3 LLNs; O-TT3 hFIX, human FIX mRNA formulated O-TT3 LLNs). (c) Human FIX protein activity. hFIX activity is 304 mIU/mL (0.55 mg/kg) and 791 mIU/mL (1.1 mg/kg) in O-TT3 hFIX LLNs treated group, while no detectable activity was found in O-TT3 FLuc LLNs treated FIX-knockout mice. (n = 3; **, P < 0.01; ***, P < 0.001; t test, double-tailed).
In summary, we have developed a new type of lipid-like compounds N1,N3,N5-tris(2-aminoethyl)benzene-1,3,5-tricarboxamide derivatives (TT2–TT8), composed of a phenyl ring, three amide linkers, and three amino lipid chains. We utilized an orthogonal experimental design for formulation optimization, which is an efficient approach to the rapid evaluation of multiple formulation parameters and the identification of the best formulation ratio with significantly reduced experimental numbers (32 out of 512). More importantly, this approach enabled us to improve the transfection efficiency over 2 orders of magnitude after two rounds of optimization. In addition, the experimental array obtained through orthogonal methods represents the independency of all formulation combinations, thus offering a series of reliable data to perform correlation analysis. The analysis results indicate that entrapment efficiency of mRNA plays a key role for cellular transfection. Zeta potential may correlate with mRNA delivery efficiency for certain LLNs formulations. These results validate the usefulness of the orthogonal experiment design in order to optimize multicomponent nanoparticles for mRNA delivery in future material development. Meanwhile, we show that PEGylation of TT3 LLNs improved particle stability and reduced particle size but hindered delivery efficiency, which is consistent with observations of other polymer-based drug delivery systems reported in the literature.29,30 After the ratio of DMG-PEG2000 was optimized, O-TT3 LLNs showed over 110-fold higher delivery efficiency compared to the start-point TT3-DSPC LLNs, and O-TT3 LLNs were stable for over 2 weeks. Moreover, O-TT3 LLNs showed much higher delivery efficiency of FLuc mRNA both in vitro and in vivo, compared to a C12-200-DSPC LLNs reported previously.15 Lastly, we evaluated the delivery efficiency of O-TT3 LLNs for a therapeutically relevant mRNA encoding hFIX (1662 nucleotides), which possesses comparable length to mRNA-encoding luciferase (1929 nucleotides). In the case that O-TT3 LLNs are utilized to deliver mRNAs with significantly different length, the formulation ratio needs to be further tuned and optimized. Our results showed that O-TT3 LLNs efficiently delivered hFIX mRNA in both wild-type and FIX-knockout mice. Most importantly, O-TT3 LLNs fully recovered the level of hFIX (791 mIU/mL at 1.1 mg/kg) to normal physiological values (500–1500 mIU/mL) in FIX-knockout mice. These results demonstrate that O-TT3 LLNs are a promising mRNA delivery system with the potential for broad therapeutic applications, including protein replacement, gene engineering, and immunotherapy.
Experimental Details
Materials
mRNAs encoding Firefly luciferase (FLuc mRNA) and human factor IX mRNA (hFIX mRNA) were purchased from TriLink Biotechnologies, Inc. (San Diego, CA). Alexa fluor 488 conjugate of wheat germ agglutinin, NucBlue Fixed cell ready probes DAPI, ProLong diamond antifade mountant reagent, Ribogreen reagent, and fetal bovine serum (FBS) were purchased from Life Technologies (Grand Island, NY). Alexa-Fluor 647-labeled RNA was purchased from Integrated DNA Technologies. 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) were purchased from Avanti Polar Lipids, Inc. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide(MTT) was purchased from Amresco (Solon, OH). Bright-Glo luciferase assay substrate was from Promega (Madison, WI). Buffered formaldehyde (10%, pH 7.4) was purchased from Ricca Chemical (Arlington, TX). Goat anti-hFIX HRP antibody was purchased from Enzyme Research Laboratories (South Bend, IN). O-Phenylenediaminedihydrochloride (15 mg substrate per tablet), cholesterol, human factor IX, monoclonal anti-human factor IX antibody, and other chemicals were purchased from Sigma-Aldrich.
Synthesis of TT2–TT8
To a suspension of compound 1 (0.1 mmol) in 10 mL of anhydrous tetrahydrofuran was added triethylamine (0.4 mmol) under nitrogen protection. The mixture was stirred for 30 min at RT. After adding dodecyl aldehyde (0.9 mmol) and NaBH(OAc)3, the reaction mixture was stirred at RT for 48 h. After the solvent was removed, the residue was purified by column chromatography using a CombiFlash Rf system with a RediSep Gold Resolution silica column (Teledyne Isco) with gradient elution from 100% CH2Cl2 to CH2Cl2/MeOH/NH4OH (75/22/3 by volume) to give TT2–TT8.
Formulation of mRNA-loaded TT LLNs
TT2–TT8 were formulated with the helper lipid (1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), 2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) or 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE)), cholesterol, 1,2-dimyristoyl-sn-glycerol, methoxypolyethylene glycol (DMG-PEG2000) (molar ratio 50/10/38.5/1.5 or based on the orthogonal design table, Supporting Information), and FLuc mRNA via pipetting for in vitro studies or via a microfluidic based mixing device (Precision NanoSystems) for in vivo studies.23 After formulation, the freshly formed mRNA-LLNs were used immediately for cell transfection. For in vivo studies, the freshly prepared LLNs were then dialyzed against PBS buffer using Slide-A-Lyzer dialysis cassettes (3.5 K MWCO, Life Technologies, Grand Island, NY). Particle size and zeta potential of LLNs were measured using a NanoZS Zetasizer (Malvern, Worcestershire, U.K.) at a scattering angle of 173° and a temperature of 25 °C. Entrapment efficiency of LLNs was determined using the Ribogreen assay reported previously.15,21
TT LLNs-Mediated Luciferase Transfection Assay
The human hepatocellular carcinoma cell line Hep3B was purchased from American Type Culture Collection (Manassas, VA) and maintained at 37 °C with 5% CO2 in Eagle’s Minimum Essential Medium (EMEM) supplemented with 10% heat inactivated FBS. Hep3B cells were seeded (2 × 104 cells per well) into each well of white 96-well plates in 150 μL of culture medium, allowed to attach overnight in growth medium, and transfected by addition of 20 μL of FLuc mRNA-loaded TT LLNs to growth medium. Transfections were performed in triplicate. After 6 h of transfection, culture medium containing TT LLNs was carefully removed, and 50 μL of serum-free EMEM and 50 μL of Bright-Glo luciferase substrate were mixed and added to each well. Five minutes later, the relative luminescence intensity was measured with the SpectraMax M5 microplate reader (Molecular Devices, LLC., Sunnyvale, CA). Free FLuc mRNA served as a negative control.
Orthogonal Array Experimental Design
In order to identify optimal molar ratio of the formulation components, an orthogonal array experiment design was utilized. Four formulation components were assigned in the following orthogonal experiments with a fixed TT/mRNA ratio (10/1). TT3-DOPE LLNs 1–1 to 1–16 were prepared according to the orthogonal array design table L16(4)4 and used to transfect Hep3B cells (Supporting Information Table S1a). The average luminescence intensity (Kn) of each factor in the same level (n = 1, 2, 3, and 4) and the difference (ΔK) between the highest and lowest values of each factor were used to evaluate the impact of the levels and factors to the transfection efficiency, respectively. On the basis of the result of the first round optimization, the levels of each factor were further fine-tuned, and the second round orthogonal experiment with 16 formulations TT3-DOPE LLNs 2–1 to 2–16 was conducted (Supporting Information Table S2a). A similar analysis was performed as described above. Lastly, the best formulation ratio was predicted, which was validated by the luciferase transfection assay.
Cryo-Transmission Electron Microscopy (Cryo-TEM)
Cryo-TEM samples were prepared by applying a small aliquot (3 μL) of O-TT3 LLNs to a specimen grid. After blotting away excess liquid, the grid was immediately plunged into liquid ethane to rapidly form a thin film of amorphous ice using Vitrobot Mark IV system (FEI, Hillsboro, OR). The grid was transferred under liquid nitrogen in cryo-transfer station to a Gatan 626 cryo-transfer holder (Gatan, Pleasanton, CA). Cryo-transfer holder was loaded to a Tecnai F20 S/TEM (FEI, Hillsboro, OR) and maintained at −173 °C. Cryo-TEM images were recorded under low dose conditions at a magnification of 18 500× on a postcolumn 1k × 1k CCD camera.
Cytotoxicity Assay
Hep3B cells were grown in 96-well plates in 150 μL of medium 24 h prior to treatment at a density of 2 × 104 cells/well. After 6 h incubation with free FLuc mRNA or LLNs, 17 μL of MTT (5 mg/mL solution in PBS) was added to each well and the cells were then incubated for 4 h at 37 °C. Then the medium was removed, and 150 μL of dimethyl sulfoxide was added. After shaking for 10 min, the absorbance at a wavelength of 570 nm was measured on a SpectraMax M5 microplate reader. Cell viability of TT LLNs was normalized by untreated cells.
Cellular Uptake of O-TT3 LLNs
Cells were plated on sterile glass coverslips (22 mm) in a 6-well plate at 8 × 104 cells/well. After overnight culture, cells on the coverslips were treated with PBS, Alexa-Fluor 647-labeled RNA or O-TT3-LLN containing FLuc mRNA and Alexa-Fluor 647-labeled RNA (weight ratio 1/1) for 3 h. Cells were then rinsed three times with PBS and fixed with 4% formaldehyde for 10 min at RT. After washing twice with PBS, cells were incubated with Alexa-Fluor 488 conjugate of wheat germ agglutinin (1 μg/μL) and NucBlue fixed cell ready probes DAPI at RT for 10 min to stain membranes and nucleus. The cells were finally mounted onto glass slides (75 × 25 mm) with a drop of ProLong diamond antifade mountant reagent. All images were acquired using an ECLIPSE Ti inverted fluorescence microscopy (Nikon, Japan).
Biodistribution of O-TT3 LLNs
All procedures used in animal studies conducted at The Ohio State University were approved by the Institutional Animal Care and Use Committee (IACUC) and were also consistent with local, state, and federal regulations as applicable. C57BL/6 mice (6–8 weeks old from the Jackson Laborator) were administered intravenously via tail vein injection of free FLuc mRNA, C12-200-DSPC LLNs, TT3-DSPC LLNs, or O-TT3 LLNs at a dose of 0.5 mg/kg (mRNA concentration, n = 3). After 6 h, mice were i.p. injected with 150 μL of the D-luciferin substrate (30 mg/mL) and euthanized in a CO2 chamber 8 min after injection. Bioluminescence signals in the dissected liver, spleen, kidney, heart, and lung were immediately measured using a Xenogen IVIS imaging system (Caliper, Alameda, CA), and signal strength of individual tissues was normalized against tissue weight.
In Vivo hFIX Expression and Histological Analysis
38C57BL/6 mice were administered intravenously via tail vein injection with free hFIX mRNA, O-TT3 FLuc (an irrelevant mRNA-loaded LLNs as a control), or O-TT3 hFIX at the indicated dosage. Six hours post administration, blood samples were collected and mixed with an anticoagulant solution (3.2% sodium citrate anticoagulant containing 0.17 mg/mL of corn trypsin inhibitor) in a ratio of 9:1, which was then centrifuged for 15 min at 2500 g. hFIX protein level was measured by enzyme-linked immunosorbent (ELISA) assay. Briefly, a 96-well immunoplate was coated overnight at 4 °C with 50 μL/well of mouse anti-hFIX antibody (1:1000 dilution) in 100 mM of bicarbonate/carbonate coating buffer (pH 9.2). After blocking with 200 μL of 6% BSA at RT for 3 h, each well was incubated with 50 μL of diluted mouse serum at RT for 2h. Human FIX bounded to the wells was detected by incubating with 100 μL goat anti-hFIX HRP antibody diluted 1:4000 at RT for 1 h. After that, 100 μL of substrate O-phenylenediaminedihydrochloride at 2 mg/mL was added at RT for 8 min and then the reaction was immediately stopped by adding 50 μL of 3 M H2SO4. The absorbance at 492 nm was determined using the SpectraMax M5 microplate reader. Levels of human FIX were calculated through a standard curve generated from the standard human FIX. The same protocol was used for FIX-knockout mice. Additionally, hFIX bioactivity was determined by using a chromogenic kit according to the manufacturer’s protocol (Rossix, Mölndal, Sweden). Histopathology on the heart, kidney, liver, lung, and spleen were processed, stained with hematoxylin and eosin (H&E), and imaged using an ECLIPSE Ti inverted fluorescence microscopy (Nikon, Japan).
Supplementary Material
Acknowledgments
This work was supported by the Early Career Investigator Award from Bayer Hemophilia Awards Program, Research Reward from Trilink Biotechnologies, as well as the startup-fund from the College of Pharmacy at the Ohio State University. We acknowledge Dr. Mark A. Kay, Dr. Adi Barzel, Dr. Inder M. Verma, Dr. Mingjun Zhang, Dr. Dennis Bong, Dr. Zhun Zhou for providing assistance in material characterization and mouse models. We acknowledge the use of the core facility provided by the Comparative Pathology and Mouse Phenotyping Shared Resource (CPMPSR) at the Ohio State University (NCI Grant P30 CA016058). We acknowledge Precision NanoSystems, Inc. for providing technical support for material formulation.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nano-lett.5b03528.
General synthetic procedures, spectral data, physicochemical properties of LLNs, correlation analysis between transfection efficiency of LLNs and physicochemical parameters, bioluminescence distribution, and histological analysis. (PDF)
Notes
The authors declare the following competing financial interest(s): B.A.K. is on advisory boards for Novo Nordisk, Bayer Healthcare US, and Baxalta and receives research support from CSL Behring. A.L.D. serves as a consultant for Baxalta, Bayer, Biogen, CSL Behring, Novo Nordisk, Pfizer and has received honoraria from Bayer, Baxalta, CSL Behring, NovoNordisk and Pfizer. A.L.D. also receives research support from Baxalta, Bayer, Biogen, Novo Nordisk and CSL Behring.
References
- 1.Pascolo S. Handb Exp Pharmacol. 2008;183:221–235. doi: 10.1007/978-3-540-72167-3_11. [DOI] [PubMed] [Google Scholar]
- 2.Sahin U, Kariko K, Tureci O. Nat Rev Drug Discovery. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 3.Andries O, Kitada T, Bodner K, Sanders NN, Weiss R. Expert Rev Vaccines. 2015;14:313–331. doi: 10.1586/14760584.2015.997714. [DOI] [PubMed] [Google Scholar]
- 4.Phua KK, Nair SK, Leong KW. Nanoscale. 2014;6:7715–7729. doi: 10.1039/c4nr01346h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tavernier G, Andries O, Demeester J, Sanders NN, De Smedt SC, Rejman J. J Controlled Release. 2011;150:238–247. doi: 10.1016/j.jconrel.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Weide B, Carralot JP, Reese A, Scheel B, Eigentler TK, Hoerr I, Rammensee HG, Garbe C, Pascolo S. J Immunother. 2008;31:180–188. doi: 10.1097/CJI.0b013e31815ce501. [DOI] [PubMed] [Google Scholar]
- 7.Weide B, Pascolo S, Scheel B, Derhovanessian E, Pflugfelder A, Eigentler TK, Pawelec G, Hoerr I, Rammensee HG, Garbe C. J Immunother. 2009;32:498–507. doi: 10.1097/CJI.0b013e3181a00068. [DOI] [PubMed] [Google Scholar]
- 8.Rittig SM, Haentschel M, Weimer KJ, Heine A, Muller MR, Brugger W, Horger MS, Maksimovic O, Stenzl A, Hoerr I, Rammensee HG, Holderried TA, Kanz L, Pascolo S, Brossart P. Mol Ther. 2011;19:990–999. doi: 10.1038/mt.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Su HH, Yang Y, Hu Y, Zhang L, Blancafort P, Huang L. Mol Ther. 2013;21:358–367. doi: 10.1038/mt.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phua KK, Leong KW, Nair SK. J Controlled Release. 2013;166:227–233. doi: 10.1016/j.jconrel.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su X, Fricke J, Kavanagh DG, Irvine DJ. Mol Pharmaceutics. 2011;8:774–787. doi: 10.1021/mp100390w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K, Cu Y, Beard CW, Brito LA, Krucker T, O’Hagan DT, Singh M, Mason PW, Valiante NM, Dormitzer PR, Barnett SW, Rappuoli R, Ulmer JB, Mandl CW. Proc Natl Acad Sci USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng C, Convertine AJ, Stayton PS, Bryers JD. Biomaterials. 2012;33:6868–6876. doi: 10.1016/j.biomaterials.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, Bogorad RL, Yin H, Chen Y, Vegas AJ, Alabi CA, Sahay G, Olejnik KT, Wang W, Schroeder A, Lytton-Jean AK, Siegwart DJ, Akinc A, Barnes C, Barros SA, Carioto M, Fitzgerald K, Hettinger J, Kumar V, Novobrantseva TI, Qin J, Querbes W, Koteliansky V, Langer R, Anderson DG. Proc Natl Acad Sci USA. 2014;111:3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, Frank-Kamenetsky M, Yip KN, Alvarez R, Sah DWY, de Fougerolles A, Fitzgerald K, Koteliansky V, Akinc A, Langer R, Anderson DG. Proc Natl Acad Sci USA. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, de Fougerolles A, Dorkin JR, Narayanannair Jayaprakash K, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev KG, Sah DW, Soutschek J, Toudjarska I, Vornlocher HP, Zimmermann TS, Langer R, Anderson DG. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Pelet JM, Heller DA, Dong Y, Chen D, Gu Z, Joseph BJ, Wallas J, Anderson DG. Adv Mater. 2013;25:4641–4645. doi: 10.1002/adma.201301917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, Fenton OS, Zhang Y, Olejnik KT, Yesilyurt V, Chen D, Barros S, Klebanov B, Novobrantseva T, Langer R, Anderson DG. Nat Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broaders KE, Pastine SJ, Grandhe S, Frechet JM. Chem Commun (Cambridge, UK) 2011;47:665–667. doi: 10.1039/c0cc04190d. [DOI] [PubMed] [Google Scholar]
- 20.Dong Y, Eltoukhy AA, Alabi CA, Khan OF, Veiseh O, Dorkin JR, Sirirungruang S, Yin H, Tang BC, Pelet JM, Chen D, Gu Z, Xue Y, Langer R, Anderson DG. Adv Healthcare Mater. 2014;3:1392–1397. doi: 10.1002/adhm.201400054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, Jeon A, Dong Y, Whitehead KA, Anderson DG. J Am Chem Soc. 2012;134:6948–6951. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 22.Cui WG, Li XH, Zhou SB, Weng J. J Appl Polym Sci. 2007;103:3105–3112. [Google Scholar]
- 23.Marks JR, Placone J, Hristova K, Wimley WC. J Am Chem Soc. 2011;133:8995–9004. doi: 10.1021/ja2017416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan D, Papamichail D. ACS Synth Biol. 2013;2:237–244. doi: 10.1021/sb300086d. [DOI] [PubMed] [Google Scholar]
- 25.Zhan JY, Zheng KY, Zhu KY, Bi CW, Zhang WL, Du CY, Fu Q, Dong TT, Choi RC, Tsim KW, Lau DT. J Agric Food Chem. 2011;59:6091–6098. doi: 10.1021/jf200728e. [DOI] [PubMed] [Google Scholar]
- 26.Meng HN, Zhang ZZ, Zhao FX, Qiu T, Yang JD. Appl Surf Sci. 2013;280:679–685. [Google Scholar]
- 27.Liu L, Zhang XQ, Zhang Y, Pu YP, Yin LH, Tang M, Liu H. J Nanosci Nanotechnol. 2013;13:8137–8143. doi: 10.1166/jnn.2013.7925. [DOI] [PubMed] [Google Scholar]
- 28.Lv H, Zhang S, Wang B, Cui S, Yan J. J Controlled Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 29.Mishra S, Webster P, Davis ME. Eur J Cell Biol. 2004;83:97–111. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 30.Otsuka H, Nagasaki Y, Kataoka K. Adv Drug Delivery Rev. 2003;55:403–419. doi: 10.1016/s0169-409x(02)00226-0. [DOI] [PubMed] [Google Scholar]
- 31.Coppola A, Di Capua M, Di Minno MN, Di Palo M, Marrone E, Ierano P, Arturo C, Tufano A, Cerbone AM. J Blood Med. 2010;1:183–195. doi: 10.2147/JBM.S6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monahan PE, White GC., 2nd Curr Opin Hematol. 2002;9:430–436. doi: 10.1097/00062752-200209000-00007. [DOI] [PubMed] [Google Scholar]
- 33.Sabatino DE, Nichols TC, Merricks E, Bellinger DA, Herzog RW, Monahan PE. Progress in molecular biology and translational science. 2012;105:151–209. doi: 10.1016/B978-0-12-394596-9.00006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barzel A, Paulk NK, Shi Y, Huang Y, Chu K, Zhang F, Valdmanis PN, Spector LP, Porteus MH, Gaensler KM, Kay MA. Nature. 2014;517:360–364. doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Zoppe M, Hackeng TM, Griffin JH, Lee KF, Verma IM. Proc Natl Acad Sci U S A. 1997;94:11563–11566. doi: 10.1073/pnas.94.21.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Hylckama Vlieg A, van der Linden IK, Bertina RM, Rosendaal FR. Blood. 2000;95:3678–3682. [PubMed] [Google Scholar]
- 37.Srivastava A, Brewer AK, Mauser-Bunschoten EP, Key NS, Kitchen S, Llinas A, Ludlam CA, Mahlangu JN, Mulder K, Poon MC, Street A, Treatment Guidelines Working Group on Behalf of The World Federation Of, H Haemophilia. 2013;19:e1–47. doi: 10.1111/j.1365-2516.2012.02909.x. [DOI] [PubMed] [Google Scholar]
- 38.Day SM, Reeve JL, Myers DD, Fay WP. Thromb Haemostasis. 2004;92:486–494. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


