Abstract
Rbfox proteins regulate tissue-specific splicing by targeting a conserved GCAUG sequence within pre-mRNAs. We report here that sequence-specific binding of the conserved Rbfox RRM to miRNA precursors containing the same sequence motif in their terminal loops, including miR-20b and miR-107, suppresses their nuclear processing. The structure of the complex between precursor miR-20b and Rbfox RRM shows the molecular basis for recognition, and reveals changes in the stem-loop upon protein binding. In mammalian cells, Rbfox2 downregulates mature miR-20b and miR-107 levels and increases the expression of their downstream targets PTEN and Dicer, respectively, suggesting that Rbfox2 indirectly regulates many more cellular miRNAs. Thus, some of the widespread cellular functions of Rbfox2 protein are attributable to regulation of miRNA biogenesis, and might include the mis-regulation of miR-20b and miR-107 in cancer and neurodegeneration.
INTRODUCTION
MicroRNAs (miRNAs, miRs), small (∼20–22 nt) non-coding RNAs, play key post-transcriptional regulatory roles by base pairing primarily to the 3′-untranslated region (UTR) of target mRNAs and repressing translation or inducing mRNA degradation (1,2). Because base pair complementarity need not be perfect, each miRNA can regulate the expression of multiple genes and each gene can be regulated by multiple miRNAs (1). Thus, even relatively small changes in expression levels of individual miRNAs can combinatorially result in large phenotypic effects. With more than 2000 miRNAs recognized in humans (3), miRNAs target at least 60% of human protein-coding genes and participate in all fundamental cellular processes (4). Moreover, the mis-regulation of their expression is associated with many human diseases (5,6).
MicroRNA expression is regulated transcriptionally as well as post-transcriptionally (7,8). MiRNAs are transcribed as long precursors (primary miRNA, pri-miRNA) and cleaved by the Drosha/DGCR8 microprocessor complex in the nucleus (9–11) to release the precursor miRNAs (pre-miRNAs), a stem-loop of 60–70 nt which embeds the mature miRNA. Following export to the cytoplasm, pre-miRNAs are excised by Dicer into ∼22 nt miRNA:miRNA* duplexes (10). One strand of the duplex, the mature miRNA, is loaded onto Argonaute (AGO)-containing RNA-induced silencing complexes (RISC) to guide the repression or degradation of target mRNAs (10), while the second strand is generally but not always degraded. Several RNA-binding proteins (12–14) have been found to regulate miRNA processing either positively or negatively by binding to miRNA precursors often within the apical portion of the stem-loops (15). These studies suggest that the presence of a binding site for an RNA-binding protein within a precursor miRNA hairpin can confer the ability to regulate the expression of a miRNA and its downstream targets.
The Rbfox family of RNA-binding proteins, conserved tissue-specific alternative splicing factors (16), consists in mammals of Rbfox1 (A2BP1), Rbfox2 (RBM9) and Rbfox3 (NeuN). High levels of mammalian Rbfox1 protein are expressed in neurons and in both skeletal and cardiac muscle cells (17,18), whereas Rbfox3 protein is only detected in neurons (19). Rbfox2 is more broadly distributed, expressed in neurons and muscles as well as in stem cells, hematopoetic cells and in the embryo (16,18). It is the only Rbfox protein expressed in embryonic stem cells, where it is required for their proliferation. Rbfox proteins contain a highly conserved nuclear localizing signal (NLS) in the C-terminal region, and most Rbfox1/2 isoforms localize to the nucleus (20,21). Rbfox3 also localizes predominantly to the nuclei of neurons (19). Mutations or abnormal expression of Rbfox1 have been found in patients with epilepsy, mental retardation, autism and heart disease (16). Moreover, expression of Rbfox2 protein is downregulated in ovarian cancer and its splicing is altered in breast cancer samples, suggesting a role of Rbfox2 in cancer proliferation (22).
Recently, Rbfox3 protein was found to directly regulate the expression of many miRNAs in neuron in a manner that is independent of the presence of a cognate binding site (23). This result, though, was very surprising, considering that, unlike most other splicing factors, the highly conserved RRM of Rbfox proteins targets RNA specifically through the GCAUG motif (21,24), a well-known cis-element required for regulation of alternative splicing. The binding of Rbfox2 protein is strongly enriched around the conserved (U)GCAUG motif in transcripts found in mouse embryonic stem cells (25). In a systematic study of binding specificity, Rbfox2 protein was found to cross-link in cells to sites containing the cognate sequence with high specificity (26); furthermore, binding and activity of Rbfox proteins were determined by the intrinsic binding preferences of the proteins and largely driven by RNA-binding affinity (26). Thus, we searched computationally for the GCAUG motif within human precursor miRNA hairpin sequences and identified >120 human miRNAs, including the precursor miR-20b (pre-miR-20b) and miR-107 (pre-miR-107), which contain the exact sequence motif in their apical loops, where it is accessible for protein binding. Because the apical loop is an important site for regulation of miRNA biogenesis (10), we hypothesized that Rbfox proteins would specifically bind to the sequence motif within those miRNA precursors, in doing so, would regulate their mature miRNA levels and affect expression of downstream targets.
We found that the RRM of Rbfox proteins (given the identical RNA specificity in all three family members, we refer to it with a single name) binds to stem loop structures of miR-20b and -107 precursors with high specificity. The nuclear magnetic resonance (NMR) structure of the complex of pre-miR-20b with the Rbfox RRM, together with SHAPE chemistry, demonstrates that protein binding differentially alters the conformation of the two miRNA hairpins. This interaction represses processing of the pri-miRNA in vitro. Importantly, Rbfox2 downregulates the production of mature miR-20b in mammalian cells and increases expression of endogenous PTEN (Phosphatase and TEnsin Homolog) protein, a tumor suppressor with a critical role in cell growth and proliferation (27); up-regulation of mature miR-107 by Rbfox2 knockdown reduces cellular levels of Dicer enzyme.
The newly identified role for Rbfox family proteins in regulation of miRNAs maturation, directly, through the Rbfox binding sites, and indirectly, through Dicer, links their abnormal expression, mutation and altered splicing forms to changes in the levels of specific miRNAs in cancer and brain diseases.
MATERIALS AND METHODS
Computationally screening miRNA hairpin sequences
Human miRNA hairpin sequences were downloaded from miRBase (release 21) (http://www.mirbase.org/ftp.html) (28,29). The desired Rbfox binding sites (GCAUG or GCACG) were searched and grouped according to their locations within the hairpin (terminal stem loop, upper stem or lower stem) (Supplemental Figure S1A).
Protein and RNA preparation
Full-length Rbfox1 and Rbfox2 cDNA clones were obtained from Open Biosystems. DNA encoding the full-length Rbfox1 (418 amino acids, transcript variant 1) and Rbfox2 proteins (380 amino acids, transcript variant 1) as well as the RRM of Rbfox1 alone (residues 109–208, Swissprot Q9NWB1 and various other extensions as discussed in the text) were amplified by polymerase chain reaction (PCR) and cloned into a pET28a (with N-terminal His-tag) vector. Since it is identical in sequence to the RRM of Rbfox2, it will be referred to as Rbfox RRM, The full-length proteins could not be expressed in Escherichia coli (E. coli) and were in vitro translated instead using the S30 T7 High-Yield Protein Expression System (Promega) and purified by the MagneHis™ Protein Purification System (Promega) according to the manufacturer's protocol. The Rbfox RRM domain was expressed in transformed BL21 (DE3) E. coli and purified by two-steps of nickel-affinity and heparin affinity column chromatography (24). Purified proteins were then dialyzed and concentrated against assay buffer of 20 mM Tris–Cl (pH7.5) and 50 mM NaCl. Protein concentrations were measured by UV absorbance at 280 nm and confirmed by Bradford assay.
Different lengths of pre-miR-20b and pre-miR-107 for SHAPE and NMR analysis were prepared by standard in vitro T7 RNA transcription (30) using DNA templates ordered form Integrated DNA Technologies (IDT).
The paramagnetic spin labeling of the RNA is described in Supplementary Material.
Gel-shift protein–RNA binding assays
Rbfox proteins were assayed for their RNA-binding activity by using synthetic RNAs (IDT) containing the apical stem-loop sequences of the pre-miRNAs. Shorter constructs as well as the full-length pre-miR-20b and pre-miR-107 derived from miRBase were used in the binding assay (Supplementary Figure S1C and D). The RNAs were 5′-end-labeled using γ32-ATP (adenosine triphosphate). The proteins were incubated with 20 pM of radiolabeled RNAs at room temperature for 1 h. Samples were run on 10% native polyacrylamide gels and the radioactive signals were detected on a GE Storm phosphor imagine scanner.
Isothermal titration Calorimetry (ITC)
MicroCal iTC200 was used to examine the thermodynamics of binding between various constructs of the Rbfox RRMs and pre-miR-20b. The proteins and pre-miR-20b were dialyzed into the same buffer containing 10 mM NaH2PO4 (pH 6.5) and 100 mM NaCl. The titration was carried out at 293 K, with pre-miR-20b in the cell as titrate (10 uM) and RRM protein in the syringe as titrant (100 uM). Data analysis was carried out using Origin 7 provided by MicroCal. All measurements were performed more than two times.
In vitro primary and precursor miRNA processing assays
The DNA segments containing pri-miRNA sequences of miR-20b and miR-107 were amplified from genomic DNA by PCR, then cloned into a pUC19 vector downstream of a T7 promoter. The plasmids were linearized and gel purified. The internally labeled pri-miRNAs were transcribed in vitro using T7 RNA polymerase (Promega) in the presence of [α-32P]-CTP, and purified with QIAGEN RNeasy Mini kit. HeLa cell nuclear extracts from ProteinOne were used for the pri-miRNA processing assays. Processing assays were carried out (typically using 25 μg cell nuclear extract per 20 μl reaction) at 30°C for 1 h. Samples were then phenol/chloroform extracted, precipitated and re-suspended in RNA loading dye prior to denaturing polyacrylamide gelelectrophoresis separation. All the assays were preformed at least in triplicate.
Processed pri-miRNA products were resolved on 12.5% acrylamide-8 M urea gels together with Molecular weight markers of radiolabeled RNA oligonucleotides (Ambion® Decade™ Marker System). After electrophoresis, the gels containing the separated RNAs were exposed to a phosphor-screen and visualized by autoradiography using a Storm PhosphorImager (GE Life Sciences). Band intensities were quantified using ImageJ software (31).
Selective 2′OH acylation analyzed by primer extension (SHAPE)
We performed SHAPE chemical probing (32) on pre-miRNA-20b and pre-miR-107 in both free and Rbfox-RRM bound forms. Prior to conducting SHAPE chemistry, RNAs were denatured by heating at 95°C for 3 min, then snap-cooled on ice to refold the RNA. Three 9-μl aliquots of free precursor miRNAs or protein–RNA complexes (100 nM each) were each combined with 1 μl of either Dimethyl sulfoxide ( DMSO) (control), or 65 mM N-Methylisatoic Anhydride (NMIA) (dissolved in DMSO), or 130 mM NMIA (in DMSO). Reactions were incubated at 37°C for 40 min, then ethanol precipitated on dry ice. After pelleting at 4°C by centrifugation, the RNA was allowed to air dry before re-suspension in 10 μl water.
The DNA primer was 5′-end labeled with γ-[32P]-ATP using T4 polynucleotide kinase and purified. Modified RNA (∼0.5 pmol) was annealed with 1.5 pmol 32P-labeled DNA primer. Reverse transcription was performed using Superscript III (Invitrogen) by incubation at 52°C for 10 min, and stopped by degrading the RNA with 200 mM NaOH at 95°C for 5 min. Each sequencing and modification reaction was analyzed by gel electrophoresis. The fixed gels were dried and exposed overnight onto a storage phosphor screen. Autoradiographs were imaged with a Typhoon laser scanner and manipulated with Semi-Automated Footprinting Analysis (SAFA) software (33). All of the SHAPE analyses were replicated at least three times. Secondary structure analysis was conducted with RNAstructure (34) and MC-Fold (35).
NMR spectroscopy
Constructs of various length corresponding to the apical part of the pre-miR-20b stem-loop were prepared by standard in vitro T7 RNA transcription using unlabeled or 13C, 15N labeled NTPs (36). Following purification through denaturing polyacrylamide gel electrophoresis, the RNA was desalted, lyophilized and re-suspended in NMR buffer (10 mM NaH2PO4, 20 mM NaCl, pH 6.5). RNA samples were annealed by heating at 80°C for 5 min followed by snap cooling on ice.
Unlabeled or 13C, 15N labeled Rbfox RRM (residues 109–208 of Rbfox1) was prepared from E. coli grown from LB or minimal M9 medium supplemented with 1 g/l 15NH4Cl and 2 g/l 13C-glucose. After His-trap and Heparin column purification, protein was buffer exchanged into the same NMR buffer as the RNA and concentrated to 1 mM for later use. The complex was formed either by titrating unlabeled protein into 13C, 15N labeled RNA, monitored by1H-13C HSQC (Heteronuclear Single-Quantum Correlation) of the RNA or, vice versa, by titrating unlabeled RNA into 13C, 15N labeled protein, monitored by1H-15N HSQC of the protein. NMR experiments were performed on Bruker Avance 600 and Avance 800 spectrometers equipped with HCN cryogenic probes and single axis pulse field gradients. NMR data were processed with NMRPipe (37) and analyzed with CCPNMR (38). Resonance assignments and structural determination of the free RNA and protein–RNA complexes were carried out using standard NMR spectroscopy (details described in Supplementary Material).
Structural modeling
Full-length pre-miR-20b free or in complex with the Rbfox RRM was modeled by using the MC-Fold and MC-Sym pipeline (35) with SHAPE and NMR data as restraints. Two structures were merged in the modeling process. The experimentally-determined apical stem loop structure of pre-miR-20b (U21 to A39), as determined by NMR with or without the Rbfox RRM, and the structure of full-length pre-miR-20b modeled by MC-Fold and MC-Sym pipeline using SHAPE data as restraints for the secondary structure, as well as base pairs identified in the analysis of the 46 nt RNA construct of pre-miR-20b. These two fragments were merged to generate a single hybrid model which was minimized using Amber with force field ff99 (39).
Cell culture, transfection and siRNA
HeLa, HEK293, SHSY5Y, NSC-34 and MCF7 cells from ATCC were grown in Dulbecco's modified Eagle's medium (DMEM, GIBCO) plus 10% fetal bovine serum (FBS) with penicillin/streptomycin supplement. MCF7 and mda-mb-231 cells were grown in DMEM plus 10% FBS, supplemented with 2 mM Glutamine, 0.1 mM non-essential amino acids and 1× penicillin/streptomycin. Plasmids were transfected using Trans IT®-LT1 Transfection Reagent (Mirus). In co-transfection experiments, cells were transfected with 2.5 μg of FLAG-Rbfox2 and 2.5 μg of miR-20b expression plasmids. In the controls, an equivalent amount of the empty expression plasmid was utilized. SiRNAs against Rbfox2 protein were purchased from Life Technologies (Invitrogen) (Silencer® Select Pre-Designed siRNA to human Rbfox2 #4392420, siRNA ID: s24047, Silencer® Select Pre-Designed siRNA to mouse Rbfox2 #4390771, siRNA ID: s96621 Negative Control No. 1 siRNA #4390843). Si-RNA transfections were performed using INTERFERin™ siRNA trasfection reagent (Polyplus transfection) according to the manufactures instructions. Forty-eight hours after transfection, cells were harvested for protein and RNA extraction.
MiRNA expression plasmids for primary miR-20b and miR-107 were obtained from OriGene. Full-length Rbfox1 (Fox-1, NM_145891) and Rbfox2 (Fox-2, RBM9, NM_001031695) proteins and relevant fragments, as described in the main text, were cloned into pCMV-Tag2B vector. The expressed proteins have N-terminal Flag-tags for western-blot detection.
Preparation of cell extracts, immunoblot analysis and antibodies
Cell extracts were prepared with radioimmunoprecipitation assay (RIPA) buffer (Sigma) supplemented with a protease-inhibitor cocktail (Roche). For immunoblot analysis protein samples were resolved on a 10% polyacrylamide Bis-Tris gel and transferred onto a nitrocellulose membrane. The following primary antibodies were used: rabbit polyclonal anti-Rbfox1 (1:1000, Abcam), mouse monoclonal anti-Rbfox2 (1:1000, Abcam), mouse monoclonal α-actinin (1:1000, Santa Cruz Biotechnology), rabbit polyclonal anti-DICER (1:1000, Santa Cruz Biotechnology), mouse monoclonal E-Cadherin (1:1000), mouse monoclonal ß-tubulin (1:1000, Santa Cruz Biotechnology), mouse monoclonal anti-actinin (1:2000, Santa Cruz Biotechnology) and mouse monoclonal anti-FLAG M2 antibody (1:1000, Sigma). Goat anti-rabbit IgG-HRP (1:3000, Santa Cruz Biotechnology) and goat anti-mouse IgG-HRP (1:3000, Santa Cruz Biotechnology) were used as secondary Abs. All Western blots were analyzed with the ChemiDoc XRS+ System (Bio-Rad).
Immunofluorescence microscopy
Cell staining and immunofluorescence microscopy were carried out as described previously (40). Briefly, MCF7 cells transfected with an FLAG-Rbfox2 expression plasmid were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 in phosphate-buffered saline. Fixed cells were incubated with primary mouse monoclonal anti-FLAG M2. Alexa-488–conjugated goat antibody against mouse IgG (1:500, Molecular Probes) was used as secondary Ab. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI, 5 μg/ml). Microscopy analysis was performed using the Zeiss Observer Z.1 microscope implemented with the Zeiss ApoTome device. Pictures were acquired using AxioVision imaging software package (Zeiss) and assembled with Adobe Photoshop CS3. Images were not modified other than for adjustments of levels, brightness and magnification.
RNA preparation, RT-PCR and qRT-PCR
Total RNA was extracted with the miRNeasy Mini kit (Qiagen). One microgram of total RNA was used in the retro-transcription reaction using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). qRT-PCR was performed in triplicate with TaqMan Universal Master Mix II, no UNG (Applied Biosystems). Relative differences in RNA levels were calculated according to the 2−ΔΔCt method and normalized against RNU48 small-nucleolarRNA. Statistical analysis from three cell cultures was performed by ANOVA followed by Bonferroni's multiple comparisons test. The TaqMan probes were 5′-CAAAGUGCUCAUAGUGCAGGUAG-3′ for mature hsa-miR-20b-5p (ID: 001014, Life Technologies); 5′-AGCAGCAUUGUACAGGGCUAUCA-3′ for mature hsa-miR-107 (ID: 000443, Life Technologies), snoRNA234 (mouse) and RNU48 (human) TaqMan Controls miRNA Assay (Life Technologies).
RESULTS
Rbfox targets the GCAUG sequence found in pre-miR-20b, pre-miR-107 and >100 other precursor miRNA hairpins
A recent manuscript reported that Rbfox3 protein regulates miRNA biogenesis in a manner that is independent of the presence of its known target sequence within the pre-miRNA (23). However, Rbfox proteins are known to target pre-mRNAs by sequence specific recognition of the GCAUG motif by their RRM and to cross-link to cellular RNAs at or near sites carrying that canonical signal (24,26). Furthermore, their function in splicing regulation is strongly correlated and dependent on their binding to the cognate motif (26). In fact, we were already investigating if the presence of Rbfox target sites within a precursor miRNA hairpin would lead to regulation of processing by the Drosha and/or Dicer, and computationally searching human precursor miRNA hairpin sequences for the presence of the GCAUG motif. Approximately 120 pre-miRNAs (from 1881 entries in miRBase release 21) contain this sequence (Supplementary Table S1), while >40 additional pre-miRNAs contain the related GCACG sequence (26) (Supplementary Table S2).
Most of the candidate miRNAs of Supplementary Tables S1 and 2 are poorly annotated with uncertain cellular abundance or functions. Thus, we focused our investigation on two well-studied miRNAs, miR-20b and miR-107, which contain the Rbfox target sequence within the terminal loop (TL) of their precursor hairpins, a hotspot for regulation of miRNA processing (15). Rbfox proteins participate in cancer and neurological disorders (16,41), conditions where abnormal levels of miR-20b and miR-107 have also been found. Finally, the TL sequence of precursor miR-107 is highly conserved throughout vertebrates (15), although the TL sequence of pre-miR-20b is conserved only among primates (Supplementary Figure S1A and B).
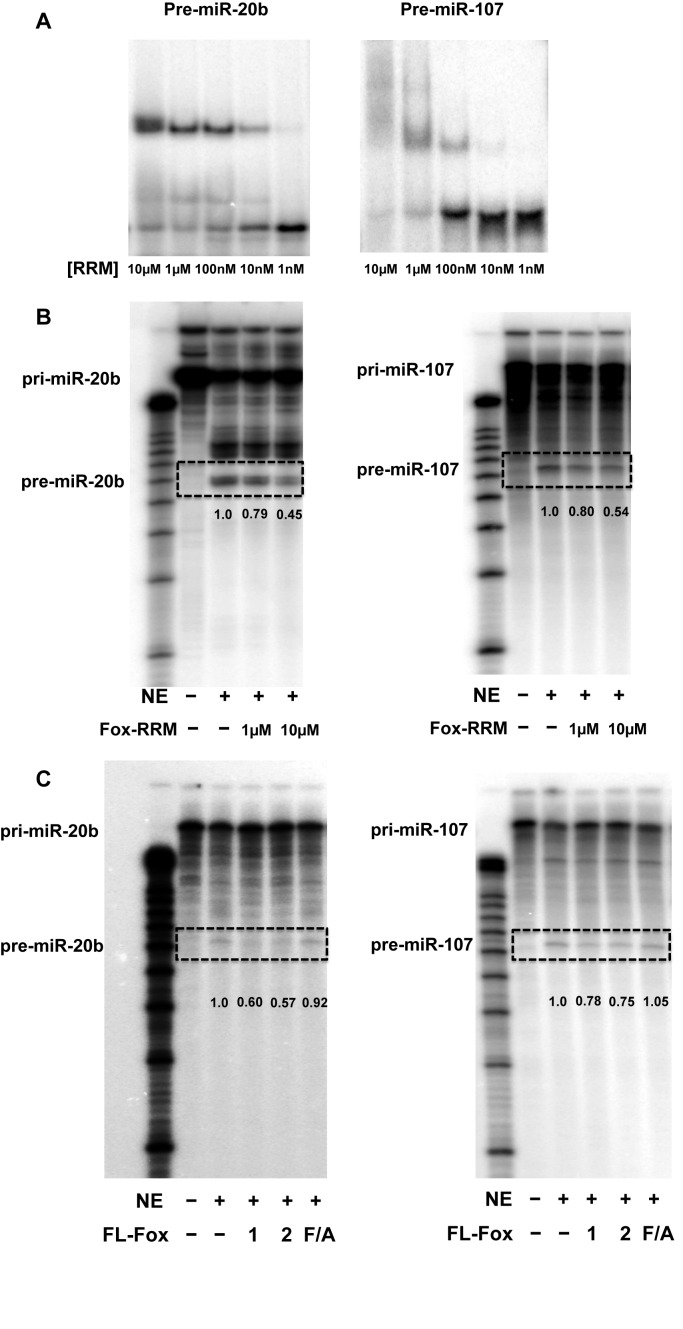
The RRM domain of human Rbfox1 protein has the same sequence as Rbfox2, and differs from the Rbfox3 RRM in only four positions which are not involved in RNA binding; thus, all three RRMs bind to RNA in the same way. We refer to the identical Rbfox1 and Rbfox2 RRMs as the Rbfox RRM. Electrophoretic mobility shift assay experiments were used to determine the binding affinity of Rbfox RRM against pre-miR-20b and pre-miR-107. This assay visualizes the presence of multiple complexes or smears indicative of non-specific binding. The protein binds to both the complete and the truncated terminal stem-loop of pre-miR-20b with a binding affinity (Kd) of ∼50 nM (Figure 1A; Supplemental Figure S1C and D). The difference in affinity compared to the single-stranded RNA is not unexpected, because secondary structure formation inhibits binding (26) and the (G)GCAUG sequence is sub-optimal compared to (U)GCAUG (24). The Rbfox RRM binds to pre-miR-107 more weakly (Kd of ∼1 μM) (Figure 1A, right side). Binding is highly specific as well: mutation of GCAUG to ACAUA within both pre-miR-20b and -107 reduced Rbfox RRM binding to only a smear in the gel at high (10 μM) protein concentration, indicating Kd >10 μM (data not shown).
Figure 1.
Rbfox proteins specifically bind to miR-20b and -107 precursor hairpins and repress their processing in vitro. (A) Electrophoretic mobility shift assay analysis of the Rbfox RRM binding to pre-miR-20b and -107. Truncated constructs of pre-miR-20b and -107 were used in these figures but the same results were obtained with the complete pre-miRNA stem-loops (Supplementary Figure S1C and D). (B) In vitro Drosha processing of pri-miR-20b and -107 in HeLa nuclear extract (NE) is inhibited by the Rbfox RRM at the indicated protein concentrations. Pri-miRNAs are uniformly radiolabeled. (C) In vitro Drosha processing of pri-miR-20b and -107 in HeLa nuclear extract is inhibited by full-length Rbfox1 and Rbfox2 proteins, but not by the Rbfox2 F/A mutant that abolishes binding to RNA. Quantification of the gel bands for pre-miRNAs in Drosha processing assays was carried out using ImagineJ and normalized to NE only.
Binding of the Rbfox RRM to pre-miR-20b and pre-miR-107 hairpins inhibit their processing in vitro by Drosha
Because of the mainly nuclear localization of Rbfox proteins (16), we investigated whether binding of the Rbfox RRM to pri-miR-20b and pri-miR-107 affects nuclear processing by Drosha. A typical primary miRNA consists of a stem-loop corresponding to the pre-miRNA, and single-stranded RNA segments at both 5′- and 3′- sides (Supplementary Figure S1A). We found that the pri-miR-20b and -107 were processed in vitro in the presence of HeLa nuclear extracts, generating products corresponding to the predicted pre-miR-20b (∼60 nt) and pre-miR-107 (∼65 nt). When the Rbfox RRM was added to the reaction, we observed reduced cleavage efficiency in a dose-dependent manner (Figure 1B). Consistent with our hypothesis that inhibition is driven by direct protein–RNA recognition, the effects correlate with binding affinity. Namely, processing of miR-20b is more strongly affected, compared to miR-107, at equivalent protein concentrations. No difference in the size of the cleavage products was observed in the presence or absence of the Rbfox RRM, suggesting that the accuracy of cleavage is not affected by protein binding. When we repeated the processing assays using recombinant full-length Rbfox1 and Rbfox2 proteins (Figure 1C), we found the same inhibitory effects as for the Rbfox RRM on both pri-miR-20b and -107 processing.
In order to demonstrate that inhibition is specific and requires binding to RNA, we expressed a mutant full-length Rbfox2 protein (Rbfox2-F/A) where all three phenylalanine residues (F126, F158, F160) on the RNA-binding protein surface are replaced with alanine. This construct abolishes RNA binding since each single mutant F126/A, F158/A and F160/A reduces RNA-binding affinity by ∼1500-, 700- and 30 000-fold, respectively (24). Processing of pri-miR-20b and -107 by Drosha in the presence of full-length Rbfox2-F/A protein is similar to the processing reactions in the absence of Rbfox proteins (Figure 1C). Taken together, our data show that sequence-specific binding of Rbfox protein to the TLs of primary and precursor miR-20b and -107 inhibit their processing by Drosha microprocessor complex, and that binding to RNA is required for this to occur.
Binding of Rbfox alters the conformation of pre-miR-20b and pre-miR-107
In order to understand mechanistically how Rbfox protein binds to pre-miRNAs and affects their processing, we investigated the effect of this protein on the structures of the pre-miR-20b and -107 hairpins. We used both NMR and SHAPE chemistry (42), which provides information on local nucleotide dynamics and accessibility in RNA and RNA–protein complexes (43).
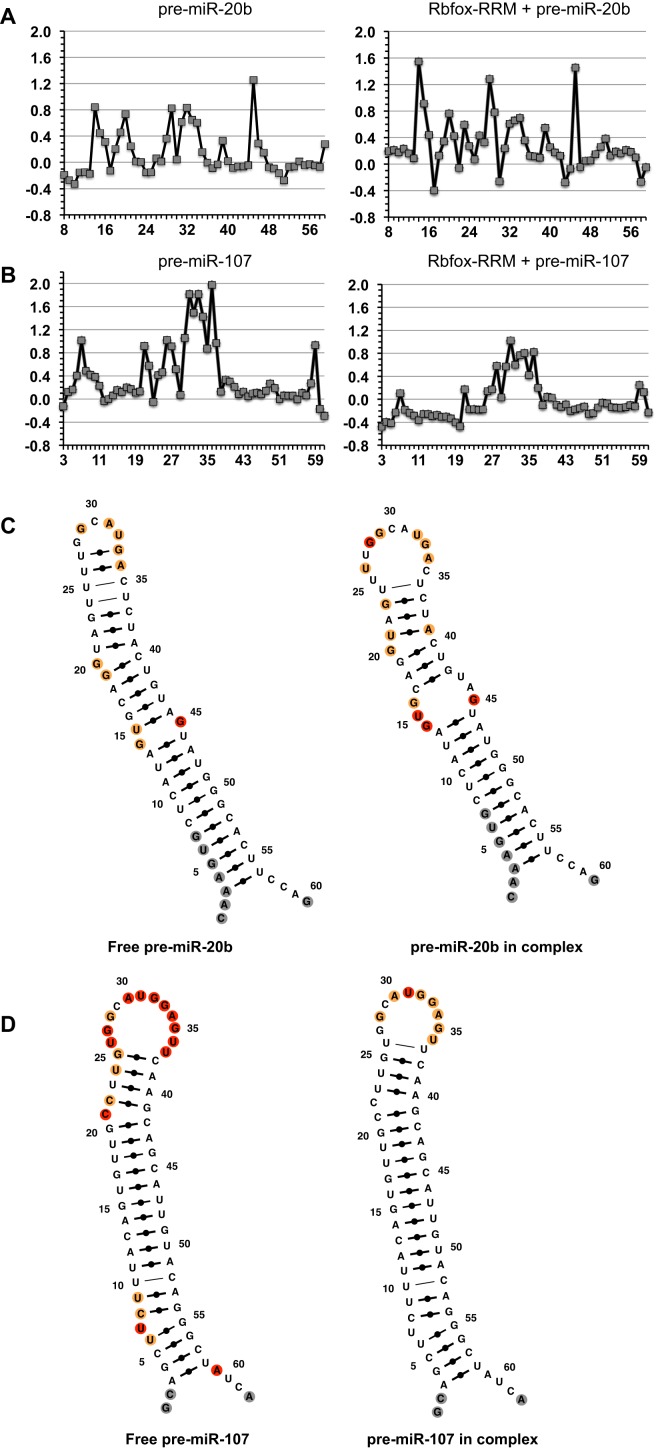
The SHAPE data for free pre-miR-20b (Figure 2A) indicates that the secondary structure computationally predicted from miRBase is incorrect. Instead of forming a 2 × 3 internal loop (G20U21 and C37U38A39, Supplemental Figure S1C), pre-miR-20b forms a much longer apical helix through the formation of a non-canonical U24·U36 base pair and a well-ordered U25·C35 mismatch (Figure 2C). Formation of this helix reduces the size of the TL to 5 nt, but leaves the Rbfox targeting sequence GCAUG exposed.
Figure 2.
Conformational changes of pre-miR-20b and -107 stem loops upon Rbfox-RRM binding. SHAPE reactivity traces for pre-miR-20b (A) and pre-miR-107 (B) in free and complex forms (protein/RNA ratio is 1:1); reactivity is normalized to untreated samples. Secondary structures of pre-miR-20b (C) and -107 (D) in their free and Rbfox RRM-bound forms calculated using MC-Fold and MC-Sym by incorporating information provided by SHAPE and NMR analyses (35). Nucleotides are colored according to their normalized SHAPE reactivity with no data as gray, low reactivity (<0.4) as black, medium reactivity (0.4–0.85) as orange and high reactivity (>0.85) as red. Canonical and non-canonical base pairs are shown by bold lines with black dots and non-bold lines, respectively.
These unexpected observations are supported by the NMR structure of the apical portion of pre-miR-20b (Figure 3A), a 23-nt stem-loop (G20-C40) containing the GCAUG target sequence whose secondary structure and NMR signals match exactly that of a much longer construct (46 nt, G7-C52; starting from the miRBase 5′-end of the precursor) covering most of the structured region of full-length pre-miR-20b. The secondary structures and protein binding of the two RNAs (G20-C40 versus G7-C52) are identical except for the bottom G20-C40 pair, as judged from the comparison of the resonances of the two constructs, which match perfectly and change in the same way in the complex (Supplementary Figure S2A and B). The 15N HSQCs of the Rbfox RRM in complex with the two RNAs are also identical (data not shown), but the shorter construct provides higher spectral quality for the complex (see below) and therefore improves structural resolution.
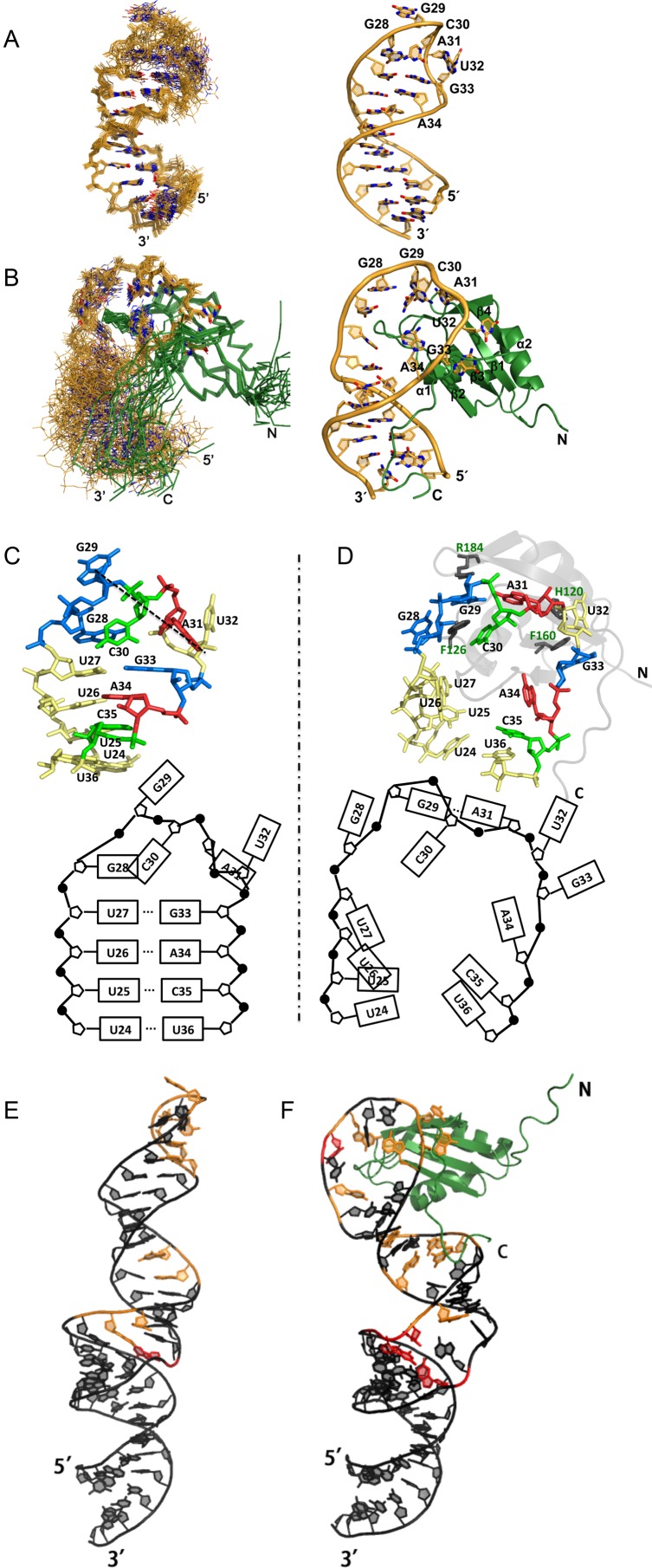
Figure 3.
Structural basis for recognition of pre-miR-20b by the Rbfox RRM: (A) superposition of NMR structures of free pre-miR-20b; (B) superposition of NMR structures pre-miR-20b (orange) in complex with Rbfox RRM (green); comparison of loop conformations in free (C) and complex (D) forms; structural models of the free full-length pre-miR-20b (E) and the complex of full-length pre-miR-20b with Rbfox RRM (109–208) (F) showing how the highly conserved C-terminal tail of the RRM can reach down into the stem-loop to provide additional contacts that are not possible with single-stranded RNA. Nucleotides are colored by their normalized SHAPE reactivity.
Not only is the apical loop of free pre-miR-20b in a closed conformation, as suggested by SHAPE, but the five unpaired nucleotides (G28-U32) within the protein targeting sequence are rigidly ordered, forming a well-defined loop structure (local RMSD 0.80 Å, comparable to that of the rigid imperfect helix 0.69 Å, Figure 3A and Supplementary Table S3). The G29, A31 and U32 bases are splayed out with their Watson–Crick face pointing outward to the solvent, while G28 continues helical stacking on top of U27 and the base of C30 faces inward toward the minor groove (Figure 3C).
This loop conformation would not allow binding of the Rbfox RRM, which must therefore reorganize the loop structure and bind to it by an induced fit mechanism (44,45). Comparison of the SHAPE reactivity for pre-miR-20b in free form and in complex with Rbfox RRM indicates that protein binding indeed opens up the structure to generate a larger unpaired apical loop. We observe increased reactivity at many nucleotides (protein binding does not decrease accessibility to the chemical probe, see below), even at sites distant from RRM binding, namely at the G20 bulge and the 1 × 1 internal loop (G14·G45). Since Rbfox RRM binds single-stranded RNA and requires at least 5 nucleotides (GCAUG) for sequence-specific interaction (24), the opening at the G20 bulge and the 1 × 1 internal loop are not likely caused by second protein binding site.
The SHAPE data for the pre-miR-107 hairpin (Figure 2B) also indicate a structure inconsistent with computational predictions from miRBase (Supplementary Figure S1C), but in a manner opposite pre-miR-20b. In the free RNA (Figure 2D), nucleotides predicted to form a short four-base-pair terminal helix, as well as those in the TL, have high SHAPE reactivity, indicating that nucleotides U26 to U37 are in a large open loop that exposes both the GCAUG motif targeted by Rbfox and the UGU motif that contributes to the accuracy of Drosha processing (46). Additional high SHAPE reactivity was observed at the C21 and U7 bulges, but the predicted 1 × 1 internal loop formed by U10 and C52 has very low SHAPE reactivity, indicating a well-ordered U10·C52 mismatch at this position, similar to the U25·C35 mismatch in pre-miR-20b. Upon Rbfox RRM binding, the pre-miR-107 hairpin has reduced SHAPE reactivity (Figure 2B), especially in the terminal stem-loop region containing the UGU motif, which becomes base-paired and rigidified (Figure 2D). This indicates an opposite effect from what we observed with pre-miR-20b, yet in both cases we observe reduction of processing efficiency. The implications of this observation are discussed later.
Structural basis for recognition of pre-miR-20b by the Rbfox RRM
We next determined the NMR structure of pre-miR-20b both free and in complex with the Rbfox RRM. Since we observed a very large number of intermolecular NOE restraints (197, a large number for a complex of this size; Supplementary Table S3), the RMS deviation of the heavy atoms for ordered parts of both RNA and protein (G29-A34 and Pro116-Arg194) is low, 0.90 Å (Figure 3B), enabling confident structural analysis.
The key structural features reported for the Rbfox1 RRM in complex with single-stranded UGCAUGU (24) are retained in our structure as well and are described in detail in the supplementary material, but the different RNA structural context adds new elements to the complex. First, several local features of the protein–RNA interface differ, as described in detail in the supplementary results (Supplementary Figure S8). Second, the β2β3 loop of the Rbfox RRM is inserted into the opened-up TL of pre-miR-20b, anchoring the RNA loop-helix junction to the protein surface in a manner reminiscent of the structure of the U1A protein–RNA complex (47), which is absent in the complex structure between Rbfox RRM with single-stranded RNA (Supplementary Figure S8). Unlike U1A, this interaction might not be optimized for binding to a specific RNA stem-loop; as a consequence, secondary structure reduces binding of Rbfox2 (26), while U1A binds much more strongly to a stem loop compared to single-stranded RNA with the same sequence (47). This feature leads to a reorganization of the RNA, which opens up the pre-structured RNA loop to allow recognition of the single-stranded nucleotides (Figure 3C and D), but also masks a critical structural element recognized by DGCR8. In the process, the base of A31 rotates about 180 degree to base pair with the base of G29, which slightly tilts inward, while the base of G33 flips outward to stack with the aromatic ring of Phe160 of Rbfox, switching from the anti to syn conformation. Third, the four base pairs in the apical stem of pre-miR-20b immediately below the loop are broken up to increase loop size drastically, which inevitably reduces the local quality of the NMR structure as reflected in the superpositions (Figure 3B).
Unexpectedly, protein binding at the TLs of pre-miR-20b and -107 cause long-range effect on the SHAPE reactivity of nucleotides further down the stem, in mismatches (G14/G45 in pre-miR-20b) and bulged nucleotide (U7 in pre-miR-107), away from the expected protein binding site (Figure 2C and D). We also found by NMR that protein recognition extends to the very end of the stem-loop, including the artificial G19·C41 closing base pair that was added to increase transcription. Intermolecular NOE connections were observed between G19 and C41 to the side chains of Tyr205 and Thr206, located at the very C-terminal end of our Rbfox RRM construct.
Sequence alignment of Rbfox family proteins revealed that sequence conservation extends beyond the canonical RRM to Ala225 (Supplementary Figure S3A), suggesting the possibility of additional interactions between the conserved C-terminal end of Rbfox RRM and the lower stem of pre-miR-20b. We compared RNA binding of Rbfox1 RRMs with variable C-terminal lengths (Supplementary Figure S3B) using ITC (Supplementary Figure S4). Among the three RRM constructs tested, the median-length Rbfox1 RRM (109–208), which we have used in the other experiments reported in the paper, binds pre-miR-20b 2-fold tighter than the RRM (109–194) alone without a C-terminal tail; by comparison, the Rbfox1 RRM (109–225) with an extended C-terminal tail binds twice as strongly as the RRM (109–208) with a much larger enthalpy change (−22 kcal·mol−1). The results suggest the formation of additional intermolecular interactions, such as stacking between protein aromatic side chains and RNA bases (48). In addition, we found that two more base pairs G23·C37 and A22·U38 were opened up upon binding of this longer Rbfox RRM (109–225) construct. Finally, paramagnetic relaxation enhancement (PRE) data confirmed the long-range contacts we predicted (Supplementary Figure S9). We conclude that the conserved C-terminus of the Rbfox RRM forms long-range interaction with the lower stem of pre-miR-20b. Structural models based on SHAPE and NMR data were generated to show how Rbfox1 RRM (109–208) interacts with full-length pre-miR-20b and its effect on RNA conformations (Figure 3E and F).
Thus, SHAPE and NMR data demonstrate that the conserved C-terminal tail of the Rbfox RRM interacts with the stem region of pre-miR-20b and affects the stability and dynamics of bulges and internal loops, in regions of the structure that affect the efficiency of Drosha cleavage. The interaction between the conserved C-terminal tail of Rbfox RRM and the stem region of pre-miR-20b also suggests that Rbfox proteins may be suited to bind to the loop sequence of stem-loop RNAs, and not just to single-stranded RNA sequences encountered within pre-mRNAs.
Rbfox2 protein inhibits miR-20b and miR-107 maturation in mammalian cells
As a pre-requisite to analyzing the effect of Rbfox proteins on miR-20b and miR-107 expression in vivo, we examined the levels of endogenous full-length Rbfox1 and Rbfox2 proteins as well as mature miR-20b (miR-20b-5p) and -107 in different cell lines by Western blot and qRT-PCR, respectively. Rbfox1 is expressed mainly in the mouse motor neuron cell line NSC-34, while Rbfox2 protein is highly expressed in HEK293 and SHSY-5Y cells but is very low in HeLa and MCF7 cells (Supplementary Figure S5A). The qRT-PCR analysis conducted in HeLa, HEK293, MCF7 and SHSY-5Y cells show that expression levels of miR-20b and miR-107 are both significantly higher in SHSY-5Y cells compared to HeLa cells (Supplementary Figure S5B). Based on these expression patterns, we focused on Rbfox2 rather than Rbfox1 in cell analysis, because it is more widely expressed, and investigated whether siRNA knock-down would alter the expression levels of mature miR-20b and miR-107 in SHSY-5Y cells. In these cells, endogenous Rbfox2 is highly expressed and, at the same time, endogenous Rbfox1 is not detected, preventing a potential cross-talk between the two proteins. MCF7 cells, where Rbfox2 is very low and Rbfox1 is absent, were used instead for protein overexpression experiments.
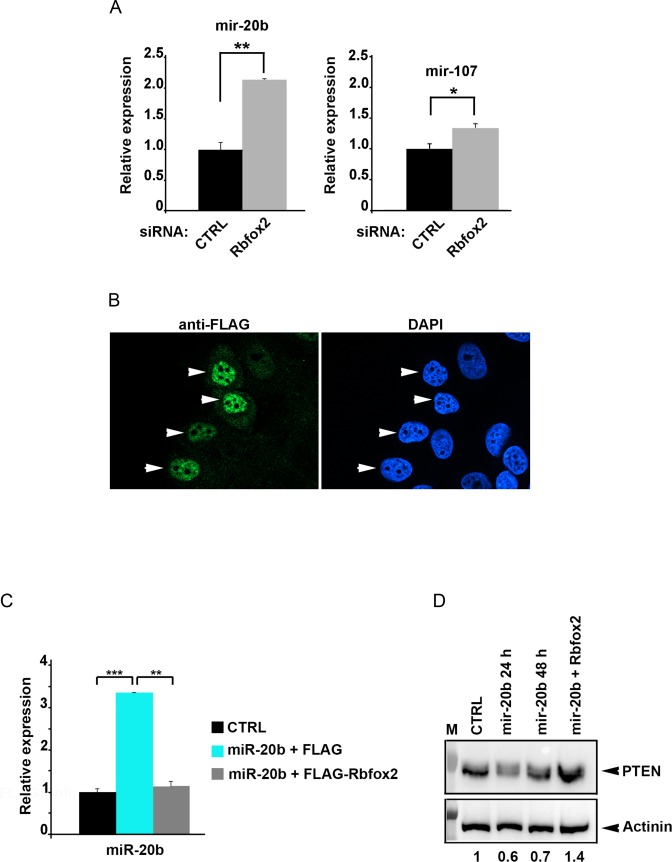
When Rbfox2 was efficiently downregulated by RNAi (compared to a scrambled control, Supplementary Figure S6A) in SHSY-5Y cells, we found significantly increased levels of miR-20b (Figure 4A). MiR-107 was also upregulated, but to a less significant level (Figure 4A). Immunoblot analysis showed that FLAG-Rbfox2 is efficiently expressed and its level is stable in MCF7 cells (Supplementary Figure S6B). When we assessed the intracellular localization of the recombinant Rbfox2 by performing immunostaining experiments, we found that the exogenously expressed FLAG-Rbfox2 localizes predominantly to the nucleus (Figure 4B), confirming that any effect on miRNA processing would occur at the nuclear step. Therefore, we transfected MCF7 with a plasmid containing primary miR-20b, not the pre-miRNA, and observed >3-fold increase in mature miR-20b levels by qRT-PCR. Over-expressing FLAG-Rbfox2 by co-transfection significantly decreased mature miR-20b levels compared with cells expressing miR-20b alone (Figure 4C). Similar results following over-expression of Rbfox2 were obtained in HeLa cells as well, using a luciferase reporter assay to monitor the effect of Rbfox2 expression on a reporter gene carrying miR-20b binding sites within the 3′-UTR (data not shown). Altogether, these data show that Rbfox2 protein interacts with pre-miRNAs carrying Rbfox binding sites within cells, confirming the in vitro data, and that mis-expression of Rbfox2 alters the biogenesis of both miR-20b and miR-107 in vivo.
Figure 4.
Rbfox2 downregulates mature miR-20b and upregulates miR-20b downstream targets (A) Quantitative RT-PCR of mature miR-20b in SHSY5Y cells. Cells were collected 48 h after siRNA transfection with anti-Rbfox2 siRNA. CTRL: cells transfected with scrambled siRNA. n = 3 (biological replicates, cell cultures), average ± s.e.m., **P < 0.01, *P < 0.05 (one way ANOVA statistical test). (B) Immunofluorescence micrographs showing nuclear localization of FLAG-Rbfox2 expressed in MCF7 cells. Exogenously expressed FLAG-Rbfox2 was detected 48 h after transfection by immunostaining as indicated. Nuclei are stained by DAPI. (C) qRT-PCR of mature miR-20b in MCF7 cells. CTRL: empty vector; miR-20b: cells collected 24 h after transfection of vector containing pri-miR-20b; miR-20b + FLAG-Rbfox2: Day 0—cells were seeded; Day 1—FLAG-Rbfox2 plasmid transfection; Day 2—pri-miR-20b vector transfection (into the same well transfected at day 1 with FLAG-Rbfox2); Day 3—cell harvest. n = 3 (biological replicates, cell cultures), average ± s.e.m., **P < 0.01, ***P < 0.001 (one way ANOVA statistical test). (D) Immunoblot analysis of endogenous PTEN protein in MCF7 cells. CTRL: empty vector; miR-20b 24 and 48 h: cells collected 24 or 48 h after transfection of vector containing pri-miR-20b; miR-20b + Rbfox2: Day 0—cells were seeded; Day 1—FLAG-Rbfox2 plasmid transfection; Day2—pri-miR-20b plasmid transfection (into the same well transfected at day 1 with FLAG-Rbfox2); Day 3—cells harvested. α-actinin serves as a loading control. The levels of PTEN were quantified by densitometry and the normalized level in the presence of control vector was set to 1.
Over-expression of Rbfox2 upregulates PTEN protein expression
One of the target mRNA for miR-20b encodes the PTEN protein, whose misregulation leads to the development of many types of human cancers (49,50). We hypothesized that the expression of PTEN, a representative downstream target of miR-20b, would be regulated by Rbfox2 through miR-20b. In order to confirm this hypothesis, we determined the expression of PTEN protein in MCF7 cells (with low endogenous levels of miR-20b and Rbfox2) by transfecting with a plasmid expressing primary miR-20b (pri-miR-20b) either alone or together with the FLAG-Rbfox2 expression plasmid. As shown in Figure 4D, miR-20b overexpression significantly decreases the levels of PTEN protein. Conversely, the expression of PTEN protein is significantly increased in cells transfected with plasmid containing primary miR-20b together with the FLAG-Rbfox2 expression plasmid, compared to cells treated only with control miRNA plasmid at 48 h post transfection. The effects on PTEN expression are much larger than for the RNA levels, but this is commonly observed, where modest (2–4-fold) effects on miRNA expression lead to much larger downstream effects. These results indicate that Rbfox2 protein regulates mature miR-20b expression levels in a manner that is inversely correlated with PTEN protein expression.
Downregulation of Rbfox2 reduces Dicer protein levels
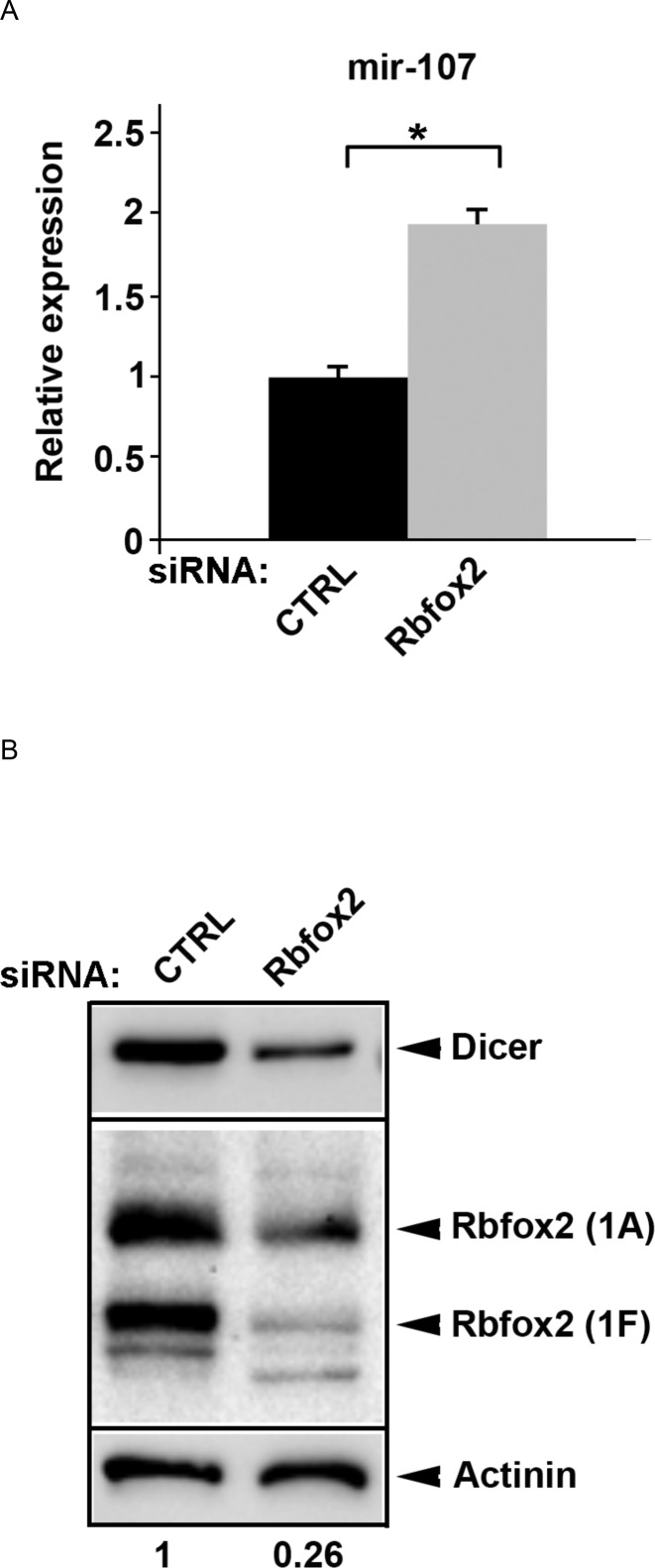
Rbfox2 expression in different cancer cells is linked to their epithelial/mesenchymal characteristics (51). In human breast cancer, high levels of miR-103/107 family miRNAs are associated with metastasis and poor outcome (52). Moreover, the miR-103/107 family globally attenuates miRNA biosynthesis by downregulating Dicer (53,54). It has been demonstrated that miR-107 directly targets the 3′UTR of Dicer mRNA (52–54). These observations prompted us to investigate whether downregulation of Rbfox2 would alter expression levels of Dicer, the downstream target of miR-107, in two cancer cell lines: a non-invasive breast cancer cell line (MCF7) and a more aggressive breast cancer cell line (mda-mb-231). By western blot, we confirmed that Rbfox2 protein expression is significantly higher in the more metastatic mda-mb-231 cell line (Supplementary Figure S7A). By qRT-PCR analysis, we observed higher expression levels of miR-107 and miR-20b in mda-mb-231 cells compared to MCF7 cells (Supplementary Figure S7B). When Rbfox2 is efficiently downregulated by RNAi compared to a scrambled control in mda-mb-231 cells, we found that knockdown of Rbfox2 protein significantly increased the level of miR-107 (Figure 5A), as compared to miR-20b which is increased to a less significant level (Supplementary Figure S7C). As we hypothesized, knockdown of Rbfox2 significantly decreased Dicer protein expression levels in mda-mb-231 cells (Figure 5B), as well as in 4TO7 mouse cell lines (Supplementary Figure S7D and E). These results demonstrate for the first time that Rbfox2 regulates Dicer expression by modulating the levels of miR-107 in a manner that correlates with the invasiveness of breast cancer cell lines.
Figure 5.
Effects of Rbfox2 on the levels of mature miR-107 and its downstream target Dicer in mda-mb-231 cells. (A) qRT-PCR of mature miR-107. Transfected siRNAs are as indicated. Rbfox2 siRNA: siRNA against Rbfox2; CTRL siRNA: non-targeting siRNA. n = 3 (biological replicates, cell cultures), average ± s.e.m., *P < 0.05 (one way ANOVA statistical test). (B) Immunoblot analysis of Dicer and Rbfox2 proteins. Rbfox2–1A and Rbfox2–1F represent two different isoforms of this protein (75). Actinin serves as a loading control.
DISCUSSION
We report herein that Rbfox family proteins regulate miRNA biogenesis by specifically targeting their cognate sequence within miRNA precursors through their conserved RRMs. Among the miRNAs directly regulated by Rbfox proteins are miR-20b and miR-107, the latter of which regulates Dicer enzyme, providing an indirect mechanism by which Rbfox proteins regulate cellular miRNA levels. Although a recent manuscript suggested that Rbfox3 contributes to miRNA biogenesis independent of the presence of an Rbfox target sequence (23), Rbfox2 crosslinks predominantly at or near sites carrying canonical target sequences and their function in vivo is strongly correlated with and dependent on their intrinsic RNA-binding activity (26). In fact, the use of negative control library in cross-linking, which was proven critical for filtering non-specific signals (55), made it possible to detect predominantly high-fidelity binding to conserved loci containing the consensus (U)GCAUG motif (25). Since the RRMs of Rbfox1, 2 and 3 have identical RNA-binding sequence specificity, we would expect the same behavior to hold for Rbfox3 as well. Consistent with the latter study, we found that abolishing binding to RNA also prevents Rbfox RRM from inhibiting processing of miR-20b and miR-107. These results indicate that at least some of the biological functions of these tissue-specific alternative splicing factors are due to their effect on miRNA processing either by directly sequence-specific binding to miRNA precursors or, indirectly, by altering Dicer levels.
A computational search of the Rbfox target sequence within human precursor miRNA hairpins identified >120 miRNAs containing this sequence motif. We have thus far investigated only miR-20b and miR107, because many of the other miRNAs identified have low cellular abundance and unclear biological role, but it is likely that more of the miRNAs listed in Supplementary Tables S1 and 2 are also regulated by Rbfox proteins. Rbfox RRM binds more strongly to pri-miR-20b compared to pri-miR-107, although the target sequence is exposed in the latter. We do not have an explanation for this observation, which would require a complete structural analysis of both complexes. However, both Rbfox RRM alone and the full-length Rbfox2 protein inhibit pri-miR-20b and -107 maturation in biochemical assays and in several cell lines.
How would binding of the Rbfox RRM, which occupies a region of the RNA that is not recognized directly by the microprocessor complex (56) suppress processing efficiency? A large and flexible TL has been proposed to promote Drosha and Dicer processing, perhaps by enhancing enzyme turnover (57), but we found that pre-miR-20b has a closed terminal hairpin structure, which is opened by the binding of Rbfox RRM. In contrast, the conformation of the pre-miR-107 TL is rigidified by the binding of RRM. Yet in both cases, we observe reduced miRNA processing, suggesting that loop size per se does not matter. The results for miR-107 can be interpreted by observing that a critical sequence motif promoting processing, the apical UGU motif (46,58) recognized by the RHED domain of DGCR8 (56,59) becomes less accessible when Rbfox binds to it (Figure 2D), as judged by SHAPE reactivity. However, the results for miR-20b processing cannot be interpreted by analysis of sequence features alone, because the stem loop of miR-20b precursor lacks the UGU and other sequence motifs required for efficient processing. Instead, it requires a more careful analysis of the structure. First, the Rbfox β2β3 loop inserts itself into the apical stem-loop junction where the RHED domain of DGCR8 binds, thus effectively competing with the RHED domain (56,59). Second, we observe both local and long-range effects on the pre-miR-20b structure and dynamics mediated both by the RRM itself and by a conserved C-terminal tail that augments the RRM. Binding of Rbfox RRM makes the stem-loop structure less regular by disrupting several base pairs and by opening up an internal loop (Figure 2C), making the RNA a less ideal substrates for Drosha/DGCR8 (58). The stem-loop structure of miRNA precursors is important for post-transcriptional regulation of miRNA processing (11,60–62), especially for miRNAs that lack positive sequence motifs like UGU and the mismatched GHG (which is paired and inaccessible in both pri-miR-20b and pri-miR-107). A less perfect double helical region, distorted and opened up by the Rbfox RRM, would interact less optimally with the double-stranded RNA-binding domain of DGCR8 (56,58).
Concerning the cellular consequences of our observations, we have shown that over-expression of Rbfox2 downregulates mature miR-20b and enhances PTEN expression (Figure 4D), a known tumor suppressor target of miR-20b that inhibits cell proliferation and migration. We chose to investigate Rbfox2 in part because it is more widely expressed than the other family members, and because it is essential to the survival of embryonic stem cells and for the progression of several cancers (16). Expression of Rbfox2 is down-regulated in ovarian cancer and its splicing is altered in breast cancer samples as well (22). The levels of miR-20b, a member of the miR-17 family of oncomirs (63,64), are significantly altered in multiple cancers as well (49–50,64). Cells are very sensitive to even subtle decreases in PTEN abundance, highlighting the importance of miRNA-mediated PTEN regulation in cancer (65) which we here show is controlled by Rbfox2, upstream of miR-20b. The role of Rbfox2 on PTEN might be even more pervasive. In addition to miR-20b, >10 other miRNAs targeting the 3′-UTR of PTEN have GCAUG or GCACG sequence motifs within their pre-miRNA hairpins (Supplementary Table S4), including some that have been validated to target PTEN.
Our discovery that Rbfox2 downregulates Dicer protein expression through miR-107 provides new insight on the role of this protein family in cancer and other diseases. MiR-107 belongs to the miR-103/107 family with identical seed sequences and very high levels of expression in brain tissue (66). Dysregulation of miR-107 is linked to cancers, obesity (through lipid metabolism) and neurological diseases (52,67–68). The miR-103/107 family attenuates miRNA biosynthesis by downregulating Dicer (53), thereby influencing cell-cycle progression, senescence, stem cell maintenance and tumorigenesis (69,70). Despite the critical function of this enzyme, the mechanisms that regulate Dicer expression are still poorly understood. Dicer mRNA transcription is positively regulated by Tap63 in mice (71), post-transcriptionally suppressed by let-7 and miR-103/107 (52,72) and by the RNA binding protein AUF1, highly expressed in several solid tumors (73,74). We show here that Rbfox2 acts upstream of miR-107 to control Dicer levels, which would have broad effects on the miRNA population in response to altered Rbfox2 expression or alternative splicing.
In summary, we report that Rbfox family proteins modulate the expression of miRNA through a direct sequence-specific interaction with pri-miRNAs carrying Rbfox binding sites. In addition, by regulating cellular levels of Dicer through miR-107, Rbfox2 might have broad influence on gene expression patterns. These functions are in addition to the known role in alternative splicing; suggest a more pervasive role of Rbfox2-mediated regulation of gene expression through its effect on miRNAs. Future work will uncover the full spectrum of regulatory and functional Rbfox2-mediated miRNA regulation.
ACCESSION NUMBERS
Atomic coordinates and related NMR restraints of pre-miR-20b and its complex with Rbfox RRM have been deposited in the Protein Data Bank as 2n7x and 2n82.
Supplementary Material
Acknowledgments
The authors thank Analytical Biopharmacy Core for using ITC in the School of Pharmacy at the University of Washington. We thank Prof. Alberto Inga for MCF7, mda-mb-231 cells and E-cadherin antibody and Prof. Stefano Piccolo for kindly providing the 4TO7 cells. The authors are grateful for Jana Mandic and other members of the laboratory for technical assistance.
Author contributions: Y. C., L. Z., P. M. and G. V. designed the project. Experiments were conducted by Y. C., L. Z., F. Y., K. G. and T. P.; and Y. C., L. Z., F. Y., P. M. and G. V. wrote the paper.
Footnotes
Present address: Yu Chen, Seattle Children's Research Institute, Seattle, WA 98101, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. National Institutes of Health [1R01 GM103834 to G.V.]; University of Trento (Progetto Biotecnologie); Autonomous Province of Trento (Madelena Project) (to P.M.). Funding for open access charge: National Institutes of Health [1R01 GM103834 to G.V.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 3.Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 4.Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman R.C., Farh K.K.H., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 8.van Kouwenhove M., Kedde M., Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 10.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 11.Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 12.Viswanathan S.R., Daley G.Q., Gregory R.I. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trabucchi M., Briata P., Garcia-Mayoral M., Haase A.D., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M.G. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michlewski G., Cáceres J.F. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michlewski G., Guil S., Semple C.a., Cáceres J.F. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell. Mol. Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibata H., Huynh D.P., Pulst S.M. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum. Mol. Genet. 2000;9:1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- 18.Underwood J.G., Boutz P.L., Dougherty J.D., Stoilov P., Black D.L. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim K.K., Adelstein R.S., Kawamoto S. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J. Biol. Chem. 2009;284:31052–31061. doi: 10.1074/jbc.M109.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakahata S., Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin Y., Suzuki H., Maegawa S., Endo H., Sugano S., Hashimoto K., Yasuda K., Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venables J.P., Klinck R., Koh C., Gervais-Bird J., Bramard A., Inkel L., Durand M., Couture S., Froehlich U., Lapointe E., et al. Cancer-associated regulation of alternative splicing. Nat. Struct. Mol. Biol. 2009;16:670–676. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- 23.Kim K.K., Yang Y., Zhu J., Adelstein R.S., Kawamoto S. Rbfox3 controls the biogenesis of a subset of microRNAs. Nat. Struct. Mol. Biol. 2014;21:901–910. doi: 10.1038/nsmb.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Auweter S.D., Fasan R., Reymond L., Underwood J.G., Black D.L., Pitsch S., Allain F.H.-T. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jangi M., Boutz P.L., Paul P., Sharp P.A. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes Dev. 2014;28:637–651. doi: 10.1101/gad.235770.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert N., Robertson A., Jangi M., McGeary S., Sharp P.A., Burge C.B. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell. 2014;54:887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song M.S., Salmena L., Pandolfi P.P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 28.Kozomara A., Griffiths-Jones S. MiRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkinson K.a., Merino E.J., Weeks K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 33.Das R., Laederach A., Pearlman S.M., Herschlag D., Altman R.B. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reuter J.S., Mathews D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parisien M., Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y., Fender J., Legassie J.D., Jarstfer M.B., Bryan T.M., Varani G. Structure of stem-loop IV of Tetrahymena telomerase RNA. EMBO J. 2006;25:3156–3166. doi: 10.1038/sj.emboj.7601195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins Struct. Funct. Genet. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 39.Cornell W.D., Cieplak P., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 40.Tenzer S., Moro A., Kuharev J., Francis A.C., Vidalino L., Provenzani A., Macchi P. Proteome-wide characterization of the RNA-binding protein RALY-interactome using the in vivo-biotinylation-pulldown-quant (iBioPQ) approach. J. Proteome Res. 2013;12:2869–2884. doi: 10.1021/pr400193j. [DOI] [PubMed] [Google Scholar]
- 41.Martin C.L., Duvall J.a., Ilkin Y., Simon J.S., Arreaza M.G., Wilkes K., Alvarez-Retuerto A., Whichello A., Powell C.M., Rao K., et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- 42.McGinnis J.L., Dunkle J.a., Cate J.H.D., Weeks K.M. The mechanisms of RNA SHAPE chemistry. J. Am. Chem. Soc. 2012;134:6617–6624. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gherghe C.M., Shajani Z., Wilkinson K.a, Varani G., Weeks K.M. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (s) in RNA strong correlation between SHAPE chemistry and the generalized NMR. J. Am. Chem. Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leulliot N., Varani G. Current topics in RNA-protein recognition: Control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 45.Williamson J.R. Induced fit in RNA-protein recognition. Nat. Struct. Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 46.Auyeung V.C., Ulitsky I., McGeary S.E., Bartel D.P. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allain F.H.T., Howe P.W.a, Neuhaus D., Varani G. Structural basis of the RNA-binding specificity of human U1A protein. EMBO J. 1997;16:5764–5774. doi: 10.1093/emboj/16.18.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLaughlin K.J., Jenkins J.L., Kielkopf C.L. Large favorable enthalpy changes drive specific RNA recognition by RNA recognition motif proteins. Biochemistry. 2011;50:1429–1431. doi: 10.1021/bi102057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou W., Shi G., Zhang Q., Wu Q., Li B., Zhang Z. MicroRNA-20b promotes cell growth of breast cancer cells partly via targeting phosphatase and tensin homologue (PTEN) Cell Biosci. 2014;4:62. doi: 10.1186/2045-3701-4-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu J., Chen L., Zou L., Yang P., Wu R., Mao Y., Zhou H., Li R., Wang K., Wang W., et al. MiR-20b, -21, and -130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Hum. Immunol. 2014;75:348–353. doi: 10.1016/j.humimm.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Venables J.P., Brosseau J.-P., Gadea G., Klinck R., Prinos P., Beaulieu J.-F., Lapointe E., Durand M., Thibault P., Tremblay K., et al. RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol. Cell. Biol. 2013;33:396–405. doi: 10.1128/MCB.01174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martello G., Rosato A., Ferrari F., Manfrin A., Cordenonsi M., Dupont S., Enzo E., Guzzardo V., Rondina M., Spruce T., et al. A microRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 53.Ristori E., Lopez-Ramirez M.A., Narayanan A., Hill-Teran G., Moro A., Calvo C.-F., Thomas J.-L., Nicoli S. A Dicer-miR-107 interaction regulates biogenesis of specific miRNAs crucial for neurogenesis. Dev. Cell. 2015;32:546–560. doi: 10.1016/j.devcel.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y., Mao L., Gao Y., Baral S., Zhou Y., Hu B. MicroRNA-107 contributes to post-stroke angiogenesis by targeting Dicer-1. Sci. Rep. 2015;5:13316. doi: 10.1038/srep13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Friedersdorf M.B., Keene J.D. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol. 2014;15:R2. doi: 10.1186/gb-2014-15-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen T.A., Jo M.H., Choi Y.-G., Park J., Kwon S.C., Hohng S., Kim V.N., Woo J.-S. Functional anatomy of the human microprocessor. Cell. 2015;161:1374–1387. doi: 10.1016/j.cell.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 57.Zhang X., Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010;38:7689–7697. doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang W., Bartel D.P. The menu of features that define primary MicroRNAs and enable de novo design of microRNA article the menu of features that define primary microRNAs and enable de novo design of microRNA genes. Mol. Cell. 2015;60:131–145. doi: 10.1016/j.molcel.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quick-Cleveland J., Jacob J.P., Weitz S.H., Shoffner G., Senturia R., Guo F. The DGCR8 RNA-binding heme domain recognizes primary microRNAs by clamping the hairpin. Cell Rep. 2014;7:1994–2005. doi: 10.1016/j.celrep.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu S., Jin L., Zhang Y., Huang Y., Zhang F., Valdmanis P.N., Kay M.A. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song L., Axtell M.J., Fedoroff N. V. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 2010;20:37–41. doi: 10.1016/j.cub.2009.10.076. [DOI] [PubMed] [Google Scholar]
- 62.Sperber H., Beem A., Shannon S., Jones R., Banik P., Chen Y., Ku S., Varani G., Yao S., Ruohola-Baker H. miRNA sensitivity to Drosha levels correlates with pre-miRNA secondary structure. RNA. 2014;20:621–631. doi: 10.1261/rna.043943.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olive V., Jiang I., He L. Mir-17–92, a cluster of miRNAs in the midst of the cancer network. Int. J. Biochem. Cell Biol. 2010;42:1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Landais S., Landry S., Legault P., Rassart E. Oncogenic potential of the miR-106–363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 65.Alimonti A., Carracedo A., Clohessy J.G., Trotman L.C., Nardella C., Egia A., Salmena L., Sampieri K., Haveman W.J., Brogi E., et al. Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilfred B.R., Wang W.X., Nelson P.T. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol. Genet. Metab. 2007;91:209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., Heim M.H., Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 68.Wang W.-X., Rajeev B.W., Stromberg A.J., Ren N., Tang G., Huang Q., Rigoutsos I., Nelson P.T. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mudhasani R., Zhu Z., Hutvagner G., Eischen C.M., Lyle S., Hall L.L., Lawrence J.B., Imbalzano A.N., Jones S.N. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J. Cell Biol. 2008;181:1055–1063. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Srikantan S., Marasa B.S., Becker K.G., Gorospe M., Abdelmohsen K. Paradoxical microRNAs: individual gene repressors, global translation enhancers. Cell Cycle. 2011;10:751–759. doi: 10.4161/cc.10.5.14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su X., Chakravarti D., Cho M.S., Liu L., Gi Y.J., Lin Y.-L., Leung M.L., El-Naggar A., Creighton C.J., Suraokar M.B., et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Forman J.J., Legesse-Miller A., Coller H.A. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ciafrè S.A., Galardi S. microRNAs and RNA-binding proteins. RNA Biol. 2014;10:934–942. doi: 10.4161/rna.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abdelmohsen K., Tominaga-Yamanaka K., Srikantan S., Yoon J.H., Kang M.J., Gorospe M. RNA-binding protein AUF1 represses Dicer expression. Nucleic Acids Res. 2012;40:11531–11544. doi: 10.1093/nar/gks930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Braeutigam C., Rago L., Rolke A., Waldmeier L., Christofori G., Winter J. The RNA-binding protein Rbfox2: an essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene. 2013;33:1082–1092. doi: 10.1038/onc.2013.50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.