Abstract
Annotation of protein-coding genes is very important in bioinformatics and biology and has a decisive influence on many downstream analyses. Homology-based gene prediction programs allow for transferring knowledge about protein-coding genes from an annotated organism to an organism of interest.
Here, we present a homology-based gene prediction program called GeMoMa. GeMoMa utilizes the conservation of intron positions within genes to predict related genes in other organisms. We assess the performance of GeMoMa and compare it with state-of-the-art competitors on plant and animal genomes using an extended best reciprocal hit approach. We find that GeMoMa often makes more precise predictions than its competitors yielding a substantially increased number of correct transcripts. Subsequently, we exemplarily validate GeMoMa predictions using Sanger sequencing. Finally, we use RNA-seq data to compare the predictions of homology-based gene prediction programs, and find again that GeMoMa performs well.
Hence, we conclude that exploiting intron position conservation improves homology-based gene prediction, and we make GeMoMa freely available as command-line tool and Galaxy integration.
INTRODUCTION
Next Generation Sequencing technologies enable rapid and cost-efficient sequencing of genomes. However, after sequencing and assembling the genome of an organism, it is important to provide annotations, especially of protein-coding genes. Annotation pipelines for newly sequenced genomes utilize three main sources of information: (i) evidence from wet-lab experiments, (ii) ab initio and (iii) homology-based gene prediction relying on (closely) related and annotated species (1,2).
Wet-lab experiments like RNA-seq (3) or Iso-seq (4) provide a wealth of information about transcripts including exon–intron boundaries and Untranslated region (UTRs), but are limited to those transcripts expressed under the studied conditions. Hence, annotations based on RNA-seq data might miss lowly or very specifically expressed transcripts. By contrast, ab initio gene prediction programs are computer programs that predict gene models without evidence from wet-lab experiments or related species.
Here, we focus on homology-based gene prediction programs that predict genes or transcripts in the newly sequenced target genomes based on the similarity to known genes from closely related and, typically, well annotated reference genomes. Given some knowledge about a gene in a specific organism, we are interested in whether a similar or, ideally, an orthologous gene exists in another organism, which could allow for transferring knowledge from one species to another.
One of the most popular tools for identifying similar genes or proteins is Basic Local Alignment Search Tool (BLAST) (5). However, BLAST does not explicitly account for the exon–intron structure of genes and, for this reason, is typically applied to find similar genes or transcripts already extracted from genomic sequence. Searching for proteins or coding sequences in complete genomes, long and variable introns might be a problem for BLAST yielding a plethora of short, similar sequences scattered over the genome. Hence, BLAST is no gene prediction program in terms of this manuscript.
To circumvent this problem, several approaches have been proposed for combining smaller, local hits of high similarity to parts of a given gene into larger, complete gene models, as for instance Genewise (6), exonerate (7), Projector (8), GeneMapper (9) and genBlastG (10). In this manuscript, we focus on exonerate and genBlastG. Exonerate is a very versatile tool allowing diverse alignments and is often used as part of genome annotation pipelines (11). GenBlastG is specially tailored to aligning proteins to genomes and especially optimized for runtime.
However, most of these tools do not utilize the known gene structures, i.e. the exon–intron boundaries and exons lengths, of the query genes while searching for target genes, although the gene structure of intron-containing orthologous genes is strongly conserved throughout the whole plant or animal kingdom and to a smaller extent even across kingdoms (12,13). By contrast, Projector (8) and GeneMapper (9) use the conservation of the gene structure in addition to the similarity of the encoded amino acid sequences. Projector uses a pairHMM approach, while GeneMapper uses bottom-up approach utilizing an alignment of codons for the exons.
Here, we propose a Gene Model Mapper approach called GeMoMa that exploits the conservation of gene structures to predict gene models in a target genome based on the gene models of a reference genome. Specifically, GeMoMa uses BLAST as a first step to align individual coding exons to the genome on the level of (translated) amino acids. For several, especially short, exons, BLAST will report multiple matches spread across the genome. To reduce computational complexity for the following steps, GeMoMa segments the genome into matching regions based on the occurrence of such exon matches. Within each region, GeMoMa uses a dynamic programming approach to create a complete gene model that joins the matching exons in this region in the correct order, but allows for intron gain and loss during this procedure. Finally, predicted exons are refined such that each exon is flanked by proper splice sites and each (coding) transcript begins with a start codon and ends with a stop codon.
Aiming at a lowly biased comparison of different tools, we extend the best reciprocal hit (BRH) approach. Two genes residing in different genomes are called BRHs or bidirectional best hits if the corresponding proteins find each other as the best hit in the opposite genome (14). Often BRHs are used to determine orthologous genes, but for several reasons this might be problematic (15). In this manuscript, we adapt the approach comparing the best hits of transcripts in different genomes. We enrich the analysis by several categories, as for instance, correct transcript, correct gene and correct gene family and measure whether the complete gene model can be predicted.
Using this extended BRH approach, we test GeMoMa for gene prediction in plants and animals, and compare it with exonerate and genBlastG. Subsequently, we utilize Sanger sequencing and demonstrate that predictions of GeMoMa for Carica papaya might allow for improving partially known gene models or predicting previously unknown gene models. Finally, we evaluate exonerate, genBlastG and GeMoMa using RNA-Seq data for Nicotiana benthamiana.
MATERIALS AND METHODS
Algorithm
The main idea for predicting gene models in a genome or genome assembly is to rely on annotated genes in other species. In addition to using the amino acid sequence, we also use the exon–intron boundaries, i.e. the split of the complete Coding Sequence (CDS) into smaller parts.
Hence, each (partially) coding exon of a transcript is translated into an amino acid sequence. Subsequently, these sequences are fed to tblastn to obtain regions potentially coding for similar amino acid sequences in the target genome (Text S1).
The tblastn results of each transcript are filtered per contig and strand. For each contig-strand combination, a dynamic programming algorithm is performed to assemble the tblastn results to an initial gene model (Text S1) returning an initial sum score. Based on this initial sum score, the contig-strand combinations are filtered obtaining promising initial gene models using the parameter ct, which specifies the percentage of the maximal initial sum score that has to be be succeeded to be used for further analysis.
For each of those contig-strand combinations, regions are identified that possibly encode for a similar transcript. In each region, coding parts of the transcript are searched that have no tblastn result. Again the dynamic programming algorithm is used that this time considers canonical splice sites and only in-frame combinations of individual parts to obtain a gene model and a corresponding score (Text S1).
Based on this score, the predictions of each region are ranked and a user-specified number of predictions is returned.
Extended best reciprocal hit approach
In order to compare the predictions of different tools, the BRH approach was extended to allow for a less biased comparison. The BRH approach can be summarized as follows: Given transcript A encoded in genome 1, we search for the best matching transcript B in genome 2. Given transcript B, we are searching for the best matching transcript C in genome 1. If A equals C, we have a BRH. Hence, the BRH approach only provides one category.
We extend this approach by introducing 8 additional categories:
A and C are the same transcript (BRH),
A and C are different transcripts of the same gene,
A and C are transcripts of a gene family,
A and C are transcripts that do not belong to a gene family,
There is no prediction of transcript A in genome 2,
There is no annotation (B) in genome 2 that overlaps with the prediction of transcript A in genome 2,
There is no prediction of transcript B in genome 1,
There is no annotation (C) in genome 1 that overlaps with the prediction of transcript B in genome 1.
Furthermore, we enrich this approach by an additional measure of confidence. The nucleotide F1 measure was computed twice, namely, between the prediction of transcript A in genome 2 and transcript B, and between the prediction of transcript B in genome 1 and transcript C (Text S4). In a nutshell, nucleotide F1 aggregates nucleotide precision and nucleotide recall into one scalar value varying between 0 and 1 corresponding to completely wrong and perfect predictions, respectively. Finally, the minimum of these two F1 values was computed and denoted as minimal F1. Predictions that fall in one of the first four categories were further characterized by the minimal F1.
Genomes and annotations
For the extended BRH approach, we downloaded the genome (assemblies) and gene annotation of the plant species Arabidopsis lyrata, Arabidopsis thaliana, Carica papaya, Oryza sativa and Solanum tuberosum from Phytozome (16), and the animal species Homo sapiens, Gallus gallus and Mus musculus from Ensembl (17) (cf. Text S3).
Additionally, we downloaded genome assembly and gene annotation of Nicotiana benthamiana v0.4.4 (18) from ftp://ftp.solgenomics.net/genomes/Nicotiana_benthamiana/assemblies/ for mapping RNA-seq data and assessing GeMoMa predictions.
For all analyses, we discarded gene models from the given annotation with missing start or stop codon, premature stop codon(s) or ambiguous nucleotide(s). In addition, we only used one representative gene model if several gene models of a gene have the same CDS, i.e. only differ in their UTRs.
Polymerase Chain Reaction (PCR) and sequencing
The mRNA was isolated from A. thaliana ecotype Col-0 and C. papaya inflorescences using the Bio & Sell RNA mini Kit (Bio&Sell e.K., Feucht, Germany). In the case of C. papaya, flowers were stored in RNAshield (Zymo research Europe GmbH, Freiburg, Germany) prior to RNA isolation. The cDNA was synthesized using an anchored oligo dT-Primer and the Maxima H Minus Reverse Transcriptase Kit (Thermo Fisher Scientific, Germany) using 2–4 μg of total RNA as template for the reverse transcriptase reaction. The genomic DNA of A. thaliana was isolated as described previously (19). The genomic DNA of C. papaya was provided by Ray Ming who was leading the original C. papaya sequencing project (University of Illinois).
For amplification of complementary and genomic DNA from A. thaliana, primer pairs were deduced and designed from the published sequence of the TAIR 10 database. For amplification of the GeMoMa predicted C. papaya exons, we designed primer pairs as near as possible to the predicted start codon (ATG) and stop codon. For primer details see Supplementary Table S7.
The PCR using cDNA of A. thaliana and C. papaya was done with an appropriate polymerization time of 1 to 2 min at 72°C depending on the size of the predicted cDNA length (from 250 to a maximum of 2037 bp). The genomic DNA of A. thaliana and C. papaya was amplified as control using the same primers and conditions adapted to the expected size (ranging from 1.2 kb up to 7 kb). The 16 kb long C. papaya gene homologous to At4g16566 was amplified in two parts covering the first 3.5 kb and the last 4 kb.
To confirm the full length sequence of the amplified cDNAs of C. papaya, we subjected them to Sanger-sequencing. The amplified cDNA and in one case (homologous gene to At4g16566) the gDNA was purified using the ‘GeneJET PCR Purification Kit’ (Thermo Scientific Germany, Braunschweig) and without cloning send to sequencing at GATC Biotech AG (Cologne, Germany). We used the PCR-amplification primer to completely sequence the cDNA from both sides. Direct sequencing was used to avoid point mutations which could have been introduced by PCR. In the case of the gene homologous to At4g38240, the papaya cDNA sequence was unreadable and therefore cloned using the ‘InsTAclone PCR Cloning Kit’ (Thermo Scientific Germany, Braunschweig). After cloning we performed amplification by M13 forward and reverse primer (cf. Supplementary Table S7) flanking the multiple cloning site of pTZ57R/T.
Next Generation Sequencing data
Three transgenic N. benthamiana plants were grown on soil under 8 h light (120 μEm−2) and 16 h dark at 20°C. The transgene, an expression cassette facilitating the expression of a stroma targeted eGFP, was introduced into N. benthamiana for the purpose of examining the plastids as part of another experiment (for details cf. (20,21)). A total of ∼100 mg of plant tissue was harvested from the third and fourth leaves of six week old plants. The Qiagen RNeasy Plant Mini Kit was used to isolate total RNA from the three plants (three biological replicates), which was then sent to MWG Eurofins (Ebersberg, Germany) for RNA sequencing with Illumina HiSeq 2000 (v3.0 chemistry). RNA-seq data have been submitted to ENA and are available under study accession number PRJEB11424.
We independently mapped the RNA-seq triplicates using TopHat2 (22) v2.0.12 to the Nicotiana benthamiana genome v0.4.4 using parameter –library-type=fr-firststrand for strand specificity. For each of the three mapping files, we assembled transcripts using Cufflinks (23) v2.2.1 using parameter –library-type fr-firststrand for strand specificity. We finally merged the three resulting transcript annotations using cuffmerge. The transcripts reported by cuffmerge were then shortened to the longest reading frame starting with a start codon and ending with a stop codon and non-coding exons are removed, since the predictions of all tools considered are based only on coding regions of the corresponding transcripts.
RESULTS
In this section, we analyze the performance of GeMoMa on plant and animal genomes. Due to the different intron size distributions in these kingdoms, we set in all studies the maximum intron length of GeMoMa for plants to 15 kb and for animals to 200 kb.
Best reciprocal hits – Benchmark
In a pilot study, we tested the performance of GeMoMa on the modified projector data set (8,9) for the prediction of mouse transcripts given the approximate regions of the mouse genome and human reference transcripts. We find that GeneMapper, which is no longer available, and GeMoMa perform comparable, while they clearly outperform the remaining tools especially for the categories exon and gene (Text S2).
Based on these promising results, we performed a large benchmark study using the extended BRH approach for predicting gene models in animal and plant genomes, including Homo sapiens (HS), Mus musculus (MM), Gallus gallus (GG), as well as Arabidopsis thaliana (AT), Arabidopsis lyrata (AL), Carica papaya (CP), Solanum tuberosum (ST) and Oryza sativa (OS). Since H. sapiens and A. thaliana have the best annotated genomes among the selected, we used these as the start and end point of the extended BRH approach.
We selected the minimal F1 as a measure of accuracy of a prediction (see Materials and Methods and Text S4). Basically, minimal F1 = 1 indicates a perfect prediction in the sense that the predicted coding exons are perfectly identical to the coding exons of an annotated transcript on the target genome, and minimal F1 = 0 indicates a prediction that does not overlap with any known exons. We may base the evaluation in the extended BRH on different thresholds on the minimal F1 values corresponding to different levels of required accuracy. For different thresholds on the minimal F1 measure, we may determine the corresponding number of predictions for several categories: correct transcript, correct gene, correct gene family and wrong prediction, whereas the remaining categories can be determined without such a threshold.
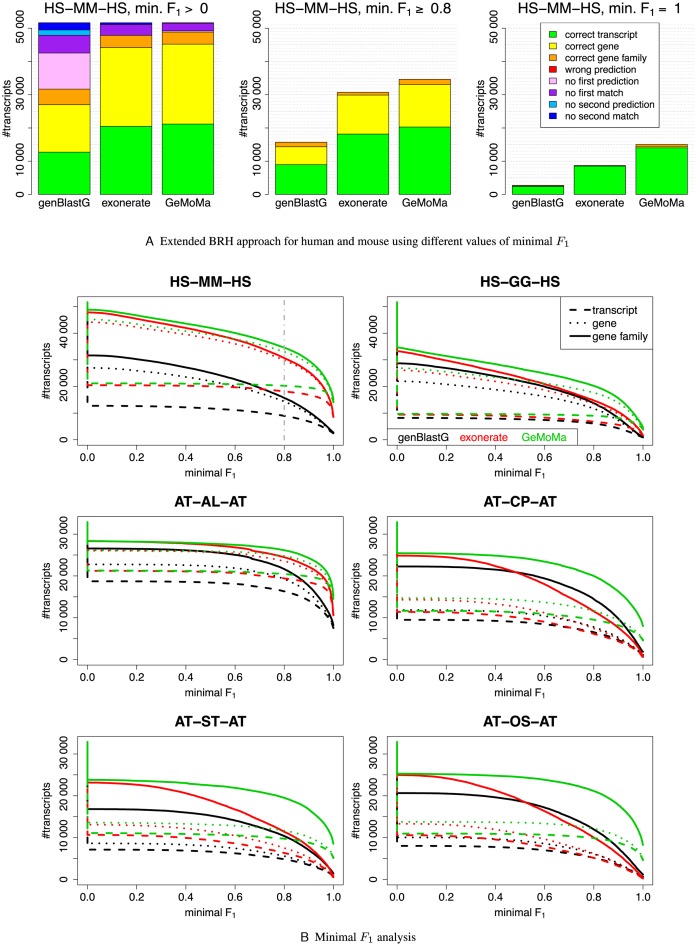
In Figure 1A, we present the results for human and mouse using the different thresholds. For the threshold of minimal F1 = 1, GeMoMa yields 14 035, 446 and 563 predictions in the categories correct transcript, correct gene and gene families, respectively. By contrast, exonerate and genBlastG yield 8526, 93 and 82 as well as 2430, 108 and 183 predictions in the corresponding categories, respectively.
Figure 1.
Results of benchmark studies using the extended BRH approach. In (A), we exemplarily visualize the results for human and mouse for the fixed minimal F1 of 0, 0.8 and 1. For a minimal F1 > 0, only the categories correct transcript, correct gene, correct gene family and wrong prediction can be evaluated. In (B), we visualize the results of the categories correct transcript, correct gene and correct gene family for animals (MM, GG) and plants (AL, CP, ST, OS) for continuous minimal F1.
However, different thresholds might give different results. Hence, we tested in total three thresholds, namely minimal F1 > 0, minimal F1 ≥ 0.8 and minimal F1 = 1. In a nutshell, we find that exonerate and GeMoMa outperform genBlastG for these three thresholds, whereas GeMoMa performs comparable to exonerate for minimal F1 > 0 and yields a larger number of correct predictions than exonerate for minimal F1 ≥ 0.8 and minimal F1 = 1.
These results indicate that the relative performance of approaches depends on the threshold on the minimal F1 values. For this reason, we plot the number of predictions for the categories correct transcript, correct gene and correct gene family against the minimal F1 in Figure 1B. For human and mouse (top left panel), we find that exonerate performs better than genBlastG and GeMoMa performs better than exonerate independent of the minimal F1.
Comparing these results with the other results for animals and plants, we find for the threshold minimal F1 = 1 that GeMoMa always predicts the highest number of BRHs of the category correct transcript (Supplementary Table S5A). The absolute numbers vary between 3807 and 14 514 for GeMoMa, whereas the numbers vary between 231 and 10 584 for exonerate and genBlastG. This is equivalent to an improvement between 37% for A. lyrata and 623% for O. sativa using GeMoMa instead of genBlastG or exonerate.
Being less conservative and using the category correct gene family and minimal F1 ≥ 0.8, GeMoMa still outperforms its competitors. However, the difference is less pronounced, varying between 7% for A. lyrata and 77% for O. sativa.
Comparing the number of predictions versus varying thresholds on the minimal F1 in Figure 1B, we find that exonerate often performs better than genBlastG, whereas GeMoMa always performs better than exonerate and genBlastG. Especially for C. papaya, S. tuberosum and O. sativa, we find that GeMoMa clearly outperforms the other tools for high values of minimal F1.
In addition, we determined for each of the three tools and each of the plant and animal genomes those predicted transcripts that do not match any annotated transcript in the target organism. If such transcripts are consistently predicted by all three tools, this might increase our confidence in those predictions. We find that the number of predicted transcripts without match in the target genome varies between 599 for M. musculus and 9607 for G. gallus. Adding the further constraint that the predictions of the three tools are located in the same genomic region, the number varies between 411 for M. musculus and 8068 for G. gallus (Supplementary Table S6). These numbers indicate that there is still a substantial potential of identifying new transcripts using tools like genBlastG, exonerate and GeMoMa even in annotated genomes.
In Figure 1, we also observe that the accuracy of predictions decreases with increasing evolutionary distance of reference and target organism. To further investigate this observation, we consider two extreme examples (A.thaliana versus Chlamydomonas reinhardtii and H. sapiens versus Drosophila melanogaster) in Supplementary Text S8. We find that, given the large evolutionary distance between these organisms, the number of perfect predictions (F1 = 1) is extremely low for all three tools. Hence, we conclude that homology-based gene prediction using any of the tools considered greatly profits from using an evolutionary related organism as reference.
For these genome-wide studies, we fixed all parameters of GeMoMa as well as genBlastG and exonerate. GeMoMa parameters that might be tuned for specific applications are (i) the maximum intron length, (ii) the parameter ct controlling the number of contigs considered and (iii) the substitution matrix. The choice of the maximum intron length has a clear impact on the prediction results only for drastic changes (e.g. 15 kb versus 200 kb for plants and animals, respectively). For the parameter ct, lower values yield a larger number of potential GeMoMa predictions but also result in an increased runtime. As we considered only the top-ranked prediction in the benchmark studies above, these are not affected by the choice of ct. We further investigated the influence of the substitution matrix on the results for A. thaliana and O. sativa and found that the performance of GeMoMa is quite stable (Text S7).
Importance of intron position conservation
We examined the characteristics of the approaches based on the predictions between A. thaliana and O. sativa. First, we scrutinized the impact of intron position conservation by running GeMoMa on protein sequences instead of exon-wise amino acid subsequences (Text S9). We find that the performance of GeMoMa without intron position is lower than that of GeMoMa using intron position conservation. This difference is especially pronounced for high values of minimal F1 corresponding to the highest-quality predictions. In summary, these results indicate that utilizing intron position conservation is a key feature of GeMoMa.
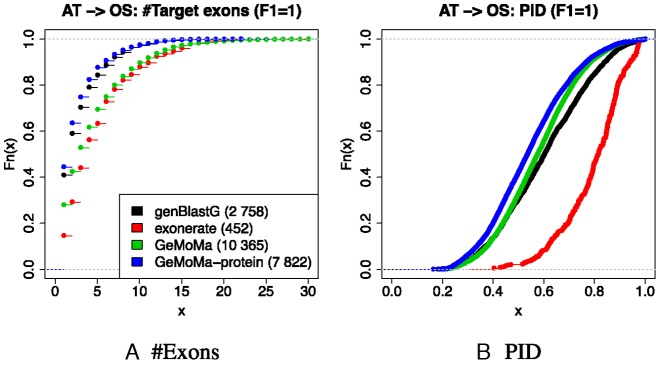
Second, we investigated the number of exons and the protein identity (PID) for the predictions with F1 = 1 of all three tools. For computing PID, we aligned the sequences globally using affine gap costs (gap opening = −11, gap extension = −1) and the BLOSUM62 substitution matrix. We determine the PID as the number of matching positions divided by the minimum of the two sequence lengths (24). We visualize the results for each tool in Figure 2.
Figure 2.
Number of exons and PID for the predictions with F1 = 1. In parentheses, we give the number of predictions with F1 = 1 for each tool. GeMoMa: GeMoMa using intron position conservation; GeMoMa-protein: GeMoMa without intron position conservation.
We find that the distribution of the number of exons for exonerate and GeMoMa with intron position conservation is broader than for genBlastG and GeMoMa without intron position conservation, whereas the distribution of PID for genBlastG and GeMoMa is broader than for exonerate. Hence, we conclude that GeMoMa with intron position conservation is able to predict transcripts with many exons and lower PID more accurately than GeMoMa without intron position conservation, genBlastG and exonerate.
Third, we visualized the difference of the number of exons of (i) reference gene and prediction, (ii) reference gene and target gene with F1 =1 and (iii) reference gene and target gene in category correct transcript with min. F1 =1 (cf. Supplementary Figure S6). We find that GeMoMa with intron position conservation yields in all three cases a similar, sharp and symmetric distribution, whereas exonerate, genBlastG and GeMoMa without intron position conservation yield a broader and asymmetric distribution for (i). This indicates that difference of the number of exons between reference gene and prediction is more similar to the desired distribution for GeMoMa with intron position conservation than for its competitors. Furthermore, the asymmetry indicates that exonerate and genBlastG might tend to miss some exons.
Despite the sharp distribution of the difference of the number of exons, we find that GeMoMa is still able to handle intron gain or loss if reasonable. Specifically, we find one intron loss/gain event in 9.6% and multiple intron loss/gain events in 1.7% of the predictions in the category correct transcript with minimal F1 = 1. Exemplarily, GeMoMa predicts LOC_Os07g09720.1 as homolog of AT5G18070.1 with seven intron gain events, LOC_Os08g43570.1 as homolog of AT2G16730.1 and LOC_Os03g63670.1 as homolog of AT5G08550.1 with five intron loss events each. Hence, we conclude that GeMoMa is able to handle intron loss or gain.
Experimental validation
To exemplarily demonstrate the quality of GeMoMa predictions that are either not annotated, i.e. do not have a match in the target genome, or which have been predicted with additional exons compared to the existing annotation, we chose 10 genes from C. papaya and performed wet-lab experiments. We designed at least two PCR primers for each gene to amplify the full length cDNA predicted by GeMoMa. As control, we also designed primer pairs for the A. thaliana genes which are used as original template for the GeMoMa prediction. The full primer list is given in Supplementary Table S7. We successfully amplified all cDNAs and genomic DNAs from A. thaliana in accordance to the annotated genes (cf. Supplementary Figure S7A and B).
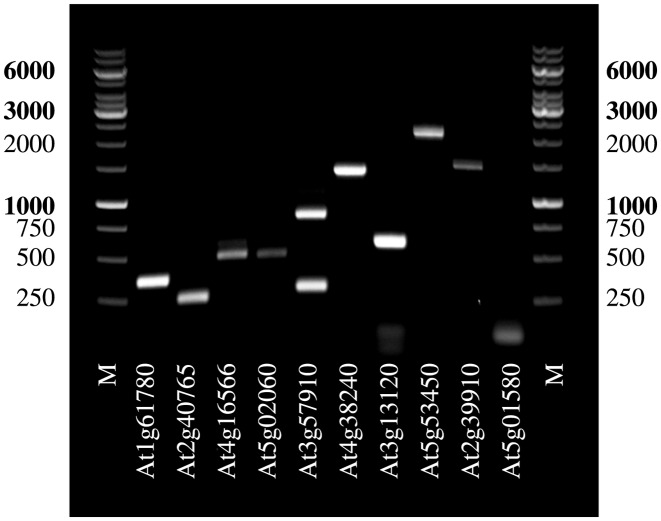
We amplified the genomic DNA of these 10 potential genes of C. papaya, but due to very large introns we had to amplify in one case (C. papaya homolog of At4g16566) two fragments of which only one is shown (Supplementary Figure S7C, At4g16566). We amplified nine out of the 10 potential genes using C. papaya cDNA (Figure 3).
Figure 3.
Gel electrophoresis image of C. papaya cDNA of 10 candidate genes indicating that the GeMoMa predictions for 9 out of these 10 seem to be reasonable.
The C. papaya gene homologous to At5g01580 was the only one which could not be amplified from cDNA (Figure 3). Despite the fact that we tested different primer pairs and PCR conditions, we were not able to get any cDNA amplification of this gene. Therefore, we have to assume that in mRNA from C. papaya flowers neither the annotated nor the predicted transcript of this gene is expressed. In one case, At3g57910, we observed two bands indicating that this gene might be alternatively spliced in C. papaya (cf. Figure 3).
We sequenced the full length cDNAs of the nine successfully amplified transcripts, aligned them to the phytozome entries and the C. papaya reference sequence, and summarized the results in Table 1. In four out of those nine cases (At1g61780, At2g40765, At4g16566, At3g57910), the prediction perfectly matches the sequenced cDNA confirming among others the alternative splicing for At3g57910. In case of At5g02060, we found a 1 bp deletion in the reference sequence leading two a 4 bp difference at the donor splice site of exon 2, which GeMoMa is conceptually unable to adjust for. In case of At4g38240, we observed minor differences due to non-canonical splice sites (AT-AC) at intron 13, and after cloning and sequencing we realized that the cDNA contained in one case out of three the unspliced intron no. 5, leading to an aberrant transcript. In case of At3g13120, we observed that the predicted three exons roughly match the two exons of the cDNA. However, due to three 1 bp indels within 80 bp of the reference sequence the first exon was split in two predicted parts. These indels additionally caused differences in the splice sites of intron 1. Finally, we found perfect matches for 2 and 1 additional predicted exons for At5g53450 and At2g39910, respectively. However due to gaps in the reference sequence and small Indels, we also observed some differences.
Table 1. Overview of the experimental validation of 10 genes which showed a different annotation in phytozome version 10.1 and GeMoMa. The table lists ten genes which show differences between the annotation and the GeMoMa prediction. Four of these genes have been missed in the official annotation and six genes have been annotated with a smaller number of exons in comparison to the GeMoMa prediction.
| AT geneID | Contig (strand) | Region in kb | Type | Experimental validation |
|---|---|---|---|---|
| At1g61780 | Sc_198 (−) | 52–57 | missing gene | as predicted |
| At2g40765 | Sc_19.43 (−) | 367–369 | missing gene | as predicted |
| At4g16566 | Sc_29.148 (−) | 1559–1575 | missing gene | as predicted |
| At5g02060 | Sc_33 (−) | 1665–1673 | missing gene | well matching |
| At3g57910 | Sc_85 (−) | 586–589 | 1 missing exon | as predicted |
| At4g38240 | Sc_28580 (−) | 4–11 | 2 missing exons | well matching |
| At3g13120 | Sc_12 (+) | 1568–1570 | 2 missing exons | roughly matching |
| At5g53450 | Sc_3 (−) | 509–514 | 4 missing exons | 2 exons as predicted |
| At2g39910 | Sc_19 (+) | 1743–1745 | 2 missing exons | 1 exon as predicted |
| At5g01580 | Sc_120 (+) | 347–349 | 1 missing exon | no amplification |
Assessment using RNA-seq data
We finally evaluated the performance of GeMoMa based on experimental RNA-seq data for Nicotiana benthamiana. N. benthamiana is a model plant for plant-microbe interactions (18,25) and widely used in transient assays and for plant molecular farming (26,27). It has a substantially larger genome (∼2.6 Gbp) than A. thaliana. The genome version used in this study (v0.4.4) comprises 140 890 scaffolds and contigs. We performed RNA-seq experiments using N. benthamiana leaves in triplicates, mapped the resulting reads to the genome accounting for splicing and derive experimentally supported transcripts from the mapped reads (see Materials and Methods). The resulting transcripts served as an experimental reference in the following evaluations.
We predicted transcripts in N. benthamiana for all A. thaliana genes also considered in the previous benchmark studies using exonerate, genBlastG and GeMoMa, and additionally included coding sequences of the official 0.4.4 genome annotation into the evaluation. N. benthamiana is an amphidiploid species (28) with parents related to Nicotiana sylvestris and Nicotiana obtusifolia (29) and, hence, can be expected to carry duplicate variants of many A. thaliana genes. For this reason, reporting a single hit for each A. thaliana transcript may not be sufficient to cover all transcripts that are present in the N. benthamiana 0.4.4 genome annotation.
Hence, we started exonerate, genBlastG and GeMoMa in two variants, (i) considering only the best prediction for each A. thaliana transcript and (ii) considering at most 10 predictions judged as reasonable by each of the 3 tools, which results in a total of 229 758, 237 633 and 347 845 predictions for GeMoMa [We ran GeMoMa with ct = 0.4 and P = 10 to obtain a similar number of predictions as genBlastG.], genBlastG and exonerate, respectively. To further illustrate potential implications of an improved prediction of gene models, we consider one specific transcript in Figure 4.
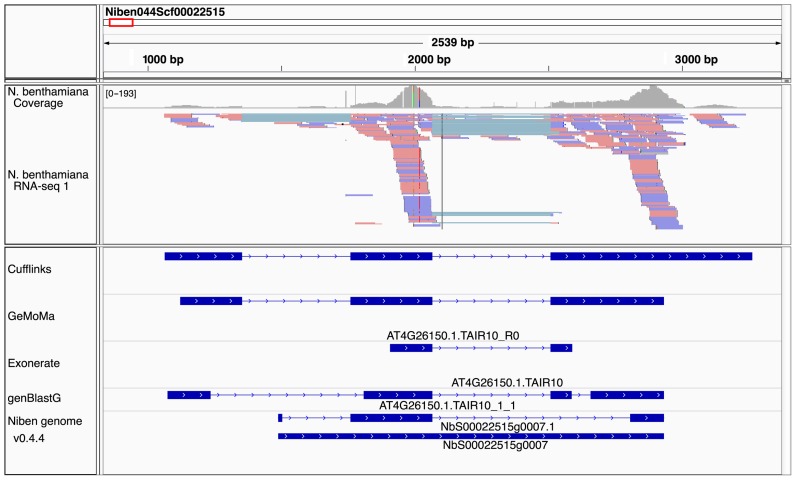
Figure 4.
Exemplary region of the N. benthamiana genome with the corresponding experimentally derived transcripts, predictions and official annotations, and mapped reads of one of the replicates.
In this region of the N. benthamiana genome exists one experimentally derived transcript with three exons. All three tools predict a transcript similar to AT4G26150.1 in this region, which codes for the transcription factor cytokinin-responsive GATA factor 1 (CGA1), while the official annotation does not list an A. thaliana match for transcript NbS00022515g0007.1. CGA1 belongs to the B-GATA subfamily with C-terminal leucine-leucine-methionine domain, which is involved in the control of germination, greening, senescence and flowering time downstream from several growth regulatory signals (30).
The prediction of GeMoMa perfectly matches the experimentally derived transcript except for the UTRs, while the prediction of exonerate and genBlastG as well as the official gene annotation show substantial differences. The prediction of exonerate misses the first exon and predicts a shorter second and third exon. GenBlastG predicts 4 exons, where all exon overlap the experimentally derived exons. However, genBlastG predicts none of the exon perfectly. The official annotation contains one transcript with three exons in this region, where only the second exon perfectly matches the experimentally derived exon. By contrast, the third exon is too short, while the first exon does not even overlap with one of the experimentally derived exons.
Further examples of specific N. benthamiana transcripts comprise additional exons only predicted by GeMoMa and genBlastG (Supplementary Figure S11), exons that are not present in the prediction of genBlastG and the official annotation (Supplementary Figure S8, Supplementary Figure S12), missing only in the official annotation (Supplementary Figure S9), or missing in the genBlastG prediction (Supplementary Figure S14). We also find predictions with experimental support that are substantially different from the official annotation and the prediction of genBlastG (Supplementary Figure S10), or that show deviation from the exonerate and genBlastG prediction, and official annotation in exon positioning and lengths (Supplementary Figure S13).
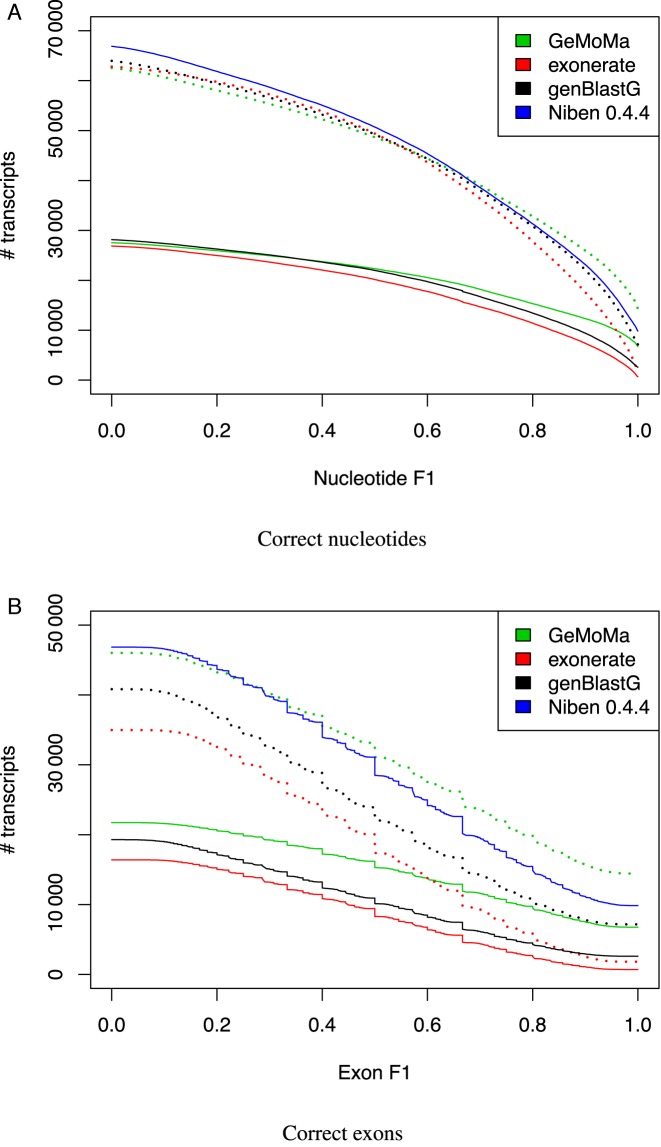
After inspecting a few examples, we aim at assessing the performance of GeMoMa, exonerate and genBlastG, and the official annotation using transcripts derived from RNA-seq data as a reference. For each of the experimentally derived transcripts, we considered for the prediction of all three tools and the official annotation only the best-matching transcript for further evaluation to avoid scoring of overlapping predictions for transcripts of multiple, different A. thaliana genes. Given a pair of experimentally derived transcript and best-matching predicted transcript, we computed the corresponding nucleotide F1 measure as before. We plot the number of matching predictions against different thresholds on these nucleotide F1 values in Figure 5A. Since the total number of experimentally derived transcripts is fixed, this number of matching predictions is proportional to sensitivity.
Figure 5.
Assessment of genBlastG, exonerate and GeMoMa predictions compared to experimentally derived transcripts in N. benthamiana. We plot the number of matching predictions using only the best prediction (solid) and at most 10 predictions (dotted) for different thresholds on the corresponding (A) nucleotide F1 values and (B) exon F1 values. As a reference, we also include the official v0.4.4 annotation.
Considering only the best prediction for each A. thaliana transcript, GeMoMa and genBlastG consistently yield a larger number of matching predictions than exonerate for all thresholds on the F1 values, where for GeMoMa, the offset is especially pronounced for F1 values between 0.5 and 1.0. GeMoMa also yields a larger number of matching predictions than genBlastG for F1 values above 0.5.
Turning to the second variant considering at most 10 predictions, we find that GeMoMa yields a larger number of matching transcripts than exonerate only for thresholds of F1 > 0.6. For values F1 < 0.6, exonerate predicts a larger number of matching transcripts than GeMoMa. However, the total number of predictions by exonerate is substantially larger than that of GeMoMa, which also increases the chance of roughly matching predictions compared to the experimentally derived transcripts. Comparing GeMoMa with genBlastG, we find a larger number of matching predictions of GeMoMa for F1 > 0.65, and the opposite for F1 < 0.65. For very stringent thresholds above F1 > 0.75, GeMoMa even yields a slightly larger number of matching predictions than the official annotation.
For an alternative perspective on prediction accuracy, we further compared the predictions of GeMoMa, genBlastG and exonerate on the exon level. To this end, we counted for each of the tools the number of correctly predicted exons for each experimentally derived transcript. We considered an exon as correctly predicted if donor and acceptor splice site are found at identical position in the transcripts derived from the RNA-seq data. Based on these counts, we computed the corresponding exon F1 values (see Materials and Methods) and, in complete analogy to Figure 5A, plot the number of matching predictions against different thresholds on the exon F1 values in Figure 5B. Considering only the best predictions, we find that GeMoMa yields a larger number of matching predictions than exonerate and genBlastG for all thresholds on the exon F1 values. In this case, the differences between the different tools are more pronounced than on the nucleotide level, which indicates that a substantial number of transcripts and corresponding exons is largely covered by the predictions of all three tools. However, the proportion of perfectly matching exons (including the exact location of splice sites) is larger for the predictions of genBlastG than for exonerate and larger for GeMoMa than for exonerate and genBlastG. This picture is widely consistent considering at most 10 predictions of all three tools. Notably, GeMoMa also yields a larger number of matching predictions than the official annotation for thresholds on the exon F1 value above 0.4.
Finally, we further examined those transcripts that are either predicted by GeMoMa using 10 predictions or present in the official annotation with perfect accuracy, i.e. with a nucleotide F1 = 1 compared with the experimentally derived transcripts. The official annotation contains 9863 and the prediction of GeMoMa contains 14 445 of such perfectly matching transcripts. The intersection of both sets contains 6660 transcripts, while 3203 transcripts are perfectly matching only for the official annotation and 7785 transcripts are perfectly matching only for the prediction of GeMoMa. Of the 6660 transcripts shared between the official annotation and the GeMoMa prediction, 3296 are also annotated with the same best-matching A. thaliana gene, whereas 542 transcripts are annotated with a putative A. thaliana homolog in the GeMoMa prediction but not in the official annotation, and the remaining 2822 transcripts are annotated with differing A. thaliana genes. Of the 7785 transcripts that are perfectly matching the experimentally derived transcript only in the GeMoMa prediction, 3668 are annotated with the same A. thaliana transcript in the official annotation, despite the apparent differences in the gene models. However, 1205 of these transcripts are annotated with a putative A. thaliana homolog only in the GeMoMa prediction.
Summarizing these results, we obtain a more accurate gene model for 7785 transcripts when complementing the official annotation with GeMoMa predictions. In addition, we gain annotations with putative homolog A. thaliana genes for 1747 N. benthamiana transcripts. Since all these GeMoMa predictions considered are perfectly matching experimentally derived transcripts, this additional information is of reasonable confidence.
DISCUSSION
We find that GeMoMa performs better than Genewise, Projector, exonerate and genBlastG on a small mouse data set using human gene models, while it performs similar to GeneMapper. In addition, GeMoMa outperforms genBlastG and exonerate in genome-wide studies for animals and plants using different categories of correctness and different thresholds for the minimal F1 measure.
Using the most conservative evaluation, we find that GeMoMa predicts between 37% and 623% more transcripts perfectly compared to genBlastG and exonerate. Despite making the evaluation less conservative, we still observe that GeMoMa performs better than its competitors by 7% to 77% using the category correct gene family and a minimal F1 ≥ 0.8. Summarizing these results, we find that GeMoMa predicts more transcripts with a higher accuracy than its competitors.
Searching for an explanation of the performance differences, we compared the predictions of genBlastG, exonerate and GeMoMa with and without intron position conservation. We find that GeMoMa with intron position conservation predicts more transcript with more exons and also with lower PID. Furthermore, the number of exons in the reference transcript and the prediction is stronger correlated using intron position conservation.
Comparing the predictions of genBlastG, exonerate and GeMoMa, we also find that hundreds to thousands of transcripts have been predicted in overlapping genomic regions, which might be promising candidates of not yet annotated transcripts. In wet-lab experiments for papaya, we show that the predictions of GeMoMa can be used to improve the official annotation identifying new transcripts or missing exons.
Finally, we use data from RNA-seq experiments to derive experimentally supported transcripts in N. benthamiana and find that a larger number of GeMoMa than exonerate and genBlastG predictions have good experimental support. We also observe several cases, where the predictions of GeMoMa are in better accordance to the experimentally derived transcripts than even the official N. benthamiana annotation.
These findings indicate that utilizing intron position conservation besides amino acid conservation might be beneficial for gene prediction in related species. In addition, it raises questions about the quality and the completeness of existing genome annotations for protein-coding genes. In several cases, we also find indels in the reference genome which affect annotations and predictions.
Despite the reasonable performance of GeMoMa, there is still room for improvement. Currently, GeMoMa only uses GT and GC as consensus donor splice sites and AG as consensus acceptor splice sites. Hence, transcripts using non-canonical splice site cannot be predicted perfectly. Further improvements might be gained by using additional intron-related features in addition to intron position conservation.
High-quality annotations of protein-coding genes are a prerequisite for many applications, as for instance, targeted resequencing including exome-capture (31). Hence, we expect that GeMoMa might be of broad interest for planing and conducting such experiments besides general genome annotation pipelines (32,33).
AVAILABILITY
RNA-seq data have been submitted to ENA and are available under study accession number PRJEB11424. GeMoMa is freely available to the community as part of the Jstacs library (34) and as Galaxy (35) integration at http://www.jstacs.de/index.php/GeMoMa.
Supplementary Material
Acknowledgments
The authors are grateful to Ray Ming for providing genomic DNA of papaya. The authors thank Thomas Berner and Katrin Schulz for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
European Regional Development Fund (to M.S. and J.E.). However, the funder had no influence on the design, analysis and interpretation of (and the decision to submit) this work.
Conflict of interest statement. None declared.
REFERENCES
- 1.Yandell M., Ence D. A beginner's guide to eukaryotic genome annotation. Nat. Rev. Genet. 2012;13:329–342. doi: 10.1038/nrg3174. [DOI] [PubMed] [Google Scholar]
- 2.Hoff K., Stanke M. Current methods for automated annotation of protein-coding genes. Curr. Opin. Insect Sci. 2015;7:8–14. doi: 10.1016/j.cois.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez-Garay M.L. Introduction to isoform sequencing using pacific biosciences technology (Iso-Seq) In: Wu J, editor. Transcriptomics and Gene Regulation. Vol. 9. Netherlands: Springer; 2016. pp. 141–160. [Google Scholar]
- 5.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Birney E., Clamp M., Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slater G., Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer I.M., Durbin R. Gene structure conservation aids similarity based gene prediction. Nucleic Acids Res. 2004;32:776–783. doi: 10.1093/nar/gkh211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterji S., Pachter L. Reference based annotation with GeneMapper. Genome Biol. 2006;7:R29. doi: 10.1186/gb-2006-7-4-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.She R., Chu J. S.-C., Uyar B., Wang J., Wang K., Chen N. genBlastG: using BLAST searches to build homologous gene models. Bioinformatics. 2011;27:2141–2143. doi: 10.1093/bioinformatics/btr342. [DOI] [PubMed] [Google Scholar]
- 11.Liang C., Mao L., Ware D., Stein L. Evidence-based gene predictions in plant genomes. Genome Res. 2009;19:1912–1923. doi: 10.1101/gr.088997.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fedorov A., Merican A.F., Gilbert W. Large-scale comparison of intron positions among animal, plant, and fungal genes. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16128–16133. doi: 10.1073/pnas.242624899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartung F., Blattner F.R., Puchta H. Intron gain and loss in the evolution of the conserved eukaryotic recombination machinery. Nucleic Acids Res. 2002;30:5175–5181. doi: 10.1093/nar/gkf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 15.Dalquen D.A., Dessimoz C. Bidirectional best hits miss many orthologs in duplication-rich clades such as plants and animals. Genome Biol. Evol. 2013;5:1800–1806. doi: 10.1093/gbe/evt132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flicek P., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bombarely A., Rosli H.G., Vrebalov J., Moffett P., Mueller L.A., Martin G.B. A draft genome sequence of nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe Interact. 2012;25:1523–1530. doi: 10.1094/MPMI-06-12-0148-TA. [DOI] [PubMed] [Google Scholar]
- 19.Kasajima I., Ide Y., Ohkama-Ohtsu N., Hayashi H., Yoneyama T., Fujiwara T. A protocol for rapid DNA extraction from Arabidopsis thaliana for PCR analysis. Plant Mol. Biol. Rep. 2004;22:49–52. [Google Scholar]
- 20.Erickson J., Ziegler J., Guevara D., Abel S., Klosgen R., Mathur J., Rothstein S., Schattat M. Agrobacterium-derived cytokinin influences plastid morphology and starch accumulation in Nicotiana benthamiana during transient assays. BMC Plant Biol. 2014;14:127. doi: 10.1186/1471-2229-14-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schattat M., Barton K., Baudisch B., Klösgen R.B., Mathur J. Plastid stromule branching coincides with contiguous endoplasmic reticulum dynamics. Plant Physiol. 2011;155:1667–1677. doi: 10.1104/pp.110.170480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raghava G., Barton G. Quantification of the variation in percentage identity for protein sequence alignments. BMC Bioinformatics. 2006;7:415. doi: 10.1186/1471-2105-7-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodin M.M., Zaitlin D., Naidu R.A., Lommel S.A. Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol. Plant Microbe Interact. 2008;21:1015–1026. doi: 10.1094/MPMI-21-8-1015. [DOI] [PubMed] [Google Scholar]
- 26.Gleba Y., Klimyuk V., Marillonnet S. Magnifection – a new platform for expressing recombinant vaccines in plants. Vaccine. 2005;23:2042–2048. doi: 10.1016/j.vaccine.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Gleba Y., Klimyuk V., Marillonnet S. Viral vectors for the expression of proteins in plants. Curr. Opin. Biotechnol. 2007;18:134–141. doi: 10.1016/j.copbio.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Sierro N., Battey J., Ouadi S., Bovet L., Goepfert S., Bakaher N., Peitsch M., Ivanov N. Reference genomes and transcriptomes of Nicotiana sylvestris and Nicotiana tomentosiformis. Genome Biol. 2013;14:R60. doi: 10.1186/gb-2013-14-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones L., Keining T., Eamens A., Vaistij F.E. Virus-induced gene silencing of argonaute genes in nicotiana benthamiana demonstrates that extensive systemic silencing requires Argonaute1-like and Argonaute4-like genes. Plant Physiol. 2006;141:598–606. doi: 10.1104/pp.105.076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Behringer C., Schwechheimer C. B-GATA transcription factors - insights into their structure, regulation and role in plant development. Front. Plant Sci. 2015;6:90. doi: 10.3389/fpls.2015.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 32.Allen J.E., Pertea M., Salzberg S.L. Computational Gene Prediction Using Multiple Sources of Evidence. Genome Res. 2004;14:142–148. doi: 10.1101/gr.1562804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zickmann F., Renard B.Y. IPred - integrating ab initio and evidence based gene predictions to improve prediction accuracy. BMC Genomics. 2015;16:134. doi: 10.1186/s12864-015-1315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grau J., Keilwagen J., Gohr A., Haldemann B., Posch S., Grosse I. Jstacs: a java framework for statistical analysis and classification of biological sequences. J. Mach. Learn. Res. 2012;13:1967–1971. [Google Scholar]
- 35.Goecks J., Nekrutenko A., Taylor J., The Galaxy Team Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.