Abstract
We evaluate the ability of hexahistidine (His6) tags on peptide and protein substrates to recruit deoxyribozymes for modifying those substrates. For two different deoxyribozymes, one that creates tyrosine-RNA nucleopeptides and another that phosphorylates tyrosine side chains, we find substantial improvements in yield, kobs, and Km for peptide substrates due to recruiting by His6/Cu2+. However, the recruiting benefits of the histidine tag are not observed for larger protein substrates, likely because the tested deoxyribozymes either cannot access the target peptide segments or cannot function when these segments are presented in a structured protein context.
Introduction
Deoxyribozymes are single-stranded DNA sequences with catalytic activity.1–5 Our laboratory seeks deoxyribozymes for covalent protein modification.6 In these efforts, a key challenge is achieving DNA-catalyzed reactions using relatively low protein concentrations that enable preparative utility. We recently described the use of azide-functionalized peptide substrates during in vitro selection, which enabled identification of deoxyribozymes with peptide Km of ~100 μM.7 However, that approach is not applicable to all DNA-catalyzed reactions. Here we pursued an alternative and potentially more general strategy: recruiting deoxyribozymes for modifying peptides or proteins using histidine tags.
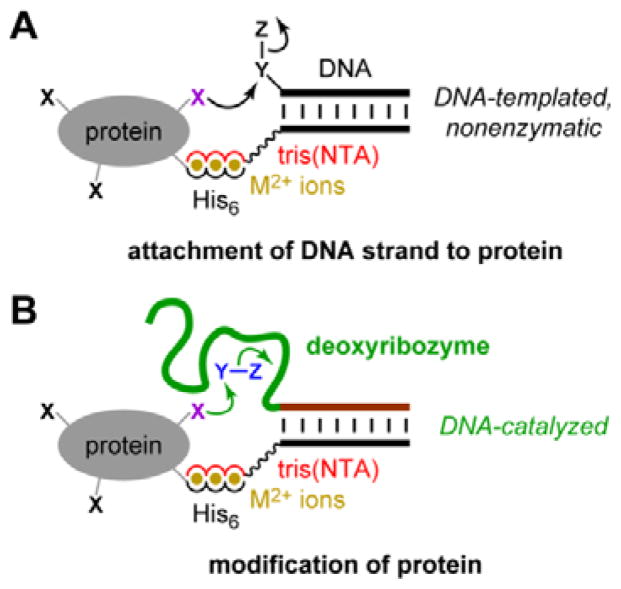
Rosen et al. recently reported that metal ion-mediated interactions between a suitably functionalized DNA oligonucleotide and a metal-binding (e.g., histidine-tagged) protein can be used to recruit the DNA oligonucleotide to a specific region on a protein surface (Fig. 1A).8 Subsequent nonenzymatic, DNA-templated reaction leads to site-specific protein modification by attachment of a DNA strand, whereas many other chemical modification approaches for proteins are not site-selective.9–23 Here, we evaluated experimentally whether a histidine tag can be used to recruit a deoxyribozyme—which in this context is simply a long DNA oligonucleotide—to a peptide or protein, thereby enhancing DNA-catalyzed covalent modification of that substrate, in principle by any modification reaction that can be catalyzed by DNA (Fig. 1B). Unlike the nonenzymatic, strictly proximity-driven approach of Fig. 1A, the strategy of Fig. 1B uses Watson-Crick base pairs for recruiting the deoxyribozyme to the peptide or protein substrate. The subsequent modification reaction still requires enzymatic catalysis by the DNA.
Fig. 1.
Use of histidine tags to recruit reagents or catalysts for peptide and protein modification. (A) The reported use of hexahistidine (His6) tags and divalent metal ions (M2+) for directing nonenzymatic, DNA-templated reaction of protein side chains.8 In one case, the authors instead used a natively metal-binding protein. NTA = nitrilotriacetic acid. (B) As evaluated in this study, the potential use of His6 tags and M2+ to recruit a deoxyribozyme for catalyzing peptide and protein modification. See Fig. 2 for details of DNA catalysis arrangement, including reaction partner for the protein (shown here as Y–Z), as well as NTA structure.
Results
Evaluating the His6- tag recruiting strategy with the 8XJ105 deoxyribozyme, which forms peptide-RNA conjugates
For evaluating the histidine tag recruiting strategy of Fig. 1B, we initially used the 8XJ105 deoxyribozyme, which attaches a 5′-triphosphorylated RNA oligonucleotide (pppRNA) to a tyrosine hydroxyl group in a peptide substrate, forming a nucleopeptide (peptide-RNA) conjugate.7 Such peptide-RNA (or peptide-DNA) conjugates are integrally involved in many key biological processes,24–32 and they are useful for various applications as well.33–40 8XJ105 was identified by an in vitro selection strategy that uses an untethered (discrete, free) peptide during each selection round. As a consequence, 8XJ105 has substantial activity with an untethered peptide only at very high peptide concentration (apparent peptide Km >1 mM). 8XJ105 tolerates a wide variety of sequence contexts for the substrate’s tyrosine residue, i.e., 8XJ105 is peptide sequence-general. Therefore, we sought to use histidine tags to recruit 8XJ105 for conjugating RNA to a tyrosine-containing peptide.
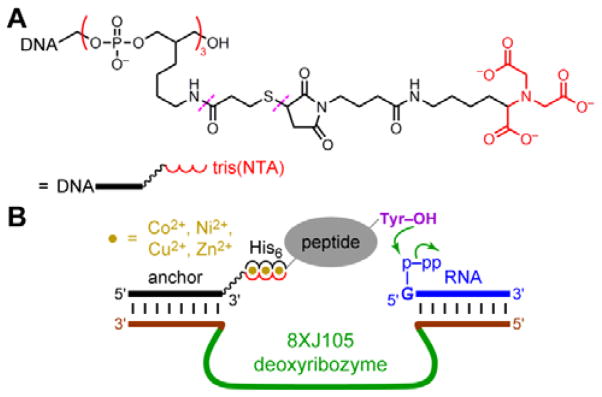
A DNA anchor oligonucleotide was synthesized, bearing at either its 3′-end or 5′-end a tris(NTA) moiety that efficiently binds certain divalent metal ions (NTA = nitrilotriacetic acid; Fig. 2A). As was done by Rosen et al.,8 we adapted the synthetic procedure from the work of Goodman et al.41 The deoxyribozyme interacts with the DNA anchor by standard Watson-Crick base pairs, such that recruiting the DNA anchor to the peptide substrate also recruits the deoxyribozyme. The peptide substrate bears at its N-terminus a hexahistidine (His6) tag. As shown in Fig. 2B, the intention is that the tris(NTA) and His6 moieties bind simultaneously to divalent metal ions (M2+), thereby recruiting the deoxyribozyme to the peptide and enabling DNA-catalyzed peptide modification.
Fig. 2.
Strategy for using histidine tags to recruit a deoxyribozyme to a peptide substrate and enable DNA-catalyzed peptide modification. (A) Tris(NTA)-modified DNA anchor oligonucleotide. The 3′-tris(NTA) moiety was connected to the DNA by a hexa(ethylene glycol) spacer. For the 5′-tris(NTA)-modified DNA anchor, the 5′-tris(NTA) moiety was connected to the DNA by a T15 oligonucleotide spacer. Each tris(NTA)-modified oligonucleotide was synthesized by a multistep procedure in which the linkages marked with dashes were created (see the Experimental section). (B) Design showing the intended interactions among the 8XJ105 deoxyribozyme that forms nucleopeptide linkages, 3′-tris(NTA)-modified DNA anchor, His6-tagged peptide substrate, and divalent metal ions. Success is revealed by either or both of increase in peptide-RNA conjugation yield and leftward shift in peptide concentration dependence.
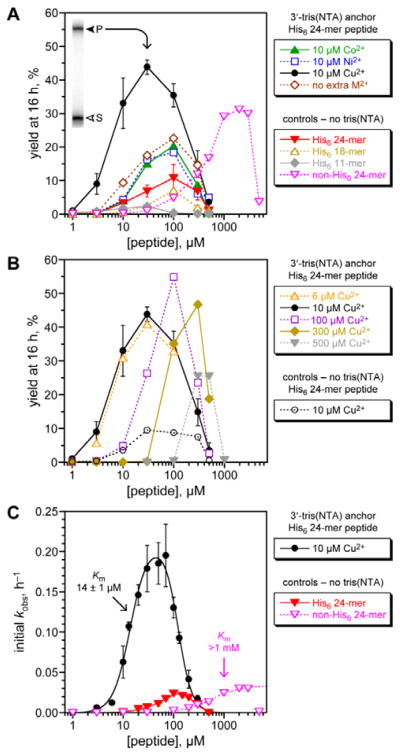
The 8XJ105 deoxyribozyme was first evaluated for its catalytic activity in control experiments with His6-tagged peptide substrates in the absence of His6/M2+ recruiting (Fig. 3A, controls). These experiments are important to establish the conditions and substrates with which the recruiting effect will then be evaluated. 8XJ105 requires 1 mM Zn2+ as a cofactor (as well as 40 mM Mg2+ and 20 mM Mn2+). Therefore, the controls used 1 mM Zn2+ and a DNA anchor oligonucleotide that lacks the tris(NTA) modification. 8XJ105 showed very little activity with the 11-mer peptide H6AAYAA (2% yield in 16 h at 10 or 30 μM peptide and <0.5% yield at 100 μM peptide). Zn2+ leads to peptide aggregation upon interaction with the His6 tag, which constitutes more than half of the peptide. Such aggregation was visible by eye as a white precipitate at high peptide concentration (1 mM). 8XJ105 was more active with two longer 18-mer and 24-mer His6-tagged peptides, each containing a single tyrosine residue. The 24-mer led to 11% yield in 16 h at 100 μM peptide, although reduced activity was still observed at even higher peptide concentrations (>100 μM). The 11% yield at 100 μM can be compared with 5% yield observed with a non-His6-tagged 24-mer peptide at the same concentration.7 The 24-mer peptide was therefore used as the substrate to assess the particular recruiting strategy of Fig. 2B, which is expected to reduce the required peptide concentration and increase the reaction yield of peptide-RNA conjugation.
Fig. 3.
Assessing histidine tag recruiting for the 8XJ105 deoxyribozyme. See the Experimental section for explanation of error bars in all panels. Conditions: 20 nM 3′-32P-radiolabeled 5′-pppRNA, 0.5 μM 8XJ105 deoxyribozyme, 2 μM 3′-tris(NTA) DNA anchor oligonucleotide, 1–5000 μM peptide, 70 mM HEPES, pH 7.5, 40 mM MgCl2, 20 mM MnCl2, 1 mM ZnCl2, 150 mM NaCl, and CoCl2, NiCl2, or Cu(NO3)2 as indicated at room temperature. CuCl2 gave equivalent outcome. (A) Yield at 16 h of peptide-RNA conjugate as a function of concentration of His6-tagged 24-mer peptide (or untagged 24-mer in one case). All experiments including the controls were performed with 1 mM Zn2+; 10 μM Co2+, Ni2+, or Cu2+ was additionally present where indicated. Inset: PAGE data at 16 h for 3′-32P-radiolabeled RNA and 30 μM His6-tagged 24-mer peptide with Cu2+ (S = substrate, P = product). See Fig. S1 (ESI†) for data with 5′-tris(NTA) moiety. 24-mer H6SAGERASAEDMARAAYAA (for non-His6, replace H6 with ASAASA); 18-mer H6SAEDMARAAYAA; 11-mer H6AAYAA. (B) Optimization of Cu2+ concentration, and demonstration that Cu2+ alone [without tris(NTA)] is not responsible for the recruiting effect. (C) Determination of peptide Km values using initial-rate kinetics. See curve fit descriptions and fit details in the Experimental section.
The third-row metal ions Co2+, Ni2+, Cu2+, and Zn2+ interact efficiently with NTA and have been used in Immobilized-Metal Affinity Chromatography (IMAC) for applications such as protein purification.42–45 We tested each of these metal ions with 8XJ105 according to the recruiting strategy of Fig. 2B. The 3′-tris(NTA)-modified DNA anchor oligonucleotide was incubated with 8XJ105, pppRNA, the His6-tagged 24-mer peptide substrate, and 10 μM of one of Co2+, Ni2+, or Cu2+, noting again that 1 mM Zn2+ is present in all experiments [Fig. 3A, 3′-tris(NTA) anchor data]. Inclusion of the tris(NTA) moiety but no extra M2+ led to 2.1-fold increase in yield (from 11% to 23%) relative to the non-tris(NTA) control at 100 μM peptide. No additional benefit arose by including 10 μM Co2+ or Ni2+. In contrast, including 10 μM Cu2+ led to 6.4-fold increase in yield (from 6.9% to 44%) relative to control at 30 μM peptide. A yield of 31% was observed for a non-His6-tagged 24-mer peptide, albeit only at 30-fold higher peptide concentration (>1 mM; yields decreased substantially at high peptide concentration, again attributed to aggregation). Therefore, a histidine tag successfully recruits the 8XJ105 deoxyribozyme to modify its peptide substrate. Among the evaluated divalent metal ions, Cu2+ was most effective, consistent with its tightest binding in metal complexes.46 The alternative 5′-tris(NTA) on the anchor oligonucleotide led to 2.8-fold yield increase relative to non-tris(NTA) control at 100 μM peptide, but none of Co2+, Ni2+, or Cu2+ provided any additional yield increase (Fig. S1, ESI†), indicating a smaller recruiting benefit from this 5′-tris(NTA) alternative.
The Cu2+ concentration was optimal at 10 μM when the 3′-tris(NTA) anchor oligonucleotide was at 2 μM, such that the metal-binding capacity is 6 μM (Fig. 3B). As an important control experiment, without the 3′-tris(NTA) moiety, 10 μM Cu2+ supported catalysis with no greater efficiency than in the absence of Cu2+.
The substantial recruiting effect of His6/Cu2+ was confirmed by evaluating the rate of DNA-catalyzed peptide modification (Fig. 3C). From initial-rate kinetics, kobs values for the His6-tagged 24-mer peptide substrate were obtained both with and without 3′-tris(NTA) on the anchor oligonucleotide. Although generic inhibition of activity at >100 μM peptide was observed, the data again reveal a strong recruiting effect, with peptide Km of 14 μM. Comparing the data for the His6-tagged 24-mer and untagged non-His6 24-mer peptides indicates >70-fold decrease in Km value due to recruiting.
Evaluating the His6-tag recruiting strategy with the 6CF134 tyrosine kinase deoxyribozyme
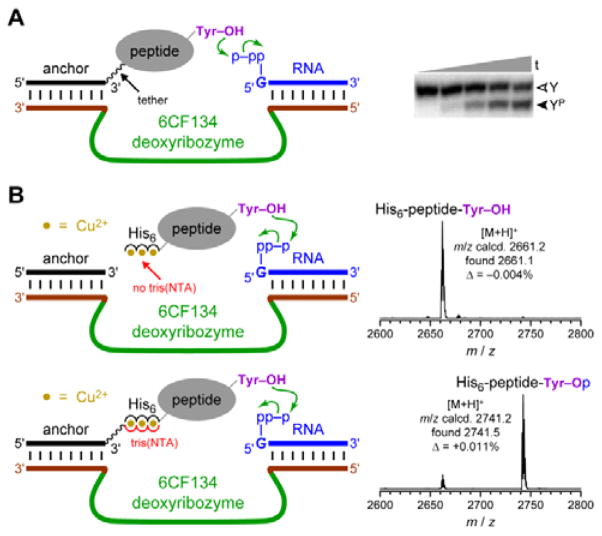
In separate experiments, we investigated the Fig. 1B recruiting strategy for DNA-catalyzed tyrosine side chain phosphorylation (kinase activity). Tyrosine phosphorylation is one of many important, natural protein post-translational modification reactions,47–52 and synthetic tyrosine kinase enzymes will have substantial utility. We previously reported the 6CF134 tyrosine kinase deoxyribozyme, which phosphorylates a short hexapeptide substrate when that substrate is covalently tethered to a DNA anchor oligonucleotide (Fig. 4A).53 However, 6CF134 does not catalyze detectable phosphorylation of a discrete, untethered peptide at any peptide concentration tested. Here we sought to recruit 6CF134 to an untethered 24-mer peptide substrate via a His6 tag. In the presence of 100 μM Cu2+, 88% conversion of phosphorylated peptide was observed by MALDI mass spectrometry when the 3′-tris(NTA) was included (Fig. 4B), compared to 50% yield for 6CF134 with a tethered peptide under similar conditions (Fig. 4A). Little or no phosphorylation was observed in negative control assays that omitted one of 3′-tris(NTA) (0.9% conversion; Fig. 4B), Cu2+ (0.3%; data not shown), or the pppRNA phosphoryl donor (<0.05%; data not shown), or when the non-His6 24-mer peptide was used (0.09%; data not shown). From these observations, we conclude that 6CF134 is inherently capable of modifying an untethered peptide, despite its strict tether requirement found in our original report.53 The His6 tag and tris(NTA) interaction functionally replaces the covalent tether and enables 6CF134 catalysis with the formally untethered peptide substrate.
Fig. 4.
The 6CF134 tyrosine kinase deoxyribozyme and its His6 recruiting to a peptide substrate. (A) Originally reported arrangement of 6CF134 and disulfide-tethered peptide substrate. No phosphorylation is observed without the tether;53 ~50% phosphorylation yield is observed with the tether. Conditions: 20 nM 5′-32P-radiolabeled DNA-anchored CAAYAA peptide, 0.5 μM 6CF134 deoxyribozyme, 5 μM 5′-pppRNA, 70 mM HEPES, pH 7.5, 40 mM MgCl2, 20 mM MnCl2, 0.5 mM ZnCl2, and 150 mM NaCl at room temperature (t = 30 s, 30 min, 2 h, 5 h, and 16 h; Y = substrate, YP = product). (B) MALDI mass spectrometry reveals successful recruiting of 6CF134 to phosphorylate the 24-mer His6-tagged peptide substrate (same substrate as in Fig. 3). Conditions: 30 μM peptide, 30 μM DNA anchor oligonucleotide with or without 3′ tris(NTA), 35 μM 6CF134 deoxyribozyme, 40 μM 5′-pppRNA, 70 mM HEPES, pH 7.5, 40 mM MgCl2, 20 mM MnCl2, 2 mM ZnCl2, 150 mM NaCl, and 100 μM Cu(NO3)2 at room temperature for 20 h.
Evaluating the His6-tag recruiting strategy with protein substrates
Rosen et al. used both His6-tagged and natively metal-binding proteins in their work.8 We evaluated the recruiting strategy of Fig. 2B for 8XJ105, using two His6-tagged proteins (lysozyme and plasminogen activator inhibitor 1 [PAI-1]) and one natively metal-binding protein (carboxypeptidase B). For all three proteins and using 3′-32P-radiolabeled pppRNA, no modification of any tyrosine residues by 8XJ105 was observed by PAGE-shift assay (Fig. S2, ESI†). Separately, we also sought to use 6CF134 with lysozyme and PAI-1, albeit unsuccessfully, as assayed by trypsin digestion and MALDI mass spectrometry (Fig. S3, ESI†).
Discussion
In this study, we performed experiments to evaluate the histidine tag recruiting strategy of Fig. 1B for enhancing DNA-catalyzed peptide and protein modification. Substantially higher peptide modification yields and kobs values were observed, along with substantial effects on peptide Km values. The His6/Cu2+ recruiting effect allowed, for the first time, successful DNA-catalyzed tyrosine phosphorylation of a discrete, untethered peptide substrate. However, where tested, DNA-catalyzed protein modification using the recruiting strategy was not observed. Unlike nonenzymatic protein modification reactions such as those studied by Rosen et al.,8 where simple proximity is responsible for the enhanced reactivity (Fig. 1A), achieving DNA-catalyzed reactions of proteins faces additional challenges, even with the benefit of histidine tag recruiting. For example, access by a deoxyribozyme to residues on a structured protein’s surface may be restricted, and the recruiting effect may not overcome this problem. In addition, a deoxyribozyme may not function well (or at all) with peptide segments on a protein’s surface, if those segments are unable to adopt a conformation suitable to allow their DNA-catalyzed modification.
For deoxyribozymes that do function well with protein substrates, the recruiting strategy should allow site-selective modification of particular surface-exposed residues, based on the geometric relationship among the tris(NTA) recruiting site, the recruited deoxyribozyme, and the side chains themselves (see Fig. 1B). This directed reactivity strategy for a sequence-general deoxyribozyme differs from our recently reported experiments in which inherently sequence-selective deoxyribozymes were identified using particular peptide sequences during the selection process, such that the resulting deoxyribozymes function only with those peptide sequences.7,54 We are currently seeking protein-modifying deoxyribozymes using various approaches, and with such DNA catalysts we intend to evaluate the histidine tag recruiting strategy to achieve site selectivity. The experiments reported here provide a encouraging framework for these future studies.
Experimental
Oligonucleotides and oligonucleotide-tris(NTA) conjugates
General considerations
DNA oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA) or prepared by solid-phase synthesis on an ABI 394 instrument using reagents from Glen Research. 5′-Triphosphorylated RNA (pppRNA) oligonucleotides were prepared by in vitro transcription using synthetic DNA templates and T7 RNA polymerase.55 All oligonucleotides were purified by 7 M urea denaturing PAGE with running buffer 1× TBE (89 mM each Tris and boric acid and 2 mM EDTA, pH 8.3) as described previously.56,57 Sequences are collected in Table S1 (ESI†).
Procedure for synthesis of tris(NTA)-modified DNA anchor
DNA oligonucleotides with three primary amino groups at the 3′-end or 5′-end were synthesized using the Uni-Link Amino Modifier (Clontech). The procedure for incorporating tris(NTA) was adapted from Goodman et al.41 A 50 μL sample containing 10 nmol of tris(amino)-modified DNA oligonucleotide in 100 mM sodium phosphate buffer, pH 7.5, 100 mM NaCl, and 25 mM SPDP [N-succinimidyl 3-(2-pyridylthio)propionate, Fisher cat. no. PI21857] was incubated at room temperature for 1 h. The SPDP was added from a freshly prepared 100 mM stock in DMF. A desalting column (Micro Bio-Spin P-6, Bio-Rad) was washed with 100 mM sodium phosphate buffer, pH 7.5, and used to remove excess SPDP from the sample. The eluted sample was treated with 6.3 μL of 100 mM TCEP [tris(carboxyethyl)phosphine hydrochloride, Chem-Impex] and incubated at room temperature for 30 min. Excess TCEP was removed with another desalting column. The eluted sample was treated with 7 μL of 100 mM maleimido-C3-NTA (Dojindo cat. no. M035-10), 1 μL of 1 M sodium phosphate buffer, pH 7.5, and 2.5 μL of 3 M NaCl, and the sample was incubated at room temperature for 1 h. The sample was purified by 20% PAGE. The product was extracted from the gel in 10 mM Tris, pH 8.0 and 300 mM NaCl (no EDTA) and precipitated with ethanol. Typical yield after gel extraction and precipitation was 20–30%. The product was analyzed by mass spectrometry on a Bruker UltrafleXtreme MALDI-TOF mass spectrometer with matrix 3-hydroxypicolinic acid in positive ion mode at the UIUC School of Chemical Sciences Mass Spectrometry Laboratory. For the 3′-tris(NTA) DNA anchor oligonucleotide used in peptide-RNA conjugation, [M+H]+ calcd. 7341.6, found 7340.6 (Δ = −0.014%). For the 5′-tris(NTA) DNA anchor oligonucleotide used in peptide-RNA conjugation, [M+H]+ calcd. 11560.3, found 11564.6 (Δ = +0.037%). For the 3′-tris(NTA) DNA anchor oligonucleotide used in peptide tyrosine phosphorylation, [M+H]+ calcd. 7343.7, found 7343.3 (Δ = −0.005%). Images of the mass spectra are shown in Fig. S4 (ESI†).
Synthesis of peptides
Peptides were prepared by solid-phase synthesis using Fmoc Rink amide MBHA resin on a Liberty Automated Microwave Peptide Synthesizer (CEM Corporation) using the instrument’s standard wash procedures. Each synthesis was performed at 0.1 mmol scale, initiated using 130 mg of Rink amide resin with a loading capacity of 0.77 mmol/g (Chem-Impex). For each Fmoc deprotection step, 20% piperidine in DMF with 0.1 M HOBt was used at 30 W of microwave power and 75 °C for 3 min. Each coupling used 5 equivalents (2.5 mL of 0.2 M in DMF, 0.5 mmol) of Fmoc-amino acid, 5 equivalents (1 mL of 0.5 M in DMF, 0.5 mmol) of HCTU (N,N,N′,N′-tetramethyl-O-(1H-6-chlorobenzotriazol-1-yl)uronium hexafluorophosphate), and 10 equivalents (0.5 mL of 2 M in N-methylpyrrolidone, 1 mmol) of DIPEA (N,N-diisopropylethylamine). All amino acid monomers except Arg and His were coupled twice at 20 W and 75 °C for 5 min. For Arg, to reduce δ-lactam formation, the first coupling was performed without microwaver power at room temperature for 25 min, followed by 25 W at 75 °C for 5 min; the second coupling was performed at 20 W and 75 °C for 5 min. For His, to reduce racemization, both couplings were performed without microwave power at room temperature for 2 min followed by 25 W at 50 °C for 4 min. After each coupling step as well as the final Fmoc deprotection step, capping was performed using 7 mL of 0.5 M acetic anhydride and 0.125 M DIPEA in DMF at 40 W and 65 °C for 2 min. The peptide was cleaved from the solid support by stirring the resin in a separate vial with a solution containing 5 mL of trifluoroacetic acid (TFA), 125 μL of water, and 50 μL of triisopropylsilane for 90 min. The liquid solution was separated from the resin by filtration. This solution was dried on a rotary evaporator, providing an oily solid. To this material was added 20 mL of cold diethyl ether, and the peptide was obtained as a white solid that was filtered and purified by HPLC. Each peptide had an N-terminal acetyl group and C-terminal amide group.
| His6 11-mer: | HHHHHHAAYAA |
| His6 18-mer: | HHHHHHSAEDMARAAYAA |
| His6 24-mer: | HHHHHHSAGERASAEDMARAAYAA |
| non-His6 24-mer: | ASAASASAGERASAEDMARAAYAA |
Single-turnover deoxyribozyme assay and mass spectrometry procedures
General considerations
Metal ions MgCl2, MnCl2, CoCl2, NiCl2, and Cu(NO3)2 were added from 10× stock solutions, which were diluted from 1 M (or 500 mM for NiCl2) solutions. ZnCl2 was added from a 10× stock solution containing 10 mM ZnCl2, 20 mM HNO3, and 200 mM HEPES at pH 7.5; this stock was freshly prepared from a solution of 100 mM ZnCl2 in 200 mM HNO3. 3′-32P-radiolabeling of 5′-pppRNA was achieved starting from the unlabeled 5′-pppRNA using 5′-32P-pCp and T4 RNA ligase.58
Peptide-RNA conjugation by the 8XJ105 deoxyribozyme
A 5 μL sample containing 20 pmol of DNA anchor oligonucleotide [with or without 3′-tris(NTA) as appropriate], one of 100 pmol of Co2+, 100 pmol of Ni2+, or 60–5000 pmol of Cu2+ if applicable, and peptide substrate was incubated in 50 mM HEPES, pH 7.5, and 150 mM NaCl at room temperature for 1 h (this preincubation step was later determined to be dispensable). The reaction was initiated by bringing the sample to 10 μL total volume containing 20 nM 3′-32P-radiolabeled 5′-pppRNA, 0.5 μM 8XJ105 deoxyribozyme, 2 μM DNA anchor oligonucleotide, 1–5000 μM peptide, 70 mM HEPES, pH 7.5, 40 mM MgCl2, 20 mM MnCl2, 1 mM ZnCl2, and 150 mM NaCl. For Co2+ or Ni2+ as the recruiting metal ion, the final concentration was 10 μM; for Cu2+, 6–500 μM. The sample was incubated at room temperature, and 2 μL aliquots were quenched at appropriate times with 5 μL of stop solution (80% formamide, 1× TBE [89 mM each Tris and boric acid, 2 mM EDTA, pH 8.3], 50 mM EDTA, and 0.025% each bromophenol blue and xylene cyanol). To each aliquot before PAGE was added 10 pmol of a decoy oligonucleotide (complementary to the deoxyribozyme’s 40 nt catalytic region along with 10 nt of binding arms on either side), to displace the deoxyribozyme from the substrate and product. Samples were separated by 20% PAGE and quantified with a PhosphorImager.
Peptide tyrosine phosphorylation by the 6CF134 deoxyribozyme
The assay of Fig. 4A was performed as described.53 The assays of Fig. 4B were performed with much higher concentrations of DNA anchor oligonucleotide, unradiolabeled 5′-pppRNA, and recruiting metal ion, to enable peptide product characterization by mass spectrometry. A 5 μL sample containing 300 pmol of DNA anchor oligonucleotide [with or without 3′-tris(NTA) as appropriate], 1 nmol of Cu(NO3)2, and 300 pmol of His6-tagged 24-mer peptide substrate was incubated in 50 mM HEPES, pH 7.5, and 150 mM NaCl at room temperature for 1 h (this preincubation step was later determined to be dispensable). The reaction was initiated by bringing the sample to 10 μL total volume containing 30 μM His6-tagged 24-mer peptide, 30 μM DNA anchor oligonucleotide, 35 μM 6CF134 deoxyribozyme, 40 μM unradiolabeled 5′-pppRNA, 70 mM HEPES, pH 7.5, 40 mM MgCl2, 20 mM MnCl2, 2 mM ZnCl2, 150 mM NaCl, and 100 μM Cu(NO3)2. The sample was incubated at room temperature for 20 h, immediately desalted by Millipore C18 ZipTip, and analyzed by MALDI mass spectrometry. Data were acquired on a Bruker UltrafleXtreme MALDI-TOF mass spectrometer with matrix 2,5-dihydroxybenzoic acid in positive ion mode.
Analysis of deoxyribozyme assay data
Data for the control experiment with the His6 24-mer (Fig. 3A and Fig. S1, ESI†) are shown as mean ± SD (n = 4). Data for the experiment with the 3′-tris(NTA) DNA anchor oligonucleotide with 10 μM Cu2+ (Fig. 3A and Fig. 3B) are shown as mean ± SD (n = 3–5). Data for the initial-rate kinetics with 10 μM Cu2+ and the control experiment with the His6 24-mer in Fig. 3C are shown as mean ± SD (n = 3); in the latter case, the error bars are smaller than the data points. All other data are n = 1.
Initial-rate kinetics data for Fig. 3C were obtained by fitting the initial linear portion of plots (yield versus time up to 2 h) to a straight line. The peptide Km values were determined by fitting the data to kobs = kmax•[Cn/(Kmn+Cn)] • [1−Cm/(Kim+Cm)], where C is the peptide concentration, kmax is the kobs at saturating peptide concentration, Km is the Km value for productive peptide binding, Ki is the inhibition constant for unproductive peptide binding, and n and m are Hill coefficients for productive and unproductive peptide binding, respectively. The Km value for the non-His6 24-mer was determined by fitting the data to kobs = kmax• [Cn/(Kmn+Cn)]. Curve fit values were as follows. For the 3′-tris(NTA) anchor with Cu2+, Km = 14 ± 1 μM, Ki = 124 ± 8 μM, n = 2.6 ± 0.4 and m = 3.2 ± 0.5. For the control experiment with the His6 24-mer, Km = 82 ± 75 μM, Ki = 218 ± 68 μM, n = 1.6 ± 0.6 and m = 3.3 ± 1.0 (these values are not necessarily interpretable due to the large errors; Km is likely to be ≫80 μM). For the control using non-His6 24-mer, Km = 579 ± 57 μM and n = 1.5 ± 0.2 (the 5 mM data point was omitted from the fit; Km is likely to be >1 mM).
Assays of 8XJ105 with protein substrates
Following the approach of Rosen et al.,6 we assayed the 8XJ105 deoxyribozyme with each of three different proteins as potential substrates. His6-lysozyme was from US Biological (cat. no. 155733, 15.2 kDa). His6-plasminogen activator inhibitor 1 (PAI-1) was from US Biological (cat. no. 170035, 45 kDa). Carboxypeptidase B, which natively binds metal ions, was from Worthington Biochemical Corporation (cat. no. LS005305, 34.5 kDa). A 5 μL sample containing 20 pmol of 3′- or 5′-tris(NTA) DNA anchor oligonucleotide, 100–3000 pmol of Cu(NO3)2, and 100–1000 pmol of protein substrate was incubated in 50 mM HEPES, pH 7.5, 150 mM NaCl, and 0.04% (v/v) Tween-20 at room temperature for 1 h. The reaction was initiated by bringing the sample to 10 μL total volume containing 20 nM 3′-32P-radiolabeled 5′-pppRNA, 0.5 μM 8XJ105 deoxyribozyme, 2 μM 3′- or 5′-tris(NTA) DNA anchor oligonucleotide, 10–100 μM protein, 70 mM HEPES, pH 7.5, 1 or 40 mM MgCl2, 5 or 20 mM MnCl2, 0.5 or 1 mM ZnCl2, 150 mM NaCl, 10–300 μM Cu(NO3)2, and 0.02% (v/v) Tween-20. The sample was incubated at room temperature for 16 h. In all three cases, 16% SDS-PAGE revealed no detectable product formation (<0.5%; Fig. S2, ESI†).
Assays of 6CF134 with protein substrates
We assayed the 6CF134 deoxyribozyme with each of lysozyme and PAI-1 as potential substrates. A 5 μL sample containing 150 or 300 pmol of 3′-tris(NTA) DNA anchor oligonucleotide, 500 or 1000 pmol of Cu(NO3)2, and 100 or 300 pmol of protein substrate was incubated in 50 mM HEPES, pH 7.5 and 150 mM NaCl at room temperature for 1 h. The reaction was initiated by bringing the sample to 10 μL total volume containing 10 or 30 μM protein, 15 or 30 μM 3′-tris(NTA) DNA anchor oligonucleotide, 20 or 35 μM 6CF134 deoxyribozyme, 25 or 40 μM unradiolabeled 5′-pppRNA, 70 mM HEPES, pH 7.5, 40 mM MgCl2, 0 or 20 mM MnCl2, 0.5 or 2 mM ZnCl2, 150 mM NaCl, and 50 or 100 μM Cu(NO3)2. The sample was incubated at room temperature for 20 or 48 h. Trypsin digestion was performed to prepare the sample for mass spectrometry. The sample was reduced and denatured by adding 10 μL of 6 M urea in 100 mM Tris-HCl, pH 8.0, and 1 μL of 250 mM DTT and incubating the sample at 37 °C for 1 h. To block cysteine residues, 1 μL of 350 mM iodoacetamide was added, and the sample was incubated at room temperature in the dark for 30 min. The trypsin digestion reaction was initiated by adding 60 μL of 50 mM Tris-HCl, pH 8.0 and 1 μL of 100 ng/μL sequencing-grade trypsin (Promega, cat. no. V511). The sample was incubated at 37 °C for 16 h, immediately desalted by Millipore C18 ZipTip, and analyzed by MALDI mass spectrometry. Data were acquired on a Bruker UltrafleXtreme MALDI-TOF mass spectrometer with matrix 2,5-dihydroxybenzoic acid in positive ion mode. Peptide fragments were calculated using ExPASy. No phosphorylation products peaks were observed for all identifiable Tyr-containing peptide fragments (Fig. S3, ESI†).
Supplementary Material
Acknowledgments
This work was supported by a grant to S.K.S. from the National Institutes of Health (R01GM065966). Mass spectrometry was performed by Kevin Tucker at the UIUC School of Chemical Sciences Mass Spectrometry Laboratory on an instrument purchased with support from NIH grant S10RR027109A. We thank Wilfred van der Donk for access to the microwave peptide synthesizer and Subha Mukherjee for guidance in its use.
Footnotes
Electronic Supplementary Information (ESI) available. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Breaker RR, Joyce GF. Chem Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 2.Schlosser K, Li Y. Chem Biol. 2009;16:311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Silverman SK. Acc Chem Res. 2009;42:1521–1531. doi: 10.1021/ar900052y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silverman SK. Angew Chem Int Ed. 2010;49:7180–7201. doi: 10.1002/anie.200906345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollenstein M. Molecules. 2015;20:20777–20804. doi: 10.3390/molecules201119730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverman SK. Acc Chem Res. 2015;48:1369–1379. doi: 10.1021/acs.accounts.5b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu C, Wong O, Silverman SK. ChemBioChem. 2014;15:1905–1910. doi: 10.1002/cbic.201402255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosen CB, Kodal AL, Nielsen JS, Schaffert DH, Scavenius C, Okholm AH, Voigt NV, Enghild JJ, Kjems J, Tørring T, Gothelf KV. Nat Chem. 2014;6:804–809. doi: 10.1038/nchem.2003. [DOI] [PubMed] [Google Scholar]
- 9.Kalia J, Raines RT. Curr Org Chem. 2010;14:138–147. doi: 10.2174/138527210790069839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephanopoulos N, Francis MB. Nat Chem Biol. 2011;7:876–884. doi: 10.1038/nchembio.720. [DOI] [PubMed] [Google Scholar]
- 11.Davis L, Chin JW. Nat Rev Mol Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 12.Rashidian M, Dozier JK, Distefano MD. Bioconjug Chem. 2013;24:1277–1294. doi: 10.1021/bc400102w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debets MF, van Hest JCM, Rutjes FPJT. Org Biomol Chem. 2013;11:6439–6455. doi: 10.1039/c3ob41329b. [DOI] [PubMed] [Google Scholar]
- 14.Takaoka Y, Ojida A, Hamachi I. Angew Chem Int Ed. 2013;52:4088–4106. doi: 10.1002/anie.201207089. [DOI] [PubMed] [Google Scholar]
- 15.Spicer CD, Davis BG. Nat Commun. 2014;5:4740. doi: 10.1038/ncomms5740. [DOI] [PubMed] [Google Scholar]
- 16.McKay CS, Finn MG. Chem Biol. 2014;21:1075–1101. doi: 10.1016/j.chembiol.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinogradova EV, Zhang C, Spokoyny AM, Pentelute BL, Buchwald SL. Nature. 2015;526:687–691. doi: 10.1038/nature15739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boutureira O, Bernardes GJL. Chem Rev. 2015;115:2174–2195. doi: 10.1021/cr500399p. [DOI] [PubMed] [Google Scholar]
- 19.ElSohly AM, Francis MB. Acc Chem Res. 2015;48:1971–1978. doi: 10.1021/acs.accounts.5b00139. [DOI] [PubMed] [Google Scholar]
- 20.Krall N, da Cruz FP, Boutureira O, Bernardes GJL. Nat Chem. 2016;8:103–113. doi: 10.1038/nchem.2393. [DOI] [PubMed] [Google Scholar]
- 21.Chudasama V, Maruani A, Caddick S. Nat Chem. 2016;8:114–119. doi: 10.1038/nchem.2415. [DOI] [PubMed] [Google Scholar]
- 22.Zhang C, Welborn M, Zhu T, Yang NJ, Santos MS, Van Voorhis T, Pentelute BL. Nat Chem. 2016;8:120–128. doi: 10.1038/nchem.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Struck AW, Bennett MR, Shepherd SA, Law BJC, Zhuo Y, Wong LS, Micklefield J. J Am Chem Soc. 2016;138:3038–3045. doi: 10.1021/jacs.5b10928. [DOI] [PubMed] [Google Scholar]
- 24.Grindley NDF, Whiteson KL, Rice PA. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 25.Ambros V, Baltimore D. J Biol Chem. 1978;253:5263–5266. [PubMed] [Google Scholar]
- 26.Rothberg PG, Harris TJ, Nomoto A, Wimmer E. Proc Natl Acad Sci USA. 1978;75:4868–4872. doi: 10.1073/pnas.75.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermoso JM, Salas M. Proc Natl Acad Sci USA. 1980;77:6425–6428. doi: 10.1073/pnas.77.11.6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bamford DH, Mindich L. J Virol. 1984;50:309–315. doi: 10.1128/jvi.50.2.309-315.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rozovics JM, Virgen-Slane R, Semler BL. PLoS ONE. 2011;6:e16559. doi: 10.1371/journal.pone.0016559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tse YC, Kirkegaard K, Wang JC. J Biol Chem. 1980;255:5560–5565. [PubMed] [Google Scholar]
- 31.Wang JC. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- 32.Samad A, Carroll RB. Mol Cell Biol. 1991;11:1598–1606. doi: 10.1128/mcb.11.3.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corey DR, Schultz PG. Science. 1987;238:1401–1403. doi: 10.1126/science.3685986. [DOI] [PubMed] [Google Scholar]
- 34.Brenner S, Lerner RA. Proc Natl Acad Sci USA. 1992;89:5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keefe AD, Szostak JW. Nature. 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baskerville S, Bartel DP. Proc Natl Acad Sci USA. 2002;99:9154–9159. doi: 10.1073/pnas.142153799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vivès E, Schmidt J, Pèlegrin A. Biochim Biophys Acta. 2008;1786:126–138. doi: 10.1016/j.bbcan.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Said Hassane F, Saleh AF, Abes R, Gait MJ, Lebleu B. Cell Mol Life Sci. 2010;67:715–726. doi: 10.1007/s00018-009-0186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Juliano RL, Ming X, Nakagawa O. Acc Chem Res. 2012;45:1067–1076. doi: 10.1021/ar2002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Josephson K, Ricardo A, Szostak JW. Drug Discov Today. 2014;19:388–399. doi: 10.1016/j.drudis.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 41.Goodman RP, Erben CM, Malo J, Ho WM, McKee ML, Kapanidis AN, Turberfield AJ. ChemBioChem. 2009;10:1551–1557. doi: 10.1002/cbic.200900165. [DOI] [PubMed] [Google Scholar]
- 42.Porath J, Carlsson J, Olsson I, Belfrage G. Nature. 1975;258:598–599. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- 43.Porath J. Protein Expr Purif. 1992;3:263–281. doi: 10.1016/1046-5928(92)90001-d. [DOI] [PubMed] [Google Scholar]
- 44.Block H, Maertens B, Spriestersbach A, Brinker N, Kubicek J, Fabis R, Labahn J, Schäfer F. Methods Enzymol. 2009;463:439–473. doi: 10.1016/S0076-6879(09)63027-5. [DOI] [PubMed] [Google Scholar]
- 45.Cheung RCF, Wong JH, Ng TB. Appl Microbiol Biotechnol. 2012;96:1411–1420. doi: 10.1007/s00253-012-4507-0. [DOI] [PubMed] [Google Scholar]
- 46.Irving H, Williams RJP. J Chem Soc. 1953:3192–3210. [Google Scholar]
- 47.Shahbazian MD, Grunstein M. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 48.Tiganis T, Bennett AM. Biochem J. 2007;402:1–15. doi: 10.1042/BJ20061548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi Y. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 50.Tarrant MK, Cole PA. Annu Rev Biochem. 2009;78:797–825. doi: 10.1146/annurev.biochem.78.070907.103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moremen KW, Tiemeyer M, Nairn AV. Nat Rev Mol Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore KE, Gozani O. Biochim Biophys Acta. 2014;1839:1395–1403. doi: 10.1016/j.bbagrm.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walsh SM, Sachdeva A, Silverman SK. J Am Chem Soc. 2013;135:14928–14931. doi: 10.1021/ja407586u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walsh SM, Konecki SN, Silverman SK. J Mol Evol. 2015;81:218–224. doi: 10.1007/s00239-015-9699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flynn-Charlebois A, Wang Y, Prior TK, Rashid I, Hoadley KA, Coppins RL, Wolf AC, Silverman SK. J Am Chem Soc. 2003;125:2444–2454. doi: 10.1021/ja028774y. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Silverman SK. Biochemistry. 2003;42:15252–15263. doi: 10.1021/bi0355847. [DOI] [PubMed] [Google Scholar]
- 58.Wong O, Pradeepkumar PI, Silverman SK. Biochemistry. 2011;50:4741–4749. doi: 10.1021/bi200585n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.