Abstract
Many neuronal groups such as dopamine-releasing (dopaminergic) neurons are functionally divergent, although the details of such divergence are not well understood. Dopamine in the nematode Caenorhabditis elegans modulates various neural functions and is released from four left-right pairs of neurons. The terminal identities of these dopaminergic neurons are regulated by the same genetic program, and previous studies have suggested that they are functionally redundant. In this study, however, we show functional divergence within the dopaminergic neurons of C. elegans. Because dopaminergic neurons of the animals were supposedly activated by mechanical stimulus upon entry into a lawn of their food bacteria, we developed a novel integrated microscope system that can auto-track a freely-moving (in actio) C. elegans to individually monitor and stimulate the neuronal activities of multiple neurons. We found that only head-dorsal pair of dopaminergic neurons (CEPD), but not head-ventral or posterior pairs, were preferentially activated upon food entry. In addition, the optogenetic activation of CEPD neurons alone exhibited effects similar to those observed upon food entry. Thus, our results demonstrated functional divergence in the genetically similar dopaminergic neurons, which may provide a new entry point toward understanding functional diversity of neurons beyond genetic terminal identification.
A variety of neuronal types exist in an animal’s brain, each of which plays specific physiological roles based on its own property for integrative brain function. Neuronal cell types are distinguished primarily on the basis of differences in gene expression as well as anatomical properties such as localization, morphology, and interneuronal connectivity. These divergences are regulated by genetic programs as well as temporal and positional interactions1,2.
Dopamine (DA) is a neurotransmitter that has been investigated intensively owing to its prominent roles in the brain functions of higher animals. It is responsible for locomotor regulation, cognition, emotion, reward, and learning, and its dysfunction causes Parkinson’s disease, schizophrenia, and drug addiction3. Despite having such extensive roles, interestingly, the cell bodies of dopaminergic (DAergic) neurons are located in limited areas of the brain such as the midbrain and hypothalamus4. Recent studies have revealed the functional divergence of DAergic neurons, such as differences in sensory information to which they respond and their response patterns5,6,7. Nonetheless, the detailed properties and mechanisms of such divergence have not been sufficiently elucidated.
Likewise, DA is released from a limited number of neurons and regulates a variety of neural functions, such as locomotion, sensory perception, and learning, in the nematode Caenorhabditis elegans8,9,10,11,12,13,14,15. Leveraging the animal’s advantages, such as its small nervous system consisting of just 302 neurons, feasibility of genetic analyses, and utility of a series of cell-specific promoters16,17, the regulatory mechanisms of DA signalling on these neural functions have been studied at the cellular and molecular levels. In hermaphrodites (the major sexual form of the animals), four left-right pairs of neurons are DAergic: CEPD (cephalic sensilla-dorsal), CEPV (cephalic sensilla-ventral), ADE (anterior deirids), and PDE (posterior deirids)18 (Fig. 1a; note that the animals’ left-right axis is vertical and their dorsal-ventral axis is horizontal when they move in the standard laboratory condition). The terminal identities of all of these DAergic neurons are tightly regulated by the same cis-regulatory elements and the same transcription factors in C. elegans as in mammals19,20. In addition, D1- and D2-type receptors function in multiple types of neurons of the animals21, a phenomenon that is analogous to DA signalling in mammals.
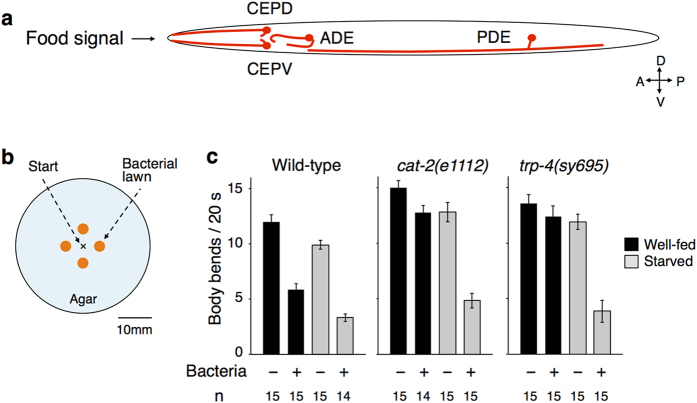
Figure 1. DAergic neurons mediate food-dependent slowing behaviour.
(a) A schematic diagram of arrangement of the four DAergic neuron pairs. Note that this is the top view of an animal on an agar surface. (b) An arrangement of patches of bacterial lawns on an agar surface. Similar experimental condition was recently reported by Hardaway et al.38. (c) Food-dependent slowing response of wild-type and mutant animals. Well-fed animals (black bars) or animals starved for 30 min (grey bars) were transferred to the centre position of the assay plate shown in b and the bending numbers were scored for 20 s after the food entry. The numbers of animals used in each condition are shown at the bottom.
Previous studies have suggested that DAergic neurons in C. elegans are activated by the presence of their food bacteria, one of the most influential environmental stimuli modulating diverse aspects of their behaviour. For instance, the animal’s locomotor speed is slowed upon entry into a bacterial lawn, which is mimicked by a mechanical stimulus from Sephadex beads8. This slowing response is mediated by DA signalling, and laser ablation of all four pairs DAergic neurons was necessary to fully suppress the slowing, suggesting that the neurons function redundantly8. Recently, the use of a genetically encoded calcium indicator22 revealed the head DAergic CEP pairs were activated by mechanical stimulation with a glass tip12. Moreover, optogenetic stimulation23,24 of all DAergic neurons caused slowing25. However, the actual relationship between bacterial presence and DAergic neuron activities remains unclear. Could the bacterial lawn provide a sufficient mechanical stimulus to activate DAergic neurons? In what manner (tonically or phasically, for example) are DAergic neurons activated? Which DAergic neurons are indeed activated by food entry? These questions should be answered to understand how food presence, the most influential information for the animals, is transformed into sensory neural activity and then transmitted to downstream target neurons to modulate multiple behavioural aspects. However, the methodology for studying this relationship has been difficult. For calcium imaging, since the bacterial presence is sensed mechanically but not chemically, the widely used microfluidic flow channel for animal’s stimulation26 is not applicable. Although auto-tracking microscope systems for in vivo calcium imaging or optogenetic stimulation of freely-moving C. elegans recently became available27,28,29,30,31,32, separately analysing the activities of individual DAergic neurons remains challenging because 3 pairs of DAergic neurons (CEPD, CEPV, and ADE) are all located in the head ganglia (Fig. 1a), and the machine-vision discrimination of these neurons has been difficult, especially during tracking.
Here we established an integrated microscope system for calcium imaging and optogenetic stimulation of individual neurons of freely-moving (in actio) C. elegans with high spatiotemporal resolution. While the terminal identities of all of the DAergic neurons are tightly regulated by the same genetic program19,20 and these neurons are considered functionally redundant8, we found functional diversification among DAergic neuron pairs in the slowing response upon food entry. In particular, the CEPD neurons were preferentially activated upon food entry compared to the CEPV neurons despite their symmetry at the gross anatomical level33 (Fig. 1a). The posterior PDE neurons did not appear to be activated. Moreover, the optogenetic activation of CEPD neurons, but not of CEPV or PDE neurons, caused a behavioural response similar to that of food entry. Thus, our results demonstrate the functional diversification of genetically and even anatomically similar DAergic neurons.
Results
Experimental setup for optophysiological analysis during food entry
Here we focused on the slowing of locomotion upon the entry of a food bacterial lawn8, one of the most prominent DA-dependent behavioural responses of C. elegans. The animals reduces the frequency of its body bending and slows down its locomotion upon food entry, which is likely because the animals migrate more quickly to find out food when they are off food, and they migrate more slowly on food to consume it. For the optophysiological analysis of the slowing response upon food entry, we optimized the experimental condition (Fig. 1b). Even with a thin layer (30–40 μm) of bacterial lawn that does not disturb fluorescent imaging, the food entry–dependent slowing response was reproduced (Fig. 1c). The slowing response was affected by mutations in tyrosine hydroxylase cat-2 or TRPN channel trp-4, which are required for DA biosynthesis or DAergic neuron activation, respectively, as previously reported8,12 (Fig. 1c, black bars). The slowing response was observed in both cat-2 and trp-4 mutants after 30 min of starvation (Fig. 1c, grey bars), which is consistent with the fact that 30-min starvation causes an enhanced slowing response that is dependent on serotonin, and not on dopamine8.
Cell-targeted calcium imaging and optogenetics system for freely-moving animals
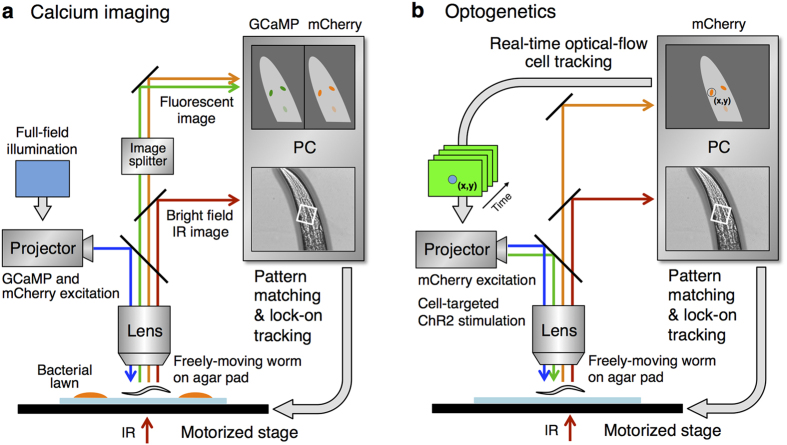
To optophysiologically analyse multiple DA neuron pairs in actio, we developed an auto-tracking microscope system featuring sophisticated image processing and machine-control techniques (Fig. 2 and Supplementary Video S1). The auto-tracking system maintains the head of a freely-moving C. elegans in the centre of the field of view using a texture pattern-matching algorithm34.
Figure 2. Schematic drawing of the OSaCaBeN system.
Setups for calcium imaging (a) and optogenetic analysis (b) are shown. The motorized stage is controlled to lock-on a part of the animal’s body at the centre of the view field. In (a), blue light is illuminated to the full view field to excite GCaMP6f and mCherry in all of the DAergic neurons. In (b), green light for mCherry is illuminated to the full view field to monitor the positions of all of the DAergic neurons, and blue light is illuminated to one of the DAergic neurons to stimulate ChR2 when necessary. The position of the blue light illumination is updated in real time because the target neuron moves in the view field due to the limitation of tracking accuracy. An IR light was used to acquire bright field images for pattern matching and tracking.
In the calcium imaging setup, the images of green fluorescence from GCaMP6f 35 and red fluorescence from mCherry36 were used to monitor the intracellular calcium concentration changes and the cell body positions of the multiple DAergic neurons, respectively. The individual signal intensities at each of the cell bodies were measured separately off-line. However, the conventional method that tracks the brightest point in the view field is not applicable. Thus, here we developed a software that individually tracks multiple fluorescent signals using the optical flow method (Supplementary Video S2). It should be noted that the measured fluorescent signal from one cell body likely reflects the sum of vertically aligned a pair of left and right neurons.
In the optogenetics setup, the optical flow method is used on-line: channelrhodopsin-2 (ChR2), a light-gated cation channel23 and mCherry are co-expressed in the DAergic neurons, and the mCherry signals were used to identify the positions of multiple DAergic neuron pairs. Blue light is illuminated only at the instantaneous position of the targeted cell using a high power video projector as a light source37 (Supplementary Video S3). With the high speed of its tracking system, this method allows for optogenetic activation of one among multiple target cells in the head ganglia of the moving C. elegans (see Discussion). We named this the Optogenetic Stimulation associated with Calcium imaging for Behaving Nematodes (OSaCaBeN or OSB) system.
Anatomically symmetrical CEPD and CEPV neurons are asymmetrically activated
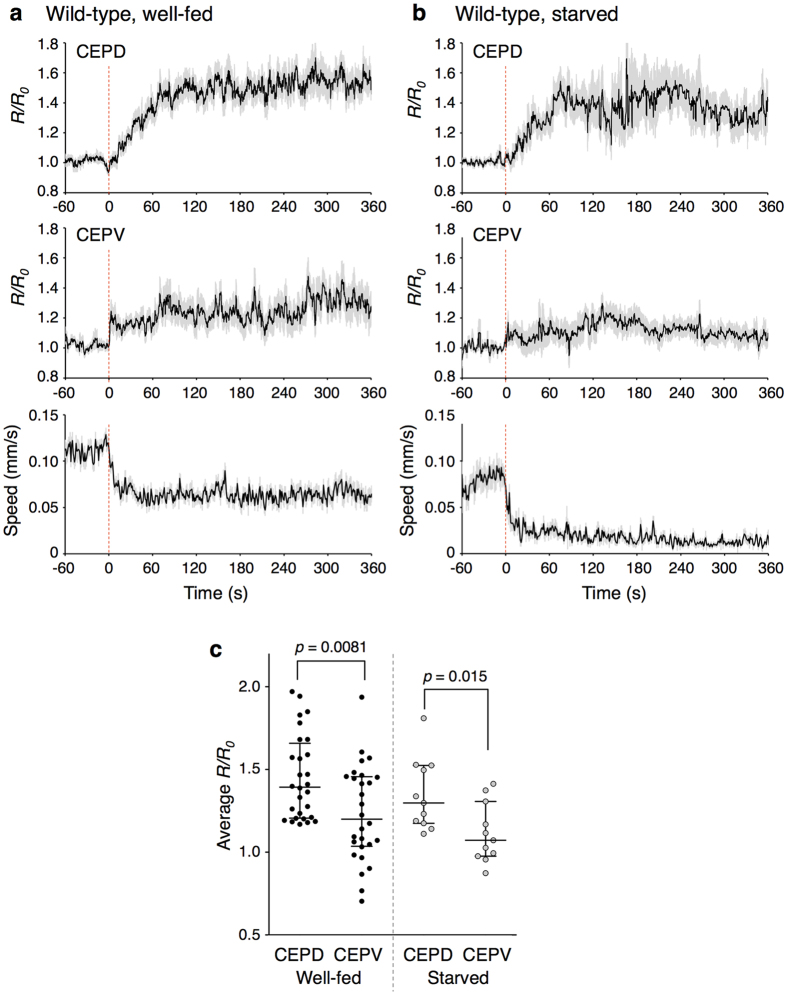
Using the calcium imaging setup, we measured the activities of the CEPD and CEPV neurons, which are symmetrical in the dorso-ventral axis at the gross anatomical level and extend their sensory endings to the anterior end of the body33 (Fig. 1a). Upon food entry, which likely produces a subtle mechanical force, CEPD and CEPV pairs were activated as reported recently38, and the activity was sustained at least for 6 minutes (Fig. 3a). Unexpectedly, however, we found that the CEPD activation was gradual and stronger, while CEPV activation was quicker and weaker (Fig. 3a,c). These results indicate that CEPD and CEPV pair activities upon food entry are asymmetrical, although these neuron pairs are considered genetically indistinguishable and functionally redundant. We also analysed the PDE pair of DAergic neurons, whose processes cover more than half of the posterior end of the body (Fig. 1a). Food entry-dependent activation of PDE pair was observed only in a few trials (Table 1, Fig. S1). The ADE neuron pair could not be analysed because of dim GCaMP fluorescence from the cells, possibly due to low intracellular calcium concentration before and after food entry. These results suggest that CEPD and CEPV neuron pairs sense the bacteria presence upon food entry, whereas ADE and PDE neuron pairs may be devoted to a different role. In trp-4 mutants, CEPD and CEPV showed little response upon food entry (Table 1, Fig. S1). This result is consistent with the fact that CEP neurons of the trp-4 mutant are defective in response to a certain type of mechanical stimulus: trp-4 mutants did not respond to gentle and repetitive press stimulus, but responded to a continuous press12.
Figure 3. CEPD and CEPV neuron pairs were asymmetrically activated upon food entry.
(a,b) Responses of CEPD (top) and CEPV (middle) as well as speed (bottom) of well-fed (a) or starved (b) wild-type animals upon food entry are shown. The time when an animal entered a bacterial lawn was determined as t = 0. The R/R0 values (average ± SEM) are shown. The responses of CEPD and CEPV were monitored from the same animals. Twenty-eight and 11 animals were analysed for well-fed and starved conditions, respectively. When the average R/R0 value was lower than 1.1 in both of CEPD and CEPV in an animal, that animal was regarded as “not responding”, and those data were excluded. (c) Scatter plot comparing averaged R/R0 values between CEPD and CEPV of well-fed (left) or starved (right) wild-type animals upon food entry. A two-tailed Mann-Whitney test was used for the statistical analysis.
Table 1. Numbers of animals that exhibited calcium response to food entry.
| Strain | Feeding status | Cell | # Tested | # Responded | % Responded |
|---|---|---|---|---|---|
| Wild-type | fed | CEPD, CEPV | n = 39 | n = 28 | 71.8% |
| Wild-type | starved | CEPD, CEPV | n = 16 | n = 11 | 68.8% |
| Wild-type | fed | PDE | n = 26 | n = 6 | 23.1% |
| trp-4(sy695) | fed | CEPD, CEPV | n = 8 | n = 3 | 37.5% |
When the average R/R0 ≥ 1.1 in either CEPD or CEPV cells of an animal after food entry, the animal was scored as “responded”.
We further measured CEPD and CEPV pair activities under starved condition. Starvation is known to modulate various aspects of C. elegans’ behaviour16,17, and DA-dependent behavioural modulations such as area-restricted search and tap habituation are generally not observable after 30 minutes of starvation10,12. In addition, after starvation, the slowing response upon food entry is enhanced in a serotonin-dependent manner8 (Fig. 1c). Regardless, it was not clear whether the responses of DAergic neurons themselves are modulated by starvation. We found the responses of CEPD and CEPV neurons to bacterial lawn were essentially similar before and after starvation (Fig. 3b,c), while the slowing response was enhanced (Fig. 3a,b, bottom). These results suggest that starvation-induced modulation of DA signalling may occur not in the DAergic neurons themselves but rather in the target neurons, such as sensory or motor neurons12,25,39.
CEPD is mainly responsible for food-dependent slowing behaviour
Calcium imaging revealed that CEPD and CEPV pairs were activated asymmetrically upon food entry. However, the relationship between this asymmetrical activation and their behavioural consequences is still unclear. To further elucidate the functional differences in DAergic neuron activities, we optogenetically activated the individual DAergic neuron pairs and analysed the effects on the slowing response. We established transgenic animals expressing ChR2 and mCherry in all of the DAergic neurons at similar levels in the cells (Fig. 4a). Blue light illumination to the animals’ whole bodies caused a slowing response similar to the one observed upon food entry as reported previously25 (Fig. 4b). The effect was observed only in the presence of all-trans-retinal (ATR), which should be supplied as a co-factor of ChR2 for the animals40. We then illuminated the blue light to a specific pair of DAergic neurons with the optogenetic setup of the OSB system (Fig. 2b). When the CEPD pair was specifically activated, a significant slowing response was observed, whereas CEPV activation did not cause significant slowing (Fig. 4b). We also tested PDE pair but observed no response. These results indicate that CEPD pair activation is mostly responsible for the slowing behaviour.
Figure 4. Activation of CEPD neuron pairs is mainly responsible for the slowing behaviour.
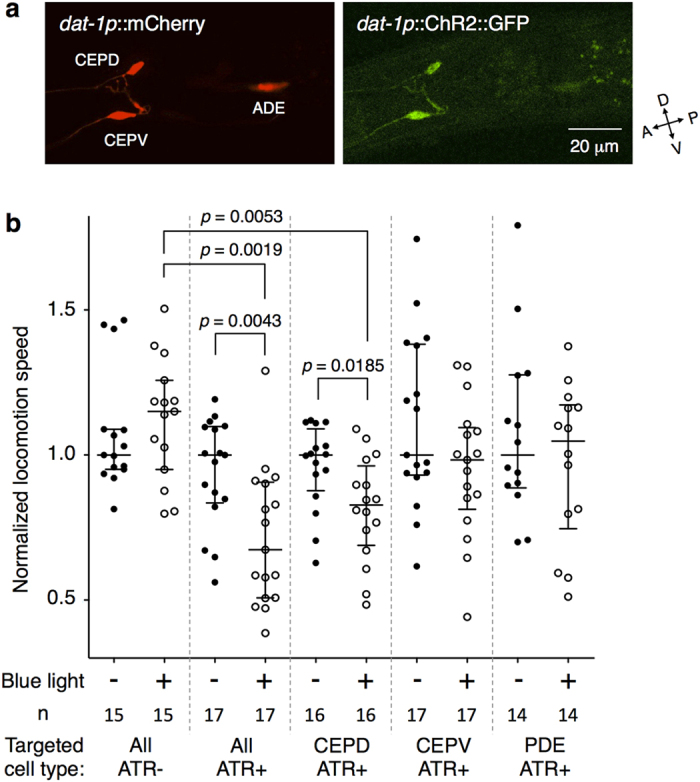
(a) Expressions of mCherry and ChR2::GFP were at similar levels between CEPD and CEPV but was lower in ADE. (b) Comparison of the effects of optogenetic stimulation on slowing. When all of the dopaminergic neurons or only CEPD were illuminated, but not CEPV or PDE, significant slowing occurred. A Mann-Whitney test was used to compare the normalized locomotion speeds before (−) and during (+) the blue light illumination within each targeted cell type, while a Kruskal-Wallis test with a post-hoc Steel-Dwass test was used to compare cell types during illumination. Details of the statistical analyses are shown in Supplementary Table S4.
Discussion
Here we revealed the functional diversification of DAergic neurons in C. elegans. To analyse the physiological properties of DAergic neurons that are activated by the mechanical stimulus of bacterial lawn, we established an integrated microscope system for in actio optophysiological analyses and found that CEPD and CEPV neuron pairs, which have been considered highly similar to each other based on their anatomical symmetry and similar gene expression patterns, are functionally asymmetrical in terms of their responsiveness to the bacterial lawn stimulus and the effect of their activation on the slowing response.
To individually analyse the food entry–dependent activation patterns of DAergic neuron pairs of C. elegans, which are located in close proximity and cannot be separated with known cell-specific promoters, we established the OSB system, which tracks freely-moving animals with high spatiotemporal resolutions and performs calcium imaging and optogenetic illumination of individual neuron pairs. Optogenetic illumination requires higher specifications than calcium imaging. For calcium imaging, multiple neuronal activities can be simultaneously captured during tracking and analysed individually afterward (i.e., off-line): individual neurons do not require separate identification during tracking. In contrast, for optogenetic illumination, individual neurons should be identified during tracking (i.e., on-line). For temporal resolution, the widely used optogenetic microscope systems for moving C. elegans track an animal with a cycle speed of 25–50 Hz29,30, while our system does the same at 200 Hz. If a system possesses a spatial resolution of 2–3 μm (the regular diameter of the animal’s neuronal cell body41), an animal moving at 150 μm/s should be tracked at >50 Hz (for details, see Stirman et al.30). For spatial resolutions, the aforementioned two systems use a 4–10× objective lens to capture the image of the animal’s entire body for tracking29,30, while our system uses a 20× or higher objective lens because it allows tracking of a part of the animal’s body by using a high-speed pattern-matching algorithm34. The two systems have been used in many laboratories for individual optogenetic analyses of multiple neurons and/or muscles that are sufficiently separated, such as those in the middle and posterior parts or on opposite sides of the body29,30,42,43,44, but not for multiple neurons within the animal’s brain. While there are a few other systems for calcium imaging or optogenetic analyses of multiple neurons in the brain of the animals27,28,31,32, our OSB system allows for both calcium imaging and optogenetic analysis and revealed the functional asymmetry of DAergic neuron pairs of moving C. elegans, which are separated by only 20–25 μm in the brain. Our system can also be used for functional dissection of other neurons that are genetically indistinguishable and located in close proximity; it could also be used with spatially and temporally more complex pattern of optogenetic illuminations to reveal dynamic relationships of activities of multiple neurons in future experiments.
We found that the CEPD and CEPV neurons are both tonically activated. Sensory neurons generally exhibit phasic, tonic or phasi-tonic responses45. Phasic response signals the changes in stimulus over a wide range, which contributes to an animal’s navigation in a chemical gradient, for example. The tonic response signals the presence/absence and strength of the stimulus over time. Tonic signalling is used to provide positional information about the body parts (proprioception) or for biologically significant chemicals such as CO245. These functions of phasic and tonic signalling are also true in C. elegans. Sensory neurons for chemotaxis or thermotaxis show phasic responses46,47,48, while those for sensing O2 show tonic responses49. As such, it is reasonable to speculate that the C. elegans DAergic neurons are tonically activated to signal the information of food presence, which modulates various aspects of the animal’s behaviour. In a previous study, CEP neurons exhibited a rapidly-adapting (i.e., phasic) response to a mechanical press with a glass probe50, suggesting that the tonic response observed in this study might be triggered by a subtle and/or fluctuating mechanical pressure brought about by the animal’s movement on a bacterial lawn. Quite interestingly, tonic signalling of DA is well known in primates and rodents: Tonic DA release enables a variety of motor, cognitive, and motivational processes, while phasic DA release signals rewards and alerting stimuli3. Thus, the role and mechanism of tonic DA signalling in the modulation of various aspects of the nervous system may be evolutionarily conserved from nematodes to mammals.
Our analysis revealed that CEPD and CEPV pairs are functionally asymmetrical. Not only were their activation patterns different, but the effects of their optogenetic activation differed: In both cases, CEPD played a more significant role. In C. elegans, there are a few precedent examples of the asymmetrical functions of anatomically symmetrical sensory neurons, including ASEL-ASER48 and AWCON-AWCOFF (ref. 51). These functional asymmetries are related to an increased repertoire of the sensory stimuli to which each sensory neuron can respond. This is likely because the number of sensory neurons is quite limited in the animals (only a few pairs of sensory neurons are used to detect tens of odorants, for example17). This may also be the case for CEP neuron pairs.
The functional asymmetry of CEP neuron pairs was unexpected because not only do they appear to be anatomically symmetrical, but DA is known to function extrasynaptically to motor39 and sensory12,25 neurons. The fact that optogenetic CEPV activation did not cause the slowing response may suggest that, at least for that particular behavioural modulation, DA function is mediated intrasynaptically. CEP neurons have 77 known postsynaptic target neurons, of which 14 neurons have synaptic inputs from both CEPD and CEPV neurons (“DV” in Table S1 according to http://wormwiring.hpc.einstein.yu.edu). Most of the remaining synaptic connections are from only one of CEP neurons (“single” in Table S1). Thus, the unshared synaptic connections from CEPD may play a significant role in the slowing response. Alternatively, the different amounts of released DA caused by similar optogenetic activations could be the reason for the different slowing response between CEPD and CEPV pairs; even in that case, DA released by CEPV is likely to have its own role.
We also found that another DAergic neuron pair, the PDEs, were not activated upon food entry and did not cause slowing with optogenetic activation. Although all of the DA pairs are considered functionally redundant, laser ablation of CEPs, but not ADEs or PDEs, significantly affected the slowing response8. Similarly, CEPs, but not ADEs or PDEs, have been recently reported to be responsible for learning of food patch size52. In addition, only CEP neuron pairs have their sensory endings at the anterior end of the body (Fig. 1a). Taken together, these data suggest that DA from CEPD plays a major role in the slowing response, while DA from other DAergic neuron pairs has different roles but still plays a minor role in the slowing response, possibly through extrasynaptic transmission. The other pairs may be activated in a much longer time-scale in the presence of food, by different stimuli, or in different contexts.
So where do the differences in DAergic neuron pairs originate? The identities of neuronal types are determined by the expression of type-specific batteries of genes, such as the ones for biosynthesis of neurotransmitters, ion channels, neurotransmitter receptors, and signalling molecules2. Such identity of DAergic neurons is tightly regulated by three types of transcription factors in C. elegans, as well as in mammals19,20. Nevertheless, the CEP pairs are functionally asymmetrical and have different synaptic partners. Thus, our results suggest there are yet-unknown mechanisms that could further determine the differences in the same neuron type beyond genetic terminal identification.
Methods
Strains
The techniques used to culture and handle C. elegans were essentially as described previously53. The C. elegans wild-type Bristol strain (N2) and the mutant strains CB1112 cat-2(e1112), TQ1101 lite-1(xu7), and TQ296 trp-4(sy695) were obtained from the Caenorhabditis Genetics Center (University of Minnesota, USA). Young adult hermaphrodites were used in all of the behavioural experiments.
Behavioural assay for bending numbers
For the food patch, the OP50 bacteria was grown in 100 mL of LB culture overnight, spun down, and suspended in 10 volumes of nematode growth medium (NGM) buffer, and 5 μL of the suspension was applied to four spots 5 mm from the centre of the NGM plate surface (Fig. 1b); this bacterial lawn was 30–40 μm thick and did not obstruct the fluorescent imaging. Five young adult worms were briefly washed with 10 μL of NGM buffer and placed on 1.7% agar NGM plates with food patches. The worms moved freely and spontaneously entered the food during video recording with a dissection microscopy SZX10 and a digital camera system DP72 (Olympus). The obtained images were manually analysed to quantify bending number as described previously8.
Molecular biology and germline transformation
For the DAergic neuron–specific expression, mCherry36, GCaMP6f 35, and codon-optimized ChR2(H134R)54 cDNAs were fused with dat-1 promoter55 using a GATEWAY system® (Thermo Fisher Scientific). ChR2 as well as mCherry were expressed in lite-1(xu7) mutant background to reduce the animal’s response to strong blue light56. Germline transformation was performed using microinjection57. Multiple transgenic lines were used for each experiment type, and the different lines produced similar results. The plasmids and transgenic lines used in this study are listed in Supplementary Tables S2 and S3, respectively.
Confocal imaging
The animals were anesthetized with 5 mM sodium azide on a 2% agarose NGM pad and covered with a cover glass (Matsunami). Images of the animals expressing dat-1p::mCherry and dat-1p::ChR2::GFP were captured with a confocal microscope LSM710 (Carl Zeiss) with a Plan Apochromat 40× 0.95 NA lens. The images were processed using Zen software (Carl Zeiss).
Behavioural tracking
An animal on an NGM plate with food patches was placed on a motorized stage HV-STU-03W (HawkVision Inc., Fujisawa, Japan) combined with a BX51WI upright microscope (Olympus). The animal was illuminated with infrared (IR) light from a halogen lamp without an IR-cut filter through a 32BP775 band-pass filter (Olympus), and the bright field image was reflected with a DF670 dichroic mirror (Semrock, USA) guided to a GRAS-03K2M-C CCD camera (Point Grey Research, Canada) with a 0.35× U-TV0.35XC-2 C-mount TV adapter (Olympus). The images were taken at 200 Hz by the camera and processed by a custom-made program for real-time pattern matching on a Linux PC (Intel Core i7-870) that regulated the motorized stage to maintain the region of interest (ROI) of a freely-moving animal in the centre of the microscope’s field of view34. In most of the frames, a ROI was set around the head ganglia. For PDE imaging, the ROI was set around the fluorescence of PDE cell body. Each animal’s speed was calculated from records of displacement of the auto-tracking stage and ROI position in the field of view.
Calcium imaging
The sample was exposed to white excitation light from a Multi-independent Light Stimulation System (MiLSS; Aska Company, Hyogo, Japan)37 through a BP460-495 bandpass-filter (Olympus) and a DM505HQ dichroic mirror (Olympus). The fluorescence was introduced into a W-View optics system (Hamamatsu, Japan) containing two dichroic mirrors (545SP and 545LP) and bandpass filters (D520/20m and 620DF35 for GCaMP and mCherry, respectively) through a 20× UPLFLN objective lens (Olympus). The GCaMP and mCherry images were split by the W-View and simultaneously captured side-by-side on an EM-CCD camera ImagEM (Hamamatsu). Images were taken at a 32.6-ms exposure time and 100-ms sampling interval with 2 × 2 binning. The calcium imaging subsystem was triggered by a TTL signal from the auto-tracking subsystem, and both subsystems ran independently thereafter in the present setup. The images were saved as uncompressed 16-bit TIFF files, the cell bodies were independently tracked with a custom-made off-line optical flow tracking program, and signal intensities of particular regions were measured by ImageJ (NIH). The signal intensity of the background was subtracted from that of the cell body. This process was performed for fluorescent images from both green (GCaMP) and red (mCherry) channels of W-View, and the ratio of fluorescent intensities of GCaMP over mCherry (R) was calculated to cancel out florescent changes caused by motion artefacts. The median ratio during 90 seconds before the food entry was defined as the baseline R0, and the normalized ratio (R/R0) was then calculated and further analysed.
Optogenetic experiments
The animals were grown in the presence or absence of ATR as described previously58 and transferred to an NGM plate on the OSB system and maintained under the 20× objective lens by auto-tracking. The sample was illuminated with green and blue lights with MiLSS through an XF3069 multi-bandpass filter (Omega) and an FF493/574-Di01 dichroic mirror (Semrock). The green light (540–570 nm; ~0.4 mW/mm2) was illuminated to the full view field for the real-time optical flow tracking of the mCherry-expressing DAergic neuron positions. The fluorescent images of mCherry were captured through the 620DF35 bandpass filter. Each DAergic neuron was identified based on vulva position from the bright-field images, and one was manually selected and then automatically targeted by the optical flow tracking. After 1 min for measuring basal locomotion rate and when the worm was moving forward, blue light (450–470 nm; ~12 mW/mm2) was overlaid for 10 s to the full view field or at the position of targeted cell body, which was updated at 31.7 Hz; because the motorized stage adjusted the position of the animal’s brain at 200 Hz, the relative neuron speed was slow enough for single cell targeting at 31.7 Hz. A locomotion speed of 10 s before and during stimulation was calculated according to the behavioural tracking of the animals and normalized with the ensemble median of averaged locomotion speed before stimulation for each targeted cell type. One animal was tested only once for the light stimulation to avoid any hysteretic effects.
To verify the spatiotemporal accuracy of our cell-targeted light stimulation, we first illuminated the full view field with green light at ~0.4 mW/mm2 to visualize all of the mCherry-expressing neuronal cell bodies and then targeted one of the cell bodies with green light at ~2.6 mW/mm2 to further excite the mCherry fluorescence on that cell only. The results (Video S3) show that the targeted light illuminated the target cell body but not any others.
Data analysis and statistics
The data were obtained over 3 days from 10–40 animals for each condition in most cases. We chose this sample number because a large-scale behavioural analysis of C. elegans concluded that a certain statistical difference can be detected in 10 animals, and 24 ± 14 (average ± SD) animals per condition were used59. After the sample acquisition, the data of some animals for Figs 3 and 4 were excluded when any of the following problems were found: (1) trials interrupted by auto-tracking errors, (2) insufficient mCherry or basal GCaMP6f fluorescence intensity for optical flow tracking, or (3) basal locomotion speed <0.06 mm/s (average speed ± SD of normal animals was 0.17 ± 0.05 mm/s). In addition, when the average of R/R0 after t = 0 was <1.1 in an animal’s CEPD and CEPV, the animal was regarded as “not responding”; those data were not included in Fig. 3 but are shown in Table 1.
Experimental conditions, such as the presence/absence of ATR, light stimulation, or different strains, were randomized on a daily basis. A Kruskal-Wallis test with a post hoc Steel-Dwass test was used for multiple comparisons in Fig. 4b, while a Mann-Whitney test was used for single comparison in Figs 3c and 4b using R (The R Project) or Prism ver. 5.0 for Mac OSX (GraphPad Software, San Diego, CA, USA).
Additional Information
How to cite this article: Tanimoto, Y. et al. In actio optophysiological analyses reveal functional diversification of dopaminergic neurons in the nematode C. elegans. Sci. Rep. 6, 26297; doi: 10.1038/srep26297 (2016).
Supplementary Material
Acknowledgments
We thank Y. Mori (HawkVision Inc., Japan) and K. Ishida (Aska Company, Japan) for their generous technical hardware support. We also thank Drs. T. Obara and M. Chen for the materials and technical support, K. Kuhara and I. Mori for the codon-optimized ChR2, and S. Suo and O. Hobert for critically reading this manuscript. Nematode strains were provided by the Caenorhabditis Genetics Center (funded by the NIH Office of Research Infrastructure Programs P40 OD010440), and neuronal information was provided by WormBase (funded by National Human Genome Research Institute grant #41 HG002223) and by WormAtlas (www.wormatlas.org). This work was supported by a Grant-in-Aid for JSPS fellows and a program for Leading Graduate Schools entitled ‘Interdisciplinary graduate school program for systematic understanding of health and disease’ (Y.T.), JSPS KAKENHI Grant Number 25249020 (K.H. and K.D.K.), KAKENHI on Innovative Areas (Comprehensive Brain Science Network), the Osaka University Life Science Young Independent Researcher Support Program, Precursory Research for Embryonic Science and Technology from MEXT, and research grants from Mitsubishi Foundation, Shimadzu Science Foundation, and Takeda Science Foundation (K.D.K.). All of the materials and study data are stored or archived in the Kimura Laboratory in the Department of Biological Sciences, Osaka University, and available upon request.
Footnotes
Author Contributions Y.T., X.F., K.H. and K.D.K. designed the experiments, Y.T., Y.G.Z. and Y.F. performed the experiments and data analyses, X.F. and K.H. developed the OSB system, and Y.T. and K.D.K. wrote the manuscript. All authors reviewed the manuscript.
References
- Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development 134, 3771–3780 (2007). [DOI] [PubMed] [Google Scholar]
- Hobert O. Regulation of Terminal Differentiation Programs in the Nervous System. Annu Rev Cell Dev Biol 27, 681–696 (2011). [DOI] [PubMed] [Google Scholar]
- Schultz W. Multiple dopamine functions at different time courses. Annu Rev Neurosci 30, 259–288 (2007). [DOI] [PubMed] [Google Scholar]
- Björklund A. & Dunnett S. B. Dopamine neuron systems in the brain: an update. Trends Neurosci 30, 194–202 (2007). [DOI] [PubMed] [Google Scholar]
- Brischoux F., Chakraborty S., Brierley D. I. & Ungless M. A. Phasic excitation of dopamine neurons in ventral VTA by noxious stimuli. Proc Natl Acad Sci USA 106, 4894–4899 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M. & Hikosaka O. Two types of dopamine neuron distinctly convey positive and negative motivational signals. Nature 459, 837–841 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z. & Davis R. L. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front Neural Circuits 3, 5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin E. R., Ranganathan R. & Horvitz H. R. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26, 619–631 (2000). [DOI] [PubMed] [Google Scholar]
- Bettinger J. C. & Mcintire S. L. State-dependency in C. elegans. Genes Brain Behav 3, 266–272 (2004). [DOI] [PubMed] [Google Scholar]
- Hills T., Brockie P. J. & Maricq A. V. Dopamine and glutamate control area-restricted search behavior in Caenorhabditis elegans. J Neurosci 24, 1217–1225 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S. et al. Dopamine modulates the plasticity of mechanosensory responses in Caenorhabditis elegans. EMBO J 23, 473–482 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindt K. S. et al. Dopamine mediates context-dependent modulation of sensory plasticity in C. elegans. Neuron 55, 662–676 (2007). [DOI] [PubMed] [Google Scholar]
- Voglis G. & Tavernarakis N. A synaptic DEG/ENaC ion channel mediates learning in C. elegans by facilitating dopamine signalling. EMBO J 27, 3288–3299 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K. D., Fujita K. & Katsura I. Enhancement of odor avoidance regulated by dopamine signaling in Caenorhabditis elegans. J Neurosci 30, 16365–16375 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Gadea A. et al. Caenorhabditis elegans selects distinct crawling and swimming gaits via dopamine and serotonin. Proc Natl Acad Sci USA 108, 17504–17509 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bono M. & Maricq A. V. Neuronal substrates of complex behaviors in C. elegans. Annu Rev Neurosci 28, 451–501 (2005). [DOI] [PubMed] [Google Scholar]
- Bargmann C. I. Chemosensation in C. elegans. WormBook 1–29, doi: 10.1895/wormbook.1.123.1 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J., Dew M. & Brenner S. Dopaminergic neurons in the nematode Caenorhabditis elegans. J Comp Neurol 163, 215–226 (1975). [DOI] [PubMed] [Google Scholar]
- Flames N. & Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature 458, 885–889 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M. et al. A combinatorial regulatory signature controls terminal differentiation of the dopaminergic nervous system in C. elegans. Genes Dev 27, 1391–1405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo S., Ishiura S. & Van Tol H. H. M. Dopamine receptors in C. elegans. Eur J Pharmacol 500, 159–166 (2004). [DOI] [PubMed] [Google Scholar]
- Palmer A. E. & Tsien R. Y. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat Protoc 1, 1057–1065 (2006). [DOI] [PubMed] [Google Scholar]
- Nagel G. et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA 100, 13940–13945 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. et al. Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639 (2007). [DOI] [PubMed] [Google Scholar]
- Ezcurra M., Tanizawa Y., Swoboda P. & Schafer W. R. Food sensitizes C. elegans avoidance behaviours through acute dopamine signalling. EMBO J 30, 1110–1122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis N., Zimmer M. & Bargmann C. I. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat Meth 4, 727–731 (2007). [DOI] [PubMed] [Google Scholar]
- Faumont S. et al. An image-free opto-mechanical system for creating virtual environments and imaging neuronal activity in freely moving Caenorhabditis elegans. PLoS ONE 6, e24666 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T. et al. An imbalancing act: gap junctions reduce the backward motor circuit activity to bias C. elegans for forward locomotion. Neuron 72, 572–586 (2011). [DOI] [PubMed] [Google Scholar]
- Leifer A. M., Fang-Yen C., Gershow M., Alkema M. J. & Samuel A. D. T. Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans. Nat Meth 8, 147–152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirman J. N. et al. Real-time multimodal optical control of neurons and muscles in freely behaving Caenorhabditis elegans. Nat Meth 8, 153–158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocabas A., Shen C.-H., Guo Z. V. & Ramanathan S. Controlling interneuron activity in Caenorhabditis elegans to evoke chemotactic behaviour. Nature 490, 273–277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Cao P., Yang J., Xu X. Z. S. & Feng Z. Calcium imaging of multiple neurons in freely behaving C. elegans. J Neurosci Methods 260, 78-82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N. & Brenner S. The Structure of the Nervous System of the Nematode Caenorhabditis elegans. Philos Trans Royal Soc B 314, 1–340 (1986). [DOI] [PubMed] [Google Scholar]
- Maru M., Igarashi Y., Arai S. & Hashimoto K. Fluorescent Microscope System to Track a Particular Region of C. elegans. IEEE/SICE Int Symp Sys Integr 347–352 (2010). [Google Scholar]
- Chen T.-W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotech 22, 1567–1572 (2004). [DOI] [PubMed] [Google Scholar]
- Sakai S., Ueno K., Ishizuka T. & Yawo H. Parallel and patterned optogenetic manipulation of neurons in the brain slice using a DMD-based projector. Neurosci Res 75, 59–64 (2013). [DOI] [PubMed] [Google Scholar]
- Hardaway J. A. et al. Glial Expression of the Caenorhabditis elegans Gene swip-10 Supports Glutamate Dependent Control of Extrasynaptic Dopamine Signaling. J Neurosci 35, 9409–9423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase D. L., Pepper J. S. & Koelle M. R. Mechanism of extrasynaptic dopamine signaling in Caenorhabditis elegans. Nat Neurosci 7, 1096–1103 (2004). [DOI] [PubMed] [Google Scholar]
- Nagel G. et al. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol 15, 2279–2284 (2005). [DOI] [PubMed] [Google Scholar]
- Schafer W. Neurophysiological methods in C. elegans: an introduction. WormBook 1–4, doi: 10.1895/wormbook.1.111.1 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly J. L. et al. Monoaminergic orchestration of motor programs in a complex C. elegans behavior. PLoS Biol 11, e1001529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Q. et al. Proprioceptive coupling within motor neurons drives C. elegans forward locomotion. Neuron 76, 750–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson S. J. et al. Microbial light-activatable proton pumps as neuronal inhibitors to functionally dissect neuronal networks in C. elegans. PLoS ONE 7, e40937 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcomyn F. Properties of sensory systems. in Foundations of Neurobiology 209–231 (W H Freeman & Company, 1998). [Google Scholar]
- Kimura K. D., Miyawaki A., Matsumoto K. & Mori I. The C. elegans thermosensory neuron AFD responds to warming. Curr Biol 14, 1291–1295 (2004). [DOI] [PubMed] [Google Scholar]
- Chalasani S. H. et al. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature 450, 63–70 (2007). [DOI] [PubMed] [Google Scholar]
- Suzuki H. et al. Functional asymmetry in Caenorhabditis elegans taste neurons and its computational role in chemotaxis. Nature 454, 114–117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K. E. et al. Tonic signaling from O2 sensors sets neural circuit activity and behavioral state. Nat Neurosci 15, 581–591 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L., Gao J., Schafer W. R., Xie Z. & Xu X. Z. S. C. elegans TRP Family Protein TRP-4 Is a Pore-Forming Subunit of a Native Mechanotransduction Channel. Neuron 67, 381–391 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes P. D. & Bargmann C. I. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature 410, 698–701 (2001). [DOI] [PubMed] [Google Scholar]
- Calhoun A. J. et al. Neural Mechanisms for Evaluating Environmental Variability in Caenorhabditis elegans. Neuron 86, 428–441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhara A., Ohnishi N., Shimowada T. & Mori I. Neural coding in a single sensory neuron controlling opposite seeking behaviours in Caenorhabditis elegans. Nat Commun 2, 355 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald P. W. et al. Vigorous motor activity in Caenorhabditis elegans requires efficient clearance of dopamine mediated by synaptic localization of the dopamine transporter DAT-1. J Neurosci 27, 14216–14227 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. L. et al. A Novel Molecular Solution for Ultraviolet Light Detection in Caenorhabditis elegans. PLoS Biol 6, e198 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D. & Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10, 3959–3970 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazoe Y., Yawo H. & Kimura K. D. A simple optogenetic system for behavioral analysis of freely moving small animals. Neurosci Res 75, 65–68 (2013). [DOI] [PubMed] [Google Scholar]
- Yemini E., Jucikas T., Grundy L. J., Brown A. E. X. & Schafer W. R. A database of Caenorhabditis elegans behavioral phenotypes. Nat Meth 10, 877–879 (2013). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.