Abstract
Embryonic stem cells (ESCs) exhibit unrestricted and indefinite, but stringently controlled, proliferation and can differentiate into any lineage in the body. In the current study, we test the hypothesis that expression of ribosomal RNA (rRNA) and ribosomal protein genes (RPGs) contribute to the ability of hESCs to proliferate indefinitely. Consistent with the accelerated growth rate of hESCs, we find that hESC lines H1 and H9 both exhibit significantly higher levels of rRNA when compared to a panel of normal and cancer human cell lines. While many RPGs are expressed at levels that comparable to other human cell lines, a few RPGs also exhibit higher expression levels. In situ nuclear run-on assays reveal that both nucleoli in hESCs actively transcribe nascent rRNA. Employing genome-wide chromatin immunoprecipitation-deep sequencing and bioinformatics approaches, we discovered that RPGs are dominantly marked by the activating H3K4me3 histone mark in the G1, M and G2 phases of the cell cycle. Interestingly, the rDNA repeats are marked by the activating H3K4me3 only in the M phase, and repressive H3K27me3 histone mark in all three cell cycle phases. Bioinformatics analyses also reveal that Myc, a known regulator of cell growth and proliferation, occupies both the rRNA genes and RPGs. Functionally, down-regulation of Myc expression by siRNA results in a concomitant decrease in rRNA levels. Together, our results show that expression of rRNA, which is regulated by the Myc pluripotency transcription factor, and of RPGs in hESCs is associated with the activating H3K4me3 modification.
Keywords: Myc, H3K4me3, H3K27me3, rRNA, Bivalency
Introduction
Cell growth, size and proliferation are intimately linked through a coordinated regulation of ribosomal RNA and protein genes (Lempiainen & Shore 2009; Mayer et al. 2006; Dez & Tollervey 2004; Moss 2004). Human embryonic stem cells (hESCs) are characterized by their ability to proliferate indefinitely and differentiate into any lineage in the body (Chen & Daley 2008; De Los Angeles et al. 2015). The higher proliferative rate of hESCs is, in part, due to a shortened G1 phase of the cell cycle (Becker, Ghule, et al. 2006; Becker et al. 2010; Kapinas et al. 2013). However, it remains to be established whether ribosomal RNA and proteins are coordinately regulated to accommodate the faster growth rate of hESCs. Ribosomal RNA makes up nearly 85% of total cellular RNA and is an essential component of ribosomes (Boisvert et al. 2007). In human cells, hundreds of tandem repeats of rRNA genes, located on 5 acrocentric chromosomes and organized as nucleoli in the interphase nucleus, account for such high levels of rRNA expression (McStay & Grummt 2008; Pechmann et al. 2013). Interestingly, in eukaryotic cells only half of the rRNA gene copies account for transcription of cellular RNA [(Conconi et al. 1989; Lucchini & Sogo 1998)]. Several studies have shown that half of the rRNA genes are inactive, as indicated by DNA methylation and repressive histone marks. Furthermore, the stoichiometry between the RNA and protein complement of ribosomes is required for functional ribosomes (Reviewed in (Woolford & Warner 1991)). Because global protein translation plays a key role in cell growth, coordinate regulation of ribosomal RNA (rRNA) and protein gene (RPGs) expression in human cells is intimately linked with growth and proliferation potential of the cell (Mayer & Grummt 2006; Lempiainen & Shore 2009).
Myc is a key oncogene that is upregulated in the majority of human cancers (Hsieh et al. 2015; van Riggelen et al. 2010). It has been shown that Myc regulates gene transcription by RNA polymerases I, II and III, thus coordinating protein synthesis at multiple levels (Kenneth & White 2009; Arabi et al. 2005; Oskarsson & Trumpp 2005; Grandori et al. 2005; Gomez-Roman et al. 2003). Interestingly, Myc is also an essential component of the Yamanaka pluripotency transcription factor cocktail, and is required for efficient induction of pluripotency in adult cells (Takahashi & Yamanaka 2006). Although pluripotency can be induced with only Nanog, Oct4 and Klf4, the presence of Myc drastically increases the efficiency of the process (Takahashi & Yamanaka 2006). However, the mechanism by which Myc increases efficiency of pluripotency is not well understood.
In this study, we tested the hypothesis that Myc plays a key role in coordinating accelerated cell cycle as well as for efficient induction of pluripotency in adult cells by transcriptionally regulating ribosomal RNA and protein genes. Our results show that rRNA expression is higher in hESCs when compared to adult human cell lines. We further show that Myc occupies and regulates the expression of rRNA genes in hESCs. Another key finding of our study is that rRNA genes are bivalently marked with activating and repressive histone marks in a cell cycle dependent manner. Interestingly, RPGs are dominantly marked by the activating H3K4me3 modification and are occupied by Myc in hESCs. These findings reveal a unique epigenetic landscape for the rRNA genes and RPGs and suggest a potential mechanism by which Myc contributes to coordinate control of rapid stem cell growth and abbreviated embryonic stem cell cycle.
Materials and Methods
hESC Culture and Differentiation
The H9 human ESC line from WiCell Research Institute (Madison, WI) was maintained on hESC-qualified Matrigel (BD Bioscience, Catalog number 354277) in mTeSR-1 medium (Stemcell Technologies, Catalog number 05850) or Essential E8 medium (Life Technologies, Catalog number A1517001), as recommended by the supplier. Cells were expanded every 5–6 days, using non-enzymatic passaging following WiCell standard protocols. To initiate undirected differentiation, hESCs were grown in mTeSR-1 medium containing 10%FBS. hESC research was approved by the Institutional Embryonic Stem Cell Research Oversight Committee at the University of Vermont.
Cell Sorting
Pure populations of cells at the G2, mitosis or G1 phases of the cell cycle were isolated by FACS, taking advantage of differences in DNA content to distinguish cells in G2/M from cells in G1, and the exclusive presence of histone H3 phosphorylated in serine 28 (H3S28p) in mitosis to discriminate between cells in G2 or M phase (Fig. 1A and C). Subsequent to cell synchronization and fixation, cells were permeabilized for 10 min using a mild permeabilization/wash buffer containing Saponin (BD Bioscience, Catalog number 51-2091KZ). For ESC isolation, staining was performed with antibodies against OCT4 (PE-conjugated, BD Bioscience, Material number 561556) and H3S28p (Alexa fluor 647-conjugated, BD Bioscience, Material number 558609) for 30 min. After staining, cells were washed with permeabilization/wash buffer, resuspended in 2% fetal bovine serum (FBS) in phosphate buffered saline (PBS) and counterstained with 1 μg/ml DAPI (Life Technologies, Catalog number D1306) for 30 min. FACS was performed on a BD FACSAria II using linear FSC and SSC scaling, followed by height and area-based doublets discrimination. Compensation was calculated using FACS Diva autocompensation algorithms, and supplemented by manual compensation to correct for autofluorescence. Cells with sub-normal DNA content were filtered out during the gating and only samples with a purity of 95% or higher were used in the following experiments.
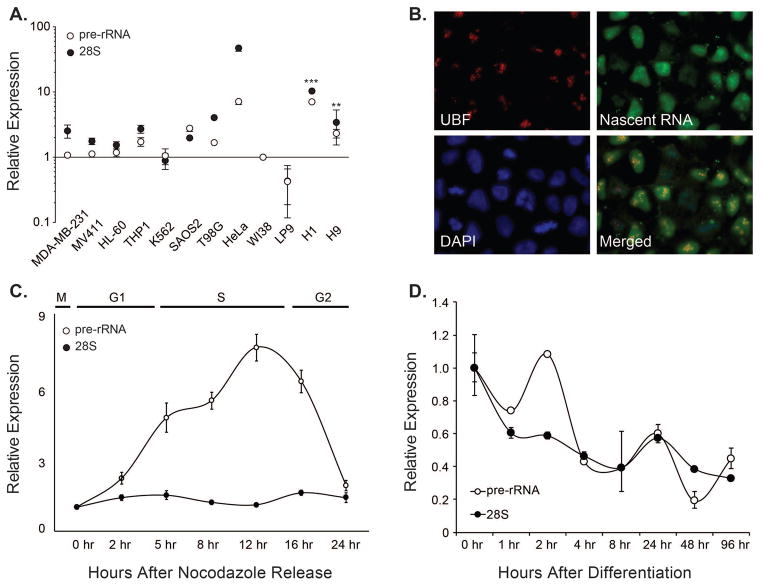
Figure 1. Expression of Ribosomal RNA genes is high in pluripotent human embryonic stem cells (hESCs), and is down-regulated during lineage commitment.
A. Comparative expression of nascent (pre) and mature (28S) ribosomal RNA in indicated human cell lines. The graph shows an average of three independent experiments, and the error bars represent standard error of means. Levels of pre- (open circle) and 28S-rRNA (close circle) in normal diploid fibroblasts (WI-38 cells) were used as a baseline. A normalizing factor was calculated from the expression of three housekeeping genes (GAPDH, Actin, and HPRT), and rRNA expression levels were normalized using this normalizing factor to account for inherent heterogeneity in growth and proliferation among the used cell lines. Significance of differential rRNA expression in hESCs is indicated, where ** represents p≤0.01 and *** represents p≤0.001. B. An in situ nuclear run-on experiment was carried out by pulse-labelling nascent RNA synthesis with Br-UTP in actively growing culture of pluripotent hESCs. Newly synthesized RNA was detected using an antibody specific for the bromo-moiety. Concurrently, cells were stained with an antibody specific for the upstream binding factor (UBF-1). A significant overlap between the signal indicates that rRNA genes are actively transcribed in hESCs. C. Levels of pre-rRNA and 28S rRNA transcripts were determined at various stages during the cell cycle. Mitotically synchronized population of hESCs was harvested at the indicated time points and the rRNA expression was determined using quantitative PCR. D. Human embryonic stem cells (H9) were subjected to undirected differentiation using fetal bovine serum and rRNA expression was determined at indicated time points after differentiation. The line graphs in Panels C and D show an average of two independent experiments (biological replicates) each, while the error bars represent standard error of the means.
Synchronization of hESC
Before every cell sorting experiment, cells were allowed to establish robust colonies. Normally, they were collected 4–5 days after plating. To improve the yield of sorting, we synchronized hESCs at either G2/M or G1 phase of the cell cycle one day before the collection, by incubation with Nocodazole (Sigma–Aldrich, Catalog number M1404-2MG) (200 ng/ml for 16 h when cells were grown in mTeSR-1). Blocking of hESCs in G2/M phase was reversible, and cells were able to resume the cell cycle and progress into G1 after withdrawing of Nocodazole from the culture medium. For cells growing in E8 medium, 25 ng/ml of Nocodazole for 16 h was enough to block most of the cells in G2/M and allowed progression into G1 after withdrawing of Nocodazole from the medium (data not shown). After synchronization, cells were collected as single cells by treatment with Acutase (MP Biomedicals, Catalog number 1000449), and subsequently fixed with 1% formaldehyde for 10 min followed by 5 min of incubation with 0.125M glycine (Sigma-aldrich, Catalog number G8790-100G). Cells were counted and stored at −80°C.
Immunofluorescence Microscopy
H9 cells were grown on coverslips previously coated with hESC-qualified Matrigel (BD Bioscience, Catalog number 354277). Normally, 3 days after plating, cells where fixed with 1% formaldehyde in PBS for 10 min and processed for immunofluorescence. Subsequently, cells were permeabilized in 0.25% Triton X-100 in PBS, and blocked 0.5% bovine serum albumin (BSA) in PBS. Cells were stained with mouse monoclonal antibodies against Myc or UBF for 1 h at room temperature. Nuclear DNA was counterstained with DAPI (0.1 μg/ml). Secondary antibodies conjugated with Alexa fluor 594 or Alexa fluor 488 (1:1000; Molecular Probes/Invitrogen) were incubated for 1 h in a humidity chamber, and at room temperature. Finally, cells were mounted in Prolong-Gold (Invitrogen).
For in situ nuclear run-on experiments, cells were pulse labelled with BrUTP for 30 minutes before processing for immunofluorescence analysis.
ChIP-seq Library Preparation
Cells at specific phases of the cell cycle, obtained from 3–4 different cell-sorting experiments, were pooled and treated as a single sample (or biological replicate). Two biological replicates were analyzed for each cell cycle phase. Cells were washed twice with PBS and subjected to extraction of nuclei according to (1) with modifications. Isolated nuclei were sonicated using a Misonix S-4000 Ultrasonic Processor (QSonica,) to obtain sheared chromatin ranging from 0.2 to 0.6 kilobases. Four μg of sheared chromatin were immunoprecipitated with either anti H3K4me3 (Abcam, ab1012) or anti H3K27me3 (Millipore, 07-449). Immunoprecipitated complexes were purified by Protein-G Dynabeads (Invitrogen), eluted, reverse crosslinked, quantified, and subjected to library preparation. Immunoprecipitated DNA or Input DNA for each cell cycle phase and biological replicate were end repaired using the End-It DNA End-Repair Kit (Epicenter), extended using a Klenow fragment (3′-5′ exo) (Epicenter), and ligated to sequencing adaptor oligos (Illumina) by following manufacturers’ recommendations. Each library was then subjected to 15 cycles of PCR amplification using PFU Ultra II Hotstart Master Mix (Agilent), and a size selected in range of 300 ± 50 bp. The final libraries were quantified by using both Qubit fluorimeter (Life Technologies) and Bioanalyzer system (Agilent Technologies), and submitted for sequencing.
ChIP-seq Analysis
Sequencing base calls were generated on the HiSeq 1500 instrument in the Advanced Genome Technologies Core Massively Parallel Sequencing Facility. For the ChIP-Seq analysis Fastq conversion and demultiplexing were done by bcl2fastq (Ilumina, v1.8.4), evaluated (Fastqc) and processed to remove low quality reads (FastX toolkit). Reads were mapped to the human genome (hg38) using STAR aligner (version 2.4) with splicing disabled (--alignIntronMax 1) (Dobin et al. 2013). Wig tracks and enriched regions (peak calls) for each replicate were generated by MACS2 (Feng et al. 2012) and replicates were then evaluated by wigCorrelate (Kent et al. 2010) and IDR (Li et al. 2011). Biological replicates were then combined and wig tracks regenerated for the combined signal. All raw data were deposited at the NCBI Gene Expression Omnibus (GSE55502).
For analysis of gene promoter regions, annotations were generated by defining the TSS for each protein coding ENSMBL gene (GENCODE release 21) and extending by +/− 1Kb. Enrichment profiles at gene promoters for each cell line was calculated by RPKMtreatment/RPKMinput where input was minimally limited to the 5th percentile of all calculated RPKM (RPKMtotal) using HTseq (Anders et al. 2014). Fold enrichment (FE) of chromosomes was determined by calculating RPKM based on total aligned reads and chromosome length, and calculating fold increase over input. Relative enrichment of histone-associated DNA at gene promoters between cell lines was defined by calculating FE (log2 B − log2 A). To ensure that signal was quantifiably different between two cell lines a FE value of > 4 was used as a minimum cutoff. For aggregate plots and heatmaps, NGSplot (version 2.47) (Shen et al. 2014) was used to generate FE profiles from alignment files. In contrast to the +/− 1Kb promoter definition used to calculate values, plot profiles were adjusted to +/− 2Kb from each TSS to better capture the pattern of histone marks at each promoter.
The resultant lists of differentially enriched gene promotors was then used to query 4914 MSigDB gene sets (Subramanian et al. 2005) in order to define over- represented gene sets using a binomial test (probability of success = MSigDB list size/21439 total unique genes in all MSigDB lists; number of trials = size of dynamically marked genes list; number of success = size of intersection of dynamically marked genes list and MSigDB list). Significance of over-representation was determined with a p-value threshold of 4.9×10−9 after Bonferroni correction.
Heatmaps of each MSigDB list were generated with a k-means method (R v 3.2.1) initialized with 4 random centers and allowed to run for 10 iterations or until convergence. Cluster centers were then individually sorted in decreasing order of total FE. Genes used to define MSigDB lists were excluded from clustering and plotted in a separate group presented at the top of each heatmap.
RNA Isolation and Quantitative PCR
Total RNAs from undifferentiated, mitotically blocked and released or FBS-differentiated H9 cells, were isolated by using Trizol (Invitrogen). Subsequently, we removed DNA contaminants by using DNA-free RNA extraction kit (Zymo Research). Following Illumina’s manual, 2 μg of DNA-free total RNAs was used to construct each paired-end RNA-Seq library with TruSeq RNA Sample Preparation Kit v2 (Illumina). Libraries were subjected to 15 cycles of amplification with pair-end PCR primers (Illumina). Libraries were quantified in Bioanalyzer and sequenced were on the HiSeq-1000 platform (Illumina) for the read length of 100 bases.
siRNA-Mediated Knockdown
ESCs were transfected with 200nM of an siRNA against Myc for 48 hours using Oligofectamine. Cells were harvested for total RNA isolation and quantitative RT-PCR as described.
Results and Discussion
Ribosomal RNA genes are highly expressed in human embryonic stem cells during S-phase and are down regulated upon differentiation
Because hESCs proliferate at a high rate, have a shortened G1 phase of the cell cycle and exhibit an open chromatin landscape (Chen & Daley 2008; Becker, Stein, et al. 2006; Becker et al. 2010; De Los Angeles et al. 2015), we hypothesized that ribosomal RNA genes are expressed at a high rate in hESCs. We initially determined levels of both pre- and 28S- rRNA in normal and in various human cancer cell lines (as indicated in Figure 1A), as well as in two different ESC lines (H01, and H09). With normal diploid WI-38 cells as baseline, both ES cell lines reproducibly exhibited higher levels of rRNA expression that were comparable to highly cancerous cell lines (Figure 1A). We controlled for inherent cell proliferation and growth variability among various cell types across triplicates by using a normalization factor calculated using multiple internal controls (Vandesompele et al. 2002). Our findings that hESCs have high levels of 28S and pre-rRNA are consistent with the notion that ESCs exhibit many characteristics of cancer cells (Monk & Holding 2001), albeit in a highly controlled and regulated manner.
We confirmed our results by carrying out in situ nuclear run-on experiments to examine active rRNA transcription. Embryonic stem cells were pulse labelled with Br-UTP, and nascent RNA molecules were identified by using antibodies against the bromo moiety to detect newly synthesized RNA and compared to the rRNA activator Upstream Binding Factor (UBF) (Figure 1B). Consistent with our RT-qPCR results (Figure 1A), hESCs exhibited a high level of new RNA synthesis in the nucleoli. These results suggest that most copies of rRNA genes are actively transcribed in human embryonic stem cells.
Because hESCs have a short G1 phase of the cell cycle, we next investigated the expression of rRNA genes during the ES cell cycle. Cells released from mitotic block were examined for the expression of pre-rRNA or total rRNA (28S) at various stages of the cell cycle. We observed an increase in pre-rRNA at the onset of S-phase that continues throughout S-phase, indicating that rRNA expression is tightly linked with the onset and progression of the S phase of the cell cycle (Figure 1C; Supplementary Figure 1A). Relative expression of 28S rRNA remained unchanged throughout the cell cycle. These findings are consistent with a highly active S-phase in embryonic stem cells (Klein & Grummt 1999), where DNA duplication, rRNA synthesis, and histone gene transcription take place concomitantly to accommodate higher cell proliferation and growth rates. We next examined the expression of rRNA genes during undirected differentiation of hESCs by culturing the cells in FBS. Our results show a progressive decrease in both pre- and 28S- rRNA levels following an initial burst in the expression of pre-rRNA at the 2 hr time point following addition of serum (Figure 1D; Supplementary Figure 1B). These results together indicate that rRNA genes are highly expressed in human embryonic stem cells during S-phase and are down regulated upon differentiation.
Ribosomal protein genes are marked by histone marks of active transcription during the embryonic stem cell cycle
Because hESCs exhibit higher expression of rRNA, we hypothesized that ribosomal protein genes (RPGs) are also expressed at higher levels in hESCs. We first determined the expression of all 78 human RPGs by RT-qPCR (Supplementary Table 1). As expected, all RPGs are expressed at high levels in hESCs (Figure 2A and B, Supplementary Figure 2 and data not shown). We also find some minor differences between the two hES cell lines that may represent slight differences in proliferation rates (Figure 2B). We next examined the genome-wide epigenetic landscape of RPGs at three different stages of the hES cell cycle for activating or repressive histone modifications. Human embryonic stem cells, sorted in G2, M or G1 phases of the cell cycle, were subjected to chromatin immunoprecipitation using antibodies against H3K4me3 and H3K27me3 marks, followed by genome-wide sequencing. We discovered that most RPGs are predominantly marked with the activating H3K4me3 mark in all three cell cycle stages. In comparison, the repressive H3K27me3 modification is detectable on RPGs at a very low level. Together, these results indicate that RPGs are actively transcribed in human embryonic stem cells. We extended our bioinformatics analyses to publicly available dataset (Varlakhanova et al. 2011), and found that that Myc, an oncogene and an activator of ribosomal RNA genes in somatic cells, occupies all of the RPGs in human embryonic stem cells (Figure 3A and B). These findings, together with higher levels of rRNA gene expression in hESCs, suggest that both the ribonucleic acid and protein components of ribosomes are readily available for synthesis to accommodate the higher growth rate of hESCs, as well as competency for differentiation into many different lineages.
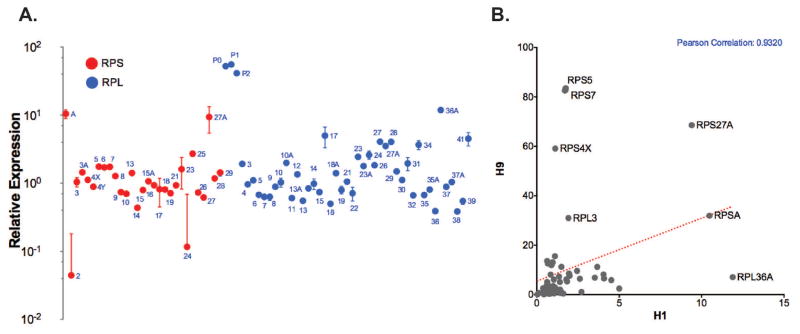
Figure 2. Ribosomal protein genes (RPGs) exhibit higher expression in hESCs.
A. Each circle represents expression of the indicated RPG for small (red circle) and large (blue circle) ribosomal subunits in pluripotent hESCs (n=3). B. RPG expression is highly correlated between the two hESC cell lines (H1 and H9), as indicated by the Pearson correlation of <0.9. Some RPGs were differentially expressed between the two cell lines, including RPS4X, a ribosomal protein that is encoded by the X chromosome and is only expressed in the female H9 cells.
Figure 3. The entire complement of ribosomal protein genes is occupied by Myc and marked by the activating H3K4me3 histone modification during cell cycle.
A. Heat maps showing occupancy of all RPGs by Myc in hESCs. Remarkably, all RPGs are predominantly marked by the activating activating histone modification H3K4me3 in G2, M and G1 phases of the cell cycle. Heat maps also reveal three distinct classes of RPGs: Class 1 (average gene length 3720 bp) contains RPGs that are marked by H3K4me3 throughout the gene body, Class 2 (average gene length 11431 bp) contains RPGs where H3K4me3 is narrowly localized to the transcription start site, while Class 3 (average gene length 7029 bp) contains RPGs with a relatively broader association of H3K4me3 around transcription start site. B. Line graphs represent cumulative occupancy of RPGs in all three classes indicating the Myc occupancy is localized to the transcription start site (tss; verticle dotted lines), and coincides with dominantly activating H3K4me3 histone modification (green line). The repressive histone mark H3K27me3 is also present on these genes, but at a very low level and remains unchanged in all three cell cycle stages. Transcription end site (tes) for RPGs is also indicated.
Ribosomal RNA genes are bivalently marked and transcriptionally regulated by Myc in human embryonic stem cells
To investigate the epigenetic landscape of rDNA repeats in hESCs, we analyzed genomic association of H3K4me3 and H3K27me3 during G2, M and G1 phases of the embryonic stem cell cycle. In contrast with RPGs that are predominantly marked by the activating H3K4me3 mark, we find that both activating and repressive marks are present on the rDNA repeats, a key characteristic of bivalently marked genes in ESCs that are poised for expression (Bernstein et al. 2006). Furthermore, we find that the H3K27me3 modification is present throughout the rDNA repeat in all three cell cycle stages, while the H3K4me3 mark is elevated in the M phase of the cell cycle (Figure 4A). Interestingly, the elevated H3K4me3 mark during the M phase is localized within known enhancer region with the intergenic spacer (IGS) of the rDNA repeat (Jacob et al. 2012). These novel findings establish intimate linkage between elevated rRNA gene expression and pluripotent state of embryonic stem cells. Furthermore, our studies reveal that rDNA repeats are bivalently marked genes in a cell cycle stage dependent manner.
Figure 4. rDNA repeats are bivalently marked during G1 phase of the cell cycle and are occupied and regulated by Myc in hESCs.
A. Schematic representation of rDNA repeat shows the 18S, 28S and 5S rRNA as well as the intergenic spacer (IGS). ChIP-seq experiments reveal that the rDNA repeats are occupied by Myc, a known transcriptional activator of rRNA genes. Bioinformatics analyses also reveal that rDNA repeats are enriched in H3K27me3 marks in all three cell cycle phases, with the activating H3K4me3 association is elevated in the M phase. B. In situ immunofluorescence showing that Myc (red) is punctately organized in the pluripotent nucleus (blue) and partially localizes to nucleoli as identified by the UBF staining (green). C. Bar graph showing that Myc plays a functional regulatory role in the expression of rRNA in hESCs. The H9 cells were transfected with a non-silencing siRNA (NS) or an siRNA against Myc (siMyc), and the total cellular RNA was examined for the expression of pre-rRNA and 28S RNA by RT-qPCR. The graph represents two independent biological replicates; significance of Myc knockdown on pre-rRNA expression is indicated (** p≤0.01).
Our bioinformatics analyses revealed that Myc is bound to transcription start sites of all RPGs in hESCs (Figure 3). Myc is a pluripotency transcription factor as well as a potent oncogene that regulates expression of gene transcripts by all three RNA polymerases (Arabi et al. 2005; Oskarsson & Trumpp 2005; Grandori et al. 2005; Gomez-Roman et al. 2003; Kenneth & White 2009). Therefore, we investigated whether Myc regulates ribosomal RNA gene expression in human embryonic stem cells. As has been shown in somatic cells (Persson & Leder 1984), Myc is localized to the embryonic stem cell nucleus (Figure 4B). We examined the occupancy of rDNA repeats by Myc in a genome-wide dataset and found that Myc dominantly occupies the regulatory regions of rDNA repeats (Figure 4A, Top Panel). These findings suggest that Myc plays a functional role in regulation of ribosomal biogenesis in hESCs through transcriptional regulation of both rRNA and ribosomal protein genes.
We directly examined the functional relevance of Myc occupancy of rRNA genes and RPGs by modulating levels of Myc protein in hESCs utilizing RNA interference- mediated knock down of the protein. We observed that rRNA gene expression is moderately decreased when Myc, a known transcriptional activator of rRNA genes, is depleted by RNA interference (Figure 4C). A modest decrease in rRNA levels upon Myc depletion suggests the existence of additional mechanisms that contribute to the regulation of ribosomal biogenesis in hESCs. Together, these findings implicate a central role for Myc in coordinate control of cell proliferation and cell growth in hESCs that exhibit restricted proliferation with stringent cell cycle control.
Conclusion
We have presented evidence that hESCs have higher expression of the components of ribosomal machinery during the abbreviated stem cell cycle. We discovered in this study that rRNA genes are bivalently marked with activating and repressive histone modifications, a key property of genes that are poised for transcription. Importantly, RPGs predominantly contain the activating histone mark. Furthermore, we show that Myc, a key pluripotency transcription factor, directly binds to and regulates rRNA as well as occupies ribosomal protein genes in hESCs. We conclude that ribosomal RNA and protein genes are competent for active transcription in hESCs and that Myc plays a central role in coordinating RNA Pol I and II gene transcription, thereby controlling accelerated cell cycle as well as cell growth in human embryonic stem cells.
Supplementary Material
Supplementary Figure 1: Western blot analysis showing down-regulation of pluripotency markers during differentiation.
Supplementary Figure 2: Heat map showing expression of RPGs, detected by gene specific primers and quantitative reverse transcription-PCR in various normal and cancer human cell lines. Red color indicates high expression, while black represents undetectable level of expression.
Supplementary Figure 3: Heatmaps showing association of Myc as well as activating H3K4me3 and repressive H3K27me3 marks with all RPGs, including pseudo genes. Inclusion of pseudo RP genes results in distribution of RPGs in 8 distinct classes that are based on differential association of H3K4me3 and H3K27me3 with genes. tss=transcription start site, tes=transcription end site.
Supplementary Figure 4: NPG plots showing various regions of human rDNA repeats individually for their association with Myc and H3K4me3 and H3K27me3 in three cell cycle stages.
Supplementary Table 1: Primers sequences used in this study for all human RPGs.
Acknowledgments
Contract Grant Sponsor: National Cancer Institute. Contract Grant Numbers: CA139322 and CA082834
References
- Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. 2014. Published online September 25, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi A, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nature Cell Biology. 2005;7(3):303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- Becker KA, et al. Human embryonic stem cells are pre-mitotically committed to self-renewal and acquire a lengthened G1 phase upon lineage programming. Journal of Cellular Physiology. 2010;222(1):103–110. doi: 10.1002/jcp.21925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KA, Ghule PN, et al. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. Journal of Cellular Physiology. 2006;209(3):883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- Becker KA, Stein JL, et al. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. Journal of Cellular Physiology. 2006;210(2):517–526. doi: 10.1002/jcp.20903. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boisvert F-M, et al. The multifunctional nucleolus. Nature Reviews Molecular Cell Biology. 2007;8(7):574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Chen L, Daley GQ. Molecular basis of pluripotency. Human Molecular Genetics. 2008;17(R1):R23–7. doi: 10.1093/hmg/ddn050. [DOI] [PubMed] [Google Scholar]
- Conconi A, et al. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57(5):753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- De Los Angeles A, et al. Hallmarks of pluripotency. Nature. 2015;525(7570):469– 478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- Dez C, Tollervey D. Ribosome synthesis meets the cell cycle. Current opinion in microbiology. 2004;7(6):631–637. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, et al. Identifying ChIP-seq enrichment using MACS. Nature Protocols. 2012;7(9):1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roman N, et al. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421(6920):290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- Grandori C, et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nature Cell Biology. 2005;7(3):311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Hsieh AL, et al. MYC and metabolism on the path to cancer. Seminars in Cell & Developmental Biology. 2015;43:11–21. doi: 10.1016/j.semcdb.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob MD, et al. Where no RNA polymerase has gone before. Nucleus. 2012;3(4):315–319. doi: 10.4161/nucl.20585. [DOI] [PubMed] [Google Scholar]
- Kapinas K, et al. The abbreviated pluripotent cell cycle. Journal of Cellular Physiology. 2013;228(1):9–20. doi: 10.1002/jcp.24104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenneth NS, White RJ. Regulation by c-Myc of ncRNA expression. Current Opinion in Genetics & Development. 2009;19(1):38–43. doi: 10.1016/j.gde.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Kent WJ, et al. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26(17):2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Grummt I. Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(11):6096–6101. doi: 10.1073/pnas.96.11.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiainen H, Shore D. Growth control and ribosome biogenesis. Current Opinion in Cell Biology. 2009;21(6):855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Li Q, et al. Measuring reproducibility of high-throughput experiments. The annals of applied statistics 2011 [Google Scholar]
- Lucchini R, Sogo JM. The dynamic structure of ribosomal RNA gene chromatin. Transcription of ribosomal RNA genes by eukaryotic … 1998 [Google Scholar]
- Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25(48):6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- Mayer C, et al. Intergenic transcripts regulate the epigenetic state of rRNA genes. Molecular Cell. 2006;22(3):351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- McStay B, Grummt I. The Epigenetics of rRNA Genes: From Molecular to Chromosome Biology. Annual Review of Cell and Developmental Biology. 2008;24(1):131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- Monk M, Holding C. Human embryonic genes re-expressed in cancer cells. Oncogene. 2001;20(56):8085–8091. doi: 10.1038/sj.onc.1205088. [DOI] [PubMed] [Google Scholar]
- Moss T. At the crossroads of growth control; making ribosomal RNA. Current Opinion in Genetics & Development. 2004;14(2):210–217. doi: 10.1016/j.gde.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Oskarsson T, Trumpp A. The Myc trilogy: lord of RNA polymerases. Nature Cell Biology. 2005;7(3):215–217. doi: 10.1038/ncb0305-215. [DOI] [PubMed] [Google Scholar]
- Pechmann S, Willmund F, Frydman J. The ribosome as a hub for protein quality control. Molecular Cell. 2013;49(3):411–421. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson H, Leder P. Nuclear localization and DNA binding properties of a protein expressed by human c-myc oncogene. Science. 1984;225(4663):718–721. doi: 10.1126/science.6463648. [DOI] [PubMed] [Google Scholar]
- Shen L, et al. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15(1):1. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nature Reviews Cancer. 2010;10(4):301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlakhanova N, Cotterman R, Bradnam K. Myc and Miz-1 have coordinate genomic functions including targeting Hox genes in human embryonic stem cells. Epigenetics : official journal of the DNA Methylation Society. 2011 doi: 10.1186/1756-8935-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL, Jr, Warner JR. The ribosome and its synthesis. The molecular and cellular biology of the yeast … 1991 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Western blot analysis showing down-regulation of pluripotency markers during differentiation.
Supplementary Figure 2: Heat map showing expression of RPGs, detected by gene specific primers and quantitative reverse transcription-PCR in various normal and cancer human cell lines. Red color indicates high expression, while black represents undetectable level of expression.
Supplementary Figure 3: Heatmaps showing association of Myc as well as activating H3K4me3 and repressive H3K27me3 marks with all RPGs, including pseudo genes. Inclusion of pseudo RP genes results in distribution of RPGs in 8 distinct classes that are based on differential association of H3K4me3 and H3K27me3 with genes. tss=transcription start site, tes=transcription end site.
Supplementary Figure 4: NPG plots showing various regions of human rDNA repeats individually for their association with Myc and H3K4me3 and H3K27me3 in three cell cycle stages.
Supplementary Table 1: Primers sequences used in this study for all human RPGs.