Abstract
Cannabis is the most widely produced and consumed illicit psychoactive substance worldwide. Occasional cannabis use can progress to frequent use, abuse and dependence with all known adverse physical, psychological and social consequences. Individual differences in cannabis initiation are heritable (40–48%). The International Cannabis Consortium was established with the aim to identify genetic risk variants of cannabis use. We conducted a meta-analysis of genome-wide association data of 13 cohorts (N=32 330) and four replication samples (N=5627). In addition, we performed a gene-based test of association, estimated single-nucleotide polymorphism (SNP)-based heritability and explored the genetic correlation between lifetime cannabis use and cigarette use using LD score regression. No individual SNPs reached genome-wide significance. Nonetheless, gene-based tests identified four genes significantly associated with lifetime cannabis use: NCAM1, CADM2, SCOC and KCNT2. Previous studies reported associations of NCAM1 with cigarette smoking and other substance use, and those of CADM2 with body mass index, processing speed and autism disorders, which are phenotypes previously reported to be associated with cannabis use. Furthermore, we showed that, combined across the genome, all common SNPs explained 13–20% (P<0.001) of the liability of lifetime cannabis use. Finally, there was a strong genetic correlation (rg=0.83; P=1.85 × 10−8) between lifetime cannabis use and lifetime cigarette smoking implying that the SNP effect sizes of the two traits are highly correlated. This is the largest meta-analysis of cannabis GWA studies to date, revealing important new insights into the genetic pathways of lifetime cannabis use. Future functional studies should explore the impact of the identified genes on the biological mechanisms of cannabis use.
Introduction
Cannabis is the most widely produced and consumed illicit psychoactive substance worldwide.1 Following initiation, occasional cannabis use can progress to frequent use, abuse and dependence. About 1 in 10 occasional users becomes dependent, which is associated with physical, psychological, social and occupational consequences.2, 3 Despite the increasing use of cannabis for medicinal purposes,4 associations with adverse health effects have been reported.5, 6 These include increased risk for psychiatric outcomes, including psychosis, schizophrenia, schizotypal personality disorder and mania.7, 8 Early cannabis use appears to moderate relationship between polygenic risk scores for schizophrenia and brain maturation.9 In view of expanding medicalization and decriminalization, the potential consequences, and the debate surrounding the benefits versus adverse consequences associated with cannabis use,10 understanding the genetics of cannabis use should be a public health priority.11
The risk of lifetime cannabis use, defined as any use of cannabis during the lifetime, varies between individuals. Previous studies have shown that individual differences in lifetime cannabis use can be partly explained by genetic differences between individuals; a meta-analysis of twin studies reported significant heritability estimates of lifetime cannabis use of 48% for males and 40% for females.12 Shared environmental factors, such as cannabis availability and parental monitoring,13, 14 also have a role accounting for 25 and 39% of the risk for males and females, respectively.12 Also, there is substantial overlap in the genetic risks underlying lifetime cannabis use and cannabis use disorder.15
Several studies have sought to identify specific genetic risk factors associated with cannabis use phenotypes. Genome-wide linkage studies have revealed suggestive evidence for linkage across many chromosomes.16, 17, 18, 19, 20 With very little consistency across studies, nearly all findings failed to meet genome-wide significance. The one study examining lifetime cannabis use16 reported a nonsignificant linkage locus on chromosome 18 (LOD score=1.97).
Candidate gene studies, including reports examining the CNR1, GABRA2, FAAH and ABCB1 genes have detected some significant associations with cannabis use but again, replication has been inconsistent.21, 22, 23 On the basis of a sample of 7452 Caucasian individuals, Verweij et al.21 found no gene-based associations between the frequency of cannabis use and 10 candidate genes identified by Agrawal and Lynskey.24 Overall, the results of candidate-gene studies are inconclusive; some associations have been replicated a few times, but failed to replicate in other studies. Moreover, findings may be further distorted due to publication bias favouring significant results.
As an alternative to the candidate-gene approach, the genome-wide association study (GWAS) is a hypothesis-free method that aims to detect novel genetic variants involved in complex traits. To date, three GWASs of cannabis use phenotypes have been published: one GWAS of cannabis dependence in 708 cannabis-dependent individuals and 2346 controls;25 a GWAS meta-analysis of lifetime cannabis use based on two studies with a combined sample size of 10 091 individuals (40.7% users);26 and a recent GWAS of lifetime cannabis use and age of cannabis use onset based on a sample of 6744 individuals (of whom 20% were users).27 None of the studies identified any genome-wide significant associations. This was likely due to the small effect sizes typical of common variants underpinning highly polygenic traits,28 thereby indicating a need for larger sample sizes. In this context, the success of larger GWASs and international consortia examining a variety of complex traits is encouraging.29 For example, multiple large meta-analyses of GWA results for number of cigarettes smoked per day have independently identified associations on chromosome 15q25 spanning the α5, α3 and β4 nicotinic receptor subunit gene clusters (CHRNA5, CHRNA3, CHRNB4).30, 31, 32
These and other recent GWA findings29 clearly illustrate the need for larger sample sizes. In response to this need, the International Cannabis Consortium was established to combine the results of multiple GWASs to identify the genetic variants underlying individual differences in cannabis use phenotypes. Our rationale for focusing on lifetime cannabis use (yes/no) is because this phenotype is heritable and shares significant genetic risks with that risk for cannabis abuse or dependence.14, 15, 33 In contrast to frequency of use or abuse and dependence, which are not commonly assessed in large-scaled genetic studies, most general population studies have assessed lifetime cannabis use, thereby increasing our sample size and power to detect associations. Currently, the combined International Cannabis Consortium sample size for lifetime cannabis is 32 330 individuals from 13 cohorts from Europe, the United States and Australia, along with four independent replication samples comprising 5627 individuals. This sample size is considerably larger than the sample size of the previous GWAS investigating lifetime cannabis use in two samples from Australia and the UK, thereby providing substantially greater power to detect genetic variants of small effect size.
The aim of the present study is to identify genetic variants associated with lifetime cannabis use by meta-analysis of the GWAS results from all contributing International Cannabis Consortium samples. The tests of association for individual genetic variants will be complemented with gene-based tests of association. In addition, we will investigate which proportion of the heritability inferred by twin studies is explained by common SNPs captured on GWAS arrays. Finally, we will estimate the genetic correlation between lifetime cannabis and smoking initiation based on the analysis of our summary statistics and those from the publicly available Tobacco Alcohol and Genetics consortium.
Materials and methods
Cohorts
We performed a meta-analysis of GWA results from 13 discovery samples from Europe, USA and Australia including a total of 32 330 individuals of European ancestry. The size of the samples ranged from 721 to 6778 individuals. The age of the participants ranged from 16 to 87 years with an average of 34 years. The percentage of females ranged from 30 to 66% with an average of 53%. Owing to the differences in recruitment strategies, cultural and temporal difference, combined with likely variation in the drug availability between countries, there was a wide range in the prevalence of lifetime use (that is, never/ever used cannabis), which varied from 1 to 92% with an average of 44.5%.
Four additional independent samples with a total of 5627 subjects were used for replication. One sample (n=2660) consisted of African American subjects. The other three included subjects of European ancestry. See Table 1 for individual sample characteristics. The procedures for data collection per sample are described in the Supplementary Information 1.
Table 1. Discovery and replication sample characteristics.
| Sample | Country | N | % Users | % Female | Mean age (range) | N SNPs |
|---|---|---|---|---|---|---|
| Discovery | ||||||
| ALSPAC | UK | 2976 | 42 | 56 | 18 (17–19) | 5 182 231 |
| BLTS | Australia | 721 | 60 | 57 | 26 (18–33) | 4 558 509 |
| CADD | USA | 853 | 79 | 30 | 25 (18–36) | 4 972 726 |
| EGCUT1 | Estonia | 2765 | 1.3 | 55 | 34 (18–66) | 6 048 479 |
| EGCUT2 | Estonia | 970 | 4.8 | 51 | 31 (18–50) | 5 171 164 |
| FinnTwin | Finland | 1029 | 27 | 52 | 23 (20–29) | 4 364 135 |
| HUVH | Spain | 981 | 20 | 30 | 36 (17–87) | 4 971 170 |
| MCTFR | USA | 6241 | 59 | 54 | 37 (18–71) | 6 304 767 |
| NTR | Netherlands | 4653 | 27 | 66 | 37 (18–60) | 4 644 238 |
| QIMR | Australia | 6778 | 51 | 54 | 45 (18–85) | 5 901 727 |
| TRAILS | Netherlands | 1226 | 51 | 47 | 19 (18–21) | 5 336 901 |
| Utrecht | Netherlands | 1173 | 54 | 54 | 21 (18–37) | 4 831 885 |
| Yale Penn EA | USA | 1964 | 92 | 40 | 38 (16–76) | 5 856 902 |
| Replication | ||||||
| Radar | Dutch | 338 | 59 | 44 | 20 (17–22) | 10 |
| SYS | Canada | 551 | 51 | 56 | 50 (36–65) | 10 |
| TwinsUK | UK | 2078 | 12 | 93 | 58 (18–86) | 10 |
| Yale Penn AA | US | 2660 | 82 | 46 | 42 (16–76) | 10 |
Abbreviations: N, sample size; N SNPs, number of SNPs used for the meta-analysis; SNP, single-nucleotide polymorphism; % female, percentage of females; % users, percentage of users that ever used cannabis.
Phenotype and covariates
For all individuals, the data were available on whether or not the subject reported having ever used cannabis during their lifetime: yes (1) versus no (0). Although phrasing of the question slightly differed between samples (see Supplementary Information 1), our unit of analysis reflected lifetime cannabis use in all the samples.
Covariates included age at the time of phenotypic assessment, sex, birth cohort and principal components (obtained from the genome-wide genotype data). Spanning 20-year intervals, birth cohort was dummy coded, with the lowest birth cohort (that is, oldest age group) used as the reference group. The details about phenotypic assessment and individual sample characteristics for the discovery and replication samples are located in Supplementary Information 1 and Supplementary Table 1.
Genotyping and imputation
Genotype imputation was based on the 1000 Genomes phase 1 reference panel.34 Allelic dosage data were used to account for genotype uncertainties. See Supplementary Table 2 for the genotyping platform, imputation program and quality control thresholds used.
Statistical analyses
GWA analysis in each discovery cohort
The GWA analyses were performed by each group separately. Associations between the binary phenotype and the genotypes were tested genome-wide using a logistic regression model including covariates (see above). For family-based samples, familial relatedness was taken into account by using a sandwich correction as implemented in PLINK.35 The analyses plan can be found in Supplementary Information 3. It should be noted that some groups did do the analyses in a slightly different manner based on the characteristics of their sample. The analyses plan that was send to the participating groups is included in Supplementary Information 3. It should be noted that some groups did do the analyses in a slightly different manner based on the characteristics of their sample. Supplementary Table 2 lists the program used by each group.
Meta-analysis of GWAS results
Before performing the meta-analysis, we applied a set of filters to each GWA results set independently. First, we removed insertions and deletions, ensuring that all base pair positions were unique and referred to the same genetic variant (that is, SNP). Second, we removed genotyped SNPs that were not in Hardy–Weinberg equilibrium (P⩽10−5). Third, we removed SNPs with minor allele frequency (MAF) <√(5/N), which under the assumption of Hardy–Weinberg equilibrium corresponded to less than five estimated individuals in the least frequent genotype group. In the EGCUT1 sample, due to very low prevalence of lifetime cannabis use (1.3%), we excluded SNPs with MAF<0.2. Fourth, regardless of the quality score type used, we excluded SNPs with imputation quality scores below 0.6. Finally, SNPs present in only one sample and SNPs with alleles or allele frequencies inconsistent with the 1000 Genomes phase I European reference panel (absolute MAF difference >0.15) were removed.
We performed a fixed-effects meta-analysis based on the cohort's effect sizes and standard errors using METAL.36 Our meta-analysis combined association summary statistics for 6 444 471 SNPs that passed all the filters. We applied the conventional threshold of 5 × 10−8 as an indication of genome-wide significance (see ref. 37). Although the combined sample size of the meta-analysis based on the discovery samples is 32 330, the sample size per SNP varies due to missingness across subsamples.
Gene-based test
Results of the GWAS were then used as part of gene-based tests of association in the Knowledge-based mining system for Genome-wide Genetic studies (KGG) software package Version 3.5.38, 39 This approach uses an extended Simes test that integrates prior functional information and the meta-analysis association results when combining the SNP P-values within a gene to obtain an overall association P-value for each entire gene. We conducted 24 576 gene-based tests of association. The genome-wide significance level according to the Knowledge-based mining system for Genome-wide Genetic studies default setting of Benjamini and Hochberg false discovery rate threshold of 0.05 (ref. 40) was 9.38 × 10−6.
Estimation of SNP-based heritability and genetic overlap with lifetime cigarette smoking
The proportion of phenotypic variance that could be explained by the SNPs was estimated using the density estimation method developed by So et al.41 The density estimation method estimates the genome-wide distribution of effect sizes based on the difference between the observed distribution of test statistics in the meta-analysis and the corresponding null distribution. Before estimation, the SNPs present in at least 25% of the meta-analysis samples were pruned for LD. We used the r2=0.15 pruning level as the primary result for consistency with other applications of this method. Additional details are located in the Supplementary Information 2. LD Score regression42, 43 was used as an alternative method to estimate the SNP-based heritability, as well as to estimate the degree of genetic covariance between lifetime cannabis use (present study) and lifetime cigarette smoking31 (see Supplementary Information 2).
Results
Meta-analysis
No genome-wide significant associations between individual SNPs and lifetime cannabis use were observed (see Manhattan plot, Supplementary Figure 1a). However, the QQ plot (Supplementary Figure 1b) reveals strong enrichment of SNPs with P<10−4. Supplementary Figures 2a–m and 3a–m illustrate the Manhattan and QQ plots for each sample. Table 2 illustrates the top 10 independent (R2<0.1) SNPs associated with lifetime cannabis use. None of these 10 SNPs were replicated in the four independent replication samples (Supplementary Table 3). In a combined meta-analysis of the 10 top SNPs (that is, discovery plus replication samples), none of the SNPs reached genome-wide significance. Local plots of the most strongly associated regions, including neighboring genes, are provided in Supplementary Figures 4a–j. The most statistically significant marker (P-value =4.6 × 10−7) was rs4984460 located on chromosome 15 (see Supplementary Figure 5 for the forest plot). The SNP is located in an intergenic region between LOC400456/LOC145820 and NR2F2 and MIR1469 genes. Supplementary Table 4 includes the 153 SNPs identified with P-values <10−5. Because not all SNPs passed the post-imputation quality control steps in all the samples, this table includes the effective sample size per SNP.
Table 2. Top 10 SNPs with meta-analysis results of discovery samples, and results of combined discovery and replication samples.
| SNP | Chr | BP (hg19) | A1 | A2 | Freq A1 |
Discovery |
Combineda |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Beta (s.e.) | P-value | Directionb | Beta (s.e.) | P-value | ||||||
| rs4984460 | 15 | 96424399 | T | G | 0.75 | −0.11 (.023) | 4.6 × 10−7 | +−−++−−−−−−−+ | −0.11 (0.023) | 2.2 × 10−6 |
| rs2099149 | 12 | 30479358 | T | G | 0.81 | −0.16 (0.032) | 9.8 × 10−7 | −−−?−??−?−−+− | −0.17 (0.034) | 5.1 × 10−7 |
| rs7675351 | 4 | 141218757 | A | C | 0.86 | −0.15 (0.031) | 1.4 × 10−6 | −−−?+−−−?−−−− | −0.13 (0.033) | 1.1 × 10−4 |
| rs4471463 | 11 | 112983595 | T | C | 0.55 | −0.09 (0.020) | 1.5 × 10−6 | −−−−+−+−−−−+− | −0.1 (0.021) | 9.0 × 10−7 |
| rs7107977 | 11 | 915764 | A | G | 0.60 | 0.27 (0.058) | 1.9 × 10−6 | ??+++?+???+?+ | 0.29 (0.064) | 6.4 × 10−6 |
| rs58691539 | 2 | 52753909 | T | G | 0.91 | −0.29 (0.062) | 2.1 × 10−6 | −????−?−????− | −0.29 (0.062) | 2.2 × 10−6 |
| rs2033867 | 2 | 175188281 | A | G | 0.06 | 0.24 (0.051) | 2.6 × 10−6 | +??????+++??+ | 0.23 (0.050) | 4.2 × 10−6 |
| rs35053471 | 3 | 47124761 | A | T | 0.38 | −0.10 (0.022) | 2.7 × 10−6 | −−−−−?+−−−−−− | −0.09 (0.022) | 9.2 × 10−5 |
| rs12518098 | 5 | 60864467 | C | G | 0.68 | 0.10 (0.022) | 3.0 × 10−6 | ++++−++++++++ | 0.09 (0.023) | 4.7 × 10−5 |
| rs73067624 | 1 | 196333461 | T | C | 0.90 | −0.18 (0.039) | 3.1 × 10−6 | −?−?−−−−?−−−− | −0.16 (0.041) | 6.3 × 10−5 |
Abbreviations: A1, allele 1; A2, allele 2; BP (hg19), location in base pairs in human genome version 19; Chr, chromosome; Freq A1, frequency of allele 1; SNP, single-nucleotide polymorphism.
The combined sample contains the discovery samples and the Radar, SYS and TwinsUK replication samples.
Direction per sample: allele A1 increases (+) or decreases (−) liability for cannabis use, or sample did not contribute to this SNP because it did not pass the post-imputation quality control (?). Order of samples: ALSPAC, BLTS, CADD, EGCUT1, EGCUT2, FinnTwin, HUVH, MCTFR, NTR, QIMR, TRAILS, Utrecht, Yale Penn EA. Sample information can be found in Table 1.
SNPs are displayed when not in linkage disequilibrium (R2<0.1. For SNPs with R2⩾0.1, only the most significant SNP is shown in the top 10).
Gene-based tests
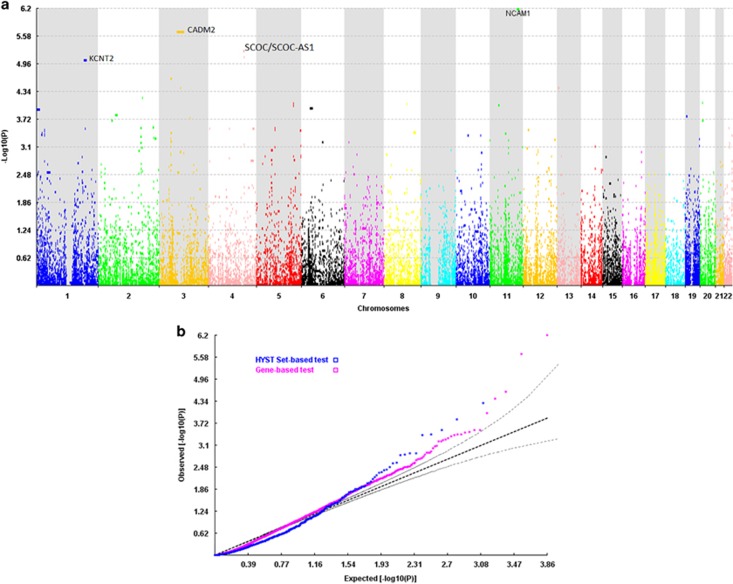
The gene-based tests of associations were run on 24 576 genes/genetic regions (see ‘Materials and Methods' section for details). The Manhattan and QQ plot for this test are shown in Figures 1a and b. Results for the top 100 genes can be found in Supplementary Table 5. As shown in Table 3, four genes and one intergenic noncoding RNA region were significantly (false discovery rate-corrected P<0.05) associated with lifetime cannabis use: (i) neural cell adhesion molecule 1 (NCAM1, on 11q23); (ii) cell adhesion molecule 2 (CADM2, on 3p12); (iii) short coiled-coil protein (SCOC) and (iv) SCOC antisense RNA1 (SCOC-AS1, both located on 4q31); and (v) potassium channel, subfamily T, member 2 (KCNT2, on 1q31). Regional plots44 of these top genes are located in Supplementary Figure 6.
Figure 1.
The Manhattan (a) and the QQ plot (b) based on results of the gene-based analysis performed in the discovery sample using HYST (hybrid set-based test).
Table 3. Top five genes from the gene-based tests of association with corrected P-values (Benjamini and Hochberg) based on the meta-analytic discovery and replication samples.
| Gene | Chr | Start position (hg19) | BP length | N SNPS | Nominal P-values discovery | Corrected P-values discovery | Nominal P-values EU replication samples | Nominal P-values replication African Americans |
|---|---|---|---|---|---|---|---|---|
| NCAM1 | 11 | 112831968 | 303 952 | 400 | 6.26 × 10−7 | 0.015 | 0.381 | 0.302 |
| CADM2 | 3 | 85008132 | 1 115 448 | 978 | 2.13 × 10−6 | 0.026 | 0.744 | 0.112 |
| SCOC-AS1 | 4 | 141204879 | 89 668 | 81 | 5.76 × 10−6 | 0.046 | 0.681 | 0.044 |
| SCOC | 4 | 141264614 | 39 097 | 111 | 7.85 × 10−6 | 0.046 | 0.636 | 0.027 |
| KCNT2 | 1 | 196194909 | 382 653 | 237 | 9.38 × 10−6 | 0.046 | 0.269 | 0.201 |
Abbreviations: BP length, base pair length; chr, chromosome; hg19, human genome version 19; N SNPs, number of SNPs used for the meta-analysis; SNP, single-nucleotide polymorphism.
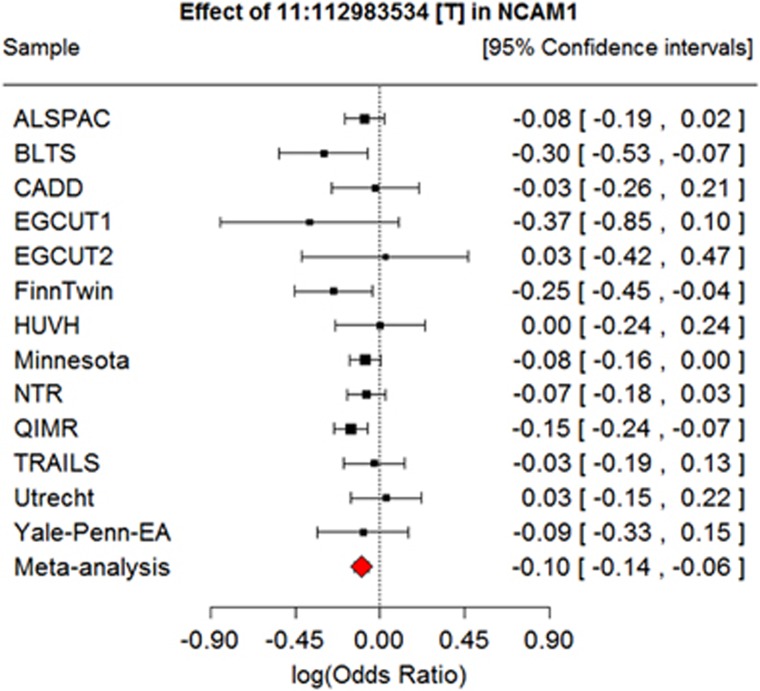
The smallest gene-based P-value was found for the NCAM1 gene. Within this gene, rs4471463 had the lowest SNP P-value, and was also among the top 10 associated SNPs. The forest plot in Figure 2 illustrates the effect of this SNP in each sample. In most samples, the effect is in the same direction, such that the major (T) allele is associated with a decreased risk of lifetime cannabis use. The forest plot for two SNPs with lowest P-values in the other significant gene regions can be found in Supplementary Figure 5.
Figure 2.
Forest plot for the top-SNP rs4471463 in the NCAM1 gene on chromosome 11. SNP, single-nucleotide polymorphism.
Of the five genes included in our replication analyses, none were replicated in two of the independent replication samples (see Table 3). In the African American replication sample, suggestive associations with SCOC-AS1 (P=0.044) and SCOC (P=0.027) were found.
SNP-based heritability and genetic overlap with lifetime cigarette smoking
Using the density estimation method (see ‘Materials and Methods' section for a description), all the SNPs available in at least 25% of the samples when combined explained 20% of the total variance in lifetime cannabis use (P<0.001). Alternative estimation with LD score regression also yielded a significant heritable component of 13% (h2LD=0.13, s.e.=0.02, P=1.4 × 10−7). These variance estimates were robust across pruned sets with similar r2 thresholds (see Supplementary Table 6). Stricter LD pruning (that is, r2=0.05), or restricting analyses to SNPs present in all studies substantially decreased the estimate of variance explained. Both SNP heritability estimates confirmed that lifetime cannabis use has a significant heritable component (13–20%), indicating that GWAS should be able to identify these common SNPs (but effect sizes are small and large sample sizes are thus required). However, because these estimates are only based on common SNPs, the total heritability of lifetime cannabis use is likely to be higher.
The LD score regression analyses revealed a strong and highly significant genetic correlation (rg=0.83, s.e.=0.15, P=1.85 × 10−8) between lifetime cannabis use and lifetime cigarette smoking (based on the Tobacco Alcohol and Genetics consortium31 summary results), implying that SNPs for lifetime cannabis use and lifetime cigarette smoking are highly correlated.
Discussion
To date, this is the largest GWA study of lifetime cannabis use. We performed meta-analysis of the GWA results based on a discovery sample comprising 32 330 individuals from 13 cohorts, and a replication sample comprising 5627 subjects from four cohorts (including one African American cohort). There were no genome-wide significant SNP associations. However, heritability analyses revealed that between 13 and 20% of the variation in lifetime cannabis use could be explained by common SNPs. Moreover, gene-based tests of association identified four protein-coding genes and one intergenic region significantly associated with lifetime cannabis use including NCAM1, which has previously been linked to substance use.45, 46, 47, 48 Finally, we revealed that the genetic liability to lifetime cannabis use correlated to a large extent (r=0.83) with the genetic liability to lifetime cigarette smoking. Our results are consistent with the hypothesis that lifetime cannabis use is a highly polygenic trait, comprising many SNPs each with small effects contributing to lifetime risk. Moreover, portions of the genetic risk in lifetime cannabis use likely correlates with other substances including cigarette smoking.
Our top gene associated with lifetime cannabis use was NCAM1, a known candidate for nicotine dependence.45 The role of NCAM1 is to regulate pituitary growth hormone secretion as a membrane-bound glycoprotein that mediates cell–cell contact by hemophilic interactions.46 NCAM1 is part of the NCAM1–TTC12–ANKK1–DRD2 (NTAD) gene cluster, which is related to neurogenesis and dopaminergic neurotransmission. Importantly, the NTAD cluster has been associated with smoking behavior and nicotine dependence,45, 47, 48, 49, 50, 51, 52 alcohol dependence,53, 54 heroin dependence,55 as well as other substance use disorders.54 Although it is plausible that NCAM1 is capturing pleiotropic risks underpinning the liability to licit and illicit substance use in general, we note that NCAM1 was not identified either by the Tobacco Alcohol and Genetics consortium or other consortia for cigarette smoking.30, 31, 32 The functions of the putative variants responsible for the associations in the candidate-gene studies remain to be determined.
The second gene, CADM2, is a synaptic cell adhesion molecule (SynCAM family) belonging to the immunoglobulin (Ig) superfamily. Variants in the CADM2 gene have been previously associated with body mass index,56 processing speed57 and autism disorders.58 Interestingly, these phenotypes were associated with cannabis use in previous studies,59, 60, 61 which together suggest that CADM2 can be considered an important gene related to a variety of complex traits. It is possible that the association with lifetime cannabis use may be driven, for example, by differences in personality rather than as a direct relationship with lifetime use.
The third gene, SCOC, encodes a short coiled-coil domain-containing protein that localizes to the Golgi apparatus. Many coiled-coil-type proteins are involved in important biological functions such as the regulation of gene expression through the regulation of transcription factor binding.62 The function of SCOC is largely unknown and no previous association studies have linked SCOC to cannabis or other substance use phenotypes. The SCOC antisense RNA1 gene is located in the same chromosomal region.
Finally, KCNT2 encodes a potassium voltage-gated channel (subfamily S, member 2). The sodium-activated potassium channels Slack and Slick are encoded by KCNT1 (potassium channel, subfamily T, member 1) and KCNT2, respectively, which are found in neurons throughout the brain. Suggestive association for SNPs near KCNT2 have previously been found for cocaine dependence and for early-onset, highly comorbid, heavy opioid use.63, 64 This suggests that potassium signaling may have a role in addiction.
The lack of genome-wide significant associations for individual SNPs is consistent with previous GWA studies of lifetime cannabis use26, 27 and cannabis dependence.25 The difficulty of identifying specific SNPs for lifetime cannabis use may be attributable to several reasons. First, complex traits are known to be influenced by many variants, each with very small effect sizes. Although power calculations reveals suitable power (96%) to detect odds ratios of 1.15 based on common SNPs (MAF=0.2), the power to detect smaller effect sizes remains lower. For example, there is only 28% power to detect effect sizes with odds ratio of 1.1 and MAF=0.2. Therefore, our data suggest that the effect sizes of single variants contributing to lifetime cannabis use are likely to be smaller than 1.15. Combining variants within larger units (that is, genes) did however reveal four significant genes associated with lifetime cannabis use implying that these genes are appropriate targets for future functional studies of cannabis use. Unfortunately, our gene-based results were not replicated in the replication samples, probably due to low sample sizes and therefore low power. In the African American replication sample, we did find suggestive association with SCOC-AS1 and SCOC.
On the basis of twin studies, the heritability of lifetime cannabis use is estimated at 40–50%.12 In our study, all common SNPs combined explained 13–20% of the variance in the liability to use cannabis depending on the method used. Stricter LD pruning (that is, r2=0.05) or restricting to SNPs observed (genotyped or imputed) in all the analyses, substantially reduces the estimate of variance explained. Speculatively, this may indicate that much of the variance explained comes from SNPs located in the regions of weak LD. Such effects are likely to be poorly tagged for the estimation of variance explained after strict LD pruning, and are likely to be more difficult to impute owing to a lack of strongly correlated genotyped SNPs (and thus missing from some studies). Our SNP-based heritability estimates lie in between two previous heritability estimates for lifetime cannabis use based on the Genome-wide Complex Trait Analysis65 software package. Verweij et al.26 estimated that 6% of the variance in lifetime cannabis use is explained by aggregated common SNPs (MAF>0.05). Minică et al.27 found an estimate of 25%. Provided that the current sample is much larger than the samples used in the previous studies, we conclude that approximately one-third to half of the heritability is explained by common SNPs captured on a GWAS array. Other sources of variation may explain the discrepancy between SNP- and twin-based heritability estimates. For example, age-related genetic differences, non-additive genetic variance, interactions between genetic variants and environmental risk factors, epistasis and/or rare mutations may also have a role.
Our results indicate a very high genetic overlap (r=0.83) between our measure of lifetime cannabis use and lifetime cigarette use when based on the SNP panel. Twin studies have shown moderate to high genetic correlations of 0.59–0.74 between lifetime cannabis and nicotine use.66 Kendler et al.67 also reported significant biometrical genetic correlations between the levels of cannabis, nicotine and alcohol use, which were increasingly influenced by common genetic risks detectable in early adulthood.
Our findings should be interpreted in the context of at least four potential limitations. First, our study was underpowered to detect very small effects of individual variants. Power analyses revealed that a twofold increase in sample size is required to detect SNP effect sizes with odds ratios of 1.1. Second, lifetime cannabis use is a dichotomous measure combining single lifetime, regular and chronic users. Consequently, our sample may compromise heterogeneous patterns of use, which has the potential to reduce the power to detect genetic association.68 Third, prevalences of lifetime cannabis use varied between 1% (EGCUT1) and 92% (Yale Penn EA). This was likely due to differences in the sample characteristics, recruitment strategies and the political differences between countries. Despite these differences, the forest plots of the key SNPs (see Figure 2; see also Supplementary Figure 5) revealed that the 95% confidence intervals surrounding the effect estimates typically included the estimated meta-analytic effect, which tends to overlap across studies. This indicates that the input samples were representative of the same population of users. Finally, the average age of participants varied between 18 (ALSPAC) and 45 (QIMR) years. Consequently, some younger participants might have initiated cannabis use at a later age, but have been classified as ‘never users' in the current study. This can decrease power, but does not invalidate our results. In addition, we note that the average age of each sample did not correlate with sample prevalences (r=−0.04, P=0.91).
On the basis of our observations, the following recommendations for future studies can be made. We have identified four genes significantly associated with cannabis use, which are candidates for follow-up functional studies. In particular, the role of NCAM1 can be examined to determine the functional role of this gene, possibly in combination with other genes in the same gene cluster (NCAM1–TTC12–ANKK1–DRD2).
The next goal of the International Cannabis Consortium is to perform a meta-analysis on GWA studies investigating the age at first cannabis use. Our rationale is based on the observation that early initiation of cannabis use is associated with rapid progression towards cannabis abuse and dependence, polysubstance use and other substance use disorders.69, 70, 71 Methods other than GWASs may also be used to reveal the biological pathways of cannabis use, such as rare variant association analyses. The environmental risk factors may be incorporated to investigate gene × environment interactions. Hopefully, the combination of advanced technologies and novel statistical approaches with larger samples will further contribute to our understanding of the genetic architecture of cannabis use.
Conclusion
We have performed the largest meta-analysis to date of GWASs investigating cannabis use phenotypes. With a sample of over 32 000 individuals, our results implicate four genes as involved in lifetime cannabis use: NCAM1, CADM2, SCOC and KCNT2. Our results illustrated that lifetime cannabis use is under the influence of many common genetic variants. The combined SNPs explained 13–20% of the phenotypic variation, and revealed a high degree of genetic sharing (r=0.83) with lifetime cigarette smoking. Future studies should investigate the impact of these genes on the biological mechanisms leading to lifetime cannabis use.
Acknowledgments
JMV, CCM and HM are supported by the European Research Council (Beyond the Genetics of Addiction ERC-284167, PI JMV). SS and EMD are supported by the Foundation Volksbond Rotterdam. KJHV is supported in part by the Netherlands Organization for Health Research and Development (ZonMW 31160212) and in part by a 2014 NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation. NAG is supported by US National Institutes of Health, National Institute on Drug Abuse R00DA023549. GHL is supported by NIDA R37 DA-018673 and NSF BCS-1229450. RW is supported by NIH U01 MH094432 and NSF BCS-1229450. Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara and financially supported by the Netherlands Organization for Scientific Research (NWO 480-05-003 PI: Posthuma) along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam. The study site acknowledgments are as follows: ALSPAC—We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council and the Wellcome Trust (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. GWAS data were generated by Sample Logistics and Genotyping Facilities at the Wellcome Trust Sanger Institute and LabCorp (Laboratory Corportation of America) using support from 23andMe. AIS is supported by a Wellcome Trust 4-year PhD studentship in molecular, genetic and lifecourse epidemiology (WT083431MA). JJW is supported by a Postdoctoral Research Fellowship from the Oak Foundation. JJW and MRM are members of the MRC Integrative Epidemiology Unit at the University of Bristol, funded by the UK Medical Research Council (MC_UU_12013/6) and the University of Bristol. JJW and MRM are also members of the UK Centre for Tobacco and Alcohol Studies, a UKCRC Public Health Research: Centre of Excellence. Funding from British Heart Foundation, Cancer Research UK, Economic and Social Research Council, Medical Research Council and the National Institute for Health Research, under the auspices of the UK Clinical Research Collaboration is gratefully acknowledged. BLTS—The BLTS was supported by grants from the United States National Institute on Drug Abuse (R00DA023549) awarded to NAG, by the Australian Research Council to MJW (Nos. DP0343921, DP0664638 and DP1093900) and by Australian National Health and Medical Research Council Australia Fellowships awarded to Ian Hickie (No. 464914) and GWM (No. 619667). We acknowledge and thank the following project staff: AKH, Leanne Wallace and Lisa Bowdler for the laboratory processing, genotyping and QC; Soad Hancock as Project Coordinator; Lenore Sullivan as Research Editor; our research interviewers Pieta-Marie Shertock and Jill Wood; and David Smyth for IT. We also thank the twins and their siblings for their willing cooperation. SEM was supported by an ARC future fellowship FT110100548. CADD—The Center on Antisocial Drug Dependence (CADD) data reported here were funded by grants from the National Institute on Drug Abuse (P60 DA011015, R01 DA012845, R01 DA021913, R01 DA021905, R01 DA035804). EGCUT—received financing by FP7 grants (278913, 306031, 313010), Center of Excellence in Genomics (EXCEGEN) and University of Tartu (SP1GVARENG). We acknowledge EGCUT technical personnel, especially Mr V Soo and S Smit. Data analyses were carried out in part in the High Performance Computing Center of University of Tartu. FinnTwin—We warmly thank the participating twin pairs and their family members for their contribution. We express our appreciation to the skilled study interviewers A-M Livonen, K Karhu, H-M Kuha, U Kulmala-Gråhn, M Mantere, K Saanakorpi, M Saarinen, R Sipilä, L Viljanen and E Voipio. Anja Häppölä and Kauko Heikkilä are acknowledged for their valuable contribution in recruitment, data collection and data management. Phenotyping and genotyping of the Finnish twin cohorts has been supported by the Academy of Finland Center of Excellence in Complex Disease Genetics (grants 213506, 129680), the Academy of Finland (grants 100499, 205585, 118555, 141054, 265240, 263278 and 264146 to JK), National Institute of Alcohol Abuse and Alcoholism (grants AA-12502, AA-00145, and AA-09203 to RJR and AA15416 and K02AA018755 to DM Dick), Sigrid Juselius Foundation (to JK), and the Welcome Trust Sanger Institute, UK. Antti-Pekka Sarin and Samuli Ripatti are acknowledged for genotype data quality controls and imputation. GWAS analyses were run at the ELIXIR Finland node hosted at CSC—IT Center for Science for ICT resources. HUVH—Financial support was received from ‘Instituto de Salud Carlos III-FIS' (PI11/00571, PI11/01629, PI12/01139), ‘Plan Nacional Sobre Drogas' (PNSD#2011-0080), ‘Agència de Gestió d'Ajuts Universitaris i de Recerca-AGAUR, Generalitat de Catalunya ‘ (2014SGR1357) and ‘Departament de Salut', Government of Catalonia, Spain. MR is a recipient of a Miguel de Servet contract from the ‘Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación', Spain. MCTFR—This research was supported in part by USPHS Grants from the National Institute on Alcohol Abuse and Alcoholism (AA09367 and AA11886), the National Institute on Drug Abuse (DA05147, DA13240 and DA024417) and the National Institute on Mental Health (MH066140). NTR—We thank the Netherlands Twin Register participants whose data we analyzed in this study. This work was supported by grants from the Netherlands Organization for Scientific Research (ZonMW Addiction 31160008; ZonMW 940-37-024; NWO/SPI 56-464-14192; NWO-400-05-717; NWO-MW 904-61-19; NWO-MagW 480-04-004; NWO-Veni 016-115-035), the European Research Council (Beyond the Genetics of Addiction ERC-284167; Genetics of Mental Illness: ERC-230374), the Centre for Medical Systems Biology (NWO Genomics), Netherlands Bioinformatics Center/BioAssist/RK/2008.024. We acknowledge the EMGO+ Institute for Health and Care Research, the Neuroscience Campus Amsterdam, BBMRI-NL (184.021.007: Biobanking and Biomolecular Resources Research Infrastructure), the Avera Institute, Sioux Falls, South Dakota (USA) for support. Genotyping was funded in part by grants from the National Institutes of Health (4R37DA018673-06, RC2 MH089951), Rutgers University Cell and DNA Repository cooperative agreement (National Institute of Mental Health U24 MH068457-06), and the National Institutes of Health (NIH R01 HD042157-01A1, MH081802, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995) and the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health. The statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) which is supported by the Netherlands Scientific Organization (NWO 480-05-003), the Dutch Brain Foundation and the Department of Psychology and Education of the VU University Amsterdam. QIMR—This is supported by National Institutes of Health Grants AA07535, AA0758O, AA07728, AA10249, AA13320, AA13321, AA14041, AA11998, AA17688, DA012854, DA018267, DA018660, DA23668 and DA019951; by Grants from the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, 552498 and 628911); by Grants from the Australian Research Council (A7960034, A79906588, A79801419, DP0770096, DP0212016 and DP0343921); and by the 5th Framework Programme (FP-5) GenomEUtwin Project (QLG2-CT-2002-01254). This research was further supported by the Centre for Research Excellence on Suicide Prevention (CRESP—Australia). We thank AKH, Richard Parker, Soad Hancock, Judith Moir, Sally Rodda, Pieta-Maree Shertock, Heather Park, Jill Wood, Pam Barton, Fran Husband, Adele Somerville, Ann Eldridge, Marlene Grace, Kerrie McAloney, Lisa Bowdler, Alexandre Todorov, Steven Crooks, David Smyth, Harry Beeby and Daniel Park. Last, we thank the twins and their families for their participation. RADAR—We thank all the adolescents and their families and friends for their participation. Moreover, we thank the various assistants that helped in recruiting participants as well as collecting and cleaning the data. The research was funded partly by the Netherlands Organisation for Scientific Research (Brain and Cognition, 056-21-010). Data of the RADAR study were used. RADAR has been financially supported by main grants from the Netherlands Organisation for Scientific Research (GB-MAGW 480-03-005), and Stichting Achmea Slachtoffer en Samenleving (SASS) and various other grants from the Netherlands Organisation for Scientific Research, the VU University Amsterdam and Utrecht University. ACH is supported by the Netherlands Organization for Health Research and Development, ZonMW 31160212. Saguenay Youth Study—The Canadian Institutes of Health Research and the Heart and Stroke Foundation of Canada fund the SYS (TP, ZP). TP is the Tanenbaum Chair in Population Neuroscience (University of Toronto) and the Dr John and Consuela Phelan Scholar (Child Mind Institute). TRAILS—TRAILS (TRacking Adolescents' Individual Lives Survey) is a collaborative project involving various departments of the University Medical Center and University of Groningen, the Erasmus University Medical Center Rotterdam, the University of Utrecht, the Radboud Medical Center Nijmegen and the Parnassia Bavo group, all in the Netherlands. TRAILS has been financially supported by grants from the Netherlands Organization for Scientific Research NWO (Medical Research Council program grant GB-MW 940-38-011; ZonMW Brainpower grant 100-001-004; ZonMw Risk Behavior and Dependence grant 60-60600-97-118; ZonMw Culture and Health grant 261-98-710; Social Sciences Council medium-sized investment grants GB-MaGW 480-01-006 and GB-MaGW 480-07-001; Social Sciences Council project grants GB-MaGW 452-04-314 and GB-MaGW 452-06-004; NWO large-sized investment grant 175.010.2003.005; NWO Longitudinal Survey and Panel Funding 481-08-013); the Dutch Ministry of Justice (WODC), the European Science Foundation (EuroSTRESS project FP-006), Biobanking and Biomolecular Resources Research Infrastructure BBMRI-NL (CP 32), the participating universities and Accare Center for Child and Adolescent Psychiatry. We are grateful to all the adolescents, their parents and teachers who participated in this research and to everyone who worked on this project and made it possible. Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003) along with a supplement from the Dutch Brain Foundation. TwinsUK—The study was funded by the Wellcome Trust; European Community's Seventh Framework Programme (FP7/2007-2013). The study also receives support from the National Institute for Health Research (NIHR)- funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London. SNP Genotyping was performed by The Wellcome Trust Sanger Institute and National Eye Institute via NIH/C. Utrecht—We are grateful to Chris Schubart and Willemijn van Gastel and numerous students for their work in the study. Foremost, we thank our study participants. This study was financially supported by a grant of the NWO (Netherlands Organization for Scientific Research), grant no. 91207039. The study was performed at the University Medical Centre Utrecht, The Netherlands. Yale Penn—Genotyping services for a part of our GWAS study were provided by the Center for Inherited Disease Research (CIDR) and Yale University (Center for Genome Analysis). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (contract number N01-HG-65403). This study was supported by the National Institutes of Health grants RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, R01 AA017535, and the VA Connecticut and Philadelphia VA MIRECCs.
HRK is a consultant or Advisory Board Member for Alkermes, Lilly, Lundbeck, Otsuka, Pfizer, Roche; member of the American Society of Clinical Psychopharmacology's Alcohol Clinical Trials Initiative, supported by AbbVie, Ethypharm, Lilly, Lundbeck and Pfizer. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- United Nations Office on Drugs and Crime. 2010 World Drug Report, New York, USA, 2010.
- Hall W, Solowij N. Adverse effects of cannabis. Lancet 1998; 352: 1611–1616. [DOI] [PubMed] [Google Scholar]
- Hall W, Babor TF. Cannabis use and public health: assessing the burden. Addiction 2000; 95: 485–490. [DOI] [PubMed] [Google Scholar]
- Aggarwal SK, Carter GT, Sullivan MD, ZumBrunnen C, Morrill R, Mayer JD. Medicinal use of cannabis in the United States: historical perspectives, current trends, and future directions. J Opioid Manag 2009; 5: 153–168. [DOI] [PubMed] [Google Scholar]
- Hall W. What has research over the past two decades revealed about the adverse health effects of recreational cannabis use? Addiction 2014; 110: 19–35. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Compton WM, Weiss SR. Adverse health effects of marijuana use. N Engl J Med 2014; 371: 879. [DOI] [PubMed] [Google Scholar]
- Gibbs M, Winsper C, Marwaha S, Gilbert E, Broome M, Singh SP. Cannabis use and mania symptoms: a systematic review and meta-analysis. J Affect Disord 2014; 171c: 39–47. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan R, Wilkinson ST, D'Souza DC. Gone to pot: a review of the association between cannabis and psychosis. Front Psychiatry 2014; 5: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French L, Gray C, Leonard G, Perron M, Pike GB, Richer L et al. Early cannabis use, polygenic risk score for schizophrenia and brain maturation in adolescence. JAMA Psychiatry 2015; 72: 1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting PF, Wolff RF, Deshpande S, Di Nisio M, Duffy S, Hernandez AV et al. Cannabinoids for medical use: a systematic review and meta-analysis. JAMA 2015; 313: 2456–2473. [DOI] [PubMed] [Google Scholar]
- Cressey D. The cannabis experiment. Nature 2015; 524: 280–283. [DOI] [PubMed] [Google Scholar]
- Verweij KJH, Zietsch BP, Lynskey MT, Medland SE, Neale MC, Martin NG et al. Genetic and environmental influences on cannabis use initiation and problematic use: a meta-analysis of twin studies. Addiction 2010; 105: 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie NA, Lubke GH, Gardner CO, Neale MC, Kendler KS. Two-part random effects growth modeling to identify risks associated with alcohol and cannabis initiation, initial average use and changes in drug consumption in a sample of adult, male twins. Drug Alcohol Depend 2012; 123: 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie NA, Neale MC, Kendler KS. Pathways to cannabis abuse: a multi-stage model from cannabis availability, cannabis initiation and progression to abuse. Addiction 2009; 104: 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Neale MC, Jacobson KC, Prescott CA, Kendler KS. Illicit drug use and abuse/dependence: modeling of two-stage variables using the CCC approach. Addict Behav 2005; 30: 1043–1048. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Morley KI, Hansell NK, Pergadia ML, Montgomery GW, Statham DJ et al. Autosomal linkage analysis for cannabis use behaviors in Australian adults. Drug Alcohol Depend 2008; 98: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Pergadia ML, Saccone SF, Lynskey MT, Wang JC, Martin NG et al. An autosomal linkage scan for cannabis use disorders in the nicotine addiction genetics project. Arch Gen Psychiatry 2008; 65: 713–722. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Gizer IR, Wilhelmsen KC. Heritability and a genome-wide linkage analysis of a Type II/B cluster construct for cannabis dependence in an American Indian community. Addict Biol 2009; 14: 338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Gizer IR, Vieten C, Wilhelmsen KC. Linkage analyses of cannabis dependence, craving, and withdrawal in the San Francisco Family Study. Am J Med Genet B Neuropsychiatr Genet 2010; 153B: 802–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfer CJ, Lessem JM, Hartman CA, Stallings MC, Cherny SS, Corley RP et al. A genome-wide scan for loci influencing adolescent cannabis dependence symptoms: evidence for linkage on chromosomes 3 and 9. Drug Alcohol Depend 2007; 89: 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij KJH, Zietsch BP, Liu JZ, Medland SE, Lynskey MT, Madden PAF et al. No association of candidate genes with cannabis use in a large sample of Australian twin families. Addict Biol 2012; 17: 687–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughey HM, Marshall E, Schacht JP, Louis A, Hutchison KE. Marijuana withdrawal and craving: influence of the cannabinoid receptor 1 (CNR1) and fatty acid amide hydrolase (FAAH) genes. Addiction 2008; 103: 1678–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Agrawal A, Montgomery GW, Heath AC, Martin NG et al. The role of GABRA2 in alcohol dependence, smoking, and illicit drug use in an Australian population sample. Alcohol Clin Exp Res 2008; 32: 1721–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT. Candidate genes for cannabis use disorders: findings, challenges and directions. Addiction 2009; 104: 518–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT, Hinrichs A, Grucza R, Saccone SF, Krueger R et al. A genome-wide association study of DSM-IV cannabis dependence. Addict Biol 2011; 16: 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij KJH, Vinkhuyzen AAE, Benyamin B, Lynskey MT, Quaye L, Agrawal A et al. The genetic aetiology of cannabis use initiation: a meta-analysis of genome-wide association studies and a SNP-based heritability estimation. Addict Biol 2013; 18: 846–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minică CC, Dolan CV, Hottenga J-J, Pool R, Fedko IO, Mbarek H et al. Heritability, SNP-and gene-based analyses of cannabis use initiation and age at onset. Behav Genet 2015; 45: 503–513, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ et al. Finding the missing heritability of complex diseases. Nature 2009; 461: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 2012; 13: 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet 2010; 42: 448–U135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furberg H, Kim Y, Dackor J, Boerwinkle E, Franceschini N, Ardissino D et al. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 2010; 42: 441–U134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet 2010; 42: 366–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Karkowski LM, Corey LA, Prescott CA, Neale MC. Genetic and environmental risk factors in the aetiology of illicit drug initiation and subsequent misuse in women. Br J Psychiatry 1999; 175: 351–356. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE et al. An integrated map of genetic variation from 1092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Purcell SM. Statistical power and significance testing in large-scale genetic studies. Nat Rev Genet 2014; 15: 335–346. [DOI] [PubMed] [Google Scholar]
- Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet 2011; 88: 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MX, Kwan JS, Sham PC. HYST: a hybrid set-based test for genome-wide association studies, with application to protein-protein interaction-based association analysis. Am J Hum Genet 2012; 91: 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 1995; 57: 289–300. [Google Scholar]
- So HC, Li M, Sham PC. Uncovering the total heritability explained by all true susceptibility variants in a genome-wide association study. Genet Epidemiol 2011; 35: 447–456. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015; 47: 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 2015; 47: 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw 2010; 36: 1–48. [Google Scholar]
- Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ et al. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum Mol Genet 2006; 15: 3498–3507. [DOI] [PubMed] [Google Scholar]
- Rubinek T, Yu R, Hadani M, Barkai G, Nass D, Melmed S et al. The cell adhesion molecules N-cadherin and neural cell adhesion molecule regulate human growth hormone: a novel mechanism for regulating pituitary hormone secretion. J Clin Endocrinol Metab 2003; 88: 3724–3730. [DOI] [PubMed] [Google Scholar]
- Ducci F, Kaakinen M, Pouta A, Hartikainen AL, Veijola J, Isohanni M et al. TTC12-ANKK1-DRD2 and CHRNA5-CHRNA3-CHRNB4 influence different pathways leading to smoking behavior from adolescence to mid-adulthood. Biol Psychiatry 2011; 69: 650–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidwell LC, McGeary JE, Gray JC, Palmer RH, Knopik VS, MacKillop J. NCAM1-TTC12-ANKK1-DRD2 variants and smoking motives as intermediate phenotypes for nicotine dependence. Psychopharmacology 2015; 232: 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Weiss R, Brady K, Poling J, Krauthammer M et al. Genomewide linkage scan for nicotine dependence: identification of a chromosome 5 risk locus. Biol Psychiatry 2007; 61: 119–126. [DOI] [PubMed] [Google Scholar]
- Laucht M, Becker K, Frank J, Schmidt MH, Esser G, Treutlein J et al. Genetic variation in dopamine pathways differentially associated with smoking progression in adolescence. J Am Acad Child Adolesc Psychiatry 2008; 47: 673–681. [DOI] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PAF et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet 2007; 16: 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo M, Clark T, Johnstone E, Murphy M, Walton R. The genetic basis for smoking behavior: a systematic review and meta-analysis. Nicotine Tob Res 2004; 6: 583–597. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao HY, Gruen JR, Luo XG, Gelernter J. Association of haplotypic variants in DRD2, ANKK1, TTC12 and NCAM1 to alcohol dependence in independent case-control and family samples. Hum Mol Genet 2007; 16: 2844–2853. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Haplotypic variants in DRD2, ANKK1, TTC12, and NCAM1 are associated with comorbid alcohol and drug dependence. Alcohol Clin Exp Res 2008; 32: 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand FL et al. ANKK1, TTC12, and NCAM1 polymorphisms and heroin dependence: importance of considering drug exposure. JAMA Psychiatry 2013; 70: 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010; 42: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim-Verbaas CA, Bressler J, Debette S, Schuur M, Smith AV, Bis JC et al. GWAS for executive function and processing speed suggests involvement of the CADM2 gene. Mol Psychiatry 2015; 21: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JP, Magalhaes T, Conroy JM, Regan R, Shah N, Anney R et al. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Genet 2012; 131: 565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayatbakhsh MR, O'Callaghan MJ, Mamun AA, Williams GM, Clavarino A, Najman JM. Cannabis use and obesity and young adults. Am J Drug Alcohol Abuse 2010; 36: 350–356. [DOI] [PubMed] [Google Scholar]
- Kelleher LM, Stough C, Sergejew AA, Rolfe T. The effects of cannabis on information-processing speed. Addict Behav 2004; 29: 1213–1219. [DOI] [PubMed] [Google Scholar]
- De Alwis D, Agrawal A, Reiersen AM, Constantino JN, Henders A, Martin NG et al. ADHD symptoms, autistic traits, and substance use and misuse in adult Australian twins. J Stud Alcohol Drugs 2014; 75: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. Chembiochem 2004; 5: 170–176. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H et al. Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biol Psychiatry 2014; 76: 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR et al. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry 2014; 19: 717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JA, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011; 88: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Silberg JL, Lynskey MT, Maes HH, Eaves LJ. Mechanisms underlying the lifetime co-occurrence of tobacco and cannabis use in adolescent and young adult twins. Drug Alcohol Depend 2010; 108: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Schmitt E, Aggen SH, Prescott CA. Genetic and environmental influences on alcohol, caffeine, cannabis, and nicotine use from early adolescence to middle adulthood. Arch Gen Psychiatry 2008; 65: 674–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchia M, Cullis J, Turecki G, Rouleau GA, Uher R, Alda M. The impact of phenotypic and genetic heterogeneity on results of genome wide association studies of complex diseases. PLoS One 2013; 8: e76295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JD, Lynskey MT, Scherrer JF, Agrawal A, Heath AC, Bucholz KK. A cotwin-control analysis of drug use and abuse/dependence risk associated with early-onset cannabis use. Addict Behav 2010; 35: 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT, Pergadia ML, Bucholz KK, Heath AC, Martin NG et al. Early cannabis use and DSM-IV nicotine dependence: a twin study. Addiction 2008; 103: 1896–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynskey MT, Vink JM, Boomsma DI. Early onset cannabis use and progression to other drug use in a sample of Dutch twins. Behav Genet 2006; 36: 195–200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.