The transcription factors Mesp1 and Mesp2 are equally efficient at promoting specification, EMT, and differentiation of early multipotent cardiovascular progenitors. However, only Mesp1 promotes the speed, polarity, and directionality of cell migration, explaining how Mesp1 coordinates progenitor fate decision and migration during development.
Abstract
During embryonic development, Mesp1 marks the earliest cardiovascular progenitors (CPs) and promotes their specification, epithelial–mesenchymal transition (EMT), and cardiovascular differentiation. However, Mesp1 deletion in mice does not impair initial CP specification and early cardiac differentiation but induces cardiac malformations thought to arise from a defect of CP migration. Using inducible gain-of-function experiments during embryonic stem cell differentiation, we found that Mesp2, its closest homolog, was as efficient as Mesp1 at promoting CP specification, EMT, and cardiovascular differentiation. However, only Mesp1 stimulated polarity and directional cell migration through a cell-autonomous mechanism. Transcriptional analysis and chromatin immunoprecipitation experiments revealed that Mesp1 and Mesp2 activate common target genes that promote CP specification and differentiation. We identified two direct Mesp1 target genes, Prickle1 and RasGRP3, that are strongly induced by Mesp1 and not by Mesp2 and that control the polarity and the speed of cell migration. Altogether, our results identify the molecular interface controlled by Mesp1 that links CP specification and cell migration.
Introduction
Mammalian heart development begins during gastrulation when Mesp1, the earliest marker of the cardiovascular lineages, is expressed in prospective cardiovascular progenitors (CPs) within the primitive streak (PS; Saga et al., 1999; Kitajima et al., 2000; Bondue and Blanpain, 2010). Mesp1-positive CPs then leave the PS and migrate anterolaterally to form the initial cardiac structure, known as the cardiac crescent (Buckingham et al., 2005; Abu-Issa and Kirby, 2007). Different retrospective and prospective clonal analyses have demonstrated that the different heart regions arise from distinct populations of CPs, specified at different time points during development (Kelly et al., 2001; Meilhac et al., 2004; Lescroart et al., 2014). The early Mesp1 CPs contribute to the first heart field derivatives and give rise to cardiomyocytes (CMs), endothelial cells (ECs), and epicardial-derived cells. In contrast, the later Mesp1 CPs contribute to the development of the second heart field derivatives, which include the atria, the right ventricle, and the outflow and inflow tract regions, and give rise to CMs, ECs, and smooth muscle cells (Bondue et al., 2011; Lescroart et al., 2014). The existence of distinct pools of Mesp1 progenitors that are specified at different times during gastrulation and contribute to the morphogenesis of different heart regions requires that CP specification and migration be tightly regulated. The mechanisms that control and couple these two key cellular processes during mammalian heart development are currently unknown.
Mesp1 expression induces cardiovascular progenitor specification and differentiation by directly promoting the expression of key transcription factors that control cardiovascular cell fate (Bondue et al., 2008, 2011; David et al., 2008; Lindsley et al., 2008). Inactivation of Mesp1 in mice results in severe cardiac malformations thought to arise from a defect of early CP migration (Saga et al., 1999, 2000; Kitajima et al., 2000). The persistence of early CP specification and early cardiac differentiation in Mesp1-null mice suggests that other genes may compensate for Mesp1 function during these processes, whereas Mesp1 presents a unique and nonredundant function during CP migration. However, the functional demonstration that other genes compensate for Mesp1 function during the early step of CP specification remains elusive, and the unique mechanisms regulated by Mesp1 to control CP migration are currently unknown. Mesp2, the closest Mesp1 homolog, has been hypothesized to compensate for Mesp1 function during the early step of cardiogenesis (Kitajima et al., 2000; Saga et al., 2000). In the absence of Mesp1, Mesp2 is up-regulated at the time of CP specification (Kitajima et al., 2000). However, Mesp2-knockout (KO) mice do not present cardiac malformation but rather major defects in somitogenesis and segmentation (Saga et al., 1997, 2000; Morimoto et al., 2005). Inactivation of both Mesp1 and Mesp2 induces a profound defect of gastrulation, leading to the absence of mesoderm formation and consequently heart development, precluding the assessment of the redundant function of Mesp1 and Mesp2 during CP specification and differentiation (Kitajima et al., 2000; Saga et al., 2000).
Here, we investigate whether Mesp2 compensates for Mesp1 function during CP specification and differentiation and what unique mechanisms are regulated by Mesp1 during CP migration. Using inducible gain-of-function experiments during embryonic stem cell (ESC) differentiation, we found that Mesp2 is as potent as Mesp1 in promoting CP specification, epithelial–mesenchymal transition (EMT), and cardiovascular lineage differentiation. However, only Mesp1 promotes cell migration and polarity of CPs by a cell-autonomous mechanism. We identified Prickle1, a core component of the planar cell polarity (PCP) pathway, and the Ras guanyl releasing protein 3 (RasGRP3) as two unique Mesp1 target genes that control polarity and speed of CP migration, linking progenitor specification and migration during cardiac development.
Results
Mesp2 promotes robust cardiovascular differentiation
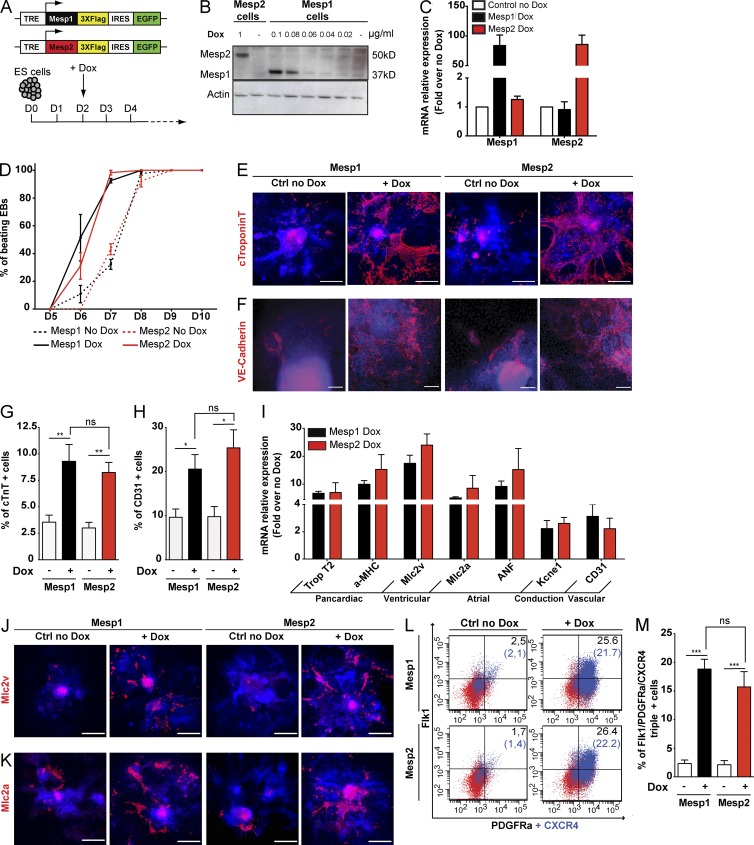
To assess whether Mesp2 promotes cardiovascular cell fate similarly to Mesp1 (Bondue et al., 2008; David et al., 2008; Lindsley et al., 2008), we generated inducible ESC lines allowing overexpression of Mesp2 after doxycycline (Dox) addition (Fig. 1 A). To compare side-by-side in the same experimental system the ability of Mesp1 and Mesp2 to promote CP specification and cardiovascular differentiation, we titrated the dose of Dox in each cell line to induce similar levels of Mesp1 and Mesp2 expression as determined by quantitative RT-PCR (qPCR) and Western blot experiments (Fig. 1, B and C). We found that 0.08 and 1 µg/ml Dox allowed similar levels of transgene expression, respectively, in Mesp1- and Mesp2-inducible ESC lines (Fig. 1, B and C). The difference in Mesp1 and Mesp2 transgene expression was observed in three different independent cell lines for each construct (not depicted), showing that this effect was caused by intrinsic differences between Mesp1 and Mesp2 sequences.
Figure 1.
Mesp1 and Mesp2 equally promote CP specification and differentiation. (A) Schematic representation of Dox-inducible Mesp1 and Mesp2 constructs (top). Experimental design for Dox-inducible Mesp1 or Mesp2 overexpression during EB differentiation (bottom). (B) Western blot analysis of Mesp1-Flag and Mesp2-Flag expression after administration of different concentrations of Dox. (C) qPCR quantification of Mesp1 and Mesp2 expression 24 h after Dox administration. 0.08 and 1 µg/ml Dox were used to stimulate, respectively, Mesp1- and Mesp2-inducible cell lines. Data are normalized to the relative mRNA expression in the absence of Dox and represent mean ± SEM of three biologically independent experiments. (D) Quantification of beating EBs at different times in control conditions and after Dox administration in Mesp1- and Mesp2-inducible ESCs. Data represent mean ± SEM of three biologically independent experiments. At least 60 EBs for each condition were counted. (E and F) Cardiac and vascular differentiation after Mesp1 or Mesp2 overexpression. Immunostaining of EBs at day 8 of EB differentiation, 6 d after Dox addition, using anti-cTnT antibody, a specific marker for cardiomyocytes (E), and anti–VE-cadherin antibody, an EC marker (F). (G and H) FACS quantification of cells positive for cTnT (G) and CD31 (EC marker; H) at day 8 of EB differentiation. Data represent mean ± SEM of at least three biologically independent experiments. (I) qPCR quantification of different cardiovascular markers at day 8 of EB differentiation. Data represent mean ± SEM of three biologically independent experiments. (J and K) Immunostaining of EBs with anti-Mlc2v antibody, a specific marker for ventricular cells (J), and anti-Mlc2a antibody, a marker for atrial cells and immature CMs (K) at day 8 of EB differentiation. (L and M) FACS quantification of Flk1, PDGFRa, and CXCR4 triple-positive CPs at day 3, 24 h after Mesp1 or Mesp2 induction, in control and stimulated cells. Percentage of Flk1/PDGFRa-positive cells and Flk1/PDGFRa/CXCR4-positive cells (in blue and in parentheses) are shown. Data represent mean ± SEM of at least four biologically independent experiments. E, F, J, and K are mosaic reconstructions of several microscopic images generated using a 10% overlap between each single acquisition. Western blots and all immunostainings are representative images of at least three independent experiments. Bars: (E, J, and K) 500 µm; (F) 100 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Induced Mesp2 expression during embryonic body (EB) differentiation accelerated the appearance and enhanced the number of beating areas with an efficiency similar to that of Mesp1 (Fig. 1 D). Immunostaining and FACS quantification revealed that both Mesp1 and Mesp2 strongly and equally promoted CM (cardiac troponin T [cTnT]) and EC (CD31 and vascular endothelial [VE]-cadherin) differentiation (Fig. 1, E–H). qPCR and immunostaining for different cardiac, conduction system, and EC markers (Fig. 1, I–K) showed that Mesp1 and Mesp2 promote the differentiation of the different cardiovascular derivatives with a similar efficiency.
Mesp1-expressing CPs coexpress KDR/Flk1, PDGFRa, and CXCR4 cell-surface markers during both ESC differentiation and embryonic development (Bondue et al., 2011; Lescroart et al., 2014). Mesp1 overexpression during ESC differentiation rapidly promotes CP specification and the appearance of a cell population coexpressing these three cell-surface markers (Bondue et al., 2011). To assess whether Mesp2 promotes CP specification as efficiently as Mesp1, we used flow cytometry to quantify the presence of Flk1+, PDGFRa+, and CXCR4+ cells, which mark CPs (Bondue et al., 2011; Lescroart et al., 2014) 24 h after Dox administration. Similarly to Mesp1, Mesp2 overexpression increased the proportion of Flk1+, PDGFRa+, and CXCR4+ cells, from 2.1% in control cells to 15.7% within 24 h after Dox administration (Fig. 1, L and M), indicating that Mesp2 overexpression strongly promotes CP specification as efficiently as Mesp1. Altogether, these data demonstrate for the first time that Mesp2 promotes CP specification and cardiovascular differentiation in a manner similar to Mesp1, both qualitatively and quantitatively.
Mesp1 promotes the speed of cell migration by a cell-autonomous mechanism
Mesp1-null embryos exhibit severe cardiac malformations, including so-called cardia bifida, because of a failure of ventral fusion of the cardiac mesoderm, attributed to a defect of the prospective cardiac mesoderm migration upon Mesp1 deletion (Saga et al., 1999, 2000; Kitajima et al., 2000). Indeed, analysis of the Mesp1LacZ/- embryo (corresponding to the Mesp1-null embryo but allowing the tracking of Mesp1 progenitors) revealed that Mesp1-deficient CPs present defects in migration, initially accumulating in the PS and then migrating to the anterior region of the embryo with some delay (Saga et al., 1999, 2000). These data suggest that Mesp1 controls CP migration and that the up-regulation of Mesp2 in the cardiac mesoderm of Mesp1-KO embryos is not sufficient to compensate for the promigratory function of Mesp1.
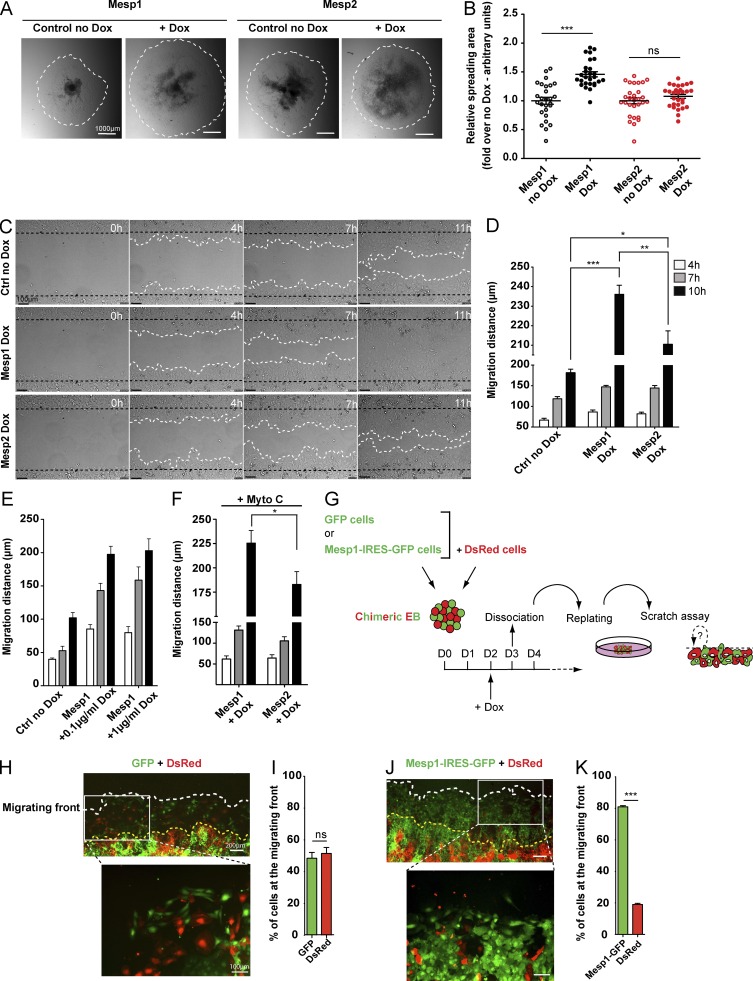
To directly address this hypothesis, we assessed the migration properties of cells after Mesp1- or Mesp2-induced expression during ESC differentiation. We first assessed the rapidity of EB spreading after replating the hanging drops to a gelatin-coated plate. 48 h after replating, the spreading area of the EBs was significantly increased after Mesp1 overexpression, and not after Mesp2 overexpression, compared with their respective controls (Fig. 2, A and B). Using an in vitro scratch wound assay on a monolayer of cells, obtained after the plating of dissociated EBs 24 h after Mesp1- or Mesp2-induced expression, at day 3 of EB differentiation, we measured the migration of these cells by time-lapse microscopy (Fig. 2, C and D). Analysis of the different time-lapse experiments showed that whereas Mesp2 slightly accelerated cell migration compared with control cells, Mesp1-expressing cells migrated much faster compared with both control and Mesp2-expressing cells, and higher doses of Dox did not impair Mesp1-induced migration (Fig. 2, C–E). Importantly, the greater promotion of cell migration by Mesp1 was not the consequence of increase in cell proliferation, as Mesp1-overexpressing cells still migrated faster than Mesp2-overexpressing cells after mitomycin C treatment (Fig. 2 F). Altogether, these results show that Mesp1 exhibits nonredundant promigratory functions during CP specification.
Figure 2.
Mesp1 promotes rapid cell migration by a cell-autonomous mechanism. (A and B) Mesp1- and Mesp2-inducible EBs were replated on a gelatin-coated plate at day 3, 24 h after Dox induction, and the cell spreading areas were measured by bright field microscopy 48 h later. Data represent the relative spreading area of EB ± SEM of at least 27 EBs from four biologically independent experiments. (C) Cell migration measured by in vitro scratch wound assay. Time-lapse microscopy images were recorded every 5 min during 11 h. Data show cell migration 0, 4, 7, and 11 h after the wound. (D) Quantification of the distance of cell migration 4, 7, and 10 h after wounding. Data represent the mean migration distance ± SEM of six biologically independent experiments. Control (no Dox) in C and D represent Mesp1 cells without Dox. No difference was observed between Mesp1 and Mesp2 no Dox cells. (E) Migration distance of control, Mesp1 0.1 µg/ml, and Mesp1 1 µg/ml after 4, 7, and 10 h after Dox addition (n = 3 independent experiments performed in duplicate). (F) Migration distance of Mesp1- and Mesp2-expressing cells after mitomycin C (Mito) treatment (n = 3). (G) Experimental strategy to assess the cell-autonomous function of Mesp1 in the promotion of cell migration. Chimeric EBs were generated by aggregating similar numbers of Mesp1-IRES-GFP–expressing and control DsRed-expressing cells. (H–K) Fluorescence microscopy analysis and quantification of the relative chimerism of GFP- and DsRed-positive cells at the migrating front, 24 h after wounding in chimeric EBs containing control GFP (H and I) or Mesp1-IRES-GFP (J and K) cell lines. Graphs (I and K) represent the mean chimerism and SEM of three independent experiments. At least 150 cells for each condition were counted. Bars: (A) 1,000 µm; (C) 100 µm; (H and J, lower magnification) 200 µm; (H and J, higher magnification) 100 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
To determine whether cell migration induced by Mesp1 is mediated by the secretion of soluble proteins or through an intrinsic and cell-autonomous mechanism, we assessed the relative migration of Mesp1-IRES-GFP–expressing cells and control wild-type (WT) cells expressing the red fluorescent protein DsRed, differentiated in chimeric EBs, containing equal proportions of both inducible cells (Fig. 2 G). As controls, we used chimeric EBs composed of GFP- and DsRed-expressing cells (Fig. 2 G). The chimeric EBs were stimulated with Dox, and in vitro scratch wound assays were performed (Fig. 2 G). In the control condition, equal numbers of green and red cells were present at the edge of the migrating front, 24 h after the scratch wound (Fig. 2, H and I). In contrast, the proportion of Mesp1-IRES-GFP–expressing cells was massively enriched at the edge of the wound compared with control DsRed cells (Fig. 2, J and K). These results demonstrate that Mesp1 promotes cell migration through a cell-autonomous mechanism.
Mesp1 but not Mesp2 promotes oriented cell migration
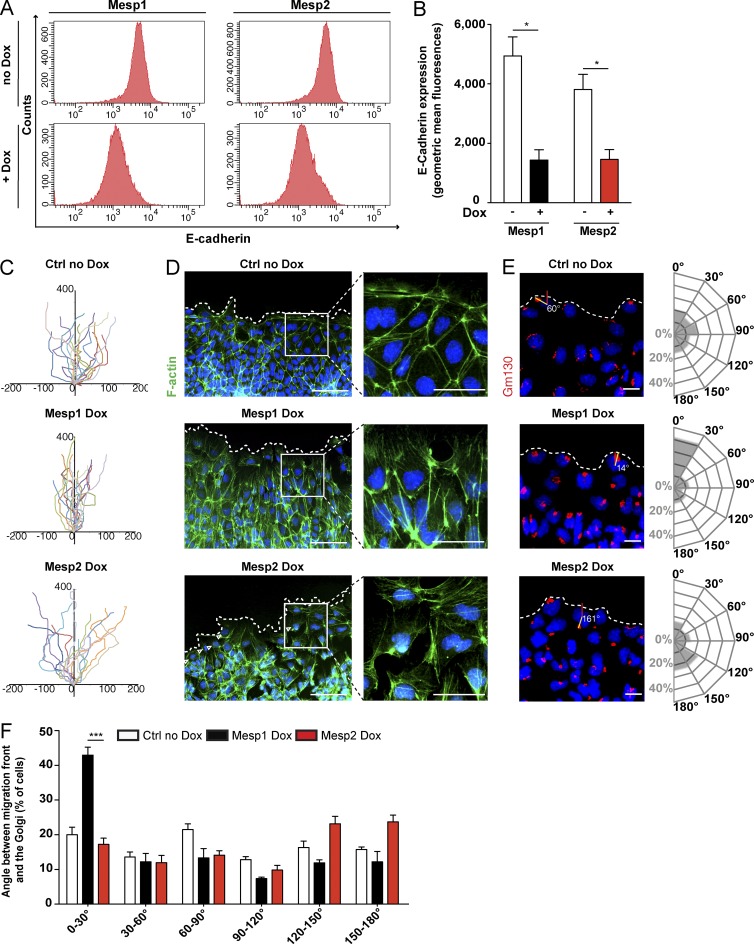
During mouse gastrulation, EMT and cytoskeleton remodeling are two critical processes, allowing the mesodermal cells to leave the PS and migrate to their specific destination (Burgess and Schroeder, 1979; Sugihara et al., 1998; Savagner, 2001; Nakaya and Sheng, 2008; Solnica-Krezel and Sepich, 2012). We and others have recently shown that Mesp1 promotes EMT through the direct up-regulation of EMT transcription factors such as Snail1 (Lindsley et al., 2008; Bondue et al., 2011; Lescroart et al., 2014). To determine whether the more rapid migration induced by Mesp1 is the consequence of its greater ability to promote EMT, we assessed the expression of E-cadherin in Mesp1- and Mesp2-inducible ESCs 48 h after Dox administration. FACS quantification of E-cadherin expression showed that Mesp2 down-regulated the expression of E-cadherin as efficiently as Mesp1 (Fig. 3, A and B), suggesting that the greater promigratory function of Mesp1 is not correlated with its ability to promote EMT.
Figure 3.
Mesp1 promotes oriented cell migration. (A) FACS quantification of E-cadherin expression in Mesp1- and Mesp2-overexpressing cells, 48 h after Dox addition, and their respective control cells. (B) Geometric mean fluorescence of E-cadherin expression in the different cell lines after Dox addition. Data represent mean ± SEM of three biologically independent experiments. (C) Time-lapse microscopy of individual migrating cells. Cells were tracked every 5 min during 11 h, and their paths were overlaid. Paths are oriented with respect to their starting point and the leading edge. The cell tracks shown are representative of five biologically independent experiments. (D) F-actin staining with Alexa Fluor 488–phalloidin (green) in the migrating front of control cells and Mesp1- and Mesp2-induced cells. Note the number of actin fibers oriented toward the migrating front in Mesp1-induced cells. (E) Cell polarity assessed by the orientation of the Golgi (stained with anti-GM130) with respect to the migrating front. Circular histograms show the proportion of cells at the migrating front with a given angle of polarization. Data represent the polarization angle of more than 300 cells counted in three biologically independent experiments. (F) Histogram showing the percentage of cells with different polarization in control conditions and Mesp1- and Mesp2-induced cells. Data represent mean ± SEM of three biologically independent experiments. At least 180 cells were counted for each condition. Control (no Dox) in C–F represents Mesp1 cells without Dox. No difference was observed between Mesp1 and Mesp2 no Dox cells. Bars: (D, left) 100 µm; (D, right) 50 µm; (E) 20 µm. *, P < 0.05; ***, P < 0.001.
At the single-cell level, the oriented migration of the prospective cardiac mesoderm involves a polarized process, in which cytoskeletal remodeling and cell protrusion, driven by actin polymerization, allow the directional cell motility (Elric and Etienne-Manneville, 2014). To further define the migrating properties induced by Mesp1 and Mesp2, we monitored the precise movement of single cells during time-lapse microscopy. Cells were tracked every 5 min from 0 to 11 h after scratch wound, and their paths were reconstructed (Fig. 3 C). This analysis revealed that although control cells and Mesp2-overexpressing cells exhibited several directional changes during their migratory path, Mesp1 cells migrated in a unidirectional manner toward the wound edge (Fig. 3 C).
To define the cellular mechanisms involved in the more efficient cell migration induced by Mesp1, we assessed the consequence of Mesp1- and Mesp2-induced expression on F-actin cytoskeleton. In control cells, F-actin was mainly restricted to the plasma membrane (Fig. 3 D). In contrast, Mesp1 expression induced a dramatic increase in the number of actin stress fibers oriented toward the leading edge (Fig. 3 D). In Mesp2-expressing cells, these actin stress fibers had no particular direction, and some cells presented almost no actin stress fiber (Fig. 3 D, arrowheads). These data show that Mesp1 induces a profound reorganization of the actin cytoskeleton, with the formation of multiple stress fibers oriented toward the leading edge of migrating cells.
To assess whether the unidirectional migration induced by Mesp1 is related to a change in cell polarity, we analyzed cell polarity by measuring the position of the Golgi (Gm130 immunostaining) with respect to the migration front of individual cells (Bershadsky and Futerman, 1994). The majority of Mesp1-expressing cells were polarized (Golgi apparatus between 0 and 30°) toward the migrating front, whereas in control and Mesp2-overexpressing cells, cell polarity was not oriented in a particular direction (Fig. 3, E and F). Altogether, these results demonstrate that Mesp1, but not Mesp2, promotes unidirectional migration by promoting cell polarity and actin cytoskeleton remodeling in migrating cells.
Common and unique Mesp1 target genes
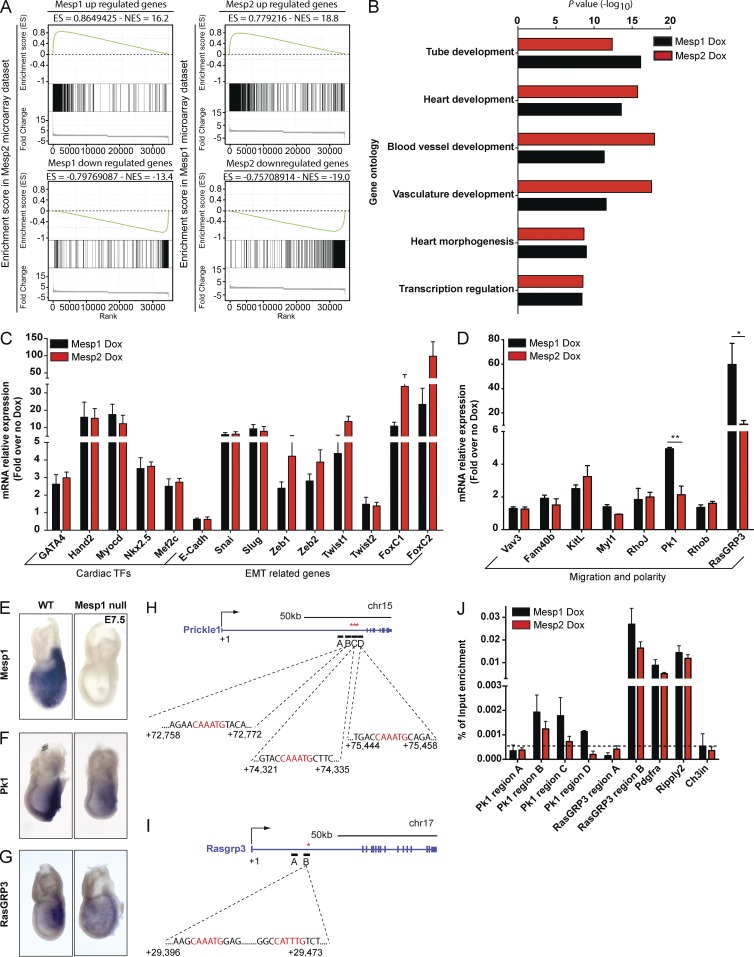
To uncover the unique molecular mechanisms by which Mesp1 promotes oriented and polarized cell migration, we compared the genes regulated after Mesp1 (Bondue et al., 2008) and Mesp2 overexpression. We first determined which genes displayed a change in expression of ≥1.5-fold after Dox administration in two separate biological replicates. Mesp2 overexpression increased the expression of 696 genes, whereas 535 genes were down-regulated, showing that both Mesp1 (Bondue et al., 2008) and Mesp2 positively and negatively regulated gene expression. Gene set enrichment analysis (GSEA) comparing Mesp1 and Mesp2 transcriptional profiles revealed that Mesp1 and Mesp2 positively and negatively controlled a common set of a large number of genes (Fig. 4 A). Gene ontology analysis showed that Mesp1- and Mesp2-overexpressing cells were highly and similarly enriched for genes controlling tube morphogenesis, heart morphogenesis and development, blood vessel and vasculature development, and transcriptional regulation, providing the molecular basis for the important functional redundancy of Mesp1 and Mesp2 during CP specification and cardiovascular differentiation (Fig. 4 B). qPCR for key cardiac transcription factors (such as GATA4, Hand2, Myocd, Nkx2.5, and Mef2c) and EMT-related regulators (such as Snai1, Slug, Zeb1, Zeb2, Twist1, Twist2, FoxC1, and FoxC2) showed that these genes were similarly up-regulated by Mesp1 and Mesp2 (Fig. 4 C), in good accordance with the functional experiments.
Figure 4.
Common and unique Mesp1 and Mesp2 target genes. (A) GSEA showing the common up-regulated and down-regulated genes induced by Mesp1 and Mesp2. The highly significant enrichment score (ES) and normalized enrichment score (NES) are shown for each analysis. (B) Comparison of gene ontology enrichment in Mesp1-overexpressing (black bars) and Mesp2-overexpressing (red bars) cells. (C and D) qPCR quantification of mRNA expression of cardiac and EMT-related transcription factors and genes involved in migration and cell polarity, 24 h after Mesp1-induced (black bars) or Mesp2-induced (red bars) expression. Results are normalized to the gene expression of the different transcripts in the absence of Dox. (E–G) In vivo in situ hybridization of Mesp1, Prickle1, and RasGRP3 transcripts, at E7.5, in WT and Mesp1-null embryos. Each in situ hybridization is representative of at least three independent hybridizations realized on at least three different litters. (H–J) ChIP-qPCR analysis of Mesp1 and Mesp2 on Prickle1 and RasGRP3 regulatory regions. (H and I) Schematic representation of regulatory regions of Prickle1 (H) and RasGRP3 (I) based on Mesp1-3HA ChiP-Seq data (unpublished data). The E-box sites are located in the first intron of Prickle1 and RasGRP3 and are indicated in red. In Prickle1, ChiP-Seq identified three regions (B, C, and D) containing Mesp1-binding sites (H). In RasGRP3, region B contains a Mesp1-binding site (I). In both genes, region A does not contain a Mesp1-binding site (negative control). Additional controls included Pdgfra and Ripply2, two genes similarly up-regulated by Mesp1 and Mesp2 (positive controls) and Ch3in (region in Chr3 not containing genes or Mesp1-binding sites used as negative control). (J) ChiP-qPCR of Mesp1 and Mesp2. Data represent mean and SEM of the relative enrichment over the input, recovered after ChIP with 3HA antibody in two biologically independent ChIP experiments. P-values for all GSEA comparisons equal 0. *, P < 0.05; **, P < 0.01.
Among the genes that were differentially regulated in Mesp1 and Mesp2 microarrays and that can potentially regulate cytoskeleton remodeling, polarity, and cell migration, we identified two genes, RasGRP3 and Prickle1, that were differently regulated by Mesp1 and Mesp2 (Fig. 4 D). RasGRP3 is a guanine nucleotide exchange factor that promotes the formation of active Ras-GTP, activates the extracellular signal-regulated kinase (ERK) signaling pathway (Yamashita et al., 2000), and is known to regulate cell motility (Brahmbhatt and Klemke, 2003; Scott et al., 2006; Mendoza et al., 2011). Prickle1 is a core component of the PCP pathway (Takeuchi et al., 2003). Both genes were much more strongly up-regulated by Mesp1 than Mesp2 (Fig. 4 D). These results suggest that, despite the high similarity between Mesp1 and Mesp2 transcriptional activities, which is consistent with their functional redundancy during cardiovascular differentiation and EMT, only Mesp1 strongly induced the expression of RasGRP3 and Prickle1, which may act downstream of Mesp1 in regulating cell polarity and directional migration.
To define whether RasGRP3 and Prickle1 are regulated by Mesp1 in vivo, we investigated the expression of these genes by in situ hybridization in WT and Mesp1-null mice at embryonic day 7.5 (E7.5). In situ hybridization of RasGRP3 and Prickle1 transcripts at E7.5 in WT mice overlapped with Mesp1 expression, and RasGRP3 and Prickle1 were both decreased in Mesp1-null embryos despite the up-regulation of Mesp2 (Kitajima et al., 2000; Fig. 4, E–G), showing that RasGRP3 and Prickle1 are both regulated by Mesp1 during embryonic development in vivo (Fig. 4, E–G).
Using chromatin immunoprecipitation followed by sequencing (ChIP-Seq), we assessed whether RasGRP3 and Prickle1 putative regulatory sequences were directly bound by Mesp1 during ESC differentiation. Mesp1 ChIP-Seq 12 h after Dox administration revealed that Mesp1 bound to one major site within the first intron of RasGRP3 and three different sites in the first intron of Prickle1 (not depicted and Fig. 4, H and I). ChIP-qPCR experiments performed on regions containing the Mesp1 binding sites identified by ChIP-Seq, which contained one or two E-boxes, were performed. Regions identified by ChiP-Seq containing one or several E-boxes were very strongly enriched after Mesp1 ChIP compared with the input DNA (Fig. 4, H–J). Interestingly, ChIP-qPCR performed under the same conditions after Mesp2 overexpression showed that whereas Mesp2 bound with the same efficiency as Mesp1 to their common induced target genes such as Pdgfra and Ripply2, Mesp2 did not bind the Prickle1 regulatory regions occupied by Mesp1 (Fig. 4, H and J), suggesting that the inability of Mesp2 to promote Prickle1 expression is related to the inability of Mesp2 to bind the Mesp1 binding sites in the regulatory regions of Prickle1. In contrast, Mesp2 bound to the RasGRP3 regulatory region almost as efficiently as Mesp1 (Fig. 4, I and J), potentially explaining the decrease but not the absence of RasGRP3 induction by Mesp2, and suggesting that other mechanisms besides the binding of Mesp2 to the regulatory region of RasGRP3 contribute to the difference in RasGRP3 up-regulation induced by Mesp1 and Mesp2. The specificity of these enriched regions was also confirmed by the absence of enrichment after Mesp1 ChIP of several other regions within the RasGRP3 and Prickle1 genes away from the E-box sites or the gene desert region in chromosome 3 (Ch3in) not bound by Mesp1 (Fig. 4, H–J). Altogether, these data show that Mesp1 directly controls the expression of RasGRP3 and Prickle1 in vitro and in vivo.
Mesp1/RasGRP3 promotes the speed of cell migration
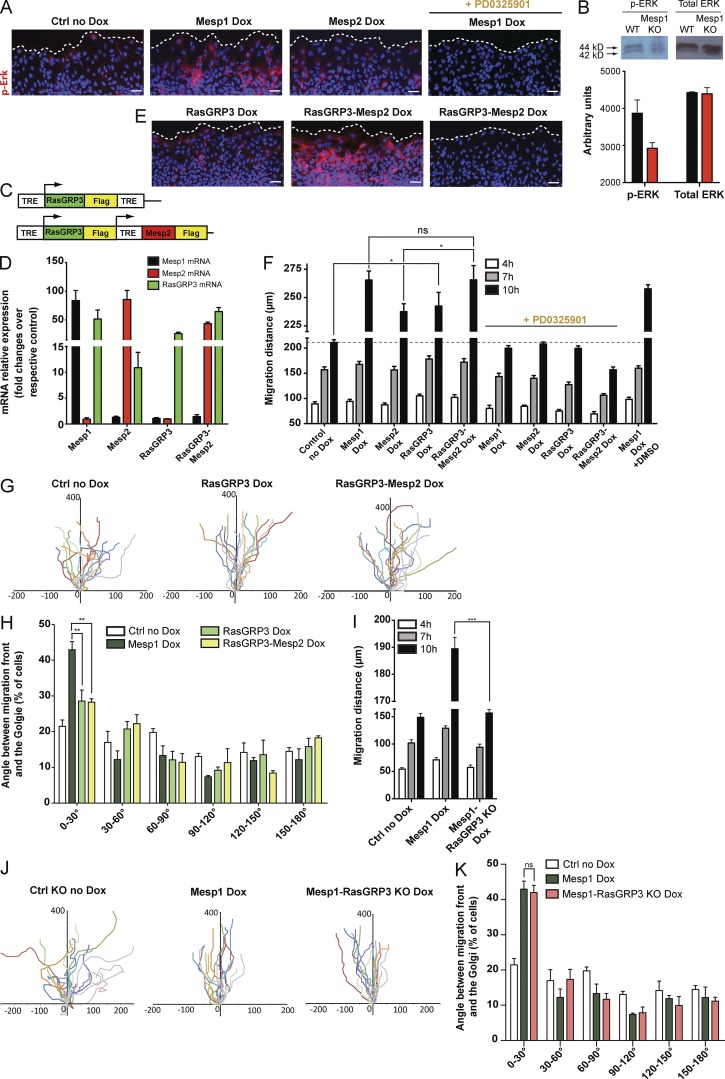
Because RasGRP3, through Ras activation, leads to ERK activation (Yamashita et al., 2000; Lorenzo et al., 2001), we assessed whether Mesp1 specifically promotes the activation of the Ras-ERK signaling pathway by monitoring phospho-ERK (p-ERK) immunostaining after Mesp1 or Mesp2 overexpression (Fig. 5 A). p-ERK immunostaining showed that Mesp1-overexpressing cells induced more robust ERK activation in migrating cells than did control or Mesp2-overexpressing cells (Fig. 5 A). ERK-specific inhibitor PD0325901 (Bain et al., 2007) abolished p-ERK immunostaining induced by Mesp1 overexpression (Fig. 5 A), showing the specificity of the assay and further demonstrating that Mesp1 specifically promotes ERK signaling in vitro. To assess whether Mesp1 modulates ERK signaling in vivo, we performed p-ERK Western blotting on whole embryos after removing the extraembryonic tissues in WT and Mesp1-KO mice, as p-ERK expression is much stronger in the extraembryonic tissues than the PS at E6.5 and E7.5 (Corson et al., 2003). Despite the absence of isolation of Mesp1-expressing cells in WT and Mesp1-null embryos, we observed a small but reproducible reduction of p-ERK in the total Mesp1-null embryos, whereas the total amount of ERK protein was unchanged (Fig. 5 B), consistent with the notion that Mesp1 promotes ERK signaling in vivo.
Figure 5.
The Mesp1/RasGRP3/ERK signaling axis controls the speed of cell migration. (A) p-ERK immunostaining at the migrating front after induced expression of Mesp1 or Mesp2 and in the presence of ERK inhibitor (PD0325901). (B) Western blot analysis of p-ERK in WT and Mesp1-KO embryos at E7.5. Western blots were scanned and quantified as described in Materials and methods. Results are represented as mean ± SEM of three biologically independent experiments. (C) Schematic representation of Dox-inducible constructs under the control of a TRE element, allowing the overexpression of RasGRP3 alone or combined with Mesp2. (D) qPCR analysis of the mRNA expression of the different transgenes, 24 h after Dox induction. (E) p-ERK immunostaining at the migrating front after induced expression of the different constructs and in the presence of PD0325901. Note the increase of p-ERK staining in Mesp1-overexpressing (A) and Mesp2/RasGRP3-overexpressing (E) cells that are abolished in the presence of ERK inhibitor. (F) Cell migration assessed by time-lapse microscopy in response to scratch wound. The migration distances were measured after 4, 7, and 10 h in the different experimental conditions. Data represent mean ± SEM of three biologically independent experiments. (G) Cells were tracked as described earlier, and all their paths were overlaid. The cell tracks shown are representative of three biologically independent experiments. (H) Cell polarity assessed by the orientation of the Golgi (stained with anti-GM130) with respect to the migrating front. Histogram showing the percentage of cells with different polarization in the different experimental conditions. Data represent mean ± SEM of three biologically independent experiments. At least 180 cells were counted for each condition. Control represents Mesp2/RasGRP3 in the absence of Dox cells. No difference was observed between the different cell lines in the absence of Dox. (I) Migration distance after Mesp1 overexpression in WT and RasGRP3-null cells at 4, 7, and 10 h after Dox administration. Data represent mean ± SEM of three biologically independent experiments. (J) Cells were tracked by time-lapse microscopy, and all their paths were overlaid. The cell tracks shown are representative of three biologically independent experiments. (K) Histogram showing the percentage of cells with different polarization in control, Mesp1-overexpressing, and Mesp1-overexpressing RasGRP3-deficient cells. Data represent mean ± SEM of three biologically independent experiments (minimum 150 cells counted). Bars: (A and E) 50 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
To define whether RasGRP3 promotes cell migration through ERK-signaling activation during cardiovascular specification, we generated two Dox-inducible ESC lines, allowing us to study the effect of RasGRP3 overexpression alone or in combination with Mesp2 (Fig. 5 C). Dox administration induced overexpression of the transgenes in all cell lines (Fig. 5 D). RasGRP3 alone induced a slight increase in ERK activation in the cells localized at the migration front and significantly faster migration in scratch wound assays compared with noninduced cells (Fig. 5, E and F). The small increase of p-ERK immunostaining after RasGRP3 overexpression alone compared with Mesp1 overexpression (Fig. 5 A) suggests that the activation of other signaling components is required in addition to RasGRP3 to promote the activation of ERK signaling downstream of Mesp1. Remarkably, combined overexpression of Mesp2 and RasGRP3 enhanced ERK activation and rescued the speed of cell migration of Mesp2-expressing cells to a similar extent as after Mesp1 expression (Fig. 5, E and F). These results indicate that Mesp1 controls the speed of cell migration through the induction of RasGRP3.
To determine whether RasGRP3 promotes the speed of cell migration through activation of the ERK signaling pathway, we monitored the speed of cell migration after Mesp1, Mesp2, RasGRP3, and RasGRP3/Mesp2 overexpression in the presence of ERK inhibitor PD0325901. Inhibition of ERK signaling prevented the increase in the speed of cell migration induced by Mesp1, Mesp2, RasGRP3, and RasGRP3/Mesp2 overexpression, showing that Mesp1 and its direct target gene RasGRP3 control the speed of cell migration by regulating ERK signaling (Fig. 5, A, E, and F).
To assess whether RasGRP3 also regulates cell polarity and oriented migration, we monitored the migration of individual cells by time-lapse microscopy and assessed their intrinsic polarity by assessing the position of the Golgi in respect to the migration front (Fig. 5, G and H). Overexpression of RasGRP3 alone or in combination with Mesp2 did not rescue the oriented cell migration induced by Mesp1 (Fig. 5, G and H). Likewise, overexpression of RasGRP3 alone or in combination with Mesp2 did not rescue the promotion of cell polarity induced by Mesp1 (Fig. 5 H).
To validate the data obtained after RasGRP3 overexpression, we performed clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 homozygous deletion of RasGRP3 in the context of Mesp1 gain of function. Consistent with the phenotype obtained after RasGRP3 gain of function, RasGRP3 loss of function strongly impaired the speed of cell migration induced by Mesp1 overexpression (Fig. 5 I), whereas RasGRP3 loss of function did not affect the promotion of cell polarity and unidirectional migration induced by Mesp1 (Fig. 5, J and K). These data indicate the important role of RasGRP3 downstream of Mesp1 in regulating the speed of migration, whereas other Mesp1 target genes regulate oriented cell migration.
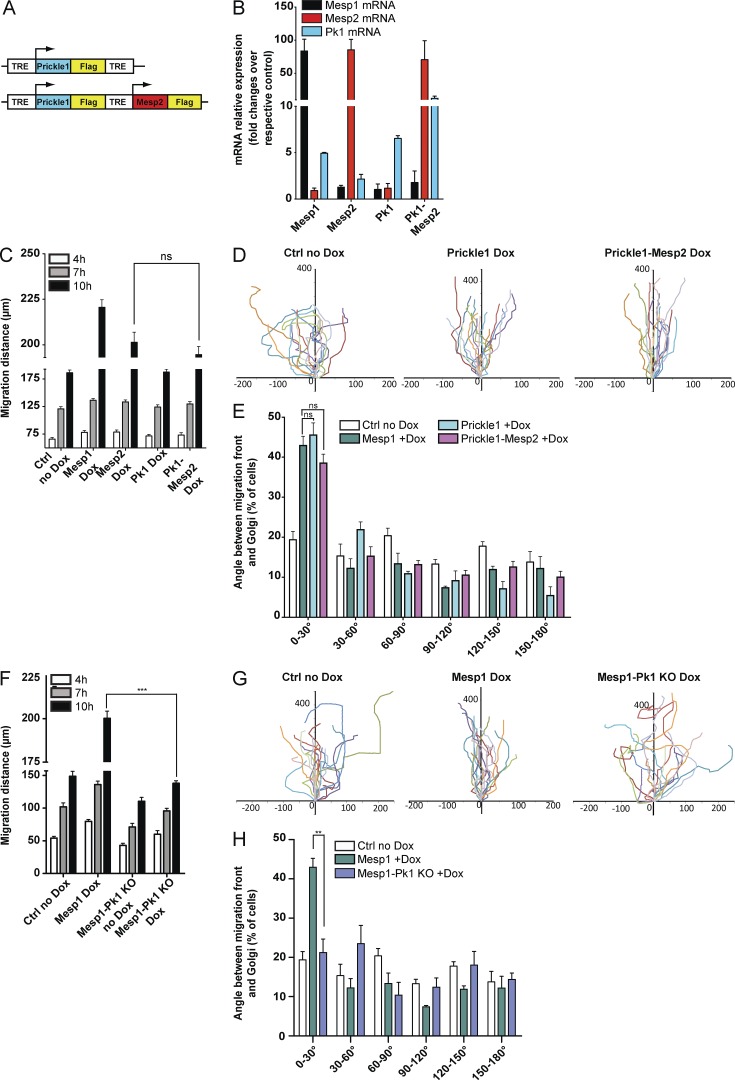
Mesp1/Prickle1 promotes oriented cell migration
Prickle1 deletion in mice leads to early critical morphological defects and tissue disorganization associated with abnormal cell polarity, leading to early lethality (Tao et al., 2009; Liu et al., 2014). To assess whether Prickle1 acts downstream of Mesp1 to control oriented cell migration, we generated Dox-inducible cell lines allowing the overexpression of Prickle1 alone or together with Mesp2, and assessed their effects on polarity and cell migration (Fig. 6, A and B). Overexpression of Prickle1 alone or in combination with Mesp2 did not influence the mean speed of cell migration (Fig. 6 C). However, cell tracking during wound scratch assays revealed that Prickle1 overexpression greatly promoted polarized cell migration, as shown by the stable orientation of the migrating cells that overexpressed Prickle1 (Fig. 6 D). Remarkably, the combined overexpression of Prickle1 and Mesp2 rescued the defect of oriented cell migration observed with Mesp2 overexpression (Fig. 6 D). In addition, Gm130 immunostaining showed that Prickle1 overexpression also rescued the polarity of migrating cells in response to Mesp2 expression (Fig. 6 E). CRISPR/Cas9 homozygous deletion of Prickle1 in Mesp1-overexpressing cells confirmed the importance of Prickle1 in the regulation of cell polarity and directional migration induced by Mesp1 (Fig. 6, F–H). Altogether, these results show that Mesp1 promotes polarized and oriented cell migration through the regulation of Prickle1 expression.
Figure 6.
Prickle1 acts downstream of Mesp1 to promote cell polarization and oriented cell migration. (A) Schematic representation of the Dox-inducible constructs under the control of a TRE element, allowing the overexpression of Prickle1 alone or in combination with Mesp2. (B) qPCR quantification of the mRNA expression of the different transgenes, 24 h after Dox induction. (C) Cell migration analyzed by time-lapse microscopy in response to scratch wound. Migration distances were measured after 4, 7, and 10 h in the different experimental conditions. Data represent mean ± SEM of three biologically independent experiments. (D) Cells were tracked as described earlier, and all their paths were overlaid. The cell tracks shown are representative of three biologically independent experiments. (E) Cell polarity assessed by the orientation of the Golgi (stained with anti-GM130) with respect to the migrating front. Histogram showing the percentage of cells with different polarization in the different experimental conditions (minimum 200 cells counted for each condition). Data represent mean ± SEM of three biologically independent experiments. Control (no Dox) shown in C represents Mesp1 cells without Dox, and in D and E it represents Prickle1/Mesp2 cells without Dox. No difference was observed between the different uninduced cells. (F) Migration distance after Mesp1 overexpression in WT and Prickle1 (Pk1)-deficient cells at 4, 7, and 10 h after Dox administration. Data represent mean ± SEM of three biologically independent experiments. (G) Cells were tracked by time-lapse microscopy, and all their paths were overlaid. The cell tracks shown are representative of three biologically independent experiments. (H) Histogram showing the percentage of cells with different polarization in control, Mesp1 overexpression in WT cells, and Mesp1 overexpression in Prickle1-deficient cells. Data represent mean ± SEM of three biologically independent experiments (minimum 150 cells counted). ***, P < 0.001; **, P < 0.01; ns, not significant.
Discussion
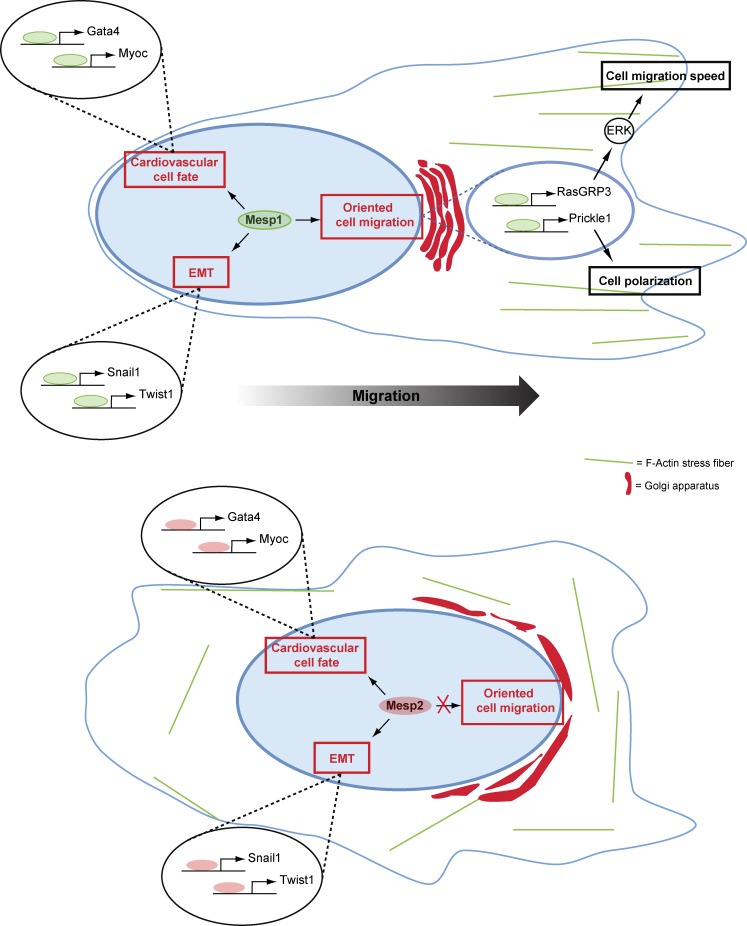
The precise migration of embryonic progenitors at the right place and at the right time is essential to coordinate the development of future tissues and organs. Defects in embryonic progenitor migration can lead to severe developmental defects (Kurosaka and Kashina, 2008; Herion et al., 2014). The heart is one of the first organs formed during embryonic development, and congenital heart diseases are the most common causes of developmental birth defects in humans (Pierpont et al., 2007). In mice embryos, CP specification and migration appear to occur concomitantly (Brand, 2003). Although extrinsic signals such as Wnt, FGF, and bone morphogenetic protein signaling pathways and EMT regulators have been shown to regulate exit from the PS and migration of cardiac progenitors (Christiaen et al., 2010; Camp and Munsterberg, 2011; Song et al., 2014), the intrinsic mechanisms driving their migration, and potentially linking CP specification and migration during mouse development, remain poorly understood. Here, we identified the molecular mechanisms by which Mesp1 acts at the interface of fate specification and cell migration, allowing coordination of CP specification and the rapid and oriented cell migration (Fig. 7).
Figure 7.
Model of Mesp1 functions during cardiovascular progenitor specification and oriented cell migration. During CP specification, Mesp1 directly controls cardiovascular progenitor cell fate decision, EMT, and oriented cell migration through the regulation of different sets of target genes, allowing the coordination of CP specification and migration. Whereas Mesp2 efficiently promotes the same Mesp1 target genes that control cardiovascular progenitor specification, EMT, and cardiovascular differentiation, only Mesp1 (and not Mesp2) controls the speed and orientation of progenitor cell migration.
By comparing Mesp1 and Mesp2 functions, we uncovered redundant roles of Mesp1 and Mesp2 in promoting CP specification, EMT, and cardiovascular differentiation. However, our study reveals that Mesp1 has a unique role in regulating cell polarity, speed, and directionality of cell migration and in coordinating CP specification and migration (Fig. 7), similar to what has been proposed for its ancestral gene Mesp during development of the early chordate Ciona (Christiaen et al., 2008). In contrast, Mesp2 was not as efficient as Mesp1 in increasing the speed of cell migration. In addition, the polarity and directionality of cell migration are promoted only by Mesp1 overexpression and not by Mesp2 (Fig. 7).
Molecular profiling of Mesp1- and Mesp2-overexpressing cells reveals that these two transcriptional factors induce a very similar transcription program and identifies a set of common target genes up-regulated by both Mesp1 and Mesp2, including many previously known direct Mesp1 target genes, such as transcription factors that promote cardiovascular development (such as Gata4, Hand2, or Myocardin) or EMT (such as Snail1, Twist1, Zeb1, Foxc1, or Foxc2; Bondue et al., 2008; Lindsley et al., 2008; Lescroart et al., 2014; Shi et al., 2015). This molecular profiling also identified several differentially regulated genes by Mesp1 and Mesp2, such as RasGRP3 and Prickle1, that could potentially explain the increased migratory properties induced by Mesp1 compared with Mesp2.
We identified RasGRP3 as preferentially induced by Mesp1 compared with Mesp2 and showed that RasGRP3 regulates the speed of cell migration induced by Mesp1. RasGRP3 was previously shown to be expressed in the nascent mesoderm (Costello et al., 2011) in Mesp1-H2B-GFP–expressing cells at E6.5 in vivo (Lescroart et al., 2014). Here, using ChiP-Seq confirmed by ChiP-qPCR, we showed that Mesp1 directly bound to a single site in the regulatory region of RasGRP3 in vitro and that RasGRP3 expression decreased in Mesp1-null mice in vivo, consistent with the notion that Mesp1 directly regulates RasGRP3 expression in early CPs in vitro and in vivo. Mesp2 was also able to bind to the same RasGRP3 enhancer region, although Mesp2 is much less potent than Mesp1 in promoting RasGRP3 up-regulation, suggesting that other mechanisms beside their binding to the DNA control the different capacity of Mesp1 and Mesp2 to promote RasGRP3 expression. RasGRP3 was initially identified as a Ras exchange factor promoting ERK signaling (Yamashita et al., 2000; Lorenzo et al., 2001). Overexpression of RasGRP3 alone minimally activated ERK phosphorylation but synergized with Mesp1 in promoting ERK signaling, suggesting that RasGRP3 is necessary but not sufficient to promote ERK signaling downstream of Mesp1, and other factors such as PDGFRa or KDR/Flk1 act downstream of Mesp1 together with RasGRP3 to promote ERK signaling at the leading edge of migrating cells. Furthermore, specific inhibition of ERK signaling abolishes the promigratory function of Mesp1; coexpression of RasGRP3 together with Mesp2 rescues ERK signaling and the speed of migration of Mesp2-overexpressing cells, to a level similar to that of Mesp1; and finally, deletion of RasGRP3 decreases the promigratory function of Mesp1, showing the importance of the Mesp1/RasGRP3/ERK axis in the regulation of the speed of CP migration. Because different ligands, such as FGFs, VEGFs, and PDGFs, activate ERK signaling (Beh et al., 2007), the up-regulation of RasGRP3 by Mesp1 may accelerate the speed of CP migration in response to extracellular signals. Whereas RasGRP3 promotes cell migration downstream of Mesp1 in vitro, Rasgrp3-null mice can undergo gastrulation (Coughlin et al., 2005), suggesting that other compensatory mechanisms can substitute for Rasgrp3 function during mouse embryonic development.
PCP describes the collective alignment of cell polarity across a tissue, which by integrating global directional cues regulates individual cell polarity (Zallen, 2007). During gastrulation, PCP pathways regulate movements of convergence and extension, which narrow the mediolateral axis and elongate the anteroposterior axis (Yin et al., 2009). We found that only Mesp1-overexpressing cells, and not Mesp2-overexpressing cells, are strongly polarized during migration, as revealed by the shape of the cells, the direction of actin stress fibers, and the position of the Golgi, which are all polarized toward the migration front. We identified Prickle1, a member of the PCP pathway (Jenny et al., 2005; Seifert and Mlodzik, 2007), as being up-regulated specifically by Mesp1 and not by Mesp2, and which controls cell polarity and oriented cell migration downstream of Mesp1. Prickle1, which regulates the asymmetrical distribution of PCP proteins (Zallen, 2007), is expressed in the PS concomitant with Mesp1 (Saga et al., 2000; Crompton et al., 2007), and Prickle1 deletion induces defects in PS formation during mouse gastrulation (Tao et al., 2009). ChiP-qPCR showed that only Mesp1 but not Mesp2 bound the regulatory regions of Prickle1, explaining the inability of Mesp2 to regulate Prickle1 expression and the decreased expression of Prickle1 in Mesp1-null mice. Consistent with the importance of Prickle1 in regulating cell polarity in migrating cells downstream of Mesp1, overexpressing Prickle1 together with Mesp2 rescues the defect in cell polarity to a level comparable to that of Mesp1-expressing cells, and deletion of Prickle1 abolishes the promigratory function of Mesp1 in vitro. Future studies will be needed to further understand the molecular mechanisms by which Mesp1 and Prickle1 control cell polarity and migration during embryonic development.
In conclusion, our study shows the important functional redundancy between Mesp1 and Mesp2 in promoting CP specification, EMT, and cardiac differentiation and identifies a unique promigratory function of Mesp1 in regulating the speed and orientation of cell migration by regulating Prickle1 and Ras-GRP3 expression (Fig. 7). Future studies are important to delineate further the role of Mesp1 and the mechanisms by which it promotes the migration of CP in vivo during embryonic development. These results have important implications for better understanding the mechanisms underlying congenital heart defects and other organ malformations associated with defective cell migration.
Materials and methods
Tetracycline-inducible ESC lines
Mesp2, RasGRP3, and Prickle1 ORFs were amplified by PCR, cloned into the p2Lox vector (Bondue et al., 2008), and validated by Sanger sequencing. For the double constructs such as RasGRP3 or Prickle1 with Mesp2 in A2Lox cells, each ORF contained its own TRE elements (Bondue et al., 2011). For the Mesp1-triple-HA and Flag-tagged construct, the triple HA or Flag-tag was cloned downstream of Mesp1 and Mesp2 ORF in the same p2Lox vector. These constructs were electroporated with the pSalCre vector in A2Lox cells, and stable knock-in cell lines were selected as previously described (Bondue et al., 2008). For the functional characterization of the different genes, three different clones for each inducible ESC line were tested.
CRISPR/Cas9 knockout ESCs
Target sites in Prickle1 and RasGRP3 for Cas9 were designed using the CRISPR online tool (http://crispr.mit.edu/), and the vectors pX330-U6-Chimeric_BB-CBh-hSpCas9 (PX330) and pSpCas9n (BB)-2A-GFP (PX461) were obtained from Addgene (plasmids #42230 and #48140). The oligo pairs encoding the guide sequence were cloned into a modified short version of PX330 in which the Cas9 cassette was removed. The plasmids containing guide sequence together with PX461 (nickase Cas9) were transfected into the relevant ESC line using Lipofectamine 2000 (Invitrogen). 48 h after transfection, cells were dissociated into single-cell suspensions and diluted in 2i medium to a final concentration of 0.5–1 cell per 100 µl. The diluted cells were plated into at least two 96-well plates (100 µl medium per well). After culture in 2i medium for 10–14 d, single-cell clones were screened through genotyping using the PCR primers that flanked the deleted region mediated by Cas9. Prickle1 and Rasgrp3 homozygous null ESCs were selected for further functional study.
ESC culture and differentiation
ESCs were cultured on irradiated mouse embryonic fibroblasts in DMEM (Invitrogen) supplemented with 15% ESC-qualified FBS (Invitrogen), 0.1 mM nonessential amino acids (Invitrogen), 1 mM sodium-pyruvate (Invitrogen), 0.1 mM β-mercaptoethanol (Sigma-Aldrich), 100 U/ml penicillin (Invitrogen), 100 µg/ml streptomycin (Invitrogen), and 1,000 U/ml leukemia inhibitory factor (ESGRO). ESCs were replated on gelatin-coated plates without fibroblasts 2 d before differentiation, and differentiation was performed by hanging drops of 1,000 cells in 25 µl differentiation medium (the same medium without leukemia inhibitory factor but containing 0.5 mM ascorbic acid [Sigma-Aldrich]), as previously described (Bondue et al., 2008). Dox (Sigma-Aldrich) was added to hanging drops at day 2, at the indicated concentration for Mesp1- and Mesp2-inducible ESCs (0.08 and 1 µg/ml, respectively) and at 1 µg/ml for Prickle1-, Prickle1/Mesp2-, RasGRP3-, and RasGRP3/Mesp2-inducible ESCs. After 4 d in hanging drops, EBs were replated on gelatin-coated dishes for further differentiation. For chimeric EB experiments, Mesp1- or GFP-inducible ESC lines were mixed with the control DsRed-inducible ESC line in equal proportions. To inhibit specifically the ERK signaling pathway during ESC differentiation, PD0325901 (Stem Gent) was added to the differentiation medium at a final concentration of 1 µM.
Mice
Mesp1v1/+ (Mesp1 KO; Saga et al., 1999) mice were obtained from the Riken Institute. Mice colonies were maintained in a certified animal facility in accordance with European guidelines. These experiments were approved by the local ethics committee under the reference #LA1230332 (CEBEA).
Western blot
10 µg protein was loaded and separated according to size by electrophoresis on a 10% SDS-PAGE gel, transferred on a polyvinylidene difluoride membrane, blocked for 1 h with TBS + 0.1 Tween + 3% BSA (TBST), incubated with a specific primary antibody overnight at 4°C, washed three times with TBST, incubated with hypoxanthine-guanine phosphoribosyltransferase (HPRT)-coupled corresponding secondary antibody, washed three times with TBST, and revealed with a Western Lighting Plus-ECL kit (PerkinElmer). M5-flagged transgene was quantified using M5 antibody (1:1,000; Sigma-Aldrich) as primary antibody, and HPRT-coupled anti-mouse secondary antibody (1:3,000; GE Healthcare). The amount of loaded protein was normalized using an anti–β-actin antibody (1:3,000; Abcam). Phosphorylated ERK was quantified in whole WT and Mesp1-KO embryos at E7.25 using Phospho-p44-42 MAPK rabbit polyclonal antibody (1:500; Cell Signaling Technology) and HPRT-coupled anti-rabbit secondary antibody (1:30,000; GE Healthcare). At least three WT and Mesp1-KO embryos were pooled after genotyping using extraembryonic tissue. The amount of loaded protein was normalized using an antibody against total ERK p44-42 MAPK rabbit monoclonal antibody (1:1,000; Cell Signaling Technology). For quantification of Western blot, ECL signals in the linear range were scanned and quantified using ImageJ 1.48s. Images in Figs. 1 B and 5 B are representative of three biologically independent experiments.
RNA isolation, reverse transcription, and qPCR
RNA extractions were performed using an Absolutely RNA Microprep kit (Agilent Technologies), according to the manufacturer’s recommendations. For each experiment, the same amount of RNA (mean 1 µg) was used to synthesize cDNA in a 50-µl final volume using Superscript II (Invitrogen) and random hexamers (Roche). Genomic DNA contaminations were avoided by treatment with DNaseI (Absolutely RNA Microprep), and control of genomic contamination was measured by performing the same procedure without reverse transcription. qPCR experiments were performed with 5 ng cDNA per reaction, using a KAPA SYBR FAST reagent (Kapabiosystems) on an Agilent Mx3005P qPCR System. The relative expressions of each gene were normalized to their expression in the respective control condition. All primers were designed using Lasergene 7.2 software (DNAStar) and are presented in Tables S1 and S2. Analyses of the results were performed using Mx Pro-Mx3005P v4.10 and GraphPad Prism software.
Immunofluorescence analysis
For immunofluorescence experiments, cells were plated on coverslips coated with gelatin (for EBs) or fibronectin (for in vitro scratch assays), fixed with 4% paraformaldehyde for 10 min (EBs) or 5 min (cell monolayers) at 4°C, washed three times in PBS, and incubated for 1 h at RT with a blocking solution containing 1% BSA, 0.2% Triton X-100, and 5% horse serum in PBS, before overnight incubation at 4°C with the primary antibodies. Antibodies used were as follows: anti-cTnT (clone 13-11, mouse monoclonal, 1:100; NeoMarkers), anti-Mlc2v (mouse monoclonal, 1:25; Alexis Corp.), anti-Mlc2a (mouse monoclonal, 1:200; Synaptic Systems), anti–VE-cadherin (clone 11D4.1, rat monoclonal, 1:100; BD), anti-Gm130 (clone EP892Y, rabbit monoclonal, 1:500; Abcam), and anti–p-ERK (Phospho-p44-42 MAPK rabbit polyclonal antibody, 1:500; Cell Signaling Technology). For p-ERK staining, cells were incubated for 10 min in 100% methanol at −20°C and washed three times with PBS before blocking and staining. Primary antibodies were revealed with appropriate RRX-coupled secondary antibody from Jackson ImmunoResearch Laboratories, Inc. (1:400). F-actin was stained with Alexa Fluor 488–phalloidin (A12379, 1:40; Thermo Fisher Scientific). Counterstaining of nuclei was performed with Hoechst (1:2,000). All immunostainings were mounted using DABCO (Sigma-Aldrich) as mounting medium. Single images and mosaics were acquired on an Axio Imager with an Axiocam MRn camera and using Axiovision Rel. v4.8.2.0 software (ZEISS). Acquisitions were performed at RT using 10× and 20× EC Plan Neofluar objectives (10×-0.3 numerical aperture and 20×-0.4 numerical aperture; ZEISS). Mosaics were generated using a 10% overlap between each single acquisition. Each representative image has been reproduced in at least three independent experiments.
Flow cytometry
For intracellular staining, EBs were dissociated by trypsinization and permeabilized with BD Cytofix-Cytoperm kit according to the manufacturer’s recommendations. For staining of the different cell-surface markers, EBs were dissociated in 3 mM EDTA. Anti-cTnT staining (NeoMarkers) was performed for 30 min in Perm-Wash buffer (BD Cytofix-Cytoperm kit) at a final concentration of 1:100 and revealed with an anti-mouse phosphatidylethanolamine (PE) secondary antibody (BD) at a final concentration of 1:400. For staining of cell-surface markers, all antibodies were diluted in PBS-BSA 1%. Flk1 (VEGFR2) was stained using a biotinylated antibody at 1:100 (clone Avas12a1; eBioscience) revealed by a streptavidin-PE-Cy7 secondary antibody at 1:400 (BD). PDGFRa was directly coupled to PE and used at 1:75 (clone APA5; eBioscience). CXCR4 antibody was directly coupled to A647 and used at 1:100 (clone 2B11; eBioscience). CD31 expression was directly coupled to PE and used at 1:100 (clone MEC 13.3; BD). Living cells were gated by propidium iodide dye exclusion (1 µg/ml). FACS analyses were performed on a FACSFortessa or FACSCalibur (BD) using BD FACS Diva v6.2 and CellQuest Pro software, respectively. Each FACS result is representative of at least three independent experiments.
In vitro scratch wound assay
Uniform wounds were made using Culture-Insert (#80209; ibidi) on fibronectin-coated plates. This approach provides two cell culture chambers separated by a physical barrier 500 µm thick. EBs were dissociated with Accutase (Invitrogen), and after neutralization and resuspension, 70-µl cell suspensions containing 80,000 cells were seeded on each chamber of the culture insert. The physical barrier separating the two cell fields (creating the wound) was removed after cell adhesion, 2 h after replating, and the wound closure was analyzed by time-lapse microscopy during a period of at least 12 h. All images in Fig. 2 C are representative of three independent experiments.
Live-sample imaging and analysis
Time-lapse imaging was performed using a Leica DMI6000B microscope mounted with a cell culture chamber, which allowed maintenance of optimal cell culture conditions (95% relative humidity, 37°C, 5% CO2) during the acquisition process. Acquisitions were taken every 5 min during at least 12 h using a DFC365FX camera and LAS AF v2.6.0.7235 software. The different migration manual cell tracks were realized using ImageJ software. All representative cell tracks illustrated in Figs. 3 C, 5 E, and 6 D were replicated in at least three independent experiments.
Microarray analysis
For microarray analysis, Mesp2-induced cells were harvested 24 h after Dox induction, and total RNA extraction and DNase treatment were performed using an Absolutely RNA Microprep kit as described earlier. RNA isolation and microarray analysis were performed in two biologically independent replicates as previously described (Bondue et al., 2008) using mouse genome 430 2.0 arrays (Affymetrix) at Nucleomics Core, VIB facility, Flanders, Belgium. To compare Mesp2 with Mesp1 microarrays, we considered all regulated genes (fold change ≥1.5) in both arrays. Gene Set Enrichment Analysis (GSEA; Subramanian et al., 2005) was downloaded from the Broad Institute website (http://www.broadinstitute.org/gsea/). We used the GSEA preranked option with standard parameters of weighted enrichment score calculation to run the GSEA against a user-supplied fold-change–ranked list of genes. Results of the enrichment analysis were plotted using R software. A functional annotation chart of genes enriched in Mesp1- or Mesp2-induced cells was performed on DAVID bioinformatic resources v6.7 (http://david.abcc.ncifcrf.gov/).
In situ hybridization
Embryos were extracted at E7.5, fixed overnight in 4% paraformaldehyde, and processed as previously described (Lescroart et al., 2014). The hybridization signal was revealed using BM purple (Roche) for all antisense probes. Chromogenic substrate and embryos were acquired in PBST (0.1% Tween 20) with a V16 stereomicroscope (ZEISS). Acquisition data were treated with Zen blue software and exported in TIF image format. All in situ hybridizations were performed on at least three different litters in three independent experiments. Antisense riboprobes for RasGRP3 (Costello et al., 2011) and Prickle1 (Crompton et al., 2007) were synthesized from vectors provided by S. Arnold (University Medical Center, Freiburg, Germany) and T.A. Rodriguez (Medical Research Council Clinical Sciences Centre, London, UK), respectively.
Mesp1 and Mesp2 ChIP-qPCR analysis
ChIP-qPCR was performed on Mesp1-3HA–flagged and Mesp2-3HA–flagged Dox-inducible ESC lines. Approximately 1,000 EBs were collected 20 h after induction of Dox (0.1 µg/ml Dox for Mesp1 and 1 µg/ml for Mesp2), fixed directly with 1% formaldehyde for 7 min at RT, and quenched with 0.125 M glycine for 5 min. EBs were lysed according to the manufacturer’s instructions (ChIP-IT express kit; Active Motif), and cross-linked DNAs were sonicated for 10–15 cycles (30 s on/30 s off) by a Bioruptor Sonicator (Diagenode). Sheared DNAs have a mean range of ∼300 bp. ChIP was performed using Chip grade anti-3HA antibody (ab9110; Abcam) according to the manufacturer’s instructions (ChIP-IT express kit).
Statistical analysis
Statistical significance was calculated using Student’s t test.
Online supplemental material
Tables S1 and S2 show qPCR primers. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201505082/DC1.
Supplementary Material
Acknowledgments
We thank S. Brohée for his help with gene set enrichment analysis. We thank Sebastian Arnold and Tristan A. Rodriguez for kindly providing the probes for in situ hybridization. We thank Navrita Mathiah, Tatiana Trevenco, and Nadia Aghbal for their help during their internships.
G. Chiapparo and S. Chabab were supported by a fellowship of the Fonds pour la Formation à la Recherche dans l’Industrie et dans l’Agriculture. F. Lescroart is supported by the European Molecular Biology Organization long-term fellowship. X. Lin is supported by the Fonds de la Recherche Scientifique - FNRS. A. Bondue was supported by the Fonds de la Recherche Scientifique - FNRS and by the Tagnon Fund. C. Blanpain is an investigator of WELBIO. This work was supported by the Fonds de la Recherche Scientifique - FNRS, Fonds pour la Formation à la Recherche dans l’Industrie et dans l’Agriculture, the Université Libre de Bruxelles Foundation, and the Bettencourt Schueller Foundation (F. Lescroart and C. Blanpain).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ChIP
- chromatin immunoprecipitation
- CM
- cardiomyocyte
- CP
- cardiovascular progenitor
- CRISPR
- clustered regularly interspaced short palindromic repeats
- cTnT
- cardiac troponin T
- EB
- embryonic body
- EC
- endothelial cell
- EMT
- epithelial–mesenchymal transition
- ERK
- extracellular signal-regulated kinase
- ESC
- embryonic stem cell
- GSEA
- gene set enrichment analysis
- HPRT
- hypoxanthine-guanine phosphoribosyltransferase
- KO
- knockout
- PCP
- planar cell polarity
- PE
- phosphatidylethanolamine
- p-ERK
- phospho-ERK
- PS
- primitive streak
- qPCR
- quantitative RT-PCR
- VE-cadherin
- vascular endothelial cadherin
- WT
- wild-type
References
- Abu-Issa R., and Kirby M.L.. 2007. Heart field: From mesoderm to heart tube. Annu. Rev. Cell Dev. Biol. 23:45–68. 10.1146/annurev.cellbio.23.090506.123331 [DOI] [PubMed] [Google Scholar]
- Bain J., Plater L., Elliott M., Shpiro N., Hastie C.J., McLauchlan H., Klevernic I., Arthur J.S., Alessi D.R., and Cohen P.. 2007. The selectivity of protein kinase inhibitors: A further update. Biochem. J. 408:297–315. 10.1042/BJ20070797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh J., Shi W., Levine M., Davidson B., and Christiaen L.. 2007. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development. 134:3297–3305. 10.1242/dev.010140 [DOI] [PubMed] [Google Scholar]
- Bershadsky A.D., and Futerman A.H.. 1994. Disruption of the Golgi apparatus by brefeldin A blocks cell polarization and inhibits directed cell migration. Proc. Natl. Acad. Sci. USA. 91:5686–5689. 10.1073/pnas.91.12.5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondue A., and Blanpain C.. 2010. Mesp1: A key regulator of cardiovascular lineage commitment. Circ. Res. 107:1414–1427. 10.1161/CIRCRESAHA.110.227058 [DOI] [PubMed] [Google Scholar]
- Bondue A., Lapouge G., Paulissen C., Semeraro C., Iacovino M., Kyba M., and Blanpain C.. 2008. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 3:69–84. 10.1016/j.stem.2008.06.009 [DOI] [PubMed] [Google Scholar]
- Bondue A., Tännler S., Chiapparo G., Chabab S., Ramialison M., Paulissen C., Beck B., Harvey R., and Blanpain C.. 2011. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J. Cell Biol. 192:751–765. 10.1083/jcb.201007063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmbhatt A.A., and Klemke R.L.. 2003. ERK and RhoA differentially regulate pseudopodia growth and retraction during chemotaxis. J. Biol. Chem. 278:13016–13025. 10.1074/jbc.M211873200 [DOI] [PubMed] [Google Scholar]
- Brand T. 2003. Heart development: Molecular insights into cardiac specification and early morphogenesis. Dev. Biol. 258:1–19. 10.1016/S0012-1606(03)00112-X [DOI] [PubMed] [Google Scholar]
- Buckingham M., Meilhac S., and Zaffran S.. 2005. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 6:826–835. 10.1038/nrg1710 [DOI] [PubMed] [Google Scholar]
- Burgess D.R., and Schroeder T.E.. 1979. The cytoskeleton and cytomusculature in embryogenesis—an overview. Methods Achiev. Exp. Pathol. 8:171–189. [PubMed] [Google Scholar]
- Camp E., and Munsterberg A.. 2011. Ingression, migration and early differentiation of cardiac progenitors. Front. Biosci. (Landmark Ed.). 16:2416–2426. 10.2741/3863 [DOI] [PubMed] [Google Scholar]
- Christiaen L., Davidson B., Kawashima T., Powell W., Nolla H., Vranizan K., and Levine M.. 2008. The transcription/migration interface in heart precursors of Ciona intestinalis. Science. 320:1349–1352. 10.1126/science.1158170 [DOI] [PubMed] [Google Scholar]
- Christiaen L., Stolfi A., and Levine M.. 2010. BMP signaling coordinates gene expression and cell migration during precardiac mesoderm development. Dev. Biol. 340:179–187. 10.1016/j.ydbio.2009.11.006 [DOI] [PubMed] [Google Scholar]
- Corson L.B., Yamanaka Y., Lai K.M., and Rossant J.. 2003. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 130:4527–4537. 10.1242/dev.00669 [DOI] [PubMed] [Google Scholar]
- Costello I., Pimeisl I.M., Dräger S., Bikoff E.K., Robertson E.J., and Arnold S.J.. 2011. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat. Cell Biol. 13:1084–1091. 10.1038/ncb2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin J.J., Stang S.L., Dower N.A., and Stone J.C.. 2005. RasGRP1 and RasGRP3 regulate B cell proliferation by facilitating B cell receptor-Ras signaling. J. Immunol. 175:7179–7184. 10.4049/jimmunol.175.11.7179 [DOI] [PubMed] [Google Scholar]
- Crompton L.A., Du Roure C., and Rodriguez T.A.. 2007. Early embryonic expression patterns of the mouse Flamingo and Prickle orthologues. Dev. Dyn. 236:3137–3143. 10.1002/dvdy.21338 [DOI] [PubMed] [Google Scholar]
- David R., Brenner C., Stieber J., Schwarz F., Brunner S., Vollmer M., Mentele E., Müller-Höcker J., Kitajima S., Lickert H., et al. 2008. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat. Cell Biol. 10:338–345. 10.1038/ncb1696 [DOI] [PubMed] [Google Scholar]
- Elric J., and Etienne-Manneville S.. 2014. Centrosome positioning in polarized cells: Common themes and variations. Exp. Cell Res. 328:240–248. 10.1016/j.yexcr.2014.09.004 [DOI] [PubMed] [Google Scholar]
- Herion N.J., Salbaum J.M., and Kappen C.. 2014. Traffic jam in the primitive streak: The role of defective mesoderm migration in birth defects. Birth Defects Res. A Clin. Mol. Teratol. 100:608–622. 10.1002/bdra.23283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A., Reynolds-Kenneally J., Das G., Burnett M., and Mlodzik M.. 2005. Diego and Prickle regulate Frizzled planar cell polarity signalling by competing for Dishevelled binding. Nat. Cell Biol. 7:691–697. 10.1038/ncb1271 [DOI] [PubMed] [Google Scholar]
- Kelly R.G., Brown N.A., and Buckingham M.E.. 2001. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev. Cell. 1:435–440. 10.1016/S1534-5807(01)00040-5 [DOI] [PubMed] [Google Scholar]
- Kitajima S., Takagi A., Inoue T., and Saga Y.. 2000. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 127:3215–3226. [DOI] [PubMed] [Google Scholar]
- Kurosaka S., and Kashina A.. 2008. Cell biology of embryonic migration. Birth Defects Res. C Embryo Today. 84:102–122. 10.1002/bdrc.20125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescroart F., Chabab S., Lin X., Rulands S., Paulissen C., Rodolosse A., Auer H., Achouri Y., Dubois C., Bondue A., et al. 2014. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat. Cell Biol. 16:829–840. 10.1038/ncb3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley R.C., Gill J.G., Murphy T.L., Langer E.M., Cai M., Mashayekhi M., Wang W., Niwa N., Nerbonne J.M., Kyba M., and Murphy K.M.. 2008. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 3:55–68. 10.1016/j.stem.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Lin C., Gao C., May-Simera H., Swaroop A., and Li T.. 2014. Null and hypomorph Prickle1 alleles in mice phenocopy human Robinow syndrome and disrupt signaling downstream of Wnt5a. Biol. Open. 3:861–870. 10.1242/bio.20148375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo P.S., Kung J.W., Bottorff D.A., Garfield S.H., Stone J.C., and Blumberg P.M.. 2001. Phorbol esters modulate the Ras exchange factor RasGRP3. Cancer Res. 61:943–949. [PubMed] [Google Scholar]
- Meilhac S.M., Esner M., Kelly R.G., Nicolas J.F., and Buckingham M.E.. 2004. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev. Cell. 6:685–698. 10.1016/S1534-5807(04)00133-9 [DOI] [PubMed] [Google Scholar]
- Mendoza M.C., Er E.E., Zhang W., Ballif B.A., Elliott H.L., Danuser G., and Blenis J.. 2011. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol. Cell. 41:661–671. 10.1016/j.molcel.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto M., Takahashi Y., Endo M., and Saga Y.. 2005. The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature. 435:354–359. 10.1038/nature03591 [DOI] [PubMed] [Google Scholar]
- Nakaya Y., and Sheng G.. 2008. Epithelial to mesenchymal transition during gastrulation: an embryological view. Dev. Growth Differ. 50:755–766. 10.1111/j.1440-169X.2008.01070.x [DOI] [PubMed] [Google Scholar]
- Pierpont M.E., Basson C.T., Benson D.W. Jr., Gelb B.D., Giglia T.M., Goldmuntz E., McGee G., Sable C.A., Srivastava D., and Webb C.L.. 2007. Genetic basis for congenital heart defects: Current knowledge: A scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 115:3015–3038. 10.1161/CIRCULATIONAHA.106.183056 [DOI] [PubMed] [Google Scholar]
- Saga Y., Hata N., Koseki H., and Taketo M.M.. 1997. Mesp2: A novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev. 11:1827–1839. 10.1101/gad.11.14.1827 [DOI] [PubMed] [Google Scholar]
- Saga Y., Miyagawa-Tomita S., Takagi A., Kitajima S., Miyazaki Ji., and Inoue T.. 1999. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 126:3437–3447. [DOI] [PubMed] [Google Scholar]
- Saga Y., Kitajima S., and Miyagawa-Tomita S.. 2000. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc. Med. 10:345–352. 10.1016/S1050-1738(01)00069-X [DOI] [PubMed] [Google Scholar]
- Savagner P. 2001. Leaving the neighborhood: Molecular mechanisms involved during epithelial-mesenchymal transition. BioEssays. 23:912–923. 10.1002/bies.1132 [DOI] [PubMed] [Google Scholar]
- Scott M.G., Pierotti V., Storez H., Lindberg E., Thuret A., Muntaner O., Labbé-Jullié C., Pitcher J.A., and Marullo S.. 2006. Cooperative regulation of extracellular signal-regulated kinase activation and cell shape change by filamin A and beta-arrestins. Mol. Cell. Biol. 26:3432–3445. 10.1128/MCB.26.9.3432-3445.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert J.R., and Mlodzik M.. 2007. Frizzled/PCP signalling: A conserved mechanism regulating cell polarity and directed motility. Nat. Rev. Genet. 8:126–138. 10.1038/nrg2042 [DOI] [PubMed] [Google Scholar]
- Shi X., Zirbes K.M., Rasmussen T.L., Ferdous A., Garry M.G., Koyano-Nakagawa N., and Garry D.J.. 2015. The transcription factor Mesp1 interacts with cAMP-responsive element binding protein 1 (Creb1) and coactivates Ets variant 2 (Etv2) gene expression. J. Biol. Chem. 290:9614–9625. 10.1074/jbc.M114.614628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnica-Krezel L., and Sepich D.S.. 2012. Gastrulation: Making and shaping germ layers. Annu. Rev. Cell Dev. Biol. 28:687–717. 10.1146/annurev-cellbio-092910-154043 [DOI] [PubMed] [Google Scholar]
- Song J., McColl J., Camp E., Kennerley N., Mok G.F., McCormick D., Grocott T., Wheeler G.N., and Münsterberg A.E.. 2014. Smad1 transcription factor integrates BMP2 and Wnt3a signals in migrating cardiac progenitor cells. Proc. Natl. Acad. Sci. USA. 111:7337–7342. 10.1073/pnas.1321764111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara K., Nakatsuji N., Nakamura K., Nakao K., Hashimoto R., Otani H., Sakagami H., Kondo H., Nozawa S., Aiba A., and Katsuki M.. 1998. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene. 17:3427–3433. 10.1038/sj.onc.1202595 [DOI] [PubMed] [Google Scholar]
- Takeuchi M., Nakabayashi J., Sakaguchi T., Yamamoto T.S., Takahashi H., Takeda H., and Ueno N.. 2003. The prickle-related gene in vertebrates is essential for gastrulation cell movements. Curr. Biol. 13:674–679. 10.1016/S0960-9822(03)00245-8 [DOI] [PubMed] [Google Scholar]
- Tao H., Suzuki M., Kiyonari H., Abe T., Sasaoka T., and Ueno N.. 2009. Mouse prickle1, the homolog of a PCP gene, is essential for epiblast apical-basal polarity. Proc. Natl. Acad. Sci. USA. 106:14426–14431. 10.1073/pnas.0901332106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S., Mochizuki N., Ohba Y., Tobiume M., Okada Y., Sawa H., Nagashima K., and Matsuda M.. 2000. CalDAG-GEFIII activation of Ras, R-ras, and Rap1. J. Biol. Chem. 275:25488–25493. 10.1074/jbc.M003414200 [DOI] [PubMed] [Google Scholar]
- Yin C., Ciruna B., and Solnica-Krezel L.. 2009. Convergence and extension movements during vertebrate gastrulation. Curr. Top. Dev. Biol. 89:163–192. 10.1016/S0070-2153(09)89007-8 [DOI] [PubMed] [Google Scholar]
- Zallen J.A. 2007. Planar polarity and tissue morphogenesis. Cell. 129:1051–1063. 10.1016/j.cell.2007.05.050 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.