Abstract
Background
Childhood wheezing and asthma vary greatly in clinical presentation and time course. The extent to which phenotypic variation reflects heterogeneity in disease pathways is unclear.
Objective
To assess the extent to which single nucleotide polymorphisms (SNPs) associated with childhood asthma in a genome-wide association study are predictive of asthma-related phenotypes.
Methods
In 8365 children from a population based birth cohort, the Avon Longitudinal Study of Parents and Children, allelic scores were derived based on between 10 and 215,443 SNPs ranked according to inverse of the p-value for their association with physician diagnosed asthma in an independent genome-wide association study (6176 cases and 7111 controls). We assessed the predictive value of allelic scores for asthma-related outcomes at age 7-9 years (physician’s diagnosis, longitudinal wheezing phenotypes, and measurements of pulmonary function, bronchial responsiveness and atopy).
Results
Scores based on the 46 highest-ranked SNPs were associated with the symptom-based phenotypes persistent (P<10-11, area under ROC curve (AUC)=0.59) and intermediate onset (P<10-3, AUC=0.58) wheeze. Among lower-ranked SNPs (ranks 21,545-46,416), there was evidence for associations with diagnosed asthma (P<10-4, AUC=0.54) and atopy (P<10-5, AUC=0.55). We found little evidence of associations with transient early wheezing, reduced pulmonary function or non-asthma phenotypes.
Conclusion
The genetic origins of asthma are diverse and: some pathways are specific to wheezing syndromes while others are shared with atopy and bronchial hyper-responsiveness. Out study also provides evidence of aetiological differences among wheezing syndromes.
Keywords: Respiratory Sounds, Hypersensitivity, Respiratory Function Tests, Genome-Wide Association Study, Forecasting
Introduction
Childhood asthma and wheezing vary greatly in clinical presentation and time course1 and are thought to have heterogeneous aetiologies. Children with wheeze can be classified into more homogeneous phenotypes1–7, but it is unclear whether these reflect distinct disease pathways. The aetiological relationship of asthma with intermediate phenotypes such as atopy, bronchial hyper-responsiveness (BHR) and reduced pulmonary function is unclear, although twin and family studies have found evidence of shared genetic components8–15.
Genome-wide association studies (GWAS) of asthma have provided evidence for both common and distinct pathways16–19. Among loci robustly associated with asthma susceptibility16, one (ORMDL3/GSDML) was primarily associated with childhood asthma but not with atopy16, 17, while associations with eosinophil count and allergic disease have been reported for others (IL1RL1 and IL33)17, 20. The hypothesis that loci associated with lung function18, 19 also confer susceptibility to asthma was not confirmed, suggesting distinct pathways for these phenotypes16.
To address the problem of false discovery associated with multiple testing in GWAS, stringent genome-wide significance thresholds are used. As a result, it is likely that many true associations do not reach these thresholds (false negatives) and are therefore neither reported nor considered for further replication21. Genome-wide prediction – examining the combined effect of a large number of variants simultaneously rather than separately – retrieves some of the predictive information lost by dismissing false negatives22–25. Including genome-wide information, rather than only the variants reaching stringent significance thresholds, can improve prediction of disease status for a number of complex diseases24, 25. Furthermore, this approach can be used to discover shared genetic liability between different complex phenotypes25.
The work presented here aimed to construct genetic prediction scores using genome-wide data from the childhood cohorts of the GABRIEL (A Multidisciplinary Study to Identify the Genetic and Environmental Causes of Asthma in the European Community) consortium16, and use these to predict childhood asthma and a range of asthma- and non-asthma-related phenotypes in independent data from a large population-based birth cohort.
Methods
Methods are described in more detail in the Online Repository.
Study population and data collection
ALSPAC is a longitudinal population-based birth cohort study that recruited 14,541 pregnant women in Avon, UK, with expected dates of delivery April 1991-December 1992. Additional 542 eligible pregnancies were recruited when children were aged 7yrs. These 15,083 pregnancies resulted in 14,610 live births. The study has been described elsewhere26, 27. Each year from 0-8yrs, study mothers were sent child health questionnaires including detailed questions on respiratory symptoms. We included only unrelated children of European ancestry. Ethical approval for the study was obtained from the ALSPAC Law and Ethics Committee and the Local Research Ethics Committees.
Genotyping and quality control
Children of the GABRIEL consortium including ALSPAC were genotyped for >500,000 SNPs. GWAS results for asthma from all childhood cohorts of the GABRIEL consortium were downloaded from http://www.cng.fr/gabriel/16. We included SNPs satisfying the following quality control criteria in ≥1 cohort: minor allele frequency ≥5%, consistency with Hardy-Weinberg equilibrium (P>10-4) in controls, and call rate ≥97%.
Construction of genetic scores
Log odds-ratios (logORs) for childhood asthma from GABRIEL cohorts excluding ALSPAC (18 cohorts with 6176 cases and 7111 controls, referred to as ‘discovery set’) were combined using random-effects meta-analysis and SNPs were ranked in inverse order of resulting P-values. In ALSPAC, 440,524 genotyped SNPs satisfied quality control criteria and overlapped with SNPs included in the discovery set. Prediction scores were calculated using the k highest-ranking SNPs, with k=10, 22, 46, 100, 215, 464, 1000, 2154, 4642, 10000, 21544, 46416, 100000, and 215443 (multiplying successively by 100.33 to achieve an even spacing on the log-scale). Two types of scores were calculated: ‘log odds scores’ using pooled logORs as weights for the allele count, and ‘allele count scores’ using the signs of the logORs (+1 if logOR>0, -1 if logOR<0). The prediction scores were constructed using the PLINK program28 (version 1.07).
Outcomes in mid-childhood
ALSPAC children were classified according to whether they had ever received a physician diagnosis of asthma or reported use of asthma medication in the past 12 months (based on the specific question “Has asthma medication been given to your child in the last 12 months?”), at age 7-8yrs. Results of skin prick tests at ages 7-8yrs were used to define atopy (mean weal diameter ≥2mm to house dust mite, cat or grass pollen) and to classify asthma as atopic or non-atopic. Phenotypes of wheezing in the first 7yrs of life were defined using latent class analysis of parental reports of wheezing at seven time points from 6 months to 7 years. We identified six phenotypes characterised by different longitudinal trajectories of wheezing. These were classified as: persistent, late onset, intermediate onset, prolonged early, transient early and never/infrequent wheeze. The nature of these phenotypes, their associations with asthma, atopy and lung function in mid childhood, and their reproducibility in independent data, have been described elsewhere3, 29. At ages 8-9yrs, lung function was measured by spirometry (Vitalograph 2120, Maids Moreton, UK) and bronchial responsiveness to methacholine was measured using the method of Yan et al.30 To evaluate the specificity of findings we also defined the following non-asthma phenotypes: highest quintiles of systolic blood pressure at 7-8yrs, intelligence quotient (IQ) at 8-9yrs and height at 7-8yrs (Table I).
Table 1.
Defined outcomes in mid-childhood and association with physician diagnosed asthma at age 7 years
| Outcome | Definition | Age at assessment (yrs) | Physician diagnosed asthma at 7 years | P* | |
|---|---|---|---|---|---|
| No (N=4545) | Yes (N=1132) | ||||
| Atopic asthma | Physician diagnosed asthma and SPT positive** | 7-8 | 0/4545 (0.0) | 332/861 (38.6) | - |
| Non-atopic asthma | Physician diagnosed asthma and SPT negative** | 7-8 | 0/4545 (0.0) | 529/861 (61.4) | - |
| Asthma medication | Use of asthma medication in past 12 months** | 7-8 | 66/4520 (1.5) | 689/1120 (61.5) | <0.001 |
| Persistent wheeze | Wheeze phenotype 'persistent'† | 0-7 | 47/4466 (1.1) | 322/1112 (29.0) | <0.001 |
| Late onset wheeze | Wheeze phenotype 'late onset'† | 0-7 | 109/4466 (2.4) | 174/1112 (15.6) | <0.001 |
| Intermediate onset wheeze | Wheeze phenotype 'intermediate onset'† | 0-7 | 14/4466 (0.3) | 119/1112 (10.7) | <0.001 |
| Prolonged early wheeze | Wheeze phenotype 'prolonged early'† | 0-7 | 272/4466 (6.1) | 142/1112 (12.8) | <0.001 |
| Transient early wheeze | Wheeze phenotype 'transient early'† | 0-7 | 451/4466 (10.1) | 104/1112 (9.4) | 0.502 |
| SPT positive | At least 1 positive skin prick test‡ | 7-8 | 539/3484 (15.5) | 332/861 (38.6) | <0.001 |
| BHR | 3rd tertile of bronchial challenge dose-response slope§ | 8-9 | 560/2410 (23.2) | 241/524 (46.0) | <0.001 |
| Low FEV1 | FEV1 z-score <20th percentile of normal distribution | 8-9 | 593/3456 (17.2) | 199/873 (22.8) | <0.001 |
| Low FVC | FVC z-score <20th percentile of normal distribution | 8-9 | 625/3510 (17.8) | 152/882 (17.2) | 0.730 |
| High systolic BP | 5th quintile of systolic blood pressure | 7-8 | 705/3946 (17.9) | 196/984 (19.9) | 0.140 |
| High IQ | 5th quintile of IQ total score | 8-9 | 736/3680 (20.0) | 161/921 (17.5) | 0.085 |
| High body height | 5th quintile of body height | 7-8 | 727/3995 (18.2) | 146/997 (14.6) | 0.008 |
Prevalence among children with and without diagnosed asthma are reported as n/N(%). Abbreviations: SPT Skin prick test, BHR bronchial hyper-responsiveness, BP blood pressure, IQ intelligence quotient
P-value of Fisher’s exact test
Phenotypes of wheezing in the first 7yrs of life were defined using latent class analysis as reported previously3.
Mean weal diameter ≥2mm to house dust mite, cat or grass pollen.
Tertiles were formed among children with a positive dose response slope. Children with a dose response slope ≤0 were then grouped together with children in the lowest tertile.
based on parental report
Statistical analysis
We used logistic regression to examine the association of each outcome with each genetic prediction score. We quantified the predictive ability of scores using area under the ROC curve (AUC), a measure of discrimination31. We also quantified the magnitude of association using inter-quartile odds ratios (IQORs) comparing the highest and lowest risk score quartiles. Analyses were repeated using scores based on independent sets of SNPs with rankings between consecutive values of k. The R statistical software version 2.11.0 (R Foundation for Statistical Computing) was used for all analyses.
Results
Genetic data were available for 8365 unrelated white European ALSPAC children. Of these, 5748 (68.7%) replied to the questionnaire at age 7 years, and 6128 (73.3%) and 5621 (67.2%) participated in study clinics at ages 7-8 and 8-9 years respectively. Wheezing phenotypes based on repeated assessment of current wheeze throughout the first 7 years of life3 were available in 7176 (85.8%) children. Of these 506 (7.1%) had persistent, 338 (4.7%) late onset, 168 (2.3%) intermediate onset, 538 (7.5%) prolonged early, 726 (10.1%) transient early wheeze and the remaining 4900 (68.3%) never or rarely wheezed. At age 7 years, 1132/5677 (19.9%) children had ever been given a physician diagnosis of asthma. Physician diagnosed asthma was associated with most defined outcomes: associations were particularly strong (ORs>15) for use of asthma medication at age 7 and the persistent and intermediate onset wheeze phenotypes (Table I). Among the non-asthma outcomes, high IQ (P=0.085) and height (P=0.008) were negatively associated with diagnosed asthma (Table I).
Meta-analysis of GABRIEL childhood cohorts excluding ALSPAC
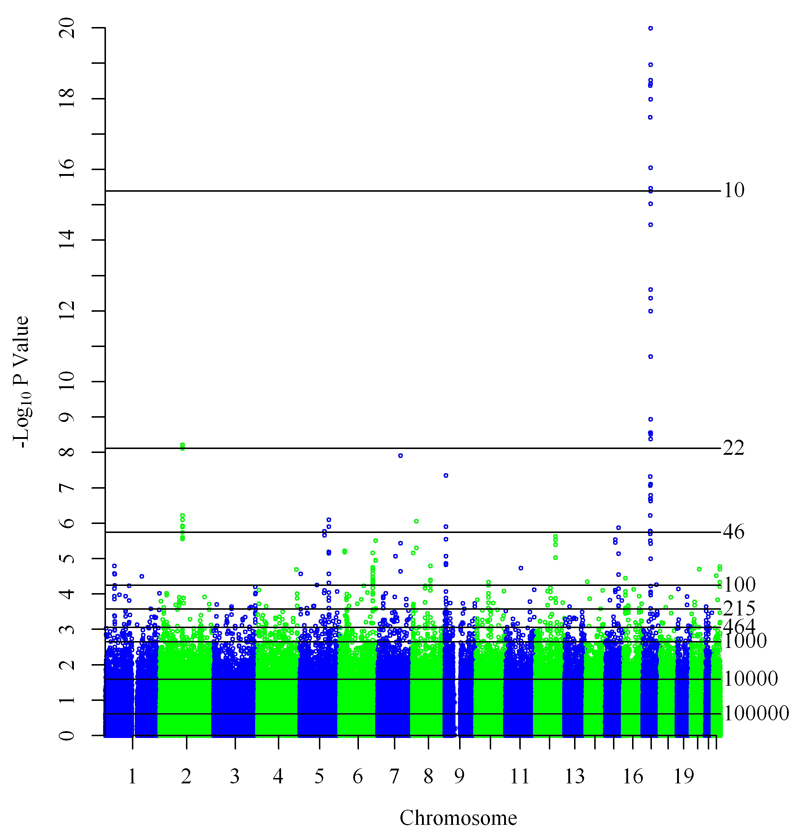
Combining GWAS results for asthma from the GABRIEL childhood cohorts excluding ALSPAC yielded similar results to the original study combining all childhood cohorts (Figure 1 in this article and supplementary Figure 2a of reference16). The top 18 SNPs, and therefore all SNPs included in the 10-SNP scores, were in the ORMDL3/GSDMB locus on chromosome 17q21, a well-established asthma susceptibility locus primarily associated with childhood-onset disease17 (Figure 1 and Table E1 in the Online Repository). Additional loci contributing to the 46-SNP scores included 6 SNPs on chromosome 2 implicating the IL1RL1 and IL18R1 genes, and 2 SNPs flanking the IL33 gene on chromosome 9. Other contributions to the 100-SNP score included loci on chromosomes 1, 5, 6, 8, 12, 15 and 22 (Figure 1 and Table E1 in the Online Repository).
Figure 1. Manhattan plot of pooled results for physician diagnosed asthma in GABRIEL childhood cohorts excluding ALSPAC.
Horizontal lines represent genetic prediction scores with varying number (shown to the right of each line) of included SNPs. All SNPs with P-values above a line are included in the respective score.
Prediction of asthma phenotypes
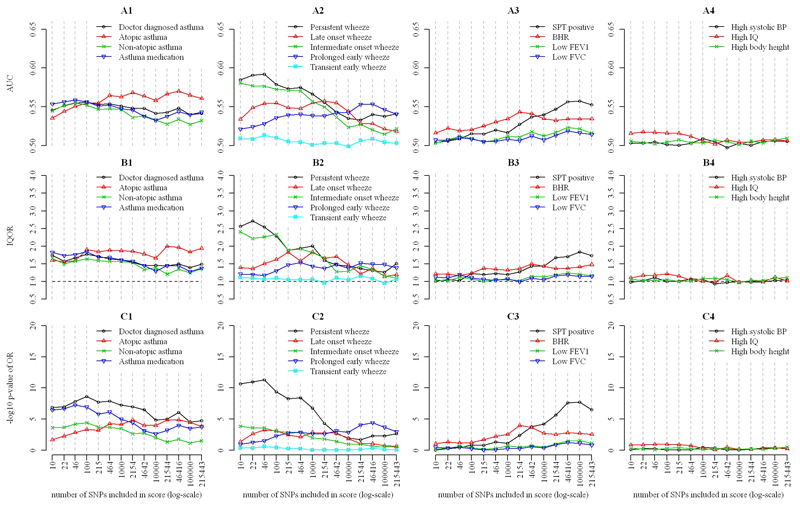
Figure 2 shows the predictive performance of log odds scores for four groups of outcomes. For the first group (asthma, atopic and non-atopic asthma, and asthma medication) there was evidence of association with genetic prediction scores (P<10-4), but discrimination was weak (AUC<0.6) (Figures 2.A1-C1). The AUC for physician-diagnosed asthma using the 10-SNP score was 0.55, while the inter-quartile OR (IQOR) was 1.75 (95%CI 1.44-2.12), and the P value for association was 1.4×10-7. Similar values of AUC and IQOR were observed for non-atopic asthma and use of asthma medication. Prediction of atopic asthma, which was weaker than for the other asthma outcomes for scores with smaller numbers of SNPs, improved with increasing numbers of SNPs: the AUC reached 0.57 for 46,416 SNPs. By contrast, the AUC for non-atopic asthma and use of asthma medication declined with increasing numbers of SNPs.
Figure 2. Comparison of predictive value of allelic log-odds scores for the defined outcomes in mid-childhood.
Data: area under ROC curve (AUC) (sub-figures A), inter-quartile odds ratio (IQOR) (B) and P-values for association (C). Outcomes: outcomes based on a physician’s diagnosis (A1-C1), phenotypes of wheeze in the first seven years of life (A2-C2), physiological measurements (A3-C3), and non-asthma outcomes (A4-C4). SNPs included in inverse order of their P-values in an independent GWAS for physician diagnosed asthma.
For the second outcome group – the early childhood wheezing phenotypes – there was evidence of association (P<10-3) for all phenotypes except transient early wheeze (P>0.25 for all scores). Discrimination and strength of association varied between phenotypes (Figures 2.A2-C2). The strongest associations, and best discrimination, were observed for persistent and intermediate onset wheeze. Prediction of persistent wheeze based on 46 SNPs resulted in an AUC of 0.59, an IQOR of 2.54 (95% CI 1.92-3.37), and P=4.9×10-12. The AUC for these two phenotypes declined to ≤0.54 for scores including 104 SNPs or more. The strongest evidence for association was with the scores including 46 SNPs (IQOR 1.51 [1.10-2.06], P=5.5×10-4) for late onset wheeze, and for the score including 46,416 SNPs (IQOR 1.49 [1.15-1.93], P=4.0×10-5) for prolonged early wheeze, but the AUC did not exceed 0.56.
Among physiological measurements (the third outcome group) BHR was associated (P<10-3) with scores including 2154 and 4642 SNPs and SPT positivity with scores including ≥4642 SNPs, while little evidence of association with any score was found for FEV1 (P>0.029) and FVC (P>0.078) (Figures 2.A3-C3). Maximum AUC values for BHR were reached with a score including 2154 SNPs (AUC=0.54) and for SPT positivity with a score including 105 SNPs (0.56) while AUC was consistently ≤0.52 for FEV1 and FVC.
There was little evidence of association between scores and the fourth group (non-asthma outcomes) (Figures 2.A4-C4). Results were closely similar when using allele count scores instead of log odds scores (Figure E1 in the Online Repository).
Using scores based on independent sets of SNPs with ranks between successive cut-offs, scores including SNPs ranked 1-10, 11-22, and 23-46 were predictive of physician diagnosed asthma (P≤10-6, AUC 0.54-0.55), asthma medication (P≤10-4, 0.54-0.55), atopic (P<0.02, 0.53-0.56) and non-atopic asthma (P<0.002, 0.54-0.55) (Figure E2 in the Online Repository). There was evidence of association (P<0.007) between scores including SNPs ranked 10001-21544 and 21545-46416 and physician diagnosed asthma, atopic asthma, and medication use, with the AUC approaching 0.56 for atopic asthma. Among early wheezing phenotypes, persistent wheeze was associated with scores including SNPs ranked 1-10, 11-22, and 23-46 (P<10-7, AUC 0.57-0.58) but there was little evidence of associations with scores based on lower ranked SNPs. A similar pattern was observed for intermediate onset and late onset wheeze though associations with the first three sets of SNPs were weaker. The score including ranks 10001-21544 was associated with prolonged early wheeze (P=3.4×10-4, AUC=0.55). Among physiological measurements notable associations were only found for scores based on lesser ranks, with SNPs ranked 21545-46416 being most predictive of SPT positivity (P=4.0×10-7, AUC=0.56) and those ranked 1001-2154 for BHR (P=2.8×10-4, AUC=0.54). There was little evidence of associations with low FEV1 and FVC, and the non-asthma outcomes.
Discussion
Main findings
We used SNPs associated with asthma in the GABRIEL GWAS16 to construct genome-wide prediction scores, and evaluated their performance in an independent population-based birth cohort study. Prediction scores were most strongly associated with the persistent and intermediate onset wheezing phenotypes, defined previously using latent class analysis3. We found associations with asthma among the lower-ranked SNPs whose p-values for association did not reach genome-wide thresholds in the GABRIEL GWAS. Although the highest-ranked SNPs were not associated with atopy, there was evidence of associations of atopy with asthma-associated SNPs that did not reach genome-wide significance. Specificity of the prediction scores for asthma-related phenotypes was confirmed by absence of associations with the non-asthma phenotypes, blood pressure, height, and IQ.
Strengths and limitations
Our study exploited the combined signal of SNPs across the genome that is usually ignored in GWAS. By constructing prediction scores based on the log odds ratios (or directions of association) from the GWAS and examining their associations with asthma phenotypes in independent data, we avoided making large numbers of multiple comparisons, and hence the need for stringent p-value thresholds. The study sample was from a well-phenotyped, large, population-based birth cohort. A limitation of the study is that, identified associations with asthma-linked SNPs with low P-value rankings did not permit identification of the specific SNPs responsible for these associations, because only the combined contribution to prediction scores of numerous SNPs, many of which are not associated with asthma, was considered. The allele scores used in our study can, by construction, only account for additive genetic effects and not for dominant allelic effects or gene by gene interaction. We did not have medical records of diagnoses, but relied on parental reports of physician diagnosed asthma. This could have resulted in misclassification of asthma, which may have attenuated the strength of associations with disease diagnosis compared with symptom-based phenotypes. Similarly, use of asthma medication was based on parental report. Although we asked specifically about asthma medication, it is possible that parents erroneously reported other treatments as being for asthma. The magnitude of any such misclassification was small, as only 6.8% of children reported asthma medication without a reported diagnosis of asthma or current symptoms.
Relationship to other genome-wide prediction studies
To our knowledge the present study is the first to use genome-wide prediction to demonstrate the existence of common genetic components of different asthma-related phenotypes. In a recent asthma GWAS, Ferreira et al. also used also used genome-wide allelic scores based on results from the GABRIEL study to predict physician diagnosed asthma in independent data32. However they did not investigate associations between these scores and other related phenotypes. Another study used a related approach to predict severe asthma exacerbations in asthmatic children33. The authors reported that the discriminative ability of the prediction score improved when they included more variants (up to 160). In two other studies using multiple genetic markers to predict asthma related phenotypes, the variants were either preselected based on whether they belonged to a particular candidate gene34, or whether they reached genome-wide significance16.
Previous studies have demonstrated the utility of genome-wide prediction in other complex diseases24, 25. Applying similar methodology, Evans et al. found that for some diseases, including bipolar disorder, coronary heart disease, and type II diabetes, the discriminative ability of genome-wide prediction scores increased as more SNPs satisfying less stringent P-value thresholds were included24. Although this increase was small and unlikely to be of diagnostic utility, it implies the existence of genetic variants contributing to disease risk that remain unidentified even in large GWAS. A genome-wide prediction score for schizophrenia was recently shown to be predictive of bipolar disorder, suggesting that these diseases share a common polygenic component25.
Interpretation
In interpreting our findings it is important to note that the predictive ability of genome-wide scores is expected to decrease as the number of number of included variants increases, unless the additional variants include loci truly associated with disease. This is because, among the lower ranked SNPs, increasing proportions are included because, by chance alone, they show evidence of association in the discovery set. These variants dilute signals originating from truly associated loci. If discrimination improves with the number of SNPs included in the prediction scores, as observed for atopy and atopic asthma, or if it is maintained, as observed for diagnosed asthma and asthma medication use, this suggests the presence of variants truly associated with these phenotypes among the lower ranking SNPs. The validity of this interpretation is supported by the fact that little or no discriminative ability was found for non-asthma phenotypes. It is likely that contamination by noise arising from sampling variation in the discovery data set partly explains the relatively poor discrimination found for all phenotypes (all AUC were <0.60). However, even in the absence of sampling variation, the AUC of genome-wide prediction scores is limited by other factors such as the prevalence and heritability of the phenotype, as discussed extensively elsewhere35.
Early wheezing phenotypes and asthma
We found that the top 46-ranking SNPs associated with childhood asthma in the GABRIEL GWAS predicted two early wheezing phenotypes - persistent wheeze and intermediate onset wheeze - better than they predicted physician diagnosed asthma (the discovery phenotype). We found little evidence that lower-ranking SNPs predicted these wheezing phenotypes, although this cannot be excluded. The first 46 SNPs had P-values <2×10-6 and included previously reported loci16 such as the ORMDL3/GSDMB locus on chromosome 17q21, the IL1RL1 and IL18R1 genes on chromosome 2, and IL33 on chromosome 9, but also some others (Table E1 in the Online Repository). Given the strong signal observed for the first 22 SNPs, 18 of which are in the 17q21 region, this locus appears to be the main single contributor to the discrimination reached for these wheezing phenotypes. Our results agree with previous findings suggesting that the 17q21 locus is primarily associated with early-onset severe asthma36–38 but not with atopy16, 36 or lung function36. Our study suggests that a symptom based phenotype defined by wheeze beginning in the first 3 years of life and persisting up to 6 years more specifically reflects the disease susceptibility conferred by this locus than physician-diagnosed asthma.
Our study provides evidence of still-undiscovered asthma susceptibility loci and adds to the evidence that asthma is a polygenic disease: we found that genome-wide prediction scores based on independent sets of SNPs that had marker specific P-values for association with childhood asthma of >0.025 (ranks >10000, Figure 1) were predictive of the asthma phenotypes, physician diagnosed asthma, atopic asthma, and medication use. This is consistent with findings by Ferreira et al. in their recent GWAS diagnosed asthma32 and with findings for other complex diseases24, 25 and suggests the presence of numerous loci of small effect. Identifying these loci will be difficult even in well-powered GWAS. Apart from the SNPs identified by the GABRIEL study, most SNPs identified as associated with asthma in recent GWAS32, 39–43 had ranks >10000 in our study (Table E2 in the Online Repository). The 11q13.5 locus (LRRC32) identified as associated with atopic status among asthmatics by Ferreira et al.32 had P-value 0.032 and rank 12607 in the present study, and may have contributed to the discrimination of atopic asthma observed for the set of SNPs ranked 10001-21544 (Figure E2 in the Online Repository).
Atopy, BHR and asthma
Our study provides evidence that childhood asthma shares common polygenic components with atopy and with BHR, suggesting that there are shared genetic pathways between these phenotypes and asthma. This has been previously been shown in twin studies which use differences in cross-trait correlations between monozygotic and dizygotic twins to infer the correlation between the heritable components (genetic correlation) underlying two traits. Several such studies have reported genetic correlation between asthma or wheeze and different measurements of allergy including self reported allergy14, skin prick test positivity10, 13, and total serum IgE10 though estimates vary widely between studies (0.15-0.71). Similar genetic correlation was found between asthma or wheeze and bronchial responsiveness (0.11-0.85)10, 13 while estimates for genetic correlation between measurements allergy and bronchial responsiveness (0.15-0.59)10, 11, 13, 15 and between FEV1 and FEV1/FVC with other asthma related traits (-0.52 to -0.11)10, 15 tended to be smaller in magnitude. The evidence provided by our study is more direct because it based on allelic information from individual SNPs. Other attempts to find a genetic overlap between asthma phenotypes based on individual SNPs have been less successful, probably because they relied on SNPs reaching stringent thresholds in GWAS. The GABRIEL study, for instance, found little overlap between loci associated with asthma and those associated with total serum IgE, concluding that elevated IgE levels have a minor role in the development of asthma and may be secondary to asthma16. Our study shows that such an overlap is detectable among variants that do not meet genome-wide significance thresholds in GWAS for asthma.
Conclusions
Our study shows that the genetic origins of asthma phenotypes are diverse. Some pathways appear to be specific to wheezing syndromes but there is also evidence for shared genetic components with atopy and BHR. Discovering the genes underlying this shared heritability will require larger sample sizes and/or more specific phenotype definitions. Among wheezing syndromes, further evidence for aetiological differences comes from the heterogeneity in patterns of genetic association with different symptom based phenotypes. Understanding these differences could be key to devising interventions that alter the natural history of disease in targeted subgroups.
Supplementary Material
Key messages.
Using genetic prediction scores based on up to 215,443 single nucleotide polymorphisms (SNPs), this study found that not only high ranked (reaching stringent significance thresholds in an independent GWAS for childhood asthma) but also lower ranked SNPs contributed to predicting asthma-related phenotypes in children.
Scores based on high ranked SNPs were most predictive for symptom-based phenotypes.
Scores based on lower ranked SNPs predicted atopy and BHR, providing evidence of shared genetic influences with asthma, while scores did not predict lung function.
Capsule summary.
This study provides evidence for shared polygenic effects between asthma, atopy, and BHR in children implicating genetic variants that did not reach genome wide significance thresholds in a GWAS for asthma alone.
Acknowledgements
We are extremely grateful to all the individuals who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. We thank the Sanger Centre, Centre National de Génotypage and 23andMe for generating the ALSPAC GWA data and John Kemp for assistance with cleaning these data.
Funding:
The UK Medical Research Council and the Wellcome Trust (Grant ref: 092731), and the University of Bristol provide core support for ALSPAC study. B.D.S is the recipient of a European Respiratory Society/Marie Curie Joint Research Fellowship - Number MC 1614-2010. The research leading to these results has received funding from the European Respiratory Society and the European Community's Seventh Framework Programme FP7/2007-2013 - Marie Curie Actions under grant agreement RESPIRE, PCOFUND-GA-2008-229571. R.G. was supported by the UK Medical Research Council (Grant no. 0401540). The GABRIEL project was supported by a contract from the European Commission (018996) and grants from the French Ministry of Research, the Wellcome Trust (WT084703MA), and Asthma UK.
Abbreviations
- ALSPAC
Avon Longitudinal Study of Parents and Children
- AUC
area under ROC curve
- BHR
bronchial hyper-responsiveness
- FEV1
forced expiratory volume in 1 s
- FVC
forced vital capacity
- GABRIEL
A Multidisciplinary Study to Identify the Genetic and Environmental Causes of Asthma in the European Community
- GWAS
genome-wide association study
- IQ
intelligence quotient
- IQOR
inter-quartile odds ratio
- OR
odds ratio
- ROC
receiver operating characteristic
- SNP
single nucleotide polymorphism
- SPT
skin prick test
References
- 1.Martinez FD, Wright AL, Taussig LM, Holberg CJ, Halonen M, Morgan WJ. Asthma and wheezing in the first six years of life. N Engl J Med. 1995;332:133–8. doi: 10.1056/NEJM199501193320301. [DOI] [PubMed] [Google Scholar]
- 2.Silverman M, Grigg J, Mc Kean M. Virus-induced wheeze in young children - A separate disease? In: Johnston S, Papadopoulos N, editors. Respiratory infections in allergy and asthma. New York: Marcel Dekker; 2002. pp. 427–71. [Google Scholar]
- 3.Henderson J, Granell R, Heron J, Sherriff A, Simpson A, Woodcock A, et al. Associations of wheezing phenotypes in the first 6 years of life with atopy, lung function and airway responsiveness in mid-childhood. Thorax. 2008;63:974–80. doi: 10.1136/thx.2007.093187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spycher BD, Silverman M, Brooke AM, Minder CE, Kuehni CE. Distinguishing phenotypes of childhood wheeze and cough using latent class analysis. Eur Respir J. 2008;31:974–81. doi: 10.1183/09031936.00153507. [DOI] [PubMed] [Google Scholar]
- 5.Henderson J, Granell R, Sterne J. The search for new asthma phenotypes. Arch Dis Child. 2009;94:333–6. doi: 10.1136/adc.2008.143636. [DOI] [PubMed] [Google Scholar]
- 6.Spycher BD, Silverman M, Kuehni CE. Phenotypes of childhood asthma: are they real? Clin Exp Allergy. 2010;40:1130–41. doi: 10.1111/j.1365-2222.2010.03541.x. [DOI] [PubMed] [Google Scholar]
- 7.Schultz A, Brand PL. Episodic Viral Wheeze and Multiple Trigger Wheeze in preschool children: A useful distinction for clinicians? Paediatr Respir Rev. 2011;12:160–4. doi: 10.1016/j.prrv.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Duffy DL, Mitchell CA, Martin NG. Genetic and environmental risk factors for asthma: a cotwin-control study. Am J Respir Crit Care Med. 1998;157:840–5. doi: 10.1164/ajrccm.157.3.9702070. [DOI] [PubMed] [Google Scholar]
- 9.Palmer LJ, Burton PR, Faux JA, James AL, Musk AW, Cookson WO. Independent inheritance of serum immunoglobulin E concentrations and airway responsiveness. Am J Respir Crit Care Med. 2000;161:1836–43. doi: 10.1164/ajrccm.161.6.9805104. [DOI] [PubMed] [Google Scholar]
- 10.Ferreira MA, O'Gorman L, Le Souef P, Burton PR, Toelle BG, Robertson CF, et al. Variance components analyses of multiple asthma traits in a large sample of Australian families ascertained through a twin proband. Allergy. 2006;61:245–53. doi: 10.1111/j.1398-9995.2005.00954.x. [DOI] [PubMed] [Google Scholar]
- 11.Thomsen SF, Ferreira MA, Kyvik KO, Fenger M, Backer V. A quantitative genetic analysis of intermediate asthma phenotypes. Allergy. 2009;64:427–30. doi: 10.1111/j.1398-9995.2008.01850.x. [DOI] [PubMed] [Google Scholar]
- 12.Thomsen SF, Kyvik KO, Backer V. Etiological relationships in atopy: a review of twin studies. Twin Res Hum Genet. 2008;11:112–20. doi: 10.1375/twin.11.2.112. [DOI] [PubMed] [Google Scholar]
- 13.Thomsen SF, Ulrik CS, Kyvik KO, Ferreira MA, Backer V. Multivariate genetic analysis of atopy phenotypes in a selected sample of twins. Clin Exp Allergy. 2006;36:1382–90. doi: 10.1111/j.1365-2222.2006.02512.x. [DOI] [PubMed] [Google Scholar]
- 14.Willemsen G, van Beijsterveldt TC, van Baal CG, Postma D, Boomsma DI. Heritability of self-reported asthma and allergy: a study in adult Dutch twins, siblings and parents. Twin Res Hum Genet. 2008;11:132–42. doi: 10.1375/twin.11.2.132. [DOI] [PubMed] [Google Scholar]
- 15.Wu T, Boezen HM, Postma DS, Los H, Postmus PE, Snieder H, et al. Genetic and environmental influences on objective intermediate asthma phenotypes in Dutch twins. Eur Respir J. 2010;36:261–8. doi: 10.1183/09031936.00123909. [DOI] [PubMed] [Google Scholar]
- 16.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–7. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 21.Williams SM, Haines JL. Correcting away the hidden heritability. Ann Hum Genet. 2011;75:348–50. doi: 10.1111/j.1469-1809.2011.00640.x. [DOI] [PubMed] [Google Scholar]
- 22.de los Campos G, Gianola D, Allison DB. Predicting genetic predisposition in humans: the promise of whole-genome markers. Nat Rev Genet. 2010;11:880–6. doi: 10.1038/nrg2898. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–9. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans DM, Visscher PM, Wray NR. Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Hum Mol Genet. 2009;18:3525–31. doi: 10.1093/hmg/ddp295. [DOI] [PubMed] [Google Scholar]
- 25.Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golding J, Pembrey M, Jones R, Team AS. ALSPAC--the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatric & Perinatal Epidemiology. 2001;15:74–87. doi: 10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- 27.Jones RW, Ring S, Tyfield L, Hamvas R, Simmons H, Pembrey M, et al. A new human genetic resource: a DNA bank established as part of the Avon longitudinal study of pregnancy and childhood (ALSPAC) European Journal of Human Genetics. 2000;8:653–60. doi: 10.1038/sj.ejhg.5200502. [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savenije OE, Granell R, Caudri D, Koppelman GH, Smit HA, Wijga A, et al. Comparison of childhood wheezing phenotypes in 2 birth cohorts: ALSPAC and PIAMA. J Allergy Clin Immunol. 2011;127:1505–12 e14. doi: 10.1016/j.jaci.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Yan K, Salome C, Woolcock AJ. Rapid method for measurement of bronchial responsiveness. Thorax. 1983;38:760–5. doi: 10.1136/thx.38.10.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steyerberg EW. Clinical Prediction Models - A Practical Approach to Development, Validation, and Updating. New York: Springer; 2009. [Google Scholar]
- 32.Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378:1006–14. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu M, Tantisira KG, Wu A, Litonjua AA, Chu JH, Himes BE, et al. Genome Wide Association Study to predict severe asthma exacerbations in children using random forests classifiers. BMC Med Genet. 2011;12:90. doi: 10.1186/1471-2350-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bureau A, Dupuis J, Falls K, Lunetta KL, Hayward B, Keith TP, et al. Identifying SNPs predictive of phenotype using random forests. Genet Epidemiol. 2005;28:171–82. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]
- 35.Wray NR, Yang J, Goddard ME, Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 2010;6:e1000864. doi: 10.1371/journal.pgen.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–94. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 37.Halapi E, Gudbjartsson DF, Jonsdottir GM, Bjornsdottir US, Thorleifsson G, Helgadottir H, et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010;18:902–8. doi: 10.1038/ejhg.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Binia A, Khorasani N, Bhavsar PK, Adcock I, Brightling CE, Chung KF, et al. Chromosome 17q21 SNP and severe asthma. J Hum Genet. 2011;56:97–8. doi: 10.1038/jhg.2010.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–93. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 41.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–92. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Doi S, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43:893–6. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noguchi E, Sakamoto H, Hirota T, Ochiai K, Imoto Y, Sakashita M, et al. Genome-wide association study identifies HLA-DP as a susceptibility gene for pediatric asthma in Asian populations. PLoS Genet. 2011;7:e1002170. doi: 10.1371/journal.pgen.1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.