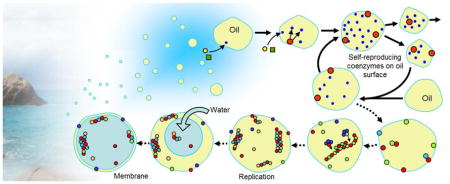
Abstract
The origin of life means the emergence of heritable and evolvable self-reproduction. However the mechanisms of primordial heredity were different from those in contemporary cells. Here I argue that primordial life had no nucleic acids; instead heritable signs were represented by isolated catalytically active self-reproducing molecules, similar to extant coenzymes, which presumably colonized surfaces of oil droplets in water. The model further assumes that coenzyme-like molecules (CLMs) changed surface properties of oil droplets (e.g., by oxidizing terminal carbons), and in this way created and sustained favorable conditions for their own self-reproduction. Such niche-dependent self-reproduction is a necessary condition for cooperation between different kinds of CLMs because they have to coexist in the same oil droplets and either succeed or perish together. Additional kinds of hereditary molecules were acquired via coalescence of oil droplets carrying different kinds of CLMs or via modification of already existing CLMs. Eventually, polymerization of CLMs became controlled by other polymers used as templates; and this kind of template-based synthesis eventually resulted in the emergence of RNA-like replicons. Apparently, oil droplets transformed into the outer membrane of cells via engulfing water, stabilization of the surface, and osmoregulation. In result, the metabolism was internalized allowing cells to accumulate free-floating resources (e.g., animoacids, ATP), which was a necessary condition for the development of protein synthesis. Thus, life originated from simple but already functional molecules, and its gradual evolution towards higher complexity was driven by cooperation and natural selection.
Keywords: coenzyme world, origin of life, lipid world, molecular heredity, molecular cooperation
Graphical Abstract
1. Introduction
Reconstruction of past evolutionary events is an exciting challenge; it requires integration of huge amounts of facts and theoretical approaches in order to filter out the most likely scenario of the origin of specific organs. It is even more difficult to approach the ultimate challenge – understanding the origin of life. Despite of the large number of publications on the origin of life (sf. reviews Ikehara, 2016; Sponer et al., 2016; Weber, 2007), the problem is rarely addressed from the systems theory perspective. Instead of defining what life is, most publications discuss the possible origins of specific cellular components and functions. These papers certainly elucidate the later steps in the evolution of cells, but they miss the origin of life itself. In particular, the RNA-world scenario (Orgel, 2004) has nothing to do with the origin of life because RNA is a product of a long earlier evolution rather than a “starting molecule” (Bernhardt, 2012). There are reasons to expect that primordial hereditary systems were organized in a radically different way than in contemporary cells and required neither nucleic acids nor proteins.
Life is often confused with self-organization or autocatalysis such as in crystal growth or dissipative structures (e.g., hurricanes, tornados, and flames) (Egel, 2012; Kauffman, 1986). Self-organization has one important feature that is common with life – the capacity to persist despite of perturbations via positive feedback driven by dissipating energy. But neither self-organization nor auto-catalysis is sufficient for life. The main difference is that self-organized systems die out without affecting future self-organizing events. In contrast, living systems are constructed by parental systems and carry unique conditions that allow them to produce the next generation of systems (Deacon et al., 2014). This feature is heredity that distinguishes life from non-life. In other words, heredity is a recursive construction (Bickhard, 2005): it is a capacity to make specific conditions (including resources, tools, scaffolds, codes) that are sufficient for both self-maintenance and making offspring systems that, in turn, carry the same capacity (“same” means that this capacity is stable in a sequence of generations1). Life creates unique self-supporting artificial structures and conditions that did not exist before. Besides stability, heredity has to support evolvability, which is a capacity to change and keep the changed state recursively across generations. Thus, the origin of life means the emergence of heredity and evolvability rather than the origin of nucleic acids or proteins (Jablonka and Szathmáry, 1995; Tessera, 2011).
The second problem with the origin of life stems from the notion of chemical evolution, which is supposed to provide an increasing diversity of organic molecules in primordial soup or inorganic compartments and supply first living systems with every resource they need (Benner et al., 2012; Ikehara, 2016; Koonin and Martin, 2005). However, nobody have described mechanisms of chemical evolution and proved that it could indeed provide these resources (e.g., sugars, aminoacids, nucleic bases, and lipids). Without heredity, past self-organizing events have no effect on the future events, and thus, chemical evolution is not “evolution” in biological sense. And if heredity indeed existed in certain chemical systems, then these systems were already alive because heredity is the essential feature of life. This is not an attempt to draw a demarcation line between life and non-life; certainly there is a gray transition zone, where heredity is weak and evolutionary potential is limited. But this transition zone already belongs to life as far as we are focused on heredity and evolution. Thus, instead of chemical evolution and soup we need to focus on early phases of biological evolution.
In this paper I first briefly discuss problems with existing models of the origin of life, and then describe an expanded version the coenzyme world model proposed earlier (Sharov, 2009). In particular, I added the discussion on the nature of primordial CLMs and their potential interaction with each other and with microelements that facilitate catalytic capacities. Also, I elaborated scenarios for transformation of surface metabolism intointernal metabolism within cells. The model is discussed in the context of recent experiments and observations related to the origin of life.
2. Problems with existing models of the origin of life
The RNA world hypothesis remains widely accepted in the field of the origin of life (Orgel, 2004; Robertson and Joyce, 2012). This model assumes that first living systems had self-replicating nucleic acids (Gilbert, 1986) or other kinds of similar heteropolymers2 (Nelson et al., 2000; Orgel, 2000). Alternatively, the RNA world is viewed as an intermediate step of evolution that preceded contemporary prokaryotic cells with DNA-based heredity and protein synthesis (Bernhardt, 2012). Bernhardt mentioned the following main objections to the RNA world as the initial stage of life: RNA is too complex to have arisen prebiotically, it is highly unstable, and is generally a weak catalyst. Another problem is the lack of resources (i.e., nucleotides) for replication of RNA. Although nucleotides can be synthesized abiogenically (Benner et al., 2012; Powner et al., 2009), they are unlikely to get concentrated in quantities sufficient for RNA replication. Even if several nucleotides appear in a close proximity to each other due to a rare coincidence and produce a complimentary RNA strain, there would be no resources left for the next round of replication. Nucleotides can be synthesized from bases and sugars by RNA-mediated catalysis (Unrau and Bartel, 1998), but these precursors are unlikely to be supplied in sufficient quantities to support the reaction. For example, in the Murchison meteorite, nucleobases and sugars are present at concentration of ~1 ppm (Callahan et al., 2011; Cooper et al., 2001). Recent discovery of alcohol and sugar on the comet Lovejoy (Biver et al., 2015) is intriguing, but it does not prove that primordial organisms used hydrocarbons of abiotic origin as resources. It is very unlikely that life originated on small comets, and if a comet landed on a planet, organic chemicals would have degraded almost immediately or become diluted. Neither chemical evolution nor primordial soup can provide resources of self-replications (Lane et al., 2010). An alternative scenario is that replicating polymers such as RNA emerged much later in evolution after primordial living systems have developed heritable mechanisms for making monomers (Sharov, 2009; Wächtershäuser, 1988).
The most appealing point of the RNA world hypothesis is the assumption that primordial replication systems were identical to the present-day nucleic acids, and thus, there is no need to reconstruct alternative mechanisms of heredity. However, self-reproduction3 can be achieved in substantially more simple systems without nucleic acids. For example, synthetic peptide-based monomers selectively self-assemble into fibers, which then reproduce via fiber elongation-and-breakage mechanism (Colomb-Delsuc et al., 2015). Heredity is a systems-level autocatalysis, where parental systems construct the same kind of descendants. However, autocatalytic synthesis (in contrast to decay) is a rare property among organic molecules. Thus, it was suggested that self-reproduction could arise more easily in multi-component mixtures of molecules with random cross-catalysis (Kauffman, 1986). In particular, Kauffman proposed that a mixture of peptides makes an autocatalytic set if the synthesis of each component is catalyzed by some other member(s) of the same set. This hypothesis has no experimental support and appears as weak as the RNA world from the theoretical point of view. It is hard to imagine a mechanism that preserves long peptide sequences over time without encoding; whereas short or random peptides (that do not need encoding) have no catalytic activity. Another problem is that autocatalytic sets easily dissipate if they are not enclosed in a membrane; but peptides are not likely to make a non-permeable membrane.
Although peptide cross-catalysis is certainly unrealistic as a model for the origin of life, the theoretical idea that autocatalytic sets can support heritable self-reproduction is valid. However, not every autocatalytic set, as defined by Kauffman, can support self-reproduction. Self-reproduction is possible only in autocatalytic sets with specific stoichiometry constraints, where a sequence of internal reactions can increase the number of all molecular species within the set (Sharov, 1991). It appears that such systems always have a unique minimum core with at least one “reproduction” reaction that generates an additional molecule within the cycle. Self-reproducing systems can propagate in space, which is similar to the growth and expansion of populations of living organisms (Gray and Scott, 1994; Tilman and Kareiva, 1997). Autocatalytic sets have two alternative steady states: “on” and “off”; thus, they represent the most simple hereditary system or memory unit (Jablonka and Szathmáry, 1995). A biochemical example of a self-reproducing autocatalytic set is a Calvin cycle (also known as a reverse citric acid cycle), which has a “reproduction” reaction: a ribulose 1 5 bisphosphate molecule divides into two molecules of 3-phosphoglycerate after acquiring carbon dioxide as a resource. It was speculated that the Calvin cycle without protein enzymes could have been the first self-reproducing system at the origin of life (Hartman, 1998; Morowitz et al., 2000). The advantage of this hypothesis is that it does not require soup or RNA world (Hartman, 1998). However, it seems unlikely that CO2 fixation was the initial source of carbon in primordial systems because the Calvin cycle requires too much energy (e.g., ATP), which was not available in simple molecular assemblies. The network modeling approach suggested that the Calvin cycle had a low probability of a random realization and the length and cost of the cycle are close to the extreme values (Zubarev et al., 2015). This indicates that the Calvin cycle is likely to be a product of adaptive evolution via selection. Because natural selection requires heredity, we have to admit that heredity existed before the emergence of the Calvin cycle. Another example of a self-reproducing molecule is prion (Griffith, 1967; Laurent, 1997), and indeed prions have been invoked in various ways in discussions on the origin of life (Maury, 2009, 2015; Steele and Baross, 2006). However, prions cannot support the synthesis of the primary (i.e., unfolded) polypeptides, and therefore they had no role in the origin of life.
Another group of theories is based on the assumption that abundant organic molecules were generated by direct reduction of CO2. In particular, Wächtershäuser (Wächtershäuser, 1988) assumed that the energy from oxidation of FeS to FeS2 at the sea floor was used for organic synthesis. He further suggested that life started from autocatalytic coenzymes similar to nicotinamide NADP+ or thiamine pyrophosphate (TPP), and that negatively-charged organic molecules were adsorbed and concentrated on positively-charged pyrite mineral surfaces. This hypothesis is close to my idea that life originated from coenzyme-like molecules, although it assumes a different source of carbon for primordial systems. Another model of the origin of life is based on the idea that formaldehyde was synthesized using reduction of CO2 via photo-catalysis mediated by ZnS (Mulkidjanian and Galperin, 2007). This model further assumed that other organic molecules and polymers were synthesized by various inorganic catalysts including Zn, preparing a soup where self-replicating systems (RNA-world) can emerge spontaneously. The common problem of these theories is that carbon fixation from CO2 is too difficult energetically and could not produce sufficient resources for the RNA world.
The next class of theories known as “lipid-world” is based on the idea that lipid-like molecules played the key role in the origin of life (Segre et al., 2001; Segre et al., 1998; Serge et al. 2001; Tessera, 2011). These models are certainly more realistic than the RNA-world or protein-world models because (1) they consider a spatially heterogeneous system of lipid compositional assemblies in water, (2) lipids do not require coding, (3) lipid surfaces and their individual molecular components have catalytic activity, and (4) the dynamics of compositional assemblies depends on affinity and binding, which are common features in lipids. In particular, the GARD (Graded Autocatalysis Replication Domain) model of the origin of life assumes that compositional assemblies grow and eventually break down into two (or more) daughter assemblies (Segre et al., 1998). As a result, these assemblies can reproduce and form discrete quasispecies. Disproportional split of components between daughter assemblies can result in a heritable variation that may occasionally give rise to new quasispecies. The lipid-world model, however, has its own problems. First, the diversity and abundance of abiotic lipid-like molecules is low and hardly can support the propagation and evolution of molecular assemblies. Lipids per se do not exist in a non-living world4, and their synthesis requires glycerol, which is a rare and unstable molecule. For example, concentration of glycerol in Murchison meteorite is only 15 ppm (Cooper et al., 2001). Finally, the catalytic activity of lipids alone is low. Thus they may require specialized catalysts to establish new covalent bonds.
Very few publications attempt to predict the functions of primordial living systems. Deacon developed a model of autocatalytic set called “autocell”, which besides cross-catalysis included self-assembly of a multi-component unit that resembles a viral capsid (Deacon, 2006; Deacon, 2011). This structure may have additional functions such as long-term survival in unfavorable conditions. Deacon further argued that the specific feature of life is “teleodynamics” which is an intrinsic tendency to create higher-order constraints that are beneficial for the self-organization of the whole system. However, Deacon admitted that the emergence and self-propagation of autocells appears highly unlikely because it requires conditions that did not exist without life, such as “(1) presence of significant quantities and concentrations of large polymers; (2) some considerable degree of structural similarity among these molecules; and (3) sufficient variety and degeneracy of their stereochemical properties to support spontaneous autocatalysis and self-assembly” (Deacon, 2006: p. 145).
It appears that the problem of the origin of life cannot be adequately addressed by focusing on one specific component of cells as a sole leading factor. It is meaningless to separate metabolism from heredity because metabolism can persist and evolve only if it is heritable, and heredity requires metabolism as a source of organic molecules and energy (Jablonka and Szathmáry, 1995). Models of prebiotic networks suggest the importance of combinations of various kinds of molecules (Caetano-Anolles and Seufferheld, 2013; Nghe et al., 2015). Thus, we need to envision primordial systems as multi-component functional units capable of capturing resources, dispersion, and heritable self-reproduction. Functions emerged and changed simultaneously with chemical and structural changes. Organic molecules in Stanley Miller’s experiments or in meteorites are not relevant for the discussion of the origin of life because these molecules are not functional alone. It is even more important to find a context/niche in which organic molecules may become functional. Heredity is the central feature of life but it can be based on other molecules than nucleic acids, and this non-genetic information was likely distributed among various molecular components of primordial systems. In summary, the next turn in the study of the origin of life should involve a system’s approach, where heredity and other functions emerge and co-evolve in multi-component systems.
3. Components, Functions, and Evolution of First Living Systems
3.1. Life on the Surface
Surfaces play an extraordinary role in living cells (Hoffmeyer, 1998). Most functional surfaces are represented by bilayer phospholipid membranes, which separate cells from the environment, prevent the dissipation of cell resources, provide a sensorial interface with the outside world, support the structural scaffold for the cell, and carry tools for cell propulsion (e.g., flagella or pseudopodia). Eukaryotic cells have additional membrane functions associated with endoplasmic reticulum, nuclear envelope, and various organelles, such as mitochondria, plastids, golgi, vacuoles, and secretion vesicles. There are two main reasons why surfaces are so abundant and enriched in various functions: (1) dimensionality effect: chemical reactions go faster on the surface because molecules encounter each other more readily in two dimensions than in three dimensions (Adam and Delbrück, 1968), and (2) continuous surfaces can separate compartments.
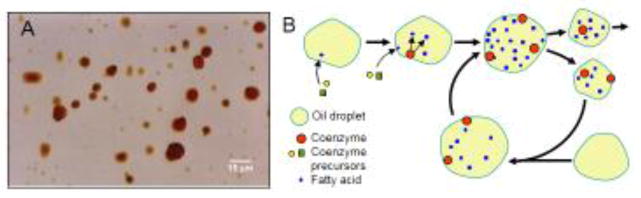
Life could not have originated in a homogeneous 3-dimensional space; thus, we need to consider what kinds of surfaces could have been important for the origin of life. Among surfaces available on Earth-like planets two kinds can be considered as most likely places for the origin of life: water–mineral and water–oil surfaces. Here, by “oil” I mean various hydrocarbons, mostly alkenes, which are the most abundant organic molecules in the universe (Deamer, 2011) and are likely to exist on early terrestrial planets (Marcano et al., 2003). These two kinds of surfaces are selected here for discussion because they both include water. Life could not originate without water because most chemical reactions in living cells require water. We do not discuss air-water surface because it is too unstable and subject to wind and radiation. In this paper we focus mostly on water-oil surfaces because (1) oil self-aggregates in water forming fine droplets or microspheres (Fig. 1A), which resemble living cells and can divide or merge, (2) oil-attached molecules are not fixed in space but can move around the surface and have a chance to interact with other oil-attached molecules, (3) oil can be used as a source of carbon for primordial life, and (4) oil droplets could be eventually transformed into a (phospho)lipid membrane (see section 4.2.).
Figure 1.
Oil (hydrocarbon) droplets in water as a potential substrate for coenzyme-like molecules. (A) Emulsion of oil/petroleum in water, from http://petrowiki.org/Oil_emulsions. (B) Scenario of coenzyme self-reproduction on oil droplets: a coenzyme molecule makes the surface hydrophilic via oxidation of hydrocarbons; this change facilitates synthesis of coenzymes from precursors on the surface. Hydrophilic oil droplets easily divide and may coalesce with new oil droplets (i.e., capture new oil resource).
Both mineral and oil surfaces can be colonized from the water side by catalytically active self-reproducing simple molecules which could be precursors of life. I call them coenzyme-like molecules (CLMs) because they may resemble coenzymes, and the model of the origin of life is therefore named “coenzyme world” (Sharov, 2009). Catalysis is necessary for self-reproduction of CLMs as well as for modification of other molecules on the surface. Because many coenzymes (e.g., ATP, NADH, and CoA) are similar to nucleotides, CLMs can be viewed as predecessors of nucleotides. Some ancient organic molecules such as carotinoids and vitamin K are oleophilic and may be relics of the primordial metabolism on the surface of oil droplets.
Let us consider a hypothetical scenario of how CLMs can colonize the surface of oil droplets in water. Assume that simple precursors of CLMs exist in water but cannot anchor to the hydrophobic oil surface. However, some droplets may include a few fatty acids with hydrophilic ends that allow the precursors to attach and make a functional CLM (Fig. 1B). This CLM can now catalyze the oxidation of outer ends of hydrocarbons in the oil droplet, making more fatty acids where additional CLMs can be assembled from precursors. Fatty acids produced by CLMs make the surface hydrophilic, and this increases the chance of a droplet to split into smaller ones. Later, small droplets can coalesce with other oil droplets, i.e., capture new oil resources (Fig. 1B). Some components of CLMs could come from the oil phase rather than from the water. These include microelements as well as organic molecules. Furthermore, some organic molecules can be obtained via degradation of oil itself. The hypothesis that oil droplets served not just as a habitat but also as a source of carbon for self-reproduction of CLMs seems very attractive because oil can provide abundant organic resources. In particular, some organic molecules could have been derived from fatty acids by pathways similar to the beta-oxidation pathway. The idea that hydrocarbons were used by primordial systems as nutrients was proposed by Oparin (1953 [1936]). Oparin assumed that hydrocarbons together with other organic molecules formed colloidal systems with spontaneous metabolism, which resembled living cells. More recently, Deemer wrote: “It seems likely that primitive cells incorporated lipid-like molecules from the environment as a nutrient, rather than undertaking the much more complex process of synthesizing complex lipids by an enzyme-catalyzed process.” (Deamer, 1999).
The emergence of self-reproducing CLMs could be facilitated by interaction of organic molecules with non-organic ions or crystals. Organic molecules produced by primordial systems were initially very simple, and simple molecules are not likely to act as strong catalysts. This limitation can be removed, however, if these small molecules acquired or modified their catalytic competence via interaction with non-organic ions or crystals. For example, aminoacids readily interact with heavy metals such as cobalt, nickel, or zinc, and some of these complexes are catalysts (Dutta et al., 2014); vanadium has effects in carbohydrate metabolism (Gruzewska et al., 2014), whereas iron and sulfur facilitate electron transfer (Kummerle et al., 2000). Oxidation of hydrocarbons can be facilitated by iron oxide. These inorganic compounds are natural contaminants in petroleum, and thus it is reasonable to assume that they may be present in abiogenic oil droplets.
Oil droplets with CLMs represent a two-level hierarchical system that includes self-reproduction at both levels: (1) level of catalytically active surface molecules and (2) level of oil droplets populated by communities of surface-bound molecules. Moreover, CLMs support self-reproduction at both levels because they change their local environment (i.e., surface properties of oil droplets) and this change enables them to capture resources, self-reproduce, and propagate to other oil droplets (Fig. 1B). As a result, CLMs play the role of hereditary signs (i.e., coding molecules) because they encode the surface properties of oil droplets (phenotype) and transfer the code to the next generation of colonized droplets in the form of additional copies of the same molecule type (Sharov, 2009). Note, that CLMs are not resources, and do not have to exist in large quantities in the environment to support the primordial metabolism. A single molecule is sufficient to initiate a wave of self-reproduction that can then spread over the surface of an individual oil droplet and over the large population of oil droplets.
Besides oil droplets, CLMs can also colonize mineral surfaces (Wächtershäuser, 1988); however minerals cannot be used as a source of carbon. Thus, Wächtershäuser assumed that CO2 was the source of carbon for the synthesis of organic molecules. However, carbon fixation from CO2 was hardly possible at the early stages of the origin of life (see section 2). A more realistic habitat for primordial molecules could be minerals covered with a film of oil. Then, self-reproducing CLMs can spread along such surfaces and possibly travel on oil droplets to colonize other patches of oleophylic minerals. Thus, mineral surfaces and oil droplets could have been two alternative niches for the same population of self-reproducing CLMs.
3.2. Evolutionary potential of the coenzyme world
The next important condition for primordial life is a capacity to evolve adaptively towards more complex functional molecules and pathways. Self-reproduction of a single kind of molecules (e.g., autocatalysis or crystal growth) is not sufficient for evolution because there is no alternative way to reproduce. Without heritable variation there is no Darwinian selection and no adaptive evolution. In rare cases, crystallization may follow multiple alternative pathways that provide a limited potential for change (Cairns-Smith, 1982). But the number of variations is always small and not sufficient to support the long-term evolution.
The only conceivable way for primordial self-reproducing molecules to increase their evolutionary potential was to establish cooperation with other self-reproducing molecules. Such cooperation can be facilitated by group selection, where groups of molecules either propagate or dissipate together. For example, if the interaction between two kinds of coding molecules (i.e., self-reproducing CLMs) facilitated the rates of self-reproduction of the jointly colonized oil droplets, then the proportion of droplets that harbor both kinds of coding molecules will increase with time. Thus, oil droplets likely harbored a community of self-reproducing molecules with cooperative and possibly conflicting relations. Mineral surfaces are probably less effective in supporting group selection as compared to oil droplets because their existence does not depend on the presence of CLMs. However, minerals can loose an oil film as a consequence of CLM activity and then become unsuitable for colonization; this would be functionally equivalent to the dissipation of an oil droplet.
The term “cooperation” in relation to molecules means that interaction (or simply coexistence) of two or more kinds of self-reproducing molecules is beneficial for each kind of molecules, where benefits are measured by the increase in the rate of reproduction. Cooperation is more than mutual catalysis (Conrad, 1982; Kauffman, 1986) because mutual catalysis is static: it is either present or not, whereas the notion of cooperation is meaningful only in the context of continuous evolutionary change. Continuous evolution appears very unlikely to happen in a system with small coding molecules. In particular, we cannot expect that CLMs easily change their chemical structure and keep the competence for self-reproduction. Thus, instead of looking for changes in the chemical structure of CLMs we should consider changes in relations between molecules or changes in their local environment. By relations I mean weak or transient interactions that do not cause irreversible change in molecule structure, as for example, in catalysis. Relations can be mediated by modification of local environments (e.g., surface properties of oil droplets). Potential relations between even simple molecules are very diverse: two molecules may turn relative to each other, approach each other from different sides, or interact via intermediate molecules that serve as signals or resources. There are many more potential cooperative relations as compared to the number of potential heritable modifications of a single molecule. These relations may become heritable signs if they reinforce each other, or appear self-reinforced. Thus, heredity is supported not just by coding molecules but also by coding relations between molecules, and in fact by the whole network of relations. Note that the autopoiesis theory always emphasized the self-renewal of relations in addition to the self-renewal of components (Maturana and Varela, 1980).
Among all kinds of relations between molecules, the relations mediated by local environment (e.g., by surface properties of oil droplet or abundance of resources) seem most important in evolution. Each kind of self-reproducing CLMs performs some function, such as capturing resources, storing energy, or catalyzing a reaction; and these effects may appear beneficial not only for this particular kind of molecules but also for other kinds of self-reproducing CLMs on the same oil droplet. If these beneficial effects are reciprocal and increase the rate of self-reproduction of the entire molecular community, then we can say that molecules have a cooperative relation. The main advantage of molecular cooperation compared to modification of single molecules is in the increase of potential variability of outcomes. Modification of oil droplets can be viewed as “niche construction” and was necessary for boosting the evolutionary potential of primordial systems. In summary, the evolvability of primordial molecular communities was supported mainly by subtle changes such as modification of local environment and cooperative relations between species of molecules. Cooperative molecular communities in perishable local environments (e.g., oil droplets) can be viewed as precursors of organisms/cells.
Evolvable primordial systems cross the threshold, where physical and chemical processes become integrated into potentially endless and branching chains of events that constitutes life. Thus, we need a term that represents this potentially immortal and evolvable component of life, and this term is sign. Historically, the doctrine of signs (or semiotics) was applied almost exclusively to human communication. Biologists however noticed that meaningful signs exist in the behavior and communication of animals (Sebeok, 1972; Uexküll, 1982). Discovery of DNA and genetic code showed, however, that molecular processes in living cells follow programs encoded in a genome, which bring the experience of ancestral generations to new-formed organisms and help them to develop, survive, and reproduce in changing environments. Thus, the genome carries a set of inter-related signs that encode meaningful information. A few decades ago, a new discipline of biosemiotics emerged whose aim is integration of theoretical biology with semiotics (Sebeok, 2001, Hoffmeyer 2008). The main challenge of biosemiotics is to depart from the anthropocentric interpretation of signs and replace it with an evolutionary approach, where signs and their meanings evolved from simple ones (as in bacteria) to more complex (as in animals and humans) (Sharov et al., 2015). Primitive sign process, or protosemiosis, do not require mind; instead signs are associated with specific actions of molecular agents (e.g., ribosome or RNA polymerase) or cells (Sharov and Vehkavaara, 2015)). Protosemiosis may exist in very simple systems, such as molecular communities on the surface of oil droplets, where CLMs play the role of signs that encode surface properties of oil and future catalytic actions, and disseminate this capacity to the progeny. Thus, the notion of protosemiosis is important for understanding the origin of life.
3.3. Diversification of molecular communities
The major criterion of progress in biological evolution at its largest scale is the increase of functional complexity (Sharov, 2006). Thus, the increase in the number of components and relations in molecular communities is the main path towards full-fledged organisms. The core of the primordial self-reproducing system is a minimal subset of irreplaceable coding components and coding relations. If any of these components or relations is lost entirely, then it cannot be recovered based on other components and relations. Establishment of new coding molecules depends on their capacity to self-reproduce as well as on their compatibility with already existing molecules. If new molecules strongly compete with other components for resources then the self-reproduction capacity of the whole system may appear compromised.
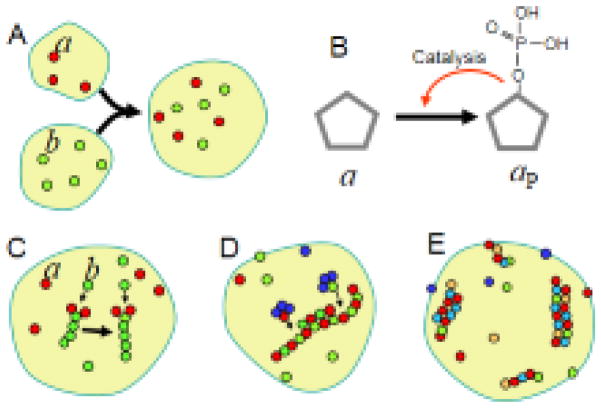
The first path for the emergence of new coding molecules is de-novo acquisition, similar to the emergence of the first self-reproducing CLMs. Such acquisition can be facilitated by the coalescence of droplets carrying different coding molecules. Consider that some population of oil droplets has coding molecules of type a, and another partially separated population has coding molecules of type b. If droplets from different populations coalesce, they make a new kind of droplets with two types of coding molecules: a and b (Fig. 2A). If the new droplet splits up due to mechanical agitation then the progeny droplets are likely to carry both kinds of coding molecules because multiple copies of each are present in the parental system. Thus, combinations of coding molecules can be inherited and make their own evolutionary lineages (Sharov, 2009). The combination is beneficial, if droplets with both coding molecules reproduce faster than droplets with just one kind of coding molecules.
Figure 2.
Emergence of new coding molecules. (A) Coalescence of oil droplets carrying different self-reproducing coenzymes. (B) Phosphorylation of molecule a, which is stably produced within the chemical community on the surface of oil droplets, results in the emergence of a new coding molecule aP if this reaction is autocatalytic. (C) Polymerization of molecules b catalyzed by self-reproducing molecules a. (D) Simple repetitive polymers facilitate polymerization of identical polymers aligned on the surface of oil droplets. (E) Template-based universal synthesis (i.e., replication) of coding polymers with any aperiodic sequence of monomers.
Initially, coding molecules were not connected and were transferred to offspring systems in different combinations at random. The probability of transferring the full set of coding molecules to descendants by pure chance may be problematic especially if droplets carry too many kinds of coding molecules and some of them are present in a small number of copies. This combinatorial problem can be partially meliorated by the “stochastic corrector” mechanism, which is a preferential propagation of systems with a full set of coding molecules (Szathmáry, 1999). Systems with an incomplete set of coding molecules are more likely to fail in surviving and reproduction because some of their functions appear missing. This kind of stochastic correction is a primordial version of the purifying selection; and like the purifying selection, it reduces the overall reproduction rate of the population.
An important consequence of the emergence of new kinds of coding molecules is the diversification of the downstream molecules. For example, if coding molecule A generates some products from available resources, then catalytic activities of the additional coding molecule B can be applied to the products of molecule A. Similarly, catalytic activities of A can be applied to the products of B. Thus, the increase of the number of coding molecules results in a much faster combinatorial increase in the diversity of downstream molecules. This diversity could in turn increase the chances of acquiring new coding molecules through paths discussed below.
The second path towards the emergence of new kinds of coding molecules is the modification of existing coding and downstream molecules (Fig. 2B). It is important that new coding molecules facilitate their self-reproduction. For example, if molecules of type a are already produced in the systems and the phosphorylated modification of a (i.e., aP) facilitates the phosphorylation of a, then a single phosphorylated molecule aP initiates a chain reaction that converts a into aP. This reaction may initially be harmful due to a decrease in the number of molecules a, and weakening of functions supported by a. However, the molecular community may eventually develop protecting mechanisms such as shielding some molecules a from phosphorylation or inhibiting the catalytic activity of aP if it becomes too abundant. As a result, the system acquires a new kind of coding molecules aP, which can self-reproduce and support a new chemical function (i.e., phosphorylation). This molecule is heritable because the descendant systems are likely to get copies of aP after the division of oil droplets.
Group selection at the level of oil droplets is likely to facilitate cooperation of newly acquired coding molecules with the previously existing molecular community. However, group selection is effective only if the exchange of members between groups is rare compared to the rate of self-reproduction of groups. This condition is required for any kind of symbiogenesis (Guerrero et al., 2013). Frequent exchange of members between groups is beneficial for the emergence of molecular “parasites” that self-reproduce at the expense of other members in molecular community inhabiting oil droplets. Thus, primordial systems should have a selection pressure to develop mechanisms that reduce the chances of molecule exchange with other systems. For example, certain molecules produced within the system could inhibit the coalescence with droplets that already carry some CLM molecules on the surface.
Finally, the third path towards the emergence of new coding molecules is the establishment of novel relations, such as polymerization (Sharov, 2009). Polymers may initially appear as downstream products of non-polymeric coding molecules. For example, a coding molecule a could catalyze the polymerization of molecules b, and in this way encode the formation of long polymers b–b–b–b–... (Fig. 2C), which could cover the surface of droplets and modify their physical properties. Some polymers may then become coding molecules if they could facilitate the synthesis of the same kind of polymers (Fig. 2D).
4. Evolution from oil droplets to LUCA
4.1. Template-based replication
The main factor that restricted the rate of primordial evolution was the absence of universal methods for producing new hereditary molecules. Transformation of coding molecules via change of functional groups or polymerization increased the evolvability of primordial systems to some extent (Sharov, 2009). But the ultimate solution of this problem came only with the invention of template-based (or digital) replication. Replication can be viewed as a special case of autocatalysis applied to hetero-polymers, where each monomer is added to the newly-constructed polymer sequentially based on a uniform simple rule applied to each monomer in another linearly aligned polymer that serves as a template (Szathmáry, 1999). Digital replication is universal in the sense that the rule of replication works for polymers of various lengths and for any sequence of monomers from the allowed set. Using the terminology of Kauffman (2014), we can say that the invention of digital replication provided “enabling constraints” that expanded the boundaries of heritable “adjacent possible”, and in this way, increased the evolutionary potential of life.
However, the notion of universal coding should be interpreted with caution because true universal properties exist only in mathematics. In the real world, there are always some limitations even within a universal coding system. For example, replication of the lagging DNA strand requires different molecular agents than replication of the leading strand. Replication and elongation of telomeres requires additional mechanisms that are not equivalent to simple template-based copying. Molecular machinery which is sufficient for replicating short DNA fragments (200–1000 bp) may not work for long sequences (e.g., >1 Mb).
The starting point for the origin of template-based replication was the presence of hetero-polymeric coding molecules with repetitive or partially random sequence. Such polymers may initially stick to each other to perform some other functions (e.g., to increase mechanical stiffness of the surface or facilitate other reactions). Then, the shorter strand in a paired sequence can become elongated by adding monomers that weakly matched the longer strand (Fig. 2D). Natural selection may have supported the increase in fidelity of this process, which helped to produce better copies of existing polymers. Template-based replication probably started with copying short repeats (e.g., telomere-like), and then progressed into copying longer repeats and entirely aperiodic sequences (Sharov, 2009) (Fig. 2E). First replicating polymers were probably similar to nucleic acids; however, the sugar-phosphate backbone of RNA does not support anchoring to the surface of oil droplet. In contrast, peptide nucleic acids (PNAs) with a pseudopeptide backbone are able to absorb at the lipid-water surface (Weronski et al., 2007). Thus, PNA-like molecules could have been evolutionary predecessors of RNA (Nelson et al., 2000). Invention of digital replication was the turning point in the origin of life that substantially increased the evolutionary potential of primordial living systems (Jablonka and Szathmáry, 1995; Sharov, 2009).
4.2. Bilayer membrane
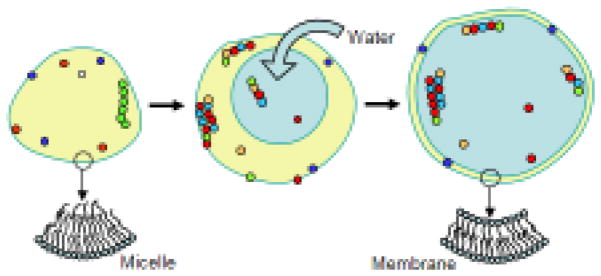
The surfaces of oil droplets provide plentiful local niches for the emerging life. However, subsequent evolution was constrained by the limited supply of oil, and by the two dimensions of a droplet surface. The lack of hydrocarbons slowed the rates of growth in primordial organisms, which in turn, affected negatively the rates of metabolism, which was surface-dependent. Although surface metabolism is helpful for the emergence of life, two-dimensional surfaces do not provide enough capacity for storing resources. Also, the mobility of polymers in a two-dimensional space is restricted because long molecules cannot easily pass each other.
To overcome these limitations primordial systems presumably transformed oil droplets into an outer membrane via engulfing water (Fig. 3). Agitation of oil emulsion in water easily makes “nested” droplets, but they are unstable. Cell-like organization is unlikely to yield any functional advantage if the membrane breaks easily. Thus, the membrane has to be strong enough to sustain mechanical disturbances, and osmoregulation is needed to prevent bursting. Membranes can be stabilized by replacing fatty acids with lipids and phospholipids. Lipids require glycerol, which has to be synthesized within the system (it is not available in the environment). Thus, the emergence of metabolic pathways for glycerol synthesis was necessary for making stable membranes. Glycerol could have been used also to make sugars that are suitable for storing energy, regulating osmosis, and making nucleic acids. It is not clear if template-based replication of coding polymers appeared before or after the formation of stable cell membrane. But in any case, these two evolutionary events produced cells with ribozyme-based catalysts that match to the RNA-world model (Gilbert, 1986).
Figure 3.
The origin of bilayer membranes and transition from surface metabolism to intra-cellular metabolism.
4.3. Chromosomes
As the number of coding molecules increased in evolution, the stochastic correction mechanism became less effective in keeping all coding components together. Stochastic corrector has a significant fitness load because daughter cells with incomplete set of coding molecules are not fully viable. This load increased with each additional type of coding molecules, which limited the increase in overall complexity of the system. In addition, the stochastic correction mechanism requires abundant copies of each type of coding molecules, so that at least one copy is transferred to each daughter cell. As a result, cells had to spend valuable resources for making redundant copies of coding molecules. These problems were resolved in evolution by concatenating heterogeneous coding molecules (e.g., RNAs) into one or very few large units (e.g., chromosomes) and by developing mechanisms of controlled transfer of chromosomes to descendent cells.
4.4. Protein synthesis
Protein synthesis (translation) presumably emerged as a modified type of self-replication (Root-Bernstein and Root-Bernstein, 2015). Apparently, peptides existed before ribosomes and were synthesized individually by specific ribozymes. Because first peptides were likely short and included only a few kinds of aminoacids, their functional capacities were limited. According to Root-Bernstein and Root-Bernstein, the evolutionary fate of peptides changed when they were used to assist the self-replication of RNA. As a result, natural selection supported not only self-replication of the RNA itself but also the production of helper peptides. In particular, peptides became longer and included a more specific sequence of aminoacids. The diversity of animoacids increased in evolution, and especially important was the emergence of aminoacids that are currently found in catalytically active portions of proteins: lysine, cysteine, serine, treonine, aspartate, histidine, and thyrosine (Holliday et al., 2009). The first four of them are rather simple and probably appeared earlier in evolution that the latter three. The evolution of peptide-assisted RNA replication also included the emergence of efficient catalysis of peptide bond formation and tRNA for transporting aminoacids. This hypothesis is supported by the fact that the ribosomal RNA of bacteria Escherichia coli carries remnants of the entire set of tRNAs for all aminoacids together with fragments of coding sequences for ribosomal proteins, polymerases, ligases, synthetases, and phosphatases (Root-Bernstein and Root-Bernstein, 2015). Thus, ribosomal RNA apparently originated from a primordial genome that encoded a self-organizing and self-replicating molecular assembly.
The correspondence between the triplets of nucleotides in the RNA molecule and corresponding aminoacids in the encoded protein is determined by the genetic code. Although genetic code is nearly universal in all living organisms, it is not likely to appear initially in its current form. It is possible that the initial code was based on nucleotide doublets rather than triplets, and encoded a smaller set of aminoacid types (Patel, 2005; Travers, 2006). It is also likely that genetic code evolved towards lower rates of translation errors (Novozhilov et al., 2007).
Reconstructions of the Last Universal Common Ancestor (LUCA) of all known living organisms (Doolittle, 2000; Theobald, 2010) indicate, that LUCA was far more complex than RNA-world systems described above. The hypothetical LUCA had a DNA-based genome and a fully developed machinery for programmed synthesis of proteins. The integrity of the DNA was maintained by a group of diverse enzymes, including DNA helicase, topoisomerase, ligase, and DNA repair proteins. The RNA-polymerase complex was used to synthesize mRNA copies of each gene. LUCA-type cells synthesized fatty acids, and thus were no longer dependent on the supply of abiotic oil. The outer membrane of LUCA included ion pumps, receptors, and other proteins. Although the genome of LUCA is a product of phylogenetic reconstruction, it can be viewed as a bacteria- or archea-like organism with the number of genes in the range from 500 to 1000 (Koonin, 2003, 2009). The difference in complexity between early cells with first encoded proteins and LUCA-type cells is hard to overestimate, and it is comparable in scope to the difference between prokaryotes and multicellular eukaryotes. Emergence of each new function, and/or new gene, opened previously unavailable possibilities for subsequent evolution (Sharov 2014). This positive feedback supported the exponential increase of the overall complexity in most successful lineages of organisms (Sharov, 2006). These later evolutionary events, however, are not relevant for the problem of the origin of life (and for this paper) because the backbone of hereditary mechanisms and chemical construction networks has been finalized with the advent of protein synthesis.
5. Discussion
Proposed scenario of the origin of life from catalytically active self-reproducing molecules (coenzymes) on the surface of oil droplets in water has multiple advantages as compared to alternative models. First, surface metabolism increases the chances of molecular interaction as compared to the open 3D space; an additional advantage of oil surfaces is that functional molecules can be covalently attached to oil (hydrocarbon) molecules and still float around. Second, surfaces provide ready access to the outside resources including both chemicals and energy. Third, there is no need for either pre-biotic soup or energetically-expensive and biochemically complex pathway of carbon fixation from CO2; instead, oil was used as a source of carbon. Fourth, coenzymes played the role of hereditary signs that encoded surface properties of oil droplets. And fifth, the emergence of advanced features of life, such as template-based replication, bilayer outer membrane, and carbohydrate metabolism, is not required at the origin of life; instead these features appeared much later in evolution. According to this model, novel chemical structures, functions, and heredity emerged simultaneously by reinforcing each other with each cycle of reproduction. Thus, it does not fit into traditional categories such as “replication first” or “metabolism first”. The model also assumes that evolution of primordial systems was driven by natural selection from the very start of life, and long before the emergence of template-based replication of polymers.
For comparison I will consider an alternative model of the origin of life in inorganic compartments within hydrothermal vents (Koonin, 2009; Koonin and Martin, 2005). Authors correctly reasoned that biochemical processes could have hardly evolved in free solution; but instead of considering surface metabolism they selected another option – inorganic compartments. Indeed, compartments prevent free diffusion and can keep molecules together. In the presence of catalysts (e.g., FeS), simple organic molecules (e.g., formaldehyde) can become concentrated within compartments. However, there is no evidence that more complex organic molecules can be generated in sufficient amounts in such systems. Even the formose reaction is hardly possible due to the low concentration of formaldehyde. The model further assumes the spontaneous emergence of self-replicating nucleic acids and even protein synthesis. As we argued in sections 1–2, self-replicating polymers were unlikely to emerge in evolution before the development of heritable pathways for producing monomers. Also, it is questionable if cooperation between different kinds of replicons could emerge in such systems. Isolated compartments do not allow the spread of replicons, whereas connected compartments cannot support group selection. In contrast to inorganic compartments with mostly static connections (pores), oil droplets provide a highly dynamic habitat for primordial life: they are fully isolated, and yet in certain conditions they may either coalesce or split into smaller droplets. This dynamic patchy environment supports both, propagation in space and group selection.
The origin of life is a very long multi-step process, which cannot be replicated in laboratory conditions. Thus, experimental studies should be focused on individual steps in this process and our expectations for the results of such experiments should be set lower. It would never be possible to mix inorganic chemicals and generate functional cells with a membrane, self-supporting metabolism, and nucleic acids as hereditary molecules. Experiments in artificially enriched medium with abundant complex organic molecules such as aminoacids, peptides, sugars, or phospholipids will not take us any closer to the understanding of the origin of life because these kinds of synthetic molecules have never been available in sufficient quantities in nature. More meaningful results can be expected from experiments that use nutrient-poor media, such as hydrocarbons in water supplemented with rare additional molecules that may become assembled into catalytically active units.
Analysis of the rates of increase in genomic complexity indicates that the doubling time of the non-redundant and functional fraction of the genome is about 340 ± 100 (c.i.)5 million years (Sharov, 2006). Similar estimates (360 million years) were obtained independently by Markov et al. (2010). Based on this rate, the evolution from the origin of life to the level of bacteria-like organisms would require from 4 to 8 billion years. Considering that bacteria-like organisms flourished on Earth as early as 3.48 By ago (Noffke et al., 2013), and the age of earth is only 4.5 By, it is likely that life originated on a planet orbiting another star and then was transferred to earth with comets, asteroids, or rogue planets. The cosmic journey of life may have been shorter if the Solar System originated from the remnants of an exploded parental star that had life on its planets. In this case, LUCA could represent the genome of organisms (possibly from multiple lineages) that arrived to the primordial earth. This hypothesis is further supported by the facts that there are no traces of pre-cellular life on earth6, no RNA-world organisms without protein synthesis, no alternative nucleotide composition, no alternative aminoacid composition, no alternative genetic code (minor modifications likely appeared after arrival to earth), and no free-living organisms with a genome <0.5 Mbp.
An alternative view is that primordial evolution was qualitatively different from subsequent evolution, and thus, extrapolations of the rates of complexity increase are not valid. In particular, it was suggested that the primordial evolution was unusually fast due to the lack of competition (Koonin and Galperin, 2003). However, primordial competition may have been very intensive because of the scarcity of organic resources; and there is no consensus on the inverse relationship between competition and rates of complexity increase. For example, competition was shown to increase the diversity and facilitate the emergence of new taxa in animals (Stanley 1973). Carl Woese proposed another hypothesis that primordial organisms evolved via non-Darwinian process that was much faster than regular evolution because of unusually intensive horizontal gene transfer (HGT) (Woese, 2002). However, historical rates of HGT are measured indirectly based on mathematical models of the evolutionary process and are highly sensitive to model parameters (Koonin and Galperin, 2003: p. 241–242). There is no direct evidence that HGT rates >3.5 By ago were much higher than subsequent HGT rates in bacterial lineages. Moreover, HGT is substantially more frequent in bacteria than in eukaryotes, and yet the rates of complexity increase are lower in bacteria than in eukaryotes (Sharov, 2006). Thus, there seem to be no positive association between HGT frequency and the rates of increase in genome complexity. In summary, these hypotheses failed to explain the origin of life on earth.
The most important novel component of the proposed model is that the evolvability of primordial systems was largely based on cooperation of molecules, which is the establishment of mutually beneficial relations between functional molecules colonizing the same local environment such as droplet of oil. Some relations became heritable if they reinforced each other, and their cooperative nature was supported by group selection. Thus, life originated from simple (not polymeric) but already functional molecules, and its gradual evolution towards higher complexity was driven by cooperation and natural selection.
Highlights.
The proposed scenario assumes that life originated from simple catalytically active self-reproducing molecules, similar to coenzymes, that played the role of hereditary signs.
These molecules colonized the surface of oil droplets in water and changed surface properties of droplets to enable and enhance self-reproduction of these molecules.
Evolvability of primordial systems was based on cooperation of multiple kinds of self-reproducing molecules and establishment of self-sustained relationships between them.
Transition to the RNA-world systems included polymerization of coenzyme-like molecules, emergence of template-based replication, transformation of oil droplets into the outer membrane of cells via engulfing water, transition of metabolism from the surface to the inner space of the cell, and emergence of protein synthesis.
Acknowledgments
This project was supported entirely by the Intramural Research Program of the National Institute on Aging (NIA/NIH), project Z01 AG000656-13. Funding organization had no involvement in this study.
Footnotes
Stability can be represented mathematically by an attractor in a model of branching dynamical systems, i.e., a quasispecies (Eigen and Schuster, 1979). However, any mathematical model is only an approximation, and thus, it never fully captures all details that remain conserved across generations. Hence, the notion of ‘sameness’ is not defined mathematically; instead it expands in content as we learn more about living organisms. For example, in the past biologists did not believe in epigenetic heredity and the notion of ‘sameness’ was restricted to genotypes or haplotypes.
For example, threose nucleic acid (TNA) or peptide nucleic acid (PNA).
I use the term “replication” only for template-guided synthesis (as in nucleic acids); in other cases I use the term “self-reproduction” or “reproduction”.
Rare molecules of lipids in meteorites do not count because their concentrations are very low; also they may be artifacts of heating during meteorite entry into the atmosphere.
Confidence interval was determined by sensitivity analysis.
Viruses likely arrived to earth together with their bacterial hosts. Thus, we cannot consider them as evidence of pre-cellular life.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam G, Delbrück M. Reduction of dimensionality in biological diffusion processes. In: Davidson ARaN., editor. Structural Chemistry and Molecular Biology. W.H. Freeman; New York: 1968. pp. 198–215. [Google Scholar]
- Benner SA, Kim HJ, Carrigan MA. Asphalt, water, and the prebiotic synthesis of ribose, ribonucleosides, and RNA. Acc Chem Res. 2012;45:2025–2034. doi: 10.1021/ar200332w. [DOI] [PubMed] [Google Scholar]
- Bernhardt HS. The RNA world hypothesis: the worst theory of the early evolution of life (except for all the others) Biol Direct. 2012;7:23. doi: 10.1186/1745-6150-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickhard MH. Functional scaffolding and self-scaffolding. New Ideas in Psychology. 2005;23:166–173. [Google Scholar]
- Biver N, Bockelee-Morvan D, Moreno R, Crovisier J, Colom P, Lis DC, Sandqvist A, Boissier J, Despois D, Milam SN. Ethyl alcohol and sugar in comet C/2014 Q2 (Lovejoy) Sci Adv. 2015;1:e1500863. doi: 10.1126/sciadv.1500863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anolles G, Seufferheld MJ. The coevolutionary roots of biochemistry and cellular organization challenge the RNA world paradigm. J Mol Microbiol Biotechnol. 2013;23:152–177. doi: 10.1159/000346551. [DOI] [PubMed] [Google Scholar]
- Cairns-Smith AG. Genetic takeover and the mineral origins of life. Cambridge University Press; Cambridge: 1982. [Google Scholar]
- Callahan MP, Smith KE, Cleaves HJ, 2nd, Ruzicka J, Stern JC, Glavin DP, House CH, Dworkin JP. Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases. Proc Natl Acad Sci U S A. 2011;108:13995–13998. doi: 10.1073/pnas.1106493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomb-Delsuc M, Mattia E, Sadownik JW, Otto S. Exponential self-replication enabled through a fibre elongation/breakage mechanism. Nat Commun. 2015;6:7427. doi: 10.1038/ncomms8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M. Bootstrapping model of the origin of life. Biosystems. 1982;15:209–219. doi: 10.1016/0303-2647(82)90006-5. [DOI] [PubMed] [Google Scholar]
- Cooper G, Kimmich N, Belisle W, Sarinana J, Brabham K, Garrel L. Carbonaceous meteorites as a source of sugar-related organic compounds for the early Earth. Nature. 2001;414:879–883. doi: 10.1038/414879a. [DOI] [PubMed] [Google Scholar]
- Deacon TW. Reciprocal linkage between self-organizing processes is sufficient for self-reproduction and evolvability. Biological Theory. 2006;1:136–149. [Google Scholar]
- Deacon TW. Incomplete nature: How mind emerged from matter. W. W. Norton and Company; New York: 2011. [Google Scholar]
- Deacon TW, Srivastava A, Bacigalupi JA. The transition from constraint to regulation at the origin of life. Front Biosci (Landmark Ed) 2014;19:945–957. doi: 10.2741/4259. [DOI] [PubMed] [Google Scholar]
- Deamer D. First life: Discovering the connections between stars, cells, and how life began. University of California Press; Berkley, CA: 2011. [Google Scholar]
- Deamer DW. How did it all begin? The self-assembly of organic molecules and the origin of cellular life. In: Scotchmoor J, Springer DA, editors. Evolution: Investigating the Evidence. Paleontological Society; Pittsburgh, PA: 1999. [Google Scholar]
- Doolittle WF. Uprooting the tree of life. Sci Am. 2000;282:90–95. doi: 10.1038/scientificamerican0200-90. [DOI] [PubMed] [Google Scholar]
- Dutta A, DuBois DL, Roberts JA, Shaw WJ. Amino acid modified Ni catalyst exhibits reversible H2 oxidation/production over a broad pH range at elevated temperatures. Proc Natl Acad Sci U S A. 2014;111:16286–16291. doi: 10.1073/pnas.1416381111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R. Primal eukaryogenesis: on the communal nature of precellular States, ancestral to modern life. Life (Basel) 2012;2:170–212. doi: 10.3390/life2010170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M, Schuster P. The hypercycle, a principle of natural self-organization. Springer-Verlag; Berlin, New York: 1979. [DOI] [PubMed] [Google Scholar]
- Gilbert W. The RNA world. Nature. 1986;319:618. [Google Scholar]
- Gray P, Scott SK. Chemical oscillations and instabilities: Non-linear chemical kinetics. Oxford University Press; New York: 1994. [Google Scholar]
- Griffith JS. Self-replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- Gruzewska K, Michno A, Pawelczyk T, Bielarczyk H. Essentiality and toxicity of vanadium supplements in health and pathology. J Physiol Pharmacol. 2014;65:603–611. [PubMed] [Google Scholar]
- Guerrero R, Margulis L, Berlanga M. Symbiogenesis: the holobiont as a unit of evolution. Int Microbiol. 2013;16:133–143. doi: 10.2436/20.1501.01.188. [DOI] [PubMed] [Google Scholar]
- Hartman H. Photosynthesis and the origin of life. Orig Life Evol Biosph. 1998;28:515–521. doi: 10.1023/a:1006548904157. [DOI] [PubMed] [Google Scholar]
- Hoffmeyer J. Surfaces inside surfaces. On the origin of agency and life. Cybernetics and Human Knowing. 1998;5:33–42. [Google Scholar]
- Hoffmeyer J. Biosemiotics: An examination into the signs of life and the life of signs. University of Scranton Press; Scranton, PA: 2008. [Google Scholar]
- Holliday GL, Mitchell JB, Thornton JM. Understanding the functional roles of amino acid residues in enzyme catalysis. J Mol Biol. 2009;390:560–577. doi: 10.1016/j.jmb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Ikehara K. Evolutionary steps in the emergence of life deduced from the bottom-up approach and GADV hypothesis (top-down approach) Life (Basel) 2016;6:6. doi: 10.3390/life6010006. doi:10.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E, Szathmáry E. The evolution of information storage and heredity. Trends in Ecology and Evolution. 1995;10:206–211. doi: 10.1016/s0169-5347(00)89060-6. [DOI] [PubMed] [Google Scholar]
- Kauffman SA. Autocatalytic sets of proteins. J Theor Biol. 1986;119:1–24. doi: 10.1016/s0022-5193(86)80047-9. [DOI] [PubMed] [Google Scholar]
- Kauffman SA. Prolegomenon to patterns in evolution. Biosystems. 2014;123:3–8. doi: 10.1016/j.biosystems.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- Koonin EV. On the origin of cells and viruses: primordial virus world scenario. Ann N Y Acad Sci. 2009;1178:47–64. doi: 10.1111/j.1749-6632.2009.04992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Galperin MY. Sequence - evolution - function : computational approaches in comparative genomics. Kluwer Academic; Boston: 2003. [PubMed] [Google Scholar]
- Koonin EV, Martin W. On the origin of genomes and cells within inorganic compartments. Trends Genet. 2005;21:647–654. doi: 10.1016/j.tig.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummerle R, Kyritsis P, Gaillard J, Moulis JM. Electron transfer properties of iron-sulfur proteins. J Inorg Biochem. 2000;79:83–91. doi: 10.1016/s0162-0134(99)00160-9. [DOI] [PubMed] [Google Scholar]
- Lane N, Allen JF, Martin W. How did LUCA make a living? Chemiosmosis in the origin of life. Bioessays. 2010;32:271–280. doi: 10.1002/bies.200900131. [DOI] [PubMed] [Google Scholar]
- Laurent M. Autocatalytic processes in cooperative mechanisms of prion diseases. FEBS Lett. 1997;407:1–6. doi: 10.1016/s0014-5793(97)00310-4. [DOI] [PubMed] [Google Scholar]
- Marcano V, Benitez P, Palacios-Pru E. Acyclic hydrocarbon environments >=n-C18 on the early terrestrial planets. Planet Space Sci. 2003;51:159–166. [Google Scholar]
- Markov AV, Anisimov VA, Korotaev AV. Relationship between the genome size and organismal complexity in the lineage leading from prokaryotes to mammals [in Russian] aleontological Journal. 2010;(4):1–12. [Google Scholar]
- Maturana H, Varela F. Autopoiesis and cognition: The realization of the living. D. Reidel Publishing Co; Dordecht: 1980. [Google Scholar]
- Maury CP. Self-propagating beta-sheet polypeptide structures as prebiotic informational molecular entities: the amyloid world. Orig Life Evol Biosph. 2009;39:141–150. doi: 10.1007/s11084-009-9165-6. [DOI] [PubMed] [Google Scholar]
- Maury CP. Origin of life. Primordial genetics: Information transfer in a pre-RNA world based on self-replicating beta-sheet amyloid conformers. J Theor Biol. 2015;382:292–297. doi: 10.1016/j.jtbi.2015.07.008. [DOI] [PubMed] [Google Scholar]
- Morowitz HJ, Kostelnik JD, Yang J, Cody GD. The origin of intermediary metabolism. Proc Natl Acad Sci U S A. 2000;97:7704–7708. doi: 10.1073/pnas.110153997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY, Galperin MY. Physico-chemical and evolutionary constraints for the formation and selection of first biopolymers: towards the consensus paradigm of the abiogenic origin of life. Chem Biodivers. 2007;4:2003–2015. doi: 10.1002/cbdv.200790167. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Levy M, Miller SL. Peptide nucleic acids rather than RNA may have been the first genetic molecule. Proc Natl Acad Sci U S A. 2000;97:3868–3871. doi: 10.1073/pnas.97.8.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghe P, Hordijk W, Kauffman SA, Walker SI, Schmidt FJ, Kemble H, Yeates JA, Lehman N. Prebiotic network evolution: six key parameters. Mol Biosyst. 2015;11:3206–3217. doi: 10.1039/c5mb00593k. [DOI] [PubMed] [Google Scholar]
- Noffke N, Christian D, Wacey D, Hazen RM. Microbially induced sedimentary structures recording an ancient ecosystem in the ca. 3.48 billion-year-old Dresser Formation, Pilbara, Western Australia. Astrobiology. 2013;13:1103–1124. doi: 10.1089/ast.2013.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novozhilov AS, Wolf YI, Koonin EV. Evolution of the genetic code: partial optimization of a random code for robustness to translation error in a rugged fitness landscape. Biol Direct. 2007;2:24. doi: 10.1186/1745-6150-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparin AI. The origin of life. Dover Publications; Mineola, NY: 1953 [1936]. [Google Scholar]
- Orgel L. Origin of life. A simpler nucleic acid. Science. 2000;290:1306–1307. doi: 10.1126/science.290.5495.1306. [DOI] [PubMed] [Google Scholar]
- Orgel LE. Prebiotic chemistry and the origin of the RNA world. Crit Rev Biochem Mol Biol. 2004;39:99–123. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- Patel A. The triplet genetic code had a doublet predecessor. J Theor Biol. 2005;233:527–532. doi: 10.1016/j.jtbi.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Powner MW, Gerland B, Sutherland JD. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature. 2009;459:239–242. doi: 10.1038/nature08013. [DOI] [PubMed] [Google Scholar]
- Robertson MP, Joyce GF. The origins of the RNA world. Cold Spring Harb Perspect Biol. 2012;4:a003608. doi: 10.1101/cshperspect.a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root-Bernstein M, Root-Bernstein R. The ribosome as a missing link in the evolution of life. J Theor Biol. 2015;367:130–158. doi: 10.1016/j.jtbi.2014.11.025. [DOI] [PubMed] [Google Scholar]
- Sebeok TA. Perspectives in zoosemiotics. Mouton de Gruyter; The Hague: 1972. [Google Scholar]
- Sebeok TA. Biosemiotics: its roots, proliferation and prospects. Semiotica. 2001;134:61–78. [Google Scholar]
- Segre D, Ben-Eli D, Deamer DW, Lancet D. The lipid world. Orig Life Evol Biosph. 2001;31:119–145. doi: 10.1023/a:1006746807104. [DOI] [PubMed] [Google Scholar]
- Segre D, Lancet D, Kedem O, Pilpel Y. Graded autocatalysis replication domain (GARD): kinetic analysis of self-replication in mutually catalytic sets. Orig Life Evol Biosph. 1998;28:501–514. [PubMed] [Google Scholar]
- Sharov A, Maran T, Tønnessen M. Towards synthesis of biology and semiotics. Biosemiotics. 2015;8:1–7. [Google Scholar]
- Sharov AA. Self-reproducing systems: structure, niche relations and evolution. Biosystems. 1991;25:237–249. doi: 10.1016/0303-2647(91)90022-d. [DOI] [PubMed] [Google Scholar]
- Sharov AA. Genome increase as a clock for the origin and evolution of life. Biol Direct. 2006;1:17. doi: 10.1186/1745-6150-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA. Coenzyme autocatalytic network on the surface of oil microspheres as a model for the origin of life. Int J Mol Sci. 2009;10:1838–1852. doi: 10.3390/ijms10041838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA. Evolutionary constraints or opportunities? Biosystems. 2014;123:9–18. doi: 10.1016/j.biosystems.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Sharov AA, Vehkavaara T. Protosemiosis: agency with reduced representation capacity. Biosemiotics. 2015;8:103–123. doi: 10.1007/s12304-014-9219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponer JE, Sponer J, Novakova O, Brabec V, Sedo O, Zdrahal Z, Costanzo G, Pino S, Saladino R, Di Mauro E. Emergence of the first catalytic oligonucleotides in a formamide-based origin scenario. Chemistry. 2016 Jan 25; doi: 10.1002/chem.201503906. doi:10.1002. [DOI] [PubMed] [Google Scholar]
- Stanley MS. Effects of competition on rates of evolution, with special reference to bivalve mollusks and mammals. Syst Zool. 1973;22:486–506. [Google Scholar]
- Steele A, Baross J. Are prions relevant to astrobiology? Astrobiology. 2006;6:284. [Google Scholar]
- Szathmáry E. The first replicators. In: Keller L, editor. Levels of selection in evolution. Princeton University Press; Princeton: 1999. pp. 31–52. [Google Scholar]
- Tessera M. Origin of evolution versus origin of life: a shift of paradigm. Int J Mol Sci. 2011;12:3445–3458. doi: 10.3390/ijms12063445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL. A formal test of the theory of universal common ancestry. Nature. 2010;465:219–222. doi: 10.1038/nature09014. [DOI] [PubMed] [Google Scholar]
- Tilman D, Kareiva P. Spatial Ecology: The Role of Space in Population Dynamics and Interspecific Interactions. Princeton University Press; Princeton, NJ: 1997. [Google Scholar]
- Travers A. The evolution of the genetic code revisited. Orig Life Evol Biosph. 2006;36:549–555. doi: 10.1007/s11084-006-9041-6. [DOI] [PubMed] [Google Scholar]
- Uexküll Jv. The theory of meaning. Semiotica. 1982;42:25–82. [Google Scholar]
- Unrau PJ, Bartel DP. RNA-catalysed nucleotide synthesis. Nature. 1998;395:260–263. doi: 10.1038/26193. [DOI] [PubMed] [Google Scholar]
- Wächtershäuser G. Before enzymes and templates: theory of surface metabolism. Microbiol Rev. 1988;52:452–484. doi: 10.1128/mr.52.4.452-484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber BH. Emergence of life. Zygon. 2007;42:837–856. [Google Scholar]
- Weronski P, Jiang Y, Rasmussen S. Molecular dynamics study of small PNA molecules in lipid-water system. Biophys J. 2007;92:3081–3091. doi: 10.1529/biophysj.106.097352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. On the evolution of cells. Proc Natl Acad Sci U S A. 2002;99:8742–8747. doi: 10.1073/pnas.132266999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubarev DY, Rappoport D, Aspuru-Guzik A. Uncertainty of prebiotic scenarios: the case of the non-enzymatic reverse tricarboxylic acid cycle. Sci Rep. 2015;5:8009. doi: 10.1038/srep08009. [DOI] [PMC free article] [PubMed] [Google Scholar]