Abstract
TMPRSS2-ERG gene fusions that occur frequently in human prostate cancers can be generated either through insertional chromosomal rearrangement or by intrachromosomal deletion. Genetically, a key difference between these two mechanisms is that the latter results in deletion of a ~3Mb interstitial region containing genes with unexplored roles in prostate cancer. In this study, we characterized two mouse models recapitulating TMPRSS22-ERG insertion or deletion events in the background of prostate-specific PTEN deficiency. We found that only the mice which lacked interstitial region developed prostate adenocarcinomas marked by poor differentiation and epithelial-to-mesenchymal transition. Mechanistic investigations identified several interstitial genes, including Ets2 and Bace2, whose reduced expression correlated in the gene homologs in human prostate cancer with biochemical relapse and lethal disease. Accordingly, PTEN-deficient mice with prostate-specific knockout of Ets2 exhibited marked progression of prostate adenocarcinomas that was partly attributed to activation of MAPK signaling. Collectively, our findings established that Ets2 is a tumor suppressor gene in prostate cancer and its loss along with other genes within the TMPRSS2-ERG interstitial region contributes to disease progression.
Keywords: TMPRSS2-ERG gene fusion, interstitial deletion, tumor suppressor, mouse model, ETS2
Introduction
Over half of Prostate-Specific Antigen (PSA)-screened prostate cancer (PCa) patients possess gene fusions involving members of the ETS transcription factor family (1, 2). In the United States, this translates to more than 120,000 men diagnosed with ETS fusion-positive PCa each year. The most common subtype of ETS fusion juxtaposes the androgen-regulated promoter of serine protease gene TMPRSS2 with the coding region of an ETS gene, ERG, leading to ERG overexpression. Both TMPRSS2 and ERG genes are located on human chromosome 21q22 and are separated by a 3Mb interstitial region. The TMPRSS2-ERG gene fusion is predominantly generated either through intrachromosomal deletion of this interstitial region (referred to as “deletion”) or via chromosomal rearrangement through insertion of the interstitial region (referred to as “insertion”) (3–6). As a whole, ERG rearrangements are believed to be a PCa-specific event (7, 8), yet their prognostic value to date remains controversial (9). Numerous studies found no association between TMPRSS2-ERG fusions and tumor grade, stage, Gleason score, PSA recurrence or mortality (9–12). A recent review and meta-analysis both have exhaustively summarized literature to date demonstrating the overall lack of consistent prognostic value for TMPRSS-ERG fusion in PCa (9, 12). Differences in patient cohorts, diagnosis method, and fusion detection method may account for these conflicting results. Nonetheless, it is important to note that since TMPRSS2-ERG fusions are thought to represent an early event in prostate tumorigenesis (2, 8), other oncogenic events that drive PCa progression to later stages may also contribute to differential clinical outcomes for patients with fusion-positive PCa. In addition, another important consideration is that TMPRSS2-ERG rearrangements are genetically distinct and may have different biological consequences. Various subclasses of TMPRSS2-ERG fusions have largely been grouped together in the literature, thus potentially masking their true prognostic value. For instance, it has been suggested that PCas with TMPRSS2-ERG fusions generated through deletion represent more aggressive cases than those with fusions formed via insertion (5). Furthermore, it was reported that patients with duplicated copies of deletion-generated fusions exhibited extremely poor survival (13). Lastly, in a rapid autopsy cohort, TMPRSS2-ERG fusions in fusion-positive metastases from patients with hormone-refractory PCa were uniformly generated through deletion (14). Collectively, it remains unclear whether interstitial deletion, which deletes one copy of the intervening genes between TMPRSS2 and ERG, represents a separate genetic event that contributes to prostate tumorigenesis, and whether deletion alone or in conjunction with TMPRSS2-ERG fusion expression correlates with a more aggressive subtype of fusion-positive PCa. A better understanding of this may reveal novel tumor suppressor(s) in the interstitial region and enhance our understanding of the underlying biology of aggressive PCa.
The TMPRSS2-ERG interstitial region contains ~16 known protein-encoding genes (Supplementary Fig. S1), several of which have reported tumor-suppressive roles. A gene encoding another ETS factor, ETS2, is located in this region and its ectopic expression decreased proliferation and invasion of PCa cell lines (15, 16). HMGN1, which binds to and maintains an open chromatin configuration, plays a critical role in DNA repair; its loss led to increased N-Cadherin expression (17), a gene associated with high-grade PCa (18). In addition, MX1 encodes an interferon-inducible GTPase MxA and its ectopic expression drastically reduced the motility and invasiveness of an aggressive subline of PC3 PCa cells (19). Together, these studies suggest that deletion of one or more genes within this region may lead to haploinsufficiency or loss of expression, which may promote PCa progression.
To study the role of TMPRSS2-ERG fusions in PCa, we recently generated two knockin mouse models that recapitulate either TMPRSS2-ERG insertion or deletion events (20). Notably, prostate-specific activation of both conditional knockin alleles leads to expression of the same fusion transcript as the predominant TMPRSS2-ERGα subtype originally described by Tomlins et al (1), and the only genetic difference is that one model has deletion of the interstitial region, whereas the other has no deletion. Importantly, these interstitial regions are syntenic between human and mouse with almost all known protein-encoding genes being conserved (Supplementary Fig. S1). Thus, these models offer a unique opportunity to study whether the interstitial deletion also represents a genetic event that contributes to prostate tumorigenesis. In this study, we compared the prostate phenotypes of these two models under the background of Probasin-Cre (Pb-Cre)-mediated biallelic Pten inactivation, and provided genetic evidence that reduced expression of the interstitial genes, in particular, ETS2, promotes PCa progression. We also explored associations of the interstitial genes in PCa cohorts to test their clinical relevance.
Materials and Methods
Mouse strains, procedures, and tissue preparation
T-ERG and T-3Mb-Erg knockin mice were generated previously (20). Pb-Cre (Pb-Cre4) transgenic mice were obtained from the Mouse Models of Human Cancers Consortium (MMHCC) repository. Rosa26-STOP-YFP (R26YFP) conditional Cre-reporter mice, Pten and Ets2 conditional knockout mice (PtenL/L, Ets2L/L) were obtained from The Jackson Laboratory (JAX). All mice were maintained on a mixed genetic background and housed in pathogen-free barrier environment. All mouse studies were approved by the Institutional Animal Care and Use Committee (IACUC). Prostate tissues used for immunohistochemistry (IHC) were fixed for 16 hours in 10% formalin (Fisher), dehydrated, and embedded in paraffin. Tissues used for immunofluorescent (IF) staining were fixed in 10% formalin for 1 hour, washed in PBS, then saturated in 30% sucrose overnight at 4°C. Tissues were then embedded in OCT compound (Sakura) and stored at −80°C prior to cryosectioning.
Histology, immunohistochemistry and immunofluorescent staining
Paraffin-embedded tissues were stained with H&E and reviewed by a trained rodent histopathologist. Pathology was defined as previously described (21, 22). IHC was carried out by rehydrating sections, performing antigen unmasking with Tris-EDTA buffer. Sections were blocked with 2.5% goat serum for 1 hour at room temperature and incubated with primary antibodies overnight at 4°C. Antibodies for IHC were used to detect ERG (Epitomics 2805), AR (Santa Cruz sc816), SMA (Sigma A2547), p63 (Chemicon MAB4135), E-Cadherin (Cell Signaling 3195), Vimentin (abcam 92547), ETS2 (Santa Cruz sc351), HMGN1 (abcam 5212), or BACE2 (abcam 5670). Staining was visualized using DAB substrate (Vector) and counter-stained with hematoxylin. Slides were dehydrated and sealed using Permount mounting media (Fisher). For IF staining, cryosections of prostate tissues were sectioned at 8μm, blocked in 2.5% goat serum, and incubated with primary antibodies overnight at 4°C. Antibodies for IF were used to detect YFP (abcam 13970), Vimentin (abcam 92547), K5 (Covance PRB-160P) or K8 (Covance MMS-162P). Alexa Fluor-conjugated secondary antibodies (Life Technologies) were incubated for 1 hour at room temperature. Nuclei were counterstained with DAPI and slides were sealed with Vectashield mounting media (Vector).
Laser capture microdissection and gene expression profiling
Paraffin-embedded prostates were stained with hematoxylin and excised using the ArcturusXT Laser Capture Microdissection system to collect genomic DNA or total RNA. DNA was isolated using the Arcturus PicoPure kit (Life Technnologies) and RNA was isolated using the RNeasy FFPE kit (Qiagen). Nucleic acid quality was validated on a BioAnalyzer (Agilent) and samples were processed using the Ovation RNA Amplification System (NuGEN) prior to gene expression profiling with the Affymetrix Mouse Gene 2.0 ST chip.
Prostate regeneration assays
In vivo prostate regeneration assays were performed as previously described (23). Briefly, prostate epithelial cells were dissociated and MACS-sorted (Miltenyi Biotec) to removed Lineage positive cells (CD31, CD45, Ter119). Cre-naiive cells used for PCa initiation studies were infected with CMV-Cre adenovirus (50 MOI, from University of Iowa Gene Transfer Core) for 1 hour at 37°C. Approximately 2.5 x 105 sorted cells were mixed with 2.5 x 105 Urogenital sinus mesenchyme (UGSM) cells and subcutaneously injected with Matrigel into flanks of Rag2-deficient mice. Outgrowths were collected 8 weeks post injection.
Cell culture, shRNA knockdown and Western blot
VCaP cell line was purchased from American Type Culture Collection (<5 years ago) and was authenticated again internally by Short Tandem Repeat (STR) profiling test; it was free from mycoplasma contamination and was cultured in DMEM medium with 10%FBS. VCaP cells were stably transduced by pGIPZ lentiviruses with shRNAs for ETS2 [shETS2_1: V3LHS_646035; shETS2_2: V3LHS_642164, or with control shRNA (empty vector pGIPZ shRNA, #RHS4351), from Open Biosystems]. Western blot was performed as previously described (24) on lysates were collected 72 hours post-infection. Antibodies used include, from Cell Signaling: pMET (3077), total MET (3127), pERK1/2 (4370), total ERK1/2 (9102), GAPDH (2118), and from Santa Cruz: ETS2 (sc351).
Data analysis
Hierarchical clustering and heatmaps were generated using MultiExperiment Viewer. Statistical significance was calculated using Student’s t-test. GSEA was performed as described (25). Analysis of human data was performed using cbio-portal.
For the PHS and HPFS analysis, we utilized data from a gene expression profiling study. Briefly, men were sampled from the HPFS and PHS Prostate Tumor Cohort using an extreme case design, which includes 119 men who died of their cancer or developed bony or distant metastases (“lethal”) and 282 men who lived at least 8 years after PCa diagnosis and were not diagnosed with metastases through 2012 (“indolent”). Briefly, RNA was extracted from tissue, then amplified using the WT-Ovation FFPE System V2 (NuGEN), a whole transcriptome amplification system that allows for complete gene expression analysis from archives of FFPE samples. After reverse transcription and fragmentation, the cDNA was hybridized to the Affymetrix GeneChip Human Gene 1.0 ST microarray. After normalization of the data, we mapped gene names to Affymetrix transcript cluster IDs using the NetAffx annotations as implemented in Bioconductor annotation package pd.hugene.1.0.st.v1. We compared the expression of the interstitial genes across the lethal and indolent cases using linear regression.
Accession numbers
The microarray expression profiling data set generated in this manuscript has been deposited to the GEO database under the following accession number: GSE63070.
Results
Tmprss2-Erg fusion with interstitial deletion more strongly cooperates with Pten-loss
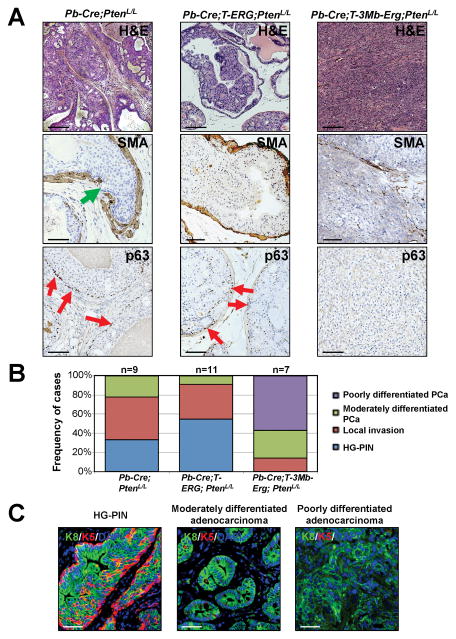
We previously reported generation and characterization of two knockin mouse models of Tmprss2-Erg fusions for PCa (20). In the insertion model, an N-terminus truncated human ERG cDNA was knocked-in to the mouse Tmprss2 locus directly (Tmprss2-STOP-ERG, Cre-mediated excision of the floxed STOP cassette leads to activation of the Tmprss2-ERG fusion allele; these are collectively referred to as “T-ERG” knockin). The second model utilized sequential gene targeting to introduce loxP sites into the mouse Tmprss2 and Erg loci, respectively, thus allowing deletion of the entire interstitial region upon Cre recombinase expression (i.e., referred to as “T-3Mb-Erg” before Cre-mediated excision of the interstitial region, and “T-Δ-Erg” after excision). In the presence of a single copy of Pten-null allele, both lines expedited formation of low-grade Prostatic Intraepithelial Neoplasia (LG-PIN) at similar rates and exhibited indistinguishable phenotypes that rarely developed into higher grade lesions, but never to frank adenocarcinoma (20). In the presence of biallelic Pten inactivation mediated by Pb-Cre, however, most Pb-Cre;T-3Mb-Erg;PtenL/L male mice developed large poorly differentiated prostate tumors (i.e., T-Δ-Erg/Pten-null tumors) in dorsolateral and ventral lobes by 12 months of age (20). To determine whether this advanced PCa phenotype is caused by Tmprss2-Erg fusion expression or interstitial deletion, or both, we similarly generated Pb-Cre;T-ERG;PtenL/L males and characterized their prostate phenotype at 12 months of age. Intriguingly, none of the Pb-Cre;T-ERG;PtenL/L males developed poorly differentiated prostate adenocarcinoma at this age (Fig. 1A–B) [compared to almost 100% penetrance for Pb-Cre;T-3Mb-Erg;PtenL/L males to develop poorly and/or moderately differentiated adenocarcinomas (Fig. 1B)]. The prostate lesions developed in these males were quite similar to those high-grade PIN (HG-PIN) lesions often observed in Pb-Cre;PtenL/L control males at this age (Fig. 1A). Furthermore, control Pb-Cre;PtenL/L males and Pb-Cre;T-ERG;PtenL/L males only displayed signs of local invasion, which was confirmed through IHC staining of smooth muscle actin (SMA) (Fig. 1A). In stark contrast, Pb-Cre;T-3Mb-Erg;PtenL/L males developed invasive prostate tumors that lacked expression of SMA and basal marker p63 (Fig. 1A). Such T-Δ-Erg/Pten-null tumors were positive for the prostate luminal epithelial marker Keratin 8 (K8), but were negative for the basal epithelial marker Keratin 5 (K5) (Fig. 1C). These Pb-Cre;T-3Mb-Erg;PtenL/L males also developed typical HG-PIN lesions composed of K8+ luminal cells surrounded by K5+ basal cells that are often observed in the control Pb-Cre;PtenL/L males (Fig. 1C), as well as localized invasive cancers with microducts mainly composed of K8+ prostate luminal cells [with almost no K5+ basal cells (Fig. 1C), and almost no SMA and p63 expression (Supplementary Fig. S2A)]. Poorly differentiated adenocarcinomas were never observed in control Pb-Cre;PtenL/L or Pb-Cre;T-ERG;PtenL/L males, whereas presence of invasive microducts consistent with moderately differentiated adenocarcinoma were only infrequently detected in these controls (Fig. 1B).
Fig. 1. Tmprss2-Erg gene fusion generated through interstitial deletion more strongly cooperates with Pten-loss.
(A) Representative haematoxylin and eosin (H&E) staining (top row), SMA IHC staining (middle row), and p63 IHC staining (bottom row) of prostate sections from mice with the indicated genotypes. Green arrow denotes discontinuous SMA staining and emergence of epithelial cells through basement membrane. Loss of basal marker p63 was used to validate adenocarcinoma. Red arrows denote p63-expressing (p63+) basal cells. Scale bars are 100μm for H&E and 50μm for SMA and p63 staining.
(B) Graphical summary of dominant histological lesions observed in aged mouse models of Tmprss2-Erg fusions with (T-3Mb-Erg) or without (T-ERG) interstitial deletion.
(C) Progressive lesions developed in Pb-Cre;T-3Mb-Erg;PtenL/L mice. IF staining for luminal marker K8 (green), basal marker K5 (red), and DAPI (blue) showing progress loss of K5+ basal cells in moderately and poorly differentiated adenocarcinomas. Scale bars are 50μm.
Adenocarcinomas and HG-PINs developed in Pb-Cre;T-3Mb-Erg;PtenL/L mice exhibit Cre-mediated interstitial deletion
By IHC staining, we confirmed that ERG protein was robustly expressed in HG-PIN lesions developed in both Pb-Cre;T-ERG;PtenL/L and Pb-Cre;T-3Mb-Erg;PtenL/L males (Supplementary Fig. S2B). We previously reported that in Pb-Cre;T-3Mb-Erg;PtenL/L male mice, moderately differentiated invasive cancers with microducts developed in their prostates often exhibited a mosaic pattern of ERG protein expression, whereas poorly differentiated invasive adenocarcinomas were largely negative for ERG (20). To rule out a possibility in which lack of ERG expression is due to reduced efficiency in generating Tmprss2-Erg fusion from Cre-mediated deletion of the large 3Mb interstitial region in the conditional T-3Mb-Erg allele, we performed laser capture microdissection to isolate epithelial cells from well-defined regions of either HG-PIN (mainly ERG positive) or poorly differentiated adenocarcinoma (tumor, mainly ERG negative) (Supplementary Fig. S3). We then extracted genomic DNA from these isolated tissues as well as from the whole prostates (i.e., containing both ERG positive and negative lesions) and by genomic DNA PCR analysis (Supplementary Fig. S4A), we verified that in both types of tissues, the Tmprss2-Erg gene fusion was generated effectively via Cre-mediated interstitial deletion (Supplementary Fig. S4B). This data suggests that advanced PCa lesions observed in the Pb-Cre;T-3Mb-Erg;PtenL/L model are most likely due to the interstitial deletion between the Tmprss2 and Erg loci.
T-Δ-Erg/Pten-null tumors exhibit an EMT phenotype
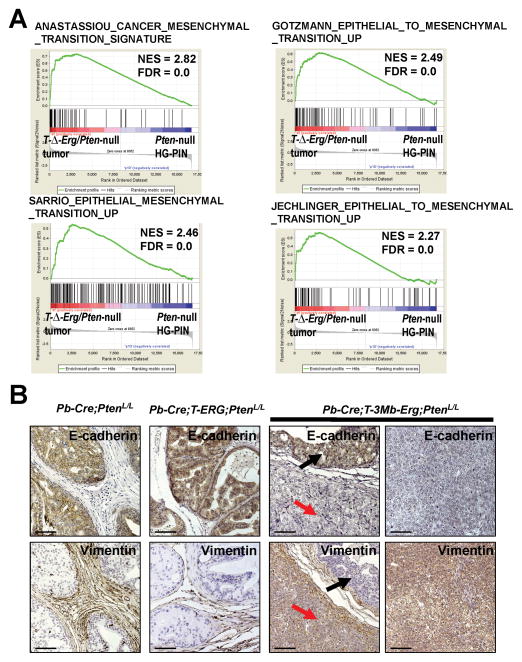
To investigate the biological mechanism underlying the more aggressive T-ΔErg/Pten-null tumors, we performed gene expression profiling using laser capture microdissected prostate epithelial cells. Only cells with round epithelial-like morphology were excised leaving behind spindle-shaped mesenchymal cells. As an internal control for stage-specific differences between cancer lesions developed in control Pb-Cre;PtenL/L males (i.e., mainly HG-PIN) and Pb-Cre;T-3Mb-Erg;PtenL/L males [i.e., poorly differentiated adenocarcinoma (tumor) and HG-PIN], we also excised HG-PIN lesions (in addition to tumors) from Pb-Cre;T-3Mb-Erg;PtenL/L mice for analysis (Supplementary Fig. S3). Among genes differentially regulated between these three groups, gene set enrichment analysis (GSEA) (25) revealed that multiple previously defined epithelial-to-mesenchymal transition (EMT) gene sets were enriched in T-Δ-Erg/Pten-null tumors compared to control Pten-null HG-PIN lesions (Fig. 2A and Supplementary Fig. S5A). Enrichment of EMT gene sets could also be found when comparing T-Δ-Erg/Pten-null HG-PIN lesions to Pten-null HG-PIN lesions (Supplementary Fig. S5B), or when comparing T-Δ-Erg/Pten-null tumors to HG-PIN lesions developed in the same mice (Supplementary Fig. S5C). These analyses suggested that the EMT signature in the T-Δ-Erg/Pten-null tumors was not simply due to a tumor stage difference (i.e., poorly differentiated adenocarcinoma versus HG-PIN), but was acquired progressively during PCa progression when under the interstitial deletion background. We validated the EMT signature using IHC for E-Cadherin and Vimentin, which display epithelial and stromal compartment-restricted expression, respectively (Fig. 2B). E-Cadherin was highly expressed in the epithelial compartment in HG-PIN lesions in Pb-Cre;PtenL/L, Pb-Cre;T-ERG;PtenL/L and Pb-Cre;T-3Mb-Erg;PtenL/L mice, but was downregulated in T-ΔErg/Pten-null tumors. Inversely, Vimentin displayed stromal-specific expression in all HG-PIN lesions yet was abundant within epithelial cells of the T-Δ-Erg/Pten-null tumors. In these Pb-Cre-based mice, a conditional Cre-reporter Rosa26-STOP-YFP (R26Y, Cre-mediated excision of a floxed STOP cassette in this allele leads to activation of the YFP reporter) was included to track Pb-Cre-expressing cells and their daughter cells (i.e., YFP+ prostate epithelial cells). The presence of EMT features was also verified in IF analyses where the epithelial marker K8 and the lineage marker YFP (for genetic marking) overlapped with Vimentin only in tumor cells but not in HG-PIN lesions (Supplementary Fig. S5D). This data suggested that although the EMT program was already upregulated in T-Δ-Erg/Pten-null HG-PIN lesions at the transcript level, changes in the expression of key EMT markers at the protein level appeared at later tumor stages. Lastly, to rule out a possibility in which the poorly differentiated tumors with mesenchymal features developed in Pb-Cre;T-3Mb-Erg;PtenL/L mice were due to a desmoplastic response in the stroma (as a response to invasive PCa developing nearby), similar to what was reported for the Pb-Cre;T-ETV1;PtenL/L mouse model (20), we stained these tumors for YFP expression and found that they were indeed YFP+ (Supplementary Fig. S5E), thus confirming that these large poorly differentiated T-ΔErg/Pten-null tumors were derived from Pb-Cre-mediated recombination and therefore of epithelial origin.
Fig. 2. T-Δ-Erg/Pten-null tumors exhibit an EMT phenotype.
(A) GSEA results showing highly significant [FDR (i.e., FDR q-val)<0.25] enrichment of multiple EMT gene sets in T-Δ-Erg/Pten-null tumors in relation to HG-PIN lesions in Pb-Cre;PtenL/L control males. The gene sets are from the c2 CGP (chemical and genetic perturbations) collection of MSigDB (http://www.broadinstitute.org/gsea/msigdb/index.jsp).
(B) IHC confirmation of EMT features in T-Δ-Erg/Pten-null tumors using E-cadherin (top rows) and Vimentin (bottom rows) staining. Control Pten-null and T-ERG/Pten-null HG-PINs (left panels), T-Δ-Erg/Pten-null HG-PIN (middle panel, black arrows), and T-Δ-Erg/Pten-null tumors (middle panel, red arrows; right panel) are shown. Scale bars are 50 μm.
T-Δ-Erg/Pten-null tumors retain partial AR signaling
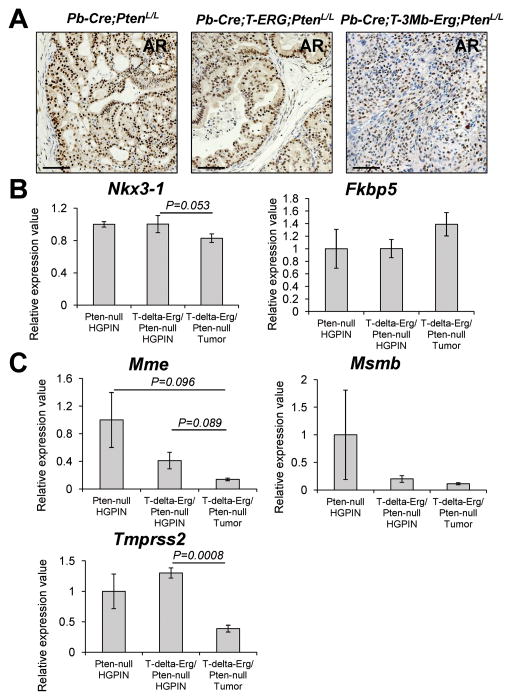
Similar to the Pb-Cre;T-3Mb-Erg;PtenL/L model, mice with Pb-Cre-induced Pten-loss and RAS/MAPK activation also develop poorly differentiated prostate tumors with an EMT phenotype (i.e., Ras/Pten tumors) (26). These Ras/Pten tumors exhibited highly heterogeneous androgen receptor (AR) expression and overall reduced expression of AR target genes (26). To determine the status of AR signaling in T-Δ-Erg/Pten-null prostate tumors, we stained them for AR protein expression and found that T-ΔErg/Pten-null tumors are largely AR positive (Fig. 3A). By analyzing microarray expression data, we found that expression of Nkx3-1, a well-known AR target and tumor suppressor gene, was not significantly changed in T-Δ-Erg/Pten-null tumors or HG-PIN lesions compared to Pten-null HG-PIN lesions; Fkbp5, a recently described AR target gene that mediates reciprocal inhibition between PTEN/PI3K and AR pathways (27, 28), was slightly upregulated in T-Δ-Erg/Pten-null tumors compared to Pten-null HG-PINs (Fig. 3B). In contrast, these two AR target genes both exhibited a trend of downregulation in Ras/Pten tumors compared to Pten-null lesions [Supplementary Fig. S6A and (26)]. However, several other AR target genes related to normal prostate function, including Mme, Msmb and Tmprss2, all exhibited a trend of downregulation in both T-Δ-Erg/Pten-null and Ras/Pten tumors in relation to Pten-null lesions [Fig. 3C, Supplementary Fig. S6A and (26)]. Of note, downregulation of Tmprss2 in T-Δ-Erg/Pten-null prostate tumors in relation to both T-Δ-Erg/Pten-null and Pten-null HG-PIN lesions may explain downregulation of Erg expression (which is under the control of the endogenous Tmprss2 promoter) in T-Δ-Erg/Pten-null tumors, but not in T-Δ-Erg/Pten-null HG-PIN lesions (20). Lastly, by GSEA using an androgen-driven signature we developed recently from human PCa cell lines (20), we found that this gene set exhibited a higher level of downregulation in Ras/Pten tumors than in T-Δ-Erg/Pten-null tumors, when compared to their corresponding Pten-null controls (Supplementary Fig. S6B–C). Collectively, these data suggest that compared to the Ras/Pten EMT tumors, T-ΔErg/Pten-null tumors retain expression of both AR and some of the AR target genes, but also exhibit downregulation of other AR targets. This is consistent with the concept of AR cistrome reprogramming in which AR binding sites change during PCa progression, when its collaborating factors are altered (e.g., due to different oncogenic events), leading to changes in the expression of select AR target genes (29).
Fig. 3. T-Δ-Erg/Pten-null tumors retain AR signaling partially.
(A) IHC staining depicting AR expression in typical prostate lesions from various PCa mouse models. Scale bars are 50μm.
(B–C) Relative expression values of AR target genes, Nkx3-1 and Fkbp5 (B), as well as Mme, Msmb and Tmprss2 (C), in T-Δ-Erg/Pten-null tumors and HG-PINs in relation to Pten-null HG-PINs (=1), based on microarray expression profiling. Data represent mean ± SEM. P-values (Student’s t-test) for pairwise comparisons that were significant (p<0.05) or marginally significant (p<0.1) were shown. All the other comparisons were not statistically significant.
Multiple interstitial genes are candidate prostate tumor suppressors
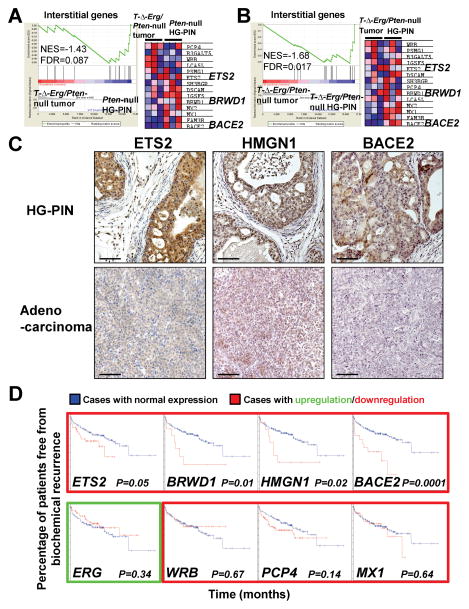
We next examined whether some interstitial genes could function as tumor suppressors during prostate carcinogenesis to explain the aggressive nature of T-ΔErg/Pten-null tumors. To do this, we first generated an “Interstitial genes” gene set composed of protein-coding genes between TMPRSS2 and ERG and then performed GSEA using this gene set. We found that it was significantly downregulated in T-ΔErg/Pten-null tumors when compared to either Pten-null HG-PINs (Fig. 4A) or T-ΔErg/Pten-null HG-PINs (Fig. 4B). Among the downregulated interstitial genes, Ets2 is abundantly expressed in prostate tissues [whereas several other ETS factor genes, such as Ets1, Erg, and Etv1, are not (Supplementary Fig. S7)] and its overexpression was shown previously to decrease proliferation and invasion of PCa cell lines (15, 16); however, other interstitial genes, such as Bace2 and Brwd1, have so far not been implicated in PCa development. We analyzed expression of interstitial genes Ets2, Hmgn1, and Bace2 in T-Δ-Erg/Pten-null HG-PIN and adenocarcinoma at the protein level (Fig. 4C). Bace2 was included as it is the most significantly downregulated gene in the above GSEA analysis (Fig. 4A–B). Although Hmgn1 failed to show up in the GSEA analysis (due to absence of Hmgn1 probe conversion), we also included it in our validation, as it has been implicated in PCa as a potential tumor suppressor (17, 18). Expression at the protein level from these genes was abundantly detected in nuclei and cytoplasm of epithelial cells in HG-PIN lesions. In advanced tumors, expression levels were dramatically decreased, although weak cytoplasmic staining could still be observed (Fig. 4C). Interestingly, in our GSEA analysis, although the “Interstitial genes” gene set was not significantly enriched in HG-PIN lesions from Pb-Cre;T-3Mb-Erg;PtenL/L mice to those from Pb-Cre;PtenL/L control mice, the above-described interstitial genes such as Ets2, Bace2 and Brwd1 all exhibited a trend toward slight downregulation in T-ΔErg/Pten-null HG-PINs (Supplementary Fig. S8 and Table S1).
Fig. 4. Multiple interstitial genes are candidate prostate tumor suppressors.
(A–B) GSEA for the “Interstitial genes” showing significant (FDR<0.25) negative enrichment (i.e., downregulation) of this gene set in T-Δ-Erg/Pten-null tumors in relation to Pten-null HG-PINs (A) or T-Δ-Erg/Pten-null HG-PINs (B). In A and B, Enrichment plots are shown on the left, heatmaps are shown on the right. In the heatmap, red to pink to light blue to blue indicate highest to lowest expression levels of indicated genes.
(C) Expression of select interstitial genes, Ets2, Hmgn1, and Bace2 were significantly lower in adenocarcinoma (bottom row) compared to HG-PIN lesions (top row) in Pb-Cre;T-3Mb-Erg;PtenL/L mice. Scale bars are 50μm.
(D) Kaplan-Meier curves of human patient data reveals that downregulation of several interstitial genes (outlined in red) predict biochemical relapse after prostatectomy (top row), whereas some do not (bottom row). ERG overexpression (outlined in green) also did not predict recurrence in this cohort. Blue lines depict patients with normal expression while patients with deregulated expression are shown as red lines for each graph.
To determine whether reduced expression of interstitial genes is associated with PCa patient outcomes, we analyzed potential association of deletion of interstitial genes with biochemical relapse after radical prostatectomy in a previously published human PCa patient cohort (30). We found that in this cohort, only 4 of 17 interstitial genes including ETS2, BRWD1, HMGN1, and BACE2 were significant for PCa progression (when downregulated), regardless of patient fusion status (Fig. 4D). Patients possessing downregulation of the remaining set of interstitial genes did not exhibit differences in biochemical recurrence compared to those with normal expression levels. Similarly, ERG overexpression was not associated with time to failure (Fig. 4D). Lastly, we examined the potential clinical association of interstitial genes with lethal PCa in a larger study with 119 lethal PCa cases, defined as men with metastatic disease or cancer death, and 282 indolent cases with no evidence of metastatic disease [from the Physicians’ Health Study (PHS) and the Health Professionals Follow-up Study (HPFS) (12, 31)]. We found that at least one-third of the interstitial genes, including ETS2 and BACE2, are expressed at significantly lower levels in lethal PCa cases than indolent cases (Table 1). Collectively, these analyses suggest that multiple genes within the TMPRSS2-ERG interstitial region are associated with lethal PCa (when downregulated) and may function as tumor suppressors for PCa.
Table 1.
Association of select interstitial genes with lethal prostate cancer, Physicians’ Health Study and Health Professionals Follow-up Study.
| Gene name | P value | Expression lower in lethal or indolent? |
|---|---|---|
| ERG | 0.482 | -- |
| ETS2 | 7.51E-05 | Lethal |
| PSMG1 | 0.01 | Indolent |
| BRWD1 | 0.07 | Indolent |
| BRWD1-IT2 (NCRNA00257) | 0.204 | -- |
| HMGN1 | 0.059 | Indolent |
| WRB | 0.656 | -- |
| LCA5L | 0.152 | -- |
| SH3BGR | 0.052 | Lethal |
| C21orf88 | 0.924 | -- |
| B3GALT5 | 0.041 | Lethal |
| IGSF5 | 0.173 | -- |
| PCP4 | 0.378 | -- |
| DSCAM | 0.128 | -- |
| BACE2 | 0.01 | Lethal |
| PLAC4 | 0.00056 | Lethal |
| FAM3B | 0.0017 | Lethal |
| MX2 | 0.875 | -- |
| MX1 | 0.132 | -- |
| TMPRSS2 | 0.538 | -- |
--no association
Reduced gene dosage of Ets2 contributes to prostate cancer progression
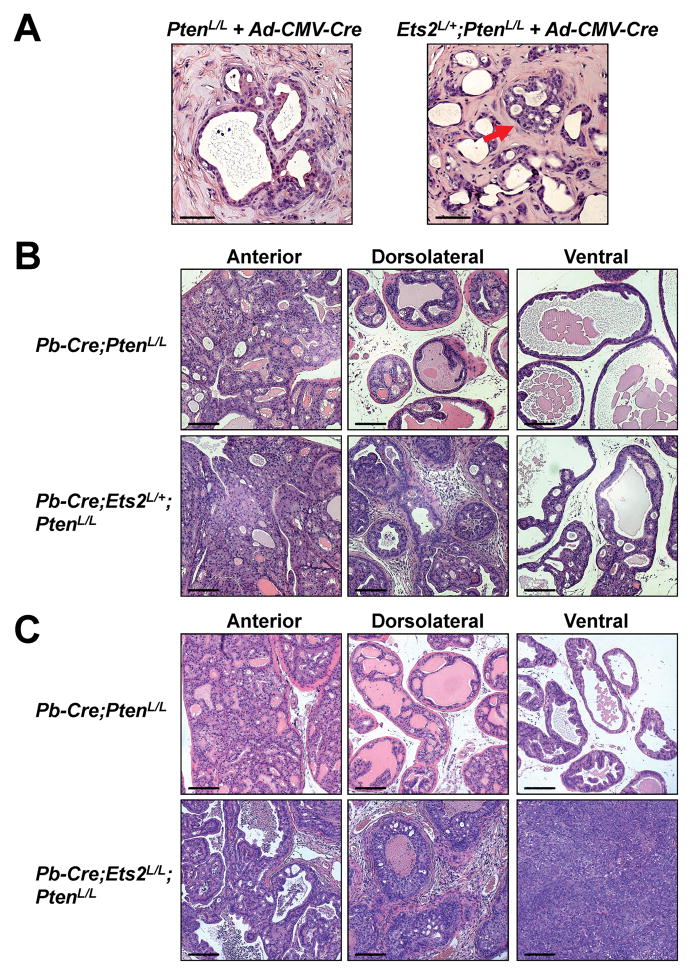
Interstitial genes exhibiting significantly lower expression levels in advanced PCa (e.g., ETS2, BACE2) can either directly contribute to prostate tumorigenesis as a tumor suppressor or serve as a biomarker for lethal PCa, or both. To distinguish between these possibilities, we focused on ETS2, which is strongly downregulated in prostate tumors compared to benign tissues [(32) and in our PHS/HPFS study (P-value=8.4x10−14)] and is also the most significantly downregulated interstitial gene in lethal PCas (Table 1). We crossed mice carrying a conditional knockout allele of Ets2 (Ets2L) (33) with PtenL/L mice to generate Ets2L/+;PtenL/L male mice. We then performed the regeneration assay upon ex vivo exposure of their prostate cells to CMV-Cre adenovirus (Ad-CMV-Cre). Ad-CMV-Cre-infected PtenL/L control cells formed largely normal ducts with occasional areas of hyperplasia (Fig. 5A). In contrast, Ad-CMV-Cre-infected Ets2L/+;PtenL/L prostate cells formed small proliferating lesions invading into the lumen, consistent with HG-PIN (Fig. 5A). Next, we generated Pb-Cre;Ets2L/+;PtenL/L, Pb-Cre;Ets2L/L;PtenL/L, and matched control Pb-Cre;PtenL/L male mice. At 6–9 months of age, we found that although the anterior lobes of all these mice developed HG-PIN lesions of comparable severity, the phenotypes from the dorsolateral and ventral lobes of the Pb-Cre;Ets2L/+;PtenL/L and Pb-Cre;Ets2L/L;PtenL/L males were notably stronger. Loss of one copy of Ets2 resulted in development of larger HG-PIN lesions in both the dorsolateral and ventral lobes and the dorsolateral lobes also contained significantly more stromal proliferation and inflammatory infiltrate (Fig. 5B). Importantly, loss of both copies of Ets2 led to emergence of poorly differentiated prostate adenocarcinoma in the ventral lobes (Fig. 5C). Collectively, these data suggest that Ets2 is a tumor suppressor in PCa and its reduced dosage in prostate epithelial cells promotes PCa progression.
Fig. 5. Reduced dosage of Ets2 contributes to PCa progression under the Pten-null background.
(A) Prostate regeneration assay using PtenL/L or Ets2L/+;PtenL/L prostate cells infected with Ad-CMV-Cre adenovirus prior to implantation. Red arrow denotes a lesion resembling HG-PIN. Scale bars are 50μm.
(B) H&E staining showing enhanced HG-PIN phenotype in a 6-month old Pb-Cre;Ets2L/+;PtenL/L male compared to its age-matched Pb-Cre;PtenL/L control male in the dorsolateral and ventral lobes. Scale bars are 100μm.
(C) H&E staining showing invasive PCa and poorly differentiated prostate adenocarcinoma phenotype in a 9-month old Pb-Cre;Ets2L/L;PtenL/L male compared to its age-matched Pb-Cre;PtenL/L control male in the dorsolateral and ventral lobes. Scale bars are 100μm.
ETS2-loss leads to activation of the MAPK pathway
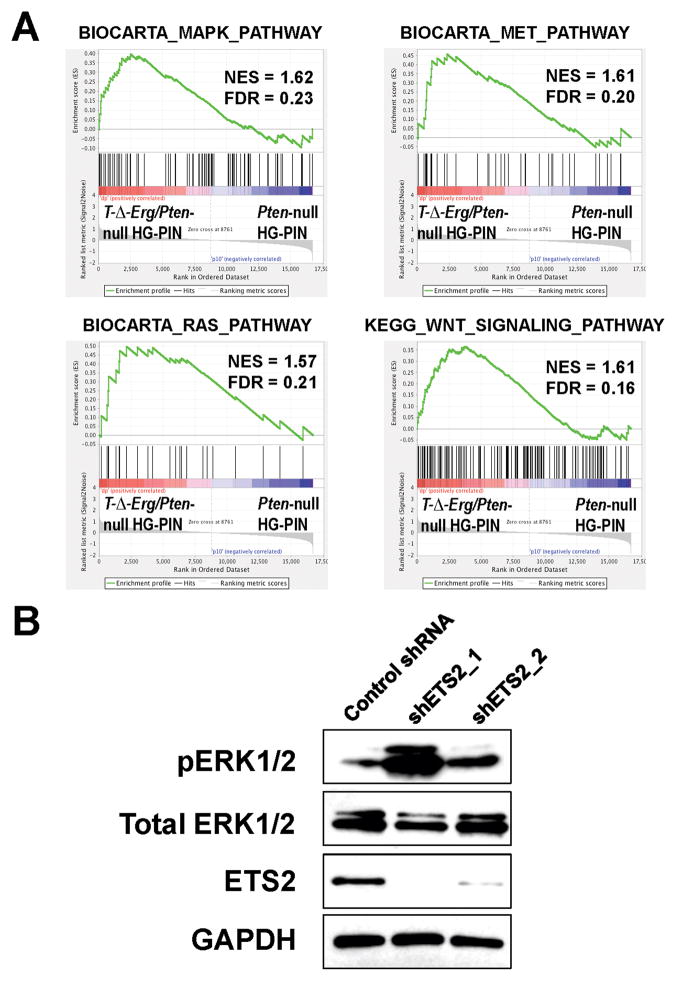
To determine how reduced Ets2 dosage contributes to PCa progression, we further analyzed the microarray data from our animal models, focusing on comparing T-Δ-Erg/Pten-null HG-PINs to Pten-null HG-PINs (thus stage-matched). By GSEA, we found that in addition to the above-described EMT-related gene sets (Supplementary Fig. S5B), gene sets related to the RAS/MAPK pathway, HGF/MET pathway, and WNT signaling, were also significantly enriched in T-Δ-Erg/Pten-null HG-PIN lesions (Fig. 6A and Supplementary Table S2). Interestingly, a recent study examining a tumor suppressor role of ETS2 in non-small cell lung cancer demonstrated association of ETS2-loss with activation of both HGF/MET and MAPK signaling, as well as induction of EMT-related genes (34). To determine whether ETS2-loss could induce similar pathway activation in PCa cells, we used shRNAs to knockdown ETS2 expression in VCaP cells, which carry the TMPRSS2-ERG fusion without the interstitial deletion (1). We found that upon ETS2 knockdown, the level of phosphorylated ERK1/2 (pERK1/2) was notably increased (Fig. 6B). Due to a low level of phosphorylated MET (pMET) in VCaP cells, we were unable to definitively determine whether reduced ETS2 expression promotes activation of the HGF/MET pathway (data not shown). Collectively, our data from both animal models and human PCa cells suggest that ETS2-loss causes activation of at least the MAPK pathway, which cooperates with Pten-loss, leading to development of advanced PCa.
Fig. 6. ETS2-loss leads to activation of the MAPK pathway.
(A) GSEA results showing significant (FDR<0.25) enrichment of gene sets related to RAS/MAPK, HGF/MET and WNT signaling in T-Δ-Erg/Pten-null HG-PIN lesions in relation to HG-PIN lesions in Pb-Cre;PtenL/L control males. Also see Supplementary Table S2 for a complete list of significantly enriched pathway-related gene sets.
(B) Western blot showing increased levels of pERK1/2 in VCaP cells upon knockdown of ETS2 by two independent shRNAs. Levels of total ERK1/2 and GAPDH were shown as loading controls.
Discussion
Our unique mouse models have allowed examination and comparison of the biological consequences of Tmprss2-Erg fusions generated by insertion or deletion. ERG overexpression in the context of Pten-loss is sufficient for early initiation events as both models similarly expedited PIN frequency and severity in the context of a single copy of Pten-loss (20). However, the difference between these two models became obvious in aged mice when under the Pten-null background. Prostate lesions in Pb-Cre;T-ERG;PtenL/L males (i.e., no deletion) were histologically similar to those in control Pb-Cre;PtenL/L males and development of invasive PCa in either line was a relatively rare event. In stark contrast, Pb-Cre;T-3Mb-Erg;PtenL/L mice (i.e., with deletion) consistently developed poorly differentiated adenocarcinomas that exhibited EMT features and partial AR signaling. Together these data suggest that interstitial deletion may program prostate cells for development of more aggressive PCa than ERG overexpression alone. However, an important consideration is the prolonged delay for the phenotypic difference between Pb-Cre;T-ERG;PtenL/L and Pb-Cre;T-3Mb-Erg;PtenL/L males to emerge. This suggests that additional mutations or epigenetic events may be required for the full T-Δ-Erg/Pten-null phenotype to manifest. Another consideration is that ERG protein expression is largely lost in the poorly differentiated T-Δ-Erg/Pten-null tumors (20), whereas the majority of human PCas with TMPRSS2-ERG fusions retain ERG overexpression (35–38). However, a small subset of human fusion-positive castration-resistant cases (e.g., small cell/neuroendocrine PCa) were also found to be negative for ERG protein staining (39). Whether our T-Δ-Erg/Pten-null model recapitulates this subset of human PCa requires further investigation. Of note, a recent mouse model of prostate-specific ERG overexpression resulted in aggressive PCa (40), rather than HG-PIN lesions as observed in our Pb-Cre;T-ERG;PtenL/L model. This phenotypic difference may be due to a difference in how ectopic ERG expression is controlled: ERG overexpression in that mouse model is controlled by the constitutive Rosa26 locus, whereas ERG overexpression in our T-ERG mouse model is driven by the endogenous Tmprss2 control region, which is under regulation by both androgen and estrogen (20, 41, 42).
Our study provided definitive genetic evidence to support ETS2 as a prostate tumor suppressor gene in the interstitial region. This is consistent with our epidemiological study demonstrating a significant association of ETS2 downregulation with lethal PCa (Table 1), as well as findings from PCa genomes in which both focal deletion and point mutation of ETS2 were identified in lethal castration-resistant PCa (16). Of note, ETS2 has also been shown as a tumor suppressor in other cancer types [e.g., colon cancer (43), lung cancer (34)]. Since both ERG and ETS2 belong to the ETS family of transcription factors that share the same or similar DNA-binding motif (44), it is possible that ectopic ERG expression may interfere with the normal function of ETS2 in prostate epithelial cells by competing with ETS2 for DNA binding. Thus, ETS factors (e.g., ETS2) normally expressed in prostate epithelial cells may serve as prostate tumor suppressors, whereas those ETS factors (e.g., ERG, ETV1, Supplementary Fig. S7) that are not expressed in prostate epithelial cells may serve as oncogenes (once ectopically expressed), by potentially interfering with functions of ETS factors normally expressed in prostate epithelial cells. This notion is consistent with the idea of an ETS transcriptional network in prostate tumorigenesis (45). Mechanistically, we showed that ETS2-loss might promote PCa progression in part via activation of the MAPK pathway. This is supported by the similar aggressive PCa phenotype (e.g., EMT) observed in both the T-Δ-Erg/Pten-null model and other mouse models with both Pten-loss and RAS/MAPK activation (26, 46).
Although our T-Δ-Erg/Pten-null model and the Ras/Pten model (26) both involve activation of the MAPK pathway, leading to development of prostate tumors with features of EMT, the EMT tumors developed in these models also exhibit difference at the molecular level (e.g., different AR activity and targets expression). Other deregulated pathways (e.g., due to ETS2-loss and/or loss of other tumor suppressor genes in the interstitial region) may contribute to this difference. In addition, although our Pb-Cre;T-3Mb-Erg;PtenL/L mice had only one copy of each interstitial gene deleted by Cre-mediated excision, the remaining alleles might also be additionally silenced through unidentified mechanisms. This might explain the lost expression of select interstitial genes (e.g., Ets2, Hmgn1, Bace2) in advanced PCa. Furthermore, recent study suggested that combinations of hemizygous deletions of multiple tumor suppressors might be required to produce maximal cancer phenotypes (47). This provides another potential mechanism for loss of multiple interstitial genes to contribute to prostate tumorigenesis jointly. Overall, although we have validated ETS2 as a major tumor suppressor in this region, we cannot exclude that other interstitial genes (e.g., BACE2) may have tumor suppressor roles as well. Future characterization of clinical samples with careful dissection of the individual and combinatorial contribution of the loss of these candidate interstitial genes, coupled with mouse modeling, may reveal additional tumor suppressor(s) in this interstitial region. Lastly, our study also suggests that clinical prognostic studies may need to consider more carefully distinguishing TMPRSS2-ERG fusions based on deletion or insertion, as they may contribute to distinct clinical outcomes.
Supplementary Material
Acknowledgments
Financial support: This research was supported by NIH grant R01 CA136578 (to L.A.M.), and by a Career Development Award from Dana-Farber/Harvard Cancer Center Prostate Cancer SPORE (P50 CA090381) and Idea Development Awards from Department of Defense (W81XWH-11-1-0329, W81XWH-15-1-0546) (to Z.L.).
We would like to thank Xin Zhang for her assistance in maintaining and genotyping some of the mouse models and Drs. Esther Baena and Ying Xie for technical assistance with experimental assays.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 2.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teixeira MR. Chromosome mechanisms giving rise to the TMPRSS2-ERG fusion oncogene in prostate cancer and HGPIN lesions. Am J Surg Pathol. 2008;32:642–4. doi: 10.1097/PAS.0b013e31815b6056. author reply 4. [DOI] [PubMed] [Google Scholar]
- 4.Hermans KG, van Marion R, van Dekken H, Jenster G, van Weerden WM, Trapman J. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer. Cancer Res. 2006;66:10658–63. doi: 10.1158/0008-5472.CAN-06-1871. [DOI] [PubMed] [Google Scholar]
- 5.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, et al. TMPRSS2:ERG Fusion-Associated Deletions Provide Insight into the Heterogeneity of Prostate Cancer. Cancer Res. 2006;66:8337–41. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 6.Yoshimoto M, Joshua AM, Chilton-Macneill S, Bayani J, Selvarajah S, Evans AJ, et al. Three-color FISH analysis of TMPRSS2/ERG fusions in prostate cancer indicates that genomic microdeletion of chromosome 21 is associated with rearrangement. Neoplasia. 2006;8:465–9. doi: 10.1593/neo.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheble VJ, Braun M, Beroukhim R, Mermel CH, Ruiz C, Wilbertz T, et al. ERG rearrangement is specific to prostate cancer and does not occur in any other common tumor. Mod Pathol. 2010;23:1061–7. doi: 10.1038/modpathol.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park K, Dalton JT, Narayanan R, Barbieri CE, Hancock ML, Bostwick DG, et al. TMPRSS2:ERG gene fusion predicts subsequent detection of prostate cancer in patients with high-grade prostatic intraepithelial neoplasia. J Clin Oncol. 2014;32:206–11. doi: 10.1200/JCO.2013.49.8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu B, Chevarie-Davis M, Chevalier S, Scarlata E, Zeizafoun N, Dragomir A, et al. The prognostic role of ERG immunopositivity in prostatic acinar adenocarcinoma: a study including 454 cases and review of the literature. Hum Pathol. 2014;45:488–97. doi: 10.1016/j.humpath.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Gopalan A, Leversha MA, Satagopan JM, Zhou Q, Al-Ahmadie HA, Fine SW, et al. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–6. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minner S, Enodien M, Sirma H, Luebke AM, Krohn A, Mayer PS, et al. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011;17:5878–88. doi: 10.1158/1078-0432.CCR-11-1251. [DOI] [PubMed] [Google Scholar]
- 12.Pettersson A, Graff RE, Bauer SR, Pitt MJ, Lis RT, Stack EC, et al. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21:1497–509. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–63. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehra R, Tomlins SA, Yu J, Cao X, Wang L, Menon A, et al. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 2008;68:3584–90. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foos G, Hauser CA. Altered Ets transcription factor activity in prostate tumor cells inhibits anchorage-independent growth, survival, and invasiveness. Oncogene. 2000;19:5507–16. doi: 10.1038/sj.onc.1203946. [DOI] [PubMed] [Google Scholar]
- 16.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubinstein YR, Furusawa T, Lim JH, Postnikov YV, West KL, Birger Y, et al. Chromosomal protein HMGN1 modulates the expression of N-cadherin. Febs J. 2005;272:5853–63. doi: 10.1111/j.1742-4658.2005.04980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaggi M, Nazemi T, Abrahams NA, Baker JJ, Galich A, Smith LM, et al. N-cadherin switching occurs in high Gleason grade prostate cancer. Prostate. 2006;66:193–9. doi: 10.1002/pros.20334. [DOI] [PubMed] [Google Scholar]
- 19.Mushinski JF, Nguyen P, Stevens LM, Khanna C, Lee S, Chung EJ, et al. Inhibition of tumor cell motility by the interferon-inducible GTPase MxA. J Biol Chem. 2009;284:15206–14. doi: 10.1074/jbc.M806324200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baena E, Shao Z, Linn DE, Glass K, Hamblen MJ, Fujiwara Y, et al. ETV1 directs androgen metabolism and confers aggressive prostate cancer in targeted mice and patients. Genes Dev. 2013;27:683–98. doi: 10.1101/gad.211011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R, et al. Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res. 2013;73:2718–36. doi: 10.1158/0008-5472.CAN-12-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JH, Walls JE, Galvez JJ, Kim M, Abate-Shen C, Shen MM, et al. Prostatic intraepithelial neoplasia in genetically engineered mice. Am J Pathol. 2002;161:727–35. doi: 10.1016/S0002-9440(10)64228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukacs RU, Goldstein AS, Lawson DA, Cheng D, Witte ON. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat Protoc. 2010;5:702–13. doi: 10.1038/nprot.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Bragt MP, Hu X, Xie Y, Li Z. RUNX1, a transcription factor mutated in breast cancer, controls the fate of ER-positive mammary luminal cells. Elife. 2014;3:e03881. doi: 10.7554/eLife.03881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012;72:1878–89. doi: 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011;19:792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mills IG. Maintaining and reprogramming genomic androgen receptor activity in prostate cancer. Nat Rev Cancer. 2014;14:187–98. doi: 10.1038/nrc3678. [DOI] [PubMed] [Google Scholar]
- 30.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettersson A, Lis RT, Meisner A, Flavin R, Stack EC, Fiorentino M, et al. Modification of the association between obesity and lethal prostate cancer by TMPRSS2:ERG. J Natl Cancer Inst. 2013;105:1881–90. doi: 10.1093/jnci/djt332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rostad K, Mannelqvist M, Halvorsen OJ, Oyan AM, Bo TH, Stordrange L, et al. ERG upregulation and related ETS transcription factors in prostate cancer. Int J Oncol. 2007;30:19–32. [PubMed] [Google Scholar]
- 33.Tynan JA, Wen F, Muller WJ, Oshima RG. Ets2-dependent microenvironmental support of mouse mammary tumors. Oncogene. 2005;24:6870–6. doi: 10.1038/sj.onc.1208856. [DOI] [PubMed] [Google Scholar]
- 34.Kabbout M, Garcia MM, Fujimoto J, Liu DD, Woods D, Chow CW, et al. ETS2 mediated tumor suppressive function and MET oncogene inhibition in human non-small cell lung cancer. Clin Cancer Res. 2013;19:3383–95. doi: 10.1158/1078-0432.CCR-13-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–8. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falzarano SM, Zhou M, Carver P, Tsuzuki T, Simmerman K, He H, et al. ERG gene rearrangement status in prostate cancer detected by immunohistochemistry. Virchows Arch. 2011;459:441–7. doi: 10.1007/s00428-011-1128-4. [DOI] [PubMed] [Google Scholar]
- 37.Chaux A, Albadine R, Toubaji A, Hicks J, Meeker A, Platz EA, et al. Immunohistochemistry for ERG expression as a surrogate for TMPRSS2-ERG fusion detection in prostatic adenocarcinomas. Am J Surg Pathol. 2011;35:1014–20. doi: 10.1097/PAS.0b013e31821e8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gopalan A, Leversha MA, Dudas ME, Maschino AC, Chang J, Al-Ahmadie HA, et al. TMPRSS2-ERG rearrangement in dominant anterior prostatic tumours: incidence and correlation with ERG immunohistochemistry. Histopathology. 2013;63:279–86. doi: 10.1111/his.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gsponer JR, Braun M, Scheble VJ, Zellweger T, Bachmann A, Perner S, et al. ERG rearrangement and protein expression in the progression to castration-resistant prostate cancer. Prostate Cancer Prostatic Dis. 2014;17:126–31. doi: 10.1038/pcan.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med. 2013;19:1023–9. doi: 10.1038/nm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Setlur SR, Mertz KD, Hoshida Y, Demichelis F, Lupien M, Perner S, et al. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst. 2008;100:815–25. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munera J, Cecena G, Jedlicka P, Wankell M, Oshima RG. Ets2 regulates colonic stem cells and sensitivity to tumorigenesis. Stem Cells. 2011;29:430–9. doi: 10.1002/stem.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. Embo J. 2010;29:2147–60. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kunderfranco P, Mello-Grand M, Cangemi R, Pellini S, Mensah A, Albertini V, et al. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PLoS ONE. 2010;5:e10547. doi: 10.1371/journal.pone.0010547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aytes A, Mitrofanova A, Kinkade CW, Lefebvre C, Lei M, Phelan V, et al. ETV4 promotes metastasis in response to activation of PI3-kinase and Ras signaling in a mouse model of advanced prostate cancer. Proc Natl Acad Sci U S A. 2013;110:E3506–15. doi: 10.1073/pnas.1303558110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solimini NL, Xu Q, Mermel CH, Liang AC, Schlabach MR, Luo J, et al. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science. 2012;337:104–9. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.