Abstract
The immunological synapse is an excellent example of cell-cell communication, where signals are exchanged between two cells, resulting in a well-structured line of defense during adaptive immune response. This process has been the focus of several studies that aimed at understanding its formation and subsequent events and has led to the understanding that it relies on a well-orchestrated molecular program that only occurs when specific requirements are met. The development of more precise and controllable T cell activation systems has led to new insights including the role of mechanotransduction in the process of formation of the immunological synapse and T cell activation. Continuous advances in our understanding of the immunological synapse formation, particularly in the context of T cell activation and differentiation, as well the development of new T cell activation systems are being applied to the establishment and improvement of immune therapeutical approaches.
Keywords: adaptive response, activation, T cell, mechanotransduction, pMHC, signaling
I. OVERVIEW
Our body responds to pathogenic challenges (caused by viruses, bacteria, fungi or parasites) not only by efficiently clearing invading pathogens but also by developing long-lasting immunity against infectious diseases. This defense against infection can be divided into innate and adaptive immune response on the basis of specificity of recognition (Janeway and Medzhitov, 2002). Whereas innate response is readily available to fight a range of pathogens in an evolutionarily pre-programmed manner, the adaptive immune response results from a selection of novel specificities during infection with a pathogen and often leads to long-term protection to re-infection by that specific pathogen. These two arms of immune response work hand in hand to fight infections, prevent autoimmunity and also to resolve the immune response when infection has cleared. The main cellular players of the adaptive immune response include antigen-presenting cells (APCs) which process and display on their surface foreign antigens that subsequently interact with B cells or T cells, leading to an adaptive immune response (Trombetta and Mellman, 2005). While APC sounds like a genereal term, this is actually highly specific to a group of cells that express high levels of MHC class II, such as B cells and dendritic cells (DC). Almost all nucleated cells expression MHC class I, but there are not generally called APC, or professional APC, if they lack MHC class II expression. Whereas B cell activation leads to the production of antibodies that fight extracellular pathogens, T cells are crucial in cell-mediated immune responses for pathogens replicating inside cells (Banchereau and Steinman, 1998). In the latter, the antigen presented by the APC triggers the T cell receptor (TCR) and leads to the formation of the “immunological synapse” (IS) (Fooksman et al., 2010; Grakoui et al., 1999), the interface between the APC and T cell, establishing the communication between the two cells and ultimately leading to the T cell activation through a signaling network. The stable IS serves a variety of functions including sustaining contact dependent receptor engagement with appropriate topology, polarization of molecules in the partner cells and confinement of secreted products (Angus and Griffiths, 2013; Choudhuri et al., 2005). Numerous studies have contributed to our understanding of how at the cellular and molecular level the IS is formed and how this reorganization and molecular architecture of the membrane interface between two cells impacts the response to an invading pathogen (Egen et al., 2008; Harris et al., 2012; Waite et al., 2011; Zinselmeyer et al., 2013). The study of the IS provides opportunities for basic investigations in cell biology and valuable insights into immunopathologies and immunotherapy.
II. THE IMMUNOLOGICAL SYNAPSE
Among the APCs, DCs are the main players in the initiation of the adaptive immune response because they coordinate antigen processing and presentation capacity with migration. Multiple likes of myeloid development converge on the DC phenotypes, making definitive fate mapping a challenge. “Immature” DC reside near barriers and in the marginal zone of the spleen where they sample self antigens and benign foreign antigens as part of maintaining self-tolerance and prevention of inappropriate response to foreign antigens such as allergies (Steinman et al., 2003). Upon encounter with foreign antigens in combination with innate activation the DC initiate a program of migration into lymphatics and into lymph nodes or across the marginal zone into white pulp (Edelson et al., 2011; Ohl et al., 2004), The migration program is coordinated with “maturation” of the DC, which involves up-regulation of major histocompatibility complex (MHC) class II complexes and costimulatory molecules in which type I interferons play an important role (Longhi et al., 2009). Within the T cell zones of lymph nodes or the spleen white pulp the DC scan thousands of naïve T cells per hour to bring MHC-peptide complexes together with the 1 in one million T cells that has appropriate specificity (Miller et al., 2004).
During this initial key step, the digested antigenic peptides, associated with the MHC on the surface of the DC, together with co-stimulatory molecules, trigger the cell receptors on the naïve T cell, leading to cytoskeleton remodeling and the formation of the IS at the interface of these two cells, ultimately leading to activation of the T cell (Celli et al., 2007). CD4+ T cells that are activated by dendritic cells can then go on to help B cells through synapse formation at the junction between the T and B cell zones and later in germinal centers (Allen et al., 2007; Victora et al., 2010).
Types of IS
There is a remarkable diversity of synaptic interactions in immune cells. Synapse- like contact interfaces are observed in many immune cells, and these cells communicate with a variety of cellular partners in the steady state and during an immune response. Although the contact between T cell-APC and cytotoxic T cell- and their targets remains the most characterized form of synapse, recent studies in other immune cell types have also characterized stable contacts that involve B cells, NK T cells, NK cells and even phagocytes (Fooksman et al., 2010; Goodridge et al., 2011). In addition, “synapse”-like contacts have also been described between T cell subtypes such as between CD4 and CD8 T cells. While most of the studies characterizing the higher order structure of non-T cell synapses have been carried out in vitro using controlled activation conditions, in vivo dynamics of the these interactions has also been investigated in specific tissue contexts, and has confirmed the specificities of these interactions (Breart et al., 2008; Mempel et al., 2004; Ruocco et al., 2012). Diverse synapse types between different immune cells, although exhibiting variations in the overall organization of molecules at the cell-cell interface, follow some common themes as studied in T cell-APC synapses. This includes the activation and spatial segregation of immunoreceptors at the cell surface, regulation of this process by co-receptors and adhesion molecules, phosphorylation of intracellular immunoreceptor tyrosine-based motifs (ITAM) and activation of intracellular signaling and eventually Ca2+ flux (Skokos et al., 2007). Most forms of synapse also exhibit cytoskeleton remodeling and specialized organization of actin cytoskeleton, required for cell polarization and optimal synapse function (Burkhardt et al., 2008).
B cells form synapses with various APCs- macrophages, DCs and follicular DCs (Harwood and Batista, 2011). The antigen displayed on the APC interacts with the B cell receptor (BCR) on the B cell surface, and this engagement process triggers the formation of BCR microclusters and B cell- APC synapse. Activated B cells internalize BCR-antigen complexes and process the antigen to present it in the context of MHC to T cells. Thus B cells relay the antigen from APCs to T cells via synapse formation (Batista et al., 2001). Actin cytoskeleton is thought to play a negative role in B cell synapse, where plasma membrane BCR is restricted in cortical actin meshwork present underneath the membrane, and antigen engagement releases this diffusion trap (Harwood and Batista, 2011). Further actin-dependent spreading of B cell facilitates formation of a larger contact area and BCR microclusters. A stable synapse is established when the B cell interface contracts following expansion, mobilizing BCR microclusters to the center of the contact. The process of BCR internalization after the contraction phase is currently under investigation in search of precise molecular players that regulate the endocytic process.
T cell -APC synapse is the most extensively characterized form of immune cell synapse. T cells can form synapses with antigen loaded B cells or antigen-loaded DCs. The TCR recognizes peptides bound to MHC on APCs and undergoes activation. TCR triggering then activates a series of signaling events inside the T cell leading to its activation. TCR-stimulated T cells then serve a variety of functions such as cytolysis of target cells by CD8 T cells and production of effector cytokines by CD4 T cells. Though less characterized, T cells can form synapses with non-APCs. A good example of this class of interaction is the synapse between T cells. Antigen CD8 T cells can form stable conjugates with CD4 T cells and this interaction can lead to amplification of the CD8 T cell response (Chaudhri et al., 2009). Alternatively, a synapse between activated CD4 T cells can also form, that can be utilized for sharing of paracrine cytokines such as IL-2 and IFN-γ, thus consolidating CD4 T cell activation response at population level (Gerard et al., 2013; Sabatos et al., 2008). The molecular players involved in these synaptic interactions are not known; however a critical role for integrin LFA-1 has been established in homotypic T cell synapses. In additional to the above-mentioned synapse, effector T cells can also form conjugates with regulatory T cells and NK cells, however, the precise cell biology of these interactions has not been characterized.
Unlike T cells where the presence of cognate antigen-MHC on the APC is sufficient to trigger activation, NK cells utilize a combination of a positive and negative surface receptor signal on the DC to regulate their cytolytic response. NK cells are innate cells that express germline-encoded activating and inhibitory receptors. The activating receptors, mainly NKG2D and NCRS, recognize a variety of molecules expressed on infected or transformed target cells. The inhibitory receptors CD94/NKG2A and KIRs recognize non-classical and classical MHC molecules (Barreira da Silva and Munz, 2011). The relative signaling of these two functionally distinct types of signaling receptors decides the nature of the NK cell synapse, i.e. activating or inhibitory. Actin cytoskeleton plays a crucial role in the strength of the contact and has been proposed to mediate polarized and localized secretion of cytolytic granules (Rak et al., 2011).
Another interesting variation of typical recognition-specificity based synapse is the synapse formed by the NK-T cells. These cells are innate-like cells and the majority of them express a semi-invariant TCR consisting of invariant α-chain along with diverse β chain, and exhibit features of both NK cells and T cells. These cells are thought to represent a link between the innate and adaptive immune system and likewise, can interact with a variety of innate and adaptive immune cells. The TCR on the NK-T cell recognizes self and non-self lipid antigens presented on the CD1d molecule. CD1d is a MHC-like molecule expressed by a variety of APCs, including macrophages, DCs, B cells, CD4 CD8 thymocytes and epithelial cells (Joyce et al., 2011). Upon recognition of lipid antigen/CD1d on the APC, NK-T cells establish a synapse with the APC. Stable synapse formation and polarization of lytic granules accompanies the optimal effector response.
T cell IS
The T cell IS has been the best studied as having a close contact is indisputably critical for antigen recognition by a T cell (Grakoui et al., 1999).
The naïve T cell is initially primed by a pathogen-activated DCs in the lymph node. The formation of an IS between these two cells results in the activation of the T cell which clonally expands ≥ 10 generations for each T cell the is activated by a DC (Moon et al., 2007). These progeny differentiate into effector cells that exit the lymph node or spleen and in turn patrol the whole body scanning for matching MHC peptide complexes on the surface of APCs.
Once a T cell encounters an APC, several conditions have to be met for an IS to form (Shaw and Dustin, 1997). These requirements were formalized over 15 years ago, but new appreciation of the function of actin networks necessitates updating these:
1) Cell membrane proximity and cell-cell adhesion
The APC and T cell have to be close enough to enable the molecules on each cell’s membrane to interact efficiently. One obstacle in reaching proximity between the two cells is the layer of glycocalyx (consisting of large glycoproteins) that covers both the T cell and APC and that creates repulsion between the two cells (Cyster et al., 1991). This effect is partially overcome by the integrin-based adhesion between the ICAM-1, on the APC and Lymphocyte function associated antigen-1 (LFA-1, a member of the integrin family of receptors), on the T cell, an antigen-nonspecifc interaction that is driven in part by chemokines (Alon and Dustin, 2007). Although LFA-1-ICAM-1 based adhesion brings the two cells’ membranes within ~45 nm of each other (Chen et al., 2012), the distance between the two cells (APC and T cell) has to be brought to less than 15 nm so that the pMHC complex on the APC can bind to the TCR on the T cell. To achieve the optimal intercellular distance, and reduce it from 45 nm to 15 nm, actin polymerization-based protrusions (invasive pseudopodia) of the T cell reach the APC, allowing the T cell to sample a large number of pMHC complexes at the surface of each APC to scan for a specific peptide. Such “invadosome-like protusions” (ILP) have been recently visualized at high resolution using endothelial cells to provide a thin, flat APC with excellent properties for imaging (Sage et al., 2012).
2) Recognition of matching TCR-pMHC
If the TCR (on the T cell) does not recognize a specific matching antigen (pMHC complex), the T cell will separate from the APC to continue sampling. Conversely, upon recognition and binding of the specific pMHC by the TCR, the 15 nm optimal distance for interaction between the two cell membranes is sustained allowing formation of a microcluster over a period of several seconds (Campi et al., 2005; Varma et al., 2006).
On a longer time scale, binding of the TCR to the specific antigen (pMHC complex) leads to the formation an IS composed of supramolecular activation clusters (SMACs) (Grakoui et al., 1999; Monks et al., 1998).
3) Co-stimulatory system
In addition to the TCR-pMHC and adhesion (LFA-1—ICAM-1) systems, the interaction of CD28 (on the surface of the T cell) with a B7 ligand on the APC (CD80 or CD86) provides a co-stimulatory signal that enhances TCR triggering of naïve T cells (Levine et al., 1997).
Several cell surface molecules are therefore required for T cell and APC to interact:
T Cell :: APC
TCR :: pMHC (class I, CD8 T cells; class II, CD4 T cells)
CD28 :: B7 molecules (CD80, CD86)
LFA-1 :: ICAM-I
Organizational architecture of the IS
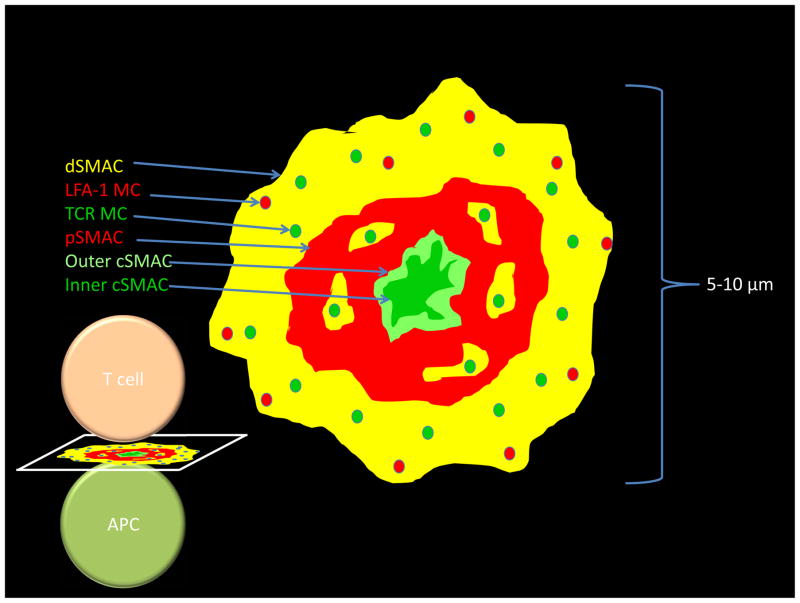
The IS is a well-organized structure that consists of three concentric rings which differ in their receptor composition and density. The different surface receptors that are part of the IS are segregated into distinct areas in the form of clusters. The TCR is initially present at the T cell surface in clusters of 5–10 molecules of TCR in “confinement zones” or “protein islands”, possibly held by an underlying actin meshwork and/or anchored to the cortical actin (Lillemeier et al., 2010). Upon engagement of the TCR, microclusters start forming in small contact areas between the T cell and APC membranes and their number increases as the contact area between the two cells increases with T cell spreading. The TCR microclusters form in the periphery of the synapse in the distal supra molecular activation cluster (dSMAC) and within a few minutes of cluster formation, they translocate centripetally, through the peripheral supra molecular activation cluster (pSMAC) eventually fusing into the central supra molecular activation cluster (cSMAC) zone. It is at the cSMAC, where the TCR clusters co-localize with markers of protein degradation (Varma et al., 2006) and components of the ubiquitin pathway (Vardhana et al., 2010), that signaling is thought to be terminated, with the TCR-pMHC being internalized possibly by phagocytosis (Alarcon et al., 2011; Coombs et al., 2002; Liu et al., 2000). As the TCR microclusters form, in the pSMAC, they associate with the signaling molecules that will eventually trigger T cell activation (described below), however, as the microclusters migrate towards the cSMAC, these associations are lost, thus, despite the large number of TCRs present in the cSMAC, signaling does not occur in this central part of the IS.
Contrary to TCR clusters, LFA-1 clusters are organized in the pSMAC - a highly contractile zone - and do not cross the cSMAC-pSMAC border. As molecules engage and TCR microclusters are formed at the pSMAC, these start signaling and continue to do so as they translocate to the cSMAC at the center of the IS. After the TCR microclusters are delivered to the cSMAC TCR signaling is downregulated, possibly as a way to control signaling strength at the synapse (Varma et al., 2006).
Studies that aimed at investigating the involvement of actin polymerization and remodeling in the formation of the IS and segregation of its main components have shown that actin is dynamically remodeled during this process. The observation that inhibition of actin polymerization inhibits T cell activation implies that actin is a key player. In an initial phase actin rapidly polymerizes as the T cell spreads, with expansion of lamellipodia-like edges. At this stage the IS presents three distinct actin organization zones: an actin-enriched area at the periphery of the IS (dSMAC), an actomyosin-rich area (pSMAC), and a central F-actin poor zone (cSMAC). Following this stage, the T cell lamellipodia start contracting leading to a retrograde flow of actin towards the cSMAC at the same time as the TCR translocates towards the cSMAC. Several studies have suggested that actin is indeed coupled to the translocation of TCR microclusters (the fact that sometimes translocation is slower than actin flow has been suggested to occur by occasional slippage). Another model suggests that the coupling between microclusters and actin is transient (“frictional coupling mechanism”) through weak links that form and break at different times producing a frictional force (DeMond et al., 2008; Smoligovets et al., 2012; Yu et al., 2010). Such studies on the role of actin in microclusters translocation and organization of the IS include the measurement of the velocity of TCR microclusters and actin speckles when tracking actin-GFP in Jurkat T cells (Kaizuka et al., 2007) and the use of chrome barriers that obstruct TCR-pMHC translocation but not the actin flow in supported lipid bilayers (SLB) (DeMond et al., 2008). The mechanism by which microclusters are coupled to F-actin is however still not clear. Experiments using myosin II activity inhibitors or depletion of myosin II also resulted in defective microcluster translocation suggesting Myosin II activity to be involved in TCR migration towards the cSMAC in an additive manner to actin polymerization (Yi et al., 2012),(Ilani et al., 2009),(Kumari et al., 2012),(Babich et al., 2012). This myosin II activity appears not to be required for TCR translocation in motile T cells, whereas actin depolymerization is (Beemiller et al., 2012). Recent studies have also implied the involvement of microtubule cytoskeleton in the TCR microcluster motility towards the cSMAC as TCR microclusters seem to be able to interact with the microtulule motor dynein (Hashimoto-Tane et al., 2011).
The final step of the IS, i.e. the separation of T cell and APC, is as critical, as in order for the T cell to move on to scan for its cognate pMHC, it must disengage from the IS. This stage of the IS is referred to as “kinapse” (Dustin, 2008). The mechanism underlying the synapse-kinapse transition is not yet fully understood.
Functional consequences of the IS
Upon antigen presentation to the T cell and recognition of the pMHC and TCR engagement, downstream signaling pathways are triggered, leading to a series of events that can result in different outcomes.
TCR signaling
As the TCR recognizes and binds the cognate antigen on the APC, the ITAM motifs of the cytosolic tail of the TCR-associated CD3 chains - that is thought to be associated with the lipids in the inner leaflet of the plasma membrane in the inactive conformation – undergo phosphorylation by the lymphocyte-specific protein tyrosine kinase (Lck).
It is not clear whether the release of the cytosolic TCR-associated CD3 zeta chains, protected from Lck, is the cause (“safety model”) (Xu et al., 2008),(Gagnon et al., 2012),(Shi et al., 2013) or the consequence (Zhang et al., 2011) of the ITAM motifs’ phosphorylation.
The “safety model”, which proposes that the TCR/CD3 cytoplasmic domains are associated with the membrane to prevent phosphorylation of the ITAM motifs, has been recently questioned, mainly due to the observation that inhibition of the association of the CD3 cytoplasmic domain with the membrane did not enhance CD3 phosphorylation, and reduced it instead, and that the ITAM motifs phosphorylation was required for the TCR/CD3 chains dissociation from the membrane (DeFord-Watts et al., 2011; Zhang et al., 2011). At the IS, the phosphorylated TCR-associated and the inhibitory phosphatase CD45 (which localizes to the p and dSMAC) are mutually exclusive, which indicates that this is a requirement for the receptor phosphorylation (Leupin et al., 2000; Varma et al., 2006). In fact, ITAM motifs can have a low level of phosphorylation in resting T cells, as they are constantly being phosphorylated, by Lck, and dephosphorylated by CD45; given the short distance between T cell and APC when TCR and pMHC bind (15 nm), glycoproteins of larger size are excluded from the TCR contact area and are localized to the pSMAC, allowing prolonged phosphorylation of the TCR-CD3 ITAM motifs and subsequent TCR triggering (Choudhuri et al., 2009). The phosphorylation of the ITAM motifs allows the recruitment of the Syk-kinase zeta chain-associated protein of 70 kDa (ZAP70) (Wang et al., 2010), which is then also phosphorylated, and activated, by Lck. The phosphorylated ZAP70 (pZAP70) phosphorylates the transmembrane protein Linker of Activated T cells (LAT). Whereas some have proposed that LAT and TCR are segregated in sub-micrometer “protein islands” surrounded by a protein-poor “lipid sea” (Lillemeier et al., 2010) - a barrier that would have to be overcome for signaling to occur - others have suggested that LAT molecules are not laterally recruited but that there is an intracellular pool of LAT sufficient to drive signaling (Williamson et al., 2011). LAT has been shown to exist in sub-synaptic vesicles that could either be phosphorylated in trans or instead, tethered or fused with the plasma membrane. The LAT “protein island” and “LAT in vesicles” models are not mutually exclusive and may reflect different experimental conditions, such as time points, or may be related to different signaling outcomes. Once phosphorylated, LAT phosphorylates and docks SH2 domain-containing leukocyte protein of 76 kDa (SLP76), Phospholipase C γ (PLCγ), phosphatidylinositol 3-kinase (PI3K), growth factor receptor-bound protein 2 (Gb2) and Grb2-homologous adaptor (GADS) (Cruz-Orcutt and Houtman, 2009; Liu et al., 2001; Wu and Koretzky, 2004; Zhang et al., 2011). The phosphorylation of LAT (4 seconds after TCR-pMHC engagement) and subsequent docking of PLCγ regulate Ca2+ influx (6–7 seconds post TCR-pMHC engagement). PLCγ activity leads to an increase in cytosolic Ca2+ through the generation of IP3, which releases Ca2+ from endoplasmic reticulum (store-operated Ca2+ entry, SOCE) then triggering the influx of Ca2+ across the plasma membrane (Ca2+ release-activated Ca2+, CRAC) channels (reviewed in (Shaw et al., 2012)). One of the main consequences of the Ca2+ influx is the relocation of the transcription factor Nuclear Factor of Activated T cells (NFAT) from the cytosol to the nucleus, through dephosphorylation by the Ca2+dependent phosphatase calcineurin (Hogan et al., 2003), eventually resulting in cytokine transcription and secretion (IL2-2, IL-4, IL-17, IFNγ and TNF) by the T cell. SLP76, phosphorylated by LAT, plays a role as scaffold for several actin effectors. Both the guanine-nucleotide-exchange factor (GEF) Vav1 (that appears to associate with the LAT-SLP76 complex (Pauker et al., 2012; Sylvain et al., 2011)) and Wiskott-Aldrich syndrome protein WASp, F-actin nucleation and remodeling regulators have been reported to be recruited to the TCR-pMHC sites and regulate actin polymerization (Barda-Saad et al., 2005; Miletic et al., 2009; Miletic et al., 2006; Sasahara et al., 2002; Tybulewicz, 2005; Yokosuka et al., 2005; Zeng et al., 2003).
The formation of the IS can have different functional consequences to provide an appropriate response in a context-dependent manner. The role of the immunological synapse is multifaceted: to aid the interaction of T cells with APC, efficient antigen-MHC sampling, to form stable conjugate with cognate pMHC, and to activate and down-regulate TCR signaling (which is as important to achieve an optimal and controlled T cell response). The IS allows for the development of the appropriate response ranging from cytokine secretion, proliferation and effector functions. Though cytokine secretion and proliferation have been thought of as a singular pathway, initiated by TCR triggering, ITAM phosphorylation, Ca2+ influx and transcription activation, Guy et al. have recently shown that proliferation and cytokine secretion are non-overlapping pathways (Guy et al., 2013). The TCR-CD3 complex has ten functional ITAM motifs - more than the two motifs that have been believed to be required to activate trigger the TCR signaling. Guy et al. have found that even though low multiplicity of TCR-CD3 ITAM motifs (only two to four functional ITAM motifs) is indeed sufficient to engage canonical TCR signaling events that lead to cytokine secretion, such as interleukin 2 (IL-2) and interferon-γ, a high multiplicity of TCR-CD3 ITAM motifs is required for TCR-induced proliferation (Guy et al., 2013). The proliferation pathway was shown to rely on the formation of a compact cSMAC, on Notch1 and up-regulation of c-Myc after ligation of TCR-CD3 to the pMHC (Guy et al., 2013). Having distinct pathways driving different functional responses, such as secretion of certain cytokines and proliferation, may be biologically relevant as it may enable the T cell to respond differently under different conditions. Indeed, Guy et al. have shown that T cells presented with superagonists have an enhanced expression and translocation of c-Myc and a greater proliferation than when presented with a weak agonist.
Polarized secretion
Formation of the IS and subsequent induction of cell polarization are required for directed secretion by CD8 cytotoxic T cells (CTL) of specialized secretory lisossomes (or lytic granules) containing lytic pore-forming perforin, which enables entry of granzymes into virally-infected target cells or tumor cells, ultimately triggering apoptosis (reviewed in (Stinchcombe and Griffiths, 2007)). Movement of the centrosome to the membrane at the edge of the cSMAC is a key step in IS-induced cell polarization, which has been observed in CTLs (Stinchcombe et al., 2006) as well as in CD4 cells (Ueda et al., 2011), NK and NKT cells (Stinchcombe et al., 2011), indicating that docking of the centrosome to the plasma membrane is a common event in direct secretion. The cytoskeleton and motor proteins such as dynein, as well as diacyl-glycerol (DAG) accumulation and restriction to the IS and other TCR signaling players, have been suggested to play a role in inducing the movement of the centrosome (Gharbi et al., 2011; Quann et al., 2009). Several candidate proteins have been suggested to also be involved at the centrosome polarization (reviewed in (Angus and Griffiths, 2013).) Other TCR signaling players, such as Lck, have been implicated specifically in the docking of the centrosome to the plasma membrane (Tsun et al., 2011), essential for granule delivery to the IS. The movement and docking of the centrosome - the microtubule organizing center (MTOC) of the cell – to the IS, reorganizes the intracellular organizing cytoskeleton and is thought to allow polarized and precise secretion of lytic granules to the IS, inducing cell death of the target cell. Though there have been advances in the study of polarized secretion in T cells regarding centrosome movement and exocytosis of the secretory vesicles at the IS (reviewed in (Griffiths et al., 2010)), much remains to be understood. Besides directing secretion of lytic granules, the IS can also induce polarized secretion of cytokines (such as IL-2, IL-4, IL-5 and IFN-γ) towards the APC (Kupfer et al., 1991; Poo et al., 1988). In addition it has been proposed that also the cytokine receptors can be selectively recruited to the synapse, adding a new regulation layer (Maldonado et al., 2009).
T cell differentiation
One important aspect of the immune response is the ability to generate the appropriate type of T cell for a suitable response. The IS plays a role in T cell differentiation by leading to changes in transcription factor programs initiated by TCR triggering and ultimately the activation of transcription factors such as NFAT, Activator protein 1 (AP-1) and Nuclear Factor-kB (NF-kB), which in turn bind to regulatory elements in genes that encode cytokines and other relevant transcription factors (reviewed in (Padhan and Varma, 2010)).
Depending on whether an immediate response or long-lasting memory is needed, the organism can generate two different subsets of cells: effector or memory T cells, respectively. The mechanism underlying T cell differentiation into effector or memory fate remains controversial and is still to be fully understood. While some studies support the hypothesis that memory cells can differentiate in a linear developmental fashion directly from effector cells (Bannard et al., 2009), it has also been proposed that the progeny of activated T cells is not homogenous compositionally leading to differences in fate potential. The latter hypothesis is supported by the evidence that following antigen presentation by the APC, the mitotic T cell shows an asymmetric distribution of polarity proteins – such as aPKC and Par3 – resulting in its asymmetric division and an uneven segregation of molecular cell fate determinants between the daughter cells, ultimately leading to two daughter cells with two different fate potentials: effector and memory (Chang et al., 2007; Oliaro et al., 2010).
The IS therefore plays a role in T cell differentiation by modulating the trafficking of transcription factors between nucleus and cytoplasm and by regulating the asymmetric division of the T cell.
Signal modulation and termination
Though it has been proposed that the cSMAC plays a role in the temporal regulation of the synapse (Varma et al., 2006), with TCR signaling down-regulation and termination taking place at the cSMAC through internalization of the TCR-pMHC, it has also been suggested that the cSMAC may enhance the stimulatory effect of a weak TCR agonist, and thus that the balance between signaling and termination is dependent on the quality of the specific antigen (Cemerski et al., 2008; Lee et al., 2003). A potential pathway to resolution of this controversy is the observation that there are actually two compartments within the cSMAC, a TCR enriched inner cSMAC compartment that is dependent upon ubiquitin recognition for signal termination (Vardhana et al., 2010) and a CD28 enriched outer cSMAC compartment that is active in TCR and CD28 signaling (Tseng et al., 2008; Yokosuka et al., 2008). It is possible that the use of weak TCR ligands results in generation of an outer cSMAC without an inner cSMAC due to low ubiquitination.
The continued TCR signaling through new microcluster formation or an active, outer cSMAC compartment is important for T cell “deceleration” or the “stop” signal of the motile T cell, to ensure the engagement of a longer duration with the APC (Dustin et al., 1997; Sims et al., 2007; Skokos et al., 2007). Cytoplasmic Ca2+ increases is required for rapid deceleration in some conditions in vivo (Skokos et al., 2007; Waite et al., 2013).
Mechanotransduction
Other factors appear to play a role in T cell activation. Mechanical forces have been shown to play an important role in several biological processes such as cell differentiation, mitosis and migration of certain cell types (reviewed in (Eyckmans et al., 2011)) and the T cell appears to be no exception (Kim et al., 2012). Several groups have shown that the T cell is responsive to mechanical forces that can trigger T cell activation: (Husson et al., 2011; Judokusumo et al., 2012; Kim et al., 2009; Li et al., 2010). The application of lateral, but not normal, forces to the TCR using a monoclonal anti-CD3 antibody that is poorly stimulatory even when presented on a particle, resulted in an increase in cytoplasmic Ca2+ levels, suggesting that the TCR receptor is an anisotropic mechanosensor (Kim et al., 2009). Li et al. have used an APC system expressing an elongated ligand against CD3 that in spite of binding to the TCR does not induce signaling under static conditions, making it a good model to test whether physical forces can trigger TRC signaling (Li et al., 2010). They observed an increase in intracellular Ca2+ levels when a force was applied normal to the T cell surface. The direction of activation of force is distinct in this study compared to the Kim et al study above. The same effect was observed when T cells were pulled away from the APC (Li et al., 2010). Husson et al. have also shown, using a biomembrane force probe, that T cells do generate pushing and pulling forces when their TCR is engaged by anti-CD3 (Husson et al., 2011). In a physiological setting, such mechanical forces could easily be exerted on the TCR as it binds pMHC during migration and scanning of APC surfaces in search for the cognate pMHC and T cell detachment, which can create tensile mechanical forces as the T cell moves in an actin-dependent manner with respect to the APC (receptor deformation model), however, there is no experimental support for this (Ma and Finkel, 2010; Ma et al., 2008). It is possible that mechanical forces applied to the TCR may play a role in dislodging the ITAM motifs from the inner leaflet of the plasma membrane making the ITAM motifs more accessible to be phosphorylated, initiating the signaling cascade, with or without the participation of actin (Xu et al., 2008). More studies will be required to address these hypotheses. The lateral transport of microclusters towards the center of the synapse may also generate force on the TCR and LFA-1 (Kaizuka et al., 2007; Zhu et al., 2008).
Additional recent studies have aimed at integrating mechanical stimuli in T cell culture, by manipulating the substrate elastic modulus (stiffness), as a way to direct T cell response, using mouse (Judokusumo et al., 2012) and human (O’Connor et al., 2012) T cells. These groups have shown that there is indeed a correlation between the substrate stiffness and the T cell response. Mouse CD4+ T cells cultured on polyacrylamide gels with increasing elastic modulus coated with antibodies against CD3 and CD28 (mimicking the APC) secreted more IL-2. The observed difference in levels of IL-2 secretion by T cells cultured in gels of different stiffness was abolished by blebbistatin, which suggests that actomyosin cytoskeleton is involved in the T cell response to mechanical stimuli. In this study it was also reported that when the anti-CD28 antibody tethered to the polyacrylamide substrate of different elastic modulus and the anti-CD3 antibody was immobilized onto beads (of constant rigidity) the difference in IL-2 secretion was no longer observed (unlike the inverse situation), supporting Li et al. hypothesis that the mechanical force acts through the TCR. Consistently with the increased levels of IL-2 produced by T cells cultured on stiffer substrates, also clusters of phosphorylated ZAP70 (pZAP70), phosphorylated Src family kinase proteins (Lck and Fyn) and phosphorylated proline rich tyrosine kinase-2 (pPYK2) were observed at the T cell-substrate interface, contrary to what was observed in cells cultured in softer substrates where no (pZAP70 and pSFK) or only a small number of clusters (pPYK2) was detected. The observation that blebbistatin induced a small decrease in levels of pPYK2 across substrates of different rigidities suggests that PYK2 responds to cell contractility and may contribute to T cell mechanosensing (Judokusumo et al., 2012). One important aspect to take into account is that a different cellular response may not necessarily imply a mechanical response, but may result from a difference in chemistry of ligand presentation among substrates of different elastic modulus (Trappmann et al., 2012). Interestingly, PKY2, which is structurally related to the mechanotransducer focal adhesion kinase (FAK), appears to be critical for CD8 T cell short-lived effector fate, as its deficiency results in the impairment of T cell activation and the loss of this type of cell CD8+ effector cells, supporting mechanotransduction as an important player in T cell signaling (Beinke et al., 2010).
An approach similar to the ones developed by Judokusumo et al. and O’Connor et al., was used to investigate the effect of cell culture antigen presenting substrates of different stiffness (polyacrylamide gels) on B cells and led to the conclusion that B cells are also affected by the substrate stiffness: mouse B cells cultured in stiffer substrates showed a stronger activation (Wan et al., 2013). Another study also on B cells suggested that the B cells mechanically test the strength of antigen binding discriminating antigen affinities for development of high-affinity antibodies (Natkanski et al., 2013). Besides substrate stiffness, other parameters such as topology and protein distribution and molecular motility remain to be accessed regarding their role on the IS and T cell activation.
T cell activation systems and imaging tools
A good understanding of the spatial organization of the signaling molecules and how they are distributed in the plasma membrane and connected to downstream pathways is crucial to understand the underlying mechanisms of T cell activation.
A number of tools has been developed and optimized and has been increasingly used to study the IS.
The development of planar TCR activation systems has allowed for the visualization of the dynamic protein distribution during the IS formation and T cell activation.
Supported lipid bilayers (SLBs)
SLBs consist of a self-assembled lipid bilayers on a solid substrate reconstituted with an adhesion molecule like ICAM-1, antigenic pMHC or with anti-CD3 antibody, aiming at mimicking the phospholipid bilayer of the APC membrane (Dustin et al., 2007; Groves and Dustin, 2003). SLB can be formed on planar substrates, which is ideal for imaging, or on glass beads, which is useful for functional assays (Gay et al., 1986). This system was utilized sparingly through the 1980’s for studies on Fc receptors and the T cell receptor (McConnell et al., 1986), but the use of lipid anchored proteins that are mobile in bilayers with fluorescence microscopy has lead to expanded use (Dustin et al., 1996). SLBs capture the lateral mobility of natural ligands that are involved in the IS allowing T cells to conjugate with activating proteins, as if recognizing an APC, and forming an IS. The mobility of this systems allows the reproduction of the IS reorganization as it enables the translocation of ligated protein clusters. SLBs provide not only a planar orientation for better visualization of the synapse but also allows for the control of antigen density for TCR triggering and study of its effect in IS formation.
Others have aimed at mimicking T cell-APC interactions by seeding T cells on planar multicomponent protein substrates with lithographically defined patterns of tethered TCR ligands (anti-CD3 antibody) and the adhesion molecule ICAM-1 attempting to reproduce the microscale organization of the IS (Doh and Irvine, 2006). This approach offers the possibility of gaining control over the IS geometry and has been expanded so that in addition to the distribution of anti-CD3, also that of the costimulatory CD28 ligand could be controlled, against a background of ICAM-1 through multiple rounds of microcontact printing. This approach has allowed the study of how T cells respond differently to different patterns and how the costimulatory signal can affect this response (Shen et al., 2008).
Mobility of proteins on the SLB, or restriction of their mobility when immobilized onto substrates, may not represent the physiological conditions of the APC/T cell interface and may influence signaling events considerably. In order to control lateral mobility of molecules in the supported membrane, Some studies have used fabricated barriers onto the underlying substrate. These engineered supported membranes can lead to new insights into our understanding of the IS. Mossman et al. have imposed geometric constraints on the formation of the IS by using supported bilayer membranes with nanometer-scale structures onto the underlying substrate and have shown restriction of the motion of TCR clusters to the center of the IS enhanced TCR signaling, i.e. TCR microclusters mechanically trapped in the periphery of the IS resulted in prolonged signaling from the TCR clusters, which is consistent with the hypothesis that translocation of the TCR clusters towards the the cSMAC acts as a signal regulation mechanism (Mossman et al., 2005). Lemond et al. have used SLB reconstituted substrates patterned with molecular mazes of chromium, which enabled the study of TCR dynamics. In this system, microclusters were not permanently trapped by the barriers and displayed long range movement on the cell surface, leading to insightful results described above. Conversely, the use of grids instead of maze lines also affected the mobility of TCR but caused TCR to be confined in a fixed position (DeMond et al., 2008). Other groups have tried to engineer supported membranes introducing patterns to control molecule diffusion (reviewed in (Yu and Groves, 2010)).
Other model systems that aim at mimicking the APCs include non-planar surfaces such as biomimetic droplets – liquid colloids in the form of droplets grafted with specific molecules. This system enabled the visualization of the binding molecules on the T cell and the droplet and has been shown to trigger the T cell signaling cascade (Bourouina et al., 2012).
The introduction into non-immune cells of genes encoding the TCR and other proteins required for regulation of its phosphorylation and conjugation of these cells with an APC (James and Vale, 2012) and red blood cells directly coated with pMHC via biotin-streptavidin coupling, serving as an artificial APC (Huang et al., 2010), have also been used as reconstituted T cell activation systems to study initial TCR triggering events (and also as force sensors in the latter case).
Because the IS is based on the communication between two cells and localized to their contact interface, visualization of the IS and subsequent events in the cells relies on the use of specific imaging techniques.
Total internal reflection fluorescence microscopy (TIRF)
TIRF offers the advantage of a much reduced background fluorescence by illuminating only 100<200nm into the cell, enabling the generation of high-resolution images of the plane between the T cell and APC, and has thus been used to study the organization of the IS. TIRF is routinely used to study the T cell interface with SLBs, where the well-defined planar interface facilitates high-resolution imaging. TIRF can also be combined with the techniques described below.
Other imaging techniques that allow for visualization at a molecular nanometer scale have been developed and extremely useful in gaining new insights in the field; these include photoactivated localization microscopy (PALM)(Betzig et al., 2006) and stochastic optical reconstruction microscopy (STORM)(Rust et al., 2006), which can be used for detection and precise localization of single molecules, and fluorescent resonance energy transfer (FRET)(Joo et al., 2008; Moerner, 2007) that enables single molecule tracking and provide high-resolution information on dynamic molecular processes.
Advances in super-resolution fluorescence microscopy applied to the IS have been key in gaining further insights into several aspects of TCR signaling. This tool has been particularly relevant in discerning between different models for 1) LAT distribution upon TCR engagement, in membrane “protein islands” (Lillemeier et al., 2010) versus cytoplasmic pool (Williamson et al., 2011)) and arrangement of other signaling molecules (Hsu and Baumgart, 2011; Sherman et al., 2011), 2) role of ITAM phosphorylation in the coordination of distinct TRC-driven pathways (Guy et al., 2013), 3) clustering mechanism of Lck driven by conformational changes of Lck following TCR activation (Rossy et al., 2013a), 4) mechanism for release of intracellular domains of the CD3 from the inner leaflet of the plasma membrane (Gagnon et al., 2012; Shi et al., 2013; Zhang et al., 2011) (reviewed in (Rossy et al., 2013b)).
One of the challenges in imaging the IS is to have it positioned in the horizontal imaging plane so that the microcluster localization can be properly resolved. To manipulate the T cell-APC conjugates and to allow for vertical cell orientation, laser tweezers or laser trap have been successfully used (Oddos et al., 2008). This system however presents some limitations such as cellular phototoxicity complications and/or thermotoxicity due to heating of the medium, and does not allow for high-throughput analysis due to its laborious nature.
To facilitate high-resolution imaging of the IS overcoming these limitations, Biggs et al. developed microfabricated substrates (micropit arrays in PDMS) to sequester single T cell-APC conjugates, not substrate-adhered or influenced by interaction with adjacent cells, so that the IS forms in the horizontal imaging plane. This experimental design made use of K562-based APCs (K562 cells transduced to express an array of stimulatory and costimulatory molecules used to activate and expand T cells) that were initially sequestered in the fabricated micropits, followed by loading of T cells. This system allowed for large-scale imaging analysis of the IS interface of T cell-APC conjugates both in fixed and live samples, as multiple cell conjugates can be analyzed within a single imaging field (Biggs et al., 2011).
The development of artificial substrates to mimic the T cell-APC interface has become a main focus in the IS field, with the generation of minimal systems with greater control and high resolution. The use of such substrates will provide a system that allows dissection of the molecular mechanism underlying the IS, in particular the requirements regarding the surface molecules’ density, precise distribution and mechanical and topological characteristics of the surface where the APC surface molecules lay. Future approaches will aim at improving the antigen presenting substrate regarding the resolution of surface molecules as well as the substrate’s fluidity, so that the dynamics of the IS can be accurately reproduced.
Molecular imaging techniques continue to improve and will undoubtedly lead to a better understanding of the IS – its formation and subsequent T cell activation downstream consequences. The ability to analyze single molecules will enable a more accurate capture of the dynamics throughout these events not only in fixed but also living cells.
CONCLUDING REMARKS AND PERSPECTIVES
IS evolution
The two broad arms of the immune system, i.e. innate and adaptive, appear to have arisen at different timescales during evolution (Cooper and Alder, 2006). While all multi-cellular organisms have innate defense mechanisms and innate-like cells, the adaptive system is found primarily in vertebrates. Adaptive immune cell antigen receptors exhibit selective reactivity to various antigens and these utilize somatic recombination mechanisms to generate a diversity of receptor recognition specificities. Needless to say that the cell-cell conjugate formation involving the mechanism of recognition or the classical “immunological synapse” has thus been studied extensively in vertebrate systems. Although theoretically the IS can exist between any two immune cells, the antigen sampling by an adaptive immune cell via interaction with another immune cell has been the focus of extensive research during the past decade (Flajnik and Kasahara, 2010).
The components of the innate immune system such as innate cells expressing pattern recognition receptors are found in the entire animal kingdom. The evidence of adaptive immune system emerged first in jawless vertebrates, such as lamprey and hagfish, when the immunization with specific antigens resulted in production of specific agglutinins. Interestingly, instead of Ig-domain rich immunoreceptors, as found in lymphocytes of higher vertebrates, lymphoid cells in these organisms have Variable Lymphocyte Receptors (VLRs) rich in Leucine containing repeats (Boehm et al., 2012). Lamprey has two VLR genes – VLRA and VLRB - which exhibit allelic exclusion and encode membrane-bound and secreted forms of the VLRs, respectively. The adaptive immune system in higher vertebrates includes cells expressing BCR, TCR and MHC molecules, where these molecules participate in specialized mechanism of antigen recognition and synapse formation. It will be interesting to know if mechanotransduction is common to both the LRR and Ig-superfamily antigen receptors. This could make sense in that lymphocytes in both systems are freely migrating cells derived from mesenchyme. Cells from this tissue lineage utilize integrins to measure the physical properties of the environment and apply forces that are important in formation of solid tissues. Thus, a key mechanism in immune triggering maybe a common sensory module handed down to immune cells through evolution (Dobereiner et al., 2006).
IS and disease
Cellular-based immunotherapy relies on providing a robust source of responsive cells to target the disease, adoptive transfer being an important approach (June, 2007a). One advantage of applying adoptive T cell therapy in cancer is that T cells can have the ability to specifically target tumor cells through recognition of tumor proteins presented on the target cell surface. Provided that relevant tumor-specific antigens are identified, antigen-specific T cells can be generated and expanded ex vivo in a bioreactor system. This approach, though specific can be costly and labor intensive. An alternative approach is based on the activation and expansion of polyclonal T cells and relies on the presence of tumor-specific T cells within the polyclonal pool - technically more rapid and feasible but depends on the number of T cells within the expanded population that are tumor-specific and responsive to tumor-antigens presented by the APCs in vivo (reviewed in (June, 2007b). Adoptive immunotherapy can be based on allogeneic donor leukocytes, which through graft-versus-leukemic effects can be successful in patients with myeloid leukemia (Loren and Porter, 2006). To resolve the complications associated with graft-versus-host disease, autologous immunotherapy is a promising approach. This approach relies on the use of patient cells that need to be expanded in culture systems ex vivo to allow the T cells to proliferate before being reintroduced into the same patient. Though APCs could be used for this purpose they may not be controllable enough to achieve optimal differentiation of the patient T cells, whose long-term persistence and efficacy seem to be key. Therefore, the current protocol for expansion of patient T cells ex vivo, used in early-phase clinical trials, is based on the use of cell-size magnetic beads coated with antibodies against CD3 and CD28 (Brentjens et al., 2011; Kalos et al., 2011; Levine et al., 1997; Rapoport et al., 2005; Scholler et al., 2012). Efforts to replace the natural APC and reproduce the IS details in a carefully designed APC-mimetic system are being made not only to gain a better understanding of the IS but also to better control ex vivo T cell expansion, directing or enhancing T cell function of patients under certain pathological conditions. Engineered surfaces have already been used to show that T cells can respond to microscale geometry of the IS, using controled ligand localization (Shen et al., 2008), and to stiffness (Judokusumo et al., 2012; O’Connor et al., 2012), and appear to be superior to the currently used anti-CD3/CD28 antibody coated beads, suggesting that these fabricated surfaces may mimic the natural T cell:APC interface more closely.
T cells also have the capability of being genetically engineered during ex vivo manipulation using TCRs or chimeric antigen receptors (CARs) to confer tumor specificity. This is achieved by linking a tumor recognition domain, most commonly the antigen-specific domain of antibodies, to molecules involved in signaling T cell effector function, leading to T cell granule and release and cytotoxicity, cytokine production and proliferation. It is crucial that CAR-engineered T cells selectively and efficiently target the malignant tissue without harming normal cells in vivo. One major advantage of this approach is that CARS are not MHC-restricted and can be applied to different patients expressing the same target molecule. Lentiviral and retroviral transduction has been used to introduce CARs with costimulatory domains (to increase antitumor activity in vivo) into T cells that are expanded for adoptive transfer. Important factors for the optimization of CAR design include costimulation, density of target molecules in the tumor cell for effective T cell signaling, accessibility, and affinity of the engineered T cell and CAR molecules to the target tumor cell membrane (reviewed in (Riddell et al., 2013)). This approach has proven promising in pilot clinical trials for treatment of patients with chronic myeloid leukemia (CLL), where T cells are removed from the patient, modified to express the CAR, expanded, and reinfused into the patient (Kalos et al., 2011), (Porter et al., 2011). Some of the current studies on CARs include the development of safety strategies through incorporation of a conditional suicide gene or regulated expression of the tumor-targeting receptor, and the study of the effect of composition of CAR T cell products (also reviewed in (Riddell et al., 2013)).
A better understanding of the IS and how it is formed in healthy individuals can be extremely insightful in the study and design of appropriate treatment of certain pathologies, where the IS may show defects. T cells of CLL patients show defects in the formation of the IS with APCs – this synapse dysfunction appears to result both from CLL cells having poor APC function and defective actin polymerization in T cells from patients with CLL (Ramsay et al., 2008). Ramsay et al. observed, using cells of CLL patients, that the conjugation of T cells with APCs and subsequent polarization of F-actin, and recruitment of key regulatory proteins to the IS contact site (TCRs, adhesion molecules and actin cytoskeleton proteins) were inhibited. These defects were observed both in T cells from CLL patients and T cells from healthy individuals that had been in contact with CLL cells, these defects could be reduced using the immunomodulating drug lenalidomide that appears to repair F-actin polymerization and signaling at the IS. This finding can have implications in the treatment of CLL patients both through autologous and allogeneic immunotherapy approaches and indicates that the repair of IS defects can be an important step in improving cancer immunotherapy.
As mentioned above, granule exocytosis-mediated cytotoxicity is the major effector function of CD8 T cells in adaptive immunity. Upon association of the CTL with the target cell and TCR engagement, lytic granules move to the IS and ultimately fuse with the effector cell plasma membrane to release cytolytic proteins. This lytic activity has been shown to be defective in human cancer and mouse human models (Frey and Monu, 2008). Lee et al. have reported that tumor-derived gangliosides inhibit lytic function in CD8 CTLs by impeding the process of TCR-induced lytic granule release (Lee et al., 2012).
In the context of HIV infection, components of the IS may facilitate latency as well as active infection. In addition, the asymmetric cell division in response to T cell stimulation by the IS could be a mechanism by which HIV infection can be maintained in a latent state and at the same time generating progeny that actively express HIV virus. This may suggest that manipulation of the IS may lead to establishment and maintenance of latency of the virus (reviewed in (Kulpa et al., 2013)).
Several immune diseases - autoimmune diseases - result from the recognition of self-pMHC by the organism’s T cells. These T cells, with self-reactive TCRs, manage to escape negative selection and yet are able to respond to self-antigen with enough strength in the target organ. ISs formed by self-reactive T cells of patients with active autoimmune pathologies (such as multiple sclerosis and type 1 diabetes), though unusual and showing a reduced formation of the cSMAC and reduced pMHC recruitment, still appear to be able to sustain active TCR signaling (Schubert et al., 2012), possibly due to sustained motility of the T cells, and similarly to ISs formed with a mutant weak pMHC agonist (Vardhana et al., 2010). The observation that T cells with altered IS can have pathogenic potential strongly supports the need for a better understanding of the IS, both in healthy individuals and immune disease patients.
Future perspectives
There are still a number of fundamental questions about all levels of organization in the IS. It is unclear how TCR triggering works and its now more and more likely that there will be multiple ways this can be initiated. It will be interesting to know if TCR microclusters are a common pathway that are formed regardless of the triggering mechanism. More studies will be needed to address questions such as the relevance of synapse stability in the T cell response, the molecular determinants involved, the cross-talk in TCR signaling pathways that will lead to diversification of T cell functional responses, and other open questions. Moreover, the combination of both the understanding of the molecular players and the development of new T cell activation systems will enable the generation of platforms that can ultimately be used to manipulate T cell response, to optimize T cell expansion and differentiation into a phenotype of choice. The ability to manipulate the immune response is particularly important in designing effective immunotherapy strategies. Cellular immunotherapy is becoming a preferred approach in the treatment of patients with cancer, thus all efforts that may improve our understanding of the events that occur from the formation of the IS to the T cell functional response will be key in the development and refinement of these therapeutic strategies.
Figure 1.
Components of the immunological synapse. The mature immunological synapse is composed of three SMACs and at least two types of microclusters (MC). The dSMAC (outer yellow) is f-actin rich and is the location in which TCR (green) and LFA-1 (red) microclusters form. The pSMAC is a perforated network of engaged LFA-1 (red) that contains some TCR microclusters in the perforations. The outer cSMAC (pale green) contains a small amount of TCR and accumulated all the CD28 (over time). It is active in signaling. The inner cSMAC (green) is dependent upon ubiquitin recognition and is not active in signaling. There is inward movement of MCs, inward movement of LFA-1 in the pSMAC (unpublished observations) and there is inward movement of TCR from the outer to the inner cSMAC.
Contributor Information
Silvia Curado, Email: silvia.curado@med.nyu.edu.
Michael L. Dustin, Email: michael.dustin@kennedy.ox.ac.uk.
References
- Alarcon B, Mestre D, Martinez-Martin N. Immunology. 2011;133:420–5. doi: 10.1111/j.1365-2567.2011.03458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen CD, Okada T, Tang HL, Cyster JG. Science. 2007;315:528–31. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- Alon R, Dustin ML. Immunity. 2007;26:17–27. doi: 10.1016/j.immuni.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Angus KL, Griffiths GM. Curr Opin Cell Biol. 2013;25:85–91. doi: 10.1016/j.ceb.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babich A, Li S, O’Connor RS, Milone MC, Freedman BD, Burkhardt JK. J Cell Biol. 2012;197:775–87. doi: 10.1083/jcb.201201018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J, Steinman RM. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Bannard O, Kraman M, Fearon DT. Science. 2009;323:505–9. doi: 10.1126/science.1166831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barda-Saad M, Braiman A, Titerence R, Bunnell SC, Barr VA, Samelson LE. Nat Immunol. 2005;6:80–9. doi: 10.1038/ni1143. [DOI] [PubMed] [Google Scholar]
- Barreira da Silva R, Munz C. Cell Mol Life Sci. 2011;68:3505–18. doi: 10.1007/s00018-011-0801-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista FD, Iber D, Neuberger MS. Nature. 2001;411:489–94. doi: 10.1038/35078099. [DOI] [PubMed] [Google Scholar]
- Beemiller P, Jacobelli J, Krummel MF. Nat Immunol. 2012;13:787–95. doi: 10.1038/ni.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinke S, Phee H, Clingan JM, Schlessinger J, Matloubian M, Weiss A. Proc Natl Acad Sci U S A. 2010;107:16234–9. doi: 10.1073/pnas.1011556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642–5. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Biggs MJ, Milone MC, Santos LC, Gondarenko A, Wind SJ. J R Soc Interface. 2011;8:1462–71. doi: 10.1098/rsif.2011.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T, McCurley N, Sutoh Y, Schorpp M, Kasahara M, Cooper MD. Annu Rev Immunol. 2012;30:203–20. doi: 10.1146/annurev-immunol-020711-075038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourouina N, Husson J, Hivroz C, Henry N. Langmuir. 2012;28:6106–13. doi: 10.1021/la300398a. [DOI] [PubMed] [Google Scholar]
- Breart B, Lemaitre F, Celli S, Bousso P. J Clin Invest. 2008;118:1390–7. doi: 10.1172/JCI34388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentjens RJ, Riviere I, Park JH, Davila ML, Wang X, Stefanski J, Taylor C, Yeh R, Bartido S, Borquez-Ojeda O, Olszewska M, Bernal Y, Pegram H, Przybylowski M, Hollyman D, Usachenko Y, Pirraglia D, Hosey J, Santos E, Halton E, Maslak P, Scheinberg D, Jurcic J, Heaney M, Heller G, Frattini M, Sadelain M. Blood. 2011;118:4817–28. doi: 10.1182/blood-2011-04-348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt JK, Carrizosa E, Shaffer MH. Annu Rev Immunol. 2008;26:233–59. doi: 10.1146/annurev.immunol.26.021607.090347. [DOI] [PubMed] [Google Scholar]
- Campi G, Varma R, Dustin ML. J Exp Med. 2005;202:1031–6. doi: 10.1084/jem.20051182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli S, Lemaitre F, Bousso P. Immunity. 2007;27:625–34. doi: 10.1016/j.immuni.2007.08.018. [DOI] [PubMed] [Google Scholar]
- Cemerski S, Das J, Giurisato E, Markiewicz MA, Allen PM, Chakraborty AK, Shaw AS. Immunity. 2008;29:414–22. doi: 10.1016/j.immuni.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, Killeen N, Orange JS, Russell SM, Weninger W, Reiner SL. Science. 2007;315:1687–91. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- Chaudhri G, Quah BJ, Wang Y, Tan AH, Zhou J, Karupiah G, Parish CR. Proc Natl Acad Sci U S A. 2009;106:14984–9. doi: 10.1073/pnas.0906554106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Lou J, Evans EA, Zhu C. J Cell Biol. 2012;199:497–512. doi: 10.1083/jcb.201201091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri K, Parker M, Milicic A, Cole DK, Shaw MK, Sewell AK, Stewart-Jones G, Dong T, Gould KG, van der Merwe PA. J Biol Chem. 2009;284:26096–105. doi: 10.1074/jbc.M109.039966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA. Nature. 2005;436:578–82. doi: 10.1038/nature03843. [DOI] [PubMed] [Google Scholar]
- Coombs D, Kalergis AM, Nathenson SG, Wofsy C, Goldstein B. Nat Immunol. 2002;3:926–31. doi: 10.1038/ni838. [DOI] [PubMed] [Google Scholar]
- Cooper MD, Alder MN. Cell. 2006;124:815–22. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Cruz-Orcutt N, Houtman JC. Mol Immunol. 2009;46:2274–83. doi: 10.1016/j.molimm.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Cyster JG, Shotton DM, Williams AF. EMBO J. 1991;10:893–902. doi: 10.1002/j.1460-2075.1991.tb08022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFord-Watts LM, Dougall DS, Belkaya S, Johnson BA, Eitson JL, Roybal KT, Barylko B, Albanesi JP, Wulfing C, van Oers NS. J Immunol. 2011;186:6839–47. doi: 10.4049/jimmunol.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMond AL, Mossman KD, Starr T, Dustin ML, Groves JT. Biophys J. 2008;94:3286–92. doi: 10.1529/biophysj.107.119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobereiner HG, Dubin-Thaler BJ, Hofman JM, Xenias HS, Sims TN, Giannone G, Dustin ML, Wiggins CH, Sheetz MP. Phys Rev Lett. 2006;97:038102. doi: 10.1103/PhysRevLett.97.038102. [DOI] [PubMed] [Google Scholar]
- Doh J, Irvine DJ. Proc Natl Acad Sci U S A. 2006;103:5700–5. doi: 10.1073/pnas.0509404103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin ML. Annu Rev Cell Dev Biol. 2008;24:577–96. doi: 10.1146/annurev.cellbio.24.110707.175226. [DOI] [PubMed] [Google Scholar]
- Dustin ML, Bromley SK, Kan Z, Peterson DA, Unanue ER. Proc Natl Acad Sci U S A. 1997;94:3909–13. doi: 10.1073/pnas.94.8.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin ML, Ferguson LM, Chan PY, Springer TA, Golan DE. J Cell Biol. 1996;132:465–74. doi: 10.1083/jcb.132.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin ML, Starr T, Varma R, Thomas VK. Curr Protoc Immunol. 2007;Chapter 18(Unit 18):13. doi: 10.1002/0471142735.im1813s76. [DOI] [PubMed] [Google Scholar]
- Edelson BT, Bradstreet TR, Hildner K, Carrero JA, Frederick KE, Kc W, Belizaire R, Aoshi T, Schreiber RD, Miller MJ, Murphy TL, Unanue ER, Murphy KM. Immunity. 2011;35:236–48. doi: 10.1016/j.immuni.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egen JG, Rothfuchs AG, Feng CG, Winter N, Sher A, Germain RN. Immunity. 2008;28:271–84. doi: 10.1016/j.immuni.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyckmans J, Boudou T, Yu X, Chen CS. Dev Cell. 2011;21:35–47. doi: 10.1016/j.devcel.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fooksman DR, Vardhana S, Vasiliver-Shamis G, Liese J, Blair DA, Waite J, Sacristan C, Victora GD, Zanin-Zhorov A, Dustin ML. Annu Rev Immunol. 2010;28:79–105. doi: 10.1146/annurev-immunol-030409-101308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey AB, Monu N. Immunol Rev. 2008;222:192–205. doi: 10.1111/j.1600-065X.2008.00606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon E, Schubert DA, Gordo S, Chu HH, Wucherpfennig KW. J Exp Med. 2012;209:2423–39. doi: 10.1084/jem.20120790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay D, Coeshott C, Golde W, Kappler J, Marrack P. J Immunol. 1986;136:2026–32. [PubMed] [Google Scholar]
- Gerard A, Khan O, Beemiller P, Oswald E, Hu J, Matloubian M, Krummel MF. Nat Immunol. 2013;14:356–63. doi: 10.1038/ni.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi SI, Rincon E, Avila-Flores A, Torres-Ayuso P, Almena M, Cobos MA, Albar JP, Merida I. Mol Biol Cell. 2011;22:4406–14. doi: 10.1091/mbc.E11-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, Wolf AJ, Bose N, Chan AS, Magee AS, Danielson ME, Weiss A, Vasilakos JP, Underhill DM. Nature. 2011;472:471–5. doi: 10.1038/nature10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. Science. 1999;285:221–7. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- Griffiths GM, Tsun A, Stinchcombe JC. J Cell Biol. 2010;189:399–406. doi: 10.1083/jcb.201002027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves JT, Dustin ML. J Immunol Methods. 2003;278:19–32. doi: 10.1016/s0022-1759(03)00193-5. [DOI] [PubMed] [Google Scholar]
- Guy CS, Vignali KM, Temirov J, Bettini ML, Overacre AE, Smeltzer M, Zhang H, Huppa JB, Tsai YH, Lobry C, Xie J, Dempsey PJ, Crawford HC, Aifantis I, Davis MM, Vignali DA. Nat Immunol. 2013;14:262–70. doi: 10.1038/ni.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TH, Banigan EJ, Christian DA, Konradt C, Tait Wojno ED, Norose K, Wilson EH, John B, Weninger W, Luster AD, Liu AJ, Hunter CA. Nature. 2012;486:545–8. doi: 10.1038/nature11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood NE, Batista FD. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Tane A, Yokosuka T, Sakata-Sogawa K, Sakuma M, Ishihara C, Tokunaga M, Saito T. Immunity. 2011;34:919–31. doi: 10.1016/j.immuni.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A. Genes Dev. 2003;17:2205–32. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- Hsu CJ, Baumgart T. PLoS One. 2011;6:e23586. doi: 10.1371/journal.pone.0023586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, Zhu C. Nature. 2010;464:932–6. doi: 10.1038/nature08944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson J, Chemin K, Bohineust A, Hivroz C, Henry N. PLoS One. 2011;6:e19680. doi: 10.1371/journal.pone.0019680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilani T, Vasiliver-Shamis G, Vardhana S, Bretscher A, Dustin ML. Nat Immunol. 2009;10:531–9. doi: 10.1038/ni.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JR, Vale RD. Nature. 2012;487:64–9. doi: 10.1038/nature11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA, Jr, Medzhitov R. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Annu Rev Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- Joyce S, Girardi E, Zajonc DM. J Immunol. 2011;187:1081–9. doi: 10.4049/jimmunol.1001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judokusumo E, Tabdanov E, Kumari S, Dustin ML, Kam LC. Biophys J. 2012;102:L5–7. doi: 10.1016/j.bpj.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- June CH. J Clin Invest. 2007a;117:1466–76. doi: 10.1172/JCI32446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- June CH. J Clin Invest. 2007b;117:1204–12. doi: 10.1172/JCI31446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizuka Y, Douglass AD, Varma R, Dustin ML, Vale RD. Proc Natl Acad Sci U S A. 2007;104:20296–301. doi: 10.1073/pnas.0710258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, June CH. Sci Transl Med. 2011;3:95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Shin Y, Brazin K, Mallis RJ, Sun ZY, Wagner G, Lang MJ, Reinherz EL. Front Immunol. 2012;3:76. doi: 10.3389/fimmu.2012.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Takeuchi K, Sun ZY, Touma M, Castro CE, Fahmy A, Lang MJ, Wagner G, Reinherz EL. The alphabeta T cell receptor is an anisotropic mechanosensor. J Biol Chem. 2009:31028–37. doi: 10.1074/jbc.M109.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa DA, Brehm JH, Fromentin R, Cooper A, Cooper C, Ahlers J, Chomont N, Sekaly RP. Immunol Rev. 2013;254:305–25. doi: 10.1111/imr.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Vardhana S, Cammer M, Curado S, Santos L, Sheetz MP, Dustin ML. Front Immunol. 2012;3:230. doi: 10.3389/fimmu.2012.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer A, Mosmann TR, Kupfer H. Proc Natl Acad Sci U S A. 1991;88:775–9. doi: 10.1073/pnas.88.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Wondimu A, Liu Y, Ma JS, Radoja S, Ladisch S. J Immunol. 2012;189:3521–7. doi: 10.4049/jimmunol.1201256. [DOI] [PubMed] [Google Scholar]
- Lee KH, Dinner AR, Tu C, Campi G, Raychaudhuri S, Varma R, Sims TN, Burack WR, Wu H, Wang J, Kanagawa O, Markiewicz M, Allen PM, Dustin ML, Chakraborty AK, Shaw AS. Science. 2003;302:1218–22. doi: 10.1126/science.1086507. [DOI] [PubMed] [Google Scholar]
- Leupin O, Zaru R, Laroche T, Muller S, Valitutti S. Curr Biol. 2000;10:277–80. doi: 10.1016/s0960-9822(00)00362-6. [DOI] [PubMed] [Google Scholar]
- Levine BL, Bernstein WB, Connors M, Craighead N, Lindsten T, Thompson CB, June CH. J Immunol. 1997;159:5921–30. [PubMed] [Google Scholar]
- Li YC, Chen BM, Wu PC, Cheng TL, Kao LS, Tao MH, Lieber A, Roffler SR. J Immunol. 2010;184:5959–63. doi: 10.4049/jimmunol.0900775. [DOI] [PubMed] [Google Scholar]
- Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. Nat Immunol. 2010;11:90–6. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Rhodes M, Wiest DL, Vignali DA. Immunity. 2000;13:665–75. doi: 10.1016/s1074-7613(00)00066-2. [DOI] [PubMed] [Google Scholar]
- Liu SK, Berry DM, McGlade CJ. Oncogene. 2001;20:6284–90. doi: 10.1038/sj.onc.1204771. [DOI] [PubMed] [Google Scholar]
- Longhi MP, Trumpfheller C, Idoyaga J, Caskey M, Matos I, Kluger C, Salazar AM, Colonna M, Steinman RM. J Exp Med. 2009;206:1589–602. doi: 10.1084/jem.20090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loren AW, Porter DL. Curr Opin Oncol. 2006;18:107–14. doi: 10.1097/01.cco.0000208781.61452.d3. [DOI] [PubMed] [Google Scholar]
- Ma Z, Finkel TH. Trends Immunol. 2010;31:1–6. doi: 10.1016/j.it.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Sharp KA, Janmey PA, Finkel TH. PLoS Biol. 2008;6:e43. doi: 10.1371/journal.pbio.0060043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado RA, Soriano MA, Perdomo LC, Sigrist K, Irvine DJ, Decker T, Glimcher LH. J Exp Med. 2009;206:877–92. doi: 10.1084/jem.20082900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell HM, Watts TH, Weis RM, Brian AA. Biochim Biophys Acta. 1986;864:95–106. doi: 10.1016/0304-4157(86)90016-x. [DOI] [PubMed] [Google Scholar]
- Mempel TR, Henrickson SE, Von Andrian UH. Nature. 2004;427:154–9. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- Miletic AV, Graham DB, Sakata-Sogawa K, Hiroshima M, Hamann MJ, Cemerski S, Kloeppel T, Billadeau DD, Kanagawa O, Tokunaga M, Swat W. PLoS One. 2009;4:e6599. doi: 10.1371/journal.pone.0006599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miletic AV, Sakata-Sogawa K, Hiroshima M, Hamann MJ, Gomez TS, Ota N, Kloeppel T, Kanagawa O, Tokunaga M, Billadeau DD, Swat W. J Biol Chem. 2006;281:38257–65. doi: 10.1074/jbc.M608913200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Hejazi AS, Wei SH, Cahalan MD, Parker I. Proc Natl Acad Sci U S A. 2004;101:998–1003. doi: 10.1073/pnas.0306407101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerner WE. Proc Natl Acad Sci U S A. 2007;104:12596–602. doi: 10.1073/pnas.0610081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Nature. 1998;395:82–6. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Immunity. 2007;27:203–13. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman KD, Campi G, Groves JT, Dustin ML. Science. 2005;310:1191–3. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- Natkanski E, Lee WY, Mistry B, Casal A, Molloy JE, Tolar P. Science. 2013;340:1587–90. doi: 10.1126/science.1237572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor RS, Hao X, Shen K, Bashour K, Akimova T, Hancock WW, Kam LC, Milone MC. J Immunol. 2012;189:1330–9. doi: 10.4049/jimmunol.1102757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddos S, Dunsby C, Purbhoo MA, Chauveau A, Owen DM, Neil MA, Davis DM, French PM. Biophys J. 2008;95:L66–8. doi: 10.1529/biophysj.108.143198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohl L, Mohaupt M, Czeloth N, Hintzen G, Kiafard Z, Zwirner J, Blankenstein T, Henning G, Forster R. Immunity. 2004;21:279–88. doi: 10.1016/j.immuni.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Oliaro J, Van Ham V, Sacirbegovic F, Pasam A, Bomzon Z, Pham K, Ludford-Menting MJ, Waterhouse NJ, Bots M, Hawkins ED, Watt SV, Cluse LA, Clarke CJ, Izon DJ, Chang JT, Thompson N, Gu M, Johnstone RW, Smyth MJ, Humbert PO, Reiner SL, Russell SM. J Immunol. 2010;185:367–75. doi: 10.4049/jimmunol.0903627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhan K, Varma R. Immunology. 2010;129:322–8. doi: 10.1111/j.1365-2567.2009.03241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauker MH, Hassan N, Noy E, Reicher B, Barda-Saad M. Sci Signal. 2012;5:rs3. doi: 10.1126/scisignal.2002423. [DOI] [PubMed] [Google Scholar]
- Poo WJ, Conrad L, Janeway CA., Jr Nature. 1988;332:378–80. doi: 10.1038/332378a0. [DOI] [PubMed] [Google Scholar]
- Porter DL, Levine BL, Kalos M, Bagg A, June CH. N Engl J Med. 2011;365:725–33. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quann EJ, Merino E, Furuta T, Huse M. Nat Immunol. 2009;10:627–35. doi: 10.1038/ni.1734. [DOI] [PubMed] [Google Scholar]
- Rak GD, Mace EM, Banerjee PP, Svitkina T, Orange JS. PLoS Biol. 2011;9:e1001151. doi: 10.1371/journal.pbio.1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay AG, Johnson AJ, Lee AM, Gorgun G, Le Dieu R, Blum W, Byrd JC, Gribben JG. J Clin Invest. 2008;118:2427–37. doi: 10.1172/JCI35017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport AP, Stadtmauer EA, Aqui N, Badros A, Cotte J, Chrisley L, Veloso E, Zheng Z, Westphal S, Mair R, Chi N, Ratterree B, Pochran MF, Natt S, Hinkle J, Sickles C, Sohal A, Ruehle K, Lynch C, Zhang L, Porter DL, Luger S, Guo C, Fang HB, Blackwelder W, Hankey K, Mann D, Edelman R, Frasch C, Levine BL, Cross A, June CH. Nat Med. 2005;11:1230–7. doi: 10.1038/nm1310. [DOI] [PubMed] [Google Scholar]
- Riddell SR, Jensen MC, June CH. Biol Blood Marrow Transplant. 2013;19:S2–5. doi: 10.1016/j.bbmt.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossy J, Owen DM, Williamson DJ, Yang Z, Gaus K. Nat Immunol. 2013a;14:82–9. doi: 10.1038/ni.2488. [DOI] [PubMed] [Google Scholar]
- Rossy J, Pageon SV, Davis DM, Gaus K. Curr Opin Immunol. 2013b;25:307–12. doi: 10.1016/j.coi.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Ruocco MG, Pilones KA, Kawashima N, Cammer M, Huang J, Babb JS, Liu M, Formenti SC, Dustin ML, Demaria S. J Clin Invest. 2012;122:3718–30. doi: 10.1172/JCI61931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Nat Methods. 2006;3:793–5. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatos CA, Doh J, Chakravarti S, Friedman RS, Pandurangi PG, Tooley AJ, Krummel MF. Immunity. 2008;29:238–48. doi: 10.1016/j.immuni.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage PT, Varghese LM, Martinelli R, Sciuto TE, Kamei M, Dvorak AM, Springer TA, Sharpe AH, Carman CV. J Immunol. 2012;188:3686–99. doi: 10.4049/jimmunol.1102594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasahara Y, Rachid R, Byrne MJ, de la Fuente MA, Abraham RT, Ramesh N, Geha RS. Mol Cell. 2002;10:1269–81. doi: 10.1016/s1097-2765(02)00728-1. [DOI] [PubMed] [Google Scholar]
- Scholler J, Brady TL, Binder-Scholl G, Hwang WT, Plesa G, Hege KM, Vogel AN, Kalos M, Riley JL, Deeks SG, Mitsuyasu RT, Bernstein WB, Aronson NE, Levine BL, Bushman FD, June CH. Sci Transl Med. 2012;4:132ra53. doi: 10.1126/scitranslmed.3003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert DA, Gordo S, Sabatino JJ, Jr, Vardhana S, Gagnon E, Sethi DK, Seth NP, Choudhuri K, Reijonen H, Nepom GT, Evavold BD, Dustin ML, Wucherpfennig KW. J Exp Med. 2012;209:335–52. doi: 10.1084/jem.20111485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AS, Dustin ML. Immunity. 1997;6:361–9. doi: 10.1016/s1074-7613(00)80279-4. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Qu B, Hoth M, Feske S. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Thomas VK, Dustin ML, Kam LC. Proc Natl Acad Sci U S A. 2008;105:7791–6. doi: 10.1073/pnas.0710295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman E, Barr V, Manley S, Patterson G, Balagopalan L, Akpan I, Regan CK, Merrill RK, Sommers CL, Lippincott-Schwartz J, Samelson LE. Immunity. 2011;35:705–20. doi: 10.1016/j.immuni.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Bi Y, Yang W, Guo X, Jiang Y, Wan C, Li L, Bai Y, Guo J, Wang Y, Chen X, Wu B, Sun H, Liu W, Wang J, Xu C. Nature. 2013;493:111–5. doi: 10.1038/nature11699. [DOI] [PubMed] [Google Scholar]
- Sims TN, Soos TJ, Xenias HS, Dubin-Thaler B, Hofman JM, Waite JC, Cameron TO, Thomas VK, Varma R, Wiggins CH, Sheetz MP, Littman DR, Dustin ML. Cell. 2007;129:773–85. doi: 10.1016/j.cell.2007.03.037. [DOI] [PubMed] [Google Scholar]
- Skokos D, Shakhar G, Varma R, Waite JC, Cameron TO, Lindquist RL, Schwickert T, Nussenzweig MC, Dustin ML. Nat Immunol. 2007;8:835–44. doi: 10.1038/ni1490. [DOI] [PubMed] [Google Scholar]
- Smoligovets AA, Smith AW, Wu HJ, Petit RS, Groves JT. J Cell Sci. 2012;125:735–42. doi: 10.1242/jcs.092825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM, Hawiger D, Nussenzweig MC. Annu Rev Immunol. 2003;21:685–711. doi: 10.1146/annurev.immunol.21.120601.141040. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JC, Griffiths GM. Annu Rev Cell Dev Biol. 2007;23:495–517. doi: 10.1146/annurev.cellbio.23.090506.123521. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM. Nature. 2006;443:462–5. doi: 10.1038/nature05071. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JC, Salio M, Cerundolo V, Pende D, Arico M, Griffiths GM. BMC Biol. 2011;9:45. doi: 10.1186/1741-7007-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvain NR, Nguyen K, Bunnell SC. Sci Signal. 2011;4:ra14. doi: 10.1126/scisignal.2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trappmann B, Gautrot JE, Connelly JT, Strange DG, Li Y, Oyen ML, Cohen Stuart MA, Boehm H, Li B, Vogel V, Spatz JP, Watt FM, Huck WT. Nat Mater. 2012;11:642–9. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Mellman I. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- Tseng SY, Waite JC, Liu M, Vardhana S, Dustin ML. J Immunol. 2008;181:4852–63. doi: 10.4049/jimmunol.181.7.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun A, Qureshi I, Stinchcombe JC, Jenkins MR, de la Roche M, Kleczkowska J, Zamoyska R, Griffiths GM. J Cell Biol. 2011;192:663–74. doi: 10.1083/jcb.201008140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tybulewicz VL. Curr Opin Immunol. 2005;17:267–74. doi: 10.1016/j.coi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Ueda H, Morphew MK, McIntosh JR, Davis MM. Proc Natl Acad Sci U S A. 2011;108:17099–104. doi: 10.1073/pnas.1113703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhana S, Choudhuri K, Varma R, Dustin ML. Immunity. 2010;32:531–40. doi: 10.1016/j.immuni.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. Immunity. 2006;25:117–27. doi: 10.1016/j.immuni.2006.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite JC, Leiner I, Lauer P, Rae CS, Barbet G, Zheng H, Portnoy DA, Pamer EG, Dustin ML. PLoS Pathog. 2011;7:e1001326. doi: 10.1371/journal.ppat.1001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite JC, Vardhana S, Shaw PJ, Jang J-E, McCarl CA, Cameron TO, Feske S, Dustin ML. Eur J Immunol. 2013 doi: 10.1002/eji.201243255. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z, Zhang S, Fan Y, Liu K, Du F, Davey AM, Zhang H, Han W, Xiong C, Liu W. J Immunol. 2013;190:4661–75. doi: 10.4049/jimmunol.1202976. [DOI] [PubMed] [Google Scholar]
- Wang H, Kadlecek TA, Au-Yeung BB, Goodfellow HE, Hsu LY, Freedman TS, Weiss A. Cold Spring Harb Perspect Biol. 2010;2:a002279. doi: 10.1101/cshperspect.a002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson DJ, Owen DM, Rossy J, Magenau A, Wehrmann M, Gooding JJ, Gaus K. Nature immunology. 2011;12:655–62. doi: 10.1038/ni.2049. [DOI] [PubMed] [Google Scholar]
- Wu JN, Koretzky GA. Semin Immunol. 2004;16:379–93. doi: 10.1016/j.smim.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Xu C, Gagnon E, Call ME, Schnell JR, Schwieters CD, Carman CV, Chou JJ, Wucherpfennig KW. Cell. 2008;135:702–13. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Wu XS, Crites T, Hammer JA., 3rd Mol Biol Cell. 2012;23:834–52. doi: 10.1091/mbc.E11-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka T, Kobayashi W, Sakata-Sogawa K, Takamatsu M, Hashimoto-Tane A, Dustin ML, Tokunaga M, Saito T. Immunity. 2008;29:589–601. doi: 10.1016/j.immuni.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka T, Sakata-Sogawa K, Kobayashi W, Hiroshima M, Hashimoto-Tane A, Tokunaga M, Dustin ML, Saito T. Nat Immunol. 2005;6:1253–62. doi: 10.1038/ni1272. [DOI] [PubMed] [Google Scholar]
- Yu CH, Groves JT. Med Biol Eng Comput. 2010;48:955–63. doi: 10.1007/s11517-010-0634-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CH, Wu HJ, Kaizuka Y, Vale RD, Groves JT. PLoS One. 2010;5:e11878. doi: 10.1371/journal.pone.0011878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R, Cannon JL, Abraham RT, Way M, Billadeau DD, Bubeck-Wardenberg J, Burkhardt JK. J Immunol. 2003;171:1360–8. doi: 10.4049/jimmunol.171.3.1360. [DOI] [PubMed] [Google Scholar]
- Zhang H, Cordoba SP, Dushek O, van der Merwe PA. Proc Natl Acad Sci U S A. 2011;108:19323–8. doi: 10.1073/pnas.1108052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Luo BH, Xiao T, Zhang C, Nishida N, Springer TA. Mol Cell. 2008;32:849–61. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinselmeyer BH, Heydari S, Sacristan C, Nayak D, Cammer M, Herz J, Cheng X, Davis SJ, Dustin ML, McGavern DB. J Exp Med. 2013;210:757–74. doi: 10.1084/jem.20121416. [DOI] [PMC free article] [PubMed] [Google Scholar]