Abstract
The fibroblast growth factors (FGFs) are a family of cell intrinsic regulatory peptides that control a broad spectrum of cellular activities. The family includes canonic FGFs that elicit their activities by activating the FGF receptor (FGFR) tyrosine kinase and non-canonic members that elicit their activities intracellularly and via FGFR-independent mechanisms. The FGF signaling axis is highly complex due to the existence of multiple isoforms of both ligands and receptors, as well as cofactors that include the chemically heterogeneous heparan sulfate (HS) cofactors, and in the case of endocrine FGFs, the Klotho coreceptors. Resident FGF signaling controls embryonic development, maintains tissue homeostasis, promotes wound healing and tissue regeneration, and regulates functions of multiple organs. However, ectopic or aberrant FGF signaling is a culprit for various diseases, including congenital birth defects, metabolic disorder, and cancer. The molecular mechanisms by which the specificity of FGF signaling is achieved remain incompletely understood. Since its application as a druggable target has been gradually recognized by pharmaceutical companies and translational researchers, understanding the determinants of FGF signaling specificity has become even more important in order to get into the position to selectively suppress a particular pathway without affecting others to minimize side effects.
It has been gratifying for early basic researchers on fibroblast growth factors (FGF) that their Cinderella in the growth factor arena is now drawing so much attention as a druggable target by pharmaceutical companies and translational researchers. The first two prototype FGFs, FGF1 and FGF2, discovered in the early seventies, were designated acidic and basic FGF (aFGF and bFGF) based on their activity to stimulate fibroblast proliferation and their isoelectric point (1, 2). Subsequently 20 more FGF homologues have been identified as the family members in mammals (3–20). Genes coding for a large number of FGFs were cloned based on homology in the amino acid sequence. It was soon found that the name “fibroblast growth factor” was not the best name to describe the diverse functions of the family members and their receptors since many FGFs do not even have receptors expressed in fibroblasts and elicit no activity in fibroblasts. In addition, many FGFs induce diverse cellular responses beyond growth promoting signals in different target cells. Despite being misleading to some degrees, the name “fibroblast growth factor” followed by a number (FGF1, 2, 3, 4, etc.) has been preserved and replaced numerous other names used to describe either tissue origin, target, function, or properties of the FGF molecule. FGF signaling has long not been a favorite of pharmaceutical companies largely because of the diversity of both ligands and receptors in the family, its wide range of target cell types, diverse functions, and complexity of FGF signals that intersect either directly or indirectly with multiple pathways. The complexity of the multi-subunit transmembrane FGF signaling complex in both the extracellular and the intracellular portions has also been a major factor. Several cofactors are integral regulatory components of the FGF signaling complex. These include the chemically heterogeneous heparan sulfate (HS) cofactors, and in the case of endocrine FGFs, the Klotho coreceptors. These cofactors and coreceptors not only participate in FGF receptor-binding specificity and affinity, but also in specifying signaling activities. Therefore, a full understanding of the molecular mechanisms underlying the specificity of FGF signaling is important for therapeutic usage of FGFs.
1. FGF signaling axis
FGFs
The FGFs are single chain polypeptides that are tissue regulatory molecules controlling a broad spectrum of cellular processes in both embryonic and adult tissues. The polypeptides have one conserved domain flanked by non-conserved extensions (Fig. 1A). Most FGFs have an N-terminal signal peptide that facilitates secretion through classical mechanisms. However, several FGFs, including FGF1 and FGF2, do not have a cleavable signal peptide and are secreted in a non-conventional manner. Seven FGF subfamilies have been defined based on their sequence homology and function (Fig. 1B). These FGF subfamilies can also be divided into two general groups, the canonical FGFs comprising paracrine or autocrine-acting FGF1–10, FGF16–18, FGF20, and FGF22 and the endocrine-acting FGFs, FGF15 (mouse)/FGF19 (human), FGF21, and FGF23; and the non-canonical FGFs comprising FGF11–14. The canonical FGFs elicit regulatory functions through high affinity binding to and activating FGF receptors (FGFR). An autocrine canonical FGF acts on the cells of origin as a self-stimulator, and a paracrine FGF is secreted by one cell and acts on another locally within tissues. In contrast, the endocrine FGF originates at a distal organ site and reaches the target through the blood circulation in a classical endocrine mode of action. The non-canonical FGFs do not bind to the FGFR but elicit their activities intracellularly, such as through interaction with voltage-gated sodium channels and calcium channels (21–23).
Fig. 1. The FGF family.

A. Schematic of the FGF. Red box, signal peptide; open boxes, non-conserved, N- and C-terminal domains; solid box, conserved core domain. B. FGF sub-families. The 22 FGFs are grouped into 7 subfamilies based on sequence homology and function.
FGFRs
The FGFR is a single chain transmembrane tyrosine kinase that consists of a ligand binding extracellular domain, a single transmembrane domain, and an intracellular tyrosine kinase domain that is separated into two parts by an insertion domain (Fig. 2). The mammalian FGFR is encoded by four highly homologous genes (24–27). Except for the Fgfr4 gene for which only one splice isoform occurs naturally (28), other three Fgfrs have been found to encode multiple splice variants. These splice variants generate diversity of sequence and function in the ligand-binding extracellular domain and the intracellular substrate-binding and kinase domains (29). It has been speculated that the combination of FGFR1 splice variation sequences can potentially encode up to 256 splice isoforms (29). FGFR3 and FGFR4 have 3 immunoglobin (Ig)-like domains in the extracellular domains. As a consequence of alternative splicing, the extracellular domain of both FGFR1 and FGFR2 can contain either 2 or 3 Ig-like loops. The presence of the first Ig-loop modulates the affinity for both FGF and FGFR-binding heparin/heparan sulfate (30–32). Two major isoforms generated by alternative splicing in the second half of Ig-loop III, namely IIIb and IIIc in FGFR1, FGFR2, and FGFR3 have been reported. This variation defines ligand-binding affinity and specificity of FGFR1–3 (33, 34). Several other splice variations at the extracellular domain have been found in FGFR2, although the functional significance of these variants remains unknown (35). The role of the alternatively spliced dipeptide VT (valine-threonine) in the intracellular juxtamembrane domain of FGFR is controversial. It has been shown that the presence of VT is required for FGFR to bind FRS2α and FRS2β and therefore contributes to signaling specificity (36, 37). However, other reports show that the dipeptide is dispensable for the binding of FRS2α and FRS2β to FGFR1 even though it enhances the binding affinity between substrate and the receptor kinase (38). The variations in the kinase domain and C-terminal tail following the kinase domain of FGFRs have only been found in cancer cells (29, 39). Although the kinase domains of the four FGFR isotypes are highly homologous (>80%) in the primary amino acid sequence and share common tyrosine phosphorylation sites (Fig.2), the four FGFRs elicit receptor-, cofactor-, coreceptor-, and cell type-specific activities in cells (40–43). Seven major tyrosine autophosphorylation sites have been identified in the FGFR1 kinase domain (44–47). Y (tyrosine) 653 is predominant in activation (derepression) of the receptor kinase activity (44–47) while Y654 contributes to maximal activation (48). Phosphorylation of Y766 is required for recruiting phospholipase Cγ (PLCγ) via its SH2 domains to the FGFR1 kinase (49–51). Y463 is a binding site for the adapter proteins CRK and CRK-like (52–54) and phosphorylated Y730 is a binding site for the 85 kDa regulatory subunit alpha of phosphatidylinositol 3-kinase (PI3K) (45, 49, 55). The function of other phosphorylation sites, including Y583, and Y585 has not been clearly established, despite some evidence that they contribute to the intensity and extent of FGFR signaling (48). An FGFR2 splice variant that lacks exon 16 has been reported in prostate epithelial cells, which does not have the PLCγ-binding site (56). The significance of this splice variant remains to be elucidated.
Fig. 2. Topology of a prototypical FGF receptor tyrosine kinase.
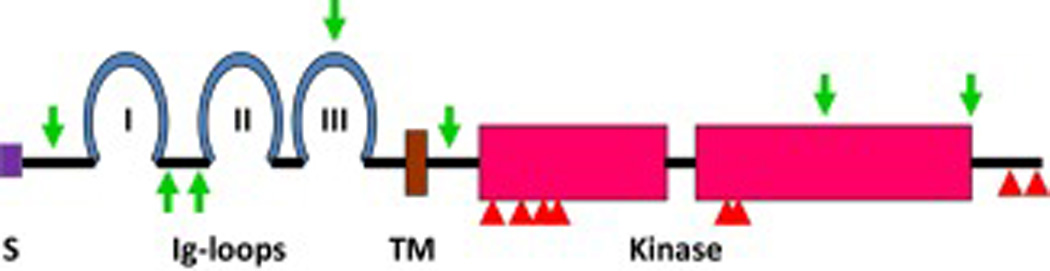
S, signal peptide; I, II, III, immunoglobulin-like domain 1, 2, and 3; TM, transmembrane domain. Red box, tyrosine kinase domain that is separated by a kinase insertion sequence; green arrows, alternative splice sites; triangles, tyrosine phosphorylation sites.
Heparan sulfate (HS) cofactors
HS is the glycan component of proteoglycans in the pericellular matrix and on the cell surface. It is a highly heterogeneous glycosaminoglycan (46, 57–59). Variations in degrees and patterns of sulfation on HS motifs affect their interaction with FGFs and FGFRs and have been shown to play a role in determination of ligand-binding and downstream signaling specificity of FGFR complexes (60–65). Although HS motifs with high affinity for FGFs and FGFRs are normally sulfated, emerging evidence shows that the affinity is not simply proportional to total charge density and degree of sulfation. Instead, these high-affinity motifs are often less than fully sulfated and have unique sulfation patterns (66, 67). Because of affinity for both FGF and FGFR, HS in the tissue environment largely plays two general roles that impact overall FGFR signaling. Tissue matrix HS acts as an FGFR-independent depot and stabilizes influence for canonical paracrine/autocrine FGFs (68, 69). It limits access of FGF that are generally long-lived and at considerable concentrations in the matrix to cell membrane FGFR except when needed (46). The second role is as an integral part of the FGF signaling complex through a distinct HS-binding domain in the extracellular domain of FGFR. Motifs within this class of HS are thought to be less abundant and potentially more specific than matrix HS (66, 67). FGFR-bound HS interacts concurrently with both FGFR and FGF within the FGF-FGFR-HS signaling complex.
Both our early models based on protein mutagenesis and in silico modeling (70, 71) and the crystal structure (72) of the FGF2-FGFR1c-HS complex show a 2-2-2 complex of FGF-FGFR-HS, in which one single heparan sulfate chain may contact Ig-loop II of one FGFR, the inter-Ig-loop connector sequence and Ig-loop III of the same FGFR, and extend to Ig-loop III of the adjacent FGFR in the FGFR dimer. Ig loops II and III cooperate both within monomers and across dimers with cellular HS to confer cell type-dependent specificity of the FGFR complex for FGF ligands (73). It is unclear whether FGF stabilizes proximity of random interactions of monomeric units or activates a pre-existing inactive oligomeric complex of HS-FGFR via conformation changes (46, 61, 66, 67). However, emerging data pose that together with HS, the FGFR forms a dimer constitutively in the absence of FGF (70, 71, 74). Interaction with HS restrains FGFR dimers in an inactive conformation and FGF binding converts the HS-FGFR complex from the inactive repressed conformation to an active arrangement that allows an initial trans-phosphorylation between the kinase domains of FGFR dimers. Structural studies indicate that trans-phosphorylation of tyrosine 653 and then 654 (in FGFR1) changes conformation of an autoinhibitory loop within the kinase domain that normally restricts access of substrates to the active site of the FGFR kinases (48, 75).
Klothos
Unlike paracrine/autocrine FGFs, the endocrine FGFs, FGF15/FGF19, FGF21, and FGF23 have little or no affinity for HS (76). This property permits their endocrine circulation and movement through tissue matrices without being trapped and stored prior to reaching distal target cells. The specificity of endocrine FGF signaling at the cellular level is directed by a family of membrane-anchored proteins that include αKlotho (αKL) and βKlotho (KLB). Although endocrine FGFs signal through the same FGFR as canonical FGFs, they have little affinity for FGFR in the absence of Klothos. KLB interacts with FGFR independent of FGF and with FGF independent of FGFR (77–79). Although αKL binds the extracellular domain of FGFR1, it poorly interacts with FGF23. Instead, binding of αKL with FGFR1 forms a de novo site generated at the composite FGFR1c-αKL interface, which binds the C-terminal domain of FGF23 (80). Thus, the major role of αKL and KLB is to facilitate high affinity binding and subsequent activation of FGFRs by endocrine FGFs. αKL specifically facilitates binding of FGF23 to its receptor and controls mineral metabolism via a vitamin D controlled bone to kidney axis where αKL is expressed (81). Inactivation of αKL induces hyperphosphatemia in mice that highly express FGF23 (82). The cofactor KLB facilitates high affinity binding and signaling of FGF19 (mouse FGF15) and FGF21 and controls cholesterol/bile acid, lipid, and glucose metabolism in the liver and adipocytes (83).
Expression of neither KLB nor FGFR4 alone affects cell population dynamics in KLB- and FGFR4-deficient cells. However, co-expression of KLB and FGFR4 restricts cell population growth via apoptosis in an endocrine FGF19 or FGF1 dependent manner (84). This indicates that the KLB interaction with the FGFR4 tyrosine kinase complex not only serves to confer high affinity for endocrine FGF19, but also plays a role in directing signaling of the FGFR4 complex independent of the activating FGF (84, 85).
The FGF signaling pathways
The signaling cascade downstream of the transmembrane receptor, including PLC-γ, MAP kinase, and PI3K pathways have been implicated in all four FGFR kinases (Fig. 3). Among them, PLC-γ binds to a specific phosphorylated tyrosine residue at the C-terminal tail of the FGFR kinases (50, 51). However, the MAP kinase and PI3K pathways need to be recruited to the FGFR by a membrane-anchored adaptor protein, FRS2α (FGF receptor substrate 2α), which undergoes an extensive pattern of tyrosine and serine/ threonine phosphorylation upon FGFR activation (86–93). In addition, CRK has also been proposed to serve as a functional adaptor that binds to the FGFR (52), which has been reported to further link the ERK pathway to FGFR1. The activation of the ERK pathway by the FGFR tyrosine kinase is tightly regulated by both positive and negative feedback loops at both transcriptional and post translational levels (Fig. 3). Sprouty (SPRY) proteins, which comprise four conserved members, SPRY1–4, present a feedback regulator of the FGF pathway at the posttranslational level (94). Tyrosine phosphorylation of SPRY creates a decoy site that binds the docking molecule GRB2 and prevents translocation of SOS to the plasma membrane to activate RAS. SEF (similar expression of FGF) inhibits binding of FRS2α to the FGFR and prevents activation of ERK, and, therefore, negatively regulates the RAS-MAPK pathway (95, 96). Activation of the ERK and PI3K/AKT pathways has been implicated in most FGFR regulatory functions. Deletion of the PLCγ binding site on FGFR1 does not affect FGFR1-elicited cellular responses, which include mitogenesis, neuronal differentiation, mesoderm induction, induction of urokinase-type plasminogen activator, and chemotaxis. However, the PLC-γ binding site is required for FGFR1 to induce benign prostate cancer cells to acquire the proliferative response to FGFR1, although it appears not to be required for the mitogenic response (97). This suggests that pathways linked to FGFR1 Y766 contribute to prostate cancer progression rather than playing a direct role in cell cycle and mitogenesis. Experiments with purified recombinant FGFRs in vitro or when overexpressed at high levels in cell lines, such as Sf9 insect cells or COS7 mammalian cells, indicate that the four FGFR isotypes exhibit similar if not identical substrate phosphorylation patterns (Wang, unpublished results). However, in the experiments with moderate expression levels in cells, the results are not consistent. In some experimental systems, the four FGFR isotypes elicit similar and redundant effects on cell phenotypes, and in others, exert different effects (40). Overexpression of the FGFR kinase at levels far beyond the minute normal cellular levels likely homogenizes and masks receptor and cell type specific effects of the different FGFR kinases. More sensitive analytical approaches in situ as well as robust and controlled experimental systems that hold promise of revealing and dissecting such differences are needed for developing FGFR isotype- and cell type-specific inhibitors or activators.
Fig. 3. Signaling pathways downstream of FGFR tyrosine kinase.
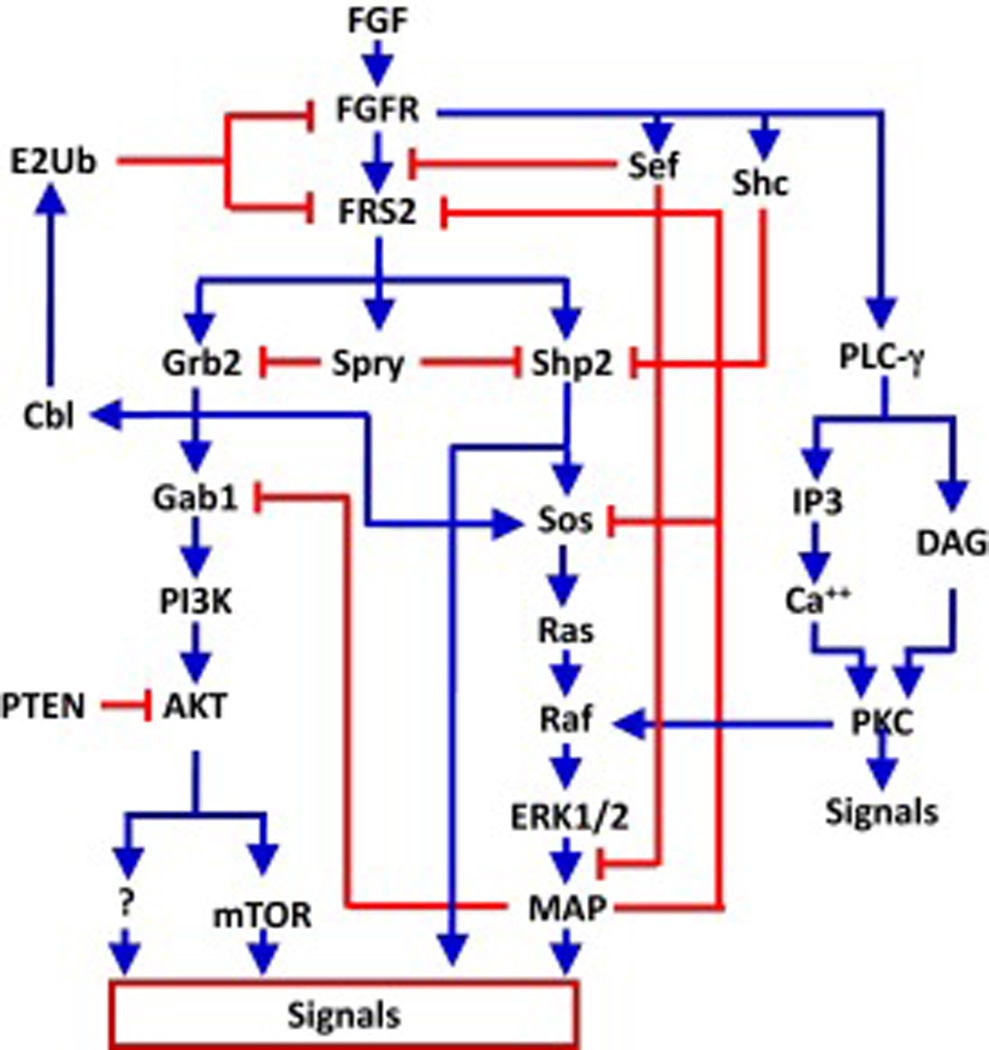
Shown is a wiring diagram with blue lines indicating positive effects, and red lines indicating negative effects.
The FRS2 family is composed of two highly homologous members, FRS2α and FRS2β, which belong to a category of adaptor proteins that have binding sites for molecules both upstream and downstream of it in signaling networks. It physically presents downstream molecules to the upstream molecules. Depletion of FRS2α abrogates the ability of FGFR kinases to activate the MAP kinase and PI3K/AKT pathways (Wang, unpublished data). Ablation of Frs2α in mice causes severe defects in embryonic development and results in early embryonic lethality at E7.0–7.5 (98). Although it is not clear whether the two FRS2 members are functionally redundant, expression of FRS2β in FRS2α-deficient cells restores the ability of FGFR1 to activate both the MAPK and PI3K/AKT pathways (99). In addition, FRS2α is also engaged in the feedback regulation of the FGF signaling pathway (100, 101). As illustrated in Fig. 3, FRS2 appears to be the key adaptor protein in the FGFR signaling cascade that mediates multiple downstream pathways of the FGFR, as well as control of the amplitude of the signaling intensity. However, whether FRS2 is also involved in the receptor and cell type specificity of signaling elicited by the FGFR kinases remains to be elucidated.
2. Translational application of the FGFs and their signaling pathways
Aberrant FGF signaling in diseases
In embryos, the FGF, FGFR kinase, and heparan sulfate components of the FGF signaling complex are expressed in a spatiotemporally- and cell-specific pattern that changes constantly as development proceeds. In adult organs, components of the FGF signaling axis are expressed in a cell type-specific mode and are important in the mediation of external signals and communication within compartments that maintain tissue homeostasis and function. Abnormal expression of FGF and FGFR and aberrant activation of the FGF signaling axis are frequently found associated with various adult tissue-specific pathologies and cause developmental disorders (40, 46, 53, 102"–110). The subversion of the homeostasis-promoting activity of resident epithelial FGFR2 in a variety of tissues (40, 41, 46, 111) and concurrent ectopic expression of normally mesenchymal FGFR1IIIc in epithelial cells (40, 112–115) is often found associated with tumor progression. Changes in core protein expression of HS proteoglycans as well as sulfation patterns have been reported to contribute to progression of premalignant tumor cells to malignancy (46). Currently, extensive efforts have been taken to explore the translational application of manipulation of the FGF signaling activities, both using the FGF directly and chemical agonists or antagonists as in the areas summarized hereafter. As a heparin-binding protein, delivery of heparin-binding FGF through the circulation and the tissue matrix remains a challenging issue. Treating large traumatic tympanic membrane perforation with FGF2 improved closure rates compared with the control group (116). Recently, new technologies, including multi-walled carbon nanotubes, have been used to deliver FGF2, which improves bone regeneration in animal models (117, 118).
Wound healing
As potent mitogenic factors, both FGF1 and FGF2 have been extensively explored for their potential in wound healing. FGF1 induces cell proliferation in the wounded area and promotes the cells to produce cytokines and other growth factors that induce migration of macrophages and monocytes towards the wounded area to remove damaged or dead cells (119, 120). FGF1 also induces epithelial cells and vessel endothelial cells to migrate toward the healing tissues. True to its original name, FGF promotes growth and differentiation of fibroblasts, and induces cells to release collagenase and plasminogen activators to promote angiogenesis in the wounded tissues (121, 122). In addition, FGF1 also down regulates αI procollagen expression and suppresses collagen production and deposit in fibroblasts and therefore prevents scar formation (123, 124).
Recombinant FGF1 and 2 (rFGF1 and rFGF2) have been developed for clinical trials in several countries. Since 1992, recombinant FGF2 has been used in several hospitals in China, and the results show that recombinant FGF2 improves healing in burn trauma, skin flap grafts, intractable cerebrospinal fluid rhinorrhea, intractable skin ulcer, postoperative mastoid cavity problems, pressure ulcers, chronically ischaemic tissue and traumatic ulcers, bone fracture, periodontitis-induced damage of human periodontal tissue, diabetic gangrene, diabetes-related chronic ulcers, peripheral artery disease, and gastric ulcers (124–149). The outcomes of the clinical trials demonstrate that FGF2 can be used as an agent to accelerate the healing of fresh and chronic wounds and improve the quality of healing of wounds of diverse types. In addition, FGF1 has been shown to elicit modest nerve regeneration after spinal cord injury (150). FGF7 acts exclusively through a subset of FGF receptor isoforms (FGFR2b) and has been developed by Amgen (palifermin) to prevent and speed up the healing of severe sores in the mouth and throat caused by chemotherapy and radiation therapy, which are used to treat cancers of the blood or bone marrow (151–156).
Cardiac protection
Heart failure also called congestive heart failure or congestive cardiac failure occurs when the heart fails to pump sufficient blood to meet the needs of the body. It remains a major cause of morbidity and mortality and causes critical health problems especially in Western societies. Reduction in the efficiency of the myocardium through overloading or damage leads to cardiac hypertrophy and fibrosis, which subsequently progresses to heart failure. Several members of the FGF family, including FGF1, FGF2, FGF5, FGF16, FGF21, and FGF23 have been shown to play roles in the heart. FGF signaling is essential for cardiomyocyte homeostasis through phosphorylating connexin 43 (Cx43), which is required for the maintenance of gap junctions (157). FGF1 is released from the myocardial tissue into pericardial fluid during severe myocardial ischemia (158).
Treatment with biodegradable hydrogel microspheres containing FGF2 improved left ventricle function and inhibited left ventricle remodeling by angiogenesis in pigs with chronic myocardial infarction (159). Intramyocardial injection of FGF-2 plus heparin suppresses the progression of cardiac failure in rat models (160). Similarly, treating pigs with adenovirus carrying FGF5 cDNA improves wall-thickening and cardiac function (161). In humans, FGF treatment has likewise shown cardioprotective effects: a single intracoronary infusion of rFGF2 shows trends toward symptomatic improvement of angina and myocardial function in patients with advanced coronary artery disease (162). Treatment of patients with a bicistronic VEGF/FGF2 plasmid improves cardiac function with respect to exercise tolerance and clinical symptoms (163). Intracoronary administration FGF-2 in patients with severe ischemic heart disease increases regional wall thickening and reduces the extent of the ischemic area (164). Treatment with Ad5-FGF4 results in favorable anti-ischemic effects (165). All these initial small and unblinded studies with FGF proteins or encoding cDNAs were encouraging and demonstrated both clinical improvement and evidence of angiogenesis. However, subsequent double-blind placebo-controlled trials did not confirm the initial high efficacy observed in the small trials (166). Future larger trials are needed to confirm whether FGF treatment is efficacious, safe, and practical for the heart failure patients.
Metabolic disorders
The FGFs are best known for their diverse roles in mediating cellular homeostasis through short-range cell-to-cell communication within tissues (167). However, FGF19 (or mouse FGF15), FGF21, and FGF23, have been identified as endocrine hormones since they originate in tissues distal to the metabolic organs they target and are transported through the circulation (17, 168–171). A diurnal physiologic role of the ileal FGF19-hepatocyte FGFR4 axis in bile acid metabolism during normal feeding has been established (169, 172). FGF21 regulates energy homeostasis mainly through activating the FGFR1/KLB complex in adipocytes (173), and represents a novel target for the development of therapies for the treatment of obesity, diabetes, and cardiovascular diseases.
Expression of FGF21 is controlled by a complex network of transcriptional regulators, which modulate FGF21 expression in response to a wide array of physiological stimuli or pharmacologic agents (174). The function of FGF21, if any, in normally fed mice is not clear. Generally, the liver FGF21-adipocyte FGFR1 signaling axis appears to come into effect only after prolonged starvation, when it uncouples lipid metabolism between the adipocytes and hepatocytes, prolonging the supply of lipid fuels to maintain lifesaving glucose levels as long as possible until a feeding opportunity arises (175). When administered, both FGF19 and FGF21 dramatically reverse obesity and its associated symptoms, including type 2 diabetes (176–178). Studies with tissue-specific Fgfr1-knockout animals have revealed that the adipocyte, via FGFR1, is the specific target of FGF19 and FGF21 that alleviates obesity and allied symptoms (173, 175, 179). Although several reports show that FGF21 controls ketogenic and triglyceride clearance in the liver (180–183), unlike FGF19, FGF21 is unable to bind FGFR4-KLB complex with affinity comparable to FGFR1-KLB (83, 170, 184", 185). Therefore, at physiological concentrations, FGF21 is unlikely to signal in the liver where FGFR4-KLB predominantly resides. It has been shown that the metabolic effects of FGF21 on glucose homeostasis and insulin sensitivity are mediated in part by controlling adiponectin production and release in adipocytes (186, 187). More recently, it has been shown that FGF21 is produced by a variety of tissues other than liver under other than extreme metabolic conditions as starvation or obesity (188). The common features of the conditions that elicit FGF21 in organisms cause diverse tissue and cellular stress. Thus it has been proposed that FGF21 is largely a stress hormone that calls on adipocytes and its metabolic and hormonal secretory products (adipokines) to alleviate diverse tissue stresses (188). Although numerous reports (189) suggest FGF21, which requires FGFR1-KLB, may directly target other tissues than adipocytes that are very low in KLB, the significance relative to adipocytes on overall FGF21 action remains to be determined.
Endocrine FGF19 and FGF21 act on the same FGFR1 that also mediates the effects of paracrine/autocrine FGFs on cellular homeostasis in developing and mature organs as well as driving numerous proliferative pathologies, such as cancer. However, the canonical cellular activities of FGF19 and FGF21, most prominently their mitogenicity, are prevented by the transmembrane co-factor KLB, which participates directly in the FGFR signaling complex and redirects its output to metabolic signaling (79, 84, 176). Co-expression of KLB directs FGFR4 signaling from growth-controlling to apoptosis-promoting, which may explain why FGFR4 elicits specific cellular context control of cell population expansion and tumor suppression rather than tumor promotion (84). Breast tumor progression in Fgfr4 null mice is delayed rather than accelerated (190). This correlates with a fortuitous chronic compensatory elevation of ileal FGF19 and an unexpected chronic elevation of circulating FGF21 in the FGFR4-knockout model (191), indicating that persistently elevated FGF19 and FGF21 has a tumor suppressive effect. The persistent elevation of FGF21 in cancer is consistent with its overall role as a stress hormone since cancer is a major source of stress on the organism affecting many tissues.
Obesity and its associated aspects of metabolic syndrome are strong promoters for several cancers, which include breast and prostate cancer. Normally, FGF19/FGF21 serves to maintain normal metabolic homeostasis between adipocytes and other tissues, primarily hepatocytes. Antitumorigenic effects of FGF21 may occur systemically through fat tissues and locally distributed adipocytes, via regulating release of their metabolites and adipokines, which affect tumor cells directly or indirectly by changing the tumor microenvironment (Fig. 4). Although FGF19 directly regulates specific aspects of hepatic contribution to metabolic homeostasis, FGF21 has exquisite specificity for adipocytes via FGFR1/KLB without direct effects on hepatic FGFR signaling. This very narrow physiologic role of FGF21 and adipocyte target specificity makes it especially attractive as a pharmacologic antiobesity, antidiabetic, and now antitumor agent for which few side effects are predicted.
Fig. 4. Endocrine FGFs suppress prostate tumor progression and metastasis by regulating adipokine secretion.
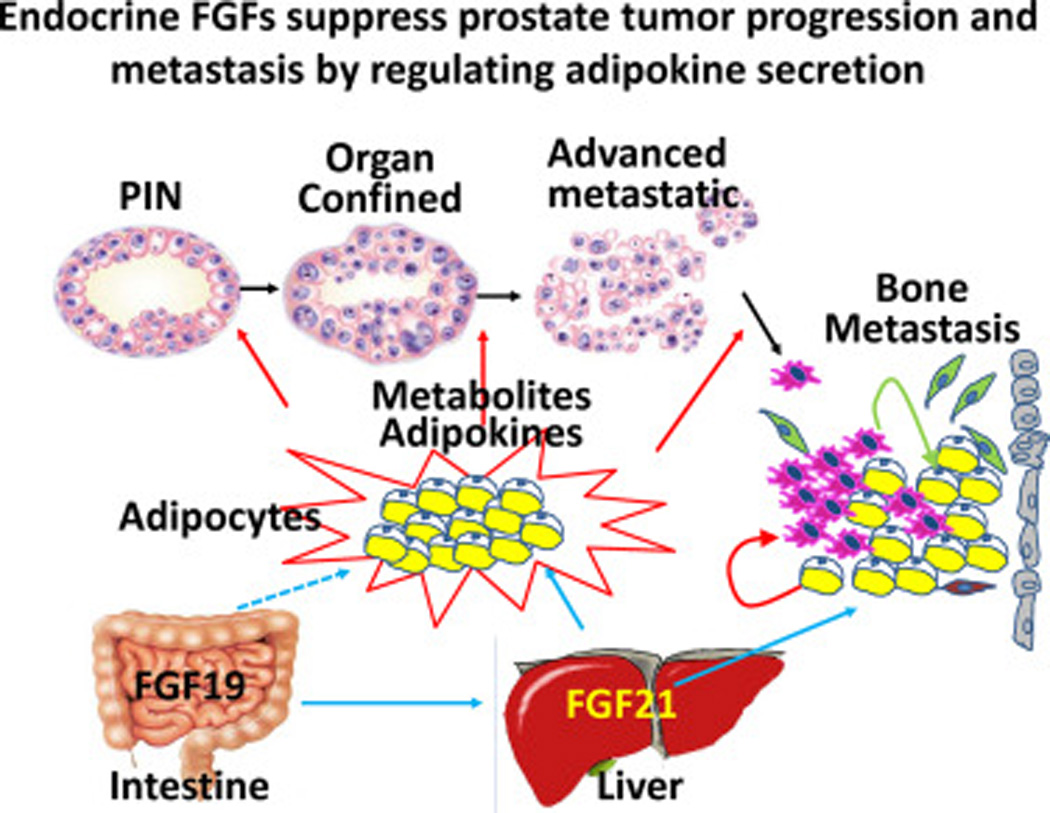
FGF21 produced by hepatocyte affects prostate cancer progression via controlling adipokine productions in both adipose tissues and local adipocytes in the tumor microenvironment.
Aberrant FGF signaling in cancer
Ectopic expression of FGF ligand or receptor, as well as mutations in the FGFR that cause activation of the FGF/FGFR signaling axis is common in many epithelial cancers including hepatocellular carcinoma, melanoma, lung, breast, bladder, endometrial, head and neck, and prostate cancers (192). Point mutations causing constitutive activation of FGFR3 have been detected in more than 60% of non-muscle invasive urothelial carcinomas (193), and a point mutation in the transmembrane domain of Fgfr4 has been reported in human prostate carcinoma (194). Gene amplification and mutations in the intronic sequence leading to overexpression of FGFR tyrosine kinases is also a mechanism underlying excessive FGF signaling in cancer. For example, amplification of chromosomal region 8p11–12, which encompasses Fgfr1, is frequently found in human prostate carcinoma (195) and approximately 10% of breast carcinomas (192), a point mutation in intron 2 of the Fgfr2 alleles has been found associated with breast cancer (196). Alternative splicing of FGFR resulting in variants with altered ligand specificities constitutes the third mechanism leading to aberrant FGF signaling in cancer (192). In addition, downregulation of feed-back controllers of FGF signaling, such as Spry or SEF can also contribute to oncogenic activity of FGF signaling (197).
Aberrant FGF signaling has been implicated in prostate carcinoma development and progression (40, 112, 198", 199). Elevated production of FGF ligands by prostatic secretory epithelial cells creates an autocrine signaling loop stimulating aberrant epithelial growth and cellular dysplasia and promoting independence from stromal regulation. Upregulation of FGF family members in primary prostate cancer correlates with higher grades of cancer and clinical stage (197, 200). FGF8 is expressed at low levels in normal prostate. However, FGF8 and its cognate receptors are overexpressed in human samples of prostatic intraepithelial neoplasia (PIN) and prostate carcinoma (201–204). Furthermore, overexpression of FGF8 is associated with decreased patient survival (205). Exogenous FGF1 induces expression of matrix metalloproteinases and promotes tumor metastasis in prostate carcinoma cells (206). Overexpression of FGF9 augments reactive stroma formation and promotes cancer progression in mouse models of prostate carcinoma (207). Attenuating FGF2 activity inhibits cell proliferation, migration, and invasion in cell culture (208, 209). Consistent with this, ablation of FGF2 inhibits prostate tumor progression in the TRAMP transgenic mouse model of prostate cancer (210). In addition, hyperactivation of the FRS2α-mediated pathway increases tumor angiogenesis and predicts poor outcomes of prostate carcinoma patients (211).
FGFs also have a role in the development of bone metastases, which occur in approximately 80% of patients with advanced prostate cancer (212). These metastases often abnormally express FGF8 and/or FGF9, which promote osteoblast proliferation/differentiation in culture (213, 214). Forced expression of FGF8 promotes bone growth of prostate carcinoma in a mouse model (215). Advanced prostate cancer is frequently resistant to castration. Multiple FGFs have been reported to be aberrantly expressed in castration-resistant (205, 216, 217) or chemotherapy resistant prostate cancer (218, 219). Inhibition of FGF8 and FGF9 signaling has an antitumor effect in mouse models of castrate-resistant prostate cancer (213, 220).
Dysregulated expression of FGFRs has also been associated with prostate cancer. Overexpression of FGFR1 has been found in human prostate cancer and accelerates tumor progression of rat premalignant prostate epithelial cells (40, 115). Exposure to aberrant FGFR1 signaling leads to dosage- and time-dependent lesions of prostate, ranging from low-grade PIN to carcinoma in situ of the prostate, invasive carcinoma, and metastasis. Forced expression of constitutively active mutants of FGFR1 leads to development of high-grade PIN lesions (221, 222). JOCK-1 is a transgenic mouse model overexpressing an FGFR1 kinase construct, iFGFR1, which contains the membrane anchored FGFR1 intracellular kinase domain in frame fused with a 12 kDa FK506 binding protein (FKBP12) at the C-terminus. Treating the mice with FK506, a dimer inducer to activate the FGFR1 kinase, causes the mice to develop invasive prostate carcinoma and metastasis (223). On the other hand, deletion of Frs2α or Fgfr1 in prostate epithelial cells inhibits the initiation and progression of prostate cancer in the transgenic adenocarcinoma of the mouse prostate (TRAMP) model (224, 225). Tissue recombination experiments in vitro also show that ectopic FGFR1 is required for prostate cancer initiation and progression (226). Epithelial-mesenchymal transition (EMT) is a process whereby polarized epithelial cells lose epithelial characteristics and acquire mesenchymal features, including enhanced migratory capacity, invasiveness, and elevated resistance to apoptosis (227). Shifts in alternative splicing of FGFR1 and FGFR2 from IIIb (epithelial) to IIIc (mesenchymal) isoforms are associated with EMT in prostate and other types of cancer (228). In contrast, downregulation of epithelial cell resident FGFR2 is associated with prostate cancer progression (40, 114). Together, these data suggest that aberrant activation of FGFR1 signaling is sufficient to disrupt prostate tissue homeostasis leading to over-proliferation of prostate cells, and contribute to initiation and progression of the lesion to malignancy in mouse models of prostate cancer.
In contrast to ectopic FGFR isoforms, however, resident FGFR signaling in prostate cells maintains tissue homeostasis, communication with stromal cells, and mediates androgen signaling. The stromal FGF7/FGF10 to epithelial FGFR2 signaling axis maintains prostate tissue homeostasis and mediates androgen signaling in the epithelial cells. Therefore, both FGF7 and FGF10 have been called andromedins (112, 199). Although ablation of Fgfr2 was insufficient to cause full progression to carcinoma, it leads to development of low-grade PIN (Wang and McKeehan, unpublished data). The epithelial FGF9 to stromal FGFR3 signaling axis is engaged in communication between epithelial and stromal cells in the prostate and is lost in advanced prostate cancer. Reinstatement of this signaling axis in advanced rat prostate cancer cells restores the interactions between cancer cells and stromal cells and induces prostate cancer cell differentiation in the Dunning R3327 rat prostate cancer model (216).
FGF pathway inhibitions in cancer treatment
A number of targeted agents that inhibit FGF/FGFR signaling have been developed, which include tyrosine kinase inhibitors (TKIs), monoclonal antibodies, and FGF ligand traps. The TKIs include both ATP binding site and non ATP binding site molecules. With the exception of AZD4547 that is relatively FGFR specific (229, 230), most ATP binding site inhibitors, including dovitinib (TKI258), nintedanib (BIBF 1120), lenvatinib, brivanib, orantinib, and PD173074, cross-inhibit multiple receptor tyrosine kinases (231–237). Non-ATP binding site inhibitors of FGFR kinase may exhibit a better specificity than the ATP-binding site inhibitors. Several non-ATP binding site inhibitors have been developed, which include L6123, Aea4, Aea25, A114, and A117 (238–240). All these non-ATP binding site inhibitors exhibit highly FGFR kinase specific inhibitory activities and suppress cancer cell proliferation, migration, and induce cell apoptosis. It remains to be determined, however, whether these non-ATP binding site inhibitors are safe and efficacious for use on cancer patients.
In addition to kinase inhibitors, several strategies have also been developed to block ligand-receptor binding, which includes antibodies against FGF or FGFR, ligand traps, and small peptides that compete with FGF for binding to the receptors. MFGR1877S is a monoclonal antibody against FGFR3 that is currently undergoing phase 1 testing for patients with advanced solid tumors (241). Both the GP369 antibody that specifically blocks FGFR2 IIIb isoform and the 1A6 antibody that neutralizes FGF19 activities are currently in preclinical development (242, 243). The fusion protein HGS1036 (FP-1039) comprises the extracellular domain of FGFR1c fused with the Fc portion of IgG1, is expected to trap FGF ligands for the FGFR1IIIc isoform and functions as a decoy receptor. It inhibits tumor cell proliferation and blocks angiogenesis and suppresses growth of patient-derived xenograft tumor models of various tissue origins (244). It has been shown to cause shrinkage of prostate cancer in a phase 1 clinical trial (245). Two short peptides, P8 (PLLQATAGGGS) that binds to FGF2 and P7 (LSPPRYP) that binds to FGFR1 were identified by screening a phage display library using FGF2 and FGFR1 as the bait, respectively (246, 247). Both peptides exhibit activities to suppress FGF2–FGFR1 binding and block FGF2-induced cell proliferation activity without cytotoxic effect in multiple cell lines. The clinical application of these two peptides in cancer treatment is currently being explored.
3. Perspective
Soon after its discovery in the early seventies, the FGF family was recognized to elicit a broad spectrum of regulatory activities. There are few tissues where no members of this large family are expressed or have an impact on some tissue response marker. Often multiple members of the family, both ligands and receptor isotypes, are co-expressed although they are most commonly cell-specific and compartmented when examined more closely. Cell culture analyses in the absence of physiological restrictions often indicate a considerable redundancy among the family members. However, as FGF family member expression and associated activities have been more closely dissected under physiological conditions, results have indicated an increasing degree of receptor isoform- tissue and cell type contextual specificity. Aberrant and ectopic expression of FGF signaling has been reported as a causal factor for multiple diseases, including cancer. Yet, the determinants of FGF isotype and cell type signaling specificity are poorly understood. It is particularly challenging to understand how the same FGFR isotype can have diverse and often opposing biological endpoints. Sometimes the endpoints are temporally dependent on the point at which activation begins and how long it is sustained. Little is known with respect to kinase-substrate specificity of the FGFR tyrosine kinases, although emerging evidence shows that FGFR elicits isoform-specific activities. The role of co-factors and co-receptors, such as heparan sulfate proteoglycans and Klothos, in FGF signaling specificity should not be ignored. In fact, it has been reported that with or without co-expression of KLB, FGFR4 elicits different activities in 293 cells (84). In addition, heparan sulfates affect ligand binding specificity of the FGFR, and it remains to be investigated whether heparan sulfates also contribute to signaling specificity at the substrate level. Understanding the signaling specificity of the FGF signaling axis will provide new strategies for developing drugs that will selectively suppress a particular pathway to minimize side effects. As new technologies emerge, unraveling the “gaitou” of FGF signaling specificity is no longer a dream. Therefore, future applications of FGF, the new focus in the pharmaceutical arena, for improving wound healing, alleviating damages of cardiovascular diseases, controlling obesity and diabetes, and suppressing cancer progression and metastasis are visible on the horizon.
Acknowledgments
This work was supported in part by the National Institutes of Health CA96824 and DE023106 to FW, CA140388 to WLM and FW, The Cancer Prevention and Research Institution of Texas CPRIT110555 to FW and WLM, TAMU1400302 to FW, the Natural Science Foundation of Zhejiang Province of China Y2110492, and the National Natural Science Foundation of China 81101712, 31371470, and 81270761 to CW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Rererences
- 1.Armelin HA. Pituitary extracts and steroid hormones in the control of 3T3 cell growth. Proceedings of the National Academy of Sciences of the United States of America. 1973;70(9):2702–2706. doi: 10.1073/pnas.70.9.2702. Epub 1973/09/01. PubMed PMID: 4354860; PubMed Central PMCID: PMC427087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gospodarowicz D. Localisation of a fibroblast growth factor and its effect alone and with hydrocortisone on 3T3 cell growth. Nature. 1974;249(453):123–127. doi: 10.1038/249123a0. Epub 1974/05/10. PubMed PMID: 4364816. [DOI] [PubMed] [Google Scholar]
- 3.Huebner K, Ferrari AC, Delli Bovi P, Croce CM, Basilico C. The FGF-related oncogene, K-FGF, maps to human chromosome region 11q13, possibly near int-2. Oncogene research. 1988;3(3):263–270. Epub 1988/01/01. PubMed PMID: 3060803. [PubMed] [Google Scholar]
- 4.Taira M, Yoshida T, Miyagawa K, Sakamoto H, Terada M, Sugimura T. cDNA sequence of human transforming gene hst and identification of the coding sequence required for transforming activity. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(9):2980–2984. doi: 10.1073/pnas.84.9.2980. Epub 1987/05/01. PubMed PMID: 2953031; PubMed Central PMCID: PMC304784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakamoto H, Mori M, Taira M, Yoshida T, Matsukawa S, Shimizu K, et al. Transforming gene from human stomach cancers and a noncancerous portion of stomach mucosa. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(11):3997–4001. doi: 10.1073/pnas.83.11.3997. Epub 1986/06/01. PubMed PMID: 3459165; PubMed Central PMCID: PMC323652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marics I, Adelaide J, Raybaud F, Mattei MG, Coulier F, Planche J, et al. Characterization of the HST-related FGF.6 gene, a new member of the fibroblast growth factor gene family. Oncogene. 1989;4(3):335–340. Epub 1989/03/01. PubMed PMID: 2649847. [PubMed] [Google Scholar]
- 7.Rubin JS, Osada H, Finch PW, Taylor WG, Rudikoff S, Aaronson SA. Purification and characterization of a newly identified growth factor specific for epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(3):802–806. doi: 10.1073/pnas.86.3.802. Epub 1989/02/01. PubMed PMID: 2915979; PubMed Central PMCID: PMC286565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka A, Miyamoto K, Minamino N, Takeda M, Sato B, Matsuo H, et al. Cloning and characterization of an androgen-induced growth factor essential for the androgen-dependent growth of mouse mammary carcinoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(19):8928–8932. doi: 10.1073/pnas.89.19.8928. Epub 1992/10/01. PubMed PMID: 1409588; PubMed Central PMCID: PMC50037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyamoto M, Naruo K, Seko C, Matsumoto S, Kondo T, Kurokawa T. Molecular cloning of a novel cytokine cDNA encoding the ninth member of the fibroblast growth factor family, which has a unique secretion property. Molecular and cellular biology. 1993;13(7):4251–4259. doi: 10.1128/mcb.13.7.4251. Epub 1993/07/01. PubMed PMID: 8321227; PubMed Central PMCID: PMC359975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamasaki M, Miyake A, Tagashira S, Itoh N. Structure and expression of the rat mRNA encoding a novel member of the fibroblast growth factor family. The Journal of biological chemistry. 1996;271(27):15918–15921. doi: 10.1074/jbc.271.27.15918. Epub 1996/07/05. PubMed PMID: 8663172. [DOI] [PubMed] [Google Scholar]
- 11.Smallwood PM, Munoz-Sanjuan I, Tong P, Macke JP, Hendry SH, Gilbert DJ, et al. Fibroblast growth factor (FGF) homologous factors: new members of the FGF family implicated in nervous system development. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9850–9857. doi: 10.1073/pnas.93.18.9850. Epub 1996/09/03. PubMed PMID: 8790420; PubMed Central PMCID: PMC38518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyake A, Konishi M, Martin FH, Hernday NA, Ozaki K, Yamamoto S, et al. Structure and expression of a novel member, FGF-16, on the fibroblast growth factor family. Biochemical and biophysical research communications. 1998;243(1):148–152. doi: 10.1006/bbrc.1998.8073. PubMed PMID: 9473496. [DOI] [PubMed] [Google Scholar]
- 13.Hoshikawa M, Ohbayashi N, Yonamine A, Konishi M, Ozaki K, Fukui S, et al. Structure and expression of a novel fibroblast growth factor, FGF-17, preferentially expressed in the embryonic brain. Biochemical and biophysical research communications. 1998;244(1):187–191. doi: 10.1006/bbrc.1998.8239. Epub 1998/03/26. PubMed PMID: 9514906. [DOI] [PubMed] [Google Scholar]
- 14.Ohbayashi N, Hoshikawa M, Kimura S, Yamasaki M, Fukui S, Itoh N. Structure and expression of the mRNA encoding a novel fibroblast growth factor, FGF-18. The Journal of biological chemistry. 1998;273(29):18161–18164. doi: 10.1074/jbc.273.29.18161. Epub 1998/07/11. PubMed PMID: 9660775. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura T, Utsunomiya Y, Hoshikawa M, Ohuchi H, Itoh N. Structure and expression of a novel human FGF, FGF-19, expressed in the fetal brain. Biochimica et biophysica acta. 1999;1444(1):148–151. doi: 10.1016/s0167-4781(98)00255-3. Epub 1999/02/05. PubMed PMID: 9931477. [DOI] [PubMed] [Google Scholar]
- 16.Ohmachi S, Watanabe Y, Mikami T, Kusu N, Ibi T, Akaike A, et al. FGF-20, a novel neurotrophic factor, preferentially expressed in the substantia nigra pars compacta of rat brain. Biochemical and biophysical research communications. 2000;277(2):355–360. doi: 10.1006/bbrc.2000.3675. PubMed PMID: 11032730. [DOI] [PubMed] [Google Scholar]
- 17.Nishimura T, Nakatake Y, Konishi M, Itoh N. Identification of a novel FGF, FGF-21, preferentially expressed in the liver. Biochimica et biophysica acta. 2000;1492(1):203–206. doi: 10.1016/s0167-4781(00)00067-1. Epub 2000/06/. PubMed PMID: 10858549. [DOI] [PubMed] [Google Scholar]
- 18.Nakatake Y, Hoshikawa M, Asaki T, Kassai Y, Itoh N. Identification of a novel fibroblast growth factor, FGF-22, preferentially expressed in the inner root sheath of the hair follicle. Biochimica et biophysica acta. 2001;1517(3):460–463. doi: 10.1016/s0167-4781(00)00302-x. Epub 2001/05/09. PubMed PMID: 11342227. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita T, Yoshioka M, Itoh N. Identification of a novel fibroblast growth factor, FGF-23, preferentially expressed in the ventrolateral thalamic nucleus of the brain. Biochemical and biophysical research communications. 2000;277(2):494–498. doi: 10.1006/bbrc.2000.3696. [DOI] [PubMed] [Google Scholar]
- 20.McWhirter JR, Goulding M, Weiner JA, Chun J, Murre C. A novel fibroblast growth factor gene expressed in the developing nervous system is a downstream target of the chimeric homeodomain oncoprotein E2A-Pbx1. Development. 1997;124(17):3221–3232. doi: 10.1242/dev.124.17.3221. Epub 1997/10/06. PubMed PMID: 9310317. [DOI] [PubMed] [Google Scholar]
- 21.Hennessey JA, Wei EQ, Pitt GS. Fibroblast growth factor homologous factors modulate cardiac calcium channels. Circulation research. 2013;113(4):381–388. doi: 10.1161/CIRCRESAHA.113.301215. PubMed PMID: 23804213; PubMed Central PMCID: PMCPMC3813963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, Wang C, Hoch EG, Pitt GS. Identification of novel interaction sites that determine specificity between fibroblast growth factor homologous factors and voltage-gated sodium channels. The Journal of biological chemistry. 2011;286(27):24253–24263. doi: 10.1074/jbc.M111.245803. PubMed PMID: 21566136; PubMed Central PMCID: PMCPMC3129206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen SK, Garbi M, Zampieri N, Eliseenkova AV, Ornitz DM, Goldfarb M, et al. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. The Journal of biological chemistry. 2003;278(36):34226–34236. doi: 10.1074/jbc.M303183200. PubMed PMID: 12815063. [DOI] [PubMed] [Google Scholar]
- 24.Lee PL, Johnson DE, Cousens LS, Fried VA, Williams LT. Purification and complementary DNA cloning of a receptor for basic fibroblast growth factor. Science. 1989;245(4913):57–60. doi: 10.1126/science.2544996. Epub 1989/07/07. PubMed PMID: 2544996. [DOI] [PubMed] [Google Scholar]
- 25.Bottaro DP, Rubin JS, Ron D, Finch PW, Florio C, Aaronson SA. Characterization of the receptor for keratinocyte growth factor. Evidence for multiple fibroblast growth factor receptors. The Journal of biological chemistry. 1990;265(22):12767–12770. Epub 1990/08/05. PubMed PMID: 2165484. [PubMed] [Google Scholar]
- 26.Keegan K, Johnson DE, Williams LT, Hayman MJ. Characterization of the FGFR-3 gene and its gene product. Ann N Y Acad Sci. 1991;638:400–402. doi: 10.1111/j.1749-6632.1991.tb49052.x. Epub 1991/01/01. PubMed PMID: 1723860. [DOI] [PubMed] [Google Scholar]
- 27.Stark KL, McMahon JA, McMahon AP. FGFR-4, a new member of the fibroblast growth factor receptor family, expressed in the definitive endoderm and skeletal muscle lineages of the mouse. Development. 1991;113(2):641–651. doi: 10.1242/dev.113.2.641. Epub 1991/10/01. PubMed PMID: 1723680. [DOI] [PubMed] [Google Scholar]
- 28.Takaishi S, Sawada M, Morita Y, Seno H, Fukuzawa H, Chiba T. Identification of a novel alternative splicing of human FGF receptor 4: soluble-form splice variant expressed in human gastrointestinal epithelial cells. Biochemical and biophysical research communications. 2000;267(2):658–662. doi: 10.1006/bbrc.1999.2010. PubMed PMID: 10631118. [DOI] [PubMed] [Google Scholar]
- 29.Hou JZ, Kan MK, McKeehan K, McBride G, Adams P, McKeehan WL. Fibroblast growth factor receptors from liver vary in three structural domains. Science. 1991;251(4994):665–668. doi: 10.1126/science.1846977. [DOI] [PubMed] [Google Scholar]
- 30.Vainshenker Iu I, Kalinina OV, Nuralova IV, Ivchenko IM, Meliucheva LA, Tsinzerling VA. Low-manifest infections in children and adolescents with consequences of perinatal damage of nervous system. Zh Mikrobiol Epidemiol Immunobiol. 2012;(5):77–80. PubMed PMID: 23163043. [PubMed] [Google Scholar]
- 31.Wang F, Kan M, Yan G, Xu J, McKeehan WL. Alternately spliced NH2-terminal immunoglobulin-like Loop I in the ectodomain of the fibroblast growth factor (FGF) receptor 1 lowers affinity for both heparin and FGF-1. The Journal of biological chemistry. 1995;270(17):10231–10235. doi: 10.1074/jbc.270.17.10231. PubMed PMID: 7730327. [DOI] [PubMed] [Google Scholar]
- 32.Kalinina J, Dutta K, Ilghari D, Beenken A, Goetz R, Eliseenkova AV, et al. The alternatively spliced acid box region plays a key role in FGF receptor autoinhibition. Structure. 2012;20(1):77–88. doi: 10.1016/j.str.2011.10.022. PubMed PMID: 22244757; PubMed Central PMCID: PMCPMC3378326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. The Journal of biological chemistry. 2006;281(23):15694–15700. doi: 10.1074/jbc.M601252200. PubMed PMID: 16597617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Y, Lu W, Mohamedali KA, Jang JH, Jones RB, Gabriel JL, et al. The glycine box: a determinant of specificity for fibroblast growth factor. Biochemistry. 1998;37(47):16506–16515. doi: 10.1021/bi9816599. PubMed PMID: 9843417. [DOI] [PubMed] [Google Scholar]
- 35.Yan G, Wang F, Fukabori Y, Sussman D, Hou J, McKeehan WL. Expression and transforming activity of a variant of the heparin- binding fibroblast growth factor receptor (flg) gene resulting from splicing of the alpha exon at an alternate 3'-acceptor site. Biochemical and biophysical research communications. 1992;183(2):423–430. doi: 10.1016/0006-291x(92)90498-a. PubMed PMID: 1312829. [DOI] [PubMed] [Google Scholar]
- 36.Hoch RV, Soriano P. Context-specific requirements for Fgfr1 signaling through Frs2 and Frs3 during mouse development. Development. 2006;133(4):663–673. doi: 10.1242/dev.02242. PubMed PMID: 16421190. [DOI] [PubMed] [Google Scholar]
- 37.Burgar HR, Burns HD, Elsden JL, Lalioti MD, Heath JK. Association of the signaling adaptor FRS2 with fibroblast growth factor receptor 1 (Fgfr1) is mediated by alternative splicing of the juxtamembrane domain. The Journal of biological chemistry. 2002;277(6):4018–4023. doi: 10.1074/jbc.M107785200. PubMed PMID: 11729184. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, McKeehan K, Lin Y, Zhang J, Wang F. Fibroblast growth factor receptor 1 (FGFR1) tyrosine phosphorylation regulates binding of FGFR substrate 2alpha (FRS2alpha) but not FRS2 to the receptor. Molecular endocrinology. 2008;22(1):167–175. doi: 10.1210/me.2007-0140. PubMed PMID: 17901128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hattori Y, Odagiri H, Nakatani H, Miyagawa K, Naito K, Sakamoto H, et al. K-sam, an amplified gene in stomach cancer, is a member of the heparin-binding growth factor receptor genes. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(15):5983–5987. doi: 10.1073/pnas.87.15.5983. PubMed PMID: 2377625; PubMed Central PMCID: PMC54454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng S, Wang F, Matsubara A, Kan M, McKeehan WL. Fibroblast growth factor receptor 2 limits and receptor 1 accelerates tumorigenicity of prostate epithelial cells. Cancer research. 1997;57(23):5369–5378. PubMed PMID: 9393762. [PubMed] [Google Scholar]
- 41.Matsubara A, Kan M, Feng S, McKeehan WL. Inhibition of growth of malignant rat prostate tumor cells by restoration of fibroblast growth factor receptor 2. Cancer research. 1998;58(7):1509–1514. PubMed PMID: 9537256. [PubMed] [Google Scholar]
- 42.Luo Y, Yang C, Jin C, Xie R, Wang F, McKeehan WL. Novel phosphotyrosine targets of FGFR2IIIb signaling. Cellular signalling. 2009;21(9):1370–1378. doi: 10.1016/j.cellsig.2009.04.004. Epub 2009/05/05. doi: S0898-6568(09)00149-1 [pii] 10.1016/j.cellsig.2009.04.004. PubMed PMID: 19410646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xian W, Schwertfeger KL, Rosen JM. Distinct roles of fibroblast growth factor receptor 1 and 2 in regulating cell survival and epithelial-mesenchymal transition. Molecular endocrinology. 2007;21(4):987–1000. doi: 10.1210/me.2006-0518. PubMed PMID: 17284663. [DOI] [PubMed] [Google Scholar]
- 44.Mohammadi M, Dikic I, Sorokin A, Burgess WH, Jaye M, Schlessinger J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Molecular and cellular biology. 1996;16(3):977–989. doi: 10.1128/mcb.16.3.977. Epub 1996/03/01. PubMed PMID: 8622701; PubMed Central PMCID: PMC231080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocrine-related cancer. 2000;7(3):165–197. doi: 10.1677/erc.0.0070165. PubMed PMID: 11021964. [DOI] [PubMed] [Google Scholar]
- 46.McKeehan WL, Wang F, Kan M. The heparan sulfate-fibroblast growth factor family: diversity of structure and function. Prog Nucleic Acid Res Mol Biol. 1998;59:135–176. doi: 10.1016/s0079-6603(08)61031-4. PubMed PMID: 9427842. [DOI] [PubMed] [Google Scholar]
- 47.Hou J, McKeehan K, Kan M, Carr SA, Huddleston MJ, Crabb JW, et al. Identification of tyrosines 154 and 307 in the extracellular domain and 653 and 766 in the intracellular domain as phosphorylation sites in the heparin-binding fibroblast growth factor receptor tyrosine kinase (flg) Protein Sci. 1993;2(1):86–92. doi: 10.1002/pro.5560020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lew ED, Furdui CM, Anderson KS, Schlessinger J. The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Sci Signal. 2009;2(58):ra6. doi: 10.1126/scisignal.2000021. Epub 2009/02/20. doi: 2/58/ra6 [pii] 10.1126/scisignal.2000021. PubMed PMID: 19224897; PubMed Central PMCID: PMC2755185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi E, Kan M, Xu J, Wang F, Hou J, McKeehan WL. Control of fibroblast growth factor receptor kinase signal transduction by heterodimerization of combinatorial splice variants. Molecular and cellular biology. 1993;13(7):3907–3918. doi: 10.1128/mcb.13.7.3907. PubMed PMID: 8321198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohammadi M, Dionne CA, Li W, Li N, Spivak T, Honegger AM, et al. Point mutation in FGF receptor eliminates phosphatidylinositol hydrolysis without affecting mitogenesis. Nature. 1992;358(6388):681–684. doi: 10.1038/358681a0. [DOI] [PubMed] [Google Scholar]
- 51.Peters KG, Marie J, Wilson E, Ives HE, Escobedo J, Del Rosario M, et al. Point mutation of an FGF receptor abolishes phosphatidylinositol turnover and Ca2+ flux but not mitogenesis. Nature. 1992;358(6388):678–681. doi: 10.1038/358678a0. [DOI] [PubMed] [Google Scholar]
- 52.Larsson H, Klint P, Landgren E, Claesson-Welsh L. Fibroblast growth factor receptor-1-mediated endothelial cell proliferation is dependent on the Src homology (SH) 2/SH3 domain- containing adaptor protein Crk. The Journal of biological chemistry. 1999;274(36):25726–25734. doi: 10.1074/jbc.274.36.25726. PubMed PMID: 10464310. [DOI] [PubMed] [Google Scholar]
- 53.Klint P, Claesson-Welsh L. Signal transduction by fibroblast growth factor receptors. Front Biosci. 1999;4:D165–D177. doi: 10.2741/klint. PubMed PMID: 9989949. [DOI] [PubMed] [Google Scholar]
- 54.Seo JH, Suenaga A, Hatakeyama M, Taiji M, Imamoto A. Structural and functional basis of a role for CRKL in a fibroblast growth factor 8-induced feed-forward loop. Molecular and cellular biology. 2009;29(11):3076–3087. doi: 10.1128/MCB.01686-08. Epub 2009/03/25. doi: MCB.01686-08 [pii] 10.1128/MCB.01686-08. PubMed PMID: 19307307; PubMed Central PMCID: PMC2681998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Francavilla C, Rigbolt KT, Emdal KB, Carraro G, Vernet E, Bekker-Jensen DB, et al. Functional proteomics defines the molecular switch underlying FGF receptor trafficking and cellular outputs. Mol Cell. 2013;51(6):707–722. doi: 10.1016/j.molcel.2013.08.002. PubMed PMID: 24011590. [DOI] [PubMed] [Google Scholar]
- 56.Yan G, McBride G, McKeehan WL. Exon skipping causes alteration of the COOH-terminus and deletion of the phospholipase C gamma 1 interaction site in the FGF receptor 2 kinase in normal prostate epithelial cells. Biochemical and biophysical research communications. 1993;194(1):512–518. doi: 10.1006/bbrc.1993.1849. [DOI] [PubMed] [Google Scholar]
- 57.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. PubMed PMID: 10872465. [DOI] [PubMed] [Google Scholar]
- 58.Esko JD, Lindahl U. Molecular diversity of heparan sulfate. The Journal of clinical investigation. 2001;108(2):169–173. doi: 10.1172/JCI13530. PubMed PMID: 11457867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park PW, Reizes O, Bernfield M. Cell surface heparan sulfate proteoglycans: selective regulators of ligand-receptor encounters. The Journal of biological chemistry. 2000;275(39):29923–29926. doi: 10.1074/jbc.R000008200. PubMed PMID: 10931855. [DOI] [PubMed] [Google Scholar]
- 60.Ye S, Luo Y, Lu W, Jones RB, Linhardt RJ, Capila I, et al. Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs. Biochemistry. 2001;40(48):14429–14439. doi: 10.1021/bi011000u. Epub 2001/11/29. doi: bi011000u [pii]. PubMed PMID: 11724555. [DOI] [PubMed] [Google Scholar]
- 61.Kan M, Wu X, Wang F, McKeehan WL. Specificity for fibroblast growth factors determined by heparan sulfate in a binary complex with the receptor kinase. The Journal of biological chemistry. 1999;274(22):15947–15952. doi: 10.1074/jbc.274.22.15947. PubMed PMID: 10336501. [DOI] [PubMed] [Google Scholar]
- 62.Kreuger J, Salmivirta M, Sturiale L, Gimenez-Gallego G, Lindahl U. Sequence analysis of heparan sulfate epitopes with graded affinities for fibroblast growth factors 1 and 2. The Journal of biological chemistry. 2001;276(33):30744–30752. doi: 10.1074/jbc.M102628200. Epub 2001/06/15. M102628200 [pii]. PubMed PMID: 11406624. [DOI] [PubMed] [Google Scholar]
- 63.Guimond SE, Turnbull JE. Fibroblast growth factor receptor signalling is dictated by specific heparan sulphate saccharides. Curr Biol. 1999;9(22):1343–1346. doi: 10.1016/s0960-9822(00)80060-3. Epub 1999/11/27. doi: bb9v54 [pii]. PubMed PMID: 10574766. [DOI] [PubMed] [Google Scholar]
- 64.Ostrovsky O, Berman B, Gallagher J, Mulloy B, Fernig DG, Delehedde M, et al. Differential effects of heparin saccharides on the formation of specific fibroblast growth factor (FGF) and FGF receptor complexes. The Journal of biological chemistry. 2002;277(4):2444–2453. doi: 10.1074/jbc.M108540200. Epub 2001/11/21. M108540200 [pii]. PubMed PMID: 11714710. [DOI] [PubMed] [Google Scholar]
- 65.Powell AK, Fernig DG, Turnbull JE. Fibroblast growth factor receptors 1 and 2 interact differently with heparin/heparan sulfate. Implications for dynamic assembly of a ternary signaling complex. The Journal of biological chemistry. 2002;277(32):28554–28563. doi: 10.1074/jbc.M111754200. Epub 2002/05/30. M111754200 [pii]. PubMed PMID: 12034712. [DOI] [PubMed] [Google Scholar]
- 66.Luo Y, Ye S, Kan M, McKeehan WL. Control of fibroblast growth factor (FGF) 7- and FGF1-induced mitogenesis and downstream signaling by distinct heparin octasaccharide motifs. The Journal of biological chemistry. 2006;281(30):21052–21061. doi: 10.1074/jbc.M601559200. Epub 2006/05/27. doi: M601559200 [pii] 10.1074/jbc.M601559200. PubMed PMID: 16728399. [DOI] [PubMed] [Google Scholar]
- 67.Luo Y, Ye S, Kan M, McKeehan WL. Structural specificity in a FGF7-affinity purified heparin octasaccharide required for formation of a complex with FGF7 and FGFR2IIIb. J Cell Biochem. 2006;97(6):1241–1258. doi: 10.1002/jcb.20724. Epub 2005/11/30. PubMed PMID: 16315317. [DOI] [PubMed] [Google Scholar]
- 68.Kan M, Shi EG, McKeehan WL. Identification and assay of fibroblast growth factor receptors. Methods Enzymol. 1991;198:158–171. doi: 10.1016/0076-6879(91)98017-z. [DOI] [PubMed] [Google Scholar]
- 69.Kan M, DiSorbo D, Hou JZ, Hoshi H, Mansson PE, McKeehan WL. High and low affinity binding of heparin-binding growth factor to a 130- kDa receptor correlates with stimulation and inhibition of growth of a differentiated human hepatoma cell. The Journal of biological chemistry. 1988;263(23):11306–11313. [PubMed] [Google Scholar]
- 70.Kan M, Wang F, To B, Gabriel JL, McKeehan WL. Divalent cations and heparin/heparan sulfate cooperate to control assembly and activity of the fibroblast growth factor receptor complex. The Journal of biological chemistry. 1996;271(42):26143–26148. doi: 10.1074/jbc.271.42.26143. PubMed PMID: 8824259. [DOI] [PubMed] [Google Scholar]
- 71.Wang F, Kan M, McKeehan K, Jang JH, Feng S, McKeehan WL. A homeo-interaction sequence in the ectodomain of the fibroblast growth factor receptor. The Journal of biological chemistry. 1997;272(38):23887–23895. doi: 10.1074/jbc.272.38.23887. PubMed PMID: 8792155. [DOI] [PubMed] [Google Scholar]
- 72.Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, et al. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6(3):743–750. doi: 10.1016/s1097-2765(00)00073-3. Epub 2000/10/13. doi: S1097-2765(00)00073-3 [pii]. PubMed PMID: 11030354. [DOI] [PubMed] [Google Scholar]
- 73.Uematsu F, Kan M, Wang F, Jang JH, Luo Y, McKeehan WL. Ligand binding properties of binary complexes of heparin and immunoglobulin-like modules of FGF receptor 2. Biochemical and biophysical research communications. 2000;272(3):830–836. doi: 10.1006/bbrc.2000.2872. PubMed PMID: 10860838. [DOI] [PubMed] [Google Scholar]
- 74.Belov AA, Mohammadi M. Molecular mechanisms of fibroblast growth factor signaling in physiology and pathology. Cold Spring Harbor perspectives in biology. 2013;5(6) doi: 10.1101/cshperspect.a015958. Epub 2013/06/05. PubMed PMID: 23732477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen H, Ma J, Li W, Eliseenkova AV, Xu C, Neubert TA, et al. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol Cell. 2007;27(5):717–730. doi: 10.1016/j.molcel.2007.06.028. PubMed PMID: 17803937; PubMed Central PMCID: PMCPMC2094128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goetz R, Beenken A, Ibrahimi OA, Kalinina J, Olsen SK, Eliseenkova AV, et al. Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Molecular and cellular biology. 2007;27(9):3417–3428. doi: 10.1128/MCB.02249-06. Epub 2007/03/07. doi: MCB.02249-06 [pii] 10.1128/MCB.02249-06. PubMed PMID: 17339340; PubMed Central PMCID: PMC1899957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin BC, Wang M, Blackmore C, Desnoyers LR. Liver-specific activities of FGF19 require Klotho beta. The Journal of biological chemistry. 2007;282(37):27277–27284. doi: 10.1074/jbc.M704244200. Epub 2007/07/14. PubMed PMID: 17627937. [DOI] [PubMed] [Google Scholar]
- 78.Micanovic R, Raches DW, Dunbar JD, Driver DA, Bina HA, Dickinson CD, et al. Different roles of N- and C- termini in the functional activity of FGF21. Journal of cellular physiology. 2009;219(2):227–234. doi: 10.1002/jcp.21675. Epub 2009/01/01. PubMed PMID: 19117008. [DOI] [PubMed] [Google Scholar]
- 79.Goetz R, Ohnishi M, Ding X, Kurosu H, Wang L, Akiyoshi J, et al. Klotho coreceptors inhibit signaling by paracrine fibroblast growth factor 8 subfamily ligands. Molecular and cellular biology. 2012;32(10):1944–1954. doi: 10.1128/MCB.06603-11. PubMed PMID: 22451487; PubMed Central PMCID: PMCPMC3347405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goetz R, Nakada Y, Hu MC, Kurosu H, Wang L, Nakatani T, et al. Isolated C-terminal tail of FGF23 alleviates hypophosphatemia by inhibiting FGF23-FGFR-Klotho complex formation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(1):407–412. doi: 10.1073/pnas.0902006107. PubMed PMID: 19966287; PubMed Central PMCID: PMCPMC2806769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, et al. Regulation of fibroblast growth factor-23 signaling by klotho. The Journal of biological chemistry. 2006;281(10):6120–6123. doi: 10.1074/jbc.C500457200. Epub 2006/01/27. PubMed PMID: 16436388; PubMed Central PMCID: PMC2637204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakatani T, Ohnishi M, Razzaque MS. Inactivation of klotho function induces hyperphosphatemia even in presence of high serum fibroblast growth factor 23 levels in a genetically engineered hypophosphatemic (Hyp) mouse model. FASEB J. 2009;23(11):3702–3711. doi: 10.1096/fj.08-123992. PubMed PMID: 19584304; PubMed Central PMCID: PMCPMC2775000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kurosu H, Choi M, Ogawa Y, Dickson AS, Goetz R, Eliseenkova AV, et al. Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. The Journal of biological chemistry. 2007;282(37):26687–26695. doi: 10.1074/jbc.M704165200. Epub 2007/07/12. doi: M704165200 [pii] 10.1074/jbc.M704165200. PubMed PMID: 17623664; PubMed Central PMCID: PMC2496965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luo Y, Yang C, Lu W, Xie R, Jin C, Huang P, et al. Metabolic regulator betaKlotho interacts with fibroblast growth factor receptor 4 (FGFR4) to induce apoptosis and inhibit tumor cell proliferation. The Journal of biological chemistry. 2010;285(39):30069–30078. doi: 10.1074/jbc.M110.148288. Epub 2010/07/27. PubMed PMID: 20657013; PubMed Central PMCID: PMC2943257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goetz R, Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nature reviews Molecular cell biology. 2013;14(3):166–180. doi: 10.1038/nrm3528. PubMed PMID: 23403721; PubMed Central PMCID: PMCPMC3695728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McDougall K, Kubu C, Verdi JM, Meakin SO. Developmental expression patterns of the signaling adapters FRS-2 and FRS-3 during early embryogenesis. Mechanisms of development. 2001;103(1–2):145–148. doi: 10.1016/s0925-4773(01)00337-9. PubMed PMID: 11335123. [DOI] [PubMed] [Google Scholar]
- 87.Rabin SJ, Cleghon V, Kaplan DR. SNT, a differentiation-specific target of neurotrophic factor-induced tyrosine kinase activity in neurons and PC12 cells. Molecular and cellular biology. 1993;13(4):2203–2213. doi: 10.1128/mcb.13.4.2203. PubMed PMID: 7681142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ong SH, Goh KC, Lim YP, Low BC, Klint P, Claesson-Welsh L, et al. Suc1-associated neurotrophic factor target (SNT) protein is a major FGF- stimulated tyrosine phosphorylated 90-kDa protein which binds to the SH2 domain of GRB2. Biochemical and biophysical research communications. 1996;225(3):1021–1026. doi: 10.1006/bbrc.1996.1288. PubMed PMID: 8780727. [DOI] [PubMed] [Google Scholar]
- 89.Arman E, Haffner-Krausz R, Gorivodsky M, Lonai P. Fgfr2 is required for limb outgrowth and lungbranching morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(21):11895–11899. doi: 10.1073/pnas.96.21.11895. PubMed PMID: 10518547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin HY, Xu J, Ischenko I, Ornitz DM, Halegoua S, Hayman MJ. Identification of the cytoplasmic regions of fibroblast growth factor (FGF) receptor 1 which play important roles in induction of neurite outgrowth in PC12 cells by FGF-1. Molecular and cellular biology. 1998;18(7):3762–3770. doi: 10.1128/mcb.18.7.3762. PubMed PMID: 9632759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, et al. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89(5):693–702. doi: 10.1016/s0092-8674(00)80252-4. PubMed PMID: 9182757. [DOI] [PubMed] [Google Scholar]
- 92.Xu H, Lee KW, Goldfarb M. Novel recognition motif on fibroblast growth factor receptor mediates direct association and activation of SNT adapter proteins. The Journal of biological chemistry. 1998;273(29):17987–17990. doi: 10.1074/jbc.273.29.17987. PubMed PMID: 9660748. [DOI] [PubMed] [Google Scholar]
- 93.Ong SH, Lim YP, Low BC, Guy GR. SHP2 associates directly with tyrosine phosphorylated p90 (SNT) protein in FGF-stimulated cells. Biochemical and biophysical research communications. 1997;238(1):261–266. doi: 10.1006/bbrc.1997.7272. PubMed PMID: 9299490. [DOI] [PubMed] [Google Scholar]
- 94.Guy GR, Wong ES, Yusoff P, Chandramouli S, Lo TL, Lim J, et al. Sprouty: how does the branch manager work? Journal of cell science. 2003;116(Pt 15):3061–3068. doi: 10.1242/jcs.00652. PubMed PMID: 12829736. [DOI] [PubMed] [Google Scholar]
- 95.Torii S, Kusakabe M, Yamamoto T, Maekawa M, Nishida E. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev Cell. 2004;7(1):33–44. doi: 10.1016/j.devcel.2004.05.019. PubMed PMID: 15239952. [DOI] [PubMed] [Google Scholar]
- 96.Xiong S, Zhao Q, Rong Z, Huang G, Huang Y, Chen P, et al. hSef inhibits PC-12 cell differentiation by interfering with Ras-mitogen-activated protein kinase MAPK signaling. The Journal of biological chemistry. 2003;278(50):50273–50282. doi: 10.1074/jbc.M306936200. PubMed PMID: 12958313. [DOI] [PubMed] [Google Scholar]
- 97.Wang F, McKeehan K, Yu C, McKeehan WL. Fibroblast growth factor receptor 1 phosphotyrosine 766: molecular target for prevention of progression of prostate tumors to malignancy. Cancer research. 2002;62(6):1898–1903. PubMed PMID: 11912171. [PubMed] [Google Scholar]
- 98.Hadari YR, Gotoh N, Kouhara H, Lax I, Schlessinger J. Critical role for the docking-protein FRS2 alpha in FGF receptor-mediated signal transduction pathways. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(15):8578–8583. doi: 10.1073/pnas.161259898. PubMed PMID: 11447289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gotoh N, Laks S, Nakashima M, Lax I, Schlessinger J. FRS2 family docking proteins with overlapping roles in activation of MAP kinase have distinct spatial-temporal patterns of expression of their transcripts. FEBS letters. 2004;564(1–2):14–18. doi: 10.1016/S0014-5793(04)00287-X. PubMed PMID: 15094036. [DOI] [PubMed] [Google Scholar]
- 100.Wong A, Lamothe B, Lee A, Schlessinger J, Lax I. FRS2 alpha attenuates FGF receptor signaling by Grb2-mediated recruitment of the ubiquitin ligase Cbl. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(10):6684–6689. doi: 10.1073/pnas.052138899. PubMed PMID: 11997436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lax I, Wong A, Lamothe B, Lee A, Frost A, Hawes J, et al. The Docking Protein FRS2alpha Controls a MAP Kinase-Mediated Negative Feedback Mechanism for Signaling by FGF Receptors. Mol Cell. 2002;10(4):709–719. doi: 10.1016/s1097-2765(02)00689-5. PubMed PMID: 12419216. [DOI] [PubMed] [Google Scholar]
- 102.Faham S, Linhardt RJ, Rees DC. Diversity does make a difference: fibroblast growth factor-heparin interactions. Curr Opin Struct Biol. 1998;8(5):578–586. doi: 10.1016/s0959-440x(98)80147-4. PubMed PMID: 9818261. [DOI] [PubMed] [Google Scholar]
- 103.Ornitz DM. FGFs, heparan sulfate and FGFRs: complex interactions essential for development. Bioessays. 2000;22(2):108–112. doi: 10.1002/(SICI)1521-1878(200002)22:2<108::AID-BIES2>3.0.CO;2-M. PubMed PMID: 10655030. [DOI] [PubMed] [Google Scholar]
- 104.McIntosh I, Bellus GA, Jab EW. The pleiotropic effects of fibroblast growth factor receptors in mammalian development. Cell Struct Funct. 2000;25(2):85–96. doi: 10.1247/csf.25.85. PubMed PMID: 10885578. [DOI] [PubMed] [Google Scholar]
- 105.Xu X, Weinstein M, Li C, Deng C. Fibroblast growth factor receptors (FGFRs) and their roles in limb development. Cell and tissue research. 1999;296(1):33–43. doi: 10.1007/s004410051264. PubMed PMID: 10199963. [DOI] [PubMed] [Google Scholar]
- 106.Kannan K, Givol D. FGF receptor mutations: dimerization syndromes, cell growth suppression, and animal models. IUBMB Life. 2000;49(3):197–205. doi: 10.1080/713803609. PubMed PMID: 10868910. [DOI] [PubMed] [Google Scholar]
- 107.Kato S, Sekine K. FGF-FGFR signaling in vertebrate organogenesis. Cell Mol Biol (Noisy-le-grand) 1999;45(5):631–638. PubMed PMID: 10512194. [PubMed] [Google Scholar]
- 108.Burke D, Wilkes D, Blundell TL, Malcolm S. Fibroblast growth factor receptors: lessons from the genes. Trends Biochem Sci. 1998;23(2):59–62. doi: 10.1016/s0968-0004(97)01170-5. PubMed PMID: 9538690. [DOI] [PubMed] [Google Scholar]
- 109.Webster MK, Donoghue DJ. FGFR activation in skeletal disorders: too much of a good thing. Trends Genet. 1997;13(5):178–182. doi: 10.1016/s0168-9525(97)01131-1. PubMed PMID: 9154000. [DOI] [PubMed] [Google Scholar]
- 110.Muenke M, Schell U. Fibroblast-growth-factor receptor mutations in human skeletal disorders. Trends Genet. 1995;11(8):308–313. doi: 10.1016/s0168-9525(00)89088-5. PubMed PMID: 8585128. [DOI] [PubMed] [Google Scholar]
- 111.Kwabi-Addo B, Ropiquet F, Giri D, Ittmann M. Alternative splicing of fibroblast growth factor receptors in human prostate cancer. The Prostate. 2001;46(2):163–172. doi: 10.1002/1097-0045(20010201)46:2<163::aid-pros1020>3.0.co;2-t. PubMed PMID: 11170144. [DOI] [PubMed] [Google Scholar]
- 112.Yan G, Fukabori Y, Nikolaropoulos S, Wang F, McKeehan WL. Heparin-binding keratinocyte growth factor is a candidate stromal-to- epithelial-cell andromedin. Molecular endocrinology. 1992;6(12):2123–2128. doi: 10.1210/mend.6.12.1491693. PubMed PMID: 1491693. [DOI] [PubMed] [Google Scholar]
- 113.Gowardhan B, Douglas DA, Mathers ME, McKie AB, McCracken SR, Robson CN, et al. Evaluation of the fibroblast growth factor system as a potential target for therapy in human prostate cancer. British journal of cancer. 2005;92(2):320–327. doi: 10.1038/sj.bjc.6602274. PubMed PMID: 15655558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Freeman KW, Gangula RD, Welm BE, Ozen M, Foster BA, Rosen JM, et al. Conditional activation of fibroblast growth factor receptor (FGFR) 1, but not FGFR2, in prostate cancer cells leads to increased osteopontin induction, extracellular signal-regulated kinase activation, and in vivo proliferation. Cancer research. 2003;63(19):6237–6243. PubMed PMID: 14559809. [PubMed] [Google Scholar]
- 115.Sahadevan K, Darby S, Leung H, Mathers M, Robson C, Gnanapragasam V. Selective over-expression of fibroblast growth factor receptors 1 and 4 in clinical prostate cancer. J Pathol. 2007 doi: 10.1002/path.2205. PubMed PMID: 17607666. [DOI] [PubMed] [Google Scholar]
- 116.Lou Z. Healing large traumatic eardrum perforations in humans using fibroblast growth factor applied directly or via gelfoam. Otology & neurotology : official publication of the American Otological Society, American Neurotology Society [and] European Academy of Otology and Neurotology. 2012;33(9):1553–1557. doi: 10.1097/JES.0b013e31826f5640. Epub 2012/11/15. PubMed PMID: 23150095. [DOI] [PubMed] [Google Scholar]
- 117.Hirata E, Menard-Moyon C, Venturelli E, Takita H, Watari F, Bianco A, et al. Carbon nanotubes functionalized with fibroblast growth factor accelerate proliferation of bone marrow-derived stromal cells and bone formation. Nanotechnology. 2013;24(43):435101. doi: 10.1088/0957-4484/24/43/435101. Epub 2013/10/01. PubMed PMID: 24077482. [DOI] [PubMed] [Google Scholar]
- 118.Kwan MD, Sellmyer MA, Quarto N, Ho AM, Wandless TJ, Longaker MT. Chemical control of FGF-2 release for promoting calvarial healing with adipose stem cells. The Journal of biological chemistry. 2011;286(13):11307–11313. doi: 10.1074/jbc.M110.180042. Epub 2011/01/26. PubMed PMID: 21262969; PubMed Central PMCID: PMC3064187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang H, Issekutz AC. Down-modulation of monocyte transendothelial migration and endothelial adhesion molecule expression by fibroblast growth factor: reversal by the anti-angiogenic agent SU6668. The American journal of pathology. 2002;160(6):2219–2230. doi: 10.1016/S0002-9440(10)61169-8. PubMed PMID: 12057924; PubMed Central PMCID: PMCPMC1850845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Matuszewska B, Keogan M, Fisher DM, Soper KA, Hoe CM, Huber AC, et al. Acidic fibroblast growth factor: evaluation of topical formulations in a diabetic mouse wound healing model. Pharm Res. 1994;11(1):65–71. doi: 10.1023/a:1018993610801. PubMed PMID: 7511240. [DOI] [PubMed] [Google Scholar]
- 121.Mellin TN, Cashen DE, Ronan JJ, Murphy BS, DiSalvo J, Thomas KA. Acidic fibroblast growth factor accelerates dermal wound healing in diabetic mice. J Invest Dermatol. 1995;104(5):850–855. doi: 10.1111/1523-1747.ep12607026. PubMed PMID: 7537778. [DOI] [PubMed] [Google Scholar]
- 122.Xie L, Zhang M, Dong B, Guan M, Lu M, Huang Z, et al. Improved refractory wound healing with administration of acidic fibroblast growth factor in diabetic rats. Diabetes Res Clin Pract. 2011;93(3):396–403. doi: 10.1016/j.diabres.2011.05.016. PubMed PMID: 21641072. [DOI] [PubMed] [Google Scholar]
- 123.Bennett NT, Schultz GS. Growth factors and wound healing: biochemical properties of growth factors and their receptors. Am J Surg. 1993;165(6):728–737. doi: 10.1016/s0002-9610(05)80797-4. PubMed PMID: 8506974. [DOI] [PubMed] [Google Scholar]
- 124.Ma B, Cheng DS, Xia ZF, Ben DF, Lu W, Cao ZF, et al. Randomized, multicenter, double-blind, and placebo-controlled trial using topical recombinant human acidic fibroblast growth factor for deep partial-thickness burns and skin graft donor site. Wound repair and regeneration : official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2007;15(6):795–799. doi: 10.1111/j.1524-475X.2007.00307.x. Epub 2007/11/22. PubMed PMID: 18028126. [DOI] [PubMed] [Google Scholar]
- 125.Akita S, Akino K, Imaizumi T, Hirano A. A basic fibroblast growth factor improved the quality of skin grafting in burn patients. Burns : journal of the International Society for Burn Injuries. 2005;31(7):855–858. doi: 10.1016/j.burns.2005.04.008. Epub 2005/10/04. PubMed PMID: 16199295. [DOI] [PubMed] [Google Scholar]
- 126.Fu X, Shen Z, Chen Y, Xie J, Guo Z, Zhang M, et al. Randomised placebo-controlled trial of use of topical recombinant bovine basic fibroblast growth factor for second-degree burns. Lancet. 1998;352(9141):1661–1664. doi: 10.1016/S0140-6736(98)01260-4. Epub 1998/12/16. PubMed PMID: 9853438. [DOI] [PubMed] [Google Scholar]
- 127.Fu X, Shen Z, Chen Y, Xie J, Guo Z, Zhang M, et al. Recombinant bovine basic fibroblast growth factor accelerates wound healing in patients with burns, donor sites and chronic dermal ulcers. Chinese medical journal. 2000;113(4):367–371. Epub 2002/01/05. PubMed PMID: 11775238. [PubMed] [Google Scholar]
- 128.Nie K, Li P, Zeng X, Sun G, Jin W, Wei Z, et al. Clinical observation of basic fibroblast growth factor combined with topical oxygen therapy in enhancing burn wound healing. Zhongguo xiu fu chong jian wai ke za zhi = Zhongguo xiufu chongjian waike zazhi = Chinese journal of reparative and reconstructive surgery. 2010;24(6):643–646. Epub 2010/07/17. PubMed PMID: 20632489. [PubMed] [Google Scholar]
- 129.Akita S, Akino K, Imaizumi T, Hirano A. Basic fibroblast growth factor accelerates and improves second-degree burn wound healing. Wound repair and regeneration : official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2008;16(5):635–641. doi: 10.1111/j.1524-475X.2008.00414.x. Epub 2009/01/09. PubMed PMID: 19128258. [DOI] [PubMed] [Google Scholar]
- 130.Akita S, Akino K, Imaizumi T, Tanaka K, Anraku K, Yano H, et al. The quality of pediatric burn scars is improved by early administration of basic fibroblast growth factor. Journal of burn care & research : official publication of the American Burn Association. 2006;27(3):333–338. doi: 10.1097/01.BCR.0000216742.23127.7A. Epub 2006/05/09. PubMed PMID: 16679903. [DOI] [PubMed] [Google Scholar]
- 131.Akita S, Akino K, Yakabe A, Tanaka K, Anraku K, Yano H, et al. Basic fibroblast growth factor is beneficial for postoperative color uniformity in split-thickness skin grafting. Wound repair and regeneration : official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2010;18(6):560–566. doi: 10.1111/j.1524-475X.2010.00620.x. Epub 2010/09/28. PubMed PMID: 20868384. [DOI] [PubMed] [Google Scholar]
- 132.Kubo S, Inui T, Hasegawa H, Yoshimine T. Repair of intractable cerebrospinal fluid rhinorrhea with mucosal flaps and recombinant human basic fibroblast growth factor: technical case report. Neurosurgery. 2005;56(3):E627. doi: 10.1227/01.neu.0000154708.18963.26. discussion E. Epub 2005/02/26. PubMed PMID: 15730594. [DOI] [PubMed] [Google Scholar]
- 133.Nakada T, Saito Y, Chikenji M, Koda S, Higuchi M, Kawata K, et al. Therapeutic outcome of hyperbaric oxygen and basic fibroblast growth factor on intractable skin ulcer in legs: preliminary report. Plastic and reconstructive surgery. 2006;117(2):646–651. doi: 10.1097/01.prs.0000197206.48963.60. discussion 52–3. Epub 2006/02/08. PubMed PMID: 16462352. [DOI] [PubMed] [Google Scholar]
- 134.Kakigi A, Sawada S, Takeda T. The effects of basic fibroblast growth factor on postoperative mastoid cavity problems. Otology & neurotology : official publication of the American Otological Society, American Neurotology Society [and] European Academy of Otology and Neurotology. 2005;26(3):333–336. doi: 10.1097/01.mao.0000169763.62679.86. discussion 6. Epub 2005/05/14. PubMed PMID: 15891629. [DOI] [PubMed] [Google Scholar]
- 135.Robson MC, Hill DP, Smith PD, Wang X, Meyer-Siegler K, Ko F, et al. Sequential cytokine therapy for pressure ulcers: clinical and mechanistic response. Annals of surgery. 2000;231(4):600–611. doi: 10.1097/00000658-200004000-00020. Epub 2000/04/05. PubMed PMID: 10749622; PubMed Central PMCID: PMC1421038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Robson MC, Phillips LG, Lawrence WT, Bishop JB, Youngerman JS, Hayward PG, et al. The safety and effect of topically applied recombinant basic fibroblast growth factor on the healing of chronic pressure sores. Annals of surgery. 1992;216(4):401–406. doi: 10.1097/00000658-199210000-00002. discussion 6–8. Epub 1992/10/01. PubMed PMID: 1417189; PubMed Central PMCID: PMC1242638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Uhl E, Barker JH, Bondar I, Galla TJ, Leiderer R, Lehr HA, et al. Basic fibroblast growth factor accelerates wound healing in chronically ischaemic tissue. The British journal of surgery. 1993;80(8):977–980. doi: 10.1002/bjs.1800800812. Epub 1993/08/01. PubMed PMID: 8402094. [DOI] [PubMed] [Google Scholar]
- 138.Yao C, Yao P, Wu H, Zha Z. Acceleration of wound healing in traumatic ulcers by absorbable collagen sponge containing recombinant basic fibroblast growth factor. Biomedical materials. 2006;1(1):33–37. doi: 10.1088/1748-6041/1/1/005. Epub 2008/05/07. PubMed PMID: 18458383. [DOI] [PubMed] [Google Scholar]
- 139.Yamanaka K, Inaba T, Nomura E, Hurwitz D, Jones DA, Hakamada A, et al. Basic fibroblast growth factor treatment for skin ulcerations in scleroderma. Cutis. 2005;76(6):373–376. Epub 2006/01/28. PubMed PMID: 16438426. [PubMed] [Google Scholar]
- 140.Kawaguchi H, Oka H, Jingushi S, Izumi T, Fukunaga M, Sato K, et al. A local application of recombinant human fibroblast growth factor 2 for tibial shaft fractures: A randomized, placebocontrolled trial. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2010;25(12):2735–2743. doi: 10.1002/jbmr.146. Epub 2010/06/10. PubMed PMID: 20533373. [DOI] [PubMed] [Google Scholar]
- 141.Kitamura M, Akamatsu M, Machigashira M, Hara Y, Sakagami R, Hirofuji T, et al. FGF-2 stimulates periodontal regeneration: results of a multi-center randomized clinical trial. Journal of dental research. 2011;90(1):35–40. doi: 10.1177/0022034510384616. Epub 2010/11/10. PubMed PMID: 21059869. [DOI] [PubMed] [Google Scholar]
- 142.Asai J, Takenaka H, Ichihashi K, Ueda E, Katoh N, Kishimoto S. Successful treatment of diabetic gangrene with topical application of a mixture of peripheral blood mononuclear cells and basic fibroblast growth factor. The Journal of dermatology. 2006;33(5):349–352. doi: 10.1111/j.1346-8138.2006.00081.x. Epub 2006/05/17. PubMed PMID: 16700668. [DOI] [PubMed] [Google Scholar]
- 143.Richard JL, Parer-Richard C, Daures JP, Clouet S, Vannereau D, Bringer J, et al. Effect of topical basic fibroblast growth factor on the healing of chronic diabetic neuropathic ulcer of the foot. A pilot, randomized, double-blind, placebo-controlled study. Diabetes care. 1995;18(1):64–69. doi: 10.2337/diacare.18.1.64. Epub 1995/01/01. PubMed PMID: 7698050. [DOI] [PubMed] [Google Scholar]
- 144.Uchi H, Igarashi A, Urabe K, Koga T, Nakayama J, Kawamori R, et al. Clinical efficacy of basic fibroblast growth factor (bFGF) for diabetic ulcer. European journal of dermatology : EJD. 2009;19(5):461–468. doi: 10.1684/ejd.2009.0750. Epub 2009/07/30. PubMed PMID: 19638336. [DOI] [PubMed] [Google Scholar]
- 145.Takagi G, Miyamoto M, Tara S, Takagi I, Takano H, Yasutake M, et al. Controlled-release basic fibroblast growth factor for peripheral artery disease: comparison with autologous bone marrow-derived stem cell transfer. Tissue engineering Part A. 2011;17(21–22):2787–2794. doi: 10.1089/ten.tea.2010.0525. Epub 2011/08/04. PubMed PMID: 21810028. [DOI] [PubMed] [Google Scholar]
- 146.Konturek SJ, Brzozowski T, Majka J, Szlachcic A, Bielanski W, Stachura J, et al. Fibroblast growth factor in gastroprotection and ulcer healing: interaction with sucralfate. Gut. 1993;34(7):881–887. doi: 10.1136/gut.34.7.881. Epub 1993/07/01. PubMed PMID: 8344573; PubMed Central PMCID: PMC1374219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hull MA, Knifton A, Filipowicz B, Brough JL, Vautier G, Hawkey CJ. Healing with basic fibroblast growth factor is associated with reduced indomethacin induced relapse in a human model of gastric ulceration. Gut. 1997;40(2):204–210. doi: 10.1136/gut.40.2.204. Epub 1997/02/01. PubMed PMID: 9071932; PubMed Central PMCID: PMC1027049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fu X, Shen Z, Chen Y. Basic fibroblast growth factor (bFGF) and wound healing: a multi-centers and controlled clinical trial in 1024 cases. Zhongguo xiu fu chong jian wai ke za zhi = Zhongguo xiufu chongjian waike zazhi = Chinese journal of reparative and reconstructive surgery. 1998;12(4):209–211. Epub 1999/08/07. PubMed PMID: 10437068. [PubMed] [Google Scholar]
- 149.Fu XB, Guo ZR, Sheng ZY. Effects of basic fibroblast growth factor on the healing of cutaneous chronic wounds. Zhongguo xiu fu chong jian wai ke za zhi = Zhongguo xiufu chongjian waike zazhi = Chinese journal of reparative and reconstructive surgery. 1999;13(5):270–272. Epub 2002/06/26. PubMed PMID: 12080816. [PubMed] [Google Scholar]
- 150.Wu JC, Huang WC, Tsai YA, Chen YC, Cheng H. Nerve repair using acidic fibroblast growth factor in human cervical spinal cord injury: a preliminary Phase I clinical study. Journal of neurosurgery Spine. 2008;8(3):208–214. doi: 10.3171/SPI/2008/8/3/208. Epub 2008/03/04. PubMed PMID: 18312071. [DOI] [PubMed] [Google Scholar]
- 151.Li E, Trovato JA. New developments in management of oral mucositis in patients with head and neck cancer or receiving targeted anticancer therapies. American journal of health-system pharmacy : AJHP : official journal of the American Society of Health-System Pharmacists. 2012;69(12):1031–1037. doi: 10.2146/ajhp100531. Epub 2012/05/31. PubMed PMID: 22644979. [DOI] [PubMed] [Google Scholar]
- 152.Finch PW, Rubin JS. Keratinocyte growth factor/fibroblast growth factor 7, a homeostatic factor with therapeutic potential for epithelial protection and repair. Advances in cancer research. 2004;91:69–136. doi: 10.1016/S0065-230X(04)91003-2. Epub 2004/08/26. PubMed PMID: 15327889. [DOI] [PubMed] [Google Scholar]
- 153.Weigelt C, Haas R, Kobbe G. Pharmacokinetic evaluation of palifermin for mucosal protection from chemotherapy and radiation. Expert opinion on drug metabolism & toxicology. 2011;7(4):505–515. doi: 10.1517/17425255.2011.566556. Epub 2011/03/23. PubMed PMID: 21417820. [DOI] [PubMed] [Google Scholar]
- 154.Fliedner M, Baguet B, Blankart J, Davies M, Henriques E, Leather A, et al. Palifermin for patients with haematological malignancies: shifting nursing practice from symptom relief to prevention of oral mucositis. European journal of oncology nursing : the official journal of European Oncology Nursing Society. 2007;11(Suppl 1):S19–S26. doi: 10.1016/S1462-3889(07)70004-2. Epub 2007/06/15. PubMed PMID: 17540295. [DOI] [PubMed] [Google Scholar]
- 155.Radtke ML, Kolesar JM. Palifermin (Kepivance) for the treatment of oral mucositis in patients with hematologic malignancies requiring hematopoietic stem cell support. Journal of oncology pharmacy practice : official publication of the International Society of Oncology Pharmacy Practitioners. 2005;11(3):121–125. doi: 10.1191/1078155205jp159oa. Epub 2006/01/05. PubMed PMID: 16390600. [DOI] [PubMed] [Google Scholar]
- 156.Palifermin: AMJ 9701, KGF-Amgen, recombinant human keratinocyte growth factor, rHu-KGF. Drugs in R&D. 2004;5(6):351–354. doi: 10.2165/00126839-200405060-00008. PubMed PMID: 15563240. [DOI] [PubMed] [Google Scholar]
- 157.Sakurai T, Tsuchida M, Lampe PD, Murakami M. Cardiomyocyte FGF signaling is required for Cx43 phosphorylation and cardiac gap junction maintenance. Experimental cell research. 2013;319(14):2152–2165. doi: 10.1016/j.yexcr.2013.05.022. Epub 2013/06/08. PubMed PMID: 23742896; PubMed Central PMCID: PMC3783259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Iwakura A, Fujita M, Ikemoto M, Hasegawa K, Nohara R, Sasayama S, et al. Myocardial ischemia enhances the expression of acidic fibroblast growth factor in human pericardial fluid. Heart and vessels. 2000;15(3):112–116. doi: 10.1007/pl00007264. Epub 2001/04/06. PubMed PMID: 11289498. [DOI] [PubMed] [Google Scholar]
- 159.Sakakibara Y, Tambara K, Sakaguchi G, Lu F, Yamamoto M, Nishimura K, et al. Toward surgical angiogenesis using slow-released basic fibroblast growth factor. European journal of cardiothoracic surgery : official journal of the European Association for Cardio-thoracic Surgery. 2003;24(1):105–111. doi: 10.1016/s1010-7940(03)00159-3. discussion 12. Epub 2003/07/11. PubMed PMID: 12853053. [DOI] [PubMed] [Google Scholar]
- 160.Yajima S, Ishikawa M, Kubota T, Moroi M, Sugi K, Namiki A. Intramyocardial injection of fibroblast growth factor-2 plus heparin suppresses cardiac failure progression in rats with hypertensive heart disease. International heart journal. 2005;46(2):289–301. doi: 10.1536/ihj.46.289. Epub 2005/05/07. PubMed PMID: 15876812. [DOI] [PubMed] [Google Scholar]
- 161.Suzuki G, Lee TC, Fallavollita JA, Canty JM., Jr Adenoviral gene transfer of FGF-5 to hibernating myocardium improves function and stimulates myocytes to hypertrophy and reenter the cell cycle. Circulation research. 2005;96(7):767–775. doi: 10.1161/01.RES.0000162099.01268.d1. Epub 2005/03/12. PubMed PMID: 15761196. [DOI] [PubMed] [Google Scholar]
- 162.Simons M, Annex BH, Laham RJ, Kleiman N, Henry T, Dauerman H, et al. Pharmacological treatment of coronary artery disease with recombinant fibroblast growth factor-2: double-blind, randomized, controlled clinical trial. Circulation. 2002;105(7):788–793. doi: 10.1161/hc0802.104407. Epub 2002/02/21. PubMed PMID: 11854116. [DOI] [PubMed] [Google Scholar]
- 163.Kukula K, Chojnowska L, Dabrowski M, Witkowski A, Chmielak Z, Skwarek M, et al. Intramyocardial plasmid-encoding human vascular endothelial growth factor A165/basic fibroblast growth factor therapy using percutaneous transcatheter approach in patients with refractory coronary artery disease (VIF-CAD) American heart journal. 2011;161(3):581–589. doi: 10.1016/j.ahj.2010.11.023. Epub 2011/03/12. PubMed PMID: 21392615. [DOI] [PubMed] [Google Scholar]
- 164.Laham RJ, Chronos NA, Pike M, Leimbach ME, Udelson JE, Pearlman JD, et al. Intracoronary basic fibroblast growth factor (FGF-2) in patients with severe ischemic heart disease: results of a phase I open-label dose escalation study. Journal of the American College of Cardiology. 2000;36(7):2132–2139. doi: 10.1016/s0735-1097(00)00988-8. Epub 2000/12/29. PubMed PMID: 11127452. [DOI] [PubMed] [Google Scholar]
- 165.Grines CL, Watkins MW, Helmer G, Penny W, Brinker J, Marmur JD, et al. Angiogenic Gene Therapy (AGENT) trial in patients with stable angina pectoris. Circulation. 2002;105(11):1291–1297. doi: 10.1161/hc1102.105595. Epub 2002/03/20. PubMed PMID: 11901038. [DOI] [PubMed] [Google Scholar]
- 166.Kastrup J. Gene therapy and angiogenesis in patients with coronary artery disease. Expert review of cardiovascular therapy. 2010;8(8):1127–1138. doi: 10.1586/erc.10.95. Epub 2010/07/31. PubMed PMID: 20670190. [DOI] [PubMed] [Google Scholar]
- 167.McKeehan WL, Wang F, Luo Y. The fibroblast growth factor (FGF) signaling complex. In: Bradshaw R, Dennis E, editors. Handbook of Cell Signaling. 2dn. New York: Academic/Elsevier Press; 2009. [Google Scholar]
- 168.Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. Journal of biochemistry. 2011;149(2):121–130. doi: 10.1093/jb/mvq121. Epub 2010/10/14. PubMed PMID: 20940169; PubMed Central PMCID: PMC3106964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, et al. Fibroblast growth factor. 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell metabolism. 2005;2(4):217–225. doi: 10.1016/j.cmet.2005.09.001. Epub 2005/10/11. PubMed PMID: 16213224. [DOI] [PubMed] [Google Scholar]
- 170.Hotta Y, Nakamura H, Konishi M, Murata Y, Takagi H, Matsumura S, et al. Fibroblast growth factor 21 regulates lipolysis in white adipose tissue but is not required for ketogenesis and triglyceride clearance in liver. Endocrinology. 2009;150(10):4625–4633. doi: 10.1210/en.2009-0119. Epub 2009/07/11. PubMed PMID: 19589869. [DOI] [PubMed] [Google Scholar]
- 171.Urakawa I, Yamazaki Y, Shimada T, Iijima K, Hasegawa H, Okawa K, et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444(7120):770–774. doi: 10.1038/nature05315. Epub 2006/11/07. PubMed PMID: 17086194. [DOI] [PubMed] [Google Scholar]
- 172.Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, et al. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. The Journal of biological chemistry. 2000;275(20):15482–15489. doi: 10.1074/jbc.275.20.15482. PubMed PMID: 10809780. [DOI] [PubMed] [Google Scholar]
- 173.Foltz IN, Hu S, King C, Wu X, Yang C, Wang W, et al. Treating diabetes and obesity with an FGF21-mimetic antibody activating the betaKlotho/FGFR1c receptor complex. Science translational medicine. 2012;4(162):162ra53. doi: 10.1126/scitranslmed.3004690. Epub 2012/12/01. PubMed PMID: 23197570. [DOI] [PubMed] [Google Scholar]
- 174.Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine fibroblast growth factors 15/19 and 21: from feast to famine. Genes & development. 2012;26(4):312–324. doi: 10.1101/gad.184788.111. PubMed PMID: 22302876; PubMed Central PMCID: PMCPMC3289879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yang C, Wang C, Ye M, Jin C, He W, Wang F, et al. Control of lipid metabolism by adipocyte FGFR1-mediated adipohepatic communication during hepatic stress. Nutrition & metabolism. 2012;9(1):94. doi: 10.1186/1743-7075-9-94. Epub 2012/10/31. PubMed PMID: 23106963; PubMed Central PMCID: PMC3545967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kharitonenkov A, Shiyanova TL, Koester A, Ford AM, Micanovic R, Galbreath EJ, et al. FGF-21 as a novel metabolic regulator. The Journal of clinical investigation. 2005;115(6):1627–1635. doi: 10.1172/JCI23606. Epub 2005/05/20. PubMed PMID: 15902306; PubMed Central PMCID: PMC1088017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fu L, John LM, Adams SH, Yu XX, Tomlinson E, Renz M, et al. Fibroblast growth factor 19 increases metabolic rate and reverses dietary and leptin-deficient diabetes. Endocrinology. 2004;145(6):2594–2603. doi: 10.1210/en.2003-1671. Epub 2004/02/21. PubMed PMID: 14976145. [DOI] [PubMed] [Google Scholar]
- 178.Kharitonenkov A, Wroblewski VJ, Koester A, Chen YF, Clutinger CK, Tigno XT, et al. The metabolic state of diabetic monkeys is regulated by fibroblast growth factor-21. Endocrinology. 2007;148(2):774–781. doi: 10.1210/en.2006-1168. Epub 2006/10/28. PubMed PMID: 17068132. [DOI] [PubMed] [Google Scholar]
- 179.Adams A, Yang C, Coskun T, Cheng CC, Cimeno R, Luo Y, et al. The breadth of FGF21's metabolic actions are governed by FGFR1 in adipose tissue. Mol Metabolism. 2012;2(1):31–37. doi: 10.1016/j.molmet.2012.08.007. Epub Feb 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Galman C, Lundasen T, Kharitonenkov A, Bina HA, Eriksson M, Hafstrom I, et al. The circulating metabolic regulator FGF21 is induced by prolonged fasting and PPARalpha activation in man. Cell metabolism. 2008;8(2):169–174. doi: 10.1016/j.cmet.2008.06.014. Epub 2008/08/06. PubMed PMID: 18680716. [DOI] [PubMed] [Google Scholar]
- 181.Berglund ED, Li CY, Bina HA, Lynes SE, Michael MD, Shanafelt AB, et al. Fibroblast growth factor 21 controls glycemia via regulation of hepatic glucose flux and insulin sensitivity. Endocrinology. 2009;150(9):4084–4093. doi: 10.1210/en.2009-0221. Epub 2009/05/28. PubMed PMID: 19470704; PubMed Central PMCID: PMC2736088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell metabolism. 2007;5(6):415–425. doi: 10.1016/j.cmet.2007.05.003. Epub 2007/06/07. PubMed PMID: 17550777. [DOI] [PubMed] [Google Scholar]
- 183.Potthoff MJ, Inagaki T, Satapati S, Ding X, He T, Goetz R, et al. FGF21 induces PGC-1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(26):10853–10858. doi: 10.1073/pnas.0904187106. Epub 2009/06/23. PubMed PMID: 19541642; PubMed Central PMCID: PMC2705613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yang C, Jin C, Li X, Wang F, McKeehan WL, Luo Y. Differential specificity of endocrine FGF19 and FGF21 to FGFR1 and FGFR4 in complex with KLB. PloS one. 2012;7(3):e33870. doi: 10.1371/journal.pone.0033870. Epub 2012/03/24. PubMed PMID: 22442730; PubMed Central PMCID: PMC3307775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Suzuki M, Uehara Y, Motomura-Matsuzaka K, Oki J, Koyama Y, Kimura M, et al. betaKlotho is required for fibroblast growth factor (FGF) 21 signaling through FGF receptor (FGFR) 1c and FGFR3c. Molecular endocrinology. 2008;22(4):1006–1014. doi: 10.1210/me.2007-0313. Epub 2008/01/12. doi: me.2007-0313 [pii] 10.1210/me.2007-0313. PubMed PMID: 18187602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lin Z, Tian H, Lam KS, Lin S, Hoo RC, Konishi M, et al. Adiponectin Mediates the Metabolic Effects of FGF21 on Glucose Homeostasis and Insulin Sensitivity in Mice. Cell metabolism. 2013;17(5):779–789. doi: 10.1016/j.cmet.2013.04.005. Epub 2013/05/15. PubMed PMID: 23663741. [DOI] [PubMed] [Google Scholar]
- 187.Holland WL, Adams AC, Brozinick JT, Bui HH, Miyauchi Y, Kusminski CM, et al. An FGF21-adiponectin-ceramide axis controls energy expenditure and insulin action in mice. Cell metabolism. 2013;17(5):790–797. doi: 10.1016/j.cmet.2013.03.019. PubMed PMID: 23663742; PubMed Central PMCID: PMCPMC3667496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Luo Y, McKeehan WL. Stressed Liver and Muscle Call on Adipocytes with FGF21. Frontiers in endocrinology. 2013;4:194. doi: 10.3389/fendo.2013.00194. PubMed PMID: 24385972; PubMed Central PMCID: PMC3866528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ohta H, Itoh N. Roles of FGFs as Adipokines in Adipose Tissue Development, Remodeling, and Metabolism. Frontiers in endocrinology. 2014;5:18. doi: 10.3389/fendo.2014.00018. PubMed PMID: 24605108; PubMed Central PMCID: PMC3932445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fuentes-Mattei E, Velazquez-Torres G, Phan L, Zhang F, Chou PC, Shin JH, et al. Effects of obesity on transcriptomic changes and cancer hallmarks in estrogen receptor-positive breast cancer. Journal of the National Cancer Institute. 2014;106(7) doi: 10.1093/jnci/dju158. PubMed PMID: 24957076; PubMed Central PMCID: PMC4110474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Luo Y, Yang C, Ye M, Jin C, Abbruzzese JL, Lee MH, et al. Deficiency of metabolic regulator FGFR4 delays breast cancer progression through systemic and microenvironmental metabolic alterations. Cancer & metabolism. 2013;1(1):21. doi: 10.1186/2049-3002-1-21. PubMed PMID: 24279986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. The Biochemical journal. 2011;437(2):199–213. doi: 10.1042/BJ20101603. PubMed PMID: 21711248. [DOI] [PubMed] [Google Scholar]
- 193.Iyer G, Milowsky MI. Fibroblast growth factor receptor-3 in urothelial tumorigenesis. Urologic oncology. 2013;31(3):303–311. doi: 10.1016/j.urolonc.2011.12.001. PubMed PMID: 22285006. [DOI] [PubMed] [Google Scholar]
- 194.Wang J, Stockton DW, Ittmann M. The fibroblast growth factor receptor-4 Arg388 allele is associated with prostate cancer initiation and progression. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10(18 Pt 1):6169–6178. doi: 10.1158/1078-0432.CCR-04-0408. PubMed PMID: 15448004. [DOI] [PubMed] [Google Scholar]
- 195.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. PubMed PMID: 20579941; PubMed Central PMCID: PMC3198787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nature genetics. 2007 doi: 10.1038/ng2075. PubMed PMID: 17529973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kwabi-Addo B, Ozen M, Ittmann M. The role of fibroblast growth factors and their receptors in prostate cancer. Endocrine-related cancer. 2004;11(4):709–724. doi: 10.1677/erc.1.00535. PubMed PMID: 15613447. [DOI] [PubMed] [Google Scholar]
- 198.Yan G, Fukabori Y, McBride G, Nikolaropolous S, McKeehan WL. Exon switching and activation of stromal and embryonic fibroblast growth factor (FGF)-FGF receptor genes in prostate epithelial cells accompany stromal independence and malignancy. Molecular and cellular biology. 1993;13(8):4513–4522. doi: 10.1128/mcb.13.8.4513. PubMed PMID: 7687739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lu W, Luo Y, Kan M, McKeehan WL. Fibroblast growth factor-10. A second candidate stromal to epithelial cell andromedin in prostate. The Journal of biological chemistry. 1999;274(18):12827–12834. doi: 10.1074/jbc.274.18.12827. PubMed PMID: 10212269. [DOI] [PubMed] [Google Scholar]
- 200.Cotton LM, O'Bryan MK, Hinton BT. Cellular signaling by fibroblast growth factors (FGFs) and their receptors (FGFRs) in male reproduction. Endocr Rev. 2008;29(2):193–216. doi: 10.1210/er.2007-0028. PubMed PMID: 18216218; PubMed Central PMCID: PMCPMC2528845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Valve EM, Nevalainen MT, Nurmi MJ, Laato MK, Martikainen PM, Harkonen PL. Increased expression of FGF-8 isoforms and FGF receptors in human premalignant prostatic intraepithelial neoplasia lesions and prostate cancer. Laboratory investigation; a journal of technical methods and pathology. 2001;81(6):815–826. doi: 10.1038/labinvest.3780291. PubMed PMID: 11406643. [DOI] [PubMed] [Google Scholar]
- 202.Leung HY, Dickson C, Robson CN, Neal DE. Over-expression of fibroblast growth factor-8 in human prostate cancer. Oncogene. 1996;12(8):1833–1835. PubMed PMID: 8622905. [PubMed] [Google Scholar]
- 203.Tanaka A, Furuya A, Yamasaki M, Hanai N, Kuriki K, Kamiakito T, et al. High frequency of fibroblast growth factor (FGF) 8 expression in clinical prostate cancers and breast tissues, immunohistochemically demonstrated by a newly established neutralizing monoclonal antibody against FGF 8. Cancer research. 1998;58(10):2053–2056. [PubMed] [Google Scholar]
- 204.Gnanapragasam VJ, Robinson MC, Marsh C, Robson CN, Hamdy FC, Leung HY. FGF8 isoform b expression in human prostate cancer. British journal of cancer. 2003;88(9):1432–1438. doi: 10.1038/sj.bjc.6600875. PubMed PMID: 12778074; PubMed Central PMCID: PMCPMC2741033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dorkin TJ, Robinson MC, Marsh C, Bjartell A, Neal DE, Leung HY. FGF8 over-expression in prostate cancer is associated with decreased patient survival and persists in androgen independent disease. Oncogene. 1999;18(17):2755–2761. doi: 10.1038/sj.onc.1202624. PubMed PMID: 10348350. [DOI] [PubMed] [Google Scholar]
- 206.Udayakumar TS, Nagle RB, Bowden GT. Fibroblast growth factor-1 transcriptionally induces membrane type-1 matrix metalloproteinase expression in prostate carcinoma cell line. The Prostate. 2004;58(1):66–75. doi: 10.1002/pros.10293. PubMed PMID: 14673954. [DOI] [PubMed] [Google Scholar]
- 207.Huang Y, Jin C, Hamana T, Liu J, Wang C, An L, et al. Overexpression of FGF9 in Prostate Epithelial Cells Augments Reactive Stroma Formation and Promotes Prostate Cancer Progression. International journal of biological sciences. 2015;11(8):948–960. doi: 10.7150/ijbs.12468. PubMed PMID: 26157349; PubMed Central PMCID: PMC4495412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wesley UV, McGroarty M, Homoyouni A. Dipeptidyl peptidase inhibits malignant phenotype of prostate cancer cells by blocking basic fibroblast growth factor signaling pathway. Cancer research. 2005;65(4):1325–1334. doi: 10.1158/0008-5472.CAN-04-1852. PubMed PMID: 15735018. [DOI] [PubMed] [Google Scholar]
- 209.Yang F, Strand DW, Rowley DR. Fibroblast growth factor-2 mediates transforming growth factorbeta action in prostate cancer reactive stroma. Oncogene. 2008;27(4):450–459. doi: 10.1038/sj.onc.1210663. PubMed PMID: 17637743. [DOI] [PubMed] [Google Scholar]
- 210.Polnaszek N, Kwabi-Addo B, Peterson LE, Ozen M, Greenberg NM, Ortega S, et al. Fibroblast growth factor 2 promotes tumor progression in an autochthonous mouse model of prostate cancer. Cancer research. 2003;63(18):5754–5760. PubMed PMID: 14522896. [PubMed] [Google Scholar]
- 211.Liu J, You P, Chen G, Fu X, Zeng X, Wang C, et al. Hyperactivated FRS2alpha-mediated signaling in prostate cancer cells promotes tumor angiogenesis and predicts poor clinical outcome of patients. Oncogene. 2015 doi: 10.1038/onc.2015.239. PubMed PMID: 26096936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bubendorf L, Schopfer A, Wagner U, Sauter G, Moch H, Willi N, et al. Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Hum Pathol. 2000;31(5):578–583. doi: 10.1053/hp.2000.6698. PubMed PMID: 10836297. [DOI] [PubMed] [Google Scholar]
- 213.Li ZG, Mathew P, Yang J, Starbuck MW, Zurita AJ, Liu J, et al. Androgen receptor-negative human prostate cancer cells induce osteogenesis in mice through FGF9-mediated mechanisms. The Journal of clinical investigation. 2008;118(8):2697–2710. doi: 10.1172/JCI33093. PubMed PMID: 18618013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Valta MP, Hentunen T, Qu Q, Valve EM, Harjula A, Seppanen JA, et al. Regulation of osteoblast differentiation: a novel function for fibroblast growth factor 8. Endocrinology. 2006;147(5):2171–2182. doi: 10.1210/en.2005-1502. PubMed PMID: 16439448. [DOI] [PubMed] [Google Scholar]
- 215.Valta MP, Tuomela J, Bjartell A, Valve E, Vaananen HK, Harkonen P. FGF-8 is involved in bone metastasis of prostate cancer. International journal of cancer Journal international du cancer. 2008;123(1):22–31. doi: 10.1002/ijc.23422. PubMed PMID: 18386787. [DOI] [PubMed] [Google Scholar]
- 216.Jin C, Wang F, Wu X, Yu C, Luo Y, McKeehan WL. Directionally specific paracrine communication mediated by epithelial FGF9 to stromal FGFR3 in two-compartment premalignant prostate tumors. Cancer research. 2004;64(13):4555–4562. doi: 10.1158/0008-5472.CAN-03-3752. PubMed PMID: 15231666. [DOI] [PubMed] [Google Scholar]
- 217.Leung HY, Mehta P, Gray LB, Collins AT, Robson CN, Neal DE. Keratinocyte growth factor expression in hormone insensitive prostate cancer. Oncogene. 1997;15(9):1115–1120. doi: 10.1038/sj.onc.1201256. PubMed PMID: 9285567. [DOI] [PubMed] [Google Scholar]
- 218.Gan Y, Wientjes MG, Au JL. Expression of basic fibroblast growth factor correlates with resistance to paclitaxel in human patient tumors. Pharm Res. 2006;23(6):1324–1331. doi: 10.1007/s11095-006-0136-6. PubMed PMID: 16741658. [DOI] [PubMed] [Google Scholar]
- 219.Song S, Wientjes MG, Gan Y, Au JL. Fibroblast growth factors: an epigenetic mechanism of broad spectrum resistance to anticancer drugs. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(15):8658–8663. doi: 10.1073/pnas.140210697. PubMed PMID: 10890892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Maruyama-Takahashi K, Shimada N, Imada T, Maekawa-Tokuda Y, Ishii T, Ouchi J, et al. A neutralizing anti-fibroblast growth factor (FGF) 8 monoclonal antibody shows anti-tumor activity against FGF8b-expressing LNCaP xenografts in androgen-dependent and -independent conditions. The Prostate. 2008;68(6):640–650. doi: 10.1002/pros.20728. Epub 2008/01/24. PubMed PMID: 18213631. [DOI] [PubMed] [Google Scholar]
- 221.Jin C, McKeehan K, Guo W, Jauma S, Ittmann MM, Foster B, et al. Cooperation between ectopic FGFR1 and depression of FGFR2 in induction of prostatic intraepithelial neoplasia in the mouse prostate. Cancer research. 2003;63(24):8784–8790. PubMed PMID: 14695195. [PubMed] [Google Scholar]
- 222.Wang F, McKeehan K, Yu C, Ittmann M, McKeehan WL. Chronic activity of ectopic type 1 fibroblast growth factor receptor tyrosine kinase in prostate epithelium results in hyperplasia accompanied by intraepithelial neoplasia. The Prostate. 2004;58(1):1–12. doi: 10.1002/pros.10311. PubMed PMID: 14673947. [DOI] [PubMed] [Google Scholar]
- 223.Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, et al. Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell. 2007;12(6):559–571. doi: 10.1016/j.ccr.2007.11.004. PubMed PMID: 18068632. [DOI] [PubMed] [Google Scholar]
- 224.Zhang Y, Zhang J, Lin Y, Lan Y, Lin C, Xuan JW, et al. Role of epithelial cell fibroblast growth factor receptor substrate 2{alpha} in prostate development, regeneration and tumorigenesis. Development. 2008;135(4):775–784. doi: 10.1242/dev.009910. PubMed PMID: 18184727. [DOI] [PubMed] [Google Scholar]
- 225.Yang F, Zhang Y, Ressler SJ, Ittmann MM, Ayala GE, Dang TD, et al. FGFR1 is essential for prostate cancer progression and metastasis. Cancer research. 2013;73(12):3716–3724. doi: 10.1158/0008-5472.CAN-12-3274. Epub 2013/04/12. PubMed PMID: 23576558; PubMed Central PMCID: PMC3686853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Memarzadeh S, Xin L, Mulholland DJ, Mansukhani A, Wu H, Teitell MA, et al. Enhanced paracrine FGF10 expression promotes formation of multifocal prostate adenocarcinoma and an increase in epithelial androgen receptor. Cancer Cell. 2007;12(6):572–585. doi: 10.1016/j.ccr.2007.11.002. PubMed PMID: 18068633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. PubMed PMID: 19487818; PubMed Central PMCID: PMCPMC2689101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Holzmann K, Grunt T, Heinzle C, Sampl S, Steinhoff H, Reichmann N, et al. Alternative Splicing of Fibroblast Growth Factor Receptor IgIII Loops in Cancer. J Nucleic Acids. 2012;2012:950508. doi: 10.1155/2012/950508. PubMed PMID: 22203889; PubMed Central PMCID: PMCPMC3238399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Gavine PR, Mooney L, Kilgour E, Thomas AP, Al-Kadhimi K, Beck S, et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer research. 2012;72(8):2045–2056. doi: 10.1158/0008-5472.CAN-11-3034. PubMed PMID: 22369928. [DOI] [PubMed] [Google Scholar]
- 230.Xie L, Su X, Zhang L, Yin X, Tang L, Zhang X, et al. FGFR2 gene amplification in gastric cancer predicts sensitivity to the selective FGFR inhibitor AZD4547. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(9):2572–2583. doi: 10.1158/1078-0432.CCR-12-3898. PubMed PMID: 23493349. [DOI] [PubMed] [Google Scholar]
- 231.Trudel S, Li ZH, Wei E, Wiesmann M, Chang H, Chen C, et al. CHIR-258, a novel, multitargeted tyrosine kinase inhibitor for the potential treatment of t(4;14) multiple myeloma. Blood. 2005;105(7):2941–2948. doi: 10.1182/blood-2004-10-3913. PubMed PMID: 15598814. [DOI] [PubMed] [Google Scholar]
- 232.Hilberg F, Roth GJ, Krssak M, Kautschitsch S, Sommergruber W, Tontsch-Grunt U, et al. BIBF 1120: triple angiokinase inhibitor with sustained receptor blockade and good antitumor efficacy. Cancer research. 2008;68(12):4774–4782. doi: 10.1158/0008-5472.CAN-07-6307. PubMed PMID: 18559524. [DOI] [PubMed] [Google Scholar]
- 233.Okamoto K, Ikemori-Kawada M, Jestel A, von Konig K, Funahashi Y, Matsushima T, et al. Distinct binding mode of multikinase inhibitor lenvatinib revealed by biochemical characterization. ACS medicinal chemistry letters. 2015;6(1):89–94. doi: 10.1021/ml500394m. PubMed PMID: 25589937; PubMed Central PMCID: PMC4291723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Bhide RS, Cai ZW, Zhang YZ, Qian L, Wei D, Barbosa S, et al. Discovery and preclinical studies of (R)- 1-(4-(4-fluoro-2-methyl-1H-indol-5-yloxy)-5- methylpyrrolo[2,1-f][1,2,4]triazin-6-yloxy)propan- 2-ol (BMS-540215), an in vivo active potent VEGFR-2 inhibitor. Journal of medicinal chemistry. 2006;49(7):2143–2146. doi: 10.1021/jm051106d. PubMed PMID: 16570908. [DOI] [PubMed] [Google Scholar]
- 235.Laird AD, Vajkoczy P, Shawver LK, Thurnher A, Liang C, Mohammadi M, et al. SU6668 is a potent antiangiogenic and antitumor agent that induces regression of established tumors. Cancer research. 2000;60(15):4152–4160. [PubMed] [Google Scholar]
- 236.Daniele G, Corral J, Molife LR, de Bono JS. FGF receptor inhibitors: role in cancer therapy. Curr Oncol Rep. 2012;14(2):111–119. doi: 10.1007/s11912-012-0225-0. PubMed PMID: 22311684. [DOI] [PubMed] [Google Scholar]
- 237.Mohammadi M, Froum S, Hamby JM, Schroeder MC, Panek RL, Lu GH, et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. Embo J. 1998;17(20):5896–5904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Chaochao X, Wulan L, Peihong Q, Yiqun X, Xiaojing D, Fen W, et al. The therapeutic potential of a novel non-ATP-competitive fibroblast growth factor receptor 1 inhibitor on gastric cancer. Anticancer drugs. 2014 doi: 10.1097/CAD.0000000000000195. PubMed PMID: 25521558. [DOI] [PubMed] [Google Scholar]
- 239.Wu J, Ji J, Weng B, Qiu P, Kanchana K, Wei T, et al. Discovery of novel non-ATP competitive FGFR1 inhibitors and evaluation of their anti-tumor activity in non-small cell lung cancer in vitro and in vivo. Oncotarget. 2014;5(12):4543–4553. doi: 10.18632/oncotarget.2122. PubMed PMID: 24980830; PubMed Central PMCID: PMC4147344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wang Y, Cai Y, Ji J, Liu Z, Zhao C, Zhao Y, et al. Discovery and identification of new non-ATP competitive FGFR1 inhibitors with therapeutic potential on non-small-cell lung cancer. Cancer letters. 2014;344(1):82–89. doi: 10.1016/j.canlet.2013.10.016. PubMed PMID: 24513267. [DOI] [PubMed] [Google Scholar]
- 241.Qing J, Du X, Chen Y, Chan P, Li H, Wu P, et al. Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice. The Journal of clinical investigation. 2009;119(5):1216–1229. doi: 10.1172/JCI38017. PubMed PMID: 19381019; PubMed Central PMCID: PMCPMC2673861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Bai A, Meetze K, Vo NY, Kollipara S, Mazsa EK, Winston WM, et al. GP369, an FGFR2-IIIb-specific antibody, exhibits potent antitumor activity against human cancers driven by activated FGFR2 signaling. Cancer research. 2010;70(19):7630–7639. doi: 10.1158/0008-5472.CAN-10-1489. PubMed PMID: 20709759. [DOI] [PubMed] [Google Scholar]
- 243.Pai R, Dunlap D, Qing J, Mohtashemi I, Hotzel K, French DM. Inhibition of fibroblast growth factor 19 reduces tumor growth by modulating beta-catenin signaling. Cancer research. 2008;68(13):5086–5095. doi: 10.1158/0008-5472.CAN-07-2325. PubMed PMID: 18593907. [DOI] [PubMed] [Google Scholar]
- 244.Harding TC, Long L, Palencia S, Zhang H, Sadra A, Hestir K, et al. Blockade of nonhormonal fibroblast growth factors by FP-1039 inhibits growth of multiple types of cancer. Science translational medicine. 2013;5(178):178ra39. doi: 10.1126/scitranslmed.3005414. PubMed PMID: 23536011. [DOI] [PubMed] [Google Scholar]
- 245.Tolcher A, Papadopoulos K, Agnew J, Marshall J, Tonzola C, Zanghi J, et al. Abstract A103: Preliminary results of a phase 1 study of FP-1039 (FGFR1:Fc), a novel antagonist of multiple fibroblast growth factor (FGF) ligands, in patients with advanced malignancies. Molecular Cancer Therapeutics. 2009;8(12 Suppl):A 103. [Google Scholar]
- 246.Fan L, Xie H, Chen L, Ye H, Ying S, Wang C, et al. A novel FGF2 antagonist peptide P8 with potent antiproliferation activity. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(10):10571–10579. doi: 10.1007/s13277-014-2356-4. PubMed PMID: 25062723. [DOI] [PubMed] [Google Scholar]
- 247.Wu X, Huang H, Wang C, Lin S, Huang Y, Wang Y, et al. Identification of a novel peptide that blocks basic fibroblast growth factor-mediated cell proliferation. Oncotarget. 2013;4(10):1819–1828. doi: 10.18632/oncotarget.1312. PubMed PMID: 24142482; PubMed Central PMCID: PMC3858566. [DOI] [PMC free article] [PubMed] [Google Scholar]


