Abstract
Protein kinase signaling along the kinetochore-centromere axis is crucial to assure mitotic fidelity, yet its spatial coordination is obscure. Here, we examined how pools of human Polo-like kinase 1 (Plk1) within this axis control signaling events to elicit mitotic functions. To do this, we restricted active Plk1 to discrete subcompartments within the kinetochore-centromere axis using chemical genetics and decoded functional and phosphoproteomic signatures of each. We observe distinct phosphoproteomic and functional roles, suggesting that Plk1 exists and functions in discrete pools along this axis. Deep within the centromere, Plk1 operates to assure proper chromosome alignment and segregation. Thus, Plk1 at the kinetochore is a conglomerate of an observable bulk pool coupled with additional functional pools below the threshold of microscopic detection/resolution. Although complex, this multiplicity of locales provides an opportunity to decouple functional and phosphoproteomic signatures for a comprehensive understanding of Plk1’s kinetochore functions.
Introduction
During mitosis, macromolecular kinetochores assemble upon centromeric chromatin to attach chromosomes to spindle microtubules. In addition to its structural role, the kinetochore generates signals to promote proper attachment of sister chromatids to opposite spindle poles (biorientation) to ensure accurate segregation into daughter cells. Several protein kinases regulate kinetochore signaling, including Aurora B, Bub1, BubR1, Mps1, Haspin and Polo-like kinase 1 (Plk1)1–3. How these mitotic kinases operate within the kinetochore is obscure, but spatial cues are clearly important. For example, one model for Aurora B function is based on a tension-generated loss of spatial access to its substrates4,5. It is unclear if the spatial distributions of mitotic kinases within the kinetochore control accessibility of substrates and functions. Here, we investigated how spatial distribution of Plk1 along the kinetochore contributes to its function and access to substrates.
Polo-like kinase 1 (Plk1) is a core regulator of mitosis required for centrosome maturation, bipolar spindle formation, chromosome arm resolution, chromosome alignment and segregation, and cytokinesis6. Plk1 localizes to several mitotic structures, including centrosomes, chromosome arms, kinetochores and the anaphase spindle midzone6,7. Its localization is established primarily via its C-terminal Polo-box domain (PBD), a phosphopeptide binding domain, which requires its direct binding partner to be primed through phosphorylation8,9. Preventing Plk1 localization disrupts its functions, resulting in failure to align and segregate chromosomes10–13 and impaired cytokinesis14.
Spatial regulation of Plk1 signaling at the kinetochore remains enigmatic due to a multiplicity of interactors and substrates located at distinct sites along the entire kinetochore-centromere axis (Supplementary Results, Supplementary Fig. 1). During mitosis, Plk1 is recruited to the outer kinetochore by Bub115, NudC16, and BubR1, where it phosphorylates BubR117,18 and CLASP19 to stabilize kinetochore-microtubule attachments, promoting chromosome alignment, and Kif2b20 to correct microtubule attachment errors, facilitating accurate chromosome segregation. At the inner kinetochore, Plk1’s role in chromosome alignment and segregation also requires recruitment by and phosphorylation of CENP-U/5021 (also called PBIP) and CENP-Q22. Surprisingly, Plk1 is also reported to function at the inner centromere, approximately 500 nm from the outer kinetochore, and normal chromosome dynamics depend on its binding to INCENP23 and phosphorylation of Survivin24,25, members of the chromosome passenger complex (CPC). Microscopically, Plk1 is visualized as a single kinetochore focus, and its kinase domain is only 5 nm in length26, so it is difficult to envision how it could span ~500 nm to access these substrates. The alternative is that multiple pools of Plk1 exist at distinct kinetochore subcompartments. If so, we would expect Plk1 kinetochore functions to be separable by precise spatial control of its activity.
A detailed understanding of Plk1 signaling at the kinetochore has remained elusive for several reasons. First, most previous studies abrogate all Plk1 activity, resulting in prometaphase-arrested cells with monopolar spindles. This prevents direct evaluation of mitotic activities that require a bipolar spindle, including chromosome alignment, segregation, and cytokinesis. Second, Plk1’s localization to multiple mitotic structures necessitates uncoupling its kinetochore activity from other locales27. Finally, phosphoproteomic studies have identified hundreds of Plk1-regulated phosphorylation sites28–31, but the abundance of substrates has impaired efficient assignment of phosphorylation events to biologic functions. To circumvent the first obstacle, we previously titrated Plk1 activity to uncouple chromosome alignment and segregation from bipolar spindle formation12. Here, we addressed the second and third obstacles through chemical genetic complementation, high-resolution microscopy, and quantitative phosphoproteomics. These data support the model that Plk1 operates in pools within the kinetochore and, surprisingly, the bulk pool is at the inner kinetochore/centromere. Moreover, Plk1 operates at chromatin and in the inner centromere in a manner that is distinct and separable from Plk1’s role in stabilizing microtubule attachments at the outer kinetochore17–19. The separability of Plk1 operation within the kinetochore provides an opportunity to match functional and phosphoproteomic signatures, and to identify phosphorylation events that regulate specific aspects of mitotic progression.
Results
Diffusible Plk1 fails to function at the kinetochore
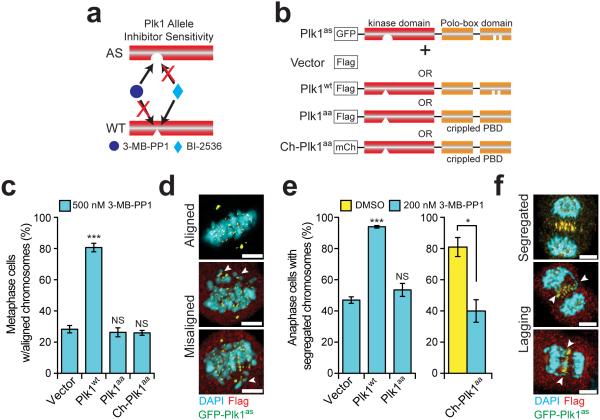
To interrogate Plk1 signaling at the kinetochore, we employed a chemical genetic system wherein each of two distinct Plk1 alleles is separately controlled by chemical inhibitors. Briefly, human RPE1 cells had the endogenous PLK1 exon 3 deleted and rescued by GFP-Plk1as, which encodes chemical sensitivity to 3-methylbenzyl-pyrazolopyrimidine (3-MB-PP1) through two point mutations32. When wild type Plk1 (Plk1wt) is co-expressed in cells with Plk1as, these complement, and either allele can execute the essential functions of this kinase. However, Plk1wt and Plk1as are separately controlled: Plk1wt is sensitive to the pharmacologic inhibitor, BI-2536 and resistant to 3-MB-PP1, whereas Plk1as is resistant to BI-253633 (Fig. 1a). This empowers exquisite spatial and temporal control of Plk1 activity along the kinetochore-centromere axis.
Figure 1. Plk1 signaling at the kinetochore requires binding via its PBD.
(a) Schematic indicating orthogonal control of Plk1 alleles. (b) Schematic of complementation strategy. RPE1 cells expressing GFP-Plk1as (Plk1as) were stably infected with an empty Flag vector (Vector), Flag-tagged wild-type Plk1 (Plk1wt), or Plk1 with a wild-type kinase domain, but crippled Polo-box domain that delocalizes the protein (Plk1aa and Ch-Plk1aa). (c-f) Delocalized Plk1 fails to restore metaphase chromosome alignment or anaphase chromosome segregation when GFP-Plk1as is inhibited with 3-MB-PP1. (c) Graph shows average percentage (± SEM) of pre-anaphase mitotic cells at metaphase with fully aligned chromosomes for each cell line challenged with 500 nM 3-MB-PP1 (n=150 cells/experiment; 3 independent experiments). (d) Representative maximal intensity projection micrographs from (c). Arrowheads indicate misaligned chromosomes. (e) Graph shows average percentage (± SEM) of anaphase cells with fully segregated chromosomes for each cell line challenged with DMSO (yellow bar) or 200 nM 3-MB-PP1 (cyan bars) (n=50 cells/experiment; 4 independent experiments). (f) Representative maximal intensity projection micrographs from (e). Arrowheads indicate lagging chromosomes. *P<.05, ***P<0.0001 by one-way ANOVA or unpaired t-test; NS, not significant (panels c,e). Scale bars, 5 μm (panels d,f).
Plk1 signaling at the kinetochore might not require localization to exert its functions. To test this, we generated lines that stably co-express GFP-Plk1as and either Plk1wt, or Plk1aa, which has a wild type kinase domain but is delocalized by virtue of a mutant PBD (Fig. 1b). The PBD of Plk1aa has two point mutations (H538A/K540M)—sufficient to disrupt Plk1 localization8,14. For the delocalized construct, we used either N-terminal Flag tag (Plk1aa) or mCherry tag (Ch-Plk1aa) (Fig. 1b). Both are expressed and exhibit catalytic activity similar to Plk1wt (Supplementary Fig. 2a-b). As expected, neither localize to kinetochores, centrosomes, or the spindle midzone (Supplementary Fig. 2c), yet both partially restore bipolar spindles (Supplementary Fig. 2d-e), consistent with previous findings10,12.
We previously titrated low inhibitor concentrations to observe Plk1 loss-of-function phenotypes in which a bipolar spindle is preserved12. Under these conditions chromosomes fail to align at the midline, presumably from impaired kinetochore function. As expected, Plk1wt restored chromosome alignment (81% vs. 28% with vector, p<0.0001) (Fig. 1c-d). With Plk1aa or Ch-Plk1aa, however, chromosome alignment remained impaired (both 26% vs. 28% for vector control). Thus, a functional PBD is required for Plk1 to align mitotic chromosomes, consistent with earlier reports10,34.
A second loss-of-function phenotype of Plk1 is lagging anaphase chromosomes, which occur at very modest losses of Plk1 activity after chromosome alignment on a bipolar spindle and silencing of the mitotic checkpoint12. We tested if delocalized Plk1 was sufficient to restore accurate anaphase chromosome segregation. As expected, Plk1wt localized and restored chromosome segregation compared to vector cells (94% vs. 47%, p<0.0001), yet both Plk1aa and Ch-Plk1aa failed to do so (54% and 40%, respectively) (Fig. 1e-f). We conclude that PBD-dependent kinase localization is required for both chromosome alignment and segregation.
Recapitulating Plk1 at the kinetochore
We next considered how Plk1 localization within the kinetochore might control function. At the extremes, Plk1 could bind each substrate directly (focal operation) or it could bind a single partner and have sufficient reach to signal throughout the kinetochore (dispersive operation). Alternatively, Plk1 could operate between these extremes—binding multiple partners with limited dispersiveness to reach adjacent substrates. To test these models, we artificially tethered Plk1 to discrete subcompartments within the kinetochore axis, by replacing the C-terminal PBD with cDNAs encoding proteins that localize to the outer kinetochore (Dsn1), to chromatin (H2B), or to the inner centromere (Kif2c) (Supplementary Fig. 3a). These partners had a diversity of localization and were superior to other tether partners in localization, expression, or activity. We selected clonal lines in which fusion proteins localized appropriately (Supplementary Fig. 3b), with low levels of delocalized extractable protein (Supplementary Fig. 3c-e). Typically, the kinetochore-tethered Plk1 constructs were expressed at lower levels than full-length Plk1 (Plk1wt) but had a higher specific activity (Supplementary Fig. 4).
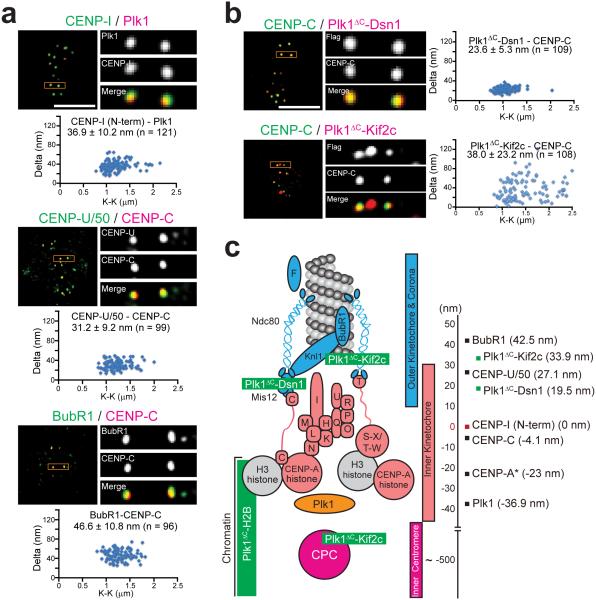
To precisely localize Plk1 and the kinetochore-tethered constructs, we employed high-resolution microscopy of metaphase cells to map mean positions relative to CENP-I/CENP-C standards as described previously35,36 (Fig. 2a-b). Surprisingly, endogenous Plk1 mapped approximately 37 nm inside of the CENP-I probe, within the inner kinetochore at the centromere periphery (Fig. 2c). This finding is unexpected because it spatially isolates bulk Plk1 from known outer kinetochore targets, such as CENP-U/50 and BubR1, which mapped approximately 64 nm and 80 nm outward from Plk1, respectively (Fig. 2a,c). This localization is not an artifact of cell type or antibody as it was similar in HeLa cells and with antibodies targeting tagged Plk1 (Supplementary Fig. 5a-d). Moreover, Plk1 localization is independent of microtubule attachment as Plk1 remains positioned interior to CENP-C in nocodazole-arrested cells (Supplementary Fig. 5e). Thus, wild type Plk1 predominantly localizes deep in the inner kinetochore, near chromatin, independent of microtubule attachment status.
Figure 2. High-resolution microscopy identifies discrete localization of endogenous Plk1 and kinetochore-tethered Plk1 constructs along the kinetochore-centromere axis.
(a) High-resolution single plane images and Delta analysis of endogenous Plk1, CENP-U/50 (also known as PBIP), and BubR1 used for kinetochore mapping in RPE1 cells. K-K indicates distances between the two centroids for CENP-I, CENP-U/50 or BubR1. (b) High-resolution single plane images and Delta analysis of kinetochore-tethered Plk1 constructs. K-K indicates distances between the two centroids for the tethered constructs. (c) Map of kinetochore with summary of Delta measurements from panels a-b set relative to the N-terminus of CENP-I (red). Endogenous proteins indicated in black and Plk1 fusion constructs in green. Positive values are outward toward the spindle microtubules and negative values are inward toward the inner centromere. *CENP-A position was reported previously35. Scale bars, 5 μm (panels a-b).
For tethered constructs, we observed two distinct pools of Plk1ΔC-Kif2c: one in the inner centromere, as expected, and a second 34 nm outside the inner kinetochore CENP-C probe, near the Ndc80 complex at the outer kinetochore (Fig. 2b-c), consistent with Kif2c at microtubule tips36–38. The Plk1ΔC-Dsn1 construct localized an average of 24 nm outside of the CENP-C probe, consistent with endogenous Dsn1 localization35,36, validating localization of the fusion constructs.
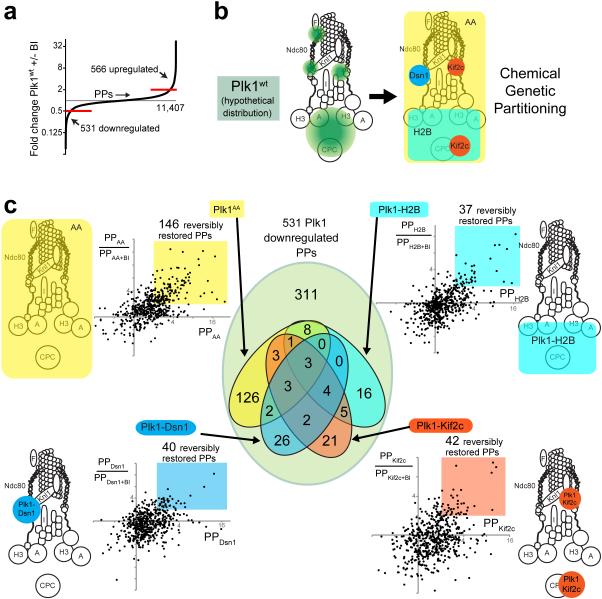
It was initially unclear whether re-localized kinases could access Plk1 substrates, and if so, whether they operate with sufficient dispersiveness to usefully separate function. To assess substrates accessed, we evaluated quantitative phosphoproteomic signatures compared to Plk1wt and Ch-Plk1aa (Fig. 3). We used 5 constructs: Plk1wt (normal localization) and Ch-Plk1aa (delocalized) controls, plus kinase tethered to Dsn1 (outer kinetochore), to H2B (chromatin), and to Kif2c (inner centromere and microtubule tips). For each, we examined phosphoproteomics with tethered/control Plk1 both on (-BI-2536) and off (+BI-2536) for a total of 5 cell lines/2 conditions using 10-plex tandem mass tags (TMT)39 (Supplementary Fig. 6a). We detected 531 phosphopeptides decreased >2x by inhibition of Plk1wt, corresponding to 396 proteins (Fig. 3a, Supplementary Fig. 6b and Supplementary Dataset 1), similar to previous Plk1 phosphoproteomic analyses28–31. Of the 531 Plk1-regulated phosphopeptides, kinetochore-tethered constructs together restored 94 (Fig. 3b-c). These 94 phosphopeptides met three criteria: (i) >2x decrease when Plk1wt was inhibited by BI; (ii) >2x increase with the tethered construct vs. Plk1wt+BI; (iii) >2x decrease when the tethered construct was inhibited with BI. Although some phosphorylation events could be indirect, 47 of these 94 phosphopeptides share the Plk1 consensus motif [D/E/N/Q-X-pS/pT]31, demonstrating many are likely direct and the reach of tethered Plk1 is significant. Additionally, there is minimal overlap in phosphopeptides reached by the 3 tethered constructs, with 63/119 unique to one construct and only 7 restored by all. Thus tethered Plk1 effectively partitions kinase signaling within kinetochore subcompartments.
Figure 3. 10-plex TMT phosphoproteomic analysis of Plk1 partitioned by locale within the KT axis.
(a) Distribution of phosphopeptides (PPs) regulated by Plk1 activity. Out of a total of 11,407 phosphopeptides encountered, 566 PPs were upregulated (> 2-fold increase +BI/−BI) and 531 downregulated (> 2-fold decrease +/−BI) with exposure to the Plk1wt inhibitor, BI-2536. (b) Hypothetical distribution of Plk1wt (green) within the KT axis and partitioning function by locale within the KT axis (Dsn1-outer KT, Kif2c-outer KT/inner centromere, H2B-chromatin) or by a delocalized control, (Ch-Plk1AA) (c) Identification of phosphopeptides (PPs) that are restored with each tethered Plk1 construct. Each dot on plot represents one of 531 physiologically Plk1-regulated PPs (downregulated >2x with Plk1wt+BI). The x-axis displays its abundance relative to control (Plk1wt+BI) with values >2 indicating that phosphorylation is restored by the construct in question. The y-axis indicates the ratio PP/PP+BI, with values >2 indicating that the restored PP is depleted with chemical inactivation of the regionally-restricted construct. Color-coded regions indicate the PPs that are reversibly restored with each regional construct. The Venn diagram at center illustrates the number of PPs that overlap by the distinct color-coded constructs.
The phosphopeptides accessed within the kinetochore are largely consistent with the localization of constructs (Supplementary Table 1). For example, Plk1ΔC-Dsn1 enhances phosphorylation of proteins that interact with microtubules and cytoskeleton, consistent with function at the outer kinetochore. Further validating outer-kinetochore activity of this construct, it restored Plk1-regulated phosphorylation of Dsn1 itself. By contrast, Plk1ΔC-H2B regulated phosphorylation of proteins involved in DNA repair and transcription. Overlapping phosphopeptides were found between Plk1ΔC-Kif2c with both Plk1ΔC-Dsn1 and with Plk1ΔC-H2B, consistent with the known localization of Kif2c at both the outer kinetochore (near Dsn1) and inner centromere (near chromatin), but no overlap was found between Plk1ΔC-Dsn1 and Plk1ΔC-H2B, demonstrating their spatial isolation. We next considered phosphopeptides from kinetochore proteins that are likely direct; they match the minimal Plk1 (D/E/N/Q-X-pS/pT) consensus (Supplementary Fig. 6c). As expected, Plk1wt reached targets at the outer kinetochore (CENPE, CENPF, CDC27), along chromatin (KIF4A, ERCC6L/PICH) and at the inner centromere (INCENP). Plk1aa successfully phosphorylated proteins both at the outer kinetochore (CENPE, CENPF) and the inner centromere (INCENP). This may occur, for example, if low levels of delocalized Plk1 activity are sufficient to phosphorylate proteins protected from phosphatases. Alternatively, delocalized Plk1 could phosphorylate soluble substrates in early mitosis, before kinetochore assembly. Surprisingly, Plk1 at the outer kinetochore (Plk1ΔC-Dsn1) failed to target kinetochore proteins distinct from Plk1aa. Plk1ΔC-Kif2c targeted some, but not all, proteins along the kinetochore-centromere axis. As expected, chromatin-bound Plk1 (Plk1ΔC-H2B) was limited to proteins along chromatin (Kif4A, USP16). We conclude that Plk1 localized to discrete sites within the kinetochore have restricted regions of activity that overlap minimally, yet are sufficiently dispersive to elicit ~40 phosphorylation events each.
Plk1 operates at chromatin and the inner centromere
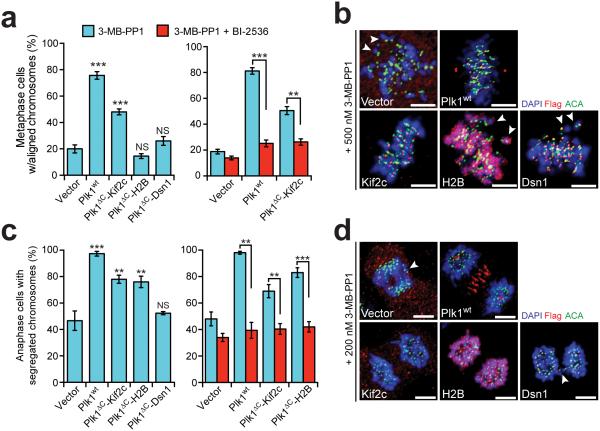
To evaluate where Plk1 operates functionally, we tested the kinetochore-tethered constructs for complementation. We evaluated chromosome alignment and segregation as in Fig. 1, by gently titrating inhibition of Plk1as. Indeed, we found that Kif2c-tethered Plk1, but not Dsn1-localized or H2B-localized kinase restored accurate chromosome alignment compared to uncomplemented (vector) cells (48% vs. 19%, p<0.0001) (Fig. 4a-b; Supplementary Fig. 7a). This rescue was substantial, but incomplete compared to Plk1wt, possibly due to insufficient activation of the requisite substrate, or perhaps, complete rescue requires additional substrate phosphorylation beyond the tethered construct’s reach. Importantly, this rescue was reversed by inhibiting tethered Plk1 with BI-2536 (Fig. 4a), demonstrating that activity of re-localized Plk1, not Kif2c expression, facilitates normal alignment. Additionally, this is unlikely to arise from a soluble pool of Plk1ΔC-Kif2c because active delocalized Plk1aa was unable to rescue (Fig. 1c). We conclude that Kif2c-tethered Plk1 rescues the chromosome alignment function, either due to activity at microtubule tips or at the inner centromere.
Figure 4. Restricting Plk1 activity along the kinetochore-centromere axis produces distinct phosphoproteomic and functional signatures.
Cells expressing GFP-Plk1as and kinetochore-tethered Plk1 constructs were challenged with 3-MB-PP1 (inhibits Plk1as) ± BI-2536 (inhibits Plk1wt/ Plk1ΔC constructs) and assayed for ability of constructs to rescue chromosome alignment during metaphase or segregation during anaphase. (a) Graphs show average percentage (± SEM) of pre-anaphase mitotic cells at metaphase with fully aligned chromosomes for each cell line (n≥100 cells/experiment; 3 independent experiments). (b) Representative maximal intensity projection micrographs from (a). Arrowheads indicate misaligned chromosomes. (c) Graph shows average percentage (± SEM) of anaphase cells with fully segregated chromosomes for each cell line (n≥40 cells/experiment; at least 3 independent experiments). (d) Representative maximal intensity projection micrographs from (c). Arrowheads indicate lagging chromosomes. **P<0.005, ***P<0.0001 by one-way ANOVA or unpaired t-test; NS, not significant (panels a,c). Scale bars, 5 μm (panels b,d).
Next, we tested anaphase chromosome segregation. As before, Plk1ΔC-Kif2c rescued accurate chromosome segregation whereas outer-kinetochore Plk1 (Plk1ΔC-Dsn1) failed to do so (Fig. 4c-d, Supplementary Fig. 7b). However, chromatin-localized Plk1 also functionally restored chromosome segregation. Again, rescue required active kinase, and was reversed with BI-2536 (Fig. 4c). Moreover, soluble Plk1aa was unable to rescue (Fig. 1e). Tethered constructs did not disrupt chromosome alignment or segregation in the absence of inhibition of Plk1as by 3-MB-PP1 (Supplementary Fig. 7c-d). Thus Plk1 acts at either chromatin or at Kif2c sites to elicit its function for proper chromosome segregation in anaphase. Moreover, the functionally distinct behavior of Plk1ΔC-H2B from Plk1ΔC-Kif2c in chromosome alignment suggests that Plk1 phosphorylates one or more unique substrates to execute the disparate functions.
Re-evaluating Plk1 function at the outer kinetochore
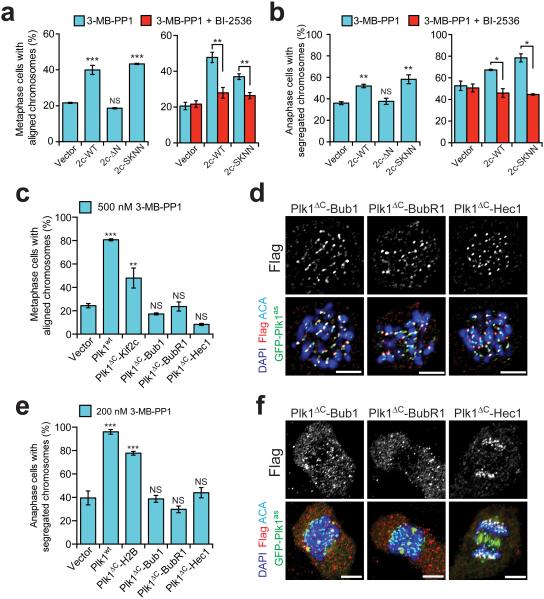
We identified kinetochore functions that arise from Plk1 signals at chromatin or the inner centromere, where the bulk of Plk1 is located. However, outer kinetochore activity could be important if Plk1ΔC-Kif2c operates at the microtubule tips, and Plk1ΔC-Dsn1 might fail to restore activity merely due to impaired access to key outer kinetochore substrates. To address the first question, we separated Plk1ΔC-Kif2c functions at the inner centromere versus outer kinetochore. The Kif2c N-terminus is important for inner centromere targeting40 and the SxIP sequence within the N-terminus promotes microtubule tip-tracking37,38 (Supplementary Fig. 8a, top). Using Kif2c localization mutant constructs (Supplementary Fig. 8a, bottom), both N-terminal deletion mutant (ΔN) and the microtubule tip-tracking mutant (SKNN) were expressed as stable Plk1 fusions and exhibited catalytic activity similar to Plk1ΔC-Kif2cwt (Supplementary Fig. 8b-c). As expected, the SKNN mutant, but not the ΔN mutant, localized to the inner centromere and the SKNN mutant failed to localize at the outer kinetochore (Supplementary Fig. 8d-e). Notably, the SKNN, but not the ΔN mutant, rescued both chromosome alignment and anaphase chromosome segregation phenotypes, and rescue depended on tethered Plk1 activity, as seen by BI-2536-dependent reversal (Fig. 5a-b; Supplementary Fig. 8f-g). Thus, Plk1ΔC-Kif2c operates in the inner centromere to execute key mitotic functions.
Figure 5. Outer kinetochore tethering of Plk1 fails to restore chromosome alignment or segregation.
(a-b) Cells expressing GFP-Plk1as and Kif2c localization mutants were challenged with 3-MB-PP1 ± BI-2536 and assayed for ability of constructs to rescue chromosome alignment during metaphase or segregation during anaphase. (a) Graphs show average percentage (± SEM) of pre-anaphase mitotic cells at metaphase with fully aligned chromosomes for each cell line (n=150 cells/experiment; 3 independent experiments). (b) Graph shows average percentage (± SEM) of anaphase cells with fully segregated chromosomes for each cell line (n=50 cells/experiment; 4 independent experiments). (c-f) Cells expressing GFP-Plk1as and outer kinetochore-tethered Plk1 constructs were challenged with 3-MB-PP1 and assayed for ability of constructs to rescue chromosome alignment during metaphase or segregation during anaphase. (c) As in panel a (n=150 cells/experiment; 3 independent experiments). (d) Representative single-plane micrographs indicating kinetochore localization of Plk1 tethers in cells failing to restore chromosome alignment. (e) As in panel b (n≥30 cells/experiment; 4 independent experiments). (f) Representative maximal intensity micrographs indicating localization of Plk1 tethers in cells failing to restore chromosome segregation. *P< 0.05, **P<0.005, ***P<0.0001 by one-way ANOVA or unpaired t-test; NS, not significant (panels a,b,c,e). Scale bars, 5 μm (panel d,f).
We next considered that Plk1ΔC-Dsn1, though localized correctly and catalytically active, might not access key outer kinetochore substrates. As a measure of Plk1 activity at the outer kinetochore, we evaluated BubR1 phosphorylation, which is detectable by a slower migrating band on SDS-PAGE18,33. Therefore, we tested BubR1 phosphorylation from mitotic extracts, +/− 3-MB-PP1 and found that, indeed, Plk1ΔC-Dsn1 did not reach or only weakly phosphorylated BubR1 (Supplementary Fig. 9a). Therefore, we generated additional constructs, tethering Plk1 to the outer kinetochore proteins Bub1, BubR1 and Hec1 (also called Ndc80). We identified stable cell lines exhibiting diverse expression levels including ones expressed at high levels or similar levels to the other tethers with comparable catalytic activity (Supplementary Fig. 9b-d). Moreover, the Plk1ΔC-Bub1 and Plk1ΔC-Hec1 constructs appeared to restore the slow-mobility p-BubR1 band (Supplementary Fig. 9a). Nevertheless, all three constructs failed to rescue either chromosome alignment or segregation (Fig. 5c-f and Supplementary Fig. 9e-f). Together, these data support the idea that outer kinetochore Plk1 activity is not required for its function in chromosome alignment or segregation. However, we do not exclude the possibility that these four constructs all fail to phosphorylate a critical outer kinetochore protein.
Spatially decoding Plk1 phosphoproteomic signatures
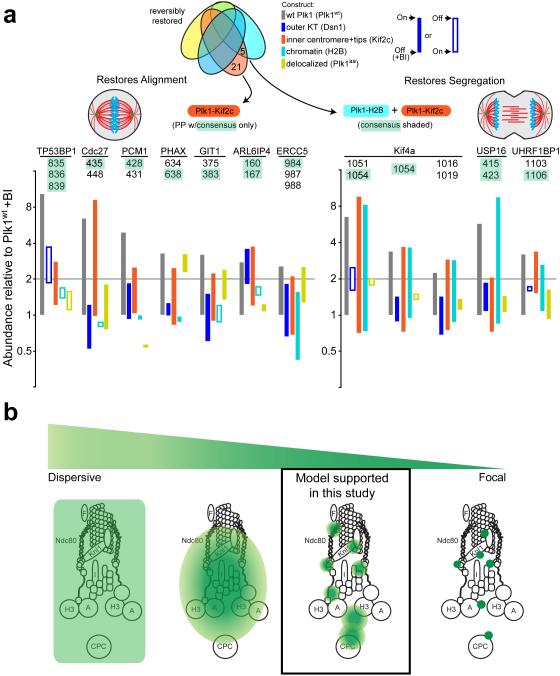
Our results afforded the opportunity to decode Plk1 function at the kinetochore by phenotypic-phosphoproteomic signature. Heretofore, phosphoproteomic analyses of pleiotropic kinases like Plk1 have discovered hundreds of new substrates, yet provided limited information to link them to cognate kinase functions. Here, we identified complex functional signatures-by-locale for Plk1-dependent phenotypes, which are expected to match a limited set of substrates (Fig. 6a). To identify potential substrates, we matched these functional signatures with those of phosphopeptides. For example, chromosome alignment is restored by Plk1 tethered to Kif2c, but is not restored by delocalized Plk1aa or Plk1 tethered to Dsn1 or H2B; only 7 of 176 Plk1-dependent phosphopeptides match this signature (Fig. 6a, left). Similarly, chromosome segregation is restored with Plk1 localized either to H2B or Kif2c, but not to Dsn1 or delocalized Plk1aa; this matches that of only 4 of 146 Plk1-dependent phosphopeptides (Fig. 6a, right). Because Plk1 operates in specific pools within the kinetochore, these data demonstrate that an enlarged set of constructs with restricted localization may effectively decode Plk1 function at the kinetochore. This match of phosphoproteomic signature with functional signature is an unbiased approach to hone in on phosphorylation events that concordantly occur with phenotypic rescue.
Figure 6. Functional/proteomic signatures and a model for Plk1 activity in the kinetochore.
(a) Phosphopeptides that are restored by constructs concordant with phenotypic rescue of chromosome alignment (left) and accurate chromosome segregation (right). Phosphopeptides (PPs) containing minimal Plk1 consensus are illustrated for each phenotype with bars illustrating difference between +/− BI-2536 for each construct. Left, phosphopeptides that are restored by Plk1wt and Kif2c only. Right, phosphopeptides that are restored with both H2B and Kif2c. (b) A model of Plk1 operation within the kinetochore could range from a highly dispersive operation where one binding partner provides access to all substrates (extreme left), versus focal operation, where Plk1 binds each substrate directly (extreme right). Our data support an intermediate model where Plk1 at each subcompartment can access elicit ~40 phosphorylation events, with minimal overlap between inner centromere, chromatin, and the outer kinetochore.
Discussion
Here, we identify the functional and phosphoproteomic signatures of Plk1 tethered to discrete locales within the centromere, chromatin, and kinetochore. A major finding is that Plk1 is predominantly located deep within the kinetochore, where it operates to ensure accurate chromosome alignment and segregation. Notably, this localization is independent of microtubule attachment, suggesting its presence during early mitosis. Previous work has reported small pools of Plk1 at the inner centromere, where it is activated41. We find that Plk1 functions at the inner centromere and at chromatin during mitosis. This finding is supported by concordant data, including high-resolution microscopy co-localizing endogenous Plk1 with centromeric chromatin, by phosphoproteomic data identifying chromatin-bound and inner centromere substrates, and by rescue of Plk1 functions when it is artificially tethered to chromatin or to the inner centromere. Although the central binding partner of Plk1 at chromatin is unclear, one known partner here is Plk1-Interacting Checkpoint Helicase (PICH)42.
We found functional rescue only with active kinase at the chromatin and inner-centromere; however, our findings do not exclude the possibility of Plk1 functions outside of this zone. Indeed, Plk1 phosphorylation at the outer kinetochore mediates its function in attaching chromosomes to the microtubule spindle17,18. Our assays do not capture the outer kinetochore functions of Plk1, likely because they are observed with partial loss-of-function12, in which residual activity may be sufficient to retain phosphorylation of BubR1 and other outer kinetochore substrates. Moreover, a number of phosphorylations are elicited with Plk1 at the outer kinetochore (Fig. 3, Supplementary Table 1). Thus, our findings do not conflict with other strong evidence that Plk1 operates at the outer kinetochore17,18,34.
A second key finding of this study is that Plk1 function at the kinetochore can be uncoupled at the inner centromere, at chromatin, and at the outer kinetochore, suggesting Plk1 operates in distinct pools within the kinetochore (Fig. 6b). A canonical model for Plk1 function is that it directly engages substrates through its C-terminal PBD9,43 (Fig. 6b, extreme right), and discovery of a new substrate is often coupled with an assay for direct binding (focal operation). Consistent with this model, Plk1 is known to bind many substrates within the kinetochore, including BubR1 and others16,18,21,42. We are aware of only one counter-example—endogenous Plk1 engages CENP-U/50 to reach CENP-Q44. Yet, our data powerfully demonstrate that direct engagement of substrates via the PBD is not only unnecessary, but that it should not be routinely expected. For example, Plk1aa lacks phosphopeptide-binding activity, yet phosphorylates 21% of the peptides reached by wild type kinase. Second, Plk1 tethered to each discrete kinetochore subcompartment restores ~40 phosphopeptides (Fig. 3c). Thus, Plk1 function at the kinetochore cannot be accounted for solely by a focal model where it engages each substrate directly.
Conversely, Plk1 operation in the kinetochore is not purely dispersive (Fig. 6b, extreme left). First, delocalized Plk1aa is incapable of rescuing chromosome alignment or segregation (Fig. 1c-f) even though it elicits phosphorylation of more peptides than any tethered construct (Fig. 3c). Indeed, the scale from inner centromere to outer kinetochore exceeds the ~5 nm width of the Plk1 kinase domain26 by two orders of magnitude, making it difficult to imagine how a single focus of kinase could reach throughout without at least limited diffusion. In addition to the theoretical considerations, we observe minimal overlap of phosphoproteomic profiles and ability to rescue Plk1 functions (Figs. 3,4). Thus, our data support a pool model, wherein Plk1 engages, perhaps, a small number of interactors that allow regional dispersiveness of phosphorylation signals within the kinetochore (Fig. 6b). Although a single dominant focus of Plk1 is observed microscopically at each kinetochore, this can be explained by limited sensitivity and diffraction limit—some pools, (e.g. outer kinetochore or inner centromere substrates) are not detected or not resolved. Thus, our data converge on a model that is complex in that it requires both a diversity of interactors and distinct requirements for operation for each pool of Plk1, depending, perhaps, on accessibility to phosphatases. A complex interplay of highly regulated kinase and phosphatase activities are consistent with recent observations that proper kinetochore attachment is regulated by access and activity of phosphatases45–47.
Our tethered Plk1 constructs cannot fully recapitulate dynamic localization, important for proper mitosis. For example, during transition from prometaphase to metaphase, CUL3-KLHL22 ubiquitylation removes Plk1 from the kinetochore48, coincident with decline of phosphorylated targets34. Furthermore, non-ubiquitylatable Plk148 and Plk1 tethered to Hec134 produce similar aberrant phenotypes—impaired kinetochore-microtubule attachments and mitotic delay—suggesting that dampened activity of Plk1 at the outer kinetochore is important for proper mitotic progression. We do not exclude a requirement for similar dynamics of Plk1 at the inner centromere and chromatin. If removal is required, the lack of dynamic activity in our tethered constructs could account for the inability to completely restore function. Alternatively, we may have inadvertently mimicked the dynamic localization of inner centromere Plk1 because Kif2c is removed at the proper time.
In principle, it is possible that rescue is afforded by phosphorylation of a non-physiologic substrate by tethered Plk1; however it is unlikely that chance phosphorylation happens to rescue a specific phenotype by a novel mechanism. It is also possible that Plk1 does not operate at the inner centromere: although we observe Kif2c at the inner centromere, another pool, not readily observable, may exist. Additionally, our conclusions from the phosphoproteomic data are limited by the small number of kinetochore peptides detected. Indeed, well-documented substrates such as BubR1 are absent from our list. It is possible that peptide abundance for these proteins were below our level of detection and different protein enrichment strategies may identify additional kinetochore targets which would better illuminate the phosphoproteomic signatures of Plk1-tethered constructs. Nevertheless, we are able to identify phosphoproteomic evidence of Plk1 function across the kinetochore.
In conclusion, concordant microscopic, phosphoproteomic, and functional evidence places Plk1 deep within the kinetochore and centromere where it executes key mitotic functions. The data best support a ‘pool model’ of Plk1 function at the kinetochore. Thus, Plk1 exists and operates at multiple locations within the kinetochore. For full function, Plk1 needs to engage the kinetochore in discrete pools via its PBD, but it need not engage each substrate individually. Moreover, such engagement is dispensable altogether for certain phosphorylations, such as on CENPE, CENPF or INCENP, which are phosphorylated well by soluble kinase. Although this complicated picture challenges our ability to decode functions of Plk1 at the kinetochore, it simultaneously provides a tool to meet the challenge—Plk1 kinase can be restricted to discrete regions of activity within the kinetochore. In principle, this approach can decode function of any spatially regulated pleiotropic kinase.
From the molecular point of view, the landscape of a kinetochore is finer than can be appreciated by conventional light microscopy. This molecular view is crucial for accurate models of molecular signaling within small, complex biologic structures.
Online Methods
Cell Line Derivation and Culture Procedures
Cell Culture
All cell lines were maintained at 37 °C and 5% CO2 in a humidified incubator and propagated in the following media supplemented with 10% fetal bovine serum and 100 units/mL penicillin-streptomycin: Phoenix retroviral packaging line, Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 4.0 mM L-Glutamine and 4500 mg/L glucose; hTERT-RPE1 derived cell lines, 1:1 mixture of DMEM and Ham’s F-12 media supplemented with 2.5 mM L-Glutamine. EGFP-Plk1as RPE1 cell lines32 and Plk1as cell lines stably expressing mCherry-Plk1 constructs with a “pincer mutant” Polo-Box domain (Ch-Plk1aa)12 were derived as previously reported.
Plasmid Construction
All DNA plasmids were cloned into pQCXIN (Clontech) retroviral vector. Human Kif2c (MHS1010-73945), Dsn1 (MHS1010-9205748), and Hec1 (MHS1010-7508250) were purchased from Open Biosystems. Bub1 and BubR1 were gifts from B.A. Weaver. Flag-tagged Plk1 constructs with a wild-type (Plk1wt) or pincer mutant Polo-Box domain (Plk1aa) were generated by PCR amplification of Plk1 from mCherry-tagged constructs and cloned into pQCXIN-Flag (pQCFIN) using standard restriction digest and ligation procedures. Plk1 fusion constructs were made using a modified USER cloning strategy (New England Biolabs). First, the Plk1 kinase domain (Plk1ΔC, 1-1056aa) was PCR amplified and inserted into pQCFIN using standard restriction digest and ligation procedures. Next, the XbaI restriction site was removed by digest with XbaI, overhang removal by T4 DNA polymerase and blunt-end ligation. A 55-base double-stranded USER acceptance sequence (Supplementary Table 2) containing XbaI and Nt.BbvCI restriction sites was inserted c-terminal to Plk1 (pQCFIN-Plk1ΔC-USER). The plasmid was then linearized by XbaI and NtBbvCI digest, creating 8-base overhangs. Kif2c, H2B and Dsn1 were PCR amplified with primers containing 8-base complementary sequences with a terminal uracil. Products were digested with USER enzyme and ligated into pQCFIN-Plk1ΔC-USER vectors according to manufacturer’s protocol. Kif2c localization mutants were made by PCR amplification of amino acids 140-726 (ΔN) or site-directed mutagenesis (SKNN) using a QuikChange protocol (Stratagene). Constructs were fully sequenced to verify integrity. See Supplementary Table 2 for primer list.
Retroviral Transgenesis
For stable retroviral transduction, constructs were co-transfected with a VSV-G envelope plasmid into Phoenix cells. Fresh medium was applied 24 h post-transfection. A further 24 h later, cells were clarified by centrifugation and filtration through a 0.45 μm membrane to remove cell debris, and diluted 1:1 with complete medium containing 10 μg/ml polybrene (Millipore). Target cells were infected at 40-60% confluence for 24 h, then selected with 0.4 mg/ml G418 for 10-14 d. Polyclonal transductants were further purified by limiting dilution to obtain individual clones.
Chemicals
Chemicals used in this study include 3-MB-PP1 (Toronto Research Chemicals), BI-2536 (Selleck), MG-132 (Enzo Life Sciences), nocodazole and thymidine (both EMD Biosciences).
Immunoblotting, Immunoprecipitation and Kinase Assays
For all experiments, cells were challenged with 0.2 μg/ml nocodazole for 19 h and mitotic cells were collected by shakeoff, pelleted, frozen down and stored at −80°C prior to use. BubR1 detection included both mitotic and non-mitotic cells after 19 h nocadazole treatment.
Immunoblotting
Cell pellets were lysed in buffer (50 mM HEPES pH 7.5, 100 mM NaCl, 0.5% NP-40, 10% glycerol) containing phosphatase inhibitors (10 mM sodium pyrophosphate, 5 mM β-glycerolphosphate, 50 mM NaF, 0.3 mM Na3VO4), 1mM PMSF, 1x protease inhibitor cocktail (Thermo-Scientific) and 1 mM dithiothreitol. Proteins were separated by SDS-PAGE, transferred to Immobilon PVDF membrane (Millipore), and blocked for 30 min in 4% milk and 0.1% Tween-20 Tris buffered saline ph 7.4 (TBST+milk). Membranes were incubated with gentle agitation for 2 h at room temperature with primary antibodies (Supplementary Table 3) diluted in TBST+milk, washed 3x with TBST, incubated for 1 h at room temperature in secondary antibodies conjugated to horse radish peroxidase diluted 1:10,000 in TBST+milk. Membranes were washed and developed with luminol/peroxide (Millipore) and visualized with film.
All results were obtained from single gels. To simultaneously probe for the protein of interest and the loading marker, the membrane was divided in two after transfer and incubated in separate antibody solutions. When identical-sized proteins prevented membrane division, the membrane was first probed for the protein of interest, stripped in an acidic glycine wash (100 mM glycine pH 2, 500 mM NaCl, 2% SDS), rinsed in deionized H2O, and then reprobed for the loading marker.
Immunoprecipitation and in vitro kinase assays
Cell lysates were prepared as above with up to 1 mg total protein used for pulldown. Protein inputs were adjusted to promote equivalent quantities after pulldown. Flag-construct lysates were immunoprecipitated with anti-Flag affinity gel (Biotool) for 1 h at 4°C. mCherry-construct lysates were incubated for 3 h with 5 μg/mL dsRed antibody, followed by immunoprecipitation with a 1:1 mixture protein A and protein G sepharose beads (GE Healthcare) for 1 h at 4°C with gentle rotation. The beads/gel were washed three times with lysis buffer and then once with kinase buffer (20 mM Tris, pH 7.4, 10 mM MgCl2, 50 mM KCl). Fifteen percent of immunoprecipitated complexes were removed for detection of bound constructs by Flag or Plk1 immunoblotting. The remainder was then divided and incubated in kinase buffer, with or without 200 nM BI-2536, plus 5 μg GST-S (substrate), 10 μM ATP, 10 mM dithiothreitol and 2.5 μCi [γ-32P] ATP for 40 min at 30°C. 32P incorporation was observed by SDS-PAGE and Typhoon TRIO imager (GE Healthcare). Three kinase assays were performed from two independent pulldowns. Signal intensity quantification was performed in ImageJ49 by calculating area underneath the curve for each lane after background subtraction. Final construct intensities were determined by subtracting signal with BI-2536 from signal without BI-2536 and expressed as a percentage of the control.
To prepare the Plk1-specific substrate (GST-S) used in the kinase assays, a single nucleic acid sequence encoding 6 known, serine-only, Plk1-specific peptide sequences identified from PhosphositePlus50 was cloned into the pGEX-6P1 vector (GE Healthcare) by Gibson assembly (New England Biolabs). Vector expression was induced in E. coli (BL21DE3) by addition of 400 μM IPTG for 3 hours at 37 °C. Bacteria were resuspended in PBS containing 250 mM NaCl, 10mM EGTA, 10 mM EDTA, 0.1% Tween-20, 1 mM dithiothrietol, 1 mM PMSF, and 1 mg/ml lysozyme prior to sonication. GST-S was purified from lysates using Glutathione Sepharose 4B beads (GE Healthcare) and eluted in 50 mM Tris-HCL pH 8.0.
Immunofluorescence Microscopy
General procedures
Cells were seeded on glass coverslips at low density in 24-well plates and allowed to grow until 80-90% confluence. For chromosome alignment experiments, cells were challenged for 2 h with 10 μM MG-132 to prevent anaphase onset and 500 nM 3-MB-PP1 ± 200 nM BI-2536. For chromosome segregation experiments, cells were challenged for 6 h with 200 nM 3-MB-PP1 ± 200 nM BI-2536. For extraction experiments, cells were challenged overnight with 0.2 μg/mL nocodazole.
For pre-extraction, coverslips were initially incubated for 15 s at room temperature (RT) in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM magnesium chloride) with 0.5% Triton X-100. Otherwise, coverslips were fixed in 4% paraformaldehyde in PHEM buffer for 10 min at RT, washed 3 times in PBS, and then blocked for 30 min at RT in 3% bovine serum albumin (BSA) and 0.1% Triton X-100 in PBS (PBSTx+BSA). Primary antibodies (Supplementary Table 3) were pooled and diluted in PBSTx+BSA. Coverslips were incubated in primary antibodies for 1 h at RT and washed 3 times in PBSTx. Alexa Fluor (Invitrogen) secondary antibodies were pooled and diluted at 1:350 in PBSTx+BSA. Coverslips were incubated in secondary antibodies for 30 min at RT and then washed twice with PBSTx. Coverslips were counterstained with DAPI and mounted on glass slides with Prolong Gold anti-fade medium (Invitrogen) and allowed to cure overnight.
Image acquisition was performed on a Nikon Eclipse Ti inverted microscope equipped with a 100x/1.4NA (Plan Apo) DIC oil immersion objective, motorized stage (Prior Scientific), and CoolSNAP HQ2 CCD camera (Photometrics). Optical sections were taken at 0.2 μm intervals and, except for extraction experiments, deconvolved using the AQI 3D Deconvolution module in Nikon Elements. Panels were cropped using Photoshop CS5 (Adobe) and assembled with overlays using Illustrator CS5 (Adobe).
During quantitation of spindle polarity, chromosome alignment and segregation phenotypes, observer blinding was performed by slide label concealment. “Bipolar” spindles were defined by tubulin staining that exhibited an oval shape, tapered at opposite ends, with pericentrin signal at each end. All other spindle types were considered “abnormal”. For chromosome alignment, all non-prophase pre-anaphase mitotic cells were included for analysis. Cells were scored as exhibiting “aligned” chromosomes if all DNA, visualized by DAPI, was present at the cell equator. All other cells were scored as “misaligned”. For chromosome segregation, anaphase cells were identified by central spindle localization of GFP-Plk1as and chromosome position was determined by DNA (DAPI) and kinetochores (ACA antibody) signals. Cells were scored as exhibiting “segregated” chromosomes if no lagging DNA or ACA signals were observed. “Lagging” chromosomes were defined if single DNA or ACA signals were observed lagging behind the segregating masses or if the two masses were not sufficiently separated despite elongation of the cytoplasm (see Fig. 5, panel f, for example).
Quantitation of cytoplasmic fluorescence intensity was performed using Nikon Elements. Images of 10 cells were acquired for each condition (‘with extraction’ vs. ‘without extraction’ prior to fixation) per Plk1 construct. Threshold levels were equally applied to all images to exclude background intensity. Average volume intensity measurements for each channel were made using a 1.5×1.5 μm box placed in the cytoplasm of each cell. To determine the quantity of construct extracted, the median intensity of the ‘with extraction’ cells was subtracted from individual intensities in the ‘without extraction’ cells. These values were then divided by the amount of GFP-Plk1as extracted to control for cell-cell variability.
Sample size was selected for cell biology experiments based on prior experience and biologically significant effect size. For immunofluorescence, the sample size was typically ~100 cells. Three biologic replicates were performed each with this sample size.
Data analysis was performed using Prism 6 (GraphPad). Statistical significance was determined using an ordinary one-way ANOVA with Dunnett’s multiple comparisons test with a single pooled variance when comparing multiple cell lines against the vector control or a two-tailed, unpaired t-test with Welch’s correction when comparing a single cell line with different chemical treatments.
High Resolution Imaging and Delta analysis
Samples were prepared as previously described35. Image acquisition was performed on a Nikon TE300 inverted microscope equipped with a Yokogawa CSU10 spinning disk confocal with image magnification yielding a 65 nm pixel size from the Orca ER cooled CCD camera and an 100X/1.4NA (Plan Apo) DIC oil immersion objective (Nikon). Sixty-five flame 3D stacks of pairs of red and green fluorescent images were obtained sequentially at 200 nm steps along the z-axis through the cell from coverslip surface using MetaMorph 6.1 software (Molecular Devices).
For each metaphase kinetochore pair, 3D centroid positions were measured with a 3D Gaussian fitting function as described previously36. The centroids of one color were projected to the axis defined by the centroids of the other color, and the Delta (average separation of the projection distance between the signals of red and green colors for theta pair) was calculated to correct for chromatic aberration. Mean Delta values were corrected for tilt of the face of the kinetochore relative to the axis between sister kinetochores.
Mass Spectrometry
Cell Culture
EGFP-Plk1as RPE1 cell lines stably expressing Flag-Plk1wt, Ch-Plk1aa, Flag-Plk1ΔC-Kif2c, Flag-Plk1ΔC-H2B, or Flag-Plk1ΔC-Dsn1 were split in half (2 chemical treatments/cell line) and cultured as above. At approximately 60% confluence, cells were synchronized in S-phase with 3 mM thymidine. After 24 h, cells were rinsed twice with Hank’s Balanced Salt Solution (HBSS), then replenished with fresh media containing 0.2 μg/ml nocodazole. After 18 h, 3-MB-PP1 (final concentration 10 mM) was added to all cells to inhibit Plk1as. To half of the cells, BI-2536 (final concentration 200 nM) was also added to inhibit the complementing Plk1wt allele. One hour later, mitotic cells were collected by shake-off, pelleted, rinsed x 1 with PBS, pelleted again, snap frozen in liquid nitrogen and stored at −80°C. A total of 10 cell pellets (5 cell lines ± BI-2536) were prepared in this manner.
MS Sample Preparation
Cell pellets were lysed in buffer (8 M urea, 50 mM Tris pH 8.0, 100 mM CaCl2) containing dissolved protease and phosphatase inhibitor tablets (Roche) and then sonicated for 20 min. Protein concentrations were determined using a BCA kit (Thermo Pierce). Cell lysates were then reduced by addition of dithiothreitol (final concentration of 5 mM), incubated at 37°C for 45 min, and alkylated by adding iodoacetamide (15 mM final concentration). Next, lysates were incubated for 45 min in the dark at room temperature, and remaining iodoacetamide was quenched by bringing each lysate back to a final 5 mM dithiothreitol concentration. Next, lysates were diluted to a 1.5 M urea concentration using a 50 mM Tris and 100 mM CaCl2 solution. Trypsin was added to each lysate in a 50:1 (protein: enzyme) ratio and digested overnight at ambient temperature. The 10 samples were desalted using 100 mg C18 Sep-Paks (Waters) and dried down using a vacuum centrifuge to obtain tryptically digested peptides. 1 mg of peptides for each of the 10 samples was incubated with 10-plex tandem mass tags (TMT) reagents (Thermo Scientific) for 3 h at room temperature. An aliquot of each sample was mixed in a 1:1 ratio and run on an Orbitrap Elite mass spectrometer (Thermo Scientific) to ensure complete TMT peptide labeling. The 10 samples were mixed in a final 1:1 ratio across all 10 TMT channels and desalted using a 500 mg C18 Sep-Pak (Waters) to produce a single pooled sample containing chemically labeled peptides from all 10 samples (5 cell lines ± BI-2536). The pooled sample was fractionated using Strong Cation Exchange (SCX) chromatography to produce 12 total peptide fractions, which were subsequently lyophilized and desalted. The resultant 12 peptide fractions were each enriched using Immobilized Metal Affinity Chromatography (IMAC) Ni-NTA magnetic agarose beads (Qiagen), leading to 12 final SCX fractionated enriched phosphopeptide and unenriched peptide fractions. Each fraction was dried down using a vacuum centrifuge and resuspended in 0.2% formic acid for mass spectrometry analyses.
Nano-LC-MS/MS Methods
Each sample was introduced to an Orbitrap Fusion mass spectrometer (Thermo Scientific) during a 90 min nano-liquid chromatography separation using a nanoAcquity UPLC (Waters). A “Top N” Fusion method was used to analyze eluting peptides, using a 60,000 resolving power survey scan followed by MS/MS scans collected at 60,000 resolving power. Peptides were fragmented using higher-energy collisional dissociation (HCD) at a normalized collision energy of 35%. Phosphopeptide fractions were analyzed with 200 msec maximum injections times for MS scans and 120 ms maximum injection times for MS/MS scans, while unenriched fractions were analyzed with 100 ms maximum injections times for MS scans and 75 ms maximum injection times for MS/MS scans. Only peptides with charge states from +2 to +8 were selected for MS/MS with an exclusion duration of 30 s. The 12 phosphopeptide samples were run in duplicate.
MS Data Analysis
Data was searched using Proteome Discoverer 1.4.1.14 (Thermo Fisher) with the Sequest search algorithm. Thermo RAW files were searched against a Homo sapiens target-decoy database (UniProt, downloaded 11/06/2014). Peptide and phosphopeptide datasets were searched using a 50 ppm precursor mass tolerance and 0.02 Da fragment tolerance for b and y ions produced by HCD fragmentation. All fractions were searched with static carbadimomethyl of cysteine residues, static TMT 10-plex modifications of peptide N-termini and lysines, dynamic methionine oxidation, and dynamic TMT 10-plex modification of tyrosine residues. Phosphopeptide fractions were searched with additional dynamic phosphorylation modifications of serine, threonine, and tyrosine residues. Resulting peptide identifications were filtered to 1% false discovery rate (FDR) and exported to tab-delimited text files compatible with the COMPASS software suite51. COMPASS was used to obtain the 10-plex TMT protein and phosphopeptide quantitation for all the 12 peptide fractions and 12 phosphopeptide fractions. Peptides were mapped back to their parent proteins using COMPASS, and Phospho RS52 was used to localize phosphorylation to amino acid residues with a fragment tolerance of 0.02 Da automatically considering neutral loss peaks for HCD and considering a maximum of 200 maximum position isoforms per phosphopeptide. The raw 10-plex reporter ion intensities of localized phosphopeptide isoforms were log2 transformed and normalized against inhibited Plk1wt (+BI-2536) to obtain the relative phosphopeptide and protein quantitation for each cell line and condition. Phosphopeptides were considered targeted by each cell line (A) if the uninhibited condition exhibited a 2-fold increase over inhibited Plk1wt (i.e. AA vs. WT_BI) and (B) if the phosphopeptide abundance decreased by half when the cell line itself was inhibited by BI-2536 (i.e. AA vs. AA_BI) (Supplementary Dataset 1).
Supplementary Material
Acknowledgements
This work was supported by NIH R01 GM097245 (to M.E.B.), NIH R01 GM080148 (to J.J.C), NIH R01 GM024364 (to E.D.S), Cancer Center Support P30 CA014520, and Effcansah.com. The authors thank I.M. Cheeseman, S.S. Taylor, and T.J. Yen for contributing reagents, and B.A. Weaver and members of the Burkard laboratory for helpful discussions.
Footnotes
Author Contributions
R.F.L. and M.E.B. designed the research. R.F.L., G.K.P., A.S., and J.M.J performed experiments. R.F.L., G.K.P., A.S., J.M.J., and M.E.B. analyzed the data. M.E.B., J.J.C., and E.D.S. supervised the research. R.F.L. and M.E.B. drafted the manuscript. All authors revised/contributed to the manuscript.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Godek KM, Kabeche L, Compton DA. Regulation of kinetochore-microtubule attachments through homeostatic control during mitosis. Nat Rev Mol Cell Biol. 2015;16:57–64. doi: 10.1038/nrm3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.London N, Biggins S. Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 2014;15:736–748. doi: 10.1038/nrm3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu D, Vader G, Vromans MJM, Lampson MA, Lens SMA. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welburn JPI, et al. Aurora B Phosphorylates Spatially Distinct Targets to Differentially Regulate the Kinetochore-Microtubule Interface. Mol. Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barr FA, Silljé HHW, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 7.Arnaud L, Pines J, Nigg EA. GFP tagging reveals human Polo-like kinase 1 at the kinetochore/centromere region of mitotic chromosomes. Chromosoma. 1998;107:424–429. doi: 10.1007/s004120050326. [DOI] [PubMed] [Google Scholar]
- 8.Elia AEH, et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 9.Elia AEH, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 10.Hanisch A, Wehner A, Nigg EA, Silljé HHW. Different Plk1 functions show distinct dependencies on Polo-Box domain-mediated targeting. Mol Biol Cell. 2006;17:448–459. doi: 10.1091/mbc.E05-08-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu D, Davydenko O, Lampson MA. Polo-like kinase-1 regulates kinetochore-microtubule dynamics and spindle checkpoint silencing. J Cell Biol. 2012;198:491–499. doi: 10.1083/jcb.201205090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lera RF, Burkard ME. High mitotic activity of polo-like kinase 1 is required for chromosome segregation and genomic integrity in human epithelial cells. J. Biol. Chem. 2012;287:42812–42825. doi: 10.1074/jbc.M112.412544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J-E, Erikson RL, Lee KS. Feed-forward mechanism of converting biochemical cooperativity to mitotic processes at the kinetochore plate. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8200–8205. doi: 10.1073/pnas.1102020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burkard ME, et al. Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol. 7. 2009:e1000111. doi: 10.1371/journal.pbio.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi W, Tang Z, Yu H. Phosphorylation- and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol Biol Cell. 2006;17:3705–3716. doi: 10.1091/mbc.E06-03-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishino M, et al. NudC Is Required for Plk1 Targeting to the Kinetochore and Chromosome Congression. Curr. Biol. 2006;16:1414–1421. doi: 10.1016/j.cub.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 17.Suijkerbuijk SJE, Vleugel M, Teixeira A, Kops GJPL. Integration of Kinase and Phosphatase Activities by BUBR1 Ensures Formation of Stable Kinetochore-Microtubule Attachments. Dev. Cell. 2012;23:745–755. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Elowe S, Hümmer S, Uldschmid A, Li X, Nigg E. a. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore-microtubule interactions. Genes Dev. 2007;21:2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maia ARR, et al. Cdk1 and Plk1 mediate a CLASP2 phospho-switch that stabilizes kinetochore-microtubule attachments. J. Cell Biol. 2012;199:285–301. doi: 10.1083/jcb.201203091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hood EA, Kettenbach AN, Gerber SA, Compton DA. Plk1 regulates the kinesin-13 protein Kif2b to promote faithful chromosome segregation. Mol. Biol. Cell. 2012;23:2264–2274. doi: 10.1091/mbc.E11-12-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang YH, et al. Self-Regulated Plk1 Recruitment to Kinetochores by the Plk1-PBIP1 Interaction Is Critical for Proper Chromosome Segregation. Mol. Cell. 2006;24:409–422. doi: 10.1016/j.molcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 22.Park CH, et al. Mammalian Polo-like Kinase 1 (Plk1) Promotes Proper Chromosome Segregation by Phosphorylating and Delocalizing the PBIP1·CENP-Q Complex from Kinetochores. J. Biol. Chem. 2015;290:8569–8581. doi: 10.1074/jbc.M114.623546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goto H, et al. Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat. Cell Biol. 2006;8:180–187. doi: 10.1038/ncb1350. [DOI] [PubMed] [Google Scholar]
- 24.Chu Y, et al. Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J. Mol. Cell Biol. 2011;3:260–267. doi: 10.1093/jmcb/mjq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colnaghi R, Wheatley SP. Liaisons between survivin and Plk1 during cell division and cell death. J. Biol. Chem. 2010;285:22592–22604. doi: 10.1074/jbc.M109.065003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kothe M, et al. Selectivity-determining residues in Plk1. Chem. Biol. Drug Des. 2007;70:540–546. doi: 10.1111/j.1747-0285.2007.00594.x. [DOI] [PubMed] [Google Scholar]
- 27.Cheerambathur DK, Desai A. Linked in: Formation and regulation of microtubule attachments during chromosome segregation. Curr. Opin. Cell Biol. 2014;26:113–122. doi: 10.1016/j.ceb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oppermann FS, et al. Combination of Chemical Genetics and Phosphoproteomics for Kinase Signaling Analysis Enables Confident Identification of Cellular Downstream Targets. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.O111.012351. O111.012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grosstessner-Hain K, et al. Quantitative phospho-proteomics to investigate the Polo-like kinase 1-dependent phospho-proteome. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.008540. M111.008540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kettenbach AN, et al. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santamaria A, et al. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.004457. M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burkard ME, et al. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4383–4388. doi: 10.1073/pnas.0701140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burkard ME, Santamaria A, Jallepalli PV. Enabling and disabling polo-like kinase 1 inhibition through chemical genetics. ACS Chem. Biol. 2012;7:978–981. doi: 10.1021/cb200551p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu D, Davydenko O, Lampson M. a. Polo-like kinase-1 regulates kinetochore-microtubule dynamics and spindle checkpoint silencing. J. Cell Biol. 2012;198:491–499. doi: 10.1083/jcb.201205090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki A, Badger BL, Wan X, DeLuca JG, Salmon ED. The Architecture of CCAN Proteins Creates a Structural Integrity to Resist Spindle Forces and Achieve Proper Intrakinetochore Stretch. Dev. Cell. 2014;30:717–730. doi: 10.1016/j.devcel.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan X, et al. Protein Architecture of the Human Kinetochore Microtubule Attachment Site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Domnitz SB, Wagenbach M, Decarreau J, Wordeman L. MCAK activity at microtubule tips regulates spindle microtubule length to promote robust kinetochore attachment. J. Cell Biol. 2012;197:231–237. doi: 10.1083/jcb.201108147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honnappa S, et al. An EB1-Binding Motif Acts as a Microtubule Tip Localization Signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 39.Phanstiel D, Unwin R, McAlister GC, Coon JJ. Peptide quantification using 8-plex isobaric tags and electron transfer dissociation tandem mass spectrometry. Anal. Chem. 2009;81:1693–1698. doi: 10.1021/ac8019202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maney T, Hunter AW, Wagenbach M, Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carmena M, et al. The chromosomal passenger complex activates Polo kinase at centromeres. PLoS Biol. 2012;10:e1001250. doi: 10.1371/journal.pbio.1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baumann C, Körner R, Hofmann K, Nigg EA. PICH, a Centromere-Associated SNF2 Family ATPase, Is Regulated by Plk1 and Required for the Spindle Checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 43.Lowery DM, et al. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J. 2007;26:2262–2273. doi: 10.1038/sj.emboj.7601683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang YH, et al. Mammalian polo-like kinase 1-dependent regulation of the PBIP1-CENP-Q complex at kinetochores. J. Biol. Chem. 2011;286:19744–19757. doi: 10.1074/jbc.M111.224105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meppelink A, Kabeche L, Vromans MJM, Compton DA, Lens SMA. Shugoshin-1 Balances Aurora B Kinase Activity via PP2A to Promote Chromosome Bi-orientation. Cell Rep. 2015;11:508–515. doi: 10.1016/j.celrep.2015.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu D, et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beck J, et al. Ubiquitylation-dependent localization of PLK1 in mitosis. Nat. Cell Biol. 2013;15:430–9. doi: 10.1038/ncb2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hornbeck PV, et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–20. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wenger CD, Phanstiel DH, Lee MV, Bailey DJ, Coon JJ. COMPASS: A suite of pre- and post-search proteomics software tools for OMSSA. Proteomics. 2011;11:1064–1074. doi: 10.1002/pmic.201000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taus T, et al. Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.