Abstract
Microtubules and actin filaments are the two main cytoskeleton networks supporting intracellular architecture and cell polarity. The centrosome nucleates and anchors microtubules and is therefore considered to be the main microtubule-organizing center. However, recurring, yet unexplained, observations have pointed towards a connection between the centrosome and actin filaments. Here we have used isolated centrosomes to demonstrate that the centrosome can directly promote actin filament assembly. A cloud of centrosome-associated actin filaments could be identified in living cells as well. Actin-filament nucleation at the centrosome was mediated by the nucleation promoting factor WASH in combination with the Arp2/3 complex. Pericentriolar material 1 (PCM1) appeared to modulate the centrosomal actin network by regulating Arp2/3 complex and WASH recruitment to the centrosome. Hence our results reveal an additional facet of the centrosome as an intracellular organizer and provide mechanistic insights into how the centrosome can function as an actin filament-organizing center.
The functional coherence between cell internal architecture and cell micro-environment depends on the accurate orchestration of cytoplasmic and peripheral polarities. This requires a tight coordination of microtubules and actin filaments in space and time 1. It is ensured by common signaling pathways co-regulating the two network dynamics 2. In addition, several cross-linkers support the physical interaction of microtubule plus ends with actin filaments at the cell periphery 3–5. However, it is worth considering that such a crosstalk could also occur at the cell center, where microtubules minus-ends are connected to the centrosome. Indeed unexplained, but recurrent, observations have highlighted the influence of the actin network on centrosome positioning 6,7.
In highly adherent cells, disassembly of actin filaments dampened centriole motion 8 while inactivation of ROCK-dependent acto-myosin contractility increased inter-centriolar distance and centriolar exploration toward cell periphery 9. In poorly adherent polymorphonuclear leukocytes, actin disassembly blocked the splitting of centriole that was associated to cell spreading in response to PKC activation 10. Similarly, at the onset of mitosis, actin filaments appeared to be involved in the splitting of duplicated centrosomes in various systems ranging from early drosophila embryos to mammalian cultured cells 11–14. Ciliogenesis is another example of the close association between the centrosome and actin filaments. It starts by centrosome migration from the center to the periphery of the cell, where it attaches to the cortical actin network 7. Actin filaments not only bind the centrosome to the cell cortex via focal-adhesion-like and stress fiber-like structures 15–17 but also regulate centrosome migration to the edge of the cell 18–20. Similarly, when cytotoxic T lymphocytes encounter a target cell, reorganization of the actin network seems to promote centrosome migration to the cell cortex where it will promote the assembly of the immune synapse 21.
The converse has also been observed and various forms of actin-network reorganizations have been described in the vicinity of centrosomes. In early Drosophila embryos, centrosomes organize and position actin-based interphase caps around them 22,23. On a different note, the inhibition of acto-myosin contractility around the sperm centrosome directs a cortical flow which further determines the one-cell stage C. elegans embryo axes 24,25. The interaction of centrosome with actin filaments seems a general feature of mitosis as dynamic actin networks at the mitotic spindle poles are involve in spindle assembly and orientation in frog embryonic cells and mammalian cultured cells 26–28. Actin network disassembly also seems to occur next to centrosome as they reach the T lymphocyte cell cortex during immune synapse formation 29.
Several physiological functions have been attributed to centrosome-actin connections, notably the regulation of centrosome attachment to the actin networks surrounding the nucleus 30,31 or spanning the cell cortex 6,15. However, and despite few examples of direct interaction between centrosome and actin filaments 15,17, microtubules were most often considered as necessary intermediates between centrosomes and the actin network. Proteomic analyses have systematically revealed the presence of actin and actin-associated proteins at the centrosome 32–35. But they were considered as contaminants because of the abundance of actin in the cytoplasm. Thus, clear evidence for a direct role of centrosomes in actin filament assembly and organization has yet to be shown.
Results
Isolated centrosomes promote the assembly of actin filaments
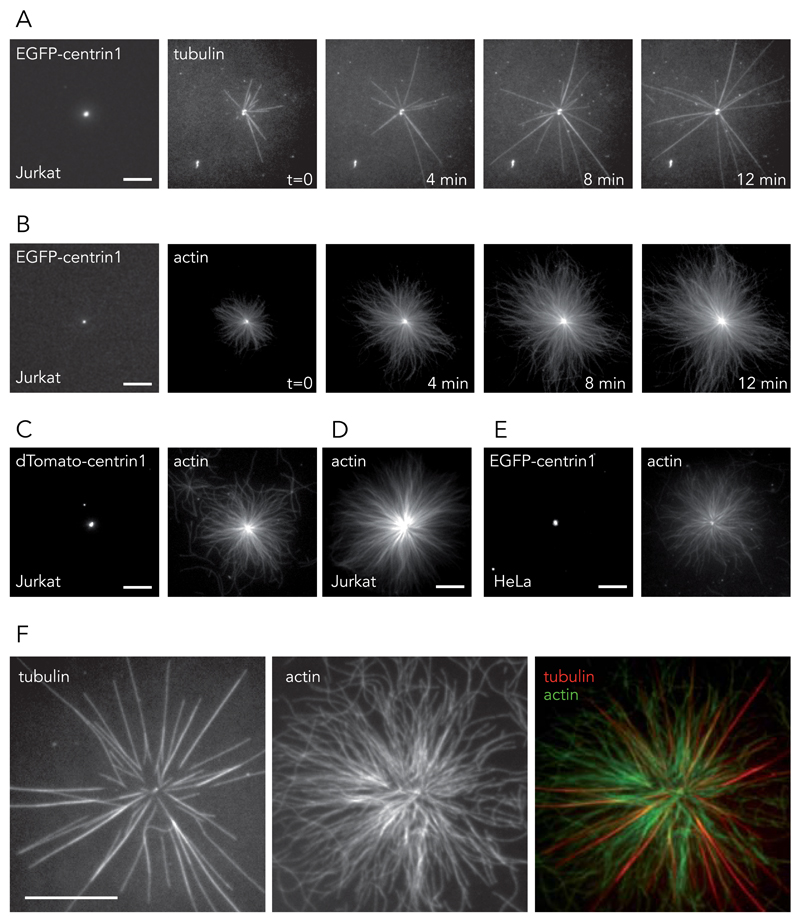
Isolated centrosomes were used to investigate a potential direct interaction between the centrosome and actin cytoskeleton. This initial in vitro approach was used in preference to an in vivo approach, to mitigate potential artifacts arising from the presence of dense cytoskeletal networks surrounding the centrosome in living cells. Centrosomes were purified from the human T lymphocyte Jurkat cell line, modified to express EGFP-centrin1, a core component of centrioles 36. Preliminary tests to validate our cytoskeleton-assembly conditions revealed that the classical buffer for the study of centrosomes and microtubules in vitro, the Brinkley buffer, impaired the nucleation of actin filaments in the presence of regulatory proteins (Supplementary Figure 1, Supplementary Video 1). This limitation was overcome by the development of a new polymerization buffer, the TicTac buffer, fully compatible with actin and microtubule assembly (see the Methods section for buffer composition and Supplementary Figure 1, Supplementary Video 1). Centrosomes, isolated in TicTac buffer and seeded on glass coverslips in the presence of 30 μM purified tubulin dimers, resulted in the classical assembly of dynamic microtubules (Fig. 1A, Supplementary Video 2). Strikingly, when seeded in the presence of 1 µM purified actin monomers, isolated centrosomes generated large radial arrays of actin filaments (Fig. 1B, Supplementary Video 3). Similar behaviours were observed with centrosomes isolated from Jurkat cells expressing dTomato-centrin1 (Fig. 1C) as well as from non-modified Jurkat cells (Fig. 1D). These results were further confirmed using centrosomes from the human epithelial HeLa cell line, modified to express EGFP-centrin1 (Fig. 1E). In vitro, 30–40% of fluorescent-centrin spots were competent to trigger actin assembly in the presence of actin monomers (n=2033 centrosomes) and 50% were competent for microtubule assembly in the presence of tubulin dimers (n= 1035 centrosomes). In the presence of a mix of actin monomers and tubulin dimers, 18 % of centrosomes (n=447) promoted the growth of heterogeneous radial arrays made of both microtubules and actin filaments (Fig. 1F, Supplementary Video 4), 33 % nucleated microtubules only and 11 % assembled actin filaments only.
Figure 1. Cytoskeleton filament assembly from isolated centrosomes.
(A and B) Centrosomes were isolated from T lymphocytes expressing EGFP-centrin1 and seeded on glass coverslips. (A) The addition of purified tubulin dimers led to the assembly of dynamic microtubules. Time is in minutes. Images are representative of 7 independent experiments. (B) The addition of purified actin monomers led to the assembly of radial arrays of actin filaments. Time is in minutes. Images are representative of 12 independent experiments. (C, D and E) Centrosomes were isolated from Jurkat cells expressing dTomato-centrin1 (5 independent experiments) (C), non-modified Jurkat cells (5 independent experiments) (D) and from HeLa cells expressing EGFP-centrin1 (8 independent experiments) (E). All centrosome preparations induced the growth of actin filaments in the presence actin monomers. (F) The assembly of both microtubules and actin filaments from isolated centrosomes. Images are representative of 5 independent experiments. Scale bars:10 μm.
Centrosomes are associated with an actin meshwork in cells
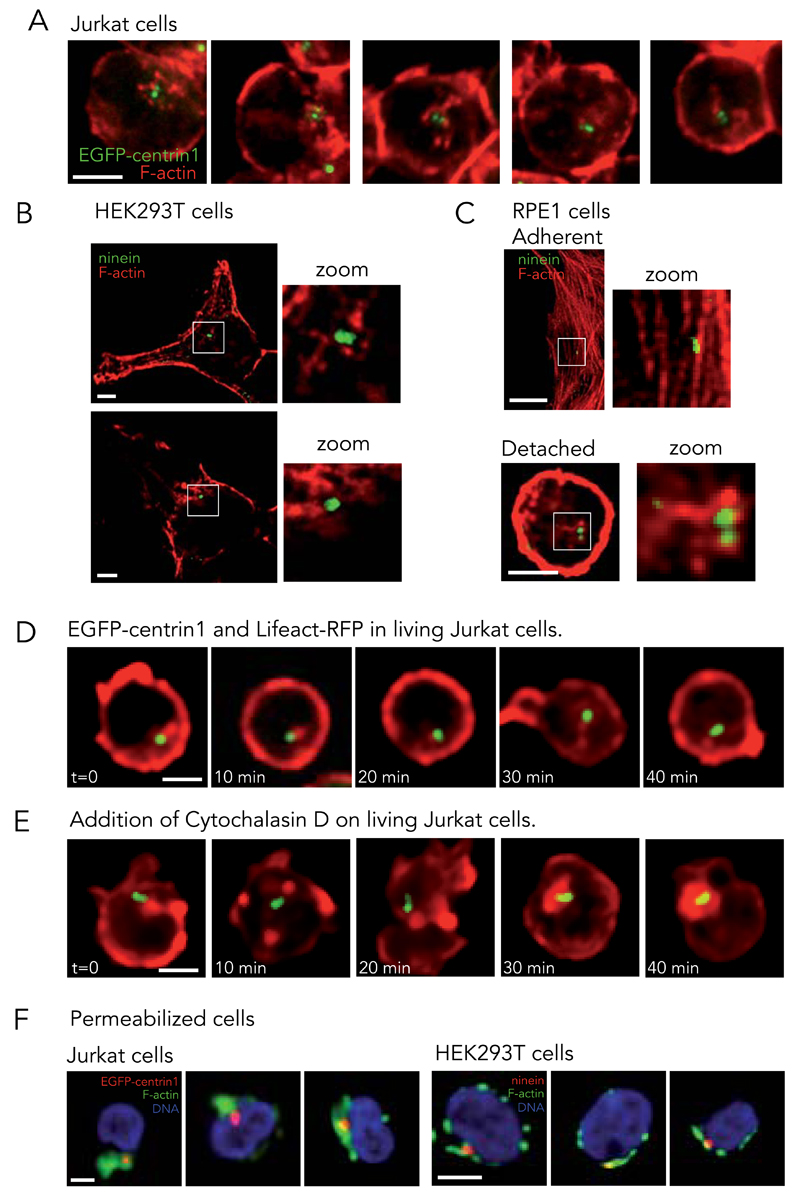
In view of these results, we closely examined the localization of actin filaments in Jurkat cells. In fixed cells, phalloidin staining revealed, as expected, an intense signal along the cell periphery emanating from the cortical actin network. However, around the centrosome, a less intense cloud-like signal was consistently observed (Fig. 2A). Similar actin clouds were also observed in human embryonic kidney derived cells HEK293T (Fig. 2B). Actin clouds could not be observed in highly adherent cell lines, such as human retinal pigment epithelial cells RPE1, in which the centrosomes were often seen close to bright actin bundles (Fig. 2C). However, an actin cloud could be detected at the centrosome when those cells were forced to detach upon trypsin treatment (Fig. 2C).
Figure 2. Association of actin filaments with centrosome in living cells.
(A) Actin filaments staining with phalloidin (red) in Jurkat cells expressing EGFP-centrin1 (green). Images are representative of 4 independent experiments. (B) F-actin (phalloidin, red) and ninein (green) in HEK293T cells. Images are representative of 2 independent experiments. (C) F-actin (phalloidin, red) and ninein (green) in RPE1 cells, adherent and in solution. Images are representative of 2 independent experiments. Cells were fixed with PFA. (D) Time-series of Jurkat cells expressing EGFP-centrin1 (green) transfected with Lifeact-RFP to visualize actin network (red). Image gamma was set to 0.65 to highlight the cytoplasmic network. Images are representative of 6 independent experiments. (E) Same as (D) except after the addition of 10 μg/ml cytochalasin D at t=0. Images are representative of 3 independent experiments. (F) DNA (blue) and F-actin (phalloidin, green) in pre-permeabilized and fixed Jurkat cells (left panel) expressing EGFP-centrin1 (red) and HEK293T cells (right panel) stained for ninein (red). Images are representative of 5 and 2 independent experiments for Jurkat and HEK293T cells respectively. Scale bars: 5 μm.
These observations were further confirmed by time-lapse microscopy. The thick cortical actin network formed dynamic protrusions deforming the roundish T lymphocytes. Meanwhile, centrosomes appeared to be continually associated with a cloud-like actin meshwork in the cytoplasm (Fig. 2D, Supplementary Video 5). This association did not appear to be disrupted by the addition of the actin polymerization inhibitor, cytochalasin D, which blocks actin filament dynamics and induces network collapse on its most steady portions 37. Within a few minutes after the addition of cytochalasin D to T lymphocytes, actin networks fragmented into dense patches all around cell periphery and at the centrosome (Fig. 2E, Supplementary Video 6). Furthermore, the association of the centrosome with the cloud-like meshwork of actin filaments was better observed in fixed T lymphocytes and HEK293T when the cells were treated with detergent prior to fixation; a process which removed the cortical-actin network and soluble cytoplasmic proteins, including actin monomers (Fig. 2F).
Centrosomes are genuine actin filament nucleators
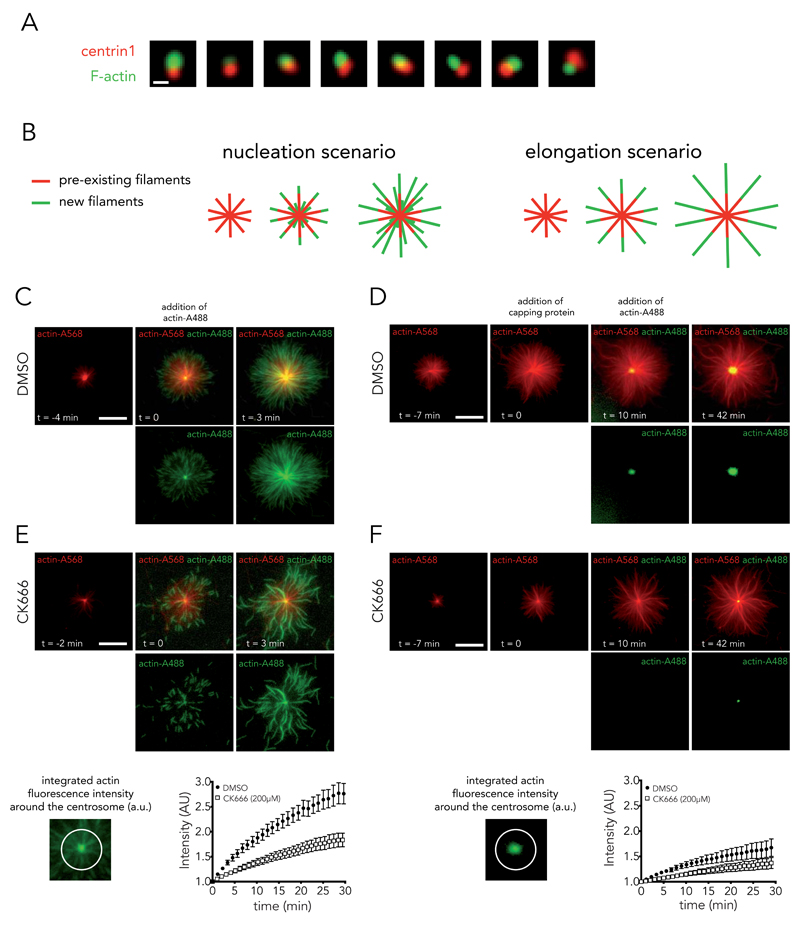
These last two experiments suggested that the pool of actin filaments at the centrosome was quite stable and may have resisted the actin-disrupting step during centrosome isolation from T lymphocytes. Indeed, phalloidin staining showed that centrosomes were bound to actin filaments after purification (Fig. 3A). It was unclear whether these filaments were already present prior to purification or they had aggregated during purification. Hence the observed actin-filament assembly on isolated centrosomes may have been due to elongation of pre-existing filaments, rather than to de novo nucleation. Therefore, to distinguish between these two scenarios (elongation versus nucleation), network assembly was performed on purified centrosomes, first by incubating with red-fluorescent actin monomers and then by incubating with green-fluorescent actin monomers (colour-switch assay). In the elongation scenario, green-fluorescent actin monomers would be added at the extremity of red filaments only. In the nucleation scenario, actin monomers would form new additional filaments at the centrosome (Fig. 3B). Using the purified centrosomes from T lymphocytes, both scenarios were observed (Fig. 3C, Supplementary Video 7). Newly assembled filaments segments were visible at the extremity of red filaments and at the centrosome (see green filaments at t=0 in Fig. 3C). The occurrence of genuine nucleation was further confirmed by the addition of capping proteins prior to initiating network assembly with green monomers in order to block the elongation of red filaments. In this situation, actin-filament assembly was then strictly initiated at the centrosome (Fig. 3D, Supplementary Video 8).
Figure 3. Isolated centrosomes nucleate actin filaments.
(A) Actin-filament staining with phalloidin (green) on isolated centrosomes (EGFP-centrin1 in red). Images are representative of 4 independent experiments. Scale bar: 2 μm. (B) Schematic representations of actin-filament nucleation and elongation hypotheses. (C, E) Time-lapse imaging of actin-filament assembly from isolated centrosomes in the presence of Alexa568-actin monomers (red) followed by addition of Alexa488-actin monomers (green). Bottom row shows green channel and top row the overlay. Growth of green actin filaments at the centrosome supported the nucleation hypothesis. Actin nucleation activity was measured at the centrosome in the presence of DMSO (full circles, mean of n = 9 actin asters) or 0.2 mM of CK666 (empty squares, mean of n = 17 actin asters); data show the results of a single experiment, representative of 2 independent experiments. Bars represent the s.d. (D, F) Same as (C, E) except that capping protein is added 10 minutes prior to Alexa488-actin monomers to block red actin filaments. Here also, growth of green actin filaments at the centrosome supported the nucleation hypothesis. DMSO and CK666 mean of n = 15 and 21 actin asters respectively; data show the results of a single experiment, representative of 3 independent experiments. Bars represent s.d. With or without the capping of older actin filaments, Arp2/3 inhibition reduced actin nucleation at the centrosome. Scale bars:10 μm.
Various molecular pathways could be involved in the assembly of actin filaments at the centrosome. They mostly differ in the polarity of the generated filaments. Formins and Ena/Vasp bind to filament barbed ends whereas the Arp2/3 complex binds to filament pointed ends 38. The addition of new monomers at the distal ends of filaments (Fig. 3C; i.e. the new filaments grew away from their nucleation sites) without signs of filament displacement as they grew (Supplementary Figure 2) suggested that filament assembly was governed by Arp2/3-based nucleation.
Actin filament nucleation at the centrosome is Arp2/3 dependent
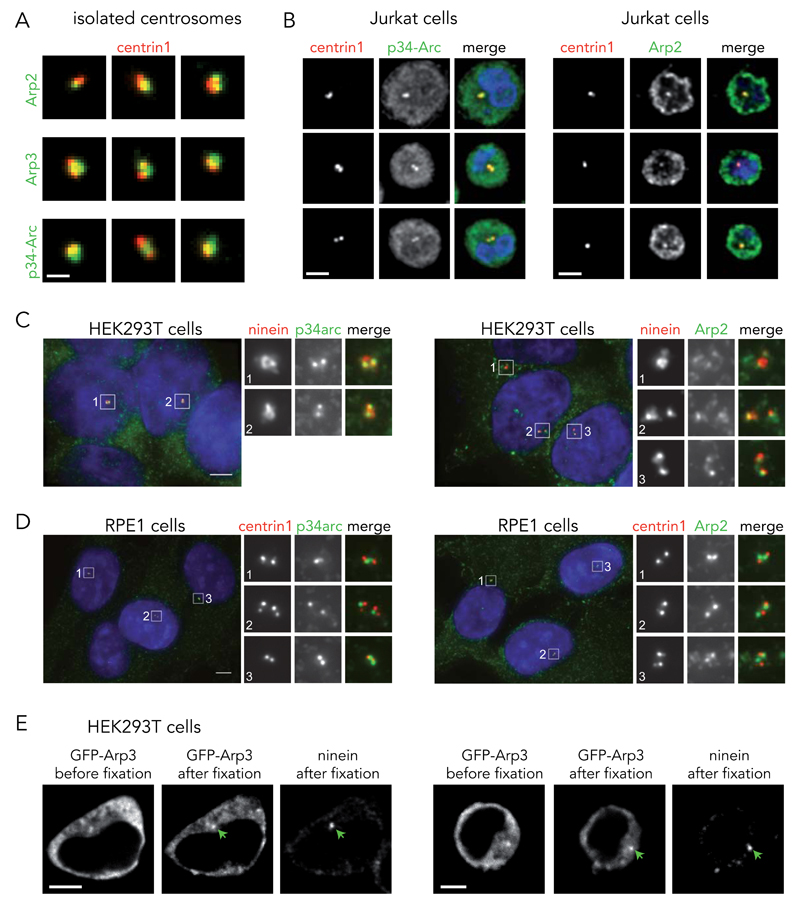
Several distinct immunostainings revealed the presence of Arp2/3 subunits on purified centrosomes (Fig. 4A and Supplementary Figure 3A). In addition, mass spectrometry of isolated centrosomes revealed the presence of five Arp2/3 complex subunits (Arp2, Arp3, p20-Arc, p21-Arc and p41-Arc) (Supplementary Table S1). We could also observe, by immunostaining, the Arp2/3 complex on the two centrioles of fixed Jurkat cells (Fig. 4B), HEK-293T (Fig. 4C) and RPE1 cells (Fig. 4D) as already reported 39. The specificity of the antibodies was confirmed by carrying out a competition assay using purified Arp2/3 complex as antigen. The level of centrosomal Arp2/3 subunit signal was progressively reduced in the presence of increasing amount of soluble Arp2/3 complexes (Supplementary Figure 4). The exogenous expression of a GFP-Arp3 construct 40 in HEK293T further confirmed the centriolar localization of the Arp2/3 complexes (Fig. 4E).
Figure 4. Arp2/3 complex localizes to the centrosome.
(A) Immunofluorescence staining of isolated centrosomes (EGFP-centrin1 in red) with antibodies against Arp2, Arp3 or p34-Arc subunits of the Arp2/3 complex (green). Images are representative of 4 independent experiments. Scale bar; 2 μm. (B) Immunofluorescence staining of T lymphocytes for p34-Arc and Arp2 after fixation with cold methanol. EGFP-centrin1 is shown in red, p34-Arc and Arp2 (2 independent experiments) in green and the DNA in blue. Scale bar: 10 µm. (C) Immunofluorescence staining of methanol-fixed HEK293T cells with antibodies to ninein (red) and p34arc or Arp2 (both green). DNA is in blue (1 experiment). Scale bar: 10 μm. (D) Methanol-fixed RPE1 cells stably expressing EGFP-centrin1 (red) stained with antibodies to p34arc or Arp2 (both green). DNA is in blue. Both p34arc and Arp2 juxtapose EGFP-centrin1 suggesting that the subunits of the Arp2/3 complex localize to or towards the proximal ends of centrioles. Images are representative of 3 independent experiments. Scale bars; 5 μm. (E) Distribution of EGFP-Arp3 in HEK293T cells before and after fixation (PFA + methanol) and ninein staining. Green arrows indicate the centrosome position. Images are representative of 2 independent experiments. Scale bar: 10 μm.
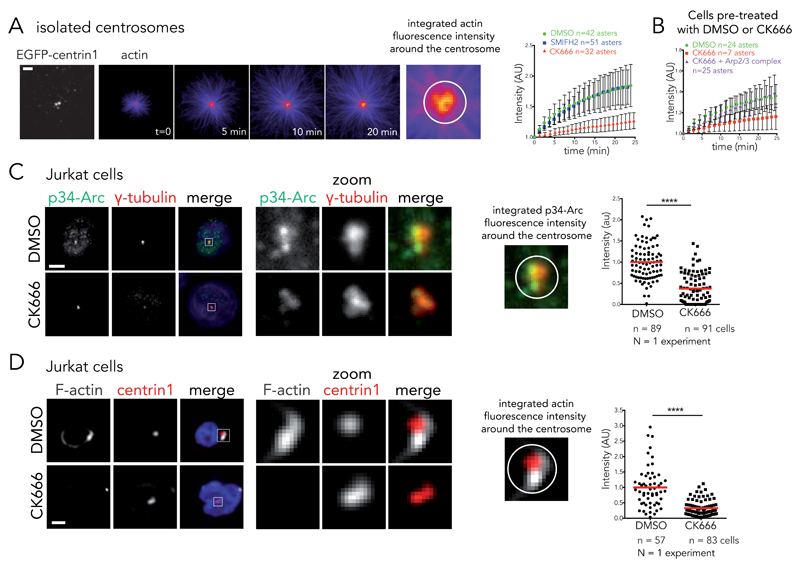
The Arp2/3 complex inhibitor CK666 41 (see activity test in Supplementary Figure 5A) was used to evaluate the contribution of the Arp2/3 complex activity on actin-filament nucleation at the centrosome. The proportion of centrosomes nucleating actin filaments in vitro was reduced in the presence of CK666 (33% +/− 3% in the presence of DMSo, 15% +/− 3% in the presence of CK666, n=3 independent experiments). The nucleation process was assessed by the increase of actin-fluorescence intensity at the centrosome over time (Fig. 5A) and by the amount of new filaments at the centrosome in the color-switch assay (Fig. 3E, F and Supplementary Videos 9 and 10). It was also strongly diminished in the presence of CK666. In addition, the weaker filament nucleation from centrosomes isolated from CK666-treated cells could be enhanced by the addition of 100 nM of purified Arp2/3 complex to the actin mix (Fig. 5B). This firmly demonstrated the existence of a genuine Arp2/3-complex-dependent nucleation of filaments at the centrosome. On the opposite, formin inhibition using SMIFH2 (see activity test Supplementary Figure 5B) had no effect on centrosomal actin nucleation (Fig. 5A).
Figure 5. Arp2/3 complex inactivation impairs actin filament nucleation at the centrosome.
(A) Time-lapse imaging of actin filament assembly from isolated centrosomes. “Fire” look-up table of actin images reveals that fluorescence intensity increases at the centrosome over time. This intensity was integrated over a 2 μm diameter circle around the centrosome and plotted against time in the presence of DMSO, 0.2 mM SMIFH2 or 0.2 mM of CK666 (right panel). Intensities were normalized with respect to initial intensity. Data show the results of a single experiment, representative of 3 independent experiments. Errors bars represent standard deviation. Scale bar: 5 μm. (B) Actin nucleation activity for centrosomes isolated from DMSO or CK666-treated cells. Addition of 100 nM purified Arp2/3 complex restores nucleation activity of CK666-treated centrosomes. Data show the results of a single experiment, representative of 3 independent experiments. Errors bars represent standard deviation. (C) Immunostaining of p34-Arc (green), γ-tubulin (red) and DNA (blue) at the centrosome of Jurkat cells incubated with DMSO or 0.2 mM CK666 and subsequently fixed with cold methanol. Right panel: p34-Arc fluorescence integrated over a 3 μm diameter circle around the centrosome for DMSO and CK666 condition. **** p ≤ 0.0001. Scale bar: 5 μm. (D) Immunostaining of F-actin (phalloidin, white) at the centrosome (EGFP-centrin1, red) of Jurkat cells incubated with DMSO or 0.2 mM CK666 and subsequently treated with detergent prior to PFA fixation. Actin fluorescence intensity was integrated over a 4 μm diameter circle around the centrosome to compare the two conditions (right panel). **** p ≤ 0.0001. Scale bar: 5 μm. Red bar indicates the mean. Unpaired t-test with Welch’s correction was used to generate p values.
In parallel, CK666 treatment of live T lymphocytes drastically reduced the concentration of Arp2/3 complexes (Fig. 5C) and the amount of polymerized actin that could be detected at the centrosome (Fig. 5D). Similar effects were observed in HEK293T (Supplementary Figure 6A) and RPE1 (Supplementary Figure 6B). Therefore both the in vitro and in vivo results supported the involvement of the Arp2/3 complex in actin filament nucleation at the centrosome.
WASH promotes actin filament nucleation at the centrosome
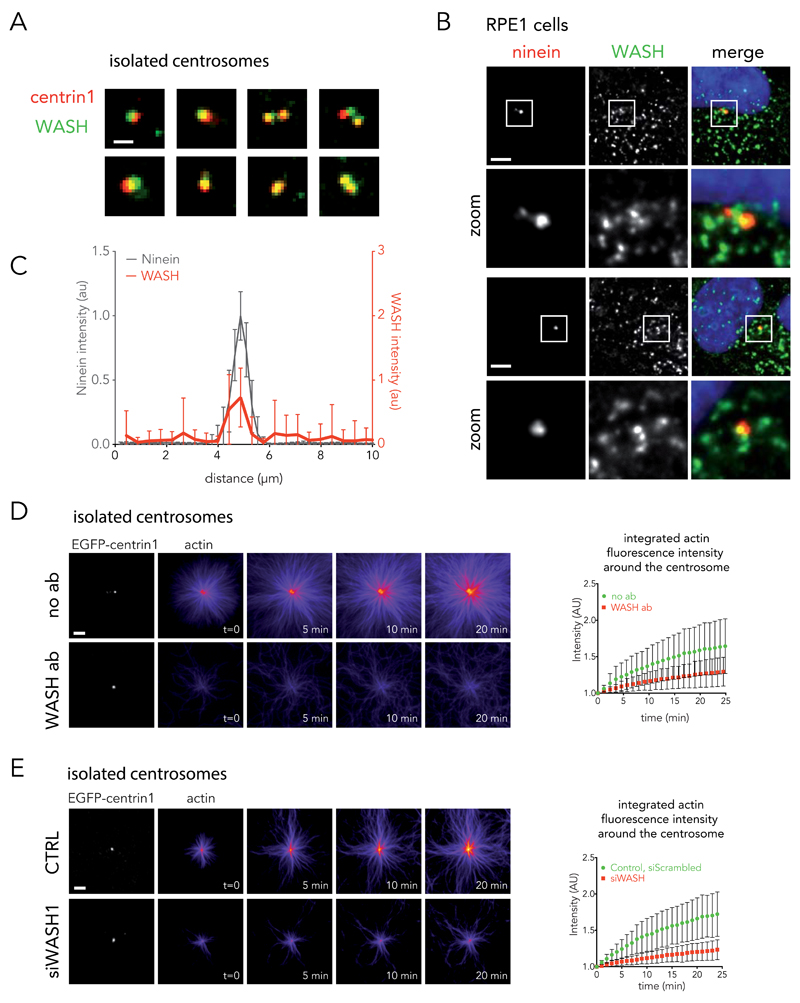
The Arp2/3 complex is constitutively inactive in the cytoplasm. Therefore, actin nucleation triggered by the Arp2/3 complex required the presence of an actin nucleation-promoting factor at the centrosome. One potential candidate is WASH; a potent nucleation-promoting factor that has been shown to be associated with the centrosome42. Indeed, KIAA1033, a WASH subdomain, was present in centrosome mass spectrometry analysis (Supplementary Table 1) and we could detect WASH on isolated centrosomes (Fig. 6A and Supplementary Figure 3C) as well as in fixed cells (Fig. 6B). The concentration of WASH was not higher on the centrosome than on the numerous endosomes in the cell cytoplasm 43. However, linescan analysis of fluorescence distribution showed that WASH was systematically localized at the centrosome (Fig. 6C). Two lines of evidence attested to a functional role for WASH at the centrosome. Anti-WASH blocking antibodies impaired the nucleation of actin filaments from isolated centrosomes (Fig. 6D). And centrosomes isolated from cells treated with siRNA against WASH1 (see Supplementary Figure 3D for the control of protein expression knock-down) displayed a lower ability to nucleate actin filaments than centrosomes isolated from cells treated with negative control siRNAs (Fig. 6E).
Figure 6. Implication of the WASH complex in actin nucleation at the centrosome.
(A) Immunostaining of WASH (green) on isolated centrosomes (EGFP-centrin1 in red). Images are representative of 2 independent experiments. Scale bar: 2 μm. (B) RPE1 cells were fixed in cold methanol and stained for ninein (left panel) and WASH (central panel). Right panel shows the overlay of WASH (green), ninein (red) and DNA (blue). Images are representative of 3 independent experiments. Scale bar: 5 μm. (C) Linescan analysis of fluorescent distribution of WASH (red line) at the centrosome (ninein, black line) (mean of n = 31 cells, data are pooled from 2 independent experiments). Bars represent the s.d. (D) Inhibition of WASH on isolated centrosomes with blocking antibodies (bottom row). Control experiments performed without antibody (no ab, top row). Data show the results of a single experiment, representative of 2 independent experiments: n = 25 and 27 actin asters for no antibody (green discs) and WASH antibody (red squares) respectively (mean ± s.d.). Scale bar: 5 μm. (E) Actin nucleation activity of centrosomes isolated from siWASH1-treated cells (bottom row) or control cells (top row). Data show the results of a single experiment, representative of 2 independent experiments: n = 20 and 26 actin asters for control (green discs) and siWASH1 (red squares) condition respectively (mean ± s.d.). Scale bar: 5 μm.
PCM1 regulates actin filament nucleation at the centrosome
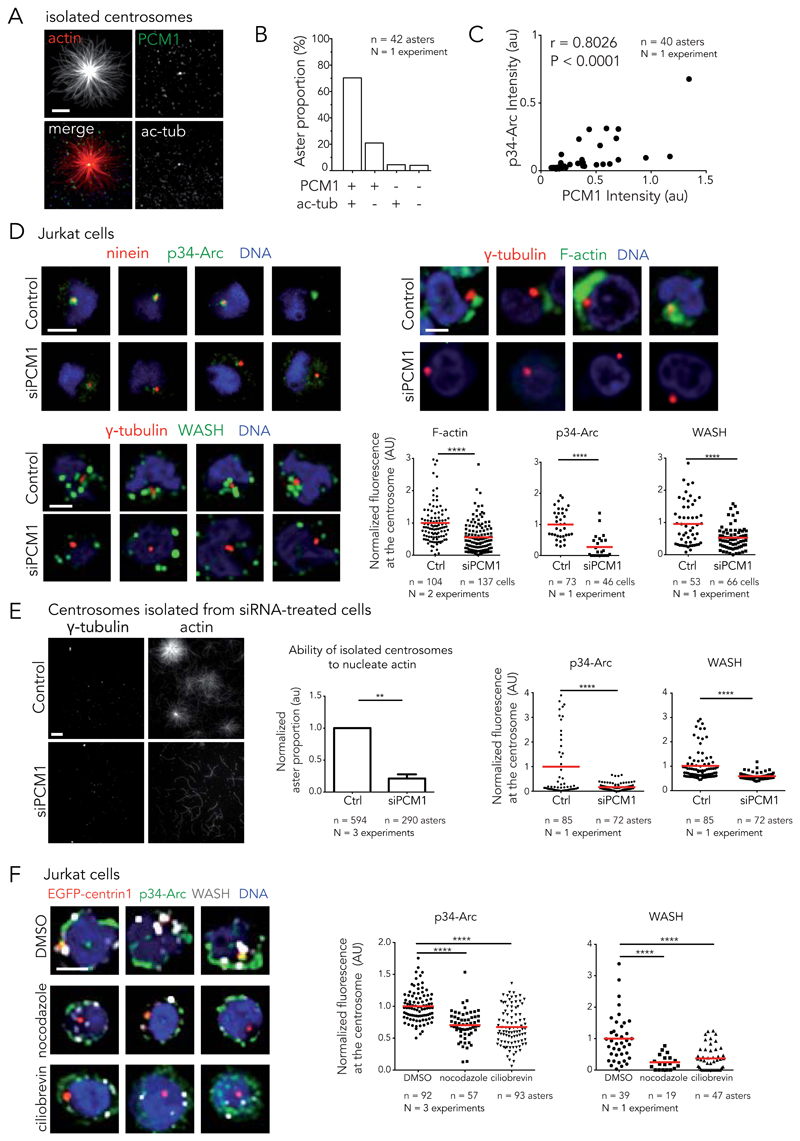
To identify centrosomal components involved in the Arp2/3 complex and WASH recruitment, we examined the biochemical composition of the various structures purified during centrosome isolation and determined their ability to nucleate actin filaments (Fig. 7A). Interestingly, we found that 70% of actin asters contained acetylated-tubulin and PCM1 (Fig. 7B). This confirmed that centrioles, which are made of acetylated microtubules, were the major scaffolds for actin filament nucleation. Interestingly, most of the remaining asters did not contain acetylated tubulin but were positive for PCM1 (Fig. 7B). In addition, the amount of PCM1 was correlated to the quantity of the Arp2/3 complex per spot (Fig. 7C). These results suggested that PCM1 could be a good candidate for actin regulation at the centrosome. Indeed, PCM1 is a key regulator of centrosome composition 44,45.
Figure 7. Regulation of actin filament assembly at the centrosome by PCM1.
(A) Immunostaining of PCM1 (green) and acetylated-tubulin (white) on isolated centrosomes in the presence of actin (red). Scale bar: 10 μm. (B) Quantification of aster proportion depending on the composition of the nucleation center (42 asters, 1 experiment). (C) Amount of p34-Arc plotted against PCM1 amount on isolated centrosomes. Pearson correlation coefficient (r) and P-value measure the correlation between the two variables. (D) Knockdown of PCM1 in Jurkat cells. Left panels: methanol fixed cells, ninein (red), p34-Arc (green) and DNA (blue) (top) and γ-tubulin (red), WASH (green) and DNA (blue) (bottom). Top right: cells treated with detergent prior to PFA fixation and stained for γ-tubulin (red), F-actin (phalloidin, green) and DNA (blue). Top row: control siRNA. Bottom row: PCM1 siRNA. Scale bars: 5 γm. Graph: fluorescence intensity at the centrosome of F-actin, p34-Arc and WASH. **** p ≤ 0.0001. (E) Left: centrosomes isolated from siPCM1 treated cells (bottom row) or control cells (top row) in the presence of actin and stained for γ-tubulin. Scale bar: 5 γm. Centre: ability of isolated centrosomes to nucleate actin calculated as ratio of the number of actin asters divided by the number of γ-tubulin spots. Error bar; s.d. ** p ≤ 0.01. Right: fluorescence intensity of p34-Arc and WASH. **** p ≤ 0.0001 . (F) Representative staining of Jurkat cells incubated with DMSO (top panel), nocodazole (central panel) and ciliobrevin D (bottom panel) and stained for DNA (blue), ninein (red), p34-Arc (green) and WASH (white). Scale bars: 5 γm. Graph: p34-Arc and WASH amount at the centrosome. **** p ≤ 0.0001. Red bar indicates the mean. Unpaired t-test with Welch’s correction was used to generate p values.
The functional implication of PCM1 in the regulation of actin filaments at the centrosome was tested using PCM1 siRNA knock-down 46. Down-regulation of PCM1 reduced the amount of Arp2/3 complex and WASH at the centrosomes of Jurkat cells (Fig. 7D) and RPE1 cells (Supplementary Figure 7A-E). It also diminished the associated cloud-like meshwork of actin filaments that was detected using the detergent-treatment prior to fixation protocol in Jurkat cells (Fig. 7D). Furthermore, centrosomes that were isolated from siRNA-treated Jurkat cells contained a lower amount of Arp2/3 complex and WASH and lost most of their ability to grow actin asters (Fig. 7E). These results pointed to a central role for PCM1 in centrosomal actin regulation.
PCM1 is the main component of centriolar satellites, which are small cytoplasmic granules that are transported on microtubules by dyneins towards the centrosome, where they accumulate and modulate centrosome composition 44,47. Both nocodazole treatment, to depolymerize microtubules, and ciliobrevin D treatment, to inhibit dyneins, reduced the concentration of the Arp2/3 complex and WASH at the centrosome in Jurkat cells (Fig. 7F) and RPE1 cells (Supplementary Figure 7F). Despite being unable to detect Arp2/3 complex and WASH on satellites by immuno-fluorescence, these results suggest they are involved in the recruitment of Arp2/3 complex and WASH to the centrosome.
Discussion
These results have identified a genuine actin filament nucleation process at the centrosome. They shed some light on numerous previous works reporting the unexplained presence of actin-binding proteins at the centrosome. Indeed, actin nucleators 21,39,42,48, focal adhesion associated proteins 15,49–51, actin regulatory proteins 52–57 and various myosins 19,26,58,59 were described to localize, at least transiently, to the centrosome. Altogether these data reveal the centrosome as a true actin-organizing center. The convergence of actin filaments and microtubules at the centrosome contributes to the view that it acts as a spatial integrator of various biochemical signals and cytoskeleton networks.
The high density of actin filaments throughout the cytoplasm precluded the clear delimitation of a specific centrosomal actin network in cells. Nonetheless, centrosome-associated actin filaments were clearly detected in live cells, in agreement with our in vitro observations. Electronic or sub-diffraction optical microscopy may shed light on the actual architecture of the centrosomal actin network. In addition, further description of the precise localization of Arp2/3 complex and WASH at the centrosome would be required to progress in this direction. At first glance, the Arp2/3 complex seems to localize to the proximal end of centrioles, where the pericentriolar material is highly structured 60,61. This suggested that actin filaments might interact there with the pericentriolar material and influence centrosome architecture and function.
Unraveling the molecular mechanism supporting the recruitment of the Arp2/3 complex and WASH to the centrosome is a critical step to further understand how the assembly of centrosomal actin network is regulated. We found that PCM1 was strongly involved in this mechanism but the operating mechanism remains to be elucidated. To our surprise, we could not detect any enrichment of Arp2/3 complex or actin filaments on PCM1-positive centriolar satellites in cells although in vitro 20% of the spots nucleating actin filaments were positive for PCM1 and negative for acetylated tubulin, strongly suggesting that they were centriolar satellites. In addition, the potential role of satellites in centrosomal actin assembly was further reinforced by the observation that microtubules and dyneins were involved in the recruitment of actin nucleators to the centrosome. This raised the question whether satellites could actually nucleate actin filaments, which would be hidden in a strong cytoplasmic background, or whether they simply carry part of the actin nucleation machinery to the centrosome. In any case, the implication of intracellular trafficking in the modulation of actin nucleation at the centrosome is of great interest as it opens a full spectrum of regulation in space and time.
Interestingly, actin nucleators could be detected at the centrosome of all tested cell types although actin filaments were observed only in the less adherent cells. Cell adhesions are central and powerful hubs in the nucleation of actin filaments. Competition between actin nucleators 62 for actin monomer has recently appeared as a central regulatory process in actin network assembly. The centrosome may thus be considered as a poor actin nucleator, from which actin filaments only grow when cell adhesion is low. This should not be considered as a weakness but instead as an increased sensitivity to external changes. Indeed, this interesting hypothesis implies that any change in cell adhesion, such as during cell mitosis or contact-induced polarization, would affect actin filament assembly at the centrosome.
Finally, how actin-filament nucleation at centrosomes contributes to physiological events involving the modulation of centrosome architecture, centrosome positioning or the interaction of the centrosome with the cell cortex or nucleus is an exciting field open to investigation.
Methods
Cell Culture
Jurkat cells, immortalized human T lymphocytes, were cultured in RPMI 1640, HeLa cells, transformed human epithelial cells, and HEK293T cells, human embryonic kidney cells, in DMEM and RPE1 cells, immortalized retinal pigment epithelial cells, in DMEM/F-12 (Gibco) at 37°C and 5% CO2. All media were supplemented with 10% foetal bovine serum and penicillin/streptomycin/antimycotic (Gibco).
Actin network disruption was performed adding 10 μg/ml cytochalasin D (Sigma Aldrich). Microtubules depolymerisation was performed adding 1 μM nocodazole (Sigma Aldrich) for 1 hour at 37°C and 5% CO2 and for 30 minutes at 4°C. Cytoplasmic dynein inhibition was performed using 50 μM ciliobrevin D (Calbiochem) for 2 hours. For the Arp2/3 complex inhibition experiments, cells were incubated with 0.2 mM CK666 (Sigma Aldrich) for 1 hour. Control experiments were performed using DMSO.
Plasmids
The plasmid pdTomato-centrin1 was created by excising the EGFP coding sequence from pEGFP-centrin1 with the restriction enzymes Age I and BsrG I (New England Biolabs) and replacing it with the dTomato coding sequence, which was amplified by PCR using Phusion DNA polymerase (Thermo Scientific) from pRSET-B-dTomato (Roger Tsien) using primers to incorporate Age I and BsrG I restriction sites at the 5′ and 3′ end of the PCR product, respectively. Sequencing of the dTomato coding sequence was carried out to verify the fidelity of the amplifying enzyme.
The plasmid pEGFP-N1-ACTR3 40 to express EGFP-Arp3 was obtained from Addgene (#8462).
Cell transfection
Stable Jurkat cell lines expressing EGFP-centrin1 were established by transfecting cells with pEGFP-centrin1 using Lipofectamine 2000 (Life Technologies) at a ratio of 2.5:1 transfection reagent (μl) to DNA (μg). Medium was supplemented with 0.75 mg/ml geneticin (Gibco) 24 hours after transfection to select for cells stably incorporating the EGFP-centrin1 sequence. After 2 weeks of selection, fluorescent-activated cell sorting was carried out to eliminate non-expressing cells from the culture. Transient transfections in Jurkat and HEK293T cells were performed using X-tremeGENE DNA transfection reagent (Roche) according to supplier’s protocol using a 3:1 ratio of transfection reagent (μl) to DNA (μg). Small interfering RNAs (siRNAs) targeting PCM1 (siPCM1, Qiagen) and ON-TARGETplus Human SMARTpool WASH1-targeting (siWASH1) siRNAs (Dharmacon, GE Healthcare) were transfected into Jurkat cells at the final concentration of 20 nM using Lipofectamine 2000 according to supplier’s protocol. PCM1-targeting siRNAs were transfected into RPE1 cells at the final concentration of 10 nM using Lipofectamine RNAiMax (Life Technologies) according to supplier’s protocol. Negative control siRNA was performed using AllStars Negative Control siRNA (Qiagen).
PCM1-target sequences were:
HS-PCM-1_1 5′-CAGUAUCACAUCUGAACUAAA-3′
HS PCM-1_2 5′-CAGGCUUUAACUAAUUAUGGA-3′
Antibodies and chemicals
For immunofluorescence staining, we used the following antibodies: rabbit anti-PCM1 (1 : 100; H-262, Santa Cruz Biotechnology), rabbit anti-γ-tubulin (1 : 500; ab11317, abcam), human anti-Ninein (1 : 100) 69, mouse anti-acetylated tubulin (1 : 1000, Sigma Aldrich T7451) mouse anti-γ-tubulin (1 : 500; T6557, Sigma-Aldrich), rabbit anti-Arp2 (1 : 500; ab47654, abcam), mouse anti-Arp3 (1 : 250 ; PA517288, ThermoFisher Scientific), mouse anti-p34-Arc 70 (undiluted) and rabbit anti-WASH 43 (5 μg/ml). For in cell competition assay, we used rabbit anti-Arp2 (1 : 500, ab47654, abcam) and mouse anti-p34-Arc 70 (1 : 8). For inhibition experiments on isolated centrosomes, we used rabbit anti-WASH antibodies 43 (2 μg/ml).
For western blot analysis, we used rabbit anti-PCM-1 (1 : 500; H-262, Santa Cruz Biotechnology), rabbit anti-GAPDH (1 : 10000; FL335, Santa Cruz Biotechnology), rabbit anti-Arp2 (1 : 250; ab47654, abcam) and rabbit anti-WASH1 (1 : 1000, ab157592, abcam). Labelled anti-mouse and anti-rabbit secondary antibodies (1 : 1000) and HRP-conjugated goat IgG anti-mouse and anti-rabbit (1 : 10000) for western blot were obtained from Jackson ImmunoResearch.
Cytochalasin D and nocodazole were purchased from Sigma-Aldrich. Ciliobrevin D was purchased from Calbiochem. PFA was purchased from Delta Microscopies.
CK666 and SMIFH2 were purchased from Sigma-Aldrich. Alexa-647-phalloidin was purchase from Life Technologies while non-labelled phalloidin was from Sigma Aldrich.
Immunofluorescence stainings in cells
Immunofluorescence staining was performed by incubating Jurkat cells on 0.1 % w/v poly-L-lysine-coated coverslip for 3 hours and RPE1 and HEK293T on clean coverslips at 37 °C and 5 % CO2.
For actin filament staining, cells were fixed with 4 % paraformaldehyde (PFA) for 20 minutes, incubated with quenching agent NH4Cl for 10 minutes and blocked with antibody blocking buffer (PBS supplemented with 1 % BSA, PBS-BSA) for 30 minutes. Permeabilization was performed with 0.2 % Triton X-100 for 1 minute. Primary and secondary antibodies were diluted in PBS-BSA and incubated on cells at room temperature for 1 hour and 30 minutes respectively. After washing, Alexa-647-phalloidin (200 nM) was incubated for 20 minutes. DNA was labeled with a 0.2 μg/ml solution of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma). The coverslips were air-dried and mounted onto glass slides using Mowiol mounting medium. Arp2/3 and WASH staining was performed by fixing cells with methanol at -20°C for 3 minutes and blocking with PBS-BSA for 30 minutes. Primary and secondary antibodies, diluted in PBS-BSA, were incubated for 1 hour and 30 minutes respectively. DNA labeling and coverslip mounting were performed as previously described. In solution RPE1 staining was performed by detaching the cells with trypsin (Life Technologies), pelleting and then fixing them with PFA (final concentration 4 %). Cells were incubated on poly-L-lysine coated coverslip for 30 minutes and permeabilized with 0.2 % Triton X-100 for 1 minute. Cells were stained as previously described. HEK293T cells expressing EGFP-Arp3 were stained under microscope. Before fixation, several z-stacks were acquired to observe the EGFP-Arp3 distribution. Fixation was performed by incubating cells with PFA for 20 minutes, washing 3 times with PBS and incubating cells with cold methanol for 3 minutes. Staining was performed as previously described.
Cell pre-extraction was carried out by pelleting Jurkat and HEK293T cells, previously detached with trypsin, and by resuspending them in 1 % Triton X-100 for 30 seconds. Fixation was performed in solution by adding PFA (final concentration 4 %). Cells were incubated on poly-L-lysine coated coverslip for 30 minutes and stained as previously described.
Competition assay in cells
We tested the specificity of the anti-Arp2 and anti-p34-Arc antibodies by performing a competition assay with blocking antigen (purified Arp2/3 complex). Briefly, primary antibodies were incubated with increasing quantity of purified Arp2/3 complex (0, 12, 24 and 48 pmol) in TicTac-BSA buffer for 1 hour. RPE1 cells were cultured on clean coverslip and fixed with methanol at -20 °C for 3 minutes. Cells were stained with antibody/purified Arp2/3 complexes as previously described by substituting the PBS-BSA buffer with the TicTac-BSA buffer.
Isolation of centrosomes
Centrosomes were isolated from Jurkat (non treated, CK666-treated or transfected with siRNA against PCM1) and HeLa cells by modifying a previously published protocol 63. In brief, cells were treated with nocodazole (0.2 μM) and cytochalasin D (1 μg/ml) followed by hypotonic lysis. Centrosomes were harvested by centrifugation onto a 60% sucrose cushion and further purified by centrifugation through a discontinuous (70%, 50% and 40%) sucrose gradient. Composition of sucrose solutions was based on an ad hoc buffer (TicTac buffer), in which the activity of tubulin, actin and actin-binding proteins is maintained: 10 mM Hepes, 16 mM Pipes (pH 6.8), 50 mM KCl, 5 mM MgCl2, 1 mM EGTA. The TicTac buffer was supplemented with 0.1% Triton X-100 and 0.1% β-mercaptoethanol. After centrifugation on gradient sucrose, supernatant was removed until only about 5 ml remained in the bottom of the tube. Centrosomes were stored at -80°C after flash freezing in nitrogen liquid.
Protein expression and purification
Tubulin was purified from fresh bovine brain by three cycles of temperature-dependent assembly/disassembly in Brinkley Buffer 80 (BRB80 buffer: 80 mM Pipes pH 6.8, 1 mM EGTA and 1 mM MgCl2) according to Shelanski 64. Fluorescent tubulin (ATTO-565-labeled tubulin) was prepared according to Hyman et al. 65. Actin was purified from rabbit skeletal-muscle acetone powder. Monomeric Ca-ATP-actin was purified by gel-filtration chromatography on Sephacryl S-300 at 4 °C in G buffer (2 mM Tris-HCl, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 1 mM NaN3 and 0.5 mM dithiothreitol (DTT). Actin was labelled on lysines with Alexa-488, Alexa-568 and Alexa-647. Recombinant human profilin, mouse capping protein, the Arp2/3 complex, GST-pWA and mDia1 were purified according to previous works 66,67.
Pyrene assay
For bulk actin polymerization assay , pyrene fluorescence at 407 nm (λex = 365 nm) was monitored over time. Experiments were performed using Brinkley buffer (supplemented with 50 mM KCl and 2.7 mM ATP) or TicTac buffer (supplemented with 2.7 mM ATP) in order to study the activity of the Arp2/3 complex. Each assay contained 2 μM actin monomers (10 % pyrene), 2 μM profilin, 50 nM Arp2/3 complex and 500 nM pWA. Elongation assay was performed with 2 μM actin monomers (10 % pyrene) from filament seeds stabilized with non-labelled phalloidin. CK666 and SMIFH2 inhibitors were tested in an actin polymerization assay. CK666 assay contained 2 μM actin monomers (10 % pyrene), 50 nM Arp2/3 complex, 500 nM pWA and 60 μM CK666. SMIFH2 inhibitor was tested using 2 μM actin monomers (10 % pyrene), 1 nM mDia1 and 0.2 mM SMIFH2.
Western blotting
Western blots were performed fractioning proteins on SDS polyacrylamide gels. Membrane blocking was carried out using 3% BSA in PBS. Primary and secondary antibodies were diluted in PBS supplemented with 1% BSA and 0.1 % Tween-20 while washing steps were performed with PBS supplemented with 1% BSA and 1% Tween-20.
Proteomic analyses
Proteomic experiments were performed as previously described 68. Briefly, proteins were stacked in the top of a 4-12% NuPAGE gel (Invitrogen) before R-250 Coomassie blue (BioRad) staining and in-gel digestion using modified trypsin (Promega, sequencing grade). Resulting peptides were analyzed by online nanoLC-MS/MS (UltiMate 3000 RSLCnano and Q-Exactive Plus, Thermo Scientific) using a 120-min gradient. Peptides and proteins were identified through concomitant searches against Uniprot (Homo sapiens taxonomy, September 2014 version) and classical contaminants database (249 sequences, homemade) and the corresponding reversed databases using Mascot (version 2.5.1). The Proline software was used to filter the results (conservation of rank 1 peptides, peptide identification FDR < 1% as calculated on peptide scores by employing the reverse database strategy, minimum peptide score of 25, and minimum of 1 specific peptide per identified protein group) before performing a compilation, grouping and comparison of the protein groups from the different samples. Proteins from the contaminants database and additional keratins were discarded from the final list of identified proteins. Only proteins identified in both biological replicates (minimum 1 specific spectral count in each replicate) and 2 spectral counts in one of them were considered as reliable purified centrosome components.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org/) via the PRIDE partner repository with the dataset identifier PXD002370.
In-vitro assays
Experiments were performed in polydimethylsiloxane (PDMS) open chambers in order to add sequentially experimental solutions when needed. PDMS (Sylgard 184 kit, Dow Corning) was mixed with the curing agent (10:1 ratio), degased, poured into a Petri dish to a thickness of 5 mm and cured for 30 minutes at 100°C on a hot plate. PDMS layer was cut to 15 mm × 15 mm and punched using a hole puncher (ted Pella) with an outer diameter of 8 mm. The PDMS chamber and clean coverslip (20 mm × 20 mm) were oxidized in an oxygen plasma cleaner for 20 s at 80 W (Femto, Diener Electronic) and brought into contact.
Isolated centrosomes were diluted in TicTac buffer (10 mM Hepes, 16 mM Pipes (pH 6.8), 50 mM KCl, 5 mM MgCl2, 1 mM EGTA) and incubated for 20 minutes. The excess of centrosomes was removed by rinsing the open chamber with large volume of TicTac buffer supplemented with 1% BSA to prevent the non-specific interactions (TicTac-BSA buffer).
Microtubules and actin assembly at the centrosome were induced by diluting tubulin dimers (labelled with ATTO-565, 30 μM final) and/or actin monomers (labelled with Alexa-488, or Alexa-568, or Alexa-647, 1 μM final) in TicTac buffer supplemented with 1 mM GTP and 2.7 mM ATP, 10 mM DTT, 20 μg/ml catalase, 3 mg/ml glucose, 100 μg/ml glucose oxidase and 0.25 % w/v methylcellulose. In addition, 3-fold molar equivalent of profilin to actin was added in the reaction mixture.
Experiments based on the Brinkley buffer, the buffer usually used for centrosome and microtubules assays, were performed as previously described for the TicTac buffer by using BRB80 buffer supplemented with 50 mM KCl, 1 mM GTP and 2.7 mM ATP, 10 mM DTT, 20 μg/ml catalase, 3 mg/ml glucose, 100 μg/ml glucose oxidase and 0.25 % w/v methylcellulose.
Antibody inhibition experiments were performed by incubating isolated centrosomes with primary antibodies (diluted in TicTac-BSA buffer) for 1 hour. The control experiment without antibodies was performed incubating isolated centrosomes for 1 hour with TicTac-BSA buffer.
Arp2/3 complex and formin homology 2 (FH2) domain inhibition experiments were performed by adding 0.2 mM CK666 and 0.2 mM SMIFH2 respectively in the reaction mixture.
Immunofluorescence staining on isolated centrosomes
Staining of F-actin on centrosomes was performed without prior fixation with 200 nM Alexa-647-phalloidin for 20 minutes. For immunofluorescence staining, isolated centrosomes were incubated with primary antibodies for 1 hour and with secondary antibodies for 30 minutes at room temperature. F-actin asters were stained under microscope without prior fixation. Numerous images of asters were acquired before the addition of primary antibodies (1 hour) and secondary antibodies (30 minutes). Phalloidin and all the antibodies were diluted in TicTac-BSA buffer. In the experiments with centrosomes isolated from siPCM1 cells, a γ-tubulin staining at the end of the movies was necessary to label the centrosome, because PCM1 depletion impairs the recruitment of EGFP-centrin to the centrosome 45. Staining was performed as previously described, without prior fixation.
Centrosomes were considered positive for a given signal when their signal exceeded two times the background signal threshold.
Image acquisition, processing and analysis
Fixed cell images were captured on a confocal microscope (Leica SP2) using a 40x 1.25 N.A. objective lens, on a spinning disk confocal microscope (Roper Scientific) equipped with an iLasPulsed system, with a confocal head (Yokogawa) and an Evolve camera (EMCCD 512 × 512, pixel = 16 μm) using a 60x N.A. 1.4 objective lens or on an Olympus BX61 upright microscope using a 100x N.A. 1.4 objective lens and Cool SNAP HQ2 camera (pixel = 6.45 μm). Live cell imaging was performed on a spinning disk confocal microscope (Roper Scientific) using a 60x N.A. 1.4 objective lens at 37 °C and 5 % CO2. 3D images processing was performed using ImageJ software. All the images were deconvolved using a theoretical point spread function (PSF). All the images show the centrosome plane, except in Figure 4D-E where maximal intensity projections are shown.
Measurement of the actin amount around the centrosome in extracted cells was performed by measuring the integrated intensity of fluorescence in a 4 μm diameter circle centered around the centrosome. Arp2, p34-Arc and WASH measurements were performed measuring the integrated fluorescence intensity in a 3 μm diameter circle centered around the centrosome. Data from separated experiments were normalized so that the average intensity in control cells was 1. Data from two distinct siRNA sequences against PCM1 (see above) were pooled.
Imaging of isolated centrosomes was performed with a Total Internal Reflection Fluorescence (TIRF) microscope (Roper Scientific) equipped by an iLasPulsed system and an Evolve camera (EMCCD 512 × 512, pixel = 16 μm) using a 60x 1.49 N.A objective lens. Aster proportion was calculated as the ratio of centrin1-positive actin asters divided by the number of centrin1 spots. For the centrosomes isolated from siPCM1 cells, aster proportion was calculated as the ratio of the number of actin asters divided by the number of γ-tubulin spots. Data from separated experiments were normalized so that the average intensity in control experiments (DMSO or control siRNA) was 1. Actin nucleation activity was quantified measuring the actin fluorescence intensity integrated over a 2 μm diameter at the center of the actin aster and normalized with respect to initial intensity over the time. Representative data for several experiments are shown.
TetraSpeck™ microspheres (Life Technologies) were used to calibrate fluorescence intensities on spinning disk and TIRF microscopes.
Statistical analyses
Statistical analyses were performed with GraphPad Prism 5 (GraphPad Software). Unpaired t-test with Welch’s correction was used to determine statistical significance. In all the graphics two-tailed P-values were calculated and we reported the following Prism convention: ns (P > 0.05), * (P ≤ 0.05), ** (P ≤ 0.01), *** (P ≤ 0.001) and **** (P ≤ 0.0001).
Supplementary Material
Acknowledgements
This work was supported by European Research Council (Starting grant 310472), the Agence Nationale pour la Recherche (ANR-12-BSV5-0004-01, ANR-10-INBS-08-01) and the “Laboratory of Excellence” (Grenoble Alliance for Integrated Structural Cell Biology). We are grateful to Michel Bornens for providing pEGFP-centrin1 plasmid, Alexey Khodjakov for RPE1 EGFP-centrin-1 cells, Roger Tsien for pRSET-B-dTomato plasmid, Alexis Gautreau for WASH antibodies and Theresia Stradal for p34-Arc antibodies. We thank Marylin Vantard and Michel Bornens for constructive discussions.
Footnotes
Authors contributions
JS, LB and MT conceived and supervised the project. FF and JS performed the experimental work. JG and CG defined the TicTac buffer. YC performed the mass spectrometry analysis. FF, JS, LB and MT analyzed the data. MT wrote the manuscript, which was edited by FF, JS and LB.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Bornens M. Organelle positioning and cell polarity. Nat Rev Mol Cell Biol. 2008;9:874–886. doi: 10.1038/nrm2524. [DOI] [PubMed] [Google Scholar]
- 2.Etienne-Manneville S. Actin and microtubules in cell motility: which one is in control? Traffic. 2004;5:470–7. doi: 10.1111/j.1600-0854.2004.00196.x. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez OC, et al. Conserved microtubule – actin interactions in cell movement and morphogenesis. Nat Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- 4.Coles CH, Bradke F. Coordinating Neuronal Actin–Microtubule Dynamics. Curr Biol. 2015;25:R677–R691. doi: 10.1016/j.cub.2015.06.020. [DOI] [PubMed] [Google Scholar]
- 5.Chesarone Ma, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 6.Vaughan S, Dawe HR. Common themes in centriole and centrosome movements. Trends Cell Biol. 2011;21:57–66. doi: 10.1016/j.tcb.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Tang N, Marshall WF. Centrosome positioning in vertebrate development. J Cell Sci. 2012;125:4951–4961. doi: 10.1242/jcs.038083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The Respective Contributions of the Mother and Daughter Centrioles to Centrosome Activity and Behavior in Vertebrate Cells 7. Cell. 2000;149:317–329. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevrier V. The Rho-associated protein kinase p160ROCK is required for centrosome positioning. J Cell Biol. 2002;157:807–817. doi: 10.1083/jcb.200203034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Euteneuer U, Schliwa M. Evidence for an involvement of actin in the positioning and motility of centrosomes. J Cell Biol. 1985;101:96–103. doi: 10.1083/jcb.101.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao J, Crest J, Fasulo B, Sullivan W. Cortical actin dynamics facilitate early-stage centrosome separation. Curr Biol. 2010;20:770–6. doi: 10.1016/j.cub.2010.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, Chen L, Ding Y, Jin J, Liao K. Centrosome separation driven by actin-microfilaments during mitosis is mediated by centrosome-associated tyrosine-phosphorylated cortactin. J Cell Sci. 2008;121:1334–43. doi: 10.1242/jcs.018176. [DOI] [PubMed] [Google Scholar]
- 13.Rosenblatt J, Cramer LP, Baum B, Mcgee KM. Myosin II-Dependent Cortical Movement Is Required for Centrosome Separation and Positioning during Mitotic Spindle Assembly. 2004;117:361–372. doi: 10.1016/s0092-8674(04)00341-1. [DOI] [PubMed] [Google Scholar]
- 14.Kunda P, Pelling AE, Liu T, Baum B. Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr Biol. 2008;18:91–101. doi: 10.1016/j.cub.2007.12.051. [DOI] [PubMed] [Google Scholar]
- 15.Antoniades I, Stylianou P, Skourides Pa. Making the connection: ciliary adhesion complexes anchor basal bodies to the actin cytoskeleton. Dev Cell. 2014;28:70–80. doi: 10.1016/j.devcel.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Pan J, You Y, Huang T, Brody SL. RhoA-mediated apical actin enrichment is required for ciliogenesis and promoted by Foxj1. J Cell Sci. 2007;120:1868–76. doi: 10.1242/jcs.005306. [DOI] [PubMed] [Google Scholar]
- 17.Kleve MG, Clark WH. Association of actin with sperm centrioles: isolation of centriolar complexes and immunofluorescent localization of actin. J Cell Biol. 1980;86:87–95. doi: 10.1083/jcb.86.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pitaval A, Tseng Q, Bornens M, Théry M. Cell shape and contractility regulate ciliogenesis in cell cycle-arrested cells. J Cell Biol. 2010;191:303–312. doi: 10.1083/jcb.201004003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong H, Kim J, Kim J. Myosin heavy chain 10 (MYH10) is required for centriole migration during the biogenesis of primary cilia. Biochem Biophys Res Commun. 2015;10:1–6. doi: 10.1016/j.bbrc.2015.04.028. [DOI] [PubMed] [Google Scholar]
- 20.Dawe HR, et al. Nesprin-2 interacts with meckelin and mediates ciliogenesis via remodelling of the actin cytoskeleton. J Cell Sci. 2009;122:2716–2726. doi: 10.1242/jcs.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez TS, et al. Formins Regulate the Actin-Related Protein 2/3 Complex-Independent Polarization of the Centrosome to the Immunological Synapse. Immunity. 2007;26:177–190. doi: 10.1016/j.immuni.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chodagam S, Royou A, Whitfield W, Karess R, Raff JW. The centrosomal protein CP190 regulates myosin function during early Drosophila development. Curr Biol. 2005;15:1308–13. doi: 10.1016/j.cub.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 23.Stevenson VA, Kramer J, Kuhn J, Theurkauf WE. Centrosomes and the Scrambled protein coordinate microtubule-independent actin reorganization. Nat Cell Biol. 2001;3:68–75. doi: 10.1038/35050579. [DOI] [PubMed] [Google Scholar]
- 24.Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cel. 2004;7:413–24. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Cowan CR, Hyman Aa. Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature. 2004;431:92–6. doi: 10.1038/nature02825. [DOI] [PubMed] [Google Scholar]
- 26.Woolner S, O’Brien LL, Wiese C, Bement WM. Myosin-10 and actin filaments are essential for mitotic spindle function. J. Cell Biol. 2008;182:77–88. doi: 10.1083/jcb.200804062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitsushima M, et al. Revolving movement of a dynamic cluster of actin filaments during mitosis. J Cell Biol. 2010;191:453–62. doi: 10.1083/jcb.201007136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fink J, et al. External forces control mitotic spindle positioning. Nat Cell Biol. 2011;13:771–8. doi: 10.1038/ncb2269. [DOI] [PubMed] [Google Scholar]
- 29.Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM. Centrosome polarization delivers secretory granules to the immunological synapse. Nature. 2006;443:462–5. doi: 10.1038/nature05071. [DOI] [PubMed] [Google Scholar]
- 30.Bornens M. Is the centriole bound to the nuclear membrane? Nature. 1977;270:80–2. doi: 10.1038/270080a0. [DOI] [PubMed] [Google Scholar]
- 31.Burakov AV, Nadezhdina ES. Association of nucleus and centrosome: magnet or velcro? Cell Biol Int. 2013;37:95–104. doi: 10.1002/cbin.10016. [DOI] [PubMed] [Google Scholar]
- 32.Bornens M, Moudjou M. Studying the composition and function of centrosomes in vertebrates. Methods Cell Biol. 1999 doi: 10.1016/s0091-679x(08)61973-1. [DOI] [PubMed] [Google Scholar]
- 33.Andersen JS, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–4. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 34.Jakobsen L, et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011;30:1520–35. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Firat-Karalar EN, Sante J, Elliott S, Stearns T. Proteomic analysis of mammalian sperm cells identifies new components of the centrosome. J Cell Sci. 2014;127:4128–33. doi: 10.1242/jcs.157008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bornens M, Paintrand M, Berges J, Marty MC, Karsenti E. Structural and chemical characterization of isolated centrosomes. Cell Motil Cytoskeleton. 1987;8:238–49. doi: 10.1002/cm.970080305. [DOI] [PubMed] [Google Scholar]
- 37.Miranda AF, Godman GC, Tanenbaum SW. Action of cytochalasin D on cells of established lines. II. Cortex and microfilaments. J Cell Biol. 1974;62:406–23. doi: 10.1083/jcb.62.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chesarone Ma, Goode BL. Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol. 2009;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubert T, Vandekerckhove J, Gettemans J. Actin and Arp2/3 localize at the centrosome of interphase cells. Biochem Biophys Res Commun. 2011;404:153–8. doi: 10.1016/j.bbrc.2010.11.084. [DOI] [PubMed] [Google Scholar]
- 40.Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nolen BJ, et al. Characterization of two classes of small molecule inhibitors of Arp2 / 3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monfregola J, Napolitano G, D’Urso M, Lappalainen P, Ursini MV. Functional characterization of Wiskott-Aldrich syndrome protein and scar homolog (WASH), a bi-modular nucleation-promoting factor able to interact with biogenesis of lysosome-related organelle subunit 2 (BLOS2) and gamma-tubulin. J Biol Chem. 2010;285:16951–7. doi: 10.1074/jbc.M109.078501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Derivery E, et al. The Arp2/3 Activator WASH Controls the Fission of Endosomes through a Large Multiprotein Complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 44.Bärenz F, Mayilo D, Gruss OJ. Centriolar satellites: busy orbits around the centrosome. Eur J Cell Biol. 2011;90:983–9. doi: 10.1016/j.ejcb.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sillibourne JE, et al. Primary ciliogenesis requires the distal appendage component Cep123. Biol Open. 2013;2:535–45. doi: 10.1242/bio.20134457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N. Centriolar Satellites: Molecular Characterization, ATP-dependent Movement Toward Centrioles and Possible Involvement in Ciliogenesis. J Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clark IB, Meyer DI. Overexpression of normal and mutant Arp1alpha (centractin) differentially affects microtubule organization during mitosis and interphase. J Cell Sci. 1999;112:3507–18. doi: 10.1242/jcs.112.20.3507. [DOI] [PubMed] [Google Scholar]
- 49.Patel H, et al. Kindlin-1 regulates mitotic spindle formation by interacting with integrins and Plk-1. Nat Commun. 2013;4:2056. doi: 10.1038/ncomms3056. [DOI] [PubMed] [Google Scholar]
- 50.Pugacheva EN, Golemis Ea. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol. 2005;7:937–46. doi: 10.1038/ncb1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fielding AB, Dobreva I, McDonald PC, Foster LJ, Dedhar S. Integrin-linked kinase localizes to the centrosome and regulates mitotic spindle organization. J Cell Biol. 2008;180:681–9. doi: 10.1083/jcb.200710074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hubert T, Van Impe K, Vandekerckhove J, Gettemans J. The actin-capping protein CapG localizes to microtubule-dependent organelles during the cell cycle. Biochem Biophys Res Commun. 2009;380:166–70. doi: 10.1016/j.bbrc.2009.01.064. [DOI] [PubMed] [Google Scholar]
- 53.Bershteyn M, Atwood SX, Woo W-M, Li M, Oro AE. MIM and cortactin antagonism regulates ciliogenesis and hedgehog signaling. Dev Cell. 2010;19:270–83. doi: 10.1016/j.devcel.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Epting D, et al. The Rac1 regulator ELMO controls basal body migration and docking in multiciliated cells through interaction with Ezrin. Development. 2015;142:174–84. doi: 10.1242/dev.112250. [DOI] [PubMed] [Google Scholar]
- 55.Sabino D, et al. Moesin Is a Major Regulator of Centrosome Behavior in Epithelial Cells with Extra Centrosomes. Curr Biol. 2015;25:879–889. doi: 10.1016/j.cub.2015.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaplan DD, Meigs TE, Kelly P, Casey PJ. Identification of a Role for β-Catenin in the Establishment of a Bipolar Mitotic Spindle. J Biol Chem. 2004;279:10829–10832. doi: 10.1074/jbc.C400035200. [DOI] [PubMed] [Google Scholar]
- 57.Ritchey L, Ottman R, Roumanos M, Chakrabarti R. A functional cooperativity between Aurora A kinase and LIM kinase1: Implication in the mitotic proce. Cell Cycle. 2012;11:296–309. doi: 10.4161/cc.11.2.18734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lemullois M, Klotz C, Sandoz D. Immunocytochemical localization of myosin during ciliogenesis of quail oviduct. Eur J Cell Biol. 1987;43:429–437. [PubMed] [Google Scholar]
- 59.Espreafico EM, et al. Localization of myosin-V in the centrosome. Proc Natl Acad Sci U S A. 1998;95:8636–8641. doi: 10.1073/pnas.95.15.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol. 2012;14:1–13. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- 61.Mennella V, et al. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol. 2012;14:1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burke TA, et al. Report Homeostatic Actin Cytoskeleton Networks Are Regulated by Assembly Factor Competition for Monomers. Curr Biol. 2014;24:579–585. doi: 10.1016/j.cub.2014.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moudjou M, Bornens M. In: Cell Biol A Lab Handb. Celis J, editor. Academic Press; 1998. pp. 111–119. [Google Scholar]
- 64.Shelanski M. Chemistry of the filaments and tubules of brain. J Histochem Cytochemistry. 1973;21:529–539. doi: 10.1177/21.6.529. [DOI] [PubMed] [Google Scholar]
- 65.Hyman A, et al. Preparation of modified tubulins. Methods Enzymol. 1991;196:478–85. doi: 10.1016/0076-6879(91)96041-o. [DOI] [PubMed] [Google Scholar]
- 66.Michelot A, et al. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr Biol. 2007;17:825–33. doi: 10.1016/j.cub.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 67.Achard V, et al. A ‘primer’-based mechanism underlies branched actin filament network formation and motility. Curr Biol. 2010;20:423–8. doi: 10.1016/j.cub.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 68.Casabona MG, Vandenbrouck Y, Attree I, Couté Y. Proteomic characterization of Pseudomonas aeruginosa PAO1 inner membrane. Proteomic. 2013;13:2419–2423. doi: 10.1002/pmic.201200565. [DOI] [PubMed] [Google Scholar]
- 69.Sillibourne JE, et al. Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol Biol Cel. 2010;21:547–61. doi: 10.1091/mbc.E09-06-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dubois T, et al. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat Cell Biol. 2005;7:353–64. doi: 10.1038/ncb1244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.