Abstract
Escherichia coli single-stranded DNA binding protein (SSB) is an essential homotetramer that binds ssDNA and recruits multiple proteins to their sites of action during genomic maintenance. Each SSB subunit contains an N-terminal globular oligonucleotide/oligosaccharide binding fold (OB-fold) and an intrinsically disordered C-terminal domain. SSB binds ssDNA in multiple modes in vitro, including the fully wrapped (SSB)65 and (SSB)56 modes, in which ssDNA contacts all four OB-folds, and the highly cooperative (SSB)35 mode, in which ssDNA contacts an average of only two OB-folds. These modes can both be populated under physiological conditions. While these different modes might be used for different functions, this has been difficult to assess. Here we used a dimeric SSB construct with two covalently linked OB-folds to disable ssDNA binding in two of the four OB-folds thus preventing formation of fully wrapped DNA complexes in vitro, although they retain a wild-type-like, salt-dependent shift in cooperative binding to ssDNA. These variants complement wild-type SSB in vivo indicating that a fully wrapped mode is not essential for function. These results do not preclude a normal function for a fully wrapped mode, but do indicate that E. coli tolerates some flexibility with regards to its SSB binding modes.
INTRODUCTION
In order for DNA replication, repair and recombination to occur enzymes must unwind the DNA double helix to form single-stranded DNA (ssDNA) intermediates. During these processes, organisms in all kingdoms of life use single-stranded DNA binding proteins (SSBs) to protect these ssDNA intermediates (1–4). SSB proteins also serve as hubs to recruit multiple enzymes and accessory proteins that act on these intermediates during genome maintenance (5). Indeed, the E. coli SSB protein (EcSSB) interacts directly with at least 14 SSB interacting proteins (SIPs) (5).
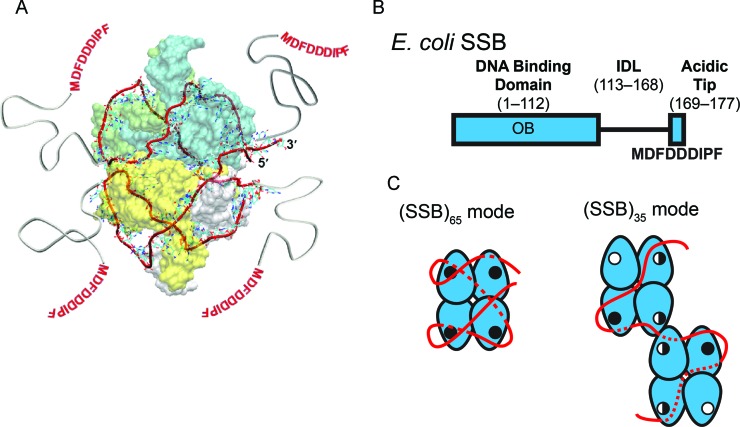
Wild-type EcSSB (wtSSB) is a D2-symmetric homotetramer (Figure 1A) (6–9), with each 177 amino acid (aa) subunit composed of two domains. The N-terminal domain, residues 1–112, forms an oligonucleotide/oligosaccharide binding (OB) fold containing an ssDNA binding site. The C-terminal domain contains an intrinsically disordered linker (IDL, residues 113–168) that connects the OB-fold to the terminal 9 aa acidic tip (MDFDDDIPF) which mediates interactions between SSB and SIPs (5) (Figure 1B).
Figure 1.
Structure and binding modes of EcSSB (A) Structural model of 65 nucleotides of ssDNA (red ribbon), wrapped around the EcSSB tetramer (8) in the (SSB)65 mode. (B) Domain organization of EcSSB, depicting the DNA binding domain (OB), the C-terminal IDL and the 9-residue acidic ‘tip.’ (C) The proposed ssDNA binding pathways of the (SSB)65 and (SSB)35 modes of EcSSB (8,12). The EcSSB tetramer is depicted in blue with the ssDNA in red. ssDNA that passes along the backside of the schematic is depicted as a dotted line. OB-fold binding sites are represented with either an open circle, for an unoccupied binding site; a half-closed circled, for a partially occupied binding site; or a closed circle, for a fully occupied binding site (see Discussion for details of this model). Two EcSSB tetramers are shown in the (SSB)35 mode to denote high cooperativity. The C-terminal tails are not depicted for clarity.
A consequence of the tetrameric structure of EcSSB is that it can bind ssDNA in multiple binding modes. The major binding modes at 25°C include the (SSB)65, (SSB)56 and (SSB)35 modes where each subscript indicates the number of nucleotides occluded by a bound SSB tetramer (10,11). In the (SSB)65 mode ssDNA interacts with all four SSB subunits with a topology that resembles the seams on a baseball (Figure 1A and C) (8,12). Conversely, in the (SSB)35 mode ssDNA interacts with an average of only two subunits of the tetramer (Figure 2C) (10,11,13,14). The (SSB)65 mode is favored at moderate monovalent salt concentrations (e.g., [NaCl] ≥ 200 mM) (10,11,15) as well as millimolar concentrations of Mg2+ and micromolar concentrations of polyamines (11,16,17). The (SSB)35 mode is stabilized at low monovalent salt concentrations ([NaCl] < 10 mM) (10,11), and by high protein binding densities (13–16,18–20). Since the properties of these different SSB binding modes differ significantly, it has been suggested that they might be used selectively in DNA replication, recombination and repair (3,5,21).
Figure 2.
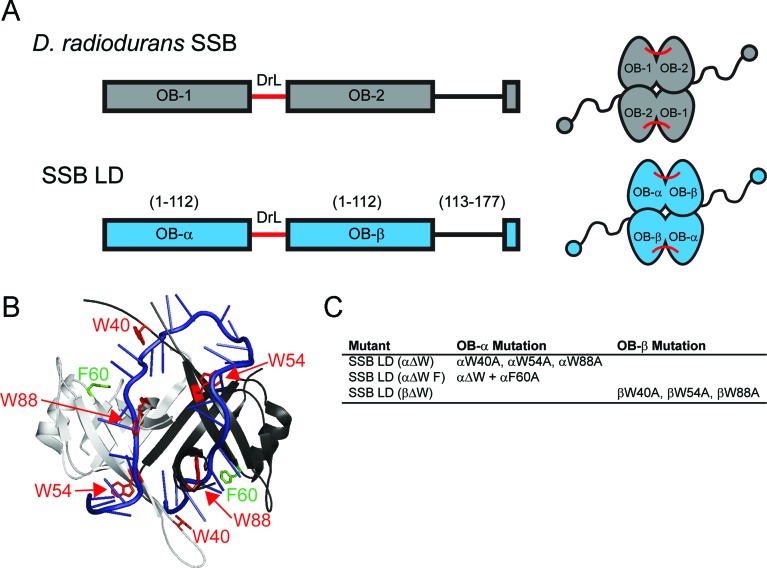
Design of SSB mutants with asymmetric ssDNA binding sites. (A) Schematic diagrams of DradSSB and SSB LD that uses the Drad linker (DrL, shown in red) to covalently link two identical EcSSB OB folds together (30). Left: Domain organization of individual subunits. Right: Final structures of folded proteins. (B) Crystal structure of two subunits of EcSSB (white and gray) bound to oligo dC (blue) (8). The positions of W40, W54 and W88 residues are indicated in red, and the positions of F60 residues are indicated in green. (C) Mutants designed to have decreased affinity for ssDNA binding in two of the four OB folds of SSB LD. For clarity, α and β prefixes are used to indicate in which OB fold the mutation(s) reside so that the amino acids could be referenced by wild-type EcSSB numbering.
Although SSB binds ssDNA with sub-nanomolar affinity depending on solution conditions (10,22), it is highly dynamic when bound to ssDNA. SSB is able to diffuse along ssDNA (12,23,24). This diffusion is functional as it provides the mechanism by which SSB can destabilize DNA hairpin structures (24,25). This ability to diffuse also stimulates RecA filament formation on natural ssDNA (24). SSB can even diffuse when bound to the RecO protein, a SIP that interacts with the C-terminal acidic tip (23). Recent studies have also suggested a further compaction of long ssDNA at higher monovalent salt concentrations beyond what is expected based on ssDNA wrapping in the (SSB)65 mode (26). In its (SSB)35 mode SSB binds with high inter-tetramer cooperativity, resulting in the formation of protein clusters on the DNA (10,11,19,27). This highly cooperative clustering requires the C-terminal IDLs and the number, length and composition of the IDL affects cooperativity (28–30). SSB in the (SSB)35 mode can also be transferred between different molecules or segments of ssDNA through a direct transfer mechanism (i.e., without a free protein intermediate) (31,32). These properties make the (SSB)35 mode an ideal candidate to function in DNA replication (32).
Under physiological conditions in vitro, all of the SSB binding modes can be populated and are in dynamic equilibrium on long ssDNA. Transitions from the fully wrapped to partially wrapped structures may function to allow other proteins, including SIPs, to transiently access the ssDNA. Since the C-terminal tails influence the transitions among SSB binding modes (28–30), interactions between the tails and SIPs may also affect the relative stabilities of the modes. In fact, recent studies suggest that the binding of two SIPs, PriA (33) and PriC (34), to a fully wrapped (SSB)65 complex promotes partial unwrapping of the ssDNA in the presence of excess SSB. These observations suggest a role for regulation of the SSB-DNA binding modes by the C-terminal tails, although the mechanism for this is not known.
To probe the functional importance of the different SSB-DNA binding modes it would be useful to have EcSSB variants that selectively inhibit one or more of the modes. Here we describe the design and characterization of SSB variants that are unable to form the (SSB)65 mode in vitro. To make these variants, we used a recently engineered SSB dimer in which each subunit possesses two covalently linked OB-folds (30) to create SSB mutants in which two of the four OB-folds have diminished affinity for ssDNA. As anticipated, these constructs are unable to form the fully wrapped (SSB)65 and (SSB)56 binding modes. These variants, however, still display a salt-dependent transition between high and low inter-tetramer cooperativity upon binding long ssDNAs. We also demonstrate that the formation of the fully wrapped modes is not essential for E. coli survival, but nonetheless are likely required for full SSB function.
MATERIALS AND METHODS
Reagents and buffers
All buffers were prepared with reagent-grade chemicals purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise noted. Water was treated with a MilliQ water purification system (Millipore; Bedford, MA, USA). Buffer T is 10 mM Tris (pH 8.1, at 25°C) and 0.1 mM Na3EDTA. TAE buffer was prepared from a 50× concentrated stock (G-Biosciences; St. Louis, MO, USA; catalog # R023) which was diluted to a 0.5× strength resulting in a solution of 20 mM Tris-Acetate (pH 8.3), 0.5 mM Na3EDTA.
Protein purification
wtSSB and the SSB LD proteins were purified as described (35) with the addition of a double-stranded DNA cellulose column to remove a potential nuclease contaminant (22) and the inclusion of 1× final concentration of a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA, Product # S8820) prior to lysing the cells. The proteins SSB LD (αΔW), SSB LD (αΔW F) and SSB LD (βΔW) were purified in the same way, except that they were loaded onto the single-stranded DNA cellulose column in [50 mM Tris (pH 8.3), 1 mM Na3EDTA, 100 mM NaCl and 10% glycerol] instead of the same buffer with 300 mM NaCl used to purify wtSSB and SSB LD. Protein concentrations were determined spectrophotometrically using ϵ280 = 1.13 × 105 M−1 (tetramer) cm−1 for wtSSB (10), ϵ280 = 1.08 × 105 M−1 (4-OB folds) cm−1 for SSB LD (30), and ϵ280 = 6.78 × 104 M−1 (4-OB folds) cm−1 for SSB LD (αΔW), SSB LD (αΔW F) and SSB LD (βΔW), as determined from their protein sequences (36).
DNA
The oligodeoxynucleotides (dT)35, (dT)70 and Cy5-(dT)68-Cy3-dT, were synthesized and purified as described (27). Poly(dT) was purchased from Midland Certified Reagent Company (Midland, TX, USA; Catalog #P-2004, Lot Number 071308) and was determined to have an average length of approximately 1000 nucleotides by analytic sedimentation velocity. M13mp18 single-stranded DNA was purchased from New England Biolabs (Ipswitch, MA, USA; catalog # N4040S). All ssDNA concentrations were determined spectrophotometrically using the extinction coefficient ϵ260 = 8.1 × 103 M−1 (nucleotide) cm−1 for oligo(dT) and poly(dT) (37), ϵ260 = 4.9 × 103 M−1cm−1 for Cy3, ϵ260 = 1.0 × 104 M−1cm−1 for Cy5 and ϵ259 = 7370 M−1cm−1 for M13 DNA (38).
Analytical sedimentation velocity
Sedimentation velocity experiments were performed using an Optima XL-A Analytical Ultracentrifuge equipped with an An50Ti rotor (Beckman Coulter; Fullerton, CA, USA) at 25°C. We measured the sedimentation properties of 750 nM (4-OB folds) of the proteins in Buffer T with both 2 mM NaCl (T2) and 300 mM NaCl (T300). The proteins were centrifuged at 42 000 rpm (25°C). The continuous sedimentation coefficient c(s) profiles, and the s20,w derived from these profiles, were calculated using the program SEDFIT (39).
Fluorescence measurements
Titrations of wtSSB, SSB LD, SSB LD (αΔW), SSB LD (αΔW F) and SSB LD (βΔW) with (dT)35 and poly(dT) were performed by monitoring quenching of the intrinsic tryptophan fluorescence of the proteins upon binding DNA using a PTI-QM-2000 spectrofluorometer (PTI Inc.; Lawrenceville, NJ, USA) using the excitation wavelength of 296 nm and monitoring emission fluorescence intensity at 350 nm as described (40–42).
Isotherms of the (dT)35 titrations were analyzed using either a single-site model or a two-site sequential model. The single-site model is given by Equation (1):
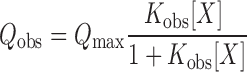 |
(1) |
Here, [X] is free ligand concentration, Qobs is the observed fluorescence tryptophan quenching at the ith titration point defined by (F0 – Fi)/F0, 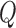 max is the fluorescence quenching at saturation of the macromolecule with ligand [in this case, the macromolecule is our SSB variant and the ligand is (dT)35] and Kobs is the observed association equilibrium constant. The concentration of free (dT)35,
max is the fluorescence quenching at saturation of the macromolecule with ligand [in this case, the macromolecule is our SSB variant and the ligand is (dT)35] and Kobs is the observed association equilibrium constant. The concentration of free (dT)35, 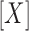 , is determined from the mass conservation given in Equation (2):
, is determined from the mass conservation given in Equation (2):
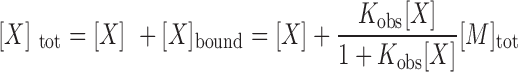 |
(2) |
Where [X]tot and [M]tot are the total concentrations of (dT)35 and SSB, respectively.
The two-site sequential model is described by Equation (3):
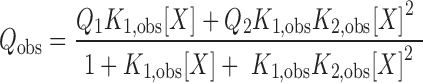 |
(3) |
Where Q1 and Q2 are the fluorescence quenching corresponding to one and two molecules of (dT)35 bound, respectively. K1,obs and K2,obs are the observed stepwise macroscopic association equilibrium constants for the first and second binding events. The concentration of free ligand was determined from Equation (4):
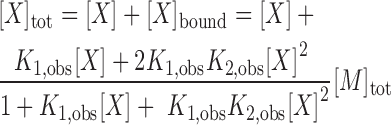 |
(4) |
Binding parameters were obtained from non-linear least squares fittings of the isotherms to Equations (1)–(4), performed using SCIENTIST (Micromath, St. Louis, MO, USA). Model-independent binding isotherms for the binding of SSB LD (αΔW F) to Cy5-(dT)68-Cy3-dT were performed and analyzed as described (42,43).
Isothermal titration calorimetry
ITC experiments were performed using a VP-ITC microcalorimeter (GE Inc.; Piscataway, NJ, USA) by titrating our SSB variants in the cell [0.93–2.96 μM (4-OB folds protein)] with DNA in the syringe [14.8–27.7 μM (molecules (dT)35)] (44,45). Prior to the experiment, the protein and DNA were extensively dialyzed into buffer T with NaCl concentrations as indicated. The reference heats were measured by titrating DNA into the appropriate buffer in the cell.
The ITC titration curves were fit either to a one-site model or a two-site sequential model to determine binding parameters. In the one-site model ligand [X = (dT)35] is bound to the macromolecule [M = SSB variant] to determine the the association equilibrium constant, Kobs, and binding enthalpy, ΔHobs. In the two-site sequential model, the binding events are described by the macroscopic association constants, K1,obs and K2,obs, and corresponding enthalpies, ΔH1,obs and ΔH2,obs. The data were fit using software provided by the ITC manufacturer as described (44,45).
ssDNA wrapping
To monitor the wrapping of ssDNA around our SSB variants, we titrated Cy5-(dT)68-Cy3-dT with SSB in buffer T with 300 mM NaCl at 25°C. The Cy3 fluorophore was excited at 515 nm and the Förster resonance energy transfer (FRET) to the Cy5 fluorophore emission was monitored at 670 nm as a function of SSB concentration. The resulting curves were analyzed with Equations (1)–(4). In this case, 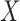 is the free SSB concentration,
is the free SSB concentration, 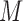 is the Cy5-(dT)68-Cy3-dT and the
is the Cy5-(dT)68-Cy3-dT and the 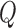 terms are the normalized Cy5 fluorescence enhancement, where Qobs = (Fi – F0)/F0.
terms are the normalized Cy5 fluorescence enhancement, where Qobs = (Fi – F0)/F0.
Agarose gel electrophoresis
Cooperativity of SSB-M13 ssDNA complexes was assessed by agarose gel electrophoresis as described (46) with one change (noted below). Briefly, increasing concentrations of protein were added to a constant amount of M13 ssDNA under the solution conditions indicated in the text. Electrophoresis in a 0.5% agarose gel was carried out in 0.5 × TAE buffer at room temperature (22°C) with constant voltage (∼1 V cm−1) for 3.5–4 h. The ssDNA was then stained for 30 min in 0.5 × TAE buffer with ethidium bromide (2 μg/ml) then destained for 30–60 min in 0.5 × TAE. We omitted a high-salt SSB dissociation step prior to ethidium bromide staining and imaged the gel with a Typhoon TRIO, Variable Mode Imager (GE Healthcare Life Sciences; Piscataway, NJ, USA) using the green (532 nm) excitation laser and the 610 BP 30 emission filter.
In vivo complementation experiments
SSB complementation (‘bumping’) experiments were performed as described (47). RPD317 is an E. coli strain with a KanR cassette in place of the wtssb gene deleted from the chromosome. Since SSB is an essential protein, it is necessary to express wtSSB ectopically from a helper plasmid with a TetR marker in order for RDP317 to survive. Plasmids expressing the SSB mutants of interest and containing an AmpR marker were transformed into RDP317. We selected transformants that grew on the LB agar plates with ampicillin (Amp, 50 μg ml−1) and kanamycin (Kan, 50 μg ml−1) and grew them overnight at 37°C in 2 ml LB media containing Amp and Kan. We then diluted this culture 1:1000 for growth the following day in fresh media. This was repeated at least four times; after the final passage the cultures were diluted and spread onto LB agar plates with either Kan and Amp or Kan and tetracycline (Tet, 10 μg ml−1). In this assay, SSB mutants that can complement loss of wtSSB, and therefore ‘bump’ the helper plasmid, display the TetS phenotype. Finally, mutant SSB expression plasmids from successfully bumped strains were sequenced to verify that no compensatory mutations had been made to the SSB mutant gene during the passages. Bumping experiments were performed at least twice for each variant.
SSB western blots
RDP317 cells containing the aforementioned mutants of interest from the bumping experiments were grown overnight in LB (supplemented with Amp, 50 μg ml−1 and Kan, 50 μg ml−1) at 37°C with shaking. To 100 μl of these saturated cultures was added 50 μl of 3× SDS PAGE loading buffer. The samples were boiled for 5 min then resolved on an Any kD Mini-PROTEAN TGX gel (Bio-Rad, Catalog #456-9036). Protein was then electrotransferred to a nylon membrane (Magna; Westboro, MA, USA; catalog No. N00HYB0010) and blocked with 5% (w:v) non-fat dry milk powder in TBST (50 mM Tris–HCl, 150 mM NaCl, 0.05% Tween-20, pH 7.4). The membranes were then labeled with a 1:15 000 dilution (in TBST 5% non-fat dry milk) of rabbit antibodies raised against EcSSB, then a 1:15 000 dilution of anti-rabbit antibodies conjugated with horseradish peroxidase (GE Healthcare Life Sciences, Catalog No. NA934). The bands were visualized using Immobilon Western Chemiluminescent HRP Substrate (Catalog No. WBKLS0050).
UV sensitivity measurements
Strains expressing the mutants of interest that resulted from the bumping experiments were tested for UV sensitivity. Overnight cultures were grown from freshly streaked plates and were spotted in 5 μl aliquots in serial dilutions onto LB plates supplemented with the appropriate antibiotics and were allowed to dry into the plate. The plates were then exposed to UV light from a Mineral Light Lamp (Model: UVGL-25, set to 254 nm) placed 4 inches above the surface of the plate. UV doses were calculated from the exposure time and the applied power of the 1.8 Wm−2 measured at this distance. After exposure, the plates were incubated overnight at 37°C.
Methyl methanesulfonate (MMS) sensitivity measurements
Sensitivities to chronic exposure of MMS were assessed as for UV sensitivity, except the overnight cultures were spotted onto plates that were dosed with the indicated concentrations of MMS and were then incubated overnight at 37°C.
RESULTS
Design of SSB variants that reduce ssDNA affinity to only two OB folds
To eliminate DNA binding in only two of the four ssDNA binding sites within the four OB-fold homotetrameric wtSSB we used an SSB construct in which two of the OB-folds are covalently linked. This construct, referred to as SSB linked dimer (SSB LD), was used previously to examine how many C-terminal tails are required for SSB function in vivo (30). SSB LD is based on the structure of the Deinococcus radiodurans SSB (DradSSB) that, like EcSSB, has four OB-folds, but is a homodimer rather than a homotetramer (Figure 2A) (48,49). Thus, while the OB-folds of EcSSB and DradSSB share significant structural homology (7–9,48,49) each DradSSB subunit contains two covalently linked OB-folds, but only one C-terminal tail (Figure 2A). In our SSB LD construct, one EcSSB N-terminal OB-fold is covalently linked to a full-length EcSSB subunit by a Drad C-terminal linker. This produces a subunit possessing two OB-folds with identical sequences that differ only in their proximity to the C-terminal tail. The N-terminal OB fold (α-OB) is attached directly to the Drad linker by its C-terminal end, while the C-terminal OB-fold (β-OB) maintains a wild-type C-terminal tail (Figure 2A) (30). The SSB LD variant complements wtSSB function in vivo (30).
The ssDNA binding site within one OB-fold of EcSSB includes a number of aromatic and basic amino acids (8). Residues of particular importance are: W40, W54, W88, F60, R3, R84, R86, K43, K62, K73 and K87. Individual mutations of these residues reduce SSB binding to ssDNA (50–54). W40 and W54 are observed to form stacking interactions with DNA bases in a co-crystal structure (8) and have been implicated in stacking interactions via spectroscopic methods (51,52). Mutation of W54 to serine produces a protein that complements loss of wild-type SSB in vivo but results in a strain that is impaired in growth and more sensitive to UV irradiation (55,56). SSB W54S forms a stable homotetramer in solution but has a biased preference for the (SSB)35 mode in vitro (53,57). While W88 is not implicated in stacking interactions, mutations of W88 influence binding stoichiometries with poly(dT), at least in the presence of a W54 mutation (53). In addition, a Δssb strain expressing the ssbW88T gene on a plasmid is hypersensitive to UV irradiation, underscoring its functional importance (55). F60 is positioned to interact with ssDNA (8) and has been shown to be involved with ssDNA binding via crosslinking (58) and mutational studies (50,59).
Mutants in which ssDNA binding is inhibited in two OB folds bind only one molecule of (dT)35
To eliminate ssDNA binding in only two OB-folds within SSB LD, we introduced three mutations, W40A, W54A, W88A, in either the α-OB folds, SSB LD (αΔW) or the β-OB folds, SSB LD (βΔW), (Figure 2B and C). We note that a triple mutant (W40T, W54S and W88T) present in all four OB folds of SSB binds weakly to poly(dT) in 0.3 M NaCl (53). In the second variant, we combined the three ΔW mutations with an F60A mutation in the α OB-fold to create SSB LD (αΔW F) (Figure 2B and C). We overexpressed and purified wtSSB, SSB LD, SSB LD (αΔW), SSB LD (βΔW) and SSB LD (αΔW F) (Supplementary Figure S1). Sedimentation velocity studies at [4-OB folds] = 750 nM, in Buffer T under both low salt (2 mM NaCl) and high salt (300 mM NaCl) conditions at 25°C indicate that all variants have hydrodynamic properties similar to SSB LD and wtSSB (Table 1). Hence, these linked-dimer mutants form homodimers in solution with wild-type-like configurations of their DNA binding cores.
Table 1. Hydrodynamic analysis of SSB variants at 25°C in buffer T.
| Protein | 300 mM NaCl | 2 mM NaCl | ||
|---|---|---|---|---|
| s20,w | f/f0 | s20,w | f/f0 | |
| SSB | 4.23 | 1.51 | 4.51 | 1.49 |
| SSB LD | 4.13 | 1.43 | 4.31 | 1.42 |
| SSB LD (αΔW) | 4.02 | 1.43 | 4.25 | 1.42 |
| SSB LD (αΔW F) | 3.81 | 1.61 | 4.15 | 1.39 |
| βΔW | 4.00 | 1.43 | 4.24 | 1.45 |
Analysis was performed via analytical ultracentrifugation sedimentation velocity at [4-OB] = 750 nM. Each variant predominately formed the 4-OB fold species yielding molecular weights within 10% of their expected values in moderate salt and in low salt. Most experiments performed on variants of SSB LD had some amount of a high molecular weight species present that corresponded to the MW of a dimer of the 4-OB fold species. This high molecular weight species never accounted for more than 4% of the total signal in the experiment.
The four OB folds in the wtSSB tetramer can bind two molecules of (dT)35 at saturation (13,14,20,45). Binding of the first (dT)35 occurs with high affinity while the second (dT)35 binds with lower affinity due to intra-tetramer negative cooperativity that becomes more pronounced at lower salt concentrations (13,14,20). Therefore, the second molecule of (dT)35 binds more readily at higher salt concentrations that favor the fully wrapped (SSB)65 mode, than at lower salt concentrations that favor the (SSB)35 mode. If the SSB variants successfully eliminate ssDNA binding to two OB folds, we expect to observe binding of only one (dT)35 under all solution conditions, even at high salt concentrations that promote exclusive binding of wtSSB in the (SSB)65 mode.
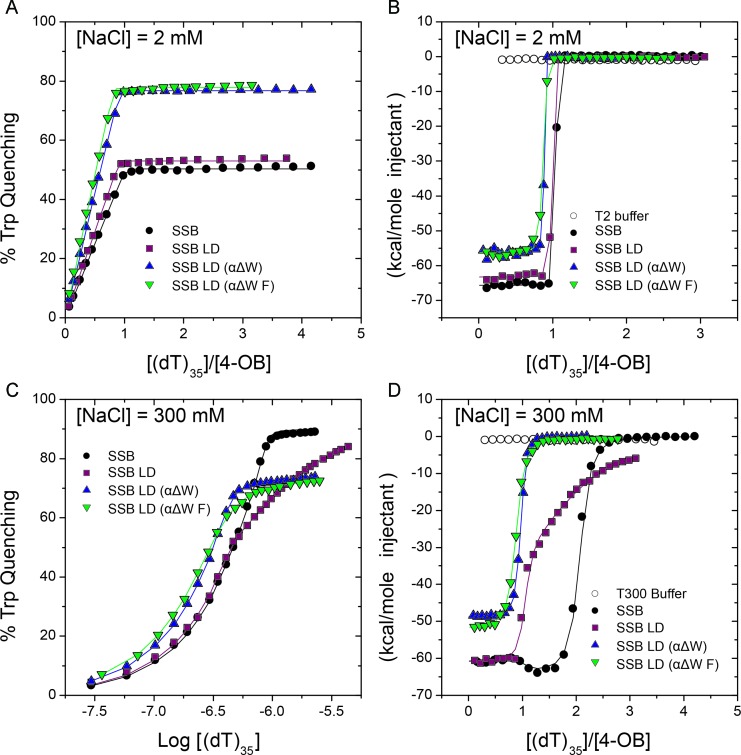
We first performed titrations of (dT)35 into wtSSB, SSB LD, SSB LD (αΔW) and SSB LD (αΔW F) under low salt (2 mM NaCl) conditions and monitored binding by the quenching of the intrinsic SSB tryptophan fluorescence (Figure 3A, Table 2). Under these conditions, we observe only a single, high affinity binding event with wtSSB (K1,obs ≥ 1010 M−1), consistent with previous reports (13,14,20), as well as with SSB LD and SSB LD (αΔW) and SSB LD (αΔW F) (Figure 3A and Table 2). SSB LD (αΔW) and SSB LD (αΔW F) show maximum Trp quenchings of 76.8 ± 0.1% and 77.8 ± 0.3%, respectively, upon binding one (dT)35. The higher extent of quenching is consistent with a higher fraction of Trp residues affected by binding.
Figure 3.
SSB variants with two-mutated ssDNA binding sites are able to only bind one molecule of (dT)35. (A) Intrinsic Trp fluorescence quenching titrations show that under (SSB)35 stabilizing conditions ([NaCl] = 2 mM) SSB, SSB LD, SSB LD (αΔW) and SSB LD (αΔW F) are only able to bind one molecule of (dT)35. (B) ITC experiments titrating (dT)35 into SSB mutants in 2 mM NaCl independently demonstrate the conclusions in (A). Further, this analysis suggests that binding of a single (dT)35 to SSB LD (αΔW) and SSB LD (αΔW F) is less enthalpically favorable than binding to SSB or SSB LD. (C) Fluorescence titrations under (SSB)65 stabilizing conditions ([NaCl] = 300 mM). SSB and SSB LD are able to bind two molecules of (dT)35 with a decrease in affinity for binding to the second molecule of (dT)35 (negative cooperativity). Isotherms of SSB LD (αΔW) and SSB LD (αΔW F), however, show that these mutants are only able to bind one molecule of (dT)35 under these conditions at saturation. In these titrations, and the titrations in (A), DNA was titrated into 0.5 μM of protein. See Table 2 for the binding parameters for (A) and (C). (D) ITC experiments performed in 300 mM NaCl agree with fluorescence titrations shown in (C). In this series, the changes in enthalpy associated with binding to (dT)35 suggest sequential binding of two molecules (dT)35 to SSB and SSB LD with negative cooperativity for binding of the second (dT)35. Isotherms of SSB LD (αΔW) and SSB LD (αΔW F) demonstrate that these proteins only bind one molecule of (dT)35. See Table 3 for the binding parameters for (B) and (D).
Table 2. Intrinsic tryptophan quenching titrations show that SSB variants with two-OB folds mutated to block ssDNA binding can only bind to one molecule of (dT)35.
| Protein | Model | [NaCl] (mM) | Q1,obs (%) | K1,obs (M−1) | Q2,obs (%) | K2,obs (M−1) |
|---|---|---|---|---|---|---|
| wtSSB | One to one | 2 | 50.3 ± 0.1 | ≥1010 | ||
| wtSSB | Two to one | 300 | 50† | ≥1010 | 90† | (2.1 ± 0.8) × 108 |
| SSB LD | One to one | 2 | 53.1 ± 0.1 | ≥1010 | ||
| SSB LD | Two to one | 300 | 53.1 ± 0.7 | ≥109 | 89.1 ± 0.7 | (1.6 ± 0.2) × 106 |
| SSB LD (αΔW) | One to one | 2 | 76.8 ± 0.1 | ≥1010 | ||
| SSB LD (αΔW) | One to one | 300 | 73.2 ± 0.1 | (1.7 ± 0.2) × 108 | ||
| SSB LD (αΔW F) | One to one | 2 | 77.8 ± 0.3 | ≥1010 | ||
| SSB LD (αΔW F) | One to one | 300 | 73.2 ± 0.2 | (3.8 ± 0.2) × 107 |
Since mutation of the Trp residues in the two OB folds in SSB LD (αΔW) and SSB LD (αΔW F) could render binding of a second (dT)35 insensitive to Trp quenching, we also examined (dT)35 binding using ITC (45). At 2 mM NaCl, all variants showed binding of one (dT)35 (Figure 3B and Table 3). Consistent with our fluorescence titrations, the binding of (dT)35 to wtSSB, SSB LD and SSB LD (αΔW) are all stoichiometric (Kobs ≥ 1010 M−1), whereas the binding of SSB LD (αΔW F) is slightly weaker [Kobs = (1.6 ± 0.3) × 109 M−1]. The values of ΔH for the binding of one (dT)35 were similar for wtSSB (−65.7 ± 0.1 kcal mol−1) and SSB LD (−63.3 ± 0.1 kcal mol−1), but lower in magnitude for SSB LD (αΔW) (−56.2 ± 0.2 kcal mol−1) and SSB LD (αΔW F) (−56.2 ± 0.3 kcal mol−1). These differences in ΔH suggest that the mutations partially disrupt the binding to (dT)35 or that the interface for (dT)35 binding is different than for wtSSB.
Table 3. Isothermal titration calorimetry demonstrates that only one molecule of (dT)35 can bind with SSB variants that have two of the four OB folds mutated to block ssDNA binding.
| Protein | [NaCl] (mM) | Model | n(1,2) Sites | K(1,2) M−1 | ΔH(1,2) kcal mol−1 |
|---|---|---|---|---|---|
| wtSSB | 2 | One Site | 0.99 ± 0.01 | ≥1010 | (−65.7 ± 0.1) |
| wtSSB | 300 | Two Sites | 1† | ≥1010 | (−60.7 ± 0.5) |
| 2† | (1.30 ± 0.19) × 108 | (−63.9 ± 0.7) | |||
| SSB LD | 2 | One Site | 0.95 ± 0.01 | ≥1010 | (−63.3 ± 0.1) |
| SSB LD | 300 | Two Sites | 1† | ≥109 | (−59.6 ± 0.7) |
| 2† | (2.0 ± 0.5) × 106 | (−43.4 ± 0.4) | |||
| SSB LD (αΔW) | 2 | One Site | 0.86 ± 0.01 | ≥1010 | (−56.2 ± 0.2) |
| SSB LD (αΔW) | 300 | One Site | 0.93 ± 0.01 | (2.36 ± 0.10) × 108 | (−48.8 ± 0.1) |
| SSB LD (αΔW F) | 2 | One Site | 0.83 ± 0.01 | (1.6 ± 0.3) × 109 | (−56.2 ± 0.3) |
| SSB LD (αΔW F) | 300 | One Site | 0.85 ± 0.01 | (8.7 ± 0.5) × 107 | (−52.0 ± 0.2) |
At high salt (300 mM NaCl) wtSSB exclusively forms a fully wrapped (SSB)65 complex with (dT)70, and due to the decrease in negative cooperativity it can readily bind two molecules of (dT)35 (13,14,20). By Trp fluorescence quenching (Figure 3C and Table 2), wtSSB binds tightly to the first (dT)35 (K1,obs ≥ 1010 M−1) but with lower affinity to the second (dT)35, K2,obs = (2.1 ± 0.8) × 108 M−1, consistent with previous studies (13,14,20). SSB LD also binds two molecules of (dT)35, the first one tightly, although the affinity for the second (dT)35 is decreased almost two orders in magnitude comparing to the wtSSB, K2,obs = (1.6 ± 0.2) × 106 M−1, (Figure 3C and Table 2).
In contrast, SSB LD (αΔW) binds only one molecule of (dT)35 with Kobs = (1.7 ± 0.2) × 108 M−1, with no evidence for binding of a second (dT)35. In addition, while Trp quenching is ∼50% for wtSSB and SSB LD for binding the first (dT)35 and near 90% for binding the second (dT)35 (Figure 3C, Table 2), the Trp quenching of SSB LD (αΔW) for binding one (dT)35 is 73.2 ± 0.1%. This degree of quenching, and 1:1 stoichiometry at 300 mM NaCl, is similar to that observed for (dT)35 binding to SSB LD (αΔW) in 2 mM NaCl (Figure 3A and Table 2). SSB LD (αΔW F) also shows 1:1 binding to (dT)35 with essentially the same degree of quenching, 73.2 ± 0.2%, but with a slightly lower affinity of K1,obs = (3.8 ± 0.2) × 107 M−1 (Figure 3A and Table 2).
ITC titrations in 300 mM NaCl yielded binding stoichiometries and affinities that agree well with the fluorescence titrations (Figure 3D and Table 3). While we cannot accurately measure the affinity of the first molecule of (dT)35 to wtSSB (K1,obs ≥ 1010 M−1), we can obtain the binding enthalpy [ΔH1 = (−60.7 ± 0.5) kcal mol−1]. The binding enthalpy of the second (dT)35 is ΔH2 = (−63.9 ± 0.7) kcal mol−1, consistent with previous studies (45). The binding enthalpy for the first (dT)35 to SSB LD is essentially the same [ΔH1 −59.6 ± 0.7 kcal mol−1] as for wtSSB, and the affinity is still high [K1,obs ≥ 109 M−1]. Binding of the second (dT)35 to SSB LD is less enthalpicaly favorable [ΔH2 = (−43.4 ± 0.4) kcal mol−1] with a much lower affinity (K2,obs = 2.0 ± 0.5) × 106 M−1) relative to wtSSB, which is also consistent with fluorescence data (see Table 2). The ΔH for binding of one (dT)35 to SSB LD (αΔW) [ΔHobs = (−48.8 ± 0.1) kcal mol−1] and SSB LD (αΔW F) [ΔHobs = (−52.0 ± 0.2) kcal mol−1] are less favorable than for binding to wtSSB and SSB LD. This as well as decreasing affinities suggest that the DNA binding interface is somewhat altered in these mutations. However, these results indicate that SSB LD (αΔW) and SSB LD (αΔW F) bind only one molecule of (dT)35 at both low and high NaCl concentrations as expected if binding to two of the OB-folds has been eliminated.
We also performed titrations of (dT)35 into SSB LD (βΔW) in buffer T with 2 and 300 mM NaCl using both tryptophan fluorescence quenching and ITC methods. This construct gave inconsistent results (not shown) and was thus excluded from further study.
Inhibiting ssDNA binding in two OB folds prevents formation of a fully wrapped (SSB)65 complex
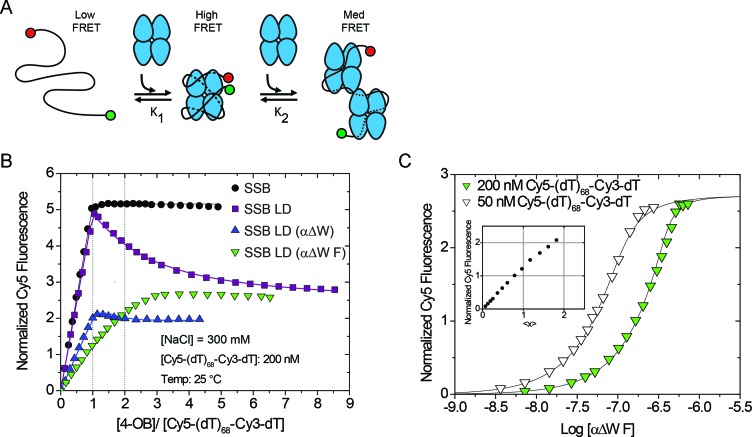
We next examined whether SSB LD (αΔW) and SSB LD (αΔW F) can form a fully wrapped (SSB)65 complex using a Förster resonance energy transfer (FRET) based assay (15,30,60,61) (Figure 4A). This assay uses a (dT)68 labeled with a Cy5 fluorophore (the FRET acceptor) on its 5' end and a Cy3 fluorophore (the FRET donor) on its 3' end. The (dT)68 is long enough to bind only one SSB tetramer in the (SSB)65 mode. When Cy5-(dT)68-Cy3-dT binds wtSSB in the fully wrapped (SSB)65 mode, the Cy3 and Cy5 fluorophores are brought in close proximity and display a high FRET signal (Figure 4A). If conditions favor the (SSB)35 mode then two SSB tetramers can bind resulting in a Cy5 enhancement that is intermediate between free DNA and the fully wrapped complex (Figure 4A).
Figure 4.
Mutants with only two wild-type-like OB folds do not form a fully wrapped complex with ssDNA. (A) Reaction scheme of SSB constructs titrated into Cy5-(dT)68-Cy3-dT, a substrate that is sensitive to the degree of wrapping and binding density of SSB:ssDNA complexes. (B) Results of equilibrium titrations of protein titrated into 200 nM Cy5-(dT)68-Cy3-dT in Buffer T + 300 mM NaCl at 25°C plotted as normalized Cy5 fluorescence versus the ratio of total protein to DNA concentrations. Under these conditions, wtSSB binds tightly with Cy5-(dT)68-Cy3-dT (Kobs ≥ 1010 M−1) in a one-to-one complex with a maximum normalized enhancement of 5.16 ± 0.01. The proteins SSB LD, and SSB LD (αΔW) first bind tightly to Cy5-(dT)68-Cy3-dT in a one-to-one complex (Kobs ≥ 5 × 1010 M−1) then a second SSB tetramer binds more weakly [(3.8 ± 0.3) × 106 M−1 and (2 ± 2) × 108 M−1, respectively]. Under these conditions, the SSB LD (αΔW F) construct does not clearly transition through a simple one-to-one complex. (C) To measure the binding of SSB LD (αΔW F) more rigorously, we also performed the titration into 50 nM 5′-Cy5-(dT)68-Cy3-dT-3′ and globally fit the data to a two-site binding model. The first molecule of SSB LD (αΔW F) binds with Kobs = (1.4 ± 0.7) × 108 M−1 with a normalized Cy5 fluorescence of 1.18 ± 0.06 and the second molecule binds with Kobs = (4 ± 1) × 107 M−1 with a normalized fluorescence of 2.71 ± 0.06. Inset shows the results of a model independent binding analysis which plots the normalized Cy5 fluorescence against the number of SSB LD (αΔW F) tetramers bound per ssDNA (<x>).
Under conditions that exclusively populate the (SSB)65 mode (300 mM NaCl) wtSSB binds tightly to Cy5-(dT)68-Cy3-dT with a 1:1 binding stoichiometry (Figure 4B). Furthermore, upon excitation of the Cy3 donor fluorescence, the Cy5 fluorescence reaches a maximum normalized enhancement of 5.16 ± 0.01 at saturation. Upon further addition of wtSSB, the enhancement remains at the maximum indicating formation of a stable, fully wrapped complex that does not subsequently form a 2:1 complex at higher SSB concentrations. Conversely, although SSB LD binding to Cy5-(dT)68-Cy3-dT also shows a maximum Cy5 fluorescence at a 1:1 binding stoichiometry, this is followed by a decrease in Cy5 fluorescence upon further addition of protein. The similar maximum fluorescence enhancement observed for wtSSB and SSB LD suggests that SSB LD forms a fully wrapped complex similar to wtSSB. The subsequent decrease in enhancement, to 2.34 ± 0.05, indicates that a second molecule of SSB LD can bind at higher protein concentrations under these conditions. This is consistent with the observation that the removal of two C-terminal tails shifts the binding mode transition toward the (SSB)35 mode (30). Titration of Cy5-(dT)68-Cy3-dT with SSB LD (αΔW) yields a maximum Cy5 fluorescence, 2.24 ± 0.04, at a 1:1 binding stoichiometry, followed by only a slight decrease in Cy5 fluorescence upon further addition of protein. Significantly, the maximum Cy5 enhancement (2.24 ± 0.04) is only ∼40% of that observed for wtSSB suggesting that the ends of Cy5-(dT)68-Cy3-dT are not brought as close together as in the fully wrapped wtSSB and SSB LD complexes. Hence, SSB LD (αΔW) does not form a fully wrapped ssDNA complex in 300 mM NaCl.
SSB LD (αΔW F) displays weaker binding to Cy5-(dT)68-Cy3-dT (50 nM DNA) (Figure 4B). Hence, in order to estimate binding parameters we performed a second titration at a higher concentration of DNA (200 nM) and fit the data globally to a two-site binding model (Figure 4C). This analysis shows that two moles of SSB LD (αΔW F) can bind to one Cy5-(dT)68-Cy3-dT and that binding of the first SSB LD (αΔW F) results in a Cy5 fluorescence enhancement of 1.18 ± 0.06, or ∼25% of the maximum enhancement observed for wtSSB and SSB LD. This value agrees with the fluorescence enhancement determined using a model independent analysis (43) (Figure 4C, inset). As with SSB LD (αΔW), the lower fluorescence signal indicates that the one-to-one complex formed between SSB LD (αΔW F) and Cy5-(dT)68-Cy3-dT is not a fully wrapped complex under these conditions. Of note, the Cy5 fluorescence signal of 2.71 ± 0.06 upon binding two SSB LD (αΔW F) to Cy5-(dT)68-Cy3-dT is similar to the plateau signal observed for SSB LD at high protein concentrations. This suggests that these proteins can form two-to-one complexes with similar configurations. These results indicate that SSB LD (αΔW) and SSB LD (αΔW F) do not form fully wrapped complexes with (dT)68 under conditions that exclusively populate the (SSB)65 mode for wtSSB, consistent with these mutations having eliminated binding of ssDNA to two of the four OB-folds.
SSB LD (αΔW) and SSB LD (αΔW F) do not form fully wrapped complexes with poly(dT)
We next examined binding of SSB LD (αΔW) and SSB LD (αΔW F) to poly(dT). At 25°C, wtSSB transitions through three binding modes on poly(dT) as a function of salt concentration that display three different occluded site sizes. At low [NaCl] (< 10 mM) wtSSB has an occluded site size of 35 nts/tetramer while at moderate-to-high [NaCl] (> 200 mM) it has a site size of 65 nts/tetramer. A third occluded site size of 56 nts occurs at intermediate [NaCl] (10,11,30,61). The site-size transitions of SSB LD follow a similar pattern at 25°C, but the transitions are shifted to higher [NaCl]; the ∼35 nt mode forms at [NaCl] < 10 mM and the 65 nt mode forms at [NaCl] ≥ 700 mM (30).
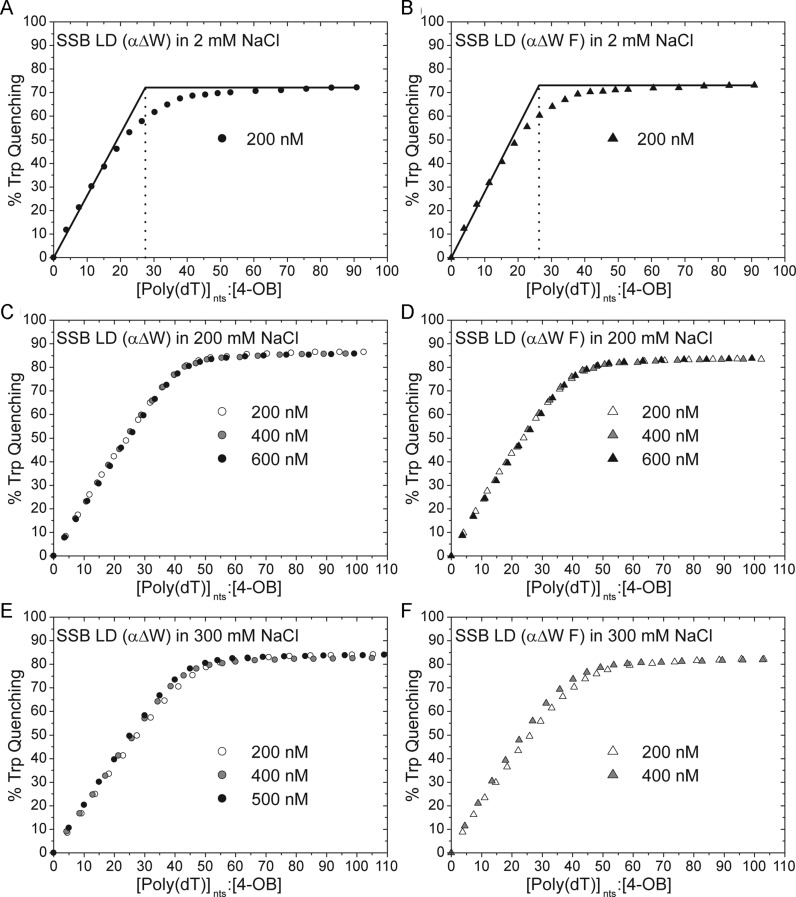
Under conditions that stabilize wtSSB in the (SSB)35 mode ([NaCl] = 2 mM), SSB LD (αΔW) displays an occluded site size of ∼28 nucleotides with a maximum tryptophan fluorescence quenching of 72% (Figure 5A). Similarly, SSB LD (αΔW F) displays a site size of ∼26 nts with 73% tryptophan quenching (Figure 5B). Both of these occluded site sizes are less than the 33–35 nt site size observed for wtSSB and SSB LD suggesting that the variants may not form some of the contacts that form in the wtSSB (SSB)35 mode. This is consistent with our observation that SSB LD (αΔW) and SSB LD (αΔW F) bind to (dT)35 with slightly different binding enthalpies under these conditions (Figure 3B).
Figure 5.
Occluded site sizes of SSB LD (αΔW) and SSB LD (αΔW F) on poly(dT). (A and B) Titrations performed in T buffer with 2 mM NaCl. (A) The site size and Qmax of SSB LD (αΔW) under these conditions are 27.5 nts and 72.2%, respectively (B) The same parameters for SSB LD (αΔW F) are 26.3 nts and 73.1%. (C and D) Titrations performed in 200 mM NaCl as a function of protein concentration (listed in the figure). (C) The averaged occluded site size and Qmax for SSB LD (αΔW) is 41.5 ± 0.7 nts and 86.1 ± 0.6%, respectively, where the error is calculated as the standard deviation of the three titrations. (D) For SSB LD (αΔW F), these values are 38.9 ± 0.8 nts and 85 ± 1%. (E) The occluded site size of SSB LD (αΔW) decreases with increasing protein concentration from 46.4 nts, to 43.3 nts, to 41.9 nts at 200 nM, 400 nM and 500 nM protein, respectively. In the same order, the Qmax for these titrations are 84.3, 85.8 and 84.0%. (F) Similarly, the occluded site size of SSB LD (αΔW F) decreases with increasing protein concentration. At 200 nM protein, the site size is 42.1 nts while at 400 nM protein it is 38.9 nts; the Qmax for these titrations are 82.3 and 82.0%, respectively.
Under moderate salt concentrations (200 mM NaCl), the observed occluded site sizes of both SSB LD (αΔW) and SSB LD (αΔW F) increase to 41 and 38 nts, respectively at 200 nM (4-OB fold) protein, with maximum Trp fluorescence quenchings of 87 and 84%, respectively (Figure 5C and D). To determine if the affinities of SSB LD (αΔW) and SSB LD (αΔW F) for poly(dT) are high enough under these conditions to obtain accurate site size estimates, we also performed titrations at 400 and 600 nM protein. All three binding curves overlap indicating that these represent accurate site size estimates (Figure 5C and D).
Upon increasing the [NaCl] to 300 mM, the apparent occluded site size for SSB LD (αΔW) increases to 46 nts at 200 nM protein. Additional titrations at higher protein concentrations, however, give non-overlapping isotherms. As the concentration of SSB LD (αΔW) is increased to 400 nM and 500 nM the site size decreases to 43 and 42 nts, respectively, indicating that 42 nucleotides is the maximum estimate of the site size at 300 mM NaCl. As expected, the poly(dT) titrations performed with different concentrations of SSB LD (αΔW F) in 300 mM NaCl are not stoichiometric. In these experiments, the apparent occluded site size of SSB LD (αΔW F) decreases from ∼42 nts at 200 nM protein, to ∼39 nts at 400 nM protein. These results suggest that the occluded site sizes of SSB LD (αΔW) and SSB LD (αΔW F) on poly(dT) are ∼40 nts at both 200 mM and 300 mM NaCl. Even though there is an increase in occluded site size from ∼26-29 nts at 2 mM NaCl to ∼40 nts at 200 mM NaCl, these are well below the fully wrapped site sizes of 56 and 65 nts indicating that neither SSB LD (αΔW) nor SSB LD (αΔW F) bind to poly(dT) in the fully wrapped (SSB)56 or (SSB)65 modes even at high salt concentrations.
The apparent differences in the occluded site size measured at low salt (2 mM NaCl, 26–29 nts) and moderate salt (200 mM NaCl, 40 nts) could result from differences in cooperative binding of these proteins to poly(dT). A protein with the same site size will show a slightly higher apparent site size if it binds with low cooperativity due to its inability to eliminate DNA gaps between bound proteins even at high protein binding densities (62). This possibility is supported by experiments discussed below.
SSB LD (αΔW) and SSB LD (αΔW F) display salt-dependent transitions between highly cooperative and low cooperative binding to ssDNA
In its (SSB)35 mode, wtSSB binds to long ssDNA with high cooperativity to form protein clusters on ssDNA (19,27,29,46). This highly cooperative binding is eliminated at higher salt concentrations that favor the (SSB)65 mode (29,46). We therefore examined whether the SSB LD (αΔW) and SSB LD (αΔW F) variants retain the ability to bind with high cooperativity to ssDNA. For example, if a specific ssDNA binding pathway is required for high cooperativity and the SSB LD (αΔW) and SSB LD (αΔW F) mutations block its formation, this might inhibit highly cooperative binding. Conversely, destabilization of the fully wrapped (SSB)65 mode might facilitate highly cooperative binding even under high salt conditions.
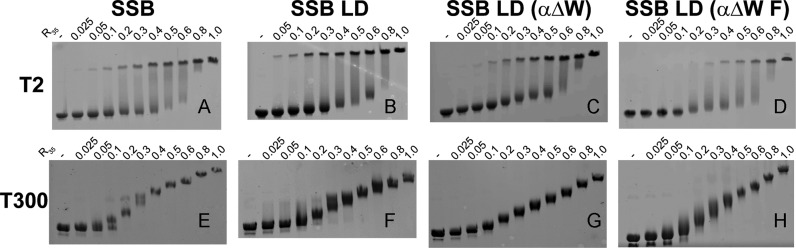
We examined cooperative binding to M13 mp18 phage ssDNA (∼7.25 kilobases) using an electrophoretic mobility shift assay (EMSA) (46). In this assay, non-cooperative, or limited-cooperative binding of SSB to the DNA results in a random binding distribution of SSB molecules such that all ssDNA molecules have essentially the same amount of SSB bound. The resulting population of ssDNA migrates as a single diffuse band at all SSB to DNA ratios. On the other hand, at sub-saturating concentrations of SSB, highly cooperative binding displays a biphasic population of ssDNA: one population that is nearly saturated with protein and another population with little bound protein (46).
Increasing concentrations of protein were mixed with M13 ssDNA in buffer T with 2 mM NaCl, allowed to equilibrate for an hour at 22°C and were then subjected to electrophoresis in an agarose gel in 0.5× TAE buffer (see Materials and Methods). While the electrophoresis buffer does not match the buffer used to form the complexes, and therefore may influence the binding properties of the complexes, we and others have found good qualitative agreement between these EMSAs and similar observations of SSB-M13 complexes made with analytical sedimentation velocity experiments where the buffers are constant (29,46). Consistent with previous studies (29,46), wtSSB and SSB LD form a bimodal banding pattern that is indicative of highly cooperative binding (Figure 6A and B). A similar bimodal distribution is also observed for SSB LD (αΔW) and SSB LD (αΔW F) under these conditions (Figure 6C and D). Conversely, when the salt concentration is raised to 300 mM NaCl all four proteins showed a more random distribution of SSB on the ssDNA substrate indicating a low level of cooperativity (Figure 6E–H). Hence, upon increasing NaCl concentration SSB LD (αΔW) and SSB LD (αΔW F) retain the wild-type-like transition from high cooperativity to low cooperativity. This suggests that the apparent increase in occluded site size on poly(dT) from 27 ± 2 nts at 2 mM NaCl to ∼40 at 200–300 mM NaCl may be partly due to a loss of highly cooperative binding. Thus, the 40 nt site size observed at 200–300 mM NaCl should be viewed as a maximum estimate.
Figure 6.
Cooperativity of SSB variants bound with ssDNA is sensitive to salt concentration. EMSAs of SSB–M13 mp18 ssDNA complexes in Buffer T as a function of protein concentration (R35 is the binding ratio of [4-OB]tot:[M13 (35 nts)]tot). (A–D) EMSAs performed T buffer with 2 mM NaCl. (E–H) EMSAs performed in T buffer with 300 mM NaCl.
Complementation and recovery from genotoxic stress of the SSB variants in vivo
Our studies indicate that the SSB LD (αΔW) and SSB LD (αΔW F) proteins are unable to form the fully wrapped (SSB)56 or (SSB)65 modes in vitro. We next determined if the fully wrapped states are essential for SSB function in vivo. Because wtSSB is essential for E. coli viability, we used an established ‘bumping’ assay (63) to determine if the variants could complement loss of wtSSB. This assay uses an E. coli strain, RDP317 (63) in which the wtSSB gene has been deleted from the chromosome and thus survival is dependent on expressing a viable SSB protein ectopically from a plasmid with a tetracycline resistance marker (63). The SSB variants to be tested are expressed under the control of the native ssb promoter from a second, low-copy number plasmid with an ampicillin resistance marker. E. coli strains containing both plasmids are then grown for a period of several days in the presence of ampicillin but in the absence of tetracycline. If the SSB variant is able to complement loss of wtSSB the bacteria will eventually lose the helper plasmid encoding wtSSB rendering the strain sensitive to tetracycline. The plasmid expressing the variant SSB that successfully ‘bumped’ the wtSSB plasmid is then purified, and the gene sequenced to determine if any compensatory mutations occurred during the growth phase. Finally, since it is possible that the ssb gene may be inserted elsewhere in the genome, or a portion of the unsequenced plasmid, we performed a western blot on whole cell extracts of these strains using antibodies raised against EcSSB (Supplementary Figure S2). We found no evidence for expression of wtSSB in these bumped strains.
When we performed this series of assays plasmids expressing wtSSB and SSB LD were able to complement loss of the wtSSB helper plasmid as demonstrated previously (30). In addition, both SSB LD (αΔW) and SSB LD (αΔW F) were able to complement loss of wtSSB. This suggests that the ability to form a fully wrapped SSB-ssDNA complex is not essential for E. coli survival.
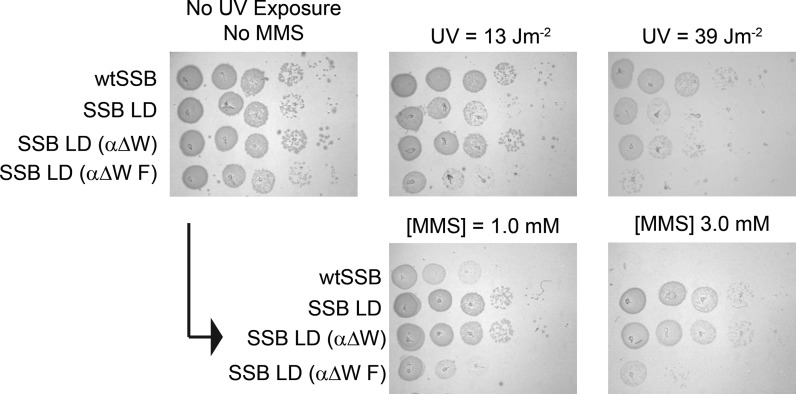
We also tested the ability of E. coli expressing either the SSB LD (αΔW) or SSB LD (αΔW F) variants to recover from challenges from genotoxic agents. For these experiments we spotted serial dilutions of the appropriate ‘bumped’ strains onto LB agar and either exposed the strains transiently to UV light or chronically to MMS (Figure 7). Exposure to UV irradiation causes DNA breaks and the formation of base crosslinks (64). As previously demonstrated, E. coli RDP317 expressing SSB LD shows a similar response to UV irradiation as E. coli RDP317 expressing wtSSB (30). In the current study, SSB LD (αΔW) also has a similar response to UV irradiation while SSB LD (αΔW F) is more sensitive (Figure 7) suggesting that the additional mutation of αF60A may interfere with DNA repair pathways. Interestingly, however, when the strains are exposed to MMS, an alkylating agent, SSB LD and SSB LD (αΔW) show hyper-resistance to the toxin relative to the wtSSB control (Figure 7). SSB LD (αΔW F) is also hyper-resistant to MMS but to a lesser degree (Figure 7). In a previous study, we demonstrated that a strain expressing SSB LD grows faster than a strain expressing wtSSB (30). It was hypothesized that the increased growth rate was due to a decrease in the ability of SSB LD to support DNA repair during replication. This was supported by the demonstration that SSB LD is better able to survive a challenge from rifampicin than wtSSB suggesting that SSB LD accumulates more compensatory spurious mutations. It is likely that the enhanced resistance of SSB LD (αΔW) and SSB LD (αΔW F) to MMS is due to a similar mechanism.
Figure 7.
In vivo UV and MMS sensitivities of SSB variants. Relative sensitivities of complementing SSB strains when challenged with UV irradiation or chronic exposure to MMS.
DISCUSSION
Due to their central roles in DNA metabolism and genome maintenance, SSB proteins are ubiquitous throughout all domains of life. The vast majority of SSBs use multiple OB-folds to interact with ssDNA (65) with two known exceptions (66,67). The oligomeric states and number of OB-folds across the SSB family are variable, however. Bacteriophages and viruses often use SSBs with one or two OB folds. For example, gene 32 protein from T4 bacteriophage is a monomeric SSB with one OB-fold (67) while gene 2.5 protein from T7 bacteriophage forms a homodimer with two OB-folds (68). The major eukaryotic SSB, replication protein A (RPA) (4), forms a heterotrimer with six OB-folds, four of which are principal ssDNA-binding domains (65,69,70). The bacterial SSBs from E. coli and D. radiodurans both contain four OB-folds, but form a homotetramer and homodimer, respectively (3,7–9,48). The presence of multiple ssDNA binding sites per SSB enables it to bind ssDNA in different binding modes. Indeed, in addition to EcSSB (3) similar behavior has been shown for yeast RPA (71), human RPA (25), DradSSB (72) and Thermus aquaticus SSB (73). However, an understanding of how these modes are used in their biological context is lacking. We are using the EcSSB protein to probe these questions due to the extensive studies that have been undertaken on its different ssDNA binding modes. Toward this goal, we have begun to create a library of EcSSB variants that are biased toward one binding mode at equilibrium. In addition to providing clues to molecular mechanisms, these variants can also be used to examine whether the different binding modes are essential for EcSSB function in vivo.
In our current study, we constructed two SSB variants designed to inhibit formation of the fully wrapped (SSB)56 and (SSB)65 binding modes in which ssDNA contacts all four OB folds. Based on the extensive characterization reported here, the SSB LD (αΔW) and the SSB LD (αΔW F) variants have this property. Under physiological salt concentrations in vitro, wtSSB is in a dynamic equilibrium populating all of its binding modes. Under these conditions, the protein to DNA stoichiometry largely determines the distribution of SSB binding modes. One can force SSB–DNA complexes to form homogeneous populations of the (SSB)65 or (SSB)35 binding modes by going to extremes of monovalent salt concentrations, > 300 mM NaCl versus < 10 mM NaCl, respectively (10,11,13,14). Although neither of these conditions is physiologically relevant, we used these extremes of [NaCl] to simplify characterization of the SSB mutants described in our study. The rationale is that if a mutant SSB protein is unable to form the fully wrapped (SSB)65 mode at high salt (300 mM NaCl), conditions under which wtSSB exclusively forms this mode, then it will not populate this mode at physiological conditions, at least at equilibrium. Although we have used high [NaCl] (300 mM) as a simple means to exclusively populate the (SSB)65 mode in vitro, this mode can also be populated in vitro under conditions that are more physiological. For example, low concentrations of Mg2+ (5–10 mM), and even micromolar concentrations of polyamines, such as spermine, that are physiologically relevant, both favor formation of the (SSB)65 binding mode in vitro (11,17). Hence, wtSSB is likely to populate all of its binding modes, including the (SSB)65 binding mode, in vivo.
Interestingly, the low salt modes for SSB LD (αΔW) or SSB LD (αΔW F) appear to be slightly altered relative to those formed by wtSSB since they possess occluded site sizes that are smaller than the 33–35 nts observed for wtSSB and SSB LD. When combined with our ITC data, these results suggest that the ssDNA contacts for these variants in the low salt modes are altered relative to wtSSB. This is consistent with the proposed pathway for ssDNA binding of wtSSB in the (SSB)35 mode where ssDNA makes full contact with one subunit and only partial contact with two other subunits for an average of two subunits (Figure 1C) (8,12). This model for ssDNA binding in the (SSB)35 mode is consistent with ssDNA passing between tetramers in such a configuration. If ssDNA normally makes at least partial contact with three OB-folds in the (SSB)35 mode, disrupting DNA binding in two of the four OB-folds would likely partially disrupt this binding pathway.
The subtle salt-dependent changes in apparent occluded site size observed for SSB LD (αΔW) and SSB LD (αΔW F) may also be related to the loss of highly cooperative binding as the salt concentration is increased. The ‘unlimited’ inter-tetramer positive cooperativity of wtSSB in its (SSB)35 mode can be regulated by the length, composition and number of intrinsically disordered C-terminal tails (29). In addition, the acidic tips of these C-terminal tails can interact weakly with the ssDNA binding sites (28,74). These observations suggested a model for highly cooperative ssDNA binding in the (SSB)35 mode in which an SSB tetramer binds ssDNA using only an average of two OB-folds and can interact via its C-terminal tails with the two unoccupied OB-folds in an adjacent SSB tetramer (29). Although the SSB LD (αΔW) or SSB LD (αΔW F) variants are unable to form fully wrapped ssDNA complexes, they still show a salt-dependent transition between highly cooperative binding at low [NaCl] and low cooperativity at higher [NaCl]. This indicates that formation of a partially wrapped SSB-ssDNA complex does not ensure highly cooperative ssDNA binding. The loss of highly cooperative binding at increased NaCl concentrations does indicate, however, an electrostatic component to this cooperativity and is consistent with the suggestion that it may involve interactions between the acidic tip (MDFDDDIPF) from a C-terminal tail on one tetramer and an unoccupied basic ssDNA binding site on an adjacent tetramer (29). Further, even though the mutations of W40, W54, W88 and F60 disrupt ssDNA binding, the mutated ssDNA binding sites still contain Lys and Arg residues that may facilitate electrostatic interactions with the acidic tip. SSB variants related to those described in this study with mutations to the basic residues in two of the OB-folds can be used to test this hypothesis.
The more difficult question is whether the different binding modes and inter-tetramer cooperativities affect biological function. The most basic question we can ask is whether SSB variants that are biased toward specific binding properties are able to complement wtSSB function in vivo. In this study we demonstrate that E. coli strains expressing only SSB LD (αΔW) or SSB LD (αΔW F) can survive demonstrating that full ssDNA wrapping is not an essential property for SSB function. We note, however, that this does not preclude a non-essential use of this mode in vivo.
The results reported here add to our prior observations concerning the functional roles of the SSB C-terminal tails (29,30). One study showed that an SSB possessing only one C-terminal tail does not complement wtSSB in vivo and shows defects in DNA replication and replication restart in vitro (29). In another study, we produced two SSB variants to examine the effects of the length and composition of the C-terminal tails on SSB function (28). The first variant was a C-terminal truncation mutant, SSB-GG, in which 56 aa of the IDL domain are removed leaving only two glycines to connect the N-terminal core to the C-terminal acidic tip (29). This mutant is able to transition from the (SSB)35 mode to an (SSB)65-like mode in low and moderate salt, respectively (29). It does not, however, display the high cooperativity that is characteristic of wtSSB in the low salt binding mode (29). Surprisingly, this protein can complement loss of wtSSB in vivo, demonstrating that the highly cooperative binding property of the (SSB)35 mode is not essential for EcSSB function.
A second SSB variant, EcPfEc, was constructed with the N-terminal DNA-binding core of EcSSB connected to the C-terminal acidic tip of EcSSB with the intrinsically disordered linker of Plasmodium falciparum SSB (PfSSB) (29). PfSSB shares structural (75) and functional (61) homology with EcSSB but is unable to form a stable, highly cooperative (SSB)35 mode in vitro (61). These properties appear to be transferred to EcPfEc variant because this protein is unable to bind poly(dT) with a (SSB)35-like site size, and is also unable to form a highly cooperative complex on ssDNA (29). Interestingly, this variant is also able to complement loss of wtSSB in vivo, suggesting that neither the (SSB)35 mode nor the highly cooperative state are essential for EcSSB function.
Our current study showed that a strain expressing SSB LD (αΔW) shares a similar UV-irradiation phenotype with wtSSB and SSB LD. This result suggests that the fully wrapped complex is not a requirement for UV damage repair. On the other hand, SSB LD (αΔW F) is hypersensitive to UV-irradiation relative to wtSSB making the interpretation of this wrapping data more nuanced. It is possible that certain contacts between SSB and ssDNA are required to achieve the wild-type phenotype in UV-irradiation survival. The small differences between SSB LD (αΔW F) and SSB LD (αΔW) in ssDNA binding could straddle this threshold. Alternatively, the hypersensitivity to UV damage of a strain expressing SSB LD (αΔW F) could be unrelated to the wrapping phenotype. Another interesting observation is that SSB LD and SSB LD (αΔW), and to a smaller degree SSB LD (αΔW F), share a hyper-resistant phenotype when exposed to MMS. The close comparison between SSB LD and SSB LD (αΔW) indicates that the inability to form the fully wrapped binding mode does not appear to affect this outcome.
Our results suggest that neither the high ‘unlimited’ inter-tetramer cooperativity nor either the (SSB)65 or the (SSB)35 modes are essential for SSB function. This may reflect the fact that SSB can function using any of its DNA binding modes. However, we note that our tests of the ability of the SSB variants that we have created to complement loss of wtSSB were performed under optimal growth conditions for E. coli. Hence, the possibility exists that a subset of the properties exhibited by the different binding modes may be essential for E. coli survival under non-optimal or stressed conditions. A further complication arises from the fact that SSB is involved in so many different aspects of genome maintenance. Further studies will be required to determine which properties of SSB are required for proper function in any particular pathway.
Supplementary Material
Acknowledgments
We thank T. Ho for synthesis and purification of the oligodeoxynucleotides, and Dr. C. Brosey and J. Obermann for assistance with the western blots.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM030498 to T.M.L., in part]. Funding for open access charge: National Institutes of Health [GM030498].
Conflict of interest statement. None declared.
REFERENCES
- 1.Chase J.W., Williams K.R. Single-stranded DNA binding proteins required for DNA replication. Annu. Rev. Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 2.Meyer R.R., Laine P.S. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohman T.M., Ferrari M.E. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 4.Wold M.S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 5.Shereda R.D., Kozlov A.G., Lohman T.M., Cox M.M., Keck J.L. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ollis D., Brick P., Abdel-Meguid S.S., Murthy K., Chase J.W., Steitz T.A. Crystals of Escherichia coli single-strand DNA-binding protein show that the tetramer has D2 symmetry. J. Mol. Biol. 1983;170:797–800. doi: 10.1016/s0022-2836(83)80134-x. [DOI] [PubMed] [Google Scholar]
- 7.Raghunathan S., Ricard C.S., Lohman T.M., Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc. Natl Acad. Sci. USA. 1997;94:6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raghunathan S., Kozlov A.G., Lohman T.M., Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 9.Savvides S.N., Raghunathan S., Futterer K., Kozlov A.G., Lohman T.M., Waksman G. The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA. Protein Sci. 2004;13:1942–1947. doi: 10.1110/ps.04661904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lohman T.M., Overman L.B. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 11.Bujalowski W., Lohman T.M. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25:7799–7802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 12.Suksombat S., Khafizov R., Kozlov A.G., Lohman T.M., Chemla Y.R. Structural dynamics of E. coli single-stranded DNA binding protein reveal DNA wrapping and unwrapping pathways. eLife. 2015;4:e08193. doi: 10.7554/eLife.08193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bujalowski W., Lohman T.M. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. II. Salt, temperature and oligonucleotide length effects. J. Mol. Biol. 1989;207:269–288. doi: 10.1016/0022-2836(89)90455-5. [DOI] [PubMed] [Google Scholar]
- 14.Bujalowski W., Lohman T.M. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. I. Evidence and a quantitative model. J. Mol. Biol. 1989;207:249–268. doi: 10.1016/0022-2836(89)90454-3. [DOI] [PubMed] [Google Scholar]
- 15.Roy R., Kozlov A.G., Lohman T.M., Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J. Mol. Biol. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bujalowski W., Overman L.B., Lohman T.M. Binding mode transitions of Escherichia coli single strand binding protein-single-stranded DNA complexes. Cation, anion, pH, and binding density effects. J. Biol. Chem. 1988;263:4629–4640. [PubMed] [Google Scholar]
- 17.Wei T.F., Bujalowski W., Lohman T.M. Cooperative binding of polyamines induces the Escherichia coli single-strand binding protein-DNA binding mode transitions. Biochemistry. 1992;31:6166–6174. doi: 10.1021/bi00141a029. [DOI] [PubMed] [Google Scholar]
- 18.Chrysogelos S., Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc. Natl Acad. Sci. USA. 1982;79:5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffith J.D., Harris L.D., Register J. 3rd. Visualization of SSB-ssDNA complexes active in the assembly of stable RecA-DNA filaments. Cold Spring Harbor Symp. Quant. Biol. 1984;49:553–559. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- 20.Lohman T.M., Bujalowski W. Negative cooperativity within individual tetramers of Escherichia coli single strand binding protein is responsible for the transition between the (SSB)35 and (SSB)56 DNA binding modes. Biochemistry. 1988;27:2260–2265. doi: 10.1021/bi00407a002. [DOI] [PubMed] [Google Scholar]
- 21.Lohman T.M., Bujalowski W., Overman L.B., Wei T.F. Interactions of the E. coli single strand binding (SSB) protein with ss nucleic acids. Binding mode transitions and equilibrium binding studies. Biochem. Pharmacol. 1988;37:1781–1782. doi: 10.1016/0006-2952(88)90443-1. [DOI] [PubMed] [Google Scholar]
- 22.Bujalowski W., Lohman T.M. Monomer-tetramer equilibrium of the Escherichia coli ssb-1 mutant single strand binding protein. J. Biol. Chem. 1991;266:1616–1626. [PubMed] [Google Scholar]
- 23.Zhou R., Kozlov A.G., Roy R., Zhang J., Korolev S., Lohman T.M., Ha T. SSB functions as a sliding platform that migrates on DNA via reptation. Cell. 2011;146:222–232. doi: 10.1016/j.cell.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy R., Kozlov A.G., Lohman T.M., Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen B., Sokoloski J., Galletto R., Elson E.L., Wold M.S., Lohman T.M. Diffusion of human replication protein A along single-stranded DNA. J. Mol. Biol. 2014;426:3246–3261. doi: 10.1016/j.jmb.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell J.C., Liu B., Kowalczykowski S.C. Imaging and energetics of single SSB-ssDNA molecules reveal intramolecular condensation and insight into RecOR function. eLife. 2015;4:e08646. doi: 10.7554/eLife.08646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrari M.E., Bujalowski W., Lohman T.M. Co-operative binding of Escherichia coli SSB tetramers to single-stranded DNA in the (SSB)35 binding mode. J. Mol. Biol. 1994;236:106–123. doi: 10.1006/jmbi.1994.1122. [DOI] [PubMed] [Google Scholar]
- 28.Kozlov A.G., Cox M.M., Lohman T.M. Regulation of single-stranded DNA binding by the C termini of Escherichia coli single-stranded DNA-binding (SSB) protein. J. Biol. Chem. 2010;285:17246–17252. doi: 10.1074/jbc.M110.118273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozlov A.G., Weiland E., Mittal A., Waldman V., Antony E., Fazio N., Pappu R.V., Lohman T.M. Intrinsically disordered C-terminal tails of E. coli single-stranded DNA binding protein regulate cooperative binding to single-stranded DNA. J. Mol. Biol. 2015;427:763–774. doi: 10.1016/j.jmb.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antony E., Weiland E., Yuan Q., Manhart C.M., Nguyen B., Kozlov A.G., McHenry C.S., Lohman T.M. Multiple C-terminal tails within a single E. coli SSB homotetramer coordinate DNA replication and repair. J. Mol. Biol. 2013;425:4802–4819. doi: 10.1016/j.jmb.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee K.S., Marciel A.B., Kozlov A.G., Schroeder C.M., Lohman T.M., Ha T. Ultrafast redistribution of E. coli SSB along long single-stranded DNA via intersegment transfer. J. Mol. Biol. 2014;426:2413–2421. doi: 10.1016/j.jmb.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozlov A.G., Lohman T.M. Kinetic mechanism of direct transfer of Escherichia coli SSB tetramers between single-stranded DNA molecules. Biochemistry. 2002;41:11611–11627. doi: 10.1021/bi020361m. [DOI] [PubMed] [Google Scholar]
- 33.Bhattacharyya B., George N.P., Thurmes T.M., Zhou R., Jani N., Wessel S.R., Sandler S.J., Ha T., Keck J.L. Structural mechanisms of PriA-mediated DNA replication restart. Proc. Natl Acad. Sci. USA. 2014;111:1373–1378. doi: 10.1073/pnas.1318001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wessel S.R., Marceau A.H., Massoni S.C., Zhou R., Ha T., Sandler S.J., Keck J.L. PriC-mediated DNA replication restart requires PriC complex formation with the single-stranded DNA-binding protein. J. Biol. Chem. 2013;288:17569–17578. doi: 10.1074/jbc.M113.478156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohman T.M., Green J.M., Beyer R.S. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 36.Gill S.C., von Hippel P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 37.Kowalczykowski S.C., Lonberg N., Newport J.W., von Hippel P.H. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. I. Characterization of the binding interactions. J Mol Biol. 1981;145:75–104. doi: 10.1016/0022-2836(81)90335-1. [DOI] [PubMed] [Google Scholar]
- 38.Berkowitz S.A., Day L.A. Molecular weight of single-stranded fd bacteriophage DNA. High speed equilibrium sedimentation and light scattering measurements. Biochemistry. 1974;13:4825–4831. doi: 10.1021/bi00720a022. [DOI] [PubMed] [Google Scholar]
- 39.Dam J., Schuck P. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Methods Enzymol. 2004;384:185–212. doi: 10.1016/S0076-6879(04)84012-6. [DOI] [PubMed] [Google Scholar]
- 40.Ferrari M.E., Lohman T.M. Apparent heat capacity change accompanying a nonspecific protein-DNA interaction. Escherichia coli SSB tetramer binding to oligodeoxyadenylates. Biochemistry. 1994;33:12896–12910. doi: 10.1021/bi00209a022. [DOI] [PubMed] [Google Scholar]
- 41.Lohman T.M., Mascotti D.P. Nonspecific ligand-DNA equilibrium binding parameters determined by fluorescence methods. Methods Enzymol. 1992;212:424–458. doi: 10.1016/0076-6879(92)12027-n. [DOI] [PubMed] [Google Scholar]
- 42.Kozlov A.G., Galletto R., Lohman T.M. SSB-DNA binding monitored by fluorescence intensity and anisotropy. Methods Mol. Biol. 2012;922:55–83. doi: 10.1007/978-1-62703-032-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lohman T.M., Bujalowski W. Thermodynamic methods for model-independent determination of equilibrium binding isotherms for protein-DNA interactions: spectroscopic approaches to monitor binding. Methods Enzymol. 1991;208:258–290. doi: 10.1016/0076-6879(91)08017-c. [DOI] [PubMed] [Google Scholar]
- 44.Kozlov A.G., Lohman T.M. SSB binding to ssDNA using isothermal titration calorimetry. Methods Mol. Biol. 2012;922:37–54. doi: 10.1007/978-1-62703-032-8_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozlov A.G., Lohman T.M. Calorimetric studies of E. coli SSB protein-single-stranded DNA interactions. Effects of monovalent salts on binding enthalpy. J. Mol. Biol. 1998;278:999–1014. doi: 10.1006/jmbi.1998.1738. [DOI] [PubMed] [Google Scholar]
- 46.Lohman T.M., Overman L.B., Datta S. Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J. Mol. Biol. 1986;187:603–615. doi: 10.1016/0022-2836(86)90338-4. [DOI] [PubMed] [Google Scholar]
- 47.Porter R.D., Black S., Pannuri S., Carlson A. Use of the Escherichia coli SSB gene to prevent bioreactor takeover by plasmidless cells. Bio/Technol. 1990;8:47–51. doi: 10.1038/nbt0190-47. [DOI] [PubMed] [Google Scholar]
- 48.Bernstein D.A., Eggington J.M., Killoran M.P., Misic A.M., Cox M.M., Keck J.L. Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc. Natl Acad. Sci. USA. 2004;101:8575–8580. doi: 10.1073/pnas.0401331101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.George N.P., Ngo K.V., Chitteni-Pattu S., Norais C.A., Battista J.R., Cox M.M., Keck J.L. Structure and cellular dynamics of Deinococcus radiodurans single-stranded DNA (ssDNA)-binding protein (SSB)-DNA complexes. J. Biol. Chem. 2012;287:22123–22132. doi: 10.1074/jbc.M112.367573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bayer I., Fliess A., Greipel J., Urbanke C., Maass G. Modulation of the affinity of the single-stranded DNA-binding protein of Escherichia coli (E. coli SSB) to poly(dT) by site-directed mutagenesis. Eur. J. Biochem./FEBS. 1989;179:399–404. doi: 10.1111/j.1432-1033.1989.tb14567.x. [DOI] [PubMed] [Google Scholar]
- 51.Khamis M.I., Casas-Finet J.R., Maki A.H., Murphy J.B., Chase J.W. Role of tryptophan 54 in the binding of E. coli single-stranded DNA-binding protein to single-stranded polynucleotides. FEBS Lett. 1987;211:155–159. doi: 10.1016/0014-5793(87)81427-8. [DOI] [PubMed] [Google Scholar]
- 52.Khamis M.I., Casas-Finet J.R., Maki A.H., Murphy J.B., Chase J.W. Investigation of the role of individual tryptophan residues in the binding of Escherichia coli single-stranded DNA binding protein to single-stranded polynucleotides. A study by optical detection of magnetic resonance and site-selected mutagenesis. J. Biol. Chem. 1987;262:10938–10945. [PubMed] [Google Scholar]
- 53.Curth U., Greipel J., Urbanke C., Maass G. Multiple binding modes of the single-stranded DNA binding protein from Escherichia coli as detected by tryptophan fluorescence and site-directed mutagenesis. Biochemistry. 1993;32:2585–2591. doi: 10.1021/bi00061a016. [DOI] [PubMed] [Google Scholar]
- 54.Chen J., Smith D.L., Griep M.A. The role of the 6 lysines and the terminal amine of Escherichia coli single-strand binding protein in its binding of single-stranded DNA. Protein Sci. 1998;7:1781–1788. doi: 10.1002/pro.5560070813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carlini L.E., Porter R.D., Curth U., Urbanke C. Viability and preliminary in vivo characterization of site-directed mutants of Escherichia coli single-stranded DNA-binding protein. Mol. Microbiol. 1993;10:1067–1075. doi: 10.1111/j.1365-2958.1993.tb00977.x. [DOI] [PubMed] [Google Scholar]
- 56.Carlini L.E., Porter R.D. Analysis of ssb mutations in vivo implicates SSB protein in two distinct pathways of SOS induction and in recombinational DNA repair. Mol. Microbiol. 1997;24:129–139. doi: 10.1046/j.1365-2958.1997.3431694.x. [DOI] [PubMed] [Google Scholar]
- 57.Ferrari M.E., Fang J., Lohman T.M. A mutation in E. coli SSB protein (W54S) alters intra-tetramer negative cooperativity and inter-tetramer positive cooperativity for single-stranded DNA binding. Biophys. Chem. 1997;64:235–251. doi: 10.1016/s0301-4622(96)02223-5. [DOI] [PubMed] [Google Scholar]
- 58.Merrill B.M., Williams K.R., Chase J.W., Konigsberg W.H. Photochemical cross-linking of the Escherichia coli single-stranded DNA-binding protein to oligodeoxynucleotides. Identification of phenylalanine 60 as the site of cross-linking. J. Biol. Chem. 1984;259:10850–10856. [PubMed] [Google Scholar]
- 59.Casas-Finet J.R., Khamis M.I., Maki A.H., Chase J.W. Tryptophan 54 and phenylalanine 60 are involved synergistically in the binding of E. coli SSB protein to single-stranded polynucleotides. FEBS Lett. 1987;220:347–352. doi: 10.1016/0014-5793(87)80844-x. [DOI] [PubMed] [Google Scholar]
- 60.Kozlov A.G., Lohman T.M. Stopped-flow studies of the kinetics of single-stranded DNA binding and wrapping around the Escherichia coli SSB tetramer. Biochemistry. 2002;41:6032–6044. doi: 10.1021/bi020122z. [DOI] [PubMed] [Google Scholar]
- 61.Antony E., Kozlov A.G., Nguyen B., Lohman T.M. Plasmodium falciparum SSB tetramer binds single-stranded DNA only in a fully wrapped mode. J. Mol. Biol. 2012;420:284–295. doi: 10.1016/j.jmb.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGhee J.D., von Hippel P.H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 63.Porter R.D., Black S. The single-stranded-DNA-binding protein encoded by the Escherichia coli F factor can complement a deletion of the chromosomal ssb gene. J. Bacteriol. 1991;173:2720–2723. doi: 10.1128/jb.173.8.2720-2723.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bonura T., Smith K.C. Enzymatic production of deoxyribonucleic acid double-strand breaks after ultraviolet irradiation of Escherichia coli K-12. J. Bacteriol. 1975;121:511–517. doi: 10.1128/jb.121.2.511-517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murzin A.G. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paytubi S., McMahon S.A., Graham S., Liu H., Botting C.H., Makarova K.S., Koonin E.V., Naismith J.H., White M.F. Displacement of the canonical single-stranded DNA-binding protein in the Thermoproteales. Proc. Natl Acad. Sci. USA. 2012;109:E398–E405. doi: 10.1073/pnas.1113277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shamoo Y., Friedman A.M., Parsons M.R., Konigsberg W.H., Steitz T.A. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376:362–366. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 68.Hollis T., Stattel J.M., Walther D.S., Richardson C.C., Ellenberger T. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc. Natl Acad. Sci. USA. 2001;98:9557–9562. doi: 10.1073/pnas.171317698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fanning E., Klimovich V., Nager A.R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bastin-Shanower S.A., Brill S.J. Functional analysis of the four DNA binding domains of replication protein A. The role of RPA2 in ssDNA binding. J. Biol. Chem. 2001;276:36446–36453. doi: 10.1074/jbc.M104386200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumaran S., Kozlov A.G., Lohman T.M. Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry. 2006;45:11958–11973. doi: 10.1021/bi060994r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kozlov A.G., Eggington J.M., Cox M.M., Lohman T.M. Binding of the dimeric Deinococcus radiodurans single-stranded DNA binding protein to single-stranded DNA. Biochemistry. 2010;49:8266–8275. doi: 10.1021/bi100920w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Witte G., Fedorov R., Curth U. Biophysical analysis of Thermus aquaticus single-stranded DNA binding protein. Biophys. J. 2008;94:2269–2279. doi: 10.1529/biophysj.107.121533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Su X.C., Wang Y., Yagi H., Shishmarev D., Mason C.E., Smith P.J., Vandevenne M., Dixon N.E., Otting G. Bound or free: interaction of the C-terminal domain of Escherichia coli single-stranded DNA-binding protein (SSB) with the tetrameric core of SSB. Biochemistry. 2014;53:1925–1934. doi: 10.1021/bi5001867. [DOI] [PubMed] [Google Scholar]
- 75.Antony E., Weiland E.A., Korolev S., Lohman T.M. Plasmodium falciparum SSB tetramer wraps single-stranded DNA with similar topology but opposite polarity to E. coli SSB. J. Mol. Biol. 2012;420:269–283. doi: 10.1016/j.jmb.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.