Abstract
As B cells engage in the immune response they express the deaminase AID to initiate the hypermutation and recombination of immunoglobulin genes, which are crucial processes for the efficient recognition and disposal of pathogens, However, AID must be tightly controlled in B cells to minimize off-targeting mutations, which can drive chromosomal translocations and the development of B cell malignancies, such as lymphomas. Recent genomic and biochemical analyses have begun to unravel the crucial question of how AID-mediated deamination is targeted outside immunoglobulin genes. Here, we discuss the transcriptional and topological features that are emerging as key drivers of AID promiscuous activity.
When B cells migrate out of the bone marrow as naïve lymphocytes, they carry substantial alterations at their B cell receptor genes. It is estimated that at the conclusion of RAG-mediated V(D)J recombination in the bone marrow, B cells have diversified their immunoglobulin gene repertoire to the extent that they can recognize an astronomical 5 × 1013 different molecules1. Yet this primary repertoire only represents a fraction of the further diversification that occurs in mature B cells and is mediated by the somatic hypermutation (SHM) of variable (V) domains, which increases the affinity of antibody molecules for the immunogen2. In addition to SHM, activated B cells replace their immunoglobulin heavy chain (IgH) constant Cµ domain (IgM isotype) for the constant (C) domain of downstream immunoglobulin isotypes (Cγ, Cα or Cε) which encode the C region for IgG, IgA or IgE respectively. This class switch recombination (CSR) process imparts antibodies with different means to eliminate pathogens and antigens.
Both CSR and SHM are initiated by activation-induced cytidine deaminase (AID; encoded by AICDA), which converts deoxycytidines into deoxyuridines upon recruitment to V and switch (S) recombination sequences3, 4. The uracil base in DNA following deamination engages the activity of base excision repair (BER) and mismatch repair (MMR) pathways, which create nicks and double-strand breaks (DSBs) that initiate SHM and CSR2.
AID activity is predominantly restricted to immunoglobulin genes. To a lesser extent however, AID promiscuously targets a subset of transcriptionally active genes, including the proto-oncogenes B cell lymphoma 6 (BCL6) and MYC5–7. Frequent lesions at these loci can result in mutations or chromosomal translocations, ultimately leading to the dysregulation of the proto-oncogene and B cell tumorigenesis7. Because of the importance of AID in both humoral immunity and lymphomagenesis, the molecular basis for the specificity of AID targeting has attracted considerable attention in the past 5 years.
In this review we summarize recent findings that help explain the affinity of AID for immunoglobulin and non-immunoglobulin loci. We first describe deep-sequencing techniques that were developed to assess AID activity across the genome. We follow with a description of the functional and topological features associated with these sites and the potential ways by which they facilitate AID off-targeting. We end with a discussion of some of the mechanisms B cells have evolved to reduce the probability of oncogenic transformation by AID.
AID off-target hypermutation
The process of SHM was initially thought to be restricted to the immunoglobulin loci. However, the BCL6 proto-oncogene was often found mutated at promoter proximal sequences in follicular lymphoma and diffuse large B cell lymphoma (DLBCL)8, 9. A priori, these mutations could be the result of tumor instability and selection. However, BCL6 mutations were also observed in primary lymphocytes from healthy donors. Furthermore, the mutations were mostly restricted to BCL6 intron 1 (which coincides with the SHM targeting area at immunoglobulin loci10) and displayed the mutation spectrum that is characteristic of immunoglobulin SHM5, 6. Following this initial discovery, additional genes including CD95, CD79A, CD79B, PIM1, MYC, RHOH and PAX5 were also found to be hypermutated in tumors and primary B cells7.
Repair pathways
The BER and MMR pathways faithfully repair U-G mismatches downstream of AID. In their absence (for example in Msh2−/−Ung−/− mice), replication over unrepaired uracils leads to the accumulation of C to T (and G to A) transition mutations11. Correspondingly, a mutation analysis of selected genes in Msh2−/−Ung−/− Peyer’s patches B cells identified additional AID off-targets12. Intriguingly, this study showed that the protection against AID-mediated mutations conferred by the BER and MMR pathways was locus specific. For instance, whereas Bcl6 mutations were evident in both Msh2−/−Ung−/− and wild-type cells, Myc was only mutated when BER and MMR were impaired12. Why protection against AID-mediated attack by repair pathways varies from gene to gene remains an interesting problem in the field.
Cataloguing the range of AID off-targets
To unravel the true nature of AID’s promiscuous activity, one must first catalogue the full range of AID off-targets. This task is complicated by the fact that DNA occupancy by AID does not predict DNA damage13, 14. Furthermore, early studies only measured SHM by Sanger sequencing at sites of interest. Conversely, the high error rate of high-throughput technologies (~1 miscalled base per 100 nucleotides sequenced15) has for the most part precluded the use of deep-sequencing to measure SHM genome-wide. A recent method, termed mutational analysis by paired-end deep-sequencing (MutPE-Seq)16, makes use of long paired-end reads to offset this ‘mutation’ background. However, similar to conventional methods, this technique can only measure mutations at pre-defined sites that are amplified by PCR. An alternative strategy, termed SHM-Seq, involves the microsequencing of cell subclones from SHM-proficient B cell lines, where acquired mutations are present at frequencies similar to single nucleotide polymorphisms (SNPs)17. In a recent study using this SHM-Seq approach, mutations were measured in DNA associated with trimethylated histone H3 Lys4 (H3K4me3), which is an epigenetic mark that closely overlaps with AID activity. The assay also involved long-term inhibition of MSH2 and UNG and overexpression of AID in the human Ramos–Burkitt lymphoma line, which undergoes trace levels of constitutive hypermutation. Furthermore, the accumulation of mutations was facilitated by enhanced expression of AID under the control of an enhancer-promoter cassette from the immunoglobulin κ-chain gene (Igκ) (first described in REF.18). This study17 confirmed that AID mediates the hypermutation of genes implicated in B cell transformation, including BCL6, MYC, BCL7A, MSH6, MIR142 and ID3, and identified several new features of AID off-targeting (discussed below).
Mapping AID off-target DNA breaks and translocations
Long before the discovery of AID, genetic studies revealed chromosomal translocations involving immunoglobulin genes in human B cell tumors7. These rearrangements altered the expression of proto-oncogenes due to their juxtaposition to potent immunoglobulin enhancers. Given that DNA is physiologically remodeled at immunoglobulin loci, it was logical to regard DNA breaks intermediate to CSR as substrates for translocations in lymphoma precursors. This hypothesis was tested in BALB/c mice19–21, in which intraperitoneal injection of pristane oil or transgenic overexpression of IL-6 causes Myc–Igh rearrangements and neoplasia22. The requirement for AID-mediated breaks in these translocations was demonstrated by their substantial reduction in Aicda−/− or Ung−/− mice (in the absence of Ung, mutations are not converted into DNA breaks)19–21.
Still unanswered was whether AID is also responsible for the DSBs at translocating oncogenes. To explore this, restriction sites for the I-SceI endonuclease were knocked-in at Myc and Igh, and artificial DSBs were induced at either locus in B cells undergoing CSR. In this setting, Myc translocation to Igh was detected only in AID-proficient cells23. Since Myc is only one of many damaged oncogenes in B cell malignancies, two high-throughput technologies (TC-Seq and HTGTS) were developed to systematically identify translocation hotspots involving AID-dependent breaks24, 25. The techniques were applied to ex vivo-activated B cells24, 25 and more recently to germinal center B cells from Plasmodium chabaudi-infected mice16. Altogether, the experiments revealed that AID targets >100 genes, including at oncogenes that are frequently translocated in lymphoma.
TC-Seq and HTGTS revealed translocation hotspots caused by AID. Detecting the actual AID-mediated breaks behind the translocations was more challenging. For instance, attempts to ChIP-Seq non-homologous end joining (NHEJ) factors, such as Nbs1 or γH2AX, did not uncover reproducible or robust hotspots of AID-mediated DNA damage26, 27. NHEJ proteins typically accumulate as foci of >100Kb in size around DSBs28. Although, such foci are clearly visible by microscopy in single cells, the extent of damage at off-target sites appears to be too infrequent in wild-type cells to be detected in the millions of cells used by ChIP-Seq assays. Consequently, the only bona fide (>100Kb) repair focus visualized by γH2AX ChIP-Seq was at Igh26. This is consistent with the fact that AID lesions at Igh are considerably more frequent than at off-target sites.
To map AID-induced DSBs across the genome the RPA-Seq technique was then developed. In this assay, AID was overexpressed from an Igκ promoter-enhancer cassette18, and the DSBs were visualized in H2AH and 53BP1 deficient B cells13, 14. In the H2ax−/− or 53bp1−/− genetic background NHEJ is crippled, so that AID-mediated lesions occurring in G1 transit unrepaired to the S-G2M phases of the cell cycle. DSBs are then resected and exposed ssDNA recruit massive amounts of homologous recombination repair factors, such as RPA and RAD51, which are then visualized by ChIP-Seq26. Under these more sensitive conditions, 235 high-confidence AID targets were identified17.
In summary, the development of TC-Seq, HTGTS, SHM-Seq, and RPA-Seq were instrumental at defining the full spectrum of AID off-targets in the mouse and human B cell genomes (see BOX 2). As discussed in detail below, recent studies from the International Cancer Genome Consortium unexpectedly uncovered a new activity for AID outside the antibody gene loci. Notably, essentially all of the AID targets identified by the new studies overlap with those catalogued by the aforementioned deep-sequencing methods.
Box 2: Deep-sequencing techniques.
To characterize AID activity across the genome, a variety of techniques based on deep-sequencing have been implemented. MutPE-Seq (mutational analysis by pair-end sequencing) measures SHM at specific genomic sites16. The domain to be interrogated is amplified by PCR and sequenced from both ends with a MiSeq 600 machine. Mutations found in both pair-end reads are considered bona fide nucleotide changes. SHM-Seq17 reveals mutations on promoter and enhancer DNA pulled down by immunoprecipitation of H3K4me3+ chromatin (ChIP-Seq). This global approach is most sensitive when repair pathways that counteract AID activity (BER and MMR) are deleted, so that AID-induced mutations accumulate over cell division. Translocations have been measured by two very similar approaches, translocation-capture sequencing (TC-Seq24) and high-throughput genomic translocation sequencing (HTGTS25). Both techniques rely on the constitutive induction of DNA breaks by the endonuclease I-SceI in cells carrying I-SceI restriction sites. AID-mediated or random DNA breaks that translocate to the I-SceI-mediated lesions are captured by PCR and deep-sequencing. The DNA breaks intermediate to translocations are detected by RPA-Seq. In this approach NHEJ proteins are deleted so that DSBs in G1 transit unrepaired to S and G2M stages of the cell cycle where they are resected by HR repair enzymes. ssDNA recruits massive amounts of HR factors RPA and Rad51, which are readily detected by ChIP-Seq. Finally, the potential function of AID in DNA methylation has been assessed by mapping the methylation status of AID-targeted genes in resting and activated B cells (R.C., unpublished observations). The technique used was Bi-Seq43, an approach that relies on bisulfite conversion of cytidines into uracils. As methylated cytidines are resistant to conversion, genome sequencing of bisulfite treated DNA can readily discriminate between methylated and unmethylated cytidines.
Kataegis: a new AID-induced lesion?
Whole genome sequencing of a wide range of human tumors uncovered a new type of off-targeting activity for AID and related deaminases. By means of rainfall plots, which display intermutation distance per chromosome, the studies revealed multiple mutation clusters of <10Kb in size in most of the tumors analyzed29, 30. Several features distinguished these ‘mutation storms’ (termed kataegis) from random substitutions, which are typically scattered across tumor genomes at a distance of ~0.1-1Mb from each other. First, kataegic mutations were predominantly (>70%) C-to-T transitions. Second, mutations at kataegis were largely fixed in the same DNA strand, a sign of catalytic processivity. Third, kataegis were frequently associated with genomic rearrangements, including localized chromothripsis, which is a phenomenon first described in cancer cells where extensive DNA damage leads to the shattering and reassembly of entire chromosomes31.
Based on these features, kataegis was proposed to be the result of processive cytidine deamination of ssDNA exposed by the resection of DSBs during repair32. As most mutated cytidines were found within TCX trinucleotides, the responsible factors were proposed to be apolipoprotein-B mRNA-editing catalytic subunit 3A (APOBEC3A) and APOBEC3B, which preferentially deaminate such triplets in vitro and in vivo29, 33. However, in contrast to non-B cell tumors, mutations in human lymphomas derived from germinal center B cells occurred preferentially within the context of the WRCY hypermutation hotspots, providing evidence that AID is responsible for the presence of kataegis in these cells29. A comparison of DLBCL and breast tumors revealed additional differences between B and non-B cell kataegis17. Whereas >80% of DLBCL kataegis were associated with transcription start sites (TSSs), only ~5% of them were found near promoters in breast cancer. Furthermore, kataegis in DLBCL were recurrent at immunoglobulin loci and mouse orthologue AID off-target sites. Conversely, in non-B cell tumors kataegis were randomly distributed across the genome, mostly at intergenic domains. AID was thus implicated in the etiology of targeted kataegis in human lymphomas, whereas APOBEC3A and APOBEC3B were proposed to generate kataegis at random sites in non-B cell tumors.
A key question is whether kataegis represent a mechanism that is distinct from and independent of SHM. Four lines of evidence suggest that this may very well be the case. One, with the exception of a few genes (e.g. BCL6), high levels of SHM are typically not observed outside the immunoglobulin loci12. In contrast, AID-mediated kataegis are characterized by large numbers of clustered mutations near promoters of off-target genes17. Two, during kataegis, substitutions are introduced in a processive manner in the same DNA strand. Conversely, although AID can act processively in vitro34, AID-mediated mutations accumulate in vivo in a stepwise manner, with only a few unlinked mutations being fixed per cell division2. Three, about half of SHM substitutions are transition mutations, compared to >70% in kataegis. This disparity fits well with the proposal that kataegis likely occurs in the S phase of the cell cycle32, where uracils are mostly replicated over into thymidines or adenines (C>T and G>A transition mutations). Four, in yeast, deletion of ung1 impairs the formation of kataegis35. At the same time, non-clustered hypermutation increases in the absence of ung1 in yeast cells35, and in Ung−/− B cells36. This suggests that kataegis occurs downstream of DSBs, in contrast to SHM, where DNA breaks are not obligate intermediates2.
Based on the above considerations it is plausible that kataegis in human lymphomas engages AID activity at two separate stages (Figure 1). Analogous to SHM, AID might first deaminate ssDNA at off-targets exposed by transcription in G1. The processing of deoxyuridine lesions by BER and MMR would then lead to occasional DSBs, which in S or G2M would be resected by the homologous recombination pathway. During or following this resection, AID may deaminate the exposed ssDNA in a processive manner, leading to long stretches of kataegic mutations. The model predicts that this second step would be independent of transcription, which is blocked following DNA damage37, 38. Another important prediction is that, in contrast to AID, APOBEC enzymes should induce kataegis in non-B cell tumors by deaminating ssDNA predominately downstream of random lesions. A resolution of these questions awaits additional biochemical and genetic experiments.
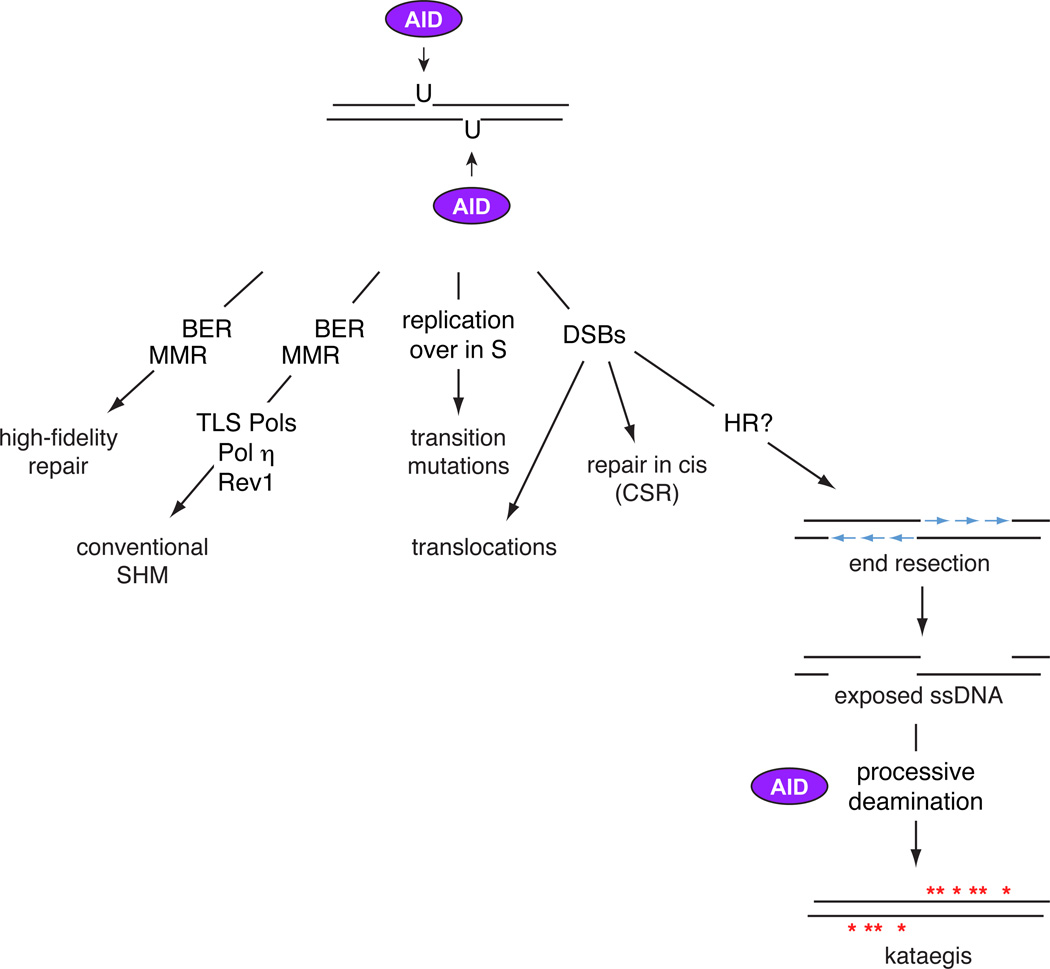
Figure 1. Differential processing of AID lesions.
The deamination of genomic DNA by AID can be processed in at least 5 distinct modes. a) High-fidelity repair by BER and MMR factors can revert deoxyuridines to deoxycitidines. b) The BER and MMR pathways, in combination with translesion polymerases such as Rev1 and Polη, carry out conventional SHM. c) DNA replication over deoxyuridines in the S phase of the cell cycle leads to transition mutations. d) The formation of staggered DSBs are either repaired in cis by NHEJ leading to CSR, or in trans leading to chromosomal translocations. e) If DNA breaks are unrepaired as the cell moves to the S or G2M stages of the cell cycle, attempts to initiate homologous recombination promotes the resection of DNA ends. The resulting ssDNA are potential targets to AID, which by processive deamination may generate kataegis.
Does promiscuous AID activity play a physiological role?
Before the DNA demethylation function of TET proteins was firmly established, there was no clear mechanism driving DNA demethylation in vertebrates. Several possibilities were thus put forward, including the deamination of CpGs by AID and related enzymes39. This was an attractive idea because deamination could in theory lead to the replacement of methylated cytidines with unmethylated nucleotides. However, although several reports have attempted to link AID to DNA demethylation (reviewed by39), the theory faces a number of key challenges. First and foremost, AID-deficient mice do not show any obvious developmental defects, as would be predicted for a factor involved in such a critical function. Second, AID expression is confined to the B cell compartment and is not expressed by germline cells at physiological levels. Third, it is unclear how AID would access methylated CpGs in the first place, since they are by and large transcriptionally silent. Fourth, no statistically significant differences in gene expression have been reported between Aicda−/− and wild-type B cells40–42. Fifth, of the 235 AID targets identified in mouse B cells, only 32 (13%) are significantly demethylated during B cell activation, as measured by high-coverage Bi-Seq43 (R.C., unpublished observations). Thus, a role for AID in DNA demethylation seems unlikely.
It is also important to point out that if AID promiscuous activity was in any way purposeful or had a useful role, one would expect AID targets to be evolutionarily conserved between different species. Yet, the overlap between AID off-targets in mouse and human B cells is less than 50%17. Interestingly, a recent report showed that AID is also expressed in self-reactive bone marrow B cells, and proposed that, when combined with RAGs, AID genotoxic activity might help remove autoreactive clones from the B cell compartment44. The idea is supported on the observation that B cells treated with shRNAs against AID fail to undergo central tolerance when they recombine self-specificities in humanized mice44. We note that under this scenario the precise genes where off-targeting damage occurs become irrelevant, so long as the damage induces apoptosis.
Thus, the available evidence so far argues against the idea that oncogenes may somehow benefit from AID off-targeting activity, but raises the intriguing possibility that the organism perhaps does employ AID promiscuity to delete autoreactive lymphocytes from the B cell repertoire.
What recruits AID activity to off-target sites?
Super-enhancers
Genomic studies that simultaneously measured DBSs, translocations, and nuclear interactions in stimulated B cells showed that interchromosomal contacts cannot explain the extent or the location of AID-induced translocations45. Instead, oncogenes are rearranged in a manner directly proportional to the frequency of AID-mediated damage. Such findings challenged long-held ideas that Igh preferentially interacts with its translocating partners in the activated B cell nucleus46. Thus, other properties intrinsic to off-target genes might enable the recruitment of AID activity. Several such properties were recently uncovered by analyzing AID off-targets within the context of transcription, chromatin and nuclear architecture. The key finding was that AID activity is mostly confined to super-enhancer domains17, 47, 48. As discussed below, the topological and functional properties of these domains fit well with the characteristics that have long been associated with AID activity.
Super-enhancers are composed of large arrays of interconnected promoters and enhancers that display unusually high levels of transcription and epigenetic accessibility43, 49. In particular, the interconnectivity between regulatory elements in super-enhancers mediates transcriptional synergy49, consistent with the observation that promoter potency increases proportionally to the number of associated enhancers43.
The interconnectivity and transcriptional synergy at super-enhancers help explain the unusual distribution of AID targets. Rather than being scattered randomly across the genome, AID-induced breaks are often found clustered into groups of 2 or 3 targets linked by long-range chromatin interactions17. Both promoter–promoter and promoter–enhancer clusters are observed, consistent with the finding that AID-mediated damage extends to transcriptionally active enhancers tethered to a targeted promoter17. The implication is that once recruited to a super-enhancer, AID deaminates topologically linked elements undergoing high levels of RNA synthesis.
Super-enhancers predominantly control transcription of genes that regulate the cell cycle and apoptosis, as well as genes that feature prominently in cell identity. These characteristics accurately describe the kinds of genes recurrently translocated in human lymphomas and mouse B cell tumors. By the same token, the transcriptional, regulatory, and architectural features of super-enhancers are shared between AID off-targets and Igh, Igκ, or Igλ loci. The key implication is that rather than mutating a specific set of genes, AID is summoned by a well-defined nuclear microenvironment, the immunoglobulin loci being its prototype (Figure 2).
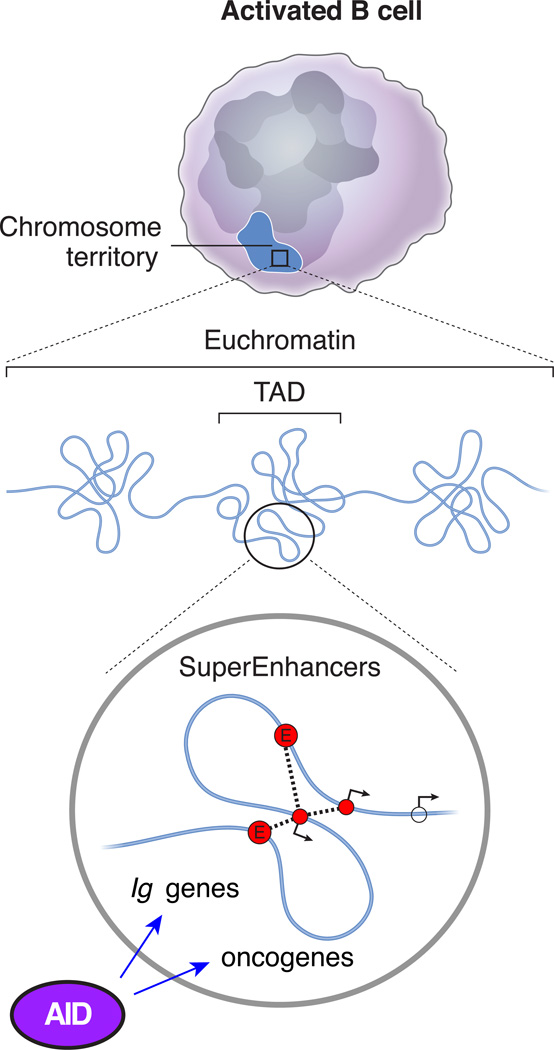
Figure 2. Super-enhancers are preferred targets of AID activity.
Topological, transcriptional, and epigenetic features that render super-enhancer domains ideal targets for AID activity, both at immunoglobulin loci and at selected oncogenes in B cells, as well as in somatic cells when AID is ectopically expressed.
In support of this model, ectopic expression of AID in mouse fibroblasts leads to chromosomal translocations predominantly between genomic sites embedded within super-enhancers17, 48. More importantly, the set of genes translocated in mouse fibroblasts and B cells is vastly different, owing to the fact that genes that define cellular identity (and are thus regulated by super-enhancers) differ in the two cell types. As in B cells, AID translocations in fibroblasts are associated with highly transcribed genes that display polymerase stalling at promoter areas, a key requirement for AID targeting of immunoglobulin genes50. In addition, engagement by AID occurs at sites that share a set of epigenetic marks, including H3K27Ac and H3K36me348. These chromatin modifications provide nucleosome accessibility and might thus contribute to the overall targeting of AID.
An analysis that includes super-enhancers, high levels of transcription, epigenetic accessibility and interconnectivity can predict AID off-targets in a given cell type with ~90% accuracy43. However, the 10% false discovery rate indicates that additional contributing factors are likely at play. Potential candidates include transcription factors, such as E2A, NF-κB and PAX5, which have long been implicated in the targeting of SHM in B cells51. However, the idea that these factors are essential for AID recruitment is challenged by at least two observations. First, as aforementioned, ectopically expressed AID targets super-enhancers in fibroblasts17, 48, where B cell factors are either not expressed or are likely functionally irrelevant. Second, AID is active and appears to contribute to cell transformation when aberrantly expressed in non-hematopoietic cells52. Most likely then, although hematopoietic transcription factors create an AID permissive microenvironment in B cells, analogous factors must do the same in other cell types. Cell-identity transcription factors might thus recruit AID by their capacity to assemble super-enhancers.
Other factors that associate with AID and might facilitate its recruitment to super-enhancers are the RNA Pol II complex53, the Pol II-associated factors SPT550 and SPT654, the Pol II elongation PAF complex55, and chromatin modifiers, such as KAP156. As discussed in detail in the next two subsections, the RNA exosome complex57 and the splicing machinery might also play a role58, 59.
The RNA exosome
CSR and translocations require nicks on both strands of DNA to produce DSBs11. Notably, in in vitro deamination assays, or when ectopically expressed in bacteria, AID predominantly targets the non-template strand60, 61. Conversely, AID can mutate both DNA strands at sites of stalled polymerases in yeast and mammalian B cells59, 62. The obvious inference is that an unknown mechanism renders the template strand accessible to AID in eukaryotes. A biochemical screen was developed to identify such a mechanism62. It revealed multiple subunits of the RNA exosome complex that associate with AID and facilitate SHM on both DNA strands (Figure 3A).
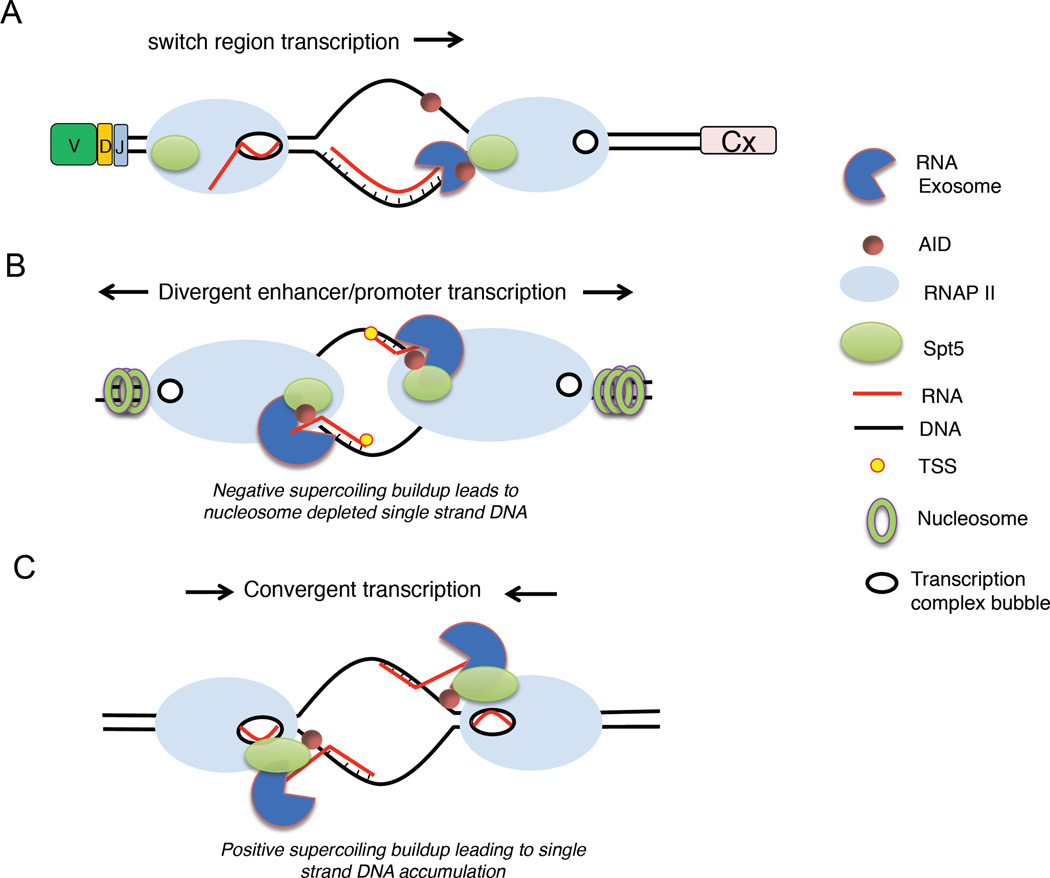
Figure 3. Potential mechanisms whereby the RNA exosome facilitates AID targeting.
(A) Transcription of S regions creates R-loops that cause stalling of RNA pol II, early transcription termination and RNA exosome recruitment. (B) Divergent transcription at enhancers and promoters create nucleosome-free DNA and ssDNA structures where cognate RNAs may associate and become substrates for exosome degradation. These activities are proposed to increase accessibility to AID. (C) Convergent transcription mediated by pol II may lead to the formation of RNA exosome substrates by the buildup of positive DNA supercoiling. SPT5, transcription elongation factor SPT5; TSS, transcription start site.
The exosome is a 3’-5’ RNA exonuclease complex that degrades non-coding RNAs. In the cytoplasm, it targets mRNAs that are not translated and in the nucleus it degrades prematurely terminated transcripts near promoter and enhancer transcriptional start sites (xTSS-RNAs63–65). Compared to silent loci, sites associated with xTSS-RNAs are in general more fragile in nature, due to the frequent presence of secondary DNA structures (e.g. R-loops or G-quartets), dynamic binding of regulatory factors, extensive chromatin remodeling, and topoisomerase activity24, 25. The exosome can thus be thought of as a caretaker of RNA-mediated genomic instability, by facilitating DNA repair at TSS-proximal sequences.
xTSS-RNAs are largely transcribed in a divergent orientation by polymerases moving in opposite directions63, 64 (Figure 3B). At enhancers, both sense and antisense transcripts are degraded by the exosome with equal efficiency, whereas antisense transcripts are preferentially targeted at promoters66. This is due to the fact that most sense transcripts enter productive elongation and undergo early transcription termination less frequently. In addition, the exosome degrades antisense RNAs synthesized from internal TSSs at genes such as Myc and at the S domains of Igh63. Since these genes also produce sense transcripts, the sites display convergent transcription originating from head-to-head TSSs47 (Figure 3C).
The in vivo role of the exosome in relation to AID was assessed by conditional deletion of EXOSC3, one of its 11 core-subunits. Exosome-deficient B cells displayed a reduction in CSR and SHM63, 64, implying that exosome-mediated degradation of Igh RNAs facilitates AID activity. The current model57 posits that convergent and divergent transcription elicit supercoilicity on TSS-proximal DNA67, leading to nucleosome eviction. Either the xTSS-RNAs or the exosome may then help stabilize the exposed ssDNA leading to AID attack (Figure 3). In this model it is important not to confuse, as is sometimes done, the ssDNA targeted by AID with the 8–22bp transcription bubble that is formed within the polymerase complex68. The latter can be accessed by nucleotides during the transcription reaction but not by proteins.
In the context of CSR, the creation of ssDNA targets might be enhanced by the highly repetitive nature of S regions, which upon transcription form long stretches of ssDNA structures69, extensive Pol II stalling70, 71, and xTSS-RNA synthesis63. It is important to point out however that this model does not account for AID targeting of immunoglobulin V sequences. These genes are not repetitive in nature, lack the polymerase-stalling capacity of S sequences, and do not seem to engage convergent or divergent transcription. How the exosome might then facilitate V gene hypermutation is thus unclear. Intriguingly, recent studies in ex-vivo cultures and in germinal center B cells clearly showed that V and S domains are simultaneously targeted by AID and, most importantly, at comparable frequencies72. Thus, the long-held notion that SHM and CSR are mechanistically or spatiotemporally uncoupled might not be correct.
Outside the immunoglobulin loci, exosome activity partially overlaps with AID off-targeting. A comparative analysis between the transcriptomes of wild-type and EXOSC3-deficient B cells reveal a genome-wide stabilization of xTSS-RNAs at a fraction of promoters and enhancers, some of which are AID off-targets47, 63. Furthermore, xTSS-RNAs-associated genes are highly transcribed and often associated with super-enhancers64. The key question however is whether these are correlative features or whether the exosome directly facilitates AID mistargeting. In support of the latter, deletion of sequences at the 5’-end of Pim1 and Cd83 TSSs reduces mutation of these genes in CH12 cells overexpressing AID63.
In conclusion, the data indicate the exosome facilitates targeting of AID to both DNA strands at S domains during CSR. It remains to be determined to what extent SHM of V genes and of-target sites rely on the exosome.
Targeting AID by mRNA splicing
In this section we summarize the potential role of splicing in targeting AID to the Igh locus and potentially to off-target sites. Unlike Igκ or Igλ, Igh constant domains are independent transcriptional units, with isotype-specific promoters that drive sterile transcription of intervening (I) exons, intronic S regions, and CH exons. S regions span 1–12kb of DNA and are composed of tandem repeats of AID-hotspot RGYW motifs. Upon transcription, S repeats form R-loops (e.g. G-quadruplexes), which enables the formation of RNA-DNA hybrids between the G-rich transcripts and the template strand69, 73 (Figure 4). These conformations are believed to expose or stabilize ssDNA for AID attack74, 75.
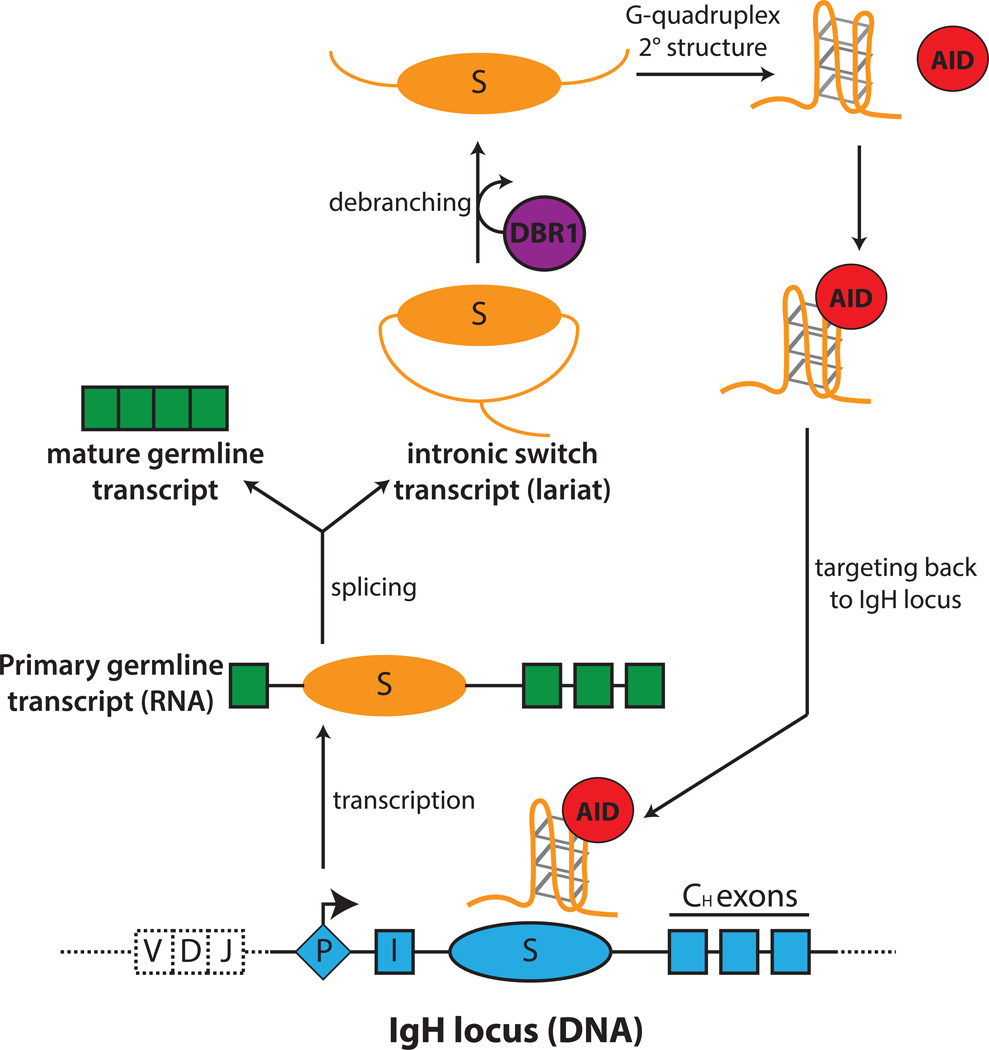
Figure 4. RNA-mediated targeting of AID to switch-region DNA.
When B cells are stimulated to undergo CSR, transcription occurs at each of the recombining switch (S) regions to produce primary switch transcripts. Primary transcripts are spliced to generate a mature germline transcripts and intronic switch region transcripts (lariat intermediate). Debranching enzyme 1 (DBR1) catalyzes the release of the lariat from the spliceosome and debranches the switch transcript into its linear form. The linear switch transcript, free of exonic sequences, can function as a guide RNA by forming a G-quadruplex structure, which allows its association with AID. AID, bound to the guide RNA, is targeted specifically by sequence information provided by the guide RNAs to the complementary S region DNA.
During splicing, sterile mRNAs are partitioned into mature transcripts and lariat intronic S region transcripts (Figure 4). Early studies suggested a critical role for this splicing event in CSR, as deletion of the Iγ1 splice donor markedly reduced recombination76, 77. This finding implied that either the spliced transcripts or the splicing machinery were crucial for efficient CSR. A functional link between AID and the splicing machinery has been further suggested since by the frequent coimmunoprecipitation of AID with splicing factors, including CTNNBL1, heterogeneous nuclear ribonucleoproteins (hnRNPs), nucleolin, and PTBP278–80. More recently, AID was also found to associate with sense S region RNA58. Biophysical analysis showed that S region RNA folds into G-quadruplexes, and that AID associates with these structures in a glycine 133-dependent manner58. Without the ability to bind S region RNA, AID G133V mutants fail to localize to S regions and support CSR58. The post-transcriptional process of lariat debranching, which catalyzes the formation of linear RNA from branched spliced introns, is an additional requirement for RNA-guided AID targeting. Knockdown of the debranching enzyme DBR1 decreased the localization of AID to S regions, and mice deficient in DBR1 displayed reduced CSR. Critically, in DBR1-depleted B cells, AID localization to S regions and CSR could be rescued by the ectopic expression of S transcripts, but only in the sense orientation58.
These findings support a model in which spliced, debranched sense S RNA forms G-quadruplexes, recruit AID, and target AID–RNA complexes back to S regions to enable CSR (Figure 4). Conversely, anti-sense S transcripts do not form G-quadruplexes and are largely dispensable for CSR81. An important implication is that unspliced transcripts either cannot form G-quadruplex structures due to steric constraints or are unable to efficiently act as an AID guide. Furthermore, the observation that deletion of the core Sµ region still allows substantial CSR82 suggests that the G-quadruplex RNA generated from the residual Sµ region can efficiently target AID to DNA.
A key challenge is to identify the precise biochemical mechanism that targets AID-RNA complexes to S regions. One possibility, analogous to CRISPR-Cas9 targeting, is that sequence complementarity between S region RNA and S region DNA mediates the interaction. This scenario would require either the displacement or the collapse of R-loops, because S region RNA-DNA hybrids are exceptionally stable and unlikely to be outcompeted for access to S region DNA73.
IgV regions and most gene transcripts do not form G-quadruplexes or R-loops. Hence, a link between splicing and SHM or off-targeting is unclear. It is also noteworthy that A:T-rich S regions from Xenopus laevis can serve as CSR substrates when inserted into mouse B cells, suggesting that additional mechanisms can target AID to these regions83. Nevertheless, mRNA splicing might play a role in AID mistargeting of at least a subset of genes. For instance, some reports have correlated AID activity at non-immunoglobulin loci to G-richness84. Furthermore, transcription of the immunoglobulin-translocation partners MYC, BCL6 and RHOH in lymphoma cells appears to induce the formation of G-loops85. This raises the possibility that G-quadruplexes, whether at the DNA or RNA level, could enhance AID accessibility to oncogenes and other loci. The G133V AID mutant, which cannot localize to S regions, provides an opportunity to explore such ideas.
In summary, the recent findings cement a role for mRNA splicing in AID targeting during CSR. It now remains to be determined whether this mechanism also facilitates SHM of V genes or off-targeted oncogenes.
Keeping AID off-targeting activity at bay
B cells have developed a plethora of mechanisms that tightly control AID mRNA and protein abundance, nuclear access, preferential targeting to immunoglobulin loci, and catalytic activity. In addition, DNA repair pathways eliminate DSBs and off-targeting SHM and actively remove cells bearing AID-induced translocations. The picture emerging from the available data suggests that these mechanisms ensure an optimal equilibrium between the ability to mount efficient antibody responses and the risk of oncogenic transformation, but do not play a major role in defining the specificity of AID targeting.
Transcriptional and post-transcriptional regulation
In mammals, Aicda transcription is maximal in activated or germinal center B cells, where T cell-derived cytokines (e.g. IL-4 and pro-inflammatory cytokines) promote transcription factor binding to upstream regulatory elements within the Aicda locus4, 43, 86–90. Aicda is also induced by T cell-independent factors, including BAFF, APRIL, Toll-like receptor (TLR) signaling, and female sex hormones87, 91–93.
Both general and B cell-specific transcription factors activate Aicda, including NF-κB, PAX5, STAT6, IRF4, C/EBP, E-proteins and FOXO189, 90, 94–97. Conversely, ID2 and BLIMP1 repress Aicda and promote B cell differentiation to the plasma cell stage87, 96, 97. Inflammatory cues also promote ectopic Aicda expression in non-germinal center B cells upon viral infection87, 98.
Post-transcriptionally, Aicda expression is regulated by the microRNAs miR-155 and miR-181b, which downmodulate AID protein levels and activity99–101. Conversely, in AID+ chronic myeloid leukaemia and acute B lymphoblastic leukaemia cells, miR-155 is not expressed, a feature that may exacerbate the pathological role of AID in those malignancies102.
The extent of antibody diversification and chromosomal translocations are directly proportional to Aicda expression levels. In Aicda+/− mice, SHM, CSR and chromosomal translocations are reduced approximately by half103, 104. Correspondingly, B cells lacking the miRNAs that regulate AID display increased CSR but also tumor-inducing translocations99–101, 105. Interestingly, although enforced overexpression of Aicda can be oncogenic18, AICDA levels in cancer cells are usually similar to or lower than those in normal B cells (reviewed in REF. 106). This suggests that AID off-targeting does not require overexpression. Indeed, low Aicda expression in pre-B cells or normal AID levels in germinal center B cells are sufficient to generate oncogenic lesions107, 108.
Protein compartmentalization and stability
AID is a nuclear–cytoplasmic shuttling protein109–111. Under steady state conditions, ~90% of AID is cytoplasmic, where its half-life is substantially longer than in the nucleus112. The mechanisms that regulate AID subcellular localization and protein stability establish a dynamic equilibrium that might help reduce AID pathological activity while allowing efficient antibody diversification.
In the cytoplasm, AID is stabilized by the HSP90 chaperone pathway113, which includes HSP90 itself and the farnesylated co-chaperone DNAJA1, one of 42 DnaJ proteins in vertebrates114. The HSP90–DNAJA1 complex regulates AID protein levels for optimal antibody diversification. Indeed, activated B cells from DnaJa1−/− mice exhibit a 50% reduction in AID protein levels and a proportional decrease in SHM and CSR114. Similarly, pharmacological inhibition of HSP90 results in a dose-dependent decrease in AID protein and activity113, 115. As such, these inhibitors could be valuable to reduce AID-dependent clonal evolution in B cell malignancies 115.
In addition to HSP90, cytoplasmic AID exists within a high molecular weight complex containing the translation elongation factor EEF1A116. The AID–HSP90 and AID–EEF1A complexes are physically and functionally distinct in that they both regulate AID half-life by different pathways117. HSP90 stabilizes immature AID, while EEF1A keeps AID out of the nucleus113, 117, where AID is actively targeted to the proteasome via the adaptor REGγ and an unknown E3 ubiquitin ligase112, 118. AID shuttling in and out of the nucleus is controlled by at least three mechanisms: one, a nuclear export signal (NES) at the C-terminal of AID is recognized by the exportin CRM1, which shuttles proteins and RNAs out of the nucleus in eukaryotes119. The association of AID with CRM1 is likely regulated by the RAS-related nuclear protein (RAN)–GTP/GDP differential on either side of the nuclear membrane. Irreversible inhibition of CRM1 by leptomycin B moderately increases the proportion of nuclear AID109–111, 117. Two, AID’s molecular mass of 24 kDa is below the maximum size for passive nuclear diffusion through the nuclear pore. In spite of this, AID is purposely imported into the nucleus by a process that probably involves karyopherin α and karyopherin β79, 120. Three, a proactive nuclear import is required for AID because it is sequestered in the cytoplasm by a multiprotein complex containing a transfer RNA-free version of EEF1A, which functions in a manner unrelated to its role in protein biosynthesis116, 117, 120. AID interacts with the EEF1A complex via a domain that partially overlaps with, but is distinct from, its NES. The stoichiometry of the association of AID with EEF1A suggests that this complex is the major cytoplasmic reservoir of AID121. Molecular modeling and structure–function analysis suggest that the EEF1A–binding motif and the NES of AID reside on opposite sides of an amphipathic helix. If confirmed, this feature explains how the small C-terminal domain of AID mediates both cytoplasmic retention and nuclear import117, 120.
Despite its accumulation in the cytoplasm, AID does not seem to play a role in this compartment. However, it is possible that this partitioning is a relict of an ancient AID role against viral infections, played now by the APOBEC enzymes, which evolved from AID122. Cytoplasmic retention might have later evolved to minimize AID off-targeting activity in the nucleus.
Post-translational modifications
AID is phosphorylated in at least five residues. PKA phosphorylates AID at Ser38 apparently only within the context of Igh chromatin123, 124. This modification enhances AID enzymatic activity during CSR and SHM123–125. An obvious implication of constraining AID Ser38 phosphorylation to the Igh is that it might help prevent the formation of chromosomal translocations, although this idea has not been directly tested. Three other phosphorylation events at Ser3, Ser27, and Thr140 have been shown to either inhibit or enhance AID enzymatic activity, but only Thr140 has a known physiological role so far, in that it potentiates SHM and CSR126–128. Finally, Tyr184 at the C-terminal of AID is phosphorylated in vivo but no role in AID compartmentalization or activity is apparent129.
Figure 5 integrates the various mechanisms that regulate AID expression and activity into a single model. HSP90 stabilizes metastable AID in the cytoplasm, probably until AID adopts a conformation that permits its association with the EEF1A complex113, 117. Nuclear export and cytoplasmic retention act in parallel to exclude AID from the nucleus117. Disruption of the AID–EEF1A interaction facilitates CSR but also increases the likelihood of chromosomal translocations117. Inhibiting HSP90 reduces both antibody diversification and off-target effects113, 115, whereas releasing AID from EEF1A increases both CSR and translocations117. These conflicting observations might reflect the fact that when associated with HSP90, AID has not yet acquired a stable conformation, whereas the opposite is true when AID is complexed with EEF1A117. Similar to transcriptional regulation, neither of these mechanisms controls AID targeting specificity but rather they determine the magnitude of its activity. Intriguingly, CRM1 inhibition does not affect CSR, despite increasing AID nuclear levels117. Thus, nuclear export and cytoplasmic retention may not be functionally equivalent. Once in the nucleus, AID is destabilized by REGγ and ubiquitylation112, 118. REGγ-deficient B cells display increased levels of nuclear AID leading to higher CSR but, interestingly, they do not display an increase in chromosomal translocations118. This is in contrast to inhibition of EEF1A and may reflect the fact that REGγ regulates AID at a latter step, perhaps following targeting of the immunoglobulin gene loci, whereas when released from EEF1A AID is capable of targeting the entire genome.
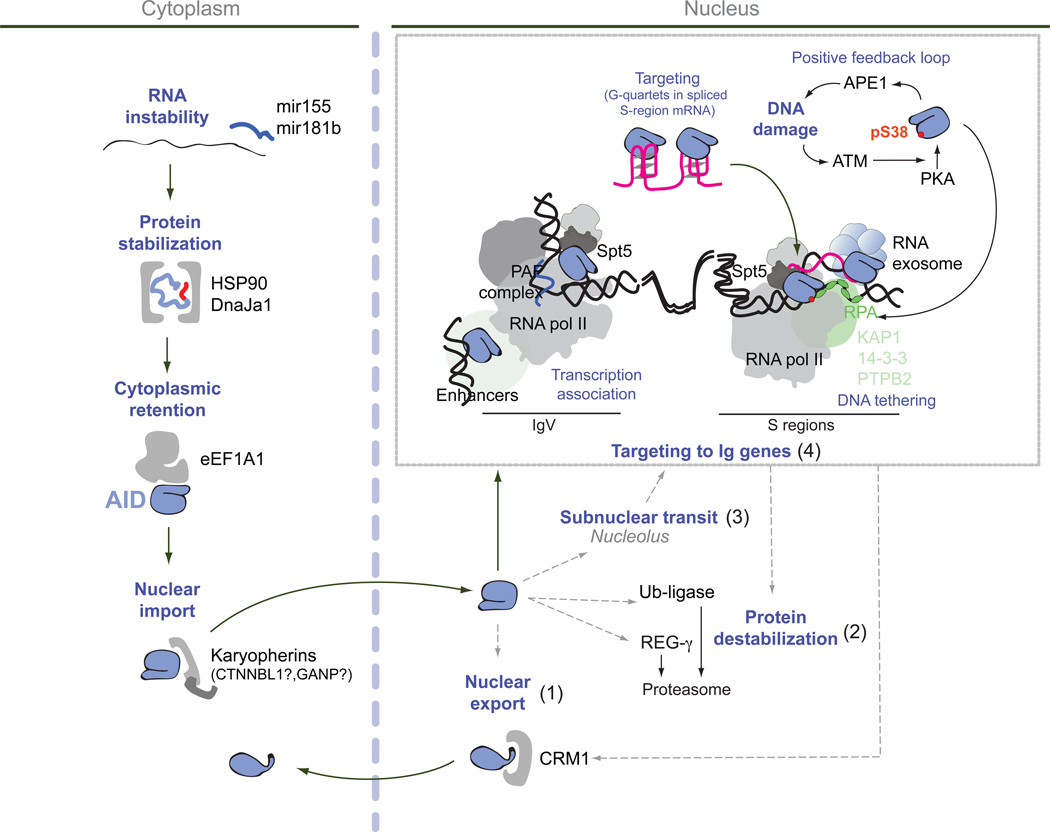
Figure 5. Integrative scheme of the mechanisms that regulate AID activity.
AID transits through HSP90- and eEF1A–containing complexes in the cytoplasm that control its functional maturation before being imported into the nucleus. Whether nuclear import of AID is regulated or stochastic is unknown. In the nucleus AID has multiple possible destinations. It can be: one, exported back to the cytoplasm, two, destabilized by proteasomal degradation, three, associate with the nucleolus and four, be recruited to the chromatin of immunoglobulin variable (IgV) and/or S regions as well as to off-target genes. It remains to be determined to what extent these various destinations are sequential while AID travels to the Ig loci, or compete for AID or synergize to minimize off-targeting. Targeting of AID to the IgV or S-regions requires strong transcription by RNA polymerase II associated with Spt5 and the PAF complex in the context of a specific chromatin microenvironment that includes abundant topological associations of multiple enhancers. Transcription can lead to DNA supercoiling and expose ssDNA that is stabilized by RPA. The repetitiveness of S-regions facilitates the formation of R-loops, exposing the untranscribed DNA strand. The exosome helps recruit AID to the template strand of R loops by degrading R-loop associated transcripts. AID is also recruited by genomic regions displaying extensive convergent or divergent transcription. A number of chromatin associating factors might help tether AID to chromatin, at least at S regions. These include KAP1, 14-3-3 and PTBP2. Once recruited, AID initiates a series of events leading to the formation of DNA breaks (after excision of uracil by UNG and the DNA nicking activity of the endonuclease APE1), which activates the DNA damage response kinase ATM. ATM promotes PKA-mediated phosphorylation of AID at serine 38. This phosphorylation increases AID activity and allows AID association to RPA. In addition, AID binds to G-quartet structures formed in the debranched intron of the S-region sterile transcript, which presumable pairs back with the DNA and helps targeting AID for CSR. Black arrows indicate mechanisms thought to act in chronological order and/or that are directly linked. Dashed grey arrows indicate possible but not yet empirically demonstrated connections.
DNA repair mechanisms both curb and facilitate AID pathological activity. First, as previously discussed, BER and MMR pathways reverse off-targeting hypermutation and the extent of this activity appears to differ depending on the genomic locus12. At the same time, it should be noted that BER and MMR generate DNA nicks and gaps, which are either filled in by DNA polymerases to allow for the full spectrum of SHM, or result in DSBs that promote CSR and chromosomal translocations21, 130. Finally, multiple components of the DNA damage response, including ATM, NBS1 and p53 eliminate cells carrying AID-induced chromosomal translocations130. This mechanism does not regulate the targeting specificity of AID but is key to prevent AID-induced B cell lymphomas 18.
In conclusion, the accumulation of AID in the B cell nucleus is controlled by the interplay between nuclear import and export, cytoplasmic retention, AID protein turnover, and possibly cell cycle regulation (Box 1; FIG. 5). DNA repair pathways provide an additional layer of control. The expectation is that the cross-talk between these regulatory pathways ultimately dictates the extent of SHM and CSR, as well as off-target deamination and damage. However, it is important to point out that these mechanisms limit the magnitude but do not control the specificity of AID off-targeting activity.
Box 1: Regulation of AID activity during the cell cycle.
AID-mediated deamination and its processing by BER or MMR is constrained to the G1 phase of the cell cycle131, 132. Likewise, downstream DSBs appear to be rapidly repaired in G126, 133, 134. B cells lacking factors necessary for NHEJ or homologous recombination accumulate AID-mediated breaks in S and G2/M26, 135, These lesions might represent delayed repair or in some cases active deamination at S and G2M. Some mechanisms that might restrict AID activity to G1 include stage-specific regulation of AID protein localization and/or stability136, 137, phosphorylation, availability of co-factors, or the regulation of nuclear AID stability by cyclins, as has been recently proposed138. To date, this topic remains largely unexplored.
Moving forward
Driven by advances in genomics, AID off-targeting activity has been narrowed down in the past few years to a nuclear microenvironment characterized by the presence super-enhancers, extensive interconnectivity between regulatory elements, high levels of convergent and divergent transcription, and a high level of RGYW accessibility. The remaining task is to elucidate the factors rendering these microenvironments preferred AID targets. A large number of co-immunoprecipitation and genetic studies have consistently implicated three general mechanisms in AID activity: splicing, transcription (mediated by Pol II-associated proteins and transcription factors), and RNA degradation. However, demonstrating the role of the isolated factors in AID targeting has been complicated by the fact that such activities are often essential to cell proliferation, which is in turn required for AID activity. Furthermore, knockdown experiments are by definition incomplete and do not always replicate under different culture conditions or in conditional knockout settings. Thus, new techniques that enable acute and transient depletion of proteins of interest are needed to overcome such shortcomings. Solving these challenges will be key to achieving a full understanding of the mechanisms that facilitate the high affinity of AID for antibody genes, and its lesser but pathological off-targeting of selected oncogenes.
Glossary Terms
- V(D)J recombination
Somatic rearrangement of variable (V), diversity (D) and joining (J) regions of the genes that encode antibody and T cell receptor proteins. The combinatorial nature of V(D)J recombination and the distribution of recombining genes in the vertebrate genome creates repertoire diversity of T and B cell surface receptors
- Somatic hypermutation (SHM)
A unique mutation mechanism that is targeted to the variable regions of rearranged immunoglobulin gene segments. Combined with selection for B cells that produce high-affinity antibody, SHM leads to affinity maturation of B cells in germinal centres
- Class-switch recombination (CSR)
A recombinational process that replaces the immunoglobulin heavy chain constant region Cµ (which encodes the Fc portion of IgM) for that of a downstream isotype Cγ Cα or Cε which encode the constant region of IgG, IgA and IgE, respectively
- DNA deamination
Removal of an amine group from pyrimidine or purine nucleic-acid bases. Deamination of cytosine and adenosine yields uracil and inosine, respectively
- Base-excision repair (BER)
A DNA-repair pathway that removes uridine nucleotides from DNA, that arise by spontaneous of purposeful deamination of cytidines. Repair is initiated by the DNA glycosylase UNG that excises the uracil base, followed by cleavage of the abasic site by the apurinic apyrimidinic endonuclease 1 (APE1)
- Mismatch repair (MMR)
A repair pathway that removes mismatched base pairs from DNA that result from errors made by replicative DNA polymerases or from deamination by AID and APOBEC deaminases. Repair involves the removal of a tract of DNA including the mismatch, and re-copying of the complementary strand. This pathway is mediated by proteins MSH2 and MSH6 among others
- Chromosomal translocations
Aberrant joining of DNA breaks from heterologous chromosomes that do not normally pair during mitosis or meiosis
- AID off-targeting activity
Promiscuous AID-mediated cytidine deamination of genomic sites other than immunoglobulin gene loci
- Enhancer
A regulatory DNA element that recruits transcription factors and influences the rate of gene expression. Enhancers function in an orientation- and position-independent manner (that is, they can function either upstream or downstream of the associated gene, or in an intron). They believe to associate to promoters via long-range chromatin interactions
- Non-homologous DNA end joining (NHEJ)
A repair pathway that joins broken DNA ends without depending on extended homology. Components of this pathway include the proteins Ku70, Ku80, ARTEMIS, X-ray repair cross-complementing protein 4 (XRCC4), DNA ligase IV, and the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs)
- Kataegis
Clusters of mutations (mostly transitions) in the same DNA strand introduced in tumor genomes by cytidine deaminases: APOBEC enzymes in non-B cell tumors and AID in B cell lymphomas
- Chromothripsis
Clustered and massive chromosomal rearrangments in one or several chromosomes of primary or transformed cells. This process occurs as a result of a catastrophic event in the history of the cells and promotes tumor development and congenital diseases
- Transition mutations
Base changes in DNA in which a cytidine (C) or thymidine (T) is replaced by a T or a C, respectively. A to G and G to A mutations are also transitions
- Homologous recombination
DNA repair pathway that makes use of homologous sequences (e.g. homologous chromosomes) as template to repair a DSB. The process involves resection of DNA ends, recruitment of RPA and Rad proteins, strand invasion of the intact sequence, DNA synthesis, ligation, and resolution
- TET proteins
A family of proteins that catalyze the conversion of methylated cytidines to hydroxymethylated cytidines. This step initiates a series of catalytic events that leads to DNA demethylation
- Super-enhancer
A cluster of transcriptional regulatory elements (promoters and enhancers) associated by long-range chromatin loops. They tend to modulate gene expression as a unit
- RNA exosome complex
Multiprotein intracellular complex that degrades short RNA molecules in the 3’-5’ orientation
- xTSS-RNAs
Transcription start site (TSS)-associated antisense transcripts that can exceed 500 base pairs in length and are transcribed divergently from cognate coding genes. These RNAs are mostly degrated by the exosome complex
- G-quadruplexes
Non-B DNA structures that form at G rich sequences. By means of Hoogsteen hydrogen bonding guanines create square planar structures known as tetrads. Two or three tetrads can stack on top of each other to form a quadruplex
Footnotes
Competing interests statement
There is no competing interest.
References
- 1.Pieper K, Grimbacher B, Eibel H. B-cell biology and development. J Allergy Clin Immunol. 2013;131:959–971. doi: 10.1016/j.jaci.2013.01.046. [DOI] [PubMed] [Google Scholar]
- 2.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 3.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2)[see comments] Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu M, et al. Class switch recombination hypermutation require activation-induced cytidine deaminase (AID), a potential RNAediting enzyme [see comments] Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 5.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 6.Pasqualucci L, et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci U S A. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- 8.Migliazza A, et al. Frequent somatic hypermutation of the 5’ noncoding region of the BCL6 gene in B-cell lymphoma. Proc Natl Acad Sci U S A. 1995;92:12520–12524. doi: 10.1073/pnas.92.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaidano G, et al. Frequent mutation of the 5’ noncoding region of the BCL-6 gene in acquired immunodeficiency syndrome-related non-Hodgkin’s lymphomas. Blood. 1997;89:3755–3762. [PubMed] [Google Scholar]
- 10.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 11.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Liu M, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 13.Yamane A, et al. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakim O, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lou DI, et al. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc Natl Acad Sci U S A. 2013;110:19872–19877. doi: 10.1073/pnas.1319590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robbiani DF, et al. Plasmodium Infection Promotes Genomic Instability and AID-Dependent B Cell Lymphoma. Cell. 2015;162:727–737. doi: 10.1016/j.cell.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian J, et al. B Cell Super-Enhancers and Regulatory Clusters Recruit AID Tumorigenic Activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robbiani DF, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramiro AR, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Kovalchuk AL, et al. AID-deficient Bcl-xL transgenic mice develop delayed atypical plasma cell tumors with unusual Ig/Myc chromosomal rearrangements. J Exp Med. 2007;204:2989–3001. doi: 10.1084/jem.20070882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovalchuk AL, et al. Mouse model of endemic Burkitt translocations reveals the long-range boundaries of Ig-mediated oncogene deregulation. Proc Natl Acad Sci U S A. 2012;109:10972–10977. doi: 10.1073/pnas.1200106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potter M. Neoplastic development in plasma cells. Immunol Rev. 2003;194:177–195. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- 23.Robbiani DF, et al. Activation Induced Deaminase is Required for the Chromosomal Translocations in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein IA, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamane A, et al. RPA accumulation during class switch recombination represents 5’-3’ DNA-end resection during the S-G2/M phase of the cell cycle. Cell Rep. 2013;3:138–147. doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staszewski O, et al. Activation-induced cytidine deaminase induces reproducible DNA breaks at many non-Ig Loci in activated B cells. Mol Cell. 2011;41:232–242. doi: 10.1016/j.molcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakofsky CJ, et al. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7:1640–1648. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burns MB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 35.Taylor BJ, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife (Cambridge) 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue K, Rada C, Neuberger MS. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J Exp Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kruhlak M, et al. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- 38.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hogenbirk MA, et al. Differential programming of B cells in AID deficient mice. PLoS One. 2013;8:e69815. doi: 10.1371/journal.pone.0069815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dominguez PM, et al. DNA Methylation Dynamics of Germinal Center B Cells Are Mediated by AID. Cell Rep. 2015;12:2086–2098. doi: 10.1016/j.celrep.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fritz EL, et al. A comprehensive analysis of the effects of the deaminase AID on the transcriptome and methylome of activated B cells. Nat Immunol. 2013;14:749–755. doi: 10.1038/ni.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kieffer-Kwon K-R, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantaert T, et al. Activation-Induced Cytidine Deaminase Expression in Human B Cell Precursors Is Essential for Central B Cell Tolerance. Immunity. 2015;43:884–895. doi: 10.1016/j.immuni.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hakim O, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aiden EL, Casellas R. Somatic Rearrangement in B Cells: It’s (Mostly) Nuclear Physics. Cell. 2015;162:708–711. doi: 10.1016/j.cell.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng FL, et al. Convergent Transcription at Intragenic Super-Enhancers Targets AID-Initiated Genomic Instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q, et al. Epigenetic targeting of activation-induced cytidine deaminase. Proc Natl Acad Sci U S A. 2014;111:18667–18672. doi: 10.1073/pnas.1420575111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pavri R, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michael N, et al. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity. 2003;19:235–242. doi: 10.1016/s1074-7613(03)00204-8. [DOI] [PubMed] [Google Scholar]
- 52.Takai A, et al. A novel mouse model of hepatocarcinogenesis triggered by AID causing deleterious p53 mutations. Oncogene. 2009;28:469–478. doi: 10.1038/onc.2008.415. [DOI] [PubMed] [Google Scholar]
- 53.Nambu Y, et al. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- 54.Begum NA, Stanlie A, Nakata M, Akiyama H, Honjo T. The histone chaperone Spt6 is required for activation-induced cytidine deaminase target determination through H3K4me3 regulation. J Biol Chem. 2012;287:32415–32429. doi: 10.1074/jbc.M112.351569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willmann KL, et al. A role for the RNA pol II-associated PAF complex in AID-induced immune diversification. J Exp Med. 2012;209:2099–2111. doi: 10.1084/jem.20112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeevan-Raj BP, et al. Epigenetic tethering of AID to the donor switch region during immunoglobulin class switch recombination. J Exp Med. 2011;208:1649–1660. doi: 10.1084/jem.20110118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pefanis E, Basu U. RNA Exosome Regulates AID DNA Mutator Activity in the B Cell Genome. Adv Immunol. 2015;127:257–308. doi: 10.1016/bs.ai.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng S, et al. Non-coding RNA Generated following Lariat Debranching Mediates Targeting of AID to DNA. Cell. 2015;161:762–773. doi: 10.1016/j.cell.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor BJ, Wu YL, Rada C. Active RNAP pre-initiation sites are highly mutated by cytidine deaminases in yeast, with AID targeting small RNA genes. Elife. 2014;3:e03553. doi: 10.7554/eLife.03553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 61.Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- 62.Basu U, et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pefanis E, et al. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014;514:389–393. doi: 10.1038/nature13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pefanis E, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–789. doi: 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim TK, Shiekhattar R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell. 2015;162:948–959. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 68.Barnes CO, et al. Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble. Mol Cell. 2015;59:258–269. doi: 10.1016/j.molcel.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 70.Rajagopal D, et al. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med. 2009;206:1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J Exp Med. 2009;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yeap LS, et al. Sequence-Intrinsic Mechanisms that Target AID Mutational Outcomes on Antibody Genes. Cell. 2015;163:1124–1137. doi: 10.1016/j.cell.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reaban ME, Lebowitz J, Griffin JA. Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin alpha switch region. J Biol Chem. 1994;269:21850–21857. [PubMed] [Google Scholar]
- 74.Chaudhuri J, et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 75.Shinkura R, et al. The influence of transcriptional orientation on endogenous switch region function. Nat Immunol. 2003;4:435–441. doi: 10.1038/ni918. [DOI] [PubMed] [Google Scholar]
- 76.Lorenz M, Jung S, Radbruch A. Switch transcripts in immunoglobulin class switching. Science. 1995;267:1825–1828. doi: 10.1126/science.7892607. [DOI] [PubMed] [Google Scholar]
- 77.Hein K, et al. Processing of switch transcripts is required for targeting of antibody class switch recombination. J Exp Med. 1998;188:2369–2374. doi: 10.1084/jem.188.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nowak U, Matthews AJ, Zheng S, Chaudhuri J. The splicing regulator PTBP2 interacts with the cytidine deaminase AID and promotes binding of AID to switch-region DNA. Nat Immunol. 2011;12:160–166. doi: 10.1038/ni.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu Y, et al. A combined nuclear nucleolar localization motif in activation-induced cytidine deaminase (AID) controls immunoglobulin class switching. J. Mol. Biol. 2013;425:424–443. doi: 10.1016/j.jmb.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 80.Hu W, Begum NA, Mondal S, Stanlie A, Honjo T. Identification of DNA cleavage- and recombination-specific hnRNP cofactors for activation-induced cytidine deaminase. Proc Natl Acad Sci U S A. 2015;112:5791–5796. doi: 10.1073/pnas.1506167112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haddad D, et al. Sense transcription through the S region is essential for immunoglobulin class switch recombination. EMBO J. 2011;30:1608–1620. doi: 10.1038/emboj.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luby TM, Schrader CE, Stavnezer J, Selsing E. The mu switch region tandem repeats are important, but not required, for antibody class switch recombination. J Exp Med. 2001;193:159–168. doi: 10.1084/jem.193.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zarrin AA, et al. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat Immunol. 2004;5:1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- 84.Duquette ML, Pham P, Goodman MF, Maizels N. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 2005;24:5791–5798. doi: 10.1038/sj.onc.1208746. [DOI] [PubMed] [Google Scholar]
- 85.Duquette ML, Huber MD, Maizels N. G-rich proto-oncogenes are targeted for genomic instability in B-cell lymphomas. Cancer Res. 2007;67:2586–2594. doi: 10.1158/0008-5472.CAN-06-2419. [DOI] [PubMed] [Google Scholar]
- 86.Dedeoglu F, Horwitz B, Chaudhuri J, Alt FW, Geha RS. Induction of activation-induced cytidine deaminase gene expression by IL-4 and CD40 ligation is dependent on STAT6 and NFkappaB. Int Immunol. 2004;16:395–404. doi: 10.1093/intimm/dxh042. [DOI] [PubMed] [Google Scholar]
- 87.Crouch EE, et al. Regulation of AID expression in the immune response. J Exp Med. 2007;204:1145–1156. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 89.Huong LT, et al. In vivo analysis of Aicda gene regulation: a critical balance between upstream enhancers and intronic silencers governs appropriate expression. PLoS ONE. 2013;8:e61433–e61433. doi: 10.1371/journal.pone.0061433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tran TH, et al. B cell-specific and stimulation-responsive enhancers derepress Aicda by overcoming the effects of silencers. Nat Immunol. 2010;11:148–154. doi: 10.1038/ni.1829. [DOI] [PubMed] [Google Scholar]
- 91.Castigli E, et al. TACI and BAFF-R mediate isotype switching in B cells. J. Exp. Med. 2005;201:35–39. doi: 10.1084/jem.20032000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He B, Qiao X, Cerutti A. CpG DNA induces IgG class switch DNA recombination by activating human B cells through an innate pathway that requires TLR9 and cooperates with IL-10. J. Immunol. 2004;173:4479–4491. doi: 10.4049/jimmunol.173.7.4479. [DOI] [PubMed] [Google Scholar]
- 93.Pauklin S, Petersen-Mahrt SK. Progesterone inhibits activation-induced deaminase by binding to the promoter. J. Immunol. 2009;183:1238–1244. doi: 10.4049/jimmunol.0803915. [DOI] [PubMed] [Google Scholar]
- 94.Dengler HS, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9:1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gonda H, et al. The balance between Pax5 and Id2 activities is the key to AID gene expression. J Exp Med. 2003;198:1427–1437. doi: 10.1084/jem.20030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 97.Sciammas R, et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 98.Gourzi P, Leonova T, Papavasiliou FN. Viral induction of AID is independent of the interferon and the Toll-like receptor signaling pathways but requires NF-kappaB. J. Exp. Med. 2007;204:259–265. doi: 10.1084/jem.20061801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.de Yébenes VG, et al. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J. Exp. Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dorsett Y, et al. MicroRNA-155 Suppresses Activation-Induced Cytidine Deaminase-Mediated Myc-Igh Translocation. Immunity. 2008;28:1–9. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Teng G, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Klemm L, et al. The B Cell Mutator AID Promotes B Lymphoid Blast Crisis and Drug Resistance in Chronic Myeloid Leukemia. Cancer Cell. 2009;16:232–245. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sernández IV, de Yébenes VG, Dorsett Y, Ramiro AR. Haploinsufficiency of activation-induced deaminase for antibody diversification and chromosome translocations both in vitro and in vivo. PLoS ONE. 2008;3:e3927. doi: 10.1371/journal.pone.0003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takizawa M, et al. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J. Exp. Med. 2008;205:1949–1957. doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang M, Yang Z, Rada C, Neuberger MS. AID upmutants isolated using a high-throughput screen highlight the immunity/cancer balance limiting DNA deaminase activity. Nat. Struct. Mol. Biol. 2009;16:769–776. doi: 10.1038/nsmb.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Orthwein A, Di Noia JM. Activation induced deaminase: how much and where? Semin Immunol. 2012;24:246–254. doi: 10.1016/j.smim.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 107.Swaminathan S, et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol. 2015;16:766–774. doi: 10.1038/ni.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pasqualucci L, et al. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 109.Brar SS, Watson M, Diaz M. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J Biol Chem. 2004;279:26395–26401. doi: 10.1074/jbc.M403503200. [DOI] [PubMed] [Google Scholar]
- 110.Ito S, et al. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci U S A. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Aoufouchi S, et al. Proteasomal degradation restricts the nuclear lifespan of AID. J Exp Med. 2008;205:1357–1368. doi: 10.1084/jem.20070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Orthwein A, et al. Regulation of activation-induced deaminase stability and antibody gene diversification by Hsp90. J Exp Med. 2010;207:2751–2765. doi: 10.1084/jem.20101321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Orthwein A, et al. Optimal functional levels of activation-induced deaminase specifically require the Hsp40 DnaJa1. EMBO J. 2012;31:679–691. doi: 10.1038/emboj.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Montamat-Sicotte D, et al. HSP90 inhibitors decrease AID levels and activity in mice and in human cells. Eur. J. Immunol. 2015 doi: 10.1002/eji.201545462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hasler J, Rada C, Neuberger MS. Cytoplasmic activation-induced cytidine deaminase (AID) exists in stoichiometric complex with translation elongation factor 1α (eEF1A) Proc. Natl. Acad. Sci. U.S.A. 2011;108:18366–18371. doi: 10.1073/pnas.1106729108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Methot SP, et al. Consecutive interactions with HSP90 and eEF1A underlie a functional maturation and storage pathway of AID in the cytoplasm. J Exp Med. 2015;212:581–596. doi: 10.1084/jem.20141157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Uchimura Y, Barton LF, Rada C, Neuberger MS. REG-γ associates with modulates the abundance of nuclear activation-induced deaminase. J. Exp. Med. 2011;208:2385–2391. doi: 10.1084/jem.20110856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hutten S, Kehlenbach RH. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 2007;17:193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 120.Patenaude A-M, et al. Active nuclear import cytoplasmic retention of activation-induced deaminase. Nat. Struct. Mol. Biol. 2009;16:517–527. doi: 10.1038/nsmb.1598. [DOI] [PubMed] [Google Scholar]
- 121.Hasler J, Rada C, Neuberger MS. Cytoplasmic activation-induced cytidine deaminase (AID) exists in stoichiometric complex with translation elongation factor 1alpha (eEF1A) Proc Natl Acad Sci U S A. 2011;108:18366–18371. doi: 10.1073/pnas.1106729108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Moris A, Murray S, Cardinaud S. AID and APOBECs span the gap between innate and adaptive immunity. Front Microbiol. 2014;5:534. doi: 10.3389/fmicb.2014.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McBride KM, et al. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc Natl Acad Sci U S A. 2006;103:8798–8803. doi: 10.1073/pnas.0603272103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vuong BQ, et al. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat Immunol. 2009;10:420–426. doi: 10.1038/ni.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Basu U, et al. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 126.Vaidyanathan B, Yen WF, Pucella JN, Chaudhuri J. AIDing Chromatin and Transcription-Coupled Orchestration of Immunoglobulin Class-Switch Recombination. Front Immunol. 2014;5:120. doi: 10.3389/fimmu.2014.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Demorest ZL, Li M, Harris RS. Phosphorylation directly regulates the intrinsic DNA cytidine deaminase activity of activation-induced deaminase and APOBEC3G protein. J Biol Chem. 2011;286:26568–26575. doi: 10.1074/jbc.M111.235721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McBride KM, et al. Regulation of class switch recombination and somatic mutation by AID phosphorylation. J Exp Med. 2008 doi: 10.1084/jem.20081319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Patenaude AM, Di Noia JM. The mechanisms regulating the subcellular localization of AID. Nucleus. 2010;1:325–331. doi: 10.4161/nucl.1.4.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ramiro AR, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Faili A, et al. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat Immunol. 2002;3:815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- 132.Sharbeen G, Yee CW, Smith AL, Jolly CJ. Ectopic restriction of DNA repair reveals that UNG2 excises AID-induced uracils predominantly or exclusively during G1 phase. J Exp Med. 2012;209:965–974. doi: 10.1084/jem.20112379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Petersen S, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schrader CE, Guikema JEJ, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J. Immunol. 2007;179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- 135.Hasham MG, et al. Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination. Nat Immunol. 2010;11:820–826. doi: 10.1038/ni.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lackey L, Law EK, Brown WL, Harris RS. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle. 2013;12:762–772. doi: 10.4161/cc.23713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Le Q, Maizels N. Cell Cycle Regulates Nuclear Stability of AID and Determines the Cellular Response to AID. PLoS Genet. 2015;11:e1005411. doi: 10.1371/journal.pgen.1005411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.He M, Cortizas EM, Verdun RE, Severinson E. Cyclin-dependent kinases regulate Ig class switching by controlling access of AID to the switch region. J Immunol. 2015;194:4231–4239. doi: 10.4049/jimmunol.1402146. [DOI] [PMC free article] [PubMed] [Google Scholar]