Abstract
Amelotin (AMTN) and kallikrein-4 (KLK4) are secreted proteins specialized for enamel biomineralization. We characterized enamel from wild-type, Amtn−/−, Klk4−/−, Amtn+/−Klk4+/− and Amtn−/−Klk4−/− mice to gain insights into AMTN and KLK4 functions during amelogenesis. All of the null mice were healthy and fertile. The mandibular incisors in Amtn−/−, Klk4−/− and Amtn−/−Klk4−/− mice were chalky-white and chipped. No abnormalities except in enamel were observed, and no significant differences were detected in enamel thickness or volume, or in rod decussation. Micro-computed tomography (µCT) maximum intensity projections localized the onset of enamel maturation in wild-type incisors distal to the first molar, but mesial to this position in Amtn−/−, Klk4−/− and Amtn−/−Klk4−/− mice, demonstrating a delay in enamel maturation in Amtn−/− incisors. Micro-CT detected significantly reduced enamel mineral density (2.5 and 2.4 gHA/cm3) in the Klk4−/− and Amtn−/−Klk4−/− mice respectively, compared with wild-type enamel (3.1 gHA/cm3). Backscatter scanning electron microscopy showed that mineral density progressively diminished with enamel depth in the Klk4−/− and Amtn−/−Klk4−/− mice. Knoop hardness of Amtn−/− outer enamel was significantly reduced relative to the wild-type and was not as hard as the middle or inner enamel. Klk4−/− enamel hardness was significantly reduced at all levels, but the outer enamel was significantly harder than the inner and middle enamel. Thus the hardness patterns of the Amtn−/− and Klk4−/− mice were distinctly different, while the Amtn−/−Klk4−/− outer enamel was not as hard as in the Amtn−/− and Klk4−/− mice. We conclude that AMTN and KLK4 function independently, but are both necessary for proper enamel maturation.
Keywords: Amelotin, kallikrein-4, Knoop hardness, µT, null mice, ameloblasts
Introduction
Dental enamel forms in two major stages [1–3]. During the secretory stage, thin ribbons of amorphous calcium phosphate initiate and lengthen at a mineralization front along the secretory surfaces of the ameloblasts’ distal membranes and these ribbons transform into biological apatite [4]. During the subsequent maturation stage the crystals grow in width and thickness, which hardens the enamel layer [5]. Some maturation occurs during the secretory stage and this growth in crystal width and thickness increases with depth toward the dentino-enamel junction (DEJ) [6,7].
The formation of dental enamel is a biological process that originated in lobe-finned fish with lungs. Lungfish deposit on the surface of incipient dentin, thin mineral ribbons that extend at a specialized mineralization front along the secretory surfaces of ameloblasts [8], which is the familiar pattern for amelogenesis in tetrapods. Many of the genes encoding key extracellular components specifically associated with dental enamel formation in mammals, such as the SCPP (secretory calcium-binding phosphoprotein) proteins amelogenin (AMEL), enamelin (ENAM), ameloblastin (AMBN), amelotin (AMTN), as well as the secretory stage enamel protease MMP20 are found in the coelacanth genome but have no clear orthologs in teleosts, which cover dentin with enameloid that forms in a dentin- like collagenous extracellular matrix [9]. The SCPP basal lamina protein SCPPPQ1 that is expressed late in the maturation stage [10,11] arose in reptiles [12]. The organization of thin crystal ribbons into rod and interrod structures is an innovation that occurred in mammals before the divergence of marsupials and may be associated with evolutionary changes in amelotin structure and a delay in its expression until completion of the secretory stage [13,14,12]. The secretion of kallikrein 4 (KLK4) to facilitate the maturation of dental enamel was a late addition evolutionarily, which occurred in placental mammals and is associated with earlier eruption of the permanent (secondary) dentition [15].
AMTN and KLK4 are both expressed by the ameloblast lineage starting in the transition between secretory and maturation stages [16–20]. Both genes show only trace expression in other tissues. The human expressed sequence tag (EST) database (which does not include developing teeth) lists only 2 AMTN and 10 KLK4 ESTs out of a total of 3,328,811 ESTs for normal tissues. Amtn and Klk4 both appear to be functionally specific for dental enamel formation and are pseudogenized in species that have lost the ability to make teeth or dental enamel during evolution [21,15,22]. The Klk4−/− mouse display severe maturation stage enamel malformations in the absence of systemic abnormalities [23,24] and defects in KLK4 cause an autosomal recessive, non-syndromic form of amelogenesis imperfecta (AI) [25–27]. Mutations in AMTN have not yet been shown to cause AI, but recently the Amtn knockout mouse was reported to exhibit a subtle enamel phenotype that primarily affects the outer and surface enamel layers [28].
The function of AMTN is poorly understood. During the maturation stage of amelogenesis the secreted AMTN protein localizes in the extracellular space adjacent to the distal membrane of the ameloblast. Immunogold labeling characterized by transmission electron microscopy showed that early in the maturation stage AMTN penetrates into the enamel surface and seems to concentrate in the less-mineralized crypts left behind as the ameloblasts retracted their Tomes’ processes at the end of the secretory stage [20]. By late maturation AMTN localizes specifically to the ameloblast-enamel interface and then disappears beneath the late, reduced enamel epithelium [20], only to reappear at the cell-enamel interface beneath the junctional epithelium following tooth eruption [29,30] where it is believed to facilitate adherence of the junctional epithelium to the tooth surface [31]. AMTN interacts with itself and with ODAM [32] and colocalizes with ODAM in the basal lamina beneath maturation stage ameloblasts and junctional epithelium.
The structure and function of KLK4 is well-characterized [33,34]. Klk4 is a glycosylated serine protease [35] that is secreted by maturation stage ameloblasts [36] and degrades enamel proteins [37] to facilitate their reabsorption by maturation stage ameloblasts. In the absence of KLK4, enamel mineralization is normal up to early maturation after which the enamel retains proteins [38] and is unable to mature beyond 85% mineral by weight. The outer enamel is hard, but the inner and middle enamel is soft and contains much more protein than normal [24]. The enamel fails after eruption, as it tends to fracture in the inner enamel near the dentin surface [23,39].
The purpose of this study was to obtain a more detailed understanding of the enamel defects in the newly engineered Amtn knock-out/lacZ knock-in mouse model [28] and to combine it with the well-defined Klk4 knock-out/lacZ knock-in mouse model [23,24] to determine if AMTN likely participates along with KLK4 to facilitate the removal of enamel proteins or if AMTN acts independently or complementarily to KLK4 to enhance enamel mineralization deposition.
Results
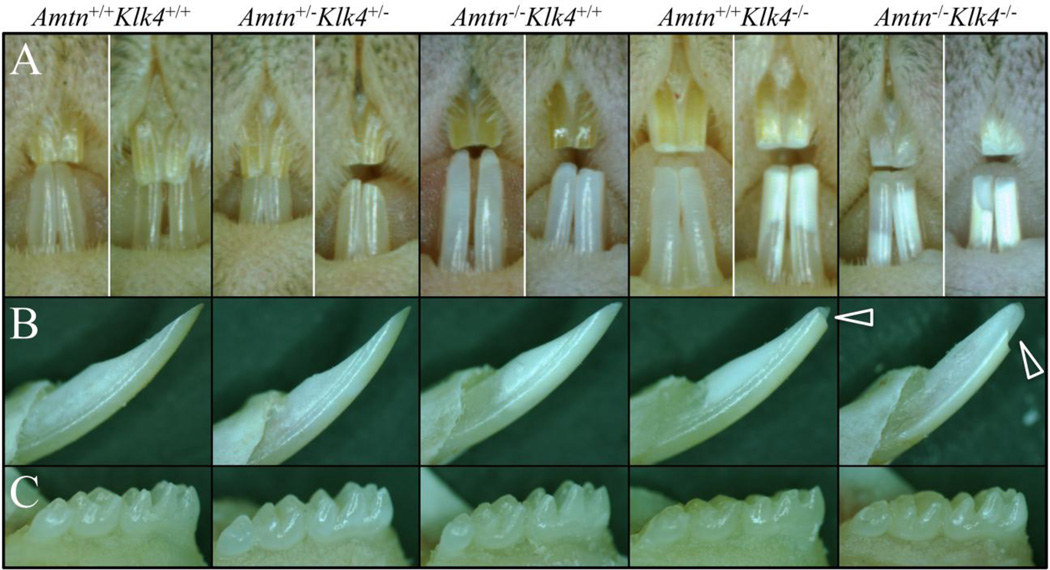
7-week old wild-type (Amtn+/+Klk4+/+), Amtn−/−, Klk4−/−, double heterozygous (Amtn+/−Klk4+/−) and double null (Amtn−/−Klk4−/−) mouse incisors and molars were examined under a dissecting microscope (Fig. 1). The Amtn null mouse (Amtn−/−Klk4+/+) mandibular incisors were chalky-white and sometimes chipped at their tips, especially if the mice were fed standard hard chow. Amtn−/− mouse mandibular incisors rarely chipped when the animals were maintained on a soft diet. The Klk4 null (Amtn+/+Klk4−/− or Klk4−/−) mouse mandibular incisors were usually chalky-white and chipped at their edges, even when maintained on soft chow. The Amtn−/−Klk4−/− mouse incisors were usually deeply chalky and showed significant attrition. The deep chalky appearance was thought to be caused by partial separation of the enamel from the underlying dentin, probably due to mechanical (occlusal) stresses. The wild-type and Amtn+/−Klk4+/− mice were similar in appearance and could not be distinguished from each other based upon their enamel phenotype.
Fig. 1.
Dissecting Microscope Photographs of 7-Week Mandibular Incisors and Molars. A-B: The wild-type (Amtn+/+Klk4+/+) and double heterozygous (Amtn+/−Klk4+/−) mice are similar in appearance and cannot be distinguished. The Amtn null mice (Amtn−/−Klk4+/+ or Amtn−/−) are chalky in color and occasionally show a chip at the edge of the mandibular incisor, especially if the mice are provided unsoftened chow. The Klk4 null (Amtn+/+Klk4−/− or Klk4−/−) incisors are chalky or deeply chalky, and always show chipping of the incisal edges (arrowheads). The AmtnKlk4 double null (Amtn−/−Klk4−/−) incisors are chalky-white and always show chipping of the incisor enamel. C: The molar crowns of the wild-type, double heterozygous and Amtn null mice are similar to each other and do not undergo rapid attrition. Extensive attrition is always observed on the molars of the Klk4−/− and Amtn−/−Klk4−/− mice.
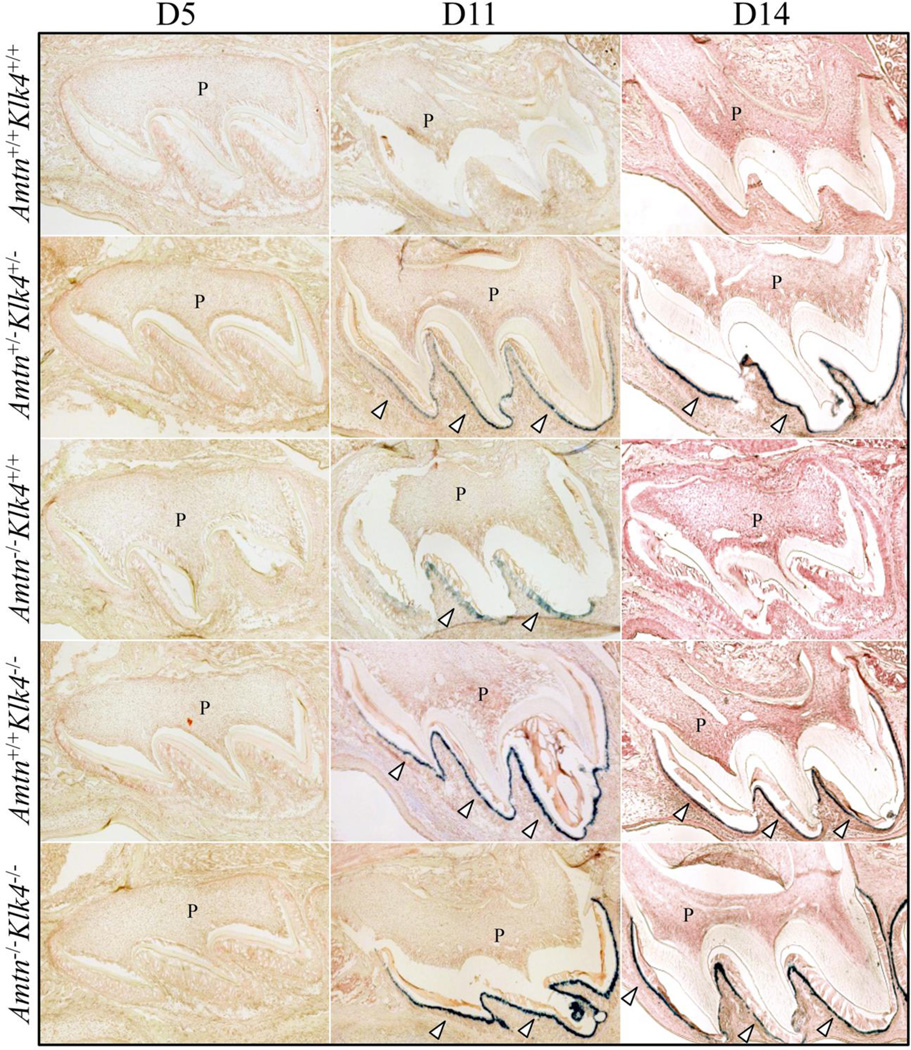
The Amtn and Klk4 genes were both knocked-out by replacing the natural translation initiation codon and downstream coding sequence with a reporter gene encoding β-galactosidase (lacZ) fused to a nuclear localization signal (NLS). We performed lacZ histochemistry on developing maxillary first molars from D5, D11, and D14 to observe reporter expression and provide additional data on the temporal and spatial patterns of these genes during amelogenesis (Fig. 2). No X-gal staining was observed in wild-type sections (negative control). Nor was any X-gal staining observed in the D5 molars from any of the genotypes, demonstrating that reporter expression was below detection in secretory stage ameloblasts. LacZ reporter expression was detected in D11 maturation stage ameloblasts in the Amtn+/−Klk4+/−, the Amtn−/−, the Klk4−/−, and the Amtn−/−Klk4−/− mice. Reporter expression in the Amtn−/− mice was weak. X-gal signal was specifically detected in D14 ameloblasts immediately prior to molar eruption in the Klk4−/−, and the Amtn+/−Klk4+/− and Amtn−/−Klk4−/− mice, but was not detected in the Amtn−/− mouse molars. These results are consistent with previous studies showing specific expression of Klk4 in maturation stage ameloblasts [40,17,39,41]. The Amtn reporter was much weaker than anticipated and did not appear to be a sensitive reporter for normal Amtn expression. RT-PCR analyses of wild-type and knockin Amtn transcripts demonstrated that expression of the reporter gene in Amtn−/− mice sometimes results in specific deletion of the modified Exon 2 during RNA splicing, which likely weakens of lacZ reporter in the Amtn knockin mice (Fig. 3). Therefore, the Amtn-lacZ reporter is not ideal for characterizing the temporal and spatial pattern of Amtn expression. The transcript formed by the skipping of Exon 2 lacks the code for the translation initiation codon, the signal peptide, and the N-terminal Leu-Pro that is part of a tripeptide motif necessary for efficient trafficking of the protein out of the endoplasmic reticulum [42], rendering it a nonfunctional transcript.
Fig. 2.
LacZ Histostaining of Cryosectioned D5, D11, and D14 Mandibular First Molars. The Amtn−/− and Klk4−/− null mice both have lacZ knockin genes introduced into the first coding exon (Exon 2). Note there is no X-gal (blue) staining in any of the D5 molars, as ameloblasts at D5 are in the secretory stage of amelogenesis prior to the onset of Amtn or Klk4 expression. The absence of X-gal staining in the wild-type mice is due to the absence of endogenous β-galactosidase expression in these tissues. Blue staining is observed in maturation stage ameloblasts in the D11 Amtn+/−Klk4+/− double heterozygous, Amtn−/− and Klk4−/− single nulls and Amtn−/−Klk4−/− double null mice (arrowheads). The blue staining in the Amtn−/− mice is specific for ameloblasts, but weaker than anticipated, and below detection in the D14 molars. The blue staining in maturation stage ameloblasts is strong in D14 mice expressing the Klk4 knockin gene.
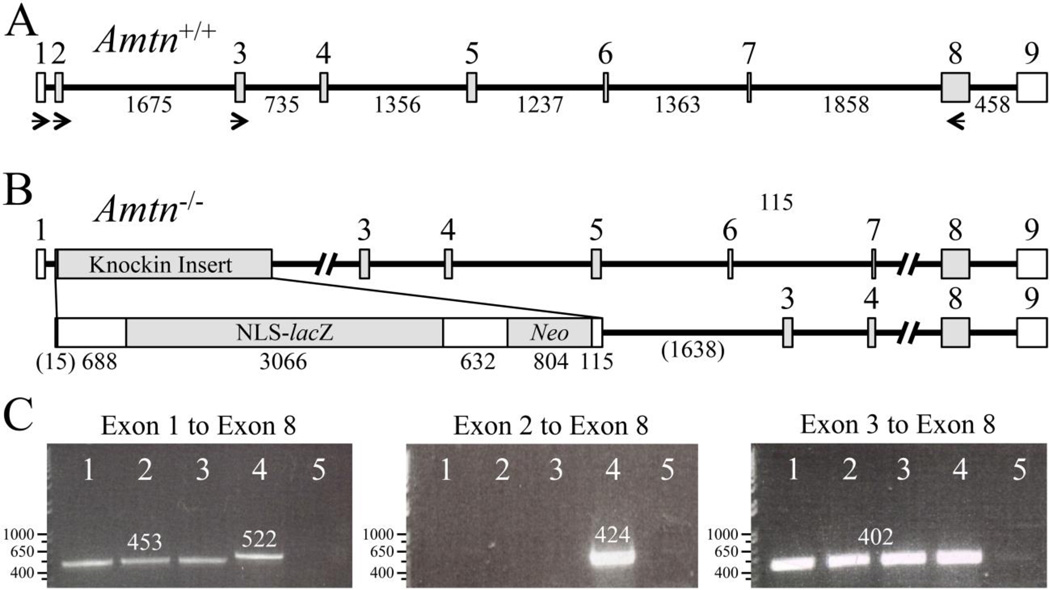
Fig. 3.
RT-PCR Assay for Amtn Expression in Amtn Null Mice. A: The wild-type Amtn gene (Amtn+/+) has 9 exons. Translation initiates early in Exon 2 and terminates early in Exon 9. The numbered boxes are exons. The horizontal lines are intron, with the number of bp below. B: The Amtn knockin gene deletes 91 bp in Amtn, starting with the Amtn translation initiation codon in Exon 2 and extending 37 bp into Intron 2. The 91 bp are replaced with 5305 bp knockin insert containing a 5’ untranslated region, NLS-lacZ code, downstream untranslated region containing a polyadenylation signal (AATAAA), neomycin phosphotransferase code, and a short 3’ untranslated region with a polyadenylation signal. C: RT-PCR using RNA template isolated from D11 (maturation stage) mandibular first molar enamel organ epithelia. Agarose gels stained with ethidium bromide; lanes 1–3 are from different Amtn−/− pups; lane 4 is wild-type (WT); and lane 5 is a negative control. The number of bp in each band is shown above the band. Left: Amtn−/− mice express an Amtn transcript that skips Exon 2 and lacks the code for translation initiation and the signal peptide. This finding explains the weakness of the lacZ reporter, despite complete loss of AMTN protein. Middle: Only WT mice express an Amtn transcript containing Exon 2. Right: Amtn+/+ and Amtn−/− mice splice the downstream Exon 3 to Exon 8 segment to remove all introns.
Enamel Thickness, Rod Decussation, and Mineral Density
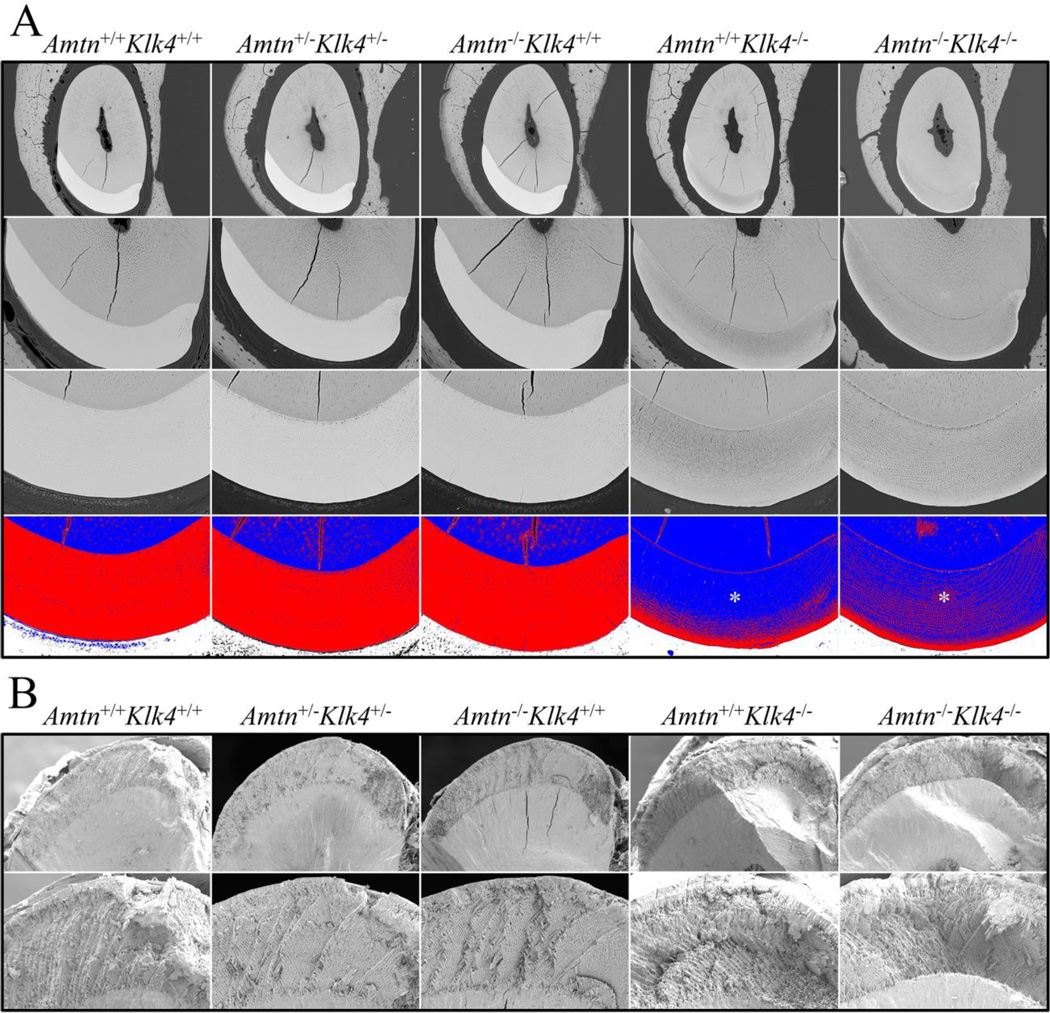
The final thickness of the incisor enamel layer, which is the distance perpendicular from the dentino-enamel junction (DEJ) to the enamel surface) and the decussation (X-shaped pattern) of adjacent rows of enamel rods, are important features of dental enamel and affect its ability to withstand occlusal forces. Enamel thickness and rod decussation have already been established by the end of the secretory stage. Mineral density however, increases substantially during the subsequent maturation stage [5]. Enamel thickness can be precisely measured by cross-sectioning and polishing 7-week mandibular incisors at the level of the alveolar crest, before this portion of the continuously growing incisor has erupted into the oral cavity (Fig. 4A). Backscatter SEM images of the polished cross-sections are used to measure enamel thickness and to evaluate mineral density. Enamel decussation is assessed at the same position as enamel thickness (the level of the alveolar crest) but using fractured surfaces (to show the alternating rows of rods), rather than polished ones (Fig. 4B).
Fig. 4.
7-Week Mandibular Incisor Cross-Sections at the Level of the Alveolar Crest. A: Backscattered scanning electron micrographs (bSEMs) of polished surfaces at 200X (top row) and 500X (second and third rows) magnifications. Images such as these were used to measure the thickness of the enamel layer (which was the same in all genotypes) and to assess the degree of enamel mineralization (lighter is more highly mineralized than darker gray). In row 3 pseudocolorization of bSEM images using ImageJ (Red is above threshold; blue is below threshold) emphasizes the similar degree of enamel mineralization in the Amtn+/+Klk4+/+Amtn+/−Klk4+/−, and Amtn−/−Klk4+/+ mice, and the decreasing level of enamel mineralization with depth (toward the DEJ) in the Amtn+/+Klk4−/− and Amtn−/−Klk4−/− mice (asterisks). B: SEM images of the enamel layer fractured at the level of the alveolar crest. Images such as these were used to assess the pattern of enamel rod decussation. The enamel rods in alternative rows showed the familiar X (decussation) pattern in all genotypes.
The thickness of the enamel layer was the same in all genotypes (Fig. 5). Normal decussation of enamel rods was also apparent in all genotypes (Fig. 4B). Thus the two major secretory stage features of dental enamel (final enamel thickness and rod decussation patterns) are unaltered by the absence of AMTN, KLK4, or both, which is consistent with previous observations that these proteins are not expressed by secretory stage ameloblasts. In contrast, the density of the enamel layer (a maturation stage feature) was equivalent in the wild-type, Amtn+/−Klk4+/−, and Amtn−/− mice and visibly reduced in both the Klk4−/− and Amtn−/−Klk4−/− mice (Fig. 4A). An interesting detail in these results was that the enamel was more highly mineralized at the enamel surface and became increasingly less-mineralized with depth in the Klk4−/− and Amtn−/−Klk4−/− mice.
Fig. 5.
Enamel Thickness. The distance from the DEJ to the enamel surface was measured in mandibular incisor cross-sections at the level of the alveolar crest. Each measurement was made at the thickest part of the enamel layer at the centro-mesial side of the incisor cross-section. ImageJ was calibrated using the magnification bar provided by the CAMECA (Cameka Instruments, Inc. Madison, WI, USA). The thickness values for each genotype were averaged and standard deviations were calculated using SPSS. The average measurements and standard deviations are listed on the left and plotted on the right. No significant differences in enamel thickness were found (t-test, p>0.05) between any of the genotypes (n = 5 per genotype).
Enamel Hardness
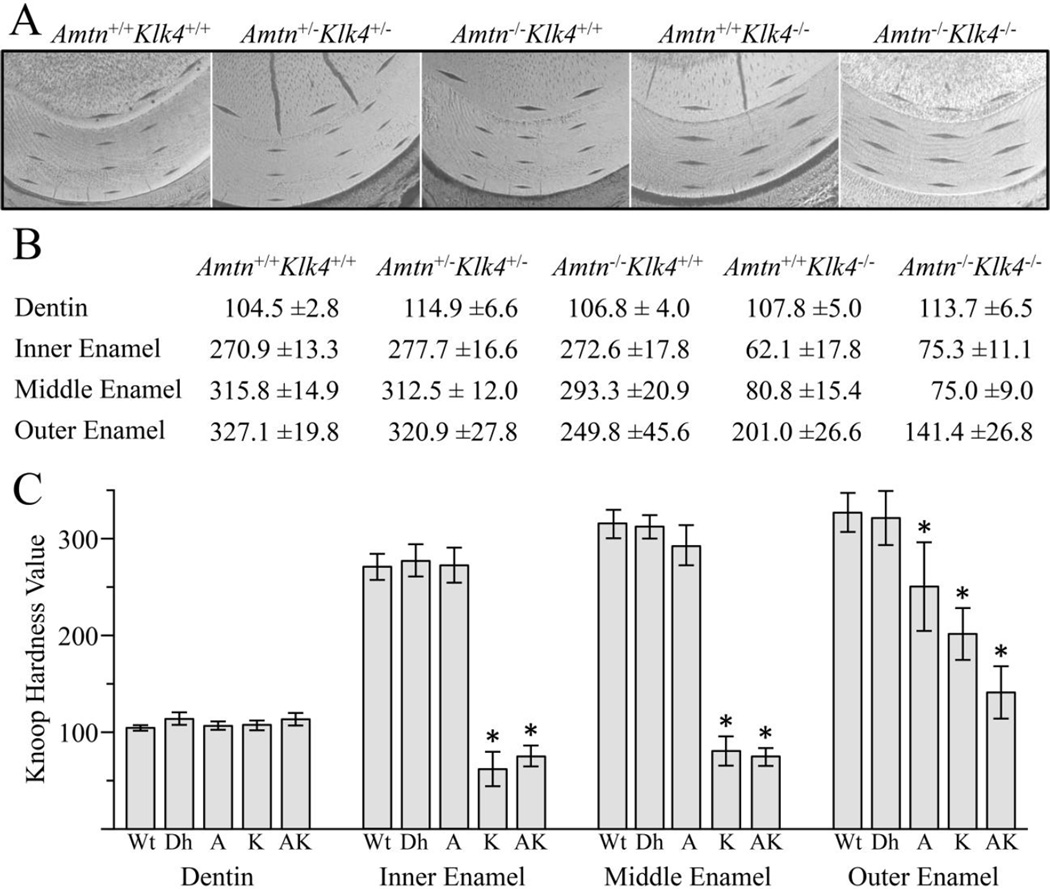
In addition to enamel thickness, architecture, and mineral density, the hardness of enamel is another parameter that relates to the ability of dental enamel to avoid failure during functional or parafunctional activities. Knoop microhardness tests quantify the resistance of a material to plastic deformation. A pyramidal diamond point was pressed with a load of 25 grams for 10 seconds onto the polished tooth surface, producing a diamond-shaped indentation that was measured to calculate Knoop hardness. Uneven or asymmetric indentations or indentations near cracks were excluded from the calculations. Indentations were placed in the outer dentin, and the inner, middle, and outer enamel, with 3 series of indentations per incisor (Fig. 6A). The points for each target position were averaged to obtain a single value per genotype per location (Fig 6B-6C).
Fig. 6.
Knoop Hardness Values. A: Indentations were made in dentin, and the inner, middle, and outer enamel of 7-week mandibular incisor sections at the level of the alveolar ridge (sample images shown). The indentations in dentin and the inner enamel were targeted between 15–20 µm from the DEJ. The outer enamel was targeted 15–20 µm from the enamel surface. The middle enamel indentation was targeted halfway between the outer and inner enamel indents. Three series of indentations were generated per incisor. The size of the indentations inversely correlates with hardness, so the larger the indentation, the softer the enamel. B: All values are expressed as mean Knoop hardness values ± standard deviation (N=6 animals per genotype). C: Plots of the average hardness measurements. Key: Wt, wild-type; Dh, double heterozygous (Amtn+/−Klk4+/−); A, Amtn−/−Klk4+/+; K, Amtn+/+Klk4−/−; AK, Amtn−/−Klk4−/−. No significant differences in the hardness of dentin were observed between any of the genotypes. No significant differences were observed between the wild-type and double heterozygous mice. The outer enamel of the Amtn−/− mouse was significantly softer than that of the wild-type. The enamel of the Klk4−/− and Amtn−/−Klk4−/− mice was softer than the wild-type enamel at all levels. Significant differences with wild-type enamel are indicated by an asterisk (t-test, p<0.001). The outer enamel of the Amtn−/−Klk4−/− mice was significantly softer than both the Amtn−/− and the Klk4−/− single nulls (t-test, p<0.001).
There were no significant differences in the hardness of dentin between any of the genotypes, indicating that Amtn and Klk4 are not critical for dentinogenesis. There were also no significant differences between the wild-type and the Amtn+/−Klk4+/− mice in any of the enamel locations (inner, middle, and outer enamel), indicating that half of the normal amount of these proteins is sufficient for normal amelogenesis. A most interesting finding was that the Amtn−/− mice produced inner and middle enamel of normal hardness, while the outer enamel was significantly reduced in hardness relative to the wild-type outer enamel (t-test, p<0.001). Furthermore, the outer Amtn−/− enamel was not as hard as the middle or inner enamel, which potentially relates to the observed chalky appearance of the incisor. In contrast, the Klk4−/− enamel was significantly reduced in hardness at all levels relative to the wild-type, but the outer enamel was significantly harder than the inner and middle enamel layers. Thus the enamel hardness patterns of the Amtn and Klk4−/− mice were distinctly different. In the absence of AMTN, the hardness of the outer enamel was reduced the most, whereas in the absence of KLK4 the inner and middle enamel was significantly softer than the outer enamel. Furthermore, the hardness of the Amtn−/−Klk4−/− outer enamel was significantly reduced compared to the outer enamel of the Amtn and Klk4 single nulls (t-test, p<0.001), suggesting that AMTN and KLK4 function independently during enamel maturation to harden the enamel layer.
Micro-CT Analyses
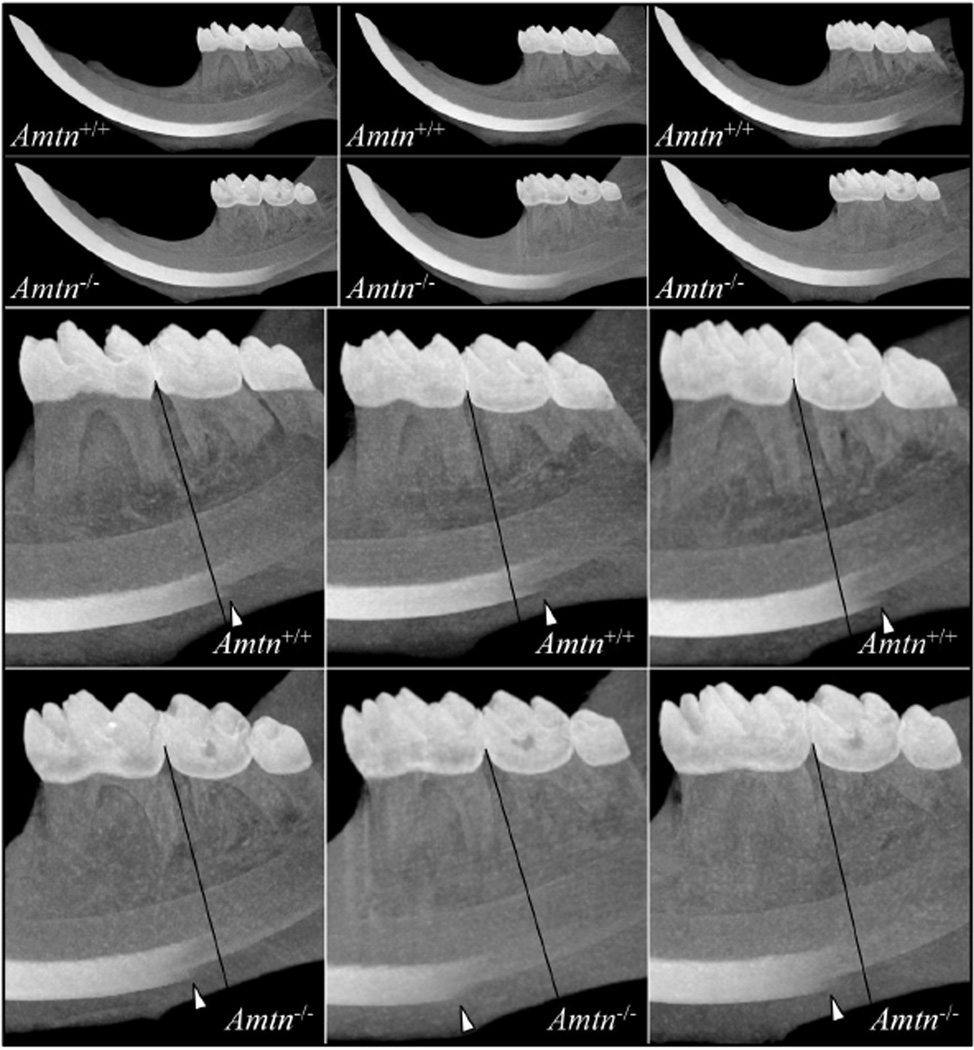
Maximum intensity projections (MIPs) allow the visualization of high density structures surrounded by lower density media (spaces that are non-homogeneously filled) and were used to compare enamel density along the length of the incisors in the hemi-mandibles between the wild-type and Amtn−/− mice (Fig. 7) and between the Klk4−/− and Amtn−/−Klk4−/− mice (Fig. 8). This method renders 3D data into a 2D image by showing the highest density that falls in the path orthogonal to the plane of projection. Histologically, the onset of the maturation stage in the continuously growing mandibular incisor is inferior to the bifurcation of the second mandibular molar [43]. The MIPs of the wild-type mice showed higher density enamel became visible just distal to a line separating the first and second mandibular molars (Fig. 7). In the MIPs of the Amtn-null mice, higher density enamel became visible mesial to this line, suggesting that enamel maturation was delayed in Amtn−/− mice. A similar delay in the onset of enamel maturation was usually observed in the Klk4−/− incisors, while in the Amtn−/−Klk4−/− mice, higher density enamel was consistently first apparent mesial to the bifurcation of the root of the mandibular first molar (Fig. 8). These findings demonstrate that Amtn plays a critical role in the maturation of dental enamel.
Fig. 7.
Micro-CT Maximum Intensity Projections for Wild-Type and Amtn Null Hemi-mandibles. Maximum intensity projections of three wild-type (Amtn+/+) and three Amtn−/− hemi-mandibles are shown at the top. Higher magnification views of these projections discovered that early maturation of the enamel layer is significantly delayed in the Amtn null mice relative to the wild-type. A line marking the midpoint between the first and second mandibular molars was drawn as a positional reference. Arrowheads mark the earliest point where the labial enamel is sufficiently dense to be projected. The arrowheads are all positioned distal (to the right) of the reference line in the wild-type and mesial (to the left) of the reference line in the Amtn−/− mice.
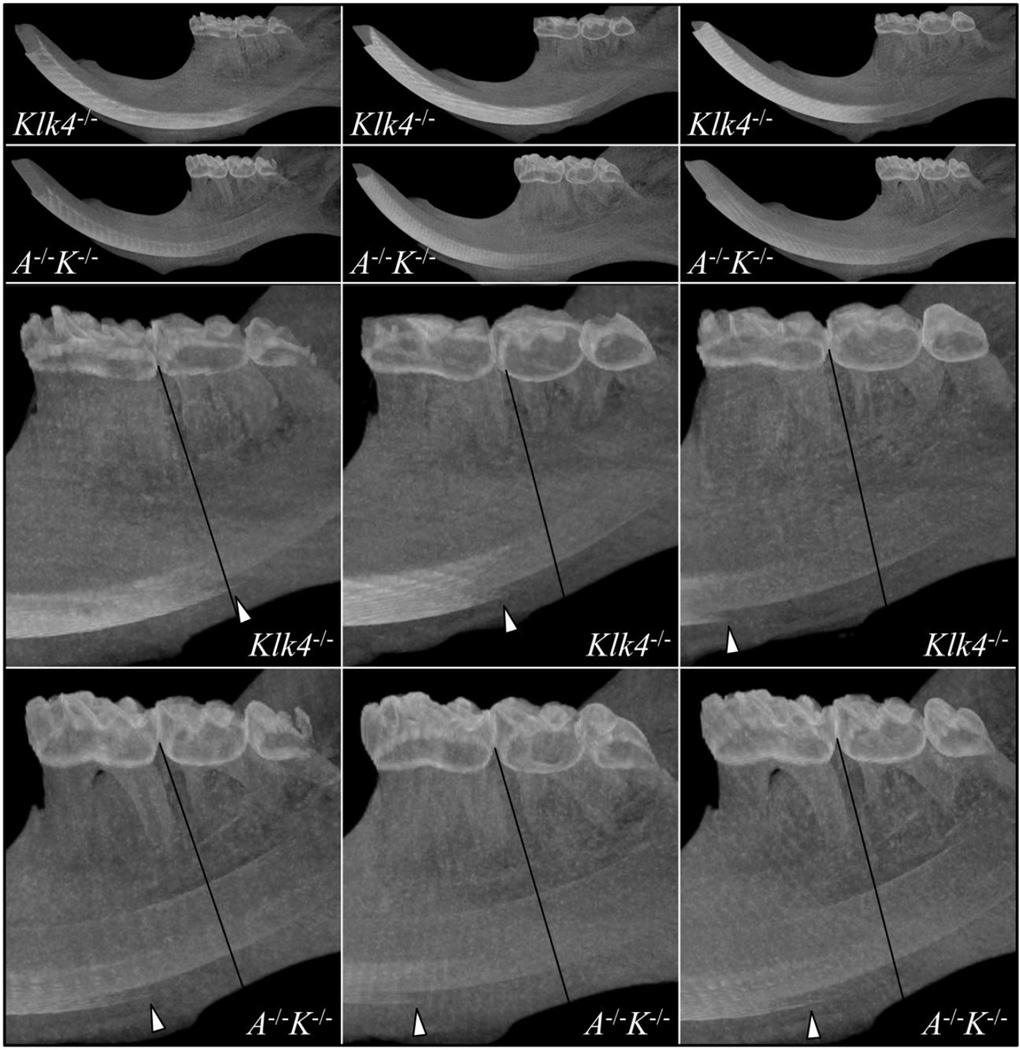
Fig. 8.
Micro-CT Maximum Intensity Projections for Klk4−/− and Amtn/Klk4 Double Null (A−/−K−/−) Hemi-mandibles. The Klk4−/− mice vary in the location of where higher density enamel starts to become visible, which indicates the progression of enamel maturation (growth of enamel crystals in width and thickness). In the Amtn−/−Klk4−/− mice the higher density enamel is consistently first observed mesial to reference line between the first and second molars and is initially very thin, suggesting that maturation of the enamel away from the surface is especially delayed.
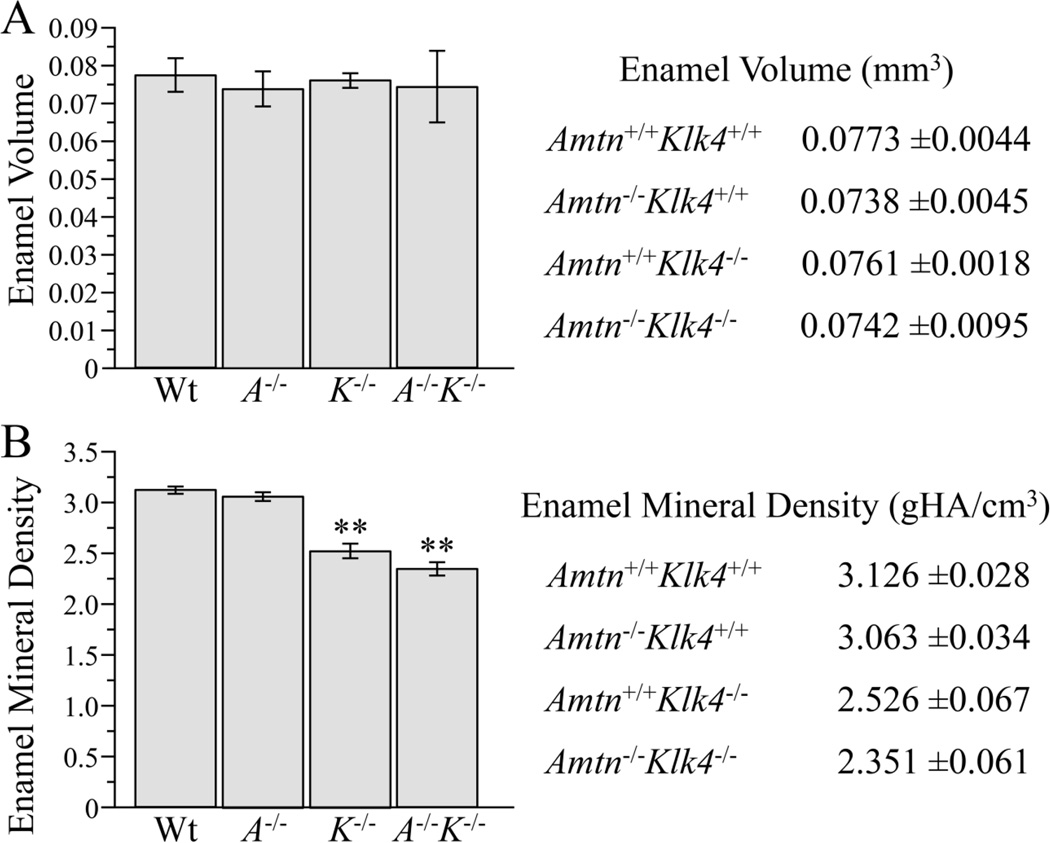
Micro-CT analyses were used to quantify enamel volume and mineral density in wild-type, Amtn−/−, Klk4−/−, and Amtn−/−Klk4−/− mandibular incisors (n=3 hemi-mandibles per group). The total enamel volume of the mandibular incisors from all genotypes was similar (t-test, p>0.05) (Fig. 9A). Given that enamel thickness was similar in all genotypes, the finding of equivalent enamel volumes in all genotypes provided additional evidence that Amtn and Klk4 are not performing functions necessary for the secretory stage of amelogenesis. By using multiple phantoms of known density to obtain a linear attenuation curve to assign mineral density values to the scanned samples based on gray-scale values, we were able to quantify the enamel mineral density in the mandibular incisors in each genotype (Fig. 9). The mineral density for the wild-type samples were similar to values reported previously for the same region [44]. The Amtn−/− enamel density was slightly lower than that of the wild-type, but this difference was not statistically significant. The enamel density measured by this method covers the entire thickness of enamel, so that a decrease in enamel mineral density only near the surface (as might be expected based upon the results of the hardness testing) was not resolvable. Enamel mineral density was similar between Klk4−/− and Amtn−/−Klk4−/− mice, and significantly lower than wild-type and Amtn-null enamel.
Fig. 9.
Micro-CT Determinations of Enamel Volume and Density. Enamel volume was the same in all genotypes. Enamel mineral density was significantly reduced in the Klk4−/− and Klk4−/−Amtn−/− mice relative to the wild-type and Amtn−/− mice. The values shown here were determined following a 1 pixel peel to diminish artifacts due to possible surface effects.
Discussion
The null mice in this study appeared to be healthy and fertile, and no abnormalities outside of the enamel phenotype were observed. No significant differences were detected in enamel thickness, enamel volume, or in rod decussation. These results are consistent with EST data showing only trace expression of Amtn and Klk4 in non-dental tissues, the decay of these genes in vertebrates that no longer make enamel [21,15,22], and the onsets of Amtn and Klk4 expression in transition ameloblasts [16–20], following the secretory stage when enamel thickness, volume, and rod architecture are established. Taken together, these results strongly support the conclusion that AMTN and KLK4 are not essential for biological processes outside of the dentition or during the secretory stage of amelogenesis. Both KLK4 and AMTN proved to be essential for the maturation of dental enamel, a process that requires the removal of extracellular matrix proteins and the deposition of ions on the sides of enamel crystallites.
KLK4 is a secreted protease that degrades extracellular enamel proteins. These proteins need to be removed to allow the thin enamel crystallites initially deposited and elongated during the secretory stage to grow in width and thickness and to interlock with adjacent crystals [23]. In the absence of KLK4, the enamel is less mineralized and shows reduced hardness throughout, while the inner and middle enamel is more severely altered than the outer enamel layer. This pattern suggests that the endocytosis mechanism, in the absence of KLK4, is still able to reabsorb much of the protein near the enamel surface, but the protein away from the surface is unable to work its way back to the cell for reabsorption. Interestingly, a pathological accumulation of enamel proteins also occurs in Wdr72−/− mice, whose enamel shows more severe reductions in mineral density and hardness than those observed in Klk4−/− mice [45,46]. WDR72 is an intracellular protein and defects in WDR72 contribute to the etiology of autosomal recessive amelogenesis imperfecta [47,48]. Identification of proteins that co-immunoprecipitated with WDR72 suggested that WDR72 is involved in endocytosis or vesicle trafficking [45,46]. These findings, and the discovery that Klk4 is a recent (evolutionary) addition to the process of enamel maturation, suggest that endocytosis is the primary mechanism for the removal of enamel matrix proteins and that secretion of the KLK4 enzyme is a later refinement that facilitates that process by degrading extracellular proteins and thereby allowing proteins in the deeper layers to more rapidly diffuse to the enamel surface and be reabsorbed by ameloblasts.
If AMTN and KLK4 were both contributing to the same process (facilitating the removal of enamel proteins), we expected the severity of the enamel defects in the double knockout mouse to be equivalent to those in the more severely affected single null mouse (which is Klk4−/−). This was not observed. We found that the pathology caused by the absence of both AMTN and KLK4 was more severe than when either protein was lacking individually. This was true for both enamel hardness and the delay in the onset of enamel maturation. These results suggest that both of these proteins are necessary for proper enamel maturation, but that they likely work independently of each other (in separate processes). We also observed no enamel phenotype in the single heterozygous (Amtn+/−Klk4+/+ or Amtn+/+Klk4+/−) mice or in the double heterozygotes (Amtn+/−Klk4+/−). This demonstrates that, at least in the mouse, loss of function defects in Amtn and Klk4 are manifested by a recessive pattern of inheritance. KLK4 defects follow a recessive pattern of inheritance in humans as well. We suspect that enamel defects caused by loss of function mutations in AMTN will also follow a recessive pattern of inheritance in humans, making them rare, which helps explain why mutations in AMTN have not yet been shown to cause AI. Many genes involved in the etiology of AI remain to be discovered. Known candidate genes account for half or fewer than half of all AI cases [49,50]. Only two ameloblastin (AMBN) [51,52], and three KLK4 [25–27] AI-causing mutations have be reported to date, despite the known importance of these genes for normal enamel development in humans and their routine inspection when searching for the genetic etiology of AI in patients. The current lack of evidence that AMTN mutations cause AI should not suggest that it doesn’t, based upon the enamel phenotype in Amtn−/− mice.
During evolution, important structural and regulatory changes have occurred in the Amtn gene [13,9]. Exon 2b encoding a Golgi casein kinase phosphorylation site and the 3’ part of Exon 8 encoding an RGD integrin binding site were lost, and expression of Amtn, previously initiating early during the secretory stage, was delayed to the transition to maturation [13,14]. These changes are associated with the emergence of prismatic enamel in mammals, and suggest that AMTN function became modified in stem mammals. Expression of Amtn during the secretory stage seems to be incompatible with formation of the Tomes’ process [53], so the restriction of Amtn expression to the maturation stages may have been permissive for the evolution of enamel rod and interrod organization. The loss of the RGD integrin binding motif may have loosened the association of AMTN with the basal lamina and allowed the protein to penetrate into the superficial enamel to promote mineralization [54]. These hypotheses are interesting and consistent with current observations, but the findings of delayed enamel maturation and reduced surface hardness in Amtn−/− enamel will require continued research to understand fully.
Experimental Procedures
Animal Protocol
Wild-type (WT), Amtn−/−, Klk4−/−, Amtn+/−Klk4+/−, and Amtn−/−Klk4−/− mice were maintained on Diet Gel® (Portland, ME, USA), and on 12-hour light/dark cycles. Genotyping of mice was done by tail biopsy using the Qiagen DNeasy blood and tissue kit (cat. 69506, Valencia, CA, USA) following manufacturer directions. Mouse photographs were taken using a Nikon DXM1200 camera on a Nikon Eclipse E600 dissecting microscope (Mitsubishi Group, Tokyo, Japan).
Klk4 Targeting Construct and Genotyping
The targeting construct for the Klk4−/− mouse was previously described [23]. Mice were generated on a C57BL/6 background. Briefly, the construct was designed to insert a NLS-lacZ (β-galactosidase) reporter containing a mouse nuclear localization signal (NLS) replacing the entire mouse Klk4 coding sequence starting with the initiation codon in exon 2 and ending downstream of the last coding exon (exon 6). Mice were genotyped using two primer pairs. To detect the wild-type Klk4 gene, we used primers that annealed to Intron 3 (5’-AACCTAAGGGACAGGGCAGT) and Exon 5 (5’-TGAGGTGGTACACAGGGTCA; 550-bp amplicon). The Klk4 knockin (null) gene was detected with a primer pair that annealed within the NLS-lacZ sequence: (5’-TGCCTCCAACCAGATAGGTC and 5’-GACAGTATCGGCCTCAGGAA; 595-bp amplicon). PCR conditions were: 94 °C/5 min, followed by 35 cycles at 94 °C/30 s, 58 °C/30 s, 74 °C/60 s with a final elongation at 72 °C for 7 min.
Amtn Targeting Construct and Genotyping
Amtn−/− mice were generated on a C57BL/6 background [28]. The Amtn knockout has a lacZ (β-galactosidase) reporter and a downstream neomycin (Neo) selection marker inserted at the Amtn translation initiation site in Exon 2. This insert is 5305 bp in length. It replaces the 54 bp of Amtn Exon 2 coding sequence and 37 bp (out of 1675 bp) at the 5’ end of Intron 2. Two primer pairs were used for genotyping. To detect the Amtn wild-type gene, the 5’ primer annealed to Amtn Exon 1 (5’-GTGGAGAAGTCATCTGGATTTCG), which is found on both the wild-type and Amtn knockin genes, and was paired with a primer that annealed to the 3’ end of Exon 2 (5’-ACTGGTAATGACTGGGCTGAT; 226-bp amplicon), which is only found on the Amtn wild-type gene. To detect the Amtn knockout/lacZ knockin gene, the 5’ primer (5’-ATTGCTGAAGAGCTTGGCGGCG) annealed to the Neo cassette downstream of the lacZ knockin, while the 3’ primer (5’-CACTCTGTCCCTCAGGGCTGCTA; 466-bp amplicon) annealed within Amtn intron 2. PCR conditions were as follows: 94 °C/5 min, followed by 35 cycles at 94 °C/30 s, 58 °C/ 30 s, 72 °C/60 s with a final elongation at 72 °C for 7 min.
Breeding Strategy
To generate Amtn−/−Klk4−/− mice, Amtn−/− mice were bred with Klk4−/− mice to generate Amtn+/−Klk4+/− mice. This cage was also used to maintain a supply of heterozygous control mice. The Amtn+/−Klk4+/− littermates were then bred with each other to obtain Amtn−/−Klk4−/− mice, which were expected to appear with a frequency of 1/16. Pups were genotyped by tail biopsy for Amtn and Klk4 as described above. Once male and female Amtn−/−Klk4−/− mice were obtained, double null breeding pairs were used to establish the Amtn−/−Klk4−/− colony.
β-Galactosidase Expression Assay
Mouse heads were collected from postnatal days 5 (secretory stage), 11 (maturation stage), and 14 (late maturation stage), sectioned in half, and fixed in 4% paraformaldehyde (PFA) overnight at 4 °C. The following morning, heads were washed in phosphate-buffered saline (PBS, 137 mM NaCl, 10 mM phosphate, 2.7 mM KCl, pH 7.4) (3 × 30 minutes). Mouse heads were then transferred to 4.13% EDTA (ethylenediaminetetraacetic acid), pH 7.4 at 4 °C with constant agitation for decalcification (D5 for 8 days; D11 for 21 days; D14 for 30 days) with a change of fresh solution every 2–3 days. Decalcified tissues were washed in PBS (3 × 30 minutes) and embedded in Tissue-Tek® O.C.T. (optimum cutting temperature) Compound (Sakura®Fintek Radnor, PA, USA). Blocks were cryosectioned at 10-µm thickness, post-fixed for 5 minutes in 0.5% glutaraldehyde, washed in PBS (3 × 5 minutes), and incubated for 5 h in freshly prepared X-gal staining buffer, pH 8.0, containing 1 mg/mL X-gal, 100 mM HEPES, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 1 mM MgCl2, 2% Triton X-100, and 1 mM dithiothreitol as described previously [23]. Tissue sections were rinsed in PBS and counterstained with hematoxylin. Images were taken using a Nikon Eclipse TE 300 microscope (Mitsubishi Group, Tokyo, Japan), Nikon DXM1200 camera (Mitsubishi Group, Tokyo, Japan), and Act1 imaging software (Mager Scientific, Dexter, MI).
RT-PCR and DNA Sequencing Analyses
RNA was extracted from the first molars of 11-day old pups using Qiagen RNeasy mini kit (cat. 74106, Qiagen, Valencia CA, USA) and analyzed with Superscript III One-Step RT-PCR system with Platinum Taq (Invitrogen, Carlsbad, CA, USA). Reactions of 50 µL containing about 200 ng of RNA were set up following manufacturer instructions. Amtn gene-specific primers were designed using Primer 3 on the Web (http://frodo.wi.mit.edu/primer3/). The forward primer annealed to Amtn Exon 1 (5’-GTGGAGAAGTCATCTGGATTTCG-3’), exon 2 (5’-GGATCAGCCCAGTCATTACC-3’), or Exon 3 (5’-AGCAGCTTAACCCTGCTTCG-3’), and was paired with a common reverse primer that annealed to Amtn Exon 8 (5’-TCTGCACATCTGGTTTAGTGC-3’). The predicted product sizes of these amplifications using the wild-type Amtn cDNA as template were: Exon 1 to Exon 8, 522-bp; Exon 2 to Exon 8, 424-bp; and Exon 3 to Exon 8, 402-bp. Control reactions were conducted using Gapdh gene-specific primers (forward 5’-ACTCCACTCACGGCAAATTC-3’ and reverse 5’-CACATTGGGGGTAGGAACAC-3’) as a loading control. RT-PCR conditions were as follows: 55 °C/30 min, 94 °C/2 min, followed by 35 cycles at 94 °C/30 s, 58 °C/30 s, 68 °C/30 s with a final elongation at 68 °C for 5 min. PCR reactions were resolved on 1% agarose gels and stained with ethidium bromide. DNA bands were removed from the gel with a scalpel and DNA was extracted using the QIAquick gel extraction kit (cat. 28706, Qiagen, Valencia, CA, USA). Samples were then sent to the University of Michigan DNA sequencing core for analysis.
Scanning Electron Microscopy – Fractured Incisors
7-week old Amtn+/+Klk4+/+, Amtn+/+Klk4−/−, Amtn−/−Klk4+/+, Amtn+/−Klk4+/−, and Amtn−/−Klk4−/− mice were anesthetized with isofluorane. The mandibles were removed and split at the mental symphysis, and the soft tissue was removed with a dental scaler. Each hemi-mandible was dehydrated with a series of graded ethanol (30 min each of 30%, 50%, 70%, 80%, 90%, 100% × 2), air dried, and fractured at the level of the alveolar crest. To create the fracture, incisors were notched in dentin on the concave portion of the tooth. The two ends of the incisor were then bent outwards, causing the fracture to propagate along the dentin and enamel. Fractured incisors were mounted on metallic stubs using conductive carbon tape and sputter coated with an Au-Pd film. Samples were imaged at the University of Michigan Microscopy and Imaging Analysis Laboratory (Ann Arbor, MI, USA) using an Amray EF 1910 Scanning Electron Microscope operating at an accelerating voltage of 5 kV.
Backscatter Scanning Electron Microscopy (bSEM)
The contralateral hemi-mandible was cleaned of soft tissue and embedded in Epon resin (EMbed812 cat#14120, Electron Microscopy Sciences, Hatfield, PA) following graded acetone dehydration (30%, 50%, 70%, 80%, 90%, 95%, 100% × 2 for 30 min each). After polymerization at 65 °C, the incisors were cut transversely (cross-section) with a model 650 low speed diamond wheel saw (South Bay Technology Inc., San Clemente, CA, USA) at the level of the crest of the alveolar bone close to where the incisor erupts into the mouth, about 8 mm from the apex of the incisor. The sectioned hemi-mandibles were then re-embedded in Castolite AC (Eager polymers, Chicago, IL, USA) using 25-mm SteriForm molds (Struers Inc., Westlake, OH, USA) with the cutting plane face down and allowed to harden overnight. The transversely sectioned faces of the mandibular incisors were polished sequentially with a syntron polisher on 400, 800, and 1200 grit waterproof silicon carbide papers followed by 1 µm diamond polishing paste (South Bay Technology Inc., San Clemente, CA, USA). Surfaces were carbon coated for backscatter SEM. The polished surfaces were examined at various magnifications in a CAMECA SX-100 electron microprobe analyzer using the backscatter mode at 15 kV and a calibration wedge to ensure that the range of intensities in each image recorded spanned a similar range of gray level intensities. The calibration wedge was based on a 0–255 pixel shade range where pure white is 255 and pure black is 0. The wild-type enamel was set at 190, dentin and bone at 160, and Epon resin at 40. Pseudo-colored image mapping was done using ImageJ on Tiff images that were normalized to have the same average gray level intensities for mineralized dentin, given that dentin and bone show no significant changes. For pseudo-colored images, gray levels 1–30 were assigned as black, 31–75 as white, 76–173 as blue, and 174–255 as red. These settings were saved in a lookup table (LUT) as described previously [55] and applied to the selected bSEM images.
Micro-Computed Tomography
7-week old mice of 4 different genotypes (wild-type, Amtn−/−, Klk4−/−, and Amtn−/−Klk4−/−; n=3 for each) were used for this analysis. Mice were anesthetized with isofluorane and euthanized by intracardiac perfusion using 5% glutaraldehyde in 0.8M sodium cacodylate with 0.05% calcium chloride, pH 7.4. Hemi-mandibles were removed and dissected of soft tissue, washed in 1X-PBS, placed in a vial and immersed in 70% ethanol. Samples were scanned in a SkyScan 1172 (Bruker SkyScan, Aartselaar, Belgium) system. Each specimen was positioned inside of the scanning tube with the ramus inferiorly and incisal tip superiorly positioned and supported with agarose. The tube was sealed with Parafilm (American National Can Company, Greenwich, CT, USA). Samples were scanned at 60 kV, 167 µA beam intensity, 0.7° rotation step, 4 frame average, 2000 × 1336 CCD, 700-millisecond exposure and 5-µm voxel size. Scan time was 35 min per entire hemi-mandible. The images were reconstructed with NRecon (Bruker SkyScan, Aartselaar, Belgium) with a Feldkamp cone-beam algorithm [56]. HA phantoms 0.25 and 0.75 g/cm3 (SkyScan), and 2.927 g/cm3 (Himed, Bethpage, NY, USA) were used for calibration prior to mineral density analyses.
The analysis of enamel volume was restricted to a region of the incisor where the enamel layer had reached its final thickness. The scanned area was chosen as the first section where the incisor is fully surrounded by alveolar bone, about 7.5 mm from the apex of the incisor. The volume of interest (VOI) started 10 slices distally from where the enamel is fully enclosed in bone. The VOI continued distally for 1.0 mm (about 200 slices). The enamel tissue was then isolated manually based on density. The manual tracing (appearing in red, but turning light blue when laid over high density areas of enamel) followed the DEJ and was then refined to exclude any areas that were not enamel, such as the air space next to the outer enamel surface. Prior to analysis, images were reoriented by aligning the image along the length of the incisor. The incisor was aligned along its length as straight as possible, given that the incisor tends to curve in two different dimensions. The cross-section of the incisor was also used to align it vertically along the midline of the incisor.
Microhardness Testing
Incisors were prepared as described for bSEM. After imaging, samples were polished with 1-micron MicroDi Permanent Diamond Suspension (South Bay Technology Inc., San Clemente, CA) to remove the carbon coating used for bSEM and air dried. Six animals of each genotype (wild-type, Amtn−/−, Klk4−/−, Amtn+/−Klk4+/−, and Amtn−/−Klk4−/−) were tested on a microhardness tester (LM247AT, s/n XM0326, Leco Corp.). Microhardness testing was performed with a load of 25 grams for 10 s with a Knoop tip to obtain a Knoop Hardness Number (KHN). Measurements were made at 500X magnification to increase precision. Indentations were placed in the outer, middle, and inner enamel as well as the dentin as a control reading for a total of four indentations per row. This series was performed 3X in each animal, for a total of 12 points per animal. Outer enamel is defined as 15 µm from the outer surface of the enamel layer. Inner enamel is defined as within 15–20 µm from DEJ on the enamel side. Middle enamel was defined as halfway between the outer and inner enamel indentation, with a range of 50–60 µm from the outer enamel surface. The dentin indentation was placed 20 µm away from the DEJ on the dentin side. Points that landed in cracks or damaged portion of the enamel or dentin were not read. Statistical significance was determined by pairwise t-tests using SPSS (software package for social sciences, Armonk, NY, USA) analysis software.
Study approval of animal use
The described animal experiments were done according to the basic protocols reviewed and approved by the Institutional Animal Care and Use Committees (IACUC) at the University of Michigan and in an agreement with the U.S. Public Health Service Policy on Human Care and Use of Laboratory Animals.
Highlights.
Amelotin nulls (Amtn−/−) exhibit delayed enamel maturation and softer outer enamel.
Klk4−/− enamel retains protein and is increasingly soft with depth.
Both AMTN and KLK4 are necessary for proper dental enamel maturation.
Amtn−/−Klk4−/− outer enamel is softer than Klk4−/− and Amtn−/− outer enamel.
The additive phenotype suggests these proteins serve independent functions.
Acknowledgments
This study was supported by NIDCR/NIH research grants DE019775 and DE015846, NIDCR/NIH career development grant DE022800, as well as a shared-instrumentation grant for the Carlisle Center for Bone and Mineral Research at the University of Texas Health Science Center at San Antonio (RR025687). It was also supported by an Operating grant (MOP-119310) from the Canadian Institutes of Health Research (CIHR) and a Discovery grant (RGPIN 403292-11) from the Natural Sciences and Engineering Research Council (NSERC) of Canada.
Abbreviations used
- DEJ
dentino-enamel junction
- HA
hydroxyapatite
- gHA
grams of HA
- MIP
maximum intensity projection
- SCPP
secretory calcium-binding phosphoprotein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
J.C-C.H, JPS, S.M.N. and BG designed the project and prepared the figures and the first draft of the manuscript. S.M.N. bred and genotyped the mice, performed the RT-PCR analyses, and microhardness testing. Y.H. and S.M.N performed the SEM and bSEM experiments. A.S.R. performed the X-gal histochemistry. J.E.S. R.F., and Y-H.P.C performed the µCT experiments and analyses. J.Y. prepared specimens for dissection microscopy. All authors read, contributed to the discussion and approved the final manuscript.
Competing financial interests
The authors declare no competing financial interests.
Contributor Information
Stephanie M. Nunez, Email: nunezste@einstein.edu.
Yong-Hee P. Chun, Email: chuny@uthscsa.edu.
Bernhard Ganss, Email: b.ganss@utoronto.ca.
Yuanyuan Hu, Email: yyhu@umich.edu.
Amelia S Richardson, Email: ramelia@umich.edu.
James E. Schmitz, Email: schmitzj@uthscsa.edu.
Roberto Fajardo, Email: fajardor@uthscsa.edu.
Jie Yang, Email: denyj78@163.com.
Jan C-C. Hu, Email: janhu@umich.edu.
James P. Simmer, Email: jsimmer@umich.edu.
References
- 1.Hu JC, Chun YH, Al Hazzazzi T, Simmer JP. Enamel formation and amelogenesis imperfecta. Cells Tissues Organs. 2007;186:78–85. doi: 10.1159/000102683. [DOI] [PubMed] [Google Scholar]
- 2.Reith EJ. The stages of amelogenesis as observed in molar teeth of young rats. J Ultrastruct Res. 1970;30:111–151. doi: 10.1016/s0022-5320(70)90068-7. [DOI] [PubMed] [Google Scholar]
- 3.Weinmann JP, Wessinger GD, Reed G. Correlation of chemical and histological investigations on developing enamel. J Dent Res. 1942;21:171–182. [Google Scholar]
- 4.Beniash E, Metzler RA, Lam RS, Gilbert PU. Transient amorphous calcium phosphate in forming enamel. J Struct Biol. 2009;166:133–143. doi: 10.1016/j.jsb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 6.Daculsi G, Kerebel B. High-resolution electron microscope study of human enamel crystallites: Size, shape, and growth. J Ultrastruct Res. 1978;65:163–172. doi: 10.1016/s0022-5320(78)90053-9. [DOI] [PubMed] [Google Scholar]
- 7.Miake Y, Shimoda S, Fukae M, Aoba T. Epitaxial overgrowth of apatite crystals on the thin-ribbon precursor at early stages of porcine enamel mineralization. Calcif Tissue Int. 1993;53:249–256. doi: 10.1007/BF01320910. [DOI] [PubMed] [Google Scholar]
- 8.Satchell PG, Shuler CF, Diekwisch TG. True enamel covering in teeth of the australian lungfish neoceratodus forsteri. Cell Tissue Res. 2000;299:27–37. doi: 10.1007/s004419900132. [DOI] [PubMed] [Google Scholar]
- 9.Kawasaki K, Amemiya CT. Scpp genes in the coelacanth: Tissue mineralization genes shared by sarcopterygians. J Exp Zool B Mol Dev Evol. 2014;322:390–402. doi: 10.1002/jez.b.22546. [DOI] [PubMed] [Google Scholar]
- 10.Moffatt P, Smith CE, Sooknanan R, St-Arnaud R, Nanci A. Identification of secreted and membrane proteins in the rat incisor enamel organ using a signal-trap screening approach. Eur J Oral Sci. 2006;114(Suppl 1):139–146. doi: 10.1111/j.1600-0722.2006.00318.x. [DOI] [PubMed] [Google Scholar]
- 11.Moffatt P, Wazen RM, Dos Santos Neves J, Nanci A. Characterisation of secretory calcium-binding phosphoprotein-proline-glutamine-rich 1: A novel basal lamina component expressed at cell-tooth interfaces. Cell Tissue Res. 2014;358:843–855. doi: 10.1007/s00441-014-1989-3. [DOI] [PubMed] [Google Scholar]
- 12.Kawasaki K, Buchanan AV, Weiss KM. Biomineralization in humans: Making the hard choices in life. Annu Rev Genet. 2009;43:119–142. doi: 10.1146/annurev-genet-102108-134242. [DOI] [PubMed] [Google Scholar]
- 13.Gasse B, Chiari Y, Silvent J, Davit-Beal T, Sire JY. Amelotin: An enamel matrix protein that experienced distinct evolutionary histories in amphibians, sauropsids and mammals. BMC Evol Biol. 2015;15:47. doi: 10.1186/s12862-015-0329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasse B, Liu X, Corre E, Sire JY. Amelotin gene structure and expression during enamel formation in the opossum monodelphis domestica. PLoS One. 2015;10:e0133314. doi: 10.1371/journal.pone.0133314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawasaki K, Hu JC, Simmer JP. Evolution of klk4 and enamel maturation in eutherians. Biol Chem. 2014;395:1003–1013. doi: 10.1515/hsz-2014-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu JC, Ryu OH, Chen JJ, Uchida T, Wakida K, Murakami C, Jiang H, Qian Q, Zhang C, Ottmers V, Bartlett JD, Simmer JP. Localization of emsp1 expression during tooth formation and cloning of mouse cdna. J Dent Res. 2000;79:70–76. doi: 10.1177/00220345000790011301. [DOI] [PubMed] [Google Scholar]
- 17.Hu JC, Zhang C, Sun X, Yang Y, Cao X, Ryu O, Simmer JP. Characterization of the mouse and human prss17 genes, their relationship to other serine proteases, and the expression of prss17 in developing mouse incisors. Gene. 2000;251:1–8. doi: 10.1016/s0378-1119(00)00203-1. [DOI] [PubMed] [Google Scholar]
- 18.Iwasaki K, Bajenova E, Somogyi-Ganss E, Miller M, Nguyen V, Nourkeyhani H, Gao Y, Wendel M, Ganss B. Amelotin--a novel secreted, ameloblast-specific protein. J Dent Res. 2005;84:1127–1132. doi: 10.1177/154405910508401207. [DOI] [PubMed] [Google Scholar]
- 19.Moffatt P, Smith CE, St-Arnaud R, Simmons D, Wright JT, Nanci A. Cloning of rat amelotin and localization of the protein to the basal lamina of maturation stage ameloblasts and junctional epithelium. Biochem J. 2006;399:37–46. doi: 10.1042/BJ20060662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somogyi-Ganss E, Nakayama Y, Iwasaki K, Nakano Y, Stolf D, McKee MD, Ganss B. Comparative temporospatial expression profiling of murine amelotin protein during amelogenesis. Cells Tissues Organs. 2012;195:535–549. doi: 10.1159/000329255. [DOI] [PubMed] [Google Scholar]
- 21.Gasse B, Silvent J, Sire JY. Evolutionary analysis suggests that amtn is enamel-specific and a candidate for ai. J Dent Res. 2012;91:1085–1089. doi: 10.1177/0022034512460551. [DOI] [PubMed] [Google Scholar]
- 22.Meredith RW, Zhang G, Gilbert MT, Jarvis ED, Springer MS. Evidence for a single loss of mineralized teeth in the common avian ancestor. Science. 2014;346:1254390. doi: 10.1126/science.1254390. [DOI] [PubMed] [Google Scholar]
- 23.Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC. Hypomaturation enamel defects in klk4 knockout/lacz knockin mice. J Biol Chem. 2009;284:19110–19121. doi: 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith CE, Richardson AS, Hu Y, Bartlett JD, Hu JC, Simmer JP. Effect of kallikrein 4 loss on enamel mineralization: Comparison with mice lacking matrix metalloproteinase 20. J Biol Chem. 2011;286:18149–18160. doi: 10.1074/jbc.M110.194258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, Wright JT. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41:545–549. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seymen F, Park JC, Lee KE, Lee HK, Lee DS, Koruyucu M, Gencay K, Bayram M, Tuna EB, Lee ZH, Kim YJ, Kim JW. Novel mmp20 and klk4 mutations in amelogenesis imperfecta. J Dent Res. 2015;94:1063–1069. doi: 10.1177/0022034515590569. [DOI] [PubMed] [Google Scholar]
- 27.Wang SK, Hu Y, Simmer JP, Seymen F, Estrella NM, Pal S, Reid BM, Yildirim M, Bayram M, Bartlett JD, Hu JC. Novel klk4 and mmp20 mutations discovered by whole-exome sequencing. J Dent Res. 2013;92:266–271. doi: 10.1177/0022034513475626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama Y, Holcroft J, Ganss B. Enamel hypomineralization and structural defects in amelotin-deficient mice. J Dent Res. 2015;94:697–705. doi: 10.1177/0022034514566214. [DOI] [PubMed] [Google Scholar]
- 29.Nishio C, Wazen R, Kuroda S, Moffatt P, Nanci A. Expression pattern of odontogenic ameloblast-associated and amelotin during formation and regeneration of the junctional epithelium. Eur Cell Mater. 2010;20:393–402. doi: 10.22203/ecm.v020a32. [DOI] [PubMed] [Google Scholar]
- 30.Nishio C, Wazen R, Moffatt P, Nanci A. Expression of odontogenic ameloblast-associated and amelotin proteins in the junctional epithelium. Periodontol 2000. 2013;63:59–66. doi: 10.1111/prd.12031. [DOI] [PubMed] [Google Scholar]
- 31.Nishio C, Wazen R, Kuroda S, Moffatt P, Nanci A. P44-expression pattern of apin and amelotin during formation and regeneration of the junctional epithelium. Bull Group Int Rech Sci Stomatol Odontol. 2010;49:111–112. [PubMed] [Google Scholar]
- 32.Holcroft J, Ganss B. Identification of amelotin- and odam-interacting enamel matrix proteins using the yeast two-hybrid system. Eur J Oral Sci. 2011;119(Suppl 1):301–306. doi: 10.1111/j.1600-0722.2011.00870.x. [DOI] [PubMed] [Google Scholar]
- 33.Bartlett JD, Simmer JP. Kallikrein-related peptidase-4 (klk4): Role in enamel formation and revelations from ablated mice. Front Physiol. 2014;5:240. doi: 10.3389/fphys.2014.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Y, Papagerakis P, Yamakoshi Y, Hu JC, Bartlett JD, Simmer JP. Functions of klk4 and mmp-20 in dental enamel formation. Biol Chem. 2008;389:695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamakoshi Y, Yamakoshi F, Hu JC, Simmer JP. Characterization of kallikrein-related peptidase 4 glycosylations. Eur J Oral Sci. 2011;119(Suppl 1):234–240. doi: 10.1111/j.1600-0722.2011.00863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simmer JP, Richardson AS, Smith CE, Hu Y, Hu JC. Expression of kallikrein-related peptidase 4 in dental and non-dental tissues. Eur J Oral Sci. 2011;119(Suppl 1):226–233. doi: 10.1111/j.1600-0722.2011.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryu O, Hu JC, Yamakoshi Y, Villemain JL, Cao X, Zhang C, Bartlett JD, Simmer JP. Porcine kallikrein-4 activation, glycosylation, activity, and expression in prokaryotic and eukaryotic hosts. Eur J Oral Sci. 2002;110:358–365. doi: 10.1034/j.1600-0722.2002.21349.x. [DOI] [PubMed] [Google Scholar]
- 38.Yamakoshi Y, Richardson AS, Nunez SM, Yamakoshi F, Milkovich RN, Hu JC, Bartlett JD, Simmer JP. Enamel proteins and proteases in mmp20 and klk4 null and double-null mice. Eur J Oral Sci. 2011;119(Suppl 1):206–216. doi: 10.1111/j.1600-0722.2011.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simmer JP, Hu Y, Richardson AS, Bartlett JD, Hu JC. Why does enamel in klk4-null mice break above the dentino-enamel junction? Cells Tissues Organs. 2011;194:211–215. doi: 10.1159/000324260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu JC, Sun X, Zhang C, Liu S, Bartlett JD, Simmer JP. Enamelysin and kallikrein-4 mrna expression in developing mouse molars. Eur J Oral Sci. 2002;110:307–315. doi: 10.1034/j.1600-0722.2002.21301.x. [DOI] [PubMed] [Google Scholar]
- 41.Simmer JP, Sun X, Yamada Y, Zhang CH, Bartlett JD, Hu JC-C. Enamelysin and kallikrein-4 expression in the mouse incisor. In: Kobayashi I, Ozawa H, editors. Biomineralization: Formation, diversity, evolution and application proceedings of the 8th international symposium on biomineralization; sept 25–28, 2001; niigata, jpn. Hadano, Jpn: Tokai University Press; 2004. pp. 348–352. [Google Scholar]
- 42.Nam AS, Yin Y, von Marschall Z, Fisher LW. Efficient trafficking of acidic proteins out of the endoplasmic reticulum involves a conserved amino terminal ileproval (ipv)-like tripeptide motif. Connect Tissue Res. 2014;55(Suppl 1):138–141. doi: 10.3109/03008207.2014.923852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith CE, Hu Y, Richardson AS, Bartlett JD, Hu JC, Simmer JP. Relationships between protein and mineral during enamel development in normal and genetically altered mice. Eur J Oral Sci. 2011;119(Suppl 1):125–135. doi: 10.1111/j.1600-0722.2011.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitz JE, Teepe JD, Hu Y, Smith CE, Fajardo RJ, Chun YH. Estimating mineral changes in enamel formation by ashing/bse and microct. J Dent Res. 2014;93:256–262. doi: 10.1177/0022034513520548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katsura KA, Horst JA, Chandra D, Le TQ, Nakano Y, Zhang Y, Horst OV, Zhu L, Le MH, DenBesten PK. Wdr72 models of structure and function: A stage-specific regulator of enamel mineralization. Matrix Biol. 2014;38:48–58. doi: 10.1016/j.matbio.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang SK, Hu Y, Yang J, Smith CE, Nunez SM, Richardson AS, Pal S, Samann AC, Hu JC, Simmer JP. Critical roles for wdr72 in calcium transport and matrix protein removal during enamel maturation. Mol Genet Genomic Med. 2015;3:302–319. doi: 10.1002/mgg3.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Sayed W, Parry DA, Shore RC, Ahmed M, Jafri H, Rashid Y, Al-Bahlani S, Al Harasi S, Kirkham J, Inglehearn CF, Mighell AJ. Mutations in the beta propeller wdr72 cause autosomal-recessive hypomaturation amelogenesis imperfecta. Am J Hum Genet. 2009;85:699–705. doi: 10.1016/j.ajhg.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee SK, Seymen F, Lee KE, Kang HY, Yildirim M, Tuna EB, Gencay K, Hwang YH, Nam KH, De La Garza RJ, Hu JC, Simmer JP, Kim JW. Novel wdr72 mutation and cytoplasmic localization. J Dent Res. 2010;89:1378–1382. doi: 10.1177/0022034510382117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan HC, Estrella NM, Milkovich RN, Kim JW, Simmer JP, Hu JC. Target gene analyses of 39 amelogenesis imperfecta kindreds. Eur J Oral Sci. 2011;119(Suppl 1):311–323. doi: 10.1111/j.1600-0722.2011.00857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright JT, Torain M, Long K, Seow K, Crawford P, Aldred MJ, Hart PS, Hart TC. Amelogenesis imperfecta: Genotype-phenotype studies in 71 families. Cells Tissues Organs. 2011;194:279–283. doi: 10.1159/000324339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poulter JA, Murillo G, Brookes SJ, Smith CE, Parry DA, Silva S, Kirkham J, Inglehearn CF, Mighell AJ. Deletion of ameloblastin exon 6 is associated with amelogenesis imperfecta. Hum Mol Genet. 2014;23:5317–5324. doi: 10.1093/hmg/ddu247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prasad MK, Geoffroy V, Vicaire S, Jost B, Dumas M, Le Gras S, Switala M, Gasse B, Laugel-Haushalter V, Paschaki M, Leheup B, Droz D, Dalstein A, Loing A, Grollemund B, Muller-Bolla M, Lopez-Cazaux S, Minoux M, Jung S, Obry F, Vogt V, Davideau JL, Davit-Beal T, Kaiser AS, Moog U, Richard B, Morrier JJ, Duprez JP, Odent S, Bailleul-Forestier I, Rousset MM, Merametdijan L, Toutain A, Joseph C, Giuliano F, Dahlet JC, Courval A, El Alloussi M, Laouina S, Soskin S, Guffon N, Dieux A, Doray B, Feierabend S, Ginglinger E, Fournier B, de la Dure Molla M, Alembik Y, Tardieu C, Clauss F, Berdal A, Stoetzel C, Maniere MC, Dollfus H, Bloch-Zupan A. A targeted next-generation sequencing assay for the molecular diagnosis of genetic disorders with orodental involvement. J Med Genet. 2015 doi: 10.1136/jmedgenet-2015-103302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lacruz RS, Nakayama Y, Holcroft J, Nguyen V, Somogyi-Ganss E, Snead ML, White SN, Paine ML, Ganss B. Targeted overexpression of amelotin disrupts the microstructure of dental enamel. PLoS One. 2012;7:e35200. doi: 10.1371/journal.pone.0035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abbarin N, San Miguel S, Holcroft J, Iwasaki K, Ganss B. The enamel protein amelotin is a promoter of hydroxyapatite mineralization. J Bone Miner Res. 2015;30:775–785. doi: 10.1002/jbmr.2411. [DOI] [PubMed] [Google Scholar]
- 55.Shin M, Hu Y, Tye CE, Guan X, Deagle CC, Antone JV, Smith CE, Simmer JP, Bartlett JD. Matrix metalloproteinase-20 over-expression is detrimental to enamel development: A mus musculus model. PLoS One. 2014;9:e86774. doi: 10.1371/journal.pone.0086774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodet T, Noo F, Defrise M. The cone-beam algorithm of feldkamp, davis, and kress preserves oblique line integrals. Med Phys. 2004;31:1972–1975. doi: 10.1118/1.1759828. [DOI] [PubMed] [Google Scholar]