Abstract
The stem cells in the shoot apical meristem (SAM) are the origin of all above ground tissues in plants. In Arabidopsis thaliana, shoot meristem stem cells are maintained by the homeobox transcription factor gene WUS (WUSCHEL) that is expressed in cells of the organizing center underneath the stem cells. In order to identify factors that operate together with WUS in stem cell maintenance, we performed an EMS mutant screen for modifiers of the hypomorphic wus-6 allele. We isolated the oberon3-2 (obe3-2) mutant that enhances stem cell defects in wus-6, but does not affect the putative null allele wus-1. The OBE3 gene encodes a PHD (Plant Homeo Domain) protein that is thought to function in chromatin regulation. Single mutants of OBE3 or its closest homolog OBE4 do not display any defects, whereas the obe3-2 obe4-2 double mutant displays broad growth defects and developmental arrest of seedlings. Transcript levels of WUS and its target gene in the stem cells, CLAVATA3, are reduced in obe3-2. On the other hand, OBE3 and OBE4 transcripts are both indirectly upregulated by ectopic WUS expression. Our results suggest a positive feedback regulation between WUS and OBE3 that contributes to shoot meristem homeostasis.
Introduction
Postembryonic growth and iterative organ formation of higher plants rely on the activity of pluripotent stem cells in organogenic centers, the meristems. The shoot meristem that will give rise to the above ground organs has been extensively studied in the model plant Arabidopsis thaliana. The homeodomain transcription factor WUS is expressed in the organizing center (OC) underneath the stem cells [1] where it directly represses cytokinin response inhibitors [2] and, after moving into the overlying stem cells [3, 4], represses cell differentiation and activates expression of the signal peptide CLV3 [4–6]. CLV3 in turn represses WUS transcription via CLV1/CLV2-CRN receptor-like kinases to delimit the size of the OC [6–8]. This negative feedback loop balances stem cell maintenance and differentiation [7]. The WUS/CLV3 loop also functions to maintain stem cells of the floral meristems [6, 7]. In contrast to the indeterminate shoot meristem, WUS in the determinate floral meristem also activates the gene encoding the MADS domain protein AGAMOUS (AG) that in turn terminates WUS expression and thus floral meristem growth [9–11]. In addition to its function in stem cell regulation, WUS is also required for the development of the female and male gametes [12–14]. However, CLV3 signaling does not appear to be targeted by WUS in these cases.
Although in the recent years, many studies identified further components affecting WUS/CLV3 homeostasis [3, 15–23], how WUS maintains stem cells remains enigmatic.
In order to find hitherto undiscovered factors involved in the WUS-mediated stem cell regulation, we used a sensitized mutant screen for genetic modifiers of the hypomorphic wus-6 allele [21, 24]. Here we report the isolation of the wus enhancer 9 (wen9) mutant that enhances stem cell defects in wus-6. We show by positional cloning that wen9 is an allele of the OBE3 gene, and characterize its function together with its closest homologue OBE4 in the shoot meristem.
Results
wen9 enhances inflorescence shoot meristem defects of wus-6
The putative null allele wus-1 causes premature termination of stem cells in the primary shoot meristem during embryogenesis, resulting in a flat apex of partially differentiated cells at the seedling stage [6]. Consequently, seedlings lack any true leaves at 10 days after germination (Fig 1B). Postembryonically initiated shoot meristems terminate after the formation of a few leaves, resulting in a stop-and-go phenotype (Fig 1C), and the seldom formed floral meristems give rise to 4 sepals, 4 petals, and a single stamen before premature termination (Fig 1D). The intermediate wus-6 allele causes reduced WUS expression levels, and the primary seedling shoot meristem and floral meristem prematurely terminate indistinguishably to wus-1 (Fig 1B and 1D; [21, 24]. In contrast to wus-1, however, postembryonically initiated wus-6 shoot meristems grow indeterminately and give rise to many floral meristems (Fig 1C; Tables 1 and 2). The wus-7 allele carries a missense mutation in the homeodomain and represents the weakest known wus allele [25]. wus-7 seedlings form several rosette leaves before the primary shoot meristem terminates (Fig 1B) and axillary shoot meristems form indeterminate shoots carrying complete flowers (Fig 1C and 1D; Table 2).
Fig 1. OBE3 gene structure and mutant phenotypes.
(A) Structure of the OBE3 gene. The upstream region used for the complementation is shown in green. (B-D) Phenotypes of the denoted genotypes of 10-day-old seedlings (B), shoots (C), and flowers (D). Scale bars: 1 mm (B, D), 2 cm (C).
Table 1. obe3-2 enhances the meristem defects of weak and intermediate wus alleles.
| % of phenotype (of germinated seeds) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| seedling (10 DAG) | shoot (51DAG) | ||||||||||
| genotype of mother plant | n | ng | wt-like | wus-7 like | wus-1 like | retard1 | wt-like | wus-6 like | wus-1 like | disorganized leaves, no stem | arrest2 |
| Ler | 244 | 42 | 99.5 | 0.0 | 0.0 | 0.5 | 99.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| obe3-2 | 51 | 2 | 98.0 | 0.0 | 0.0 | 2.0 | 98.0 | 0.0 | 0.0 | 0.0 | 2.0 |
| wus-1/+ | 202 | 3 | 69.9 | 0.0 | 28.1 | 2.0 | 68.8 | 1.5 | 24.6 | 4.0 | 1.1 |
| obe3-2 wus-1/+ | 205 | 2 | 75.9 | 0.0 | 20.7 | 3.4 | 69.5 | 3.9 | 1.5 | 19.7 | 5.4 |
| wus-6/+ | 245 | 0 | 73.5 | 0.0 | 25.3 | 1.2 | 73.1 | 25.7 | 0.0 | 0.0 | 1.2 |
| obe3-2 wus-6/+ | 212 | 1 | 74.4 | 0.0 | 24.2 | 1.4 | 74.4 | 0.0 | 0.0 | 23.7 | 1.9 |
| wus-7/+ | 237 | 2 | 94.0 | 5.6 | 0.0 | 0.4 | 79.6 | 19.5 | 0.0 | 0.0 | 0.9 |
| obe3-2 wus-7/+ | 228 | 4 | 93.8 | 4.9 | 0.4 | 0.9 | 75.4 | 19,2 | 0.0 | 4.5 | 0.9 |
Seedling phenotype classes: wt-like, shoot meristem forming a rosette of leaves; wus-7-like; shoot meristem termination after true leaves have been formed, wus-1-like: shoot meristem termination without any leaves.
Shoot phenotype classes: wt-like, rosette, indeterminate inflorescence; wus-6-like, disorganized leaves, indeterminate inflorescence; wus-1-like, disorganized leaves, stop-and-go inflorescence rarely forming flowers.
DAG, days after germination; n, number of plants analyzed; ng, not germinated;
1, retard: small whitish seedlings with retarded growth and cotyledons only.
2, arrest: plants stopped development at seedling stage.
Chi-square test results for the seedling phenotype difference between:
wus-1/+ vs obe3-2 wus-1/+, p>0.05, not significant
wus-6/+ vs obe3-2 wus-6/+, p>0.05, not significant
wus-7/+ vs obe3-2 wus-7/+, p>0.05, not significant
Chi-square test results for the shoot phenotype difference between:
wus-1/+ vs obe3-2 wus-1/+, p<0.0001, highly significant
wus-6/+ vs obe3-2 wus-6/+, p<0.0001, highly significant
wus-7/+ vs obe3-2 wus-7/+, 0.01<p<0.05, significant
Table 2. Flower phenotypes of obe3-2 wus-1, obe3-2 wus-7 and obe3-2 wus-6.
| Genotype | n flowers | stamens | carpels |
|---|---|---|---|
| wild type (Ler) | 20 | 6.0±0.0 | 2.0±0.0 |
| obe3-2 | 20 | 6.0±0.0 | 1.8±0.6 |
| wus-1 | 8 | 1.0±0.0 | 0.0±0.0 |
| obe3-2 wus-1 | 3 | 1.0±0.0 | 0–0±0.0 |
| wus-6 | 20 | 1.0±0.0 | 0.0±0.0 |
| obe3-2 wus-6 | 3 | 1.0±0.0 | 0.0±0.0 |
| wus-7 | 15 | 5.9±0.6 | 1.6±0.8 |
| obe3-2 wus-7 | 20 | 4.0±0.0 | 0.2±0.6 |
At 79 DAG (except the obe3-2 wus-6 at 100DAG), opened flowers were taken from the genotyped plants and the organ numbers were counted. Organ numbers in first and second whorls were 4 sepals and 4 petals, respectively, for all genotypes.
In order to identify factors that cooperate with WUS in stem cell maintenance, we searched for EMS mutants that modify the stem cell defects of the intermediate allele wus-6. One of the isolated enhancers, wus enhancer 9 (wen9), was mapped to a 97 kb region between position 5001124 and 5098789 on chromosome 1, and a nonsense mutation was identified in the predicted first exon of locus AT1G14740. The encoded protein OBERON3 (OBE3) [26], also named TITANIA1 (TTA1) [27], is 733 amino acids in size and contains a potential PHD (plant homeo domain) DNA binding domain (Fig 1A). The wen9 mutation introduces a stop codon after the PHD domain at amino acid position 518. OBE3 transcript levels are reduced in wen9 to about half of the wild-type level (S1 Fig). Transformation with a 4.6 kb genomic OBE3 fragment (Fig 1A) suppressed the enhanced phenotype of wen9 wus-6 plants (S1 Table; S4 Fig) and two independent T-DNA insertion mutants in the OBE3 locus enhance wus-6 similar to wen9, albeit to a weaker extent (S2 Table; S5 Fig). We thus conclude that the mutation in OBE3 caused the enhanced phenotype and assigned wen9 as obe3-2.
To investigate the genetic interaction between WUS and OBE3, we analyzed double mutants between different wus alleles and the obe3-2 mutant. Development of the homozygous obe3-2 single mutant is indistinguishable from wild type (Figs 1B–1D and 2A). The obe3-2 mutation does not affect the seedling phenotypes of wus-1, -6, or -7 (Fig 1B; Table 1). However, it strongly reduces the postembryonic formation of inflorescence stems in all combinations (Fig 1C; Table 1) and causes the formation of leaves in a disorganized (= wuschel-like) pattern. In cases where flowers are made, obe3-2 does not further enhance the already early termination of wus-1 and wus-6 floral meristems, but causes premature termination of wus-7 floral meristems (Fig 1D; Table 2).
Fig 2. Genetic combinations of obe3-2 and obe4-2.
(A) Plants of obe3-2 and obe4-2 single mutants are indistinguishable from wild type. (B) obe3-2 obe4-2 seedlings that did not develop further. (C) obe3-2 obe4-2/+ plants displayed two phenotypic classes at 60 DAG. (D) obe4-2 wus-6 plant displayed wus-6-like shoot formation at 60 DAG. (E) obe4-2 wus-6 plant produced complete flowers and wus-1 like flowers. Scale bars: 2 cm (A, C and D), 1 mm (B and E).
In summary, residual WUS activity in hypomorphic wus alleles requires OBE3 for maintenance of inflorescence meristems (wus-6 and wus-7) and floral meristems (wus-7).
OBE3 functions redundantly with OBE4
Because the obe3-2 single mutant does not display any developmental defect, we asked whether related genes might mask its function. To this end, we isolated an insertion mutant in the closest OBE3 homolog OBE4, also named TITANIA2 (TTA2) [27]. The obe4-2 (SAIL_827_F11) mutation disrupts exon1, suggesting that it is a severe loss of function allele (S2 Fig). OBE4 transcript levels are reduced in obe4-2 to about half of the wild-type level (S1 Fig). obe4-2 single mutants are indistinguishable from the Col wild type (Fig 2A).
In the segregating progeny of an obe3-2/+ obe4-2/+ mother plant, we found 2.8% (n = 143) very small seedlings with partially fused cotyledons and without any true leaves, did not develop further, and were genotyped as homozygous double mutants by PCR (Fig 2B). By contrast, all obe3-2 obe4-2/+ plants identified by PCR (9.8%, n = 143) formed true leaves, but display severe growth retardation (Fig 2C), whereas the remaining segregating sibling plants look like wild type. We independently confirmed these results with the obe3-1 obe4-1 combination (S3 Fig). Thus, OBE3 and OBE4 are redundantly required for plant growth.
obe4-2 wus-6 double mutants are indistinguishable from wus-6, with the exceptions that obe4-2 wus-6 inflorescences produced less than 10 siliques (data not shown) and that all double mutant plants (n = 6) were mosaics carrying both, wild-type-like complete flowers (40/170) and wus-1-like incomplete flowers (130/170; Fig 2D and 2E). Thus, in contrast to obe3-2, the obe4-2 mutation restores carpel and seed development in wus-6, suggesting that in floral meristems, WUS and OBE4 act oppositely.
Mutual expression regulation between WUS and OBE3
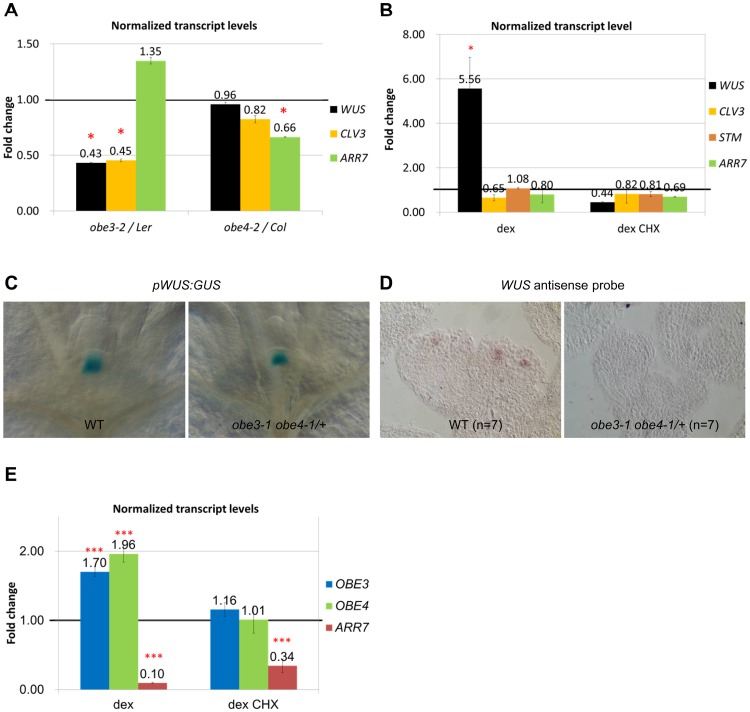
In order to investigate whether the expression levels of WUS and CLV3 genes are altered, we performed qRT-PCR with the 7 day-old obe3-2 and obe4-2 seedlings. WUS and CLV3 mRNA levels are significantly reduced in obe3-2 (0.43 and 0.45 fold, respectively) compared to the Ler wild type, whereas ARR7 mRNA levels appear increased (Fig 3A). In a converse experiment, WUS mRNA level is increased after induction of p35S:cOBE3-GR expression and this effect is suppressed in the presence of cycloheximide, whereas mRNA levels of CLV3, STM, and ARR7 are not significantly changed (Fig 3B). In contrast to obe3-2, expression levels of WUS or CLV3 genes are not significantly changed in obe4-2 (Fig 3A), but the ARR7 mRNA level is reduced (Fig 3A). In summary, OBE3 is required for normal WUS and CLV3 expression.
Fig 3. Changes of transcripts in obe3-2 and obe4-2.
(A) Transcript levels of 7-day-old seedlings as indicated. Error bars represent SE. (B) After induction of OBE3 overexpression, mRNA levels of WUS are increased, whereas mRNA levels of CLV3, STM, and ARR7 are unchanged in 7-day-old seedling. Error bars represent SD. (C) pWUS:GUS expression in 6-day-old obe3-1 obe4-1/+ seedlings is confined to the OC as in the wild type. (D) WUS mRNA is undetectable by in situ hybridization in obe3-1 obe4-1/+ floral meristems of 30-day-old plants. (E) WUS overexpression upregulates OBE3 and OBE4 mRNA levels in 7-day-old seedlings. ARR7 expression is used as a control. Error bars represent SD. Relative mRNA levels compared to the mock control are shown.*,p<0.05, calculated from Cp’ values; ***, p<0.001, calculated from Cp’ values.
Because double mutant plants are severely retarded, we analyzed the WUS expression pattern in obe3-1 obe4-1/+ plants. In 6-day-old seedlings, expression of the pWUS:GUS (Fig 3C) reporter is confined to the OC of obe3-1 obe4-1/+ plants as in the wild type. However, in 20-day-old inflorescences, WUS expression is not detectable in obe3-1 obe4-1/+ (genotyped by PCR) inflorescence and floral meristems unlike in the wild type (Fig 3D).
In order to address whether WUS affects OBE3/OBE4 transcript levels, we analyzed the effects of inducible WUS activity. After induction of p35S:WUS-GR plants with dexamethasone, mRNA levels of OBE3 and OBE4 are upregulated, and this effect is suppressed in the presence of cycloheximide (Fig 3E). The direct WUS target in the shoot meristem, ARR7 [2], is used as a control for WUS-GR induction. Upregulation of OBE3 and OBE4 expression by WUS is also suggested by published microarray data (S3 Table). Thus, WUS activity is sufficient to induce OBE3/OBE4 expression by an indirect mechanism. However, we did not detect any abnormal phenotype in p35S:cOBE3 or p35S:cOBE3-GR plants.
Overexpression of OBE3 alleviates shoot meristem but not floral meristem defects of wus-1
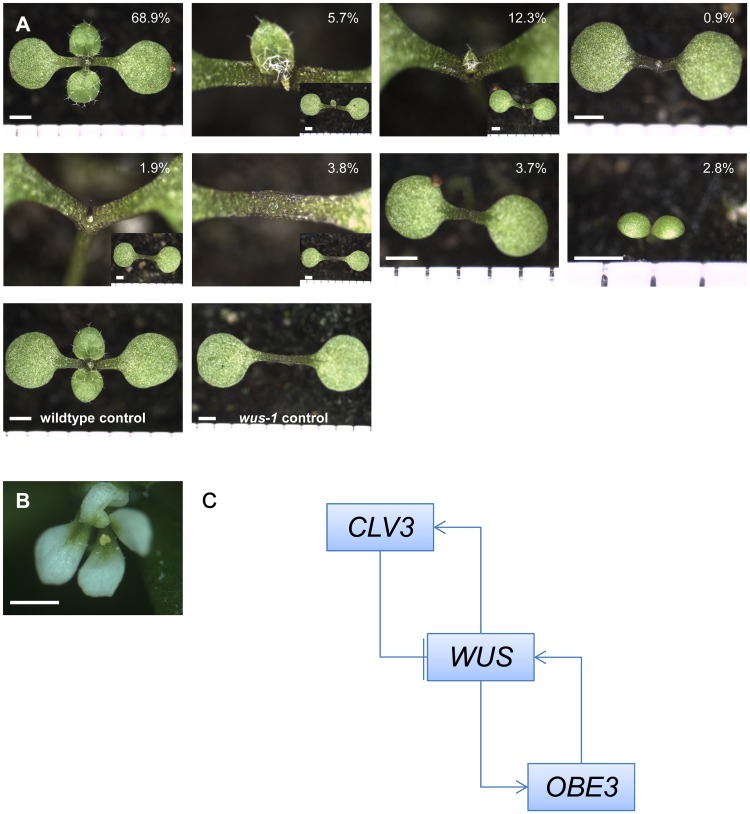
Because WUS overexpression can upregulate OBE3, we asked whether forced expression of OBE3 can overcome the absence of WUS activity and expressed p35S:cOBE3 in wus-1. When comparing segregating p35S:cOBE3 wus-1/+ plants with wus-1 empty vector controls, we find that the number of seedlings lacking the shoot meristem are reduced (10.3% vs. 28.5%) whereas seedlings with weak shoot meristem defects are increased (20.8% vs. 6.0%; Fig 4A, S4 Table). Furthermore, postembryonic shoot formation is increased (29.2% vs. 14.7%; S4 Table). The difference between p35S:cOBE3 wus-1/+ and empty vector wus-1/+ control is highly significant in both seedling and shoot stages (both Chi square, p<0.0001). By contrast, defective wus-1 flower development is not altered by p35S:cOBE3 (Fig 4B). Thus, OBE3 activity can partially replace WUS activity in seedling and inflorescence shoot meristems, but not in the floral meristem.
Fig 4. p35S:cOBE3 expression partially suppresses wus-1 defects.
(A) Phenotypes of segregating seedlings in the progeny of a p35S:cOBE3 wus-1/+ mother plant. (B) p35s:cOBE3 wus-1 plants produce wus-1-like flowers. (C) Model for WUS-OBE3 interaction. Scale bars: 1 mm.
Discussion
Stem cell homeostasis requires the balanced activities of a complex network of regulatory factors. Despite strong advances, our knowledge of regulatory pathways is still fragmentary with many components unknown. This is due in part to the fact that a limited number of mutants display informative stem cell phenotypes. Furthermore, many other essential stem cell factors may remain undiscovered due to genetic redundancy or pleiotropic mutant phenotypes. Here we used a modifier screen to overcome this problem and discovered the obe3-2 mutant as an enhancer of the hypomorphic wus-6 allele.
What is the developmental nature of the WUS-OB3 interaction?
The role of WUS in stem cell maintenance can be observed at several developmental stages. Mature wus-1 embryos and seedlings lack shoot meristem stem cells and display differentiated cells instead. Postembryonically formed adventitious meristems terminate prematurely after forming a few leaves. Only occasionally an inflorescence is formed, but it terminates prematurely after formation of 1–3 flowers, which in turn terminate prematurely after a single first anther. Whereas seedling and floral meristems appear to absolutely require WUS activity, the occasional formation of inflorescences suggests that at this stage other factors can sustain stem cells for some time [21]. Although OBE3 is ubiquitously expressed [27], the obe3-2 mutation enhances only the premature vegetative shoot meristem termination in wus-1 and thus represents one of these additional factors. In the hypomorphic allele wus-6, obe3-2 causes premature termination of the inflorescence meristem indistinguishable to wus-1. Finally, in combination with wus-7, which as a single mutant displays higher floral meristem activity compared to wus-1 and wus-6, obe3-2 enhances premature termination of the floral meristem. These results indicate that in addition to the seedling phase, OBE3 is required for residual WUS activity of hypomorphic wus alleles in inflorescence and floral meristems. By contrast, obe3-2 does not enhance wus defects in embryonic shoot meristem formation.
Curiously, despite their redundancy in the shoot meristem, obe3 enhances wus flower termination whereas obe4 mutant suppresses it. One possible reason for this particular behavior might be that in obe3-1 and obe3-2 mutants, the C-terminal region of OBE3 is disrupted and the PHD domain is still intact, whereas in obe4-1 and obe4-2 mutants the PHD domain is disrupted. Alternatively, both wild-type proteins might have divergent functions specifically in floral meristems.
Considering the ubiquitous expression of OBE3, it is noteworthy that the obe3-2 mutation reduces the organ number only of the two inner whorls of wus-7 but does not affect the perianth. A plausible explanation is that WUS-mediated stem cell maintenance is only required to provide the cells for the inner two whorls, whereas perianth organs appear to consume the cells present in the initial floral meristem formed independently of WUS, as described previously [6].
What is the genetic nature of the WUS-OB3 interaction?
Based on our mutant analysis and expression studies, OBE3 appears to act downstream of WUS. On the other hand, WUS expression levels are reduced in obe3-2 mutants and increased by OBE3 overexpression from the ubiquitous 35S promoter. The reduction of pWUS:GUS expression in the shoot meristem of obe3-1 obe4-1/+ mutants and the requirement of OBE3 in wus hypomorphs suggest that this is also the case in shoot meristem regulation. One plausible interpretation of this data is that WUS and OBE3 reinforce each other's expression in a positive feedback loop (Fig 4C), albeit this effect seems moderate.
OBE3 is a member of a small group of related proteins and, together with its closest homolog OBE4, is redundantly required for plant growth, consistent with previous observations [27]. The seedling lethality of obe3-2 obe4-2 double mutants suggests that both genes are involved in several processes other than shoot meristem regulation. Likewise, obe1 obe2 [28] displays seedling lethality, but not any other obe double mutant combinations, indicating two pairs of redundant functions in this group, OBE1,2 and OBE3,4. In contrast to obe3-2 wus-6 plants, obe4-2 wus-6 double mutants have wus-6-like inflorescences and partially restored floral organs and seed production. This indicates that OBE3 and OBE4, in addition to their redundant functions in general growth control, have opposite roles at least in floral meristems.
What is a possible molecular basis of interaction?
The upregulation of OBE3 mRNA levels by WUS and vice versa is abolished by the presence of the protein inhibitor cycloheximide, suggesting that intermediate components are necessary. OBEs are PHD domain containing proteins, which originally were found by their homology to the Potyvirus VPg-interacting protein [29]. The PHD domain is reported to bind to potentially activating H3K4me2/3 modifications [30–34]. Further experiments are necessary to address how OBE affects WUS expression and vice versa and whether the enhancement of hypomorphic wus mutant defects by obe3-2 can be attributed to the reduction of WUS expression levels.
Material and Methods
Plant materials and growth conditions
The obe3-2 mutant was isolated from EMS-mutagenized populations in a wus-6 background in Ler [25]. The insertion alleles obe3-3SALK_078036, obe3-4SALK_042597c and obe4-2SAIL_827_F11 in the Col background were identified from the SALK collection [35] of T-DNA tagged lines and the SAIL collection [36], respectively. All other mutant alleles and transgenic lines used in this study are listed in S5 Table. For segregation analysis, entire siliques from genotyped mother plants were harvested and the seeds were sown in randomized schemes [37]. Plant growth conditions were as previously described [6]
Mapping, genetic analysis and PCR genotyping
The wen9 wus-6/+ x Col cross was performed for map-based cloning. Among 12113 F2 plants from F1 parents genotyped as wus-6/+, we identified 446 plants (3.7%) with an enhanced phenotype. The wen9 mutation was mapped with SSLP and dCAPS markers to a 97 kb region in chromosome 1 (between SNP CER465614 and CER424346). Sequencing of all 23 candidate loci in this region detected a mutation only in the OBE3 gene. The identified G1554>A “stop” mutation is in exon1 of the predicted reading frame of locus AT1G14740. The primers wen9-F (5’- CAGAGATGTTTGGATTCGTTAAGGATGTTTTTGTGTGTTGCGCTAAGAATCG-3’) and wen9-R (5’-GAAATTGTGATAAGAGAAGG-3’) were used for genotyping PCRs (Ta 55°C). After TaqI restriction cleavage, the wild type displays a 300bp band, and wen9 displays a 250bp and a 50bp band. Genotyping primers of other mutants including T-DNA lines used in this study are listed in S6 Table.
Preparation of constructs and selection of transformants
The genomic fragment including the intergenic region of OBE3 was amplified by PCR from Ler and cloned as pOBE3:gOBE3 in a pGPTV-HPT-based vector. The RALF11-03K20 cDNA clone from RIKEN BRC was used to amplify the full length cDNA by PCR for construct preparation. The cDNA fragments starting from ATG to the end of gene, with or without the stop codon, were cloned as: p35S:cOBE3, and p35S:cOBE3-GR respectively, in a pGreenII-based vector. Arabidopsis plants were transformed by floral dip, and T1 seeds were selected on plates with the respective antibiotics.
Quantitative RT-PCR analysis
Arabidopsis seeds were surface-sterilized with 1% hypochlorite for 10 minutes and washed two times with 70% ethanol. Sterilized seeds were sown on 1/2 MS plates, stratified for 3 days in the dark at 4°C and then grown in a Percival growth cabinet with constant illumination for 7 or 10 days. For all qRT-PCR experiments, 3 biological replicates with two technical replicates each were done.
For experiments without further treatment, seedlings were collected from the plates and frozen in liquid nitrogen immediately. For the induction experiments, dexamethasone (5 μM), with or without cycloheximide (50 μM) were applied by spraying the plates, and flooding the seedlings for 15 minutes. After removal of the liquid, seedlings were returned to the Percival growth cabinet for 4 hours before sample collection.
Total RNA was extracted from whole seedlings using the RNeasy® Mini kit (QIAGEN), followed by RQ1 RNase-Free DNase (Promega) treatment and reverse-transcribed with SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen). Quantitative PCR was performed with the LightCycler® 480 system (Roche) coupled with SYBR Green I Master (Roche). For each qPCR reaction, 25 ng of cDNA was used. Transcript level analysis was carried out according to a published protocol [38]. For statistical analysis, ANOVA or t-tests were performed on Cp’ values. The Cp’ values were calculated after normalizing the Cp values with three independent reference genes, which passed the geNorm v3.5 [39] test. For graphic presentation, Normalized Relative Quantity (NRQ) was first rescaled by setting NRQ of mock treated wild-type samples as 1, then adjusted for the unspecific DEX effect and the transgene effect sequentially in order to calculate the transcript fold change. Fold changes were plotted as bar graphs. Primers used for quantitative PCR are listed in S7 Table.
Supporting Information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank SALK, ABRC, NASC and RIKEN BRC for providing seeds and cDNA clones. We thank members of the Laux laboratory for discussions. We thank Dr. Edwin Groot for proofreading the manuscript and help in statistical analysis. This research was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan to M.A. and from the Deutsche Forschungsgemeinschaft (SFB592 and BMBF-FRISYS) to T.L. The article processing charge was funded by the German Research Foundation (DFG) and the Albert Ludwigs University Freiburg in the funding programme Open Access Publishing.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was partially supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (http://www.mext.go.jp/english/) to M.A. and mainly by grants from Deutsche Forschungsgemeinschaft [SFB592 (http://www.sfb592.uni-freiburg.de/)] and BMBF-FRISYS (http://www.bioinf.uni-freiburg.de/Research/bmbf-frisys.html) to T.L. The article processing charge was funded by the German Research Foundation (DFG) and the Albert Ludwigs University Freiburg in the funding programme Open Access Publishing. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mayer KFX, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. Role of WUSCHEL in Regulating Stem Cell Fate in the Arabidopsis Shoot Meristem. Cell. 1998;95(6):805–15. [DOI] [PubMed] [Google Scholar]
- 2.Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438(7071):1172–5. . [DOI] [PubMed] [Google Scholar]
- 3.Daum G, Medzihradszky A, Suzaki T, Lohmann JU. A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc Natl Acad Sci U S A. 2014;111(40):14619–24. Epub 2014/09/24. 10.1073/pnas.1406446111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yadav RK, Perales M, Gruel J, Girke T, Jonsson H, Reddy GV. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011;25(19):2025–30. Epub 2011/10/08. 10.1101/gad.17258511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci U S A. 2009;106(12):4941–6. 10.1073/pnas.0900843106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laux T, Mayer KF, Berger J, Jurgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122(1):87–96. . [DOI] [PubMed] [Google Scholar]
- 7.Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100(6):635–44. . [DOI] [PubMed] [Google Scholar]
- 8.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–9. [DOI] [PubMed] [Google Scholar]
- 9.Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, et al. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105(6):793–803. Epub 2001/07/07. S0092-8674(01)00384-1 [pii]. . [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Kim YJ, Muller R, Yumul RE, Liu C, Pan Y, et al. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell. 2011;23(10):3654–70. Epub 2011/10/27. tpc.111.091538 [pii] 10.1105/tpc.111.091538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenhard M, Bohnert A, Jürgens G, Laux T. Termination of Stem Cell Maintenance in Arabidopsis Floral Meristems by Interactions between WUSCHEL and AGAMOUS. Cell. 2001;105(6):805–14. [DOI] [PubMed] [Google Scholar]
- 12.Deyhle F, Sarkar AK, Tucker EJ, Laux T. WUSCHEL regulates cell differentiation during anther development. Dev Biol. 2007;302(1):154–9. . [DOI] [PubMed] [Google Scholar]
- 13.Groß-Hardt R, Lenhard M, Laux T. WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes & Development. 2002;16(9):1129–38. 10.1101/gad.225202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieber D, Lora J, Schrempp S, Lenhard M, Laux T. Arabidopsis WIH1 and WIH2 genes act in the transition from somatic to reproductive cell fate. Curr Biol. 2011;21(12):1009–17. Epub 2011/06/11. S0960-9822(11)00542-2 [pii] 10.1016/j.cub.2011.05.015 . [DOI] [PubMed] [Google Scholar]
- 15.Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, et al. Hormonal control of the shoot stem-cell niche. Nature. 2010;465(7301):1089–92. 10.1038/nature09126 [DOI] [PubMed] [Google Scholar]
- 16.Kwon CS, Chen C, Wagner D. WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 2005;19(8):992–1003. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieffer M, Stern Y, Cook H, Clerici E, Maulbetsch C, Laux T, et al. Analysis of the Transcription Factor WUSCHEL and Its Functional Homologue in Antirrhinum Reveals a Potential Mechanism for Their Roles in Meristem Maintenance. Plant Cell. 2006;18(3):560–73. 10.1105/tpc.105.039107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucker MR, Hinze A, Tucker EJ, Takada S, Jurgens G, Laux T. Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development. 2008;135(17):2839–43. 10.1242/dev.023648 [DOI] [PubMed] [Google Scholar]
- 19.Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci U S A. 2009;106(38):16529–34. 10.1073/pnas.0908122106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yadav RK, Tavakkoli M, Reddy GV. WUSCHEL mediates stem cell homeostasis by regulating stem cell number and patterns of cell division and differentiation of stem cell progenitors. Development. 2010;137(21):3581–9. Epub 2010/09/30. dev.054973 [pii] 10.1242/dev.054973 . [DOI] [PubMed] [Google Scholar]
- 21.Graf P, Dolzblasz A, Wurschum T, Lenhard M, Pfreundt U, Laux T. MGOUN1 encodes an Arabidopsis type IB DNA topoisomerase required in stem cell regulation and to maintain developmentally regulated gene silencing. Plant Cell. 2010;22(3):716–28. 10.1105/tpc.109.068296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr Biol. 2011;21(5):345–52. 10.1016/j.cub.2011.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chickarmane VS, Gordon SP, Tarr PT, Heisler MG, Meyerowitz EM. Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc Natl Acad Sci U S A. 2012;109(10):4002–7. Epub 2012/02/22. 1200636109 [pii] 10.1073/pnas.1200636109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamada S, Onouchi H, Tanaka H, Kudo M, Liu Y-G, Shibata D, et al. Mutations in the WUSCHEL gene of Arabidopsis thaliana result in the development of shoots without juvenile leaves. the Plant Journal. 2000;24(1):91–101. [DOI] [PubMed] [Google Scholar]
- 25.Graf P. Genetic modifiers of WUSCHEL function in Arabidopsis stem cell maintenance [Inaugural-Dissertation zur Erlangung der Doktorwürde]. Freiburg im Breisgau: Albert-Ludwigs-Universität; 2007.
- 26.Thomas CL, Schmidt D, Bayer EM, Dreos R, Maule AJ. Arabidopsis plant homeodomain finger proteins operate downstream of auxin accumulation in specifying the vasculature and primary root meristem. the Plant Journal. 2009;59:426–36. 10.1111/j.1365-313X.2009.03874.x [DOI] [PubMed] [Google Scholar]
- 27.Saiga S, Möller B, Watanabe-Taneda A, Abe M, Weijers D, Komeda Y. Control of embryonic meristem initiation in Arabidopsis by PHD-finger protein complexes. Development. 2012;139(8):1391–8. 10.1242/dev.074492 [DOI] [PubMed] [Google Scholar]
- 28.Saiga S, Furumizu C, Yokoyama R, Kurata T, Sato S, Kato T, et al. The Arabidopsis OBERON1 and OBERON2 genes encode plant homeodomain finger proteins and are required for apical meristem maintenance. Development. 2008;135:1751–9. 10.1242/dev.014993 [DOI] [PubMed] [Google Scholar]
- 29.Dunoyer P, Thomas C, Harrison S, Revers F, Maule A. A Cysteine-Rich Plant Protein Potentiates Potyvirus Movement through an Interaction with the Virus Genome-Linked Protein VPg. Journal of Virology. 2004;78(5):2301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature, Letters. 2006;442:91–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellor J. It Takes a PHD to Read the Histone Code. Cell. 2006;126:22–4. [DOI] [PubMed] [Google Scholar]
- 32.Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature, Letters. 2006;442:100–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature, Letters. 2006;442:96–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature, Letters. 2006;442:86–90. [DOI] [PubMed] [Google Scholar]
- 35.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–7. . [DOI] [PubMed] [Google Scholar]
- 36.Sessions A, Burkea E, Prestinga G, Auxb G, McElverb J, Pattonb D, et al. A High-Throughput Arabidopsis Reverse Genetics System. The Plant Cell. 2002;14:2985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urbaniak GC, Plous S. Research Randomizer (Version 3.0)[Computer software]. Available: http://www.randomizer.org/. 2009. Epub 3.
- 38.Rieu I, Powers SJ. Real-Time Quantitative RT-PCR: Design, Calculations, and Statistics. The Plant Cell. 2009;21:1031–3. 10.1105/tpc.109.066001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandesompele J, Preter KD, Pattyn F, Poppe B, Roy NV, Paepe AD, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.