Summary
We identify inherited genetic variants associated with telomere length that may also confer risk for childhood cancers. Analyses reveal that genetic predisposition to longer telomere length increased risk of neuroblastoma, and potentially risk of osteosarcoma and acute lymphoblastic leukemia.
Abstract
Aberrant telomere lengthening is an important feature of cancer cells in adults and children. In addition to somatic mutations, germline polymorphisms in telomere maintenance genes impact telomere length. Whether these telomere-associated polymorphisms affect risk of childhood malignancies remains largely unexplored. We collected genome-wide data from three groups with pediatric malignancies [neuroblastoma (N = 1516), acute lymphoblastic leukemia (ALL) (N = 958) and osteosarcoma (N = 660)] and three control populations (N = 6892). Using case–control comparisons, we analyzed eight single nucleotide polymorphisms (SNPs) in genes definitively associated with interindividual variation in leukocyte telomere length (LTL) in prior genome-wide association studies: ACYP2, TERC, NAF1, TERT, OBFC1, CTC1, ZNF208 and RTEL1. Six of these SNPs were associated (P < 0.05) with neuroblastoma risk, one with leukemia risk and one with osteosarcoma risk. The allele associated with longer LTL increased cancer risk for all these significantly associated SNPs. Using a weighted linear combination of the eight LTL-associated SNPs, we observed that neuroblastoma patients were predisposed to longer LTL than controls, with each standard deviation increase in genotypically estimated LTL associated with a 1.15-fold increased odds of neuroblastoma (95%CI = 1.09–1.22; P = 7.9×10−7). This effect was more pronounced in adolescent-onset neuroblastoma patients (OR = 1.46; 95%CI = 1.03–2.08). A one standard deviation increase in genotypically estimated LTL was more weakly associated with osteosarcoma risk (OR = 1.10; 95%CI = 1.01–1.19; P = 0.017) and leukemia risk (OR = 1.07; 95%CI = 1.00–1.14; P = 0.044), specifically for leukemia patients who relapsed (OR = 1.19; 95%CI = 1.01–1.40; P = 0.043). These results indicate that genetic predisposition to longer LTL is a newly identified risk factor for neuroblastoma and potentially for other cancers of childhood.
Introduction
The etiology of cancer in children and adolescents is largely unknown, although recent reports suggest that 5–10% of cases may result from an underlying germline mutation in a gene linked to autosomal dominant cancer predisposition syndromes (1). Carcinogenesis in the remaining patients is believed to result from the combination of low-penetrance risk alleles, environmental factors and stochastic processes (2). Genome-wide association studies (GWAS) have identified a number of low-penetrance risk alleles that are associated with four of the most common cancers of childhood: acute lymphoblastic leukemia (ALL) (3), osteosarcoma (4), Wilms tumor (5) and neuroblastoma (6). Furthermore, the combination of targeted fine mapping and resequencing, combined with functional genomics approaches, have successfully identified the causative alleles underlying several of the associations observed in GWAS of ALL and neuroblastoma (7–9).
GWAS of adult cancers have shown that inherited single nucleotide polymorphisms (SNPs) in telomere-related genes are associated with numerous malignancies (10–13). Human telomeres, composed of a tandem hexanucleotide repeat (TTAGGG), are many kilobases long in newborns but shorten an average of 20–40 base-pairs annually (14,15). Very large GWAS analyses have identified eight genes that are reproducibly associated with interindividual variation in leukocyte telomere length (LTL), including SNPs in: ACYP2, TERC, NAF1, TERT, OBFC1, CTC1, ZNF208 and RTEL1 (16,17). Recent Mendelian randomization studies indicate that these LTL-associated SNPs influence adult cancer risk, with genetic predisposition to longer telomere length associated with increased risk of adult glioma, melanoma and lung cancer (18–20).
Telomeres are repetitive DNA sequences that cap and protect chromosomes. These repeats are lost with each somatic cellular division. When a chromosome’s telomeres become depleted, future mitoses will result in the loss of integral genomic DNA, inducing replicative senescence and apoptosis (21). Telomere attrition helps to prevent cancer by limiting the replicative potential of somatic cells. However, if adequate oncogenic mutations are acquired before reaching replicative senescence, unlimited proliferation may result. Thus, long telomeres may increase the risk of cancer by allowing more time and more divisions in which a cell can accumulate mutations before it reaches this critical apoptotic checkpoint (22).
Unlike adult cancers, GWAS have not implicated polymorphisms in telomere-maintenance genes in pediatric cancer predisposition (23). While it is possible that telomere biology does not play a significant role in pediatric cancer predisposition, it is also possible that the effect sizes are smaller than those observed in adult cancers and, therefore, are less amenable to detection by GWAS. Because GWAS of pediatric cancers typically have smaller sample sizes than those of adult cancers, the power to detect risk loci of moderate effect size is additionally limited. Furthermore, real SNP associations may be restricted to specific molecular subtypes of cancer, as previously observed for ALL risk loci (24,25). While the effect of individual alleles in telomere-maintenance genes is likely small, the combined effect of numerous such polymorphisms could potentially be quite large and could help identify the ‘missing heritability’ of pediatric cancers (26). In the context of GWAS, missing heritability refers to the inability of SNP associations to account for the heritability of the disease under study. The source of this missing heritability is a current conundrum in human genetics, but is most often attributed to rare genetic variants, gene × environment interactions, epistasis and very low-penetrance variants that GWAS are underpowered to detect.
To determine whether common genetic variants associated with interindividual variation in telomere length confer risk for cancers of childhood and adolescence, we analyzed eight independent SNPs in patients with ALL, osteosarcoma, neuroblastoma and a shared set of controls (N = 3134 total cases, 6892 controls). These eight SNP have previously been definitively associated with interindividual variation in LTL in a GWAS of 37 684 individuals of European ancestry conducted by the ENGAGE Consortium Telomere Group (16). In addition to single variant analyses, we also constructed a weighted linear combination of subject genotype at the eight SNPs to create a summary score that quantifies genotypic contributions to differences in LTL across patients and controls. Because SNP genotypes are present since birth, case–control comparisons are not confounded by the effect that age, chemotherapy or other factors may have on both telomere length and cancer risk, eliminating the possibility of reverse causation. This Mendelian randomization approach for examining the relationship between telomere length and cancer risk has been previously applied to adult cancers, but not to cancers of childhood (18–20).
Methods
Ethics statement
The genome-wide meta-analysis of mean LTL obtained approval by local ethics committees, as outlined previously (16). All other genomic data were obtained from dbGaP, Illumina’s iControlDB database and the Wellcome Trust Case-Control Consortium following approval from the respective agency and local approval for data security policies and procedures.
Pediatric cancer patients and controls
The ALL Relapse GWAS dataset used for the analyses described in this study were obtained from dbGaP study accession phs000638.v1.p1 (Genome-Wide Association Study of Relapse of Childhood acute lymphoblastic leukemia) and include a total of 958 non-Hispanic white children (age < 21) diagnosed with ALL. Children were recruited under COG protocols P9904 and P9905, as described previously (27,28). The osteosarcoma GWAS dataset used for the analyses described in this study were obtained from dbGaP study accession phs000734.v1.p1 (A Genome-wide Association Study (GWAS) of Risk for Osteosarcoma) and contain 660 non-Hispanic white patients diagnosed with osteosarcoma, as described previously (4). These patients were predominantly children and adolescents (aged < 21), although the dbGaP dataset did not provide individual-level age data and did not restrict to a specific age range. The neuroblastoma GWAS data described in this study were obtained from dbGaP study accession phs000124.v2.p1 (Neuroblastoma Genome-Wide Association Study). Neuroblastoma cases include 1513 non-Hispanic white children and adolescents (age < 19) diagnosed with neuroblastoma, identified through the COG Neuroblastoma Tumor Bank and the Children’s Hospital of Philadelphia.
The Geisinger control GWAS dataset used for the analyses described in this study were obtained from dbGaP study accession phs000381.v1.p1 (eMERGE Geisinger eGenomic Medicine (GeM)—MyCode Project Controls) and includes 1167 non-Hispanic white control subjects provided by the Geisinger MyCode® Project. An additional 3166 non-Hispanic white Illumina iControls were included in analyses, as described previously (18). Control genotype data for a third set of 2559 European-ancestry control samples were obtained from the Wellcome Trust Case–Control Consortium (WTCCC). WTCCC samples have previously been described in detail (29). Although control populations included both children and adults, adjustment for age differences is unnecessary in the gene-based case–control analyses conducted here.
Genotyping and imputation
Genotype data for case and control subjects were downloaded from dbGaP, Illumina’s iControlDB database and The Wellcome Trust Consortium. All subjects were genotyped on a standard commercial genome-wide SNP array, including: the Affymetrix 6.0 array (ALL patients, Wellcome Trust controls), the Illumina OmniExpress array (osteosarcoma patients, Geisinger controls) and the Illumina HumanHap550 (neuroblastoma patients, Illumina iControls). SNP array data for each case set and each control set were cleaned individually using Plink (30). SNPs with call rates < 0.98 were removed from analyses. Following removal of poorly performing SNPs, subjects with genotyping call rates < 0.97 were removed. Within each case and control dataset, ancestry-informative principal components were calculated using Eigenstrat and HapMap reference samples (31). Mean values of the first five principal components were calculated among HapMap CEPH samples. Subjects that fell more than 3 SDs from the mean CEPH values were excluded from further analyses. This successfully removed all subjects that self-identified as non-European in each dataset, and also removed additional subjects showing evidence of non-European ancestry. Following removal of subjects with non-European ancestry, SNPs with Hardy–Weinberg equilibrium P < 0.0001 among controls were removed from the three control datasets. SNP data from the three case groups and three control groups were subsequently merged using 43 664 genotyped SNPs common to all panels to identify and exclude duplicate subjects and cryptically related individuals (proportion of genome identical by descent > 0.20).
Within the three cancer patient datasets and the three control datasets, we imputed 100kb regions centered on eight SNPs previously associated with LTL in GWAS (16,17,32,33): rs11125529 (ACYP2), rs10936599 (TERC), rs7675998 (NAF1), rs2736100 (TERT), rs9420907 (OBFC1), rs3027234 (CTC1), rs8105767 (ZNF208) and rs755017 (RTEL1). Imputation was performed using the Impute2 v2.1.2 software and its standard Markov chain Monte Carlo algorithm and default settings for targeted imputation (34). All 1000 Genomes Phase 3 haplotypes were provided as the imputation reference panel (35). All SNPs had imputation quality (info) scores >0.90 and posterior probabilities >0.95. Individuals with imputed genotype probabilities <0.80 were excluded from analyses to prevent allele misclassification and minimize the effect of SNP imputation on case–control analyses. In the Affymetrix 6.0 datasets (ALL, Wellcome Trust controls), the top LTL SNP was directly genotyped on-array for ACYP2, TERT, OBFC1, CTC1 and ZNF208 and was imputed for the other three genes. In the Illumina OmniExpress datasets (osteosarcoma, Geisinger controls), the top LTL SNP was directly genotyped on-array for TERC, TERT and RTEL1 and was imputed for the remaining genes. In the Illumina HumanHap550 datasets (neuroblastoma, Illumina iControls), the top LTL SNP was directly genotyped on-array for TERC, TERT and OBFC1 and was imputed for the remaining genes.
Statistical analyses
For single locus SNP associations, allele frequencies in cancer patients were compared to those in the pooled control dataset using logistic regression in SNPTESTv2 under an allelic additive model (36), adjusting for the first five ancestry-informative principal components from Eigenstrat. To account for potential errors in imputation, a missing data likelihood score-test was applied to produce standard errors which account for the additional uncertainty inherent in the analysis of imputed genotypes. Additionally, sensitivity analyses were conducted wherein cases were compared only to controls genotyped on the same genotyping platform to account for potential differences in genotyping or imputation across arrays.
To investigate the combined effect of the eight LTL-associated SNPs, we created a weighted linear combination by summing the number of ‘long LTL’ alleles that an individual possesses and weighting each allele by its effect size in data from the ENGAGE Consortium Telomere Group (16). The effect size used for weighting was expressed as the number of additional base-pairs of telomere length associated with each allele, adjusted for age and sex, as calculated and previously reported by the ENGAGE Consortium Telomere Group (16,18). The number of base-pairs was used because the model can be interpreted as the relative difference in estimated LTL across individuals. The weighted model assigns a value of ‘0’ to an individual who possesses 0 of the alleles associated with longer LTL, while an individual possessing all sixteen alleles (two alleles at each of eight SNPs) would have a value of ‘1215’. These differences in genotypically estimated LTL were compared between pediatric cancer patients and controls using logistic regression, adjusted for the first five ancestry-informative principal components from Eigenstrat (31). Odds ratios correspond to the change in cancer risk relative to a one standard deviation (131.8bp) increase in genotypically estimated LTL, with the standard deviation defined among the pooled controls. To control for the family-wise error rate, null hypotheses for the weighted linear combination were tested using the Bonferroni–Holm approach and an alpha of 0.05.
Results
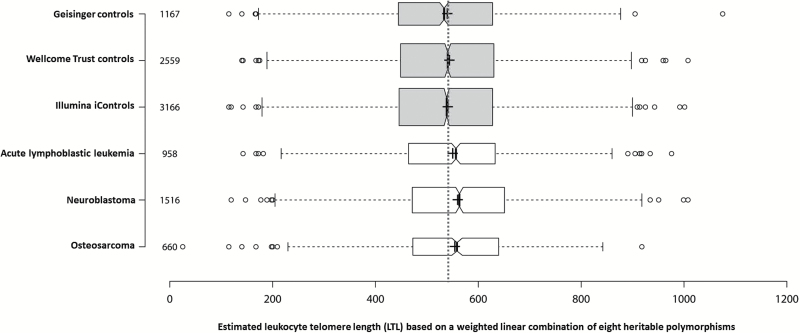
After removing subjects with poor call rates, duplicate samples, cryptically related individuals and subjects with non-European ancestry, a total of 10 026 subjects had complete data for all eight LTL-associated SNPs (3134 cases, 6892 controls). The cancer cases included 958 ALL patients, 1516 neuroblastoma patients and 660 osteosarcoma patients. The 6892 controls included 1167 Geisinger controls, 3166 Illumina iControls and 2559 Wellcome Trust controls. The weighted linear combination of the 8 LTL-associated SNP (16 LTL-associated alleles) ranged from a minimum value of 115bp to a maximum value of 1075bp in controls, representing a 960bp range in estimated LTL across individuals. Because the weighted sum estimates LTL differences using unlinked autosomal SNPs, present since birth, there was no association between the weighted sum and either subject age or sex among controls. The mean value of the weighted sum was similar in all three control groups (Geisinger controls = 540bp, Illumina iControls=541bp, Wellcome Trust controls = 544bp).
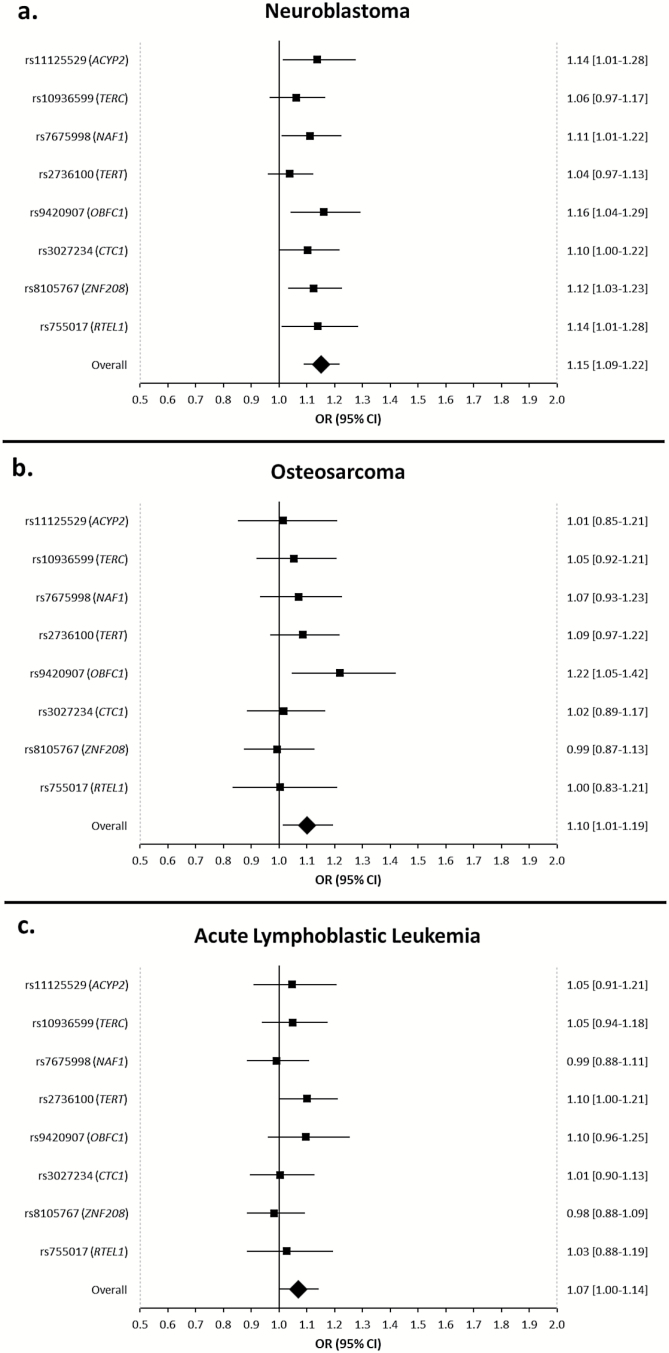
When comparing SNP genotypes in neuroblastoma cases and controls, the allele associated with longer LTL was associated with increased neuroblastoma risk for all eight SNPs (Figure 1a). For six of the eight SNPs, this association had uncorrected P < 0.05, including: rs11125529 in ACYP2 (OR = 1.14; 95%CI = 1.01–1.28; P = 0.028), rs7675998 in NAF1 (OR = 1.11; 95%CI = 1.01–1.22; P = 0.033), rs9420907 in OBFC1 (OR = 1.16; 95%CI = 1.04–1.29; P = 0.0066), rs3027234 in CTC1 (OR = 1.10; 95%CI = 1.00– 1.22; P = 0.049), rs8105767 in ZNF208 (OR = 1.13; 95%CI = 1.03–1.23; P = 0.0071) and rs755017 in RTEL1 (OR = 1.14; 95%CI = 1.01–1.28; P = 0.034).
Figure 1.
Forest plot showing the effect of alleles previously associated with longer leukocyte telomere length on risk of (A) neuroblastoma (N = 1516), (B) osteosarcoma (N = 660) and (C) acute lymphoblastic leukemia (N = 958) compared with 6892 controls. Odds ratios > 1.0 indicate that the allele associated with longer telomere length is associated with an increased risk of cancer. Allelic odds ratios are plotted with 95% confidence intervals. The overall estimate is for the combined effect of all eight SNPs, where the odds ratio relates to the change in cancer risk for a one standard deviation increase in genotypically estimated leukocyte telomere length, with standard deviation determined in the controls.
The weighted linear combination of the 8 LTL-associated SNPs was significantly greater in neuroblastoma cases than controls (560 versus 542bp, Figure 2) after adjusting for multiple comparisons using the Bonferroni–Holm procedure (P < 0.05/3). Each standard deviation increase in genotypically estimated LTL (131.8bp) was associated with a 1.15-fold increase in the odds of neuroblastoma (95%CI = 1.09–1.22; P = 7.9×10−7) (Figure 1a). Results were similar when neuroblastoma cases were exclusively compared to Illumina iControls, also genotyped on the Illumina HumanHap550 platform (OR = 1.16; 95%CI = 1.09–1.23).
Figure 2.
Boxplots of genotypically estimated leukocyte telomere length (LTL) in three pediatric cancer datasets (acute lymphoblastic leukemia, neuroblastoma and osteosarcoma) and three control datasets. The vertical dotted line shows the average value in the pooled control sets (542bp). Crosses show sample means; center lines show sample medians. Box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquartile range (IQR) from the 25th and 75th percentiles. Outliers are represented by dots. Height of the boxes is proportional to the square root of the sample size. Sample sizes in each dataset are listed on the left. The notches in boxes are defined as ±1.58*IQR/sqrt(n) and give roughly 95% confidence intervals for sample medians. Estimated LTL was significantly longer in leukemia patients (P = 0.044), neuroblastoma patients (7.9×10−7) and osteosarcoma patients (P = 0.017) than in combined control samples.
Although the effect size of each individual SNP was small in neuroblastoma case–control comparisons (allelic ORs < 1.20), the combined effect of all 8 SNPs was substantially larger (Figure 1a). Dividing the weighted linear combination into quintiles, individuals in the fifth quintile of genotypically estimated LTL (>653bp) had a 1.50-fold increased risk of neuroblastoma compared to individuals in the first quintile (<432bp) (95%CI = 1.25–1.80; P = 1.43×10−5). Risk of neuroblastoma increased with each quintile in an approximately linear fashion (r = 0.94; P trend = 0.017).
In case-only analyses, the weighted sum did not differ when patients were stratified by ploidy (diploid versus hyperdiploid), MYCN amplification, International Neuroblastoma Staging System tumor stage (1, 2a, 2b, 3, 4, 4s) or Shimada Histology Index (favorable versus unfavorable), but did differ according to age at diagnosis (P trend = 0.024). When case-control comparisons were stratified according to the patient’s age at diagnosis (<18 months, 18 months–12 years, 12–19 years), the effect of each standard deviation increase in genotypically estimated LTL was largest in the adolescent group (OR = 1.46; 95%CI = 1.03–2.08), followed by children (OR = 1.15; 95%CI = 1.07–1.24), then infants (OR = 1.13; 1.05–1.23).
In analyses of osteosarcoma patients and controls, one of the eight SNPs was associated with osteosarcoma risk at P < 0.05: rs9420907 in OBFC1 (OR = 1.22; 95%CI = 1.05–1.42; P = 0.0029). For seven of the eight SNPs, including rs9420907, the allele associated with longer LTL was associated with increased osteosarcoma risk (Figure 1b). The weighted linear combination of the eight SNPs was significantly greater in osteosarcoma cases than controls (554 versus 542bp, Figure 2) after adjusting for multiple comparisons using the Bonferroni–Holm procedure (P < 0.05/2). Each 1 standard deviation increase in genotypically estimated LTL was associated with a 1.10-fold increase in the odds of osteosarcoma (95%CI = 1.01–1.19; P = 0.017). Results were similar when analyses were limited to patients and controls genotyped on the Illumina OmniExpress (OR = 1.09; 95%CI = 0.99–1.21).
In case–control analyses of ALL risk, the allele associated with longer LTL was associated with increased ALL risk at six of the eight SNPs that were modeled. With the exception of rs2736100 in TERT (OR = 1.10; 95%CI = 1.00–1.21; P = 0.047), these associations has uncorrected P > 0.05 (Figure 1c). When the effect of all eight LTL-associated SNPs were combined into a single summary variable, the weighted linear combination was significantly greater in ALL cases than controls (550 versus 542bp, Figure 2) after adjusting for multiple comparisons using the Bonferroni–Holm procedure (P < 0.05/1). One standard deviation increase in genotypically estimated LTL was associated with a 1.07-fold increase in the odds of ALL (95%CI = 1.00–1.14; P = 0.044). Results were similar when analyses were limited to patients and controls genotyped on the Affymetrix 6.0 platform (OR = 1.07; 95%CI = 0.98–1.15). The effect of the weighted linear combination was stronger in the 15% of ALL patients that experienced a relapse (OR = 1.19; 95%CI = 1.01–1.40; P = 0.043), suggesting that longer LTL may be a risk factor for more refractory ALL subtypes. Unfortunately, molecular subtype data were unavailable for these analyses.
Discussion
Our results indicate that a genetic predisposition to longer telomere length may be a risk factor for several pediatric cancers, including ALL, osteosarcoma and neuroblastoma. The effect appears strongest in neuroblastoma, where the allele associated with longer telomeres was more common in neuroblastoma patients than in control subjects for all eight SNPs analyzed. Six of these eight SNPs were associated with neuroblastoma risk at P < 0.05, suggesting that many genetic loci of relatively weak effect may combine to impact telomere biology and neuroblastoma risk. The two SNPs that were not associated with neuroblastoma risk at P < 0.05 are located in TERT and TERC, genes which encode the two components of telomerase. Telomerase, an enzyme capable of synthesizing telomeric repeats onto the ends of the chromosomes, can counteract replicative telomere attrition. Heritable genetic variation in TERT and TERC is strongly associated with telomere length in adults, but it is unknown whether it impacts telomere length in children. Although telomere length in healthy tissues is known to differ between newborns (14), future work is needed to understand the role of inherited genetic variation in establishing telomere length at baseline versus maintaining telomere length throughout life.
The issue of ‘missing heritability’ is often raised in discussions of GWAS approaches for cancer research (26), referring to the inability of SNP associations to account for the heritability of the disease under study. A potential source of this missing heritability is very low-penetrance variants that GWAS are underpowered to detect. We observe strong evidence that low-penetrance variants in telomere-related genes contribute to neuroblastoma risk. In case–control analyses of neuroblastoma, all allelic odds ratios were <1.20 and P values ranged from 0.0066 to 0.26. However, when all eight SNPs were combined into a single summary variable, this variable was much more significantly associated with neuroblastoma risk (P = 7.9×10−7). This reflects a major strength of our approach: the ability to combine numerous SNPs into a single variable that directly informs on the biology of the disease. Intriguingly, a recent study of germline mutations in pediatric cancer revealed that patients with neuroblastoma had the lowest prevalence of high-penetrance germline mutations (4.0%) (1), suggesting that common low-penetrance variants, such as SNPs in telomere-related genes, may underlie a large proportion of neuroblastoma cases.
Mendelian randomization is an epidemiologic technique wherein genetic variants known to influence an exposure of interest (e.g. LTL) are used as surrogate markers to investigate the effect of that exposure on a disease (37). Although each of the LTL-associated variants explains only a small proportion of the total variance in telomere length across individuals (16), a summary variable made by combining the eight SNPs accounted for a 960-bp difference in LTL. Assuming an annual telomere attrition rate of ~30bp/year in leukocytes, this translates to more than 30 years of age-related telomere attrition (15). Although Mendelian randomization reduces confounding and bias, study results can be influenced by linkage disequilibrium, population stratification and pleiotropy (38). Because we analyzed unlinked SNPs on separate autosomes, linkage disequilibrium is unlikely to influence our results. By carefully excluding individuals with non-European ancestry and adjusting for ancestry-informative principal components in all analyses, inflation of test statistics due to population stratification is unlikely. There remains a possibility that pleiotropy could underlie the association between telomere length variants and cancer risk observed in our data. Additional functions of the ACYP2, TERC, NAF1, TERT, OBFC1, CTC1, ZNF208 and RTEL1 genes merit additional study.
We observe a strong association between common genetic variants associated with telomere length and risk for neuroblastoma. This effect was largest in adolescent cases (aged 12–19), but was also significant in infants and children. Although GWAS have not previously implicated SNPs in telomere maintenance genes in predisposition to pediatric cancers, frequent TERT gene rearrangements were recently identified in high-risk neuroblastoma tumor genomes (39,40). These somatic mutations reactivate telomerase, thereby enabling cancer cells to lengthen their telomeres and escape replicative senescence. It will be valuable to investigate whether LTL-associated alleles confer different risk for TERT-mutated versus TERT wild-type neuroblastoma. This has been previously observed in adult glioma (41), suggesting that inherited and acquired variants in telomere-maintenance genes may confer cancer risk synergistically (42).
Both neuroblastoma and osteosarcoma harbor somatic mutations in ATRX, leading to telomere extension through a telomerase-independent mechanism known as alternative lengthening of telomeres (43,44). Unfortunately, valuable molecular data for the ALL and osteosarcoma patients were not available for analysis. However, the observation that genotypically estimated LTL conferred greater risk in ALL patients that experienced a relapse suggests that longer LTL may be a risk factor for more refractory molecular subtypes.
Somatic alterations of TERT and ATRX have not been reported in ALL, although somatic mutation of ACD, a shelterin-complex gene involved in telomere protection, has been observed to lengthen telomeres in childhood leukemia cells (45). These prior observations suggest that telomere biology is important in several malignancies of childhood, in-line with observations from adult cancers. Our analyses support this conclusion by identifying inherited genetic variants associated with longer telomere length that confer risk for neuroblastoma, and potentially, for additional cancers of childhood.
Supplementary material
Supplementary Data can be found at http://carcin.oxfordjournals.org/
Funding
This work was financially supported by The Alex’s Lemonade Stand Foundation (K.M.W., AJd.S., S.S.F.), The Pediatric Brain Tumor Foundation (K.M.W.), NCI R01CA194189 (K.M.W., J.L.W.), NCI R01CA155461 (J.L.W.), NIEHS and EPA P01ES018172 (J.L.W., T.P.W., C.M.), The British Heart Foundation (V.C., N.J.S.) and The Sontag Foundation (K.M.W.). Work at University of Leicester was undertaken under the framework of European Union Framework 7 ENGAGE Project (HEALTH-F4-2007-201413).
Supplementary Material
Acknowledgements
This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113 and 085475. Samples and data in this study were provided by the Geisinger MyCode® Project. Funding for the MyCode® Project was provided by a grant from Commonwealth of Pennsylvania and the Clinic Research Fund of Geisinger Clinic. Funding support for the genotyping of the MyCode® cohort was provided by a Geisinger Clinic operating funds and an award from the Clinic Research Fund. Datasets used for the analyses described in this study were obtained from dbGaP study accession phs000381.v1.p1 [eMERGE Geisinger eGenomic Medicine (GeM) - MyCode Project Controls]. Additional controls were accessed from Illumina’s iControlDB database. The ALL Relapse GWAS dataset used for the analyses described in this manuscript were obtained from dbGaP study accession phs000638.v1.p1 (Genome-Wide Association Study of Relapse of Childhood Acute Lymphoblastic Leukemia). The ALL Relapse GWAS dataset was generated at St. Jude Children’s Research Hospital and by the Children’s Oncology Group, supported by NIH grants CA142665, CA21765, CA158568, CA156449, CA36401, CA98543, CA114766, CA140729 and U01GM92666, Jeffrey Pride Foundation, the National Childhood Cancer Foundation, and by ALSAC. Datasets used for the analyses described in this manuscript were obtained from dbGaP study accession phs000734.v1.p1 [A Genome-wide Association Study (GWAS) of Risk for Osteosarcoma]. Datasets used for the analyses described in this manuscript were obtained from dbGaP study accession phs000124.v2.p1 [Neuroblastoma Genome-Wide Association Study (NBL-GWAS)]. The Neuroblastoma genome-wide association study is made possible through a collaboration between the Children’s Hospital of Philadelphia (CHOP), the University of Pennsylvania and the National Cancer Institute-funded Children’s Oncology Group (COG; U10-CA98543). Cases were identified through the COG Neuroblastoma Tumor Bank, and controls recruited at CHOP. This work was funded by NIH grant R01-CA124709, the Giulio D’Angio Endowed Chair at CHOP, the CHOP Research Endowment, the Alex’s Lemonade Stand Foundation, the Evan Dunbar Foundation, the Rally Foundation, Andrew’s Army Foundation and the Abramson Family Cancer Research Institute.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ALL
acute lymphoblastic leukemia
- GWAS
genome-wide association studies
- LTL
leukocyte telomere length
- SNP
single nucleotide polymorphisms
References
- 1. Zhang J., et al. (2015) Germline mutations in predisposition genes in pediatric cancer. N. Engl. J. Med., 373, 2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tomasetti C., et al. (2015) Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science, 347, 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moriyama T., et al. (2015) Inherited genetic variation in childhood acute lymphoblastic leukemia. Blood, 125, 3988–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Savage S.A., et al. (2013) Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat. Genet., 45, 799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Turnbull C., et al. (2012) A genome-wide association study identifies susceptibility loci for Wilms tumor. Nat. Genet., 44, 681–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bosse K.R., et al. (2016) Advances in the translational genomics of neuroblastoma: from improving risk stratification and revealing novel biology to identifying actionable genomic alterations. Cancer, 122, 20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wiemels J.L., et al. (2015) A functional polymorphism in the CEBPE gene promoter influences acute lymphoblastic leukemia risk through interaction with the hematopoietic transcription factor Ikaros. Leukemia. Accepted article preview 16 September 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walsh K.M., et al. (2015) A heritable missense polymorphism in CDKN2A confers strong risk of childhood acute lymphoblastic leukemia and is preferentially selected during clonal evolution. Cancer Res., 75, 4884–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oldridge D.A., et al. (2015) Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature, 528, 418–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walsh K.M., et al. ; ENGAGE Consortium Telomere Group (2014) Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat. Genet., 46, 731–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Law M.H., et al. ; GenoMEL Consortium; Essen-Heidelberg Investigators; SDH Study Group; Q-MEGA and QTWIN Investigators; AMFS Investigators; ATHENS Melanoma Study Group (2015) Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nat. Genet., 47, 987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Speedy H.E., et al. (2014) A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia. Nat. Genet., 46, 56–60. [DOI] [PubMed] [Google Scholar]
- 13. Mocellin S., et al. (2012) Telomerase reverse transcriptase locus polymorphisms and cancer risk: a field synopsis and meta-analysis. J. Natl. Cancer Inst., 104, 840–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Okuda K., et al. (2002) Telomere length in the newborn. Pediatr. Res., 52, 377–381. [DOI] [PubMed] [Google Scholar]
- 15. Daniali L., et al. (2013) Telomeres shorten at equivalent rates in somatic tissues of adults. Nat. Commun., 4, 1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Codd V., et al. (2013) Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet., 45, 422–7, 427e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mangino M., et al. (2012) Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum. Mol. Genet., 21, 5385–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walsh K.M., et al. ; ENGAGE Consortium Telomere Group (2015) Longer genotypically-estimated leukocyte telomere length is associated with increased adult glioma risk. Oncotarget, 6, 42468–42477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iles M.M., et al. ; AMFS Investigators; IBD investigators; QMEGA and QTWIN Investigators; SDH Study Group; GenoMEL Consortium (2014) The effect on melanoma risk of genes previously associated with telomere length. J. Natl. Cancer Inst., 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang C., et al. ; GECCO and GAME-ON Network: CORECT, DRIVE, ELLIPSE, FOCI, and TRICL (2015) Genetic determinants of telomere length and risk of common cancers: a Mendelian randomization study. Hum. Mol. Genet., 24, 5356–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blackburn E.H. (2011) Walking the walk from genes through telomere maintenance to cancer risk. Cancer Prev. Res. (Phila), 4, 473–475. [DOI] [PubMed] [Google Scholar]
- 22. Walsh K.M., et al. (2013) Genetic variants in telomerase-related genes are associated with an older age at diagnosis in glioma patients: evidence for distinct pathways of gliomagenesis. Neuro. Oncol., 15, 1041–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hindorff L.A., et al. (2015) A catalog of published genome-wide association studies. Available at: www.genome.gov/gwastudies (16 November 2015, date last accessed).
- 24. Walsh K.M., et al. (2013) Novel childhood ALL susceptibility locus BMI1-PIP4K2A is specifically associated with the hyperdiploid subtype. Blood, 121, 4808–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perez-Andreu V., et al. (2013) Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat. Genet., 45, 1494–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zaitlen N., et al. (2012) Heritability in the genome-wide association era. Hum. Genet., 131, 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang J.J., et al. (2012) Genome-wide association study identifies germline polymorphisms associated with relapse of childhood acute lymphoblastic leukemia. Blood, 120, 4197–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang J.J., et al. (2011) Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat. Genet., 43, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. The Wellcome Trust Case Control Consortium. (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature, 447, 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Purcell S., et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Price A.L., et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet., 38, 904–909. [DOI] [PubMed] [Google Scholar]
- 32. Codd V., et al. ; Wellcome Trust Case Control Consortium (2010) Common variants near TERC are associated with mean telomere length. Nat. Genet., 42, 197–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levy D., et al. (2010) Genome-wide association identifies OBFC1 as a locus involved in human leukocyte telomere biology. Proc. Natl. Acad. Sci. USA, 107, 9293–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Howie B.N., et al. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet., 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abecasis G.R., et al. ; International HapMap 3 Consortium (2010) A map of human genome variation from population-scale sequencing. Nature, 467, 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marchini J., et al. (2010) Genotype imputation for genome-wide association studies. Nat. Rev. Genet., 11, 499–511. [DOI] [PubMed] [Google Scholar]
- 37. Smith G.D., et al. (2003) ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol., 32, 1–22. [DOI] [PubMed] [Google Scholar]
- 38. Jansen H., et al. (2014) Mendelian randomization studies in coronary artery disease. Eur. Heart J., 35, 1917–1924. [DOI] [PubMed] [Google Scholar]
- 39. Peifer M., et al. (2015) Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature, 526, 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Valentijn L.J., et al. (2015) TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat. Genet., 47, 1411–1414. [DOI] [PubMed] [Google Scholar]
- 41. Eckel-Passow J.E., et al. (2015) Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N. Engl. J. Med., 372, 2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Walsh K.M., et al. (2015) Telomere maintenance and the etiology of adult glioma. Neuro. Oncol., 17, 1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen X., et al. ; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project (2014) Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep., 7, 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheung N.K., et al. ; St Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project (2012) Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA, 307, 1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spinella J.F., et al. (2015) A novel somatic mutation in ACD induces telomere lengthening and apoptosis resistance in leukemia cells. BMC Cancer, 15, 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.