Abstract
Background
Trastuzumab showed survival benefit for Her2-positive gastroesophageal cancers (GEC). Immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) currently determine eligibility for trastuzumab-based therapy. However, these low-throughput assays often produce discordant or equivocal results.
Methods
We developed a targeted proteomic assay based on selected reaction monitoring mass spectrometry (SRM-MS) and quantified levels (amol/ug) of Her2-SRM protein in cell lines (n=27) and GEC tissues (n=139). We compared Her2-SRM protein expression with IHC/FISH, seeking to determine optimal SRM protein expression cut-offs to identify HER2 gene amplification.
Results
After demonstrating assay development, precision, and stability, Her2-SRM protein measurement was observed to be highly concordant with HER2/CEP17 ratio, particularly in a multivariate regression model adjusted for SRM-expression of Met, Egfr, Her3, and HER2-heterogeneity covariates, and their interactions (cell lines r2=0.9842; FFPE r2=0.7643). In GEC tissues, Her2-SRM protein was detected in 71.2% of cases. ROC curves demonstrated Her2-SRM protein levels to have high specificity (100%) at an upper-level cut-off of >750 amol/μg and sensitivity (75%) at lower-level cut-off of <450 amol/ug to identify HER2 FISH amplified tumors. An ‘equivocal-zone’ of 450-750 amol/ug of Her2-SRM protein was analogous to ’IHC2+#x2019;, but represented fewer cases (9-16% of cases versus 36-41%).
Conclusions
Compared to IHC, targeted SRM-Her2 proteomics provided more objective and quantitative Her2 expression with excellent HER2/CEP17 FISH correlation and fewer equivocal cases. Along with the multiplex capability for other relevant oncoproteins, these results demonstrated a refined HER2 protein expression assay for clinical application.
Keywords: Her2 expression, HER2(ERBB2) amplification, gastric, esophageal, gastroesophageal adenocarcinoma, stomach cancer, SRM-MS, selected reaction monitoring mass spectrometry, companion diagnostic, clinical biomarker assay, multiplex protein expression analysis in FFPE tissue
Introduction
The human epidermal growth factor receptor-2 (HER2, ERBB2 ) is a receptor tyrosine kinase promoting cell development, differentiation and survival.[1, 2] Aberrant HER2 activity due to gene amplification and consequent protein overexpression results in a HER2-driven oncogenic phenotype.[1, 2] HER2 is amplified/overexpressed in various cancers including breast (~20%), gastroesophageal (GEC) (~10-15%) and endometrial cancers (~12%).[3] HER2 positivity is higher in esophageal/esophagogastric adenocarcinomas (~15%), compared to distal gastric adenocarcinomas (~10%).[4, 5] The ‘Trastuzumab in the treatment Of GAstric cancer’ (‘ToGA’) trial reported a survival benefit among ‘HER2-positive’ GEC patients treated with trastuzumab-based therapy in comparison to standard chemotherapy, with ensuing widespread incorporation of immunohistochemistry (IHC) and/or fluorescence in situ hybridization (FISH) testing into routine GEC care (Supplementary Figure 1).[4]
‘ToGA’ trial eligibility defined ‘HER2 positivity’ as either a positive FISH score (with any IHC score) or by an IHC score of 3+ (with any FISH score). However, concordance between FISH and IHC is often variable. Patients whose tumors tested FISH positive/IHC negative (0-1+) comprised 22% of those enrolled; these patients derived no benefit from trastuzumab which suggests that binary gene amplification status is imperfectly correlated with protein overexpression (Supplementary Figure 2). Consequently, Her2 IHC was subsequently validated in an independent cohort of GEC samples and the clinical definition of HER2-positive changed to either IHC3+ or IHC2+/FISH-positive (Supplementary Figure 1,2).[6-9] However, discordance between IHC and FISH results continue to affect anti-HER2 clinical trials. Ensuing Her2-selective phase III GEC trials in the first line (‘LOGiC’)[10] and second line (‘TyTAN’)[11] metastatic settings evaluated chemotherapy plus either the HER2/Egfr specific oral tyrosine kinase inhibitor, lapatinib, or placebo. Both trials were negative for the primary endpoint overall survival in the intention-to-treat populations. Interestingly, ‘TyTAN’ enrolled 261 patients of which 31% were FISH-positive/IHC 0-1+.[11] Notably, the IHC3+/FISH-positive subset demonstrated a survival advantage (14 vs 7.6 months, HR 0.59, p=0.0175).[11] Recently, a report suggested that the degree of HER2 amplification/expression may better predict therapeutic benefit from anti-HER2 therapy.[12] These observations suggest the need for revised HER2 criteria/diagnostics, and also implications regarding optimal therapeutic strategies within classic ‘HER2+#x2019; groups.[13, 14]
Despite noted utility of HER2 IHC and FISH, various reports detail numerous limitations.[15-20] IHC is semi-quantitative attempting to incorporate staining intensity and extensity into a ‘0-3+#x2019; scoring system. IHC is notoriously subjective, and sensitive to antigen instability in formalin fixed paraffin embedded (FFPE) unstained-sections, as recently demonstrated.[21-24] ‘HER2-equivocal’ (IHC2+) scores require reflex FISH analysis – accounting for almost 30% (159/584) of FISH-positive cases enrolled cases in ‘ToGA’ (Supplementary Figure 2A), not including undocumented IHC2+/FISH- negative screen failures. Reflex FISH testing is laborious, may be time-consuming (especially serially after IHC), costly if multiple genes are assessed, and remains operator dependent/subjective, particularly in molecularly heterogeneous cases.[7, 25-28] Both assays are low-throughput and result in delayed results and a less-than-economical use of limited tissue samples.[13, 15, 19, 26] Refinement of HER2 diagnostic methods is welcomed.
Mass spectrometry-based selected-reaction-monitoring (SRM-MS) targeted proteomics has gained broad acceptance as a specific and sensitive technology for quantifying levels of specific protein targets. [29-31] However, the ability to apply this technology to FFPE tissues had been technically challenging until recently. A multiplexed and quantitative Liquid Tissue-SRM method to quantify proteins in FFPE tissues based on unique peptide sequences does not have the same technical limitations as IHC/FISH.[21-24]
We sought to evaluate a clinical role of quantitative Her2-SRM protein expression for GEC. Herein we describe application of the Her2-SRM protein assay to 27 cell lines and uniquely to 139 FFPE GEC tumors. After testing the precision and temporal reproducibility of the assay, we assessed correlation of Her2-SRM protein levels with Her2 IHC/FISH scores, along with multivariate modeling to account for HER2-heterogeneity and SRM-expression of other relevant GEC oncoproteins Met, Egfr, and Her3. We determined optimal Her2-SRM expression cut-off values correlating with HER2/CEP17 FISH ratio for clinical application using ROC curves. Finally, clinical cases are presented to substantiate advantages of the ‘GEC-plex’ assay with the capability of quantifying multiple oncoproteins simultaneously, addressing issues surrounding the current molecular profiling hurdles of inter- and intra-patient molecular heterogeneity.
Materials and Methods (See Supplementary Materials for details)
GEC Clinical Samples and Cell Lines
Patient GEC samples and cell lines were obtained from the University of Chicago (Chicago, IL) under preapproved protocols (Supplementary Tables 3,6).[23, 32]
Cell line mixing studies with HER2 amplified OE-19 and HER2 non-amplified MKN-1 to demonstrate dilutional effects of molecular subclones were performed in six lysate conditions with varying ratios (0/100, 20/80, 40/60, 60/40, 80/20, 100/0).
Sample Preparation and Her2-SRM Assay Development
Laser microdissection isolated cells were obtained from FFPE tumor sections as previously described.[21-24] Total protein content for lysates was measured using Micro-BCA assay (Thermo Fisher Scientific Inc, Rockford, IL). Her2-SRM assay development followed previously described methods.[21-24]
Her2-SRM Assay Precision & Temporal Reproducibility
Assay precision and temporal stability of the Her2-SRM assay was performed as previously described.[23, 24]
Quantitative Analysis and Validation of Her2 in Clinical GEC Tissues and Cell lines
Her2-SRM for 139 GEC FFPE samples and 27 cell lines was calculated from the ratio of area under the curve (AUC) for the endogenous and isotopically-labeled standard peptide multiplied by the known amount of isotopically-labeled standard peptide spiked into the sample before analysis, as previously described.[21-24]
HER2 Fluorescence in situ hybridization (FISH)
FISH results were obtained through routine clinical testing for HER2/CEP17 ratio. The majority of samples which had clinical FISH testing were those with an initial Her2 IHC 2+ score, per routine standards. FISH was retrospectively performed on available samples missing FISH results, as previously described.[32, 33]
HER2 Heterogeneity: FISH HER2 heterogeneity (hetero+) was defined as 10-50% of enumerated nuclei having HER2/CEP17 ratio ≥2.[26] HER2 negative (HER2-) was defined as <10% of scored nuclei having ratio ≤2, and HER2+ (non-heterogeneous) was defined as >50% of scored nuclei having ratio ≥2.[26]
Her2 Immunohistochemistry (IHC)
IHC Her2 scores were obtained per routine clinical care using the Hercept Test kit from DAKO.[6] (Supplementary Figure 1,2). For samples without clinical Her2 IHC (eg. archived curative-intent resections), when tissue was available, DAB-labeled dextrose-based polymer complex bound to secondary antibody (Leica Microsystems Inc., Buffalo Grove, IL) was performed as previously described.[34]
Statistical Methods
To examine association between Her2-SRM and HER2 gene copy number (GCN) or HER2/CEP17 ratio in cell lines and tissues, we used univariate and multivariate linear regression models with Her2-SRM as the independent and HER2 GCN or HER2/CEP17 ratio as the dependent variable. Multivariate models included SRM expression for Met, Egfr, and Her3, and their interaction terms, due to putative influence of these proteins on HER2 signaling. Presence of HER2 FISH heterogeneity was additionally included in the models. To compare IHC to either FISH or Her2-SRM in GEC tissues, we included indicators for IHC2+ and IHC3+ (IHC0/1+ reference category) in the regression model. To assess the most effective cutoff value for Her2-SRM to identify HER2/CEP17 ratio, we computed a receiver-operating-characteristic (ROC) curve. All analyses were performed using R software (www.r-project.ort), version 3.0.1.
Results
SRM Assay Development
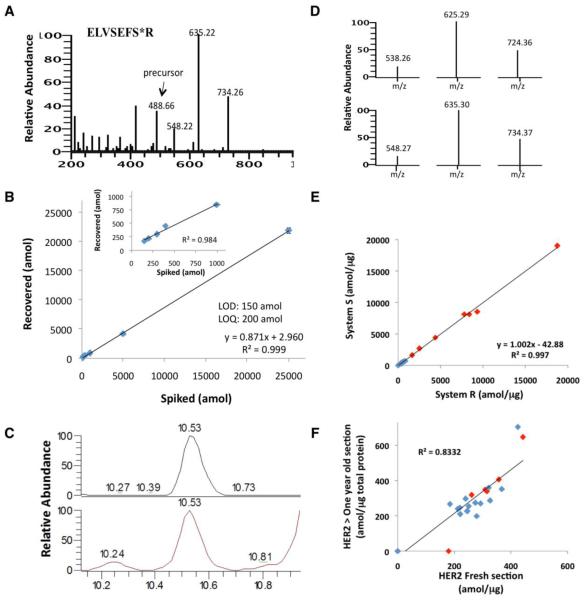
For development of the quantitative Her2-SRM assay, multiple peptides obtained from a tryptic digest of recombinant Her2 were measured using MS. The resulting three candidate peptides, SLTEILK, VLQGLPR, and ELVSEFSR, were then extensively screened in multiple formalin-fixed cell lines and FFPE clinical samples. The peptide ELVSEFSR provided the most reproducible peak heights, retention times, chromatographic ion intensities, clean elution profile, and distinctive/reproducible transition ion ratios; therefore, this peptide was selected for clinical assay development. SRM transitions used for the quantification of Her2 was selected based on the representative MS fragmentation spectrum for peptide ELVSEFSR [13C6,15N4] (Figure 1A). A calibration curve was built in formalin-fixed PC3 cell lysates to assess the linearity and the limits of detection and quantitation (LOD, LOQ) of the assay (Figure 1B). Coefficients of variation (CV’s) ranged from 2.20-14.04% for samples (5 replicates). The LOD/LOQ was 150/200 amol, respectively, with a linear regression value of r2 = 0.9998. The linearity and tight %CV over the concentration range demonstrated the accuracy, reproducibility, and quantitative resolution of the assay. The total ion chromatograms for the light/heavy isotopically-labeled peptides (Figure 1C), and the transition ions (Figure 1D) are shown. Fgfr2-SRM and Her3-SRM are detailed in Supplementary Figure 3.
Figure 1. Development of HER2 SRM assay.
(A) The fragmentation spectrum for heavy ELVSEFSR peptide and (B) the standard curve generated in human PC3 cell lysate; inset: the standard curve generated without the highest two spiking points (5000 and 25000 amol). Each point injected contained 5000 amol heavy (ELVSEFSR [13C6,15N4]) on column. (C) The total ion chromatograms for the light and heavy isotopically labeled peptides, with (D) the transition ions used to identify and quantitate each peptide. (E) Precision assessment for measuring Her2 level in 8 breast cancer (red) and 11 GEC (blue) FFPE tissues. (F) Temporal reproducibility of FFPE sections processed and analyzed using LT-SRM at two time points over one year apart (blue, GEC (n=18); red, NSCLC (n=9)).
Precision of Her2-SRM Assay
To test assay precision, Her2 protein was measured in eight human breast cancer and eleven human GEC tissues. Using two different LC-MS systems and operators, all breast cancer samples expressed Her2, ranging 395.1-18896.7 amol/ug, CVs ranging 3.7-10.4%. Nine of eleven GEC samples expressed Her2 (≥LOD) ranging 306-767.7 amol/ug, CVs ranging 7.5-14.6%. The two operating systems showed very good concordance, r2 = 0.9978, corroborating previous findings with other peptides (Figure 1E, Supplemental Table 1).[21, 23]
Temporal Reproducibility of Her2-SRM Assay
To test the assay’s temporal reproducibility, two sections from 18 GEC and 9 NSCLC samples were processed 13 months apart. Very good correlation (r2=0.8332) supported the reproducibility of Her2-SRM results in archival FFPE sections (Figure 1F, Supplemental Table 2).
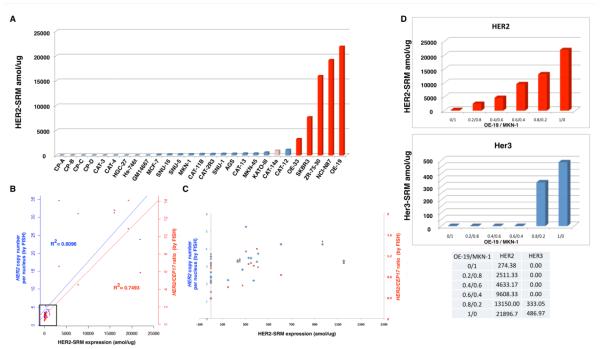
Her2-SRM Correlation with FISH in Cell Lines & HER2-heterogeneity Mixing Studies
The correlation of Her2-SRM expression and HER2 GCN or ratio was assessed in 27 GEC and breast cancer lines. (Figure 2, Supplemental Table 3).Her2-SRM ranged from <150-21896.7 amol/ug (Figure 2A). Her2-SRM results correlated well with HER2 GCN and ratio in univariate analyses (r2 = 0.6096 and 0.7493, respectively) (Figure 2B,C). Adjusting for SRM-expressions for Met, Egfr, and Her3, the multivariate regression model resulted in an improved and very good correlation between Her2-SRM and HER2 GCN or ratio (r2 = 0.8829 and 0.9842, respectively) (Supplementary Figure 4, Supplementary Table 5). Using a preliminary cut-off of 1175 amol/ug, derived from these data, Her2-SRM discerned cell lines with HER2 amplification versus non-amplification with 100% sensitivity (5/5) and specificity (22/22).
Figure 2. Her2 expression levels using LT-SRM and correlation with HER2 gene amplification (by FISH) in 27 cell lines.
(A) Quantification of Her2-SRM (amol/ug) for 27 cell lines including GM15677 lymphoblast control. HER2 amplified cell lines (HER2/CEP7 ratio ≥2) indicated in dark red, and heterogenous HER2 amplification in pink (see methods for hetero+ scoring). (B) Her2-SRM and FISH gene copy number (GCN) univariate correlations: The left y-axis (blue plus sign) represents the mean HER2 GCN per nucleus and the right y-axis (red triangle) indicates HER2:CEP17 ratio (see text for multivariate analysis including Egfr-, Her3-, and Met-SRM coexpression, GCN R2=0.8829 and Ratio R2= 0.9824) and (C) scatter plot of HER2 GCN/ratio (by FISH) (blue diamond, red square, respectively) and Her2-SRM expression in samples where Her2 expressions are < 1500 amol/ug represented by the black box in (3B). A preliminary Her2-SRM cut-off, from these cell line data, correlating with HER2 amplification was determined to be ≥1150 amol/ug. (D) Her2-SRM (red) and Her3-SRM (blue) levels in a cell line mixing study (OE-19: HER2 amplified / MKN-1: HER2 non-amplified), modeling intra-tumor clonal heterogeneity.
One cell line, CAT-14a, was observed to be ‘HER2+/hetero+#x2019;, having 30% of nuclei with ratio ≥2; Her2-SRM level was 969.33amol/ug, below the established cut-off of 1175 amol/ug. To demonstrate effects of stromal elements and clonal sub-populations within tumor masses,[14, 23, 25-28, 34] a mixing study of HER2-amplified and non-amplified cell lines (OE-19/MKN-1, respectively) exemplified a dilutional effect of HER2 expression upon decreasing OE-19 concentrations, likely recapitulating consequences of intra-tumoral HER2-heterogeneity, such as within CAT-14a (Figure 2D).
SRM, IHC and FISH on FFPE Samples
Her2-SRM results were obtained for 139 GEC samples. Among this cohort, Her2 IHC was available for 122 samples. Among these 122 IHC cases, 51 had FISH HER2/CEP17 ratio results, and 42 with absolute GCN scores. (see Supplementary Sample Flow Chart)
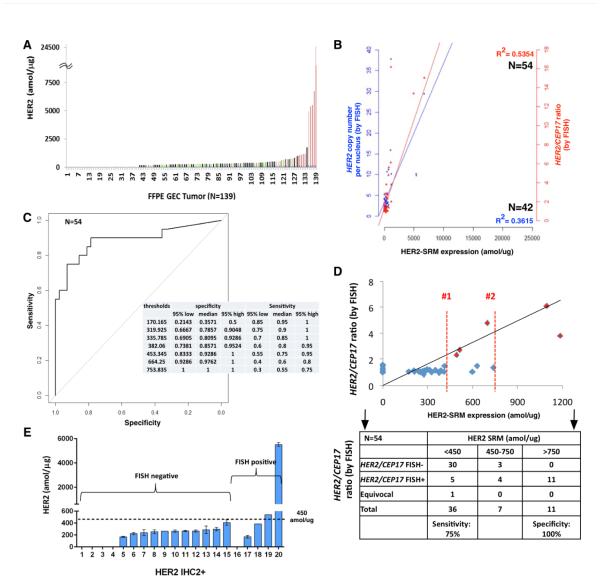
Her2-SRM Expression in FFPE Samples & Comparison to HER2 FISH
Her2-SRM levels were quantitated in 139 GEC tumors (Supplemental Table 4), and expression was above the LOD in 99/139 (71.2 %) samples, ranging 150-24,671amol/μg (Figure 3A).
Figure 3. Absolute levels of Her2 in GEC tissues and correlation of Her2-SRM levels with HER2 gene amplification.
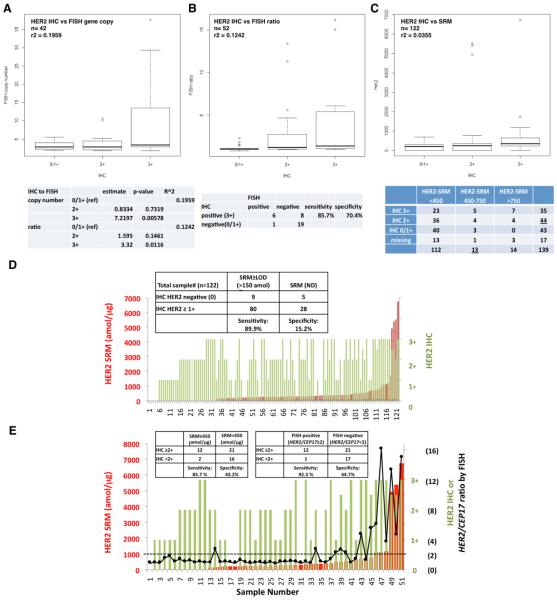
(A) Her2-SRM analysis of clinical FFPE GEC tissues (n=139) ranging <150-24617 amol/ug. Red highlighted samples are verified by FISH to be HER2 amplified (HER2/CEP17 ratio≥2); non-amplified samples are labeled as green, and samples in black were not FISH tested. (B) Univariate correlation of Her2-SRM and HER2 FISH GCN (n=42, blue, r2 = 0.3615) and ratio (n=54, red, r2=0.5354). Multivariate analysis revealed stronger correlations when incorporating Met-SRM, Egfr-SRM, Her3-SRM and HER2 FISH heterogeneity in the model: Her2-SRM:HER2 GCN r2 = 0.7345 and Her2-SRM:HER2/CEP17 ratio r2=0.7643 (54 Her2-SRM cases had absolute FISH ratio available). (C) Optimal Her2-SRM cutoff values determined by receiver operating characteristics (ROC) curve with respect to HER2/CEP17 ratio ≥2 . Using one cut-off level, a value of 450 amol/ug was 75% sensitive and 93% specific to identify ‘amplification’; alternatively, a cut-off level of 750 amol/ug was 55% sensitive and 100% specific. (D) Using two cut-points (analogous to IHC 0/1+ = Her2 negative, and IHC 3+ = Her2 positive), with values in between (analogous to IHC 2+) = ‘equivocal’, an upper SRM level bound of 750 amol/ug and lower bound of 450 amol/ug created an equivocal range 450-750 amol/ug. The two red lines (1 and 2) represent the Her2-SRM expression falling into this equivocal range (n=9/54, 16%). (54 cases were available with binary FISH ratio data (≥2 or <2). Currently, it is recommended that these SRM-equivocal cases undergo confirmatory FISH testing. (E) Among tumors exhibiting Her2 IHC 2+ with FISH results (n=20), 15 tumors (75%) were FISH- and 5 (25%) were FISH+. Her2-SRM expression levels are superimposed, demonstrating that the majority (18, 90%) of these IHC2+ samples were below the 450 amol/ug SRM cut-off.
Among cases having both Her2-SRM and either FISH GCN (n=42) or FISH ratio (n=54) results, univariate analyses demonstrated fair/moderate correlation (r2 = 0.3615 and r2 = 0.5354, respectively) (Figure 3B). After incorporating HER2 FISH heterogeneity along with SRM co-expression of Met, Egfr, and Her3 (Supplementary Figure 4, Supplementary Table 5) there was improvement in the fit of the regression model, and good correlation between Her2-SRM and FISH GCN (r2 = 0.7345) or FISH ratio (r2 = 0.7643) was observed.
Optimal Her2-SRM cutoffs corresponding with HER2/CEP17 ratio ≥2 were determined using a ROC curve (Figure 3C). Exploring various cut-offs, 450amol/ug was 92.86% specific [95%CI 83.33-100] and 75% sensitive [95%CI 55-95] to identify HER2 amplification by FISH; alternatively, a cut-off level of 750 amol/ug was 100% specific [95%CI 100-100] and 55% sensitive [95% CI 30-75].
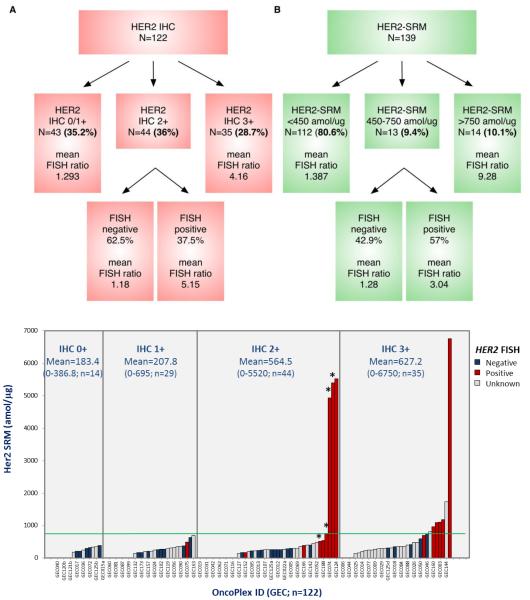
While most tumors demonstrated Her2-SRM levels < 450 amol/ug (112/139 (80.1%)), a few samples had values >750 amol/μg (14/139 (10.1%)). Eleven of 14 samples >750amol/ug that were available for FISH testing, all (100%) were FISH-positive (mean ratio 9.28) - a positive predictive value (PPV) of 100%. A ‘double cut-off’ level, or ‘equivocal’ zone, was applied to better identify marginal HER2 positive cases (not unlike IHC2+ ‘equivocal’). Within the identified SRM ‘equivocal zone’ of 450-750 amol/ug, there were 13 (9.4%) samples (Figure 3C, 5B). In terms of identifying FISH ratio ≥2, the performance of the upper/lower boundaries of this equivocal zone was evaluated on the 54 cases having both Her2-SRM and FISH ratio results (Figure 3D, 5B). Of 7 samples (7/54, 12.9%) between 450-750 amol/ug, there were 3/7 (42.9%) that were deemed FISH-negative (mean FISH ratio 1.28). Therefore, the PPV was 4/7 (57.1%). A lower mean ratio (3.04) was noted for these FISH-positive samples falling within the Her2-SRM equivocal zone than samples with Her2-SRM >750amol/ug (Figure 5B). Of 36 samples <450amol/ug (mean ratio 1.387), 5 (13.8%) were FISH-positive (mean ratio 2.758), one (2.78%) was FISH ‘equivocal’, and the remaining 30 samples were FISH-negative, demonstrating a negative predictive value (NPV) of 83.3%. Depicting IHC, SRM and FISH results sorted by IHC explicitly demonstrates the wide range of SRM expression within the IHC 2+ and 3+ categories, and the large proportion of samples with very low Her2-SRM expression within these two groups, potentially leading to better predictive capacity with respect to benefit from anti-HER2 therapy (Figure 5C). In summary, using the two SRM expression boundaries, the sensitivity of the lower boundary was 75% and the specificity of the upper boundary was 100% to discriminate HER2 FISH ratio≥2, and values within the Her2-SRM equivocal zone showed a FISH-positve PPV of 57.1%. (Figure 3D table)
Figure 5. Her2 status assessment of GEC cases by IHC (A) and Her2-SRM (B) to identify HER2 ‘positive’ and ‘negative’ cases, as determined by underlying FISH HER2/CEP17 ratio.
Both assays resulted in an equivocal zone in identifying underlying HER2 amplification (by ratio>2). However, the incidence of the Her2-SRM equivocal zone (450-750amol/ug) was much lower (9.4%) compared to IHC2+ (36%). Within these Her2-SRM equivocal cases, the PPV was relatively high (57%) compared to IHC2+ (PPV 37.5%). Her2-SRM cases >750amol/ug demonstrated a high mean HER2/CEP17 ratio (9.28) and all were FISH amplified, compared to IHC3+ (4.16); mean ratio for Her2-SRM equivocal cases that were FISH positive was 3.04, compared to 5.15 in IHC2+/FISH+. Fewer equivocal cases requiring less reflex FISH testing along with better stratification of HER2/CEP17 ratio demonstrated superiority of Her2-SRM over IHC. (C) Depicting these same IHC, SRM and FISH results now sorted by IHC category demonstrated the wide range of SRM expression within each IHC category, particularly the large proportion of samples with very low SRM expression within IHC2+ and IHC3+ groups, potentially leading to better predictive capacity of Her2-SRM with respect to benefit from anti-HER2 therapy. Four cases were scored clinically as IHC2-3+ cases (represented with ‘*) (Supplementary Table 4), and all four had FISH ratio >2, with three cases having Her2-SRM >750 amol/ug and one case between 450-750 amol/ug; these cases were included in the IHC 2+ category given the pathologist’s uncertainty of scoring (ie equivocal) and requirement for reflex FISH.
Comparison of Her2 IHC2+ Status to FISH Ratio and Her2-SRM in Tissues
Among tumors with HER2 testing by all three methods, there were 20/54 (37%) that were IHC 2+, of which 15 (75%) were FISH-negative and 5 (25%) were FISH-positive, demonstrating a PPV of 25% (Figure 3E). By Her2-SRM, 18 (90%) of these IHC2+ samples were <450 amol/ug. Among all IHC samples, 44/122 (36%) were IHC2+, with PPV for FISH-positive of 37.5% (Figure 5A). The PPV for Her2-SRM was comparatively higher (57%) within the 450-750amol/ug ‘equivocal’ zone (Figure 3D, 5B), suggesting that Her2-SRM better discriminated FISH amplification status compared to IHC.
Comparison of IHC to FISH or Her2-SRM in Tissues
Among samples having both Her2 IHC and HER2 FISH GCN (n=42) or HER2/CEP17 FISH ratio (n=52), poor correlation was observed (r2=0.1959 and r2=0.1242, respectively) (Figure 4A,B). While IHC2+ (as referenced to IHC0/1+) was not associated with either FISH GCN or ratio, only IHC3+ was significantly associated (p=0.00578 and p=0.016, respectively).
Figure 4. Her2 IHC correlations with FISH assay or SRM.
Correlation of Her2 IHC to (A) FISH GCN in 42 GEC tumors (r2=0.1959), (B) HER2/CEP17 ratio in n=52 GEC tumors (r2=0.1242) (52 cases with IHC results had absolute FISH ratio results); and (C) Her2-SRM (amol/μg) in 122 GEC tumors (r2=0.0355). (D) IHC compared to Her2-SRM with inset 2X2 table; and (E) three-way comparison of Her2-IHC, Her2-SRM and HER2/CEP17 ratio by FISH in 51 GEC tumors where all three values were available. Inset tables demonstrate the comparison of the lower boundary for each assay (left, IHC2+ versus SRM 450amol/ug) and IHC2+ versus FISH (right). Red bar-SRM, green bar-IHC, and black dot-HER2/CEP17 ratio by FISH. Inset tables assess sensitivity/specificity of IHC assuming SRM (D,E) and FISH (B,E) as the comparative standards.
The subset of GEC samples (n=122) having both Her2 IHC (categorical variable) and Her2-SRM (linear variable) demonstrated very poor correlation, r2=0.0355 (Figure 4C). However, when comparing IHC to SRM both as categorical variables, there was noted dependence, Chi Square p=0.02219 (Figure 4C table). To demonstrate the differences in sensitivity, specificity, and resolution between Her2 IHC and Her2-SRM, we compared the LODs for IHC (≥1+) versus Her2-SRM (≥150amol/ug) (Figure 4D). Of 122 cases, 108 (88.5%) were ≥IHC 1+, while 89 (73%) samples were ≥150amol/ug. IHC was 89.9% sensitive in identifying cases ≥150amol/ug, but only 15.2% specific in discerning SRM-negative cases. A score of ‘IHC2+#x2019; was observed in 36% of all cases (44/122) (Figure 5A), or 41% (21/51) of cases available for a three-way comparison of IHC, SRM and FISH ratio. These results revealed that Her2-SRM better correlated with FISH ratio compared to IHC, with better sensitivity and specificity in identifying FISH ratio ≥2 (Figure 4E).
Multivariate regression model with HER2 heterogeneity and multi-plex SRM analysis of oncoproteins
To test whether the correlation between Her2-SRM expression and HER2 FISH would improve after adjusting for the covariates ‘HER2-hetero+#x2019;, Met-SRM, Egfr-SRM, and Her3-SRM for both the cell line and tissue analyses, we evaluated a multivariate regression model (Supplementary Figure 4A,B, Supplementary Table 5). Interactions were observed with Her3-SRM and Egfr-SRM, (positive-interactions) as well as Met-SRM and HER2-hetero+ (negative-interactions) on the association of Her2-SRM with FISH GCN as well as with FISH ratio. These interactions were observed when either FISH status or Her2-SRM was the outcome variable.
Clinical Correlation with Patient Vignettes
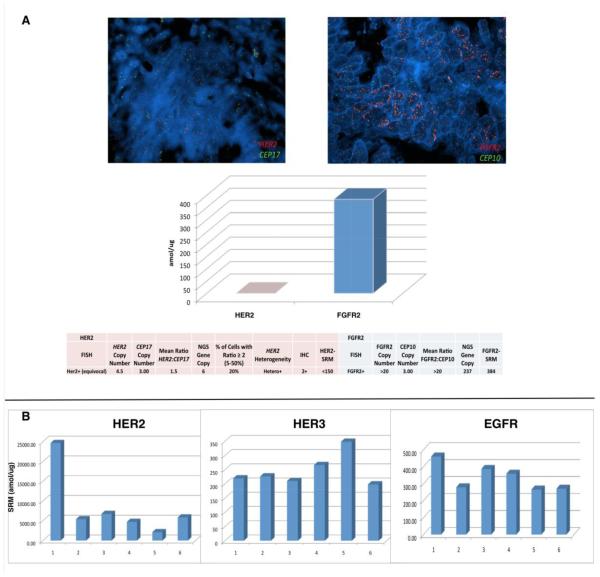
Patient 1, GEC181 (Figure 6A), was diagnosed clinically as stage IV HER2-positive gastric cancer, yet Her2-SRM demonstrated no baseline expression (<150amol/ug, Supplementary Table 4 sample #2) and extremely high Fgfr2-SRM. Upon rapid progression on anti-Her2 therapy, the patient responded to second line Fgfr-specific tyrosine kinase inhibition.
Figure 6. (A). Molecular profiling and a call for treatment prioritization based on degree of genomic/proteomic aberration.
A GEC patient diagnosed clinically as ‘HER2+#x2019; based on IHC (2+) and FISH, yet a more appropriate drug pairing was suggested by SRM multiplex testing (Fgfr2). Her2-SRM was observed to be <150 amol/ug (the range observed in all preclinical/clinical samples to date is <150-26170 amol/ug) while Fgfr2-SRM was observed to be 384 amol/ug (above the 95 percentile of documented Fgfr2 expression). NGS, next generation sequencing. (B) Serial Her2-SRM, Her3-SRM, and Egfr-SRM levels over time/treatment of a Her2 overexpressing and HER2 gene amplified esophageal tumor of a 39 year old man. Segment 1 to 2: First line cisplatin/irinotecan-trastuzumab 6 cycles (D1,D8 q 21 days) not well tolerated; RT to lumbar metastases; Second line FOLFIRI – trastuzumab 12 doses, SD but increasing tumor markers/bleeding; Segment 2 to 3: Embolization to bleeding primary tumor; Third Line –anti-PD1 antibody (PDL1+ IHC), 3 biweekly doses then PD (new multiple liver metastases); Segment 3 to 4: Fourth Line docetaxel-trastuzumab 10 cycles, then recurrent primary tumor bleeding but SD; Segment 4 to 5: RT 20Gy 1/2014-2/2014 to bleeding primary tumor. Fifth Line lapatinib/paclitaxel plus trastuzumab (15 biweekly cycles then PD on CT as well as PD clinically and by tumor markers); Segment 5 to 6: Sixth line FOLFOX-trastuzumab/pertuzumab – stable disease on CT with slight progression of tumor markers. Segment 6 to present: FOLFIRINOX-trastuzumab/pertuzumab for three cycles, response in tumors markers and PR on CT. Biopsy time points (all via EGD of primary tumor, except #6 – liver core biopsy): 1) 5/22/2012 2) 6/10/2013 3) 10/30/2013 4) 1/14/2014 5) 10/7/2014 6) 12/17/15. Diagnosed with symptoms 1/2012, ultimate tissue diagnosis 5/2012 with stage IV disease (bone, M1 lymph nodes) - now almost 36 months from onset of symptoms, 33 months from biopsy (as of 2/2015).
Patient 2, GEC159 (Figure 6B), was diagnosed stage IV HER2+ esophago-gastric cancer, with extremely high baseline Her2-SRM levels observed (24671 amol/ug, Supplementary Table 4 sample #139). Upon interval treatments, serial primary tumor biopsies revealed interesting SRM-expression evolutionary patterns. Upon initial trastuzumab exposure, a dramatic decrease (-78.1%) in Her2-SRM was noted at first tumor progression. After treatment with trastuzumab and lapatinib, elevation of Her3-SRM (+30.8%) was observed (+58.9% from baseline). Subsequently, addition of pertuzumab led to the most recent clinical and biochemical responses with improved dysphagia/tumor markers. The patient is maintained on this therapy 36 months from diagnosis.
Discussion
Current HER2 diagnostics have recognized limitations, and there is urgent need for more objective, expedient, and ‘tissue economic’ assays in order to optimize clinical outcomes for patients.[8, 13, 16, 20, 23, 35] We developed a Her2-SRM assay within a multiplex proteomic quantification assay, and demonstrated its precision, stability, and reproducibility in cell lines and clinical FFPE samples, along with correlation with IHC/FISH and other relevant oncoproteins (Her3/Egfr/Met).
We observed a wide range of Her2-SRM expression not only within the entire GEC cohort (N=139), but also within the subgroups of ‘IHC3+#x2019; or ‘FISH+#x2019; samples (ranging <150-21896.7 amol/ug). Her2-SRM correlated well with HER2 FISH amplification status, reliably identifying highly amplified samples. The degree of HER2 amplification (HER2/CEP17 ratio) linearly correlated with Her2-SRM, as was previously reported,[36] just as the absolute HER2/CEP17 ratio was recently shown to correlate with the degree of anti-Her2 therapeutic benefit.[12] One criticism of SRM technology in FFPE tissue is the low sensitivity to identify very low expression levels (ie <LOD). However, when considering gene amplified proteins, it was evident that the expression levels were dramatically higher than non-amplified expression levels, and therefore this limitation does not appear to be relevant when applying the technology to identify gene amplified tumors. We have noted this across various genes/proteins of interest.[22, 23]
Most IHC2+ cases demonstrated little Her2-SRM expression. The ‘equivocal’ zone that we defined for Her2-SRM expression (450-750 amol/ug), which lacked good correlation with HER2/CEP17 ratio, represented approximately 10-15% of cases, substantially lower than semi-quantitative IHC2+ scoring (~35-40%). Within respective ‘equivocal zones’, the PPV for Her2-SRM was 57% compared to 25-37.5% with IHC2+; others have demonstrated a PPV within IHC2+ as low as 13%.[36] Our IHC2+ rate is similar to that observed in the ‘TOGA’ trial, particularly if including their IHC2+/FISH-undocumented cases. These IHC2+ rates represent the current experience in routine clinical care. Evaluating the performance of IHC versus Her2-SRM, as contrasted in Figure 5, demonstrated the superiority of SRM over IHC in identifying truly FISH-positive HER2 samples, importantly relying less on reflex FISH testing.
In previous years, HER2 cut-offs for IHC and FISH rendering eligibility for anti-Her2 therapy erred towards lower thresholds, likely intending to avoid missing potential benefit of anti-Her2 therapies in patients who would otherwise be given standard cytotoxics alone. However, lack of benefit for these low-expressing subgroups is now recognized.[4, 11] Patient 1 was deemed clinically ‘HER2+#x2019; yet Her2-SRM was low (<450) - this ultimately predicted lack of benefit from anti-Her2 therapy. The ability to further stratify using Her2-SRM within currently clinically accepted HER2+ patients may have significant treatment implications by judiciously assigning anti-HER2 therapy to only those patients most likely to benefit, while sparing those likely to not benefit from both the clinical and financial toxicity of such therapy. Further prospective validation is required in independent datasets which is presently underway, Moreover, multiplex SRM-testing to globally survey biomarkers may allow for optimal treatments towards most-likely tumor ‘drivers’ to be administered as early as possible. In patient 1, very high Fgfr2 expression (consistent with FGFR2 amplification) might have trumped borderline/low Her2 expression. Trials testing prioritized personalized treatment algorithms based on higher-throughput molecular profiling, including SRM-MS are ongoing.[14, 37]
Spatial intratumoral heterogeneity of FISH/IHC resulted in an observed dilutional Her2-SRM measurement, similar to the exemplary cell line mixing study. It is likely that the identified Her2-SRM ‘cut-off’ for cell lines was higher than that of tissues (>1175 versus >750 amol/ug) due to less subclonal and stromal influences. Supporting this, the CAT-14a cell line, which demonstrated HER2-heterogeneity, possessed Her2-SRM levels (969.33 amol/ug) lower than the 1175 amol/ug cell line cut-off. As such, the Her2-SRM assay on FFPE samples inherently captured intra-tumoral HER2 clonal heterogeneity by effectively providing an objective aggregate Her2 expression level representing all the invasive tumor sampled via microdissection using standard H&E staining. As hypothesized, an improved correlation between Her2-SRM level and HER2/CEP17 ratio was observed when including the HER2-heterogeneity status into the multivariate linear regression model. The improved correlation between SRM and FISH after adjusting for FISH-heterogeneity was likely due FISH scores reflecting certain select areas of tumor, while SRM selected all H&E invasive tumor indiscriminately. A significant negative interaction between Her2-SRM level and presence of HER2 FISH heterogeneity was therefore demonstrated (lower-than-expected Her2-SRM level with presence of HER2-heterogeneity).
Her2 functional interactions with Met, Her3, and Egfr have been described. [26, 36, 38-43] After adjusting for these three covariates (Met/Her3/Egfr-SRM co-expression), interestingly, a stronger linear correlation between Her2-SRM and FISH HER2/CEP17 ratio was observed. Although the mechanisms for each of these interactions are not clearly defined, ultimately, gene amplification is a surrogate marker for protein overexpression. Specifically, the relationship between gene amplification and protein expression is likely multifactorial, and may be influenced by the expression of other key oncoproteins within the cell. HER2 amplified tumors tended to have relatively lower Her2-SRM levels if they were also highly expressing SRM-Met, compared to HER2 amplified tumors that were not highly expressing SRM-Met. Supporting our findings, Met overexpression has been linked with resistance to anti-Her2 therapy for HER2 amplified tumors, and vice-versa.[38, 40] On the other hand, Her3- and Egfr-SRM levels were observed to be positively associated with Her2-SRM levels, and both receptors have been implicated in signal transduction of HER2 amplified tumors. Regardless, further work to understand these associations more clearly is required. Notwithstanding, it is possible that incorporating these co-expression covariates, such as is feasible with SRM-MS multiplex technology, while assessing clinical outcome with anti-Her2 therapies will better identify most-likely responders, and may also direct better future multi-drug targeted regimens.[38, 40, 42, 44] This is currently being assessed prospectively in a clinically linked independent dataset.
The ability to evaluate molecular heterogeneity longitudinally through time after treatment was demonstrated using the SRM-multiplex assay in Patient 2. Extremely high Her2-SRM levels appeared to portend for prolonged benefit from anti-Her2 trastuzumab therapy - approximately 12 months before first progression, twice the median progression free survival in the ToGA study (Figure 6B Segment 1-2). At trastuzumab-progression, Her2-SRM levels were ~5-fold lower, offering a potential mechanism of resistance by down-regulating receptor expression - yet it remained well above IHC, FISH, and SRM cut-offs for HER2 positivity. Her2-SRM expression increased slightly after withdrawing trastuzumab for brief anti-PD1 therapy (Segment 2-3), providing more evidence of continued ‘HER2-addiction’. This expression trend, along with previous evidence that maintaining therapeutic inhibition beyond progression upon a persistent oncogenic-driver, provided rationale for resuming trastuzumab-based therapy (Segment 3-4).[14, 45-48] After an initial response to reintroduction of trastuzumab-based therapy, but upon further progression, ‘vertical inhibition’ with lapatinib/trastuzumab led to continued response (Segment 4-5).[48, 49] Finally, evolution towards higher Her3-SRM expression suggested another mechanism of resistance; pertuzumab-based therapy was then introduced with clinical benefit (Segments 5-6-present).[36, 43, 50] Serial testing in order to ‘re-target’ therapies based on ‘real-time’ molecular profiles merits further testing in ongoing novel prospective clinical trial designs.[14]
Compared to IHC, SRM-MS provided more quantitative Her2 expression with better HER2 FISH correlation, and a narrower ‘equivocal zone’. Ultimately, FISH testing for HER2 amplification is a surrogate for Her2 protein overexpression, and we showed that this expression level is influenced by several factors, including not only absolute HER2/CEP17 ratio, but also HER2-heterogeneity within the sample, along with co-expression levels of various other critical oncoproteins. Therefore, a single Her2-SRM expression cut-off in the context of the SRM-MS ‘GEC-plex’ may in the future better predict anti-HER2 therapeutic benefit without any reliance on FISH or IHC. This is the subject of ongoing evaluation and validation in a large cohort of clinically-linked samples. Along with the multiplex capability of quantifying other protein biomarkers, these results demonstrate a refined Her2 expression assay for clinical application.
Supplementary Material
Acknowledgements
DVTC would like to thank Drs. Hedy Kindler, Ravi Salgia, Funmi Olopade and Mitchell Posner for continued support.
All authors would like to thank Dr. Michael F. Press (University of Southern California) for thoughtful and critical review of this manuscript.
Grant Support: This work was supported by NIH K12 award (CA139160-01A), NIH K23 award (CA178203-01A1), UCCCC (University of Chicago Comprehensive Cancer Center) Award in Precision Oncology- CCSG (Cancer Center Support Grant) (P30 CA014599), Cancer Research Foundation Young Investigator Award, ALLIANCE for Clinical Trials in Oncology Foundation Young Investigator Award, Oncoplex Dx Collaborative Research Agreement, LLK (Live Like Katie) Foundation Award, and the Sal Ferrara II Fund for PANGEA (to D.V.T.C).
Nomenclature
- HER2
reference to the pathway in general
- HER2
reference to the gene
- Her2
reference to the protein
Key Abbreviations
- SRM
selection reaction monitoring (mass spectrometry)
- IHC
immunohistochemistry
- FISH
fluorescence in situ hybridization
Footnotes
Ethics Statement: All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later versions. Informed consent or substitute for it was obtained from all patients for being included in the study.
Disclosures:
WLL, ST, KB, JU, MD, DBK, FC, AB, TH, and JB are/were paid employees and stock owners at Oncoplex Dx and/or NantOmics, LLC. DVTC received collaborative research funding from Oncoplex Dx.
References
- 1.Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Natali PG, Nicotra MR, Bigotti A, et al. Expression of the p185 encoded by HER2 oncogene in normal and transformed human tissues. International journal of cancer. Journal international du cancer. 1990;45:457–461. doi: 10.1002/ijc.2910450314. [DOI] [PubMed] [Google Scholar]
- 3.Slamon DJ, deKernion JB, Verma IM, Cline MJ. Expression of cellular oncogenes in human malignancies. Science. 1984;224:256–262. doi: 10.1126/science.6538699. [DOI] [PubMed] [Google Scholar]
- 4.Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 5.Sehdev A, Catenacci DV. Gastroesophageal cancer: focus on epidemiology, classification, and staging. Discovery medicine. 2013;16:103–111. [PubMed] [Google Scholar]
- 6.Hofmann M, Stoss O, Shi D, et al. Assessment of a HER2 scoring system for gastric cancer: results from a validation study. Histopathology. 2008;52:797–805. doi: 10.1111/j.1365-2559.2008.03028.x. [DOI] [PubMed] [Google Scholar]
- 7.Ruschoff J, Dietel M, Baretton G, et al. HER2 diagnostics in gastric cancer-guideline validation and development of standardized immunohistochemical testing. Virchows Archiv : an international journal of pathology. 2010;457:299–307. doi: 10.1007/s00428-010-0952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruschoff J, Hanna W, Bilous M, et al. HER2 testing in gastric cancer: a practical approach. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2012;25:637–650. doi: 10.1038/modpathol.2011.198. [DOI] [PubMed] [Google Scholar]
- 9.Bartley AN, Christ J, Fitzgibbons PL, et al. Template for Reporting Results of HER2 (ERBB2) Biomarker Testing of Specimens From Patients With Adenocarcinoma of the Stomach or Esophagogastric Junction. Archives of pathology & laboratory medicine. 2014 doi: 10.5858/arpa.2014-0395-CP. [DOI] [PubMed] [Google Scholar]
- 10.Hecht JR, Bang YJ, Qin S, et al. Lapatinib in combination with capecitabine plus oxaliplatin (CapeOx) in HER2-positive advanced or metastatic gastric, esophgael, or gastroesophageal adenocarcinoma (AC): The TRIO-013/LOGiC Trial. J Clin Oncol. 2013;31 doi: 10.1200/JCO.2015.62.6598. abstr LBA4001. [DOI] [PubMed] [Google Scholar]
- 11.Satoh T, Xu RH, Chung HC, et al. Lapatinib plus paclitaxel versus paclitaxel alone in the second-line treatment of HER2-amplified advanced gastric cancer in Asian populations: TyTAN--a randomized, phase III study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:2039–2049. doi: 10.1200/JCO.2013.53.6136. [DOI] [PubMed] [Google Scholar]
- 12.Gomez-Martin C, Plaza JC, Pazo-Cid R, et al. Level of HER2 gene amplification predicts response and overall survival in HER2-positive advanced gastric cancer treated with trastuzumab. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:4445–4452. doi: 10.1200/JCO.2013.48.9070. [DOI] [PubMed] [Google Scholar]
- 13.Khoury JD, Catenacci DV. Next-Generation Companion Diagnostics: Promises, Challenges, and Solutions. Archives of pathology & laboratory medicine. 2014 doi: 10.5858/arpa.2014-0063-ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catenacci DVT. Next-generation clinical trials: Novel strategies to address the challenge of tumor molecular heterogeneity. Molecular Oncology. 2014 doi: 10.1016/j.molonc.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allison M. The HER2 testing conundrum. Nature biotechnology. 2010;28:117–119. doi: 10.1038/nbt0210-117. [DOI] [PubMed] [Google Scholar]
- 16.Carlson B. HER2 TESTS: How Do We Choose? Biotechnology healthcare. 2008;5:23–27. [PMC free article] [PubMed] [Google Scholar]
- 17.Cho EY, Srivastava A, Park K, et al. Comparison of four immunohistochemical tests and FISH for measuring HER2 expression in gastric carcinomas. Pathology. 2012;44:216–220. doi: 10.1097/PAT.0b013e3283513e8b. [DOI] [PubMed] [Google Scholar]
- 18.Buza N, English DP, Santin AD, Hui P. Toward standard HER2 testing of endometrial serous carcinoma: 4-year experience at a large academic center and recommendations for clinical practice. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26:1605–1612. doi: 10.1038/modpathol.2013.113. [DOI] [PubMed] [Google Scholar]
- 19.McCullough AE, Dell'orto P, Reinholz MM, et al. Central pathology laboratory review of HER2 and ER in early breast cancer: an ALTTO trial [BIG 2-06/NCCTG N063D (Alliance)] ring study. Breast cancer research and treatment. 2014;143:485–492. doi: 10.1007/s10549-013-2827-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Hurley G, Sjostedt E, Rahman A, et al. Garbage in, garbage out: a critical evaluation of strategies used for validation of immunohistochemical biomarkers. Molecular oncology. 2014;8:783–798. doi: 10.1016/j.molonc.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hembrough T, Thyparambil S, Liao WL, et al. Application of selected reaction monitoring for multiplex quantification of clinically validated biomarkers in formalin-fixed, paraffin-embedded tumor tissue. The Journal of molecular diagnostics : JMD. 2013;15:454–465. doi: 10.1016/j.jmoldx.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Hembrough T, Thyparambil S, Liao WL, et al. Selected Reaction Monitoring (SRM) Analysis of Epidermal Growth Factor Receptor (EGFR) in Formalin Fixed Tumor Tissue. Clinical proteomics. 2012;9(5) doi: 10.1186/1559-0275-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Catenacci DV, Liao WL, Thyparambil S, et al. Absolute quantitation of Met using mass spectrometry for clinical application: assay precision, stability, and correlation with MET gene amplification in FFPE tumor tissue. PloS one. 2014;9:e100586. doi: 10.1371/journal.pone.0100586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hembrough T, Henderson L, Rambo B, et al. Quantification of HER2 from Gastroesophageal Cancer (GEC) FFPE Tissue by Mass Spectrometry (MS) J Clin Oncol. 2014;32(suppl 3) abstr 17. [Google Scholar]
- 25.Seol H, Lee HJ, Choi Y, et al. Intratumoral heterogeneity of HER2 gene amplification in breast cancer: its clinicopathological significance. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2012;25:938–948. doi: 10.1038/modpathol.2012.36. [DOI] [PubMed] [Google Scholar]
- 26.Lee HE, Park KU, Yoo SB, et al. Clinical significance of intratumoral HER2 heterogeneity in gastric cancer. European journal of cancer. 2013;49:1448–1457. doi: 10.1016/j.ejca.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 27.Arena V, Pennacchia I, Vecchio FM, Carbone A. HER-2 intratumoral heterogeneity. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26:607–609. doi: 10.1038/modpathol.2012.147. [DOI] [PubMed] [Google Scholar]
- 28.Lee HJ, Park SY. Reply to 'Intratumoral heterogeneity of HER2 gene amplification in breast cancer: its clinicopathological significance'. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26:610–611. doi: 10.1038/modpathol.2013.38. [DOI] [PubMed] [Google Scholar]
- 29.Nilsson T, Mann M, Aebersold R, et al. Mass spectrometry in high-throughput proteomics: ready for the big time. Nature methods. 2010;7:681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- 30.Addona TA, Abbatiello SE, Schilling B, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nature biotechnology. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whiteaker JR, Lin C, Kennedy J, et al. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nature biotechnology. 2011;29:625–634. doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Catenacci DV, Cervantes G, Yala S, et al. RON (MST1R) is a novel prognostic marker and therapeutic target for gastroesophageal adenocarcinoma. Cancer biology & therapy. 2011;12:9–46. doi: 10.4161/cbt.12.1.15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Catenacci DV, Henderson L, Xiao SY, et al. Durable complete response of metastatic gastric cancer with anti-Met therapy followed by resistance at recurrence. Cancer Discov. 2011;1:573–579. doi: 10.1158/2159-8290.CD-11-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Catenacci D, Polite B, Henderson L, et al. Towards personalized treatment for gastroesophageal adenocarcinoma (GEC): Strategies to address tumor heterogeneity - PANGEA. J Clin Oncol. 2014 GI ASCO 2014, Abstr 60. [Google Scholar]
- 35.Chia S. Testing for discordance at metastatic relapse: does it matter? Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:575–576. doi: 10.1200/JCO.2011.36.6385. [DOI] [PubMed] [Google Scholar]
- 36.Yoon HH, Sukov WR, Shi Q, et al. HER-2/neu gene amplification in relation to expression of HER2 and HER3 proteins in patients with esophageal adenocarcinoma. Cancer. 2014;120:415–424. doi: 10.1002/cncr.28435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abrams J, Conley B, Mooney M, et al. Am Soc Clin Oncol Educ Book. 2014. National Cancer Institute's Precision Medicine Initiatives for the new National Clinical Trials Network; pp. 71–76. [DOI] [PubMed] [Google Scholar]
- 38.Chen CT, Kim H, Liska D, et al. MET activation mediates resistance to lapatinib inhibition of HER2-amplified gastric cancer cells. Molecular cancer therapeutics. 2012;11:660–669. doi: 10.1158/1535-7163.MCT-11-0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yonesaka K, Zejnullahu K, Okamoto I, et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Science translational medicine. 2011;3:99ra86. doi: 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulson AK, Linklater ES, Berghuis BD, et al. MET and ERBB2 are coexpressed in ERBB2+ breast cancer and contribute to innate resistance. Molecular cancer research : MCR. 2013;11:1112–1121. doi: 10.1158/1541-7786.MCR-13-0042. [DOI] [PubMed] [Google Scholar]
- 41.Shattuck DL, Miller JK, Laederich M, et al. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Molecular and cellular biology. 2007;27:1934–1946. doi: 10.1128/MCB.00757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rusnak DW, Alligood KJ, Mullin RJ, et al. Assessment of epidermal growth factor receptor (EGFR, ErbB1) and HER2 (ErbB2) protein expression levels and response to lapatinib (Tykerb, GW572016) in an expanded panel of human normal and tumour cell lines. Cell proliferation. 2007;40:580–594. doi: 10.1111/j.1365-2184.2007.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang XL, Yang YS, Xu DP, et al. Comparative study on overexpression of HER2/neu and HER3 in gastric cancer. World journal of surgery. 2009;33:2112–2118. doi: 10.1007/s00268-009-0142-z. [DOI] [PubMed] [Google Scholar]
- 44.Yoon HH, Shi Q, Sukov WR, et al. Adverse prognostic impact of intratumor heterogeneous HER2 gene amplification in patients with esophageal adenocarcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:3932–3938. doi: 10.1200/JCO.2012.43.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verma S, Miles D, Gianni L, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. The New England journal of medicine. 2012;367:1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Minckwitz G, du Bois A, Schmidt M, et al. Trastuzumab beyond progression in human epidermal growth factor receptor 2-positive advanced breast cancer: a german breast group 26/breast international group 03-05 study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:1999–2006. doi: 10.1200/JCO.2008.19.6618. [DOI] [PubMed] [Google Scholar]
- 47.Geyer CE, Forster J, Lindquist D, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. The New England journal of medicine. 2006;355:2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- 48.Blackwell KL, Burstein HJ, Storniolo AM, et al. Overall survival benefit with lapatinib in combination with trastuzumab for patients with human epidermal growth factor receptor 2-positive metastatic breast cancer: final results from the EGF104900 Study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:2585–2592. doi: 10.1200/JCO.2011.35.6725. [DOI] [PubMed] [Google Scholar]
- 49.Moasser MM. Two dimensions in targeting HER2. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:2074–2077. doi: 10.1200/JCO.2014.55.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baselga J, Cortes J, Kim SB, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. The New England journal of medicine. 2012;366:109–119. doi: 10.1056/NEJMoa1113216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.