Abstract
DNA-protein crosslinks (DPCs) arise from a wide range of endogenous and exogenous chemicals, such as chemotherapeutic drugs and formaldehyde. Importantly, recent identification of aldehydes as endogenous genotoxins in Fanconi anemia has provided new insight into disease causation. Due to their bulky nature, DPCs pose severe threats to genome stability, but previous methods to measure formaldehyde-induced DPCs were incapable of discriminating between endogenous and exogenous sources of chemical. In this study, we developed methods that provide accurate and distinct measurements of both exogenous and endogenous DPCs in a structurally-specific manner. We exposed experimental animals to stable isotope-labeled formaldehyde ([13CD2]-formaldehyde) by inhalation and performed ultrasensitive mass spectrometry to measure endogenous (unlabeled) and exogenous (13CD2-labeled) DPCs. We found that exogenous DPCs readily accumulated in nasal respiratory tissues, but were absent in tissues distant to the site of contact. This observation together with the finding that endogenous formaldehyde-induced DPCs were present in all tissues examined suggests that endogenous DPCs may be responsible for increased risks of bone marrow toxicity and leukemia. Furthermore, the slow rate of DPC repair provided evidence for persistence of DPCs. In conclusion, our method for measuring endogenous and exogenous DPCs presents a new perspective for the potential health risks inflicted by endogenous formaldehyde, and may inform improved disease prevention and treatment strategies.
Keywords: DNA-Protein crosslinks, Endogenous, Exogenous, Formaldehyde, Mass spectrometry
Introduction
DNA damage is a major culprit in many diseases, including cancer and aging. Toxic chemicals originating from a wide range of endogenous and exogenous sources continuously damage DNA (1). It is well known that our DNA is not pristine and endogenous DNA damage occurs at a high frequency compared with exogenous damage, having more than 40,000 lesions in every cell in our body (2). The “Endogenous Exposome” was first explored by our laboratory from the concept of the “Exposome” that emphasized the importance of understanding relationships between human disease and lifetime exposures to both environmental and internal chemicals (2). Recent advances have identified endogenous aldehydes as possible genotoxic agents that cause severe bone marrow failure (BMF) and leukemia in mice with mutations in both Fanconi anemia group D2 (fancd2) and aldehyde dehydrogenase 2 (Aldh2) (3). Notably, Aldh2 is mutated in ~1 billion people, most frequently observed in Southeast Asians due to inherited genetic mutations (4). Deficiency of Aldh2 has been demonstrated to dramatically accelerate BMF in Japanese Fanconi anemia patients (5). In addition, endogenous formaldehyde has been shown to have greater genotoxicity than acetaldehyde (6–8). These findings raise new challenges for understanding the cellular environments involved in disease causation. Linear extrapolation of risk down to zero remains a common approach used by regulators, despite the fact that such models use no biology and that when biology exists, linear extrapolation will over estimate risks (9). Thus, better understanding of endogenous and exogenous DPC data are important for advancing science-based risk assessments.
DNA-protein crosslinks (DPCs) strongly disrupt normal DNA-protein interactions and interfere with DNA replication, transcription, and repair, which ultimately threatens genomic integrity and cell viability (10). DPCs can originate from exposure to environmental agents such as ionizing radiation, UV light, formaldehyde and transition metal ions (11). Furthermore, anticancer drugs have been developed based on their ability to form DPCs, such as nitrogen mustards, platinum containing agents (i.e.: cisplatin), and 5-aza-2’deoxycytidine. Notably, DPCs can also arise upon exposure to endogenous carcinogens, such as reactive aldehydes that are produced during various cellular processes (2,12). Due to their bulky nature, DPCs impact practically all aspects of genomic activity, and may therefore result in genomic instability or cell death. Thus, DPC formation and repair are crucial for genomic integrity in humans and consequently for understanding and preventing carcinogenesis (1,10,13–15). Surprisingly, however, DPC repair mechanisms have not been fully elucidated (15).
Until now, accurate and distinct measurements of both endogenous and exogenous DPCs have not been studied, which significantly limits our understanding of the biological effects and repair of DPCs. Accurate measurement of DPCs is complicated by two critical issues. First, covalent DPCs must be completely isolated from non-covalent DNA-protein complexes, since the latter are present in a clear excess over the former throughout the genome (16). Secondly, DPCs need to be measured with structural specificity, given that there are numerous lesions that complicate the background of DPCs resulting from both endogenous and exogenous electrophiles. However, such stringent measurements of DPCs have not been possible using previously available DPC detection techniques, such as SDS/KCl precipitation, phenol-chloroform extraction, nitrocellulose filter binding, comet assay, alkaline elution, and CsCl density gradient centrifugation, all of which are nonselective (16,17). Recently, mass spectrometry-based methods have emerged as powerful tools for chemical analysis of digested DPCs (11). Elegant work using mass spectrometry has comprehensively characterized and ultra-sensitively quantified the DPCs induced by bifunctional electrophiles, such as nitrogen mustards (18,19), cisplatin and diepoxybutane (20).
Formaldehyde is classified as an animal and human carcinogen. Humans are exposed to formaldehyde originating from environmental sources due to its broad range of applications and formation from various chemical processes, as well as internal sources due to its role as an essential intermediate of various cellular processes (21). As a strong electrophile, formaldehyde is a well-known crosslinking agent that is cytotoxic, resulting in increased cell proliferation, mutagenesis and nasal carcinomas in rats exposed by inhalation (22). Formaldehyde has been widely used to elucidate the cellular pathways of DPC repair (8,10,23,24). Furthermore, endogenous formaldehyde is an important source of DNA damage (6,8). It thus poses unique challenges for understanding the risks associated with exposure. One major question that must be addressed is whether inhaled formaldehyde can increase the risk for leukemia. A critical dilemma has been to determine whether exogenous formaldehyde reaches bone marrow. To this end, we focused on the tissue distribution of endogenous and exogenous formaldehyde-induced DPC formation in rats and nonhuman primates (NHP) at exposure concentrations that had been studied in earlier DPC experiments, as well as in carcinogenicity studies in rats (25–27). For the first time, the incorporation of stable isotope labeled formaldehyde ([13CD2]-formaldehyde) in animal exposures permitted readily distinguishable measurements of endogenous and exogenous DPCs, based on their mass differences through the use of ultrasensitive and selective liquid chromatography-mass spectrometry. These measurements of endogenous and exogenous DPCs provide critical new data that advances science-based risk assessment, as well as providing improved methods for understanding pathways for DPC repair, such as the Fanconi anemia pathways that may lead to better disease treatment.
Materials and Methods
Chemicals and materials
2’-deoxyguanosine, reduced L-glutathione (GSH), sodium phosphate monobasic (BioXtra), sodium phosphate dibasic (BioXtra), magnesium chloride solution (1 M), calcium chloride solution (1 M), sucrose, ammonium acetate, acetic acid, formic acid, formaldehyde-DNPH standards, sodium cyanoborohydride (NaCNBH3), pronase, leucine aminopeptidase M, carboxypeptidase Y, prolidase, alkaline phosphatase, and phosphodiesterase were all purchased from Sigma (St Louis, MO). DNAzol and Turbo DNase were obtained from Life Technologies (Grand Island, NY). Methanol, acetonitrile, high-performance liquid chromatography (HPLC) grade water, and formaldehyde solution (37%, w/w) were all purchased from Thermo Fisher Scientific (Pittsburgh, PA). Formaldehyde solution was used to synthesize the dG-Me-Cys standard. [15N5, 13C10]-deoxyguanosine, [15N5]-deoxyguanosine, [13CD2]-paraformaldehyde (≥98% purity), and [13CD2]-formaldehyde solution (20% w/w in D2O) were ordered from Cambridge Isotope Laboratories, Inc. (Cambridge, MA). [15N5]-deoxyguanosine and [13CD2]-formaldehyde solution were used to synthesize the [15N5]-dG-[13CD2]-Me-Cys (Internal Standard). 2,4-Dinitrophenylhydrazine (DNPH) cartridges were ordered from Waters (Milford, Mass). Proteinase K (20 mg/ml) was obtained from 5 PRIME, Inc. (Gaithersburg, MD). Amicon Ultra Centrifugal Filters (0.5 ml, 3K) were purchased from EMD Millipore (Darmstadt, Germany). Nanosep Centrifugal Devices (MWCO 3K) were purchased from Pall Life Sciences (Ann Arbor, MI).
Synthesis of dG-Me-Cys standard and internal standard
A synthesized standard, dG-Me-Cys, was prepared by digestion of dG-Me-Glutathione according to our previous study (28), using the experimental details described in the Supplementary Information. As demonstrated in a previous study, dG-Me-Cys showed a very close UV absorbance spectrum to that of dG (28). Thus, quantification of dG-Me-Cys was based on a dG standard calibration curve created using a HPLC-UV method with a detection wavelength at 254 nm. For the preparation of internal standard, [15N5]-deoxyguanosine and [13CD2]-formaldehyde were used to synthesize [15N5]-dG-[13CD2]-Me-Glutathione. Specifically, glutathione (100 mM) was incubated with [13CD2]-formaldehyde (60 mM) in 1.0 ml sodium phosphate buffer (100 mM, pH 7.2) at 37 °C for 3 h. [15N5]-dG was then added at a final concentration of 5 mM, and the crosslinking reaction was performed at 37 °C for 14 h. The same methods described in our previous study were applied for the purification and digestion of [15N5]-dG-[13CD2]-Me-Glutathione (28). The isolated [15N5]-dG-[13CD2]-Me-GSH was digested in 0.4 ml sodium phosphate buffer (40 mM, pH 6.0) by carboxypeptidase Y (50 µg/ml), and leucine aminopeptidase M (250 µg/ml) in the presence of MgCl2 (10 mM) and CaCl2 (10 mM) at room temperature for 15 h. The internal standard, [15N5]-dG-[13CD2]-Me-Cys, was purified and quantified by the same method as described in Supplementary Information.
DNA-Protein Crosslink isolation and digestion
Animal exposure was described in the Supplementary Information. DPCs were isolated from animal tissue samples using DNAzol from Life Technologies (Carlsbad, CA) following the manufacturer’s instructions with some modifications. First, animal tissues (30–50 mg) were homogenized in 1 ml of sucrose buffer (20 mM sodium phosphate, 250 mM sucrose, pH 7.2). The nuclei were isolated from homogenate solution by centrifugation at 1,500 × g and 4 °C for 10 min. The nuclear pellets were washed with 1 ml of 20 mM sodium phosphate buffer (pH 7.2). The washed nuclear pellets were dissolved in 50 µl of water, followed by the addition of 800 µl of DNAzol and 20 µl of proteinase K (20 mg/ml). After protein digestion at room temperature for 15 hours, DPCs were precipitated by adding 600 µl of 100% ethanol. The precipitated DPCs were washed with 800 µl of 75% ethanol 4 times. The isolated DPCs were dissolved in 450 µl digestion buffer (20 mM ammonium acetate, pH 6.0) containing MgCl2 (5 mM), CaCl2 (5 mM), DNase I (40 U/ml), alkaline phosphatase (4 U/ml), phosphodiesterase I (0.001 U/ml), and pronase (0.5 mg/ml). DNA digestion was performed at room temperature for 20 hours, resulting in the formation of nucleoside-peptide crosslinks. The enzymes and undigested DNA were removed using a Nanosep Centrifugal Device (MWCO, 3K) at 10,000 × g and 4 °C for 40 min. The resultant filtrate was combined with 30 µl of an enzyme mixture containing carboxypeptidase Y (0.3 mg/ml), aminopeptidase M (0.6 mg/ml), and prolidase (0.3 mg/ml) to further digest peptides to amino acids. Following a 20 hour incubation at room temperature, the reaction was terminated with the addition of 4 µl of 30% acetic acid. After adding 8 fmol of internal standard (4 µl, 2 nM), the final reaction mixtures were subjected to centrifugation at 10,000 × g and 4 °C for 40 min in a Nanosep Centrifugal Device to remove enzymes prior to HPLC purification. In order to eliminate matrix interferences, digestion enzymes were washed before use as described in the Supplementary Information.
HPLC purification and fractionation
The target analyte, dG-Me-Cys, was purified from the reaction mixture on an Agilent 1200 series UV HPLC system by using two C18 reverse phase columns connected in series (Waters Atlantis T3, 3 µm, 150 mm × 4.6 mm, Milford, MA). Following digestion, there were two products detected by HPLC-UV, both of which had similar retention times to that of DPCs. The use of two HPLC columns connected in series was found to greatly increase the resolution of HPLC separation between the dG-Me-Cys and the other digestion products. This increased the detection sensitivity of the DPC using nano-LC-ESI-MS/MS. The detection wavelength and column temperature were set at 254 nm and 15 °C, respectively. The mobile phases consisted of 0.05% acetic acid in water (A) and pure acetonitrile (B). The flow rate was 0.45 ml/min and elution gradient conditions were set as follows: 0 min, 1% B; 3 min, 1% B; 42 min, 3% B; 86 min, 3.5% B; 86.5 min, 80% B; 95 min, 80% B; 95.5 min, 1% B; 110 min, 1% B. dG-Me-Cys was eluted at a retention time of 81.5 min. The fractions containing target compounds were combined and concentrated to approximately 10–20 µl using a vacuum concentrator. The amount of digested dG in each sample was quantitated by the UV peak area (λ = 254 nm) at the corresponding retention time using a calibration curve ranging from 4 to 80 nmole dG on column.
Nano-LC-ESI-MS/MS analysis
Nano-LC-ESI-MS/MS analysis of dG-Me-Cys was performed on a TSQ Quantum Ultra triple-stage quadrupole mass spectrometer (Thermo Scientific, San Jose, CA) in positive-mode electrospray ionization. Selected reaction monitoring (SRM) mode was used to detect and quantify dG-Me-Cys. Sample introduction and separation was accomplished on a nano-Acquity Ultra Performance Liquid Chromatography system from Waters Corporation (Milford, MA). Following injection, the compounds were retained on two Symmetry C18 trap columns (5 µm, 20 × 0.18 mm) connected in series, followed by sample separation on a HSS T3 analytical column (1.8 µm, 100 × 0.1 mm) from Waters Corporation (Milford, MA) at room temperature. Mobile phases consisted of water with 0.05% acetic acid (A) and acetonitrile (B). Analytes were first retained on two trap columns with a flow rate of 1.8 µl/min of 1% mobile phase B for 3 min, followed by transfer to the analytical column. The flow rate was 0.4 µl/min and elution gradient conditions were set as follows: 0 min, 1% B; 3 min, 1% B; 16 min, 50% B; 20 min, 1% B; 35 min, 1% B. Mass spectrometer conditions were set as the following: source voltage, 2200 V; temperature of ion transfer tube, 280 °C; skimmer offset, 0; scan speed, 75 ms; scan width, 0.7 m/z; Q1 and Q3 peak width, 0.7 m/z; collision energy, 31 eV; collision gas (argon), 1.5 arbitrary units. A sample volume of 5 µl was injected for analysis. Endogenous DPC (dG-Me-Cys) and exogenous DPC (dG-[13CD2]-Me-Cys) were quantified by using the transition of m/z 401.1 to m/z 164.1, and m/z 404.1 to m/z 167.1, respectively. The transition of m/z 409.1 to m/z 172.1 was monitored for the internal standard ([15N5]-dG-[13CD2]-Me-Cys). Linear calibration curves were obtained using the ratio of integrated peak area of the analytical standard over that of the internal standard.
Method Validation for LC-ESI-MS/MS analysis
Standard curves were established by plotting the peak area ratios of solutions containing a fixed amount of internal standard ([15N5]-dG-[13CD2]-Me-Cys, 8.0 fmol) and increasing amounts of dG-Me-Cys from 0.15 to 6 fmol. The limit of quantification (LOQ) was determined as the amount of standard that produced a signal-to-noise ratio >10. To evaluate method accuracy and precision, analytical standard was spiked into the digested mixture of rat liver samples at three different levels (0.8, 2.0, and 5.0 fmol) followed by HPLC purification and concentration using a vacuum concentrator.
Artifact determination
The digestion process of DPCs isolated from animal tissues has the potential to generate some chemically reactive species, including nucleotides containing a hydroxymethyl group, hydroxymethyl-dG, dG, and peptides containing cysteine. These products could potentially crosslink to form artifactual dG-Me-Cys during sample preparation. Control studies were performed to determine the potential for artifactual formation of dG-Me-Cys. The extent of possible artifacts was evaluated in two ways. The first technique involved spiking isotope-labeled [13C10, 15N5]-dG into the DPC solution to determine the potential for extrinsic formation of [13C10, 15N5]-dG-Me-Cys. Specifically, [13C10, 15N5]-deoxyguanosine (4 mM) was treated with 600 µl of 0.5 M NaCNBH3 for 48 hours at 37 oC. The treated [13C10, 15N5]-dG was spiked into an equal amount of dG produced from DNA digestion. This spiked sample was subjected to DNA and protein digestion, followed by HPLC purification and LC-ESI-MS/MS analysis as described above. The potential artifact, [13C10, 15N5]-dG-Me-Cys, was monitored using the corresponding transition of m/z 416.1 to m/z 174.1. The second technique to determine artifact formation employed NaCNBH3 to reduce active hydroxymethyl groups in DNA and protein during the isolation of DPCs. Specifically, all solutions used in the isolation of DPCs, including homogenate buffer, wash solution and DNazol, contained 20 mM NaCNBH3. For control samples, NaCNBH3 was excluded in all solutions used for DPC isolation.
Results
Isolation and detection of formaldehyde specific DPCs
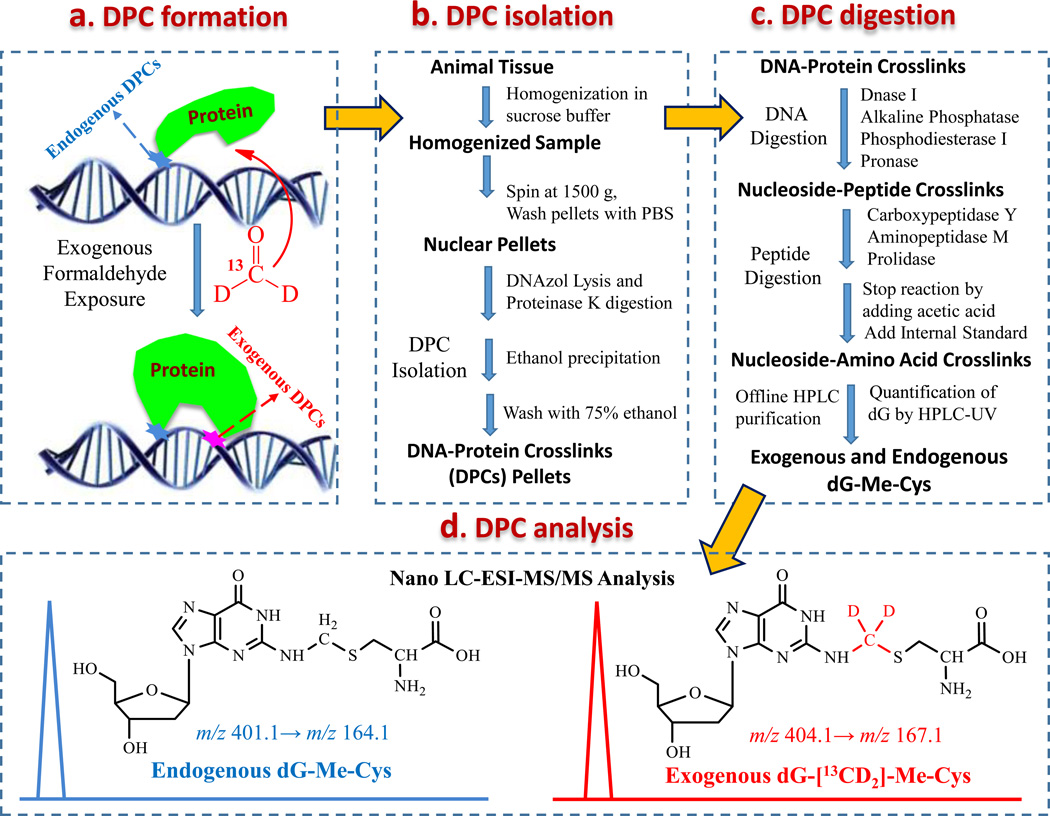
To study the chemical identity of formaldehyde induced-DPCs, we investigated the in vitro crosslinking reaction between nucleosides and amino acids in the presence of formaldehyde (29). Our results demonstrated that dG and cysteine cross-linked in the presence of formaldehyde to form dG-Me-Cys. We hypothesized that this crosslink would serve as a specific DPC biomarker of formaldehyde exposure based on its relatively high stability and abundance. To test this hypothesis, we examined formaldehyde-induced DPCs, specifically dG-Me-Cys, in both rats and NHPs that were exposed to [13CD2]-formaldehyde using nose-only and whole body inhalation chambers, respectively (Fig. 1a). The use of [13CD2]-formaldehyde permitted the differential measurement of endogenous and exogenous dG-Me-Cys based on their mass difference (+3 m/z) by mass spectrometry. A challenging step in structurally specific analysis of DPCs is the digestion or hydrolysis of bulky DPCs into small molecules suitable for MS/MS analysis. Many DPCs are unstable and chemical hydrolysis is generally too harsh to preserve the DPC linkages for their chemical identities. To this end, we optimized a mixture of enzymes to digest large DPCs into small nucleoside-amino acid crosslinks (dG-Me-Cys) under mild conditions (pH 6.0 and room temperature), thereby enabling preservation of the DPC chemical identity before MS analysis (Fig.1b and 1c). After complete digestion, formaldehyde induced-DPCs were isolated by offline HPLC fraction collection along with the quantification of digested dG using UV absorbance at 256 nm. Using authentic standards, the isolated endogenous and exogenous dG-Me-Cys were differentially quantified by mass spectrometry (Fig. 1d).
Figure 1.
Experimental workflow for distinct measurement of endogenous and exogenous DPCs. (a) Animals were exposed to [13CD2]-formaldehyde by inhalation. The use of [13CD2]-formaldehyde allowed the differentiation of endogenous and exogenous dG-Me-Cys crosslinks having the same chemical identity, but different masses. (b) DPCs were isolated from animal tissues using a DNAzol method. (c) DPCs were digested to nucleoside-amino acid crosslinks by DNA and protein enzymes, followed by offline HPLC purification of formaldehyde-DPCs, along with quantification of digested dG; (d) Endogenous and exogenous formaldehyde-DPCs were differentially quantified as the corresponding dG-Me-Cys using Nano LC-MS/MS. The normalization of DPC concentration was achieved by calculating the ratio of the amount of dG-Me-Cys to digested dG.
Nano UPLC-MS/MS method development for DPC analysis
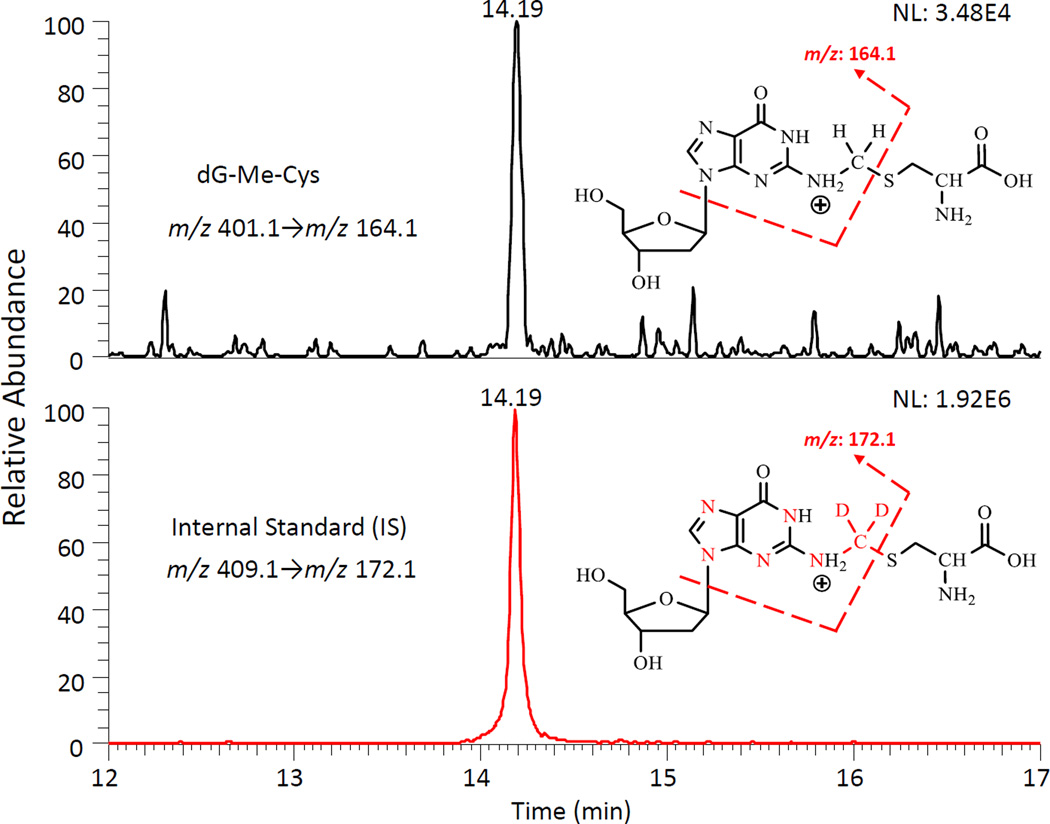
Given the inherent advantage of improved sensitivity, we adopted a nanospray UPLC-ESI-MS/MS methodology to measure dG-Me-Cys. The validated method exhibits ultra-sensitivity, as well as excellent accuracy and precision. Both the standard (dG-Me-Cys) and the internal standard (IS) were detected at the same retention time (Fig. 2). The limit of quantification was 37.5 amol on the column (S/N = 10). Good linearity was observed in the concentration range from 0.15 to 6.0 fmol with an R2 value of 0.9993 (Supplementary Fig. S1). The accuracy of this method was greater than 93.4%, with precision being less than 11.2% RSD using three spiking concentrations: 0.8, 2.0 and 5 fmol (Supplementary Table S1).
Figure 2.
Typical Nano-LC-ESI-MS/MS-SRM chromatogram and calibration curve of dG-Me-Cys. Nano-LC-ESI-MS/MS-SRM chromatograms of dG-Me-Cys (0.0375 fmol) and Internal Standard [15N5]-dG-[13CD2]-Me-Cys (0.5 fmol). NL = normalized spectrum to largest peak in particular chromatogram.
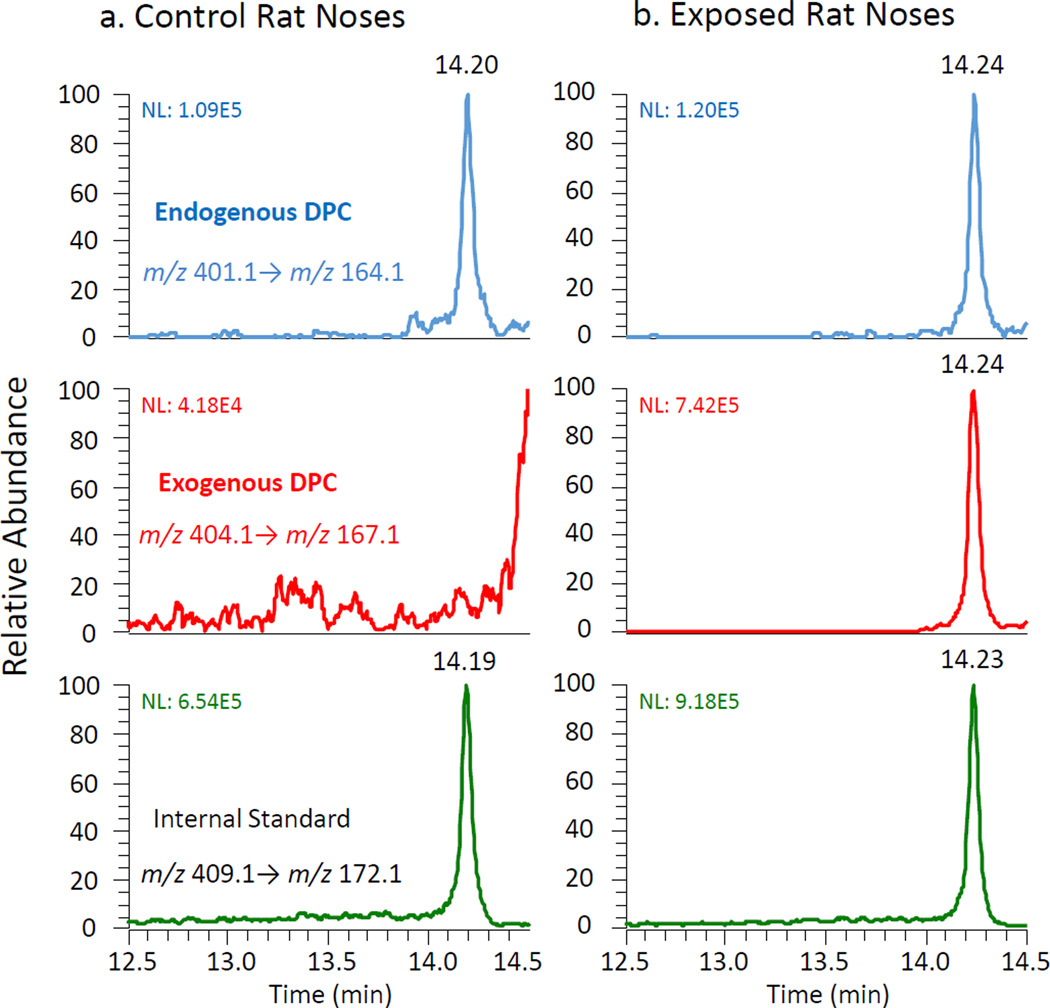
The validated method was then utilized to identify endogenous and exogenous formaldehyde-specific DPCs in rat nasal tissue (respiratory epithelium) as shown in Fig. 3. Only the endogenous peak (dG-Me-Cys) corresponding to the specific transition of m/z 401.1 to m/z 164.1 was detected in control rat nasal tissue at the same retention time as the internal standard at 14.20 min (Fig. 3a). Both endogenous and exogenous crosslinks were detected in [13CD2]-formaldehyde exposed rat nasal tissue at the same chromatographic retention time (Fig. 3b). The exogenous DPC fragment ions retained the stable isotope label, demonstrating the predicted isotope mass shift (+3 m/z) and were clearly distinguished from fragment ions of endogenous origin.
Figure 3.
Distinct detection of endogenous and exogenous dG-Me-Cys in control and exposed rat noses by Nano LC-MS/MS. NL, normalized spectrum to largest peak in this particular chromatogram.
Artifact determination for the validated method
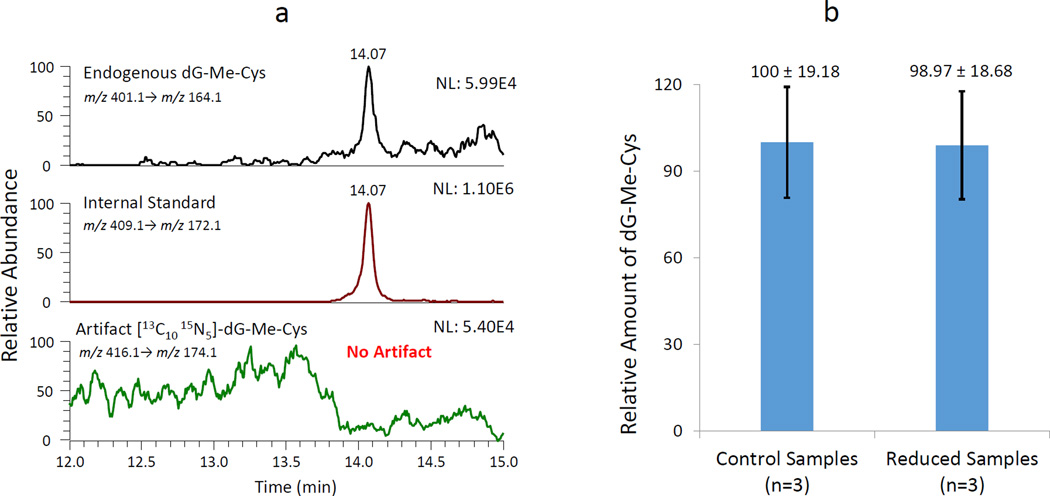
The potential for artifactual dG-Me-Cys formation during sample processing was investigated in two ways. First, stable isotope-labeled [13C10, 15N5]-dG was spiked into the reaction mixture to investigate whether free dG from the DNA digestion could form the crosslink. The potential formation of artifact, [13C10, 15N5]-dG-Me-Cys, was monitored using the corresponding transition of m/z 416.1 to m/z 174.1. The results showed that endogenous dG-Me-Cys and internal standard were observed as shown in the top and middle panels of Fig. 4a, respectively. On the contrary, no peak corresponding to the artifactual formation of [13C10, 15N5]-dG-Me-Cys was found in the same sample (Fig. 4a, bottom panel). This evidence demonstrated that the digestion of dG does not lead to artifactual formation of dG-Me-Cys during sample preparation. The second technique employed sodium cyanoborohydride (NaCNBH3) to reduce active hydroxymethyl groups that could possibly form artifactual dG-Me-Cys (Fig. 4b). The results demonstrated that the reduction of hydroxymethyl groups did not change the amount of dG-Me-Cys in rat liver samples. These data imply that the digested dG and other products do not generate artifacts that interfere with the detection of DPCs.
Figure 4.
Evaluation of artifactual formation of dG-Me-Cys. (a) Artifact determination in a rat liver sample spiked with isotope labeled [13C10, 15N5]-dG. (b) Relative amount of endogenous dG-Me-Cys in rat liver samples treated without (Control Sample) and with NaCNBH3 (Reduced Sample). Error bars represent the standard deviation of three individual experiments.
Distribution of formaldehyde-induced DPCs in rats and monkeys
Using this sensitive method, endogenous and exogenous formaldehyde-induced DPCs were distinctly quantified in selected tissues (nasal epithelium, peripheral blood mononuclear cells (PBMC), bone marrow, and liver) of animals exposed to either filtered air only or [13CD2]-formaldehyde. Consistent results from rat and NHP studies clearly demonstrate that exogenous DPCs were only found in the nasal epithelium of animals exposed to varying amounts of [13CD2]-formaldehyde. Specifically, exogenous DPCs were detected at 1.36 ± 0.20 crosslinks/108 dG in NHP nasal tissues exposed to [13CD2]-formaldehyde with a targeted aerosol concentration of 6.0 ppm for 2 consecutive days (6h per day) (Table 1). A much higher number of exogenous DPCs were measured at 18.18 ± 7.23 crosslinks/108 dG in the rat nasal tissues from animals exposed to [13CD2]-formaldehyde at a targeted concentration of 15 ppm for 4 consecutive days (6h per day) (Table 2). In contrast to the exogenous DPCs, endogenous DPCs were present in all examined tissues in both air control and exposed animals. Surprisingly, endogenous DPCs were present at higher amounts, compared to exogenous DPCs in exposed NHP nasal tissue (Table 1). More strikingly, the number of endogenous DPCs (15.46 ± 1.98 crosslinks/108 dG) in air control NHP livers, was close to the highest amounts of exogenous DPCs (18.18 ± 7.23 crosslinks/108 dG) observed in the rat nasal tissues exposed to 15 ppm [13CD2]-formaldehyde for 4 consecutive days, a highly carcinogenic exposure concentration in the rat cancer bioassays (30–32).
Table 1.
Formaldehyde-induced dG-Me-Cys in nose, peripheral blood mononuclear cells (PBMC), bone marrow, and liver of primates exposed to air control vs. 6 ppm of [13CD2]-formaldehyde (6 h per day).
| Tissue | Targeted [13CD2]- Formaldehyde Concentration (ppm) |
Exposure period (days) |
dG-Me-Cys (crosslink/108 dG) | |
|---|---|---|---|---|
| Endogenous | Exogenous | |||
| Nose | Air Control | 2 | 3.59 ± 1.01 (n=5) | ND* |
| 6 ppm | 2 | 3.76 ± 1.50 (n=5) | 1.36 ± 0.20 | |
| PBMC | Air Control | 2 | 1.34 ± 0.25 (n=5) | ND |
| 6 ppm | 2 | 1.57 ± 0.58 (n=4) | ND | |
| Bone Marrow |
Air Control | 2 | 2.30 ± 0.30 (n=4) | ND |
| 6 ppm | 2 | 1.40 ± 0.46 (n=5) | ND | |
| Liver | Air Control | 2 | 15.46 ± 1.98 (n=6) | ND |
| 6 ppm | 2 | 11.80 ± 2.21 (n=6) | ND | |
ND, Not Detected.
Table 2.
Formaldehyde-induced dG-Me-Cys in nasal tissue, peripheral blood mononuclear cells (PBMC), and bone marrow of rats exposed to Air Control verses 15 ppm of [13CD2]-formaldehyde (6 h per day).
| Tissue | Targeted [13CD2]- Formaldehyde Concentration (ppm) |
Exposure period (days) |
dG-Me-Cys (crosslink/108 dG) | |
|---|---|---|---|---|
| Endogenous | Exogenous | |||
| Nasal | 0 - Air Control | 4 | 6.50 ± 0.30 (n=5) | ND |
| 15.0 | 1 | 4.42 ± 1.10 (n=6) | 5.52 ± 0.80 | |
| 15.0 | 2 | 4.28 ± 2.34 (n=6) | 4.69 ± 1.76 | |
| 15.0 | 4 | 3.67 ± 0.80 (n=6) | 18.18 ± 7.23 | |
| PBMC | 0 - Air Control | 4 | 4.98 ± 0.61 (n=5) | ND |
| 15.0 | 1 | 3.26 ± 0.73 (n=4) | ND | |
| 15.0 | 2 | 3.00 ± 0.98 (n=5) | ND | |
| 15.0 | 4 | 7.19 ± 1.73 (n=5) | ND | |
| Bone Marrow |
0 - Air Control | 4 | 1.64 ± 0.49 (n=4) | ND |
| 15.0 | 1 | 1.80 ± 0.47 (n=4) | ND | |
| 15.0 | 2 | 1.84 ± 0.61 (n=4) | ND | |
| 15.0 | 4 | 1.58 ± 0.38 (n=4) | ND | |
ND, Not Detected.
Formation and elimination of formaldehyde-induced DPCs in rat nasal tissues
To monitor the induction and elimination of DPCs, we applied our method to investigate the accumulation and persistence of formaldehyde induced-DPCs in rat nasal tissues. As shown in Tables 2 and 3, both high and low exposure concentrations of inhaled formaldehyde gave rise to accumulation of exogenous DPCs in rat nasal tissues. At a high exposure concentration of 15 ppm [13CD2]-formaldehyde, similar amounts of exogenous DPCs were detected in 1-day and 2-day exposed rat nasal tissues (5.52 ± 0.80 and 4.69 ± 1.76 crosslinks/108 dG), while 4-day exposed rat nasal tissues showed a dramatic increase in the formation of exogenous DPCs (18.18 ± 7.23 crosslinks/108 dG) (Table 2). In contrast to the large changes in exogenous DPC formation, less difference in the amounts of endogenous DPCs were present in the same samples, suggesting that the endogenous DPCs were at steady-state concentrations. Most importantly, exogenous DPCs showed little change in the animals exposed for 28 days with either a 24 or 168 hour (7 day) post exposure recovery period (Table 3), indicating high stability and long-term persistence of DPCs with the dG-Me-Cys linkage. The discovery of accumulation and persistence of formaldehyde-DPCs highlights the need for a better understanding of DPC stability and repair.
Table 3.
Formaldehyde-induced dG-Me-Cys in nose of rats exposed to 2 ppm of [13CD2]-formaldehyde (6 h per day) for up to 28 days..
| Exposure period (day) | dG-Me-Cys (crosslink/108 dG) | |
|---|---|---|
| Endogenous | Exogenous | |
| 7 days | 4.78 ± 0.64 (n=4) | 0.96 ± 0.17 |
| 28 days | 4.51 ± 1.48 (n=3) | 2.46 ± 0.44 |
| 28 days + 24h post exposure |
3.78 ± 0.69 (n=4) | 2.12 ± 1.00 |
| 28 days + 168h post exposure |
3.51 ± 0.16 (n=3) | 2.14 ± 1.02 |
Discussion
Formaldehyde was first shown to be an animal carcinogen in 1980, causing squamous cell carcinomas in the nasal passages of exposed rats at concentrations at or above 6 ppm (30,31). Recent epidemiological studies have suggested that exposure to formaldehyde vapors may lead to the development of hematopoietic cancers such as leukemia, however these findings remain under debate (28,33–37). While leukemia has been a major finding in epidemiology studies of inhaled formaldehyde, no mechanism for leukemia has been established. The International Agency for Research on Cancer (IARC) working group also was not in full agreement on the evaluation of formaldehyde causing leukemia in humans (22,38). A recent paper by Coggon et al., did not find any leukemias in one of the largest cohorts of formaldehyde workers, even though some of those workers were exposed to inhaled formaldehyde at concentrations much higher than 2 ppm (37). In addition, the induction of leukemia by inhaled formaldehyde exposures has not been supported in rat carcinogenicity studies (39). Previous studies in rats and NHP also have not found any evidence that inhaled [13CD2]-formaldehyde reached the bone marrow as exogenous DNA adducts (28,34).
Endogenous formaldehyde is produced as an essential metabolic intermediate by enzymatic and nonenzymatic pathways, and as a detoxification product of xenobiotics during cellular metabolism (40). It is well understood that endogenous formaldehyde originates from numerous sources including one-carbon pool metabolism, amino acid metabolism, methanol metabolism, lipid peroxidation, cytochrome P450–catalyzed demethylation, and histone demethylation reactions (34,41,42). Endogenous formaldehyde has been assumed to be present in all aqueous body fluids because of its water solubility. Its half-life in humans is estimated to be 1–1.5 min (43). In particular, the concentration of endogenous formaldehyde was detected at ~0.1 mM in the blood of rats, monkeys, and humans (22,44–46). In addition, endogenous formaldehyde was found to be 2–4 times higher in the liver and nasal mucosa than in the blood of a rat (47). Endogenous formaldehyde is also formed in cellular nuclei, secondary to demethylation of histone III (42). These formaldehyde molecules are released in close proximity to DNA, providing an important source for bone marrow DPC and formaldehyde monoadducts (42). Thus, with endogenous formaldehyde-induced DNA adducts and DPC present in all tissues examined, but a complete lack of exogenous DNA adducts and DPC in tissues distant to the portal of entry (6,28), the key issue that must be addressed by risk assessors and epidemiologists is whether or how exogenous formaldehyde exposure could increase cancer risks at distal sites.
Due to their significant biological consequences, formaldehyde-induced DPCs have long been recognized as a highly mutagenic form of formaldehyde DNA damage. The amounts of formaldehyde-induced DPCs are considered to represent a good molecular biomarker of formaldehyde exposure (6,48). However, the lack of robust DPC measurements have limited our understanding of the genotoxic activity of formaldehyde. Previous studies using chloroform/iso-amyl alcohol/phenol extraction based DPC isolation following inhaled formaldehyde exposures in rats and NHPs found increased amounts of DPC formation in the nasal tissue but not in tissues distant from the portal of entry (25,27). In contrast, increased numbers of DPCs were detected in circulating lymphocytes of workers occupationally exposed to formaldehyde, and in remote tissues, such as bone marrow, liver, kidney, and testes of mice exposed to inhaled formaldehyde using SDS/KCl precipitation based DPC isolation (49,50). This scientific dispute may be due to the major disadvantage of previous DPC assays that were unable to provide chemical identity for specific and accurate measurements of DPCs. Furthermore, it is has been impossible to distinguish endogenous from exogenous formaldehyde-induced DPCs prior to the use of stable isotopes.
In particular, the National Research Council (NCR) committee considered our earlier work using sensitive and distinct measurements between exogenous and endogenous formaldehyde-induced DNA adducts as being highly informative and should be incorporated in the EPA’s draft Integrated Risk Information System (IRIS) assessment (34,48). Moreover, the NRC committee suggested that DPCs play a more important role in formaldehyde genotoxicity and carcinogenicity compared to DNA adducts (48). This study provides accurate measurements that discriminate between endogenous and exogenous DPCs through the use of stable isotope formaldehyde exposures and ultrasensitive mass spectrometry. Our data demonstrate that exogenous DPCs were only found in nasal samples of exposed animals, but not in sites remote to the portal of entry. Consistently, our previous studies reported that inhaled formaldehyde-induced DNA adducts and protein adducts were only detected in rat and NHP nasal tissues, but not in other tissues such as lung, liver, bone marrow and PBMC (28,34,51). These findings suggest that there is no additional dose to the bone marrow beyond that from endogenous formaldehyde. The lack of exogenous DPCs in remote tissues does not support a plausible relationship between inhaled formaldehyde and increased risks of leukemia. On the other hand, with high amounts of endogenous formaldehyde always present in all cells, one would expect the induction of disease states by this highly reactive aldehyde. Our previous work identified specific DNA adducts (N2-Me-dG) and DNA-DNA crosslinks (dG-Me-dG) induced by endogenous formaldehyde (34). This study provides the first evidence to the existence of endogenous formaldehyde-induced DPCs in all examined tissues. Furthermore, the recent paper by our lab and KJ Patel’s group clearly demonstrated that endogenous formaldehyde is a hematopoietic genotoxin and a metabolic carcinogen (6). Additionally, as recently reported, as many as 1/3 of the acute myeloid leukemia patients have deficiencies in aldehyde dehydrogenases that play important roles in preventing DNA damage by detoxifying endogenous aldehydes (52). Thus, recent medical research has provided strong data that better explain the formaldehyde epidemiology findings.
Formaldehyde-induced DPCs had been considered to be unstable in cell and animal studies. A rapid decline of the formaldehyde-induced DPCs was reported in cell culture, and there was no accumulation of DPCs observed in rat nasal tissues after repeated exposure (16,53,54). However, the present study demonstrated that exogenous DPCs accumulated with exposure time and showed little change after one week post exposure. Previous inconsistent results may be due to the use of non-structurally specific measurements which are highly susceptible to the interference of non-covalent DNA protein complexes. Although DPC repair mechanisms have not been well understood, proteolysis of the protein components of the DPC to small peptides has been reported as a protease-based DNA repair pathway specific for DPCs (10,11,15,55). The present study measures single amino acid-nucleoside crosslinks, specifically dG-Me-Cys. The small change in the amount of dG-Me-Cys measured one week post exposure does not rule out the possibility that the protein components of DPCs as a whole, do not undergo proteolytic degradation to small peptides. In a recent study, synthesized DPCs, including dG-Me-Cysteine, dG-Me-Glutathione, and dG-Me-Peptide crosslinks, were found to undergo rapid hydrolysis in vitro (28). The long term persistence of DPCs observed in vivo is likely due to the steric hindrance and localized hydrophobic conditions of DNA and the associated protein that greatly increase DPC stability.
The success of distinctly measuring endogenous and exogenous formaldehyde-DPCs has demonstrated the importance of structurally specific DPC data. The lack of exogenous DPCs in remote tissues away from the portal of entry suggest that the effects of inhaled formaldehyde may have been overemphasized, while endogenous formaldehyde has been underappreciated as a source of exposure leading to the induction of leukemia. The finding of exogenous DPC accumulation during inhalation exposure and its minimal repair following one week post exposure, provides the first evidence of long-term persistence of DPCs. The discovery of endogenous DPCs in all tissues examined raises an important perspective on the potential health risks posed by endogenous formaldehyde. Considering the high background of endogenous DPCs, it would be impossible to measure the induction, distribution, and elimination of exogenous DPCs without exogenous DPC measurements, particularly at low levels of exposure.
Future applications of this method include, but are not limited to 1) investigation of DPC repair mechanism(s) in vivo using formaldehyde-induced DPCs; 2) understanding whether and how the Fanconi anemia genes are involved in aldehyde induced-DPC repair; 3) application of such data for science-based risk assessment of aldehydes in a manner that contributes to the EPA’s IRIS program assessment of formaldehyde and aldehydes in general; 4) utilizing structurally specific methodologies to determine the makeup of DPCs induced by various endogenous and exogenous agents such as malondialdehyde, 4-hydroxynonenal, acrolein and aldehydic lesions present on abasic sites. Likewise, DPCs associated with exogenous anticancer drugs can be better understood, contributing to further advancements of chemotherapy. In summary, this study provides a new approach to accurately determine the roles of endogenous and environmental DPCs to form mutagenic DNA damage as factors that contribute to disease and provide more accurate data that can improve cancer risk assessments.
Supplementary Material
Acknowledgments
The authors would like to thank Dean Kracko, Raul Romero, and Dr. Melanie Doyle-Eisele at Lovelace Respiratory Research Institute for providing technical assistance and guidance in setting up and conducting the animal exposures. The costs of animal exposures at Lovelace Respiratory Research Institute were generously provided by Formacare and the Research Foundation for Health and Environmental Effects. The authors would also like to thank Leonard B. Collins for his assistance with HPLC purification and nano-UPLC-MS-MS.
Financial Support: This work was supported by the National Institutes of Environmental Health Sciences (NIEHS) Superfund Basic Research Program (P42 ES005948), NIEHS Center for Environmental Health and Susceptibility (P30 ES010126), the Texas Commission for Environmental Quality (582-12-21861) and Formacare.
Footnotes
Disclosure of Potential Conflicts of Interest:
The authors declare no competing financial interests.
Authors’ Contributions
Y.L., B.C.M., and J.A.S. conceptualized the study and designed the experiments; Y.L. performed experiments; W.M.B, and J.A.S. contributed new reagents or analytical tools; Y.L., R.Y., H.J.H., B.C.M., and J.A.S. performed data analyses; Y.L., R.Y., H.J.H., B.C.M., and J.A.S. wrote the manuscript.
References
- 1.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle. Proc Natl Acad Sci U S A. 2010;107(52):22475–22480. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamura J, Mutlu E, Sharma V, Collins L, Bodnar W, Yu R, et al. The endogenous exposome. DNA repair. 2014;19:3–13. doi: 10.1016/j.dnarep.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489(7417):571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Miller S, Younus H, Vanam R, Chen CH, Mochly-Rosen D, Hurley TD. Alda-1 is an agonist and chemical chaperone for the common human aldehyde dehydrogenase 2 variant. Nat Struct Mol Biol. 2010;17(2):159–164. doi: 10.1038/nsmb.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hira A, Yabe H, Yoshida K, Okuno Y, Shiraishi Y, Chiba K, et al. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood. 2013;122(18):3206–3209. doi: 10.1182/blood-2013-06-507962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pontel Lucas B, Rosado Ivan V, Burgos-Barragan G, Garaycoechea Juan I, Yu R, Arends Mark J, et al. Endogenous Formaldehyde Is a Hematopoietic Stem Cell Genotoxin and Metabolic Carcinogen. Mol Cell. 2015;60(1):177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duxin JP, Walter JC. What is the DNA repair defect underlying Fanconi anemia? Curr Opin Cell Biol. 2015;37:49–60. doi: 10.1016/j.ceb.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat Struct Mol Biol. 2011;18(12):1432–1434. doi: 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- 9.Crump KS. The linearized multistage model and the future of quantitative risk assessment. Hum Exp Toxicol. 1996;15(10):787–798. doi: 10.1177/096032719601501001. [DOI] [PubMed] [Google Scholar]
- 10.Stingele J, Schwarz MS, Bloemeke N, Wolf PG, Jentsch S. A DNA-dependent protease involved in DNA-protein crosslink repair. Cell. 2014;158(2):327–338. doi: 10.1016/j.cell.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 11.Tretyakova NY, Groehler A, Ji S. DNA–Protein Cross-Links: Formation, Structural Identities, and Biological Outcomes. Acc Chem Res. 2015;48(6):1631–1644. doi: 10.1021/acs.accounts.5b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voulgaridou GP, Anestopoulos I, Franco R, Panayiotidis MI, Pappa A. DNA damage induced by endogenous aldehydes: current state of knowledge. Mutat Res. 2011;711(1–2):13–27. doi: 10.1016/j.mrfmmm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Reardon JT, Sancar A. Repair of DNA-polypeptide crosslinks by human excision nuclease. Proc Natl Acad Sci U S A. 2006;103(11):4056–4061. doi: 10.1073/pnas.0600538103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano T, Morishita S, Katafuchi A, Matsubara M, Horikawa Y, Terato H, et al. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol Cell. 2007;28(1):147–158. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 15.Duxin JP, Dewar JM, Yardimci H, Walter JC. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell. 2014;159(2):346–357. doi: 10.1016/j.cell.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoulkamy MI, Nakano T, Ohshima M, Hirayama R, Uzawa A, Furusawa Y, et al. Detection of DNA-protein crosslinks (DPCs) by novel direct fluorescence labeling methods: distinct stabilities of aldehyde and radiation-induced DPCs. Nucleic Acids Res. 2012;40(18):e143. doi: 10.1093/nar/gks601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ide H, Shoulkamy MI, Nakano T, Miyamoto-Matsubara M, Salem AM. Repair and biochemical effects of DNA-protein crosslinks. Mutat Res. 2011;711(1–2):113–122. doi: 10.1016/j.mrfmmm.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Loeber R, Michaelson E, Fang Q, Campbell C, Pegg AE, Tretyakova N. Cross-linking of the DNA repair protein Omicron6-alkylguanine DNA alkyltransferase to DNA in the presence of antitumor nitrogen mustards. Chem Res Toxicol. 2008;21(4):787–795. doi: 10.1021/tx7004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michaelson-Richie ED, Ming X, Codreanu SG, Loeber RL, Liebler DC, Campbell C, et al. Mechlorethamine-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells. J Proteome Res. 2011;10(6):2785–2796. doi: 10.1021/pr200042u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gherezghiher TB, Ming X, Villalta PW, Campbell C, Tretyakova NY. 1,2,3,4-Diepoxybutane-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells. J Proteome Res. 2013;12(5):2151–2164. doi: 10.1021/pr3011974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swenberg JA, Lu K, Moeller BC, Gao L, Upton PB, Nakamura J, et al. Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment. Toxicol Sci. 2011;120(Suppl 1):S130–S145. doi: 10.1093/toxsci/kfq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.IARC. Lyon, France: World Health Organization; 2006. Formaldehyde, 2-Butoxyethanol and 1-tert-Butoxypropan-2-ol; pp. 1–287. [PMC free article] [PubMed] [Google Scholar]
- 23.de Graaf B, Clore A, McCullough AK. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair (Amst) 2009;8(10):1207–1214. doi: 10.1016/j.dnarep.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, Arakawa H, et al. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67(23):11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- 25.Casanova-Schmitz M, Heck HD. Effects of formaldehyde exposure on the extractability of DNA from proteins in the rat nasal mucosa. Toxicol Appl Pharmacol. 1983;70(1):121–132. doi: 10.1016/0041-008x(83)90185-0. [DOI] [PubMed] [Google Scholar]
- 26.Swenberg JA, Barrow CS, Boreiko CJ, Heck HD, Levine RJ, Morgan KT, et al. Non-Linear Biological Responses to Formaldehyde and Their Implications for Carcinogenic Risk Assessment. Carcinogenesis. 1983;4(8):945–952. doi: 10.1093/carcin/4.8.945. [DOI] [PubMed] [Google Scholar]
- 27.Casanova M, Morgan KT, Steinhagen WH, Everitt JI, Popp JA, Heck HD. Covalent binding of inhaled formaldehyde to DNA in the respiratory tract of rhesus monkeys: pharmacokinetics, rat-to-monkey interspecies scaling, and extrapolation to man. Fundam Appl Toxicol. 1991;17(2):409–428. doi: 10.1016/0272-0590(91)90230-2. [DOI] [PubMed] [Google Scholar]
- 28.Yu R, Lai Y, Hartwell HJ, Moeller BC, Doyle-Eisele M, Kracko D, et al. Formation, accumulation and hydrolysis of endogenous and exogenous formaldehyde induced DNA damage. Toxicol Sci. 2015;146(1):170–182. doi: 10.1093/toxsci/kfv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu K, Ye W, Zhou L, Collins LB, Chen X, Gold A, et al. Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J Am Chem Soc. 2010;132(10):3388–3399. doi: 10.1021/ja908282f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swenberg JA, Kerns WD, Mitchell RI, Gralla EJ, Pavkov KL. Induction of Squamous Cell Carcinomas of the Rat Nasal Cavity by Inhalation Exposure to Formaldehyde Vapor. Cancer Res. 1980;40(9):3398–3402. [PubMed] [Google Scholar]
- 31.Kerns WD, Pavkov KL, Donofrio DJ, Gralla EJ, Swenberg JA. Carcinogenicity of formaldehyde in rats and mice after long-term inhalation exposure. Cancer Res. 1983;43(9):4382–4392. [PubMed] [Google Scholar]
- 32.Monticello TM, Swenberg JA, Gross EA, Leininger JR, Kimbell JS, Seilkop S, et al. Correlation of Regional and Nonlinear Formaldehyde-induced Nasal Cancer with Proliferating Populations of Cells. Cancer Res. 1996;56(5):1012–1022. [PubMed] [Google Scholar]
- 33.Zhang L, Steinmaus C, Eastmond DA, Xin XK, Smith MT. Formaldehyde exposure and leukemia: a new meta-analysis and potential mechanisms. Mutat Res. 2009;681(2–3):150–168. doi: 10.1016/j.mrrev.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Lu K, Collins LB, Ru H, Bermudez E, Swenberg JA. Distribution of DNA adducts caused by inhaled formaldehyde is consistent with induction of nasal carcinoma but not leukemia. Toxicol Sci. 2010;116(2):441–451. doi: 10.1093/toxsci/kfq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehman-McKeeman L. Paracelsus and Formaldehyde 2010: The Dose to the Target Organ Makes the Poison. Toxicol Sci. 2010;116(2):361–363. doi: 10.1093/toxsci/kfq164. [DOI] [PubMed] [Google Scholar]
- 36.Beane Freeman LE, Blair A, Lubin JH, Stewart PA, Hayes RB, Hoover RN, et al. Mortality from lymphohematopoietic malignancies among workers in formaldehyde industries: the National Cancer Institute Cohort. J Natl Cancer Inst. 2009;101(10):751–761. doi: 10.1093/jnci/djp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coggon D, Ntani G, Harris EC, Palmer KT. Upper airway cancer, myeloid leukemia, and other cancers in a cohort of British chemical workers exposed to formaldehyde. Am J Epidemiol. 2014;179(11):1301–1311. doi: 10.1093/aje/kwu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.IARC. Lyon, France: World Health Organization; 2012. Formaldehyde, A review of human carcinogens. Part F: Chemical agents and related occupations; pp. 401–435. [Google Scholar]
- 39.NTP. 13th Report on Carcinogens. Washington, DC: US Department of Health and Human Services; 2014. [Google Scholar]
- 40.ATSDR. Toxicological profile for Formaldehyde. Atlanta, GA: U.S. Department of Health and Human Services; 1999. [Google Scholar]
- 41.Dhareshwar SS, Stella VJ. Your prodrug releases formaldehyde: should you be concerned? No! J Pharm Sci. 2008;97(10):4184–4193. doi: 10.1002/jps.21319. [DOI] [PubMed] [Google Scholar]
- 42.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan J, Krieger G. Clinical Environmental Health and Toxic Exposures. Philadelphia: Lippincott Williams & Wilkins; 2001. Formaldehyde. [Google Scholar]
- 44.Heck HD, Casanova-Schmitz M, Dodd PB, Schachter EN, Witek TJ, Tosun T. Formaldehyde (CH2O) concentrations in the blood of humans and Fischer-344 rats exposed to CH2O under controlled conditions. Am Ind Hyg Assoc J. 1985;46(1):1–3. doi: 10.1080/15298668591394275. [DOI] [PubMed] [Google Scholar]
- 45.Casanova M, Heck HD, Everitt JI, Harrington WW, Jr, Popp JA. Formaldehyde concentrations in the blood of rhesus monkeys after inhalation exposure. Food Chem Toxicol. 1988;26(8):715–716. doi: 10.1016/0278-6915(88)90071-3. [DOI] [PubMed] [Google Scholar]
- 46.NTP. Final Report on Carcinogens Background Document for Formaldehyde. Washington, DC: US Department of Health and Human Services; 2010. [Google Scholar]
- 47.Heck HD, White EL, Casanova-Schmitz M. Determination of formaldehyde in biological tissues by gas chromatography/mass spectrometry. Biomed Mass Spectrom. 1982;9(8):347–353. doi: 10.1002/bms.1200090808. [DOI] [PubMed] [Google Scholar]
- 48.NRC. Review of the environmental protection agency's draft IRIS assessment of formaldehyde. The National Academies Press; 2011. [PubMed] [Google Scholar]
- 49.Shaham J, Bomstein Y, Gurvich R, Rashkovsky M, Kaufman Z. DNA–protein crosslinks and p53 protein expression in relation to occupational exposure to formaldehyde. Occup Environ Med. 2003;60(6):403–409. doi: 10.1136/oem.60.6.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye X, Ji Z, Wei C, McHale CM, Ding S, Thomas R, et al. Inhaled formaldehyde induces DNA-protein crosslinks and oxidative stress in bone marrow and other distant organs of exposed mice. Environ Mol Mutagen. 2013;54(9):705–718. doi: 10.1002/em.21821. [DOI] [PubMed] [Google Scholar]
- 51.Edrissi B, Taghizadeh K, Moeller BC, Kracko D, Doyle-Eisele M, Swenberg JA, et al. Dosimetry of N(6)-formyllysine adducts following [(1)(3)C(2)H(2)]-formaldehyde exposures in rats. Chem Res Toxicol. 2013;26(10):1421–1423. doi: 10.1021/tx400320u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith C, Gasparetto M, Humphries K, Pollyea DA, Vasiliou V, Jordan CT. Aldehyde dehydrogenases in acute myeloid leukemia. Ann N Y Acad Sci. 2014;1310:58–68. doi: 10.1111/nyas.12414. [DOI] [PubMed] [Google Scholar]
- 53.Grafstrom RC, Fornace A, Jr, Harris CC. Repair of DNA damage caused by formaldehyde in human cells. Cancer Res. 1984;44(10):4323–4327. [PubMed] [Google Scholar]
- 54.Casanova M, Morgan KT, Gross EA, Moss OR, Heck HA. DNA-protein cross-links and cell replication at specific sites in the nose of F344 rats exposed subchronically to formaldehyde. Fundam Appl Toxicol. 1994;23(4):525–536. doi: 10.1006/faat.1994.1137. [DOI] [PubMed] [Google Scholar]
- 55.Stingele J, Habermann B, Jentsch S. DNA-protein crosslink repair: proteases as DNA repair enzymes. Trends Biochem Sci. 2015;40(2):67–71. doi: 10.1016/j.tibs.2014.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.