Abstract
Aptamers are valuable tools that provide great potential to develop cost-effective diagnostics and therapies in the biomedical field. Here, we report a novel DNA aptamer that folds into an unconventional G-quadruplex structure able to recognize and enter specifically into human Burkitt's lymphoma cells. We further optimized this aptamer to a highly versatile and stable minimized version. The minimized aptamer can be easily equipped with different functionalities like quantum dots, organic dyes, or even a second different aptamer domain yielding a bi-paratopic aptamer. Although the target molecule of the aptamer remains unknown, our microscopy and pharmacological studies revealed that the aptamer hijacks the clathrin-mediated endocytosis pathway for its cellular internalization. We conclude that this novel class of aptamers can be used as a modular tool to specifically deliver different cargoes into malignant cells. This work provides a thorough characterization of the aptamer and we expect that our strategy will pave the path for future therapeutic applications.
Keywords: Aptamers, G-cuadruplex, Burkitt́s lymphoma, RAMOS, bi-paratopic
Introduction
Targeting strategies for cellular delivery of potential therapeutic agents, e.g., toxins,1 microRNAs,2 siRNAs,3 and antagomirs4,5 represent an emerging research field.6 Typically, most of the molecules used for targeting approaches are antibodies, peptides, and lipids. However, aptamers represent a true versatile alternative in this regimen, as they are generated in vitro through appropriate selection procedures against various target structures, being either homogenous or complex.7,8,9 Most studies reporting the use of aptamers as targeting ligands employ the highly characterized aptamer A10, a 2′-fluoro-2′-deoxy pyrimidine modified RNA molecule, which recognizes prostate-specific membrane-antigen on cancer cells.10 The A10 aptamer has extensively been tested as a delivery tool. For instance, it was covalently attached to nanoparticles with encapsulated chemotherapeutics and used to treat tumors in mice.11 Importantly, A10 not only binds its target molecule on the cell-surface of prostate tumors, but it also gets internalized into them. Thereby, A10 was shown to deliver its toxic nanoparticle cargo. Subsequently, the same A10 aptamer was shown to be also capable of carrying conjoined siRNA molecules into tumor cells, even in vivo, enabling specific mRNA knockdown activity.12 These findings nurtured the idea of using aptamers as common and generic targeting and delivering modules. Besides A10 few aptamers have been used for delivery purposes, e.g., the nucleolin-binding aptamer AS1411.13 Likewise toxins14 and nucleic acid-type15 of therapeutics, Levy and co-workers recently demonstrated that aptamers binding to DEC205 on dendritic cells are enable to induce T-cells responses by delivering tumor-antigens.16 However, turning an in vitro selected aptamer into an in vivo functioning component can be a laborious task.17 From previous studies, it became evident that aptamers should meet certain requirements to qualify for delivery approaches.18 First, they should be short to be affordable at reasonable costs. Second, the structure should be stable over a broader range of temperatures and environmental conditions. Third, the aptamers should bear a certain flexibility to allow the introduction of modifications (e.g., to optimize stability and half-life in vivo) and to equip them with other functionalities like imaging modalities and/or siRNA molecules without losing their binding and uptake capabilities.
Herein, we report on a short DNA aptamer that recognizes selectively Burkitt's lymphoma cells and bears an unusual G-quadruplex architecture. The aptamer provides a compact and stable structure, which allows its assembly with other aptamers, dyes, and nanoparticles, noteworthy without loss of function. We further show that the target cells specifically took up the aptamer via a clathrin-mediated endocytosis with a relatively fast kinetic, which opens the door for its utilization as a new modular delivery tool.
Results
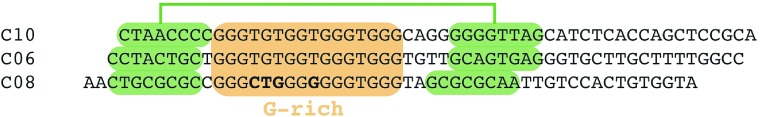
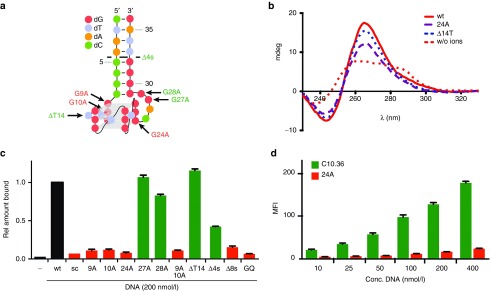
We identified a series of aptamers recognizing Burkitt's lymphoma cells.19,20 Here we provide an in-depth characterization of one of the motifs found in the enriched DNA library, exemplified by the aptamers C10, C06, and C08.20 This motif bears a G-rich segment (Figure 1), which suggests that the aptamer folds into a G-quadruplex structure. Besides, the G-rich segment, the motif comprises flanking sequences built from two complementary nucleotide stretches (Figure 1), which most likely serve as clamp (Figure 2a). To analyze this, we generated a variant of one of the aptamers (C10) made of only 36 nucleotides. This variant was named C10.36 and essentially covers the proposed G-quadruplex and clamp-structure (Figure 2a). We performed circular dicroism (CD) spectroscopy to investigate whether C10.36 indeed resumes a G-quartet containing structure. These studies reveal that C10.36 most likely folds into a parallel G-quadruplex structure when compared to the control condition in the absence of ions (Figure 2b). We next used flow cytometry and demonstrated that fluorescently-conjugated C10.36 has preserved binding properties and recognizes Burkitt's lymphoma cells, whereas a scrambled control sequence (C10.36sc) showed no binding (Figure 2c). To further delineate our findings, we synthesized point mutants of C10.36 (Figure 2a) and tested their impact on the aptamer's cell recognition properties. Based on our structural data and theoretical model, we introduced the following point mutations: G9A, G10A, G24A, G27A, and G28A (Figure 2a). The former three are expected to form part of the three-layer G-quadruplex structure, whereas the latter two not. Using flow cytometry and Atto647N-labeled aptamers, we observed a drastic reduction in binding of the G-quartet mutants (G9A, G10A, G24A). However, mutations of G-residues outside of the proposed G-quadruplex (G27A, G28A) retained the aptamer's cell-binding properties (Figure 2c). Even though CD-spectroscopy revealed formation of a G-quadruplex structure of mutant G24A (Figure 2b), concentration-dependent measurements clearly indicate that compared to C10.36 this mutant lacks Burkitt's lymphoma cell recognition properties (Figure 2d). Based on these results, we propose that C10.36 comprises a three-layer G-quadruplex structure arranged in a parallel orientation, as it has been described for other aptamers targeting the IL6R21,22 and the telomere ends of chromosomal DNA.23 According to this model, nucleotide T14 might be bulged out to avoid interference with G-quartet formation (Figure 2a). Such an unusual arrangement of G-quadruplexes has been described recently.24,25 To further prove the impact of nucleotide T14 on G-quadruplex formation and aptamer binding, we synthesized a deletion mutant (ΔT14). Flow cytometry and CD measurements on ΔT14 revealed that the lack of this thymidine-residue does neither interfere with quadruplex formation (Figure 2b) nor with aptamer binding to Burkitt's lymphoma cells (Figure 2c).
Figure 1.
Sequence analysis depicted a G-rich motif (coloured orange) flanked by two complementary strands (coloured green). Variant nucleotides within the G-rich motif are depicted in bold.
Figure 2.
The shortened aptamer C10.36 interacts with Burkitt's lymphoma cells and folds into a G-quadruplex structure. (a) Predicted structure of C10.36, folding into a parallel G-quadruplex. Nucleotides are depicted as symbols as indicated. G9A and G24A (red) indicate point mutants that lead to loss of affinity, whereas point mutants G28A and G27A (green) show preserved binding capabilities (see also Supplementary Figure S1). (b) CD-spectrometry analysis of C10.36 (red curve) compared to HD1 (black curve), C10.36 G24A (purple curve, dashed), C10.36 ΔT14 (blue curve, dashed) and C10.36 (red curve, dashed) in pure H2O. (c) Interaction analysis using flow cytometry of 5′-Atto647N-labeled C10.36 mutants [200 nmol/l] (mutations as indicated) with Burkitt's lymphoma cells compared to C10.36 (black bar, wt) and the non-binding control sequences C10.36 scramble (sc). D4s: truncated variant covering the sequence from nt 5 to 32. D8s: truncated variant covering the sequence from nt 9 to 28. GQ: truncated variant covering the sequence from nt 9 to 24 (G-quadruplex only). (d) Interaction analysis of 5′-Atto647N-labeled C10.36 (green bars) and the 5́-Atto647N labeled mutant G24A (orange bars) with Burkitt's lymphoma cells. MFI, mean fluorescence intensity.
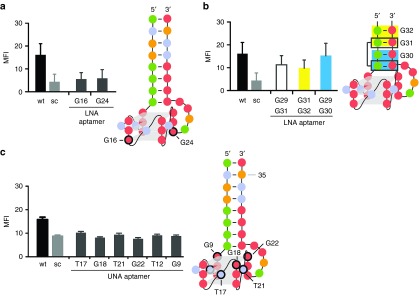
In parallel to all mutation and deletions aforementioned, we synthesized variants of C10.36 bearing either locked nucleic acid (LNA)- or unlocked nucleic acid (UNA)-building blocks.26,27 All modified variants were equipped with a 5′-fluorescein moiety to enable flow cytometry-based interaction analysis. C10.36 variants bearing LNA moieties embedded in the G-quadruplex structure (G16, G24) were found to no longer interact with the target cells (Figure 3a). This finding is in agreement with previous studies that report the loss of aptamer activity upon incorporation of LNA building blocks at positions that are engaged in G-quadruplex formation.28 We next extended our analysis toward three new aptamer variants bearing two LNA base pairs each, in the clamp region (G29,31; G31,32 and G29,30; Figure 3b). We also truncated the aptamer by shortening the stem size resulting in either four (G29,31; G31,32) or two remaining base-pairs (G29,30) (Figure 3b). These LNA-containing variants were found to retain binding to Burkitt's lymphoma cells (Figure 3b). Notably, G29,30 represents the shortest variant of C10.36 (24 nuclotides) but still capable of binding to cells. These findings indicate that the terminal stem structure is important for aptamer activity, but most likely it plays an important structural role rather than interacting directly with its target on the cell surface. In contrast to LNA, UNA-nucleotides provide a higher degree of flexibility at the nucleic acid backbone. Therefore, we investigated if UNA nucleotides embedded at different positions within the G-quadruplex domain of C10.36 also affect the structure and activity of the aptamer. We replaced, one-by-one, the bridging T-residues (T12, T17 and T21) of the quadruplex by UNA-nucleotides, as well as G9, G18, or G22 that seems to be involved in G-quadruplex formation. As shown in Figure 3c, all UNA-modified aptamer variants revealed loss of cell binding. This result indicates that the conformation of the G-quadruplex is essential for C10.36 recognition properties and besides residue T14, which is abdicable no variations in this region are tolerable. However, T14 points toward possible insertions of other entities, e.g., twisted-intercalating nucleic acids,29 which might allow to further modulate the aptamer's activity.
Figure 3.
Impact of LNA And UNA moieties on C10.36 binding to Burkitt's lymphoma cells. (a) Replacement of residues G16 and G24 (black circled) by LNA-variants leads to loss of binding. 5′-FAM labeled aptamer variants were used during flow cytometry analysis to detect cell binding. (b) Generation of truncated aptamer-variants with two LNA-moieties embedded in the stem structure (see also Figure 2a). Double modified variants G29,G31 (black box, no fill) and G31,G32 (yellow box) comprise 28 nucleotides whereas double modified variant G29,G30 (blue box) comprises 24 nucleotides. (c) Interaction analysis of 5′-FAM labeled C10.36 variants with UNA moieties at the indicated (black circled) T- or G-positions. MFI, mean fluorescence intensity. UNA, unlocked nucleic acid; LNA, locked nucleic acid. Each variant was used at a concentration of 400 nmol/l.
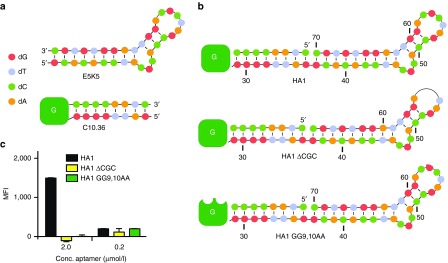
To this end, we describe a compact novel DNA aptamer, built from 36 nucleotides that recognize Burkitt's lymphoma cells. Due to its compact and well-defined structure, this aptamer might represent a valuable starting point for generating modular aptamer-directed delivery systems. For instance, combining the cellular binding activity of C10.36 with another aptamer or with different nanoparticles. To investigate this, we started an endeavor to create an hetero-bivalent aptamer: C10.36 conjoined to a second DNA aptamer domain that binds streptavidin.30 The selection and characteristics of a streptavidin (SA)-binding aptamer are described in Supplementary Figure S1.30 The minimal motif of the SA-binding aptamer, named E5K5, bears 34 nucleotides (Figure 4a). We conjoined this aptamer with the 3′-end of the cell binding aptamer C10.36 yielding a hetero-bivalent DNA aptamer, termed HA1 (Figure 4b). As controls, we synthesized hetero-bivalent variants of HA1 bearing mutations either in the SA- (HA1 ΔCGC) or in the cell-binding (HA1 GG9-10AA) domain (Figure 4b). HA1 preserved binding properties of both domains, shown by filter retention analysis of radioactively labeled HA1 to SA and competition experiments using fluorescently labeled C10.36 and increasing concentrations of HA1 (Supplementary Figure S1d,e, respectively). The synthesized control molecules behave as predicted and reveal either loss of cell- (HA1 GG9-10AA) or SA-binding (HA1 ΔCGC) (Supplementary Figure S1). We next investigated the properties of HA1 to simultaneously interact with SA and cells. Therefore, we incubated HA1 with Burkitt's lymphoma cells in the presence of fluorescently labeled SA (SA-488) and monitored binding by flow cytometry. These experiments show that HA1 simultaneously interacts with both target structures (Figure 4c). The control molecules did not reveal binding, clearly indicating the specificity of HA1 (Figure 4c). Having shown that HA1 performs in a predictable fashion, we went a step forward and shortened the bivalent aptamer further. We generated HA1.62 and HA1.54, which still were found to interact with SA, but to a lesser extend than HA1 (Supplementary Figure S1d). These data strongly suggest that due to its compact and stable structure C10.36 can be combined with other aptamers to generate bi-functional molecules capable of binding simultaneously to different targets.
Figure 4.
Assembly of hetero-bivalent aptamers. (a) Secondary structure of the DNA aptamer E5K5 (see also Supplementary Figure S1) that interacts with streptavidin (SA) and a schematic of the Burkitt's lymphoma cell-binding aptamer C10.36. The green box with white G indicates the G-quadruplex. Nucleotides are depicted by color code, as indicated. (b) Assembly of the hetero-bivalent aptamer HA1 built from C10.36 and E5K5 (top) and the control aptamers HA1 ΔCGC (middle, loss of nucleotides 55-57) and HA1 GG9,10AA (bottom, double point mutants indicated by bites in green box). (c) Interaction analysis of HA1 (black bar), HA1 ΔCGC (yellow bar) and HA1 GG9,10AA (green bar) with Burkitt's lymphoma cells in the presence of SA-488 at indicated concentrations.
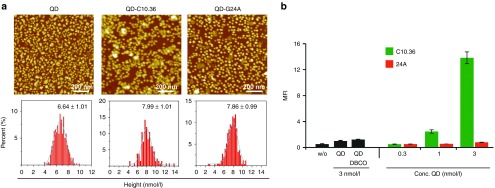
We extended our “lego-block”-approach toward chemical coupling methods and generated C10.36 or a nonbinding control oligodeoxynucleotide (G24A) equipped with quantum dots (QD). To covalently attach C10.36 to the QD-surface, we employed copper-free click chemistry.31 Briefly, C10.36 was synthesized bearing a 5′-azide moiety, which was then reacted with dibenzylcyclooctyne-derivatized QD (Figure 5a and Supplementary Figure S2). These QD were incubated with Burkitt's lymphoma cells and binding was detected by flow cytometry. As shown in Figure 5b, QD equipped with C10.36 recognize the target cells, whereas QD derivatized with the mutant G24A exhibited only background binding. These data further indicate the versatility of C10.36 being compatible to different modifications and, thus, applications without loss of cell-recognition properties. Advantageously, one QD is equipped with several aptamers, thus leading to a low detection limit of 1 nmol/l compared to monovalent interactions most likely reflecting the multivalency effect (Figure 5b).
Figure 5.
(a) Atomic force microscopy images (top) and height distributions (bottom) of QD, QD-C10.36, and QD-G24A. (b) Interaction analysis using flow cytometry of QDs derivatized with C10.36 (green bars) or C10.36 G24A (24A, red bars) with Burkitt's lymphoma cells at different QD concentrations (0.3 - 3 nmol/l).
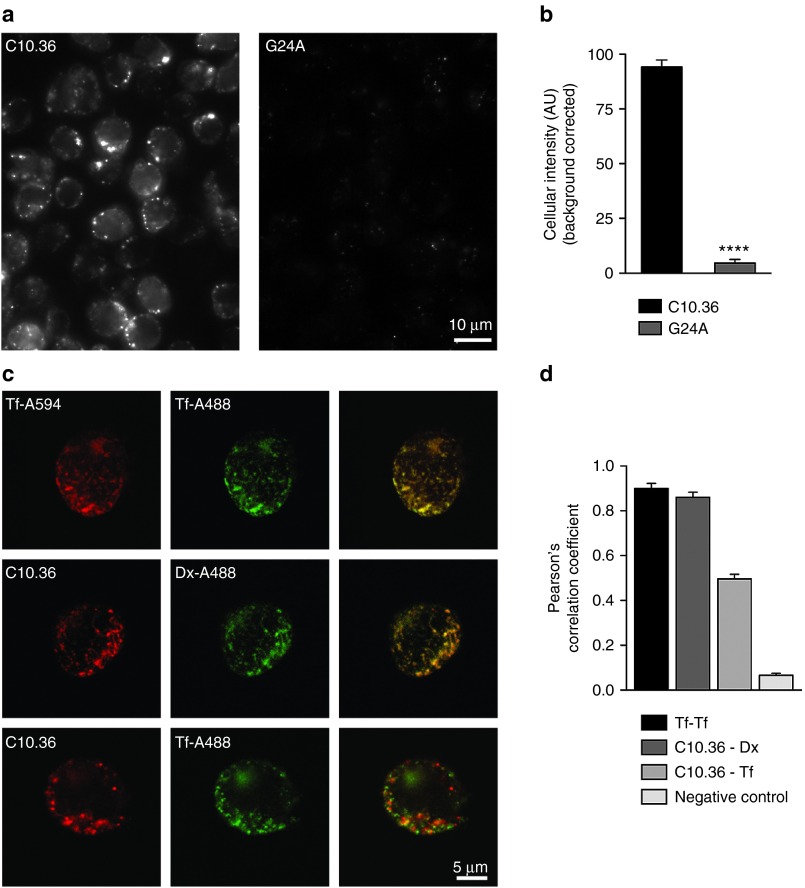
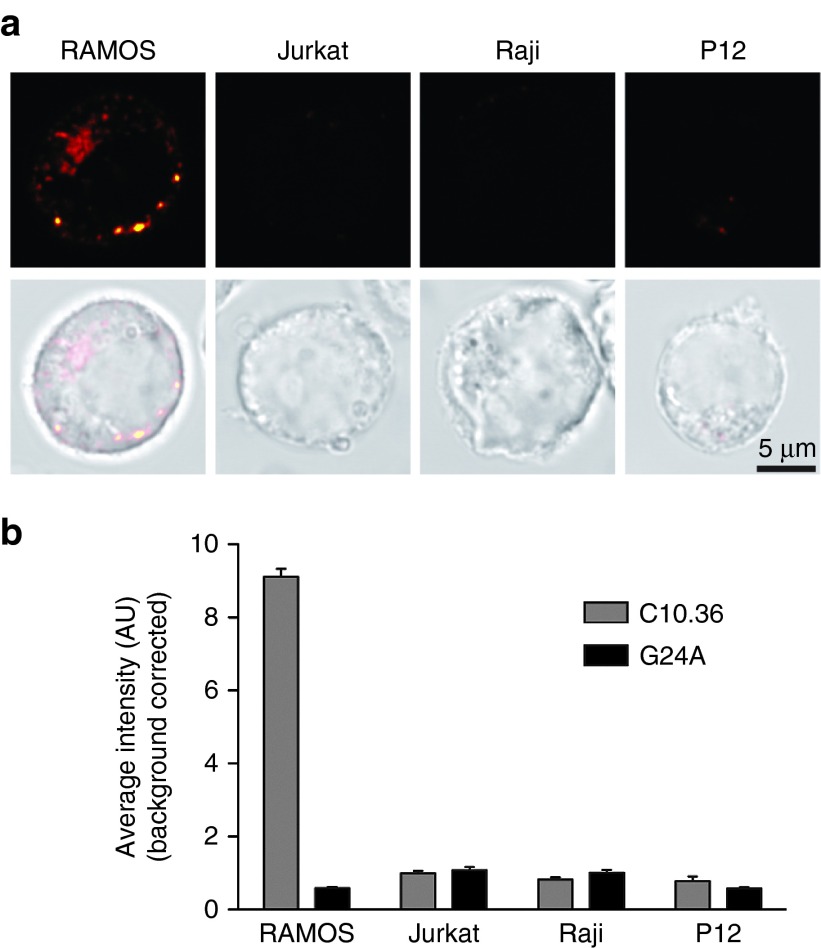
We next investigated the fate of C10.36 upon target cell recognition. We therefore performed microscopy studies, employing Atto674N-modifed C10.36 or the nonbinding control (G24A) to visualize association of the aptamer with Burkitt's lymphoma cells. After incubation of C10.36 with cells, a clear uptake of the fluorescent aptamer but not of the nonbinding control G24A was observed (Figure 6a,b). The kinetics of aptamer-uptake was shown to be rapid, with a maximum yield being achieved after only 15 minutes (Supplementary Figure S3). To elucidate the underlying endocytotic mechanism by which C10.36 is entering the cells, we performed colocalization studies with fluorescently labled transferrin and dextran. In these studies, we coincubated Burkitt's lymphoma cells with Atto647N-labeled C10.36 and Alexa488-labeled transferrin or dextran. The C10.36 aptamer colocalizes with dextran and transferrin (Figure 6c,d). The uptake of transferrin receptors occurs via clathrin-mediated endocytosis.32 However, dextran molecules can enter via clathrin-mediated endocytosis but also using caveolin-mediated endocytosis, phagocytosystosis, or macropinocytosis depending on the cell type.33 Therefore, we incubated the cells with C10.36 and the clathrin-mediated endocytosis-blocker Dynasore,34 which resulted in a significant reduction of aptamer internalization (Supplementary Figure S3). This further supports the hyposthesis that C10.36 enters into cells predominantly via clathrin-mediated endocytosis. Finally, to corroborate the specificity of C10.36 for Burkitt's lymphoma cells, we tested it on different lymphoma-derived cell lines. In these experiments, C10.36 was found to be highly specific, as it internalized only in Ramos Burkitt's lymphoma cells and under the applied conditions no significant uptake was observed in Raji, Jurkat, or P12 cells (Figure 7).
Figure 6.
Cell staining and the endocytic pathway used by C10.36 aptamer. (a) Epifluorescence images of RAMOS cells live stained with Atto647N labeled C10.36 or control aptamer G24A (see methods). (b) Quantification of cellular fluorescence after staining with Atto647N labeled C10.36 or the G24A mutant aptamer. Error bars represent the s.e.m of 3 independent experiments (**** = P < 0.0001, Student́s t-test). (c) Confocal images of Ramos cells co-incubated with fluorescent transferrin (Tf-A594 or Tf-A488), dextran (Dx-A488) and Atto647N labeled C10.36. (d) Pearson's correlation coefficient was used to evaluate the coincidence detection between the coincubated molecules in c). Coincubation of Tf-A594 and Tf-A488 (Tf-Tf) provide the maximal expected correlation (positive control). Negative control was obtained by inverting one of the images resulting in random Pearson's correlation values. Error bars represent the s.e.m of 3 independent experiments.
Figure 7.
Cellular specificity of the C10.36 aptamer. (a) Examples of confocal images obtained from different cell lines. The upper row of images shows the fluorescent signal of cells stained for 1 hour at 37 °C with 200 nmol/l of C10.36-Atto647N (see Methods for details). The lower row of images shows the overlay of fluorescence signal and transmission light. Note that the C10.36 signal in RAMOS cells is clearly intracellular. (b) Quantification of C10.36 and control aptamer signal for the different cell lines. Error bars represent the s.e.m from three independent experiments (~10 cell per experiment).
Discussion
The described aptamer C10.36 is a short single-chained DNA molecule that recognizes and internalizes specifically into Burkitt́s lymphoma cells (Ramos cell line). Colocalization and toxin studies revealed that the aptamer most likely hijacks a clathrin-mediated endocytotic pathway, via a yet unknown cell surface molecule. In this way, C10.36 represents a potential delivery vehicle for cargo transport; here we provide evidence that it can be coupled to fluorescent small molecules, QD and a second SA-binding aptamer. If this strategy can be adapted towards delivering toxins or nucleic acid-type of therapeutics,13 e.g., siRNAs and antagomirs, will be examined in follow up studies. This type of delivery strategies has been performed using mainly the prostate-specific membrane-antigen-specific aptamer A10, which is also taken up through a clathrin-dependent endocytotic mechanism.12 Alternatively, some other aptamers were developed to serve as delivery vehicles, e.g., a transferrin receptor-specific aptamer,35 T-cell recognizing aptamers15 and AS1411 (ref. 36), a pan-tumor cell acting homodimeric DNA molecule. However, in most cases, the delivery efficiency, required for activtiy of associated cargo-RNA, is limited and far from optimal yet.4 Typically, endocytosis leads to the trapping of the cargo-molecule in the endosomal lumen and, thus, diminishing their potential activity.37 In this regard, endosomal escape strategies may be needed, which have not been established using aptamer-based delivery systems to date.38
The C10.36 aptamer folds into a compact G-quadruplex structure bearing an uncommon topology. This type of topology has been recently described where the formation of quadruplexes was facilitated by out-bulged residues.25 In aggrement with such report, our study demonstrates that a simple repetitive run of G-residues is not the sole criteria for defining G-quadruplexes and that this is also true for G-quadruplexes found in aptamers, as shown in this study for the first time. The compact structure of C10.36 enables its modular assembly with other aptamers or nanoparticles; exemplified by the combination of C10.36 with a streptavidin-binding aptamer and quantum dots. Assemblies of aptamers with other molecules and particles represent valuable tools for generating higher order architectures and multifunctional molecules. The “lego-block”-type assembly, in which components are conjoined and exchanged modularly, is beneficial for rapid confirmation strategies alleviating cumbersome optimization processes. Especially a combination with SA-binding domains allows to exchnage cargos rapidly without further engineering efforts, as biotinylated molecules can be easily assembled with the complex. Other investigations generated these assemblies by using adaptor oligonucleotides to which different modules can hybridize.39,40,41 However, this method relies on a noncovalent interaction, thus less stable assembly with greater synthesis and generation efforts. In either case aptamers that have a compact, stable, and short sequence length, as revealed by C10.36 are optimal. C10.36 was also tolerant toward implementation of LNA building blocks, as long as they do not interfere with G-quartet formation. Modification of known aptamers by introduction of LNA nucleotides has been performed in attempts to improve aptamer properties. For instance, a Tenascin-C binding aptamer demonstrated increased stabilization of the aptamer in serum and maintained strong binding to Tenascin-C of the LNA-containing aptamer.42 While implementation of LNA improves serum stability, our study delineates that LNA building blocks allows further truncation of the aptamer's stem structure to yield a shorter but still active aptamer. Likewise, UNA has demonstrated utility in aptamer structures.26 Due to its more flexible nature, UNA has the potential to alleviate strain in structures with loops as confirmed by UNA modification of the loops of a thrombin binding quadruplex aptamer. However, incorporation of UNA abolished C10.36 cell-targeting properties, at least in respect of the nucleotide positions investigated.
The adaptability of C10.36 to different modifications and combinations with other components opens a multitude of downstream applications. For example, quantum dots equipped with C10.36 may be useful as diagnostic tools. We could demonstrate in this study that QD-C10.36 conjugates enable the sensitive detection of Burkitt's lymphoma cells. This likewise opens the path toward multiplex analysis in combination with other available aptamers.43 However, it remains to be determined whether C10.36 also recognizes Burkitt's lymphoma cells derived from patient samples and in respect of therapeutic endeavors reveals in vivo activity. To this end, we provide a thorough characterization of C10.36, its utility as valuable tool for detection, delivery and assembly of multifunctional devices for cell-targeting approaches. Albeit its molecular target is not known to date and which will be unraveled in ongoing studies using aptamer-based affinity labeling,44 we believe that C10.36 represents an useful tool for biomedical research, diagnostics, and developing targeted therapies.
Materials and methods
Cells. Human Burkitt's lymphoma cell line RAMOS was kindly provided by Professor Marianne Hokland from the Institute of Biomedicin, Aarhus University. RAMOS cells were cultured at 37 °C and 5% CO2 in RPMI 1640 media supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.
Flow cytometry. Flow cytometry results were obtained from a BD FACSCanto cytometer. For binding experiments, 2 × 105 Ramos cells were incubated with aptamers in the respective concentrations in 100 µl of binding buffer (RPMI 1640 supplemented with 10% fetal bovine serum and 1 mg/ml of sheared sperm salmon DNA (sssDNA)) for 15 minutes at 37 °C and 5% CO2. The cells were then washed three times with Hank′s Balance salt solution (Gibco Life Technologies, Karlsruhe, Germany) using centrifugation at 200 g for 5 minutes at room temperature. For the measurement, the cell pellet was resuspended in 200 µl Hank′s Balance salt solution and ~104 cells were analyzed in the flow cytometer. The data were postanalyzed using FlowJo software. Flow cytometry for aptamers and QD-derivatized aptamers were done according to the follwing protocol: Approximately 200,000 Ramos cells were suspended in 150 µl media in 48-well plates and incubated with different amounts of aptamer conjugated QDs for 4 hours in cell culture medium (0.33, 1, or 3 nmol/l QD). The cells were then centrifuged, washed with phosphate buffer saline (PBS), and the cells were resuspended in PBS containing 1% formaldehyde. Flow cytometry was done using a Gallios Flow Cytometer from Beckman Coulter. The cell count was 5,000 and the FL4 channel was used for detection of the QD signal. Data were analyzed using Kaluza Analysis Software. Results are represented as mean fluorescence intensity and standard deviation, normalized to unmodified QD, n =3.
UNA and LNA oligonucleotide synthesis. Oligonucleotides were synthesized in 1.0 µmol scale using standard phosphoramidite chemistry on an automated nucleic acid synthesizer. LNA phosphoramidites were purchased from Exiqon (www.exiqon.com) and UNA phosphoramidites from RiboTask (www.ribotask.com). The synthesis conditions used were as follows: trichloroacetic acid in CH2Cl2 (3:97) as detritylation reagent; 0.25 M 4,5-dicyanoimidazole in CH3CN as activator; acetic anhydride in Tetrahydrofuran (THF) (9:91, v/v) as cap A solution; N-methylimidazole in THF (1:9, v/v) as cap B solution. As an oxidizing solution 0.02 M iodine in H2O/pyridine/THF was used for phosphate linkages whereas a thiolation solution 0.02 M PADS (phenylacetyl disulfide) in 3-picoline/acetonitrile (1:1, v/v) was used for phosphorothioate linkages. Coupling times used were 12 minutes for incorporation of LNA, UNA, and DNA monomers. The stepwise coupling yields (~99% per step) were determined from the absorbance of the dimethoxytrityl cations (DMT+) released after each coupling step. Cleavage from the support was carried out by using 32% aqueous ammonia solution, 12 hours at 55 °C. All oligonucleotides were purified by reversed phase HPLC (RP-HPLC) using a Waters 600 system equipped with an XBridge OST C18 (2.5 µm, 19 × 100 mm) column and an XBridge Prep C18 (5 µm, 10 × 10 mm) precolumn. After removal of the DMT-group, oligonucleotides were characterized (purity >90%) by ion exchange HPLC (IE-HPLC) on a Dionex system HPLC (VWR) and their composition verified by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF) on a Microflex Maldi (Bruker instruments, Leipzig, Germany).
SELEX. Selection of streptavidin (SA)-binding DNA aptamer was done according to the previous experiments.30,45,46 Briefly, a DNA library was incubated with SA-derivatized magentic beads (Dynal) and after removal of nonbound sequences, the bound sequences were recovered by heat denaturation followed by polymerase chain reaction and single strand generation by λ exonuclease digestion. As selection buffer PBS, supplemented with 3 mmol/l MgCl2 was used. After 10 selection cycles, the enriched library was cloned and sequenced. We employed the following aptamer DNA library: 5′-AGCATAGAGACATCTGCTATTGGTAGCACA-N25-TGTGCAACCG TAGACTCCAGACTTCAGGTA-3′ and for polymerase chain reaction, the following primer molecules were used: ADMA4.20 (fw) 5′-AGC ATA GAG ACA TCT GCT AT-3′; ADMA4.20.Phos (rev) 5′-Phos-TAC CTG AAG TCT GGA GTC TA-3′.
Filter retention analysis. To assess apatmer biding to streptavidin, we employed filter retention analysis using 5′-radioactively labeled DNA molecules (32-P) according to a previously reported protocol.47
Azide modification of aptamers. Aptamer sequences C10.36 and G24A containing a 5′-amino-modifier-C6 were synthesized by DNA Technology A/S. 40 nmol C10.36 or G24A (1 mmol/l stock), 80 µl 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer (0.1 M, pH 8.0), and 2 µmol azido-dPEG8-NHS (Quanta Biodesign) were mixed and incubated overnight at room temperature on a shaking plate at 500 rpm. Not coupled azido-dPEG8-NHS was removed from the supernantant after aptamer was precipitated. For ethanol precipitation of the aptamer, the concentration of sodium acetate was adjusted to 0.3 M and a 2.5-fold volume of ethanol (99.9%) added to the sample. The sample was stored at room temperature for 30 minutes followed by 30 minutes on dry ice. The sample was then centrifuged at 14,000 rpm for 15 minutes at 4 °C and the supernatant was discarded. Two hundred microliters of of 70% ethanol were added and the sample was centrifuged at full speed for 5 minutes. The supernatant was removed and the aptamer was dissolved in PBS supplemented with 0.5 mmol/l MgCl2. The azido-modified aptamer was stored at −20 °C.
Aptamer conjugation to QDs. First, dibenzylcyclooctyne (DBCO) modified QDs were made by mixing 200 pmol Qdot 705 ITK amino (Polyethylene glycol) quantum dots (QD, Life Technologies) (8 µmol/l solution in 50 mmol/l borate, pH 8.3) with 200 nmol DBCO-NHS ester (Jena Biosciences) in 30% dimethyl sulfoxide overnight on a shaking plate (500 rpm) at room temperature. Buffer exchange was performed with a Nanosep 100 K Omega centrifugal devices (Pall Corporation) and the QDs were washed four times with PBS containing 0.5 mmol/l MgCl2 to finally resuspend them in 100 µl PBS containing 0.5 mmol/l MgCl2. The DBCO functionalized QDs were then mixed with C10.36 or G24A bearing a 5′-azide moiety in a total volume of 50 µl PBS containing 0.5 mmol/l MgCl2 and 10% dimethyl sulfoxide. The reaction was incubated overnight at 50 °C (2 nmol aptamer/17 pmol QD-DBCO for the cell binding study and 2 nmol aptamer/29 pmol QD-DBCO for the atomic force microscopy (AFM) study). Enreachment and washing of aptamer-conjugated QD were achieved using Nanosep 100 KDa (MWCO). Final aptamer-functionalized QD were resuspended in PBS + 0.5 mmol/l MgCl2. Samples were then centrifuged 2 minutes at 10,000 rpm to remove aggregates and the samples were stored at 4 °C. The QD concentration was determined by fluorescence spectroscopy using a Fluorimax 3 (Jobin Yvon) by comparing to a standard curve made from different concentrations of unmodified QDs in PBS. The samples were excited at 530 nm and emission measured at 705 nm.
AFM. All AFM images were recorded under ambient conditions with a commercial AFM MultiMode VIII (Bruker, Santa Barbara). Experiments were performed in tapping mode at a scan frequency of 1 Hz with optimized feedback parameters. Ultrasharp silicon cantilevers (OMCL-AC160TS-R3; Olympus) with a nominal spring constant of 26 Nm−1 were used. Muscovite mica, which was freshly cleaved by adhesive tape before each experiment, was used as the substrate. All the images and size distributions were analyzed by using the commercial software Scanning Probe Image Processor (Image Metrology, Denmark).
Aptamer staining. Ramos cells were cultured as descried above. Cells were tipically incubated at 37 °C for 1 hour with 200 nmol/l of Atto647N coupled C10.36 or G24A. All incubation were performed in complete medium (RPMI medium (PAA Laboratories GmbH, Paching, Austria) with 5% fetal calf serum (PAA), 2 mmol/l L-Glutamine and 100 U/ml of penicillin and streptomycin) supplemented with 1 mg/ml of sheared sperm salmon DNA (Invitrogen). For the colocalization studies (Figure 6c,d), 200 nmol/l of C10.36 aptamer was coincubated at 37 °C with 50 µg/ml of transferrin-Alexa488 for 20 minutes or 100 µg/ml of dextran-Alexa488 (10 kDa) for 20 minutes. Positive control was performed by coincubation of 50 µg/ml of each transferrin-Alexa488 and transferrin-Alexa594 for 20 minutes. For the inhibition of clathrin-mediated endocytosis, cells were pretreated with 80 µmol/l dynasore (Sigma-Aldrich) and 1 mg/ml of sssDNA in complete medium for 15 minutes inside the incubator (37 °C and 5% CO2). After one washing step, cells were incubated with 200 nmol/l of aptamer (C10.36 or G24A) in presence of 80 µmol/l dynasore and 1 mg/ml of sheared sperm salmon DNA in complete medium for 20 minutes. After the staining incubations, cells were washed thoroughly in Dulbecco's PBS (DPBS) and fixed in ice-cold 4% paraformaldehyde for 30 minutes while centrifuging (300 g at 4 °C) on poly-L-lysine-coated glass coverslips. After washing with DPBS, the remaining active formaldehyde groups were quenched by incubating the cells in DPBS supplemented with 0.1 M glycine and 0.1 M NH4Cl for 15 minutes at room temperature in the dark. After a brief washing with DPBS, cells were mounted on glass slides using Mowiöl solution (6 g glycerol; Merck Millipore, 6 ml deionized water, 12 ml 0.2 M Tris buffer pH 8.5, Merck Millipore, 2.4 g Mowiöl 4–88; Merck Millipore).
Microscopy. Conventional epifluorescence images were obtained (Figure 6a and Supplementary Figure S3) with an Olympus IX71 microscope equipped with 0.75 NA/40×, 1.35 NA/60×, and 1.45 NA/100× objectives. Images were captured with an Olympus F-View II CCD camera (all from Olympus, Hamburg, Germany).
Confocal images (Figures 6c and 7) were obtained using a True Confocal System STED SP5 fluorescence microscope (Leica Microsystems GmbH, Mannheim, Germany), equipped with a 1.4 NA/100× STED objective (Leica). Images were scanned unidirectionally at 1 kHz with the pinhole set to one Airy unit.
Image analysis and statistics. Data analysis was performed by custom-written procedures, in Matlab (The Mathworks, Natick, MA). Cell intensities (Figure 6b and Supplementary Figure S3) were assessed by manually selecting the background and cell. All values are given in mean ± standard error of the mean. Graphs and statistical analysis were carried out with GraphPad Prism 5.0 (GraphPad Software, San Diego, CA).
SUPPLEMENTARY MATERIAL Figure S1. Characterisation of a streptavidin binding aptamer and its conjunction to C10.36. Figure S2. Reaction scheme showing aptamer functionalization of QDs by copper-free click chemistry. Figure S3. Internalization kinetics of C10.36 and the effect of the endocytosis inhibitor dynasore.
Acknowledgments
This work has been made possible by funds from ERA-NET Euronanomed2 (META consortium) and the Collaborative Research Center 704 (SFB704) from the German Research Council (DFG) to G.M. J.W. greatly appreciates support from the European Research Council under the European Union's Seventh Framework program (FP7/2007–2013)/ERC Grant Agreement No. 268776. The Lundbeck Foundation Nanomedicine Centre for Individual Management of Tissue Damage and Regeneration (LUNA). We greatly thank Qiang Li for AFM measurements.
Supplementary Material
References
- Bagalkot, V, Farokhzad, OC, Langer, R and Jon, S (2006). An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem Int Ed Engl 45: 8149–8152. [DOI] [PubMed] [Google Scholar]
- Esposito, CL, Cerchia, L, Catuogno, S, De Vita, G, Dassie, JP, Santamaria, G et al. (2014). Multifunctional aptamer-miRNA conjugates for targeted cancer therapy. Mol Ther 22: 1151–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, TC, Twu, KY, Ellington, AD and Levy, M (2006). Aptamer mediated siRNA delivery. Nucleic Acids Res 34: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pofahl, M, Wengel, J and Mayer, G (2014). Multifunctional nucleic acids for tumor cell treatment. Nucleic Acid Ther 24: 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catuogno, S, Rienzo, A, Di Vito, A, Esposito, CL and de Franciscis, V (2015). Selective delivery of therapeutic single strand antimiRs by aptamer-based conjugates. J Control Release 210: 147–159. [DOI] [PubMed] [Google Scholar]
- Xu, X, Ho, W, Zhang, X, Bertrand, N and Farokhzad, O (2015). Cancer nanomedicine: from targeted delivery to combination therapy. Trends Mol Med 21: 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H, Li, J, Zhang, XB, Ye, M and Tan, W (2015). Nucleic acid aptamer-mediated drug delivery for targeted cancer therapy. ChemMedChem 10: 39–45. [DOI] [PubMed] [Google Scholar]
- Zhou, J and Rossi, JJ (2014). Cell-type-specific, Aptamer-functionalized Agents for Targeted Disease Therapy. Mol Ther Nucleic Acids 3: e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibiel, A, Quang, NN, Gombert, K, Thézé, B, Garofalakis, A and Ducongé, F (2014). From ugly duckling to swan: unexpected identification from cell-SELEX of an anti-Annexin A2 aptamer targeting tumors. PLoS One 9: e87002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupold, SE, Hicke, BJ, Lin, Y and Coffey, DS (2002). Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res 62: 4029–4033. [PubMed] [Google Scholar]
- Farokhzad, OC, Cheng, J, Teply, BA, Sherifi, I, Jon, S, Kantoff, PW et al. (2006). Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci USA 103: 6315–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, JO 2nd, Andrechek, ER, Wang, Y, Viles, KD, Rempel, RE, Gilboa, E et al. (2006). Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol 24: 1005–1015. [DOI] [PubMed] [Google Scholar]
- Yan, AC and Levy, M (2009). Aptamers and aptamer targeted delivery. RNA Biol 6: 316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruspe, S and Hahn, U (2014). An aptamer intrinsically comprising 5-fluoro-2'-deoxyuridine for targeted chemotherapy. Angew Chem Int Ed Engl 53: 10541–10544. [DOI] [PubMed] [Google Scholar]
- Takahashi, M, Burnett, JC and Rossi, JJ (2015). Aptamer-siRNA chimeras for HIV. Adv Exp Med Biol 848: 211–234. [DOI] [PubMed] [Google Scholar]
- Wengerter, BC, Katakowski, JA, Rosenberg, JM, Park, CG, Almo, SC, Palliser, D et al. (2014). Aptamer-targeted antigen delivery. Mol Ther 22: 1375–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassie, JP, Liu, XY, Thomas, GS, Whitaker, RM, Thiel, KW, Stockdale, KR et al. (2009). Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol 27: 839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Z and Farokhzad, OC (2012). Aptamer-functionalized nanoparticles for medical applications: challenges and opportunities. ACS Nano 6: 3670–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, G, Ahmed, MS, Dolf, A, Endl, E, Knolle, PA and Famulok, M (2010). Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat Protoc 5: 1993–2004. [DOI] [PubMed] [Google Scholar]
- Raddatz, MS, Dolf, A, Endl, E, Knolle, P, Famulok, M and Mayer, G (2008). Enrichment of cell-targeting and population-specific aptamers by fluorescence-activated cell sorting. Angew Chem Int Ed Engl 47: 5190–5193. [DOI] [PubMed] [Google Scholar]
- Magbanua, E, Zivkovic, T, Hansen, B, Beschorner, N, Meyer, C, Lorenzen, I et al. (2013). d(GGGT) 4 and r(GGGU) 4 are both HIV-1 inhibitors and interleukin-6 receptor aptamers. RNA Biol 10: 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, C, Eydeler, K, Magbanua, E, Zivkovic, T, Piganeau, N, Lorenzen, I et al. (2012). Interleukin-6 receptor specific RNA aptamers for cargo delivery into target cells. RNA Biol 9: 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, G, Kröck, L, Mikat, V, Engeser, M and Heckel, A (2005). Light-induced formation of G-quadruplex DNA secondary structures. Chembiochem 6: 1966–1970. [DOI] [PubMed] [Google Scholar]
- Mukundan, VT, Do, NQ and Phan, AT (2011). HIV-1 integrase inhibitor T30177 forms a stacked dimeric G-quadruplex structure containing bulges. Nucleic Acids Res 39: 8984–8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukundan, VT and Phan, AT (2013). Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J Am Chem Soc 135: 5017–5028. [DOI] [PubMed] [Google Scholar]
- Campbell, MA and Wengel, J (2011). Locked vs. unlocked nucleic acids (LNA vs. UNA): contrasting structures work towards common therapeutic goals. Chem Soc Rev 40: 5680–5689. [DOI] [PubMed] [Google Scholar]
- Wahlestedt, C, Salmi, P, Good, L, Kela, J, Johnsson, T, Hökfelt, T et al. (2000). Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci USA 97: 5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacio, L, Church, FC and Jarstfer, MB (2008). Effect of locked-nucleic acid on a biologically active g-quadruplex. A structure-activity relationship of the thrombin aptamer. Int J Mol Sci 9: 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrbach, F, Fatthalla, MI, Kupper, T, Pötzsch, B, Müller, J, Petersen, M et al. (2012). Chemical maturation of a bivalent aptamer by single domain variation. Chembiochem 13: 631–634. [DOI] [PubMed] [Google Scholar]
- Bing, T, Yang, X, Mei, H, Cao, Z and Shangguan, D (2010). Conservative secondary structure motif of streptavidin-binding aptamers generated by different laboratories. Bioorg Med Chem 18: 1798–1805. [DOI] [PubMed] [Google Scholar]
- Codelli, JA, Baskin, JM, Agard, NJ and Bertozzi, CR (2008). Second-generation difluorinated cyclooctynes for copper-free click chemistry. J Am Chem Soc 130: 11486–11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaroli, DM, Bellvé, KD, Standley, C, Lifshitz, LM, Cardia, J, Lambright, D et al. (2012). Rabenosyn-5 defines the fate of the transferrin receptor following clathrin-mediated endocytosis. Proc Natl Acad Sci USA 109: E471–E480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustylnikov, S, Sagar, D, Jain, P and Khan, ZK (2014). Targeting the C-type lectins-mediated host-pathogen interactions with dextran. J Pharm Pharm Sci 17: 371–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen, T, Macia, E and Pelish, HE (2008). Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol 438: 77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilner, SE, Wengerter, B, Maier, K, de Lourdes Borba Magalhães, M, Del Amo, DS, Pai, S et al. (2012). An RNA alternative to human transferrin: a new tool for targeting human cells. Mol Ther Nucleic Acids 1: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Reyes, EM, Teng, Y and Bates, PJ (2010). A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res 70: 8617–8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkouhi, AK, Scholte, M, Storm, G and Haisma, HJ (2011). Endosomal escape pathways for delivery of biologicals. J Control Release 151: 220–228. [DOI] [PubMed] [Google Scholar]
- Nguyen, J and Szoka, FC (2012). Nucleic acid delivery: the missing pieces of the puzzle? Acc Chem Res 45: 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollins, CM, Nair, S, Boczkowski, D, Lee, J, Layzer, JM, Gilboa, E et al. (2008). Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem Biol 15: 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, JO, Kolonias, D, Pastor, F, Mittler, RS, Chen, L, Giangrande, PH et al. (2008). Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J Clin Invest 118: 376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J, Soontornworajit, B, Martin, J, Sullenger, BA, Gilboa, E and Wang, Y (2009). A hybrid DNA aptamer-dendrimer nanomaterial for targeted cell labeling. Macromol Biosci 9: 831–835. [DOI] [PubMed] [Google Scholar]
- Schmidt, KS, Borkowski, S, Kurreck, J, Stephens, AW, Bald, R, Hecht, M et al. (2004). Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res 32: 5757–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallikaratchy, P, Tang, Z, Kwame, S, Meng, L, Shangguan, D and Tan, W (2007). Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt's lymphoma cells. Mol Cell Proteomics 6: 2230–2238. [DOI] [PubMed] [Google Scholar]
- Vinkenborg, JL, Mayer, G and Famulok, M (2012). Aptamer-based affinity labeling of proteins. Angew Chem Int Ed Engl 51: 9176–9180. [DOI] [PubMed] [Google Scholar]
- Müller, J, Isermann, B, Dücker, C, Salehi, M, Meyer, M, Friedrich, M et al. (2009). An exosite-specific ssDNA aptamer inhibits the anticoagulant functions of activated protein C and enhances inhibition by protein C inhibitor. Chem Biol 16: 442–451. [DOI] [PubMed] [Google Scholar]
- Mayer, G, Wulffen, B, Huber, C, Brockmann, J, Flicke, B, Neumann, L et al. (2008). An RNA molecule that specifically inhibits G-protein-coupled receptor kinase 2 in vitro. RNA 14: 524–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, G and Höver, T (2009). In vitro selection of ssDNA aptamers using biotinylated target proteins. Methods Mol Biol 535: 19–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.