Abstract
Hepatitis C virus (HCV) NS3 protease inhibitors (PIs) are important components of novel HCV therapy regimens. Studies of PI resistance initially focused on genotype 1. Therefore, knowledge about the determinants of PI resistance for the highly prevalent genotypes 2 to 6 remains limited. Using Huh7.5 cell culture-infectious HCV recombinants with genotype 1 to 6 NS3 protease, we identified protease positions 54, 155, and 156 as hot spots for the selection of resistance substitutions under treatment with the first licensed PIs, telaprevir and boceprevir. Treatment of a genotype 2 isolate with the newer PIs vaniprevir, faldaprevir, simeprevir, grazoprevir, paritaprevir, and deldeprevir identified positions 156 and 168 as hot spots for resistance; the Y56H substitution emerged for three newer PIs. Substitution selection also depended on the specific recombinant. The substitutions identified conferred cross-resistance to several PIs; however, most substitutions selected under telaprevir or boceprevir treatment conferred less resistance to certain newer PIs. In a single-cycle production assay, across genotypes, PI treatment primarily decreased viral replication, which was rescued by PI resistance substitutions. The substitutions identified resulted in differential effects on viral fitness, depending on the original recombinant and the substitution. Across genotypes, fitness impairment induced by resistance substitutions was due primarily to decreased replication. Most combinations of substitutions that were identified increased resistance or fitness. Combinations of resistance substitutions with fitness-compensating substitutions either rescued replication or compensated for decreased replication by increasing assembly. This comprehensive study provides insight into the selection patterns and effects of PI resistance substitutions for HCV genotypes 1 to 6 in the context of the infectious viral life cycle, which is of interest for clinical and virological HCV research.
INTRODUCTION
With more than 100 million chronic infections causing approximately 500,000 deaths annually, hepatitis C virus (HCV) is a major global health and economic burden (1, 2). The six epidemiologically important genotypes differ in ∼30% of their sequence and in their sensitivity to antiviral regimens (3–6). In Europe, the Americas, Asia, and Australasia, genotypes 1, 2, and 3 are most prevalent. While genotypes 4, 5, and 6 are more restricted to specific geographical regions in Africa and Asia, they account for 20% of global HCV infections and have spread beyond these primary geographical locations (1, 2, 7).
The development of directly acting antivirals (DAAs) has revolutionized HCV therapy. The main components of interferon-free regimens introduced in the clinic are inhibitors of the HCV nonstructural (NS) proteins NS3 protease (NS3P), NS5A, and NS5B (4–6, 8, 9). Even though DAA-based therapy regimens could cure most patients in clinical trials, failure rates of 5 to 10% are to be expected in real life, due primarily to the development of DAA resistance (4, 8). Given the large number of HCV-infected individuals who will be treated, DAA resistant HCV variants will be common in the future. Treating patients with DAA resistant variants and avoiding DAA resistance will be aided by understanding the determinants and the molecular virology of resistance. The selection of specific resistance substitutions is thought to depend on several factors such as the specific DAA, the level of resistance conferred by the resistance substitution, the genetic barrier to resistance of the HCV isolate, and the fitness of the resistant variant (4, 8, 10). Of note, additional substitutions might compensate for fitness impairment caused by resistance substitutions (10).
Currently, the NS3 protease inhibitors (PIs) telaprevir, boceprevir, simeprevir, asunaprevir, paritaprevir, vaniprevir, and grazoprevir have been licensed (4–6). The first licensed PIs, telaprevir and boceprevir, have linear structures and covalently bind the NS3P active site. Newer PIs have either linear or macrocyclic structures and do not form covalent bonds (9). However, since all PIs target the NS3P active site, substitutions conferring cross-resistance to different PIs have been identified (4, 8, 10). As with most DAAs, PIs were initially developed to target genotype 1 and have only recently been used to treat other genotypes. Thus, most available data relate to genotype 1, while for other genotypes, only comparatively limited data on the determinants of PI resistance are available (4, 8).
Together with its cofactor NS4A, NS3P processes the HCV polyprotein by cleavage of the junctions between NS3, NS4A, NS4B, NS5A, and NS5B, which is essential for viral replication (9, 11). NS3P also interferes with innate immune responses by cleaving adaptor proteins of cellular pathways necessary for interferon production (9). However, given the multifunctional nature of the NS3–NS4A complex, targeting NS3P with PIs or modification of NS3P by substitutions might influence different steps of the viral life cycle. NS4A and the NS3 helicase (NS3H) are both thought to mediate viral replication and assembly (9, 12–15). Changes in the activity and/or conformation of NS3P might have an impact on NS3H and NS4A functions (16). Further, NS3P activity has been shown to influence the phosphorylation state of NS5A (17), which is thought to regulate the functions of NS5A in viral replication as well as assembly (12, 18). Lastly, NS3 interacts with other nonstructural proteins, such as NS2, which is important for the coordination of assembly (12).
In this study, we aimed to identify and characterize determinants of PI resistance in the context of the complete viral life cycle by using infectious HCV recombinants with the NS3P of genotypes 1 to 6 from prototype strains (19). We aimed to identify putative resistant variants that emerged under treatment with PIs and to use reverse genetics to characterize changes in fitness and PI sensitivity caused by the substitutions identified. Finally, we studied the effect of PI treatment as well as the effects of resistance substitutions and of fitness-compensating substitutions on different steps of the viral life cycle.
MATERIALS AND METHODS
HCV genotype 1 to 6 recombinants.
The genotype 2a J6/JFH1 virus (20) (referred to below as genotype 2a [isolate JFH1]) and J6/JFH1-based recombinants with genotype (isolate) 2a (J6)-, 3a (S52)-, 3a (452)-, 5a (SA13)-, and 6a (HK6a)-specific NS3P/NS4A have been described elsewhere (21). We also used a recombinant with a genotype 4a (isolate ED43)-specific component (from the 5′ untranslated region to NS5A) and a genotype 2a (isolate JFH1)-specific component (from NS5B to the 3′ untranslated region) (22). For genotype 1 escape experiments, we used a genotype 1a (isolate TN) recombinant available at the study outset (full-length TN with F1464L, A1672S, D2979G, Y2981F [LSGF], A1226G, and Q1773H) (23); for single-cycle production assays, we used a further adapted recombinant (TNcc) (23). For a graphical presentation of the HCV recombinants, see Fig. S1 in the supplemental material. Virus stocks were generated from supernatants of infected Huh7.5 cell cultures and were sequenced as described previously (21–23).
Culturing of Huh7.5 cells, immunostaining, and infectivity titration.
Culturing of Huh7.5 cells (24) has been described elsewhere (25). Viral spread was monitored by estimating the percentage of HCV antigen-positive cells, using primary mouse antibody 9E10 against HCV NS5A (20) and Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) (Invitrogen) as a secondary antibody as described previously (25). HCV infectivity titers were determined as focus-forming units (FFU) per milliliter after infection of three replicate cultures with serially diluted supernatants. Infected cells were stained after 48 h with anti-HCV NS5A antibody 9E10 and with ECL anti-mouse IgG (horseradish peroxidase-conjugated whole antibody; GE Healthcare Amersham) as a secondary antibody, and FFU were counted automatically (25–27). For genotype 1a (isolate TN) and genotype 4a (isolate ED43), a combination of anti-HCV NS5A antibody 9E10 and mouse anti-HCV core antibody C7-50 (Abcam) was used for immunostaining (23).
Induction of viral escape in PI-treated HCV-infected cultures.
Huh7.5 cells (3.5 × 105/well) were plated in a 6-well plate, incubated overnight, and infected with stocks of HCV genotype 1 to 6 recombinants for 24 h. PIs purchased from Acme Bioscience were dissolved in dimethyl sulfoxide (Sigma) (21, 28). Treatment with telaprevir (VX-950) or boceprevir (SCH 503034) at 0.75, 1, 2, 4, 16, or 64 times the 50% effective concentration (EC50) was initiated when ∼10% to 60% of cells were infected, as estimated by immunostaining. Treatment of genotype 2a (isolate JFH1) with 4, 16, or 64 times the EC50 of vaniprevir (MK-7009), simeprevir (TMC435350), or grazoprevir (MK-5172) or with 1, 2, 4, 16, or 64 times the EC50 of faldaprevir (BI 201335), paritaprevir (ABT-450), or deldeprevir (ACH-2684) was initiated when ∼1% to 20% cells were infected. Cultures were retreated with the PI upon cell splitting every 2 to 3 days; cells plated on a chamber slide after treatment and incubated overnight were immunostained as described above in order to evaluate viral spread. Nontreated control cultures were followed until HCV spread to ≥80% of cells. Treated cultures were followed until a peak in the percentage of infected cells was observed, potentially representing viral escape, or until viral suppression, defined as the absence of HCV-positive cells in six consecutive immunostainings, was observed. Cytotoxic effects were observed when genotype 3a (isolate 452) was treated with telaprevir at 4 times the EC50 (41,572 nM). Therefore, treatment with telaprevir at doses exceeding this concentration (treatment of genotype 2a [isolate J6], genotype 4a [isolate ED43], and genotype 5a [isolate SA13] with 64 times the EC50 and of genotype 3a [isolate S52] with ≥16 times the EC50) was not carried out.
Identification of putative PI resistance substitutions in HCV.
The NS3Ps of genotype 2 to 6 viruses from two time points following escape from telaprevir or boceprevir were analyzed; for genotype 2a (isolate JFH1) treated with newer PIs, the NS3P from at least one time point was analyzed. RNA extraction, reverse transcription, and PCR were carried out as described elsewhere (25), using the primers and cycling conditions specified in Table S1 in the supplemental material. Amplicons were sequenced (Macrogen) to obtain consensus sequences. In most cases with potentially combined substitutions, amplicons were TOPO TA cloned (Invitrogen), and individual clones were sequenced. Analysis was carried out with Sequencher (Gene Codes) and BioEdit. Numbers in the H77 (GenBank accession no. AF009606) reference sequence were determined using European and Los Alamos HCV databases (29).
Reverse genetic studies to evaluate the viral fitness and genetic stability of NS3P variants.
The putative resistance substitutions identified were introduced into the recombinants in which they were selected by using chemically synthesized DNA fragments (GenScript) and standard cloning procedures. Substitutions that occurred in nontreated cultures were not studied. Most recombinants encoding the R155K and A156S substitutions have been published (28). For final plasmid preparations (Qiagen Plasmid Maxi kit or Plasmid Midi kit), the complete HCV genome was sequenced (Macrogen).
For transfections, 3.5 × 105 Huh7.5 cells/well in 6-well plates were incubated overnight. HCV RNA transcripts generated with T7 polymerase (Promega) (25) were DNase treated for 30 min at 4°C using 1 U of RQ1 DNase (Promega) per μg of template DNA, purified (Qiagen RNeasy minikit), and quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). Eighteen micrograms of RNA transcripts was incubated with 5 μl Lipofectamine 2000 in 500 μl of Opti-MEM (Invitrogen) for 20 min at room temperature. Cells were incubated overnight with transfection complexes in Opti-MEM serum-free medium and were split first on day 1 posttransfection and then every 2 to 3 days.
First-passage virus stocks were generated by inoculating Huh7.5 cells with 1 ml of the supernatant from the peak of infection in transfection experiments; 106 Huh7.5 cells were plated in a T25 flask 1 day prior to inoculation. The cultures were split every 2 to 3 days from day 1 postinfection and were amplified to several triple-layer T175 flasks. To evaluate genetic stability, the NS3P sequences of variants in culture supernatants were determined at the peak of infection as described above.
The percentage of HCV-infected Huh7.5 cells and supernatant infectivity titers in transfection/infection experiments were evaluated as described above. Peak supernatant infectivity titers were defined as the highest representative titers obtained after transfection. In transfection experiments, the difference between the infectivity titer (expressed in log10 FFU per milliliter) of each variant and that of the original recombinant was calculated by subtracting the peak infectivity titer of the original virus from the infectivity titer of the variant on the same day on which the original virus achieved its peak titer. In cases where the NS3P variant reached its peak infectivity titer before the original recombinant, the difference was calculated as follows: (peak infectivity titer of variant) − (titer of the original virus on the same day). For these comparisons, the original and variant viruses were always studied in the same transfection experiment.
High-throughput treatment assay for determination of the EC50.
Briefly, Huh7.5 cells were plated in 96-well plates and were infected the following day with stock viruses of either the original or the mutated recombinant for 24 h. Infected cells were treated once with serial dilutions of the PI for 48 h and were then immunostained as described above. Each concentration was tested in triplicate. For each virus and compound, a concentration-response curve was generated from which the EC50 was determined as described previously (21, 27). Fold resistance was calculated by relating the EC50 obtained for the variant to the EC50 obtained for the original virus included in the same experiment, as described by us recently (28).
Single-cycle HCV production assay.
Single-cycle HCV production assays were carried out using CD81-deficient S29 cells (30) in 6-well plates as described previously (31). For genotype 5a (isolate SA13) recombinants, transfections were scaled up to T25 cell culture flasks, and transfection reagents were replaced with 3 ml of fresh medium 4 h after transfection. When specified, cells were kept in a PI-containing medium from 4 h after transfection until the termination of the assay at 48 h posttransfection. Intracellular (IC) and extracellular (EC) HCV core protein levels and infectivity titers were determined as described previously (31). IC infectivity titers were determined as FFU per well (where “well” refers to 1 well of the 6-well plates of the S29 assay). IC infectivity titration was carried out using serially diluted cell lysates for infection of two replicate cultures, which were immunostained followed by automated FFU counting as described above. EC infectivity titers were determined as FFU per milliliter as described above. IC and EC infectivity titers that were below the cutoff of the automated counting procedure (1.5 log10 FFU/well and 2.3 log10 FFU/ml, respectively) (27) were determined manually. For manual counting, the cutoffs were 0.9 log10 FFU/well and 1.6 log10 FFU/ml for IC and EC infectivity titers, respectively, based on a limit of detection of 2 FFU per well in the infectivity titration assay.
RESULTS
NS3P substitutions in escape variants of HCV genotypes 1 to 6 during treatment with PIs in vitro.
To identify potential PI resistance substitutions, Huh7.5 cells infected with stock viruses of HCV recombinants with the NS3P/NS4A of genotype (isolate) 1a (TN), 2a (JFH1 and J6), 3a (S52 and 452), 4a (ED43), 5a (SA13), or 6a (HK6a) were treated with different concentrations of telaprevir or boceprevir (Fig. 1; see also Fig. S1 and S2 in the supplemental material). Similarly, we induced the escape of genotype 2a (isolate JFH1) treated with the newer PIs vaniprevir, faldaprevir, simeprevir, grazoprevir, paritaprevir, and deldeprevir (Fig. 2; see also Fig. S3 in the supplemental material). Compared to that for nontreated control cultures, treatment with lower PI concentrations typically delayed viral spread, while higher PI concentrations resulted in an initial decrease in the percentage of HCV-positive culture cells (data not shown). This initial decrease was followed either by viral spread to ≥50% of culture cells, suggesting viral escape, or by a further decrease until no HCV-positive cells were detected, suggesting viral suppression. The highest telaprevir or boceprevir concentrations resulted in viral suppression for most viruses. However, among newer PIs, only treatment with the highest concentration of paritaprevir resulted in viral suppression. Overall, the HCV recombinants were able to escape treatment with several concentrations of PIs.
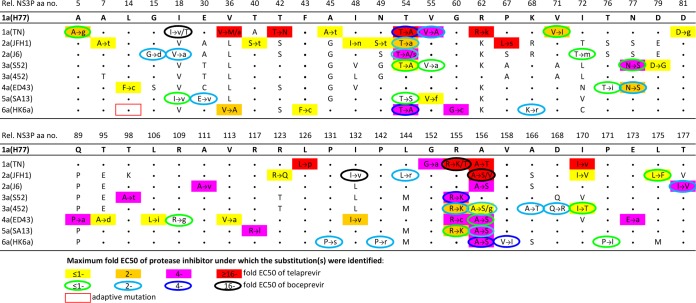
FIG 1.
Substitutions identified in the NS3Ps of HCV genotype 1 to 6 escape viruses during treatment with telaprevir or boceprevir. Huh7.5 cells were infected with viruses with the NS3P/NS4A of genotype (isolate) 1a (TN), 2a (JFH1 or J6), 3a (S52 or 452), 4a (ED43), 5a (SA13), or 6a (HK6a) (see Fig. S1 in the supplemental material), treated with telaprevir or boceprevir, and followed as described in Materials and Methods and in Fig. S2 in the supplemental material. At the time of viral escape, the NS3Ps of viruses recovered at two different time points were amplified using reverse transcription-PCR and were directly sequenced. NS3P positions at which a substitution for at least one of the viruses treated with telaprevir or boceprevir was identified are shown. Rel. NS3P aa no., NS3 protease amino acid number relative to that in the genotype 1a reference strain H77 (GenBank accession no. AF009606); the H77 amino acid residues are indicated. For each of the viruses studied, a dot indicates that the amino acid residue at the respective position is identical to that in strain H77, and a single letter neither shaded nor circled indicates the amino acid identity at a nonidentical residue. Substitutions occurring at one or more time points under PI treatment are indicated by colored filled rectangles for telaprevir and by colored circle outlines for boceprevir. The color of the rectangle or the circle outline indicates the highest multiple of the EC50 of the PI at which the substitution was identified. The letter to the left of each arrow represents the original amino acid, while the letter(s) to the right indicates the substitution(s) identified in escape variants. Different substitutions identified at the same position are separated by a slash. The substitutions estimated to be present in at least 50% of viral genomes are indicated by capital letters, while substitutions estimated to be present in a minor percentage are indicated by lowercase letters. Substitutions that were also identified in nontreated cultures are not shown. The genotype 6a (isolate HK6a) recombinant contains the cell culture-adaptive substitution V14L (boxed in red). For further details, see Tables S2 to S17 in the supplemental material.
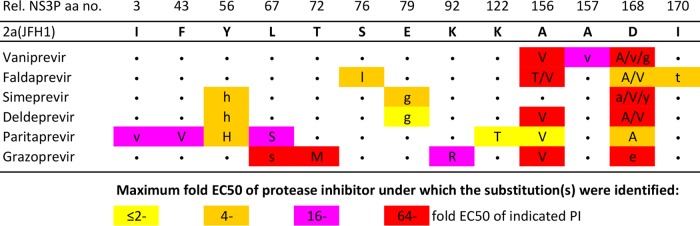
FIG 2.
Substitutions identified in the NS3Ps of HCV genotype 2a escape viruses during treatment with newer PIs. Huh7.5 cells were infected with the genotype 2a (isolate JFH1) virus, treated with vaniprevir, faldaprevir, simeprevir, deldeprevir, paritaprevir, or grazoprevir, and followed as described in Materials and Methods and in Fig. S3 in the supplemental material. At the time of viral escape, the NS3Ps of viruses were amplified using reverse transcription-PCR and were directly sequenced. Only NS3P positions at which a substitution was identified for at least one of the PIs are shown. Rel. NS3P aa no., NS3 protease amino acid number relative to that in the genotype 1a reference strain H77 (GenBank accession no. AF009606). The original genotype 2a (isolate JFH1) virus amino acid residues are used as references in this alignment. For each of the PIs studied, a dot indicates that the amino acid residue at the respective position did not change. Substitutions occurring under PI treatment are indicated by colored rectangles. The color of the rectangle indicates the highest multiple of the EC50 of the PI at which the substitution was identified. The letter(s) in each colored rectangle indicates the substitution(s) identified in escape variants, and different substitutions identified at the same position are separated by a slash. The substitutions estimated to be present in at least 50% of viral genomes are indicated by capital letters, while those estimated to be present in a minor percentage of viral genomes are indicated by lowercase letters. Substitutions that were also identified in nontreated cultures are not shown. For further details, see Tables S18 to S23 in the supplemental material.
We identified NS3P substitutions in viruses recovered during telaprevir or boceprevir treatment (Fig. 1; see also Tables S2 to S17 in the supplemental material). In genotype 1a (isolate TN), substitutions associated with in vivo genotype 1 resistance to telaprevir or boceprevir, including V36(M/A), T54A, V55A, R155(K/T), A156T, V36M plus T54A, T54A plus R155K, or V36M plus T54A plus R155K, were observed (4, 8, 10, 32, 33). Thus, the HCV in vitro escape assay permitted the selection of clinically relevant PI resistance substitutions. For genotype 2 to 6 recombinants, NS3P amino acid positions 54, 155, and 156 were hot spots for the selection of substitutions. The T54A substitution emerged in genotype (isolate) 2a (JFH1 and J6), 3a (S52), and 6a (HK6a), and the T54S substitution in 2a (J6) and 5a (SA13). The R155K substitution was selected in genotype (isolate) 3a (S52 and 452) and 5a (SA13), while the R155C substitution emerged in 4a (ED43). Finally, the A156S substitution emerged in genotype (isolate) 2a (JFH1 and J6), 3a (452), 4a (ED43), 5a (SA13), and 6a (HK6a), while the A156V and A156G substitutions emerged in 2a (JFH1) and 3a (452), respectively. In addition, for genotypes 2 to 6, substitutions at positions 18, 55, 77, 132, and 170 were acquired by two recombinants. Substitutions at other NS3P positions were acquired by a single recombinant. We also identified combinations of substitutions. In most cases, a substitution at position 54 was combined with a substitution at position 155 or 156, or a substitution at one of these positions was combined with another substitution.
For genotype 2a (isolate JFH1) treated with newer PIs, NS3P positions 156 and 168 were substitution hot spots (Fig. 2; see also Tables S18 to S23 in the supplemental material). The A156V substitution was selected with all newer PIs except simeprevir. Substitutions at position 168 emerged for all the newer PIs; changes to A or V were most frequent, while changes to G, E, or Y were also observed. Among 13 positions at which genotype 2a (isolate JFH1) acquired substitutions when treated with newer PIs, 3 (amino acids [aa] 67, 156, and 170) also acquired substitutions under treatment with telaprevir or boceprevir, and 6 others (aa 43, 72, 76, 79, 168, and 170) showed changes in other recombinants treated with telaprevir or boceprevir (Fig. 1). Thus, the patterns of substitutions selected under telaprevir or boceprevir and newer PIs partially overlapped. The main difference was the almost exclusive selection of substitutions at position 168 with newer PIs, whereas substitutions at position 54 were selected only with telaprevir or boceprevir. Of note, the Y56H substitution, close to position 54, was selected under treatment with three newer PIs.
Fitness of genotype 2 to 6 viruses with putative PI resistance substitutions.
We studied the effects of substitutions emerging under telaprevir or boceprevir treatment (Fig. 1) on the fitness of the recombinants in which they were identified. We examined 60 recombinants with single NS3P substitutions, including previously tested R155K and A156S recombinants (28), and 17 recombinants encoding combinations of NS3P substitutions (Fig. 3). Differences in viral spread kinetics were determined by comparison of the infectivity titers of variants and original recombinants after Huh7.5 cell transfections. The genetic stability of NS3P variants was tested by NS3P sequencing following viral passage to naïve cells (Fig. 3). While all except 3 engineered variants encoding single NS3P substitutions were viable, the substitutions often had a negative effect on HCV fitness (Fig. 3). Large fitness decreases were seen for 13 variants; among these, the substitutions reverted in 8 variants, while 1 acquired an additional NS3P substitution. Moderate fitness decreases were found in 15 variants, all of which maintained the engineered substitutions; 3 acquired other NS3P substitutions. Twenty-four variants had levels of fitness comparable to those of the original recombinants and maintained the engineered substitutions; 1 had an additional NS3P substitution. Finally, 5 variants had fitness increases and were genetically stable. Genotype 2a (isolate JFH1) recombinants with substitutions of A, G, E, or V for D168, selected in 2a (JFH1) treated with newer PIs (Fig. 2), showed different degrees of fitness impairment but were all genetically stable (28). Overall, viral fitness depended on the NS3P position, the specific substitution, and the recombinant (Fig. 3).
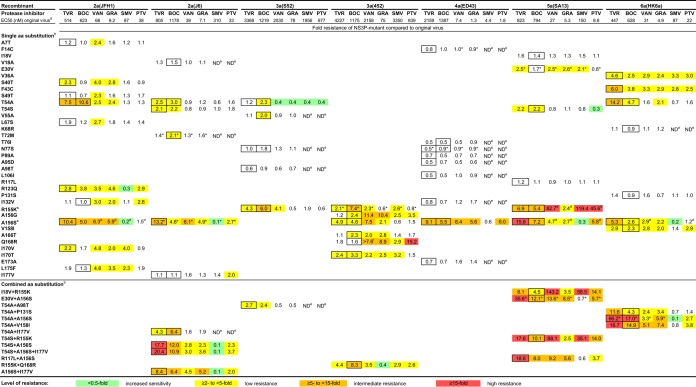
FIG 3.
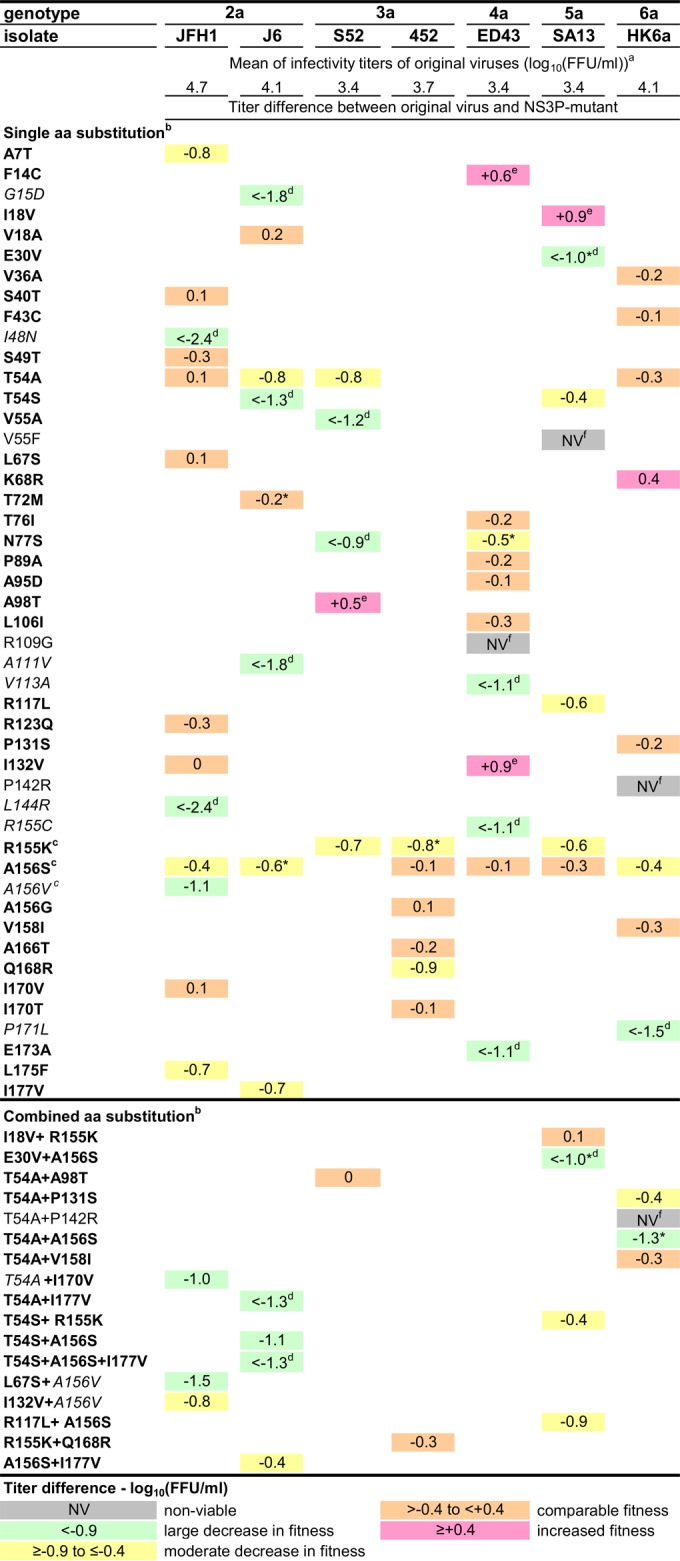
Fitness and genetic stability of HCV genotype 2 to 6 recombinants with engineered NS3P substitutions identified in escape variants. Huh7.5 cells were transfected with HCV RNA transcripts of the original and variant recombinants. For each variant, the difference in the infectivity titer (expressed as log10 FFU per milliliter) was calculated as described in Materials and Methods. Titer differences are color-coded. First-passage experiments were carried out by infecting naïve Huh7.5 cells with supernatants derived from the peak of infection in transfection experiments. The genetic stability of the engineered recombinants was investigated by direct sequencing of the NS3Ps of viruses in supernatants obtained from the peak of infection of first-passage cultures. Substitutions shown in boldface were maintained, while substitutions shown in italics reverted. a, the means of peak infectivity titers obtained for the original viruses in transfection experiments are given in log10 FFU per milliliter; the standard error of the mean ranged from 0.03 to 0.15. b, the numbering for engineered amino acid substitutions is relative to that for the reference strain H77 (GenBank accession no. AF009606). c, except for genotype 3a (isolate 452) R155K, the fitness and genetic stability of variants with single R155K, A156S, and A156V substitutions have also been reported previously (28). d, the infectivity titer of the variant was <2.3 log10 FFU/ml, the lower cutoff of the assay. e, the NS3P variant reached its peak infectivity titer before the original recombinant. f, recombinants for which no HCV-specific immunostaining was observed for at least 2 weeks posttransfection were considered nonviable in vitro. *, the following additional amino acid substitutions, estimated to be present in at least 50% of viral genomes, were identified in the NS3P sequences of the indicated viruses derived at the peak of infection in first-passage experiments: I177V in genotype 2a (isolate J6) T72M, P86L in genotype 2a (isolate J6) A156S, Q168K in genotype 3a (isolate 452) R155K, N29K and L127M in genotype 4a (isolate ED43) N77S, I17M in genotype 5a (isolate SA13) E30V and in genotype 5a (isolate SA13) E30V plus A156S, and A116P in genotype 6a (isolate HK6a) T54A plus A156S.
Most variants with combinations of NS3P substitutions showed decreased fitness from that of the original recombinants; one variant was nonviable, and three others did not maintain one of the substitutions introduced (Fig. 3). Most variants with combinations of substitutions did not have greater fitness than variants with single substitutions. However, for genotype 5a (isolate SA13), the I18V substitution compensated for the fitness cost of the R155K substitution, and for genotype 3a (isolate S52), the A98T substitution compensated for the fitness cost of the T54A substitution. The I18V and A98T substitutions increased the fitness of the respective recombinants when tested individually. The fitness of genotype 3a (isolate 452) R155K plus Q168R was unchanged, while variants with single substitutions showed fitness impairment. Thus, certain combinations of substitutions apparently had fitness-compensatory functions.
Effects of the substitutions identified on HCV resistance to telaprevir and boceprevir.
Variants that maintained engineered substitutions following viral passage (Fig. 3) (28) were assayed for sensitivity to telaprevir or boceprevir (Fig. 4). For each variant, concentration-response curves were used to determine EC50 values, and fold resistance was calculated by relating the EC50 values of variant viruses to those of the corresponding original viruses. Among 49 recombinants with single substitutions, 27 had ≥2-fold resistance to telaprevir and/or boceprevir, while 22 had susceptibilities similar to those of their respective original recombinants (Fig. 4). Resistant variants were 2.1- to 15.6-fold and 2.0- to 10.6-fold less sensitive to telaprevir and boceprevir, respectively. Only substitutions at positions 54, 155, and 156 conferred resistance on two or more genotypes; all 16 such variants were resistant to telaprevir and/or boceprevir. The 13 variants with combined NS3P substitutions had 2.7- to 66.2-fold and 2.4- to 17.0-fold decreased sensitivity to telaprevir and boceprevir, respectively (Fig. 4). The combination of a substitution at position 54 with changes at position 155, 156, or 158 conferred the highest resistance level relative to that of single substitutions.
FIG 4.
Levels of resistance of HCV genotype 2 to 6 recombinants with engineered NS3P substitutions to telaprevir (TVR), boceprevir (BOC), vaniprevir (VAN), grazoprevir (GRA), simeprevir (SMV), and paritaprevir (PTV). The level of resistance is expressed as the fold change in the EC50, which was calculated by comparing the EC50 of a variant to the EC50 of the original virus of the same genotype and isolate, as described in Materials and Methods. Fold resistance values are color-coded. The substitutions tested were selected under treatment with TVR and/or BOC; a boxed value indicates under which of these two PIs a particular amino acid substitution was selected. a, the indicated mean EC50 values (expressed as nanomolar concentrations) were determined previously for each original recombinant (28), and similar EC50 values were obtained for the original viruses in the current study. b, the numbering for engineered amino acid substitutions is relative to that for the reference strain H77 (GenBank accession no. AF009606). c, except for genotype 3a (isolate 452) R155K, the fold resistance of R155K and A156S variants has been determined previously (28) and is in agreement with the values obtained in the current study. d, the fold resistance was obtained from reference 28. e, ND, not determined. Resistance to simeprevir and paritaprevir was determined only for substitutions that either (i) conferred ≥2-fold resistance to vaniprevir and/or grazoprevir as single substitutions, (ii) conferred ≥2-fold resistance to vaniprevir and/or grazoprevir in combination with other NS3P substitutions, or (iii) were identified at position 54, 155, or 156. f, the indicated variant was <50% inhibited by the highest concentration of vaniprevir applied (15,000 nM). Asterisks indicate the following additional substitutions in NS3P acquired by the indicated first-passage stocks: I177V in genotype 2a (isolate J6) T72M, P86L in genotype 2a (isolate J6) A156S, Q168K in genotype 3a (isolate 452) R155K, N29K and L127M in genotype 4a (isolate ED43) N77S, I17M in genotype 5a (isolate SA13) E30V and in genotype 5a (isolate SA13) E30V plus A156S, and A116P in genotype 6a (isolate HK6a) T54A plus A156S. Similarly, the genotype 4a (isolate ED43) F14C second-passage stock acquired G90A.
The NS3P substitutions identified mediated cross-resistance.
We investigated whether substitutions selected under telaprevir or boceprevir treatment conferred cross-resistance to newer PIs (Fig. 4). Of the 49 recombinants with single substitutions, 26 showed decreased sensitivity to vaniprevir (2.0- to 82.7-fold) and 22 had decreased sensitivity to grazoprevir (2.0- to 10.4-fold). Among the subset of 35 recombinants tested, 10 showed decreased sensitivity to simeprevir (2.1- to 119.4-fold) and 12 had decreased sensitivity to paritaprevir (2.0- to 45.6-fold). Thus, while cross-resistance between telaprevir or boceprevir and newer PIs was observed, there were several notable exceptions. In general, substitutions selected at position 155 or 156 conferred the greatest level of cross-resistance to newer PIs, but with important differences among the drugs (Fig. 4). For example, genotype 5a (isolate SA13) R155K had high resistance to vaniprevir, simeprevir, and paritaprevir but low resistance to grazoprevir. Also, all A156S variants displayed no resistance or increased sensitivity to simeprevir. Substitutions at position 54 conferred limited cross-resistance to newer PIs. Most other substitutions conferring low to intermediate boceprevir or telaprevir resistance showed low resistance to certain of the four newer PIs. Conversely, several substitutions that did not confer resistance to telaprevir or boceprevir conferred low resistance to certain newer PIs, especially vaniprevir. Of note, the D168(A/G/E/V) substitution, selected in genotype 2a (isolate JFH1) with newer PIs (Fig. 2), conferred low to high resistance on 2a (JFH1) against these PIs but not against telaprevir or boceprevir (28). Thus, substitutions at positions 155 and 156 conferred the highest level of cross-resistance to newer PIs. Substitutions at position 168 conferred resistance to newer PIs, in contrast to substitutions at position 54, which conferred resistance only to telaprevir or boceprevir.
Most variants with combinations of substitutions showed low to intermediate resistance to vaniprevir and grazoprevir (Fig. 4). For simeprevir, increased sensitivity to high resistance was found in accordance with resistance levels for single substitutions. Four of the six A156S variants showed increased sensitivity to simeprevir, while two showed comparable sensitivity. For paritaprevir, most variants showed low resistance. Most combinations including R155K in genotype 5a (isolate SA13) conferred higher resistance to vaniprevir, simeprevir, and paritaprevir, but lower resistance to grazoprevir, than to telaprevir or boceprevir, as observed for genotype 5a (isolate SA13) R155K. However, variants with combinations including A156S and especially those with T54A or T54S had lower resistance to the newer PIs than to telaprevir or boceprevir. Thus, several combinations conferred cross-resistance to newer PIs. In general, the level of resistance to newer PIs was lower than the level of resistance to telaprevir or boceprevir.
Influence of PI-induced substitutions and PI treatment on the HCV life cycle.
We performed single-cycle production assays in CD81-deficient S29 cells (30), where the intracellular (IC) HCV core levels indicate the efficacy of viral replication/translation (summarized as replication), and the IC and extracellular (EC) infectivity titers allow conclusions on the efficacy of viral assembly and release.
The fitness costs of key NS3P resistance substitutions were due primarily to impaired viral replication.
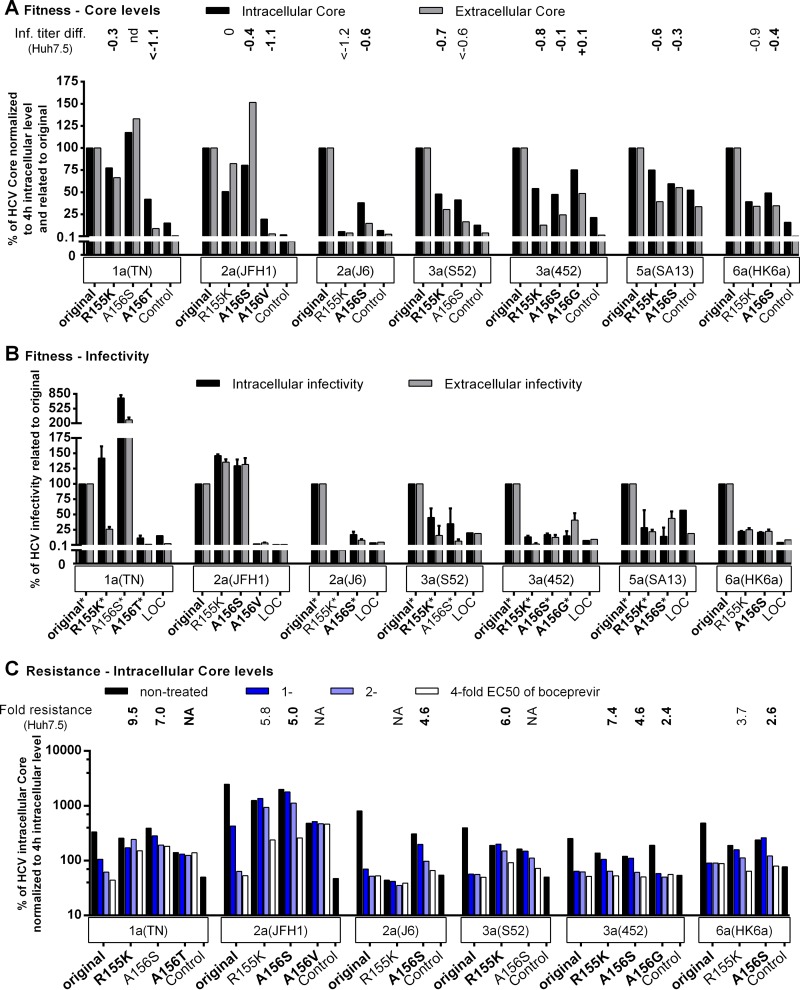
We studied the mechanisms of fitness impairment induced by the R155K and A156S substitutions across genotypes (Fig. 5A and B). Compared to those of the original recombinants, most R155K and A156S variants had decreased IC and EC core levels and infectivity titers, suggesting impairment of replication. Exceptions were genotype 1a (isolate TN) A156S, genotype 2a (isolate JFH1) R155K, and genotype 2a (isolate JFH1) A156S, for which the NS3P substitutions might have a positive impact on replication and/or assembly. Other PI-induced substitutions at position 156—A156T in genotype 1a (isolate TN), A156V in genotype 2a (isolate JFH1), and A156G in genotype 3a (isolate 452)—had a negative impact on replication, suggested by decreased IC core levels. For genotype 1a (isolate TN) A156T and genotype 2a (isolate JFH1) A156V, the decrease in replication was greater than for 1a (TN) and 2a (JFH1) R155K and A156S variants, in agreement with studies reporting high fitness costs of A156(T/V) (Fig. 3) (28, 34–37).
FIG 5.
Fitness impairment and PI resistance associated with key resistance substitutions at NS3P positions 155 and 156 affected viral replication across genotypes. RNA transcripts from the indicated genotype 1a (isolate TN), 2a (isolates JFH1 and J6), 3a (isolates S52 and 452), 5a (isolate SA13), and 6a (isolate HK6a) HCV recombinants were transfected into S29 cells. (A and B) For nontreated replicates, IC and EC core concentrations (A) and infectivity titers (B) were determined as described in Materials and Methods. (C) For the replicates treated 4 h posttransfection with boceprevir at the indicated multiple of the EC50, only the IC core concentration is shown. The corresponding IC and EC infectivity titers and the EC core concentration obtained under treatment are shown in Fig. S4 in the supplemental material. The low levels of replication of genotype 4a (isolate ED43) and genotype 5a (isolate SA13) recombinants in S29 cells have prevented or limited their use in this assay. To account for possible differences in transfection efficiency, IC and EC core concentrations at 48 h were normalized to IC core concentrations at 4 h (A and C). To determine the effects of the indicated NS3P substitutions on viral fitness, the normalized core values (A) and infectivity titers (B) of variant recombinants were related to the values for the respective original recombinants (original). In panels A and C, “Control” indicates the replication-deficient genotype 2a (isolate JFH1) negative-control virus with the mutation of the NS5B RNA polymerase active site (GlyAspAsp to GlyAsnAsp [GND]) (20), which was included in each transfection experiment. For this control, IC core concentrations at 48 h ranged from 9.4 × 103 to 25.0 × 103 fmol/liter, and EC core concentrations at 48 h ranged from 17.5 to 131.8 fmol/liter. The values shown for the control were normalized to the IC core concentrations at 4 h. In panel A, the values were also related to those obtained for the respective original recombinants (original) as was described for the NS3P variants. Transfections of recombinants of the same genotype (isolate) shown in the same graph were carried out in the same experiment; each of these experiments included the genotype 2a (isolate JFH1) GND negative-control virus. In panel B, LOC indicates the lower cutoff of the infectivity titration assay. For automated counting of FFU, the LOCs for IC and EC infectivity titers were 1.5 log10 FFU/well and 2.3 log10 FFU/ml, respectively. In instances where low replication efficiency in S29 cells precluded automated counting, FFU were counted manually, and the resulting titers are indicated by asterisks. The LOC for infectivity titers derived from manually counted FFUs was 0.9 log10 FFU/well for IC titers and 1.6 log10 FFU/ml for EC titers. LOC values were related to the values obtained for the respective original recombinants (original) as described for NS3P variants. The NS3P substitutions in boldface were specifically selected in the indicated virus under PI treatment in this study (Fig. 1 and 2). For comparison, the difference in the infectivity titer (Inf. titer diff.) observed following transfection of Huh7.5 cells (A) or the fold resistance to boceprevir (C) is indicated above each variant. Differences in infectivity titers and fold resistance were calculated as described in Materials and Methods. Values determined in this study (Fig. 3 and 4) are indicated in boldface; other values either were reported previously (28) or were calculated on the basis of infectivity titers determined in reference 28. The infectivity titer of genotype 1a (isolate TN) A156S was not determined, preventing calculation of the titer difference in Huh7.5 cells (nd, not determined). Instances in which fold resistance values could not be determined because the specified recombinant could not be grown in Huh7.5 cells (Fig. 3) (28) are indicated by NA (not applicable).
Fitness-compensatory substitutions act at different steps of the viral life cycle.
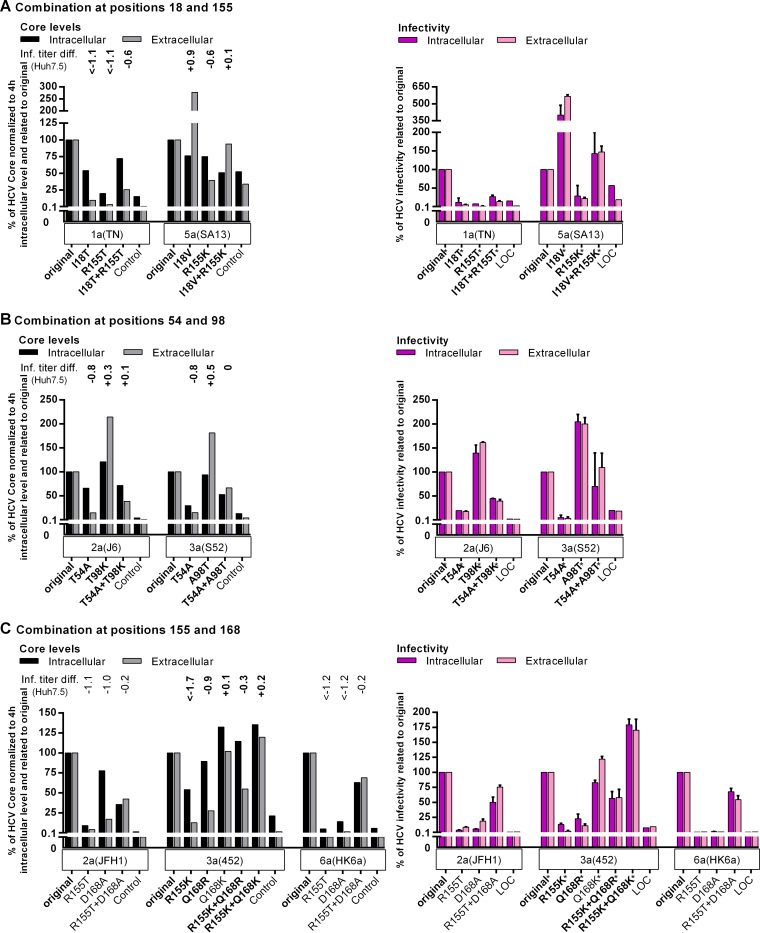
The fitness cost induced by PI resistance substitutions could be rescued by compensating NS3P substitutions (Fig. 3) (10, 28). We selected identified combinations of substitutions to study mechanisms of fitness compensation. In S29 cells, I18V increased the fitness of genotype 5a (isolate SA13) and of genotype 5a (isolate SA13) R155K by increasing assembly efficacy, as suggested by a strong increase in IC infectivity, combined with a minimal decrease in the IC core level (Fig. 6A). For genotype 1a (isolate TN), I18T and R155T individually decreased replication, while combining the substitutions partially rescued replication, as indicated by an increased IC core level (Fig. 6A). Thus, although changes at NS3P residue 18 had differential fitness effects, they compensated for the fitness cost induced by resistance substitutions at position 155 by increasing viral replication or assembly.
FIG 6.
Substitutions compensating for fitness impairment acted at different steps of the viral life cycle. The effects of combinations of substitutions at positions 18 and 155 (A), positions 54 and 98 (B), and positions 155 and 168 (C) on the HCV life cycle were studied by transfection of RNA transcripts from the indicated recombinants into S29 cells, followed by the determination of IC and EC core concentrations and infectivity titers as described in Materials and Methods. To account for possible differences in transfection efficiency, IC and EC core concentrations at 48 h were normalized to IC core concentrations at 4 h. To determine the effects of the indicated NS3P substitutions on viral fitness, normalized core values and infectivity titers of variant recombinants were related to the values of the respective original recombinants (original). Transfections of recombinants of the same genotype (isolate) were carried out in the same experiment. “Control” indicates the replication-deficient genotype 2a (isolate JFH1) GND negative-control virus. For this control, IC core concentrations at 48 h ranged from 9.4 × 103 to 25.0 × 103 fmol/liter, and EC core concentrations at 48 h ranged from 17.5 to 131.8 fmol/liter. The values shown for the control were normalized to the IC core concentrations at 4 h and were related to the values obtained for the respective original recombinants (original) as described for the NS3P variants. LOC indicates the lower cutoff of the infectivity titration assay. For automated counting of FFU, the LOCs for IC and EC infectivity titers were 1.5 log10 FFU/well and 2.3 log10 FFU/ml, respectively. In instances where low replication efficiency in S29 cells precluded automated counting, FFU were counted manually, and the resulting titers are indicated by asterisks. The LOCs for infectivity titers derived from manually counted FFU were 0.9 log10 FFU/well for IC titers and 1.6 log10 FFU/ml for EC titers. LOC values were related to the values obtained for the respective original recombinants (original) as described for NS3P variants. The NS3P substitutions in boldface were specifically selected in the indicated virus under PI treatment in the current study (Fig. 1; see also Tables S4, S5, S12, S14, and S16 in the supplemental material). For comparison, titer differences (Inf. titer diff.) observed following transfection of Huh7.5 cells are indicated above each variant. Titer differences were calculated as described in Materials and Methods. With some exceptions, titer differences were calculated using the titers determined in this study (Fig. 3) and are indicated in boldface. Titer differences for the genotype 2a (isolate JFH1) and genotype 6a (isolate HK6a) recombinants in panel C were calculated from infectivity titers determined in reference 28. For the genotype 3a (isolate 452) R155K recombinant, the titer difference was calculated on the basis of infectivity titers obtained from a transfection other than that for Fig. 3. The higher titer difference observed in this transfection might be due to slower acquisition of a compensatory NS3P substitution by the genotype 3a (isolate 452) R155K recombinant in this transfection or to reversion of the R155K substitution, as observed previously (28). The results obtained for genotype 1a (isolate TN) with V36M and R155K, previously suggested as a combination with compensatory effects, are shown in Fig. S5 in the supplemental material.
For genotype 2a (isolate J6) and genotype 3a (isolate S52), T98K and A98T, respectively, increased fitness and compensated for the T54A-induced fitness impairment. The T54A substitution impaired the replication of both recombinants, as suggested by decreased IC core levels (Fig. 6B). However, substitutions at position 98 increased the fitness of the original viruses and T54A variants primarily by increasing the efficacy of assembly, as suggested by increased IC infectivity (Fig. 6B). However, effects on replication efficiency cannot be excluded.
In Huh7.5 cells, genotype 3a (isolate 452) R155K plus Q168R had greater fitness than variants with individual substitutions (Fig. 3 and 6C). Additionally, R155T variants of genotype 2a (isolate JFH1) and genotype 6a (isolate HK6a) acquired D168A, which compensated for the R155T-induced fitness impairment (28). In S29 cells, the R155T substitution induced large decreases in replication (Fig. 6C). While Q168K had no effect on replication, other substitutions at position 168 decreased replication. Variants with combinations of substitutions at positions 155 and 168 had higher IC core levels than R155 variants. Therefore, substitutions at position 168 compensated for the replication cost imposed by substitutions at position 155.
For genotype 1, V36M was previously suggested to compensate for the fitness impairment induced by R155K (38). However, for genotype 1a (isolate TN), V36M alone or in combination with R155K resulted in decreased fitness in Huh7.5 cells and decreased replication in S29 cells (see Fig. S5 in the supplemental material).
Key PI resistance substitutions rescued PI inhibition of viral replication.
Following the transfection of S29 cells, the original genotype 1a, 2a, 3a, and 6a viruses and their R155K and A156S variants were treated with different concentrations of boceprevir (Fig. 5C; see also Fig. S4 in the supplemental material). For the original recombinants, PI treatment resulted in large decreases in IC and EC core levels and infectivity titers, suggesting that boceprevir inhibited replication. R155K and A156S variants had less-pronounced concentration-dependent decreases in IC core levels, corresponding to the resistance levels in Huh7.5 cells (Fig. 5C). For genotype 1a (isolate TN) A156T and genotype 2a (isolate JFH1) A156V, whose effects on resistance could not be determined in Huh7.5 cells due to genetic instability, IC core levels remained stable under increasing boceprevir concentrations, indicating high boceprevir resistance (Fig. 5C). This is in agreement with the findings of previous studies using enzymatic assays and replicons, which have associated A156T and A156V with high levels of PI resistance (4, 8, 10). The changes observed in IC core values (Fig. 5C) were reflected by changes in IC and EC infectivity titers and EC core levels (see Fig. S4 in the supplemental material). Thus, substitutions conferred boceprevir resistance by rescuing replication.
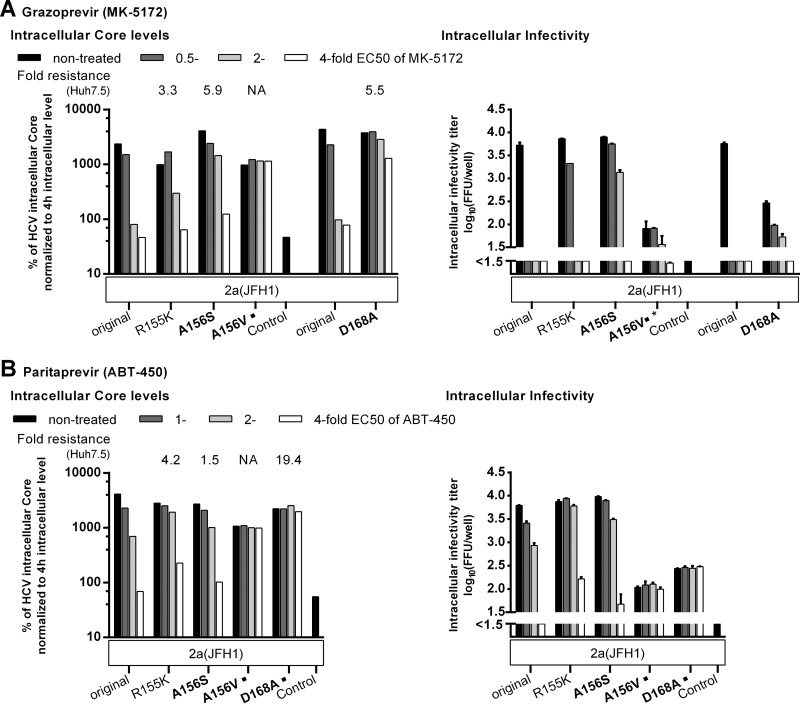
We obtained similar results when treating genotype 2a (isolate JFH1) R155K, A156S, A156V, or D168A variants with grazoprevir or paritaprevir (Fig. 7; see also Fig. S6 in the supplemental material). The A156V substitution was induced under treatment with both PIs, and the D168A substitution was selected with paritaprevir (Fig. 2; see also Tables S22 and S23 in the supplemental material). Both PIs inhibited the replication of the original recombinants. However, after treatment with 0.5 times the EC50 of grazoprevir, an additional effect on assembly was apparent, since IC core levels were minimally affected, while a pronounced decrease in IC infectivity was observed. As with boceprevir, the substitutions conferred PI resistance by rescuing replication. Most changes conferred only partial resistance to paritaprevir and grazoprevir. However, the A156V and D168A substitutions conferred total paritaprevir resistance, with stable IC and EC core levels and infectivity titers under increasing PI concentrations. In contrast, IC core levels for A156V or D168A variants were relatively stable under increasing grazoprevir concentrations, with comparatively large effects on IC and EC infectivity. Thus, these substitutions conferred only partial resistance to grazoprevir, probably due to an effect on assembly. Of the three PIs tested in this assay, grazoprevir was the most efficient against the highly resistant A156V variant.
FIG 7.
Key resistance substitutions at NS3P positions 155, 156 and 168 rescued the replication of genotype 2a (isolate JFH1) under treatment with newer PIs. RNA transcripts from the indicated genotype 2a (isolate JFH1) recombinants were transfected into S29 cells, and 4 h later, cultures were treated with the indicated concentrations of grazoprevir (MK-5172) (A) or paritaprevir (ABT-450) (B). IC core concentrations and infectivity titers were determined as described in Materials and Methods. To account for possible differences in transfection efficiency, IC core concentrations at 48 h were normalized to IC core concentrations at 4 h. The EC core concentrations and infectivity titers determined in these experiments are shown in Fig. S6 in the supplemental material. Transfections of recombinants treated with paritaprevir were carried out in the same experiment. Transfections of recombinants treated with grazoprevir were carried out in two different experiments. For each experiment, the original genotype 2a (isolate JFH1) recombinant was included. “Control” indicates the replication-deficient genotype 2a (isolate JFH1) GND negative-control virus. For this control, IC core concentrations at 48 h ranged from 9.4 × 103 to 25.0 × 103 fmol/liter. The values shown were normalized to 4-h IC core values as described for the NS3P variants. The break in the y axis indicates the lower cutoff (LOC) of the infectivity titration assay. For automated counting of FFU, the LOC for IC infectivity titers was 1.5 log10 FFU/well. For genotype 2a (isolate JFH1) A156V treated with grazoprevir, low replication efficiency in S29 cells precluded automated counting. FFU were counted manually, and the resulting titers are indicated by an asterisk. The LOC for IC infectivity titers derived from manually counted FFU was 0.9 log10 FFU/well. NS3P substitutions in boldface were specifically selected in the indicated virus under PI treatment in the current study. ■, the substitution was identified in escape variants emerging under treatment with the newer PIs (Fig. 2). For comparison, the fold resistance to grazoprevir (A) or paritaprevir (B) as determined in Huh7.5 cells (28) is indicated above each variant. Instances in which the fold resistance values could not be determined because the specified recombinant could not be grown in Huh7.5 cells (Fig. 3) (28) are indicated by NA (not applicable).
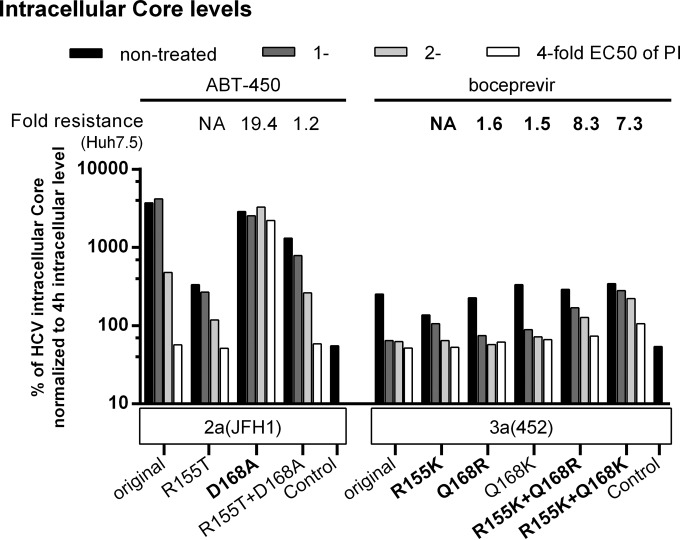
The spread of certain R155 variants required the acquisition of substitutions at NS3P residue 168. Therefore, the effects of single substitutions at residue 155 on PI resistance could not be determined in Huh7.5 cells. The S29 cell assay based on transfection of HCV transcripts allowed us to determine the effects of single or combined substitutions at positions 155 and 168 on resistance (Fig. 8; see also Fig. S7 in the supplemental material). For genotype 2a (isolate JFH1), D168A conferred a higher level of resistance to paritaprevir than R155T or the combination of R155T and D168A. For genotype 3a (isolate 452), Q168R and Q168K did not confer boceprevir resistance, while R155K, R155K plus Q168R, and R155K plus Q168K conferred similar intermediate resistance levels. Thus, combinations of substitutions at positions 155 and 168 apparently increased viral fitness but not resistance.
FIG 8.
Combinations of substitutions at positions 155 and 168 increased viral fitness but not PI resistance. RNA transcripts from the indicated genotype 2a (isolate JFH1) and genotype 3a (isolate 452) recombinants were transfected into S29 cells. Four hours later, cultures were treated with the indicated concentrations of paritaprevir (ABT-450) or boceprevir. IC core concentrations were determined as described in Materials and Methods. To account for possible differences in transfection efficiency, IC core concentrations at 48 h were normalized to IC core concentrations at 4 h. The IC and EC infectivity titers and EC core levels determined in these experiments are shown in Fig. S7 in the supplemental material. Transfections of recombinants of the same genotype (isolate) were carried out in the same experiment. “Control” indicates the replication-deficient genotype 2a (isolate JFH1) GND negative-control virus. For this control, IC core concentrations at 48 h ranged from 9.4 × 103 to 25.0 × 103 fmol/liter. The values shown were normalized to 4-h IC core values as described for NS3P variants. The NS3P substitutions in boldface were specifically selected in the indicated virus under treatment with the indicated PI. For comparison, the fold resistance to paritaprevir or boceprevir, as determined in Huh7.5 cells, is indicated above each variant. The fold resistance values were calculated as described in Materials and Methods. Values for boceprevir determined in this study (Fig. 4) are indicated in boldface; values for paritaprevir were reported previously (28). NA (not applicable) indicates that for single R155T and R155K variants, fold resistance values in Huh7.5 cells could not be determined, because introduction of these single substitutions led to the acquisition of additional substitutions at position 168 in first-passage virus stocks used for treatment experiments (Fig. 3) (28).
DISCUSSION
We examined viral escape and resistance to PIs for different HCV genotypes, and we identified NS3P positions 54, 155, 156, and 168 as hot spots for resistance substitutions. In general, substitutions at positions 155 and 156 selected during telaprevir or boceprevir treatment conferred cross-resistance to the newer PIs vaniprevir, grazoprevir, simeprevir, and paritaprevir. However, as reported previously, simeprevir showed high efficacy against A156S variants, and grazoprevir showed relatively high efficacy against R155K variants (28, 39). Simeprevir and paritaprevir had the highest efficacy against variants with substitutions at position 54, while grazoprevir showed relatively high efficacy against the otherwise resistant A156V variant of genotype 2a (isolate JFH1). PI treatment primarily decreased viral replication; for grazoprevir, an additional effect on assembly appears likely. Resistance substitutions rescued HCV replication under PI treatment and decreased viral fitness primarily by reducing viral replication. Fitness-compensating substitutions influenced either replication or assembly. These findings for HCV genotypes 1 to 6 in the context of the complete viral life cycle increase insight into various aspects of PI resistance.
Studies of HCV fitness and PI resistance require suitable in vitro systems. The development of genotype 1 to 6 replicons has permitted replication studies (40–46). However, NS3 is also involved in later steps of the viral life cycle (12). Cell culture infectious recombinants allow PI studies of the full viral life cycle (47), but lack of a strain-matched NS3P and helicase in some recombinants used here could influence functions. Yet similar EC50s were obtained for recombinants expressing only genotype-specific NS3P or the entire NS3 protein (21, 22, 48). Using infectious cultures, we previously demonstrated different PI susceptibilities of HCV genotypes 1 to 6 (21–23, 48, 49), correlating with clinical findings, and found that substitutions associated with PI or NS5A inhibitor resistance in vivo conferred resistance in vitro (21, 27). Recently, we found that substitutions at NS3P positions 155, 156, and 168, mediating genotype 1 PI resistance, conferred resistance on genotypes 2 to 6 (28). However, these studies could not reveal whether the engineered substitutions would be selected under PI treatment. This was studied here, and we found that in vivo PI resistance substitutions were selected in genotype 1 to 6 recombinants.
The selection of PI resistance substitutions depended on the HCV strain, the specific PI, and its concentration. As in clinical and culture studies for genotype 1, we identified positions 54, 155, and 156 as hot spots for the selection of substitutions under telaprevir or boceprevir treatment, whereas positions 155, 156, and 168 are hot spots for newer PIs (4, 8, 10). Although T54(A/S) was not selected by newer PIs, the emergence of Y56H under simeprevir, deldeprevir, or paritaprevir treatment suggests a role for this region in PI resistance. Although limited data are available for genotypes 2 to 6 (4, 8), recent clinical and in vitro studies on selected genotypes and substitutions suggested that these patterns might be found across genotypes (50–65). Certain substitutions were preferentially acquired by specific recombinants, depending on the genetic barrier to resistance, as evidenced by the fact that the substitutions identified were encoded by a single nucleotide change. For example, R155K was selected only in genotype 1a, 3a, and 5a viruses, where R155K requires one mutation. In contrast, two or three mutations are required for genotype 2a, 4a, and 6a viruses. This holds true for other published isolates of these subtypes (66). Such differences likely translate into clinical differences, and higher treatment failure rates were observed for genotype 1a viruses, which require one nucleotide change for R155K, than for genotype 1b viruses, which require two changes (4). However, T54(A/S) and A156(S/T/V) substitutions were selected only in certain recombinants, even though these substitutions required only one nucleotide change in all strains tested. Therefore, additional factors determined by the genetic context might influence the acquisition of resistance substitutions. The fitness of original recombinants and resistant variants is thought to influence how readily specific substitutions are selected (4, 8, 10), and our data support this notion. For example, of the genotype 2a recombinants studied, only the highly efficient genotype 2a (isolate JFH1) recombinant acquired A156V, and the fitness costs of T54A and A156S depended on the specific recombinant. We did not study NS4A, since it does not seem to influence sensitivity to PIs (67).
Viral fitness influences not only the selection, but also the persistence and spread, of resistant variants (4, 10, 68). We found that even in variants with decreased fitness, the engineered substitutions persisted. However, several recombinants acquired other NS3P substitutions, possibly compensating for the loss of fitness induced by resistance substitutions. Some engineered changes were not maintained following viral passage and might be less prone to persist in vivo, as shown for A156V (69). Here the engineered A156V substitution reverted, even when combined with coselected NS3P substitutions (L67S or I132V) identified in genotype 2a (isolate JFH1), suggesting compensating changes outside NS3P. Such changes could be relevant for other escape variants and could explain the selection of NS3P substitutions despite their fitness cost.
Given the multifunctional nature of the NS3/NS4A complex, NS3P resistance substitutions might influence different steps of the viral life cycle. However, we found that across genotypes, the fitness decrease induced by key resistance substitutions was due primarily to impairment of viral replication. An additional impairment of viral assembly cannot be excluded, since it is not clear how much the IC infectivity titer decreases in response to a decrease in replication. Certain NS3 substitutions with opposite effects on replication and the production of infectious viruses have been described (70) which could possibly be explained by NS3 mediating a switch between replication and assembly (12). Similarly, we found that genotype 2a (isolate JFH1) R155K and genotype 2a (isolate JFH1) A156S had decreased replication and increased assembly. Shimakami and colleagues, using the infectious genotype 1a virus H77S.3, reported the effects of a panel of PI resistance substitutions on replication and virus production (37, 71). In agreement with our results, T54A, R155K, R155T, A156T, A156V, and D168A resulted in replication decreases; R155T mediated an additional decrease in virus production (37). In contrast to our results, no decrease in replication was found for V36M (37). For A156S, decreased replication and virus production were reported (37). For genotype 2 to 6 recombinants, A156S also resulted in decreased replication. However, we did not observe decreased replication or decreased virus production for genotype 1a (isolate TN) A156S, and we did not observe decreased virus production for genotype 2a (isolate JFH1) A156S. Differences from the prior study (37) might be related to the HCV strain. Also, H77S.3 contains a reporter between p7 and NS2, both of which are important for viral assembly (12), and H77S.3 is not as well adapted to culture as the genotype 2a and 1a recombinants used in this study (20, 23, 71, 72). Our study shows partial agreement with prior replicon studies, where A156T and A156V had a strong negative effect, while R155K, A156S, and T54A had either no effect or a moderate negative effect, on replication (34–36, 73–76). Differences might be explained by the subgenomic nature of replicons and/or genotype/strain-specific differences. Our study represents the first systematic investigation of the effect of PI resistance substitutions on different steps of the viral life cycle for various HCV genotypes.
The substitutions rescuing fitness functioned by different mechanisms. The combination of two substitutions, both decreasing replication, improved fitness by rescuing replication, as observed for the addition of I18T or D168A to R155T and for R155K with Q168R. In contrast, I18V, T98K, or A98T increased fitness and, combined with resistance substitutions, rescued fitness primarily by increasing viral assembly. I18 is important for the formation of the NS3P N-terminal amphipathic α-helix, which targets NS3P to intracellular membranes (77). Substitutions at position 18 might influence the positioning of the NS3P active site on the membrane and thus NS3P activity, or they might induce conformational changes influencing interactions with NS4A and/or NS3H. Substitutions might increase HCV fitness across genotypes, since changes at position 18 were observed in genotype (isolate) 1a (TN), 2a (J6), and 5a (SA13) boceprevir escape variants, in treatment-naïve HCV genotype 1 or 4 patients (78), and in HCV genotype 1 patients with linear-PI treatment failure (38, 79, 80). Further, position 98 is located between two positions coordinating the zinc ion, and changes might influence NS3P stability (9, 81, 82). A98T was identified in HCV genotype 3 patients, either treatment naïve or failing telaprevir therapy without apparent resistance (55, 78). Interactions between residues 155 and 168 have been described previously (83, 84), and substitutions at position 168 might compensate for the loss of replication induced by substitutions at position 155 by restoring such interactions.
In H77S.3, telaprevir blocked replication and assembly (85). We showed that boceprevir inhibited replication across genotypes, and paritaprevir and grazoprevir inhibited genotype 2a (isolate JFH1) replication. Resistance substitutions primarily rescued decreased replication induced by PIs. A156V restored replication under treatment with boceprevir, paritaprevir, or grazoprevir. The relatively high efficacy of grazoprevir against genotype 2a (isolate JFH1) A156V might be mediated by an additional effect on assembly.
A number of substitutions selected under telaprevir or boceprevir conferred <2-fold resistance. Among these, substitutions at positions 14, 18, 68, 98, and 132 increased viral fitness. Also, several such substitutions (A7T, S49T, L67S, I132V, L175F, and I177V) conferred low resistance to certain newer PIs. The binding of newer PIs might depend more on interactions with residues at these positions. Supporting this notion, I132V has been shown to induce minor reductions in the binding affinity of linear PIs (86). For genotype 4a (isolate ED43), many substitutions did not confer resistance. This might be explained by natural telaprevir or boceprevir resistance (22) or by conformational instability of the catalytic triad of genotype 4 NS3P (87).
Certain newer PIs were efficient against telaprevir or boceprevir resistance substitutions (e.g., R155K or A156S). A156(T/V) and position 168 substitutions might pose a treatment challenge for newer PIs. Grazoprevir showed efficacy against genotype 2a (isolate JFH1) A156V in short-term assays, but A156V was the main variant selected by grazoprevir, suggesting that it can mediate resistance to efficient newer PIs.
We investigated which NS3P substitutions were preferentially selected under PI treatment across HCV genotypes 1 to 6 in the context of the infectious viral life cycle. The selection of substitutions depended on the genetic context of the viruses. Reverse genetic studies identified increased fitness and/or PI resistance as the requirement for the selection of various substitutions, including several with unknown clinical significance. Functional studies suggested that PI treatment resulted in the inhibition of replication across genotypes and that replication could be rescued by PI resistance substitutions. This study has important clinical implications and furthers understanding of the molecular virology of PI resistance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anna-Louise Sørensen and Lotte Mikkelsen for laboratory assistance, Steen Ladelund for statistical advice, Jannick Prentoe for scientific discussions, Bjarne Ø. Lindhardt, Ove Andersen, Linda Andresen, and Jens Ole Nielsen (Hvidovre Hospital), as well as Carsten Geisler (University of Copenhagen), for support, Charles Rice (Rockefeller University) and Suzanne U. Emerson and Robert H. Purcell (National Institutes of Health, USA) for providing reagents, and CTL Europe GmbH for customized software.
We declare no conflicts of interest.
S.B.N.S., J.B., and J.M.G. contributed the study concept and design. S.B.N.S, S.B.J., L.G., D.G.H., S.R., Y.-P.L., H.K., and J.M.G. acquired the data. S.B.N.S, S.B.J., J.B., and J.M.G. analyzed and interpreted the data. S.B.N.S and J.M.G. drafted the original manuscript. S.B.N.S, S.B.J, J.B. and J.M.G. revised the manuscript. J.B. and J.M.G. supervised the study.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02929-15.
REFERENCES
- 1.Hajarizadeh B, Grebely J, Dore GJ. 2013. Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol 10:553–562. doi: 10.1038/nrgastro.2013.107. [DOI] [PubMed] [Google Scholar]
- 2.Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. 2014. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol 61:S45–S57. doi: 10.1016/j.jhep.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 3.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2014. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarrazin C. 2016. The importance of resistance to direct antiviral drugs in HCV infection in clinical practice. J Hepatol 64:486–504. doi: 10.1016/j.jhep.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Pawlotsky JM. 2015. Hepatitis C treatment: the data flood goes on—an update from the liver meeting 2014. Gastroenterology 148:468–479. doi: 10.1053/j.gastro.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Pawlotsky JM. 2014. New hepatitis C therapies: the toolbox, strategies, and challenges. Gastroenterology 146:1176–1192. doi: 10.1053/j.gastro.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Antaki N, Craxi A, Kamal S, Moucari R, Van der Merwe S, Haffar S, Gadano A, Zein N, Lai CL, Pawlotsky JM, Heathcote EJ, Dusheiko G, Marcellin P. 2010. The neglected hepatitis C virus genotypes 4, 5 and 6: an international consensus report. Liver Int 30:342–355. doi: 10.1111/j.1478-3231.2009.02188.x. [DOI] [PubMed] [Google Scholar]
- 8.Lontok E, Harrington P, Howe A, Kieffer T, Lennerstrand J, Lenz O, McPhee F, Mo H, Parkin N, Pilot-Matias T, Miller V. 2015. Hepatitis C virus drug resistance-associated substitutions: state of the art summary. Hepatology 62:1623–1632. doi: 10.1002/hep.27934. [DOI] [PubMed] [Google Scholar]
- 9.Bartenschlager R, Lohmann V, Penin F. 2013. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol 11:482–496. doi: 10.1038/nrmicro3046. [DOI] [PubMed] [Google Scholar]
- 10.Vermehren J, Sarrazin C. 2012. The role of resistance in HCV treatment. Best Pract Res Clin Gastroenterol 26:487–503. doi: 10.1016/j.bpg.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Gottwein JM, Bukh J. 2008. Cutting the Gordian knot—development and biological relevance of hepatitis C virus cell culture systems. Adv Virus Res 71:51–133. doi: 10.1016/S0065-3527(08)00002-X. [DOI] [PubMed] [Google Scholar]
- 12.Lindenbach BD. 2013. Virion assembly and release. Curr Top Microbiol Immunol 369:199–218. doi: 10.1007/978-3-642-27340-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones DM, Atoom AM, Zhang X, Kottilil S, Russell RS. 2011. A genetic interaction between the core and NS3 proteins of hepatitis C virus is essential for production of infectious virus. J Virol 85:12351–12361. doi: 10.1128/JVI.05313-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mousseau G, Kota S, Takahashi V, Frick DN, Strosberg AD. 2011. Dimerization-driven interaction of hepatitis C virus core protein with NS3 helicase. J Gen Virol 92:101–111. doi: 10.1099/vir.0.023325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohlway A, Pirakitikulr N, Barrera FN, Potapova O, Engelman DM, Pyle AM, Lindenbach BD. 2014. Hepatitis C virus RNA replication and virus particle assembly require specific dimerization of the NS4A protein transmembrane domain. J Virol 88:628–642. doi: 10.1128/JVI.02052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aydin C, Mukherjee S, Hanson AM, Frick DN, Schiffer CA. 2013. The interdomain interface in bifunctional enzyme protein 3/4A (NS3/4A) regulates protease and helicase activities. Protein Sci 22:1786–1798. doi: 10.1002/pro.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neddermann P, Clementi A, De Francesco R. 1999. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J Virol 73:9984–9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheel TK, Rice CM. 2013. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med 19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bukh J, Meuleman P, Tellier R, Engle RE, Feinstone SM, Eder G, Satterfield WC, Govindarajan S, Krawczynski K, Miller RH, Leroux-Roels G, Purcell RH. 2010. Challenge pools of hepatitis C virus genotypes 1–6 prototype strains: replication fitness and pathogenicity in chimpanzees and human liver-chimeric mouse models. J Infect Dis 201:1381–1389. doi: 10.1086/651579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 21.Gottwein JM, Scheel TK, Jensen TB, Ghanem L, Bukh J. 2011. Differential efficacy of protease inhibitors against HCV genotypes 2a, 3a, 5a, and 6a NS3/4A protease recombinant viruses. Gastroenterology 141:1067–1079. doi: 10.1053/j.gastro.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Li YP, Ramirez S, Humes D, Jensen SB, Gottwein JM, Bukh J. 2014. Differential sensitivity of 5′UTR-NS5A recombinants of hepatitis C virus genotypes 1–6 to protease and NS5A inhibitors. Gastroenterology 146:812–821. doi: 10.1053/j.gastro.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 23.Li YP, Ramirez S, Jensen SB, Purcell RH, Gottwein JM, Bukh J. 2012. Highly efficient full-length hepatitis C virus genotype 1 (strain TN) infectious culture system. Proc Natl Acad Sci U S A 109:19757–19762. doi: 10.1073/pnas.1218260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottwein JM, Scheel TK, Hoegh AM, Lademann JB, Eugen-Olsen J, Lisby G, Bukh J. 2007. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology 133:1614–1626. doi: 10.1053/j.gastro.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Gottwein JM, Scheel TK, Callendret B, Li YP, Eccleston HB, Engle RE, Govindarajan S, Satterfield W, Purcell RH, Walker CM, Bukh J. 2010. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. J Virol 84:5277–5293. doi: 10.1128/JVI.02667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheel TK, Gottwein JM, Mikkelsen LS, Jensen TB, Bukh J. 2011. Recombinant HCV variants with NS5A from genotypes 1–7 have different sensitivities to an NS5A inhibitor but not interferon-alpha. Gastroenterology 140:1032–1042. doi: 10.1053/j.gastro.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 28.Jensen SB, Serre SB, Humes DG, Ramirez S, Li YP, Bukh J, Gottwein JM. 2015. Substitutions at NS3 residue 155, 156, or 168 of hepatitis C virus genotypes 2 to 6 induce complex patterns of protease inhibitor resistance. Antimicrob Agents Chemother 59:7426–7436. doi: 10.1128/AAC.01953-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuiken C, Combet C, Bukh J, Shin I, Deleage G, Mizokami M, Richardson R, Sablon E, Yusim K, Pawlotsky JM, Simmonds P. 2006. A comprehensive system for consistent numbering of HCV sequences, proteins and epitopes. Hepatology 44:1355–1361. doi: 10.1002/hep.21377. [DOI] [PubMed] [Google Scholar]
- 30.Russell RS, Meunier JC, Takikawa S, Faulk K, Engle RE, Bukh J, Purcell RH, Emerson SU. 2008. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of hepatitis C virus. Proc Natl Acad Sci U S A 105:4370–4375. doi: 10.1073/pnas.0800422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serre SB, Krarup HB, Bukh J, Gottwein JM. 2013. Identification of alpha interferon-induced envelope mutations of hepatitis C virus in vitro associated with increased viral fitness and interferon resistance. J Virol 87:12776–12793. doi: 10.1128/JVI.00901-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kieffer TL, De Meyer S, Bartels DJ, Sullivan JC, Zhang EZ, Tigges A, Dierynck I, Spanks J, Dorrian J, Jiang M, Adiwijaya B, Ghys A, Beumont M, Kauffman RS, Adda N, Jacobson IM, Sherman KE, Zeuzem S, Kwong AD, Picchio G. 2012. Hepatitis C viral evolution in genotype 1 treatment-naive and treatment-experienced patients receiving telaprevir-based therapy in clinical trials. PLoS One 7:e34372. doi: 10.1371/journal.pone.0034372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Susser S, Vermehren J, Forestier N, Welker MW, Grigorian N, Fuller C, Perner D, Zeuzem S, Sarrazin C. 2011. Analysis of long-term persistence of resistance mutations within the hepatitis C virus NS3 protease after treatment with telaprevir or boceprevir. J Clin Virol 52:321–327. doi: 10.1016/j.jcv.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 34.Lin C, Gates CA, Rao BG, Brennan DL, Fulghum JR, Luong YP, Frantz JD, Lin K, Ma S, Wei YY, Perni RB, Kwong AD. 2005. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J Biol Chem 280:36784–36791. doi: 10.1074/jbc.M506462200. [DOI] [PubMed] [Google Scholar]
- 35.Yi M, Tong X, Skelton A, Chase R, Chen T, Prongay A, Bogen SL, Saksena AK, Njoroge FG, Veselenak RL, Pyles RB, Bourne N, Malcolm BA, Lemon SM. 2006. Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations. J Biol Chem 281:8205–8215. doi: 10.1074/jbc.M510246200. [DOI] [PubMed] [Google Scholar]
- 36.Jiang M, Mani N, Lin C, Ardzinski A, Nelson M, Reagan D, Bartels D, Zhou Y, Nicolas O, Rao BG, Muh U, Hanzelka B, Tigges A, Rijnbrand R, Kieffer TL. 2013. In vitro phenotypic characterization of hepatitis C virus NS3 protease variants observed in clinical studies of telaprevir. Antimicrob Agents Chemother 57:6236–6245. doi: 10.1128/AAC.01578-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimakami T, Welsch C, Yamane D, McGivern DR, Yi M, Zeuzem S, Lemon SM. 2011. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology 140:667–675. doi: 10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarrazin C, Kieffer TL, Bartels D, Hanzelka B, Muh U, Welker M, Wincheringer D, Zhou Y, Chu HM, Lin C, Weegink C, Reesink H, Zeuzem S, Kwong AD. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767–1777. doi: 10.1053/j.gastro.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 39.Summa V, Ludmerer SW, McCauley JA, Fandozzi C, Burlein C, Claudio G, Coleman PJ, Dimuzio JM, Ferrara M, Di Filippo M, Gates AT, Graham DJ, Harper S, Hazuda DJ, McHale C, Monteagudo E, Pucci V, Rowley M, Rudd MT, Soriano A, Stahlhut MW, Vacca JP, Olsen DB, Liverton NJ, Carroll SS. 2012. MK-5172, a selective inhibitor of hepatitis C virus NS3/4a protease with broad activity across genotypes and resistant variants. Antimicrob Agents Chemother 56:4161–4167. doi: 10.1128/AAC.00324-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohmann V, Bartenschlager R. 2014. On the history of hepatitis C virus cell culture systems. J Med Chem 57:1627–1642. doi: 10.1021/jm401401n. [DOI] [PubMed] [Google Scholar]
- 41.Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 42.Saeed M, Scheel TK, Gottwein JM, Marukian S, Dustin LB, Bukh J, Rice CM. 2012. Efficient replication of genotype 3a and 4a hepatitis C virus replicons in human hepatoma cells. Antimicrob Agents Chemother 56:5365–5373. doi: 10.1128/AAC.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng B, Yu M, Xu S, Lee YJ, Tian Y, Yang H, Chan K, Mo H, McHutchison J, Delaney W, Cheng G. 2013. Development of robust hepatitis C virus genotype 4 subgenomic replicons. Gastroenterology 144:59–61. doi: 10.1053/j.gastro.2012.09.033. [DOI] [PubMed] [Google Scholar]
- 44.Saeed M, Gondeau C, Hmwe S, Yokokawa H, Date T, Suzuki T, Kato T, Maurel P, Wakita T. 2013. Replication of hepatitis C virus genotype 3a in cultured cells. Gastroenterology 144:56–58. doi: 10.1053/j.gastro.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 45.Yu M, Peng B, Chan K, Gong R, Yang H, Delaney W, Cheng G. 2014. Robust and persistent replication of the genotype 6a hepatitis C virus replicon in cell culture. Antimicrob Agents Chemother 58:2638–2646. doi: 10.1128/AAC.01780-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wose Kinge CN, Espiritu C, Prabdial-Sing N, Sithebe NP, Saeed M, Rice CM. 2014. Hepatitis C virus genotype 5a subgenomic replicons for evaluation of direct-acting antiviral agents. Antimicrob Agents Chemother 58:5386–5394. doi: 10.1128/AAC.03534-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li YP, Ramirez S, Gottwein JM, Scheel TK, Mikkelsen L, Purcell RH, Bukh J. 2012. Robust full-length hepatitis C virus genotype 2a and 2b infectious cultures using mutations identified by a systematic approach applicable to patient strains. Proc Natl Acad Sci U S A 109:E1101–E1110. doi: 10.1073/pnas.1203829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramirez S, Li YP, Jensen SB, Pedersen J, Gottwein JM, Bukh J. 2014. Highly efficient infectious cell culture of three HCV genotype 2b strains and sensitivity to lead protease, NS5A, and polymerase inhibitors. Hepatology 59:395–407. doi: 10.1002/hep.26660. [DOI] [PubMed] [Google Scholar]
- 50.Imhof I, Simmonds P. 2010. Development of an intergenotypic hepatitis C virus (HCV) cell culture method to assess antiviral susceptibilities and resistance development of HCV NS3 protease genes from HCV genotypes 1 to 6. J Virol 84:4597–4610. doi: 10.1128/JVI.02698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Imhof I, Simmonds P. 2011. Genotype differences in susceptibility and resistance development of hepatitis C virus to protease inhibitors telaprevir (VX-950) and danoprevir (ITMN-191). Hepatology 53:1090–1099. doi: 10.1002/hep.24172. [DOI] [PubMed] [Google Scholar]
- 52.Lenz O, Verbinnen T, Lin TI, Vijgen L, Cummings MD, Lindberg J, Berke JM, Dehertogh P, Fransen E, Scholliers A, Vermeiren K, Ivens T, Raboisson P, Edlund M, Storm S, Vrang L, de Kock H, Fanning GC, Simmen KA. 2010. In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435. Antimicrob Agents Chemother 54:1878–1887. doi: 10.1128/AAC.01452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng G, Chan K, Yang H, Corsa A, Pokrovskii M, Paulson M, Bahador G, Zhong W, Delaney W. 2011. Selection of clinically relevant protease inhibitor-resistant viruses using the genotype 2a hepatitis C virus infection system. Antimicrob Agents Chemother 55:2197–2205. doi: 10.1128/AAC.01382-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foster GR, Hézode C, Bronowicki JP, Carosi G, Weiland O, Verlinden L, van Heeswijk R, van Baelen B, Picchio G, Beumont M. 2011. Telaprevir alone or with peginterferon and ribavirin reduces HCV RNA in patients with chronic genotype 2 but not genotype 3 infections. Gastroenterology 141:881–889. doi: 10.1053/j.gastro.2011.05.046. [DOI] [PubMed] [Google Scholar]
- 55.De Meyer S, Ghys A, Foster GR, Beumont M, Van Baelen B, Lin TI, Dierynck I, Ceulemans H, Picchio G. 2013. Analysis of genotype 2 and 3 hepatitis C virus variants in patients treated with telaprevir demonstrates a consistent resistance profile across genotypes. J Viral Hepat 20:395–403. doi: 10.1111/jvh.12046. [DOI] [PubMed] [Google Scholar]
- 56.Gottwein JM, Jensen SB, Li YP, Ghanem L, Scheel TK, Serre SB, Mikkelsen L, Bukh J. 2013. Combination treatment with hepatitis C virus protease and NS5A inhibitors is effective against recombinant genotype 1a, 2a, and 3a viruses. Antimicrob Agents Chemother 57:1291–1303. doi: 10.1128/AAC.02164-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lenz O, Vijgen L, Berke JM, Cummings MD, Fevery B, Peeters M, De Smedt G, Moreno C, Picchio G. 2013. Virologic response and characterisation of HCV genotype 2–6 in patients receiving TMC435 monotherapy (study TMC435-C202). J Hepatol 58:445–451. doi: 10.1016/j.jhep.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 58.Silva MO, Treitel M, Graham DJ, Curry S, Frontera MJ, McMonagle P, Gupta S, Hughes E, Chase R, Lahser F, Barnard RJ, Howe AY, Howe JA. 2013. Antiviral activity of boceprevir monotherapy in treatment-naive subjects with chronic hepatitis C genotype 2/3. J Hepatol 59:31–37. doi: 10.1016/j.jhep.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 59.Bronowicki JP, Ratziu V, Gadano A, Thuluvath PJ, Bessone F, Martorell CT, Pol S, Terg R, Younes Z, He B, Eley T, Cohen D, Yu F, Hernandez D, McPhee F, Mendez P, Hughes E. 2014. Randomized trial of asunaprevir plus peginterferon alfa and ribavirin for previously untreated genotype 1 or 4 chronic hepatitis C. J Hepatol 61:1220–1227. doi: 10.1016/j.jhep.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 60.Lawitz E, Sullivan G, Rodriguez-Torres M, Bennett M, Poordad F, Kapoor M, Badri P, Campbell A, Rodrigues L Jr, Hu Y, Pilot-Matias T, Vilchez RA. 2015. Exploratory trial of ombitasvir and ABT-450/r with or without ribavirin for HCV genotype 1, 2, and 3 infection. J Infect 70:197–205. doi: 10.1016/j.jinf.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 61.Chayama K, Notsumata K, Kurosaki M, Sato K, Rodrigues L Jr, Setze C, Badri P, Pilot-Matias T, Vilchez RA, Kumada H. 2015. Randomized trial of interferon- and ribavirin-free ombitasvir/paritaprevir/ritonavir in treatment-experienced hepatitis C virus-infected patients. Hepatology 61:1523–1532. doi: 10.1002/hep.27705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moreno C, Hézode C, Marcellin P, Bourgeois S, Francque S, Samuel D, Zoulim F, Grange JD, Shukla U, Lenz O, Ouwerkerk-Mahadevan S, Fevery B, Peeters M, Beumont M, Jessner W. 2015. Efficacy and safety of simeprevir with PegIFN/ribavirin in naive or experienced patients infected with chronic HCV genotype 4. J Hepatol 62:1047–1055. doi: 10.1016/j.jhep.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 63.Hézode C, Asselah T, Reddy KR, Hassanein T, Berenguer M, Fleischer-Stepniewska K, Marcellin P, Hall C, Schnell G, Pilot-Matias T, Mobashery N, Redman R, Vilchez RA, Pol S. 2015. Ombitasvir plus paritaprevir plus ritonavir with or without ribavirin in treatment-naive and treatment-experienced patients with genotype 4 chronic hepatitis C virus infection (PEARL-I): a randomised, open-label trial. Lancet 385:2502–2509. doi: 10.1016/S0140-6736(15)60159-3. [DOI] [PubMed] [Google Scholar]
- 64.Zeuzem S, Ghalib R, Reddy KR, Pockros PJ, Ari ZB, Zhao Y, Brown DD, Wan S, DiNubile MJ, Nguyen BY, Robertson MN, Wahl J, Barr E, Butterton JR. 2015. Grazoprevir-elbasvir combination therapy for treatment-naive cirrhotic and noncirrhotic patients with chronic hepatitis C virus genotype 1, 4, or 6 infection: a randomized trial. Ann Intern Med 163:1–13. doi: 10.7326/M15-0785. [DOI] [PubMed] [Google Scholar]
- 65.Kumada H, Sato K, Takehara T, Nakamuta M, Ishigami M, Chayama K, Toyota J, Suzuki F, Nakayasu Y, Ochi M, Yamada I, Okanoue T. 2015. Efficacy of telaprevir-based therapy for difficult-to-treat patients with genotype 2 chronic hepatitis C in Japan. Hepatol Res 45:745–754. doi: 10.1111/hepr.12416. [DOI] [PubMed] [Google Scholar]
- 66.Kuiken C, Hraber P, Thurmond J, Yusim K. 2008. The hepatitis C sequence database in Los Alamos. Nucleic Acids Res 36:D512–D516. doi: 10.1093/nar/gkm962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gottwein JM, Jensen SB, Serre SB, Ghanem L, Scheel TK, Jensen TB, Krarup H, Uzcategui N, Mikkelsen LS, Bukh J. 2013. Adapted J6/JFH1-based hepatitis C virus recombinants with genotype-specific NS4A show similar efficacies against lead protease inhibitors, alpha interferon, and a putative NS4A inhibitor. Antimicrob Agents Chemother 57:6034–6049. doi: 10.1128/AAC.01176-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Franco S, Tural C, Nevot M, Molto J, Rockstroh JK, Clotet B, Martinez MA. 2014. Detection of a sexually transmitted hepatitis C virus protease inhibitor-resistance variant in a human immunodeficiency virus-infected homosexual man. Gastroenterology 147:599–601. doi: 10.1053/j.gastro.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 69.De Meyer S, Dierynck I, Ghys A, Beumont M, Daems B, Van Baelen B, Sullivan JC, Bartels DJ, Kieffer TL, Zeuzem S, Picchio G. 2012. Characterization of telaprevir treatment outcomes and resistance in patients with prior treatment failure: results from the REALIZE trial. Hepatology 56:2106–2115. doi: 10.1002/hep.25962. [DOI] [PubMed] [Google Scholar]
- 70.Pietschmann T, Zayas M, Meuleman P, Long G, Appel N, Koutsoudakis G, Kallis S, Leroux-Roels G, Lohmann V, Bartenschlager R. 2009. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog 5:e1000475. doi: 10.1371/journal.ppat.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamane D, McGivern DR, Wauthier E, Yi M, Madden VJ, Welsch C, Antes I, Wen Y, Chugh PE, McGee CE, Widman DG, Misumi I, Bandyopadhyay S, Kim S, Shimakami T, Oikawa T, Whitmire JK, Heise MT, Dittmer DP, Kao CC, Pitson SM, Merrill AH Jr, Reid LM, Lemon SM. 2014. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat Med 20:927–935. doi: 10.1038/nm.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saeed M, Andreo U, Chung HY, Espiritu C, Branch AD, Silva JM, Rice CM. 2015. SEC14L2 enables pan-genotype HCV replication in cell culture. Nature 524:471–475. doi: 10.1038/nature14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tong X, Chase R, Skelton A, Chen T, Wright-Minogue J, Malcolm BA. 2006. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res 70:28–38. doi: 10.1016/j.antiviral.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 74.He Y, King MS, Kempf DJ, Lu L, Lim HB, Krishnan P, Kati W, Middleton T, Molla A. 2008. Relative replication capacity and selective advantage profiles of protease inhibitor-resistant hepatitis C virus (HCV) NS3 protease mutants in the HCV genotype 1b replicon system. Antimicrob Agents Chemother 52:1101–1110. doi: 10.1128/AAC.01149-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou Y, Muh U, Hanzelka BL, Bartels DJ, Wei Y, Rao BG, Brennan DL, Tigges AM, Swenson L, Kwong AD, Lin C. 2007. Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: sensitivity to telaprevir (VX-950) and interferon alpha. J Biol Chem 282:22619–22628. doi: 10.1074/jbc.M610207200. [DOI] [PubMed] [Google Scholar]
- 76.Zhou Y, Bartels DJ, Hanzelka BL, Muh U, Wei Y, Chu HM, Tigges AM, Brennan DL, Rao BG, Swenson L, Kwong AD, Lin C. 2008. Phenotypic characterization of resistant Val36 variants of hepatitis C virus NS3-4A serine protease. Antimicrob Agents Chemother 52:110–120. doi: 10.1128/AAC.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brass V, Berke JM, Montserret R, Blum HE, Penin F, Moradpour D. 2008. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc Natl Acad Sci U S A 105:14545–14550. doi: 10.1073/pnas.0807298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paolucci S, Fiorina L, Piralla A, Gulminetti R, Novati S, Barbarini G, Sacchi P, Gatti M, Dossena L, Baldanti F. 2012. Naturally occurring mutations to HCV protease inhibitors in treatment-naive patients. Virol J 9:245. doi: 10.1186/1743-422X-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Susser S, Welsch C, Wang Y, Zettler M, Domingues FS, Karey U, Hughes E, Ralston R, Tong X, Herrmann E, Zeuzem S, Sarrazin C. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718. doi: 10.1002/hep.23192. [DOI] [PubMed] [Google Scholar]
- 80.Sølund C, Krarup H, Ramirez S, Thielsen P, Røge BT, Lunding S, Barfod TS, Madsen LG, Tarp B, Christensen PB, Gerstoft J, Laursen AL, Bukh J, Weis N. 2014. Nationwide experience of treatment with protease inhibitors in chronic hepatitis C patients in Denmark: identification of viral resistance mutations. PLoS One 9:e113034. doi: 10.1371/journal.pone.0113034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim JL, Morgenstern KA, Lin C, Fox T, Dwyer MD, Landro JA, Chambers SP, Markland W, Lepre CA, O'Malley ET, Harbeson SL, Rice CM, Murcko MA, Caron PR, Thomson JA. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343–355. doi: 10.1016/S0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 82.Love RA, Parge HE, Wickersham JA, Hostomsky Z, Habuka N, Moomaw EW, Adachi T, Hostomska Z. 1996. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87:331–342. doi: 10.1016/S0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 83.Romano KP, Ali A, Aydin C, Soumana D, Ozen A, Deveau LM, Silver C, Cao H, Newton A, Petropoulos CJ, Huang W, Schiffer CA. 2012. The molecular basis of drug resistance against hepatitis C virus NS3/4A protease inhibitors. PLoS Pathog 8:e1002832. doi: 10.1371/journal.ppat.1002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lim SR, Qin X, Susser S, Nicholas JB, Lange C, Herrmann E, Hong J, Arfsten A, Hooi L, Bradford W, Najera I, Smith P, Zeuzem S, Kossen K, Sarrazin C, Seiwert SD. 2012. Virologic escape during danoprevir (ITMN-191/RG7227) monotherapy is hepatitis C virus subtype dependent and associated with R155K substitution. Antimicrob Agents Chemother 56:271–279. doi: 10.1128/AAC.05636-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McGivern DR, Masaki T, Lovell W, Hamlett C, Saalau-Bethell S, Graham B. 2015. Protease inhibitors block multiple functions of the NS3/4A protease-helicase during the hepatitis C virus life cycle. J Virol 89:5362–5370. doi: 10.1128/JVI.03188-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Welsch C, Shimakami T, Hartmann C, Yang Y, Domingues FS, Lengauer T, Zeuzem S, Lemon SM. 2012. Peptidomimetic escape mechanisms arise via genetic diversity in the ligand-binding site of the hepatitis C virus NS3/4A serine protease. Gastroenterology 142:654–663. doi: 10.1053/j.gastro.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rimmert B, Sabet S, Ackad E, Yousef MS. 2014. A 3D structural model and dynamics of hepatitis C virus NS3/4A protease (genotype 4a, strain ED43) suggest conformational instability of the catalytic triad: implications in catalysis and drug resistivity. J Biomol Struct Dyn 32:950–958. doi: 10.1080/07391102.2013.800001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.