Abstract
Bacterial genome sequencing projects routinely uncover gene clusters that are predicted to encode the biosynthesis of uncharacterized small molecules. A subset of these cryptic genetic elements appears as individual operons. Here we investigate potential single operon biosynthetic systems found in the genome of the pathogenic bacterium Burkholderia pseudomallei. Placing these operons under the control of an inducible promoter led to the production of seven new metabolites. Among the molecules we identified are inhibitors of type 4 phosphodiesterases, suggesting that previously cryptic biosynthetic operons may encode metabolites that could contribute to microbial virulence by disrupting host signaling pathways.
Graphical abstract
Bacteria rely extensively on low molecular weight organic compounds to interact with the world around them and, in the case of pathogenic bacteria, these small molecules can be key components of pathogenesis.1,2 Small molecule biosynthetic gene clusters that do not appear to encode for metabolites previously identified in fermentation based studies are routinely found in sequenced bacterial genomes.3 These cryptic biosynthetic gene clusters represent candidate pathways from which additional small molecule virulence factors might be identified in pathogenic bacteria. While the biosynthesis of many small molecules is encoded on large stretches of genomic DNA, consisting of multiple operons, the biosynthesis of some metabolites can require as little as a single gene or operon. Systematically decoupling these short cryptic biosynthetic operons from their native promoters and placing them under the control of well-studied inducible promoters in model bacterial hosts should provide a means to access compounds encoded by previously uncharacterized single operon, small molecule biosynthetic systems (Figure 1). An examination of putative cryptic biosynthetic operons found in the genome of Burkholderia pseudomallei, the etiologic agent of melioidosis,4 has led to the identification of two operons that, upon induction in Pseudomonas aeruginosa, confer the production of novel metabolites to this host (Figure 2). Melioidosis is endemic in Southeast Asia and Northern Australia and because of its historical use in biological weapons programs, B. pseudomallei is categorized as a class B select agent by the Centers for Disease Control in the United States.4
Figure 1.
It should be possible to produce metabolites encoded by tightly regulated cryptic single operon biosynthetic systems using well-controlled inducible promoters in model heterologous hosts.
Figure 2.
The positions of potential single operon biosynthetic systems examined in this study are mapped onto chromosome II of B. pseudomallei K96243. Chromosome I did not yield any metabolites and is not shown. Compounds identified in this study are shown in red. Gene clusters either known to be, or predicted to be, associated with the production of structurally characterized metabolites in B. pseudomallei are tagged with the metabolites they encode (black).5–7 A metabolite identified in heterologous expression experiments in E. coli using a previously cryptic B. pseudomallei operon appears in green.8
In total, 13 potential biosynthetic operons were selected by manually examining the genome of B. pseudomallei K96243 for uncharacterized operons containing genes predicted to encode enzymes with strong links to secondary metabolism, including polyketide synthases (PKS), non-ribosomal peptide synthetases (NRPS), acyl-CoA ligases, functional group transferases and redox-active proteins (Figure 2, Table S1). Each operon was PCR amplified from genomic DNA, cloned under the control of an IPTG-inducible Ptac promoter (pMMB67EXPH) and conjugated into P. aeruginosa for heterologous expression studies.9 P. aeruginosa was chosen as the heterologous host for this study because it is easily transformable, grows rapidly and natively produces a diverse collection of secondary metabolites.
All P. aeruginosa transformants were grown in LB broth at 30 °C (250 rpm) to an OD600 = 0.1, at which time the expression of individual operons was induced with 0.1 mM IPTG. After 24–36 h of shaking, induced cultures were extracted with an equal volume of ethyl acetate, and these extracts were then examined for the presence of clone specific molecules by silica gel TLC. Clone specific molecules were detected in extracts derived from cultures of P. aeruginosa transformed with two operons: BTH-II0204-207/BPSS0130-133 and BPSS2111-2113 (Figure 3a and 3b).10 Although subsets of the genes found in these operons are found in other bacterial genomes, both operons appear to be unique to sequenced Burkholderia genomes. NMR, HRMS and X-ray crystallography data were used to elucidate the structures of the seven clone specific metabolites isolated from ethyl acetate extracts of large-scale (6 L) cultures of P. aeruginosa expressing these operons (Figure 4).
Figure 3.
IPTG induced expression of either the BTH-II0204-207BPSS0130-133 operon (a) or the BPSS2111-2113 operon (b) leads to the production of clone specific metabolites in P. aeruginosa. In each case the control extract is derived from cultures of P. aeruginosa transformed with the empty expression vector. (c) B. thailandesis E264 grown in LB broth with and without phenylalanine.
Figure 4.
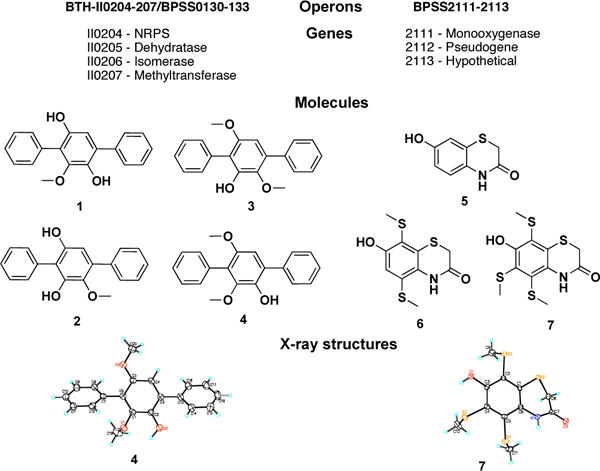
Clone-specific molecules identified in culture broths of P. aeruginosa transformed with potential cryptic biosynthetic operons from Burkholderia spp.
None of the metabolites identified in this study have been reported previously as natural products. As B. pseudomallei is a highly infectious BSL3 level pathogen, it was impractical for us to look for compounds 1 – 7 in large scale broth cultures of this organism. Burkholderia thailandensis is a close relative of B. pseudomallei that is not generally considered to be a human pathogen. At high titers however, it is infectious thus making it a good low-virulence model for studying Burkholderia pathogens in the laboratory.11,12 The BTH-II0204-207/BPSS0130-133 operon is found in both B. pseudomallei and B. thailandensis, and therefore it was possible to look for the products of this operon in cultures of B. thailandensis grown under standard conditions. In our initial exmaination of culture broth extracts from B. thailandensis grown in a variety of minimal and rich media, none of the compounds associated with this operon could be detected by HPLC-MS. We subsequently explored the possibility that the BTH-II0204-207/BPSS0130-133 operon might be induced in the presence of high substrate concentrations.
BTH-II0204-0207/BPSS0130-133 is a four-gene operon that is predicted to contain a single NRPS gene, a dehydratase, an isomerase and a methyltransferase (Figure 4, 5b). The closest experimentally defined homologs of the NRPS module are from fungi (40% identity).13 In fungal systems, this module carries out the dimerization of deaminated aromatic amino acids to produce tricyclic structures in which the central ring is a hydroxyquinone (Figure 5a).13 The NRPS module from the BTH-II0204-207/BPSS0130-133 operon likely carries out a similar dimerization of phenylananine or a phenylananine-derived precursor (Figure 5b). Cultures of B. thailandensis grown in LB broth were therefore spiked with phenylalanine (10 mM), and extracts from these cultures were examined for the presence of 1 – 4. As shown in Figure 3c compound 1 can be found easily in extracts from cultures spiked with phenylalanine. Deletion of the NRPS gene in B. thailandensis E264 by targeted mutagenesis14 abolished the production of 1, confirming the biosynthetic role of this operon (Figure S1).
Figure 5.
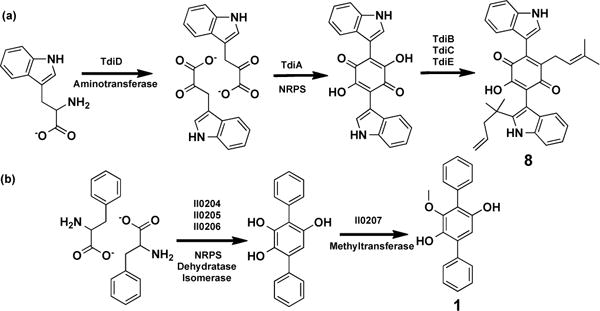
(a) Initial steps in the biosynthesis of the fungal metabolite terrequinone (8).13 (b) Predicted general biosynthetic scheme for 1. Promiscuity of the methyltransferase would account for the varied methylation patterns. The exact role of the predicted dehydratase and isomerase enzymes remains to be determined.
In cytotoxicity assays run against representative bacteria (Bacillus subtilis, Staphylococcus aureus, and Escherichia coli), Sacchromyces cereviseae and human HeLa cells, most of these metabolites were inactive. Compounds 1 and 3 show activity against B. subtilis (MICs of 11 and 33 ug/ml, respectively), with 1 also showing mild activity against S. cerevisiae (MIC of 11 ug/ml). A potential biological role for these metabolites was sought by examining the literature for known metabolites with similar structures and known biological activities. Compounds 1 – 4 resemble the Streptomyces sp. derived metabolite terferol (9, Figure 6b), which was originally identified in a screen for eukaryotic phosphodiesterase (PDE) inhibitors.10,15,16 Compounds 1 – 4 differ from terferol by either the position or the number of methyl substituents found on each metabolite.
Figure 6.
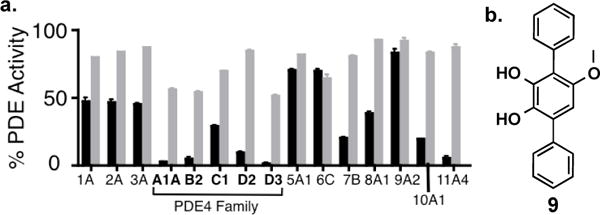
a. Percent activity of a spectrum of phosphodiesterases in the presence of either 100 μM of either compound 1 (black) or compound 3 (gray). b. Terferol (9).
PDEs terminate signaling of cyclic nucleotide second messengers by hydrolyzing cAMP and/or cGMP to the corresponding inactive 5′-mononucleotides. PDEs are divided into 11 biochemically and pharmacologically distinct enzyme families that are distinguished primarily by differences in substrate preference, structure, and inhibitor sensitivities. Compounds 1 and 3, the major clone specific metabolites produced by the BTH-II0204-207/BPSS0130-133 operon in our heterologous expression studies, were assayed for PDE inhibition activity against a panel of purified eukaryotic PDEs. At 100 μM the dimethylated 3 is largely inactive. At the same concentration the monomethylated 1 shows activity against PDE11 as well as 4 out of the 5 PDE4s that we examined (Figure 6a). The most active derivative, compound 1, is the major product we observe in cultures of wild type B. thailandensis. PDE4s have been implicated in inflammation responses across multiple immune cell-types.17 As a consequence, PDE4 inhibitors have been extensively investigated as potential therapeutics for a number of inflammatory diseases.18 It has been reported that animals treated with PDE inhibitors (rolipram or aminophylline) show impaired host defenses against pathogenic bacteria.19,20 In particular the PDE4 inhibitor rolipram was found to inhibit tumor necrosis factor-alpha production and neutrophil migration into the site of infection, resulting in earlier lethality and enhanced bacterial load in infected mice. While compound 1 is significantly less active (40 μM IC50 for PDE4A1) than optimized synthetic PDE4 inhibitors, the immediate environment adjacent to a pathogen during an infection would likely see a high effective concentration of any secreted metabolite.21 The production of PDE4 inhibitors could therefore represent a way by which pathogenic bacteria abrogate host defense mechanisms.
By decoupling individual cryptic biosynthetic operons from their native promoters and activating them with controlled inducible promoters, it should be possible to functionally access many simple, cryptic gene clusters found in bacterial genomes. When studying pathogenic bacteria like B. pseudomallei, this strategy provides the added advantage of being able to study metabolites produced by infectious bacteria without having to grow pathogens in the laboratory.
Supplementary Material
Acknowledgments
We thank Emil Lobkovsky at the Cornell X-ray crystallography facility for his assistance. This work was supported by the Northeast Biodefense Center (U54-AI057158). Phosphodiesterase assays were run by BPS Biosciences (San Diego, CA).
Footnotes
Supporting Information Available: Experimental protocols for operon cloning, molecule production and isolation as well as NMR spectra and structural characterization details for all new compounds are available free of charge at http://pubs.acs.org.
References
- 1.Bassler BL, Losick R. Cell. 2006;125:237–46. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Njoroge J, Sperandio V. EMBO Mol Med. 2009;1:201–10. doi: 10.1002/emmm.200900032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Zerikly M, Challis GL. ChemBioChem. 2009;10:625–33. doi: 10.1002/cbic.200800389. [DOI] [PubMed] [Google Scholar]; (b) Hertweck C. Nat Chem Biol. 2009;5:450–2. doi: 10.1038/nchembio0709-450. [DOI] [PubMed] [Google Scholar]; (c) Li MH, Ung PM, Zajkowski J, Garneau-Tsodikova S, Sherman DH. BMC Bioinformatics. 2009;10:185. doi: 10.1186/1471-2105-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Van Lanen SG, Shen B. Curr Opin Microbiol. 2006;9:252–60. doi: 10.1016/j.mib.2006.04.002. [DOI] [PubMed] [Google Scholar]; (e) Bode HB, Muller R. Angew Chem Int Ed. 2005;44:6828–6846. doi: 10.1002/anie.200501080. [DOI] [PubMed] [Google Scholar]; (f) Peric-Concha N, Long PF. Drug Discov Today. 2003;8:1078–84. doi: 10.1016/s1359-6446(03)02901-5. [DOI] [PubMed] [Google Scholar]
- 4.Adler NR, Govan B, Cullinane M, Harper M, Adler B, Boyce JD. FEMS Microbiol Rev. 2009;33:1079–99. doi: 10.1111/j.1574-6976.2009.00189.x. [DOI] [PubMed] [Google Scholar]
- 5.Song Y, Xie C, Ong YM, Gan YH, Chua KL. J Bacteriol. 2005;187:785–90. doi: 10.1128/JB.187.2.785-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duerkop BA, Varga J, Chandler JR, Peterson SB, Herman JP, Churchill ME, Parsek MR, Nierman WC, Greenberg EP. J Bacteriol. 2009;191:3909–18. doi: 10.1128/JB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seyedsayamdost MR, Chandler JR, Blodgett JA, Lima PS, Duerkop BA, Oinuma K, Greenberg EP, Clardy J. Org Lett. 2010;12:716–9. doi: 10.1021/ol902751x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brady SF, Bauer JD, Clarke-Pearson MF, Daniels R. J Am Chem Soc. 2007;129:12102–3. doi: 10.1021/ja075492v. [DOI] [PubMed] [Google Scholar]
- 9.Some operons present in both B. pseudomallei K96243 and its close relative B. thailandensis E264 were amplified from B. thailandensis genomic DNA.
- 10.BPSS2111–2113 was originally cloned as what was thought to be a three-gene operon. However, on closer inspection, the second ORF contains a series of mutations that results in it being a truncated pseudogene. Most sequenced B. pseudomallei genomes available in GenBank contain mutations in this gene that introduce premature stop codons. Cultures of P. aeruginosa transformed with BPSS2111-2113, BPSS2111-2112 or BPSS2111 alone produce the same set of clone specific compounds. BPSS2111, a predicted FAD dependent monooxygenese is therefore necessary and sufficient to confer the production of all three metabolites (5, 6, and 7) to P. aeruginosa. To confirm the role of this single ORF in the production of these metabolites, we introduced a BPSS2111 expression construct into a second host, Burkohlderia graminis C4D1M. As seen in P. aeruginosa, the expression of BPSS2111 alone is sufficient to generate 5, 6, and 7 in B. graminis. BPSS2111 homologs are found in a number of sequenced bacterial genomes. Additional studies are required to determine the exact biosynthetic origin of this family of metabolites. It is possible that BPSS2111 generates 5, which appears first in the culture broth, and that host enzymes add the thiomethyls-groups. None of the compounds produced by the BPSS2111–2113 operon closely resemble previously described metabolites with well-defined functions
- 11.(a) Kim HS, Schell MA, Yu Y, Ulrich RL, Sarria SH, Nierman WC, DeShazer D. BMC Genomics. 2005;6:174. doi: 10.1186/1471-2164-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yu Y, Kim HS, Chua HH, Lin CH, Sim SH, Lin D, Derr A, Engels R, DeShazer D, Birren B, Nierman WC, Tan P. BMC Microbiol. 2006;6:46. doi: 10.1186/1471-2180-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass MB, Gee JE, Steigerwalt AG, Cavuoti D, Barton T, Hardy RD, Godoy D, Spratt BG, Clark TA, Wilkins PP. J Clin Microbiol. 2006;44:4601–4. doi: 10.1128/JCM.01585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Balibar CJ, Howard-Jones AR, Walsh CT. Nat Chem Biol. 2007;3:584–92. doi: 10.1038/nchembio.2007.20. [DOI] [PubMed] [Google Scholar]; (b) Schneider P, Weber M, Hoffmeister D. Fungal Genet Biol. 2008;45:302–9. doi: 10.1016/j.fgb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Thongdee M, Gallagher LA, Schell M, Dharakul T, Songsivilai S, Manoil C. Appl Environ Microbiol. 2008;74:2985–9. doi: 10.1128/AEM.00030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakagawa F, Takahashi S, Naito A, Sato S, Iwabuchi S, Tamura C. J Antibiot (Tokyo) 1984;37:10–2. doi: 10.7164/antibiotics.37.10. [DOI] [PubMed] [Google Scholar]
- 16.Terferol has also been reported to be parasite-specific cGMP-dependent protein kinase inhibitor; Zhang C, Ondeyka JG, Herath KB, Guan Z, Collado J, Pelaez F, Leavitt PS, Gurnett A, Nare B, Liberator P, Singh SB. J Nat Prod. 2006;69:710–2. doi: 10.1021/np0505418. [DOI] [PubMed] [Google Scholar]
- 17.(a) Omori K, Kotera J. Circ Res. 2007;100:309–27. doi: 10.1161/01.RES.0000256354.95791.f1. [DOI] [PubMed] [Google Scholar]; (b) Press NJ, Banner KH. Prog Med Chem. 2009;47:37–74. doi: 10.1016/S0079-6468(08)00202-6. and references therein. [DOI] [PubMed] [Google Scholar]
- 18.Houslay MD, Schafer P, Zhang KY. Drug Discov Today. 2005;10:1503–19. doi: 10.1016/S1359-6446(05)03622-6. [DOI] [PubMed] [Google Scholar]
- 19.Soares AC, Souza DG, Pinho V, Vieira AT, Barsante MM, Nicoli JR, Teixeira M. Br J Pharmacol. 2003;140:855–62. doi: 10.1038/sj.bjp.0705517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson S, Summer WR, Jakab GJ. Am Rev Respir Dis. 1985;131:923–7. doi: 10.1164/arrd.1985.131.6.923. [DOI] [PubMed] [Google Scholar]
- 21.Wild-type PDE activities are gated by spatiotemporal regulation within cell-signaling niches, whose efficacy is normally low (Km ~ 1–10 μM; ref 17b), allowing locally concentrated PDE inhibitors the potential ability to disrupt normal PDE signaling in localized cell populations.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


