ABSTRACT
An increase in ethanol yield by yeast from the fermentation of conventional sugars such as glucose and sucrose is possible by reducing the production of a key byproduct such as cellular biomass. Previously we have reported that overexpression of PHO8 gene encoding non-specific ATP-hydrolyzing alkaline phosphatase can lead to a decrease in cellular ATP content and to an increase in ethanol yield during glucose fermentation by Saccharomyces cerevisiae. In this work we further report on 2 new successful approaches to reduce cellular levels of ATP that increase ethanol yield and productivity. The first approach is based on the overexpression of the heterologous Escherichia coli apy gene encoding apyrase or SSB1 part of the chaperon that exhibit ATPase activity in yeast. In the second approach we constructed a futile cycle by the overexpression of S. cerevisiae genes encoding pyruvate carboxylase and phosphoenolpyruvate carboxykinase in S. cerevisiae. These genetically engineered strains accumulated more ethanol compared to the wild-type strain during alcoholic fermentation.
KEYWORDS: ATPase, apyrase, ethanol, futile cycle, Saccharomyces cerevisiae
Introduction
Today, global industrial ethanol production mostly relies on the fermentation of conventional feedstocks such as glucose (derived from starch) and sucrose. The ethanol produced from these feedstocks is known as 1st generation ethanol.1 In 2014, about 93 billion liters of 1st generation ethanol was produced worldwide (http://www.afdc.energy.gov/data/10331). Today, the production of 1st generation ethanol represents the largest industrial biotechnological application of yeast. For this reason, new scientific approaches to improve ethanol yield and productivity in yeast are of great importance. Ethanol is produced from glucose or sucrose by Saccharomyces cerevisiae under anaerobic conditions as the main end product of sugar catabolism. The pathway for the production of ethanol from glucose in yeast (known as glycolysis or Embden-Meyerhof-Parnas, EMP, pathway), yields 2 moles of ATP from one mole of glucose. Some of the ATP produced by glycolysis is subsequently used to produce cellular biomass. Under anaerobic condition, 93% of the glucose used is converted to ethanol and to other low-molecular weight products while the remaining 7% is primarily converted to cell biomass. Among the other minor products of alcoholic fermentation, glycerol is the most abundant. Theoretically, it is possible to increase ethanol yield and productivity by decreasing the accumulation of cellular biomass and glycerol. There are several metabolic engineering approaches to increase ethanol yield by reducing glycerol production by yeast.1 In one approach, NADH accumulated in yeast during biomass production, is used for the reduction of acetate and to simultaneously lower the synthesis of the enzymes involved in glycerol synthesis from dihyroxyacetone-3-phosphate. By comparison to glycerol reduction, much less is known about ways to increase ethanol yield by decreasing biomass production.
There have been several attempts to reduce ATP production in yeast. One approach substituted genes of EMP pathway with Zymomonas mobilis genes encoding unique enzymes of Entner-Doudoroff (ED) pathway which is used for glucose alcoholic fermentation in this bacterium. The ED pathway in Zymomonas only yields 1 mol of ATP per mole of glucose.2,3 As a result of the lower ATP yield from glucose, Z. mobilis converts 97% of glucose to ethanol with only 3% of the sugar converted to cell biomass. Attempts to express of Z. mobilis genes encoding specific aldolase and dehydratase in S. cerevisiae were only partially successful as transformants did not express dehydratase activity.4 Other approaches to lower ATP production in yeast were based on ATP dissipation using futile cycle formed by phosphofructokinase and fructose-1,6-bisphosphatase5 or by the activation of enzymes involved in ATP degradation, such as acid phosphatase Pho56 or Fo, the subunit of membrane ATPase.7 Recently, we have found that the overexpression of the vacuolar alkaline phosphatase Pho8 leads to an increase in yield and productivity of ethanol synthesis from glucose in laboratory and in industrial strains of S. cerevisiae whereas the expression of the truncated cytosol located form is detrimental to the cells.8 Since our earlier published work, we designed and tested several other approaches to increasing ethanol yield from glucose based on the construction of alternative futile cycles which could dissipate intracellular ATP level. In this work, we report on the successful engineering of yeast cells for the reduction of cellular ATP level with an increase in ethanol yield and productivity during glucose fermentation. These approaches entail the overexpression of ATP degrading enzymes apyrase and SSB1 part of the chaperon or the construction of a futile cycle due to overexpression of S. cerevisiae genes encoding pyruvate carboxylase and phosphoenolpyruvate carboxykinase.
Results and discussion
NSSB1 and apyrase
S. cerevisiae SSB1 gene encodes ribosome associated molecular chaperon. Ssb1p is responsible for the correct folding of nascent polypeptide chain during its release from the ribosome. The energy from ATP hydrolysis is utilized to stabilize Ssb1p complex with nascent protein. The Ssb1p consists of 3 distinct domains. One of these is located in the N-terminal 44 kDa region of the protein and has ATPase activity that is repressed by 2 other C-terminal domains. The isolated 44 kDa ATPase domain was shown to have higher affinity to ATP with increased rate of reaction compared with the full length protein.9 ATP-diphosphohydrolases (apyrases) (EC 3.6.1.5), are enzymes that hydrolyze both the γ- and β-phosphates of ATP and ADP. They are distinct from other phosphohydrolases with respect to their specific activity, nucleotide substrate specificity, divalent cation requirement, and sensitivity to inhibitors.10,11 Apyrases are ubiquitously expressed in eukaryotes and have been found additionally in some prokaryotes, indicating a general role for these enzymes across these 2 major classes. In our experiments, the N-terminal region of Ssb1 protein (NSsb1) and bacterial apyrase apy from E. coli were used to decrease yeast intracellular ATP level.
The 1,233 bp fragment of SSB1 ORF encoding the N-terminal 411 amino acid residues of the Ssb1p protein (we named it NSSB1) and the apyrase gene apy from E. coli lacking the N-terminal periplasmatic targeting sequence, were functionally expressed in S. cerevisiae under the control of the galactose-inducible GAL1 promoter. The corresponding representative transformants had a slightly slower growth. Transformants that expressed ATPase activity had different growth rates with an increase of 21% for the transformant bearing apy gene (ВY4742/pYES2-apy) with no increase in growth rate for the transformant bearing NSSB1 gene (ВY4742/pYES2-NSSB). Ethanol accumulation and yield of the constructed strains, was increased by 22% and 39% for ВY4742/pYES2- NSSB strain and an increase of 17% and 28% for ВY4742/pYES2-apy, when compared to the control ВY4742/pYES2 during fermentation in the galactose containing media (1% glucose + 10% galactose)12 (Table 1).
Table 1.
Ethanol synthesis, yield and specific ATPase activity of S. cerevisiae transformants with overexpressed apy and SSB1 genes and the wild-type strain on the third day of fermentation.
| 1% glucose + 10% galactose |
5% glucose + 10% galactose |
||||
|---|---|---|---|---|---|
| Strain | Ethanol, g L−1 | Ethanol, g g−1 of biomass | Ethanol, g L−1 | Ethanol, g g−1 of biomass | ATPase, U/mg |
| ВY4742/pYES2 | 4.11 ± 0.3 | 5.73 ± 0.6 | 10.95 ± 0.9 | 14.26 ± 1.7 | 0.66 ± 0.06 |
| ВY4742/pYES2- apy | 4.80 ± 0.4 | 7.34 ± 0.8 | 10.95 ± 1.1 | 14.10 ± 1.5 | 0.80 ± 0.10 |
| ВY4742/pYES2-NSSB | 5.03 ± 0.5 | 7.94 ± 0.7 | 15.00 ± 1.3 | 16.70 ± 1.9 | 0.53 ± 0.04 |
The efficiency of galactose fermentation of the transformants with plasmid pYES2-apy, pYES2-NSSB and control plasmid pYES2 was evaluated on SD medium supplemented with mixture 1% or 5% glucose and 10% of galactose. Fermentation was carried out under semi-anaerobic (120 revolutions/min) condition for 3 d. The initial biomass concentration was 15 μg L−1. The concentration of ethanol in fermentation media was determined using alcohol oxidase/peroxidase-based enzymatic kit “Alcotest.”19 ATPase activity was measured as described elsewhere.12
Futile cycles
Phosphofructokinase and fructose-1,6-bisphosphatase
Fructose-1,6-biphosphatase (FBPase) (EC 3.1.3.11) is one of the major gluconeogenesis enzymes. This enzyme hydrolyzes D-fructose-1,6-bisphosphate to D-fructose-6-phosphate in an ATP-dependent reaction. The simultaneous action of 2 enzymes – phosphofructokinase and fructose-1,6-biphosphatase –leads to the generation of a futile cycle between D-fructose-1,6-biphosphate and D-fructose-6-phosphate with ATP dissipation. In living cells, there are however multiple regulatory mechanisms of the FBP1 gene and enzyme activity, such as catabolic repression, inactivation by ubiquitination, inhibition by AMP and fructose-2,6-biphosphate.5,13 Therefore, intracellular enzyme activity is maintained at basal level in cells grown in media containing fermentable carbon sources.
To overcome tight regulation of the yeast FBPase, the bacterial FBPase that is insensitive to fructose-2,6-biphosphate inhibition, was expressed in yeast.13 The E. coli FBPase was expressed under the control of the constitutive yeast promoter of the TPI1 gene encoding triose phosphate isomerase in the S. cerevisiae strain BY4742 using a replicative plasmid.
Fermentation experiments of the 2 independent transformants was performed using a YPD medium. Strains BY4742/fbp1_7 and BY4742/fbp1_13 produced 23.3 and 24.5 g L-1 of ethanol respectively, while control strain harboring basal plasmid pRS42H14 produced only 21.3 g L-1 (Table 2). After re-calculation of the results taking into account the increase in yeast biomass, an increase of 5.2% and 8.8% in ethanol yield was demonstrated (Table 2). The ATP level of these strains was also measured. Total ATP level of analyzed transformants was decreased by 31-39% when compared to the control strain (Table 2).
Table 2.
Ethanol synthesis, biomass, yield and ATP of S. cerevisiae transformants with increased expression of fbp1 gene of E. coli and the wild-type strain strain on the fourth day of fermentation.
| Strain | Ethanol, g L−1 | Biomass, g L−1 | Ethanol, g g−1 of biomass | ATP, µM g−1 |
|---|---|---|---|---|
| ВY4742/pRS42H | 21.3 ± 0.9 | 3.7 ± 0.2 | 5.7 ± 0.15 | 7.75 ± 0.18 |
| BY4742/fbp1_7 | 23.3 ± 1.1 | 3.9 ± 0.4 | 6.0 ± 0.12 | 4.76 ± 0.10 |
| BY4742/fbp1_13 | 24.5 ± 1.2 | 3.9 ± 0.3 | 6.2 ± 0.20 | 5.31 ± 0.12 |
Alcoholic fermentation was performed using a 10% glucose containing medium at 30°C under semi-anaerobic condition (120 revolutions/min) for 4 d as described elsewhere.8 The concentration of ethanol in fermentation media was determined using alcohol oxidase/peroxidase-based enzymatic kit “Alcotest.”19 ATP was measured by coupled enzymatic reactions with hexokinase and glucose-6-phosphate dehydrogenase as described elsewhere.20
The specific activity of fructose-1,6-bisphosphatase of the transformed strains was measured after 1 day of cultivation on a synthetic medium supplemented with 2% glucose and compared to FBPase activity of the wild type strain BY4742 grown on glucose or ethanol as a sole carbon source. The specific FBPase activity of the obtained recombinant strains was 1.5-3-folds higher than that of the parental strain BY4742. However, FBPase activity was around 2-folds lower than that of the wild-type strain cultivated on an ethanol containing medium as a sole carbon source (gluconeogenic substrates de-repress FBP1 gene expression in yeast). These results are in agreement with previously published data.5
Pyruvate kinase, pyruvate carboxylase and phosphoenolpyruvate carboxykinase
Another way to construct an alternate futile cycle is via the simultaneous activation of the enzymes, pyruvate carboxylase and phosphoenolpyruvate carboxykinase. Pyruvate carboxylase uses ATP energy to convert pyruvate into oxaloacetate, and phosphoenolpyruvate carboxykinase uses ATP energy to convert oxaloacetate into phosphoenolpyruvate. ATP is also synthesized when phosphoenolpyruvate is converted to pyruvate by pyruvate kinase. Therefore the resulting total loss in ATP is one molecule for each one turn of the cycle.
In order to increase pyruvate carboxylase activity, the promoter of the PYC2 gene was substituted by the strong constitutive TEF1 promoter. The selected recombinant strain showed a 3-5-fold increase in the specific activity of this target enzyme when compared to the initial strain.
As S. cerevisiae phosphoenolpyruvate carboxykinase activity is carefully regulated at the post-translational level, and in order to avoid this regulation, we decided to overexpress heterologous gene encoding the corresponding enzyme from E. coli. The expression module containing the E. coli pckA gene encoding phosphoenolpyruvate carboxykinase under the control of yeast ADH1 gene promoter was integrated into the genome of the previously isolated strain overexpressing PYC2 gene. The specific pyruvate carboxylase and phosphoenolpyruvate carboxykinase activities were elevated 3-4 and 6-7-fold, when compared to the parental strain BY4742, respectively. Ethanol production by the constructed recombinant strains showed a 2-fold increase over the parental strain (Fig. 1).
Figure 1.
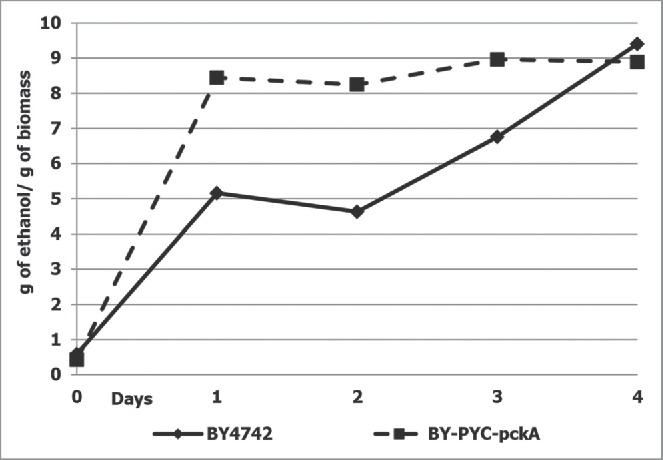
Ethanol production by strain overexpressing PYC2 and pckA genes during glucose fermentation. Diamonds represent wild-type strain (BY4742); squares represent strain overexpressing PYC2 and pckA genes. The data represent means of typical single cultivation. Alcoholic fermentation was performed in 10% glucose containing medium at 30°C under semi-anaerobic condition (120 revolutions/min) during 4 d as described elsewhere.8
We have also considered the induction of an alternative futile cycle based on the interconversion between glucose and trehalose, which theoretically could lead to ATP dissipation in cells of S. cerevisiae. The disaccharide trehalose is an essential metabolite in S. cerevisiae cells. This disaccharide, serves in yeast as one of the major stress protectants. In yeast, trehalose is synthesized from glucose-1-phosphate and UDP-glucose in 2 sequential reactions catalyzed by the enzymes, trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase. Trehalose can be catabolized to glucose by the action of acid or neutral trehalase. In yeast, acid trehalase is mainly localized in the periplasmic space, with a small fraction of activity present in the cell wall.15. The cell wall bound enzyme fraction uses extracellular trehalose as a substrate. In yeast, neutral trehalase is a cytosolic enzyme that is required for the hydrolysis of intracellular trehalose.16 In this metabolic pathway, ATP is used in the conversion of glucose into glucose-1-phosphate and UDP-glucose. To verify whether a perpetual conversion between glucose and trehalose will lead to ATP dissipation, we constructed several S. cerevisiae strains with simultaneous overexpression of the 2 genes: TPS1, encoding trehalose-6-phosphate synthase, and NTH1, encoding neutral trehalase. The over-expression of both genes was accomplished in both cases by using the strong constitutive promoter of the ADH1 gene. Biochemical analysis of these strains showed that despite the increase in the activities of both trehalose-6-phosphate synthase and neutral trehalase in transformants, when compared to the wild type strain, biomass accumulation did not decrease (data not shown).
We have also tried to induce the production of histatins in S. cerevisiae cells. Histatins constitute a group of small, cationic multifunctional proteins that are present in the saliva of human and in some other primates. It has been shown that histatin hst5 kills the fungal pathogen Candida albicans via a mechanism that involves the release of cellular ATP in the absence of cytolysis.17 We tested if the expression of codon-optimized human histatin hst5 under the control of maltose-inducible promoter MAL32 can induce ATP release and subsequent intracellular ATP depletion in S. cerevisiae. Unfortunately, the growth level of the WT and histatin-expressing strains on maltose was virtually the same, presumably due to inefficient expression of histatin gene in S. cerevisiae. Thus, the last 2 approaches turned out to be unsuccessful.
Our previous works8,12 showed that it is possible to obtain an increase in the production of 1st generation ethanol following genetic manipulation of the ATP level in S. cerevisiae. The transformed strains accumulated more ethanol due to a reduction in yeast biomass. This was achieved by decreasing intracellular ATP content following the introduction of the ATP degrading enzymes or by the activation of the futile cycles that lead to the dissipation of ATP. We report here on the construction of S. cerevisiae strains which have over-expressed genes encoding ATP degrading enzymes (NSSB1, an ATPase encoding part of the host chaperon, and the bacterial apyrase apy). Furthermore, we report on the introduction and activation of 2 futile cycles consisting of phosphofructokinase and fructose-1,6-bisphosphatase or pyruvate kinase, pyruvate carboxylase and phosphoenolpyruvate carboxykinase. All these approaches appear to be in agreement with the results reported by other authors5 on the introduction of an active futile cycle of phosphofructokinase and fructose-1,6-bisphosphatase into yeast. The new approaches that we describe are based on the overexpression of heterologous apyrase and futile cycle consisting of pyruvate kinase plus pyruvate carboxylase and phosphoenolpyruvate carboxykinase are original and have been developed and successfully applied by us for the first time. We hope these new modifications would lead to the successful improvement of yeast strains that can be used for the industrial production of 1st generation ethanol. The modifications we described here on strain improvements using gene cloning also complement other methods where we reported on the positive selection of ethanol overproducing mutants that are resistant to several antimetabolites, that are mostly known as glycolytic enzyme inhibitors.18 By combining both approaches (i.e. classical selection for antimetabolite-resistant strains and the subsequent cloning and overexpression of ATP dissipating genes in antimetabolite-resistant strains), provide promising tools for the development of more efficient yeast strains with increased yield and productivity of ethanol synthesis from glucose.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported in part by Archer Daniels Midland Company (Decatur, IL); Polish grant of National Scientific Center (NCN) DEC-2012/05/B/NZ1/01657 awarded to AAS and FEMS Fellowship grant # UA-SMU2012-2 awarded to MVS.
References
- [1].Gombert AK, van Maris AJ. Improving conversion yield of fermentable sugars into fuel ethanol in 1st generation yeast-based production processes. Curr Opin Biotechnol 2015; 33:81-6; PMID:25576737; http://dx.doi.org/ 10.1016/j.copbio.2014.12.012 [DOI] [PubMed] [Google Scholar]
- [2].Doelle HW. Bacterial metabolism, 2nd edition. New York: Academic Press; 1975. [Google Scholar]
- [3].Panesar PS, Marwaha SS, Kennedy JF. Zymomonas mobilis: an alternative ethanol producer. J Chem Technol Biotechnol 2006; 81:623-35; http://dx.doi.org/ 10.1002/jctb.1448 [DOI] [Google Scholar]
- [4].Benisch F, Boles E. The bacterial Entner-Doudoroff pathway does not replace glycolysis in Saccharomyces cerevisiae due to the lack of activity of iron-sulfur cluster enzyme 6-phosphogluconate dehydratase. J Biotechnol 2014; 171:45-55; PMID:24333129; http://dx.doi.org/ 10.1016/j.jbiotec.2013.11.025 [DOI] [PubMed] [Google Scholar]
- [5].Navas MA, Cerdan S, Gancedo JM. Futile cycles in Saccharomyces cerevisiae strains expressing the gluconeogenic enzymes during growth on glucose. Prot Natl Acad Sci USA 1993; 90:1290-4; PMID:8381962; http://dx.doi.org/ 10.1073/pnas.90.4.1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rogers DT, Szostak JW. Strains of yeast with increased rates of glycolysis. US Patent 1993; 5268285:A1 16. [Google Scholar]
- [7].Jensen P, Snoep J, Westerhoff H. Method of improving the production of biomass or a desired product from a cell. US Patent 2006; 20060094078:A1 17. [Google Scholar]
- [8].Semkiv MV, Dmytruk KV, Abbas CA, Sibirny AA. Increased ethanol accumulation from glucose via reduction of ATP level in a recombinant strain of Saccharomyces cerevisiae overexpressing alkaline phosphatase. BMC Biotechnol 2014; 14:42; PMID:24884834; http://dx.doi.org/ 10.1186/1472-6750-14-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pfund C, Huang P, Lopez-Hoyo N, Craig EA. Divergent functional properties of the ribosome-associated molecular chaperone Ssb compared with other Hsp70s. Mol Biol Cell 2001; 12:3773-82; PMID:11739779; http://dx.doi.org/ 10.1091/mbc.12.12.3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Plesner L. Ecto-ATPases: identities and functions. Int Rev Cytol 1995; 158:141-214; PMID:7721538; http://dx.doi.org/ 10.1016/S0074-7696(08)62487-0 [DOI] [PubMed] [Google Scholar]
- [11].Handa M, Guidotti G. Purification and cloning of a soluble ATPdiphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum). Biochem Biophys Res Commun 1996; 218:916-923; PMID:8579614; http://dx.doi.org/ 10.1006/bbrc.1996.0162 [DOI] [PubMed] [Google Scholar]
- [12].Dmytruk KV, Semkiv MV, Sibirny AA. Ethanol yield and reduction of biomass accumulation in the recombinant strain of Saccharomyces cerevisiae overexpressing ATPase. US Patent Application 2012; 0088290:A1. [Google Scholar]
- [13].Navas MA, Gancedo JM. The regulatory characteristics of yeast fructose-1,6-bisphosphatase confer only a small selective advantage. J Bacteriol 1996; 178: 1809-12; PMID:8606152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Taxis C, Knop M. System of centromeric, episomal, and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. Biotechniques 2006; 40:73-8; PMID:16454043; http://dx.doi.org/ 10.2144/000112040 [DOI] [PubMed] [Google Scholar]
- [15].Jules M, Guillou V, François J, Parrou JL. Two distinct pathways for trehalose assimilation in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol 2004; 70(5):2771-8; PMID:15128531; http://dx.doi.org/ 10.1128/AEM.70.5.2771-2778.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kopp M, Muller H, Holzer H. Molecular analysis of the neutral trehalase gene from Saccharomyces cerevisiae. J Biol Chem 1993; 268(7):4766-74; PMID:8444853 [PubMed] [Google Scholar]
- [17].Koshlukova S, Araujo M, Didi Baev, Edgerton M. Released ATP is an extracellular cytotoxic mediator in salivary histatin 5-induced killing of Candida albicans. Infection and Immunity 2000; 68(12):6848-56; PMID:11083804; http://dx.doi.org/ 10.1128/IAI.68.12.6848-6856.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dmytruk K, Kshanovska B, Abbas C, Sibirny A. New methods for positive selection of yeast ethanol overproducing mutants. Bioethanol 2016; 2:24-31. [Google Scholar]
- [19].Gonchar MV. Sensitive method for quantitative determination of hydrogen peroxide and oxidase substrates in biological samples. Ukr Biokhim Zh 1998; 70:157-63; PMID:10445279 [PubMed] [Google Scholar]
- [20].Ano Y, Hattori T, Kato N, Sakai Y. Intracellular ATP correlates with mode of pexophagy in Pichia pastoris. Biosci Biotechnol Biochem 2005; 69:1527-33; PMID:16116281; http://dx.doi.org/ 10.1271/bbb.69.1527 [DOI] [PubMed] [Google Scholar]


