Abstract
Eukaryotic cells initiate DNA replication from multiple origins, which must be tightly regulated to promote precise genome duplication in every cell cycle. To accomplish this, initiation is partitioned into two temporally discrete steps: a double hexameric MCM complex is first loaded at replication origins during G1 phase, and then converted to the active CMG (Cdc45, MCM, GINS) helicase during S phase. Here we describe the reconstitution of budding yeast DNA replication initiation with 16 purified replication factors, made from 42 polypeptides. Origin-dependent initiation recapitulates regulation seen in vivo. Cyclin dependent kinase (CDK) inhibits MCM loading by phosphorylating the origin recognition complex (ORC) and promotes CMG formation by phosphorylating Sld2 and Sld3. Dbf4 dependent kinase (DDK) promotes replication by phosphorylating MCM, and can act either before or after CDK. These experiments define the minimum complement of proteins, protein kinase substrates and co-factors required for regulated eukaryotic DNA replication.
The initiation of eukaryotic DNA replication origin firing is understood in outline1,2, but the process has not been reconstituted with purified proteins. MCM can be loaded onto DNA with purified ORC, Cdc6 and Cdt1·MCM3,4 and loaded MCMs can be activated to replicate in yeast extracts5–7. Mass spectrometry of complexes assembled during replication in these extracts identified previously characterised ‘firing factors’ including Sld2, Sld3, Sld7, Dpb11, Cdc45, GINS and DNA polymerase ε (pol ε), but did not identify any novel factors6, suggesting this list may be complete.
DNA replication is regulated during the cell cycle by two protein kinases, CDK and DDK8. CDK plays two roles in regulating replication: it inhibits MCM loading and it is essential for helicase activation1,2. Consequently, MCM loading can only occur during G1 phase when CDK activity is low, and origins can only fire after G1 phase when CDK levels rise. CDK phosphorylation of ORC, Cdc6 and MCM all contribute to preventing MCM loading outside G1 phase in budding yeast9. Genetic analysis has indicated that Sld2 and Sld3 are the two key CDK substrates required for helicase activation. Phosphorylation of these proteins generates binding sites for tandem BRCT repeats in Dpb1110–12. DDK is required for origin firing8 and genetic analysis has indicated that Mcm4 and 6 are the key DDK substrates13–15. Until origin firing is reconstituted with purified proteins, however, we will not know whether we have the complete inventory of essential firing factors or whether CDK and DDK have any additional important substrates.
CDK- and DDK-dependent recruitment of purified firing factors
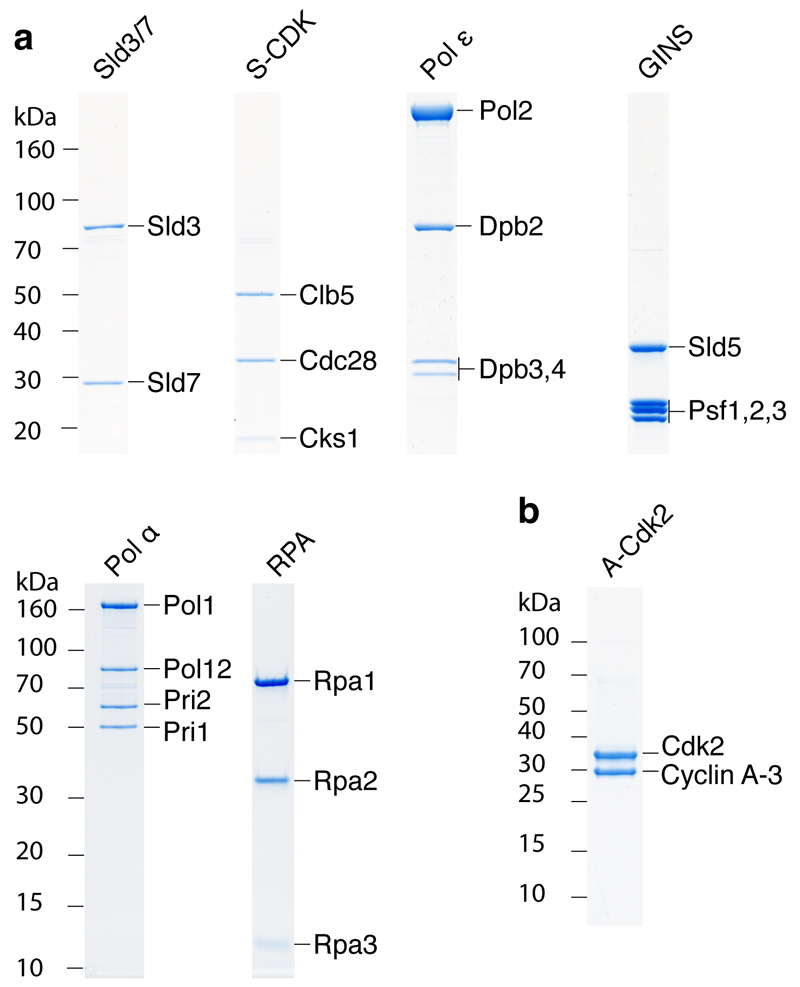
To reconstitute MCM loading and activation, we expressed and purified the thirteen replication factors shown in Fig. 1a (i) and Extended Data Fig. 1a and b. Cdc6, GINS, Mcm10 and cyclin A/Cdk2 (A-Cdk2) were expressed in Escherichia coli whilst the remaining proteins were expressed in Saccharomyces cerevisiae. In addition to A-Cdk2, budding yeast S phase CDK (S-CDK) expressed in Saccharomyces cerevisiae (Fig. 1a (i)) was used in some experiments. Details of expression and purification strategies can be found in Methods.
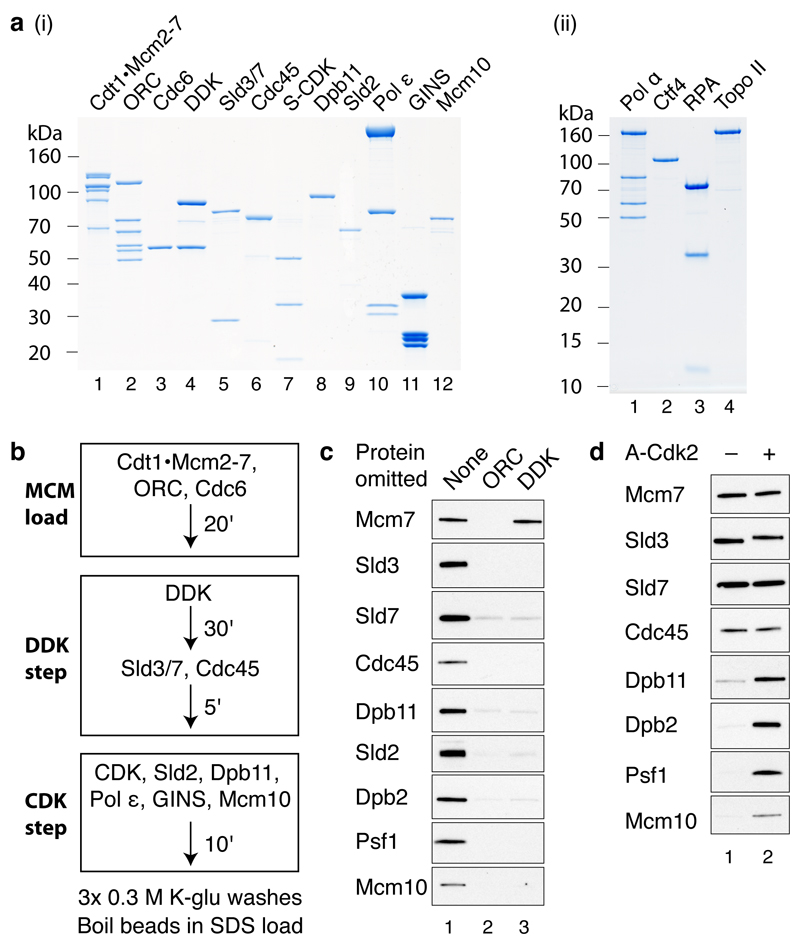
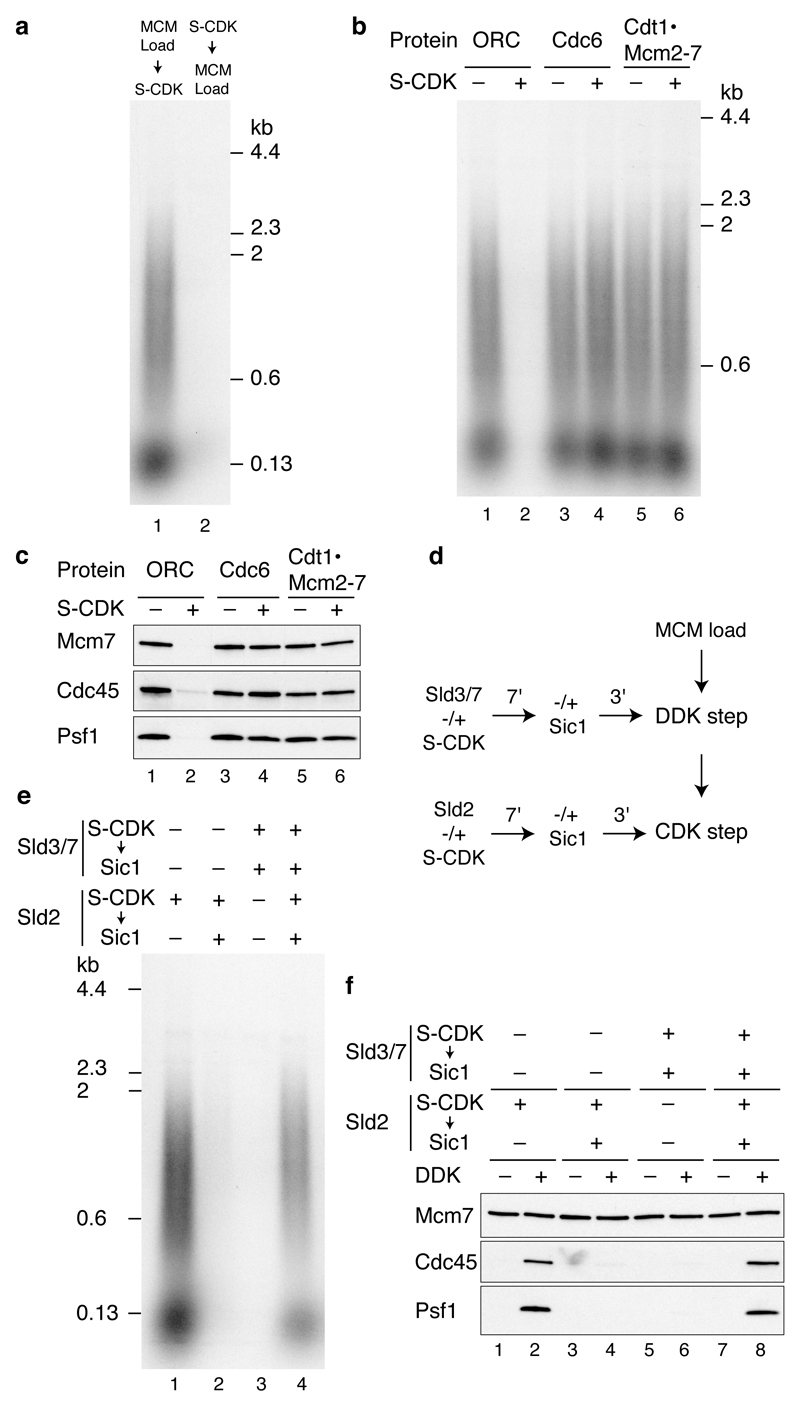
Figure 1. DDK- and CDK-dependent firing-factor recruitment with purified proteins.
a, Purified MCM loading and firing factors (i), and additional factors required for DNA replication (ii) analysed by SDS-PAGE with Coomassie staining. b, Reaction scheme for firing-factor recruitment. c, d, Immunoblots of recruitment reactions conducted as illustrated in bon ARS305 linear DNA.
We adopted the strategy outlined in Fig. 1b to assemble firing factors onto the loaded MCM in a staged manner. We first loaded MCM onto DNA attached to magnetic beads3 (‘MCM load’). The loaded MCM was phosphorylated with DDK6, beads were isolated and Sld3/7 and Cdc45 were added (‘DDK step’). Beads were again isolated and the remaining firing factors were added with A-Cdk2 (‘CDK step’). After washing the beads with a low salt buffer, we analysed bound proteins by immunoblotting. As shown in Fig. 1c, Sld3/7, Cdc45, Dpb11, Sld2, pol ε (the Dpb2 B subunit), GINS (the Psf1 subunit) and Mcm10 were all recruited in an ORC- and DDK-dependent manner. When A-Cdk2 was omitted from the final step, Sld3/7 and Cdc45 were still recruited but the remaining factors were not (Fig. 1d). Therefore, the recruitment of Sld3/7 and Cdc45 required DDK but not CDK, whilst the recruitment of the remaining firing factors required both DDK and CDK.
Stable recruitment of Cdc45 and GINS into a stable complex
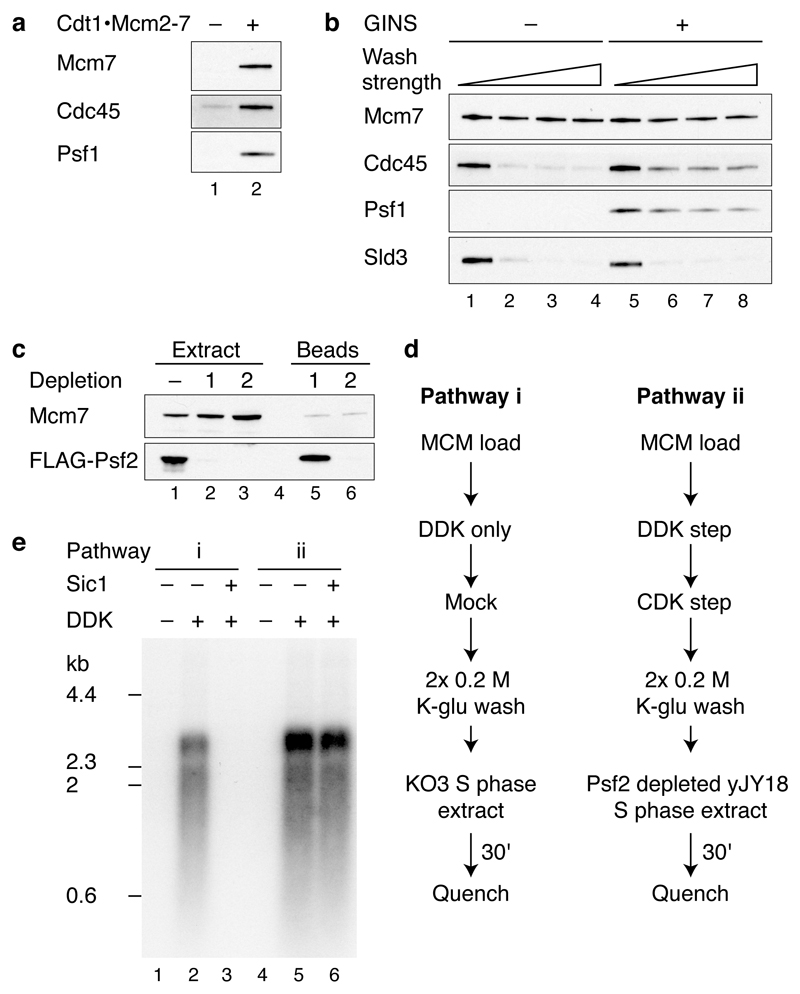
Consistent with the dependencies shown in Fig. 1c and 1d, Fig. 2a shows that the recruitment of Cdc45 did not require any of the factors acting in the CDK step (Dpb11, Sld2, pol ε, GINS, Mcm10). However, the recruitment of GINS (Psf1) required all of the other factors acting in this CDK step except for Mcm10 (i.e. Dpb11, Sld2, pol ε). CMG is salt-stable16,17, so we tested our complex under more stringent extraction conditions. Fig. 2b (lane 1) and Extended Data Fig. 2a,b show that, in addition to MCM, a fraction of Cdc45 and GINS is stable to salt extraction. In contrast to Cdc45 and GINS, Sld3 is not stabilised in this complex (Extended Data Fig. 2b). Fig 2b, lanes 2-5 shows that salt-stable Cdc45 recruitment requires Dpb11, Sld2, pol ε and GINS. Mcm10, however, is not required for this salt-stable complex (Fig. 2b, lane 6).
Figure 2. The purified firing factors are functional.
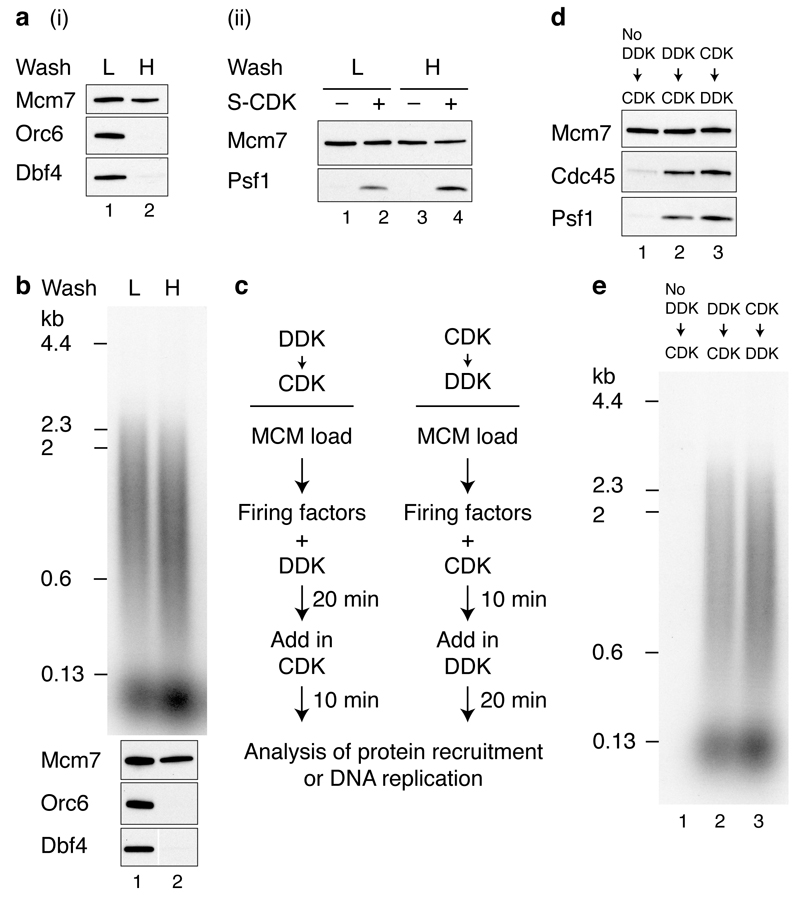
a, b, Immunoblots of recruitment reactions performed as in Fig. 1b using ARS305 linear DNA with either, a, 0.3 M K-glutamate (K-glu) or b, 0.3 M KCl washes. c, Scheme for extract-based replication reactions. Unless otherwise stated, in this and all subsequent experiments, components of the ‘CDK’ and ‘DDK’ steps are as in Fig. 1. Firing factors were recruited as illustrated in Fig. 1b. Beads were isolated, washed twice and added to an S phase extract not overexpressing firing factors (yJY18) and where the Psf2 subunit of the GINS complex was immunodepleted (Extended Data Fig. 2c). d, Reaction performed as in c. Nascent DNA was labelled by including α32P-dCTP in the extract step and products were separated through a 0.6% alkaline agarose gel.
To investigate whether the complex we assembled might be functional, we tested its ability to support DNA replication in extracts. We have previously shown that MCM loaded with purified proteins and phosphorylated with purified DDK can replicate in extracts from S phase cells made from a strain (KO3) that over-expresses Dpb11, Cdc45, Sld2, Sld3 and Sld76. It has been shown that over-expression of firing factors is required for replication in a related system7. We therefore constructed a new strain that does not over-express any firing factors (yJY18) and made S phase extracts from this strain in which GINS was also depleted (Extended Data Fig. 2c). We reasoned that addition of the complex of MCM with firing factors to this extract might support DNA replication (Fig. 2c). Fig. 2d lanes 1 and 9 shows that replication occurred under these conditions. Indeed, the replication products seen with the purified factors were equivalent to those synthesised in extracts from KO3 (Extended Data Fig. 2e). Furthermore, as shown in Fig. 2d lanes 2-8, replication was not observed if either Sld3/7, Cdc45, A-Cdk2, Dpb11, Sld2, pol ε or GINS were omitted, indicating that all of the recombinant proteins tested are functional and required for replication. This also suggested that the complex of MCM and firing factors may be functional.
Biochemical reconstitution of origin firing with purified proteins
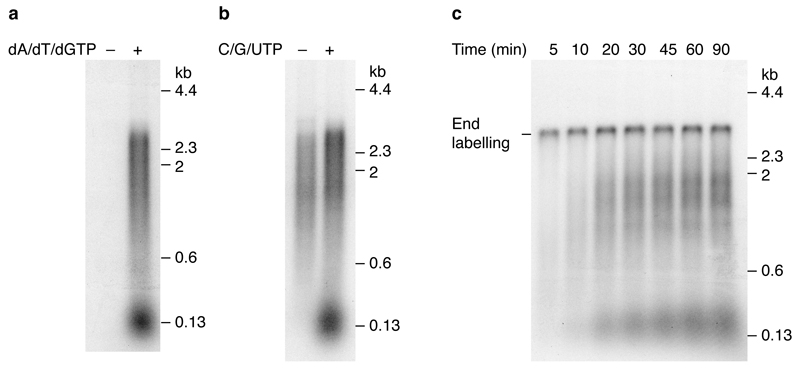
We next expressed and purified four additional proteins predicted to be important for DNA replication: DNA polymerase α - primase (pol α), Ctf4, replication protein A (RPA), and topoisomerase II (Topo II)18,19 (Fig. 1a (ii) and Extended Data Fig. 1a). Following the CDK step we isolated the beads and added a new buffer containing these four proteins along with additional Mcm10 and ribo- and deoxyribo nucleoside triphosphates (NTPs and dNTPs), as outlined in Fig. 3a. As shown in Fig. 3b (lane 1), this resulted in DNA synthesis that generated labelled products on alkaline agarose gels. Synthesis required ORC, DDK, A-Cdk2 and pol α (Fig. 3b) as well as dNTPs (Extended Data Fig. 3a), suggesting it was genuine initiation.
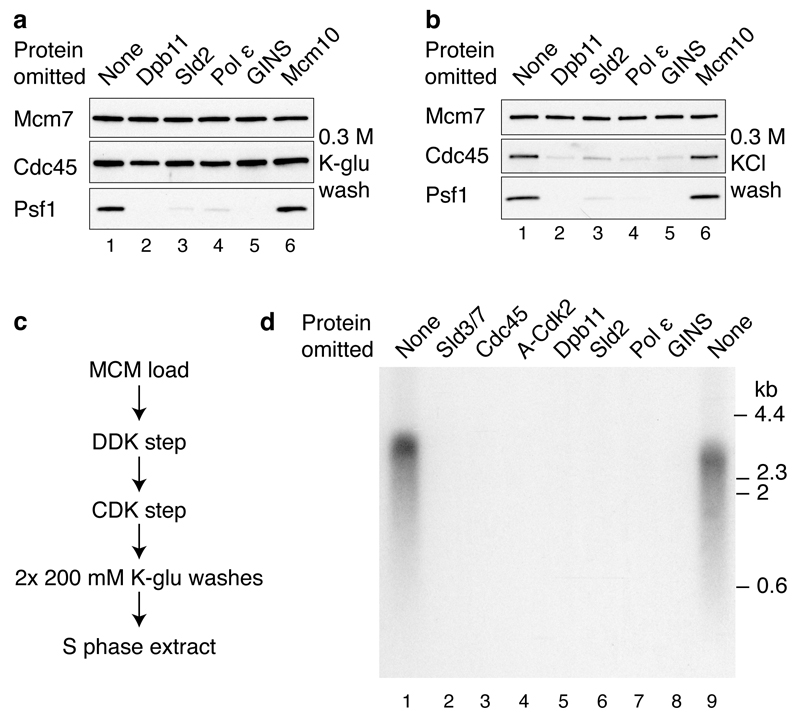
Figure 3. The initiation of DNA synthesis with purified proteins.
a, Reaction pathway for DNA replication with purified proteins. Firing factors were bound to MCM, the complex isolated, and added to a new buffer containing proteins required for DNA synthesis (Fig. 1a (i)). b, Replication reaction conducted as shown in a on ARS1 circular DNA. In this and all subsequent replication assays products were separated through 0.7% alkaline agarose gels. c, Replication on ARS1 linear templates. (i) Photocleaved DNA templates analysed by native agarose gel electrophoresis and ethidium bromide staining. (ii) Replication following the pathway shown in a. The MCM loading conditions were modified to confer origin specificity on the replication reaction (see Methods for details).
In this and subsequent experiments, two classes of labelled products were observed: a population of short products of approximately 150 nucleotides in length, and a more heterogeneous smear of larger products. The template used in these experiments was a 3.2 kbp plasmid, which had been randomly biotinylated and attached to streptavidin beads. Previous work has shown that attachment of DNA to beads interferes with the completion of DNA replication7. Since the position of the ARS1 origin relative to the site of attachment to the beads will be random, leading strand synthesis would be predicted to generate a smear of products averaging half the plasmid size (~1600 nucleotides), but ranging in size from very small to almost full plasmid length, very similar to the products observed. Synthesis of the smaller product class was more strongly affected by omission of C/G/UTP than the larger class (Extended Data Fig. 3b), suggesting that the smaller class may correspond to lagging strand synthesis, which requires repeated re-priming, but further work is needed to establish this. The residual larger products formed without C/G/UTP may be primed by RNA made from ATP alone (ARS1 is AT rich), which we cannot omit because it is required for MCM unwinding, or may be primed by inefficient dNTP use by primase. Extended Data Fig. 3c and d show full length products appear by 20 min and continue to accumulate with time; the smaller products accumulate with the same kinetics as the larger products, but appear to be less abundant, which may reflect incomplete lagging strand synthesis. A pulse-chase experiment (Extended Data Fig. 3e) shows that the smaller products are not a precursor of the larger ones, or vice versa. Approximately 10% of input plasmid is routinely replicated during replication reactions.
A template containing a functional origin yielded approximately 3.2 fold more DNA synthesis than an equivalent template containing a mutant, non-functional origin (A-B2-) (Fig. 3c (ii), compare lanes 2 and 4) indicating that replication exhibited origin preference under these conditions. The experiments in Fig. 3c and Extended Data Figure 3c-e were performed on linear DNA attached at one end to beads, and consequently, in addition to DDK-dependent replication products, there is also a DDK-independent end-dependent labelling product.
Requirements for DNA synthesis and RPA recruitment
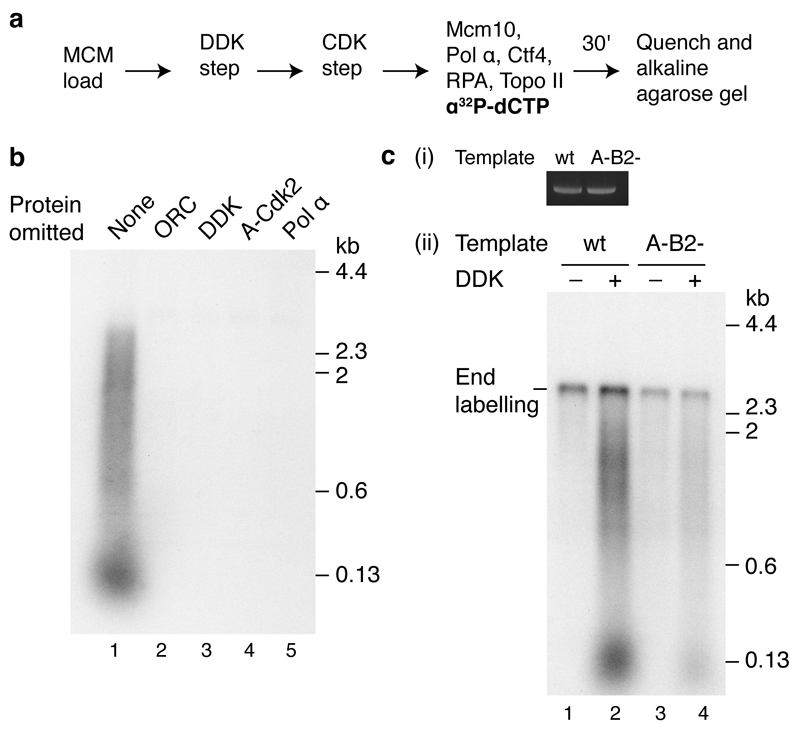
Fig. 4a shows that DNA synthesis in this system is dependent upon all of the firing factors, including Mcm10, which was not required for salt-stable association of Cdc45 and GINS (Fig. 2b). Omission of pol α also prevented DNA synthesis (Fig. 4b, lane 2) whilst Ctf4 omission had little effect on DNA synthesis, though the products were reproducibly slightly smaller (Fig. 4b, lane 3). In the absence of RPA, the small product class was completely lost whilst the larger smear was reduced in both intensity and size (Fig. 4b, lane 4). Finally, in the absence of Topo II, accumulation of products longer than 600 nucleotides was greatly reduced (Fig. 4b, lane 5).
Figure 4. Requirements for origin firing.
Reactions were performed as illustrated in Fig. 3a on circular templates unless stated. a, Firing factors required to initiate DNA synthesis. b, Protein dependencies of DNA replication for components functioning downstream of firing factor recruitment. c, Topoisomerase dependence on circular and linear ARS1 templates. d, RPA recruitment in a complete replication reaction. e, Dependence of RPA recruitment in the absence of pol α.
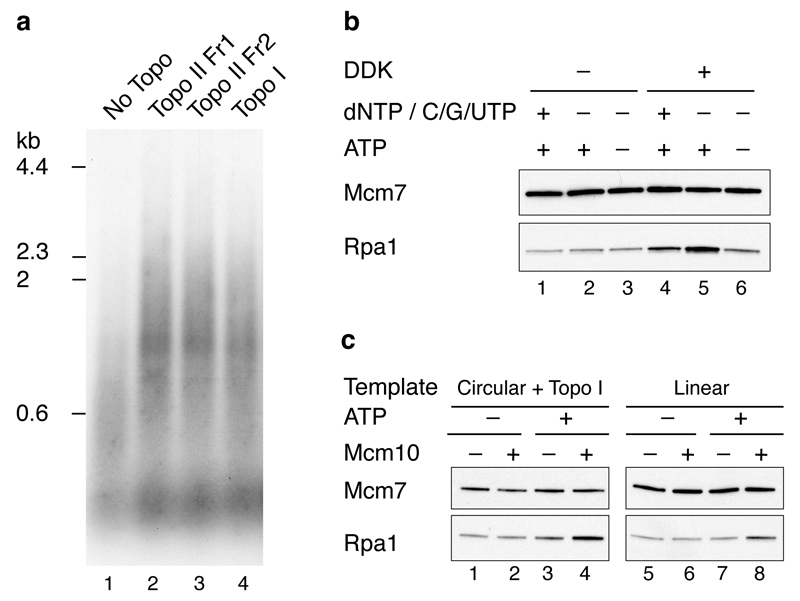
Topo II was required for accumulation of larger products from the circular template (Fig. 4b and 4c), but not from the linear template (Fig. 4c, compare lanes 2 and 5). Moreover, topoisomerase I from Vaccinia virus supported DNA replication on the circular template (Extended Data Fig. 4a). Together, these results indicate that removal of supercoils is required for the replication of circular molecules in this system. This experiment also shows that linear and circular templates generate similar labelled products (aside for the end labelled product on the linear template) and replicate with similar efficiencies (compare lanes 1 and 4).
We next examined the recruitment of RPA during the DNA synthesis reaction (Fig. 4d). RPA showed some DDK- and Mcm10-dependent recruitment (compare lanes 1-3). RPA recruitment was enhanced by omission of pol α (compare lanes 1 and 4), suggesting some uncoupling of template unwinding from DNA synthesis. This was supported by the fact that RPA recruitment was also enhanced in complete reactions lacking dNTPs and C/G/UTP (Extended Data Fig. 4b). The elevated RPA recruitment observed in the absence of pol α required GINS, Mcm10 and Topo II, but not Ctf4 (Fig. 4e). The requirement for Topo II is consistent with the idea that extensive unwinding was occurring. This is supported by the fact that enhanced RPA recruitment in the absence of Topo II could be restored by either Topo I or use of a linear template (Extended Data Fig. 4c). Although Cdc45 and GINS stably associate with loaded MCM in the absence of Mcm10 (Fig. 2b), RPA recruitment in the absence of DNA synthesis was substantially reduced without Mcm10 (Fig. 4e, lane 3 and Extended Data Fig. 4c), indicating that unwinding requires Mcm10.
Regulation of DNA replication by CDK
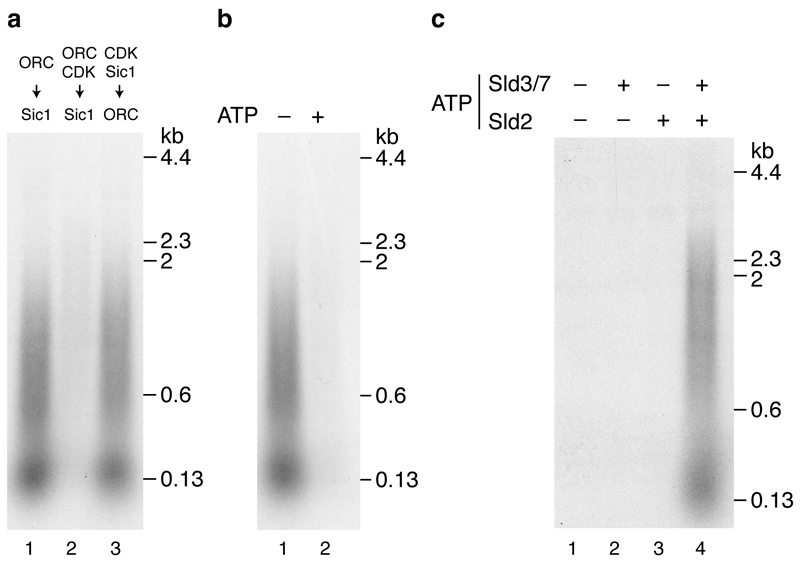
To examine the regulation of origin firing by protein kinases in more detail we used budding yeast S-CDK (Fig. 1 (i), lane 7) in place of A-Cdk2 as it can be selectively inhibited by the protein Sic1. CDK prevents MCM loading outside of G1 phase. Consistent with this, Fig. 5a (lane 2) shows that phosphorylation of MCM loading factors with S-CDK prevented DNA replication, but addition of S-CDK after MCM loading did not (lane 1). To determine which proteins were inhibited by S-CDK, we incubated each individually with S-CDK and ATP, then inhibited S-CDK with Sic1. Fig. 5b shows that treatment of ORC with S-CDK prevented DNA replication whilst treatment of Cdc6 and Cdt1·MCM with S-CDK had no effect. This is likely because, as shown in Fig. 5c, S-CDK treatment of ORC, but not Cdc6 or Cdt1·MCM, blocked MCM loading and consequently, Cdc45 and GINS recruitment. Pre-treatment of S-CDK with Sic1 before incubation with ORC prevented the inhibition of replication (Extended Data Fig. 5a, lane 3). Moreover, the inhibition of replication by S-CDK was ATP dependent (Extended Data Fig. 5b). Together, these results indicate that ORC’s ability to load MCM is inhibited by CDK-dependent phosphorylation in this system.
Figure 5. Regulation of MCM loading and origin firing by S-CDK.
a,Replication reactions where MCM loading factors were incubated with S-CDK before or after MCM loading on ARS1 circular DNA. Replication, b, and protein recruitment, c, on ARS1 circular DNA where ORC, Cdc6 and Cdt1·Mcm2-7 were individually incubated with S-CDK followed by addition of Sic1. The remaining MCM loading factors were added and reactions performed as shown in Fig. 3a (replication) and Fig. 1b (recruitment). d, Scheme to phosphorylate Sld3/7 and Sld2 in isolation from MCM and firing factors. Prior to both the DDK step and CDK step Sld3/7 and Sld2 were incubated with S-CDK before addition of Sic1. The remaining firing factors were added together with isolated DNA beads and each reaction step was performed as for standard reactions. e, Replication reaction staged as illustrated in d. f, Firing factor recruitment conducted as in d, followed by 0.3 M KCl washes.
Previous genetic analysis indicated that Sld2 and Sld3 are the two critical CDK substrates required for origin firing10–12. To examine this biochemically we phosphorylated Sld3/7 and Sld2 each individually with S-CDK then inhibited S-CDK with Sic1 prior to adding the additional proteins required for the DDK step (for Sld3/7) and CDK step (for Sld2) (Fig. 5d). Phosphorylation of either Sld2 or Sld3 individually did not support DNA replication (Fig. 5e lanes 2 and 3) or salt-stable association of Cdc45 and GINS (Fig. 5f lanes 3-6). However, when both Sld2 and Sld3 were phosphorylated separately and added in the same reaction, DNA replication (Fig. 5e lane 4) and salt-stable association of Cdc45 and GINS (Fig. 5f lanes 7 and 8) occurred efficiently. If Sic1 was omitted from the Sld2 CDK phosphorylation step, replication and salt-stable Cdc45 and GINS association occurred even when Sld3 was not pre-phosphorylated (Fig. 5e lane 1, Fig. 5f lanes 1,2), presumably because active CDK was added to the complete reaction, where it could also phosphorylate Sld3. Thus, comparison of lanes 1 and 2 in Fig. 5e and lanes 2 and 4 in Fig. 5f shows Sic1 was effective in inhibiting S-CDK. Extended Data Fig. 5c shows that ATP is essential for activation of both Sld2 and Sld3/7. Taken together, these results indicate that phosphorylation of Sld2 and Sld3/7 is necessary and sufficient for initiating DNA replication.
Regulation by DDK and the order of kinase action
High salt wash after MCM loading and DDK phosphorylation removes both ORC and DDK (Fig. 6a (i); Orc6 and Dbf4). This high salt washed MCM complex was competent for salt-stable Cdc45 and GINS association (Fig. 6a (ii)) as well as DNA replication (Fig. 6b) without further DDK addition. This indicates that MCM is the key substrate of DDK. This experiment also shows that ORC plays no essential role in initiating DNA replication after MCM loading, similar to conclusions from Xenopus egg extracts20. Previous work using yeast extracts indicated that replication did not happen if the CDK phosphorylation step was executed before the DDK step7. However, using the approach outlined in Fig. 6c with purified proteins, stable association of Cdc45 and GINS (Fig. 6d) and DNA replication (Fig. 6e) were equally efficient regardless of the order of kinase treatment.
Figure 6. DDK phosphorylation of MCM promotes origin firing either before or after S-CDK.
a, Reactions performed essentially as in Fig. 1b on circular DNA but with an additional mid-reaction wash immediately following DDK phosphorylation, in either a low salt (L) (0.3 M K-glu), or high salt (H) (0.6 M NaCl) buffer. Bound proteins immediately following the mid-reaction wash (i), and after firing factor recruitment followed by 0.3 M KCl washes (ii). b, Replication reactions with either high or low salt mid-reaction washes. c, Reaction schemes to test the order of DDK and CDK action. Firing factor recruitment was performed in a single stepwith DDK and CDK added at different times as indicated. Protein recruitment, d, and replication, e, performed with S-CDK as illustrated in c.
Discussion
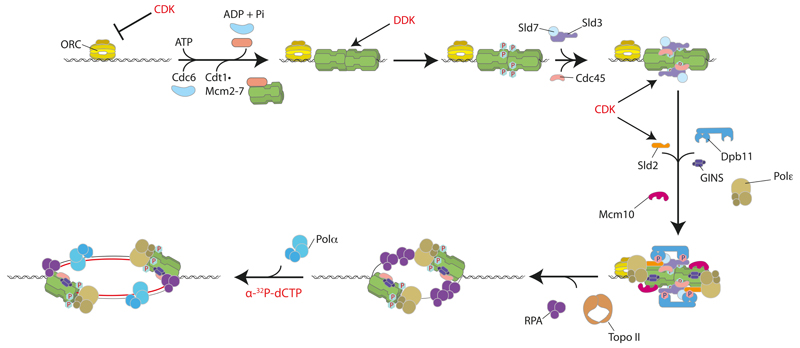
Our experiments define the minimum set of proteins and small molecule co-factors required to initiate origin-dependent DNA replication in a eukaryotic system (Extended Data Fig. 6). We show that the MCM complex assembled with purified proteins is a precursor of DNA replication initiation. Moreover, because this complex is competent for replication after high salt wash, the loading factors ORC, Cdc6 and Cdt1 play no essential role in initiation after MCM loading. Sld3/7 and Cdc45 are recruited to MCM in a DDK-dependent manner, and the remaining firing factors (Sld2, Dpb11, GINS, pol ε, Mcm10) are recruited in a DDK- and CDK-dependent manner. This set of firing factors is sufficient to promote RPA recruitment. RPA recruitment is likely due to the generation of substantial single stranded DNA because it requires topoisomerase on a circular template, and it is enhanced when DNA synthesis is prevented either by omitting either pol α or dNTPs from the reaction. Mcm10 is required for RPA recruitment, but not salt-stable association of Cdc45 and GINS, consistent with results in yeast extracts7 and in vivo21,22. Thus, a CMG-like complex can be assembled without Mcm10, but this complex is inactive. All DNA synthesis requires pol α, consistent with its role in generating primers required for DNA synthesis. Because pol ε is required for CMG formation, we cannot at present determine whether it also contributes to subsequent DNA synthesis in this system, for example, through generation of a CMGE complex23. The presence of short and long products is suggestive of leading and lagging strand synthesis. However, our replication system currently lacks DNA polymerase δ, which is responsible for lagging strand synthesis in vivo24, RFC and PCNA, which localises to lagging strands during replication in vivo25, as well as the many factors required for Okazaki fragment maturation. It also lacks other replisome-associated factors such as Mrc1 and Tof1/Csm317. Consequently, DNA synthesis in this system does not yet fully recapitulate normal coupled leading and lagging strand replication, and this is clearly an important goal for the future.
Our experiments also define the minimum set of CDK and DDK substrates required for regulated replication. CDK phosphorylation of ORC directly prevents it from loading MCM and promoting replication, consistent with previous work1,26,27. Although it is clear that CDK phosphorylation of Cdc6 and Cdt1·MCM contributes to preventing re-replication in vivo in budding yeast, our experiments show that CDK phosphorylation does not directly inhibit the ability of these proteins to load MCM. Instead, it is likely that CDK inhibits Cdc6 function by promoting its degradation in cells, and CDK inhibits Cdt1·MCM function by promoting nuclear exclusion2. Our work also confirms that Sld3/7 and Sld2 are the only two CDK substrates required for replication initiation. In contrast to conclusions from experiments in yeast extracts7, our results with purified proteins show CDK does not have any inhibitory role after MCM loading, and CDK and DDK can act in either order. In the experiments using extracts7, we suggest rapid dephosphorylation of Sld2 and Sld3 occurred after Sic16 and DDK addition, requiring one or both of these proteins to be re-phosphorylated by CDK after Sld3/7 and Cdc45 were loaded. Our experiments show that the essential DDK substrate for initiation is MCM. Further work will be required to ascertain which MCM subunits and sites are critical.
The initiation step of DNA replication has taken many years to reconstitute because of the large number of factors involved and the complicated regulation by protein kinases. Having achieved this, we are now in a position to understand how the MCM double hexamer is activated in molecular and atomic detail. Moreover, this work sets the stage for the complete reconstitution of chromosome replication, which has the potential to provide insights into many DNA replication-coupled nuclear processes.
Extended Methods
Yeast expression strain construction
All strains were constructed by transforming strain yJF127 with linearised expression vectors using standard genetic techniques (See Extended Data Tables 2 and 3 for details of vectors and strains). For expression of Sld3, Dpb11 and Sld2, affinity tags were added by transformation with PCR products following integration of the expression vector. The PCR products for Sld3 tagging were generated with oligonucleotides Sup sld3 TCP tag fwd and Sup TCP tag rev from the template pBS1539/TAPTCP 3(yTD6), and oligonucleotides Sld3SUP C-term FLAG tag fwd and Sld2SUP C-term FLAG rev using pBP83 as template (yTD11). PCR products for Sld2 and Dpb11 were generated from pBP83 (a derivative of pYM1828 modified to insert a C-terminal 3xFLAG tag associated with the NatNT2 marker) using oligonucleotides Sld2SUP C-term FLAG rev and Sld2SUP C-term FLAG rev for Sld2 and JTY10 and Cdc45 flag tag rev for Dpb11. Affinity tagged protein expression was verified by immunoblot.
Proteins and protein expression
Vaccinia virus topoisomerase I was purchased from Sigma. ORC, Cdc6, Cdt1·Mcm2-7, DDK, cyclin A/Cdk2 and Sic1 were expressed and purified as previously described6,27,29. All proteins purified in this study contained affinity tags, which were utilised in the first step of purification. For the amino acid sequences of the affinity tags and their locations see Extended Data Table 1. GINS and Mcm10 were expressed in E. coli. All other proteins were over-expressed in S. cerevisiae from bidirectional Gal1-10 promoters in a pep4Δ, bar1Δ strain background (yJF1) (see Extended Data Table 2 for details of expression strains). Expression strains for Dpb11, Sld2, Cdc45, Ctf4, RPA and Pol ε were grown at 30°C in YP + 2% raffinose to a density of ~ 2-4 × 107 cells per ml. Cells were arrested in G1 with 100 ng ml-1alpha factor and incubation was continued for 3 hours. Protein expression was induced by addition of galactose to 2% and growth continued for 3 hours at 30°C. Topo II expression was conducted essentially as described above but alpha factor was added together with galactose and growth was continued for 6 hours. For pol α expression, cells were grown to a density of ~ 2 × 107 cells per ml and expression was induced for 2 hours by addition of galactose (2%). S-CDK expressing cells were grown to 2 × 107 cells per ml and protein expression induced by addition of galactose (2%) together with nocodazole (5 μg/ml) with growth continued for a further 3 hours. In all cases, cells were collected, washed once with 25 mM HEPES-KOH pH 7.6, 1 M sorbitol and once with the appropriate initial protein purification buffer (see individual purification protocols for buffers used) lacking protease inhibitors. Cells were resuspended in 0.3 – 0.4 volumes of the initial purification buffer + protease inhibitors and the suspensions frozen drop-wise in liquid nitrogen. Frozen cells were crushed in a freezer mill (SPEX CertiPrep 6850 Freezer/Mill) with 6 cycles of 2 min at a crushing rate of 15. The resulting powders were stored at -80°C until required.
Sld2 purification
Frozen cell powder was thawed and resuspended in 3 volumes 25 mM HEPES-KOH pH 7.6, 10% glycerol, 0.02% v/v Nonidet P40 substitute (NP-40-S) (Roche #11754599001), 1 mM EDTA, 1 mM DTT, 500 mM KCl (buffer H + 500 mM KCl) + protease inhibitors (0.3 mM PMSF, 7.5 mM Benzamidine, 0.5 mM AEBSF, 1 mM Leupeptin, 1 mM Pepstatin A and 1 μg/ml Aprotinin (Sigma)). Insoluble material was cleared by centrifugation (235,000g, 4°C, 1 hour) and solid ammonium sulphate was added to the supernatant to 32% followed by gentle stirring (10 min, 4°C). Insoluble material was removed by centrifugation (27,000g, 4°C 20 min) and the ammonium sulphate concentration increased to 48% followed by stirring for 10 min. Precipitated protein was collected by centrifugation (27,000g, 4°C 20 min) and resupended in 1/3 of the original volume buffer H + 500 mM KCl + protease inhibitors. FLAG-tagged Sld2 was bound to ANTI-FLAG M2 affinity gel (Sigma) in batch for 30 min at 4°C. Resin was collected in 20 ml chromatography columns (Biorad) and washed extensively in buffer H + 500 mM KCl + protease inhibitors. The resin was resuspended in 10 column volumes (CV) buffer H without EDTA + 500 mM KCl + 10 mM magnesium acetate + 1 mM ATP and incubated for 10 min at 4°C. The flow through was discarded and the column washed with 20-40 CV buffer H + 500 mM KCl. Sld2-FLAG was eluted in 1 CV buffer H + 500 mM KCl + 0.5 mg/ml 3xFLAG peptide, followed by 2 CV buffer H + 500 mM KCl + 0.25 mg/ml 3xFLAG peptide.
The eluates were pooled, dialysed against buffer H + 280 mM KCl and applied to a 1 ml HiTrap SP FF column (GE Healthcare) equilibrated in buffer H + 250 mM KCl. Sld2-FLAG was eluted with an 18 CV gradient from 250 mM KCl to 1 M KCl in buffer H. Peak fractions were pooled and dialysed against buffer H with 40% v/v glycerol + 350 mM KCl. Protein concentration was assessed using the Bradford assay (Biorad) and this method was used for all proteins in this study.
Sld3/7 purification
To purify TCP-tagged Sld3/7, cell powder from yTD6 was thawed in buffer H + 500 mM KCl + protease inhibitors and insoluble material was cleared by centrifugation (235,000g, 4°C, 1 hour). Sld3/7 was depleted from the soluble extract by incubation for 40 min at 4°C with IgG Sepharose 6 Fast Flow (GE Healthcare). The resin was collected in a 20 ml column and washed extensively in buffer H with 0.5 mM EDTA + 500 mM KCl. Sld3/7 was cleaved from the column by incubation for 2 hours at 4°C with TEV protease (50 μg/ml) in buffer H with 0.5 mM EDTA + 500 mM KCl. His-tagged TEV protease was removed by passing the eluate over a Ni-NTA Agarose column (Qiagen). The resulting flow through was concentrated and separated on a Superdex 200 column (GE Healthcare) equilibrated in buffer H + 500 mM KCl.
The FLAG-tagged Sld3/7 used for Cdc45 depletion (Extended Data Fig. 7) was purified from yTD11 cells essentially as described above, except that Sld3/7 was depleted from the soluble extract by incubation with ANTI-FLAG M2 affinity gel for 1 hour at 4°C. Bound Sld3/7 was eluted as described for Sld2 and the eluate was concentrated and separated on a Superdex 200 column equilibrated in buffer H + 500 mM KCl.
Cdc45 purification
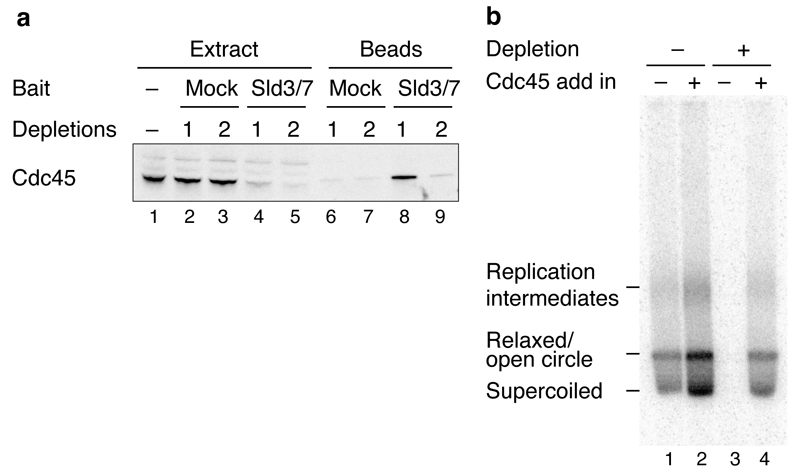
80g cell powder was thawed in 200 ml buffer H without NP-40-S + 500 mM KOAc + protease inhibitors and cell debris was removed by centrifugation (235,000g, 4°C, 1 hour). 8 ml ANTI-FLAG M2 affinity gel was added and the extract incubated for 4 hours at 4°C. Resin was collected in 20 ml columns (2 ml per column) washed extensively in buffer H without NP-40-S + 500 mM KOAc, then buffer H without NP-40-S + 300 mM KOAc. Cdc45 was eluted by incubation of the resin in 1 CV buffer H without NP-40-S + 300 mM KOAc + 0.5 mg/ml 3xFLAG peptide. The eluate was dialysed against 150 mM KOAc, 0.5 mM DTT, 10 % glycerol, 20 mM K-phosphate pH 7.4, (buffer P + 20 mM K-phosphate) and then applied to 1.5ml Hydroxyapatite column (Biorad) equilibrated in the same buffer. The column was washed with buffer P + 80 mM K-phosphate and Cdc45 was eluted with buffer P + 250 mM K-phosphate. The eluate was extensively dialysed against buffer H without NP-40-S + 300 mM KOAc prior to storage. To test the functionality of the internally FLAG-tagged Cdc45, we depleted endogenous Cdc45 from an S phase extract (Extended Data Fig. 7a). DNA replication was not detected after depletion but was restored following addition of purified Cdc45 (Extended Data Fig. 7b). The distributions of replication products following Cdc45 depletion and add back were indistinguishable from an undepleted sample (compare lanes 1 and 4) suggesting that the recombinant protein is functional.
S-CDK purification
40g cell powder was thawed in 80 ml 40 mM HEPES-KOH pH 7.6, 10% glycerol, 0.02% v/v NP-40-S, 300 mM KOAc (buffer CD + 300 mM KOAc) + protease inhibitors. Cell debris was cleared by centrifugation (235,000g, 4°C, 1 hour), calcium chloride was added to 2 mM together with Calmodulin affinity resin (Agilent) and the extract was incubated at 4°C for 1 hour. Resin was collected in a 20 ml column, washed extensively with buffer CD + 300 mM KOAc + 2 mM CaCl2 and S-CDK was eluted by incubation for 16 hours with 100 μg/ml TEV protease in buffer CD + 2 mM CaCl2. TEV protease was removed by passing the eluate over Talon resin (Clontech) and collecting the flow through, which was then applied to a Superose 6 column (GE Healthcare) equilibrated in buffer CD + 300 KOAc. Peak fractions were pooled and concentrated.
Dpb11 purification
Cell powder was resuspended in 2 volumes buffer H + 500 mM KCl and the debris removed by centrifugation (235,000g, 4°C, 1 hour). Dpb11-FLAG was bound to ANTI-FLAG M2 affinity gel for 90 min at 4°C. The resin was collected, washed extensively with buffer H + 500 mM KCl and the protein eluted in buffer H + 500 mM KCl using 3xFLAG peptide as for Sld2. The eluate was dialyased against buffer H + 150 mM KCl and applied to a 1 ml MonoS column (GE Healthcare). Dpb11-FLAG was eluted with a 30 CV gradient from 150 mM KCl to 1 M KCl in buffer H with the peak fractions then dialysed against buffer H + 300 mM KOAc.
DNA Polymerase Epsilon purification
Cell powder was slowly thawed in buffer H without NP-40-S and EDTA + 400 mM KOAc (buffer E + 400 mM KOAc) + 1 Complete, EDTA free protease inhibitor tablet (Roche) per 25 ml buffer and cell debris was removed by centrifugation (235,000g, 4°C, 1 hour). To the supernatant CaCl2 was added to 2 mM together with Calmodulin affinity resin and the solution was rotated at 4°C for 60 min. Resin was collected in a 20 ml column, washed extensively in buffer E + 400 mM KOAc + 2 mM CaCl2 and Pol epsilon was eluted in 2 ml fractions with buffer E + 400 mM KOAc + 2 mM EDTA + 2 mM EGTA. The eluate was pooled and applied directly to a 5 ml Hi-trap heparin column (GE Healthcare) equilibrated in buffer E + 400 mM KOAc. Following extensive washing with buffer E + 450 mM KOAc proteins were eluted with a 12 CV gradient from 450 mM – 1 M KOAc in buffer E. Heparin fractions containing Pol epsilon were pooled, concentrated and separated on a Superdex 200 column (GE Healthcare) equilibrated in buffer E + 500 mM KOAc.
Ctf4 purification
Ctf4 purification was based on a published method30 with modifications. Cell powder was thawed in 2 volumes 25 mM Tris-HCl pH 7.2, 10% glycerol, 1 mM DTT, 200 mM NaCl (buffer C + 200 mM NaCl) + protease inhibitors and the insoluble material removed by centrifugation (235,000g, 4°C, 1 hour). Calcium chloride was added to 2 mM together with Calmodulin affinity resin. The extract was incubated at 4°C for 90 min before the resin was collected and washed extensively with buffer C + 200 mM NaCl + 2 mM CaCl2. Proteins were eluted in buffer C + 200 mM NaCl + 2 mM EDTA + 2 mM EGTA. The NaCl concentration of the eluate was reduced to 150 mM by dilution in buffer C before application to a 1 ml MonoQ equilibrated in buffer C + 150 mM NaCl + 1 mM EDTA. Ctf4 was eluted with a 30 CV gradient from the 150 mM – 1 M NaCl in buffer C. Peak fractions were pooled, concentrated and separated on a Superdex 200 column (GE Healthcare) equilibrated in buffer C + 150 mM NaCl + 1 mM EDTA. Ctf4 containing fractions were dialysed against buffer C + 75 mM NaCl + 1 mM EDTA.
DNA Polymerase alpha–primase purification
Cell powder was thawed in buffer C + 0.02% NP-40-S + 400 mM NaCl (buffer D + 400 mM NaCl) + protease inhibitors and insoluble material was cleared by centrifugation (235,000g, 4°C, 1 hour). The NaCl concentration was reduced to 300 mM by dilution in buffer lacking NaCl, and calcium chloride was added to 2 mM together with Calmodulin affinity resin. The extract was incubated at 4°C for 90 min before the resin was collected in a 20 ml column, washed extensively with buffer D + 300 mM NaCl + 2 mM CaCl2 and proteins eluted with buffer D + 300 mM NaCl + 2 mM EDTA + 2 mM EGTA. Pooled fractions were diluted in buffer D to a conductivity equivalent to buffer D + 120 mM NaCl and were loaded onto a 1 ml MonoQ column. Bound proteins were removed with a 30 CV gradient from 120 mM – 1 M NaCl in buffer D and fractions containing pol α were pooled, concentrated to 400 μl and applied to a Superdex 200 column equilibrated in buffer D + 150 mM NaCl. The peak fractions were pooled, concentrated to ~ 0.7 mg/ml and snap frozen in liquid nitrogen.
RPA purification
40g cell powder was thawed in buffer C + 500 mM NaCl + protease inhibitors and insoluble material cleared by centrifugation (235,000g, 4°C, 1 hour). Lysate conductivity was reduced to the equivalent of buffer C + 200 mM NaCl by 2-fold dilution in buffer C + protease inhibitors, followed by 1 hour dialysis against buffer C + 200 mM NaCl. CaCl2 was added to 2 mM together with 1 ml Calmodulin affinity resin and the extract incubated at 4°C for 90 min with rotation. Resin was collected in a 20 ml column, washed with 35 CVs buffer C + 200 mM NaCl + 2 mM CaCl2 and bound proteins were eluted in 1 ml fractions of buffer C + 2 mM EDTA + 2 mM EGTA. Peak fractions were pooled, diluted 2-fold in buffer C + 1 mM EDTA, dialysed for 1 hour against buffer C + 50 mM NaCl + 1 mM EDTA and loaded onto a 1 ml Hi-trap heparin column equilibrated in the same buffer. After extensive washing proteins were eluted with a 30 CV gradient from 50 mM – 1 M NaCl in buffer C + 1 mM EDTA. Peak fractions were pooled, diluted 3-fold in buffer C + 1 mM EDTA, dialysed for 1 hour against buffer C + 150 mM NaCl + 1 mM EDTA and loaded onto a 1 ml MonoQ equilibrated in the same buffer. RPA was eluted with a 25 CV gradient from 150 mM – 1 M NaCl in buffer C + 1 mM EDTA and peak fractions were pooled and dialysed overnight in buffer C with 38% glycerol + 1 mM EDTA + 50 mM NaCl.
Topoisomerase II purification
Powder was thawed in buffer D + 300 mM NaCl + protease inhibitors and insoluble material removed by centrifugation (235,000g, 4°C, 1 hour). The soluble extract was supplemented with 2 mM CaCl2 and Calmodulin affinity resin was added and the solution incubated at 4°C for 1 hour. Resin was collected in a 20 ml column, washed extensively with buffer D + 300 mM NaCl and the CBP-tagged Topo II eluted with buffer D + 300 mM NaCl + 2 mM EDTA + 2 mM EGTA. The eluate was concentrated and applied directly to a Superdex 200 column equilibrated in buffer D + 150 mM NaCl. Topo II containing fractions were pooled and the salt concentration adjusted to ~ 100 mM by dilution prior to loading onto a 1 ml MonoQ equilibrated in buffer D + 100 mM NaCl. Topo II was eluted with a 25 CV gradient from 100 – 800 mM NaCl in buffer D and the peak fractions were pooled and dialysed against buffer D + 150 mM NaCl prior to storage.
GINS purification
Plasmid pFJD5 (Gift from K. Labib), which is a derivative of pFJD1219 in which the Psf3 subunit is expressed with an N-terminal His-tag, was used to transform Bl21 (DE3) Rosetta pLysS. Cells were grown in LB at 37°C to an A600 of 0.5 and protein expression was induced by addition of IPTG to 1 mM. Growth was continued for 2 hours before cells were harvested by centrifugation. Cell pellets were resuspended in buffer D + 400 mM NaCl + 10 mM imidazole + protease inhibitors, cells were lysed by sonication and the debris removed by centrifugation (257,000g, 4°C, 30 min). His-tagged proteins were depleted by incubation with Ni-NTA resin (Qiagen) for 1 hour at 4°C and the resin was collected in a 20 ml column. Following extensive washing with buffer D + 400 mM NaCl + 10 mM imidazole and then buffer D + 100 mM NaCl + 15 mM imidazole, GINS was eluted with buffer D + 100 mM NaCl + 200 mM imidazole. Fractions were pooled and dialysed against buffer D + 100 mM NaCl before being applied to a 1 ml MonoQ column equilibrated in the same buffer. Proteins were eluted with a 25 CV gradient from 100 mM – 500 mM NaCl in buffer D and the GINS containing fractions were pooled, concentrated and separated through a Superdex 200 column equilibrated in buffer D + 150 mM NaCl. The peak was rebound to Ni-NTA Agarose resin and the resin was washed with 50 mM HEPES-KOH pH 7.6, 10% glycerol, 0.05% NP-40-S, 10 mM Mg(OAc)2 before GINS was eluted in the same buffer supplemented with 50 mM imidazole. Finally the eluate was dialysed against buffer H + 200 mM KOAc prior to storage.
Mcm10 purification
Plasmid pET28a-Mcm10, which expresses Mcm10 with an N-terminal His/T7 tag, was used to transform BL21 (DE3). Cells were grown at 37°C to an A600 of 0.6 before the temperature was reduced to 25°C and protein expression induced by addition of IPTG to 1 mM. Growth was continued for 3 hours at 25°C before cells were harvested, washed in 50 mM Tris-HCl pH 7.6, 10 % sucrose and resuspended in 25 mM Tris-HCl pH 7.6, 10% glycerol, 0.01% NP-40-S, 500 mM NaCl (buffer M + 500 mM NaCl) + protease inhibitors. Cells were lysed by sonication and the debris removed by centrifugation (257,000g, 4°C, 30 min). The clear lysate was applied under gravity flow to a 1.5 ml Ni-NTA Agarose column. The column was washed with 15 CV buffer M + 500 mM NaCl followed by 10 CV buffer M + 200 mM NaCl + 20 mM imidazole and His-Mcm10 was eluted with buffer M + 200 mM NaCl + 200 mM imidazole. The eluate was applied to a 1 ml MonoS equilibrated in buffer M + 200 mM NaCl and proteins were eluted with a 20 CV gradient from 200 mM – 1M NaCl in buffer M. Peak Mcm10 fractions were pooled, the NaCl concentration adjusted to 200 mM by dilution in buffer M and reapplied to a 1 ml MonoS. The column was run as described for the first run and Mcm10 containing fractions were pooled and dialysed against buffer H with 0.01% NP-40-S + 200 mM K-glutamate.
S phase extracts
S phase extracts were prepared as described previously6. Psf2-FLAG (GINS complex) was depleted from the yJY18 extract by two 45 min incubations at 4°C with a 1:10 extract volume of ANTI-FLAG M2 magnetic beads (Sigma). To deplete Cdc45 from the yJY16 extract, FLAG-tagged Sld3/7 was pre-bound to ANTI-FLAG M2 magnetic beads (5 pmol per 1 μl of resin). Cdc45 was then depleted from the extract by two 30 min incubations using 2.5 μl beads per 20 μl of extract.
DNA templates
1 kbp biotinylated linear ARS305 DNA templates were generated as described3,31. For linear ARS1 templates a 2.8 kbp region surrounding ARS1 was amplified using primers ARS1_XmaI-F and ARS1_XhoI-R and cloned into pBluescript KS (+) using XhoI and XmaI to generate plasmid pCFK1. An A-B2- derivative, pCFK2, was generated by sequential rounds of site-directed mutagenesis using oligonucleotides MutA_F, MutA_R, MutB2_F, MutB2_R. 2.8 kbp biotinylated wild type and A-B2- ARS1 linear DNA templates were generated by PCR using the oligonucleotides ARS1-PC-Bio-F and ARS1-Bio-R with pCFK1 and pCFK2 as templates respectively. 3.2 kbp randomly biotinylated circular DNA templates were generated as described6. Biotinylated DNA was coupled to streptavidin M-280 magnetic DNA beads (Invitrogen) essentially as described3,31. For linear DNA templates 250 ng DNA was coupled to 5 μl bead slurry. As biotinylation efficiency was estimated only to be ~10-20% for the circular DNA templates 1 μg of DNA was coupled per 5 μl bead slurry.
Standard protein recruitment and replication reactions
Unless stated in the figure legends, protein recruitment assays were conducted using the ARS305 linear template, whereas DNA replication reactions used ARS1 circular DNA as template. All incubations were conducted with agitation (1250 rpm) in a Thermomixer (Eppendorf). The concentrations of proteins in recruitment and replication reactions were determined empirically using the concentrations of individual firing factors in S phase extracts for guidance. Sld3/7 purified from yTD6 was used in all protein recruitment and replication reactions. MCM loading reactions (‘MCM’ load) (7.5-10 μl) were performed in a buffer containing 25 mM HEPES-KOH pH 7.6, 100 mM K-glutamate, 10 mM Mg(OAc)2, 0.02% NP-40-S, 5% glycerol, 2 mM DTT, 5 mM ATP (loading buffer), 45 nM ORC, 45 nM Cdc6, 100 nM Cdt1·Mcm2-7 and either 6.25 ng/μl linear or approximately 4 ng/μl circular DNA immobilised on M-280 streptavidin magnetic beads (Invitrogen). After 20 min incubation at 25°C DDK was added directly to the reaction to a final concentration of 130 nM and incubation was continued at 25°C for 30 min. Buffer and unbound proteins were removed using a magnetic rack and were replaced by a new buffer of 2-fold the MCM loading reaction volume (15-20 μl) containing 40 mM HEPES-KOH pH 7.6, 300 mM K-glutamate, 10 mM Mg(OAc)2, 0.02% NP-40-S, 8% glycerol, 400 μg/ml BSA (NEB), 2 mM DTT, 5 mM ATP (firing-factor recruitment buffer + 300 mM K-glutamate) and 26 nM Sld3/7 and 50 nM Cdc45. Following incubation at 25°C for 5 min the supernatant was removed and replaced with the same volume (15-20 μl) of firing-factor recruitment buffer + 250 mM K-glutamate and 40 nM Dpb11, 62 nM Sld2, 30 nM Pol ε, 210 nM GINS, 2.5 nM Mcm10 and 50 nM A-Cdk2 (cyclin A/Cdk2) (Figs. 1-4 and Extended Data Figs. 2-4) or S-CDK (Clb5/Cdc28/Cks1) at a concentration of 30 nM for Fig. 6, 20 nM for Fig. 5e, f and Extended Data Fig. 5c and 25 nM for Fig. 5a-c and Extended Data Fig. 5a, b. Reactions were incubated at 25°C for 10 min (‘CDK’ Step). For protein recruitment assays that were terminated after the ‘CDK’ step, 10 ng/μl poly(dI-dC) (Sigma) was added to both the buffer containing Sld3/7 and Cdc45, and the ‘CDK’ step. After the ‘CDK’ step supernatants were again removed and replaced with the same volume of buffer containing 40 mM HEPES-KOH pH 7.6, 150 mM K-glutamate, 10 mM Mg(OAc)2, 5% glycerol, 2 mM DTT, 2 mM ATP, 200 μM CTP, GTP, UTP, 40 μM dATP, dTTP, dCTP, dGTP, 2.5 nM Mcm10, 50 nM RPA, 25 nM pol α, 30 nM Ctf4, 25 nM Topo II and, for replication reactions, 40 nM [α-32P]dCTP (Perkin Elmer). Reactions were incubated at 30°C for 30 min.
For protein recruitment assays, unless stated in the figure legends, the final reaction buffer (either after both the ‘DDK’ and ‘CDK’ steps or the final replication step) was removed and beads were washed three times in 150 μl room temperature buffer containing 40 mM HEPES-KOH pH 7.6, 300 mM K-glutamate, 10 mM Mg(OAc)2, 5% glycerol, 0.02% NP-40-S (wash buffer + 300 mM K-glutamate). Beads were resuspended in SDS-loading buffer, heated to 95°C for 3 min and proteins were separated through 4-12% Bis-Tris polyacrylamide gels (Biorad) before analysis by immunoblotting.
Replication reactions were terminated by removing the supernatant and immediately washing the beads twice with 150 μl 40 mM HEPES-KOH pH 7.6, 600 mM KCl, 5% glycerol, 0.02% NP-40-S, 5 mM EDTA before resuspending the beads in 5 mM EDTA and then adding NaOH and sucrose to 50 mM and 1% w/v respectively. Samples were incubated at room temperature for 20-30 min before the products were separated through 0.7% alkaline agarose gels in 30 mM NaOH, 2 mM EDTA for 16 hours at 25 volts. Gels were fixed with 5% cold trichloroacetic acid, dried onto chromatography paper (Whatman) and autoradiographed with Amersham Hyperfilm-MP (GE Healthcare). When gels were quantified images were scanned using a Typhoon phosphorimager (GE Healthcare) and were quantified using ImageQuant software (GE Healthcare).
Modified protein recruitment and replication reactions
For experiments where mid-reaction washes were included (Fig. 6a and b), two 150 μl washes were conducted in wash buffer containing the salts indicated in the figure legends, followed by one 150 μl wash in wash buffer + 300 mM K-glutamate. Reactions with a single firing-factor recruitment step (Figs. 6d and 6e) were conducted in firing-factor recruitment buffer + 250 mM K-glutamate for 30 min at 25°C, with 30 nM S-CDK, 65 nM DDK and all other proteins at the same concentrations as in standard reactions.
In Fig 5a lane 2, MCM loading factors (22.5 nM Orc, 45 nM Cdc6, 100 nM Cdt1·Mcm2-7) were pre-treated in loading buffer with 25 nM S-CDK for 10 min at 25°C before DNA addition for 20 min. When S-CDK was added after DNA (lane 1), MCM loading factors were incubated with DNA for 20 min and S-CDK then added for a further 10 min. Pre-treatment of individual MCM loading factors with S-CDK (Figs. 5b, c and Extended Data Fig. 5a, b) was performed under the same conditions as those used for MCM loading except that ORC was present at 22.5 nM and S-CDK and Sic1 were at 25 nM and 125 nM respectively. Proteins were incubated at 25°C with S-CDK for 10 min, S-CDK was inhibited with Sic1 (5 min) before the remaining proteins required for MCM loading were added and incubation continued for 20 min. To ensure S-CDK and Sic1 were removed from the above reactions, samples were subjected to two 150 μl washes in wash buffer + 600 mM NaCl and one 150 μl wash in wash buffer + 300 mM K-glutamate after either the MCM loading step (Fig. 5a), or after DDK phosphorylation (Fig. 5b, c and Extended Data Fig. 5a, b).
When Sld3/7 and Sld2 were pre-incubated with S-CDK (Fig. 5e, f and Extended Data Fig. 5c) the incubations were performed in the standard firing-factor recruitment buffers with 20 nM S-CDK for 7 min followed by addition of Sic1 (100 nM) for 3 min. The remaining firing factors were then added to the same concentrations as used in standard reactions.
For origin dependent replication reactions (Fig. 3c) ORC was pre-bound to DNA for 10 min at 30°C in reactions (10 μl) containing 25 mM HEPS-KOH pH 7.6, 50 mM KCl, 10 mM Mg(OAc)2, 5% Glycerol, 2 mM ATP, 1 mM DTT, 100 μg/ml BSA, 6.25 ng/μl ARS1 linear DNA beads and 2.5 nM ORC. The supernatant was removed, the beads washed twice with wash buffer + 300 mM KOAc and a new buffer (10 μl) containing 25 mM HEPES-KOH pH 7.6, 100 mM KOAc, 10 mM Mg(OAc)2, 0.02% NP-40-S, 5% glycerol, 2 mM DTT, 5 mM ATP, 45 nM Cdc6, 100 nM Cdt1·Mcm2-7 was added and the mix incubated at 30°C for 30 min. The buffer was removed and replaced with loading buffer + 130 nM DDK (10 μl) and the samples incubated at 25°C for 30 min. All downstream steps were performed as described for standard reactions.
Extract-based replication
For reactions on bead-bound DNA (Fig. 2d and Extended Data Fig. 2e), MCM and firing factors were assembled as described for standard protein recruitment and replication assays on immobilised ARS1 circular template (10 μl MCM loading reactions). Beads were washed twice with 150 μl wash buffer + 200 mM K-glutamate and resuspended in extract replication buffer (20 μl) containing 65 mM HEPES-KOH pH 7.6, 150 mM K-glutamate, 10.5 mM Mg(OAc)2, 5% glycerol, 0.5 mM EDTA, 0.5 mM EGTA, 40 mM creatine phosphate and 100 ng/μl creatine phosphokinase, 3.5 mM DTT, 2.5 mM ATP, 200 μM CTP, GTP, UTP, 100 μM dGTP, dATP, dTTP, 25 μM dCTP and 50 nM [α-32P]dCTP. Samples were incubated at 25°C for 30 min and were processed as described for standard replication reactions.
The soluble replication reaction in Extended Data Fig. 7b was performed as descried previously6 with unmodified ARS1 circular DNA as template. Products were separated through a 1% native agarose gel.
Oligonucleotides
JTY10(AAACCTATGAGACGACAGACAAGAAATCAGACAAAGGAATTAGATTCTCTGGAAGTGCTGTTTCAGGGCCCGCGTACGCTGCAGGTCGAC)
JTY32(CGATGATGAGACAATATCTAATAAAAGAGG)
JTY33(TCTTTATAGTCCTCGTCTGTGACTTCATC)
JTY40(GATGATGATGGGGACTATAAAGACGATGATGAGAC)
JTY41 (TTTATAATCCTCGTCTGTGACTTCATCGGC)
JTY92(AGCACAATAAGGCGCGCCTATAAAACAATGTTCAGGCAGTCAAAAAG) JTY100(CTTTTCCCTTCTCGAGTTAATGATTACCATTATTG)
Dpb11 fwd (AAGCTCACCGGTGATGAAGCCCTTTCAAGGAATA)
Dpb11 reverse(CTGAGGCGGCCGCTCAAGAATCTAATTCCTTTGT)
Sld3SUP C-term FLAG fwd (CTTCTAAGAGAAGAGTTAGAAGAAGATTGTTCGCTCCAGAATCTACTCTGGAAGTGCTGTTTCAGGGCCCGCGTACGCTGCAGGTCGAC)
Sld2SUP C-term FLAG fwd (GTTGCCAAAGAAGAACAGATTCTCCAACGGTAGATGGGGTAGAAGACTGGAAGTGCTGTTTCAGGGCCCGCGTACGCTGCAGGTCGAC)
Sld2SUP C-term FLAG rev (GAGAAAAGAAAAAAATTGATCTATCGATTTCAATTCAATTCAATTTAATCGATGAATTCGAGCTCG)
Sup sld3 TCP tag fwd (CTAAGAGAAGAGTTAGAAGAAGATTGTTCGCTCCAGAATCTACTGAAAACTTGTACTTCCAAGG)
Sup TCP tag rev (GGAAAGAGAAAAGAAAAAAATTGATCTATCGATTTCAATTCAATTCAATTACGACTCACTATAGGG)
Cdc45 flag tag rev(CGACTCACTATAGGGCGAATTGGAGCTCCACCGCGGTGGCGGCCGCTTAATCGATGAATTCGAGCTCG)
Cdc45 fwd(CTGGACACCGGTGATGTATTATGGAATCAGCCAG)
Cdc45 rev(AGCACGCGGCCGCTTATAACAATCCACTCAAGGT)
ARS1_XmaI-F(CGATCCCGGGGGTAGTTATAAGAAAGAGACCGAGTTAG)
ARS1_XhoI-R(CGATCTCGAGAAGAGTATTGGCGATGACGAAAC)
ARS1-PC-Bio-F(Bio-GGTAGTTATAAGAAAGAGACCGAGTTAG)
ARS1-Bio-R(AAGAGTATTGGCGATGACGAAAC)
MutA_F(GCATAAAAGATCTAAACATACCTCGAGGAAAATAACAAGATGTAAAG)
MutA_R(CTTTACATCTTGTTATTTTCCTCGAGGTATGTTTAGATCTTTTATGC)
MutB2_F(GTTATTACTGAGTAGTATTTCCTCGAGGATTGTTTGTGCACTTGCCTG)
MutB2_R(CAGGCAAGTGCACAAACAATCCTCGAGGAAATACTACTCAGTAATAAC)
Antibodies
Anti-Mcm7 (yN-19, sc-6688, Santa Cruz), anti-Orc6 (SB49). Psf2- FLAG was visualized with ANTI-FLAG M2-peroxidase (Sigma), Mcm10 was detected using the T7-tag antibody (69522, Novagen), Dbf4 was visualized with an anti-CBP tag antibody (07-482 Merck Millipore) and Rpa1 was detected with anti-scRPA (AS07 214, Agrisera). Psf1 antibodies were a gift from K. Labib. Polyclonal antibodies against Sld3, Sld7, Sld2, Cdc45 and Dpb11 were described previously6. Polyclonal antibodies against the Dpb2 subunit of pol ε were raised against full-length protein, and their specificity was confirmed using purified pol ε.
Extended Data
Extended Data Figure 1. Coomassie stained SDS-PAGE analysis of multisubunit complexes required for DNA replication.
a, Annotation of the polyacrylamide gels in Figs. 1a in which individual protein subunits have been labelled. b, Analysis of A-Cdk2. The protein complex consists of human Cdk2 and the bovine cyclin A-3 fragment32.
Extended Data Figure 2. Analysis of firing factor recruitment.
a, Immunoblots of protein recruitment conducted as in Fig. 1b but with 0.3 M KCl washes. b, Stability of recruited firing factors following washes of varying strength (lanes 1 and 5, 0.3 M K-glu; lanes 2 and 6, 0.3 M KCl; lanes 3 and 7, 0.45 M KCl; lanes 4 and 8, 0.6 M KCl). c,, Psf2-FLAG was depleted from a yJY18 S phase extract by two rounds of incubation with ANTI-FLAG M2 magnetic beads. Levels of Psf2-FLAG were determined by immunoblotting with the FLAG-M2 antibody. Soluble and bead bound protein fractions are illustrated. d, Extract-based replication reaction schemes. In pathway (i), loaded MCM is treated with DDK and added to a KO3 extract (Sld3, Sld7, Cdc45, Dpb11, Sld2 over-expression). For pathway (ii), firing factors are recruited to MCM as illustrated in Fig. 1b and the complex is added to a yJY18 extract (no firing factor over-expression) in which Psf2 (GINS complex) has been depleted. e, Replication reactions as described in d using A-Cdk2 for firing factor recruitment. Where indicated, Sic1 was added to the extract 20 min prior to replication.
Extended Data Figure 3. Characterisation of the in vitro DNA replication reaction.
a, b, Replication reactions conducted as in Fig. 3a with A-Cdk2 on ARS1 circular DNA templates for 1 hour. c, Time course of a standard replication reaction using A-Cdk2 on the ARS1 linear DNA template. d, Quantitation of a time course conducted as in c. DNA synthesis was normalised to the total DNA synthesis at 90 minutes. Small and large replication products were not quantified separately at 5 min as they are not well resolved at this time point. e, Pulse chase experiment conducted with A-Cdk2 on the ARS1 linear DNA template. For the pulse the dCTP concentration was reduced to 4 μM. Following a 10 min incubation unlabelled dCTP was added to 100 μM.
Extended Data Figure 4. Characterisation of RPA recruitment.
Unless stated reactions were conducted on ARS1 circular templates. a, Vaccinia virus topoisomerase I supports DNA replication with purified proteins. Replication reactions with either Topo II (25 nM) or Vaccinia virus topoisomerase I (0.125 units/μl). Two different Topo II fractions (Fr1 and Fr2) were used for comparison. b, Nucleotide dependence of RPA recruitment in a complete replication reaction with Topo II. c, RPA recruitment reactions were conducted on ARS1 circular template in the presence of Vaccinia virus topoisomerase I (0.125 units/μl), or on a linear template in the absence of a topoisomerase. dNTPs, C/G/UTP, pol α and Ctf4 were omitted from the final step of the reaction.
Extended Data Figure 5. Regulation of MCM loading and origin firing by S-CDK is ATP dependent.
a, Replication reaction where ORC was pre-incubated with S-CDK prior to MCM loading. When Sic1 was added before ORC the mix was incubated for 5 min and ORC was then added for 10 min. b, Pre-incubation of ORC with S-CDK in the presence or absence of ATP. After incubation with Sic1, ATP was added to the reaction lacking ATP. c, Sld3/7 and Sld2 were pre-incubated with S-CDK as illustrated in Fig. 5d in the presence or absence of ATP. Following incubation with Sic1, samples that did not contain ATP for the pre-incubation step were supplemented with ATP.
Extended Data Figure 6. Cartoon illustrating protein kinase regulated eukaryotic DNA replication origin firing with purified proteins.
Firing factors are recruited to loaded MCM in a DDK- and CDK-dependent manner. DNA synthesis is initiated once the DNA template has been unwound. CDK also functions to inhibit MCM loading by phosphorylating ORC.
Extended Data Figure 7. Internally FLAG-tagged Cdc45 supports normal DNA replication in S phase extracts.
a, The previously reported interaction between Sld3 and Cdc4533 was exploited to co-immunoprecipitate Cdc45 from yJY16 extracts (Dpb11 and Sld2 over-expression) by incubation with FLAG-Sld3/7 that was pre-coupled to ANTI-FLAG M2 magnetic beads. b, In vitro extract-based replication reaction on soluble ARS1 circular template using yJY16 extracts where endogenous Cdc45 was depleted as indicated. The extract was supplemented with purified Sld3/7 as the complex is not over-expressed in yJY16. The experiment was conducted for 30 min as described previously6 and products were separated through a 1% native agarose gel. Internally FLAG-tagged Cdc45 (52 nM) was added back as indicated. The locations of the different replication products are illustrated.
Extended Data Table 1. Affinity tag strategies for protein purification.
All N-terminal tags are located immediately upstream of original start codon. All C-terminal tags are located immediately before the original stop.
| Protein | Yeast strain | Affinity tag strategy | Affinity tag sequence |
|---|---|---|---|
| Cdc45 | yJY13 | Internal 2xFLAG tag | E197 – DYKDDDG – D198, E199Y, E200K, E202D * |
| Sld3/7 | yTD6 | C-terminal TCP tag on Sld3 | ENLYFQGEKRRWKKNFIAVSAANRFKKISSSGALDYDIPTTASKTAALAQHDEAVDNKFNKEQQNAFYEILHLPNLNEEQRNAFIQSLKDDPSQSANLLAEAKKLNDAQAPKVDNKFNKEQQNAFYEILHLPNLNEEQRNAFIQSLKDDPSQSANLLAEAKKLNGAQAPKVDANSAGKST |
| Sld3/7 | yTD11 | C-terminal 3xFLAG tag on Sld3 | LEVLFQGPRTLQVDDYKDDDDKDYKDDDDKDYKDDDDK |
| Sld2 | yTD8 | C-terminal 3xFLAG tag | LEVLFQGPRTLQVDDYKDDDDKDYKDDDDKDYKDDDDK |
| Dpb11 | yJY26 | C-terminal 3xFLAG tag | LEVLFQGPRTLQVDDYKDDDDKDYKDDDDKDYKDDDDK |
| Pol ε | yAJ2 | C-terminal CBP tag on Dpb4 | ENLYFQGEKRRWKKNFIAVSAANRFKKISSSGAL |
| S-CDK | yAE37 | N-terminal CBP tag on Clb5 | MKRRWKKNFIAVSAANRFKKISSSGALENLYFQGE |
| Pol α - primase | yJY23 | N-terminal CBP tag on Pri1 | MKRRWKKNFIAVSAANRFKKISSSGALENLYFQGE |
| Ctf4 | yAE40 | N-terminal CBP tag | MKRRWKKNFIAVSAANRFKKISSSGALENLYFQGE |
| Topo II | yAE46 | C-terminal CBP tag | ENLYFQGEKRRWKKNFIAVSAANRFKKISSSGAL |
| GINS | N/A | N-terminal 6His tag on Psf3 | MGSSHHHHHHSSGLVPRGSHMAS |
| Mcm10 | N/A | N-terminal 6His tag | MGSSHHHHHHSSGLVPRGSHMASMTGGQQMGRGSEF |
| RPA | yAE31 | N-terminal CBP tag on Rfa1 | MKRRWKKNFIAVSAANRFKKISSSGALENLYFQGE |
Amino acid residue numbers correspond to the wild type Cdc45 sequence.
Extended Data Table 2. Saccharomyces cerevisiae strains.
| Strain | Genotype | Reference |
|---|---|---|
| yTD6 |
MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100, bar1::Hyg pep4::KanMX, his3::HIS3pRS303/SLD3-TCP, GAL4, leu2::LEU2pRS305/SLD7 |
This study |
| yTD8 |
MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::Hyg pep4::KanMX his3::HIS3pRS303/Sld2-3xflag (Nat-NT2), GAL4 |
This study |
| yTD11 |
MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::Hyg pep4::KanMX his3::HIS3pRS303/SLD3-3xflag (Nat-NT2), GAL4 leu2::LEU2pRS305/SLD7 |
This study |
| yJY7 |
MATɑ
ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100, cdc7-4 pep4::KanMX |
This study |
| yJY13 |
MATa
ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::Hyg pep4::KanMX his3::HIS3pRS303/Cdc45iflag2, GAL4 |
This study |
| yJY16 |
MATɑ
ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100, cdc7-4 pep4::KanMX ura3::URA3pRS306/Dpb11 trp1::TRP1pRS304/Sld2 |
This study |
| yJY18 |
MATɑ
ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100, cdc7-4 pep4::KanMX Psf2-3xFlag (Nat-NT2) |
This study |
| yJY23 |
MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::Hyg pep4::KanMX trp1::TRP1pRS304/Pol1/Pol12 ura3::URA3pRS306/CBP-Tev-Pri1/Pri2 |
This study |
| yJY26 |
MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::Hyg pep4::KanMX his3::HIS3pRS303/Dpb11-3xflag (Nat-NT2), GAL4 |
This study |
| yAJ2 |
MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::Hyg pep4::KanMX ura3::URA3pRS306/Dpb2, Dpb3 trp1::TRP1pRS304/Pol2, Dpb4-Tev-CBP |
This study |
| yAE31 |
MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::Hyg pep4::KanMX his3::HIS3pRS303/CBP-Tev-RFA1, GAL4 ura3::URA3pRS306/RFA2, RFA3 |
This study |
| yAE37 |
MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::Hyg pep4::KanMX ura3::URA3pRS306/CKS1, CDC28 his3::HIS3pRS303/CBP-Tev-Clb5, GAL4 |
This study |
| yAE40 |
MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::Hyg pep4::KanMX his3::HIS3pRS303/CBP-Tev-Ctf4, GAL4 |
This study |
| yAE46 |
MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1::Hyg pep4::KanMX trp1::TRP1pRS304/TOP2-Tev-CBP |
This study |
Extended Data Table 3. Plasmids used to generate yeast expression strains.
Synthetic constructs were codon optimised for expression in Saccharomyces cerevisiae34. Sequences from the 5′ and 3′ end of the PGK1 gene were also added. Genes were synthesised by GeneArt Gene Synthesis (Life technologies).
| Plasmid | Original vector* | Insert | Plasmid construction |
|---|---|---|---|
| pTD2 | pJF2 | Dpb11 | PCR product from S. cerevisiae W303 genomic DNA using primers Dpb11 fwd and Dpb11 reverse cloned 5′ - SgrAI , 3′ - NotI |
| pTD5 | pJF2 | Sld2 | Synthetic construct cloned 5′ - SgrAI , 3′ - NotI |
| pTD6a | pJF2 | Sld3 | Synthetic construct cloned 5′ - SgrAI , 3′ - NotI |
| pTD6b | pJF4 | Sld7 | Synthetic construct cloned 5′ - SgrAI , 3′ - NotI |
| pRS303-Cdc45 | pJF2 | Cdc45 | PCR product cloned from S. cerevisiae W303 genomic DNA using primers cdc45 fwd and cdc45 rev 5′ - SgrAI , 3′ - NotI |
| pRS303-Cdc45iFlag1 | pJF2 | Cdc45iFlag1 | Site directed mutagenesis using oligonucleotides JTY32 and JTY33 and pRS303-Cdc45 as template |
| pRS303-Cdc45iFlag2 | pJF2 | Cdc45iFlag2 | Site directed mutagenesis using oligonucleotides JTY40 and JTY41 and pRS303-Cdc45iFlag1 as template |
| pRS304 (Pol2 + Dpb4-CBP) | pJF18 | Pol2 Dpb4-CBP |
Pol2 - synthetic construct cloned 5′ - AscI , 3′ - XhoI Dpb4-CBP - synthetic construct cloned 5′ - SgrAI , 3′ - NotI |
| pRS306 (Dpb2 + Dpb3) | pJF19 | Dpb2 Dpb3 |
Dpb2 - synthetic construct cloned 5′ - SgrAI , 3′ - NotI Dpb3 - synthetic construct cloned 5′ - AscI , 3′ - XhoI |
| pRS305/Gal-CBP-Top2 | pCG001 | Top2 | Synthetic construct cloned 5′ - AscI , 3′ - XhoI |
| pRS303/Gal-CBP-Ctf4 | pJF2 | Ctf4 | Synthetic construct cloned 5′ - SgrAI , 3′ - NotI |
| pRS303/Gal4 + CBP-Tev-Clb5 | pJF2 | Clb5 | Synthetic construct cloned 5′ - SgrAI , 3′ - NotI |
| pRS306/Cks1-Gal-Cdc28 | pJF5 | Cks1 Cdc28 |
Cks1 - synthetic construct cloned 5′ - SgrAI , 3′ - NotI Cdc28 - synthetic construct cloned 5 - AscI , 3 - XhoI |
| pRS303/Gal4 + CBP-Tev-Rfa1 | pJF2 | Rfa1 | Synthetic construct cloned 5′ - SgrAI , 3′ - NotI |
| pRS306/Rfa2-Gal-Rfa3 | pJF5 | Rfa2 Rfa3 |
Rfa2 - synthetic construct cloned 5′ - AscI , 3′ - XhoI Rfa3 - synthetic construct cloned 5′ - SgrAI , 3′ - NotI |
| pRS304/Pol1-Gal1-10-Pol12 | pJF3 | Pol1 Pol12 |
Pol1 - synthetic construct cloned 5′ - SgrAI , 3′ - NotI Pol12 - synthetic construct cloned 5′ - AscI , 3′ - XhoI |
| pRS306/CBP-Tev-Pri1-Gal1- 10-genomic Pri2 | pJF5 | Pri1 Pri2 |
Pri 1 - synthetic construct cloned 5′ - AscI , 3′ - XhoI Pri2 - PCR product from S. cerevisiae W303 genomic DNA using primers JTY92 and JTY100 cloned 5′ - AscI , 3′ - XhoI |
Supplementary Material
Acknowledgements
We are grateful to Boris Pfander and Max Douglas for the Mcm10 expression plasmid and advice on purification, Christoph Kurat for construction of plasmids used to generate ARS1 linear templates, Kenneth On and Dominik Boos for Sic1 and A-Cdk2 and Karim Labib for Psf1 antibodies and the E. coli GINS expression strain. This work was supported by Cancer Research UK, a FEBS Return-to-Europe fellowship to J.T.P.Y., a Boehringer Ingelheim studentship to A.J. and an ERC grant (249883 – EUKDNAREP) to J.F.X.D.
Footnotes
Author contributions
J.F.X.D and J.T.P.Y designed the experiments and wrote the manuscript. J.T.P.Y performed the experiments. T.D.D, A.J and A.E generated protein expression constructs and strains, and established protein purification protocols.
References
- 1.Tanaka S, Araki H. Helicase Activation and Establishment of Replication Forks at Chromosomal Origins of Replication. Cold Spring Harbor perspectives in biology. 2013 doi: 10.1101/cshperspect.a010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siddiqui K, On KF, Diffley JFX. Regulating DNA Replication in Eukarya. Cold Spring Harbor perspectives in biology. 2013 doi: 10.1101/cshperspect.a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gros J, Devbhandari S, Remus D. Origin plasticity during budding yeast DNA replication in vitro. EMBO J. 2014;33:621–636. doi: 10.1002/embj.201387278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.On KF, et al. Prereplicative complexes assembled in vitro support origin-dependent and independent DNA replication. EMBO J. 2014;33:605–620. doi: 10.1002/embj.201387369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heller RC, et al. Eukaryotic Origin-Dependent DNA Replication In Vitro Reveals Sequential Action of DDK and S-CDK Kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- 10.Zegerman P, Diffley JFX. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka S, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 12.Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- 13.Randell JC, et al. Mec1 Is One of Multiple Kinases that Prime the Mcm2-7 Helicase for Phosphorylation by Cdc7. Mol Cell. 2010;40:353–363. doi: 10.1016/j.molcel.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463:113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 18.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 19.Gambus A, et al. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowles A, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J Cell Sci. 1999;112:2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watase G, Takisawa H, Kanemaki MT. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr Biol. 2012;22:343–349. doi: 10.1016/j.cub.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 22.van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 2012;31:2195–2206. doi: 10.1038/emboj.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgescu RE, et al. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol. 2014;21:664–670. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clausen AR, et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struct Mol Biol. 2015 doi: 10.1038/nsmb.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu C, et al. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell. 2014;56:551–563. doi: 10.1016/j.molcel.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Bell SP. CDK prevents Mcm2-7 helicase loading by inhibiting Cdt1 interaction with Orc6. Genes Dev. 2011;25:363–372. doi: 10.1101/gad.2011511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frigola J, Remus D, Mehanna A, Diffley JFX. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–343. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 29.Boos D, et al. Regulation of DNA Replication through Sld3-Dpb11 Interaction Is Conserved from Yeast to Humans. Curr Biol. 2011;21:1152–1157. doi: 10.1016/j.cub.2011.05.057. [DOI] [PubMed] [Google Scholar]
- 30.Simon AC, et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature. 2014;510:293–297. doi: 10.1038/nature13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coster G, Frigola J, Beuron F, Morris EP, Diffley JFX. Origin Licensing Requires ATP Binding and Hydrolysis by the MCM Replicative Helicase. Mol Cell. 2014;55:666–677. doi: 10.1016/j.molcel.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown NR, et al. The crystal structure of cyclin A. Structure. 1995;3:1235–1247. doi: 10.1016/s0969-2126(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 33.Itou H, Muramatsu S, Shirakihara Y, Araki H. Crystal structure of the homology domain of the eukaryotic DNA replication proteins Sld3/Treslin. Structure. 2014;22:1341–1347. doi: 10.1016/j.str.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.