Abstract
Nitrogenase catalyzes the important reactions of N2-, CO- and CO2-reduction at its active cofactor site. Designated the M-cluster, this complex metallocofactor is assembled through the generation of a characteristic 8Fe-core prior to the insertion of Mo and homocitrate that completes the stoichiometry of the M-cluster. NifB catalyzes the critical step of radical SAM-dependent carbide insertion that occurs concomitant with the insertion a “9th” sulfur and the rearrangement/coupling of two 4Fe-clusters into a complete 8Fe-core of the M-cluster. Further categorization of a family of NifB proteins as a new class of radical SAM methyltransferases suggests a general function of these proteins in complex metallocofactor assembly and provides a new platform for unveiling unprecedented chemical reactions catalyzed by biological systems.
Catalysis by nitrogenase, particularly the reduction of nitrogen (N2) to ammonia (NH3) [1–3] and the reduction of carbon monoxide (CO) or carbon dioxide (CO2) to hydrocarbons under ambient conditions [4–7], bears tremendous significance to environment- and energy-related areas. The “conventional” Mo-nitrogenase utilizes a specific reductase (NifH) to deliver electrons to the active cofactor site (designated the M-cluster) of the catalytic component (NifDK), where substrate reduction takes place [1]. Arguably the most complex metallcofactor utilized by biological systems, the M-cluster consists of a [MoFe7S9C] core that can be viewed as [MoFe3S3] and [Fe4S3] subclusters bridged by three μ2-sulfides and one μ6-carbide (C4−) ion, and it is further coordinated by a homocitrate moiety at its Mo end [8–10]. Understanding the assembly mechanism of this complex metallocenter is not only important for elucidation of the structural-functional relationship of nitrogenase, but also crucial for future development of strategies to synthesize biomimetic complexes for production of valuable products.
The M-cluster: a multifaceted model for metallocofactor assembly
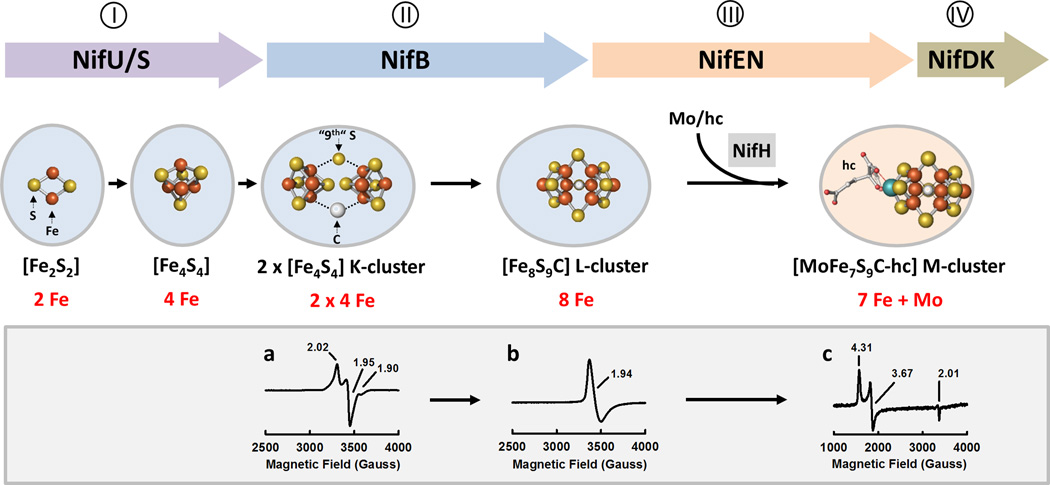
Biosynthesis of the M-cluster is launched by the concerted action of NifS and NifU, with NifS acting as a pyridoxal phosphate-dependent cysteine desulfurase, forming a protein-bound cysteine persulfide that is donated to NifU for the sequential formation of [Fe2S2] and [Fe4S4] clusters (Figure 1, I) [11–14]. Subsequently, a [Fe4S4] cluster pair (designated the K-cluster) is transferred from NifU to NifB and processed into a [Fe8S9C] cluster (designated the L-cluster), which is nearly indistinguishable from the M-cluster in structure except for the substitution of Fe for Mo/homocitrate at one end of the cluster (Figure 1, II) [15–19]. The L-cluster is then transferred from NifB to NifEN, where it is matured into an M-cluster upon NifH-mediated insertion of molybdenum and homocitrate prior to the delivery of the M-cluster to its target location in NifDK (Figure 1, III) [20–23].
Figure 1. Assembly of nitrogenase cofactor.
Actions of a series of assembly proteins lead to the sequential formation of 2Fe (on NifS/U; I), 4Fe (on NifS/U; I) and 8Fe (on NifB; II) clusters prior to the insertion of Mo and homocitrate (hc) by NifH that gives rise to a mature M-cluster (on NifEN, III) and the subsequent delivery of the M-cluster to its target location (in NifDK, IV). NifB catalyzes the K- to L-cluster conversion, which involves radical SAM-dependent carbide insertion concomitant with the insertion of a “9th” sulfur and the rearrangement/coupling of the two 4Fe units of the K-cluster into an 8Fe L-cluster. The characteristic EPR features of the (a) K-, (b) L- and (c) M-clusters are shown below the structural models.
Identification of the assembly pathway of the M-cluster was facilitated by strategic deletions of essential nif genes [24,25], which permitted the capture of biosynthetic intermediates of the M-cluster on various nif-encoded (Nif) assembly proteins (see Figure 1). Subsequent biochemical and EPR analyses demonstrated the sequential K→L→M cluster conversion, which could be monitored by the corresponding changes of the characteristic EPR features of these cluster species in this process (Figure 1, a–c) [15,20,22–24]; whereas XAS/EXAFS and crystallographic studies provided structural proofs for the identities of these biosynthetic intermediates, showing the formation of a complete 8Fe-core of the M-cluster (i.e., the L-cluster) prior to the insertion of Mo and homocitrate (Figure 1, II) [16–18] while assigning a previously-unidentified function to NifH (the reductase component of Mo-nitrogenase) as a Mo/homocitrate insertase that transforms the L-cluster into a fully-matured M-cluster (Figure 1, III) [21].
Overall, assembly of the M-cluster utilizes a fusion strategy that stepwise generates the 2Fe-, 4Fe- and 8Fe-cores. However, there are a number of variations on this theme, including the insertion of carbon and sulfur that occurs concomitant with the coupling and rearrangement of two 4Fe units of the K-cluster into an 8Fe L-cluster (see Figure 1, II), as well as the replacement of a terminal Fe atom of the L-cluster by Mo and homocitrate that leads to the formation of the M-cluster (see Figure 1, III). These variations not only render the M-cluster chemically unprecedented and functionally unique, but also establish this cluster as a multifaceted model system for the investigation of various types of chemistry that are employed for complex metallocofactor assembly. Among them, the NifB-catalyzed carbide insertion is of particular interest [26–29], as it represents a novel, radical SAM-dependent biosynthetic route to complex, bridged metalloclusters.
The NifB protein: a “radical SAM assemblase (RSA)”
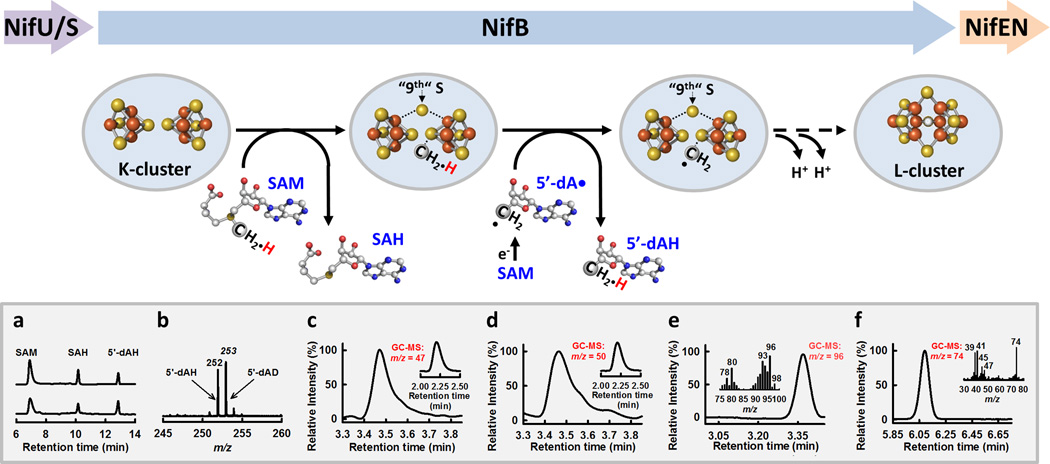
NifB carries a signature CxxxCxxC motif at its N-terminus for the coordination of an S-adenosyl-L-methionine (SAM)-binding [Fe4S4] cluster (designated the SAM-cluster), as well as additional ligands for the accommodation of the two [Fe4S4] units of the K-cluster (a biosynthetic precursor to the M-cluster) (Figure 2) [24,29]. This observation has led to the hypothesis that NifB utilizes radical SAM chemistry for the coupling of the two 4Fe units of the K-cluster into an 8Fe L-cluster. EPR analysis provided the initial proof for this hypothesis, showing the disappearance of the K-cluster-specific, S = 1/2 signal concomitant with the appearance of the L-cluster-specific, g = 1.94 signal (see Figure 1, a, b) [15] upon incubation of NifB with SAM. Subsequent HPLC analysis revealed the identities of the products of SAM cleavage by NifB as those of S-adenosyl-L-homocysteine (SAH) and 5’-deoxyadenosine (5’-dAH) (Figure 2, a) [24], suggesting the utilization of two molecules of SAM in the reaction catalyzed by NifB: one serves as the donor of a methyl group (giving rise to SAH upon removal of the methyl group); the other generates a 5’-dA• radical for hydrogen abstraction (giving rise to 5’-dAH). Isotope labeling experiments further traced the 14C label to the L-cluster when NifB was incubated with [methyl-14C] SAM [24,30] while demonstrating the formation of deuterated 5’-deoxyadenosine (5’-dAD) when NifB was incubated with [methyl-d3] SAM (Figure 2, b) [24], thereby establishing the methyl group of SAM as the source of the interstitial carbide that undergoes an initial processing step upon hydrogen abstraction by a 5’-dA• radical (see Figure 2) [24].
Figure 2. Formation of an 8Fe-core by NifB.
Proposed pathway of carbide insertion, which begins with methyltransfer from one SAM molecule to a sulfide atom of the K-cluster, followed by hydrogen abstraction from this methyl group by a 5’-dA• radical that is derived from a second SAM molecule. The resulting, cluster-bound carbon intermediate (e.g., a methylene radical) then initiates radical chemistry-based rearrangement/coupling of the two 4Fe units of the K-cluster into an 8Fe L-cluster concomitant with the insertion of a “9th” sulfur and further dehydrogenation/deprotonation of the carbon intermediate until a carbide ion appears in the center of the L-cluster. (a) HPLC profile of the standards (upper) and the actual reaction of SAM cleavage by NifB (lower); (b) LC-MS analysis of the 5’-dA species generated upon incubation of NifB with [methyl-d3] SAM; (c–f) GC-MS analyses of products generated upon acid quenching of reactions containing (d) NifB and SAM; (d) NifB and [methyl-14C] SAM; (e) Fe/Se-reconstituted NifB and SAM; and (f) NifB and ally SAM.
A recent study further refined these early steps along the carbide insertion pathway. Incubation of NifB with unlabeled SAM and [methyl-d3] SAM, respectively, resulted in the formation of methanethiol (CH3SH) and methane-d3-thiol (CD3SH), respectively, upon acid quenching, suggesting transfer of the SAM-derived methyl group to the acid-labile sulfur atom of an FeS cluster (Figure 2, c, d) [28]. Substitution of the Fe/Se-reconstituted NifB for the Fe/S-associated NifB in the same reaction led to the formation of methylselenol (CH3SeH), reaffirming the sulfide atom of the FeS precursor on NifB (i.e., the K-cluster) as the point of methyl group attachment (Figure 2, e) [28]. When ally SAM—a SAM analog containing an allyl group (-CH-CH=CH2) in place of the methyl group (-CH3)—was incubated with NifB, SAH was detected as the sole product of SAM cleavage [28]. Interestingly, acid quenching of this incubation mixture resulted in the formation of allylthiol (CH2=CH-CH-SH), suggesting the occurrence of allytransfer in the absence hydrogen abstraction by a 5’-dA• radical (Figure 2, f) [28]. This observation provides compelling evidence that methyltransfer occurs via an SN2-type mechanism prior to the abstraction of hydrogen from this group, resulting in a carbon intermediate (e.g., a methylene radical) that can be further processed into a carbide ion
While the early events of the carbide insertion process have been elucidated through these studies, the post-hydrogen-abstraction events are yet to be explored to address such questions as (i) how is the methyl-derived carbon intermediate processed into a carbide ion; and (ii) where does the “9th” sulfur originate from and how is it incorporated to complete the stoichiometry of the L-cluster? With regard to the former, it is tempting to speculate that a continuation of hydrogen abstraction from the carbon intermediate by SAM-derived 5’-dA• radicals will eventually give rise to a carbide ion, although a reaction mechanism based on acid/base chemistry could be used to accomplish the same task. With regard to the latter, one appealing scenario is that the “9th” sulfur is a “dangling sulfur” attached externally to one Fe atom of the K-cluster in an analogous manner to those observed in the cases of other radical SAM-dependent enzymes like RimO, MiaB and HydG [31–33], as well as the radical enzyme (R)-2-hydroxyisocaproyl-CoA dehydratase [34]; on the other hand, a protein origin of this sulfur atom cannot be excluded, particularly when drawing analogy to the recent observation that one “belt sulfur” of the M-cluster might be temporarily “parked” on a protein residue during the process of substrate turnover by nitrogenase [35–38]. Perhaps the most important question, however, is how radical chemistry enables the rearrangement and coupling of the two 4Fe modules of the K-cluster into an 8Fe L-cluster concomitant with the insertion of both the interstitial carbide and the “9th” sulfur. While the answer to this question remains elusive as of the moment, the capability of NifB to carry out this interesting chemistry establishes this enzyme as an efficient “radical SAM assemblase (RSA)” that specializes in complex metallocofactor assembly.
The NifB family: a new class of radical SAM methyltransferases (RSMTs)
The vast majority of our current knowledge of the NifB protein was derived from studies of this protein from a soil bacterium, Azotobacter vinelandii (designated NifBAv). Recently, two naturally “truncated” NifB homologs have been identified in two nitrogen-fixing methanogenic microorganisms: one (designated NifBMa) from the mesophilic Methanosarcina acetivorans; and the other (designated NifBMt) from the thermophilic Methanobacterium thermoautotrophicum. Unlike NifBAv, NifBMa and NifBMt lack the “NifX domain”—a sequence sharing homology with NifX, an accessory protein of unclear function in nitrogenase assembly–toward the C-termini of their primary sequences [29]. However, both proteins contain the CxxxCxxC motif for the coordination of the SAM-cluster, as well as a sufficient number of conserved Cys and His residues for the accommodation of a K-cluster. Biochemical analyses further demonstrated the functional exchangeability between NifBMa/NifBMt and NifBAv, showing the ability of the truncated NifB proteins to catalyze the same radical SAM-dependent carbide insertion that is coupled to the conversion of K- to L-cluster as their full-length counterpart [29]. This observation is important, as it establishes the minimum sequence requirement for a functional NifB protein that could facilitate mechanistic investigation of this protein in M-cluster assembly.
Subsequent BLAST search have led to the identification of a large number of proteins with high sequence homology to the three NifB proteins. Overall, this family of proteins can be divided into two categories—full-length and truncated–based on the presence or absence of the NifX domain toward the C-termini of their sequences (Figure 3). The hosts of most full-length NifB proteins are proteobacteria; whereas the hosts of the truncated NifB proteins are much more widespread across the microbial biorealm, ranging from euryarcheotes to firmicutes (Figure 3). Interestingly, many hosts of the truncated NifB proteins are not nitrogen-fixing organisms, suggesting that the NifB proteins in these organisms carry out other functions that are yet to be established [29].
Figure 3. Family of NifB proteins.
Phylogenetic analysis divides this protein family into “full-length” (left) and “truncated” (right) NifB proteins based on the presence or absence of the “NifX domain” toward the C-terminus of the sequence.
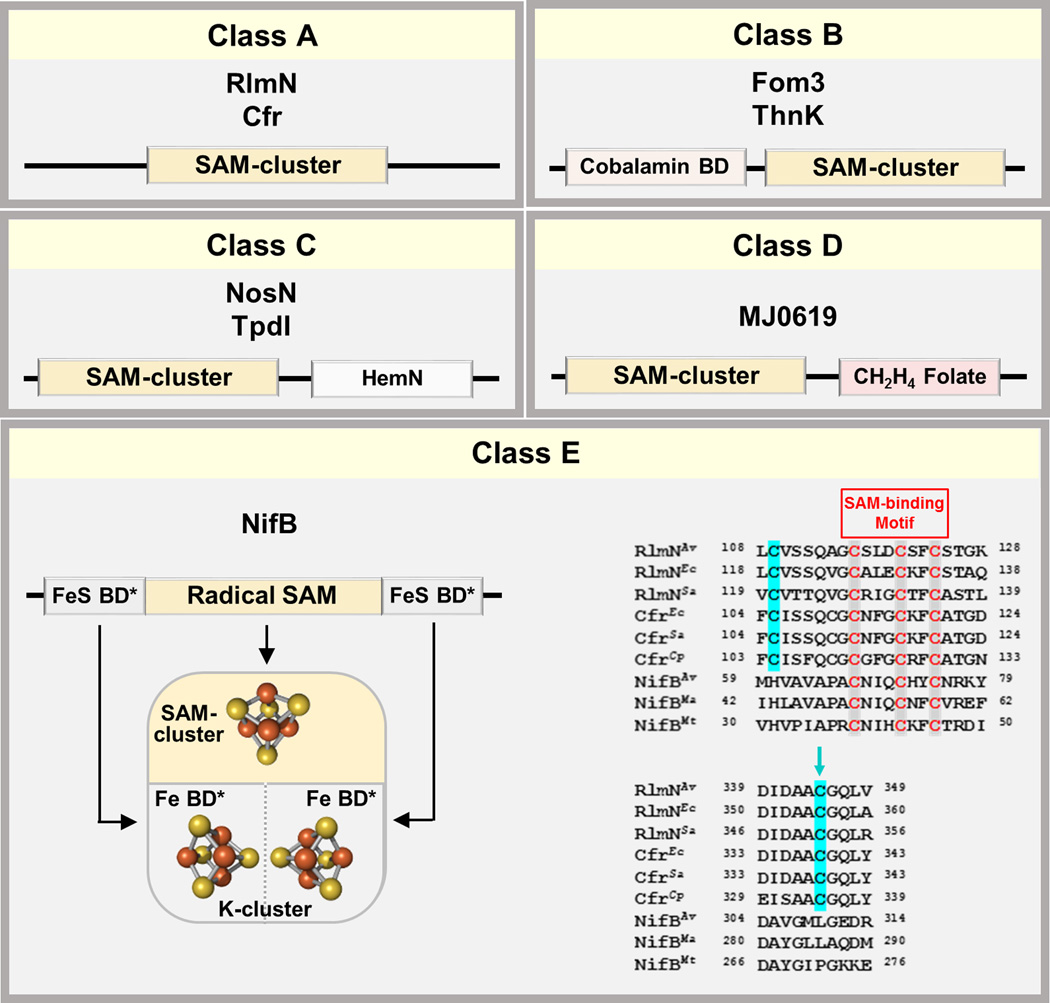
Identification of a family of NifB proteins has led to the categorization of a new class (class E) of radical SAM methyltransferases (RSMTs) (Figure. 4) [29], a large subset of radical SAM enzymes that catalyze methylation reactions using SAM or other methyl donor molecules as co-substrates. Class E RSMTs clearly differ from the previously-defined classes B, C and D of RSMTs, which carry a cobalamin-binding domain, a HemN domain and a methylenetetrahydrofolate domain, respectively, in addition to the canonical radical SAM domain (Figure 4) [39–42]. Moreover, while sharing the same byproducts of SAM cleavage (i.e., SAH and 5’-dAH), class E RSMTs do not possess a pair of conserved Cys residues in the sequences of class A RSMTs (including one that serves as the site of intermediary methylation) and, consequently, do not route methyltransfer via a protein residue (as exemplified by NifBAv, NifBMa and NifBMt) (Figure 4) [39–42]. Instead, this new class of RSMTs contain a number of conserved Cys and His residues that flank the radical SAM domain, which could potentially serve as FeS cluster-binding domains for radical SAM-based assembly of complex metallocenters (Figure 4) [29].
Figure 4. Five classes of radical SAM methyltransferases.
(Upper) Proteins of classes A–D contain a canonical radical SAM domain and two conserved Cys residues (class A), an N-terminus cobalamin binding domain (class B), a C-terminus HemN domain (class C) and a C-terminus methylenetetrahydrofolate domain (class D), respectively. (Lower) Proteins of class E contain a radical SAM domain flanked by conserved Cys and His residues that could potentially serve as FeS cluster-binding domains (left). This class of proteins (represented by NifB) do not contain a pair of conserved Cys residues of class A proteins (represented by RlmN and Cfr) based on partial sequence alignment (right). The blue arrow indicates the site of intermediary methylation in Rlm and Cfr (right). BD, binding domain.
Concluding remarks
NifB plays a pivotal role in the maturation of nitrogenase cofactor, catalyzing radical SAM-dependent carbide insertion concomitant with the generation of a characteristic [Fe8S9C] core that is nearly indistinguishable from, and hence easily convertible into, the [MoFe7C] core of a mature M-cluster. Classification of a family of NifB proteins as a distinct subset of RSMTs not only suggests a broader function of these proteins in the assembly of complex metallocofactors, but also opens up new avenues to study the structure and mechanism of this important protein family, both of which remain relatively uncharacterized and promise to reveal unprecedented chemical reactions catalyzed by biological systems.
Highlights.
The carbide of nitrogense M-cluster originates from the methyl group of SAM.
The insertion of carbide is catalyzed by NifB via a radical SAM-dependent mechanism.
The methyl group is transferred from SAM to an sulfur atom of the M-cluster precursor.
The transfer of methyl to the M-cluster precursor occurs before hydrogen abstraction.
The NifB protein family represents as a new class of radical SAM methyltransferases.
Acknowledgments
This work was supported by NIH Grant R01 GM67626 (to M.W.R.) and a Hellman Fellowship (to Y.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Burgess BK, Lowe DJ. Mechanism of molybdenum nitrogenase. Chem Rev. 1996;96:2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 2.Howard JB, Rees DC. Structural basis of biological nitrogen fixation. Chem Rev. 1996;96:2965–2982. doi: 10.1021/cr9500545. [DOI] [PubMed] [Google Scholar]
- 3.Eady RR. Structure-function relationships of alternative nitrogenases. Chem Rev. 1996;96:3013–3030. doi: 10.1021/cr950057h. [DOI] [PubMed] [Google Scholar]
- 4.Lee CC, Hu Y, Ribbe MW. Vanadium nitrogenase reduces CO. Science. 2010;329:642. doi: 10.1126/science.1191455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Y, Lee CC, Ribbe MW. Extending the carbon chain: hydrocarbon formation catalyzed by vanadium/molybdenum nitrogenases. Science. 2011;333:753–755. doi: 10.1126/science.1206883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebelein JG, Hu Y, Ribbe MW. Differential reduction of CO2 by molybdenum and vanadium nitrogenases. Angew Chem Int Ed Engl. 2014;53:11543–11546. doi: 10.1002/anie.201406863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang ZY, Moure VR, Dean DR, Seefeldt LC. Carbon dioxide reduction to methane and coupling with acetylene to form propylene catalyzed by remodeled nitrogenase. Proc Natl Acad Sci USA. 2012;109:19644–19648. doi: 10.1073/pnas.1213159109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Einsle O, Tezcan FA, Andrade SL, Schmid B, Yoshida M, Howard JB, Rees DC. Nitrogenase MoFe-protein at 1.16 Å resolution: a central ligand in the FeMo-cofactor. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 9.Spatzal T, Aksoyoglu M, Zhang L, Andrade SL, Schleicher E, Weber S, Rees DC, Einsle O. Evidence for interstitial carbon in nitrogenase FeMo cofactor. Science. 2011;334:940. doi: 10.1126/science.1214025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lancaster KM, Roemelt M, Ettenhuber P, Hu Y, Ribbe MW, Neese F, Bergmann U, DeBeer S. X-ray emission spectroscopy evidences a central carbon in the nitrogenase iron-molybdenum cofactor. Science. 2011;334:974–977. doi: 10.1126/science.1206445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng L, White RH, Cash VL, Jack RF, Dean DR. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc Natl Acad Sci USA. 1993;90:2754–2758. doi: 10.1073/pnas.90.7.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith AD, Jameson GNL, Dos Santos PC, Agar JN, Naik S, Krebs C, Frazzon J, Dean DR, Huynh BH, Johnson MK. NifS-mediated assembly of [4Fe-4S] clusters in the N- and C-terminal domains of the NifU scaffold protein. Biochemistry. 2005;44:12955–12969. doi: 10.1021/bi051257i. [DOI] [PubMed] [Google Scholar]
- 13.Yuvaniyama P, Agar JN, Cash VL, Johnson MK, Dean DR. NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc Natl Acad Sci USA. 2000;97:599–604. doi: 10.1073/pnas.97.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng LM, Dean DR. Catalytic formation of a nitrogenase iron-sulfur cluster. J Biol Chem. 1994;269:18723–18726. [PubMed] [Google Scholar]
- 15.Wiig JA, Hu Y, Ribbe MW. NifEN-B complex of Azotobacter vinelandii is fully functional in nitrogenase FeMo cofactor assembly. Proc Natl Acad Sci USA. 2011;108:8623–8627. doi: 10.1073/pnas.1102773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser JT, Hu Y, Wiig JA, Rees DC, Ribbe MW. Structure of precursor-bound NifEN: a nitrogenase FeMo cofactor maturase/insertase. Science. 2011;331:91–94. doi: 10.1126/science.1196954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fay AW, Blank MA, Lee CC, Hu Y, Hodgson KO, Hedman B, Ribbe MW. Spectroscopic characterization of the isolated iron-molybdenum cofactor (FeMoco) precursor from the protein NifEN. Angew Chem Int Ed Engl. 2011;50:7787–7790. doi: 10.1002/anie.201102724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corbett MC, Hu Y, Fay AW, Ribbe MW, Hedman B, Hodgson KO. Structural insights into a protein-bound iron-molybdenum cofactor precursor. Proc Natl Acad Sci USA. 2006;103:1238–1243. doi: 10.1073/pnas.0507853103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lancaster KM, Hu Y, Bergmann U, Ribbe MW, Debeer S. X-ray spectroscopic observation of an interstitial carbide in NifEN-bound FeMoco precursor. J Am Chem Soc. 2013;136:610–612. doi: 10.1021/ja309254g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Y, Fay AW, Ribbe MW. Identification of a nitrogenase FeMo cofactor precursor on NifEN complex. Proc Natl Acad Sci USA. 2005;102:3236–3241. doi: 10.1073/pnas.0409201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Y, Corbett MC, Fay AW, Webber JA, Hodgson KO, Hedman B, Ribbe MW. Nitrogenase Fe protein: a molybdate/homocitrate insertase. Proc Natl Acad Sci USA. 2006;103:17125–17130. doi: 10.1073/pnas.0602651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Corbett MC, Fay AW, Webber JA, Hodgson KO, Hedman B, Ribbe MW. FeMo cofactor maturation on NifEN. Proc Natl Acad Sci USA. 2006;103:17119–17124. doi: 10.1073/pnas.0602647103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshizawa JM, Blank MA, Fay AW, Lee CC, Wiig JA, Hu Y, Hodgson KO, Hedman B, Ribbe MW. Optimization of FeMoco maturation on NifEN. J Am Chem Soc. 2009;131:9321–9325. doi: 10.1021/ja9035225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribbe MW, Hu Y, Hodgson KO, Hedman B. Biosynthesis of nitrogenase metalloclusters. Chem Rev. 2014;114:4063–4080. doi: 10.1021/cr400463x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y, Ribbe MW. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. J Biol Chem. 2013;288:13173–13177. doi: 10.1074/jbc.R113.454041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wiig JA, Hu Y, Lee CC, Ribbe MW. Radical SAM-dependent carbon insertion into the nitrogenase M-cluster. Science. 2012;337:1672–1675. doi: 10.1126/science.1224603. •• This work uses radiolabeling experiments to show that the carbide of the M-cluster originates from the methyl group of SAM and that it is inserted by the assembly protein NifB via a radical SAM-dependent mechanism.
- 27.Boal AK, Rosenzweig AC. Biochemistry. A radical route for nitrogenase carbide insertion. Science. 2012;337:1617–1618. doi: 10.1126/science.1229088. [DOI] [PubMed] [Google Scholar]
- 28. Wiig JA, Hu Y, Ribbe MW. Refining the pathway of carbide insertion into the nitrogenase M-cluster. Nat Commun. 2015;6:8034. doi: 10.1038/ncomms9034. •• This work demonstrates that the methyl group of SAM is transferred to an M-cluster precursor-associated sulfur atom prior to hydrogen abstraction, thereby refining the initial steps of the carbide insertion pathway.
- 29. Fay AW, Wiig JA, Lee CC, Hu Y. Identification and characterization of functional homologs of nitrogenase cofactor biosynthesis protein NifB from methanogens. Proc Natl Acad Sci USA. 2015;112:14829–14833. doi: 10.1073/pnas.1510409112. •• This work categorizes the NifB protein family as a new class of radical SAM methyltransferases.
- 30.Wiig JA, Lee CC, Hu Y, Ribbe MW. Tracing the interstitial carbide of the nitrogenase cofactor during substrate turnover. J Am Chem Soc. 2013;135:4982–4983. doi: 10.1021/ja401698d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landgraf BJ, Arcinas AJ, Lee KH, Booker SJ. Identification of an intermediate methyl carrier in the radical S-adenosylmethionine methylthiotransferases RimO and MiaB. J Am Chem Soc. 2013;135:15404–15416. doi: 10.1021/ja4048448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forouhar F, Arragain S, Atta M, Gambarelli S, Mouesca JM, Hussain M, Xiao R, Kieffer-Jaquinod S, Seetharaman J, Acton TB, et al. Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases. Nat Chem Biol. 2013;9:333–938. doi: 10.1038/nchembio.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinis P, Suess DL, Fox SJ, Harmer JE, Driesener RC, De La Paz L, Swartz JR, Essex JW, Britt RD, Roach PL. X-ray crystallographic and EPR spectroscopic analysis of HydG, a maturase in [FeFe]-hydrogenase H-cluster assembly. Proc Natl Acad Sci USA. 2015;112:1362–1367. doi: 10.1073/pnas.1417252112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knauer SH, Buckel W, Dobbek H. Structural basis for reductive radical formation and electron recycling in (R)-2-hydroxyisocaproyl-CoA dehydratase. J Am Chem Soc. 2011;133:4342–4347. doi: 10.1021/ja1076537. [DOI] [PubMed] [Google Scholar]
- 35. Spatzal T, Perez KA, Einsle O, Howard JB, Rees DC. Ligand binding to the FeMo-cofactor: structures of CO-bound and reactivated nitrogenase. Science. 2014;345:1620–1623. doi: 10.1126/science.1256679. • This work reports the crystal structure of CO-inhibited nitrogenase MoFe-protein, which reveals that a CO molecule is bridged between Fe2 and Fe6 of the M-cluster. The μ2 binding geometry is achieved by replacing a belt-sulfur atom, highlighting the generation of a reactive iron species uncovered by the displacement of sulfur.
- 36.Lee CC, Fay AW, Weng TC, Krest CM, Hedman B, Hodgson KO, Hu Y, Ribbe MW. Uncoupling binding of substrate CO from turnover by vanadium nitrogenase. Proc Natl Acad Sci USA. 2015;112:13845–13849. doi: 10.1073/pnas.1519696112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spatzal T, Perez KA, Howard JB, Rees DC. Catalysis-dependent selenium incorporation and migration in the nitrogenase active site iron-molybdenum cofactor. Elife. 2015;4 pii:e11620. doi: 10.7554/eLife.11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Högbom M. Nitrogenase Mechanism. A dynamic tool for nitrogen reduction. Science. 2014;345:1568. doi: 10.1126/science.1260021. [DOI] [PubMed] [Google Scholar]
- 39.Hutcheson RU, Broderick JB. Radical SAM enzymes in methylation and methylthiolation. Metallomics. 2012;4:1149–1154. doi: 10.1039/c2mt20136d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujimori DG. Radical SAM-mediated methylation reactions. Curr Opin Chem Biol. 2013;17:597–604. doi: 10.1016/j.cbpa.2013.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q, van der Donk WA, Liu W. Radical-mediated enzymatic methylation: a tale of two SAMS. Acc Chem Res. 2012;45:555–564. doi: 10.1021/ar200202c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauerle MR, Schwalm EL, Booker SJ. Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation. J Biol Chem. 2015;290:3995–4002. doi: 10.1074/jbc.R114.607044. [DOI] [PMC free article] [PubMed] [Google Scholar]