Abstract
The absence of Bmal1, a core clock gene, results in a loss of circadian rhythms, an acceleration of aging, and a shortened life span in mice. To address the importance of circadian rhythms in the aging process, we generated conditional Bmal1 knockout mice that lacked the BMAL1 protein during adult life and found that wild-type circadian variations in wheel-running activity, heart rate, and blood pressure were abolished. Ocular abnormalities and brain astrogliosis were conserved irrespective of the timing of Bmal1 deletion. However, life span, fertility, body weight, blood glucose levels, and age-dependent arthropathy - which are altered in standard Bmal1 knockout mice - remained unaltered, while atherosclerosis and hair growth improved, in the conditional adult-life Bmal1 knockout mice, despite abolition of clock function. Hepatic RNA-Seq revealed that expression of oscillatory genes was dampened in the adult-life Bmal1 knockout mice, while overall gene expression was largely unchanged. Thus, many phenotypes in conventional Bmal1 knockout mice, hitherto attributed to disruption of circadian rhythms, reflect the loss of properties of BMAL1 that are independent of its role in the clock. These findings prompt re-evaluation of the systemic consequences of disruption of the molecular clock.
INTRODUCTION
Circadian rhythms are biological processes that display endogenous and entrainable oscillations of about 24 hours. They are driven by a group of clock genes that are widely conserved across plants, animals, and bacteria (1). In mammals, the core clock genes, including Bmal1, Clock, Cry, and Per, are rhythmically expressed in both the suprachiasmatic nucleus (SCN) - the master clock that resides in the hypothalamus - and almost all peripheral tissues, in which they control the expression of numerous target genes in a circadian manner, influencing many physiological and biochemical processes (2). Chronic disruption of circadian rhythms has been associated with cardiometabolic morbidity in humans (3). In mice, deletion of Bmal1 prenatally disrupts clock-dependent oscillatory gene expression and behavioral rhythmicity coincident with reduced body weight, impaired hair growth, abnormal bone calcification, eye pathologies, neurodegeneration, and a shortened life span (4–7). However, although Bmal1 is the sole nonredundant gene in the core molecular clock, the degree to which phenotypes expressed in conventional knockout mice (cKOs) reflect disruption of clock function is unknown.
In this study, we describe the characterization of inducible Bmal1 knockout (iKO) mice that express the gene during embryogenesis but not in after birth. Despite ablation of clock function in both iKO and cKO mice, we observed striking physiological differences between the two model systems, prompting re-evaluation of the systemic consequences of disruption of the molecular clock.
RESULTS
Loss of circadian rhythms
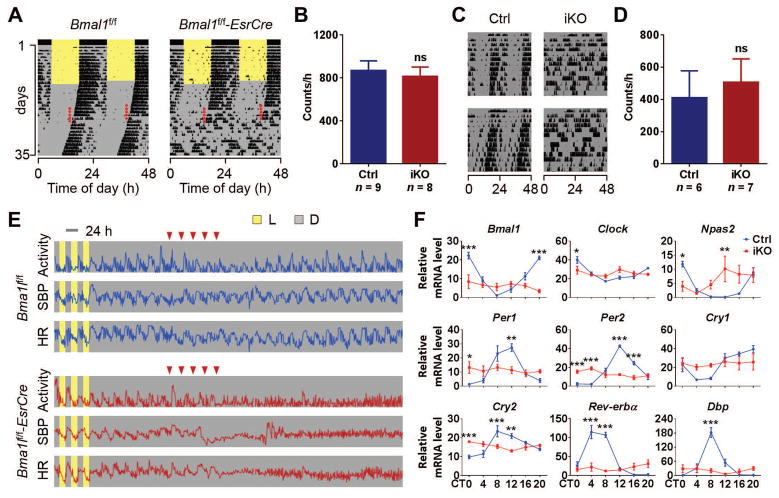
Bmal1 deletion in various tissues of the iKO mice (Bmal1f/f-EsrCre) - including brain, heart, lung, liver, kidney, spleen, skeletal muscle, and epididymal fat - was confirmed by quantitative reverse transcription–polymerase chain reaction (qRT-PCR). The iKO mice showed an 80% (liver) to 99% (brain and muscle) reduction of Bmal1 mRNA levels at Zeitgeber time 0 (ZT0) when Bmal1 expression is high (fig. S1A). Disruption of circadian behavior in iKOs was confirmed using running wheels. Before tamoxifen (TAM) treatment, the Bmal1f/f-EsrCre mice showed normal rhythmic locomotor activities under both 12 h:12 h light/dark (LD) and constant darkness (DD), which is indistinguishable from their Cre negative Bmal1f/f control littermates (Fig. 1A). When mice were administered TAM, Bmal1f/f mice (Ctrl) maintained their circadian behavior under DD, whereas all Bmal1f/f-EsrCre mice lost rhythmicity immediately after the treatment (Fig. 1A), which is similar to conventional Bmal1 KO mice (cKO) (fig. S2A). Loss of circadian behavior in iKOs was still evident 15 months after Bmal1 deletion (Fig. 1C), suggesting the permanent disruption of circadian rhythms. Interestingly, the reduction in overall locomotor activity in cKOs (8) (fig. S2B) was not recapitulated in iKOs (Fig. 1, B and D), indicating that adult-life deletion of Bmal1 does not predispose mice to the usual age-related decline of wheel running activity (9).
Fig. 1. Loss of circadian rhythms in iKO mice.
(A) Representative double-plotted actograms of wheel-running activity of 3-month-old Bmal1f/f and Bmal1f/f-EsrCre mice (red dots, TAM treatment). Similar results were obtained in N=8 to 9 mice/group. (B) The counts of wheel revolutions per hour from control mice (Ctrls) and iKO mice under conditions of constant darkness (DD) (N=8 to 9, Student’s t-test; ns, no significant difference). (C) Representative double-plotted actograms of wheel-running activity from 18-month-old Ctrl and iKO mice under DD. Similar results were obtained in N=6 to 7 mice/group. (D) The counts of wheel revolutions from 18-month-old Ctrls and iKOs under DD (N=6 to 7, Student’s t-test; ns, no significant difference). (E) Representative radiotelemetry results of locomotor activity, systolic blood pressure (SBP), and heart rate (HR) in Bmal1f/f and Bmal1f/f-EsrCre mice (red inverted triangles, TAM treatment). Similar results were obtained in N=3 mice/group. (F) Hepatic mRNA levels of canonical clock genes and clock-controlled gene Dbp, were determined by qRT-PCR [N=4/genotype/time point; x-axis, circadian time (CT); y-axis, relative mRNA levels; 2-way ANOVA; *, P<0.05; **, P<0.01; ***, P<0.001].
Consistent with disruption of core clock function, diurnal variation in heart rate (HR) and blood pressure (BP) was lost in iKOs (Fig. 1E), and circadian expression of hepatic clock genes was dampened (Fig. 1F). The variation of clock gene expression between ZT0 and ZT12 was also dampened in other tissues in iKOs (fig. S1B). Thus, behavioral, physiological, and molecular evidence for molecular clock disruption was present in the iKOs, consistent with what has previously been reported in cKOs (8, 10).
Conserved life span, weight, fertility, and blood glucose
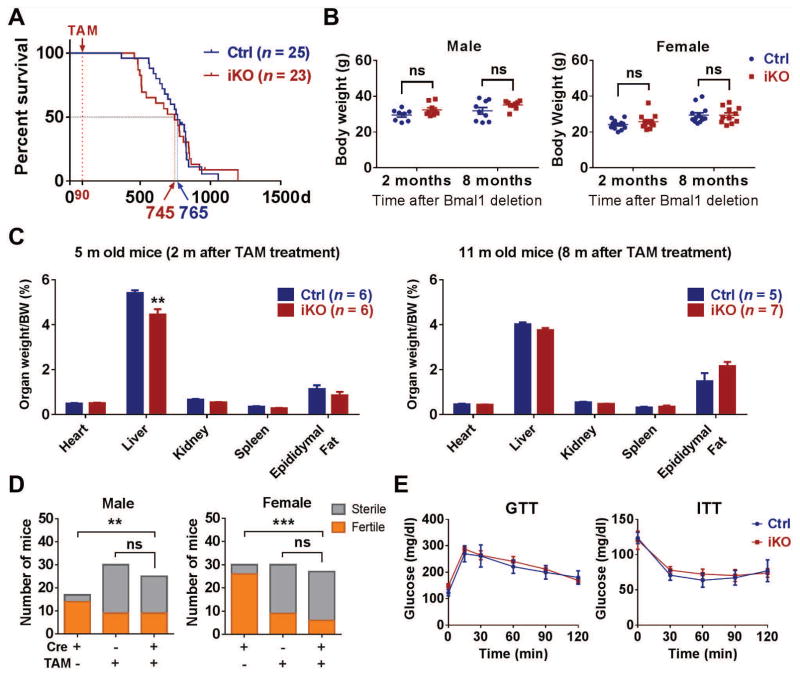
Despite permanent loss of circadian rhythmicity in iKOs, the mice possessed a normal average life span of more than 2 years (y) (Fig. 2A and fig. S3A). By contrast, the average life span of cKOs was just 9 months (Fig. S3B) (6). Except for ocular abnormalities, the iKO mice generally exhibit no gross morphological defects, and body weight was conserved in both genders (Fig. 2B and fig. S2C). Similarly, the weight of organs examined in the iKOs did not differ from controls, except for the liver at 2 months (m) after Bmal1 deletion (Fig. 2C). Although iKO mice are less fertile than normal mice (TAM untreated), the fertility percentage was comparable to their TAM-treated littermate controls (36% versus 30% in male, and 22% versus 27% in female; Fig. 2D), suggesting that the defect in fertility resulted from the TAM treatment, not the consequent gene deletion or disruption of circadian rhythms. In contrast, the cKOs were completely sterile (fig. S2D). Glucose tolerance tests (GTTs) and insulin tolerance tests (ITTs) did not differ between Ctrls and iKOs (Fig. 2E).
Fig. 2. General status of iKO mice.
(A) Life span of Ctrls and iKOs with medians of 745 days and 765 days, respectively (log-rank test; P=0.9852). (B) Body weight of Ctrls and iKOs for both genders (Student’s t-test; ns, no significant difference). (C) The ratio of organ to body weight in 5-month-old and 11-month-old old Ctrls and iKOs (multiple t-tests with Holm-Sidak correction; **, P<0.01). (D) Fertility analysis in both male and female Ctrl, iKO, and untreated Cre+ mice (χ2 test; ns, no significant difference; **, P<0.01; ***, P<0.001). (E) Blood glucose concentrations of Ctrls and iKOs that underwent GTT and ITT (N=6 to 7, 2-way ANOVA; no significant difference between Ctrls and iKOs at any time point).
Hair growth and arthropathy
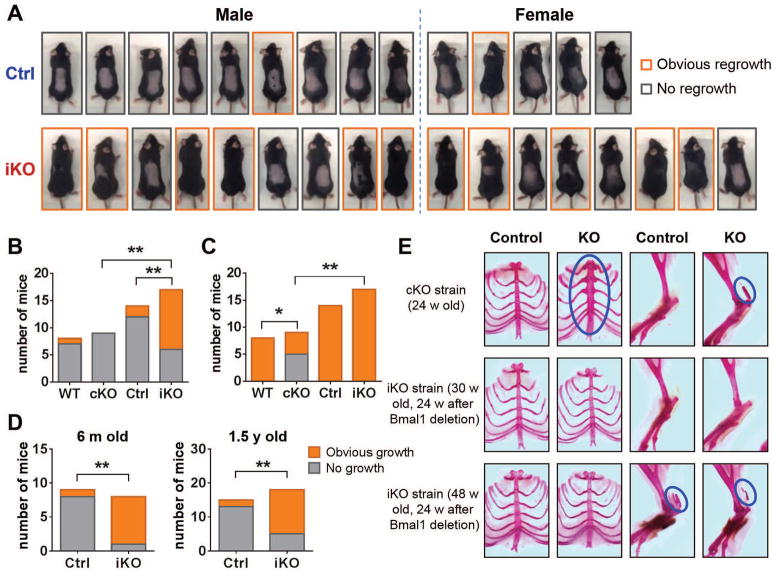
Loss, greying, and growth inactivity of hair (telogen) are hallmarks of aging (11, 12). Indeed, conventional Bmal1 and Clock mutant mice demonstrate an increase in telogen follicles compared to controls (6, 13). Here, in an assay to assess hair follicle cycling in which hair is shaved and new hair regrowth assessed (Fig. 3A), cKOs were shown not to enter the hair growth phase (anagen) as frequently as did WT mice (Fig. 3C), as previously reported. Unexpectedly, however, hair follicles in iKO mice entered anagen more frequently than did controls (Fig. 3B) even in aged (6 m and 1.5 y) mice (Fig. 3D). In young cKO mice, anagen follicles were absent 7 weeks (w) after shaving (Fig. 3B), indicating that the follicles stayed in telogen. Anagen was also rare in wild type (WT) and Ctrl mice; however, two-thirds of the iKO mice entered anagen at 7 w after shaving (Fig. 3B). Similarly, 12 w after shaving all WT, Ctrl, and iKO mice had entered anagen, but more than half of the cKOs still had not done so (Fig. 3C). Expression of genes associated with anagen (Ccnd1 and Mki67) was upregulated in iKOs compared with controls (fig. S4).
Fig. 3. Hair cycling and arthropathy.
(A) 6-week-old Bmal1f/f and Bmal1f/f-EsrCre mice were treated with tamoxifen. Meanwhile, dorsal hair was shaved. Pictures were taken 7 weeks after hair shaving. Mice with obvious hair regrowth are highlighted with orange boxes. (B to C) Contingency bar graphs show frequency distribution of hair regrowth in both cKO and iKO strains. The hair regrowth was observed 7 w (B) or 12 w (C) after hair shaving. (D) Hair regrowth assay in 6-month-old and 1.5-year-old mice. The hair regrowth was observed 7 w after hair shaving. (E) Representative photographs of alizarin red–stained ribcages and hind limbs from both cKO and iKO strains. Blue circles indicate calcification.
Accelerated age-related arthropathy has been reported in cKOs and was studied here using Aliza red–stained ribcages and hind limbs, as previously described (4, 5). As expected, we observed abnormal calcification in the costosternal junctions and calcaneal tendons of all cKOs (N=5) at 24 w of age (Fig. 3E). By contrast, abnormal calcification was not evident in any 30 w old iKOs (N=5, 24 w after Bmal1 deletion). Calcification of the calcaneal tendon was observed in some 48-w-old iKO mice (3 out of 8, 24 w after Bmal1 deletion) but was indistinguishable from that in littermate controls (4 out of 10).
Differential impact on atherogenesis
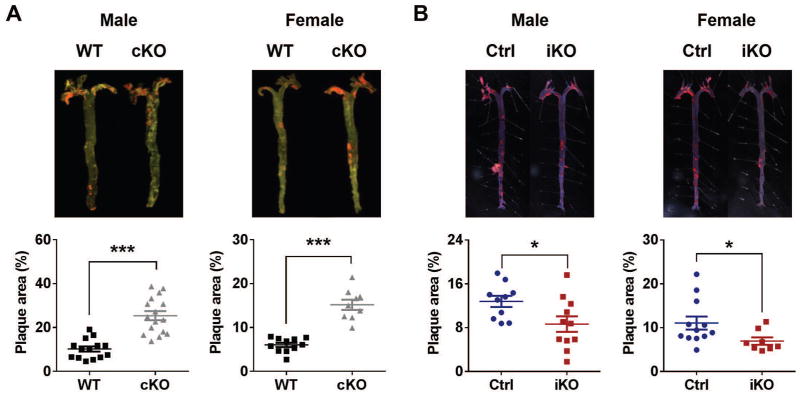
The molecular clock has been implicated in various aspects of cardiovascular function, including blood pressure homeostasis and thrombogenesis (14). Atherogenesis is accelerated in ClockΔ19/Δ19 mutant mice (15). Here, we compared conventional and adult-life Bmal1 deletion in mice on a hyperlipidemic low-density lipoprotein receptor deficient (LDLR−/−) background that were fed a high fat diet (HFD). Similar to ClockΔ19/Δ19 mutant mice, en face aortic lesion analysis revealed marked acceleration of atherogenesis in both male and female cKOs (Fig. 4A), whereas, unlike ClockΔ19/Δ19 mutant mice, cKOs show plasma total cholesterol and triglyceride levels comparable to those of WT mice (fig. S5A). By contrast, atherogenesis relative to Ctrl mice was retarded when Bmal1 was deleted in adulthood (Fig. 4B). Adult-life deletion of Bmal1 did not affect blood pressure or heart rate, at least at ZT8 to ZT9 (fig. S5B), whereas the increase in body weight in HFD-fed mice was reduced in male iKOs. Female iKOs showed a trend towards a reduction in body weight (fig. S5C). Consistently, aortae from iKOs showed reduced expression of proinflammatory genes, including CD68 (cluster of differentiation 68), MCP-1 (monocyte chemotactic protein 1), and iNOS (inducible nitric oxide synthase), compared to controls (fig. S5D). Among these genes, MCP-1 is a direct target gene of Bmal1, as revealed by ChIP-Seq in mouse liver (16).
Fig. 4. HFD-induced atherosclerosis.
Both (A) cKO and (B) iKO mice and their littermate controls were in a LDLR−/− background and fed with a HFD (42% kcal from fat) from 12 w of age. 16 w after HFD treatment, mice were euthanized and whole aortas were dissected and stained with oil red O. Plaque areas (shown in red) were quantified with Image-Pro. The percentage of plaque area relative to the entire area was calculated (Student’s t-test; *, P<0.05; **, P<0.01; ***, P<0.001).
Ocular abnormalities and astrogliosis
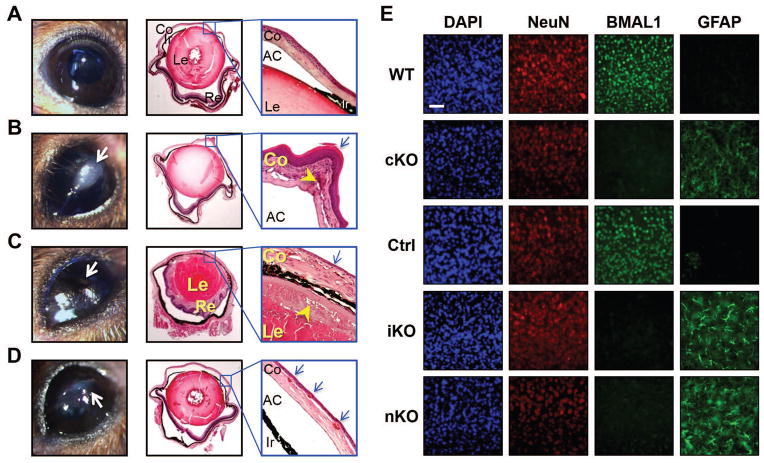
Despite the differential effects on hair regrowth, arthropathy, and atherogenesis, ocular abnormalities similar to those observed with prenatal Bmal1 depletion were evident in the iKOs (Fig. 5, A–D). By 8 m of age (5 m after Bmal1 deletion), about 80% of iKOs developed cloudy changes in one or both eyes, whereas none of their littermate controls displayed any visible abnormalities. Histological examination revealed various pathological changes, including corneal neovascularization, keratinization, and progressive inflammation in the iKOs.
Fig. 5. Ocular abnormalities and astrogliosis in iKO mice.
Representative gross images (left) and H&E-stained sections of eyes (middle and right) from (A) Ctrl and (B–D) iKOs. (A) Unremarkable globe from a Ctrl mouse. AC, anterior chamber; Co, cornea; Ir, iris; Le, lens; Re, retina. (B) Pathologic changes in a male mouse eye. Grossly, there is a leukoplakic plaque on the cornea (left, arrow). Histologically, the cornea appears thickened with keratinization of the epithelium (right, arrow) and chronic inflammation and neovascularization of the stroma (right, arrowhead). (C) In the contralateral eye of the same mouse, the corneal surface appears irregular with a flattened chamber. Histologically, the cornea appears thickened. The retina is adherent to the lens. The corneal epithelium is attenuated (right, arrow) with chronic inflammation and neovascularization in the stroma. A subcapsular anterior cataract is present (right, arrowhead). (D) A female mouse eye shows an irregular corneal surface with leukoplakia (left, arrow) and corneal neovascularization (right, arrow). (E) Immunofluorescent staining of DAPI (all nuclei), NeuN (neuronal nuclei), BMAL1 and GFAP (activated astrocytes) in the motor cortex from cKO, iKO, and nKO mouse strains. Representative images show loss of Bmal1 immunoreactivity in all KOs, which is accompanied by severe astrogliosis (GFAP panels). Similar results were obtained in N=5 mice/group. Scale bar: 150 μm.
We have reported that cKOs and neural-specific Bmal1 knockout mice (Bmal1f/f-NestinCre, nKO) exhibit severe astrogliosis (7). Here, we examined the brains of 11- to 12-m-old iKO mice and found them to be nearly indistinguishable from those of cKO mice (aged 6 m) or nKO mice (aged 11 to 12 m), with undetectable BMAL1 immunoreactivity in neurons and severe astrogliosis, which was reflected by GFAP (glial fibrillary acidic protein) staining throughout the cerebral cortex (Fig. 5E). Transcriptional analysis revealed similar changes in Gfap mRNA expression (fig. S6). However, such neurodegenerative changes did not affect the life spans of iKOs (Fig. 2A) or nKOs (fig. S3C). It is not surprising that this pathological brain phenotype did not lead to premature death, as similar levels of inflammation and synapse loss are observed in some mouse models of Amyloid-β-mediated cerebral amyloidosis, and these mice have normal life spans (17).
Loss of the circadian transcriptome
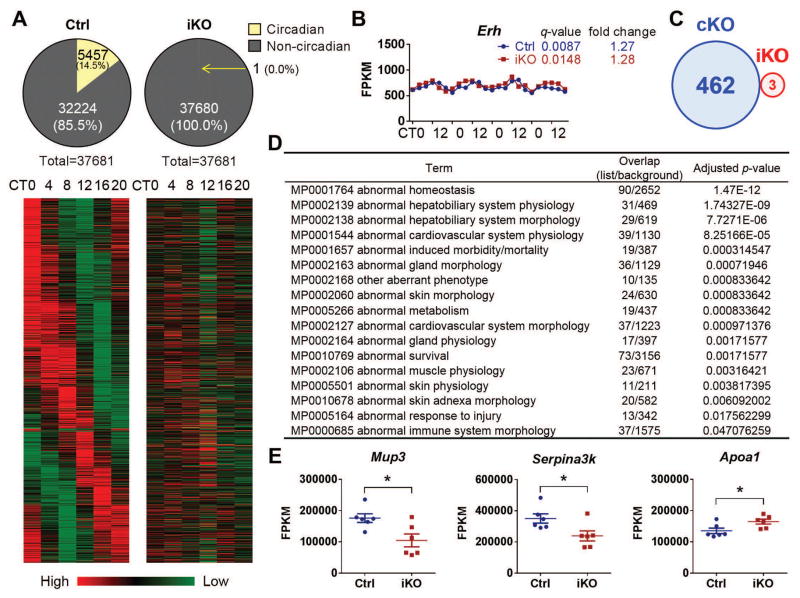
To characterize gene expression profiles, we performed RNA-Seq using samples of liver, where most circadian genes are expressed (18). As expected, all clock genes oscillated in control mice, but were expressed at relatively constant levels in iKOs (fig. S7). This expression pattern was highly consistent with the qRT-PCR results (Fig. 1F). A circadian analysis was performed using JTK(Jonckheere-Terpstra-Kendall)_CYCLE algorithm (19) to explore 24-h oscillations in transcript abundance. We used a 5% false discovery rate (FDR) threshold to detect circadian genes in control mice and found 14.5% genes (5457 among 37681, data file S1) were rhythmically expressed (Fig. 6A). By contrast, even those genes that top the list in control mice displayed no circadian expression pattern in iKOs (table S1). Only 1 gene, Erh, showed circadian variability at the mRNA level in iKOs (Fig. 6B), illustrating how few genes are rhythmically expressed independent of the classic circadian clock.
Fig. 6. Hepatic transcriptome.
RNA samples from iKOs and their littermates under DD condition were used for RNA-Seq. (A) Out of the 37,681 transcripts, 5,457 exhibited circadian variability in Ctrl mice. By contrast, only 1 gene in iKOs exhibited a circadian pattern. Heat map rendering of the temporal gene expression pattern of these 5,457 circadian genes is shown with the average gene expression level measured for each time point. (B) Erh is the only hepatic gene that shows a circadian expression pattern in iKOs, and the q-value (JTK_CYCLE analysis) and fold change (ratio of peak/trough) are shown [x-axis, circadian time; y-axis, fragments per kilobase of transcript per million mapped reads (FPKM)]. (C) A Venn diagram (sizes not to scale) depicting the number of differentially expressed genes in cKO and iKO strains. (D) MGI Mammalian Phenotype Level 3 enrichment analysis for cKO differentially expressed genes. The top 17 (adjusted P value<0.05) phenotypes related to these genes are given. Overlap, the number of appearing gene/the number of background genes. (E) Mup3, Serpina3k, and Apoa1 are the only differentially expressed genes in the iKO strain.
Phenotype enrichment analysis of differentially expressed genes
To understand the potential reasons why there is a striking phenotypic difference between iKOs and cKOs despite a shared ablation of circadian rhythms, we compared hepatic gene expression profiles between WT versus cKOs and Ctrl versus iKOs, irrespective of time (data file S2). An FDR threshold of 5% was used for detection of differentially expressed genes for both KO strains, although the specific value of this cutoff did not alter the fact that the iKOs had many fewer differentially expressed genes than did the cKOs (table S2); from these analyses, we found 462 differentially expressed genes in cKOs and 3 in iKOs (Fig. 6C). Enrichment analysis of these 462 genes revealed multiple potential phenotypes that are consistent with what might be expected, including abnormal cardiovascular function, induced morbidity and mortality, abnormal skin morphology, abnormal metabolism, abnormal survival, abnormal muscle physiology, and abnormal immune function (Fig. 6D and data file S3). However, only 3 genes in the iKOs (without overlap with the cKOs) were identified as differentially expressed (Fig. 6, C and E). Enrichment analysis showed that no category reached the cutoff (P<0.05), perhaps because of the small number of genes involved (data file S3).
DISCUSSION
Among canonical clock genes, Bmal1 is the only one whose deletion results in complete loss of circadian rhythms (8); therefore, Bmal1 knockout mice have become a valuable resource for study of the effects of circadian disruption. Other than loss of circadian rhythms, however, these mice also exhibit a greatly reduced life span and develop various features consistent with premature aging (4–7). Because of the absence of Bmal1 expression during development in cKO mice, it is not clear whether these phenotypes relate to its absence during embryogenesis or postnatal development or, indeed, whether these phenotypes reflect disruption of the molecular clock or loss of other properties of BMAL1. Here, we compared iKOs with cKOs and found that both approaches to gene deletion similarly disrupt behavioral and transcriptional indicators of clock-dependent circadian function and both types of knockout mice exhibit markers consistent with accelerated aging [such as ocular abnormalities and brain astrogliosis (table S3)]. In contrast, other biomarkers of aging and premature death, so evident in cKOs - including a markedly shortened life span, infertility, reduced body weight, organ shrinkage (6), impaired glucose homeostasis (20, 21), and abnormal bone calcification (4) - are not replicated in the iKOs. Further, two aging related phenotypes, hair growth inactivity and a predisposition to atherosclerosis, were reversed in iKOs. To characterize Bmal1 KO mice at the level of gene expression as well as to determine potential mechanisms underlying the general difference between cKO and iKO strains, we performed RNA-Seq using liver samples. Although the various phenotypes may not be explained simply by changes of the transcriptome in any given tissue, liver affords an appropriate initial opportunity to address this question, as it expresses most circadian genes (18). All time-effect genes were dampened in iKO mice without obvious changes in time-independent genes. These striking differential effects between the two strains of mice, despite a shared ablative effect on clock-dependent circadian biology, question the attribution of many phenotypes in the widely used cKOs to disruption of the molecular clock.
Circadian recruitment of clock proteins including BMAL1 to DNA binding sites is believed to contribute to the oscillation of target genes (16). However Bmal1 is expressed in mice throughout embryogenesis (22) but does not oscillate, either in the SCN (23, 24) or in peripheral tissues (25), suggesting that clock regulatory loops may not be well established until late in development. As such, Bmal1 expression might exert important non-clock functions during early development, and its deletion during this period might account for premature aging and various related phenotypes observed in the adult mice. Alternatively, the physiologic effects of Bmal1 may vary across tissues and defects in one tissue may compensate for those in another when analyzing multi tissue mutants. This possibility is not addressed in the present study. However, while shortened life span has yet to be reported in any prenatal tissue-specific Bmal1 KO mice, some do exhibit phenotypes that are not evident in adult-life global KOs. For example, impaired glucose tolerance was reported in prenatal hepatocyte (19, 25) and pancreatic β-cell (26) specific Bmal1 KO mice. This is consistent with cKOs, but is not seen in adult-life global KOs.
To our surprise, our cKO and iKO mice showed opposite phenotypes in hair growth and atherogenesis. We found increased growing follicles in all young, middle-aged, and old iKO mice as well as consistent expression of hair growth–promoting genes Ccnd1 and Mik67 in the skin. ChIP-Seq has revealed that the Ccnd1 gene contains binding sites for all core clock proteins including BMAL1 (16), which implies that Ccnd1 is directly clock controlled. However, its expression in cKOs was not changed. It is possible that this regulation is masked and hair growth is retarded by the overall premature aging phenotype. Pan et al. found impaired cholesterol metabolism and enhanced atherosclerosis in HFD-fed Clock mutant mice generated in a LDLR−/− background, implicating the circadian clock in atherogenesis (15). Consistent with their finding, we observed increased atherogenesis in Bmal1 cKOs. However, unlike the case for Clock mutant mice, Bmal1 deletion does not alter cholesterol metabolism, which suggests that disruption of the two clock-directed genes influences atherogenesis by distinct the mechanisms. By contrast, adult-life Bmal1 depletion restrains atherogenesis coincident with dampened expression of proinflammatory genes in the vasculature and a reduction in the weight gain induced by the HFD. The relative contribution of local and systemic effects to this phenotype remains to be determined. Ocular abnormalities and brain astrogliosis were preserved in iKO mice, consistent with Bmal1 exerting a direct role in the eye and neural system irrespective of developmental issues. However, it remains unknown if these phenotypes are caused by loss of circadian rhythms or loss of non-clock functions of Bmal1, because deletion of core clock genes not only disrupts rhythmicity of genes relevant to expression of these phenotypes, but may also has striking consequences on their overall expression levels (26, 27).
The apparent restraining influence of Bmal1 expression during embryogenesis on aging-related phenotypes and, indeed, life span in the adult mice, is reminiscent of the Barker hypothesis, which postulated that factors impinging on fetal development would influence the expression of aging and life span in adult humans (28). Insight into the impact of environmental factors and maternal behavior, such as smoking and alcohol consumption, on postnatal health has suggested an epigenetic basis for this hypothesis (29). Here, in the case of Bmal1, we raise the additional possibility of a direct genetic effect on such phenomena. A provocative consideration is that the anticipatory role of the clock might condition the impact of environmental insults, attenuating them at certain times of day.
In summary, despite a shared impact on circadian function, many phenotypes observed in conventional KOs are not observed when Bmal1 is deleted in adult mice. This prompts reevaluation of the systemic role of the molecular clock and its importance in the biology of aging.
MATERIALS AND METHODS
Study design
The primary objective of this study was to investigate the role of circadian rhythms in aging in adult mice. We used Bmal1-inducible KO mice in order to bypass possible issues about developmental defects in conventional KOs. For all experiments, the number of independent replicates is outlined in each figure or legend unless individually plotted. Sample sizes were based on the desired magnitude of the difference to be detected, and the variance of the estimates was based on prior data where available and setting α = 0.05 and 1-β = 0.9. Mice in all experiments were randomized into various groups. The studies were not performed under double-blind conditions. The experimenters were not blinded to the identities of individual groups. Animals were not withdrawn from the studies according to predetermined criteria in the Institutional Animal Care and Use Committee (IACUC) protocols, nor were they excluded from any of the analyses.
Mice
Both conventional Bmal1 knockout mice (cKO) and Bmal1f/f mice (30) were obtained from C. Bradfield (University of Wisconsin, Madison, WI, USA). The floxed mice were crossed with a tamoxifen-inducible universal Cre (EsrCre) mice or NestinCre mice (Jackson Laboratories, Bar Harbor, ME) to yield Bmal1f/f-EsrCre mice and Bmal1f/f-NestinCre (nKO) mice, respectively. To generate inducible Bmal1 knockout (iKO) mice, 3m old (unless specified) Bmal1f/f-EsrCre mice were treated with tamoxifen as previously described (31). Cre- Bmal1f/f littermates treated with tamoxifen served as controls (Ctrl). For atherogenesis, all mice were crossed into a LDLR−/− background. All mice were housed under 12 h:12 h LD conditions with free access to food and water unless specified. All procedures were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Wheel running activity
Young (3m, both cKO and iKO strains) and aged (18m, iKO strain only) mice were individually housed in cages equipped with running wheels (Coulbourn instruments, PA). Cages were kept within a ventilated and light-tight isolation chamber with a computer-controlled lighting system. Locomotor activity was continuously recorded and analyzed using ClockLab software (Actimetrics). For the young group, mice from both cKO (N=4/genotype) and iKO (N=8–9/genotype) strains were housed in 12:12 LD and then released into DD. After 1 week, Bmal1f/f and Bmal1f/f-EsrCre mice were treated with 5mg tamoxifen by gavage for 5 consecutive days followed by recording for 10 days.
Radiotelemetry reading
The implantation of radiotelemetry (model No. TA11PA-C20, DSI, MN) was performed as previously described (31). Following 1w of recovery, blood pressure (BP), heart rate (HR) and locomotor activity were monitored continuously in 3m old Bmal1f/f (N=3) and Bmal1f/f-EsrCre (N=3) mice using the Dataquest LabPRO Acquisition System. Mice were housed under LD and then released into DD. After 1 week, mice were treated with 5mg tamoxifen by gavage for 5 consecutive days followed by recording for 15 days.
Fertility analysis
Both genders were used for fertility analysis. 8w old Bmal1f/f and Bmal1f/f-EsrCre mice were treated with TAM. At 10w old, each iKO, control or untreated Cre+ littermate was mated with a wild type C57Bl/6 mouse. All breeding cages were monitored for birth daily for 9m.
Glucose tolerance test (GTT)
12-m-old mice (10m after TAM treatment) were fasted for 16 h before the GTT. Blood was obtained from a tail cut and was assessed for fasting glucose levels using a One-touch Ultra 2 (Lifescan, Johnson & Johnson) glucometer. The mice were then received 2g/kg body weight of a 0.2g/ml glucose solution delivered by oral gavage. At 15, 30, 60, 90 and 120 min after the administration, dried blood was quickly removed from the tail wound and fresh blood was collected again to measure the glucose concentration.
Insulin tolerance test (ITT)
20-m-old mice (18m after TAM treatment) were fasted for 4 h before ITT. After baseline blood glucose measurement, 1 U/kg insulin (Eli Lilly) was injected intraperitoneally and blood glucose was measured again at time points of 30, 60, 90, and 120 min post injection.
Hair regrowth assay
To characterize hair growth, the backs of mice were shaved and animals were monitored for hair regrowth for three months. For RNA analysis, dorsal skins from 8-w-old mice (1 w after TAM treatment for iKO strain) was collected for RNA extraction and quantitative RT-PCR analysis for Ccnd1 and Mki67.
Bone staining
Skeletons were stained using Alizarin red as previously described (5) with modifications. Briefly, skinned and eviscerated animal carcasses were submerged in 1% KOH for 4h followed by incubation in 2% KOH overnight. Carcasses were then stained in 2% KOH containing 0.004% Alizarin Red (Sigma) for 2 days. After clearing in 2:1:2 glycerin:benzyl alcohol:70% ETOH, stained skeletons were photographed.
Atherosclerosis
All mice were on a LDLR−/− background and fed HFD (42% kcal from fat, Harlan Laboratories) for 16 w from 12 w of age. Mouse aortic trees were prepared and stained and atherosclerotic lesions were quantified as described (32). Briefly, the entire aorta from the aortic root to the iliac bifurcation was collected and fixed in formalin-free fixative Prefer (Anatech LTD). Adventitial fat was removed. The aorta was opened longitudinally, stained with Sudan IV (Sigma), and pinned down on black wax to expose the intima. The extent of atherosclerosis was determined by the en face lesion area percentage to the entire intimal area.
Lipid analysis in plasma
Blood was collected from the vena cava of CO2-euthanized mice. Plasma total cholesterol and HDL levels were measured by commercial kits from Wako Chemicals.
Tail-cuff BP and HR measurement
SBP and HR were measured in conscious mice at ZT8-9 using a tail cuff system (Visitech Systems Inc.) as previously described (33).
Ocular pathology
Animals were euthanized and both eyes were harvested and fixed in 10% paraformaldehyde solution. Following gross examination, globes were routinely processed and embedded in paraffin. Sections were obtained, stained with hematoxylin-eosin (H&E), and reviewed by an ocular pathologist.
Immunostaining of brain sections
Cryosections from the prefrontal cortex to caudal hippocampus were prepared as previously described (7). Sections were washed in tris-buffered saline (TBS), blocked for 30 min in Tris-buffered saline containing 3% donkey serum and 0.25% Triton X-100, then incubated overnight in TBS plus 0.25% Triton X-100 with 1% donkey serum containing rabbit GFAP (Dako), BMAL1 (Novus), or mouse NeuN primary antibodies at 4°C. Sections were washed then incubated in TBS containing AlexFlour 488-conjugated anti-rabbit or AlexFlour 568-conjugated anti-mouse secondary antibodies for 1hr at room temperature, then washed again in TBS. Sections were mounted and imaged using an epiflourescent microscope.
Quantitative RT-PCR
Total RNA from various tissues was isolated using the Qiagen RNeasy Kit. Reverse transcription was performed using an RNA-cDNA kit (Applied Biosystems, Carlsbad, CA). Real-time PCR was performed using ABI Taqman primers and reagents on an ABI Prizm 7500 thermocycler according to manufacturer’s instructions. All mRNA measurements were normalized to beta-actin mRNA levels.
Sample preparation for RNA-Seq
24 male iKO mice aged 4–6m (2w after TAM treatment) and their littermates were maintained under 12h:12h LD conditions, then released into DD condition. Starting at 36h after release, 4 mice/genotype were killed in darkness every 4 h for 20 h (6 time points). Liver samples were quickly excised and snap-frozen in liquid nitrogen. Total RNA was isolated using Qiagen RNeasy Kit and the RNA integrity was checked using an Agilent Technologies 2100 Bioanalyzer. RNA samples were subjected to two rounds of hybridization to oligo (dT) beads. The resulting mRNA was then used as template for double stranded cDNA synthesis. After purification cDNA samples were fragmented with nebulization technique to yield fragment sizes of 100–300 bps and subjected to end-repair, adenylation, ligation of Illumina sequencing adapters, and PCR amplification. The PCR product was purified and used directly for cluster generation and sequencing (HISeq2000).
When comparing iKOs with cKOs irrespective of time points, we randomly picked one mouse/genotype/time point from iKOs and littermates. To match these samples, six male cKO mice and six littermate WT controls (one mouse/genotype/time point) were euthanized for liver collection. Samples for RNA-Seq were prepared as described above.
RNA-Seq analysis
RNA-Seq data were aligned to the mouse genome build mm9 by STAR version 2.4.2a (34). Data were normalized using a resampling strategy as described previously (35), the code is available at (https://github.com/itmat/Normalization). The normalized counts thus produced were then used for the subsequent statistical analyses. JTK_CYCLE was used to explore 24-h oscillations in transcript abundance (19). Differential expression analysis was performed by standard permutation methods to control the False Discovery Rate (FDR) (36). The list of resulting differentially expressed genes were used for enrichment analysis using Enrichr (http://amp.pharm.mssm.edu/Enrichr/). All data have been submitted to GEO (Accession GSE70479, GSE 70499 and GSE70500).
Statistical analysis
All statistical tests were two-sided. A log-rank test was used for survival curve analysis, and χ2 test was used for contingency data. For other comparisons except RNA-Seq results, Student’s t-test or 1-way ANOVA was used when a single variable was compared between two or more genotypes, and 2-way ANOVA with Bonferroni’s posttest was used when multiple variables were compared between genotypes. The cutoff for significance was P<0.05 (*). #, **, ***, and ns represent P<0.1, P<0.01, P<0.001 and no significant difference, respectively. In all figures with error bars, the graphs depict the mean ± SEM.
Supplementary Material
Acknowledgments
We thank Christopher A. Bradfield (University of Wisconsin, Madison, WI, USA) for conventional Bmal1 knockout mice and Bmal1f/f mice. We thank Eun Ji Kim, Nick Lahens, Katharina Hayer, Faith Colden, Angel Pizarro and Anand Srinivasan (University of Pennsylvania, Philadelphia, PA, USA) for their support in RNA-Seq analysis. We thank Ying Zheng (University of Pennsylvania, Philadelphia, PA, USA) for discussions about hair regrowth. We thank Weili Yan (University of Pennsylvania, Philadelphia, PA, USA) for technical assistance. We thank Amita Sehgal for reading and commenting on this manuscript. Dr FitzGerald is the McNeil Professor of Translational Medicine and Therapeutics.
Funding: This work was supported by a grant from the NIH (HL097800) and the University of Pennsylvania Genomics Frontiers Institute's Translational and Personalized Genomics Centers Initiative.
Footnotes
Author contributions: G.Y. and G.A.F. conceived the project. G.Y. and L.C. performed and analyzed most of the experiments in this study. G.P. prepared cKO samples for RNA-Seq. E.S.M. and V.L. conducted staining of brain and eye, respectively. W.S. performed atherosclerosis experiment for cKO mice. S.C.M. collected iKO organs for weight measurement. T.G designed and conducted the RNAseq experiments and G.R.G. did the data analysis. G.C. discussed the data of hair growth assay. G.R.G., E.S.M., V.L. and G.C. provided critical intellectual input in manuscript preparation of their pertinent parts. G.Y., L.C. and G.A.F. wrote the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: RNA-Seq data can be retrieved from the Gene Expression Omnibus under accession number GSE70479, GSE 70499 and GSE70500.
www.sciencetranslationalmedicine.org/cgi/content/full/7/3XX/3XXraXXX/DC1
Fig. S1. Validation of Bmal1 deletion and dampening effect of other clock genes
Fig. S2. Wheel-running activity, body weight, and fertility in cKO mice
Fig. S3. Life span of iKO, cKO, and nKO mice
Fig. S4. mRNA levels of Ccnd1 and Mki67 in skin
Fig. S5. Other parameters in HFD-fed mice
Fig. S6. iKO mice display similar Gfap induction in the brain as cKO
Fig. S7. RNA-Seq results revealed dampening effect in core clock genes in iKO mice
Table S1. Top 20 oscillating hepatic genes (JTK_CYCLE q-value) in Ctrl mice show no circadian pattern in iKO mice
Table S2. The ratio of differentially expressed gene numbers in cKO strain to the numbers in iKO strain
Table S3. Summary of phenotypes of cKO and iKO mice
Data file S1. Circadian transcriptome
Data file S2. Differentially expressed genes irrespective of time points
Data file S3. Phenotype enrichment analysis of differentially expressed genes
REFERENCES AND NOTES
- 1.Harmer SL, Panda S, Kay SA. Molecular bases of circadian rhythms. Annu Rev Cell Dev Biol. 2001;17:215–253. doi: 10.1146/annurev.cellbio.17.1.215. [DOI] [PubMed] [Google Scholar]
- 2.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 3.Ruger M, Scheer FA. Effects of circadian disruption on the cardiometabolic system. Rev Endocr Metab Disord. 2009;10:245–260. doi: 10.1007/s11154-009-9122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, Takahashi JS. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunger MK, Walisser JA, Sullivan R, Manley PA, Moran SM, Kalscheur VL, Colman RJ, Bradfield CA. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis. 2005;41:122–132. doi: 10.1002/gene.20102. [DOI] [PubMed] [Google Scholar]
- 6.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musiek ES, Lim MM, Yang G, Bauer AQ, Qi L, Lee Y, Roh JH, Ortiz-Gonzalez X, Dearborn JT, Culver JP, Herzog ED, Hogenesch JB, Wozniak DF, Dikranian K, Giasson BI, Weaver DR, Holtzman DM, Fitzgerald GA. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J Clin Invest. 2013;123:5389–5400. doi: 10.1172/JCI70317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valentinuzzi VS, Scarbrough K, Takahashi JS, Turek FW. Effects of aging on the circadian rhythm of wheel-running activity in C57BL/6 mice. Am J Physiol. 1997;273:R1957–1964. doi: 10.1152/ajpregu.1997.273.6.R1957. [DOI] [PubMed] [Google Scholar]
- 10.Ukai H, Ueda HR. Systems biology of mammalian circadian clocks. Annu Rev Physiol. 2010;72:579–603. doi: 10.1146/annurev-physiol-073109-130051. [DOI] [PubMed] [Google Scholar]
- 11.Harrison DE, Archer JR. Biomarkers of aging: tissue markers. Future research needs, strategies, directions and priorities. Exp Gerontol. 1988;23:309–325. doi: 10.1016/0531-5565(88)90034-4. [DOI] [PubMed] [Google Scholar]
- 12.Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, Park SH, Thompson T, Karsenty G, Bradley A, Donehower LA. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 13.Lin KK, Kumar V, Geyfman M, Chudova D, Ihler AT, Smyth P, Paus R, Takahashi JS, Andersen B. Circadian clock genes contribute to the regulation of hair follicle cycling. PLoS Genet. 2009;5:e1000573. doi: 10.1371/journal.pgen.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westgate EJ, Cheng Y, Reilly DF, Price TS, Walisser JA, Bradfield CA, FitzGerald GA. Genetic components of the circadian clock regulate thrombogenesis in vivo. Circulation. 2008;117:2087–2095. doi: 10.1161/CIRCULATIONAHA.107.739227. [DOI] [PubMed] [Google Scholar]
- 15.Pan X, Jiang XC, Hussain MM. Impaired cholesterol metabolism and enhanced atherosclerosis in clock mutant mice. Circulation. 2013;128:1758–1769. doi: 10.1161/CIRCULATIONAHA.113.002885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maia LF, Kaeser SA, Reichwald J, Hruscha M, Martus P, Staufenbiel M, Jucker M. Changes in amyloid-beta and Tau in the cerebrospinal fluid of transgenic mice overexpressing amyloid precursor protein. Sci Transl Med. 2013;5:194re192. doi: 10.1126/scitranslmed.3006446. [DOI] [PubMed] [Google Scholar]
- 18.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vollmers C, Schmitz RJ, Nathanson J, Yeo G, Ecker JR, Panda S. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson MH, Lim A, Fernando D, Day ML. Circadian clockwork genes are expressed in the reproductive tract and conceptus of the early pregnant mouse. Reprod Biomed Online. 2002;4:140–145. doi: 10.1016/s1472-6483(10)61931-1. [DOI] [PubMed] [Google Scholar]
- 23.Sladek M, Sumova A, Kovacikova Z, Bendova Z, Laurinova K, Illnerova H. Insight into molecular core clock mechanism of embryonic and early postnatal rat suprachiasmatic nucleus. Proc Natl Acad Sci U S A. 2004;101:6231–6236. doi: 10.1073/pnas.0401149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ansari N, Agathagelidis M, Lee C, Korf HW, von Gall C. Differential maturation of circadian rhythms in clock gene proteins in the suprachiasmatic nucleus and the pars tuberalis during mouse ontogeny. Eur J Neurosci. 2009;29:477–489. doi: 10.1111/j.1460-9568.2008.06605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolatshad H, Cary AJ, Davis FC. Differential expression of the circadian clock in maternal and embryonic tissues of mice. PLoS One. 2010;5:e9855. doi: 10.1371/journal.pone.0009855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 27.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang X, Takahashi JS, Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barker DJ. Maternal nutrition, fetal nutrition, and disease in later life. Nutrition. 1997;13:807–813. doi: 10.1016/s0899-9007(97)00193-7. [DOI] [PubMed] [Google Scholar]
- 29.Aguilera O, Fernandez AF, Munoz A, Fraga MF. Epigenetics and environment: a complex relationship. J Appl Physiol (1985) 2010;109:243–251. doi: 10.1152/japplphysiol.00068.2010. [DOI] [PubMed] [Google Scholar]
- 30.Johnson BP, Walisser JA, Liu Y, Shen AL, McDearmon EL, Moran SM, McIntosh BE, Vollrath AL, Schook AC, Takahashi JS, Bradfield CA. Hepatocyte circadian clock controls acetaminophen bioactivation through NADPH-cytochrome P450 oxidoreductase. Proc Natl Acad Sci U S A. 2014;111:18757–18762. doi: 10.1073/pnas.1421708111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang G, Jia Z, Aoyagi T, McClain D, Mortensen RM, Yang T. Systemic PPARgamma Deletion Impairs Circadian Rhythms of Behavior and Metabolism. PLoS One. 2012;7:e38117. doi: 10.1371/journal.pone.0038117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Z, Crichton I, Tang SY, Hui Y, Ricciotti E, Levin MD, Lawson JA, Pure E, FitzGerald GA. Disruption of the 5-lipoxygenase pathway attenuates atherogenesis consequent to COX-2 deletion in mice. Proc Natl Acad Sci U S A. 2012;109:6727–6732. doi: 10.1073/pnas.1115313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Y, Wang M, Yu Y, Lawson J, Funk CD, Fitzgerald GA. Cyclooxygenases, microsomal prostaglandin E synthase-1, and cardiovascular function. J Clin Invest. 2006;116:1391–1399. doi: 10.1172/JCI27540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Tibshirani R. Finding consistent patterns: a nonparametric approach for identifying differential expression in RNA-Seq data. Stat Methods Med Res. 2013;22:519–536. doi: 10.1177/0962280211428386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grant GR, Liu J, Stoeckert CJ., Jr A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics. 2005;21:2684–2690. doi: 10.1093/bioinformatics/bti407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.