Abstract
Alveolar macrophages are the main line of innate immune response against M. tuberculosis (Mtb) infection. However, these cells serve as the major intracellular niche for Mtb enhancing its survival, replication and, later on, cell-to-cell spread. Mtb-associated cytotoxicity of macrophages has been well documented, but limited information exists about mechanisms by which the pathogen induces cell necrosis. To identify virulence factors involved in the induction of necrosis, we screened 5,000 transposon mutants of Mtb for clones that failed to promote the host cell necrosis in a similar manner as the wild-type bacterium. Five Mtb mutants were identified as potential candidates inducing significantly lower levels of THP-1 cell damage in contrast to the H37Rv wild-type infection. Reduced levels of the cell damage by necrosis deficient mutants (NDMs) were also associated with delayed damage of mitochondrial membrane permeability when compared with the wild-type infection over time. Two knockout mutants of the Rv3873 gene, encoding a cell wall PPE68 protein of RD1 region, were identified out of 5 NDMs. Further investigation lead to the observation that PPE68 protein interacts and exports several unknown or known surface/secreted proteins, among them Rv2626c is associated with the host cell necrosis. When the Rv2626c gene is deleted from the genome of Mtb, the bacterium displays significantly less necrosis in THP-1 cells and, conversely, the overexpression of Rv2626c promotes the host cell necrosis at early time points of infections in contrast to the wild-type strain.
Keywords: M tuberculosis, macrophages, necrosis, PPE68, Rv2626c
Introduction
Tuberculosis is one of the most prevalent diseases on the globe that accounts for a large portion of illness and death. The infection is caused by a human-adapted bacterium, Mycobacterium tuberculosis (Mtb), that is able to infect many cell types in the host including macrophages.1,2 Mtb has the ability to subvert a number of killing processes present in macrophages possibly at every level. A fundamental virulence property of Mtb is the ability to alter the normal phagosome biogenesis2,3 and prevent trafficking to acidic, degradative lysosomes.4 Pathogen escapes killing mechanisms of toxic reactive oxygen or nitrogen intermediates by limiting phagocytic cells to generate these most effective anti-bacterial molecules.5,6 Mtb contains molecular strategies to hijack trafficking pathways to prevent autophagy and the destruction of macrophages by apoptosis7-9 that has been proposed to be the virulence characteristic of Mtb. Many manners, employed by Mtb, have been described to inhibit a macrophage self-death,7-11 leading to the conclusion that this property must be an important one for the pathogen survival outcome. The natural cycle of the infection, however, suggests that although prevention of apoptosis is achieved, exit from macrophages has an important role in establishing a definitive niche.
The ability of Mtb to trigger necrosis of infected cells has been known for quite some time, and there is increasing evidence that virulent Mtb strain leaves the macrophage via necrosis at later time point of the infection.12-14 Alveolar epithelial cells are also subjected to the same destiny when infected with Mtb.7,14 Evidence suggests that upon infection of macrophages, Mtb induces an anti-apoptotic mechanism by suppressing TNF-α-mediated extrinsic apoptosis but, concomitantly, pathogen activates the intrinsic pathway through mitochondrial damage,11 which leads to necrosis as an exit strategy.11,15 Experimental models have shown that the RD1 region, present in both M. tuberculosis and Mycobacterium marinum, is involved in the necrosis process and deficiency to secrete ESAT-6 and CFP-10 proteins results in impairment of both pathogen to exit macrophages.16 The fact is that ESAT-6 and CFP-10 proteins are secreted inside of macrophages soon after phagocytosis, but necrosis and eventual exit from the cell does not happen until several days after infection. In addition, both proteins have been associated with a number of virulence-related mechanisms17-19 and there is a possibility that knockout mutants would have additional characteristics that might depend on ESAT-6 and CFP-10 indirectly.
Our laboratory has demonstrated that Mtb partially inhibits macrophage apoptosis.7 The evidence shows that the inhibition of the extrinsic pathway is post-transcriptional, and the intrinsic pathway of apoptosis is used to trigger the cell death.11,20 The association of intrinsic pathway of apoptosis and necrosis has been recently documented.21,22 Our observation suggests that disturbance of the mitochondria membrane most likely is involved in the process of necrosis.
In this study, by adopting an unbiased screening, we identified a transport mechanism linked to secretion of protein(s) and the ability of M. tuberculosis to cause necrosis in phagocytic cells.
Results
Identification of Mtb mutants defective in necrosis but competent for intracellular growth in macrophages
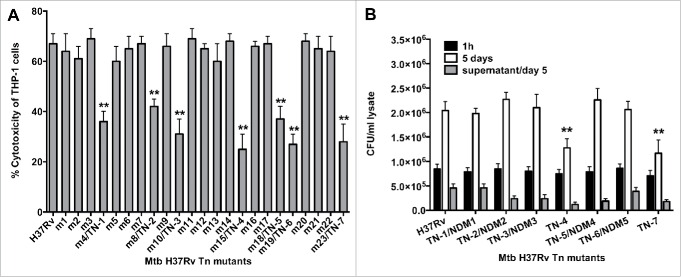
We have previously evaluated the viability of M. tuberculosis H37Rv (wild-type) infected macrophages over time, and established that at 7 day post-infection more than 70% of cells detached from the monolayer due to necrosis. We used this observation to visually screen infected monolayers with 5,000 Mtb transposon clones and to identify wells with approximately 30% fewer detached mononuclear cells. Using this unbiased approach, we observed 23 wells that clearly displayed significantly less disturbed monolayers compared with control wells infected with Mtb wild-type strain with 70% detached monolayer. These mutants were further evaluated for cytotoxicity by measuring the Lactate Dehydrogenase (LDH) enzyme levels in the supernatants of infected wells. As shown on Figure 1A, 7 mutants were identified with impaired ability to cause the cell damage when compared to the wild-type bacterium-infected macrophages at the same time points. To ensure that the significantly lower level of necrosis observed was not due to mutations in Mtb genes that would prevent intracellular replication, we examined intracellular growth rates of necrosis deficient mutants (NDMs) in THP-1 cells. Selected Mtb mutants were renamed as follow: m4/TN-1, m8/TN-2, m10/TN-3, m15/TN-4, m18/TN-5, m19/TN-6 and m23/TN-7. As shown on Figure 1B, mutants TN-4 and TN-7 invaded THP-1 cells similarly to the wild-type bacterium, but over time they showed significant impairment in intracellular growth when compared with the wild-type infection.(**, p < 0.01).
Figure 1.
(A) LDH levels was measured in THP-1 monolayers that were infected with the Mtb wild-type and transposon mutants at a MOI of 10 bacterium to 1 cell, and the percentage of necrosis was calculated according to the manufacturer's protocol. The MOI used was 10. **, p < 0.01, the significance of differences between Mtb transposon mutants and the wild-type. (B) Survival assay for Mtb necrosis deficient mutants in vitro. THP-1 cells were infected with Mtb wild-type and 7 NDMs for 1h and 5 days; CFUs were recorded from lysed macrophages as well as from the supernatants of culture medium at day 5 post-infection. Results represent means ± standard error of the mean of 2 independent experiments. **, p < 0.01, the significance of differences between Mtb transposon mutants and the wild-type.
Distinguishing apoptotic and necrotic cells during infection
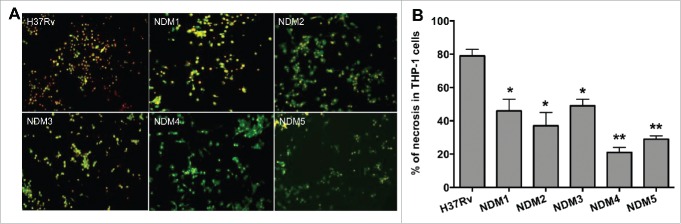
M. tuberculosis non-attenuated mutants (renamed as TN-1/NDM1, TN-2/NDM2, TN-3/NDM3, TN-5/NDM4 and TN-6/NDM5) were further assayed to quantify 2 populations of apoptotic and necrotic cells using Annexin V-FITC and propidium iodide reagents. THP-1 cells were infected with the H37Rv wild-type and necrosis deficient mutants and, after 7 days, 2 populations of dying THP-1 cells were quantified. While apoptotic cells labeled with the FITC-conjugated Annexin V are visualized in green, propidium iodide, a marker for the cell death, labels cell in red. As shown on Figure 2, monolayers that are infected with the wild-type bacterium contain significantly more propidium iodide stained cells. THP-1 macrophage monolayers infected with necrosis deficient mutants, however, demonstrated staining with both propidium iodide and Anenxin V positive cells, with majority of the cells being apoptotic. While Figure 2A displays micrographs obtained with fluorescent microscopy, Figure 2B shows percentage of necrotic cells in the wild-type M. tuberculosis and mutant infected macrophages. Two hundred cells were scored for quantification of apoptotic and necrotic cells, in both control (wild-type) and experimental (necrosis deficient mutant) wells.
Figure 2.
(A) Fluorescence micrographs of THP-1 cells at 7 days post-infection with the Mtb wild-type and NDMs. Macrophage monolayers were stained with both Annexin V-FITC and propidium iodide to visualize the early and late stage of apoptosis and/or necrosis of THP-1 cells, respectively. The early apoptotic THP-1 cells with intact membranes are seen in green; Late apoptotic cells that have already lost their membrane integrity are seen in orange (or orange to red) with green membrane fragments; Totally destroyed necrotic cells are seen in red. (B) The percentage of necrosis quantified in 2 hundred cells that was infected with either the Mtb wild-type or NDM transposon mutants. Results represent means ± standard error of the mean of 3 independent experiments. **, p < 0.01 and *, p < 0.05, the significance of differences between Mtb NDMs and the wild-type.
Necrosis deficient mutants are associated with delayed activation of the intrinsic pathway of apoptosis
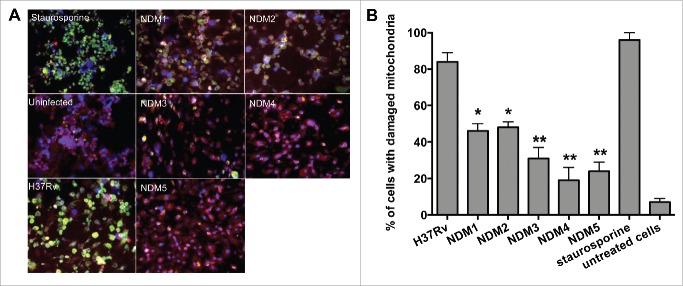
We hypothesized that the stimulation of the intrinsic apoptotic pathway by M. tuberculosis is a required strategy for the host cell necrosis outcome and virulence. We tested necrosis deficient mutants for deficiency in activation of the intrinsic pathway of apoptosis by measuring mitochondrial membrane damage. MitoLight™ assay (Chemicon), based on the lipophilic cationic dye that stains living macrophage mitochondria, was used to detect mitochondrial membrane disruption in both the wild-type- and mutant-infected macrophages. Staurosporine (Sigma) treatment was added to experiments as a positive control, Figure 3A shows that the lipophilic dye is accumulated in the mitochondria of uninfected cells (bright red fluorescence). The mitochondrial membrane damage where the dye is diffused in the cytoplasm of THP-1 cells is presented with bright green fluorescence. After quantification of 2 hundred cells, results indicate that NDM1 through NDM5-infected macrophages contain significantly lower number of cells with damaged mitochondria compared with H37Rv infection (Fig. 3B).
Figure 3.
(A) Immunofluorescence analysis of mitochondrial transmembrane disruption after Mtb wild-type and NDMs infection compared to uninfected cells and staurosporine treatment (positive control). Red scattered staining in THP-1 cells indicate undamaged mitochondria; however, cells with altered mitochondrial membrane stains green as the dye accumulates in the cytoplasm and remains in its monomeric form. (B) The percentage of necrosis quantified in 2 hundred cells that was infected with either the Mtb wild-type or NDMs at MOI of 10. **, p < 0.01 and *, p < 0.05, the significance of differences between NDMs and the wild-type.
PPE68 is involved in necrotic cell death of macrophages
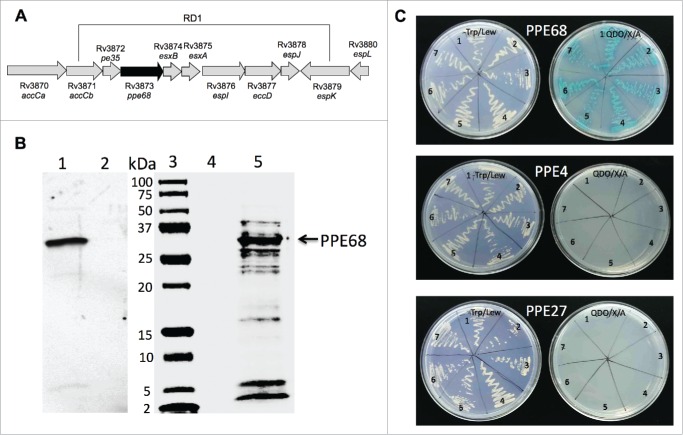
Transposon mutants were sequenced using nonspecific nested suppression PCR method. Samples were submitted to the Central Service Laboratories (CSL), Center for Gene Research and Biotechnology (CGRB), Oregon State University, Corvallis, and database search of obtained sequenced fragments was performed at the National Center for Biotechnology Information (www.ncbi.nih.gov), using BLAST network service. Sequencing results shows that NDM1 and NDM2 mutants had the same gene (Rv3873) interrupted in their genome. The Rv3873 gene is situated within the ESX-1 region and encodes PPE68 protein (Fig. 4A). The mass spectrometry analysis of M. tuberculosis lipophilic membrane- and membrane-associated proteins identified the PPE68 as a part of the cell envelop in H37Rv strain.23 Recent studies have demonstrated that PPE68 is a significant immunogenic component of RD1 and most likely plays an important role in Mtb pathogenicity.24
Figure 4.
(A) Schematic organization of the M. tuberculosis RD1 region containing the PPE68 gene. (B) His-tagged pull-down assay of Mtb proteins interacting with PPE68. The overexpressed PPE68 protein in induced (lane 1) and uninduced (lane 2) E.coli cells is visualized with anti-His antibody. Lane 3, protein marker; lane 4, the pull-down control using the uninduced E.coli cell lysate; lane 5, Mtb bound proteins to PPE68. The membrane was visualized with Li-Cor Odyssey imaging system. (C) The interactions between PPE68 and Rv0462/lpdC (1), Rv0652/rplL (2), Rv1093/glyA1 (3), Rv1388/mihF (4), Rv1837c/glcB (5), Rv2220/glnA1 (6) and Rv2626c (7) were confirmed using a yeast 2-hybrid system. To test the specificity of PPE68 interaction to target Mtb proteins, the yeast reporter strain containing the bait pGBKT7:PPE4 and pGBKT7:PPE27 constructs were screened against the selected Mtb gene constructs in pGADT7 vector; The yeast zygotes that grew on quadruple dropout medium SD/–Ade/–His/–Leu/–Trp supplemented with X-a-Gal and Aureobasidin A (QDO/X/A) and turned blue in color were considered positive. The growth of the yeast cells on the –Trp/Lew medium was used as a control that diploid yeast cells contained both bait and pray constructs.
The profile of PPE68 interacting Mtb proteins
Studies provide evidence that PPE68 interacts with multiple components of the ESX-1 machinery17,25 and results further indicate that disruption of PPE68 is associated with increased secretion of ESAT-6 effector protein, suggesting the possible role of PPE68 as a gating protein during the type VII-dependent protein secretion by mycobacterium.26 To identify if PPE68 interacts with mycobacterium secreted/surface exposed proteins, other than ESX-1 components, and, possibly, is involved in the control of translocation of these proteins, we performed the pull-down assay. The pET:PPE68 construct was expressed in E.coli and purified with the His-purification kit according the manufacturers protocol (Clontech). The recombinant PPE68 protein then was loaded into columns containing His-raisin together with H37Rv cell lysate extracted from the mid-log grown culture. The samples were incubated at 4°C with agitation for overnight. Next day, samples were washed and captured proteins were eluted, purified and submitted for mass spectrometric analysis at the Oregon Health and Science University (OHSU) Proteomics facility. Figure 4B shows the recombinant PPE68 protein (lane 1) and captured Mtb proteins (lane 5) that were bound to PPE68 during pull-down study, and the list of proteins identified by mass spectrometric analysis is presented on Table 1.
Table 1.
PPE68 interacting Mtb proteins.
| Accession | Gene | Function | Spectral counts |
|---|---|---|---|
| NP_216402.1 | fbpB | Secreted antigen 85-B FbpB | 62.9 |
| NP_215610.1 | desA2 | Possible acyl-desaturase DesA2 | 51 |
| NP_217289.1 | dapB | Dihydrodipicolinate reductase | 36 |
| NP_214662.1 | Rv0148 | Short-chain type dehydrogenase/reductase | 32.0 |
| NP_217347.1 | echA16 | Probable enoyl-CoA hydratase | 31.0 |
| NP_218321.1 | fbpA | Secreted antigen 85-a FbpA | 24.1 |
| NP_217502.1 | hupB | DNA-binding protein HU homolog | 21.0 |
| NP_216625.1 | prcA | Proteasome α subunit | 21.0 |
| NP_215155.1 | rplA | 50S ribosomal protein L1 | 20.0 |
| NP_217935.1 | groES | 10 kDa chaperonin | 184 |
| NP_215904.1 | mihF | Putative integration host factor | 149 |
| NP_214954.1 | groEL | 60 kDa chaperonin 2 | 51 |
| NP_216736.1 | glnA1 | Glutamine synthetase | 226 |
| NP_216424.1 | katG | Catalase-peroxidase-peroxynitritase T | 153 |
| NP_216547.1 | hspX | Heat shock protein | 78 |
| NP_215965.1 | tkt | Transketolase | 143 |
| NP_215228.1 | rplN | 50S ribosomal protein L14 | 23 |
| NP_217142.1 | Rv2626c | Hypoxic response protein 1/ Hrp1 | 57 |
| NP_215166.1 | rplL | 50S ribosomal protein L7/L12 | 43 |
| NP_216701.1 | TB16.3 | Conserved protein | 32 |
| NP_214864.1 | dnaK | Chaperone protein | 29 |
| NP_214972.1 | Rv0458 | Probable aldehyde dehydrogenase | 27 |
| NP_217299.1 | gpsI | Polyribonucleotide nucleotidyltransferase | 93 |
| NP_218114.1 | lsr2 | Iron-regulated H-NS-like protein | 20 |
| NP_217765.1 | sahH | Probable adenosylhomocysteinase | 44 |
| NP_218026.1 | ilvX | Probable acetohydroxyacid synthase | 65 |
| NP_215649.1 | metE | homocysteine methyltransferase | 90 |
| NP_216546.1 | Rv2030c | Conserved protein | 30 |
| NP_215267.1 | mmsA | Methylmalonate-semialdehyde dehydrogenase | 22 |
| NP_216978.1 | tig | Probable trigger factor (TF) protein | 30 |
| NP_215375.1 | fadB | Fatty oxidation protein | 28 |
| NP_216152.1 | TB15.3 | Iron-regulated universal stress protein | 22 |
| NP_218366.1 | EspR | ESX-1 transcriptional regulatory protein | 39 |
| NP_218431.1 | trxC | Thioredoxin | 23 |
| YP_177943.1 | TB9.4 | Conserved protein | 26 |
| NP_214712.1 | Zmp1 | Zinc metalloprotease | 34 |
| NP_214661.1 | Rv0147 | Aldehyde dehydrogenase (NAD+) dependent | 30 |
| YP_177746.1 | fusA1 | Probable elongation factor G | 23 |
| NP_216353.1 | glcB | Malate synthase G | 110 |
| NP_214976.1 | lpd | Dihydrolipoamide dehydrogenase | 50.0 |
| NP_216247.2 | gabD1 | Succinate-semialdehyde dehydrogenase | 41.0 |
| NP_216359.1 | guaB1 | Inosine-5′-monophosphate dehydrogenase | 29.0 |
| YP_177782.1 | cbs | Probable cystathionine β-synthase | 41.0 |
| NP_216729.1 | pepB | Aminopeptidase | 25.0 |
| YP_177787.2 | glyA | serine hydroxymethyltransferase | 25.0 |
| NP_214745.1 | fadE4 | Probable acyl-CoA dehydrogenase | 25.0 |
The interaction of identified Mtb proteins is specific to PPE68
We further confirmed and demonstrated the specific interaction between PPE68 and H37Rv Rv0462/lpdC, Rv0652/rplL, Rv1093/glyA1, Rv1388/mihF, Rv1837c/glcB, Rv2220/glnA1 and Rv2626c using the yeast 2-hybrid system. The PPE68 gene was synthesized in frame with a translational fusion of DNA-binding domain of pGBKT7 vector, whereas Mtb selected genes were constructed in fusion with GAL4 activation domain of pGADT7. In addition, to examine that the binding of identified Mtb proteins were explicit to PPE68, we randomly selected PPE4 (Rv0286) and PPE27 (Rv1790) genes for interaction analysis against lpdC, rplL, glyA1, mihF, glcB, glnA1 and Rv2626c. PPE4 and PPE27 genes were cloned into pGBKT7 similarly to PPE68 and screened on the Quadruple Dropout agar plates containing 20 mg/ml X-a-Galactosidase and 125 ng/ml Aureobasidin. As shown on Figure 4C, interaction of selected proteins were found to be specific to PPE68 but not to PPE4 and PPE27.
Overexpression of glnA1 and Rv2626c genes leads to increased cell necrosis at early time point infection
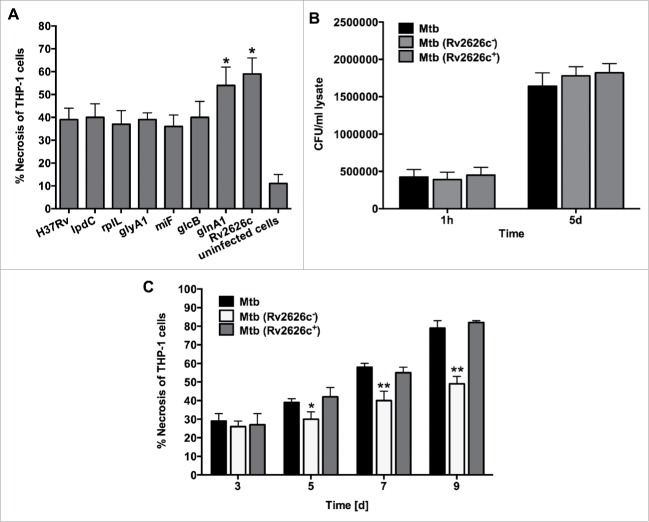
To determine if any of the identified Mtb genes were associated with the cell necrosis, we constructed overexpression clones in H37Rv, and cell death was assayed in THP-1 cells after 5 days of post-infection. Significantly higher levels of necrosis were seen in macrophages infected with either glnA1 or Rv2626c overexpressed clones when compared with control Mtb H37Rv containing empty plasmid (Fig. 5A). The cell death was also observed in macrophages infected with the wild-type bacterium control and the recombinant clones expressing lpdC, rplL, glyA1, mihF and glcB. The levels of necrosis were comparable among clones.
Figure 5.
(A) Necrosis in THP-1 cells that was infected with the Mtb wild-type and overexpressed clones of lpdC, rplL, glyA1, mihF, glcB, glnA1 and Rv2626c was measured by the LDH activity assay. The MOI used was 10. Results represent means ± standard error of the mean of 3 independent experiments. *, p < 0.05, the significance of differences between Mtb overexpressed clones and the wild-type. (B) Infection and survival assay for Mtb knockout and complemented clone of Rv2626c in vitro. THP-1 cells were infected with Mtb wild-type, Mtb (Rv2626c−) and Mtb (Rv2626c+) clones and CFUs were recorded at 1 h and 5 days post-infection. Results that represent means ± standard error of the mean of 2 independent experiments indicate that Mtb knockout and complemented clones invade and survive in macrophages similarly to the wild-type strain. (C) The Mtb (Rv2626c−) infection of THP-1 cells demonstrates significantly less necrosis when compared to Mtb wild-type and Mtb (Rv2626c+) infection. Cells were infected with an MOI of 10. **, p < 0.01 and *, p < 0.05, the significance of differences between Mtb (Rv2626c−) and the wild-type and complemented clone.
Infection with the knockout mutant of Rv2626c displays significantly less necrosis in macrophages
The knockout mutant of Rv2626c in M. tuberculosis CDC1551 was obtained from BEI resources and examined for the ability of Mtb to replicate and induce necrosis in THP-1 cells. In vitro studies revealed no differences between Mtb parental strain and Rv2626c knockout clone growth within human macrophages (Fig. 5B); However, the Rv2626c knockout clone showed significantly lower levels of cell necrosis evident at 5 days post-infection (Fig. 5C). The necrosis phenotype was recovered by complementing knockout strain with the functional Rv2626c gene.
Discussion
The ability of tubercle bacilli to control and modulate cell death pathways in infected macrophages is emerging as a mechanism of pathogenesis. The apoptotic cell death promotes microbicidal activity in phagocytic cells, and removes the intracellular niche for bacillary replication, whereas necrotic cell death causes cell lysis and facilitate release and cell-to-cell spread of the viable bacterium. Recently, it has become apparent that the timing and mode of death of infected primary host cells have decisive role in the control of M. tuberculosis infection and, later on, in the development of disease. Although, necrotic cell death is usually considered to be an “uncontrolled” death, several studies have indicated that it can follow as a controlled series of cellular and molecular events,27,28 and it has been demonstrated that virulent strains of Mtb are capable to induce necrotic death as an escape strategy from macrophages.15 Mtb evades apoptosis of host macrophages by release of TNF-R2 resulting in inactivation of TNF-α,29 and bacterium is able to modulate the induction of anti-apoptotic Bcl-2 family members7,30 Previously, we identified that H37Rv strain blocks the extrinsic pathway of apoptosis through secreting effector proteins Rv3654c and Rv3655c and interfering with caspases' post-transcriptional events.11 While Mtb inhibits the extrinsic pathway of apoptosis, it concurrently activates the intrinsic pathway, leading to the mitochondrial transmembrane damage and macrophage necrosis.11 This strategy potentially links apoptotic and non-apoptotic death pathways. M. tuberculosis, both virulent H37Rv and attenuated H37Ra strains, have been shown to disrupt the mitochondrial outer membrane, but only the virulent H37Rv strain induces the significant mitochondrial transmembrane potential loss leading to macrophage necrosis as an exit strategy for spreading of the infection.15 Our findings suggest that the pathogen uses the intrinsic pathway of apoptosis to trigger cell death, and the intrinsic mode of death at later time points has direct correlation to increased levels of necrosis of Mtb infected macrophages.11 These observations demonstrate how pathogenic Mtb utilizes concurrent stimulation and inhibition of different death modes to control the fate of its host cell.
Studies by several groups have described the ESX-1-independent31 and ESX-1-dependent necrotic cell death19,32 employed by mycobacteria. Different experimental systems identified ESAT-6 and CFP-10 proteins linked to necrosis in Mtb infected macrophages.33 ESAT-6 is a multi-functional protein with many roles in pathogenesis being identified. Importantly, ESAT-6 and CFP-10 are proteins secreted by Mtb and M. marinum when outside and inside cells.34,35 Furthermore, ESAT-6 have been associated with an array of pathogenic mechanisms of both Mtb and M. marinum, mechanisms, such as necrosis of polymorphnuclear phagocytes, lysis of lung mucosal epithelial cells, survival of the bacterium in disseminated infection, and necrosis of macrophages to cite a few.33,36,37 The secretion of ESAT-6 in macrophages has been shown at a time frame that varies from early uptake events to days following the infection.
One of the possibilities to incorporate the current information to the existing body of knowledge is to consider that many of the ESAT-6 reported functions might be only indirectly linked to the protein. The idea, that Mtb and M. marinum escape macrophages to spread the infection is supported by experiments in mice and zebrafish,37,38 raises the aspect that it is quite important for progression of the infection.
To identify virulence factors contributing to macrophage necrosis, in this study we compared the ability of a virulent H37Rv strain and its isogenic mutants to stimulate macrophage death and control balance between apoptotic and necrotic death. By screening mutants of M. tuberculosis in an unbiased manner we identified PPE68, which has been shown to interact with components of the ESX-1 secretion system.24 The inactivation of PPE68 leads to an enhancement of the secretion of ESAT-6, suggesting that PPE68 is likely to have a role and participates in the control of secretion by the type VII secretion apparatus.17 Similar observation has been made regarding the role of PPE68 protein by other groups.26 This raises an important question: why the inactivation of PPE68, associated with increased secretion of ESAT-6, decreases the necrotic cell death as observed in our assays. What is reassuring is that our screen identified PPE68 gene in multiple NDM mutants. The experimental evidence that PPE68 protein can dimerize suggests that this protein might act as a gating protein preventing premature secretion.17 Our data indicate that as a “secretion gate” PPE68 interacts with many proteins with known and many of unknown functions. Two of proteins recognized by PPE68 were glnyA1 encoding glutamine synthetase 1 and hypothetical Rv2626c protein. The glynA1 product is important in nitrogen metabolism and this gene is an essential for M. tuberculosis virulence. It has been demonstrated by several studies that the knockout mutant is attenuated for intracellular growth in THP-1 cells and in vivo.39 Our results indicate that overexpression of this protein in Mtb leads to increased levels of necrosis in macrophages. Rv2626c protein is also known as a Hypoxia-Related Protein-1 (Hrp-1), because the association with the DsoR/DosS regulon.40 Expression of Hrp-1 has been described upon exposure to concentrations of 1% oxygen as well as nitric oxide.41,42 Inactivation of Rv2626c resulted in significant decrease in macrophage necrosis. This observation brings the hypothesis that the level of hypoxia in the mycobacterial vacuole would trigger the eventual events leading to the exit from the macrophage.
What advantage does the pathogen see in lysing the host cell in which it has been living? The accepted concept would be that pathogens lyse the host cells to gain access to a new environment when the amount of nutrient becomes limited and/or the pathogens become too numerous. It is possible that the hypoxia becomes a cellular environmental signal “forcing” the bacterium to either go into latency or to exit cells for continues replication. If this hypothesis is correct, the implications are that the Dos regulon is involved in necrosis of host cells, and that additional regulation must exist to explain the “decision points.”
We did not evaluate the phenotype of Rv2626c in vivo, since there is information showing that the mutation leads to attenuation in mice.42 In vitro, however, Rv2626c inactivation did not reduce the ability of the bacterium to survive in the macrophage. It can be explained by the limited duration of the assay.(5 days).
In summary, we identified new pathway implied by Mtb during exit from infected macrophages. We also hypothesize that cell necrosis triggered by the bacterium is linked to hypoxia in the phagosome, and it possibly changes depending on unknown factors. Future studies will discern the complexities involved in the pathogen “decision making” to understand how the bacterium achieves intracellular growth while stimulating the necessary changes to drive cell-to-cell spread.
Materials and Methods
Bacterial strains and growth conditions
Mycobacterium tuberculosis H37Rv strain was purchased from the American Type Culture Collection (ATCC). The Mtb transposon mutant bank of 5,000 clones was generated in the previous study.11 Mtb CDC1551 strain and knockout mutant of the Rv2626c gene (Mycobacterium tuberculosis, Strain CDC1551, Transposon Mutant 1121 (MT2701, Rv2626c), NR-18010), were obtained through BEI Resources, NIAID, NIH. Bacteria were grown in the Middlebrook 7H9 broth supplemented with 10% OADC (Oleic, Albumin, Dextrose, Catalase, Hardy Diagnostics) at 37°C until the mid-exponential phase. Kanamycin (200μg/ml) was added to the medium, where appropriate. Bacteria were homogenized and passed through a syringe with a 24-gauge needle to remove clumps and, before any infection, viable counts of inocula were determined by plating of serial dilutions on 7H11 agar with 10% OADC.
Cell culture
The THP-1 human monocyte cell line (ATCC) was maintained in RPMI-1640 (Lonza) medium supplemented with the heat-inactivated 10% fetal bovine serum (Gemini). Fifty nanograms/ml of phorbol 12-myristate 13-acetate (PMA, Sigma Aldrich) was added to cells, to promote maturation and adherence, and then depending on the experiment, THP-1 cells were seeded at 90% confluence into 75cm2 tissue culture flasks, 24-, 96-well plates or 2-chamber glass slides. After 24h, wells were replenished with new medium and incubated for additional 48h for cell differentiation. In all experiments, THP-1 cells were infected at a multiplicity of infection (MOI) of ~10 bacteria per macrophage. After 1h infection, the wells were washed and extracellular bacteria were killed with 200 μg/ml amikacin treatment for additional 1 h. The total number of cell-associated bacteria over time was determined by plate counts.
Reagents
The anti-Mtb monoclonal antibodies against GlcB (Rv1837c), GlnA1 (Rv2220) and polyclonal antibody against Rv2626c were obtained through BEI Resources, NIAID, NIH: Monoclonal Anti-Mycobacterium tuberculosis GlcB (Gene Rv1837c), Clone α-GlcB (produced in vitro), NR-13799, Monoclonal Anti-Mycobacterium tuberculosis GlnA1 (Gene Rv2220), Clone IT-58 (CBA5) (produced in vitro), NR-13656, Polyclonal Anti-Mycobacterium tuberculosis HRP1 (Gene Rv2626c) (antiserum, Rabbit), NR-36512. The anti-His antibody was purchased from Santa Cruz Biotechnology. All chemicals utilized were obtained through Sigma-Aldrich. The CytoTox96® non-radioactive cytotoxicity assay kit was from Promega. The Annexin V-FITC assay kit was from PromoKine, and the MitoLight™ Apoptosis assay kit was purchased from Chemicon. The Yeast Two Hybrid bait, pray and control vectors, the Yeast Transformation System, yeast strains and medium were obtained from Clontech.
Mtb library screening for necrosis
Past work at the laboratory established that after 7 days post-infection, approximately 70% of macrophages infected with Mtb wild-type H37Rv undergo necrotic cell death and detach from the monolayer. To screen an M.tb transposon library of 5,000 for clones that failed to promote host cell necrosis, at first, THP-1 cells were infected with the mutant library in 96-well plates with MOI of 10, and after 7 days of infection the wells that containing approximately 30% detached cells compare with the wild-type-infected wells were visually selected. Then the selected clones were processed for the CytoTox96® non-radioactive cytotoxicity assay (Promega) to measure the Lactate Dehydrogenase (LDH) enzyme levels in the supernatants of infected THP-1 cells. Background release of LDH was obtained with untreated control cells, and maximum release of LDH with lysed uninfected cells. Percentage cytotoxicity was calculated by the formula: [release of LDH from infected cells (OD490)/maximum LDH release (OD490)] × 100.
Quantification of apoptotic and dead cells
Approximately 1 × 106 THP-1 cells grown on the 2-chamber glass slides were infected with the Mtb wild-type and mutant clones (MOI of 10) at 37°C in an atmosphere of 5% CO2. After 7 days of infection, 5 μl of Annexin V-FITC and 5 μl of propidium iodide were added to the wells and incubated at room temperature for 5 min in the dark. Samples were washed and then fixed with a freshly prepared 2% paraformaldehyde solution (in PBS, pH 7.4) for 1 h. Fixed slides were then analyzed under the fluorescence microscopy using a dual filter set for FITC and rhodamine.
Analysis of the mitochondrial transmembrane disruption
The mitochondrial membrane permeability in Mtb wild-type and mutant infected THP-1 cells (2 x 106) were detected using the MitoLight™ Apoptosis assay (Chemicon) according to the manufacturer's protocol. Briefly, 1 μl of MitoLight™ was resuspended into 900 μl of deionized water and 100 μl of 10X incubation buffer. Control (uninfected) and experimental (infected) wells were incubated with the prediluted MitoLight™ solution for 15-20 minutes at 37°C in a 5% CO2 incubator and then washed 3 times with 1X incubation buffer. As a positive control, the staurosporine (Sigma) was added to selected wells to trigger apoptosis. Samples were immediately observed under fluorescence microscopy using FITC and Texas Red channels.
Expression of PPE68 and pull-down assay
The PPE68 (Rv3873) gene was amplified with sense TTTTTAAGCTTCTGTGGCACGCAATGCCA and antisense TTTTTAAGCTTCCAGTCGTCCTCTTCGTC primers and cloned into HinDIII site of the pET6xHN-C vector (Clontech). The protein was expressed in E. coli strain BL21 (DE3) and purified by passing it through the His-column according to the manufacturer's protocol (Clontech). Protein of interest was separated on a 12% Tris–HCl gel, transferred to nitrocellulose membrane and blocked with 3% Bovine Serum Albumin (BSA). Membrane was then probed with 6xHN antibody (1:250 dilution) for 1 h and visualized with the corresponding IRDye® secondary antibody (Li-Cor Biosciences, Inc.) at a dilution of 1:5,000 for 30 min. Proteins of interest was detected using an Odyssey Imager (Li-Cor).
Alternatively, Mtb H37Rv was harvested from the liquid Middlebrook 7H9 medium at mid-log phase, lysed in PBS by mechanical disruption and cleared by centrifugation at 3,500 rpm for 20 min followed by filtration through 0.2 μm filter. The total protein extract from Mtb was incubated with 700 ng of purified 6HN:PPE68 protein for overnight at 4°C. Next day, samples were loaded into His-columns and washed according to the manufacturer's protocol (Clontech). Eluted samples were processed for electrophoresis. The PPE68 bound proteins were subjected for In-Gel Tryptic Digestion (PIERCE, Rockford, IL) and sequencing by electrospray ionization mass spectrometry (ESI-MS/MS) at OHSU facility.
The yeast 2-hybrid interaction
The PPE68 gene was fused in frame with the GAL4 DNA binding domain by inserting the PCR-generated fragment into the EcoRI and BamHI sites of pGBKT7. The resultant bait vector pGBKT7:PPE68 was transformed into Saccharomyces cerevisiae strain Y2HGold using Yeastmaker Yeast Transformation System 2, according to the manufacturer's instructions (Clontech). The following Mtb genes: Rv0462/lpdC, Rv0652/rplL, Rv1093/glyA1, Rv1388/miF, Rv1837c/glcB, Rv2220/glnA1 and Rv2626c that were identified through Mass Spectrometric analysis were selected to construct the pray vectors for 2-hybrid interaction studies. These genes were fused with the GAL4 activation domain of pGADT7 and transformed into the yeast strain Y187 (Clontech). Plasmids pGBKT7-53, pGBKT7-lam, and pGADT7-T were used as positive and negative controls for interaction studies and were obtained from Clontech. One ml of bait strain was combined with the one ml of prey strain and grown in 2xYPDA liquid medium containing 50 μg/ml kanamycin at 30°C for 24 h. The activation of AUR1-C, ADE2, HIS3, and MEL1 reporters controlled by the Gal4 promoter were screened by plating yeast zygotes on Double Dropout (SD-Leu/–Trp), Triple Dropout (SD– His/–Leu/–Trp), Quadruple Dropout (SD–Ade/–His/–Leu/–Trp) and Quadruple Dropout agar plates containing 20 mg/ml X-a-Galactosidase and 125 ng/ml Aureobasidin (QDO/X/A). The colonies that grew of blue color were identified as positive clone.
Construction of Mtb overexpression clones
Mtb clones overexpressing the Rv0462/lpdC, Rv0652/rplL, Rv1093/glyA1, Rv1388/miF, Rv1837c/glcB, Rv2220/glnA1 and Rv2626c genes were amplified from Mtb H37Rv genomic DNA and ligated at the HindIII and EcoRI restriction sites of pMV261 vector containing 6xHis. The recombinant plasmids were transformed to Mtb H37Rv. Mtb clones were used for THP-1 cell infection, and necrosis assay was performed for LDH release.
Complementation of Rv2626c
To complement the Transposon Mutant 1121, the MT2701 gene, homologous to Rv2626c, was amplified from the chromosomal DNA of Mtb CDC1551 strain. The PCR product was cloned into the SpeI site of pMV261:AprII plasmid and transformed into Rv2626c knockout strain of Mtb (Mtb (Rv2626c−). Positive clones were selected on 7H11 agar plates containing apramycin 200 μg/ml, and the complemented clone was named Mtb (Rv2626c+).
Statistics
The Student's t test was used to assess statistical significance between experimental and control groups. Experiments were carried out in duplicate and repeated at least 2 times. The p-value p < 0.05 was considered significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The research was supported by a grant from the Department of Biomedical Sciences, College of Veterinary Medicine, Oregon State University. The mass spectrometric analysis was performed by the OHSU proteomics shared resource with partial support from NIH core grants P30EY010572, P30CA069533, S10OD012246, and S10RR025571.
References
- 1.Bermudez LE, Goodman J. Mycobacterium tuberculosis invades and replicates within type II alveolar cells. Infect Immun 1996; 64:1400-6; PMID:8606107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crowle AJ, Dahl R, Ross E, May MH. Evidence that vesicles containing living, virulent Mycobacterium tuberculosis or Mycobacterium avium in cultured human macrophages are not acidic. Infect Immun 1991; 59:1823-31; PMID:1902198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vergne I, Chua J, Lee HH, Lucas M, Belisle J, Deretic V. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 2005; 102:4033-8; PMID:15753315; http://dx.doi.org/ 10.1073/pnas.0409716102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, Allen RD, Gluck SL, Heuser J, Russell DG. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 1994; 263:678-81; PMID:8303277; http://dx.doi.org/ 10.1126/science.8303277 [DOI] [PubMed] [Google Scholar]
- 5.Axelrod S, Oschkinat H, Enders J, Schlegel B, Brinkmann V, Kaufmann SH, Haas A, Schaible UE. Delay of phagosome maturation by a mycobacterial lipid is reversed by nitric oxide. Cellular Microbiol 2008; 10:1530-45; PMID:18363878; http://dx.doi.org/ 10.1111/j.1462-5822.2008.01147.x [DOI] [PubMed] [Google Scholar]
- 6.Colangeli R, Haq A, Arcus VL, Summers E, Magliozzo RS, McBride A, Mitra AK, Radjainia M, Khajo A, Jacobs WR, et al.. The multifunctional histone-like protein Lsr2 protects mycobacteria against reactive oxygen intermediates. Proc Natl Acad Sci U S A 2009; 106:4414-8; PMID:19237572; http://dx.doi.org/ 10.1073/pnas.0810126106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danelishvili L, McGarvey J, Li YJ, Bermudez LE. Mycobacterium tuberculosis infection causes different levels of apoptosis and necrosis in human macrophages and alveolar epithelial cells. Cellular Microbiol 2003; 5:649-60; PMID:12925134; http://dx.doi.org/ 10.1046/j.1462-5822.2003.00312.x [DOI] [PubMed] [Google Scholar]
- 8.Loeuillet C, Martinon F, Perez C, Munoz M, Thome M, Meylan PR. Mycobacterium tuberculosis subverts innate immunity to evade specific effectors. J Immunol 2006; 177:6245-55; PMID:17056554; http://dx.doi.org/ 10.4049/jimmunol.177.9.6245 [DOI] [PubMed] [Google Scholar]
- 9.Jayakumar D, Jacobs WR Jr., Narayanan S. Protein kinase E of Mycobacterium tuberculosis has a role in the nitric oxide stress response and apoptosis in a human macrophage model of infection. Cell Microbiol 2008; 10:365-74; PMID:17892498 [DOI] [PubMed] [Google Scholar]
- 10.Behar SM, Martin CJ, Booty MG, Nishimura T, Zhao X, Gan HX, Divangahi M, Remold HG. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol 2011; 4:279-87; PMID:21307848; http://dx.doi.org/ 10.1038/mi.2011.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danelishvili L, Yamazaki Y, Selker J, Bermudez LE. Secreted Mycobacterium tuberculosis Rv3654c and Rv3655c proteins participate in the suppression of macrophage apoptosis. PLoS One 2010; 5:e10474; PMID:20454556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Assis PA, Espindola MS, Paula-Silva FW, Rios WM, Pereira PA, Leao SC, Silva CL, Faccioli LH. Mycobacterium tuberculosis expressing phospholipase C subverts PGE2 synthesis and induces necrosis in alveolar macrophages. BMC Microbiol 2014; 14:128; PMID:24886263; http://dx.doi.org/ 10.1186/1471-2180-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tundup S, Mohareer K, Hasnain SE. Mycobacterium tuberculosis PE25/PPE41 protein complex induces necrosis in macrophages: Role in virulence and disease reactivation? FEBS Open Bio 2014; 4:822-8; PMID:25379378; http://dx.doi.org/ 10.1016/j.fob.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonough KA, Kress Y. Cytotoxicity for lung epithelial cells is a virulence-associated phenotype of Mycobacterium tuberculosis. Infect Immun 1995; 63:4802-11; PMID:7591139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen M, Gan H, Remold HG. A mechanism of virulence: virulent Mycobacterium tuberculosis strain H37Rv, but not attenuated H37Ra, causes significant mitochondrial inner membrane disruption in macrophages leading to necrosis. J Immunol 2006; 176:3707-16; PMID:16517739; http://dx.doi.org/ 10.4049/jimmunol.176.6.3707 [DOI] [PubMed] [Google Scholar]
- 16.Houben D, Demangel C, van Ingen J, Perez J, Baldeon L, Abdallah AM, Caleechurn L, Bottai D, van Zon M, de Punder K, et al.. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol 2012; 14:1287-98; PMID:22524898; http://dx.doi.org/ 10.1111/j.1462-5822.2012.01799.x [DOI] [PubMed] [Google Scholar]
- 17.Teutschbein J, Schumann G, Mollmann U, Grabley S, Cole ST, Munder T. A protein linkage map of the ESAT-6 secretion system 1 (ESX-1) of Mycobacterium tuberculosis. Microbiol Res 2009; 164:253-9; PMID:17433643; http://dx.doi.org/ 10.1016/j.micres.2006.11.016 [DOI] [PubMed] [Google Scholar]
- 18.Aguilo N, Marinova D, Martin C, Pardo J. ESX-1-induced apoptosis during mycobacterial infection: to be or not to be, that is the question. Front Cell Infect Microbiol 2013; 3:88; PMID:24364000; http://dx.doi.org/ 10.3389/fcimb.2013.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdallah AM, Bestebroer J, Savage ND, de Punder K, van Zon M, Wilson L, Korbee CJ, van der Sar AM, Ottenhoff TH, van der Wel NN, et al.. Mycobacterial secretion systems ESX-1 and ESX-5 play distinct roles in host cell death and inflammasome activation. J Immunol 2011; 187:4744-53; PMID:21957139; http://dx.doi.org/ 10.4049/jimmunol.1101457 [DOI] [PubMed] [Google Scholar]
- 20.Chen F, He Y. Caspase-2 mediated apoptotic and necrotic murine macrophage cell death induced by rough Brucella abortus. PLoS One 2009; 4:e6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butler RE, Brodin P, Jang J, Jang MS, Robertson BD, Gicquel B, Stewart GR. The balance of apoptotic and necrotic cell death in Mycobacterium tuberculosis infected macrophages is not dependent on bacterial virulence. PLoS One 2012; 7:e47573; PMID:23118880; http://dx.doi.org/ 10.1371/journal.pone.0047573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicotera P, Melino G. Regulation of the apoptosis-necrosis switch. Oncogene 2004; 23:2757-65; PMID:15077139; http://dx.doi.org/ 10.1038/sj.onc.1207559 [DOI] [PubMed] [Google Scholar]
- 23.Malen H, Pathak S, Softeland T, de Souza GA, Wiker HG. Definition of novel cell envelope associated proteins in Triton X-114 extracts of Mycobacterium tuberculosis H37Rv. BMC Microbiol 2010; 10:132; PMID:20429878; http://dx.doi.org/ 10.1186/1471-2180-10-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demangel C, Brodin P, Cockle PJ, Brosch R, Majlessi L, Leclerc C, Cole ST. Cell envelope protein PPE68 contributes to Mycobacterium tuberculosis RD1 immunogenicity independently of a 10-kilodalton culture filtrate protein and ESAT-6. Infect Immun 2004; 72:2170-6; PMID:15039340; http://dx.doi.org/ 10.1128/IAI.72.4.2170-2176.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh A, Mai D, Kumar A, Steyn AJ. Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc Natl Acad Sci U S A 2006; 103:11346-51; PMID:16844784; http://dx.doi.org/ 10.1073/pnas.0602817103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brodin P, Rosenkrands I, Andersen P, Cole ST, Brosch R. ESAT-6 proteins: protective antigens and virulence factors? Trends Microbiol 2004; 12:500-8; PMID:15488391; http://dx.doi.org/ 10.1016/j.tim.2004.09.007 [DOI] [PubMed] [Google Scholar]
- 27.Marshall KD, Baines CP. Necroptosis: is there a role for mitochondria? Front Physiol 2014; 5:323; PMID:25206339; http://dx.doi.org/ 10.3389/fphys.2014.00323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feoktistova M, Leverkus M. Programmed necrosis and necroptosis signalling. FEBS J 2015; 282:19-31; PMID:25327580; http://dx.doi.org/ 10.1111/febs.13120 [DOI] [PubMed] [Google Scholar]
- 29.Balcewicz-Sablinska MK, Keane J, Kornfeld H, Remold HG. Pathogenic Mycobacterium tuberculosis evades apoptosis of host macrophages by release of TNF-R2, resulting in inactivation of TNF-α. J Immunol 1998; 161:2636-41; PMID:9725266 [PubMed] [Google Scholar]
- 30.Sly LM, Hingley-Wilson SM, Reiner NE, McMaster WR. Survival of Mycobacterium tuberculosis in host macrophages involves resistance to apoptosis dependent upon induction of antiapoptotic Bcl-2 family member Mcl-1. J Immunol 2003; 170:430-7; PMID:12496428; http://dx.doi.org/ 10.4049/jimmunol.170.1.430 [DOI] [PubMed] [Google Scholar]
- 31.Lee J, Repasy T, Papavinasasundaram K, Sassetti C, Kornfeld H. Mycobacterium tuberculosis induces an atypical cell death mode to escape from infected macrophages. PLoS One 2011; 6:e18367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong KW, Jacobs WR. Critical role for NLRP3 in necrotic death triggered by Mycobacterium tuberculosis. Cell Microbiol 2011; 13:1371-84; PMID:21740493; http://dx.doi.org/ 10.1111/j.1462-5822.2011.01625.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo S, Xue R, Li Y, Wang SM, Ren L, Xu JJ. The CFP10/ESAT6 complex of Mycobacterium tuberculosis may function as a regulator of macrophage cell death at different stages of tuberculosis infection. Medical Hypotheses 2012; 78:389-92; PMID:22192908; http://dx.doi.org/ 10.1016/j.mehy.2011.11.022 [DOI] [PubMed] [Google Scholar]
- 34.Berthet FX, Rasmussen PB, Rosenkrands I, Andersen P, Gicquel B. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiol 1998; 144 (Pt 11):3195-203; PMID:9846755; http://dx.doi.org/ 10.1099/00221287-144-11-3195 [DOI] [PubMed] [Google Scholar]
- 35.Sorensen AL, Nagai S, Houen G, Andersen P, Andersen AB. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect Immun 1995; 63:1710-7; PMID:7729876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan T, Lee WL, Alexander DC, Grinstein S, Liu J. The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell Microbiol 2006; 8:1417-29; PMID:16922861; http://dx.doi.org/ 10.1111/j.1462-5822.2006.00721.x [DOI] [PubMed] [Google Scholar]
- 37.Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci U S A 2003; 100:13001-6; PMID:14557536; http://dx.doi.org/ 10.1073/pnas.2235593100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao LY, Guo S, McLaughlin B, Morisaki H, Engel JN, Brown EJ. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol Microbiol 2004; 53:1677-93; PMID:15341647; http://dx.doi.org/ 10.1111/j.1365-2958.2004.04261.x [DOI] [PubMed] [Google Scholar]
- 39.Tullius MV, Harth G, Horwitz MA. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect Immun 2003; 71:3927-36; PMID:12819079; http://dx.doi.org/ 10.1128/IAI.71.7.3927-3936.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharpe ML, Gao C, Kendall SL, Baker EN, Lott JS. The structure and unusual protein chemistry of hypoxic response protein 1, a latency antigen and highly expressed member of the DosR regulon in Mycobacterium tuberculosis. J Mol Biol 2008; 383:822-36; PMID:18640126; http://dx.doi.org/ 10.1016/j.jmb.2008.07.001 [DOI] [PubMed] [Google Scholar]
- 41.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α -crystallin. Proc Natl Acad Sci U S A 2001; 98:7534-9; PMID:11416222; http://dx.doi.org/ 10.1073/pnas.121172498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bashir N, Kounsar F, Mukhopadhyay S, Hasnain SE. Mycobacterium tuberculosis conserved hypothetical protein rRv2626c modulates macrophage effector functions. Immunology 2010; 130:34-45; PMID:20201990; http://dx.doi.org/ 10.1111/j.1365-2567.2009.03196.x [DOI] [PMC free article] [PubMed] [Google Scholar]