Abstract
Polymyxins are often last-line therapeutic agents used to treat infections caused by multidrug-resistant A. baumannii. Recent reports of polymyxin-resistant A. baumannii highlight the urgent need for research into mechanisms of polymyxin resistance. This study employed genomic and transcriptomic analyses to investigate the mechanisms of polymyxin resistance in A. baumannii AB307-0294 using an in vitro dynamic model to mimic four different clinically relevant dosage regimens of polymyxin B and colistin over 96 h. Polymyxin B dosage regimens that achieved peak concentrations above 1 mg/L within 1 h caused significant bacterial killing (~5 log10CFU/mL), while the gradual accumulation of colistin resulted in no bacterial killing. Polymyxin resistance was observed across all dosage regimens; partial reversion to susceptibility was observed in 6 of 8 bacterial samples during drug-free passaging. Stable polymyxin-resistant samples contained a mutation in pmrB. The transcriptomes of stable and non-stable polymyxin-resistant samples were not substantially different and featured altered expression of genes associated with outer membrane structure and biogenesis. These findings were further supported via integrated analysis of previously published transcriptomics data from strain ATCC19606. Our results provide a foundation for understanding the mechanisms of polymyxin resistance following exposure to polymyxins and the need to explore effective combination therapies.
The antimicrobial resistance crisis has become a significant threat to public health1. Globally, hospital outbreaks of infections caused by multi-drug resistant (MDR) Gram-negative pathogens, such as Acinetobacter baumannii, are being increasingly reported2,3. With few novel antibiotics in late-stage clinical development, clinicians may soon be left with no options for the treatment of recalcitrant infections caused by these MDR pathogens. A. baumannii has emerged as a particularly problematic pathogen, owing to its propensity to acquire resistance to most currently available antibiotics4. Polymyxins (i.e. polymyxin B and colistin) are used as a salvage therapy for A. baumannii infections where susceptibility testing suggests that carbapenems and aminoglycosides are unlikely to be effective5,6,7. Owing to their clinical introduction in the 1950s and fall from favour a decade or so later, the pharmacology of polymyxins has not been as thoroughly investigated as for modern antibiotics, until recently. While polymyxins demonstrate in vitro activity against many MDR A. baumannii bacterial isolates3,8,9, reports of polymyxin-resistant A. baumannii clinical isolates10 highlight an urgent need to investigate the influence of polymyxin dosage regimens on the emergence of resistance.
Polymyxins are cationic amphipathic compounds, containing a cyclic heptapeptide ring joined to a fatty acyl tail by a linear tripeptide. The L-2,4-diaminobutyric acid residues give rise to the cationic and hydrophilic nature of polymyxins, while the fatty acyl tail and position 6/7 amino acids of the heptapeptide ring contribute to the hydrophobicity of the compounds11. The aforementioned physicochemical properties of polymyxins are critical for their initial interaction with the negatively charged moieties and hydrophobic regions of lipid A of lipopolysaccharide (LPS) within the bacterial outer membrane (OM), leading to its permeabilisation11. While the interaction between lipid A and polymyxins is well characterised and essential for their ultimate bactericidal effect11, the mechanism of polymyxin killing following perturbation of the OM has yet to be fully elucidated12,13,14,15,16. To date, two mechanisms of polymyxin resistance have been identified in A. baumannii: modification of lipid A with phosphoethanolamine and/or galactosamine and the complete loss of LPS from the OM17,18,19,20,21. Current literature suggests that both mechanisms of resistance abolish polymyxin-induced bacterial killing by preventing the interaction of polymyxins with the OM, and are mediated by the pmrCAB operon17,21 (for lipid A modification with phosphoethanolamine), naxD20 (for modification with galactosamine) or lpx biosynthetic cluster (for LPS loss)19. There is a paucity of knowledge on the emergence and mechanism(s) of resistance in response to the polymyxin exposure profiles associated with clinically relevant dosage regimens of colistin and polymyxin B.
The two clinically used polymyxins, colistin and polymyxin B, differ in their administered forms and exhibit markedly different clinical pharmacokinetics (PK)5. Colistin is administered parenterally as the sodium salt of its inactive pro-drug colistin methanesulphonate (CMS), while polymyxin B is available in the clinic as the sulphate salt of its active form. Following administration, CMS is converted slowly to colistin while simultaneously undergoing rapid renal elimination, which leads to a delay in the attainment of target colistin concentrations22,23,24. In contrast, the administration of polymyxin B enables target concentrations to be more rapidly achieved25. Although colistin and polymyxin B are considered equivalent based upon their antimicrobial activity in vitro26, it was hypothesised that differences in their plasma concentration versus time profiles following initiation of therapy with CMS and polymyxin B, respectively, are likely to substantially affect their pharmacodynamic responses in patients. The objectives of this study were to investigate the transcriptomic profile and stability of polymyxin resistance in A. baumannii when exposed in an in vitro dynamic model to clinically relevant concentration versus time profiles of colistin and polymyxin B.
Methods
Bacterial strain and media
A. baumannii strain AB307-0294, a previously characterised polymyxin-susceptible (MIC: 1.0 mg/L) clinical isolate belonging to international clonal complex I27,28, was investigated in this study. Cation-adjusted Mueller-Hinton broth (CAMHB, Oxoid, Ca2+: 20–25 mg/L, Mg2+: 10–15 mg/L) was used in both the in vitro dynamic model and subsequent passaging. All bacterial cultures, including starter cultures and the in vitro model, were maintained at 37 °C for the duration of the experiment.
In vitro model and passaging in drug-free broth
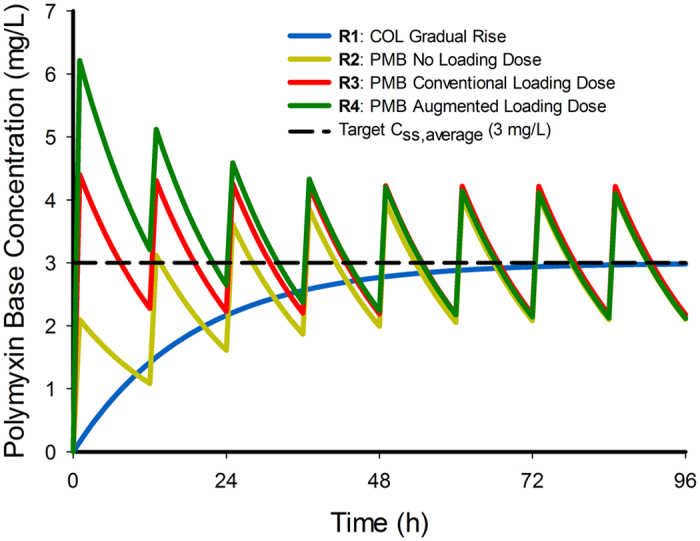
A starting inoculum of 106 CFU/mL of log-phase bacteria cultured from a single colony was introduced into a previously described in vitro one-compartment model (IVM). This model allows clinically relevant concentration versus time profiles of an antibiotic to be accurately achieved in a central reservoir inoculated with the organism of interest29. A total of 4 concentration-time profiles were simulated in the IVM (Fig. 1) with a central reservoir volume of 250 mL. These profiles corresponded to: the gradual accumulation of colistin as would be seen at the initiation of CMS therapy with no loading dose22 (regimen 1); 1-h polymyxin B infusion every 12 h without a loading dose (regimen 2); as for regimen 2 but with a loading dose to achieve the steady state immediately (regimen 3); and, regimen 2 initiated with an augmented loading dose to achieve concentrations over the first several hours higher than the eventual steady-state concentrations (regimen 4). Each regimen and the growth control were conducted in two replicates. For all regimens, an elimination half-life of 11.6 h was applied for both colistin and polymyxin B, representative of pharmacokinetic behaviour of both polymyxins in critically-ill patients22,23,25,30. For all four regimens an average steady-state concentration of 3 mg/L was simulated; for regimen 4, the augmented loading dose achieved a peak polymyxin B concentration of 6 mg/L after this initial dose and subsequently the concentrations declined to achieve the same steady-state profile as for the other two polymyxin B regimens (Fig. 1). The elimination half-life and target polymyxin concentrations were selected to mimic the disposition of polymyxin B and colistin in critically-ill patients22,25,23. Samples (1 mL) were collected from the central reservoir at 0, 1, 8, 23, 28, 47, 52, 71 and 96 h, and numbers of viable bacteria were determined by plating onto drug-free agar plates. Population analysis profiles (PAPs) were obtained at 23, 47, 71 and 96 h by counting viable bacteria after plating cultures on polymyxin B containing agar plates (2, 4 and 8 mg/L as sulphate). At the conclusion of the IVM (96 h), polymyxin-resistant bacterial cells were isolated from each of the treated reservoirs (n = 8 total) and passaged daily for a further 96 h in drug-free CAMHB. PAPs were obtained daily on polymyxin B-containing (1, 2, 4 and 8 mg/L) agar plates.
Figure 1.
Genomics and transcriptomics
Genomics samples were collected at the conclusion of passaging in drug-free CAMHB (192 h; consisting of 96 h in IVM and 96 h drug-free passaging) and DNA was prepared using a QIAamp DNA mini kit (Qiagen, USA) for high-throughput sequencing (150 bp paired-end reads). Transcriptomic profiling was performed on cultures recovered from each reservoir (n = 10; including growth control) at the conclusion of the IVM (96 h). Cultures collected for transcriptomic profiling (containing ~109 CFU per sample) were centrifuged at 9000 × g (4 °C) for 10 min and resuspended in 1 mL of RNALater (Qiagen, USA) for 10 min before a second centrifugation at 5000 × g for 10 min, with the pellet stored at −80 °C prior to sequencing. Total RNA was purified from each sample (Qiagen RNeasy; Qiagen, USA), ribosomal RNAs removed (Ribo-Zero rRNA removal kit; Illumina, UK) and libraries prepared for RNA sequencing (100 bp single-end reads) as previously described31. DNA and RNA sequencing was performed on an Illumina HiSeq (Medical Genomics Facility, Monash Health Translation Precinct, Monash University, Victoria, Australia). RNA sequencing was performed over two Illumina HiSeq lanes, with replicates for each treatment condition analysed on separate lanes.
Next-generation sequencing data analysis
Illumina HiSeq reads for both genomic and transcriptomic analyses were clipped using the Nesoni software package (Victorian Bioinformatics Consortium) before mapping to a previously published27 genome for A. baumannii AB307-0294 (Genbank accession: NC_011595) using the Short Read Mapping Package (SHRiMP 2.2.3). The average number of reads per sample was ~5 million for the genomic analysis, and ~23 million mapped reads per sample for transcriptomics analysis. Single nucleotide polymorphisms (SNPs) in the genomic and transcriptomic data were identified with Freebayes32, using the previously published genome for A. baumannii AB307-0294 as a reference. Differential gene expression analysis of transcriptomic data was performed in Degust (www.vicbioinformatics.com/degust), a visual interface for the Voom and Limma R packages33. Statistical significance of differential gene expression was calculated using the F-statistic, jointly considering all treatment groups and adjusted using the Benjamini Hochberg method to control the false discovery rate (FDR)34. Differential expression was defined as a log2 fold-change (log2FC) of >1.0 in any of the treatment groups relative to the growth control with a corresponding FDR of <0.05. Interproscan35 (version 5) was used for functional and gene ontology (GO) term annotation of the published A. baumannii AB307-0294 genome27. Principal component and GO term enrichment analyses (Fisher’s exact test) were performed in R.
Integrated gene expression analysis for comparison of data from closely related strains
In addition to the conventional differential gene expression analysis, a sparse partial least squares regression discriminant analysis36 (SPLS-DA) model was constructed to identify gene expression patterns that were (1) unique to the early stages of polymyxin exposure (≤1 h), and (2) shared between early- and late-stage polymyxin exposure (1 and 96 h). Matching of orthologous genes, defined as gene pairs with a p-value of <10−5 when clustered using OrthoMCL37, was used to merge data from the present study with a previously published transcriptomic dataset from A. baumannii strain ATCC 19606 sampled 15 and 60 min following colistin, doripenem, or colistin/doripenem combination treatment31. In total, 16 samples consisting of 5 untreated controls, 6 polymyxin-treated, 3 doripenem-treated and 2 combination-treated samples were used from the previously published dataset31. A variance stabilisation transformation was performed on the combined data in R using the DESeq238 package prior to SPLS-DA.
SPLS-DA model validation
The SPLS-DA model was subjected to k-fold (k = 2) validation to select the smallest number of genes that optimally described the biological variation within the combined dataset. The combined data from the present study and our previous paper31 were randomly partitioned into two segments (n = 13 each), with each segment used individually for model construction and the combined dataset used to determine the classification error rate (two tests per partitioning). The error rate for each candidate model was calculated as the average of 300 trials (600 tests) to account for the stochastic nature of partitioning. Candidate models contained between 10 and 150 genes (10 gene increments; 300 error rate trials per model) and were evaluated by their corresponding error rates. Once the smallest number of genes for inclusion had been determined, a second k-fold (k = 6; 6 tests per partitioning) validation was performed (50 trials; 300 tests total) to identify the inclusion rate of individual genes within models constructed on partitioned data sets.
Results
Characterisation of polymyxin activity and resistance in A. baumannii
Polymyxin B dosage regimens that rapidly attained concentrations >1 mg/L exhibited more bacterial killing compared to the simulated colistin dosing regimen (Fig. 2A). The development of polymyxin resistance was phenotypically similar across all dosage regimens (Fig. 2B). At the conclusion of polymyxin B or colistin treatment (96 h), bacterial cells isolated from the treated IVM reservoirs showed only a ~1–2 log10 CFU/mL difference between viable bacterial cells enumerated on drug-free and polymyxin-containing plates (8 mg/L), compared to the ~7 log10 CFU/mL difference seen in the controls at the same time point. Bacterial cells isolated from the control arms showed little change in polymyxin resistance profiles over the course of the IVM.
Figure 2.
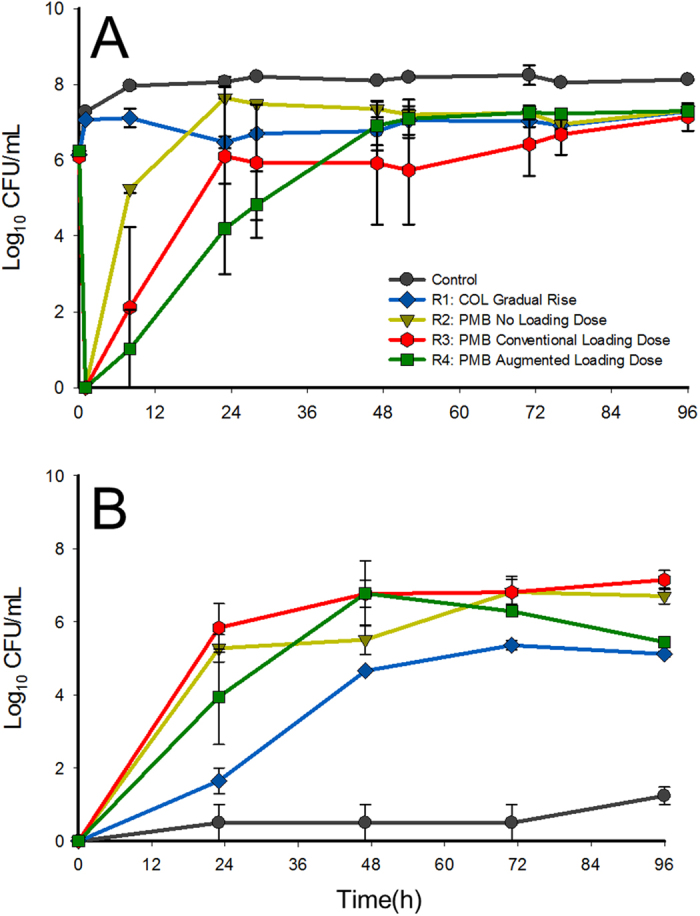
Viable counting results (mean ± SD; n = 2) for A. baumannii AB307-0294 samples grown on (A) drug-free agar plates and (B) polymyxin B containing (8 mg/L) agar plates.
Drug-free passaging of the bacterial cells isolated from the polymyxin-treated reservoirs revealed the presence of stable (n = 2; one each from regimens 2 and 3) and non-stable (n = 6) polymyxin resistance (Fig. 3). There was insufficient evidence to strongly link the development of stable polymyxin resistance with total polymyxin exposure or dosing intensity. Further, the proportion of resistant bacteria in stable resistant bacterial samples was unchanged between the commencement and conclusion of passaging (96 h) (Fig. 3A). In the case of non-stable polymyxin resistance, partial reversion to susceptibility was extensive (>2 log10 CFU/mL) but incomplete over 96 h of passaging (Fig. 3B), with a ~2–3 log10 CFU/mL increase in viable counts on polymyxin-containing (8 mg/L) plates compared to the control.
Figure 3.
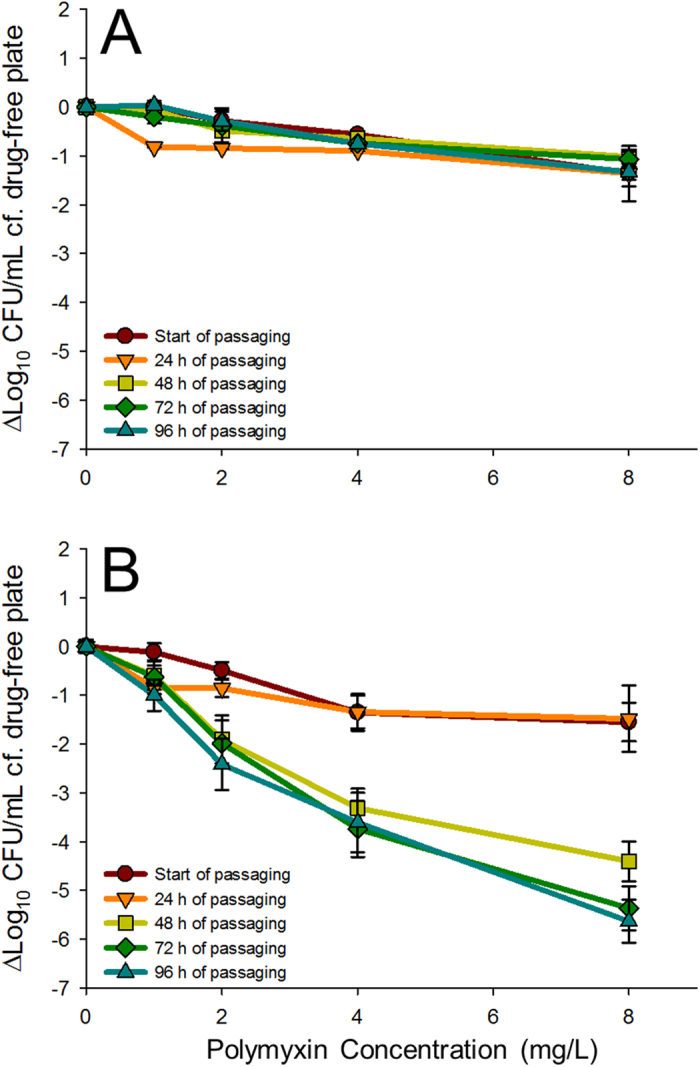
Population analysis profiles (mean ± SD; (A) n = 2; (B) n = 6) of polymyxin-treated bacterial samples during passaging in drug-free CAMHB, showing stable polymyxin resistance (A) and the partial reversion (>2 Log10 CFU/mL) to polymyxin susceptibility (B). Y-axis values reflect the difference in viable counts obtained on drug-free agar plates and polymyxin B containing agar plates at the concentrations indicated.
Genomic analysis of the stable and non-stable polymyxin resistance phenotypes
Interrogation of the genomes of the two stable polymyxin-resistant bacterial samples revealed a SNP in pmrB that led to a substitution of alanine at position 227 to valine (A227V). The same SNP was also found in the corresponding transcriptomic samples collected at the conclusion of polymyxin B or colistin treatment in the IVM, with >96% of reads covering the affected base containing the SNP. In all but one of the bacterial samples with non-stable polymyxin resistance, examination of the transcriptome yielded no evidence of the pmrB A227V mutation or any other common genomic changes across the samples. However, in the case of one non-stable polymyxin-resistant bacterial sample treated with regimen 2, 90% of transcriptomic reads covering the affected base pairs contained the pmrB A227V SNP.
Transcriptomic analysis
In bacterial samples collected after polymyxin B or colistin treatment for 96 h in the IVM (polymyxin-resistant cultures), 33 genes showed increased expression and 28 showed decreased expression relative to the untreated control across the four different regimens (Tables 1 and 2). Substantially increased expression (>2-fold) of AdeA and AdeB (ABBFA_001707 and ABBFA_001708, respectively), members of the AdeABC multidrug efflux system, was observed in all polymyxin-treated samples. Similarly, increased expression of the genes encoding components of the Lol lipoprotein transport complex (ABBFA_000739 and ABBFA_000869) and the TolQRA transmembrane complex (ABBFA_000889, ABBFA_000888 and ABBFA_000382) was also evident. GO term enrichment analysis of genes exhibiting over-expression showed a statistically significant (FDR < 0.05) over-representation of GO terms GO:0016020 (Cellular component: Membrane), GO:0005215 (Molecular function: Transporter activity), and GO:0006810 (Biological process: Transport). For the genes showing reduced expression in the presence of polymyxins, a common pattern of gene functions was less apparent, and was further confounded by the proportion of genes (12 out of 28 genes) identified as being hypothetical proteins. However, reduced expression was observed for six transcriptional regulators that have yet to be fully characterised in A. baumannii. No GO terms were found to be over-represented in the down-regulated gene set. Principal components analysis of transcriptomic profiles obtained at 96 h from the IVM revealed a high degree of separation between the control and polymyxin-treated (polymyxin B and colistin) samples (Fig. 4). However, a clear relationship between the dosage regimen used and the transcriptomic profile observed was not evident.
Table 1. Genes up-regulated in A. baumannii AB307-0294 following polymyxin treatment for 96 h in the IVM.
| Locus Tag | Published Annotation | Annotation by Similarity Searching (Hypothetical Proteins) | Dosage Regimen (log2 Fold-Change) |
||||
|---|---|---|---|---|---|---|---|
| Colistin (R1) | PMB No Loading Dose (R2) | PMB with Loading Dose (R3) | PMB with Augmented Loading Dose (R4) | FDR | |||
| ABBFA_000201 | Succinate semialdehyde dehydrogenase | – | 1.64 | 1.20 | 0.73 | 1.37 | 0.050 |
| ABBFA_000261 | Hypothetical protein | Signal peptide | 3.48 | 2.05 | 2.11 | 3.62 | 0.036 |
| ABBFA_000382 | Protein TolA | – | 3.43 | 1.21 | 1.92 | 2.71 | 0.044 |
| ABBFA_000413 | Hypothetical protein | Toluene tolerance protein Ttg2E | 2.15 | 0.79 | 1.11 | 1.56 | 0.049 |
| ABBFA_000570 | TonB dependent receptor family protein | – | 1.69 | 1.16 | 0.64 | 0.88 | 0.044 |
| ABBFA_000739 | Outer membrane lipocarrier protein LolA | – | 3.43 | 1.18 | 2.01 | 2.74 | 0.044 |
| ABBFA_000816 | Multidrug resistance protein mexB | – | 1.64 | 0.48 | 0.76 | 1.18 | 0.044 |
| ABBFA_000869 | Lipoprotein-releasing system transmembrane protein lolE | – | 1.78 | 0.62 | 0.80 | 1.46 | 0.037 |
| ABBFA_000870 | Lipoprotein releasing system, ATP-binding protein | – | 1.83 | 0.82 | 0.80 | 1.41 | 0.036 |
| ABBFA_000885 | Peptidoglycan-associated lipoprotein | – | 1.25 | 1.00 | 0.65 | 1.08 | 0.050 |
| ABBFA_000888 | Protein TolR | - | 1.29 | 0.52 | 0.63 | 1.08 | 0.044 |
| ABBFA_000889 | Protein TolQ | – | 1.12 | 0.21 | 0.41 | 0.91 | 0.050 |
| ABBFA_000904 | Hypothetical protein | No matches | 1.76 | 1.06 | 0.82 | 1.42 | 0.044 |
| ABBFA_001022 | Hypothetical protein | Putative signal peptide | 2.07 | 0.97 | 1.17 | 1.72 | 0.050 |
| ABBFA_001302 | Biofilm PGA synthesis protein pgaA precursor | – | 2.58 | 0.93 | 1.65 | 2.11 | 0.044 |
| ABBFA_001303 | Biofilm PGA synthesis lipoprotein pgaB precursor | – | 2.43 | 1.01 | 1.40 | 1.95 | 0.044 |
| ABBFA_001304 | IcaA | – | 2.25 | 1.11 | 1.30 | 1.85 | 0.044 |
| ABBFA_001629 | HTH-type transcriptional repressor Bm3R1 | – | 1.30 | 0.58 | 0.38 | 1.22 | 0.044 |
| ABBFA_001662 | HlyD family secretion family protein | – | 1.23 | 1.19 | 0.58 | 0.93 | 0.050 |
| ABBFA_001663 | Multidrug resistance protein Y | – | 1.84 | 1.84 | 1.17 | 1.52 | 0.037 |
| ABBFA_001707 | Acriflavine resistance protein E precursor | – | 3.26 | 3.42 | 2.07 | 3.26 | 0.028 |
| ABBFA_001708 | AcrB protein | – | 2.59 | 2.59 | 1.36 | 2.45 | 0.036 |
| ABBFA_001709 | Outer membrane protein oprM precursor | – | 1.51 | 1.63 | 0.62 | 1.29 | 0.050 |
| ABBFA_001779 | UTRA domain protein | – | 0.01 | 1.33 | 1.09 | 0.41 | 0.050 |
| ABBFA_002407 | Hypothetical protein | Putative signal peptide | 1.62 | 1.54 | 1.00 | 1.62 | 0.036 |
| ABBFA_002498 | Putative phospholipid-binding domain protein | – | 1.46 | 0.57 | 0.79 | 1.36 | 0.050 |
| ABBFA_002880 | Hypothetical protein | No matches | 0.75 | 1.64 | 1.15 | 0.61 | 0.050 |
| ABBFA_003020 | Outer membrane protein oprM precursor | – | 2.51 | 1.46 | 1.11 | 1.78 | 0.036 |
| ABBFA_003050 | phosphogluconate dehydratase | – | 1.45 | 0.88 | 0.61 | 1.29 | 0.050 |
| ABBFA_003147 | 50S ribosomal protein L31 type B | – | 2.07 | 1.71 | 1.20 | 1.39 | 0.037 |
| ABBFA_003406 | AMP-binding enzyme family protein | – | 1.59 | 0.89 | 0.87 | 1.88 | 0.050 |
| ABBFA_003500 | Hypothetical protein | Lipoprotein | 3.28 | 0.64 | 2.07 | 2.69 | 0.050 |
| ABBFA_003501 | Hypothetical protein | Lipoprotein | 2.93 | 1.35 | 1.94 | 3.22 | 0.037 |
Table 2. Genes down-regulated in A. baumannii AB307-0294 following polymyxin treatment for 96 h in the IVM.
| Locus Tag | Published Annotation | Annotation by Similarity Searching (Hypothetical Proteins) | Dosage Regimen (log2 Fold-Change) |
||||
|---|---|---|---|---|---|---|---|
| Colistin (R1) | PMB No Loading Dose (R2) | PMB with Loading Dose (R3) | PMB with Augmented Loading Dose (R4) | FDR | |||
| ABBFA_000133 | Hypothetical protein | Ribosomal subunit interface protein | −1.05 | −1.84 | −1.38 | −1.87 | 0.044 |
| ABBFA_000218 | Hypothetical protein | No matches | −1.47 | −1.99 | −1.48 | −1.28 | 0.036 |
| ABBFA_000426 | Rrf2 family protein (transcriptional regulator) family protein | – | −1.70 | −2.32 | −1.45 | −1.81 | 0.045 |
| ABBFA_000555 | Virulence sensor protein bvgS precursor | – | −1.60 | −1.51 | −1.36 | −1.80 | 0.037 |
| ABBFA_000602 | Hemolysin-3 | – | −1.56 | −2.07 | −1.15 | −1.73 | 0.038 |
| ABBFA_000699 | PaaX-like family protein | – | −1.14 | −1.82 | −1.06 | −1.28 | 0.044 |
| ABBFA_000748 | Hypothetical protein | No matches | −1.04 | −1.29 | −1.19 | −1.56 | 0.044 |
| ABBFA_000934 | Hypothetical protein | No matches | −1.14 | −1.91 | −1.17 | −1.02 | 0.038 |
| ABBFA_000972 | HTH-type transcriptional regulator gltR | – | −2.04 | −2.22 | −1.36 | −1.89 | 0.050 |
| ABBFA_000981 | CRISPR-associated protein Cas1 | – | −1.90 | −2.65 | −1.07 | −1.27 | 0.047 |
| ABBFA_000982 | CRISPR-associated helicase Cas3 | – | −1.53 | −2.07 | −1.12 | −1.28 | 0.036 |
| ABBFA_000983 | Hypothetical protein | CRISPR-associated protein, Csy1 family | −1.12 | −1.52 | −0.96 | −1.09 | 0.050 |
| ABBFA_001178 | Hypothetical protein | Sulphur transfer protein SirA | −1.32 | −1.32 | −0.67 | −1.51 | 0.044 |
| ABBFA_001409 | Benzoate membrane transport protein | – | −0.94 | −1.77 | −1.12 | −1.44 | 0.050 |
| ABBFA_001469 | Nitrogen regulation protein ntrB | – | −0.80 | −1.29 | −0.76 | −1.24 | 0.038 |
| ABBFA_001575 | Hypothetical protein | Transporter component | −1.58 | −1.32 | −1.47 | −1.55 | 0.039 |
| ABBFA_001576 | Hypothetical protein | Transporter component | −1.53 | −1.44 | −1.52 | −1.69 | 0.047 |
| ABBFA_001979 | Hypothetical protein | No matches | −0.42 | −0.99 | −0.80 | −1.30 | 0.045 |
| ABBFA_002011 | Tautomerase enzyme family protein | – | −0.81 | −1.70 | −0.92 | −1.21 | 0.037 |
| ABBFA_002314 | Hypothetical protein | Lipoprotein | −1.08 | −1.23 | −1.01 | −1.11 | 0.050 |
| ABBFA_002503 | Arginine exporter protein argO | – | −0.73 | −1.54 | −0.71 | −1.01 | 0.044 |
| ABBFA_002926 | Hypothetical protein | Protein FilA | −0.88 | −1.14 | −0.86 | −1.14 | 0.050 |
| ABBFA_003141 | Oxygen-independent coproporphyrinogen III oxidase-like protein yggW | – | −0.89 | −1.02 | −0.90 | −1.16 | 0.044 |
| ABBFA_003359 | Hypothetical protein | TetR/AcrR family transcriptional regulator | −1.90 | −2.19 | −1.35 | −1.77 | 0.044 |
| ABBFA_003360 | Transcription regulatory protein opdE | – | −2.62 | −2.41 | −1.73 | −2.40 | 0.050 |
| ABBFA_003361 | AraC family transcriptional regulator | – | −2.35 | −2.42 | −1.50 | −2.18 | 0.036 |
| ABBFA_003470 | Linoleoyl-CoA desaturase (Delta(6)-desaturase) | – | −3.91 | −3.67 | −2.08 | −3.84 | 0.037 |
| ABBFA_003471 | Flavohemo (Hemoglobin-like protein) | – | −4.32 | −3.92 | −2.30 | −3.98 | 0.044 |
Figure 4. Principal components plots constructed from the transcriptomes of bacterial samples collected at the conclusion of the IVM.
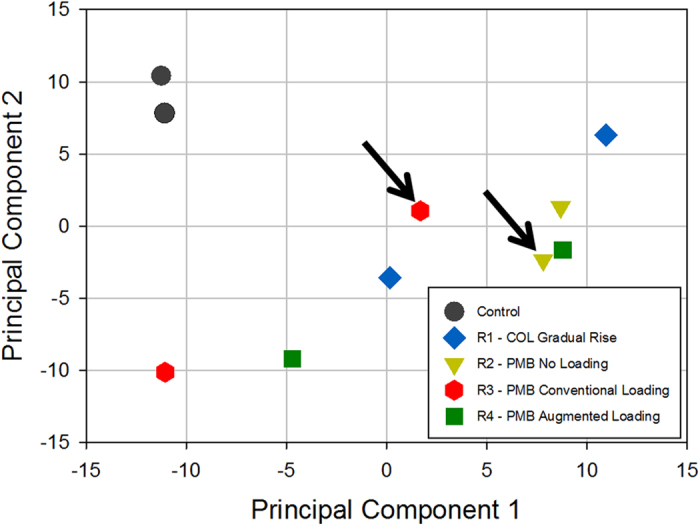
Arrows point to the samples exhibiting stable polymyxin resistance.
SPLS-DA of multiple gene expression data sets
The transcriptomic data from published experiments in A. baumannii ATCC 1960631 were successfully merged and analysed with data from the current study. The validated model contained two components that included 10 and 50 genes, respectively. The early-stage transcriptomic response to polymyxin exposure was described by the first component of the SPLS model (Table 3), while commonalities between early- and late-stage responses to polymyxin exposure were characterised by the second model component (Table 4). Important predictors of early-stage polymyxin exposure in the SPLS model included genes involved in cellular metabolism (ABBFA_002620: Polyphosphate kinase, ABBFA_003493: NADPH-dependent FMN reductase family protein, ABBFA_002755: NAD+ synthetase) and protein mis-folding (ABBFA_002915: Peptidase C13 family protein). The genes common to early- and late-stage polymyxin exposure included seven efflux transporters (Resistance-Nodulation-Division [RND] family efflux transporters and multidrug resistance proteins A, B, and Y). Although these data are consistent with outer membrane perturbation, GO term enrichment analysis pointed to a statistically significant over-representation of only GO term GO:0009306 (Biological process: Protein secretion).
Table 3. Genes and their inclusion frequencies (10 most frequently included shown; 300 trials) in component 1 of the SPLS-DA model and corresponding expression levels.
|
A. baumannii locus tags |
Frequency of inclusion in SPLS-DA model | Annotated Product | log2-Fold Change vs Control |
|||
|---|---|---|---|---|---|---|
| AB307-0294 | ATCC 19606 | Early-Stage | Late-Stage | FDR | ||
| ABBFA_002620 | HMPREF0010_01526 | 63.7% | Polyphosphate kinase | −1.10 | 0.80 | 1.79E-09 |
| ABBFA_003493 | HMPREF0010_03371 | 57.3% | NADPH-dependent FMN reductase family protein | 1.67 | −0.40 | 1.90E-10 |
| ABBFA_000791 | HMPREF0010_02052 | 56.0% | FtsJ-like methyltransferase family protein | 0.68 | −0.33 | 1.68E-08 |
| ABBFA_003446 | HMPREF0010_03271 | 50.0% | Phosphomannomutase (PMM) | 1.65 | −0.31 | 3.70E-11 |
| ABBFA_002750 | HMPREF0010_01358 | 42.7% | Hypothetical protein | 1.13 | −0.59 | 4.00E-10 |
| ABBFA_002755 | HMPREF0010_01353 | 42.7% | Probable glutamine-dependent NAD(+) synthetase | 1.00 | −0.13 | 9.09E-10 |
| ABBFA_002915 | HMPREF0010_01204 | 33.7% | Peptidase C13 family protein | 0.74 | −0.28 | 1.79E-08 |
| ABBFA_003004 | HMPREF0010_01698 | 32.3% | Hypothetical protein | 1.12 | −0.12 | 1.14E-09 |
| ABBFA_002591 | HMPREF0010_01555 | 32.3% | Protein hupE precursor | −0.52 | 0.83 | 1.16E-08 |
| ABBFA_000299 | HMPREF0010_02497 | 30.7% | Cobalt-zinc-cadmium resistance protein czcD | 0.62 | −0.44 | 1.97E-07 |
Component 1 of the SPLS-DA model described gene expression profiles unique to early-stage polymyxin exposure.
Table 4. Genes and their inclusion frequencies (Genes included in >50% of models shown; 300 trials) in component 2 of the SPLS-DA model (over 300 trials) and corresponding expression levels.
|
A. baumannii Locus Tags |
Frequency of Inclusion in SPLS-DA Model | log2-Fold Change vs Control |
||||
|---|---|---|---|---|---|---|
| AB307-0294 | ATCC 19606 | Annotated Product | Early-Stage | Late-Stage | FDR | |
| ABBFA_003136 | HMPREF0010_01842 | 78.7% | Long-chain-acyl-CoA synthetase | 1.13 | 1.51 | 1.68E-08 |
| ABBFA_001663 | HMPREF0010_00531 | 77.3% | Multidrug resistance protein Y | 1.30 | 1.73 | 4.11E-08 |
| ABBFA_002953 | HMPREF0010_01645 | 77.3% | Acetyl-CoA carboxylase, carboxyl transferase, alpha subunit | −1.61 | −1.73 | 9.23E-09 |
| ABBFA_000610 | HMPREF0010_02771 | 76.7% | AraC family transcriptional regulator | −1.10 | −1.42 | 2.34E-08 |
| ABBFA_000999 | HMPREF0010_02246 | 76.7% | Linoleoyl-CoA desaturase (Delta(6)-desaturase) | −1.77 | −1.93 | 1.71E-07 |
| ABBFA_001662 | HMPREF0010_00530 | 75.7% | HlyD family secretion family protein | 0.72 | 1.27 | 2.10E-06 |
| ABBFA_000369 | HMPREF0010_02566 | 74.7% | CobQ/CobB/MinD/ParA nucleotide binding domain protein | −0.75 | −1.00 | 2.38E-06 |
| ABBFA_002706 | HMPREF0010_01403 | 74.7% | Multidrug resistance protein B | 1.51 | 1.89 | 1.16E-06 |
| ABBFA_002838 | HMPREF0010_01276 | 73.3% | HlyD family secretion family protein | 1.36 | 1.58 | 1.83E-06 |
| ABBFA_003296 | HMPREF0010_01991 | 71.7% | Response regulator | −0.84 | −1.17 | 2.81E-07 |
| ABBFA_003340 | HMPREF0010_02034 | 71.3% | Hypothetical protein | 1.57 | 2.20 | 7.38E-05 |
| ABBFA_001460 | HMPREF0010_00336 | 70.3% | Hypothetical protein | −1.20 | −1.42 | 1.41E-06 |
| ABBFA_001969 | HMPREF0010_00851 | 69.0% | Biotin synthase | −1.11 | −1.75 | 2.84E-08 |
| ABBFA_002943 | HMPREF0010_01635 | 68.3% | Transcriptional regulator OhrR | −0.85 | −1.10 | 5.16E-06 |
| ABBFA_000374 | HMPREF0010_02571 | 68.0% | Hypothetical protein | −1.50 | −2.12 | 2.84E-07 |
| ABBFA_002707 | HMPREF0010_01402 | 68.0% | Multidrug resistance protein A | 1.55 | 1.97 | 5.16E-06 |
| ABBFA_001415 | HMPREF0010_00288 | 67.3% | 3-ketoacyl-(acyl-carrier-protein) reductase | 1.91 | 1.77 | 1.94E-07 |
| ABBFA_002751 | HMPREF0010_01357 | 66.3% | Beta-ketoacyl synthase, N-terminal domain protein | −1.17 | −1.74 | 1.16E-07 |
| ABBFA_003341 | HMPREF0010_02035 | 63.3% | Flavoprotein wrbA (Trp repressor-binding protein) | 1.78 | 1.76 | 5.88E-05 |
| ABBFA_000033 | HMPREF0010_03386 | 62.7% | Efflux transporter, RND family, MFP subunit | 1.49 | 1.57 | 3.91E-06 |
| ABBFA_000205 | HMPREF0010_02441 | 62.3% | Hypothetical protein | −1.30 | −2.46 | 6.42E-09 |
| ABBFA_002898 | HMPREF0010_01218 | 61.7% | Sorbitol dehydrogenase | 1.24 | 1.60 | 2.86E-05 |
| ABBFA_002836 | HMPREF0010_01278 | 60.3% | TetR family regulatory protein | 2.28 | 1.71 | 5.54E-08 |
| ABBFA_001549 | HMPREF0010_00427 | 57.7% | 2,4-dienoyl-CoA reductase [NADPH](2,4-dienoyl coenzymeA reductase) | 1.03 | 2.25 | 7.05E-06 |
| ABBFA_002613 | HMPREF0010_01533 | 54.3% | Hypothetical protein | −0.87 | −1.66 | 7.02E-07 |
| ABBFA_000601 | HMPREF0010_02763 | 52.0% | MFS transporter, metabolite:H+ symporter (MHS) family protein | −0.59 | −0.69 | 5.28E-05 |
| ABBFA_000867 | HMPREF0010_02122 | 52.0% | Hypothetical protein | −0.43 | −0.71 | 3.25E-05 |
| ABBFA_000206 | HMPREF0010_02442 | 51.7% | Methyltransferase domain protein | −0.83 | −1.45 | 6.02E-06 |
| ABBFA_003085 | HMPREF0010_01789 | 51.7% | Coniferyl aldehyde dehydrogenase (CALDH) | −1.23 | −1.60 | 3.22E-06 |
| ABBFA_000998 | HMPREF0010_02245 | 51.3% | Phthalate dioxygenase reductase (PDR) | −2.06 | −1.88 | 2.64E-06 |
Component 2 of the SPLS-DA model described gene expression profiles common to early- and late-stage polymyxin exposure.
Discussion
Given the worsening antimicrobial resistance crisis, there is an urgent need to further our understanding of the emergence of polymyxin resistance in A. baumannii and the association with the polymyxin exposure profile. A. baumannii AB307-0294, a multidrug-resistant clinical isolate from a bloodstream infection, has been genomically characterised27 and represents an ideal model organism for mechanistic studies into polymyxin activity and resistance in A. baumannii. The present study indicates that the rapid and extensive bacterial killing associated with polymyxins was dependent on rapidly attaining therapeutic concentrations (Fig. 2). Extensive polymyxin resistance was a common finding across all dosage regimens and reversion to susceptibility was substantial but incomplete during drug-free passaging in non-stable polymyxin-resistant bacterial samples (Fig. 3). Notably, there was insufficient evidence in the transcriptomic profiles to identify a clear link between the dosage regimen employed and the transcriptomic responses associated with polymyxin resistance. Complex multi-level regulatory networks are likely involved in the development of polymyxin resistance and limit the utility of transcriptomics in isolation (i.e. in the absence of metabolomic and proteomic studies) to characterise the mechanisms that give rise to non-stable polymyxin resistance. Further, the clinical implications of these findings will require additional investigations using in vivo infection models and clinical studies that adequately account for the role of immune response in infection39,40. The in vitro data in the present study highlight the possible limitations of polymyxin monotherapy and the need for other strategies (e.g. combination therapy) for preventing the widespread emergence of polymyxin resistance41.
In the present study, stable polymyxin resistance was caused by a previously documented pmrB A227V mutation42, which is hypothesized to constitutively up-regulate pmrCAB operon expression17,21. The notable loss of polymyxin resistance during drug-free passaging in a sample found to contain the same pmrB A227V mutation implicates a fitness cost associated with the mutation. This is supported by previously published studies in pmrB A227V mutants of A. baumannii strain ATCC 19606, which discovered that the mutant displayed a slower growth rate compared to wild-type strains42. However, the development of non-stable resistance highlights that genetic mutations are unlikely to be the sole driver of polymyxin resistance. Our findings that the transcriptomic profiles of bacteria exhibiting stable and non-stable polymyxin resistance were highly similar indicate that the stable and non-stable resistance may share a common mechanism involving pmrB-mediated modification of lipid A. This finding is striking in light of the conspicuous absence of the PhoPQ – PmrD signal transduction pathway in A. baumannii strains AB307-0294 and ATCC 19606. In other Gram-negative organisms, the PhoPQ two-component system is known to sense the presence of polymyxins and interfaces with PmrB via PmrD43. To date, an orthologous polymyxin sensing mechanism in A. baumannii has not been identified; there remains a pressing need to understand the role of polymyxin sensing and non-stable polymyxin resistance in determining the pharmacodynamics of polymyxin treatment in critically-ill patients.
While this study was limited to the examination of the transcriptome of A. baumannii strain AB307-0294, transcriptomic data from this study was successfully combined with previously published data characterising the early-stage responses to polymyxin exposure in A. baumannii ATCC 1960631. The combination of orthologous protein matching with SPLS-DA analysis enabled the integrated analysis of gene expression data between closely-related bacterial strains. SPLS-DA is a methodology developed to improve the analysis of high-dimensional omics datasets, combining multivariate statistics, dimension-reduction and feature selection36. This novel framework maximises the information gained from transcriptomics experiments by incorporating prior transcriptomic data. In this study, the fitted SPLS-DA model implicated the involvement of polyphosphate kinase (PPK; ABBFA002620) in the bacterial response to polymyxin exposure, a finding supported by studies in Salmonella that reported increased polymyxin susceptibility in ΔPPK mutants44. From these results, it can be hypothesised that the accumulation of inorganic polyphosphates is critical to the initial response to polymyxin exposure. Collectively, the altered expression of genes involved in cellular metabolism (ABBFA_003493: NADPH-dependent FMN reductase family protein, ABBFA_002755: NAD(+) synthetase) and PPK suggests that in the early stages of polymyxin exposure, the intracellular redox reactions are either directly disrupted by polymyxins or essential to the initial stress response following the exposure. Evidence of these disruptions has also been found in the metabolomes of colistin-treated A. baumannii45. Confirmation of the importance of these metabolic pathways on polymyxin activity and resistance using molecular techniques across a broader collection of A. baumannii strains will be crucial for identifying potential targets for novel antimicrobial agents.
GO term enrichment analysis, SPLS-DA, and a conventional analysis of gene expression showed remodelling of the OM to be a key aspect of polymyxin resistance; transcriptomic profiles obtained from A. baumannii strains AB307-0294 and ATCC19606 contained evidence of compensatory adaptations associated with OM remodelling. Increased expression of (RND) efflux transporter proteins (AdeABC and HlyD family) was a common finding across all analysis methodologies and stages of polymyxin exposure. This up-regulation of efflux transporters, observed in concert with over-expression of protein complexes involved in membrane homeostasis, supports previously published findings31 that point to the diminished integrity and barrier function of the remodelled OM in polymyxin-treated A. baumannii. Polymyxins have been shown to exhibit synergistic activity in combination with other antibiotics such as carbapenems and chloramphenicol46,47,48, and understanding polymyxin-induced OM remodelling will facilitate the development of rational antibiotic combination regimens that maximise bacterial killing and minimise the emergence of resistance.
Conclusions
To our knowledge, this is the first study to investigate both the genomic and transcriptomic profiles of polymyxin resistance in A. baumannii following exposure to clinically relevant dosage regimens over an extended period. Unlike previous investigations into polymyxin resistance in Gram-negative organisms which focused on bacterial isolates that exhibit stable resistance, the present study reveals that both stable and non-stable polymyxin-resistant phenotypes are selected during treatment. Further, a framework for the integrative analysis of prior transcriptomic data from closely related bacterial strains revealed new insights into responses to polymyxins in A. baumannii. It remains to be elucidated the extent to which non-stable polymyxin resistance affects clinical outcomes. Our findings provide a foundation for understanding the mechanistic drivers of polymyxin resistance during polymyxin exposure resulting from clinically relevant dosage regimens, and highlight the importance of exploring optimised combination therapy in addressing the antimicrobial resistance crisis.
Additional Information
How to cite this article: Cheah, S.-E. et al. Polymyxin Resistance in Acinetobacter baumannii: Genetic Mutations and Transcriptomic Changes in Response to Clinically Relevant Dosage Regimens. Sci. Rep. 6, 26233; doi: 10.1038/srep26233 (2016).
Acknowledgments
This study was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (Award Numbers R01AI079330 to R.L.N., J.L., B.T.T. and J.B.B. and R01AI111990 to B.T.T., J.L., R.L.N., J.D.B. and J.B.B.). J.L. is an Australian National Health and Medical Research Council (NHMRC) Senior Research Fellow. J.B.B. is the recipient of an NHMRC Career Development Fellowship (APP1084163). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Author Contributions S.-E.C. performed the experiments and data analysis described in this manuscript. M.D.J. and Y.Z. assisted with data interpretation. B.T.T., A.F. and J.B.B. assisted with the preparation of the manuscript and data interpretation. J.L. and R.L.N. conceived the study with S.-E.C. and J.D.B. and assisted with manuscript preparation and data interpretation.
References
- Walker B. et al. Environment. Looming global-scale failures and missing institutions. Science 325, 1345–1346, doi: 10.1126/science.1175325 (2009). [DOI] [PubMed] [Google Scholar]
- Gales A. C., Castanheira M., Jones R. N. & Sader H. S. Antimicrobial resistance among Gram-negative bacilli isolated from Latin America: results from SENTRY Antimicrobial Surveillance Program (Latin America, 2008–2010). Diagn Microbiol Infect Dis 73, 354–360, doi: 10.1016/j.diagmicrobio.2012.04.007 (2012). [DOI] [PubMed] [Google Scholar]
- Sader H. S., Farrell D. J., Flamm R. K. & Jones R. N. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009–2011). Diagn Microbiol Infect Dis, doi: 10.1016/j.diagmicrobio.2013.11.025 (2013). [DOI] [PubMed] [Google Scholar]
- Peleg A. Y., Seifert H. & Paterson D. L. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21, 538–582, doi: 10.1128/CMR.00058-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nation R. L., Velkov T. & Li J. Colistin and polymyxin B: peas in a pod, or chalk and cheese? Clin Infect Dis 59, 88–94, doi: 10.1093/cid/ciu213 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nation R. L. & Li J. Colistin in the 21st century. Curr Opin Infect Dis 22, 535–543, doi: 10.1097/QCO.0b013e328332e672 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavascki A. P., Goldani L. Z., Li J. & Nation R. L. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemother 60, 1206–1215, doi: 10.1093/jac/dkm357 (2007). [DOI] [PubMed] [Google Scholar]
- Jones R. N. et al. Susceptibility rates in Latin American nations: report from a regional resistance surveillance program (2011). Braz J Infect Dis 17, 672–681, doi: 10.1016/j.bjid.2013.07.002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanel G. G. et al. Antimicrobial susceptibility of 22746 pathogens from Canadian hospitals: results of the CANWARD 2007-11 study. J Antimicrob Chemother 68 Suppl 1, i7–22, doi: 10.1093/jac/dkt022 (2013). [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Shin J., Shin S. Y. & Ko K. S. Characteristics of carbapenem-resistant Enterobacteriaceae isolates from Korea. Diagn Microbiol Infect Dis 76, 486–490, doi: 10.1016/j.diagmicrobio.2013.04.006 (2013). [DOI] [PubMed] [Google Scholar]
- Velkov T., Thompson P. E., Nation R. L. & Li J. Structure-activity relationships of polymyxin antibiotics. J Med Chem 53, 1898–1916, doi: 10.1021/jm900999h (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson T. R. et al. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob Agents Chemother 56, 5642–5649, doi: 10.1128/AAC.00756-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deris Z. Z. et al. A secondary mode of action of polymyxins against Gram-negative bacteria involves the inhibition of NADH-quinone oxidoreductase activity. J Antibiot (Tokyo) 67, 147–151, doi: 10.1038/ja.2013.111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale J. D. & Hancock R. E. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev Anti Infect Ther 5, 951–959, doi: 10.1586/14787210.5.6.951 (2007). [DOI] [PubMed] [Google Scholar]
- Hancock R. E. & Chapple D. S. Peptide antibiotics. Antimicrob Agents Chemother 43, 1317–1323 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Dhillon P., Yan H., Farmer S. & Hancock R. E. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob Agents Chemother 44, 3317–3321, doi: 10.1128/aac.44.12.3317-3321.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo L. A. et al. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother 55, 3743–3751, doi: 10.1128/AAC.00256-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt J. H. et al. Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob Agents Chemother 55, 3022–3024 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt J. H. et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54, 4971–4977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. Y., Gregg K. A., Napier B. A., Ernst R. K. & Weiss D. S. A PmrB-regulated deacetylase required for lipid A modification and polymyxin resistance in Acinetobacter baumannii. Antimicrob Agents Chemother, doi: 10.1128/AAC.00515-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. K., Choi J. Y., Shin D. & Ko K. S. Correlation between overexpression and amino acid substitution of the PmrAB locus and colistin resistance in Acinetobacter baumannii. Int J Antimicrob Agents 37, 525–530 (2011). [DOI] [PubMed] [Google Scholar]
- Plachouras D. et al. Population pharmacokinetic analysis of colistin methanesulfonate and colistin after intravenous administration in critically ill patients with infections caused by gram-negative bacteria. Antimicrob Agents Chemother 53, 3430–3436, doi: 10.1128/AAC.01361-08 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garonzik S. M. et al. Population pharmacokinetics of colistin methanesulfonate and formed colistin in critically ill patients from a multicenter study provide dosing suggestions for various categories of patients. Antimicrob Agents Chemother 55, 3284–3294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaiskos I. et al. Colistin population pharmacokinetics after application of a loading dose of 9 MU colistin methanesulfonate (CMS) in critically ill patients. Antimicrob Agents Chemother, doi: 10.1128/AAC.00554-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri A. M. et al. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis 57, 524–531, doi: 10.1093/cid/cit334 (2013). [DOI] [PubMed] [Google Scholar]
- Gales A. C., Reis A. O. & Jones R. N. Contemporary assessment of antimicrobial susceptibility testing methods for polymyxin B and colistin: review of available interpretative criteria and quality control guidelines. J Clin Microbiol 39, 183–190, doi: 10.1128/JCM.39.1.183-190.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M. D. et al. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190, 8053–8064, doi: 10.1128/JB.00834-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diancourt L., Passet V., Nemec A., Dijkshoorn L. & Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5, e10034, doi: 10.1371/journal.pone.0010034 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen P. J. et al. Comparison of once-, twice- and thrice-daily dosing of colistin on antibacterial effect and emergence of resistance: studies with Pseudomonas aeruginosa in an in vitro pharmacodynamic model. J Antimicrob Chemother 61, 636–642, doi: 10.1093/jac/dkm511 (2008). [DOI] [PubMed] [Google Scholar]
- Mohamed A. F. et al. Application of a loading dose of colistin methanesulfonate in critically ill patients: population pharmacokinetics, protein binding, and prediction of bacterial kill. Antimicrob Agents Chemother 56, 4241–4249, doi: 10.1128/aac.06426-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry R. et al. The transcriptomic response of Acinetobacter baumannii to colistin and doripenem alone and in combination in an in vitro pharmacokinetics/pharmacodynamics model. J Antimicrob Chemother 70, 1303–1313, doi: 10.1093/jac/dku536 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E. & Marth G. Haplotype-based variant detection from short-read sequencing. Preprint at arXiv:1207.3907v2 [q-bio.GN] (2012).
- Ritchie M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47, doi: 10.1093/nar/gkv007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 57, 289–300 (1995). [Google Scholar]
- Jones P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240, doi: 10.1093/bioinformatics/btu031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cao K. A., Boitard S. & Besse P. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC bioinformatics 12, 253, doi: 10.1186/1471-2105-12-253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J. Jr. & Roos D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome res 13, 2178–2189, doi: 10.1101/gr.1224503 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drusano G. L. et al. Interaction of drug- and granulocyte-mediated killing of Pseudomonas aeruginosa in a murine pneumonia model. J Infect Dis 210, 1319–1324, doi: 10.1093/infdis/jiu237 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drusano G. L. et al. Saturability of granulocyte kill of Pseudomonas aeruginosa in a murine model of pneumonia. Antimicrob Agents Chemother 55, 2693–2695, doi: 10.1128/AAC.01687-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nation R. L. et al. Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect Dis 15, 225–234, doi: 10.1016/s1473-3099(14)70850-3 (2015). [DOI] [PubMed] [Google Scholar]
- Adams M. D. et al. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 53, 3628–3634, doi: 10.1128/AAC.00284-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas M. E., Rafailidis P. I. & Matthaiou D. K. Resistance to polymyxins: Mechanisms, frequency and treatment options. Drug Resist Updat 13, 132–138, doi: 10.1016/j.drup.2010.05.002 (2010). [DOI] [PubMed] [Google Scholar]
- Kim K. S., Rao N. N., Fraley C. D. & Kornberg A. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc Natl Acad Sci USA 99, 7675–7680, doi: 10.1073/pnas.112210499 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maifiah M. H. M. et al. In interscience conference of antimicrobial agents and chemotherapy (San Diego, 2015).
- Abdul Rahim N. et al. Synergistic killing of NDM-producing MDR Klebsiella pneumoniae by two ‘old’ antibiotics-polymyxin B and chloramphenicol. J Antimicrob Chemother 70, 2589–2597, doi: 10.1093/jac/dkv135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Antibiograms of multidrug-resistant clinical Acinetobacter baumannii: promising therapeutic options for treatment of infection with colistin-resistant strains. Clin Infect Dis 45, 594–598, doi: 10.1086/520658 (2007). [DOI] [PubMed] [Google Scholar]
- Deris Z. Z. et al. The combination of colistin and doripenem is synergistic against Klebsiella pneumoniae at multiple inocula and suppresses colistin resistance in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother 56, 5103–5112, doi: 10.1128/AAC.01064-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]


