Chloride is ubiquitous in physiology but understood to provide ionic strength for tissue function. The authors discover a molecular function of chloride whereby the ion signals the assembly of collagen IV, establishing a microenvironment on the outside of cells.
Abstract
Basement membranes are defining features of the cellular microenvironment; however, little is known regarding their assembly outside cells. We report that extracellular Cl− ions signal the assembly of collagen IV networks outside cells by triggering a conformational switch within collagen IV noncollagenous 1 (NC1) domains. Depletion of Cl− in cell culture perturbed collagen IV networks, disrupted matrix architecture, and repositioned basement membrane proteins. Phylogenetic evidence indicates this conformational switch is a fundamental mechanism of collagen IV network assembly throughout Metazoa. Using recombinant triple helical protomers, we prove that NC1 domains direct both protomer and network assembly and show in Drosophila that NC1 architecture is critical for incorporation into basement membranes. These discoveries provide an atomic-level understanding of the dynamic interactions between extracellular Cl− and collagen IV assembly outside cells, a critical step in the assembly and organization of basement membranes that enable tissue architecture and function. Moreover, this provides a mechanistic framework for understanding the molecular pathobiology of NC1 domains.
Introduction
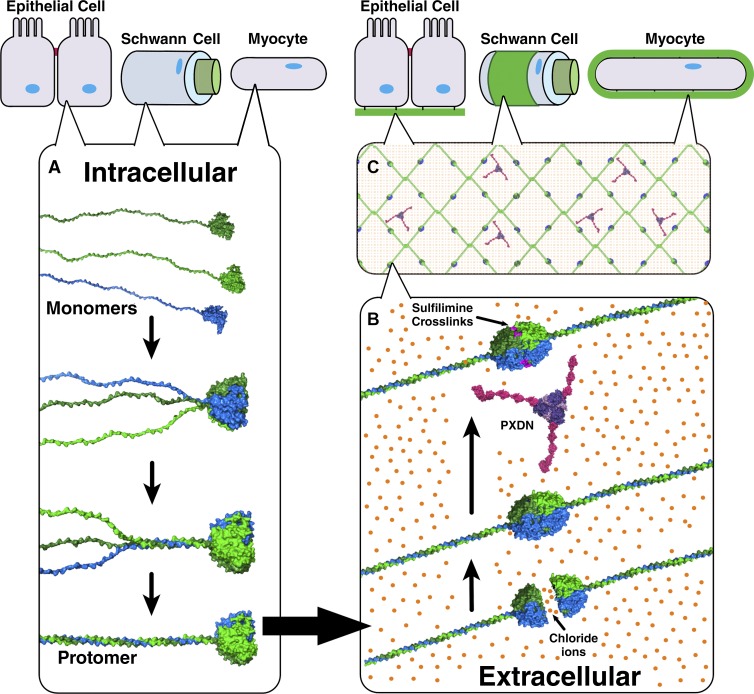
Basement membranes (BMs) are hallmarks of the cellular microenvironment of epithelial (Hagios et al., 1998; Lu et al., 2012), endothelial (Rhodes and Simons, 2007), muscle (Campbell and Stull, 2003; Sanes, 2003), fat (Sillat et al., 2012), Schwann (Court et al., 2006), and decidua cells (Fig. 1; Wewer et al., 1985; Farrar and Carson, 1992). BMs are supramolecular scaffolds that perform numerous functions, including compartmentalizing and reinforcing tissue architecture, organizing growth factor gradients, guiding cell migration and adhesion, delineating apical-basal polarity modulating cell differentiation, orchestrating cell behavior in tissue repair after injury, and guiding organ regeneration (Vracko, 1974; Pöschl et al., 2004; Wang et al., 2008; Hynes, 2009; Pastor-Pareja and Xu, 2011; Song and Ott, 2011; Yurchenco, 2011; Daley and Yamada, 2013).
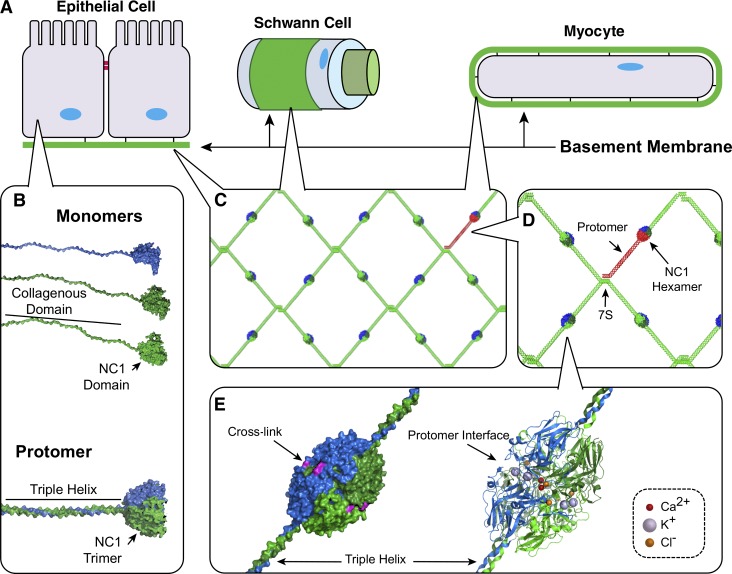
Figure 1.
NC1 domain is a primary junction point in collagen IV network assembly in BMs. (A) BMs interact directly with most eukaryotic cell types enabling tissue functions. (B) Heterotrimeric collagen IV protomers are composed of three α chain monomers; however, their assembly mechanisms remain unknown. (C) Within the BM, collagen IV networks act as scaffolds to tether ECM molecules and provide strength. (D) In network assembly, two protomers self-associate at their NC1 domains, whereas four collagen IV protomers associate at their 7S domains. (E) Crystal structures reveal multiple ion binding sites along the NC1 interprotomer interface.
A principle component of BMs is collagen IV, a complex network of triple helical molecules. The network functions as a scaffold that provides tensile strength to epithelial tissues and tethers diverse macromolecules, including laminin and proteoglycan, and growth factors and binds integrins (Fig. 1 B; Emsley et al., 2000; Wang et al., 2008; Parkin et al., 2011; McCall et al., 2014). Disrupting collagen IV networks causes BM destabilization and tissue dysfunction in mice, flies, zebrafish, and nematodes (Borchiellini et al., 1996; Gupta et al., 1997; Pöschl et al., 2004; Fidler et al., 2014). Clinically, collagen IV mutations cause Alport’s syndrome and are reported in some cases of hemorrhagic stroke (Hudson et al., 2003; Kuo et al., 2012).
The assembly of collagen IV networks includes two distinct stages of α-chain oligomerization (Fig. 1). Within cells, three α-chains associate to form triple helical molecules, termed protomers, which harbor binding sites for other BM macromolecules and integrin receptors. On the outside of cells, these protomers oligomerize into networks, whereby two protomers associate via their trimeric noncollagenous 1 (NC1) domains at the C terminus forming a NC1 hexamer at the junction, and four protomers associate at their N termini forming tetrameric 7S domains. NC1 hexamers are structurally reinforced with sulfilimine cross-links (Fig. 1 C) formed by peroxidasin (PXDN) and Br− ions (Vanacore et al., 2009; Bhave et al., 2012; McCall et al., 2014). Exemplifying the importance of NC1 reinforcement, loss of sulfilimine cross-links disrupts BM and tissue architecture, whereas cross-linked collagen IV networks enable Eumetazoa tissue development (Bhave et al., 2012; Fidler et al., 2014; McCall et al., 2014).
As recently articulated (Inman et al., 2015; Sherwood, 2015), a paramount knowledge gap in cell biology is those mechanisms outside cells that build functional and dynamic cellular microenvironments. To this end, we investigated the molecular basis of collagen IV network formation with a focus on the function of NC1 domains. Noting that x-ray structures of the NC1 hexamer contains Cl−, K+, and Ca2+ ions (Fig. 1 C), we hypothesized that protomer and network assembly is ion-dependent. Using new recombinant technology to produce hetero-triple helical molecules, we demonstrated that NC1 domains direct both protomer and network assembly and showed that Cl− triggers a molecular switch within the NC1 domain, enabling network assembly. For validation, we performed Cl− depletion studies in PFHR9 cell culture, which perturbed collagen IV network assembly and BM organization. Our discoveries provide insights into the intra- and extracellular mechanisms of collagen IV assembly in BMs.
Results
Assembly and stabilization of NC1 Hexamers requires Cl−
To examine the putative role of ions in assembly, we characterized the assembly and dissociation of native NC1 hexamers from bovine lens capsule basement membrane (LBM) as well as uncross-linked hexamers from PFHR9 cell culture (Bhave et al., 2012). LBM hexamers dialyzed from TBS into Tris-acetate buffer (TrisAc; pH 7.4) dissociated into monomers, seen by size-exclusion chromatography (SEC; Fig. 2 A). Notably, this dissociation was comparable with treatment with strong protein denaturants, including guanidine or urea (Fig. S1, A–C). Moreover, uncross-linked PFHR9 hexamers dissociated in TrisAc (Fig. S1 D), and hexamers from either source dissociated in phosphate buffer (Fig. S1, E and F), confirming that loss of Cl− ions induces hexamer dissociation.
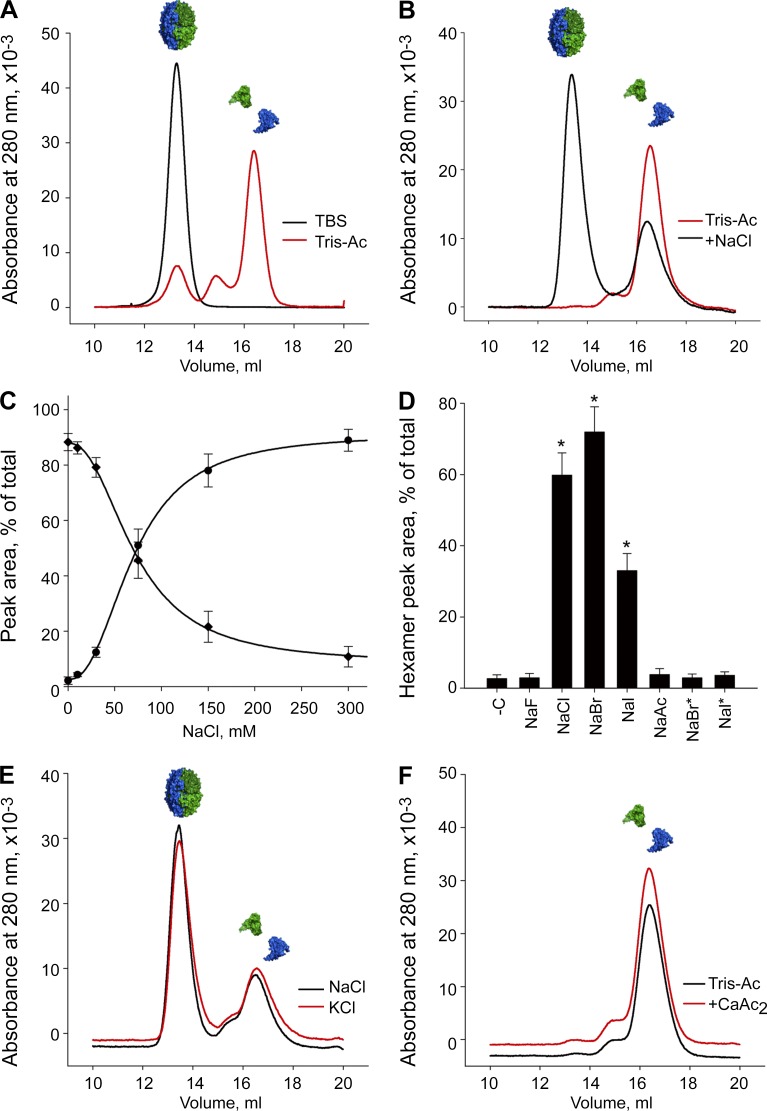
Figure 2.
Cl− is required for NC1 hexamer assembly. (A) LBM hexamers (black line) dissociate into NC1 monomers in Cl−-free TrisAc buffer (red line) by SEC. (B) LBM hexamers reassemble from monomers in the presence of 100 mM NaCl. (C) Yield of reassembled LBM hexamer depends on NaCl concentration. NC1 monomer (filled diamond) concentration decreases proportionally to NC1 hexamer formation (filled circle). (D) Effect of monovalent anions (F−, Cl−, Br−, I−, acetate, and sodium salts) at 100 mM on LBM hexamer assembly. To model physiological concentrations of Br− and I−, 100 µM NaBr (NaBr*) and 50 µM NaI (NaI*) were also tested and found to not support hexamer assembly. -C, Cl−-free control sample in 50 mM TrisAc. *, P < 0.01 versus TrisAc buffer. (E) K+ (red line) and Na+ (black line) yield similar amounts of hexamer. Cations tested at 100 mM (Cl− salt). (F) Ca2+ ions at 1 mM, does not support hexamer formation from LBM NC1 monomers. SEC profiles shown of dissociated LBM NC1 monomers in 50 mM TrisAc with (red line) and without (black line) 1 mM CaAc2.
We next asked whether Cl− triggers hexamer reassembly from dissociated monomers. We isolated LBM NC1 monomers via TrisAc-based hexamer dissociation and SEC fractionation (Fig. 2 A), incubated with 100 mM NaCl (physiological Cl− concentration), and observed the formation of an SEC peak that was identical to authentic LBM hexamer and contained α1 and α2 NC1 domains (Fig. 2 B and Fig. S1 G). Yield of reassembled hexamer was dependent on NaCl concentration, temperature, protein concentration, and time, reaching equilibrium in 24 h (Fig. 2 C and Fig. S1, H–J). PFHR9 NC1 domains also reassembled into hexamers with Cl− (Fig. S1 K).
Investigating whether hexamer assembly specifically requires Cl− ions, we screened various halides for their biochemical impact on hexamer assembly (all at 100 mM, Na+ salts), observing that Cl− and Br− strongly induced hexamer formation, whereas I− showed modest activity, F− was completely inactive, and acetate provided a negative control (Fig. 2 D). Aside from Cl−, the human physiological concentrations for all were ≤100 µM, far below the 100-mM test conditions (van Leeuwen and Sangster, 1987). We reexamined Br− and I− at more physiologically relevant concentrations (100 and 50 µM, respectively); however, neither halide induced hexamer assembly (Fig. 2 D). Thus, among the halides, Cl− is required for hexamer assembly under physiological conditions.
We did not detect any cation-specific effect on assembly. Modeling the K+ (monovalent cation) binding site from published crystal structures of the NC1 hexamers (Fig. S1, L and M; Sundaramoorthy et al., 2002; Vanacore et al., 2004), we noted that the K+ is orthogonal to the plane of the aromatic side chains (Fig. S1 M), presumably limiting the structural impact of the cation. In the assembly assay, K+ acted similarly to Na+ when tested in chloride form (Fig. 2 E and Fig. S1 N), whereas cesium and ammonium were comparable (Fig. S1 N).
Ca2+ can induce protein conformational changes (Chou et al., 2001), and the ion coordinates residues D148 and E149 in the α2 NC1 domain (Fig. S1 O). During MD simulations of the hexamer in 150 and 0 mM Cl−, the interaction between Ca2+ and the opposing D148 appeared to be enhanced by Cl−, potentially benefiting hexamer assembly (Fig. S1 P). However, physiological concentrations of Ca2+ were inactive (Fig. 2 F and Fig. S1 Q), and EDTA did not block Cl−-driven assembly (Fig. S1 R). Intriguingly, we noted increased hexamer yield in 1 mM CaCl2 plus 100 mM NaCl (Fig. S1 S), suggesting that Ca+ may potentiate the activity of Cl−.
Concluding that Cl− ions are required for hexamer assembly and seeking a mechanistic explanation, we first addressed the seminal and unanswered question of NC1 function during assembly. To this end, we developed a novel recombinant methodology to definitively determine if α1 and α2 chains can self-assemble into recombinant collagen IV α112 protomers (r-Prot) and protomer dimers, the latter comprising two recombinant protomers joined via their NC1 domains, forming an NC1 hexamer with protruding helices (Fig. 3 A). This approach enabled us to functionally distinguish the roles of NC1 domains and Cl− during assembly.
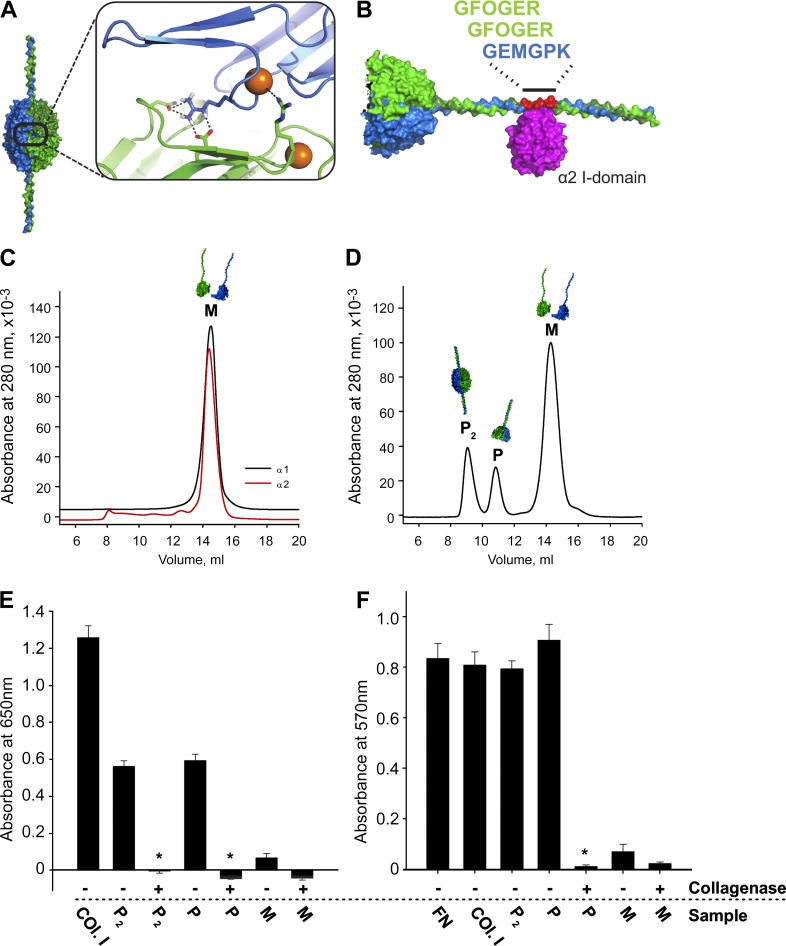
Figure 3.
Design, production, and characterization of r-Prot. (A) Model of Cl− binding site, from α 112 NC1 hexamer x-ray structure. (B) Modeled r-Prot with integrin α2β1 binding site with bound α2 I domain (magenta); 84 helical amino acids from α1 and α2 chains immediately adjacent to NC1 domains. (C) Purified α1 (black line) and α2 (red line) recombinant monomers eluted as 14.5-ml peaks by SEC. (D) SEC profile of products after in vitro assembly. Peaks identified as monomers (M, 14.5 ml), protomers (P, 11 ml), and protomer dimers (P2, 9 ml). (E) Integrin α2 I domain solid-phase binding demonstrates activity in protomer helices (P and P2) but not monomers. Digestion of helices with bacterial collagenase, to remove binding site, abolished binding activity. Collagen I (Col.I) is the positive control for binding. (F) HT-1080 cells adhere only to undigested P2 and P. Fibronectin (FN) was additional positive control. All experiments performed in triplicate. *, P < 0.01.
Production, characterization, and assembly of recombinant protomers
We designed constructs containing an NC1 domain contiguous with 28 GXY-repeats, representing truncated α1- and α2−chains of sufficient length to assemble a stable triple helix (Fig. 3 B and Fig. S2 A). We reasoned that helical tails should cause the recombinant products to elute earlier by SEC, whereas collagenase treatment would shift migration times to match native control LBM hexamers or recombinant monomers. Individual constructs were expressed as monomers (M) and purified by SEC (Fig. 3 C). Upon incubation in TBS at 2:1 ratio of α1:α2, additional 9 and 11 ml SEC peaks emerged (Fig. 3 D). Collagenase treatment shifted the 9-ml peak to match the native LBM hexamer (Fig. S2 B), and this fraction contained a 2:1 ratio of α1:α2 NC1 domains (Fig. S2 C). We concluded this peak contained protomer dimers (Fig. 3 A), and thus termed it P2. The second peak (P, 11 ml; Fig. 3 D) was converted to NC1 monomers by collagenase (Fig. S2 D), representing individual r-Prot. The remaining monomers (M, 14 ml; Fig. 3 D) were also converted to NC1 monomers by collagenase (Fig. S2 E). All peaks contained NC1 domains with expected mobility by SDS-PAGE (Fig. S2 F).
We confirmed the presence of triple helices in P and P2 through limited proteolysis with trypsin (Fig. S2 G) and circular dichroism (CD) spectroscopy, which showed a characteristic positive peak at 220 nm and melting temperature (Tm) of 30°C (Fig. S2, H and I). Importantly, the isolation of r-Prot in peak P establishes the required role of triple helix for stabilizing trimerized NC1 domains; lateral association between NC1 monomers by itself is insufficient for stabilization of NC1 trimers.
For functional testing, we engineered an α2β1 integrin binding site from the collagen IV cyanogen-bromide–derived fragment of collagen IV with integrin binding sites (CB3) region in the helical domain (Fig. 3 B and Fig. S2 A; Vandenberg et al., 1991; Eble et al., 1993). Both P and P2 exhibited Mg2+-dependent binding activity with α2 integrin I-domain, whereas monomers were inactive and collagenase treatment eliminated binding to P and P2 (Fig. 3 E). For validation, HT1080 cells adhered to either P or P2, but not to monomers, collagenase-treated P or P2, or in the presence of anti-α2β1 integrin-blocking antibodies (Fig. 3 F and Fig. S2 J). Concluding that the r-Prot displays key characteristics of native collagen IV protomers, including folded and functional NC1 trimer and helices, we turned to interrogating the assembly functions of NC1 domains and Cl−.
NC1 domains direct protomer and network assembly; Cl− signaling prompts networking
We asked if Cl− is required for assembling the P2 fraction. Indeed, the P2 peak shifted to P in TrisAc (Fig. 4 A), indicating dissociation of dimerized protomers into individual protomers with loss of Cl−. Incubation above the Tm, inducing helical unwinding, further dissociated the protomers into monomers (Fig. 4 A). Remarkably, incubation of monomers below the Tm in TrisAc reformed peak P (Fig. 4 B), indicating that r-Prot assembly relies solely on NC1 domains without Cl−. However, r-Prot only formed a P2 peak in the presence of Cl− (Fig. 4 B). Hence, NC1 domains alone direct self-trimerization of α-chains into protomers. However, joining two protomers via their NC1 domains requires Cl−. Intriguingly, Cl− concentrations are low in cells where collagen IV protomer assembly occurs and they are high extracellularly (e.g., ∼12 mM in muscle cells and ∼100 mM serum; Ziomber et al., 2008). We contend that NC1 domains alone drive protomer assembly, and once outside cells, Cl− signaling prompts the oligomerization of NC1 trimers into collagen IV networks.
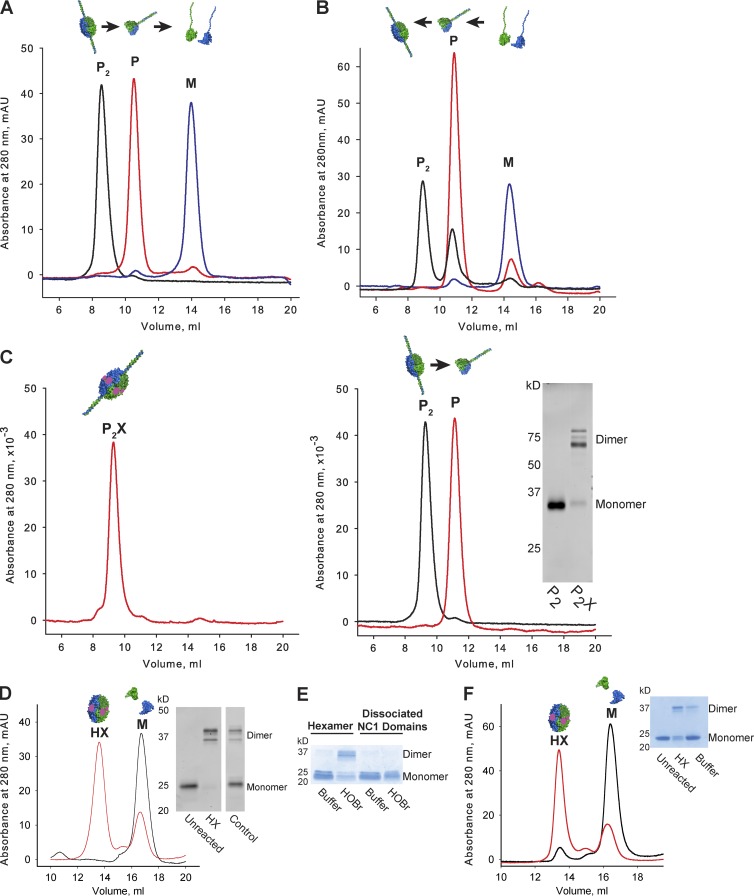
Figure 4.
Protomers self-assemble whereas network self-assembly requires Cl−. (A) Protomer dimers (P2, black line) dissociate into monomeric (M; blue line) chains through controlled steps. Protomer dimers (P2; black line) were noted in TBS, which dissociated into protomers (P; red line) in TrisAc and further dissociated into monomers (M; blue line) at 37°C. (B) Controlled reassembly of monomers into protomers (P). Monomers (blue line) spontaneously assembled into protomers (red line) in TrisAc, without Cl−. At 100 mM Cl−, protomers (P) reassembled into protomer dimers (P2; black line). (C) PXDN–cross-linked P2 (P2X) resist dissociation in TrisAc (left), whereas uncross-linked P2 dissociated into P (right). (inset) SDS-PAGE of P2 and P2X samples. (D) NC1 hexamers resist dissociation in TrisAc after cross-linking by PXDN, Br−, and H2O2 (red line). Unreacted control hexamers dissociate in TrisAc (black line). (inset) SDS-PAGE of uncross-linked (Unreacted) and cross-linked (HX) hexamers, as well as control native LBM hexamers (Control). HX and M denote cross-linked hexamer and NC1 monomer peaks by SEC, respectively. (E) Hexamer assembly precedes cross-linking. HOBr cross-links hexamer substrates but not dissociated NC1 domains by SDS-PAGE. HOBr and Buffer denote reacted and unreacted samples, respectively. (F) HOBr–cross-linking prevents LBM hexamer dissociation in guanidine-HCl (6 M, 30 min, 65°C) by SEC (red line), whereas uncross-linked controls were dissociated (black line). (inset) SDS-PAGE of unreacted (Unreacted) and HOBr–cross-linked (HX) hexamer and native LBM hexamer (Control). HX and M denote cross-linked hexamer and NC1 monomer peaks by SEC, respectively.
Cl− assembles the hexamer substrate for PXDN
Because BMs display hexamers with sulfilimine cross-links (Fidler et al., 2014), we hypothesized that Cl− signaling forms the substrate for sulfilimine cross-linking by PXDN and hypobromous acid (HOBr), the catalytic intermediate (Bhave et al., 2012; McCall et al., 2014). We thus incubated P2 with recombinant PXDN, Br−, and oxidant, noting rapid cross-linking of NC1 domains (Fig. 4 C, inset). Importantly, cross-linking protected P2 from dissociation in TrisAc, whereas uncross-linked P2 was readily dissociated (Fig. 4 C). Similarly, PXDN cross-linking enabled reassembled LBM hexamers to resist dissociation (Fig. 4 D). HOBr cross-linked NC1 hexamers, but not dissociated NC1 monomers (Fig. 4 E), indicating that hexamer assembly precedes cross-linking. HOBr–cross-linked hexamers resisted dissociation in guanidine (Fig. 4 F).
Cl−-dependent conformational switch triggers protomer–protomer assembly
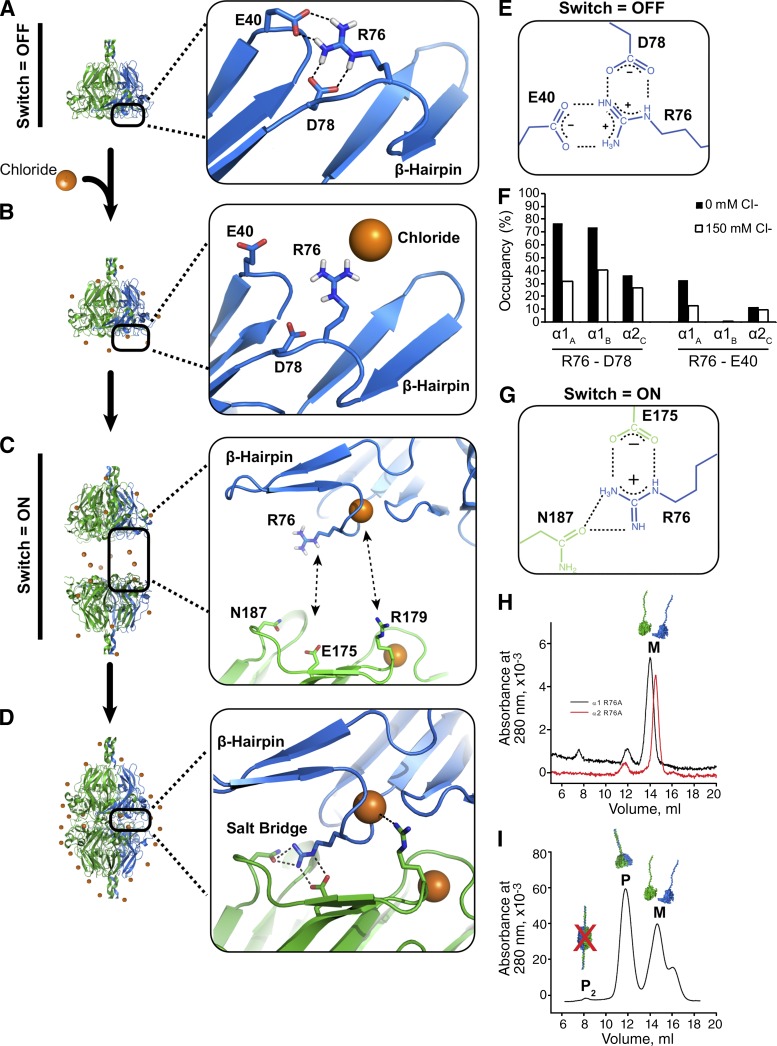
We sought to know how Cl− triggers protomer dimerization but not protomer assembly from α-chains. We previously hypothesized that β-hairpin and variable region 3 (VR3) regions on NC1 domains drive the side-to-side interactions during trimer assembly (Khoshnoodi et al., 2006b). Mechanistically, we reasoned that Cl− may act on these regions or at the trimer–trimer interface and thus modeled the surface charges of NC1 monomers and NC1 trimers. α1 and α2 NC1 monomers displayed strong electronegativity on the interior surface (relative to hexamer), negatively and positively charged patches on the exterior, and mostly neutral β-hairpin and VR3 regions (Fig. S3, A and B; and Table S1). In contrast, the α112 trimer interface has a highly electronegative core surrounded by an electrostatic recognition motif formed by residues R76 and E175 (Fig. S3, B and C) that may respond to local Cl− concentrations. Using nonlinear Poisson–Boltzmann calculations (García-García and Draper, 2003), we estimate an eightfold greater impact of salt on the binding free energy for hexamer assembly over NC1 trimer assembly (Fig. S3 D).
Having previously noted a Cl− binding site in the α112 hexamer crystal structure (Vanacore et al., 2004), we rigorously examined whether Cl− signals through this site. The binding site is formed by residues A74, S75, R76, N77, and D78 (Fig. 3 A), where Cl− coordinates the backbone amides. The R76 side chain bridges the trimer–trimer interface to form a side-on interaction with E175 and an end-on interaction with N187, altogether creating a bridging-networked salt bridge. Finally, Cl− directly coordinates with R179 from the opposing NC1 trimer (Fig. 3 A). In summary, Cl− is central to the electrostatic interactions along the trimer–trimer interface, indirectly participating in six bridging-networked salt bridges and directly engaging in up to six additional electrostatic interactions with R179.
To gain a mechanistic understanding, we used MD simulations to model residue-specific changes occurring in 150 mM Cl−. Solvent Cl− did not influence the β-hairpin motif or VR3 region (Fig. S3, E–H); therefore, we focused on understanding how Cl− triggers hexamer assembly. In monomers and the α112 NC1 trimer, R76 formed intramonomer salt bridges with D78 and E40 in 0 mM Cl− (Fig. 5 A), whereas 150 mM Cl− reduced the R76-D78 salt bridge occupancy likely via Debye–Hückel electrostatic screening (Fig. 5 B). Because R76 participates in the bridging-networked salt bridges (Fig. 5 D), disruption of the R76-D78 interactions is apparently critical to hexamer assembly (Fig. 5 C). We reason that site-specific Cl− binding within the nest restricts the available side chain conformations of R76 and D78, repositioning R76 to enable hexamer assembly (Fig. 5, C and D) by forming bridging-networked salt bridges between NC1 trimers. We view the combined residue-specific conformational changes as a “molecular switch,” triggered by Cl− that drives hexamer assembly.
Figure 5.
Cl− triggers a molecular switch enabling network assembly. (A) Without Cl−, R76 forms intramolecular salt bridge with D78 and/or E40 in MD simulations. (B) Extracellular Cl− disrupts R76-D78 salt bridge by electrostatic screening. (C) Specific binding activity of Cl− causes the ion to coordinate the R76 backbone amide of R76, thus orienting the side toward an opposing NC1 timer. (D) Each bound Cl− ion can coordinate two distinct electrostatic interactions, up to 12 such interactions per hexamer, including six bridging-networked salt bridges. (E) Molecular structure of interactions among R76, E40, and D78. (F) Occupancy plot shows simulated hydrogen bond occupancies of R76 in 0 mM Cl− (closed bars) and 150 mM Cl− (open bars), indicating that Cl− disrupts R76 intramolecular interactions. (G) Molecular structure of electrostatic interactions that comprise the bridging-networked salt bridge. (H–I) R76A mutations prevent formation of hexamers. SEC profiles of purified α1-R76A (black line) and α2R76A (red line) r-monomers (H). SEC profile after mixing and incubation of both r-monomers in 100 mM NaCl showing the formation of protomers (P) but not protomer dimers (P2 and I).
To test whether this switch indeed mediates Cl−-dependent assembly, we generated α1 and α2 recombinant proteins with R76A mutations. R76A mutants formed protomers but not P2 in 100 mM Cl− (Fig. 5, H and I; and Fig. S3, I–L), confirming that R76 is critical to in vitro hexamer assembly and demonstrating that collagen IV network assembly may be disrupted by a single NC1 domain mutation.
Switch residues identified throughout animal kingdom
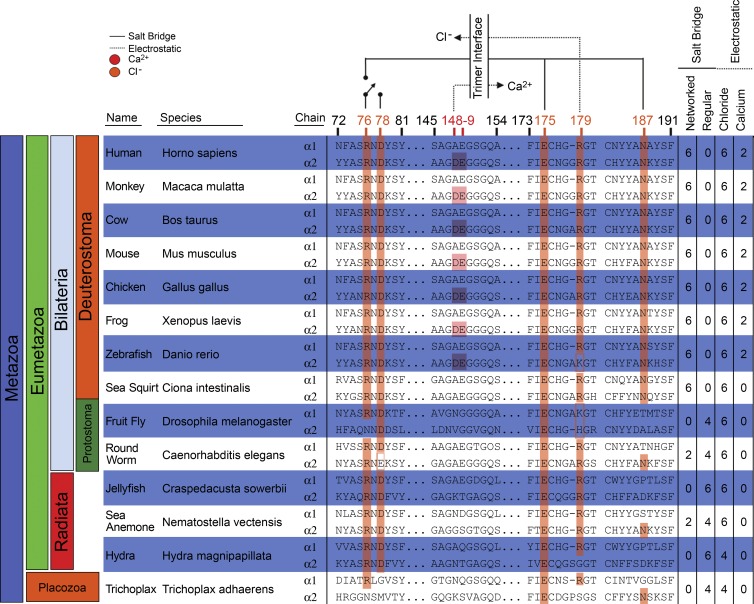
Collagen IV networks are found throughout Metazoa; therefore, we searched for the Cl−-mediated switch in NC1 protein sequences from Placozoa to human (Fig. 6). All phyla displayed the principal salt bridge residues R76 and E175 in either the α1 or α2 chain, with R179 in nearly all α1 chains and most α2 chains. Residue N187, enabling the networked interaction, is restricted to Deuterostoma. Residue D78, stabilizing the “off” switch conformation, was seen throughout Eumetazoa. Moreover, all vertebrate α3–α6 chains displayed R76, D78, and E175 (Fig. S4). Thus, we infer that the switch is a fundamental mechanism of collagen IV network assembly.
Figure 6.
Key residues of Cl−-mediated assembly switch are defining features of collagen IV. All species examined show at least one chain with residues R76 and D78 and capacity for direct electrostatic interaction with Cl−. Ca2+ binding site only in Deuterostoma. Table (right) denotes salt bridges and electrostatic interactions at the trimer–trimer interface. Salt bridge type (regular or networked) predicted by presence of N187.
Chloride is required for collagen IV network assembly in PFHR9 cell culture
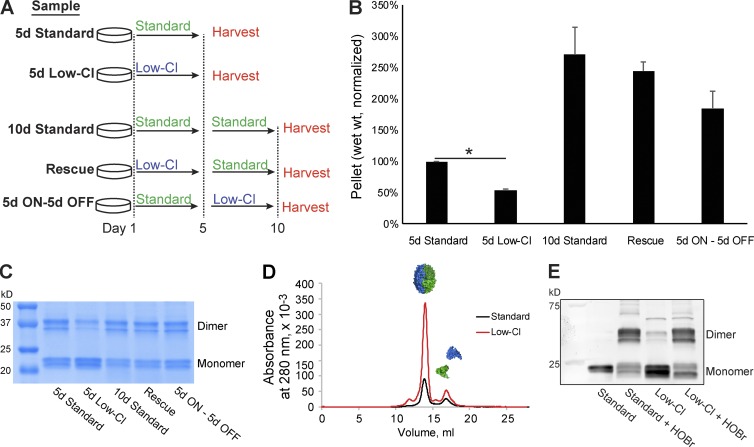
Because in vitro NC1 hexamer assembly requires Cl−, we tested whether Cl− is also required for collagen IV network assembly in cell culture by growing collagen IV matrices from PFHR9 cells for 5 d in the presence of low Cl− concentrations (∼5 mM Cl−; Fig. 7 A, 5d Low-Cl) and control cells for 5 d in standard media (Fig. 7 A, 5d Standard). Recoverable matrix (wet weight) from 5-d low-Cl conditions was significantly lower than 5-d standard cultures (Fig. 7 B) but rescued when Cl− levels were restored for an additional 5 d (Fig. 7 A, Rescue). Cell viability was not reduced in low-Cl conditions (Fig. S5 A).
Figure 7.
Chloride is required for collagen IV network assembly in PFHR9 cell culture. (A) Experimental design for culturing collagen IV networks from PFHR9 culture. (B) Low-Cl conditions reduced matrix yield after 5 d in culture. Addition of Cl− to media restored matrix yield. (C) NC1 sulfilimine cross-linking was reduced in 5-d low-Cl conditions by SDS-PAGE. (D) NC1 monomers isolated from low-Cl cultures assembled into NC1 hexamers in presence in TBS. (E) HOBr cross-linked reassembled NC1 hexamers from low-Cl cultures. Standard and low-Cl labels denote unreacted samples; standard + HOBr and low-Cl + HOBr labels denote HOBr-reacted samples. All hexamers were isolated by SEC as in D. *, P < 0.001.
We hypothesized that low-Cl treatment prevents hexamer assembly in matrices and should be evidenced by fewer sulfilimine cross-links. Indeed, cross-linking was reduced in 5-d low-Cl cultures relative to all other conditions (Fig. 7 C). Probing further, we reasoned that NC1 domains from low-Cl cultures should retain the capacity to form hexamers in normal Cl− concentrations. We thus obtained NC1 monomers from low-Cl cultures by TrisAc dissociation and SEC fractionation (Fig. S5 B). In 150 mM Cl−, these monomers yielded a hexamer peak by SEC (Fig. 7 D), indicating that low-Cl conditions reversibly block NC1 hexamer assembly. Finally, HOBr treatment cross-linked the assembled hexamers (Fig. 7 E), demonstrating that NC1 domains from low-Cl conditions can assemble in vitro into structurally authentic hexamers. Thus, we conclude that Cl− depletion in culture impairs collagen IV hexamer assembly.
NC1 domains are pivotal to collagen IV network organization
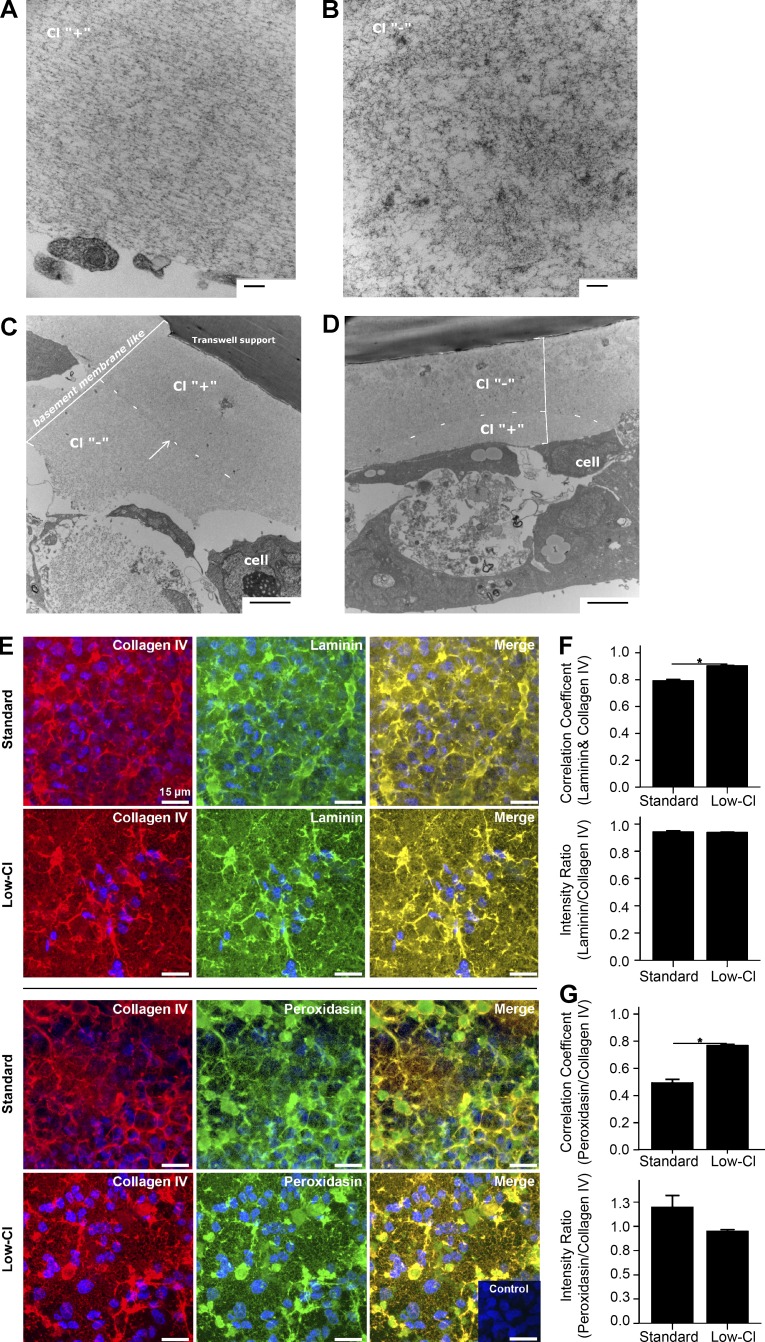
Sulfilimine cross-links are critical to BM architecture (Bhave et al., 2012; McCall et al., 2014); therefore, we examined the suprastructure of PHFR9 matrices from control and low-Cl conditions. Transmission electron microscopy of the control matrix revealed an organized structure with granular and fibrillar appearance (Fig. 8 A), whereas the low-Cl matrix was disordered with granular patches (Fig. 8 B). Matrix thickness was similar in both conditions.
Figure 8.
Disorganization of collagen IV-rich PFHR9 matrix under low-Cl conditions. (A–D) Transmission electron microscopy of the BM-like matrix from PFHR9 cells under standard (A, Cl “+”) or low-Cl (B, Cl “−”) conditions. Standard conditions yielded organized matrices (A), whereas low-Cl conditions yielded disorganized matrices with patches of accumulated granular material (B). (C and D) Transmission electron microscopy of matrix deposited in alternating standard and low-Cl conditions. (C) Cells grown in standard media (Cl “+”) for 5 d followed by 5 d in low-Cl media. An abrupt density change occurred approximately in the middle of the deposited matrix (bracketed area). (D) Cells grown in low-Cl media followed by standard conditions (5 d each) displayed a disorganized patchy matrix nearest the support membrane with more ordered structure nearest the cells (bracketed area). (E–G) Confocal microscopy reveals that low-Cl conditions stimulate the enhanced localization of laminin and PXDN to collagen IV in PFHR9 matrix. (E) Maximum intensity projection of immunofluorescent confocal z-stacks (7.2-µm projections) of matrix after 5 d in standard media or low-Cl media; collagen IV (red), laminin (green), or PXDN (green), merge where colocalization is yellow/orange, and nuclei/Hoechst stain (blue). Secondary antibody only negative control inset. Bars, 15 µm. (F and G) Collagen IV (F) and laminin or collagen IV and PXDN (G) colocalization analysis by Pearson correlation coefficient (1.0 = perfect colocalization) and ratio of green/red signal per field-of-view. B, n = 8; C, n = 4. *, P < 0.001.
To test if low-Cl causes matrix disorganization, we examined matrices where Cl− concentrations had changed in culture. In Rescue experiments, the initial deposited matrix (low Cl) appeared disorganized, whereas the later deposited matrix (standard) was well-ordered (Fig. 8 C). Conversely, when cultures were moved from standard media to low-Cl media, matrix architecture progressed from well organized to disorganized (Fig. 8 D). We conclude that Cl− signaling is required for collagen IV network assembly, pivotally influencing extracellular matrix architecture.
With confocal microscopy, we examined collagen IV networks, laminin, and PXDN in low-Cl conditions. Colocalization of laminin and PXDN to collagen IV networks was significantly higher in low-Cl matrices than controls (Fig. 8, E–G); however, signal intensity was not altered (Fig. 8, F and G), indicating that low-Cl conditions reposition BM components. Because collagen IV networks provide a structural framework for arranging and binding other BM components, we interpret this as evidence of the downstream outcomes of Cl− signaling within collagen IV networks. Namely, by enabling NC1 hexamer formation, Cl− signaling prompts the formation of collagen IV networks that provide a sophisticated array of binding sites to organize BMs.
To examine network assembly in vivo, we adopted an approach to express collagen IV transgenes in Drosophila melanogaster alongside endogenous collagen IV. We generated an α1-like collagen IV transgene with a partial NC1 sequence (Cg25CΔNC1; see Materials and methods) to destroy the NC1 architecture and thus probe the NC1 role during network formation. After injection into larvae, we noted that the Cg25CΔNC1 protein was present in circulation rather than BMs (Fig. S5, C, D, and F–I), indicating that mutational damage to NC1 domains can prevent formation of mutant-bearing collagen IV networks. In contrast, a transgene with R76A NC1 mutation (Cg25C1R76A) resulted in the mutant protein being incorporated into BMs (Fig. S5 E). Residue 76 is noncritical for NC1 trimerization to form protomers, allowing the mutant protein to incorporate with endogenous protein into the matrix. We view this as analogous to the allelic heterogeneity model of collagen IV mutations (Kuo et al., 2014), and it is consistent with low-Cl studies where collagen IV deposition continued despite incomplete network assembly. Together, we contend that these results underscore the essentiality of NC1 domains in forming collagen IV networks and BMs, and they also provide an experimental strategy to address numerous aspects of normal and defective collagen IV assembly.
Discussion
BM signaling is pivotal to cell behavior and differentiation; however, many basic assembly mechanisms are unknown (Inman et al., 2015; Sherwood, 2015). These BMs rely on collagen IV networks that function in the organization, signaling activity, and structural support of the overall matrix. We view collagen IV networks as a type of “smart scaffold” to help shape BM composition, architecture, and biology. Accordingly, the mechanisms of collagen IV network assembly can possess widespread physiological influence.
Previously hypothesizing that collagen IV NC1 domains are recognition modules for protomer and network assembly (Khoshnoodi et al., 2006a), our data now unveil an atomic-level understanding of the assembly functions for NC1 domains and Cl−, including intracellular and extracellular stages of assembly (Fig. 9). As such, Cl− has a specific signaling and structural function in assembling collagen IV networks outside of cells. Thus, Cl− is pivotal for building a collagen IV network that embeds growth factors and other signaling molecules, binds laminin and other macromolecules, and forms a complex BM scaffold that interacts with cellular receptors. Finally, these mechanisms provide a putative framework for understanding the pathobiologic mechanism underlying Alport’s syndrome and stroke in patients with certain collagen IV NC1 domain mutations.
Figure 9.
Functional NC1 domains control collagen IV protomer and network assembly. (A) Collagen IV NC1 domains nucleate protomer assembly by controlling chain stoichiometry, specificity, and registration. (B) Extracellular Cl− concentrations signal protomers to oligomerize into networks, forming NC1 hexamers at the protomer–protomer junctions. At the NC1 junctions, hexamers are reinforced with sulfilimine cross-links by PXDN and Br−. (C) Assembled collagen IV networks function as a scaffold for binding other macromolecules and growth factors in BMs.
Knowns and frontiers of Cl− activity
In vivo, Cl− is mostly found in the extracellular space with serum concentrations near 100 mM in healthy human adults. However, in muscle cells, which have a BM, intracellular Cl− concentrations are reported near 12 mM (Ziomber et al., 2008), which is inadequate for collagen IV network formation (Fig. 2 C). Although many functions are attributed to Cl−, few involve a structural role for the ion, despite its abundance in protein crystal structures. As we show, Cl− is capable of performing specific structural roles in proteins in addition to acting through ionic strength or a Hofmeister effect (Zhang and Cremer, 2010; Zhou et al., 2010).
Specific structural role for Cl− in collagen IV network assembly
Cl− signals network assembly by modulating salt bridge interactions within NC1 domains. In the hexamer, salt bridges and hydrogen bonds are crucial in defining the electrostatic complementarity across the trimer–trimer interface (Khoshnoodi et al., 2006b). Prior to hexamer formation and in a low Cl− environment, MD simulations reveal an intramonomer salt bridge between R76 and D78, likely contributing to monomer stability by decreasing conformational entropy via loop closure (Bastolla and Demetrius, 2005). However, this intramonomer salt bridge is disrupted by extracellular Cl− as a prerequisite to hexamer assembly. From its binding site in the hexamer, crystallographic Cl− coordinates the amide backbone of R76, limiting the side chain conformations and enabling R76 to form the bridging-networked salt bridges (Donald et al., 2011) that span the trimer–trimer interface. We are aware of this motif in only three other structures: Acyl-CoA oxidase (Protein Data Bank accession no. 1IS2; Nakajima et al., 2002), α-L-arabinofuranosidase (Protein Data Bank accession no. 1WD3; Miyanaga et al., 2004), and malate dehydrogenase (Protein Data Bank accession no. 1BMD; Kelly et al., 1993). However, collagen IV NC1 hexamers are the only description of this motif being biologically useful in assembling a quaternary protein structure.
Chloride as a putative regulator of extracellular matrix assembly
We view Cl− as a putative regulator of matrix assembly, acting outside of cells and just beyond collagen IV. For example, in vitro collagen I fibril formation requires physiological Cl− concentrations for proper D-spacing and fibril thickness (Harris et al., 2013). Building on this, we show that Cl− acts through a specific binding site to induce collagen IV NC1 conformational changes, remarkably by using backbone interactions in part to modulate the R76 conformation (Fig. 5, C and D). By inference, we propose that Cl− ions may also act on specific residues in other collagens, such as in type I collagen fibril formation, with broad influence over ECM assembly.
Pivotal role for NC1 domains in BM assembly
We directly demonstrate the function of NC1 domains in assembling protomers and networks. Their capacity for selective self-trimerization governs chain selectivity and registration, controlling which protomers are formed (α112, α345, or α556) and oligomerize into networks. In turn, the networks function as scaffolds for PXDN (Fig. 8, E and G), growth factors, laminins (Fig. 8, E and F), and proteoglycans (Parkin et al., 2011). Indeed, NC1 domains are key regulators of BM organization and function, and the Drosophila, cell culture, and recombinant systems described here provide powerful tools and strategies for unraveling these NC1 activities.
Framework for understanding NC1 pathobiology
NC1 domains mutations occur in some patients with Alport’s syndrome and stroke (Lemmink et al., 1993; Kuo et al., 2012); however, their underlying pathological mechanisms are unknown. In Alport’s syndrome, mutations cause loss of the α345 collagen IV network, structural alterations in the glomerular BM, and kidney failure. Our findings with recombinant protein assembly and cell culture exemplify how a point mutation or altered halide concentrations can impact NC1 functionality, thereby disrupting collagen IV network assembly and BM architecture (Fig. 5 E). Moreover, our fly studies here demonstrate the importance of NC1 structure in collagen IV network assembly, echoing studies where loss of sulfilimine cross-links disrupts tissue architecture and is embryonic lethal in Drosophila (Bhave et al., 2012; McCall et al., 2014). Collectively, these findings reveal that NC1 domains are recognition modules during BM assembly and maintenance, thus providing a framework for considering how specific NC1 mutations cause disease, namely by interfering with protomer assembly, network assembly, and BM stabilization and/or organization.
Materials and methods
Chemicals
Cell culture reagents were purchased from CellGro (Corning), and all other chemicals and reagents were purchased from Sigma-Aldrich.
Cells and tissues
HT1080 and PHFR9 cells were purchased from the ATCC. All bovine tissues were purchased frozen from Pel-Freez Biologicals.
Collagen IV NC1 hexamer purification
Cultured collagen IV
Matrix production followed established protocols (Bhave et al., 2012; McCall et al., 2014) where cells were plated at high density and maintained at confluency for 5–8 d in the presence of 50 µg/ml ascorbic acid, with media changes every 24–36 h. Cross-linking was inhibited by supplementing the culture conditions with either 1 or 10 mM of potassium iodide (KI) or 50 µM of phloroglucinol. Inhibitor and ascorbic acid treatments were initiated upon confluency.
Cultured matrix was washed in 1× PBS before being scraped in a lysis buffer containing 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1% (wt/vol) sodium deoxycholate, 0.4 mM PMSF, 1 µg/ml aprotinin, and 1 µg/ml leupeptin. Matrix was sonicated to sheer genomic DNA, and 1-ml aliquots were placed into 1.5-ml eppendorf tubes on ice and centrifuged at 14,000 rpm for 20 min at 5°C. The pellet material was washed in a high salt buffer containing 50 mM Tris-HCl, pH 7.5, and 1 M NaCl before centrifugation at 14,000 rpm for 10 min at 5°C. The insoluble material was washed in low salt buffer consisting of 10 mM Tris-HCl, pH 7.5, and stored at 4°C until use.
Isolation of NC1 domains from collagen IV matrix was accomplished by treating the matrix with bacterial collagenase enzyme (Worthington Biochemical Corporation) at 37°C in 10 mM Tris-HCl, pH 7.5, 10 mM CaCl2, 10 mM KI, 0.4 mM PMSF, 1 µg/ml aprotinin, and 1 µg/ml leupeptin. Collagenase enzyme was stored in the aforementioned buffer at –20°C until use, in 1 mg/ml aliquots, and thawed before use. Matrix was digested overnight with 0.167 µg/ml enzyme in 60 µl, typically by adding 10 µl of 1 mg/ml enzyme solution to 50 µl of buffer. Because collagenase treatment effectively solubilized NC1 domains as well as cross-linking activity, the supernatant was obtained after digestion and used directly for SDS-PAGE.
For further purification of NC1 hexamers, collagenase digest supernatant was dialyzed into 50 mM Tris-HCl, pH 7.5, before being passed over a DEAE-cellulose column. The flow-through material was collected and purified by SEC.
Low-Cl culture conditions
PFHR9 cells were seeded and grown to confluency using standard conditions, washed in low-Cl media, then cultured in low-Cl media conditions containing 50 µg/ml ascorbic acid. Low-Cl media was supplemented with 5% FBS and sterile filtered before use. The recipe for low-Cl in found in Table S2 and is derived from McCall et al. (2014).
Tissue-derived collagen IV
Bovine LBM was purified as described previously (Boutaud et al., 2000). In brief, the LBM matrix was removed from semi-thawed bovine lenses, and LBM homogenate was pelleted and washed in 1% sodium deoxycholate (wt/vol), 50 mM Tris-HCl, pH 7.4, and protease inhibitors. Samples were sonicated, subsequently washed in 50 mM Tris-HCl and 1 M NaCl, pH 7.4, and finally washed in 50 mM Tris-HCl, pH 7.4, before being digested with bacterial collagenase (Worthington Biochemical Corporation) to solubilize NC1 hexamers. Further purification of hexamers used DE-52 chromatography and SEC.
Recombinant collagen IV constructs
DNA constructs were designed to encode the wild-type collagen IV α1 and α2 monomers containing their respective NC1 domains and 84 aa residues from collagenous domain (designated α1-84 and α2-84) from residues 1249–1732 (Fig. S2 A). The QuikChange Lightening Multi Site-Directed Mutagenesis kit (Agilent Technologies) was used to incorporate integrin α2β1-binding sequence from collagen IV (Vandenberg et al., 1991) into α1-84 and α2-84 at residues 1461–1472 of the native sequence designated α1-CB3 and α2-CB3 (Fig. S2 A). Arginine-to-alanine point mutations were inserted to the α1-84 and α2-84 constructs using PCR. Constructs were subcloned into the pRcX expression vector for expression in HEK293 cells. All vectors were sequenced to confirm composition.
Recombinant expression and purification
HEK293 cells were grown in DMEM/F12 medium (Sigma-Aldrich) containing 5% FBS and 50 µg/ml ascorbic acid phosphate (Wako Pure Chemical Industries). Transfections were performed by using the calcium phosphate precipitation method and 5 µg of each plasmid DNA individually. Selection of transfected cells was started with 250 µg/ml G418 (Corning) 2 d after transfection. Resistant clones were isolated and expanded. Expression was evaluated using anti-FLAG (Sigma-Aldrich), α1 (IV) NC1, and α2 (IV) NC1-specific antibodies (H11 and H22, respectively; Sado et al., 1995).
Recombinant proteins were initially purified by affinity chromatography on anti-FLAG M2 affinity agarose (Sigma-Aldrich) according to the manufacturer’s instructions and were further purified by SEC. Recombinant products were concentrated separately using Amicon centrifugal filters (EMD Millipore). Protein concentration was determined with a Nanodrop 2000c spectrophotometer (Thermo Fisher Scientific) using the following conversion factors calculated from PROTPARAM: 1.08 A280 = 1 mg/ml for α1-84/α1-CB3 and 1.219 A280 = 1 mg/ml for α2-84/α2-CB3.
In vitro assembly of recombinant protomers
The α1-84/α1-CB3 and α2-84/α2-CB3 recombinant monomers were diluted individually to a final concentration of 100 µg/ml in buffer containing 50 mM Tris-HCl and 0.5 M NaCl and were incubated for 30 min at RT. Monomers were then combined at a 2:1 ratio of α1-84/α1-CB3 to α2-84/α2-CB3, incubated for 5 min at RT, concentrated to 1 mg/ml, and left at RT overnight. The products were then fractionated by SEC.
SEC
SEC of NC1 hexamers was conducted with a Superdex 200 10/300GL gel filtration column (GE Healthcare) in TBS, using an AKTA Purifier P-900 HPLC (GE Healthcare) at a 0.5-ml/min flow rate. Eluting proteins were monitored by A280. The area under hexamer peak was integrated using Unicorn software (GE Healthcare) and expressed as a percentage of the total peak area.
Gel electrophoresis and Western blotting
Purified proteins were separated by SDS-PAGE using 12% gels and then were either stained by Coomassie Brilliant Blue R250 or transferred to nitrocellulose membranes for probing with FLAG-, α1 (IV) NC1–, and/or α2 (IV) NC1-specific antibodies.
CD
CD spectra were measured on a spectropolarimeter (J-810; Jasco) using 1-nm steps and a 1-mm path length cuvette at 20°C in the wavelength range of 195 to 260 nm. Samples were equilibrated into PBS buffer to a concentration of 0.17 mg/ml. Thermal stability curves were recorded at 225 nm between 20 and 80°C, by increasing the temperature by 10°C/h. The conversion of the ellipticity CD signal to mean residue ellipticity was determined as previously described (Myers et al., 1997).
Solid phase–binding assays
Microtiter plates (Nunc Maxisorp; Thermo Fisher Scientific) were coated overnight at 4°C with 0.1 ml of 10 µg/ml of purified recombinant collagen IV proteins in TBS or type I collagen from rat tail (5 µg/ml; BD) in 0.02 N acetic acid. All wells were blocked for 1 h at RT with 0.2 ml of 300 µg/ml BSA in TBS. Recombinant integrin α2 I domain were diluted in two wash buffers (TBS containing 0.05% Tween-20, 30 µg/ml BSA, and either 1 mM EDTA or 2 mM MgCl2). The wells were washed with the appropriate wash buffer and then incubated for 1.5 h at RT with 0.1 ml of recombinant I domain with conjugated GST tag at 10 µg/ml. The bound α2 I domain was detected using anti-GST horseradish peroxidase conjugated antibodies (GE Healthcare). Tetramethylbenzidine substrate (Sigma-Aldrich) was added to each well, and the absorbance was read at 650 nm using a SpectraMax 190 plate reader (Molecular Devices). The nonspecific binding obtained in the presence of EDTA was subtracted for each sample, and all binding assays were a mean of three assays performed in duplicates.
Cell adhesion assays
Adhesion assays were performed by coating microtiter plates overnight at 4°C with recombinant collagen IV proteins as in the solid phase-binding assay. All wells were blocked for 1 h at RT with 1% BSA in TBS. HT1080 human fibrosarcoma cells were suspended in binding buffer (0.5% BSA, 1 mM MgCl2, and 0.2 mM MnCl2 in DMEM/F12) at 106 cells/ml and when necessary preincubated with 5 µg/ml of α2β1 integrin blocking monoclonal antibodies (MAB1998Z; EMD Millipore). After washing of the wells, 0.1 ml of cell suspension was added and allowed to adhere for 1 h at RT. Nonadherent cells were removed by washing with PBS. Adherent cells were fixed and stained with 0.1% crystal violet (Kueng et al., 1989). All wells were washed with TBS, and the bound dye was solubilized by the addition of 0.1 ml of 10% acetic acid. Absorbance was measured on a microtiter plate reader at 570 nm. All adhesion assays were performed in triplicates, and data were corrected for background binding in blank wells blocked with BSA.
Hexamer dissociation and assembly studies
Dissociation of uncross-linked NC1 hexamers occurred by dialyzing the protein from 50 mM Tris-HCl, pH 7.5, into 50 mM TrisAc, pH 7.4. Dissociation occurred with 1 mg/ml protein concentration, and dialysis was conducted at 4°C with at least one buffer change over a 2-d period. Hexamer assembly reactions occurred at 37°C in 50 mM TrisAc, pH 7.4, in 25-µl volumes. Reactions were initiated by the addition of various salts, followed by a brief centrifugation, and incubation of samples at 37°C. After the reaction, samples were centrifuged at 14,000 rpm and 5°C for 15 min and loaded onto the Superdex 200 column equilibrated in TBS. Unless otherwise specified, all experiments used standard conditions of 150 mM Cl− and 2 mg/ml NC1 at 37°C for 24 h. Assembly was quantified from SEC elution profiles after reaction as a percentage of the hexamer peak from the total peak area.
HOBr synthesis and cross-linking assay
HOBr was synthesized as previously described (Bhave et al., 2012). Briefly, reagent hypochlorite (ClO−) was mixed at 10 mM with an equal volume of 12 mM KBr and reacted for 1 min at pH >10. Subsequently, the solution was diluted into PBS, pH 7.4, to allow protonation of the hypohalite. Freshly synthesized hypohalous acid was reacted with NC1 preparations for 5 min at 37°C before quenching with 1 mM l-methionine. Cross-link formation was monitored by SDS-PAGE and Coomassie Blue R250 staining of the reaction samples.
PXDN expression, purification, and cross-linking assay
Recombinant PXDN was purified in Br-free Cl−buffers as described previously (Bhave et al., 2012; McCall et al., 2014). Briefly, constructs encoding the human PXDN were stably transfected into HEK293 cells. Confluent cultures were grown in the presence of serum-free DMEM/F12 with 12 µM hematin and 5 mM sodium butyrate, and culture medium was supplemented with protease inhibitors. Proteins were precipitated with 40% (wt/vol) ammonium sulfate and then resuspended at 1/50th the starting volume in 300 mM sucrose, 100 mM Br-Free NaCl, and 20 mM TrisAc, pH 8.5, and dialyzed against the same buffer before separation by MonoQ ion-exchange chromatography (GE Healthcare). Enzyme-containing fractions were further purified by ultracentrifugation using a 5–20% (wt/vol) sucrose gradient in 50 mM Br-free NaCl buffer, 3 mM hexadecyltrimethylammonium chloride, and 10 mM phosphate buffer, pH 7.5.
Molecular dynamics simulations
Molecular dynamics (MD) were conducted using starting coordinates derived from bovine placenta collagen IV NC1 hexamer (Protein Data Bank accession no. 1T61; Vanacore et al., 2004) with the AMBER 12 program suite and the ff99SB parameter set (Case et al., 2005; Hornak et al., 2006). Eight MD systems were constructed for analysis: NC1 (α112)2 hexamer, NC1 (α112) trimer, α1 monomer, and α2 monomer each at concentrations of either 0 or 150 mM chloride ions. Potassium, chloride, and calcium ions were placed in α112 NC1 hexamer and α112 NC1 trimer as dictated by x-ray coordinates (Vanacore et al., 2004). Monovalent ion parameter sets were used as described previously (Joung and Cheatham, 2008), whereas divalent ion parameters were generated with the xLEaP module of AMBER. Systems were solvated in a truncated octahedral box using the TIP3P water model to a distance of 8.0 Å. Chloride ions were added along a 1.0 Å coulombic potential grid. Energy minimization and solvent equilibration were achieved under periodic boundary conditions. Production calculations were conducted at constant pressure for 100 ns. Temperature was maintained at 300 K by a Langevin coupling algorithm using a collision frequency of 0.5 ps–1 (Larini et al., 2007). Electrostatic interactions were treated with the particle mesh Ewald method (Essmann et al., 1995). The SHAKE algorithm (Ryckaert et al., 1977) was used to constrain bond lengths involving hydrogen atoms. A nonbonded cutoff of 9.0 Å was set during all minimization, equilibration, and production stages.
Multiple sequence alignment
Multiple sequence alignments were generated with GENEIOUS v.4.8.5 using the blosum62matrix. The following sequences were used for alignment: human (Homo sapiens): COL4A1 (NCBI Protein database accession no. NP_001836.2), COL4A2 (NCBI Protein database accession no. NP_001837.2), COL4A3 (NCBI Protein database accession no. NP_000082.2), COL4A4 (NCBI Protein database accession no. NP_000083.3), COL4A5 (NCBI Protein database accession no. NP_000486.1), and COL4A6 (NCBI Protein database accession no. NP_001838.2); monkey (Macaca mulatta): COL4A1 (NCBI Protein database accession no. XP_001088660), COL4A2 (NCBI Protein database accession no. XP_002800878), COL4A3 (NCBI Protein database accession no. XP_001101448), COL4A4 (NCBI Protein database accession no. XP_001110249), COL4A5 (NCBI Protein database accession no. XP_001098546), and COL4A6 (NCBI Protein database accession no. XP_002806394); cow (Bos taurus): COL4A1 (NCBI Protein database accession no. NP_001159983.1), COL4A2 (NCBI Protein database accession no. XP_005194260.1), COL4A3 (NCBI Protein database accession no. NP_001160001.1), COL4A4 (NCBI Protein database accession no. XP_002685685), COL4A5 (NCBI Protein database accession no. XP_001790185), and COL4A6 (NCBI Protein database accession no. XP_601826); mouse (Mus musculus): COL4A1 (NCBI Protein database accession no. NP_034061.2), COL4A2 (NCBI Protein database accession no. NP_034062.3), COL4A3 (NCBI Protein database accession no. NP_031760.2), COL4A4 (NCBI Protein database accession no. NP_031761.1), COL4A5 (NCBI Protein database accession no. (NP_031762.2), and COL4A6 (NCBI Protein database accession no. NP_444415.2); zebrafish (Danio rerio): COL4A1 (NCBI Protein database accession no. XP_694040.5), COL4A2 (NCBI Protein database accession no. XP_687811.5), COL4A3 (NCBI Protein database accession no. XP_694013.7), COL4A4 (NCBI Protein database accession no. XP_009290110.1), COL4A5 (NCBI Protein database accession no. NP_001116702.1), and COL4A6 (NCBI Protein database accession no. XP_009301719.1); chicken (Gallus gallus): COL4A1 (NCBI Protein database accession no. NP_001155871.1) and COL4A2 (NCBI Protein database accession no. NP_001155862.1); frog (Xenopus tropicalis): COL4A1 (NCBI Protein database accession no. XP_002933064.1) and COL4A2 (NCBI Protein database accession no. NW_003163368.1); sea squirt (Ciona intestinalis): COL4A1 (NCBI Protein database accession no. XP_002120982.1) and COL4A2 (NCBI Protein database accession no. XP_002119477.1); fruit fly (D. melanogaster): COL4A1 (NCBI Protein database accession no. AAA28404.1) and COL4A2 (NCBI Protein database accession no. AAB64082.1); roundworm (Caenorhabditis elegans): COL4A1 (NCBI Protein database accession no. AAB59179.1) and COLA2 (NCBI Protein database accession no. AAA27989.1); sea anemone (Nematostella vectensis): COL4A1 (NCBI Protein database accession no. XP_001626265) and COL4A2 (NCBI Protein database accession no. XP_001626269.1); hydra (Hydra magnipapillata): COL4A1 (NCBI Protein database accession no. XP_002157001.1) and COL4A2 (NCBI Protein database accession no. XP_002164888); trichoplax (Trichoplax adhaerens): COL4A1 (NCBI Protein database accession no. EDV21329.1) and COL4A2 (NCBI Protein database accession no. EDV21231.1); and jellyfish (Craspedacusta sowerbyi): predicted protein sequences derived from RNA-Seq (GAMY01000000).
Molecular modeling and analysis
Molecular modeling was based upon the 1.5-Å resolution crystal structure of the (α112) NC1 hexamer from bovine placenta (Protein Data Bank accession no. 1T61; Vanacore et al., 2004). Molecular visualization was conducting with PYMOL (Schrodinger). Noncovalent interactions of the NC1 (α112) hexamer subdomain interfaces were analyzed using the LIGPLOT+ algorithm (Wallace et al., 1995). Domain interface surface areas were calculated using the INTERSURF algorithm (Ray et al., 2005) in the CHIMERA molecular graphics program (Pettersen et al., 2004). Structural motifs searches of 98,117 entries of the protein databank were conducted using the SUMO (Jambon et al., 2003), PINTS (Stark et al., 2003), RASMOT-3D (Magis et al., 2006), and PDBeMOTIF (Golovin and Henrick, 2008) algorithms on February 26, 2014. Nonlinear Poisson–Boltzmann calculations were used to estimate the electrostatic contributions to the binding free energy (ΔGel) of NC1 subdomains as a function of the monovalent chloride concentration with APBS (Baker et al., 2001) using the following parameters: macromolecule dielectric constant, 2; solvent dielectric constant, 78.54; probe radius, 1.4 Å; and ion exclusion radius, 2.0 Å. The input partial atomic charge and radius parameters were generated with the PDB2PQR (Dolinsky et al., 2004) webserver using the AMBER force field potential (Case et al., 2005). Conformational changes associated with binding were not modeled.
Cell viability assay
PrestoBlue (Thermo Fisher Scientific) was used to assess the viability of PFHR9 cells after 5 d in low-Cl culture, according to the manufacturer’s protocol. Briefly, PFHR9 cultures were seeded onto 96-well plates in standard media. After reaching confluence, cells were washed in PBS and grown for an additional 5 d in either low-Cl or standard media, and PrestoBlue reagent was added for 2 h at 37°C before absorbance measurements. Viability was calculated as the difference between absorbance at 570 and 600 nm, minus calculated absorbance (570–600 nm) from media-only wells. Separate controls were run for standard media as well as low-Cl media because of differences in pH indicator dye between the two media.
Confocal microscopy
There were 250,000 cells seeded per 1.7 cm2 well of four-chamber glass slides (Lab-Tek; Thermo Fisher Scientific) in standard DMEM (Thermo Fisher Scientific) or low-chloride media. After 48 h, the cells were maintained for 5 d in media supplemented with 50 µg/ml ascorbic acid with media changes every 24 h. Subsequently, the chamber walls were removed, and matrix and adherent cells were processed directly on the slide for immunohistochemistry. After a brief wash with PBS (Corning), the cells were snap-frozen, air-dried, and fixed in –20°C acetone for 10 min. Slides were washed several times with PBS and PBS/0.2% Tween (Sigma-Aldrich). Slides were treated with dissociation buffer consisting of 6 M urea in 0.1 M glycine buffer, pH 3.0, for 30 min (Ninomiya et al., 1995), followed by several washes with PBS and PBS/0.2% Tween. Slides were preincubated with 10% normal goat serum (Invitrogen) for 1 h at RT to block nonspecific binding of antibodies. The following primary antibodies were used for antigen detections: rat anti-collagen IV NC1 (1:250 dilution, JK2; from Y. Sado, Shigei Medical Research Institute, Okayama, Japan), rabbit anti-laminin (1:250 dilution; Abcam), and rabbit anti-PXDN (1:250 dilution; Bhave et al., 2012). The secondary antibodies used for immunofluorescence detection were Alexa555 goat anti–rat (1:200 dilution; Abcam) and Alexa488 goat anti–rabbit (1:200 dilution; Abcam). All antibodies were diluted in PBS/0.1% Tween and 5% normal goat serum. Slides were incubated with primary antibodies overnight at 4°C in a humidified chamber and then washed three times in PBS/0.2% Tween before incubating with secondary antibodies for 1 h at RT. Negative control slide was processed similarly to experimental slide, but without primary antibodies. After several washes, 1 µM Hoechst fluorescent dye was applied for 10 min for labeling cell nuclei. After washes in PBS/0.2% tween and PBS, sections were mounted in Prolong Gold (Thermo Fisher Scientific) and cured at RT for 2 d. Images were captured at 19 overlapping sections through 7.2-µm volume using an LSM 710 META microscope equipped with a 64 × 63×/1.40 Plan-Apochromat Oil objective (ZEISS) in Immersol 518 N (ZEISS) at RT. Voxel colocalization was quantified with Imaris software (v.7.6.0; Bitplane). Field-of-view intensity was quantified, and projection images were rendered using ImageJ (v.2.0.0-rc-46/1.50g; National Institutes of Health).
Transmission electron microscopy
PFHR9 cells were seeded and grown to confluency on Transwell Permeable Supports (Corning) in the standard DMEM high glucose medium (Gibco) containing 10% FBS. After reaching confluency, standard or low chloride DMEM media containing 50 µg/ml of ascorbic acid were used to maintain the cells for 10 d with daily media replacement. Supports were fixed in 2.5% glutaraldehyde buffered in 0.1 M sodium cacodylate buffer, pH ∼7.5, postfixed in 1% osmium tetroxide, followed by dehydration through a grade series of ethanol to 100%. Samples were further dehydrated in propylene oxide and infiltrated and embedded in Spurr’s epoxy. 70-nm ultrathin sections were collected on 300 mesh copper grids and stained in 2% uranyl acetate followed by Reynold’s lead citrate. Stained sections were examined using a T-12 electron microscope (Philips/FEI) operated at 100 kV and photographed using a 2K camera (AMT).
Drosophila collagen IV transgenes
For expression of C-terminally GFP-tagged collagen IV α1 chain (Cg25C) in Drosophila larvae, we used the GAL4-UAS binary expression system (Brand and Perrimon, 1993) under control of the Cg-GAL4 driver, expressed in fat body, the main source of collagen IV for BMs in the larva (Pastor-Pareja and Xu, 2011). The UAS-Cg25C-GFP construct and transgenic flies were previously described (Zang et al., 2015). To obtain GFP-tagged mutants UAS-Cg25CR76A-GFP (R to A mutation in aa 1626 of Cg25C) and UAS-Cg25CΔNC1-GFP (deletion of C terminal from aa 1663), we mutated donor plasmid pDONR221-Cg25C, containing the open reading frame of Cg25C, through PCR amplification with the following primers: R76A F: 5′-ACGTCTGCAACTACGCCTCCGCCAATGACAAGA-3′, R76A R: 5′-GGCGGAGGCGTAGTTGCAGACGTTATTCTGACC-3′, ΔNC1 F: 5′-TGAGGCGCCGGACCCAGCTTTCTTGTACAAAGTTG-3′, and ΔNC1 R: 5′-AAGCTGGGTCCGGCGCCTCACAAACGACGCAACGT-3′.
The resulting PCR products were incubated with DMT enzyme (Transgen Biotech) and transformed into DMT competent cells (Transgen Biotech) according to the manufacturer’s instructions. After confirmation of successful mutagenesis through sequencing, pDONR221-Cg25CR76A and pDONR221-Cg25CΔNC1 were recombined with pTWG (Carnegie Drosophila Vector Collection) using Gateway LR Clonase Enzyme Mix (Thermo Fisher Scientific) to obtain UAS-Cg25CR76A-GFP and UAS-Cg25CΔNC1-GFP constructs. Transgenic lines were obtained through standard P-element transgenesis (Rubin and Spradling, 1982).
Wing imaginal disks were dissected in PBS from third instar larvae, fixed in 4% PFA, and mounted in Vectashield-DAPI (Vector Laboratories) for imaging in an LSM 780 confocal microscope (ZEISS).
Statistical analyses
The results for all quantitative experiments are reported as mean ± SEM of at least three independent experiments. For assembly experiments, comparisons between all groups for statistical significance were assessed by analysis of variance followed by Holm–Sidak multiple comparison test. For low-Cl experiments, matrix production was compared between 5-d standard and low Cl using t test, whereas colocalization comparisons used independent variables t test.
Online supplemental material
Table S1 is a comparison of interactions stabilizing monomer–monomer and trimer–trimer associations. Table S2 shows low-Cl media composition. Fig. S1 shows chloride is required for hexamer assembly. Fig. S2 shows design and characterization of recombinant protomer (recombinant collagen IV α112 protomers). Fig. S3 shows thermodynamic and mutational analysis of hexamer assembly. Fig. S4 shows assembly switch motif is present in α3-α6 chains. Fig. S5 shows NC1 domains are required for collagen IV network assembly. Online supplemental is available at http://www.jcb.org/cgi/content/full/jcb.201510065/DC1.
Supplementary Material
Acknowledgments
We thank the Vanderbilt Center for Structural Biology for use of size exclusion chromatography multiple angle light scattering instrumentation, the Vanderbilt Center for Matrix Biology for support, and the Vanderbilt Cell Imaging Shared Resource for assistance and use of confocal and electron microscopes. Technical assistance from Neonila Danylevych is appreciated.
This work was supported by the National Institutes of Health (R01 DK18381, R01 DK065138, and P01 DK065123 to B.G. Hudson; R01 18381-38S1 to I.A. Ero-Tolliver; 5T32HL94296-06 to K.L. Brown; K08 DK097306 to G. Bhave; DK099467 to R. Vanacone; and F30 DK100094 to A.S. McCall).
C.F. Cummings and B.G. Hudson are co-founders of Sulfilatec, Inc., where B.G. Hudson is a scientific consultant and C.F. Cummings is a director and officer. B.G. Hudson and C.F. Cummings have provided consulting services to RGDI3, Inc.
The authors declare no additional competing financial interests.
Footnotes
Abbreviations used in this paper:
- BM
- basement membrane
- CD
- circular dichroism
- HOBr
- hypobromous acid
- LBM
- bovine lens capsule basement membrane
- MD
- molecular dynamics
- NC1
- noncollagenous 1
- PXDN
- peroxidasin
- r-Prot
- recombinant collagen IV α112 protomers
- SEC
- size-exclusion chromatography
- TrisAc
- Tris-acetate buffer
- VR3
- variable region 3
References
- Baker N.A., Sept D., Joseph S., Holst M.J., and McCammon J.A.. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 98:10037–10041. 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastolla U., and Demetrius L.. 2005. Stability constraints and protein evolution: the role of chain length, composition and disulfide bonds. Protein Eng. Des. Sel. 18:405–415. 10.1093/protein/gzi045 [DOI] [PubMed] [Google Scholar]
- Bhave G., Cummings C.F., Vanacore R.M., Kumagai-Cresse C., Ero-Tolliver I.A., Rafi M., Kang J.S., Pedchenko V., Fessler L.I., Fessler J.H., and Hudson B.G.. 2012. Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. Nat. Chem. Biol. 8:784–790. 10.1038/nchembio.1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchiellini C., Coulon J., and Le Parco Y.. 1996. The function of type IV collagen during Drosophila muscle development. Mech. Dev. 58:179–191. 10.1016/S0925-4773(96)00574-6 [DOI] [PubMed] [Google Scholar]
- Boutaud A., Borza D.B., Bondar O., Gunwar S., Netzer K.O., Singh N., Ninomiya Y., Sado Y., Noelken M.E., and Hudson B.G.. 2000. Type IV collagen of the glomerular basement membrane. Evidence that the chain specificity of network assembly is encoded by the noncollagenous NC1 domains. J. Biol. Chem. 275:30716–30724. 10.1074/jbc.M004569200 [DOI] [PubMed] [Google Scholar]
- Brand A.H., and Perrimon N.. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415. [DOI] [PubMed] [Google Scholar]
- Campbell K.P., and Stull J.T.. 2003. Skeletal muscle basement membrane-sarcolemma-cytoskeleton interaction minireview series. J. Biol. Chem. 278:12599–12600. 10.1074/jbc.R300005200 [DOI] [PubMed] [Google Scholar]
- Case D.A., Cheatham T.E. III, Darden T., Gohlke H., Luo R., Merz K.M. Jr., Onufriev A., Simmerling C., Wang B., and Woods R.J.. 2005. The Amber biomolecular simulation programs. J. Comput. Chem. 26:1668–1688. 10.1002/jcc.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J.J., Li S., Klee C.B., and Bax A.. 2001. Solution structure of Ca(2+)-calmodulin reveals flexible hand-like properties of its domains. Nat. Struct. Biol. 8:990–997. 10.1038/nsb1101-990 [DOI] [PubMed] [Google Scholar]
- Court F.A., Wrabetz L., and Feltri M.L.. 2006. Basal lamina: Schwann cells wrap to the rhythm of space-time. Curr. Opin. Neurobiol. 16:501–507. 10.1016/j.conb.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Daley W.P., and Yamada K.M.. 2013. ECM-modulated cellular dynamics as a driving force for tissue morphogenesis. Curr. Opin. Genet. Dev. 23:408–414. 10.1016/j.gde.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky T.J., Nielsen J.E., McCammon J.A., and Baker N.A.. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32(Web Server):W665–W667. 10.1093/nar/gkh381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald J.E., Kulp D.W., and DeGrado W.F.. 2011. Salt bridges: geometrically specific, designable interactions. Proteins. 79:898–915. 10.1002/prot.22927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eble J.A., Golbik R., Mann K., and Kühn K.. 1993. The alpha 1 beta 1 integrin recognition site of the basement membrane collagen molecule [alpha 1(IV)]2 alpha 2(IV). EMBO J. 12:4795–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley J., Knight C.G., Farndale R.W., Barnes M.J., and Liddington R.C.. 2000. Structural basis of collagen recognition by integrin alpha2beta1. Cell. 101:47–56. 10.1016/S0092-8674(00)80622-4 [DOI] [PubMed] [Google Scholar]
- Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., and Pedersen L.G.. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Farrar J.D., and Carson D.D.. 1992. Differential temporal and spatial expression of mRNA encoding extracellular matrix components in decidua during the peri-implantation period. Biol. Reprod. 46:1095–1108. 10.1095/biolreprod46.6.1095 [DOI] [PubMed] [Google Scholar]
- Fidler A.L., Vanacore R.M., Chetyrkin S.V., Pedchenko V.K., Bhave G., Yin V.P., Stothers C.L., Rose K.L., McDonald W.H., Clark T.A., et al. Aspirnauts . 2014. A unique covalent bond in basement membrane is a primordial innovation for tissue evolution. Proc. Natl. Acad. Sci. USA. 111:331–336. 10.1073/pnas.1318499111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-García C., and Draper D.E.. 2003. Electrostatic interactions in a peptide-RNA complex. J. Mol. Biol. 331:75–88. 10.1016/S0022-2836(03)00615-6 [DOI] [PubMed] [Google Scholar]
- Golovin A., and Henrick K.. 2008. MSDmotif: exploring protein sites and motifs. BMC Bioinformatics. 9:312 10.1186/1471-2105-9-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.C., Graham P.L., and Kramer J.M.. 1997. Characterization of alpha1(IV) collagen mutations in Caenorhabditis elegans and the effects of alpha1 and alpha2(IV) mutations on type IV collagen distribution. J. Cell Biol. 137:1185–1196. 10.1083/jcb.137.5.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagios C., Lochter A., and Bissell M.J.. 1998. Tissue architecture: the ultimate regulator of epithelial function? Philos. Trans. R. Soc. Lond. B Biol. Sci. 353:857–870. 10.1098/rstb.1998.0250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J.R., Soliakov A., and Lewis R.J.. 2013. In vitro fibrillogenesis of collagen type I in varying ionic and pH conditions. Micron. 49:60–68. 10.1016/j.micron.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., and Simmerling C.. 2006. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 65:712–725. 10.1002/prot.21123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson B.G., Tryggvason K., Sundaramoorthy M., and Neilson E.G.. 2003. Alport’s syndrome, Goodpasture’s syndrome, and type IV collagen. N. Engl. J. Med. 348:2543–2556. 10.1056/NEJMra022296 [DOI] [PubMed] [Google Scholar]
- Hynes R.O. 2009. The extracellular matrix: not just pretty fibrils. Science. 326:1216–1219. 10.1126/science.1176009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman J.L., Robertson C., Mott J.D., and Bissell M.J.. 2015. Mammary gland development: cell fate specification, stem cells and the microenvironment. Development. 142:1028–1042. 10.1242/dev.087643 [DOI] [PubMed] [Google Scholar]
- Jambon M., Imberty A., Deléage G., and Geourjon C.. 2003. A new bioinformatic approach to detect common 3D sites in protein structures. Proteins. 52:137–145. 10.1002/prot.10339 [DOI] [PubMed] [Google Scholar]
- Joung I.S., and Cheatham T.E. III. 2008. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 112:9020–9041. 10.1021/jp8001614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C.A., Nishiyama M., Ohnishi Y., Beppu T., and Birktoft J.J.. 1993. Determinants of protein thermostability observed in the 1.9-A crystal structure of malate dehydrogenase from the thermophilic bacterium Thermus flavus. Biochemistry. 32:3913–3922. 10.1021/bi00066a010 [DOI] [PubMed] [Google Scholar]
- Khoshnoodi J., Cartailler J.P., Alvares K., Veis A., and Hudson B.G.. 2006a Molecular recognition in the assembly of collagens: terminal noncollagenous domains are key recognition modules in the formation of triple helical protomers. J. Biol. Chem. 281:38117–38121. 10.1074/jbc.R600025200 [DOI] [PubMed] [Google Scholar]
- Khoshnoodi J., Sigmundsson K., Cartailler J.P., Bondar O., Sundaramoorthy M., and Hudson B.G.. 2006b Mechanism of chain selection in the assembly of collagen IV: a prominent role for the alpha2 chain. J. Biol. Chem. 281:6058–6069. 10.1074/jbc.M506555200 [DOI] [PubMed] [Google Scholar]
- Kueng W., Silber E., and Eppenberger U.. 1989. Quantification of cells cultured on 96-well plates. Anal. Biochem. 182:16–19. 10.1016/0003-2697(89)90710-0 [DOI] [PubMed] [Google Scholar]
- Kuo D.S., Labelle-Dumais C., and Gould D.B.. 2012. COL4A1 and COL4A2 mutations and disease: insights into pathogenic mechanisms and potential therapeutic targets. Hum. Mol. Genet. 21(R1):R97–R110. 10.1093/hmg/dds346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo D.S., Labelle-Dumais C., Mao M., Jeanne M., Kauffman W.B., Allen J., Favor J., and Gould D.B.. 2014. Allelic heterogeneity contributes to variability in ocular dysgenesis, myopathy and brain malformations caused by Col4a1 and Col4a2 mutations. Hum. Mol. Genet. 23:1709–1722. 10.1093/hmg/ddt560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larini L., Mannella R., and Leporini D.. 2007. Langevin stabilization of molecular-dynamics simulations of polymers by means of quasisymplectic algorithms. J. Chem. Phys. 126:104101 10.1063/1.2464095 [DOI] [PubMed] [Google Scholar]
- Lemmink H.H., Schröder C.H., Brunner H.G., Nelen M.R., Zhou J., Tryggvason K., Haagsma-Schouten W.A.G., Roodvoets A.P., Rascher W., van Oost B.A., and Smeets H.J.M.. 1993. Identification of four novel mutations in the COL4A5 gene of patients with Alport syndrome. Genomics. 17:485–489. 10.1006/geno.1993.1351 [DOI] [PubMed] [Google Scholar]
- Lu P., Weaver V.M., and Werb Z.. 2012. The extracellular matrix: a dynamic niche in cancer progression. J. Cell Biol. 196:395–406. 10.1083/jcb.201102147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magis C., Gasparini D., Lecoq A., Le Du M.H., Stura E., Charbonnier J.B., Mourier G., Boulain J.C., Pardo L., Caruana A., et al. 2006. Structure-based secondary structure-independent approach to design protein ligands: Application to the design of Kv1.2 potassium channel blockers. J. Am. Chem. Soc. 128:16190–16205. 10.1021/ja0646491 [DOI] [PubMed] [Google Scholar]
- McCall A.S., Cummings C.F., Bhave G., Vanacore R., Page-McCaw A., and Hudson B.G.. 2014. Bromine is an essential trace element for assembly of collagen IV scaffolds in tissue development and architecture. Cell. 157:1380–1392. 10.1016/j.cell.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanaga A., Koseki T., Matsuzawa H., Wakagi T., Shoun H., and Fushinobu S.. 2004. Crystal structure of a family 54 alpha-L-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose. J. Biol. Chem. 279:44907–44914. 10.1074/jbc.M405390200 [DOI] [PubMed] [Google Scholar]
- Myers J.K., Pace C.N., and Scholtz J.M.. 1997. Helix propensities are identical in proteins and peptides. Biochemistry. 36:10923–10929. 10.1021/bi9707180 [DOI] [PubMed] [Google Scholar]
- Nakajima Y., Miyahara I., Hirotsu K., Nishina Y., Shiga K., Setoyama C., Tamaoki H., and Miura R.. 2002. Three-dimensional structure of the flavoenzyme acyl-CoA oxidase-II from rat liver, the peroxisomal counterpart of mitochondrial acyl-CoA dehydrogenase. J. Biochem. 131:365–374. 10.1093/oxfordjournals.jbchem.a003111 [DOI] [PubMed] [Google Scholar]
- Ninomiya Y., Kagawa M., Iyama K., Naito I., Kishiro Y., Seyer J.M., Sugimoto M., Oohashi T., and Sado Y.. 1995. Differential expression of two basement membrane collagen genes, COL4A6 and COL4A5, demonstrated by immunofluorescence staining using peptide-specific monoclonal antibodies. J. Cell Biol. 130:1219–1229. 10.1083/jcb.130.5.1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin J.D., San Antonio J.D., Pedchenko V., Hudson B., Jensen S.T., and Savige J.. 2011. Mapping structural landmarks, ligand binding sites, and missense mutations to the collagen IV heterotrimers predicts major functional domains, novel interactions, and variation in phenotypes in inherited diseases affecting basement membranes. Hum. Mutat. 32:127–143. 10.1002/humu.21401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Pareja J.C., and Xu T.. 2011. Shaping cells and organs in Drosophila by opposing roles of fat body-secreted Collagen IV and perlecan. Dev. Cell. 21:245–256. 10.1016/j.devcel.2011.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pöschl E., Schlötzer-Schrehardt U., Brachvogel B., Saito K., Ninomiya Y., and Mayer U.. 2004. Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development. 131:1619–1628. 10.1242/dev.01037 [DOI] [PubMed] [Google Scholar]
- Ray N., Cavin X., Paul J.C., and Maigret B.. 2005. Intersurf: dynamic interface between proteins. J. Mol. Graph. Model. 23:347–354. 10.1016/j.jmgm.2004.11.004 [DOI] [PubMed] [Google Scholar]
- Rhodes J.M., and Simons M.. 2007. The extracellular matrix and blood vessel formation: not just a scaffold. J. Cell. Mol. Med. 11:176–205. 10.1111/j.1582-4934.2007.00031.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M., and Spradling A.C.. 1982. Genetic transformation of Drosophila with transposable element vectors. Science. 218:348–353. 10.1126/science.6289436 [DOI] [PubMed] [Google Scholar]
- Ryckaert J.-P., Ciccotti G., and Berendsen H.J.C.. 1977. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23:327–341. 10.1016/0021-9991(77)90098-5 [DOI] [Google Scholar]
- Sado Y., Kagawa M., Kishiro Y., Sugihara K., Naito I., Seyer J.M., Sugimoto M., Oohashi T., and Ninomiya Y.. 1995. Establishment by the rat lymph node method of epitope-defined monoclonal antibodies recognizing the six different alpha chains of human type IV collagen. Histochem. Cell Biol. 104:267–275. 10.1007/BF01464322 [DOI] [PubMed] [Google Scholar]
- Sanes J.R. 2003. The basement membrane/basal lamina of skeletal muscle. J. Biol. Chem. 278:12601–12604. 10.1074/jbc.R200027200 [DOI] [PubMed] [Google Scholar]
- Sherwood D.R. 2015. A developmental biologist’s “outside-the-cell” thinking. J. Cell Biol. 210:369–372. 10.1083/jcb.201501083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillat T., Saat R., Pöllänen R., Hukkanen M., Takagi M., and Konttinen Y.T.. 2012. Basement membrane collagen type IV expression by human mesenchymal stem cells during adipogenic differentiation. J. Cell. Mol. Med. 16:1485–1495. 10.1111/j.1582-4934.2011.01442.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.J., and Ott H.C.. 2011. Organ engineering based on decellularized matrix scaffolds. Trends Mol. Med. 17:424–432. 10.1016/j.molmed.2011.03.005 [DOI] [PubMed] [Google Scholar]
- Stark A., Sunyaev S., and Russell R.B.. 2003. A model for statistical significance of local similarities in structure. J. Mol. Biol. 326:1307–1316. 10.1016/S0022-2836(03)00045-7 [DOI] [PubMed] [Google Scholar]
- Sundaramoorthy M., Meiyappan M., Todd P., and Hudson B.G.. 2002. Crystal structure of NC1 domains. Structural basis for type IV collagen assembly in basement membranes. J. Biol. Chem. 277:31142–31153. 10.1074/jbc.M201740200 [DOI] [PubMed] [Google Scholar]
- Vanacore R.M., Shanmugasundararaj S., Friedman D.B., Bondar O., Hudson B.G., and Sundaramoorthy M.. 2004. The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links. J. Biol. Chem. 279:44723–44730. 10.1074/jbc.M406344200 [DOI] [PubMed] [Google Scholar]
- Vanacore R., Ham A.J., Voehler M., Sanders C.R., Conrads T.P., Veenstra T.D., Sharpless K.B., Dawson P.E., and Hudson B.G.. 2009. A sulfilimine bond identified in collagen IV. Science. 325:1230–1234. 10.1126/science.1176811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg P., Kern A., Ries A., Luckenbill-Edds L., Mann K., and Kühn K.. 1991. Characterization of a type IV collagen major cell binding site with affinity to the alpha 1 beta 1 and the alpha 2 beta 1 integrins. J. Cell Biol. 113:1475–1483. 10.1083/jcb.113.6.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F.X., and Sangster B.. 1987. The toxicology of bromide ion. Crit. Rev. Toxicol. 18:189–213. 10.3109/10408448709089861 [DOI] [PubMed] [Google Scholar]
- Vracko R. 1974. Basal lamina scaffold-anatomy and significance for maintenance of orderly tissue structure. Am. J. Pathol. 77:314–346. [PMC free article] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski R.A., and Thornton J.M.. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8:127–134. 10.1093/protein/8.2.127 [DOI] [PubMed] [Google Scholar]
- Wang X., Harris R.E., Bayston L.J., and Ashe H.L.. 2008. Type IV collagens regulate BMP signalling in Drosophila. Nature. 455:72–77. 10.1038/nature07214 [DOI] [PubMed] [Google Scholar]
- Wewer U.M., Faber M., Liotta L.A., and Albrechtsen R.. 1985. Immunochemical and ultrastructural assessment of the nature of the pericellular basement membrane of human decidual cells. Lab. Invest. 53:624–633. [PubMed] [Google Scholar]
- Yurchenco P.D. 2011. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect. Biol. 3:a004911 10.1101/cshperspect.a004911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y., Wan M., Liu M., Ke H., Ma S., Liu L.P., Ni J.Q., and Pastor-Pareja J.C.. 2015. Plasma membrane overgrowth causes fibrotic collagen accumulation and immune activation in Drosophila adipocytes. eLife. 4:e07187 10.7554/eLife.07187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., and Cremer P.S.. 2010. Chemistry of Hofmeister anions and osmolytes. Annu. Rev. Phys. Chem. 61:63–83. 10.1146/annurev.physchem.59.032607.093635 [DOI] [PubMed] [Google Scholar]
- Zhou P., Tian F., Zou J., Ren Y., Liu X., and Shang Z.. 2010. Do halide motifs stabilize protein architecture? J. Phys. Chem. B. 114:15673–15686. 10.1021/jp105259d [DOI] [PubMed] [Google Scholar]
- Ziomber A., Machnik A., Dahlmann A., Dietsch P., Beck F.X., Wagner H., Hilgers K.F., Luft F.C., Eckardt K.U., and Titze J.. 2008. Sodium-, potassium-, chloride-, and bicarbonate-related effects on blood pressure and electrolyte homeostasis in deoxycorticosterone acetate-treated rats. Am. J. Physiol. Renal Physiol. 295:F1752–F1763. 10.1152/ajprenal.00531.2007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.