Abstract
We report on PAX6 alleles associated with a clinical diagnosis of classical aniridia in 81 affected individuals representing 66 families. Allelic variants expected to affect PAX6 function were identified in 61 families (76 individuals). Ten cases of sporadic aniridia (10 families) had complete (8 cases) or partial (2 cases) deletion of the PAX6 gene. Sequence changes that introduced a premature termination codon into the open reading frame of PAX6 occurred in 47 families (62 individuals). Three individuals with sporadic aniridia (3 families) had sequence changes (1 deletion, 2 run-on mutations) expected to result in a C-terminal extension. An intronic deletion of unknown functional significance was detected in one case of sporadic aniridia (1 family), but not in unaffected relatives. Within these 61 families, single nucleotide substitutions accounted for 30/61 (49%), indels for 23/61 (38%), and complete deletion of the PAX6 locus for 8/61 (13%). In five cases of sporadic aniridia (5 families), no disease-causing mutation in the coding region was detected. In total, 27 unique variants were identified that have not been reported in the LOVD database. Within the group assessed, 92% had sequence changes expected to reduce PAX6 function, confirming the primacy of PAX6 haploinsufficiency as causal for aniridia.
Keywords: Absent Iris, Eye Abnormalities, Eye Disease, Irideremia, chromosome 11p13
INTRODUCTION
Aniridia is a congenital, progressive disorder for which the majority of cases are caused by heterozygous loss-of-function mutations of the PAX6 gene (1, 2). Aniridia occurs in approximately 1/64,000 to 1/96,000 live births and is primarily characterized by iris hypoplasia that is clinically detectable at birth (2, 3). Associated foveal hypoplasia, indicated by early infancy nystagmus, causes reduced visual acuity. The progressive nature of the disease frequently leads to multiple ocular abnormalities such as keratopathy, corneal vascularization and opacification, glaucoma, anterior chamber fibrosis, and cataracts (2, 4–7). Along with distinct ocular characteristics, the condition is associated with a number of other irregularities spanning sensory, neural, cognitive (8) and pancreatic phenotypes (9, 10). These include auditory processing deficits (11, 12), deficient pituitary function (13) and olfactory dysfunction (14). Previous studies have identified a correlation between PAX6-mediated aniridia and a number of other disorders, including diabetes (15) and autism spectrum disorders (16). Additionally, structural abnormalities in major fiber tracts and subcortical structures in the brain including the corpus callosum, anterior and posterior commissures, pineal gland and probst bundles have been observed (14, 17). A functional understanding of the PAX6 gene in context of the manifestation of aniridia-related clinical traits continues to be the focus of much research.
Functional mutations that occur only in the PAX6 gene or associated regulatory regions give rise to aniridia, which can be sporadic or familial. WAGR syndrome (Wilms tumor, aniridia, genitourinary anomalies and mental retardation) is associated with large heterozygous genomic deletions at the 11p13 chromosome that include deletion of the PAX6 locus and the WT1 locus (18, 19). In addition to clinical symptomologies associated with aniridia, patients with WAGR syndrome present with Wilms tumor and, depending on the size of the causal chromosomal deletion, often exhibit cognitive delays and disabilities in addition to developmental genitourinary defects (19). Aniridia is not comorbid with WAGR syndrome; however, it can be further classified at the cellular and molecular level by the specific mutation affecting the PAX6 gene.
The PAX6 gene encodes a transcription factor critical for normal ocular and neural development. The gene is highly conserved and is expressed in the developing eye, brain, spinal cord and pancreas (20). PAX6 is required for various aspects of anatomical and functional development. The mechanistic role of PAX6 in eye development has been investigated and its involvement has been demonstrated in initial lens development (21), cell differentiation (22–24), and cell proliferation and adhesion/migration (25). PAX6 has also been identified as having critical maintenance functions in corneal homeostasis (26, 27). Additionally, in the central nervous system PAX6 is thought to be involved in brain patterning and regionalization and the formation of neural circuits, particularly in the forebrain (25, 28–30). In these ways, PAX6 is an integral upstream regulator involved in numerous developmental gene networks. It follows that the broad developmental and progressive phenotypic outcomes caused by PAX6 mutations are a consequence of the extensive roles of the gene and its products.
The PAX6 protein is composed of four functional domains: a paired domain and a homeodomain (involved in DNA binding) joined by a linker domain, and a proline-serine-threonine (PST) rich transactivation domain (31). Should products of mutant alleles undergo translation, disruptions in the coding sequence of different functional domains will alter the efficacy of PAX6 as a transcriptional regulator, which has been shown in molecular and phenotypic studies of missense and run-on PAX6 mutations in aniridia and other related disorders (32–39). However, the majority of PAX6-mediated aniridia cases are caused by mutations that give rise to premature termination codons (PTCs) leading to haploinsufficient levels of wild-type PAX6 protein.
Prior to the current study, the Human PAX6 Allelic Variant Database (LOVD database) (http://lsdb.hgu.mrc.ac.uk/home.php?select_db=PAX6) has identified 361 unique variants of PAX6 (833 total reported). The current study sought to further examine and classify mutations of PAX6 in patients diagnosed with classical aniridia, as well as examine our data in the context of previously identified mutations of the gene. This line of investigation provides insights towards identifying locations on the PAX6 gene that are more susceptible to mutations, and seeks to explore individual-specific mutation characteristics which could benefit from new therapeutics within the aniridia population.
MATERIALS & METHODS
Participants
DNA samples were collected from 81 individuals with a clinical diagnosis of aniridia and 77 unaffected genetic relatives from 66 families (Supplementary Table 1). Participants were recruited through the Aniridia Foundation International Conferences in 2007, 2009, and 2011 through affiliation and direct physician referral for participation in the study. The study protocol was in keeping with the tenets of the Declaration of Helsinki and was reviewed, approved, and overseen by the Institutional Review Boards (IRBs) at the University of Tennessee Health Science Center and the University of Georgia. All samples were collected after written informed consent had been obtained from each participant.
DNA Preparation
Genomic DNA from individuals with aniridia and their relatives was prepared from peripheral venous whole blood. Briefly, 10mL volume blood samples were collected from each adult participant, and 2mL blood samples were collected from each infant. Buffy coat preparation was performed on each sample and genomic DNA (gDNA) prepared by use of a Gentra Puregene Blood Kit (Qiagen). Buccal cell lysates were prepared from saliva or swab samples (Oragene-DNA OG-575, DNA Genotek, Inc; buccal swabs and QuickExtract DNA extraction solution, Epicentre). All gDNA samples were stored in 10 mM Tris-HCl (pH 7.5), 1 mM EDTA buffer at −20°C.
Molecular Analysis
The PAX6 gene (11p13) [OMIM: 607108] is composed of sixteen exons (Supplementary Table 2) (20), of which eleven (4 to 13 and 5a) contribute to the protein-coding regions of the mRNA transcripts. The other exons (0, 1, 2, 3, and alpha) contribute to the non-coding regions of the mRNA transcripts. For this study, PAX6 exons 1–13 and 5a were individually amplified in a polymerase chain reaction (PCR) with primers located in the introns as previously described (40, 41). Mutation screening was performed on the PCR products by direct bi-directional sequencing using either the same PCR primers or more internal primers (40, 41). Bi-directional sequence was analyzed and compared to PAX6 reference sequences. Reference sequences for PAX6 cDNA and protein are contained in GenBank entry NM_000280 (http://www.ncbi.nlm.nih.gov/Genbank; provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD). The February 2009 human reference sequence GRCh37 produced by the Genome Reference Consortium (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/) was used as the PAX6 genomic reference after first validating against genomic sequences obtained from cosmids FAT5 and A1280 (Genbank accession numbers Z95332 and Z83307, respectively), which together encompass all PAX6 exons. Supplementary Table 2 lists intron-exon boundary sequences for human PAX6 and all exon sizes. Potential mutations were identified as differences relative to the appropriate reference sequence. Results were confirmed by repeat PCR, which in most cases was performed using an independent, second gDNA sample prepared from either blood or buccal cells. Changes from the reference sequence were identified, classified and are documented in Table 1 using the nomenclature system recommended by the Human Genome Variation Society (42), with the first base of the ATG initiation codon in the PAX6 cDNA reference sequence NM_000280 denoted as nucleotide 1.
Table 1. Identified variants in the current study.
(A) 51 sequence variants (from 51 families) sorted by genomic location and are described by type of variant, intronic/exonic location, and the predicted effect on gene products. (B) Complete and partial deletions of PAX6 from 10 families. (C) 5 families with no pathological change in PAX6 detected. Variants in bold represent those not previously identified in the LOVD database.
| Table 1A: PAX6 variants expected to cause pathology. | ||||
|---|---|---|---|---|
| FID | LOCATION | VARIANT | TYPE OF VARIANT | PREDICTED EFFECT |
| 157 | Exon 5 | c.28C>T | transition | nonsense |
|
|
||||
| 118 | Exon 5 | c.57delG | indel | frameshift deletion |
|
|
||||
| 108 | Exon 5 | c.63-70delGCCGGACT | indel | frameshift deletion |
|
|
||||
| 133 | Exon 5 | c.112delC | indel | frameshift deletion |
|
|
||||
| 101 | Exon 5 | c.112-116delCGGCC | indel | frameshift deletion |
|
|
||||
| 135 | Exon 5 | c.121_122insGCGG | indel | frameshift insertion |
|
|
||||
| 138 | Intron 5 | c.141+1G>T | transversion | splice junction disruption |
|
|
||||
| 152 | Intron 5 | c.141+2_+30delTGATCCTCCCGGCGCCGCCCCACTCGCCG | indel | splice junction disruption |
|
|
||||
| 128 | Intron 5 | c.141+18_+20delGCC | indel | Unknown effect |
|
|
||||
| 150 | Exon 6 | c.179-185delATTACGAinsCTGAT | indel | frameshift deletion/insertion |
|
|
||||
| 112 | Exon 6 | c.199A>T | transversion | nonsense |
|
|
||||
| 147 | Exon 6 | c.204delC | indel | frameshift deletion |
|
|
||||
| 155 | Exon 6 | c.332insG | indel | frameshift insertion |
|
|
||||
| 1003 | Exon 6 | c.343delG | indel | frameshift deletion |
|
|
||||
| 163 | Exon 6 & Intron 6 | c.352-357+2delCCAAGCGT | indel | frameshift deletion |
|
|
||||
| 100 | Intron 6 | c.357+1G>A | transition | splice junction disruption |
|
|
||||
| 129 | Intron 6 | c.357+1G>A | transition | splice junction disruption |
|
|
||||
| 230 | Intron 6 | c.357+1G>A | transition | splice junction disruption |
|
|
||||
| 105 | Intron 6 | c.357+1G>T | transversion | splice junction disruption |
|
|
||||
| 169 | Intron 6 & Exon 7 | c.358-3_361delCAGGTGT | indel | splice junction disruption |
|
|
||||
| 168 | Exon 7 | c.365C>A | transversion | nonsense |
|
|
||||
| 141 | Exon 7 | c.401delA | indel | frameshift deletion |
|
|
||||
| 104 | Exon 7 | c.454C>T | transition | nonsense |
|
|
||||
| 126 | Exon 7 | c.467G>A | transition | nonsense |
|
|
||||
| 127 | Exon 7 | c.480delT | indel | frameshift deletion |
|
|
||||
| 114 | Exon 7 | c.482delG | indel | frameshift deletion |
|
|
||||
| 117 | Intron 7 & Exon 8 | c.524-101_534del112bp | indel | splice junction disruption |
|
|
||||
| 102 | Exon 8 | c.607C>T | transition | nonsense |
|
|
||||
| 160 | Exon 8 | c.607C>T | transition | nonsense |
|
|
||||
| 189 | Exon 8 | c.631C>T | transition | nonsense |
|
|
||||
| 173 | Intron 9 | c.766-3C>G | transversion | splice junction disruption |
|
|
||||
| 124 | Exon 10 | c.771delG | indel | frameshift deletion |
|
|
||||
| 144 | Exon 10 | c.781C>T | transition | nonsense |
|
|
||||
| 164 | Exon 10 | c.781C>T | transition | nonsense |
|
|
||||
| 170 | Exon 10 | c.781C>T | transition | nonsense |
|
|
||||
| 232 | Exon 10 | c.781C>T | transition | nonsense |
|
|
||||
| 110 | Exon 10 | c.794G>A | transition | nonsense |
|
|
||||
| 227 | Exon 10 | c.795G>A | transition | nonsense |
|
|
||||
| 191 | Exon 10 | c.799A>T | transversion | nonsense |
|
|
||||
| 103 | Exon 10 | c.802_806delGAAGA | indel | frameshift deletion |
|
|
||||
| 120 | Exon 11 | c.949C>T | transition | nonsense |
|
|
||||
| 153 | Exon 11 | c.949C>T | transition | nonsense |
|
|
||||
| 1001 | Exon 11 | c.949C>T | transition | nonsense |
|
|
||||
| 130 | Intron 11 | c.1032+6T>G | transversion | splice junction disruption |
|
|
||||
| 122 | Intron 11 | c.1033-2A>G | transition | splice junction disruption |
|
|
||||
| 143 | Exon 12 & Intron 12 | c.1174_+6delACTTCAACAGGTGAGC | indel | frameshift deletion |
|
|
||||
| 119 | Intron 12 | c.1183+1G>A | transition | splice junction disruption |
|
|
||||
| 1004 | Intron 12 | c.1183+1G>A | transition | splice junction disruption |
|
|
||||
| 185 | Exon 13 | c.1256delC | indel | frameshift deletion; C-terminal extension |
|
|
||||
| 115 | Exon 13 | c.1268A>T | transversion | run-on, C-terminal extension |
|
|
||||
| 140 | Exon 13 | c.1268A>T | transversion | run-on, C-terminal extension |
| Table 1B: Partial and gross deletions of PAX6. | ||||
|---|---|---|---|---|
| FID | LOCATION | VARIANT | TYPE OF VARIANT | PREDICTED EFFECT |
| 228 | Deletion P1 promoter to exon 4 | indel | ||
|
|
||||
| 1002 | Deletion exons 6 and 7 | indel | ||
|
|
||||
| 161 | WT1-PAX6; 3′ extent unknown | gross deletion | loss-of-function | |
|
|
||||
| 174 | WT1-PAX6; 3′ extent unknown | gross deletion | loss-of-function | |
|
|
||||
| 180 | WT1-PAX6; 3′ extent unknown | gross deletion | loss-of-function | |
|
|
||||
| 182 | WT1-PAX6; 3′ extent unknown | gross deletion | loss-of-function | |
|
|
||||
| 214 | WT1-PAX6; 3′ extent unknown | gross deletion | loss-of-function | |
|
|
||||
| 219 | WT1-PAX6; 3′ extent unknown | gross deletion | loss-of-function | |
|
|
||||
| 225 | WT1-PAX6; 3′ extent unknown | gross deletion | loss-of-function | |
|
|
||||
| 501 | Deletion of PAX6 | gross deletion | loss-of-function | |
| Table 1C: Families with no pathological mutations of PAX6 detected. | ||||
|---|---|---|---|---|
| FID | LOCATION | VARIANT | TYPE OF VARIANT | PREDICTED EFFECT |
| 121 | No changes detected | N/A | ||
|
|
||||
| 162 | Intron 9 | c.766-12C>T | transition | Variant of unknown significance |
|
|
||||
| 166 | Intron 9 | c.766-12C>T | transition | Variant of unknown significance |
|
|
||||
| 190 | Intron 9 | c.766-12C>T | transition | Variant of unknown significance |
|
|
||||
| 231 | No changes detected | N/A | ||
Deletion/duplication testing was performed either by array-comparative genomic hybridization (CGH) analysis (43) (GeneDx or Emory Genetics Laboratory), fluorescence in situ hybridization (44), or physical mapping (45).
Individuals for which no PAX6 pathogenic mutations were detected were retested using fresh samples by GeneDx or the Denver Genetic Laboratories (University of Colorado Anschutz Medical Campus).
RESULTS
A total of 158 individuals (81 affected, 77 unaffected genetic relatives) from 66 families were evaluated in this study (Supplementary Table 1). All affected individuals had a clinical diagnosis of aniridia (Figure 1, data not shown). In the context of this study, “family” was used to group genetically related individuals, and includes instances of sporadic aniridia (a single individual within the family) or familial aniridia (multiple individuals). Of the eighty-one affected individuals, 53 were female and 28 were male. Fifty-eight cases were sporadic (44 female, 14 male), with no affected relatives at the time of birth. Of these, five females subsequently had children (2 female, 4 male) with aniridia. Fifteen additional cases (6 female, 9 male) were familial from 4 separate families. The parents for two cases (1 female, 1 male) were unknown, and so it is not possible to determine if these two individuals were sporadic or not.
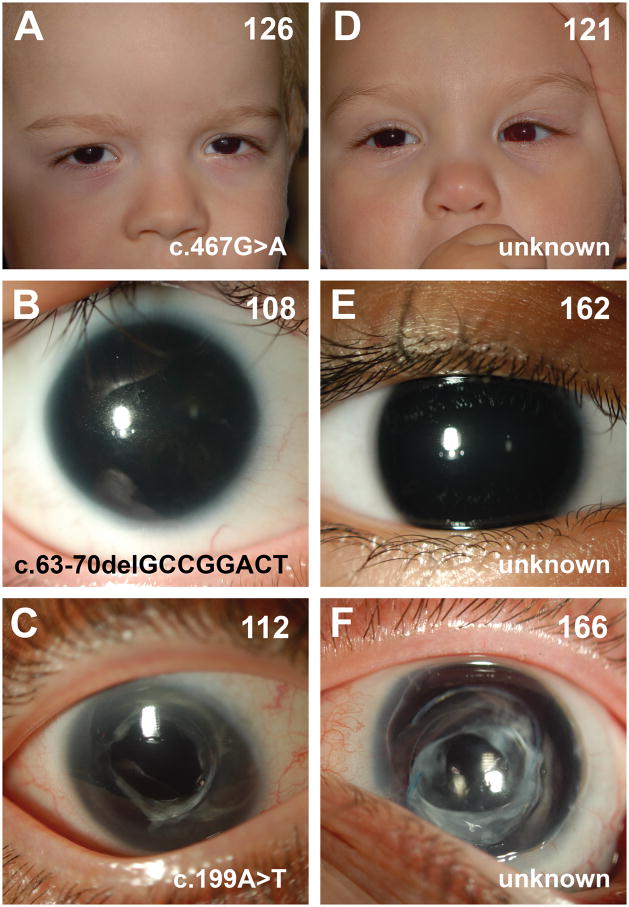
Figure 1.
Ocular phenotypes associated with individuals harboring PAX6 coding mutations (A–C) and those for whom no coding mutations were detected (D–F). Images from individuals with known PAX6 mutations were chosen to facilitate comparison with panels D–F and do not capture the full range of ocular phenotypes observed in individuals with identified mutations of the PAX6 gene. Family ID, upper right; causal sequence variant, lower right.
The PAX6 gene was characterized for all individuals. Sequence changes relative to the appropriate PAX6 reference sequence (see Methods) were identified and classified (Table 1, Supplementary Table 1). Potential aniridia-causing changes were identified as differences relative to the reference sequence and sequences from unaffected genetically related family members. Sequence variants were identified in individuals with aniridia in 64 of the 66 families (79 out of 81 individuals). One of these variants (c.766-12C>T) was detected in both affected and unaffected individuals in three families (3 individuals with sporadic aniridia) and is therefore likely to be a benign sequence change (Table 1C), as has been previously suggested (46–48). No sequence changes were detected in unaffected relatives in the remaining 74 families. Thus aniridia-specific alterations in the PAX6 gene were identified in 61 families (76 individuals).
Sequence alterations predicted to cause loss-of-function of one copy of the PAX6 gene were identified in 57 families (72 individuals). Whole gene deletions were detected in 8 cases of sporadic aniridia (8 families); seven of these deletions also included the WT1 gene. A partial deletion of PAX6 was detected in 2 cases of sporadic aniridia (2 families). In one case, the region extending from the P1 promoter to exon 4 was deleted, and in the other, a region encompassing exons 6 and 7 was deleted. Nonsense mutations were identified in 18 families (24 individuals), and frameshifting deletions or insertions were identified in 16 families (23 individuals). Changes expected to disrupt normal splicing were identified in 13 families (15 individuals). Of these, 9 were changes to the dinucleotides in the 5′ and 3′ splice sites. Also included were a C to T transition at the −3 position in the 5′ flanking sequence of exon 10 (c.766-3C>T, family 173) and a T to G transversion at the +6 position in the 3′ flanking sequence of exon 11 (c.1032+6T>G, family 130). These two variants were included because changes in these positions are known to affect splicing in other human genes (49) and there is a high degree of conservation in the sequences immediately flanking PAX6 exons between humans, dogs, mice, chickens, Xenopus tropicalis, and zebrafish (data not shown). In families 117, 143, 163, and 169, where deletions included both exonic and intronic sequences, the deletion was scored as “frameshifting” if the deletion originated within the exon and included the 3′ flanking sequence (families 143 and 163) or “splice junction” if the deletion originated in the intron and included the 5′ end of the subsequent exon (families 117 and 169).
Mutations predicted to result in a PAX6 protein with a C-terminal extension were identified in three cases of sporadic aniridia. Two cases had an A to T transversion (c.1268A>T) that converts the stop codon (TAA) to a leucine codon (TTA). This mutation is predicted to result in the addition of 14 residues to the C-terminal portion of the PAX6 protein.
One case had a single nucleotide deletion (c.1256delC) that is predicted to result in a frameshift at amino acid 419 and the addition of 105 residues to the C-terminal portion of the PAX6 protein. One case of sporadic aniridia (family 128) had a heterozygous deletion of the 18th – 20th nucleotides in the intronic sequence just 3′ to exon 5 (c.141+18_+20delGCC). This deletion was not present in unaffected family members and is, therefore, likely to be causal for aniridia in this individual. Although the molecular effect of this change is not known, its position within the intron may affect splicing or alter a regulatory element.
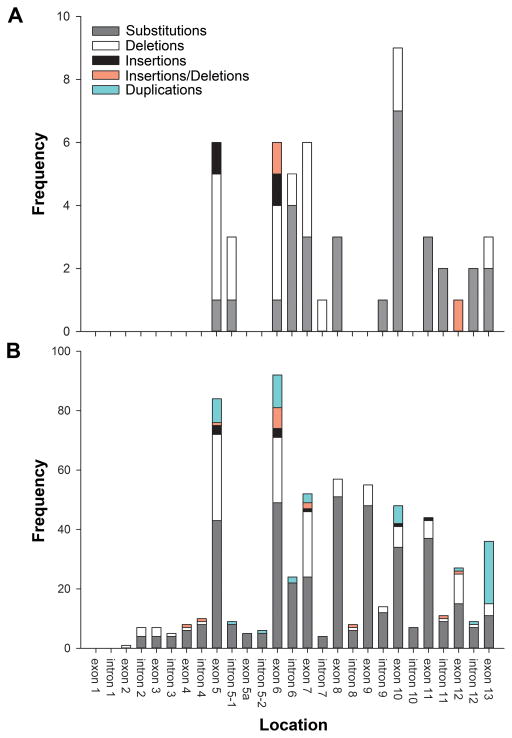
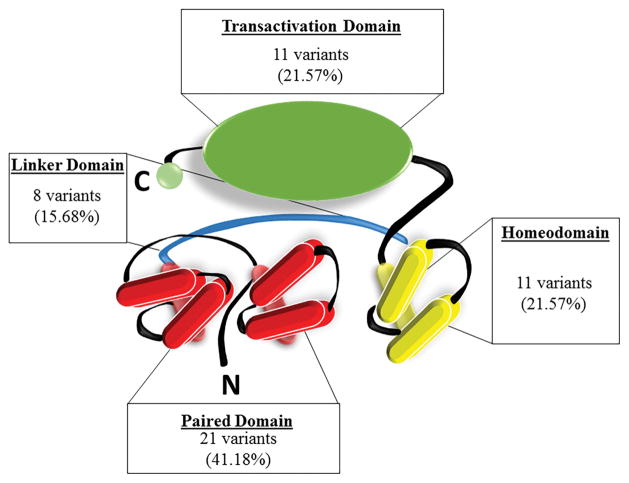
The exon-by-exon distribution of single nucleotide substitutions and small indels (≤ 150 bp) associated with aniridia that were found in 51 families is shown in Figure 2. Mutations were detected in all exons except for 4, 5a and 9. Twenty-one variants (41%) occurred within the paired domain, 11 variants (22%) occurred within the homeodomain, 8 (16%) within the linker domain, and 11 variants (22%) occurred within the C-terminal transactivation domain (Figure 3). In total, 27 unique variants were identified which have not been previously reported in the LOVD database (Table 1: variants in bold).
Figure 2.
Distribution of PAX6 variants. (A) Frequency of pathological variants identified in the current study located in each exon and intron of the PAX6 gene (N=51). (B) Frequency of variants complied from the PAX6 Allelic Variant Database as of June 2, 2015 (N=623). Variants classified based on nature of sequence alteration.
Figure 3.
Summary of allelic variants predicted to affect each protein domain. Illustration of the PAX6 protein with summary statistics of the variants in the current study predicted to affect protein structure in (A) the paired domain, (B) the linker domain (C) the homeodomain, and (D) the transactivation (PST) domain.
DISCUSSION
In the group of individuals with a diagnosis of classical aniridia analyzed here, aniridia-associated alterations in the PAX6 gene were identified in 61 out of 66 families (76 out of 81 individuals). These changes included complete (8 instances) or partial deletion (2 instances) of the PAX6 gene, introduction of a premature termination codon into the open reading frame of PAX6 (47 instances), and likely addition of residues to the C-terminal end of the PAX6 protein (3 instances). These changes are all predicted to reduce PAX6 function. Additionally, an intronic deletion of unknown functional significance was detected in one case of sporadic aniridia (1 family), but not in unaffected relatives. Overall, the distribution of the variants identified in the current study demonstrates similar characteristics when compared with the LOVD database (Figure 2a, 2b), except that the comparative frequency of frame-shifting insertions or deletions is higher in this study. These results confirm the primacy of PAX6 haploinsufficiency as causal for aniridia.
No pathological sequence changes were observed in 5 families with aniridia. At this time, these individuals appear clinically indistinguishable from others with aniridia who have known PAX6 mutation (Figure 1, data not shown). Three of these individuals are children and it is possible that clinical differences will emerge as they get older. These persons could harbor mutations in other genes involved in anterior eye development, such as FOXC1 or PITX3, among others, or they may have mutations in PAX6 regulatory elements. We are currently exploring these possibilities.
The major mutational mechanisms underlying genetic variation are single nucleotide substitutions (50–52), small DNA insertions and deletions (indels) ranging from 1 to 10,000 bp in length (53–56), and large scale (>10 kb) genome rearrangements (57–63). For the 61 families with aniridia-specific alterations in the PAX6 gene, single nucleotide substitutions accounted for 30/61 (49%), indels accounted for 23/61 (38%), and gross deletion of the PAX6 locus accounted for 8/61 (13%) of instances. These results demonstrate that single nucleotide substitutions and indels in PAX6 are a significant source of aniridia-causal mutations. Of the single nucleotide substitutions, 13/30 (43%) occurred at CpG dinucleotides, a structure known for its high mutability in the human genome (64, 65) and to be associated with significant mutation hotspots in PAX6 (66, 67). Consistent with previous reports (66, 67), nonsense mutations in exons 8, 10 and 11 occurred at CpG locations that created arginine CGA codons Arg203 (c.607C>T, 2 instances), Arg261 (c.781C>T, 4 instances) and Arg317 (c.949C>T, 3 instances), respectively. Additionally, a CpG located at the 3′ exon/intron junction of exon 6 was mutated in four families (c.357+1G>A, 3 instances; c.357+1G>T, 1 instance). The frequency of indels is notable as it is about twofold more than has been reported for the genome at large (56). Since indels result from nonhomologous end joining as a result of DNA break repair, the high frequency of indels in PAX6 suggests that the 11p13 region of human chromosome 11 may be unstable relative to other regions of the genome. Alternatively, the frequency of indels in the human genome may currently be underreported due to technical issues with detecting these types of changes throughout the genome (68). Indels have been reported to occur preferentially in the male germline (69), and may be useful in tracking inheritance.
Analysis of the different types of mutations found in this study, in conjunction with those previously reported in the LOVD database can help to differentiate possible functional implications of the nature of mutations, in the context of phenotype analysis as well as potential genetic treatments. This requires consideration of individual mutations in the context of molecular and cellular mechanisms that act on mutant gene products. Mutations that lead to a PTC give rise to mRNA transcripts, which are putative targets of nonsense-mediated mRNA decay (NMD). NMD is an RNA-surveillance pathway which recognizes and degrades transcripts with PTCs upstream of the last exon-junction complex (EJC) during the primary round of translation (70). This mechanism serves to prevent the translation of truncated proteins with possible dominant negative effects, and is expected to target all transcripts with a PTC upstream of the last EJC regardless of the nature of the causal mutation. In patients with mutations fitting these criteria, degradation of transcripts from the mutant PAX6 allele should result in PAX6 haploinsufficiency. However, PAX6 transcripts harboring mutations upstream of the coding region of PAX6, those causal for PTCs near or downstream of the last EJC, or missense mutations would likely evade NMD and could produce dysregulated or truncated dominant negative PAX6 protein products from the mutant allele. Three cases in the current study previously mentioned as causal for C-terminus extensions of the open reading frame would fall into this category, leaving the vast majority of cases subject to this RNA surveillance pathway and ultimately leading to PAX6 haploinsufficiency. Dominant negative PAX6 proteins could interfere with proteins produced from the wild-type allele, especially if the DNA binding domain structures remain intact. Future studies will seek to compare phenotypic differences at the levels of both cellular/molecular differences and clinical presentation between patients with and without mutations identified as putative NMD targets.
In addition to contributing to our understanding of PAX6 allelic differences in the context and implications of haploinsufficiency, knowledge about the different disease-causing mutations is necessary for the development of potential therapeutic approaches to treat individuals with aniridia. Approaches that could be used include: 1) those that are cell-based coupled with genome editing or manipulation; 2) those designed to increase PAX6 expression from the normal allele, through the use of microRNAs for example; and 3) those targeted towards generating functional PAX6 protein from the mutant transcripts.
Recently, a novel therapeutic known as ataluren (formerly PTC124) has been identified as a treatment for genetically-mediated diseases which are caused by mutations leading to a PTC (71). With successful clinical applications for other diseases (72–75) as well as evidence of the efficacy of ataluren in mouse models of aniridia (76), data from the current study could serve to identify potential candidates for this treatment based on specific genetic mutations of PAX6. Though ataluren has been demonstrated to effectively promote read-through of all PTCs, it shows the most significant increase in read-through when the PTC consists of the UGA nucleotide sequence (followed in significance by UAG and then UAA) (71). The efficacy of ataluren on promoting read-through of PTCs (via START drug formulation) specifically in the PAX6 gene has recently been demonstrated in a mouse model of aniridia possessing a nonsense mutation which induces a UGA in-frame premature stop codon (76). Further, administration of this drug to mice with a mutant PAX6 allele provides demonstrable evidence for partial rescue of ocular phenotypes associated with aniridia, as a product of both systemic and topical postnatal application (76). These results are promising for the treatment of human aniridia with START therapy in patients with nonsense mutations, especially those consisting of a UGA in-frame premature stop codon. The current study has identified 14 families (20 individuals) with an UGA in-frame premature stop codon who may be good candidates for this therapy, followed by 3 families (3 individuals) with an UAG in-frame premature stop and 1 family (1 individual) with an UAA in-frame premature stop.
Results of the current study have identified new variants in the human PAX6 gene causal for aniridia and, in conjunction with previously identified mutations, serve to further the understanding of PAX6 mutations in this disorder as well as inform future studies of novel pharmacological treatments which may be beneficial for the treatment of aniridia.
Supplementary Material
Supplementary Table 1. Demographic and sequence variant data for all 66 families included in the study. This includes the number of family members sequenced and affected, sex and full sequence variations.
Supplementary Table 2. Intron and exon sequences are shown in lower and upper cases, respectively. Donor and acceptor splice sites are indicated by a slash (/). The 5′ ends of transcripts corresponding to the P0, P1, and Pα promoters are indicated with forward brackets. A reverse bracket denotes the inferred 3′-most end of the untranslated sequence of exon 13 based on ten independent transcripts obtained from eye tissues and encompasses three potential polyadenylation sites located at 573 bp, 800 bp, and 967 bp, respectively past the start of exon 13.
Acknowledgments
This research was supported by National Institutes of Health grant R01 EY017841 and an Unrestricted Grant from Research to Prevent Blindness to EEG; by Dr. Barrett Haik, Department of Ophthalmology, Hamilton Eye Institute to EEG and PAN; and grants from the Sharon Stewart Aniridia Research Trust to AMB and JDL, and grants from the Children’s Glaucoma Foundation, Vision for Tomorrow, and the Department of Cellular Biology Vision Research Fund to JDL. AMB is a Franklin Foundation Fellow and an ARCS Scholar. Ms. Amita Nawathe and Ms. Brittany Fox participated in this research through the University of Georgia Center for Undergraduate Research Opportunities. M. McDougal included data reported here as part of her thesis titled Investigating Aniridia, a Human Genetic Eye Disorder, which was submitted to the Honors Council of the University of Georgia in partial fulfillment of the requirements for her B.S. degree with highest honors and CURO scholar distinction. The authors wish to thank Ms. Jill Nerby, Director and Founder of Aniridia Foundation International, and the participants in this study for their support of this research.
Footnotes
Conflict of Interest statement:
There are no conflicts of interest for any of the authors of this study. The funders of this research had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hanson IM, Seawright A, Hardman K, et al. PAX6 mutations in aniridia. Hum Mol Genet. 1993;2:915–920. doi: 10.1093/hmg/2.7.915. [DOI] [PubMed] [Google Scholar]
- 2.Parekh M, Poli B, Ferrari S, et al., editors. Aniridia: Recent Developments in Scientific and Clinical Research. Springer International Publishing; 2015. [Google Scholar]
- 3.Pozdeyeva NA, Pashtayev NP, Lukin VP, et al. Artificial iris-lens diaphragm in reconstructive surgery for aniridia and aphakia. J Cataract Refract Surg. 2005;31:1750–1759. doi: 10.1016/j.jcrs.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 4.Grant WM, Walton DS. Progressive changes in the angle in congenital aniridia, with development of glaucoma. Am J Ophthalmol. 1974;78:842–847. doi: 10.1016/0002-9394(74)90308-0. [DOI] [PubMed] [Google Scholar]
- 5.McCulley TJ, Mayer K, Dahr SS, et al. Aniridia and optic nerve hypoplasia. Eye (Lond) 2005;19:762–764. doi: 10.1038/sj.eye.6701642. [DOI] [PubMed] [Google Scholar]
- 6.Nishida K, Kinoshita S, Ohashi Y, et al. Ocular surface abnormalities in aniridia. Am J Ophthalmol. 1995;120:368–375. doi: 10.1016/s0002-9394(14)72167-1. [DOI] [PubMed] [Google Scholar]
- 7.Tsai JH, Freeman JM, Chan CC, et al. A progressive anterior fibrosis syndrome in patients with postsurgical congenital aniridia. Am J Ophthalmol. 2005;140:1075–1079. doi: 10.1016/j.ajo.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 8.Thompson PJ, Mitchell TN, Free SL, et al. Cognitive functioning in humans with mutations of the PAX6 gene. Neurology. 2004;62:1216–1218. doi: 10.1212/01.wnl.0000118298.81140.62. [DOI] [PubMed] [Google Scholar]
- 9.Ashery-Padan R, Zhou X, Marquardt T, et al. Conditional inactivation of Pax6 in the pancreas causes early onset of diabetes. Dev Biol. 2004;269:479–488. doi: 10.1016/j.ydbio.2004.01.040. [DOI] [PubMed] [Google Scholar]
- 10.Nishi M, Sasahara M, Shono T, et al. A case of novel de novo paired box gene 6 (PAX6) mutation with early-onset diabetes mellitus and aniridia. Diabet Med. 2005;22:641–644. doi: 10.1111/j.1464-5491.2005.01469.x. [DOI] [PubMed] [Google Scholar]
- 11.Bamiou DE, Musiek FE, Sisodiya SM, et al. Deficient auditory interhemispheric transfer in patients with PAX6 mutations. Ann Neurol. 2004;56:503–509. doi: 10.1002/ana.20227. [DOI] [PubMed] [Google Scholar]
- 12.Bamiou DE, Free SL, Sisodiya SM, et al. Auditory interhemispheric transfer deficits, hearing difficulties, and brain magnetic resonance imaging abnormalities in children with congenital aniridia due to PAX6 mutations. Archives of pediatrics & adolescent medicine. 2007;161:463–469. doi: 10.1001/archpedi.161.5.463. [DOI] [PubMed] [Google Scholar]
- 13.Shimo N, Yasuda T, Kitamura T, et al. Aniridia with a heterozygous PAX6 mutation in which the pituitary function was partially impaired. Internal medicine. 2014;53:39–42. doi: 10.2169/internalmedicine.53.1184. [DOI] [PubMed] [Google Scholar]
- 14.Sisodiya SM, Free SL, Williamson KA, et al. PAX6 haploinsufficiency causes cerebral malformation and olfactory dysfunction in humans. Nat Genet. 2001;28:214–216. doi: 10.1038/90042. [DOI] [PubMed] [Google Scholar]
- 15.Peter NM, Leyland M, Mudhar HS, et al. PAX6 mutation in association with ptosis, cataract, iris hypoplasia, corneal opacification and diabetes: a new variant of familial aniridia? Clin Experiment Ophthalmol. 2013;41:835–841. doi: 10.1111/ceo.12109. [DOI] [PubMed] [Google Scholar]
- 16.Davis LK, Meyer KJ, Rudd DS, et al. Pax6 3′ deletion results in aniridia, autism and mental retardation. Hum Genet. 2008;123:371–378. doi: 10.1007/s00439-008-0484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell TN, Free SL, Williamson KA, et al. Polymicrogyria and absence of pineal gland due to PAX6 mutation. Ann Neurol. 2003;53:658–663. doi: 10.1002/ana.10576. [DOI] [PubMed] [Google Scholar]
- 18.Riccardi VM, Sujansky E, Smith AC, et al. Chromosomal imbalance in the Aniridia-Wilms’ tumor association: 11p interstitial deletion. Pediatrics. 1978;61:604–610. [PubMed] [Google Scholar]
- 19.Fischbach BV, Trout KL, Lewis J, et al. WAGR syndrome: a clinical review of 54 cases. Pediatrics. 2005;116:984–988. doi: 10.1542/peds.2004-0467. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Lauderdale JD. Analysis of Pax6 expression using a BAC transgene reveals the presence of a paired-less isoform of Pax6 in the eye and olfactory bulb. Dev Biol. 2006;292:486–505. doi: 10.1016/j.ydbio.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 21.Ashery-Padan R, Gruss P. Pax6 lights-up the way for eye development. Curr Opin Cell Biol. 2001;13:706–714. doi: 10.1016/s0955-0674(00)00274-x. [DOI] [PubMed] [Google Scholar]
- 22.Kohwi M, Osumi N, Rubenstein JL, et al. Pax6 is required for making specific subpopulations of granule and periglomerular neurons in the olfactory bulb. J Neurosci. 2005;25:6997–7003. doi: 10.1523/JNEUROSCI.1435-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins MJ, Smilinich NJ, Sait S, et al. An ordered NotI fragment map of human chromosome band 11p15. Genomics. 1994;23:211–222. doi: 10.1006/geno.1994.1479. [DOI] [PubMed] [Google Scholar]
- 24.Ericson J, Rashbass P, Schedl A, et al. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- 25.Mastick GS, Davis NM, Andrew GL, et al. Pax-6 functions in boundary formation and axon guidance in the embryonic mouse forebrain. Development. 1997;124:1985–1997. doi: 10.1242/dev.124.10.1985. [DOI] [PubMed] [Google Scholar]
- 26.Koroma BM, Yang JM, Sundin OH. The Pax-6 homeobox gene is expressed throughout the corneal and conjunctival epithelia. Invest Ophthalmol Vis Sci. 1997;38:108–120. [PubMed] [Google Scholar]
- 27.Collinson JM, Chanas SA, Hill RE, et al. Corneal development, limbal stem cell function, and corneal epithelial cell migration in the Pax6(+/−) mouse. Invest Ophthalmol Vis Sci. 2004;45:1101–1108. doi: 10.1167/iovs.03-1118. [DOI] [PubMed] [Google Scholar]
- 28.Manuel M, Price DJ. Role of Pax6 in forebrain regionalization. Brain Res Bull. 2005;66:387–393. doi: 10.1016/j.brainresbull.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Simpson TI, Price DJ. Pax6; a pleiotropic player in development. Bioessays. 2002;24:1041–1051. doi: 10.1002/bies.10174. [DOI] [PubMed] [Google Scholar]
- 30.Osumi N. The role of Pax6 in brain patterning. The Tohoku journal of experimental medicine. 2001;193:163–174. doi: 10.1620/tjem.193.163. [DOI] [PubMed] [Google Scholar]
- 31.Tang HK, Singh S, Saunders GF. Dissection of the transactivation function of the transcription factor encoded by the eye developmental gene PAX6. J Biol Chem. 1998;273:7210–7221. doi: 10.1074/jbc.273.13.7210. [DOI] [PubMed] [Google Scholar]
- 32.Singh S, Chao LY, Mishra R, et al. Missense mutation at the C-terminus of PAX6 negatively modulates homeodomain function. Hum Mol Genet. 2001;10:911–918. doi: 10.1093/hmg/10.9.911. [DOI] [PubMed] [Google Scholar]
- 33.Azuma N, Nishina S, Yanagisawa H, et al. PAX6 missense mutation in isolated foveal hypoplasia. Nat Genet. 1996;13:141–142. doi: 10.1038/ng0696-141. [DOI] [PubMed] [Google Scholar]
- 34.Azuma N, Yamaguchi Y, Handa H, et al. Missense mutation in the alternative splice region of the PAX6 gene in eye anomalies. Am J Hum Genet. 1999;65:656–663. doi: 10.1086/302529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang HK, Chao LY, Saunders GF. Functional analysis of paired box missense mutations in the PAX6 gene. Hum Mol Genet. 1997;6:381–386. doi: 10.1093/hmg/6.3.381. [DOI] [PubMed] [Google Scholar]
- 36.Hanson I, Churchill A, Love J, et al. Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations. Hum Mol Genet. 1999;8:165–172. doi: 10.1093/hmg/8.2.165. [DOI] [PubMed] [Google Scholar]
- 37.Chao LY, Mishra R, Strong LC, et al. Missense mutations in the DNA-binding region and termination codon in PAX6. Human mutation. 2003;21:138–145. doi: 10.1002/humu.10163. [DOI] [PubMed] [Google Scholar]
- 38.Aggarwal S, Jinda W, Limwongse C, et al. Run-on mutation in the PAX6 gene and chorioretinal degeneration in autosomal dominant aniridia. Mol Vis. 2011;17:1305–1309. [PMC free article] [PubMed] [Google Scholar]
- 39.Maekawa M, Iwayama Y, Nakamura K, et al. A novel missense mutation (Leu46Val) of PAX6 found in an autistic patient. Neurosci Lett. 2009;462:267–271. doi: 10.1016/j.neulet.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 40.Love J, Axton R, Churchill A, et al. A new set of primers for mutation analysis of the human PAX6 gene. Human mutation. 1998;12:128–134. doi: 10.1002/(SICI)1098-1004(1998)12:2<128::AID-HUMU8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 41.Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat Genet. 1992;2:232–239. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- 42.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Human mutation. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 43.Xu S, Han JC, Morales A, et al. Characterization of 11p14-p12 deletion in WAGR syndrome by array CGH for identifying genes contributing to mental retardation and autism. Cytogenetic and genome research. 2008;122:181–187. doi: 10.1159/000172086. [DOI] [PubMed] [Google Scholar]
- 44.Crolla JA, Cawdery JE, Oley CA, et al. A FISH approach to defining the extent and possible clinical significance of deletions at the WAGR locus. J Med Genet. 1997;34:207–212. doi: 10.1136/jmg.34.3.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lauderdale JD, Wilensky JS, Oliver ER, et al. 3′ deletions cause aniridia by preventing PAX6 gene expression. Proc Natl Acad Sci U S A. 2000;97:13755–13759. doi: 10.1073/pnas.240398797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dharmaraj N, Reddy A, Kiran V, et al. PAX6 gene mutations and genotype-phenotype correlations in sporadic cases of aniridia from India. Ophthalmic genetics. 2003;24:161–165. doi: 10.1076/opge.24.3.161.15607. [DOI] [PubMed] [Google Scholar]
- 47.Villarroel CE, Villanueva-Mendoza C, Orozco L, et al. Molecular analysis of the PAX6 gene in Mexican patients with congenital aniridia: report of four novel mutations. Mol Vis. 2008;14:1650–1658. [PMC free article] [PubMed] [Google Scholar]
- 48.Kokotas H, Petersen MB. Clinical and molecular aspects of aniridia. Clin Genet. 2010;77:409–420. doi: 10.1111/j.1399-0004.2010.01372.x. [DOI] [PubMed] [Google Scholar]
- 49.Ward AJ, Cooper TA. The pathobiology of splicing. J Pathol. 2010;220:152–163. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sachidanandam R, Weissman D, Schmidt SC, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 51.International HapMap C. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 52.International HapMap C. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber JL, David D, Heil J, et al. Human diallelic insertion/deletion polymorphisms. Am J Hum Genet. 2002;71:854–862. doi: 10.1086/342727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhangale TR, Rieder MJ, Livingston RJ, et al. Comprehensive identification and characterization of diallelic insertion-deletion polymorphisms in 330 human candidate genes. Hum Mol Genet. 2005;14:59–69. doi: 10.1093/hmg/ddi006. [DOI] [PubMed] [Google Scholar]
- 55.Mills RE, Luttig CT, Larkins CE, et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mullaney JM, Mills RE, Pittard WS, et al. Small insertions and deletions (INDELs) in human genomes. Hum Mol Genet. 2010;19:R131–136. doi: 10.1093/hmg/ddq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 58.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 59.Tuzun E, Sharp AJ, Bailey JA, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 60.McCarroll SA, Hadnott TN, Perry GH, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 61.Hinds DA, Kloek AP, Jen M, et al. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 62.Conrad DF, Andrews TD, Carter NP, et al. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 63.Kidd JM, Cooper GM, Donahue WF, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988;78:151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- 66.Prosser J, van Heyningen V. PAX6 mutations reviewed. Human mutation. 1998;11:93–108. doi: 10.1002/(SICI)1098-1004(1998)11:2<93::AID-HUMU1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 67.Tzoulaki I, White IM, Hanson IM. PAX6 mutations: genotype-phenotype correlations. BMC genetics. 2005;6:27. doi: 10.1186/1471-2156-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mills RE, Pittard WS, Mullaney JM, et al. Natural genetic variation caused by small insertions and deletions in the human genome. Genome Res. 2011;21:830–839. doi: 10.1101/gr.115907.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Messer PW, Arndt PF. The majority of recent short DNA insertions in the human genome are tandem duplications. Mol Biol Evol. 2007;24:1190–1197. doi: 10.1093/molbev/msm035. [DOI] [PubMed] [Google Scholar]
- 70.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 71.Welch EM, Barton ER, Zhuo J, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 72.Sermet-Gaudelus I, Boeck KD, Casimir GJ, et al. Ataluren (PTC124) induces cystic fibrosis transmembrane conductance regulator protein expression and activity in children with nonsense mutation cystic fibrosis. Am J Respir Crit Care Med. 2010;182:1262–1272. doi: 10.1164/rccm.201001-0137OC. [DOI] [PubMed] [Google Scholar]
- 73.Bushby K, Finkel R, Wong B, et al. Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle Nerve. 2014;50:477–487. doi: 10.1002/mus.24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kerem E, Konstan MW, De Boeck K, et al. Ataluren for the treatment of nonsense-mutation cystic fibrosis: a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Respir Med. 2014;2:539–547. doi: 10.1016/S2213-2600(14)70100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilschanski M, Miller LL, Shoseyov D, et al. Chronic ataluren (PTC124) treatment of nonsense mutation cystic fibrosis. Eur Respir J. 2011;38:59–69. doi: 10.1183/09031936.00120910. [DOI] [PubMed] [Google Scholar]
- 76.Gregory-Evans CY, Wang X, Wasan KM, et al. Postnatal manipulation of Pax6 dosage reverses congenital tissue malformation defects. J Clin Invest. 2014;124:111–116. doi: 10.1172/JCI70462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Demographic and sequence variant data for all 66 families included in the study. This includes the number of family members sequenced and affected, sex and full sequence variations.
Supplementary Table 2. Intron and exon sequences are shown in lower and upper cases, respectively. Donor and acceptor splice sites are indicated by a slash (/). The 5′ ends of transcripts corresponding to the P0, P1, and Pα promoters are indicated with forward brackets. A reverse bracket denotes the inferred 3′-most end of the untranslated sequence of exon 13 based on ten independent transcripts obtained from eye tissues and encompasses three potential polyadenylation sites located at 573 bp, 800 bp, and 967 bp, respectively past the start of exon 13.