Abstract
Xenobiotic compounds undergo a critical range of biotransformations performed by the phase I, II, and III drug-metabolizing enzymes. The oxidation, conjugation, and transportation of potentially harmful xenobiotic and endobiotic compounds achieved by these catalytic systems are significantly regulated, at the gene expression level, by members of the nuclear receptor (NR) family of ligand-modulated transcription factors. Activation of NRs by a variety of endo- and exogenous chemicals are elemental to induction and repression of drug-metabolism pathways. The master xenobiotic sensing NRs, the promiscuous pregnane X receptor and less-promiscuous constitutive androstane receptor are crucial to initial ligand recognition, jump-starting the metabolic process. Other receptors, including farnesoid X receptor, vitamin D receptor, hepatocyte nuclear factor 4 alpha, peroxisome proliferator activated receptor, glucocorticoid receptor, liver X receptor, and RAR-related orphan receptor, are not directly linked to promiscuous xenobiotic binding, but clearly play important roles in the modulation of metabolic gene expression. Crystallographic studies of the ligand-binding domains of nine NRs involved in drug metabolism provide key insights into ligand-based and constitutive activity, coregulator recruitment, and gene regulation. Structures of other, noncanonical transcription factors also shed light on secondary, but important, pathways of control. Pharmacological targeting of some of these nuclear and atypical receptors has been instituted as a means to treat metabolic and developmental disorders and provides a future avenue to be explored for other members of the xenobiotic-sensing NRs.
Keywords: Nuclear receptors, drug metabolism, xenobiotics, ligand-binding domain, structural biology, coactivator, corepressor, agonist, antagonism
Introduction
A vast array of enzymes are responsible for the biotrans-formation and metabolism of xenobiotic compounds in the human body. Xenobiotic metabolism occurs by three generally accepted steps, termed phase I (functionalization), phase II (conjugation), and phase III (transporters) (Handschin and Meyer, 2003; Miller and Willson, 2001; Moore, 2001; Omiecinski et al., 2003; Sonoda and Evans, 2003; Xie et al., 2004). Key proteins involved in these steps include cytochrome p450 (CYPs), UDP-glycosyltransferases (UGTs), glutathione-S-transferases (GSTs), sulfotransferases (SULTs), organic anion transporters (OATs), multidrug-resistance proteins (MDRs), and multidrug-resistance–associated proteins (MRPs). They can be collected into three groups that mirror the phases: oxidative drug-metabolizing enzymes (CYPs) (Gonzalez, 1988); conjugative enzymes (UGTs, GSTs, SULTs) (Larrieu and galtier, 1988); and transporter proteins (OATs, MDRs, MRPs) (Scheper et al., 1992). It has been well established that endo- and xenobiotic compounds regulate the expression of these drug-metabolizing enzymes through their direct binding to nuclear receptors (NRs) (Miller and Willson, 2001; Sonoda and Evans, 2003; Xie et al., 2004; Remmer, 1972; Xie and Evans, 2001) (Table 1).
Table 1.
List of formal NRs and atypical receptor-like proteins known to be involved in drug-metabolism gene regulation.
| Formal NRs involved in drug metabolism and disposition | |||
|---|---|---|---|
| NR | Major gene types regulated | Effects upon induction | Key ligands and types |
| PXR | CYPs, UGTs, GST, SULTs, MDR, MRP, OATs | Detoxification and biotransformation of xenobiotics, regulation of homeostasis | Various drug, drug-like, and endobiotic compounds |
| CAR | CYPs, UGTs, GST, SULTs, MDR, MRP, OATs | Detoxification and biotransformation of xenobiotics, regulation of homeostasis | Small-molecule compounds |
| FXRa | CYPs, UGTs, SULTs, MRP, OATs, MDR, SHP, PPAR, PXR | Bile acid and lipid homeostasis, bile acid export and regulation of bile acid formation | Cholesterol-based compounds, farnesol metabolites, bile acid metabolites |
| VDR | CYPs, SULTs, MRP, FXR | Calcium homeostasis, cell proliferation and differentiation | 1α,25-dihydroxyvitamin D3, LCA |
| HNF4αb | CYPs, SULTs, MDR, OATs, PXR, CAR, PPAR, FXR | Liver development, lipid and bile metabolism, bile acid synthesis | Fatty acids, linoleic acid |
| PPAR | CYPs, UGTs, GSTs, SULTs, MDR, FXR, SHP | Fatty acid homeostasis, repression of bile acid synthesis, inflammation | Fatty acids, thiazolidinediones, hypolipidemic fibrates |
| GRb | CYPs, MRP, CAR, PXR, RXRα | Immunoresponse, stimulation of bile acid transport | Glucocorticoids |
| LXR | SULTs | Regulation of cholesterol synthesis and absorption, modulation of bile acid toxicity and cholestasis | Cholesterol-based compounds (oxysterols) |
| RORb | CYPs, SULTs, GSTs | Triglyceride regulation, glucose homeostasis | Cholesterol, retinoic acids, melatonin, thiazolidindiones |
| RXR | Dependent on binding Partner | Dependent on binding partner | Retinoic acids |
| Atypical NR-like proteins involved in drug metabolism and disposition | |||
|---|---|---|---|
| Receptor | Major genes regulated | Effects upon induction | Key ligands and types |
| AhR | CYPs, GSTs, UGTs, MRP, SULTs | Cell-cycle control, metabolic adaptation to xenobiotics, chemical toxicity signals | Polycyclic aromatic hydrocarbons, halogenated aromatic hydrocarbons |
| SHP | AR, ER, HNF4, LRH-1, LXR, PPAR, RAR, RXR, and other NRs | Repression of NRs through heterodimerization | Small, synthetic molecules, others unknown |
| Nrf2 | GSTs, UGTs, SULTs, MRP, AhR | Protection against oxidative and electrophilic stress | — |
Detailed for each NR (top) are the major gene types regulated, the observed effects upon receptor activation, and some of the known ligands and ligand types. Further outlined are atypical and receptor-like proteins (bottom) that have been identified as modulators of drug metabolism and formal NRs with direct effects on gene regulation. For these informal receptors, the major genes and proteins regulated, the observed effects, and key ligand types are given.
Can function with or without RXRα.
Functions as monomer or homodimer.
Pregnane X receptor (PXR) and constitutive androstane receptor (CAR) are the master xenobiotic sensors that bind a variety of ligands and modulate a number of drug-metabolizing enzymes (Kliewer et al., 2002; Moore et al., 2000; Kachaylo et al., 2011) (Table 1). Farnesoid X receptor (FXR), vitamin D receptor (VDR), hepatocyte nuclear factor 4 alpha (HNF4α), peroxisome proliferator-activated receptor (PPAR), glucocorticoid receptor (GR), liver X receptor (LXR), and RAR-related orphan receptor (ROR) are also involved in the regulation of genes critical for drug metabolism (Kang et al., 2007; Chiang, 2009; Zhang and Guan, 2007; Reschly and Krasowski, 2006; Hong and Tontonoz, 2008; Einstein et al., 2004) (Table 1).
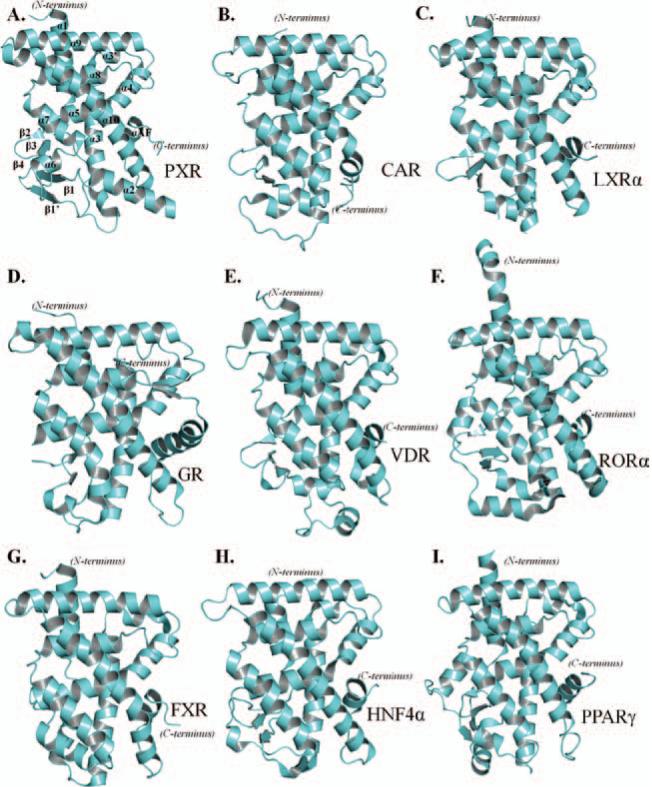
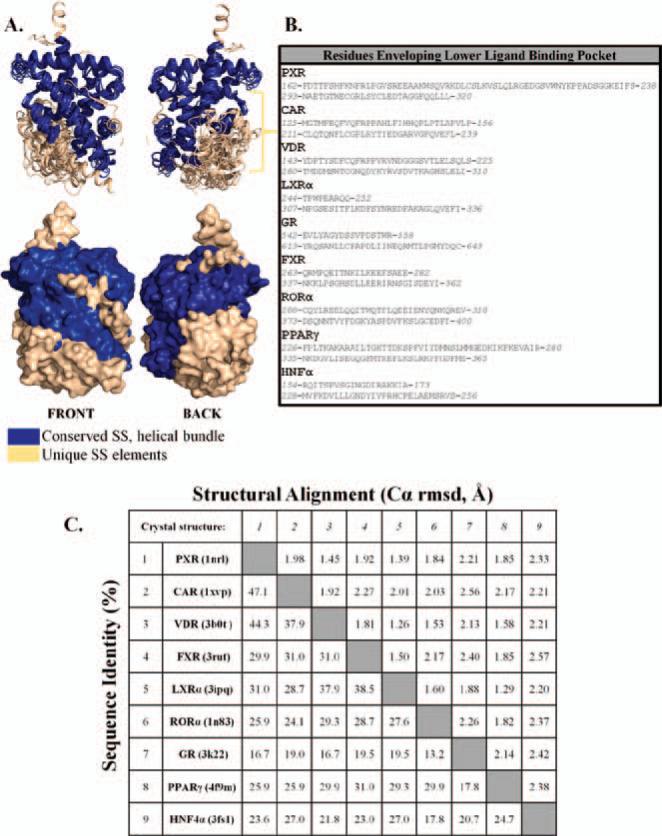
X-ray crystallographic studies focused on the ligand-binding domain (LBD) of these NRs have greatly assisted our understanding of the recognition of endo- and xeno-biotic compounds (Figure 1). Nuclear receptors form a conserved three-layered, α-helical sandwich that makes up the bulk of the LBD. This canonical make-up is composed of several α-helices (α1–10, AF) and β-sheets that cap the ligand-binding pocket (as depicted in Figure 1A) for PXR, and similar compositions are found in the other NRs (shown in Figure 1). The LBD of various NRs envelop sizeable binding cavities that are able to attract and bind to a wide variety of compounds. Each receptor contains divergent secondary structural elements in the lower left portion of the LBD (Figure 2A). Further, the amino-acid composition of this region supports the differentiation between these NRs (Figure 2B). However, examination of the domain in its entirety reveals relative conservation of both secondary structure and amino-acid composition between these nine NRs (Figure 2C) (Holm et al., 2008). A compilation of LBD crystallographic studies is presented in this review in an attempt to better understand the collaboration between the ligand-binding properties of NRs and activation of drug-metabolizing genes.
Figure 1.
(A–I) LBD crystal structures of the nuclear receptors, PXR, CAR, LXRα, GR, VDR, RORα, FXR, HNF4α, and PPARα. Each LBD contains the conserved, three-layered α-helical sandwich, along with structural elements that line the lower left portion of the ligand-binding pocket.
Figure 2.
(A) Structural overlay of all nine NR LBD crystal structures. In blue are the conserved α-helical bundles, and colored in tan are the divergent structural features that line the binding pocket. (B) Amino acids that are found to envelop the lower left ligand-binding pocket for each NR. Identities of these regions are disparate in terms of both length and chemical property. (C) Structural alignment and sequence identity performed by Dali (Holm et al., 2008) using known LBD crystal structures reveals both primary and secondary similarities.
PXR and CAR as master xenobiotic sensors
Regulation of drug-metabolizing genes
It was originally noted in the 1970s that certain pharmaceutical compounds regulated the expression of a number of enzymes capable of protecting against toxic effects of xenobiotics (Selye, 1971). The receptor capable of sensing the presence of such exogenous compounds and, ultimately, the upregulation of various metabolizing enzymes was later identified in mice as PXR (Blumberg et al., 1998; Kliewer et al., 1998). Activation was shown to be induced by natural steroids as well as synthetic corticoids. The human ortholog, originally named PXR as well as SXR (steroid and xenobiotic receptor) and PAR (pregnane-activated receptor), was found to be highly expressed in the liver and intestine, where drug metabolism and clearance is most prominent. Regulation of the phase I major drug-metabolizing enzyme, CYP, was additionally found to be linked to xenobiotic activation of PXR (Kliewer et al., 1998; Goodwin et al., 1999, 2002; Brobst et al., 2004). CAR was also identified as a xeno-biotic-activated receptor that regulates CYP enzymes through a variety of steroidal and xeno-compounds (Forman et al., 1998; Kawamoto et al., 1999; Sueyoshi et al., 1999). Similar to PXR, CAR is primarily expressed in the liver, and a multitude of ligands are known to activate CAR, though less so than PXR. A distinction of CAR is its activity in the absence of ligands in nonhepatic cells. Although PXR has well-established basal activity in in vitro reporter gene assays, it appears that PXR is as active in vivo as CAR in the absence of ligand.
Metabolic transformation by phase I drug metabolism prepares xenobiotics for subsequent reactions through oxidative mechanisms, creating sites for phase II metabolizing enzymes to act upon (Zamek-Gliszczynski et al., 2006). By utilizing phase III–based efflux pumps, such xenobiotic metabolites are then excreted from cells to complete the metabolic cycle. It is becoming increasingly accepted that xenobiotics sensors, such as PXR and CAR, coordinate the regulation of all three phases of drug metabolism. Because they are involved in the transcriptional control of UGTs, SULTs, GSTs, as well as hydrolases and acetyltransferases, CAR and PXR are capable of regulating ~90% of all phase II metabolism (Wells et al., 2004; Evans and Relling, 1999; Bélanger et al., 2003; Nowell and Falany, 2006; Glatt, 2000; Pool-Zobel, 2005; Haimeur et al., 2004; Weinshilboum, 2006). This allows for the conjugation of a wide variety of compounds, both xeno- and endobiotics. As such, PXR and CAR are able to induce detoxifying enzymes to protect against the toxicity derived from the metabolism of endogenous compounds and xenobiotics, facilitating their excretion. Several xenobiotics, acting as agonists or antagonists, interact with both PXR and CAR, either to the same effect or with inverse responses. Notably, because CAR and PXR can be activated by the same ligands, upregulation of overlapping sets of genes can create collaborative mechanisms for eliminating toxic compounds (Moore et al., 2000, 2002; Timsit and Negishi, 2007; Ekins et al., 2002; Maglich et al., 2002). PXR and CAR utilize traditional mechanisms of gene regulation, which include ligand binding, nuclear translocation, dimerization with their binding partner, retinoid X receptor (RXR), and binding to a variety of response elements in the promoter regions of regulatory enzymes (Stanley et al., 2006).
PXR as a promiscuous xenobiotic sensor
Originally characterized as critical in the role for detecting endogenous pregnanes, the types of compounds that activate PXR has, since then, been expanded to include several xenobiotics and pharmaceutically relevant compounds (Takeshita, 2001; Ricketts et al., 2007; Sonoda et al., 2005; Cheng et al., 2009). Deemed as a central xenobiotic sensor, PXR is expressed predominantly in areas of the human body along the traditional metabolic pathway (liver and gastrointestinal tract) (Kliewer et al., 1998; Lehmann et al., 1998; Bertilsson et al., 1998). PXR ligands are structurally and chemically unique, such as phenobarbital, rifampicin, dexamethasone, and hyper-forin, further supporting PXR's role as a master xenobiotic sensor (Goodwin et al., 1999; Watkins et al., 2003; Waxman and Azaroff, 1992; Shi et al., 2010) controlling a number of phase I–III metabolizing enzymes. In addition to xenobiotic activation, PXR has also been shown to be activated by a number of endogenous ligands (pregnanes, bile acids, and vitamins) (Kliewer et al., 1998; Xue et al., 2007). These endobiotics control the induction of a number of genes involved in bile acid metabolism and transport, cholesterol homeostasis, and protection from toxic endobiotics (Uppal et al., 2005). The promiscuity of PXR has been validated by ligand-binding assays, establishing that PXR is activated by the direct binding of compounds within its flexible ligand-binding cavity, a unique feature of this receptor.
PXR LBD properties
A number of crystallographic studies of the LBD of PXR have been conducted since its initial discovery as a crucial regulator of drug metabolism. Crystal structures have been elucidated of both the apo- and ligand-bound form. Some of these complexes include the cholesterol-lowering drug, SR12813, the macrocyclic antibiotic, rifampicin, hyperforin from St. John's Wort, as well as the anti-HIV (human immunodeficiency virus) drug, PNU-142721 (Watkins et al., 2003; Xue et al., 2007; Wang et al., 2008; Noble et al., 2006; Cheng and Redinbo, 2011; Watkins et al., 2001; Chrencik et al., 2005).
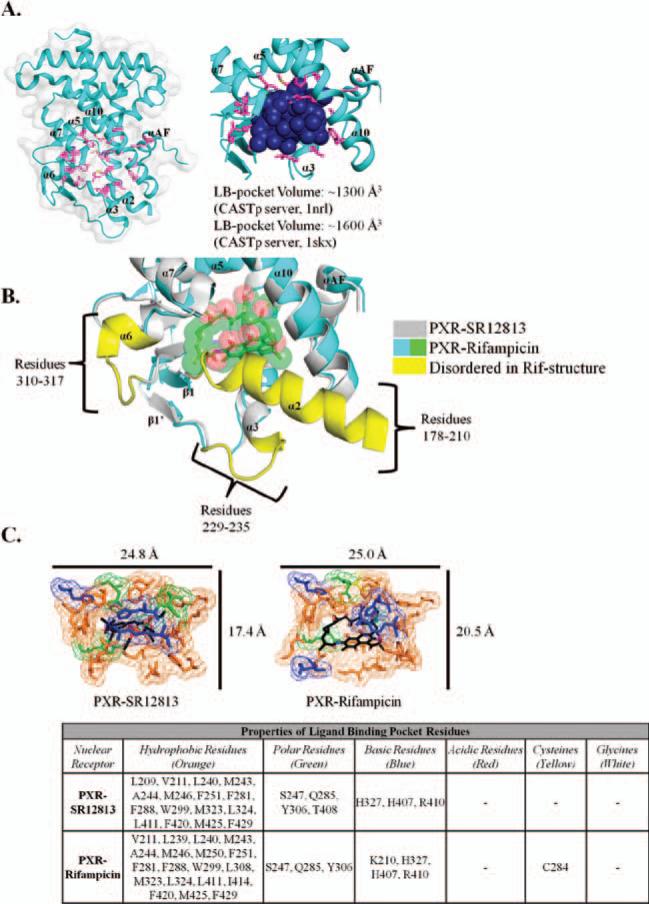
The overall structure of the PXR LBD is shown in Figure 3A and adopts the classical helical sandwich observed in other nuclear-receptor LBD structures (Figure 1A). The ligand-binding pocket residues (magenta) envelop a large volume capable of accommodating a variety of compounds. Further, PXR has the capability to expand its binding pocket to allow for larger compounds (e.g., rifampicin), as evidenced by the increase in volume of space in the binding pocket from ~1,300 Å3 for the SR12813-bound structure to ~1,600 Å3 for the rifampicin-bound structure [Figure 3A, calculated by Computed Atlas of Surface Topography of proteins (CASTp); Dundas et al., 2006; Binkowski et al., 2003]. This conformational change results in the disordering of residues 229–235, 310–317, and 178–210 (Figure 3B) (Chrencik et al.,2005). An in-depth analysis of the LBD reveals a generally hydrophobic pocket, with a few polar residues and basic residues adding additional potential interactions for ligands (Figure 3C). The overall size change of the binding pocket, when comparing different ligand sizes, can be distinguished in Figure 3C.
Figure 3.
(A) Crystal structure of the PXR LBD, with the ligand-binding pocket residues highlighted in magenta. A space-filling model of the binding pocket is shown to better understand the cavity of the pocket. PXR has the unique ability to alter its pocket to allow for the binding of different chemical structures, as exemplified by the pocket volume change from ~1,300 (SR12813 bound) to ~1,600 Å3 (rifampicin bound). (B) Overlay of the SR12813-bound (gray/yellow) and rifampicin-bound (cyan/green) PXR LBDs. Of note is the conformational change required to accommodate the large, macrocyclic compound, rifampicin, which results in three regions in the LBD becoming disordered (yellow). (C) Chemical properties of the residues that surround the ligand-binding pocket. Amino acids are colored to represent the chemical property: orange (hydrophobic); green (polar); blue (basic); red (acidic); yellow (cysteine); and white (glycine).
Several studies have shown that Ser247, Gln285, His407, Met243, Trp299, and Phe420 are consistently involved in direct protein/ligand interactions in human PXR LBD complex structures (Watkins et al., 2001, 2003; Chrencik et al., 2005; Teotico et al., 2008). It has also been observed that a number of other residues lining the pocket may also interact with the ligand or simply enclose the cavity (Figure 3C). It has also been hypothesized that cooperativity exists between coregulator binding to the AF-2 region of PXR's LBD and ligand stability in the ligand-binding pocket (Orans et al., 2005; Ingraham and Redinbo, 2005). Although not directly involved in ligand binding, PXR contains a novel secondary structural element, five-stranded antiparallel β-sheet that results in a unique homodimeric interface adjacent to the ligand-binding pocket. Although most NRs contain two to three stranded β-sheets, this element does not result in the formation of a homodimer species for other NRs, CAR included. Several studies have shown that the PXR homodimer is biologically relevant (Noble et al., 2006), and the full implications of this oligomer interface have yet to be completely elucidated.
CAR xenobiotic binding, overlap with PXR, and constitutive activity
Similar to PXR, CAR has been shown to bind to a wide variety of xeno- and endobiotic compounds that result in the induction or repression of drug-metabolizing enzymes (Kachaylo et al., 2011; Forman et al., 1998; Kawamoto et al., 1999; Sueyoshi et al., 1999; di Masi et al., 2009; Suino et al., 2004; Wei et al., 2000; Xie et al., 2000; Chang, 2009; Sakai et al., 2006; Baskin-Bey et al., 2006). CAR is known to modulate the expression of some of the same genes as PXR (CYPs, GSTs, UGTs, SULTs, and so on) (Yanagiba et al., 2009; Xu et al., 2009; Kohle and Bock, 2009; Dong et al., 2009; Buckley and Klaassen, 2009; Echchgadda et al., 2007; Chen et al., 2007). The functional overlap between PXR and CAR leads to a sophisticated method of drug clearance and detoxification through coordinated regulation of metabolic genes. Although both nuclear receptors can be activated by the same ligand, different modes of gene regulation are resultant. For example, phenobarbital is known to activate both PXR and CAR; however, activation of PXR by this compound results in increased expression of CYP3A, whereas CAR activation leads to CYP2B upregulation (Kawamoto et al., 1999; Sueyoshi et al., 1999; Waxman and Azaroff, 1992; Zelko and Negishi, 2000; Martin et al., 2003). A similar effect has also been demonstrated for rifampicin activation (Nannelli et al., 2008).
CAR is unique, relative to other NRs, in its ability to maintain constitutive activity, even in the absence of a bound ligand (Jyrkkärinne et al., 2008; Dussault et al., 2002; Windshügel and Poso, 2007; Windshügel et al., 2005, 2007; Xu et al., 2004). Alternative activation pathways are integrated into the overall function of CAR that are more indirect, independent of xenobiotic activation. Maintained in an inactive state through cytoplasmic localization by chaperone molecules (e.g., CAR cytoplasmic retention protein, heat shock protein 90) (Kawamoto et al., 1999; Kobayashi et al., 2003), CAR is then released through the action of an inducing compound, which allows the nuclear translocation of CAR and its subsequent activation of genes (Timsit and Negishi, 2007). Relatively unclear at this point, the mechanism of translocation is thought to be dependent on protein phosphatase activity, followed by response element recognition and DNA binding (Blattler et al., 2007; Shindo et al., 2007). Because of the regulation of CAR's activity by other proteins and biochemical pathways versus ligand-based activation, CAR appears to utilize a complex mode of transcriptional activation, relative to other members of the NR family.
CAR LBD crystal structures and analysis
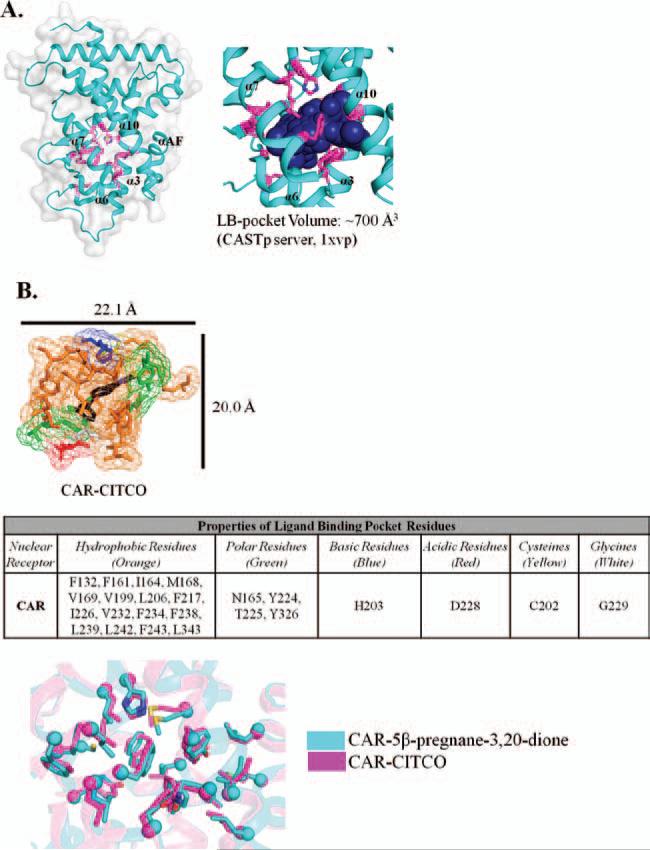
Atomic resolution studies of CAR's LBD include mouse CAR in apo form and human CAR in complex with its heterodimeric binding partner, RXRα (Shan et al., 2004; Xia and Kemper, 2005). The CAR LBD adopts similar secondary structural elements as other NRs (Figure 1B). The size of the ligand-binding pocket is smaller than that of PXR (Figure 4A), with a calculated pocket volume of ~700 Å3 for the CITCO-bound CAR LBD (calculated using CASTp). Although CAR is less promiscuous than PXR, the smaller pocket size of CAR appears to result in a more-stable ligand-bound complex than PXR, resulting in better packing of the AF-2 helix in the active conformation. The stability that coactivator recruitment provides to PXR upon ligand binding may instead be conferred to CAR through the more-compact, smaller ligand-binding pocket, with or without a bound compound.
Figure 4.
(A) Crystal structure of the CAR LBD. Ligand-binding pocket residues are highlighted in magenta. The cavity of the binding pocket is represented as a space-filled model. The CAR pocket has a calculated volume of ~700 Å3. (B) Chemical properties of the residues that surround the CAR ligand-binding pocket. Amino acids are colored to represent the chemical property: orange (hydrophobic); green (polar); blue (basic); red (acidic); yellow (cysteine); and white (glycine).
Further analysis of the amino-acid properties of the ligand-binding pocket reveals a largely hydrophobic pocket, with polar, acidic, and basic residues lining the cavity (Figure 4B). The hydrophobic nature of the binding pocket is similar to PXR in nature; however, the size limitation of the pocket, no known malleability of the pocket, as well as the incorporation of a greater variety of residue properties support the conclusion that the activity of CAR, although promiscuous, is modulated by a smaller variety of ligands, compared to the plethora that activate PXR. An examination of the two human CAR-ligand–bound structures currently deposited in the Protein Data Bank show that there is no significant conformational change or disordering of structural elements in the ligand-binding pocket (Figure 4B), as occurs in the LBD of PXR.
Splice variants and implications
The canonical sequences for PXR and CAR [PXR.1 (434 residues, Uniprot ID O75469-1) and CAR.1 (352 residues, Uniprot ID Q14994-1)] maintain the three-domain structure of all formal NRs, with a DNA-binding domain, hinge region, and the LBD. The LBD for PXR.1 consists of residues 205–434 and the LBD of CAR.1 103–352. A number of splice variants exist for each of these NRs, resulting in insertions and deletions throughout the full-length sequence. Focus will be placed on those splice variants concentrated around the LBD as well as their implication for ligand binding and gene regulation.
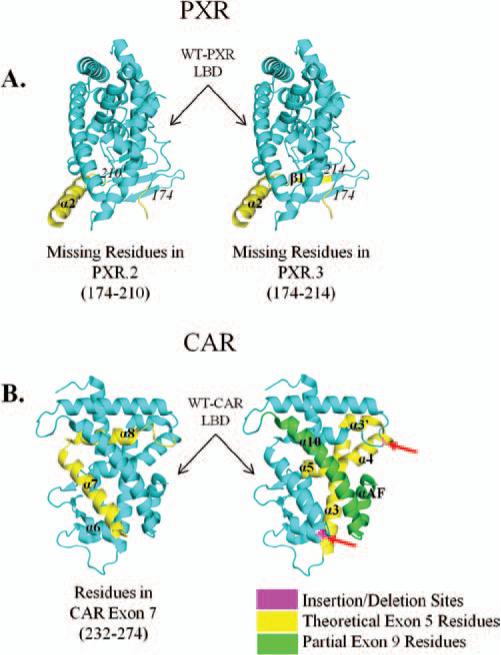
The major PXR splice variant is PXR.2, which lacks 111 nucleotides, resulting in the deletion of 37 amino acids in the LBD (Lin et al., 2009). This variant of PXR represents 6.7% of all PXR transcripts, with PXR.3 representing 0.33% (Koyano et al., 2004). Studies have discovered that PXR.2 has a reduced basal activity level as well as decreased ligand-induced (rifampicin and corticosterone) (Lamba et al., 2004; Koyano et al., 2004) transcriptional activation. Further, PXR.2 failed to activate CYP3A4 upon treatment with smaller ligands, such as deoxycholic acid, oxysterols, and bile acids (Lin et al., 2009; Lamba et al., 2004; Koyano et al., 2004; Reschly et al., 2008). The 37 residues missing from PXR.2 (Figure 5A) are a portion of the ligand-binding pocket of PXR.1, and it is to be expected that the ligand binding of properties of PXR.2 would be drastically altered. This could be a potential explanation for the loss of trans-activation upon ligand treatment. Both the canonical PXR.1 and PXR.2 are coexpressed in the liver, and it has been shown that PXR.2 regulates the overall activity of PXR.1 by repressing transactivation (Lamba et al., 2004). Indeed, it was also found that coregulator recruitment to PXR.2 is different than PXR.1; corepressors are retained by PXR.2, but coactivators show little binding (Lin et al., 2009). The two major splice variants of PXR, PXR.2 (lacking 37 residues in the LBD) and PXR.3 (lacking a 41-amino-acid deletion in the LBD) appear to be similar in predicted fold to more canonical NRs. These same residues are absent in such NRs as FXR, LXR, CAR, RXR, and ER (Ekins et al., 2002).
Figure 5.
(A) The WT PXR LBD (cyan) with residues that would be removed in the various PXR splice variants highlighted (PXR.2, left; PXR.3, right; yellow). The deletion of these residues would still maintain the canonical LBD structure, but would also result in a smaller binding pocket, which would alter PXR's ability to bind to larger xenobiotics. (B) WT CAR LBD (cyan) with residues predicted to be deleted from CAR splice variants (but still maintaining the classical LBD structure) (yellow/green). Exon 7 is a major deletion in many of CAR's variants (left, yellow) and would affect ligand binding. Shown on the right, exon 5 (predicted, yellow), exon 9 (predicted, green), and various insertion and deletion sites (magenta) are highlighted, describing additional alterations to CAR, resulting in many of the known splice variants.
There are 26 splice variants (Lamba et al., 2004) for full-length CAR, with CAR LBD variants involving the entire deletion of exon 5 and/or 7 (Lamba et al., 2004; van der Vaart and Schaaf, 2009), as well as deletions of exon 4 (which constitutes a portion of the N-terminal LBD), partial deletions of exon 9 (C-terminal end), and insertions throughout the LBD. The major splice variants of CAR involve the deletion of exon 7 (Figure 5B), which includes variants 3, 5, 8–11, and 16–21, and involve the elimination of major helices that line the ligand-binding pocket. These variants are unable to bind response-element DNA and are unable to lead to the transactivation of the major drug-metabolism enzymes, CYP2B, CYP3A4, UGT1A1, and MDR1 (Lamba et al., 2004; van der Vaart and Schaaf, 2009).
Additional CAR splice variants are created through the combination of deletion and insertions sites around exon 7 and deletions of exons 4–9 (Figure 5B). Splice variants 1, 6–7, 12, 14–15, and 23–24 incorporate an insertion on either side of exon 7. CAR variants lead to a number of consequences, resulting in altered gene expression (Auerbach et al., 2003, 2005, 2007; Arnold et al., 2004). Insertions on the N-terminal side of exon 7 result in decreased DNA binding to response elements, decreased interactions with various coactivators, and increased specificity for activation of CYP2B and MDR reporters, but decreased induction of CYP3 and UGT genes. Insertions on the C-terminal side of exon 7 lead to complete loss of response element binding and no activation of CYP2, CYP3, UGT, and MDR genes. Consequences of exon 5 and partial exon 9 deletion are varied: Removal of exon 5 results in a premature termination and loss of CAR expression; partial exon 9 deletion results in decreased DNA binding, loss of transactivation, and no interaction with coactivators.
Splice variants of PXR and CAR result in a number of deleterious or altered effects on gene transactivation, coregulator recruitment, as well as ligand binding. LBD splicing results in potentially dramatic changes in the structural elements surrounding the ligand-binding pockets and, consequentially, leads to such altered characteristics as mentioned above. Currently, no crystal structures have been solved of any PXR or CAR splice variants; however, this is perhaps not unexpected because of the predicted loss of secondary structural elements, and thus stability, within the helical sandwich that defines formal NRs. It is clear, though, that the presence of such splice variants serve to modulate the activities of PXR and CAR. The altered characteristics of the PXR and CAR variants, as described above, yield LBDs that resemble NRs not directly involved in xenobiotic sensing, such as FXR, LXR, and others. Removal of several residues surrounding the ligand-binding cavity (Figure 5) most likely results in a smaller pocket, significantly limiting ligand promiscuity and binding. This not only would reduce the ability to regulate the vast array of drug-metabolizing enzymes, but also may have a profound effect on coregulator recruitment, which is known to be linked to the type of ligands that bind to these two NRs (Zhang et al., 2011; Xu et al., 2002).
Tissue distributions
Considered as the preliminary responders to xeno- and endobiotic recognition, it is expected that PXR and CAR are highly expressed in tissues at the entry points for xenobiotic metabolism in the body. For example, these NRs are most notably expressed in the liver (both), intestinal tract (both), and kidney (PXR) (Kliewer et al., 1998; Lehmann et al., 1998; Bertilsson et al., 1998; Baes et al., 1994), key tissues involved in initial drug metabolism, transportation, and reabsorption. CAR is additionally expressed in much lower levels in the heart, skeletal muscle, brain, kidney, and lung, but the exact function in these tissues is not fully understood (Choi et al., 1997; Swales and Negishi, 2004) and may relate to its constitutive activity. As a key influencer of protective measures in the body, PXR is more widely expressed, found in the lung, stomach, blood monocytes, reproductive organs, heart, bone, and brain tissues (Lamba et al., 2004; Bauer et al., 2004). It is also clear that the expression levels of both CAR and PXR are regulated by the presence of xenobiotics (Huss and Kasper, 2000; Pascussi et al., 2000), further exemplifying their intricate involvement in biotransformation and metabolism. Other NRs also control PXR and CAR expression, suggesting secondary measures for xenobiotic sensing and metabolic regulation (Kliewer et al., 2002; Matic et al., 2007; Modica and Bellafante, 2009; Khan et al., 2009; Peet et al., 1998).
Additional nuclear receptors involved drug metabolism
LXR
The LXRs were originally identified in the 1990s and were shown to be critical regulators of cholesterol, fatty acid, and glucose homeostasis (Peet et al., 1998; Lehmann et al., 1997; Janowski et al., 1996; Willy et al., 1995). Two isoforms were identified (α and β) and were shown to be sterol sensors, activated by oxysterol cholesterol derivatives (Lehmann et al., 1997; Janowski et al., 1996). LXRα is most notably expressed in the liver, kidney, intestine, adipose, macrophages, lung, and spleen (Janowski et al., 1996). The LXRβ isoform is expressed ubiquitously, resulting in its original name, ubiquitous receptor (Song et al., 1994, 1995). As with other NRs, the LXRs form a heterodimer with RXR upon ligand activation, then binding the LXR response-element sequences (Korf et al., 2009). LXRs are known to increase cholesterol removal and to decrease endogenous cholesterol synthesis and dietary absorption; however, LXRs also have the ability to activate proteins involved in lipogenic activity (Ouvrier et al., 2009a, 2009b; Larrede et al., 2009; Zelcer et al., 2009; Delvecchio et al., 2008). More recently, the drug-metabolizing SULT enzymes have been shown to be regulated by LXRs (Uppal et al., 2007). Synthetic LXR agonists have been shown to be effective in mice models of diabetes, inflammation, and other disorders, and glucose tolerance and bile-duct–induced cholestasis are both conferred through LXR agonism (Hong et al., 2008; Dushkin et al., 2009; Cermenati et al., 2012).
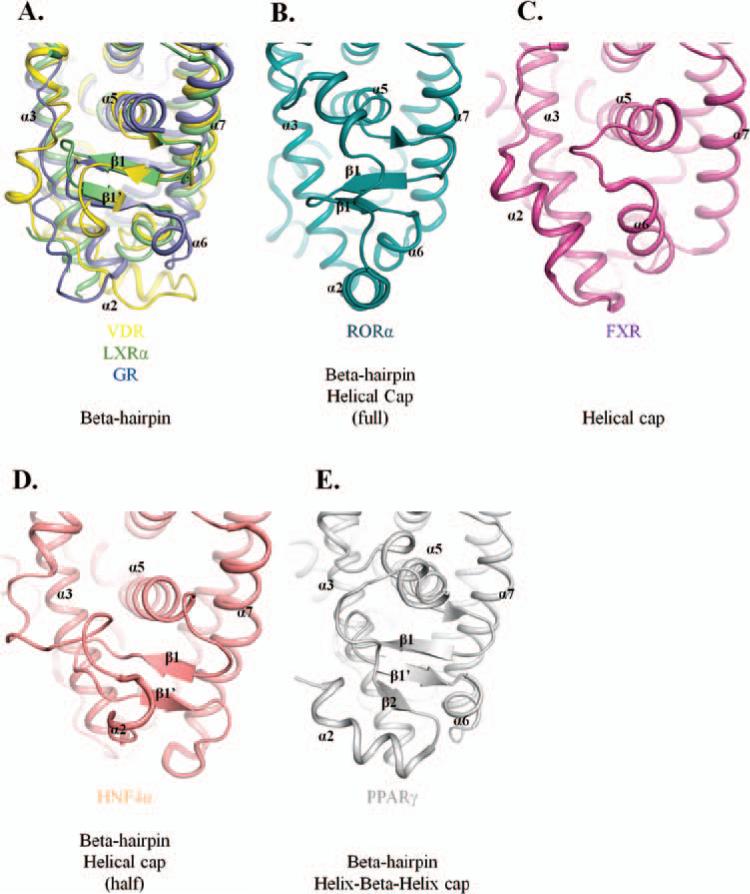
Several crystal structures of the LXRα- and β-ligand-binding domains have been elucidated, both in the apo and liganded form. The overall secondary structure is similar to other NRs (Figure 1C), with the three-layered helical sandwich, and the ligand-binding pocket in the lower portion of the LBD. In addition to the helices surrounding the binding pocket, a beta-hairpin caps the lower left side, similar to the VDR and GR (Figure 6A). The ligand-binding pocket is classified as one of the smaller ones, relative to other NRs (Figure 7A). The overall composition of the pocket residues are conserved, with a largely hydrophobic nature and a few polar residues lining the cavity (Figure 7A; Table 2). Additional basic and acidic residues are also present, resulting in a pocket that confers specificity for LXR ligands.
Figure 6.
(A) Overlay of VDR, LXRα, and GR showing secondary structural elements that cap the left portion of the ligand-binding pocket; these NRs contain a beta-hairpin cap to enclose the cavity. (B) RORα ligand-binding pocket is capped by a beta-hairpin, as with other receptors, and an additional alpha-helical cap on top of the beta-hairpin element. (C) Binding cavity of FXR is enclosed only by an alpha-helical cap, but not by a beta-hairpin. (D) HNF4α maintains the beta-hairpin structural element and incorporates half of an alpha-helical cap. (E) The PPARα ligand-binding pocket is enclosed by a beta-hairpin element found in most NRs and a unique helix-beta-helix feature to add unique specificity to this receptor.
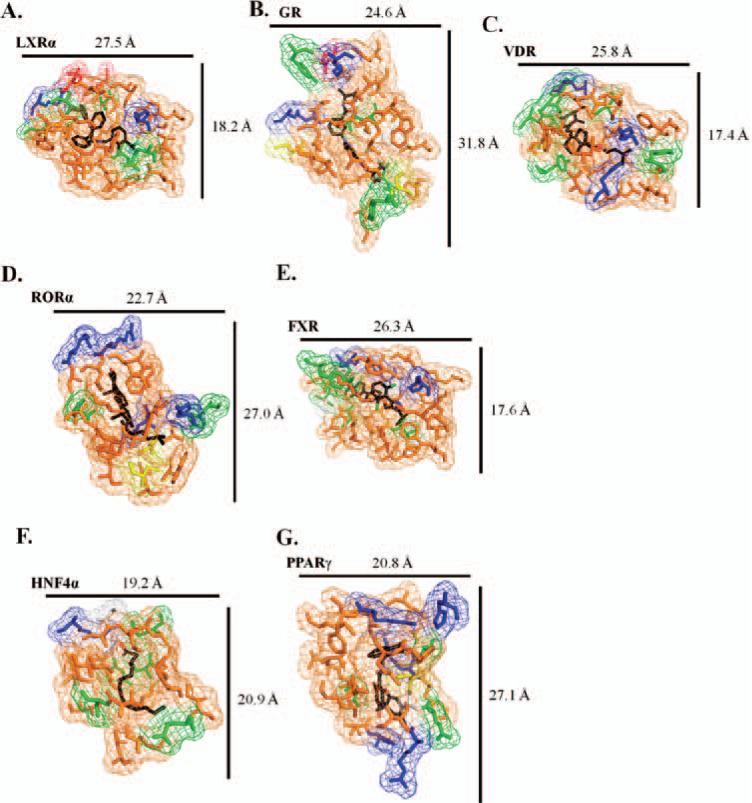
Figure 7.
(A–G) Details of the ligand-binding pockets for the nine NR LBDs. Residues are color coded based on chemical properties: orange (hydrophobic); green (polar); blue (basic); red (acidic); yellow (cysteine); and white (glycine). For each illustration, approximate measurements of binding cavities are determined to compare their relative size and shape to each other. As further detailed in Table 2, each pocket is heavily hydrophobic in nature, with various elements of polarity and charge to facilitate ligand-binding specificity.
Table 2.
Chemical properties of residues that line the ligand-binding pocket of NRs involved in drug metabolism.
| NR | Hydrophobic residues (orange) | Polar residues (green) | Basic residues (blue) | Acidic residues (red) | Cysteines (yellow) | Glycines (white) |
|---|---|---|---|---|---|---|
| GR | P541, M560, L563, L566, V571, A573, A574, W577, W600, M601, L603, M604, A605, A607, L608, F623, M646, L732, F749, L753 | N564, Q570 Y663, Y735 |
R611, K667 | E540 | C622, C736 | G567, G568 |
| LXRα | F254, F257, L260, A261, V263, I295, M298, L299, I313, F315, L316, F326, L331, F335, I336, I339, F340, V425, L428, L435, L439, W443 | T258, S264, T302, T314, Q424 | R305, H421 | E267, E301 | — | — |
| VDR | F150, L227, L230, A231, L233, V234, I268, I271, M272, W286, V300, A303, L309, L313, L404, L414, V418, F422 | Y143, Y147, S237, S275, S278, Y295, Y401 | R274, H305, H397 | — | C288 | — |
| RORα | W320, A324, I327, A330, V364, F365, M368, A371, V379, F381, F391, L394, F399, I400, V403 | Y380, Y507 R370, H484 |
K326, R367, | — | C323, C396 | — |
| FXR | M265, I273, F284, L287, M290, A291, V325, M328, F329, I335, L348, I352, I357, M365, M450, L451, W454, F461, L465, W469 | T270, T288, S332, S342, Y369 | H294, R331, H447 | — | — | G343 |
| HNF4α | V178, M182, L219, L220, A223, L234, L235, L236, V242, L249, M252, V255, I259, M342, I346 | S181, Q185, N238, S256, Q345 | R226 | — | — | G222, G237 |
| PPARγ | I262, I281, F287, A292, I326, L330, M334, V339, L340, I341, M348, L353, F363, M364, F368 | Q283, S289, S342 R288, K367, H449 |
K263, R280, | — | C285 | G284 |
In coordination with Figure 7, residues are identified as either hydrophobic, polar, basic, acidic, cysteines, or glycines. It is noted that each binding pocket is mostly hydrophobic in nature, with patches of polar and charged residues to incorporate ligand specificity.
GR
The GR is a ubiquitously expressed NR that binds to cortisol and other glucocorticoids (Wira and Munck, 1970; Svec and Rudis, 1981; Munck et al., 1972). Upon activation, GR forms a homodimer complex, which results in transactivation of various drug-disposition genes, and regulates the body's response to inflammation (Ray et al., 1995; Laue et al., 1988). The GR also serves as a repressor by binding to various other transcription factors and preventing them from activation. This particular characteristic affects other xenobiotic-sensing nuclear receptors, such as CAR, PXR, and RXR (Pascussi et al., 2000, 2003), and likely serves as another way to modulate drug-metabolism pathways. CYP2 appears to be responsive to GR activation, as well as bile acid transport mechanisms (Pascussi et al., 2003).
The clinical significance of the GR is wide reaching; the GR is involved in the central nervous system, influences endocrine systems, is involved in psychological disorders, such as Cushing's disease, and plays a role in other responses to stressors, such as inflammation (Schwabe et al., 2009; van der Akker et al., 2008; Ebisawa et al., 2008; Raddatz et al., 2004). As such, drugs targeting the GR are frequently utilized for a number of ailments. Although largely advantageous, the integration of the GR in both neural and physical networks can also lead to unwanted side effects upon prolonged glucocorticoid treatment, ranging from osteoporosis, metabolic issues, stunted development, psychosis, and depression (Kawano and Kumagai, 2007; Ng and Celermajer, 2004).
The crystal structure of the LBD of the GR has been determined in complexes with various agonists and antagonists, as well as coactivators and corepressors. Although the sequence identity of GR is low, relative to other NRs, the overall LBD structure is similar (Figure 1D). As with LXRα, the left portion of the pocket cavity is closed off by a beta-hairpin structural element (Figure 6A). Further, the ligand-binding pocket is elon-gated, when compared with other LBDs (Figure 7B). This suggests a less-stable binding pocket and, perhaps, allows for a different mode of ligand binding along the vertical axis versus the horizontal. Sugar-linked steroidal compounds would also add additional size to the base ligand, which requires a larger binding pocket to compensate. Enveloping the ligand-binding pocket is a mostly hydrophobic cavity, with polar residues, as well as charged side chains that confer specificity (Figure 7B; Table 2).
VDR
The VDR, also known as the calcitriol receptor, is an NR that binds the endogenous ligand, 1,25-dihydroxyvitamin D3 (Campbell and Adorini, 2006). The VDR is also receptive to many other ligands, such as glucocorticoids, and bile acids, such as lithocholic acid (Makishima et al., 2002). Upon ligand activation and binding to its binding partner, RXR, the VDR is known to regulate calcium homeostasis, cell proliferation and differentiation, and other immunological and microbial functions (Campbell and Adorini, 2006). VDR is most prominently found in the intestine, bone, kidney, and parathyroid glands, with limited expression in liver tissue (Gascon-Barré et al., 2003). Regulation of phase I–III drugs are known to be attributed to VDR activation and repression, such as CYP2 and CYP3 enzymes, as well as SULTs (Drocourt et al., 2002; Echchgadda et al., 2004). Further, there is an inverse relationship between FXR and VDR as regulators bile acid homeostasis; as such, calcitriol inhibits FXR activation (Honjo et al., 2006). Because CYP3 and VDR expression is high in the intestine, it should be noted that vitamin D is involved in first-pass drug metabolism. Vitamin D analogs have been utilized to treat a number of diseases, such as rickets, osteoporosis, and psoriasis (Zintzaras et al., 2006; Arita et al., 2008; Malloy et al., 2007). Preliminary studies have shown that such analogs are effective at treating immune disorders and tumor malignancies, as well (Meyer et al., 2012; Valrance et al., 2007). Similar to PXR, VDR activation by the intestinal bacterial metabolite, lithocholic acid (LCA), leads to CYP3A induction, which serves to detoxify bile acids (Makishima et al., 2002). Modulating VDR activity can alter bile acid transport and detoxification, although the clinical application of this has not been established.
Crystal structures of the LBD of VDR have been elucidated as bound to vitamin D and several analogs. The VDR maintains a high sequence and structural similarity to steroid and thyroid hormone receptors (Figure 1E). As with LXR and GR, beta-hairpin element encapsulates the ligand-binding pocket (Figure 6A) to produce a relatively small space (Figure 7C). The environment surrounding the binding cavity is mainly hydrophobic, with polar and basic residues to incorporate ligand specificity (Figure 7C; Table 2).
ROR
There are three different RORs (α, β, and γ; Jetten et al., 2001), and each is expressed in separate tissues and have distinctive functions (Carlberg et al., 1994). The exact role of RORβ is not fully understood; it is highly expressed in the brain and retina and is believed to have a critical function in rod and cone photoreceptor cell development, as well as in bipolar disorder (Carlberg et al., 1994; Stehlin-Gaon et al., 2003; André et al., 1998). There are two types of RORγ: γ1 and γt (γ2). RORγ1 is expressed in several tissues, including the liver, adipose, skeletal muscle, and kidney, whereas the expression of RORγt is restricted to certain cell types in the immune system (Medvedev et al., 1996). RORγ is known to control the development of lymphoid tissues and plays a role in thymopoiesis (Kurebayashi et al., 2000; He et al., 1998; Villey et al., 1999). RORα is found in a variety of tissues, such as the testis, kidney, adipose, and liver, and is most highly expressed in the brain (Matsui, 1997; Hamilton et al., 1996). RORα is involved in lymph node and cerebellum development, lipid metabolism, and immune responses (Matsui, 1997; Delerive et al., 2001; Sashihara et al., 1996). It remains unclear what the identity of endogenous ROR ligands are, but several studies suggest that cholesterol and sulfonated metabolites, as well as melatonin and thiazolidinediones, might induce ROR-guided responses (Kallen et al., 2002; Missbach et al., 1996; Wiesenberg et al., 1995). Additionally, there is evidence that retinoids may act as antagonists for the α and γ isoforms (Stehlin-Gaon et al., 2003). Although RORα is most directly linked to drug metabolism, mice knockout studies show that RORγ also appears to be involved (Kang et al., 2007).
RORα can bind either as monomers or homodimers to ROR DNA response elements, and the associated transcriptional activities can have positive or negative effects on the expression of drug-metabolism genes (Carlberg et al., 1994). Studies toward the pharmacological targeting of RORα appear far off, because even the identity of the endogenous RORα ligand is not certain. Additionally, ligands believed to be involved in ROR activation or repression also bind to other NRs. Thus, specifically targeting ROR may prove difficult. However, a recent structural and biochemical study of RORγ provides evidence that hydroxycholesterols lead to coactivator promotion (Jin et al., 2010; Wang et al., 2010). These results may lead to the production of synthetic agonists for further ROR testing.
The LBD structures of RORα (Figure 1F) and RORγ have been determined with a number of different ligands. As with VDR, GR, and LXR, RORα has a beta-hairpin cap on the lower left side of the binding pocket; however, this NR incorporates an additional helical cap, which would serve to not only restrict ligand binding and types, but could also provide additional ligand contacts (Figure 6B). The ligand-binding pocket of RORα is elon-gated, as with the GR, and is similarly hydrophobic, with both polar and basic contacts (Figure 7D; Table 2). This larger pocket is able to bind a variety of RORα ligands, including cholesterol-based compounds, as well as retinoids and melatonin.
FXR
Also known as the bile acid receptor, the major function of the FXR is the regulation of bile acid export and of bile acid and lipid homeostasis (Parks et al., 1999; Makishima et al., 1999; Wang et al., 1999). The first NR shown to be activated by bile acids, many natural FXR ligands are known, the most notable being chenodeoxycholic acid. FXR is highly expressed in the liver, gut, and kidney, all of which are part of enterohepatic circulation and targets of bile acids (Lu et al., 2001). In addition, though, expression has been noted in other tissues, such as the heart, eye, spleen, reproductive organs, and smooth muscle cells (Rizzo et al., 2006; Bishop-Bailey et al., 2004; Huber et al., 2002). As with many NRs, FXR forms a heterodimer with RXRα and binds to inverted and everted repeats, resulting in regulation of SULTs, MRP, and critical metabolizing genes, as well as other xenobiotic receptors (Laffitte et al., 2000; Urizar et al., 2000; Kast et al., 2002; Song et al., 2001). LCA, which activates VDR, has an inverse agonistic effect on FXR, suggesting that these two NRs cooperate in bile acid metabolism (Yu et al., 2002). It is also noteworthy that monomeric FXR activates the expression of UGTs (Barbier et al., 2003b); most formal orphan receptors similar to FXR are obligate heterodomers with RXR.
FXR is known to negatively regulate bile acid uptake systems through the receptor's interaction with small heterodimer partner (SHP). This prevents the overaccumulation of toxic bile acid metabolites, inverse to the properties of the VDR (Honjo et al., 2006; Yu et al., 2002). In addition, CYP3A, UGTs, and SULTs are shown to be effected by FXR activation or repression (Gnerre et al., 2004; Miyata et al., 2006; Lu et al., 2005). More recently, liver regeneration and liver protection from carcinogenesis has been linked to FXR (Zhang et al., 2012; Fiorucci et al., 2012). Although not a major drug target, the development of targeted compounds has the potential to precisely control bile-acid–regulatory systems and to modulate NRs directly involved in xenobiotic sensing, such as PXR and PPAR. Additionally, because FXR is closely integrated in liver protection and repair, controlling this NR's function could alter drug-treatment methods where liver damage is of concern.
The FXR LBD crystal structure has been determined as being bound to agonist ligands and coactivator peptides. Although the LBD maintains the canonical secondary structural elements of other NRs (Figure 1G), unlike the other NRs presented here, though, the left side of the ligand-binding cavity is enclosed solely by a helical cap, instead of the common beta-hairpin feature (Figure 6C). The binding pocket is similar to LXR and VDR—smaller and compact, with a nearly complete hydrophobic nature (Figure 7E; Table 2). FXR is expected to contain binding-cavity properties similar to VDR because they bind closely related or identical compounds, such as LCA.
HNF4α
Highly expressed in the liver, intestine, kidney, and pancreas, HNF4α plays an important role in organ function, particularly development and maintenance of the liver (Miquerol et al., 1994; Zhong et al., 1994). Acting as a homodimer, HNF4α can regulate gene transcription in the absence of ligands, but its activity is affected by fatty thioesters (Ladias et al., 1992; Sladek et al., 1990). Drug transporters that are suppressed by bile-acid–activated receptors FXR and SHP, like the organic cation transporters 1 and 2 and organic anion transporter 1, are instead transactivated by HNF4α, detailing a mechanism of modulation between NRs (Saborowski et al., 2006; Popowski et al., 2005). This can be extended to PXR and CAR that both contain promoters regulated by HNF4α (Kamiya et al., 2003; Ding et al., 2006). Various CYP and SULT enzymes contain HNF4α-binding sites, providing a direct link between this NR and critical drug-metabolizing enzymes (Echchgadda et al., 2007; Tirona et al., 2003; Jover et al., 2001). Pharmacological targeting of HNF4α may prove deleterious because it plays such an integral role in the regulation of many essential NRs that control xeno- and endobiotic metabolism.
There are a number HNF4α LBD crystal structures, complexed with fatty acids and coactivator fragments (Figure 1H). Secondary structural elements that surround the cavity include the helical sandwich, as well as a beta-hairpin cap (consistent with many other NRs) and an additional small helical cap further closes the lower left portion (Figure 6D). The ligand-binding pocket of HNF4α is relatively compact, coinciding with the thin nature of the fatty acid compounds it is known to bind (Figure 7F; Table 2). The environment is heavily hydrophobic, but also contains a number of polar residues, and a charged basic residue.
PPAR
The three forms of PPAR (PPARα, PPARβ, and PPARγ) are lipid sensors and controllers of lipid homeostasis, adipocyte differentiation, development, metabolism, and tumorigenesis (Brown and Plutzky, 2007; Willson et al., 2000). PPARα is expressed mainly in the heart, liver, and kidney brown fat, whereas PPARγ is expressed in those locations as well as white adipose, colon, pancreas, and spleen (Brown and Plutzky, 2007; Willson et al., 2000). PPARβ is most notably found in the brain and skin tissue (Girroir et al., 2008). PPARα and PPARγ are both known to regulate drug-metabolism genes and, as such, are regular pharmaceutical targets for different purposes. PPARα activation leads to induction of organic cation transporters, involved in drug uptake, as well as SULTs and UGTs after activation by fibrates (Luci et al., 2006; Barbier et al., 2003a; Fang et al., 2005). PPARγ is critical for adipogenesis and insulin response, and treatments with thiazolidinediones are used to regulate these factors (Szatmari et al., 2006).
Crystal structures of the PPARγ LBD (Figure 1I) consist of all three forms of PPAR and have been determined to be bound to agonists, antagonists, and coactivator and -repressor peptide fragments. PPARγ contains a beta-hairpin element that caps the lower left of the ligand-binding pocket, and an additional helix-beta-helix element further encloses the cavity (Figure 6E). The binding pocket is comparatively large, similar in size and shape to RORα or GR, and would support the binding of a variety of compounds (Figure 7G; Table 2). This is confirmed by the number of basic residues interspersed throughout the largely hydrophobic pocket.
RXRα is a binding partner for gene regulation
Obligate RXRα recruitment
Upon ligand binding, a number of NRs are complexed with a common binding partner (RXRα) (Mangelsdorf and Evans, 1995). Several studies have shown that this heterodimer complex is required for response element recognition and DNA binding (Mangelsdorf and Evans, 1995; Shulman et al., 2004; Zechel et al., 1994; Mader et al., 1993). Because the basic makeup of a formal NR includes only a single DNA-binding domain (DBD), the recruitment of RXRα is necessary to incorporate an additional DBD to bind two repeating hexameric half-sites. Understandably, RXRα is expressed in the liver, kidney, skin, and intestinal tissues, where NRs requiring heterodimerization are present (Mangelsdorf et al., 1990). NRs involved in drug metabolism requiring RXRα include PXR, CAR, FXR, VDR, PPAR, and LXR (Orlov et al., 2003; Chandra et al., 2008; Zhang et al., 2006; Cai et al., 2010; Son and Lee, 2009; Conde et al., 2008). Although RXRα assists many NRs in the sensing and binding of various response elements, it should be noted that its presence is not essential to the other receptors involved in drug metabolism. These NRs include GR, ROR, HNF4α, as well as FXR that can specifically regulate UGT enzymes in the absence of RXRα (Lu et al., 2005; Rauch et al., 2010; Cieśla, 2011). As such, RXRα appears to be critical for NRs that exhibit xenobiotic sensing properties, such as PXR and CAR, whereas NRs with a more-indirect effect on metabolism do not require heterodimerization.
Atypical receptors and receptor-like proteins
Aryl hydrocarbon receptor (AhR)
The AhR is technically classified as a member of the basic helix-loop-helix transcription factors (Fukunaga et al., 1995; Burbach et al., 1992). Although not a member of the NR family, AhR consists of four major domains, some of which are strikingly similar to that of the canonical domain composition of nuclear receptors, including a DBD and LBD (Fukunaga et al., 1995). There is an additional PAS-A domain, and a large transcriptional activation binding domain. The AhR has been shown to function in a manner reminiscent of NRs, such as being initiated by ligand binding. The AhR nuclear translocator is then recruited to AhR, and the resulting complex binds to the response elements in the promoter region of a number of drug-metabolizing genes, such as CYPs, UGTs, and GSTs (Köhle and Bock, 2007; Fujii-Kuriyama, 2005). The AhR is known to bind include many synthetic xenobiotic compounds, such as halogenated and polycyclic aromatic hydrocarbons (Denison et al., 2002; Denison and Nagy, 2003), including dioxins (environmental toxins), polychlorinated dibenzofurans (known environmental carcinogens and mutagens), polychlorinated biphenyls (found in coolants and subsequently identified as carcinogens), benzo(a)prene found in coal tar, tetracene (an organic semiconductor), and other aromatics (Denison and Nagy, 2003). With the ability to bind a wide range of xenobiotic compounds, AhR is believed to act as a detection mechanism, sensing potentially toxic foreign compounds and facilitating their eventual metabolism and elimination (Denison et al., 2002; Baba et al., 2005). However, there are no known crystal structures of the AhR.
SHP
Although classified as an NR, the SHP is unique in that it consists of the classical LBD, but lacks the conserved NR, DBD (Seol et al., 1996). The primary function of the SHP is understood to be the repression of other NRs by forming inactive heterodimer complexes (Johansson et al., 2000; Zhang et al., 2011). Crystal structures of the SHP reveal that it adopts the canonical helical sandwich motif and thus most likely forms heterodimers similarly to formal NRs (Johansson et al., 2000; Lee et al., 1998; Ortlund et al., 2005). The SHP has been shown to affect many of the key NRs that modulate the activity of drug-metabolizing genes, including the GR, FXR, LXR, HNF4α, PPARγ, PXR, and CAR (Zhang et al., 2011). Potential models of SHP repression include the blocking of the coactivator-binding site, increased recruitment of corepressors, and inhibition of DNA binding (Zhang et al., 2011). The repressive function of the SHP serves to alter the regulation of drug-metabolizing genes and thus plays a critical role in how xenobiotics are treated in the human body. Recently, activation of the SHP has been shown to be ligand induced upon interaction with certain compounds, such as retinoid-related molecules (Miao et al., 2011). This ligand-dependent activation was shown to repress certain CYP enzymes involved in bile acid regulation. Additional studies are also exposing further potential synthetic ligands that could be utilized to modulate SHP activity and its repressive functions (Xia et al., 2011, 2012).
Nuclear factor erythroid 2-related factor 2 (Nrf2)
Nrf2 is a family member of the basic leucine zipper transcription factors and is known to induce the gene expression of antioxidant enzymes (Aleksunes and Manautou, 2007). Nrf2 is most notably expressed in the kidneys, muscle, lung, heart, liver, and brain (Moi et al., 1994). Activated by reactive oxygen species and electrophiles, Nrf2 is involved in the regulation of oxidative stress (OS) through mechanisms similar to the NR family, including binding to antioxidant response elements and modulating constitutive and ligand-induced expression. Normally sequestered by the Kelch-like ECH-associated protein 1 (Keap1), influences by OS or electrophiles leads to the releases of Nrf2 and translocation to the nucleus, followed by gene transcription (Itoh et al., 1999; Kang et al., 2004). Phase II enzymes, such as GSTs, UGTs, and SULTs, are known to be effected by Nrf2 activation, as well as the drug-efflux–transporter proteins, Mrps (Itoh et al., 1997; Yueh and Tukey, 2007; Alnouti and Klaassen, 2008). Structural studies of Nrf2 have been limited to only a portion of the protein; however, sequence alignments reveal that it contains six highly conserved Nrf2/ECH homology domains, which allow for several interactions, including Maf dimerization, binding to Keap1, protein stability and transactivation, binding to cyclic adenosine monophosphate response element-binding proteins, and degradation signaling (Motohashi et al., 2004; Motohashi and Yamamoto, 2004; Nioi et al., 2005; McMahon et al., 2004). Drug targeting of Nrf2 is being investigated as a therapeutic method for OS-related ailments, as is the case with bardoxolone methyl, used to treat advanced chronic kidney disease (Reisman et al., 2012; Pergola et al., 2011).
Coregulator recruitment and involvement in metabolism
Implications of agonist versus antagonist binding
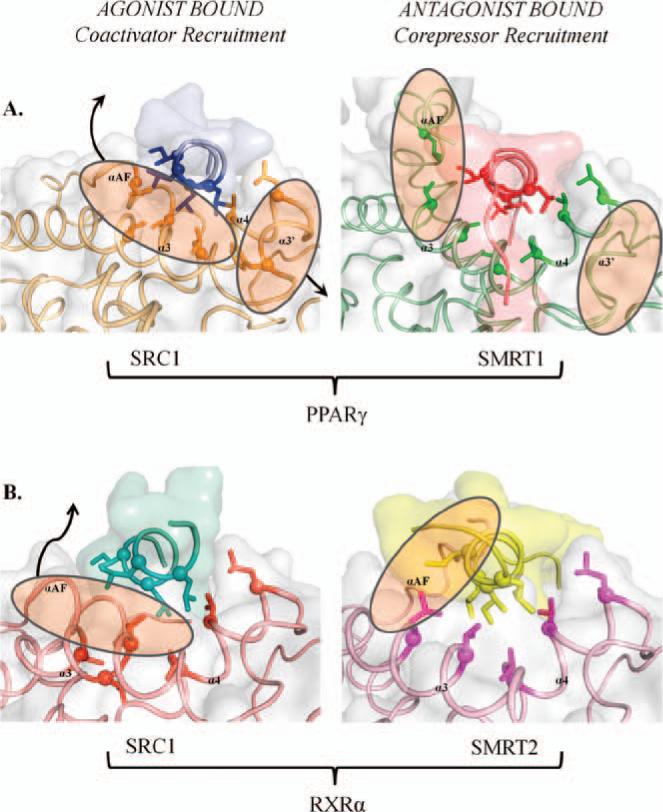
NRs have the ability to exhibit dramatically different effects dependent upon the receptor's identity, the chemical makeup of a target ligand, and the tissue involved. These effects range from agonism, antagonism, and inverse agonism (Gronemeyer et al., 2004). Agonists, such as endogenous ligands and other synthetic mimics, bind to the LBD of NRs and lead to upregulation of gene expression, whereas antagonism works to competitively block agonist binding and prevents gene induction (Brzozowski et al., 1997). A major accompanying factor to ligand binding is the location of the AF helix and the coregulator-binding surface found in formal NRs (Zhang et al., 2011; Xu et al., 2002). Agonist-bound NRs allow for favorable coactivator binding, as is the case in the “agonist-bound” state (Figure 8). Antagonism of these NRs causes a conformational change (Figure 8), preventing coactivator recruitment, but allowing for corepressors, such as silencing mediator of retinoic acid and thyroid hormone receptor 1 and 2, to bind with greater affinity. The connection between ligand-binding specificity and stability and with coregulator binding has previously been documented for a number of NRs (Xu et al., 2004; Watkins et al., 2003). The proximity of the ligand-binding pocket and the AF-2 region, where cofactors are known to bind in crystal structures, further supports consequential interplay between the two. As such, regulation of gene expression by NRs are not only dependent on ligand binding, but are also additionally further tuned by coregulator recruitment.
Figure 8.
Coregulator recruitment for agonist versus antagonist binding. (A) Crystal structures of the PPARα LBD have been solved in complex with an agonist/coactivator (left) and antagonist/corepressor (right). Comparing the two structures, it is noted that the AF helix changes conformation dramatically upon antagonist binding, producing a unique site for corepressor binding. (B) RXRα LBD crystal structures complexed with agonist/coactivator (left) and antagonist/corepressor (right) elements. As with other NRs, the consequences of antagonist/ corepressor binding are evident in the conformational change of the AF helix.
Conclusions and future directions
NR regulation of drug-metabolism gene expression has been well documented. The structural basis for endo- and xenobiotic ligand binding in the LBD reveals conserved modes of binding for certain NRs, such as LXRα, VDR, and FXR, and more-disparate modes, as observed for GR, RORα, and PPARγ. The primary xenobiotic sensor, PXR, has the unique ability to alter the shape of its pocket and the positions of neighboring elements to accommodate a wide variety of large and small compounds. Coregulator recruitment has also been directly correlated with agonist versus antagonist binding for the whole group of NRs considered. With the widespread tissue distribution of these NRs, nearly every critical tissue contains a NR known to regulate the key drug-metabolizing genes.
The discovery of agonists and antagonists has been noted for some NRs, but this does not extend to all. No synthetic, designed PXR antagonist, for example, has yet been created, though the implications of such a finding would have serious clinical applications for regulating gene expression. Additionally, more structural studies of LBD complex structures with RXRα, coupled with biological experiments, could provide a deeper understanding of coregulator recruitment to each NR and, possibly, could then be extrapolated to local or systemic gene induction or repression.
Acknowledgments
Declaration of interest
This work was funded by the National Institutes of Health (grant nos.: CA98468 and DK62229).
References
- Aleksunes LM, Manautou JE. Emerging role of Nrf2 in protecting against hepatic and gastrointestinal disease. Toxicol Pathol. 2007;35:459–473. doi: 10.1080/01926230701311344. [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. Regulation of sulfotransferase enzymes by prototypical microsomal enzyme inducers in mice. J Pharmacol Exp Ther. 2008;324:612–621. doi: 10.1124/jpet.107.129650. [DOI] [PubMed] [Google Scholar]
- André E, Gawlas K, Becker-André M. A novel isoform of the orphan nuclear receptor RORbeta is specifically expressed in pineal gland and retina. Gene. 1998;216:277–283. doi: 10.1016/s0378-1119(98)00348-5. [DOI] [PubMed] [Google Scholar]
- Arita K, Nanda A, Wessagowit V, Akiyama M, Alsaleh QA, McGrath JA. A novel mutation in the VDR gene in hereditary vitamin D-resistant rickets. Br J Dermatol. 2008;158:168–171. doi: 10.1111/j.1365-2133.2007.08232.x. [DOI] [PubMed] [Google Scholar]
- Arnold KA, Eichelbaum M, Burk O. Alternative splicing affects the function and tissue-specific expression of the human constitutive androstane receptor. Nucl Recept. 2004;2:1. doi: 10.1186/1478-1336-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach SS, Dekeyser JG, Stoner MA, Omiecinski CJ. CAR2 displays unique ligand binding and RXRalpha heterodimerization characteristics. Drug Metab Dispos. 2007;35:428–439. doi: 10.1124/dmd.106.012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach SS, Ramsden R, Stoner MA, Verlinde C, Hassett C, Omiecinski CJ. Alternatively spliced isoforms of the human constitutive androstane receptor. Nucleic Acids Res. 2003;31:3194–3207. doi: 10.1093/nar/gkg419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach SS, Stoner MA, Su S, Omiecinski CJ. Retinoid × receptor-alpha-dependent transactivation by a naturally occurring structural variant of human constitutive androstane receptor (NR1I3). Mol Pharmacol. 2005;68:1239–1253. doi: 10.1124/mol.105.013417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Mimura J, Nakamura N, Harada N, Yamamoto M, Morohashi K, et al. Intrinsic function of the aryl hydrocarbon (dioxin) receptor as a key factor in female reproduction. Mol Cell Biol. 2005;25:10040–10051. doi: 10.1128/MCB.25.22.10040-10051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol. 1994;14:1544–1522. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier O, Duran-Sandoval D, Pineda-Torra I, Kosykh V, Fruchart JC, Staels B. Peroxisome proliferator-activated receptor alpha induces hepatic expression of the human bile acid glucuronidating UDP-glucuronosyltransferase 2B4 enzyme. J Biol Chem. 2003a;278:32852–32860. doi: 10.1074/jbc.M305361200. [DOI] [PubMed] [Google Scholar]
- Barbier O, Torra IP, Sirvent A, Claudel T, Blanquart C, Duran-Sandoval D, et al. FXR induces the UGT2B4 enzyme in hepatocytes: a potential mechanism of negative feedback control of FXR activity. Gastroenterology. 2003b;124:1926–1940. doi: 10.1016/s0016-5085(03)00388-3. [DOI] [PubMed] [Google Scholar]
- Baskin-Bey ES, Huang W, Ishimura N, Isomoto H, Bronk SF, Braley K, et al. Constitutive androstane receptor (CAR) ligand, TCPOBOP, attenuates Fas-induced murine liver injury by altering Bcl-2 proteins. Hepatology. 2006;44:252–262. doi: 10.1002/hep.21236. [DOI] [PubMed] [Google Scholar]
- Bauer B, Hartz AM, Fricker G, Miller DS. Pregnane × receptor up-regulation of P-glycoprotein expression and transport function at the blood-brain barrier. Mol Pharmacol. 2004;66:413–419. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- Bélanger A, Pelletier G, Labrie F, Barbier O, Chouinard S. Inactivation of androgens by UDP-glucuronosyltransferase enzymes in humans. Trends Endocrinol Metab. 2003;14:473–479. doi: 10.1016/j.tem.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Bäckman M, et al. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci U S A. 1998;95:12208–12213. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkowski TA, Naghibzadeh S, Liang J. CASTp: Computed Atlas of Surface Topography of proteins. Nucleic Acids Res. 2003;31:3352–3355. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop-Bailey D, Walsh DT, Warner TD. Expression and activation of the farnesoid × receptor in the vasculature. Proc Natl Acad Sci U S A. 2004;101:3668–3673. doi: 10.1073/pnas.0400046101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattler SM, Rencurel F, Kaufmann MR, Meyer UA. In the regulation of cytochrome P450 genes, phenobarbital targets LKB1 for necessary activation of AMP-activated protein kinase. Proc Natl Acad Sci U S A. 2007;104:1045–1050. doi: 10.1073/pnas.0610216104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B, Sabbagh W, Jr., Juguilon H, Bolado J, Jr., van Meter CM, Ong S, et al. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brobst DE, Ding X, Creech KL, Goodwin B, Kelley B, Staudinger JL. Guggulsterone activates multiple nuclear receptors and induces CYP3A gene expression through the pregnane × receptor. J Pharmacol Exp Ther. 2004;310:528–535. doi: 10.1124/jpet.103.064329. [DOI] [PubMed] [Google Scholar]
- Brown JD, Plutzky J. Peroxisome proliferator-activated receptors as transcriptional nodal points and therapeutic targets. Circulation. 2007;115:518–533. doi: 10.1161/CIRCULATIONAHA.104.475673. [DOI] [PubMed] [Google Scholar]
- Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engström O, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- Buckley DB, Klaassen CD. Induction of mouse UDP-glucuronosyltransferase mRNA expression in liver and intestine by activators of aryl-hydrocarbon receptor, constitutive androstane receptor, pregnane × receptor, peroxisome proliferator-activated receptor alpha, and nuclear factor erythroid 2-related factor 2. Drug Metab Dispos. 2009;37:847–856. doi: 10.1124/dmd.108.024190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbach KM, Poland A, Bradfield CA. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci U S A. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai SY, He H, Nguyen T, Mennone A, Boyer JL. Retinoic acid represses CYP7A1 expression in human hepatocytes and HepG2 cells by FXR/RXR-dependent and independent mechanisms. J Lipid Res. 2010;51:2265–2274. doi: 10.1194/jlr.M005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MJ, Adorini L. The vitamin D receptor as a therapeutic target. Expert Opin Ther Targets. 2006;10:735–748. doi: 10.1517/14728222.10.5.735. [DOI] [PubMed] [Google Scholar]
- Carlberg C, Hooft van Huijsduijnen R, Staple JK, DeLamarter JF, Becker-André M. RZRs, a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Mol Endocrinol. 1994;8:757–770. doi: 10.1210/mend.8.6.7935491. [DOI] [PubMed] [Google Scholar]
- Cermenati G, Abbiati F, Cermenati S, Brioschi E, Volonterio A, Cavaletti G, et al. Diabetes-induced myelin abnormalities are associated with an altered lipid pattern: protective effects of LXR activation. J Lipid Res. 2012;53:300–310. doi: 10.1194/jlr.M021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, et al. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TK. Activation of pregnane × receptor (PXR) and constitutive androstane receptor (CAR) by herbal medicines. AAPS J. 2009;11:590–601. doi: 10.1208/s12248-009-9135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang J, Baker SM, Chen G. Human constitutive androstane receptor mediated methotrexate induction of human dehydroepiandrosterone sulfotransferase (hSULT2A1). Toxicology. 2007;231:224–233. doi: 10.1016/j.tox.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Ma X, Krausz KW, Idle JR, Gonzalez FJ. Rifampicin-activated human pregnane × receptor and CYP3A4 induction enhance acetaminophen-induced toxicity. Drug Metab Dispos. 2009;37:1611–1621. doi: 10.1124/dmd.109.027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Redinbo MR. Activation of the human nuclear xenobiotic receptor PXR by the reverse transcriptase-targeted anti-HIV drug PNU-142721. Protein Sci. 2011;20:1713–1719. doi: 10.1002/pro.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JY. Hepatocyte nuclear factor 4alpha regulation of bile acid and drug metabolism. Expert Opin Drug Metab Toxicol. 2009;5:137–147. doi: 10.1517/17425250802707342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Chung M, Tzameli I, Simha D, Lee YK, Seol W, et al. Differential transactivation by two isoforms of the orphan nuclear hormone receptor. CAR J Biol Chem. 1997;272:23565–23571. doi: 10.1074/jbc.272.38.23565. [DOI] [PubMed] [Google Scholar]
- Chrencik JE, Orans J, Moore LB, Xue Y, Peng L, Collins JL, et al. Structural disorder in the complex of human pregnane × receptor and the macrolide antibiotic rifampicin. Mol Endocrinol. 2005;19:1125–1134. doi: 10.1210/me.2004-0346. [DOI] [PubMed] [Google Scholar]
- Cieśla W. Can melatonin regulate the expression of prohormone convertase 1 and 2 genes via monomeric and dimeric forms of RZR/ROR nuclear receptor, and can melatonin influence the processes of embryogenesis or carcinogenesis by disturbing the proportion of cAMP and cGMP concentrations? Theoretic model of controlled apoptosis. Med Hypotheses. 2001;56:181–193. doi: 10.1054/mehy.2000.1137. [DOI] [PubMed] [Google Scholar]
- Conde I, Lobo MV, Zamora J, Pérez J, González FJ, Alba E, et al. Human pregnane × receptor is expressed in breast carcinomas, potential heterodimers formation between hPXR and RXR-alpha. BMC Cancer. 2008;8:174. doi: 10.1186/1471-2407-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delerive P, Monté D, Dubois G, Trottein F, Fruchart-Najib J, Mariani J, et al. The orphan nuclear receptor ROR alpha is a negative regulator of the inflammatory response. EMBO Rep. 2001;2:42–48. doi: 10.1093/embo-reports/kve007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvecchio CJ, Bilan P, Nair P, Capone JP. LXR-induced reverse cholesterol transport in human airway smooth muscle is mediated exclusively by ABCA1. Am J Physiol Lung Cell Mol Physiol. 2008;295:L949–L957. doi: 10.1152/ajplung.90394.2008. [DOI] [PubMed] [Google Scholar]
- Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- Denison MS, Pandini A, Nagy SR, Baldwin EP, Bonati L. Ligand binding and activation of the Ah receptor. Chem Biol Interact. 2002;141:3–24. doi: 10.1016/s0009-2797(02)00063-7. [DOI] [PubMed] [Google Scholar]
- di Masi A, De Marinis E, Ascenzi P, Marino M. Nuclear receptors CAR and PXR: Molecular, functional, and biomedical aspects. Mol Aspects Med. 2009;30:297–343. doi: 10.1016/j.mam.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Ding X, Lichti K, Kim I, Gonzalez FJ, Staudinger JL. Regulation of constitutive androstane receptor and its target genes by fasting, cAMP, hepatocyte nuclear factor alpha, and the coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha. J Biol Chem. 2006;281:26540–26551. doi: 10.1074/jbc.M600931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Qatanani M, Moore DD. Constitutive androstane receptor mediates the induction of drug metabolism in mouse models of type 1 diabetes. Hepatology. 2009;50:622–629. doi: 10.1002/hep.23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drocourt L, Ourlin JC, Pascussi JM, Maurel P, Vilarem MJ. Expression of CYP3A4, CYP2B6, and CYP2C9 is regulated by the vitamin D receptor pathway in primary human hepatocytes. J Biol Chem. 2002;277:25125–25132. doi: 10.1074/jbc.M201323200. [DOI] [PubMed] [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dushkin MI, Khoshchenko OM, Chasovsky MA, Pivovarova EN. The content of PPAR, LXR, and RXR and the PPAR DNA-binding activity in macrophages over the course of inflammation in mice. Bull Exp Biol Med. 2009;147:345–348. doi: 10.1007/s10517-009-0505-3. [DOI] [PubMed] [Google Scholar]
- Dussault I, Lin M, Hollister K, Fan M, Termini J, Sherman MA, et al. A structural model of the constitutive androstane receptor defines novel interactions that mediate ligand-independent activity. Mol Cell Biol. 2002;22:5270–5280. doi: 10.1128/MCB.22.15.5270-5280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisawa T, Tojo K, Tajima N, Kamio M, Oki Y, Ono K, et al. Immunohistochemical analysis of 11-beta-hydroxysteroid dehydrogenase type 2 and glucocorticoid receptor in subclinical Cushing's disease due to pituitary macroadenoma. Endocr Pathol. 2008;19:252–260. doi: 10.1007/s12022-008-9052-0. [DOI] [PubMed] [Google Scholar]
- Echchgadda I, Song CS, Oh T, Ahmed M, De La Cruz IJ, Chatterjee B. The xenobiotic-sensing nuclear receptors pregnane × receptor, constitutive androstane receptor, and orphan nuclear receptor hepatocyte nuclear factor 4alpha in the regulation of human steroid-/bile acid-sulfotransferase. Mol Endocrinol. 2007;21:2099–2111. doi: 10.1210/me.2007-0002. [DOI] [PubMed] [Google Scholar]
- Echchgadda I, Song CS, Roy AK, Chatterjee B. Dehydroepiandrosterone sulfotransferase is a target for transcriptional induction by the vitamin D receptor. Mol Pharmacol. 2004;65:720–729. doi: 10.1124/mol.65.3.720. [DOI] [PubMed] [Google Scholar]
- Einstein M, Greenlee M, Rouen G, Sitlani A, Santoro J, Wang C, et al. Selective glucocorticoid receptor nonsteroidal ligands completely antagonize the dexamethasone mediated induction of enzymes involved in gluconeogenesis and glutamine metabolism. J Steroid Biochem Mol Biol. 2004;92:345–356. doi: 10.1016/j.jsbmb.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Ekins S, Mirny L, Schuetz EG. A ligand-based approach to understanding selectivity of nuclear hormone receptors PXR, CAR, FXR, LXRalpha, and LXRbeta. Pharm Res. 2002;19:1788–1800. doi: 10.1023/a:1021429105173. [DOI] [PubMed] [Google Scholar]
- Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- Fang HL, Strom SC, Cai H, Falany CN, Kocarek TA, Runge-Morris M. Regulation of human hepatic hydroxysteroid sulfotransferase gene expression by the peroxisome proliferator-activated receptor alpha transcription factor. Mol Pharmacol. 2005;67:1257–1267. doi: 10.1124/mol.104.005389. [DOI] [PubMed] [Google Scholar]
- Fiorucci S, Zampella A, Distrutti E. Development of FXR, PXR and CAR agonists and antagonists for treatment of liver disorders. Curr Top Med Chem. 2012;12:605–624. doi: 10.2174/156802612799436678. [DOI] [PubMed] [Google Scholar]
- Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, et al. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- Fujii-Kuriyama Y, Mimura J. Molecular mechanisms of AhR functions in the regulation of cytochrome P450 genes. Biochem Biophys Res Commun. 2005;338:311–317. doi: 10.1016/j.bbrc.2005.08.162. [DOI] [PubMed] [Google Scholar]
- Fukunaga BN, Probst MR, Reisz-Porszasz S, Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor. J Biol Chem. 1995;270:29270–29278. doi: 10.1074/jbc.270.49.29270. [DOI] [PubMed] [Google Scholar]
- Gascon-Barré M, Demers C, Mirshahi A, Néron S, Zalzal S, Nanci A. The normal liver harbors the vitamin D nuclear receptor in nonparenchymal and biliary epithelial cells. Hepatology. 2003;37:1034–1042. doi: 10.1053/jhep.2003.50176. [DOI] [PubMed] [Google Scholar]
- Girroir EE, Hollingshead HE, He P, Zhu B, Perdew GH, Peters JM. Quantitative expression patterns of peroxisome proliferator-activated receptor-beta/delta (PPARbeta/delta) protein in mice. Biochem Biophys Res Commun. 2008;371:456–461. doi: 10.1016/j.bbrc.2008.04.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt H. Sulfotransferases in the bioactivation of xenobiotics. Chem Biol Interact. 2000;129:141–170. doi: 10.1016/s0009-2797(00)00202-7. [DOI] [PubMed] [Google Scholar]
- Gnerre C, Blattler S, Kaufmann MR, Looser R, Meyer UA. Regulation of CYP3A4 by the bile acid receptor FXR: evidence for functional binding sites in the CYP3A4 gene. Pharmacogenetics. 2004;14:635–645. doi: 10.1097/00008571-200410000-00001. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, Liddle C. The orphan human pregnane × receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol. 1999;56:1329–1339. doi: 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Redinbo MR, Kliewer SA. Regulation of cyp3a gene transcription by the pregnane × receptor. Annu Rev Pharmacol Toxicol. 2002;42:1–23. doi: 10.1146/annurev.pharmtox.42.111901.111051. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ. The molecular biology of cytochrome P450s. Pharmacol Rev. 1988;40:243–288. [PubMed] [Google Scholar]
- Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- Haimeur A, Conseil G, Deeley RG, Cole SP. The MRP-related and BCRP/ABCG2 multidrug resistance proteins: biology, substrate specificity and regulation. Curr Drug Metab. 2004;5:21–53. doi: 10.2174/1389200043489199. [DOI] [PubMed] [Google Scholar]
- Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, et al. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- Handschin C, Meyer UA. Induction of drug metabolism: the role of nuclear receptors. Pharmacol Rev. 2003;55:649–673. doi: 10.1124/pr.55.4.2. [DOI] [PubMed] [Google Scholar]
- He YW, Deftos ML, Ojala EW, Bevan MJ. RORgamma t, a novel isoform of an orphan receptor, negatively regulates Fas ligand expression and IL-2 production in T cells. Immunity. 1998;9:797–806. doi: 10.1016/s1074-7613(00)80645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Kääriäinen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C, Tontonoz P. Coordination of inflammation and metabolism by PPAR and LXR nuclear receptors. Curr Opin Genet Dev. 2008;18:461–467. doi: 10.1016/j.gde.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo Y, Sasaki S, Kobayashi Y, Misawa H, Nakamura H. 1,25-dihydroxyvitamin D3 and its receptor inhibit the chenodeoxycholic acid-dependent transactivation by farnesoid × receptor. J Endocrinol. 2006;188:635–643. doi: 10.1677/joe.1.06105. [DOI] [PubMed] [Google Scholar]
- Huber RM, Murphy K, Miao B, Link JR, Cunningham MR, Rupar MJ, et al. Generation of multiple farnesoid-X-receptor isoforms through the use of alternative promoters. Gene. 2002;290:35–43. doi: 10.1016/s0378-1119(02)00557-7. [DOI] [PubMed] [Google Scholar]
- Huss JM, Kasper CB. Two-stage glucocorticoid induction of CYP3A23 through both the glucocorticoid and pregnane × receptors. Mol Pharmacol. 2000;58:48–57. doi: 10.1124/mol.58.1.48. [DOI] [PubMed] [Google Scholar]
- Ingraham HA, Redinbo MR. Orphan nuclear receptors adopted by crystallography. Curr Opin Struct Biol. 2005;15:708–715. doi: 10.1016/j.sbi.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- Jetten AM, Kurebayashi S, Ueda E. The ROR nuclear orphan receptor subfamily: critical regulators of multiple biological processes. Prog Nucleic Acid Res Mol Biol. 2001;69:205–247. doi: 10.1016/s0079-6603(01)69048-2. [DOI] [PubMed] [Google Scholar]
- Jin L, Martynowski D, Zheng S, Wada T, Xie W, Li Y. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol Endocrinol. 2010;24:923–929. doi: 10.1210/me.2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson L, Båvner A, Thomsen JS, Färnegårdh M, Gustafsson A, Treuter E. The orphan nuclear receptor SHP utilizes conserved LXXLL-related motifs for interactions with ligand-activated estrogen receptors. Mol Cell Biol. 2000;20:1124–1133. doi: 10.1128/mcb.20.4.1124-1133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jover R, Bort R, Gómez-Lechón MJ, Castell JV. Cytochrome P450 regulation by hepatocyte nuclear factor 4 in human hepatocytes: a study using adenovirus-mediated antisense targeting. Hepatology. 2001;33:668–675. doi: 10.1053/jhep.2001.22176. [DOI] [PubMed] [Google Scholar]
- Jyrkkärinne J, Windshügel B, Rönkkö T, Tervo AJ, Küblbeck J, Lahtela-Kakkonen M, et al. Insights into ligand-elicited activation of human constitutive androstane receptor based on novel agonists and three-dimensional quantitative structure-activity relationship. J Med Chem. 2008;51:7181–7192. doi: 10.1021/jm800731b. [DOI] [PubMed] [Google Scholar]
- Kachaylo EM, Pustylnyak VO, Lyakhovich VV, Gulyaeva LF. Constitutive androstane receptor (CAR) is a xenosensor and target for therapy. Biochemistry (Mosc) 2011;76:1087–1097. doi: 10.1134/S0006297911100026. [DOI] [PubMed] [Google Scholar]
- Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, et al. X-ray structure of the hRORalpha LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha. Structure. 2002;10:1697–1707. doi: 10.1016/s0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Inoue Y, Gonzalez FJ. Role of the hepatocyte nuclear factor 4alpha in control of the pregnane × receptor during fetal liver development. Hepatology. 2003;37:1375–1384. doi: 10.1053/jhep.2003.50212. [DOI] [PubMed] [Google Scholar]
- Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, et al. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol Genomics. 2007;31:281–294. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- Kang MI, Kobayashi A, Wakabayashi N, Kim SG, Yamamoto M. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc Natl Acad Sci U S A. 2004;101:2046–2051. doi: 10.1073/pnas.0308347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast HR, Goodwin B, Tarr PT, Jones SA, Anisfield AM, Stoltz CM, et al. Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane × receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J Biol Chem. 2002;277:2908–2915. doi: 10.1074/jbc.M109326200. [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol Cell Biol. 1999;19:6318–6322. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano S, Kumagai S. Diagnosis and treatment of glucocorticoid-induced osteoporosis in collagen vascular disease. Nihon Rinsho. 2007;65(Suppl 9):504–507. [Article in Japanese] [PubMed] [Google Scholar]
- Khan AA, Chow EC, van Loenen-Weemaes AM, Porte RJ, Pang KS, Groothuis GM. Comparison of effects of VDR versus PXR, FXR and GR ligands on the regulation of CYP3A isozymes in rat and human intestine and liver. Eur J Pharm Sci. 2009;37:115–125. doi: 10.1016/j.ejps.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Goodwin B, Willson TM. The nuclear pregnane × receptor: a key regulator of xenobiotic metabolism. Endocr Rev. 2002;23:687–702. doi: 10.1210/er.2001-0038. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Willson TM. Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane × receptor. J Lipid Res. 2002;43:359–364. [PubMed] [Google Scholar]
- Kobayashi K, Sueyoshi T, Inoue K, Moore R, Negishi M. Cytoplasmic accumulation of the nuclear receptor CAR by a tetratricopeptide repeat protein in HepG2 cells. Mol Pharmacol. 2003;64:1069–1075. doi: 10.1124/mol.64.5.1069. [DOI] [PubMed] [Google Scholar]
- Köhle C, Bock KW. Coordinate regulation of phase I and II xenobiotic metabolisms by the Ah receptor and Nrf2. Biochem Pharmacol. 2007;73:1853–1862. doi: 10.1016/j.bcp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Köhle C, Bock KW. Coordinate regulation of human drug-metabolizing enzymes, and conjugate transporters by the Ah receptor, pregnane × receptor and constitutive androstane receptor. Biochem Pharmacol. 2009;77:689–699. doi: 10.1016/j.bcp.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Korf H, Vander Beken S, Romano M, Steffensen KR, Stijlemans B, Gustafsson JA, et al. Liver × receptors contribute to the protective immune response against Mycobacterium tuberculosis in mice. J Clin Invest. 2009;119:1626–1637. doi: 10.1172/JCI35288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano S, Kurose K, Saito Y, Ozawa S, Hasegawa R, Komamura K, et al. Functional characterization of four naturally occurring variants of human pregnane × receptor (PXR): one variant causes dramatic loss of both DNA binding activity and the transactivation of the CYP3A4 promoter/enhancer region. Drug Metab Dispos. 2004;32:149–154. doi: 10.1124/dmd.32.1.149. [DOI] [PubMed] [Google Scholar]
- Kurebayashi S, Ueda E, Sakaue M, Patel DD, Medvedev A, Zhang F, et al. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc Natl Acad Sci U S A. 2000;97:10132–10137. doi: 10.1073/pnas.97.18.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladias JA, Hadzopoulou-Cladaras M, Kardassis D, Cardot P, Cheng J, Zannis V, et al. Transcriptional regulation of human apolipoprotein genes ApoB, ApoCIII, and ApoAII by members of the steroid hormone receptor superfamily HNF-4, ARP-1, EAR-2, and EAR-3. J Biol Chem. 1992;267:15849–15860. [PubMed] [Google Scholar]
- Laffitte BA, Kast HR, Nguyen CM, Zavacki AM, Moore DD, Edwards PA. Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J Biol Chem. 2000;275:10638–10647. doi: 10.1074/jbc.275.14.10638. [DOI] [PubMed] [Google Scholar]
- Lamba J, Lamba V, Schuetz E. Genetic variants of PXR (NR1I2) and CAR (NR1I3) and their implications in drug metabolism and pharmacogenetics. Curr Drug Metab. 2005;6:369–383. doi: 10.2174/1389200054633880. [DOI] [PubMed] [Google Scholar]
- Lamba JK, Lamba V, Yasuda K, Lin YS, Assem M, Thompson E, et al. Expression of constitutive androstane receptor splice variants in human tissues and their functional consequences. J Pharmacol Exp Ther. 2004;311:811–821. doi: 10.1124/jpet.104.069310. [DOI] [PubMed] [Google Scholar]
- Lamba V, Yasuda K, Lamba JK, Assem M, Davila J, Strom S, et al. PXR (NR1I2): splice variants in human tissues, including brain, and identification of neurosteroids and nicotine as PXR activators. Toxicol Appl Pharmacol. 2004;199:251–265. doi: 10.1016/j.taap.2003.12.027. [DOI] [PubMed] [Google Scholar]
- Larrede S, Quinn CM, Jessup W, Frisdal E, Olivier M, Hsieh V, et al. Stimulation of cholesterol efflux by LXR agonists in cholesterol-loaded human macrophages is ABCA1-dependent but ABCG1-independent. Arterioscler Thromb Vasc Biol. 2009;29:1930–1936. doi: 10.1161/ATVBAHA.109.194548. [DOI] [PubMed] [Google Scholar]
- Larrieu G, Galtier P. A comparative study of some oxidative and conjugative drug metabolizing enzymes in liver, lung and kidney of sheep. Comp Biochem Physiol C. 1988;89:225–228. doi: 10.1016/0742-8413(88)90215-0. [DOI] [PubMed] [Google Scholar]
- Laue L, Kawai S, Brandon DD, Brightwell D, Barnes K, Knazek RA, et al. Receptor-mediated effects of glucocorticoids on inflammation: enhancement of the inflammatory response with a glucocorticoid antagonist. J Steroid Biochem. 1988;29:591–598. doi: 10.1016/0022-4731(88)90156-2. [DOI] [PubMed] [Google Scholar]
- Lee HK, Lee YK, Park SH, Kim YS, Park SH, Lee JW, et al. Structure and expression of the orphan nuclear receptor SHP gene. J Biol Chem. 1998;273:14398–14402. doi: 10.1074/jbc.273.23.14398. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Kliewer SA, Moore LB, Smith-Oliver TA, Oliver BB, Su JL, et al. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YS, Yasuda K, Assem M, Cline C, Barber J, Li CW, et al. The major human pregnane × receptor (PXR) splice variant, PXR.2, exhibits significantly diminished ligand-activated transcriptional regulation. Drug Metab Dispos. 2009;37:1295–1304. doi: 10.1124/dmd.108.025213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TT, Repa JJ, Mangelsdorf DJ. Orphan nuclear receptors as eLiXiRs and FiXeRs of sterol metabolism. J Biol Chem. 2001;276:37735–37738. doi: 10.1074/jbc.R100035200. [DOI] [PubMed] [Google Scholar]
- Lu Y, Heydel JM, Li X, Bratton S, Lindblom T, Radominska-Pandya A. Lithocholic acid decreases expression of UGT2B7 in Caco-2 cells: a potential role for a negative farnesoid × receptor response element. Drug Metab Dispos. 2005;33:937–946. doi: 10.1124/dmd.104.003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luci S, Geissler S, König B, Koch A, Stangl GI, Hirche F, et al. PPARalpha agonists up-regulate organic cation transporters in rat liver cells. Biochem Biophys Res Commun. 2006;350:704–708. doi: 10.1016/j.bbrc.2006.09.099. [DOI] [PubMed] [Google Scholar]
- Mader S, Chen JY, Chen Z, White J, Chambon B, Gronemeyer H. The patterns of binding of RAR, RXR and TR homo- and heterodimers to direct repeats are dictated by the binding specificites of the DNA binding domains. EMBO J. 1993;12:5029–5041. doi: 10.1002/j.1460-2075.1993.tb06196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane × receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, et al. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, et al. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- Malloy PJ, Wang J, Peng L, Nayak S, Sisk JM, Thompson CC, et al. A unique insertion/duplication in the VDR gene that truncates the VDR causing hereditary 1,25-dihydroxyvitamin D-resistant rickets without alopecia. Arch Biochem Biophys. 2007;460:285–292. doi: 10.1016/j.abb.2006.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Ong ES, Dyck JA, Evans RM. Nuclear receptor that identifies a novel retinoic acid response pathway. Nature. 1990;345:224–229. doi: 10.1038/345224a0. [DOI] [PubMed] [Google Scholar]
- Martin H, Sarsat JP, de Waziers I, Housset C, Balladur P, Beaune P, et al. Induction of cytochrome P450 2B6 and 3A4 expression by phenobarbital and cyclophosphamide in cultured human liver slices. Pharm Res. 2003;20:557–568. doi: 10.1023/a:1023234429596. [DOI] [PubMed] [Google Scholar]
- Matic M, Mahns A, Tsoli M, Corradin A, Polly P, Robertson GR. Pregnane × receptor: promiscuous regulator of detoxification pathways. Int J Biochem Cell Biol. 2007;39:478–483. doi: 10.1016/j.biocel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Matsui T. Orphan nuclear receptor ROR alpha in cerebellum. Tanpakushitsu Kakusan Koso. 1997;42:2039–2048. [Article in Japanese] [PubMed] [Google Scholar]
- McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J Biol Chem. 2004;279:31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- Medvedev A, Yan ZH, Hirose T, Giguère V, Jetten AM. Cloning of a cDNA encoding the murine orphan receptor RZR/ROR gamma and characterization of its response element. Gene. 1996;181:199–206. doi: 10.1016/s0378-1119(96)00504-5. [DOI] [PubMed] [Google Scholar]
- Meyer MB, Goetsch PD, Pike JW. VDR/RXR and TCF4/beta-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression. Mol Endocrinol. 2012;26:37–51. doi: 10.1210/me.2011-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Choi SE, Seok SM, Yang L, Zuercher WJ, Xu Y, et al. Ligand-dependent regulation of the activity of the orphan nuclear receptor, small heterodimer partner (SHP), in the repression of bile acid biosynthetic CYP7A1 and CYP8B1 genes. Mol Endocrinol. 2011;25:1159–1169. doi: 10.1210/me.2011-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RT, Willson TM. Regulation of xenobiotic metabolism by orphan nuclear receptors. Toxicol Pathol. 2001;29:3–5. doi: 10.1080/019262301301418801. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Lopez S, Cartier N, Tulliez M, Raymondjean M, Kahn A. Expression of the L-type pyruvate kinase gene and the hepatocyte nuclear factor 4 transcription factor in exocrine and endocrine pancreas. J Biol Chem. 1994;269:8944–8951. [PubMed] [Google Scholar]
- Missbach M, Jagher B, Sigg I, Nayeri S, Carlberg C, Weisenberg I. Thiazolidine diones, specific ligands of the nuclear receptor retinoid Z receptor/retinoid acid receptor-related orphan receptor alpha with potent antiarthritic activity. J Biol Chem. 1996;271:13515–13522. doi: 10.1074/jbc.271.23.13515. [DOI] [PubMed] [Google Scholar]
- Miyata M, Matsuda Y, Tsuchiya H, Kitada H, Akase T, Shimada M, et al. Chenodeoxycholic acid-mediated activation of the farnesoid × receptor negatively regulates hydroxysteroid sulfotransferase. Drug Metab Pharmacokinet. 2006;21:315–323. doi: 10.2133/dmpk.21.315. [DOI] [PubMed] [Google Scholar]
- Modica S, Bellafante E, Moschetta A. Master regulation of bile acid and xenobiotic metabolism via the FXR, PXR and CAR trio. Front Biosci. 2009;14:4719–4745. doi: 10.2741/3563. [DOI] [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DD. Regulation of drug transport by new xenobiotic receptors. Pharmacogenomics J. 2001;1:224–225. doi: 10.1038/sj.tpj.6500058. [DOI] [PubMed] [Google Scholar]
- Moore LB, Maglich JM, McKee DD, Wisely B, Willson TM, Kliewer SA, et al. Pregnane × receptor (PXR), constitutive androstane receptor (CAR), and benzoate × receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol Endocrinol. 2002;16:977–986. doi: 10.1210/mend.16.5.0828. [DOI] [PubMed] [Google Scholar]
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, et al. Orphan nuclear receptors constitutive androstane receptor and pregnane × receptor share xenobiotic and steroid ligands. J Biol Chem. 2000;275:15122–15127. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Katsuoka F, Engel JD, Yamamoto M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc Natl Acad Sci U S A. 2004;101:6379–6384. doi: 10.1073/pnas.0305902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Munck A, Wira C, Young DA, Mosher KM, Hallahan C, Bell PA. Glucocorticoid-receptor complexes and the earliest steps in the action of glucocorticoids on thymus cells. J Steroid Biochem. 1972;3:567–578. doi: 10.1016/0022-4731(72)90103-3. [DOI] [PubMed] [Google Scholar]
- Nannelli A, Chirulli V, Longo V, Gervasi PG. Expression and induction by rifampicin of CAR- and PXR-regulated CYP2B and CYP3A in liver, kidney and airways of pig. Toxicology. 2008;252:105–112. doi: 10.1016/j.tox.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Ng MK, Celermajer DS. Glucocorticoid treatment and cardiovascular disease. Heart. 2004;90:829–830. doi: 10.1136/hrt.2003.031492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nioi P, Nguyen T, Sherratt PJ, Pickett CB. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol Cell Biol. 2005;25:10895–10906. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Carnahan VE, Moore LB, Luntz T, Wang H, Ittoop OR, et al. Human PXR forms a tryptophan zipper-mediated homodimer. Biochemistry. 2006;45:8579–8589. doi: 10.1021/bi0602821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell S, Falany CN. Pharmacogenetics of human cytosolic sulfotransferases. Oncogene. 2006;25:1673–1678. doi: 10.1038/sj.onc.1209376. [DOI] [PubMed] [Google Scholar]
- Omiecinski CJ, Vanden Heuvel JP, Perdew GH, Peters JM. Xenobiotic metabolism, disposition, and regulation by receptors: from biochemical phenomenon to predictors of major toxicities. Toxicol Sci. 2011;120(Suppl 1):S49–S75. doi: 10.1093/toxsci/kfq338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orans J, Teotico DG, Redinbo MR. The nuclear xenobiotic receptor pregnane × receptor: recent insights and new challenges. Mol Endocrinol. 2005;19:2891–2900. doi: 10.1210/me.2005-0156. [DOI] [PubMed] [Google Scholar]
- Orlov I, Rochel N, Moras D, Klaholz BP. Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA. EMBO J. 2011;31:291–300. doi: 10.1038/emboj.2011.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortlund EA, Lee Y, Solomon IH, Hager JM, Safi R, Choi Y, et al. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat Struct Mol Biol. 2005;12:357–363. doi: 10.1038/nsmb910. [DOI] [PubMed] [Google Scholar]
- Ouvrier A, Cadet R, Lobaccaro JM, Drevet JR, Saez F. LXR regulate cholesterol homeostasis in the proximal mouse epididymis. Folia Histochem Cytobiol. 2009a;47:S75–S79. doi: 10.2478/v10042-009-0057-4. [DOI] [PubMed] [Google Scholar]
- Ouvrier A, Cadet R, Vernet P, Laillet B, Chardigny JM, Lobaccaro JM, et al. LXR and ABCA1 control cholesterol homeostasis in the proximal mouse epididymis in a cell-specific manner. J Lipid Res. 2009b;50:1766–1775. doi: 10.1194/jlr.M800657-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Pascussi JM, Drocourt L, Fabre JM, Maurel P, Vilarem MJ. Dexamethasone induces pregnane × receptor and retinoid × receptor-alpha expression in human hepatocytes: synergistic increase of CYP3A4 induction by pregnane × receptor activators. Mol Pharmacol. 2000;58:361–372. doi: 10.1124/mol.58.2.361. [DOI] [PubMed] [Google Scholar]
- Pascussi JM, Gerbal-Chaloin S, Drocourt L, Maurel P, Vilarem MJ. The expression of CYP2B6, CYP2C9 and CYP3A4 genes: a tangle of networks of nuclear and steroid receptors. Biochim Biophys Acta. 2003;1619:243–253. doi: 10.1016/s0304-4165(02)00483-x. [DOI] [PubMed] [Google Scholar]
- Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro JM, Hammer RE, et al. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- Pergola PE, Raskin P, Toto RD, Meyer CJ, Huff JW, Grossman EB, et al. Bardoxolone methyl and kidney function in CKD with type 2 diabetes. N Engl J Med. 2011;365:327–336. doi: 10.1056/NEJMoa1105351. [DOI] [PubMed] [Google Scholar]
- Pool-Zobel B, Veeriah S, Böhmer FD. Modulation of xenobiotic metabolising enzymes by anticarcinogens—focus on glutathione S-transferases and their role as targets of dietary chemoprevention in colorectal carcinogenesis. Mutat Res. 2005;591:74–92. doi: 10.1016/j.mrfmmm.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Popowski K, Eloranta JJ, Saborowski M, Fried M, Meier PJ, Kullak-Ublick GA. The human organic anion transporter 2 gene is transactivated by hepatocyte nuclear factor-4 alpha and suppressed by bile acids. Mol Pharmacol. 2005;67:1629–1638. doi: 10.1124/mol.104.010223. [DOI] [PubMed] [Google Scholar]
- Raddatz D, Middel P, Bockemühl M, Benöhr P, Wissmann C, Schwörer H, et al. Glucocorticoid receptor expression in inflammatory bowel disease: evidence for a mucosal down-regulation in steroid-unresponsive ulcerative colitis. Aliment Pharmacol Ther. 2004;19:47–61. doi: 10.1046/j.1365-2036.2003.01802.x. [DOI] [PubMed] [Google Scholar]
- Rauch A, Seitz S, Baschant U, Schilling AF, Illing A, Stride B, et al. Glucocorticoids suppress bone formation by attenuating osteoblast differentiation via the monomeric glucocorticoid receptor. Cell Metab. 2010;11:517–531. doi: 10.1016/j.cmet.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Ray A, Siegel MD, Prefontaine KE, Ray P. Anti-inflammation: direct physical association and functional antagonism between transcription factor NF-KB and the glucocorticoid receptor. Chest. 1995;107:139S. doi: 10.1378/chest.107.3_supplement.139s. [DOI] [PubMed] [Google Scholar]
- Reisman SA, Chertow GM, Hebbar S, Vaziri ND, Ward KW, Meyer CJ. Bardoxolone methyl decreases megalin and activates Nrf2 in the kidney. J Am Soc Nephrol. 2012;23:1663–1673. doi: 10.1681/ASN.2012050457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmer H. Induction of drug metabolizing enzyme system in the liver. Eur J Clin Pharmacol. 1972;5:116–136. [Google Scholar]
- Reschly EJ, Krasowski MD. Evolution and function of the NR1I nuclear hormone receptor subfamily (VDR, PXR, and CAR) with respect to metabolism of xenobiotics and endogenous compounds. Curr Drug Metab. 2006;7:349–365. doi: 10.2174/138920006776873526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschly EJ, Ai N, Ekins S, Welsh WJ, Hagey LR, Hofmann AF, et al. Evolution of the bile salt nuclear receptor FXR in vertebrates. J Lipid Res. 2008;49:1577–1587. doi: 10.1194/jlr.M800138-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketts ML, Boekschoten MV, Kreeft AJ, Hooiveld GJ, Moen CJ, Müller M, et al. The cholesterol-raising factor from coffee beans, cafestol, as an agonist ligand for the farnesoid and pregnane × receptors. Mol Endocrinol. 2007;21:1603–1616. doi: 10.1210/me.2007-0133. [DOI] [PubMed] [Google Scholar]
- Rizzo G, Disante M, Mencarelli A, Renga B, Gioiello A, Pellicciari R, et al. The farnesoid × receptor promotes adipocyte differentiation and regulates adipose cell function in vivo. Mol Pharmacol. 2006;70:1164–1173. doi: 10.1124/mol.106.023820. [DOI] [PubMed] [Google Scholar]
- Saborowski M, Kullak-Ublick GA, Eloranta JJ. The human organic cation transporter-1 gene is transactivated by hepatocyte nuclear factor-4alpha. J Pharmacol Exp Ther. 2006;317:778–785. doi: 10.1124/jpet.105.099929. [DOI] [PubMed] [Google Scholar]
- Sakai H, Iwata H, Kim EY, Tsydenova O, Miyazaki N, Petrov EA, et al. Constitutive androstane receptor (CAR) as a potential sensing biomarker of persistent organic pollutants (POPs) in aquatic mammal: molecular characterization, expression level, and ligand profiling in Baikal seal (Pusa sibirica). Toxicol Sci. 2006;94:57–70. doi: 10.1093/toxsci/kfl088. [DOI] [PubMed] [Google Scholar]
- Sashihara S, Felts PA, Waxman SG, Matsui T. Orphan nuclear receptor ROR alpha gene: isoform-specific spatiotemporal expression during postnatal development of brain. Brain Res Mol Brain Res. 1996;42:109–117. doi: 10.1016/s0169-328x(96)00118-0. [DOI] [PubMed] [Google Scholar]
- Scheper RJ, Broxterman HJ, Scheffer GL, Meijer CJ, Pinedo HM. Drug-transporter proteins in clinical multidrug resistance. Clin Chim Acta. 1992;206:25–32. doi: 10.1016/0009-8981(92)90005-b. [DOI] [PubMed] [Google Scholar]
- Schwabe K, Vacca G, Duck R, Gillissen A. Glucocorticoid receptor gene polymorphisms and potential association to chronic obstructive pulmonary disease susceptibility and severity. Eur J Med Res. 2009;14(Suppl 4):210–215. doi: 10.1186/2047-783X-14-S4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selye H. Hormones and resistance. J Pharm Sci. 1971;60:1. doi: 10.1002/jps.2600600102. [DOI] [PubMed] [Google Scholar]
- Seol W, Choi HS, Moore DD. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996;272:1336–1339. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- Shan L, Vincent J, Brunzelle JS, Dussault I, Lin M, Ianculescu I, et al. Structure of the murine constitutive androstane receptor complexed to androstenol: a molecular basis for inverse agonism. Mol Cell. 2004;16:907–917. doi: 10.1016/j.molcel.2004.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Yang D, Yan B. Dexamethasone transcriptionally increases the expression of the pregnane × receptor and synergistically enhances pyrethroid esfenvalerate in the induction of cytochrome P450 3A23. Biochem Pharmacol. 2010;80:1274–1283. doi: 10.1016/j.bcp.2010.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo S, Numazawa S, Yoshida T. A physiological role of AMP-activated protein kinase in phenobarbital-mediated constitutive androstane receptor activation and CYP2B induction. Biochem J. 2007;401:735–741. doi: 10.1042/BJ20061238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman AI, Larson C, Mangelsdorf DJ, Ranganathan R. Structural determinants of allosteric ligand activation in RXR heterodimers. Cell. 2004;116:417–429. doi: 10.1016/s0092-8674(04)00119-9. [DOI] [PubMed] [Google Scholar]
- Sladek FM, Zhong WM, Lai E, Darnell JE., Jr. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- Son YL, Lee YC. Molecular determinants of the interactions between LXR/RXR heterodimers and TRAP220. Biochem Biophys Res Commun. 2009;384:389–393. doi: 10.1016/j.bbrc.2009.04.131. [DOI] [PubMed] [Google Scholar]
- Song CS, Echchgadda I, Baek BS, Ahn SC, Oh T, Roy AK, et al. Dehydroepiandrosterone sulfotransferase gene induction by bile acid activated farnesoid × receptor. J Biol Chem. 2001;276:42549–42556. doi: 10.1074/jbc.M107557200. [DOI] [PubMed] [Google Scholar]
- Song C, Hiipakka RA, Kokontis JM, Liao S. Ubiquitous receptor: structures, immunocytochemical localization, and modulation of gene activation by receptors for retinoic acids and thyroid hormones. Ann N Y Acad Sci. 1995;761:38–49. doi: 10.1111/j.1749-6632.1995.tb31367.x. [DOI] [PubMed] [Google Scholar]
- Song C, Kokontis JM, Hiipakka RA, Liao S. Ubiquitous receptor: a receptor that modulates gene activation by retinoic acid and thyroid hormone receptors. Proc Natl Acad Sci U S A. 1994;91:10809–10813. doi: 10.1073/pnas.91.23.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Chong LW, Downes M, Barish GD, Coulter S, Liddle C, et al. Pregnane × receptor prevents hepatorenal toxicity from cholesterol metabolites. Proc Natl Acad Sci U S A. 2005;102:2198–2203. doi: 10.1073/pnas.0409481102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Evans R. Biological function and mode of action of nuclear xenobiotic receptors. Pure Appl Chem. 2003;75:1733–1742. [Google Scholar]
- Stanley LA, Horsburgh BC, Ross J, Scheer N, Wolf CR. PXR and CAR: nuclear receptors which play a pivotal role in drug disposition and chemical toxicity. Drug Metab Rev. 2006;38:515–597. doi: 10.1080/03602530600786232. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human. CYP2B6 gene J Biol Chem. 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- Suino K, Peng L, Reynolds R, Li Y, Cha JY, Repa JJ, et al. The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol Cell. 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Svec F, Rudis M. Glucocorticoids regulate the glucocorticoid receptor in the AtT-20 cell. J Biol Chem. 1981;256:5984–5987. [PubMed] [Google Scholar]
- Swales K, Negishi M. CAR, driving into the future. Mol Endocrinol. 2004;18:1589–1598. doi: 10.1210/me.2003-0397. [DOI] [PubMed] [Google Scholar]
- Szatmari I, Vámosi G, Brazda P, Balint BL, Benko S, Széles L, et al. Peroxisome proliferator-activated receptor gamma-regulated ABCG2 expression confers cytoprotection to human dendritic cells. J Biol Chem. 2006;281:23812–23823. doi: 10.1074/jbc.M604890200. [DOI] [PubMed] [Google Scholar]
- Takeshita A, Koibuchi N, Oka J, Taguchi M, Shishiba Y, Ozawa Y. Bisphenol-A, an environmental estrogen, activates the human orphan nuclear receptor, steroid and xenobiotic receptor-mediated transcription. Eur J Endocrinol. 2001;145:513–517. doi: 10.1530/eje.0.1450513. [DOI] [PubMed] [Google Scholar]
- Teotico DG, Bischof JJ, Peng L, Kliewer SA, Redinbo MR. Structural basis of human pregnane × receptor activation by the hops constituent colupulone. Mol Pharmacol. 2008;74:1512–1520. doi: 10.1124/mol.108.050732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit YE, Negishi M. CAR and PXR: the xenobiotic-sensing receptors. Steroids. 2007;72:231–246. doi: 10.1016/j.steroids.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, Lamba V, et al. The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat Med. 2003;9:220–224. doi: 10.1038/nm815. [DOI] [PubMed] [Google Scholar]
- Uppal H, Saini SP, Moschetta A, Mu Y, Zhou J, Gong H, et al. Activation of LXRs prevents bile acid toxicity and cholestasis in female mice. Hepatology. 2007;45:422–432. doi: 10.1002/hep.21494. [DOI] [PubMed] [Google Scholar]
- Uppal H, Toma D, Saini SP, Ren S, Jones TJ, Xie W. Combined loss of orphan receptors PXR and CAR heightens sensitivity to toxic bile acids in mice. Hepatology. 2005;41:168–176. doi: 10.1002/hep.20512. [DOI] [PubMed] [Google Scholar]
- Urizar NL, Dowhan DH, Moore DD. The farnesoid X-activated receptor mediates bile acid activation of phospholipid transfer protein gene expression. J Biol Chem. 2000;275:39313–39317. doi: 10.1074/jbc.M007998200. [DOI] [PubMed] [Google Scholar]
- Valrance ME, Brunet AH, Acosta A, Welsh J. Dissociation of growth arrest and CYP24 induction by VDR ligands in mammary tumor cells. J Cell Biochem. 2007;101:505–1519. doi: 10.1002/jcb.21263. [DOI] [PubMed] [Google Scholar]
- van den Akker EL, Koper JW, van Rossum EF, Dekker MJ, Russcher H, de Jong FH, et al. Glucocorticoid receptor gene and risk of cardiovascular disease. Arch Intern Med. 2008;168:33–39. doi: 10.1001/archinternmed.2007.41. [DOI] [PubMed] [Google Scholar]
- van der Vaart M, Schaaf MJ. Naturally occurring C-terminal splice variants of nuclear receptors. Nucl Recept Signal. 2009;7:e007. doi: 10.1621/nrs.07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villey I, de Chasseval R, de Villartay JP. RORgammaT, a thymus-specific isoform of the orphan nuclear receptor RORgamma / TOR, is up-regulated by signaling through the pre-T cell receptor and binds to the TEA promoter. Eur J Immunol. 1999;29:4072–4080. doi: 10.1002/(SICI)1521-4141(199912)29:12<4072::AID-IMMU4072>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- Wang W, Prosise WW, Chen J, Taremi SS, Le HV, Madison V, et al. Construction and characterization of a fully active PXR/SRC-1 tethered protein with increased stability. Protein Eng Des Sel. 2008;21:425–433. doi: 10.1093/protein/gzn017. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kumar N, Solt LA, Richardson TI, Helvering LM, Crumbley C, et al. Modulation of retinoic acid receptor-related orphan receptor alpha and gamma activity by 7-oxygenated sterol ligands. J Biol Chem. 2010;285:5013–5025. doi: 10.1074/jbc.M109.080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins RE, Davis-Searles PR, Lambert MH, Redinbo MR. Coactivator binding promotes the specific interaction between ligand and the pregnane × receptor. J Mol Biol. 2003;331:815–828. doi: 10.1016/s0022-2836(03)00795-2. [DOI] [PubMed] [Google Scholar]
- Watkins RE, Maglich JM, Moore LB, Wisely GB, Noble SM, Davis-Searles PR, et al. 2.1 A crystal structure of human PXR in complex with the St. John's wort compound hyperforin. Biochemistry. 2003;42:1430–1438. doi: 10.1021/bi0268753. [DOI] [PubMed] [Google Scholar]
- Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, et al. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292:2329–2333. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- Waxman DJ, Azaroff L. Phenobarbital induction of cytochrome P-450 gene expression. Biochem J. 1992;281:577–592. doi: 10.1042/bj2810577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM. Pharmacogenomics: catechol O-methyltransferase to thiopurine S-methyltransferase. Cell Mol Neurobiol. 2006;26:539–561. doi: 10.1007/s10571-006-9095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells PG, Mackenzie PI, Chowdhury JR, Guillemette C, Gregory PA, Ishii Y, et al. Glucuronidation and the UDP-glucuronosyltransferases in health and disease. Drug Metab Dispos. 2004;32:281–290. doi: 10.1124/dmd.32.3.281. [DOI] [PubMed] [Google Scholar]
- Wiesenberg I, Missbach M, Kahlen JP, Schräder M, Carlberg C. Transcriptional activation of the nuclear receptor RZR alpha by the pineal gland hormone melatonin and identification of CGP 52608 as a synthetic ligand. Nucleic Acids Res. 1995;23:327–333. doi: 10.1093/nar/23.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willson TM, Brown PJ, Sternbach DD, Henke BR. The PPARs: from orphan receptors to drug discovery. J Med Chem. 2000;43:527–550. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- Windshügel B, Jyrkkärinne J, Poso A, Honkakoski P, Sippl W. Molecular dynamics simulations of the human CAR ligand-binding domain: deciphering the molecular basis for constitutive activity. J Mol Model. 2005;11:69–79. doi: 10.1007/s00894-004-0227-4. [DOI] [PubMed] [Google Scholar]
- Windshügel B, Jyrkkärinne J, Vanamo J, Poso A, Honkakoski P, Sippl W. Comparison of homology models and X-ray structures of the nuclear receptor CAR: assessing the structural basis of constitutive activity. J Mol Graph Model. 2007;25:644–657. doi: 10.1016/j.jmgm.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Windshügel B, Poso A. Constitutive activity and ligand-dependent activation of the nuclear receptor CAR-insights from molecular dynamics simulations. J Mol Recognit. 2007;24:875–882. doi: 10.1002/jmr.1132. [DOI] [PubMed] [Google Scholar]
- Wira C, Munck A. Specific glucocorticoid receptors in thymus cells. Localization in the nucleus and extraction of the cortisol-receptor complex. J Biol Chem. 1970;245:3436–3438. [PubMed] [Google Scholar]
- Xia J, Kemper B. Structural determinants of constitutive androstane receptor required for its glucocorticoid receptor interacting protein-1-mediated nuclear accumulation. J Biol Chem. 2005;280:7285–7293. doi: 10.1074/jbc.M409696200. [DOI] [PubMed] [Google Scholar]
- Xia Z, Correa RG, Das JK, Farhana L, Castro DJ, Yu J, et al. Analogues of orphan nuclear receptor small heterodimer partner ligand and apoptosis inducer (E)-4-[3-(1-adamantyl)-4-hydroxyphenyl]-3-chlorocinnamic acid. 2. Impact of 3-chloro group replacement on inhibition of proliferation and induction of apoptosis of leukemia and cancer cell lines. J Med Chem. 2012;55:233–249. doi: 10.1021/jm2011436. [DOI] [PubMed] [Google Scholar]
- Xia Z, Farhana L, Correa RG, Das JK, Castro DJ, Yu J, et al. Heteroatom-substituted analogues of orphan nuclear receptor small heterodimer partner ligand and apoptosis inducer (E)-4-[3-(1-Adamantyl)-4-hydroxyphenyl]-3-chlorocinnamic acid. J Med Chem. 2011;54:3793–3816. doi: 10.1021/jm200051z. [DOI] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Simon CM, Pierce AM, Safe S, Blumberg B, et al. Reciprocal activation of xenobiotic response genes by nuclear receptors SXR/PXR and CAR. Genes Dev. 2000;14:3014–3023. doi: 10.1101/gad.846800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Evans RM. Orphan nuclear receptors: the exotics of xenobiotics. J Biol Chem. 2001;276:37739–37742. doi: 10.1074/jbc.R100033200. [DOI] [PubMed] [Google Scholar]
- Xie W, Uppal H, Saini SP, Mu Y, Little JM, Radominska-Pandya A, et al. Orphan nuclear receptor-mediated xenobiotic regulation in drug metabolism. Drug Discov Today. 2004;9:442–449. doi: 10.1016/S1359-6446(04)03061-2. [DOI] [PubMed] [Google Scholar]
- Xu C, Wang X, Staudinger JL. Regulation of tissue-specific carboxylesterase expression by pregnane × receptor and constitutive androstane receptor. Drug Metab Dispos. 2009;37:1539–1547. doi: 10.1124/dmd.109.026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HE, Stanley TB, Montana VG, Lambert MH, Shearer BG, Cobb JE, et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415:813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- Xu RX, Lambert MH, Wisely BB, Warren EM, Weinert EE, Waitt GM, et al. A structural basis for constitutive activity in the human CAR/RXRalpha heterodimer. Mol Cell. 2004;16:919–928. doi: 10.1016/j.molcel.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Xue Y, Moore LB, Orans J, Peng L, Bencharit S, Kliewer SA, et al. Crystal structure of the pregnane × receptor-estradiol complex provides insights into endobiotic recognition. Mol Endocrinol. 2007;21:1028–1038. doi: 10.1210/me.2006-0323. [DOI] [PubMed] [Google Scholar]
- Yanagiba Y, Ito Y, Kamijima M, Gonzalez FJ, Nakajima T. Octachlorostyrene induces cytochrome P450, UDP-glucuronosyltransferase, and sulfotransferase via the aryl hydrocarbon receptor and constitutive androstane receptor. Toxicol Sci. 2009;111:19–26. doi: 10.1093/toxsci/kfp130. [DOI] [PubMed] [Google Scholar]
- Yu J, Huang L, Zhao A, Metzger E, Adams A, Meinke PT, et al. Lithocholic acid decreases expression of bile salt export pump through farnesoid × receptor antagonist activity. J Biol Chem. 2002;277:31441–31447. doi: 10.1074/jbc.M200474200. [DOI] [PubMed] [Google Scholar]
- Yueh MF, Tukey RH. Nrf2-Keap1 signaling pathway regulates human UGT1A1 expression in vitro and in transgenic UGT1 mice. J Biol Chem. 2007;282:8749–8758. doi: 10.1074/jbc.M610790200. [DOI] [PubMed] [Google Scholar]
- Zamek-Gliszczynski MJ, Hoffmaster KA, Nezasa K, Tallman MN, Brouwer KL. Integration of hepatic drug transporters and phase II metabolizing enzymes: mechanisms of hepatic excretion of sulfate, glucuronide, and glutathione metabolites. Eur J Pharm Sci. 2006;27:447–486. doi: 10.1016/j.ejps.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Zechel C, Shen X, X. Q, Chambon P, Gronemeyer H. Dimerization interfaces formed between the DNA binding domains determine the cooperative binding of RXR/RAR and RXR/TR heterodimers to DR5 and DR4 elements. EMBO J. 1994;13:1414–1424. doi: 10.1002/j.1460-2075.1994.tb06395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325:100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelko I, Negishi M. Phenobarbital-elicited activation of nuclear receptor CAR in induction of cytochrome P450 genes. Biochem Biophys Res Commun. 2000;277:1–6. doi: 10.1006/bbrc.2000.3557. [DOI] [PubMed] [Google Scholar]
- Zhang H, Chen L, Chen J, Jiang H, Shen X. Structural basis for retinoic × receptor repression on the tetramer. J Biol Chem. 2011;286:24593–24598. doi: 10.1074/jbc.M111.245498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bae Y, Kemper JK, Kemper B. Analysis of multiple nuclear receptor binding sites for CAR/RXR in the phenobarbital responsive unit of CYP2B2. Arch Biochem Biophys. 2006;451:119–127. doi: 10.1016/j.abb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Zhang XY, Guan YF. Farnesoid × receptor (FXR): a novel regulative factor involved in metabolism. Sheng Li Ke Xue Jin Zhan. 2007;38:219–223. [Article in Chinese] [PubMed] [Google Scholar]
- Zhang Y, Ge X, Heemstra LA, Chen WD, Xu J, Smith JL, et al. Loss of FXR protects against diet-induced obesity and accelerates liver carcinogenesis in ob/ob mice. Mol Endocrinol. 2012;26:272–280. doi: 10.1210/me.2011-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hagedorn CH, Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta. 2011;1812:893–908. doi: 10.1016/j.bbadis.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Mirkovitch J, Darnell JE., Jr. Tissue-specific regulation of mouse hepatocyte nuclear factor 4 expression. Mol Cell Biol. 1994;14:7276–7284. doi: 10.1128/mcb.14.11.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zintzaras E, Rodopoulou P, Koukoulis GN. BsmI, TaqI, ApaI and FokI polymorphisms in the vitamin D receptor (VDR) gene and the risk of osteoporosis: a meta-analysis. Dis Markers. 2006;22:317–326. doi: 10.1155/2006/921694. [DOI] [PMC free article] [PubMed] [Google Scholar]