Integrative multiomics analyses of adipose and muscle tissue transcripts, S, and genotypes revealed novel genetic regulatory mechanisms of insulin resistance in African Americans.
Abstract
Context:
Compared with European Americans, African Americans (AAs) are more insulin resistant, have a higher insulin secretion response to glucose, and develop type 2 diabetes more often. Molecular processes and/or genetic variations contributing to altered glucose homeostasis in high-risk AAs remain uncharacterized.
Objective:
Adipose and muscle transcript expression profiling and genotyping were performed in 260 AAs to identify genetic regulatory mechanisms associated with insulin sensitivity (SI). We hypothesized that: 1) transcription profiles would reveal tissue-specific modulation of physiologic pathways with SI, and 2) a subset of SI-associated transcripts would be controlled by DNA sequence variants as expression quantitative traits, and these variants in turn would be associated with SI.
Design and Settings:
The cross-sectional research study was performed in a clinical research unit.
Participants:
Unrelated nondiabetic AAs were recruited for the study.
Main Outcome Measures:
SI was measured by frequently sampled iv glucose tolerance test.
Results:
The expression levels of 2212 transcripts in adipose and 145 transcripts in muscle were associated with SI. Genes involved in eIF2, eIF4-p70S6K, and mTOR signaling were modulated with SI in both tissues. Genes involved in leukocyte extravasation signaling showed adipose-specific regulation, and genes involved in oxidative phosphorylation had discordant regulation between tissues. Intersecting cis-expression quantitative trait loci results with data from transcript-SI association analysis identified cis-regulatory single nucleotide polymorphisms for 363 and 42 SI-associated transcripts in adipose and muscle, respectively. Cis-eSNPs for three SI-associated adipose transcripts, NINJ1, AGA, and CLEC10A were associated with SI. Abrogation of NINJ1 induction in THP1 macrophages modulated expression of genes in chemokine signaling, cell adhesion, and angiogenesis pathways.
Conclusion:
This study identified multiple pathways associated with SI; particularly discordant tissue-specific regulation of the oxidative phosphorylation pathway, and adipose-specific regulation of transcripts in the leukocyte extravasation signaling pathway that seem to be important in insulin resistance. Identification of single nucleotide polymorphisms associated with SI and with modulation of expression of SI-associated transcripts, including NINJ1, reveals novel genetic regulatory mechanisms of insulin resistance in AAs.
Type 2 diabetes (T2D) is a heterogeneous metabolic disorder that can result from dysfunctional interactions between complex physiological processes maintaining glucose homeostasis (1). Insulin resistance in muscle, adipose, and liver, impaired pancreatic β-cell insulin secretion, and obesity typically precede the development of T2D. Although the relative contributions and timing of these processes remain unclear (2, 3), longitudinal studies in high-risk individuals suggest that insulin resistance occurs years before glucose intolerance or β-cell failure (1, 4, 5). Investigating early pathophysiological changes may help to identify the molecular processes involved in T2D.
In healthy individuals, the relationship between insulin sensitivity (SI) and the β-cell insulin response is reciprocal and nonlinear (hyperbolic), and ethnic-specific variation is observed in its distribution (2). Compared with age-, sex-, and body mass index (BMI) –matched non-Hispanic Caucasians, African Americans (AAs) are more insulin resistant (lower SI), but show a greater acute insulin response to iv glucose (AIRG) and a higher disposition index (DI = SI × AIRG) (6, 7). Unique evolutionary histories and ancestry-specific genetic variation could contribute to ethnic-specific physiology and higher risk for T2D in AAs. Genome-wide transcript profiling analyses implicated modulation of many genes in insulin-responsive tissues, involving derangement of multiple biological pathways in insulin-resistant individuals (1). Although elucidation of these pathways advanced our understanding of altered glucose homeostasis at the molecular level, most studies did not assess individuals of African ancestry. Molecular processes and/or genetic variations that may contribute to altered glucose homeostasis in high-risk populations, including AAs, remain uncharacterized.
Thus, we systematically analyzed genome-wide transcript expression profiles of sc adipose and skeletal muscle (insulin-responsive tissues) and genome-wide SNP genotypes from a metabolically characterized cohort of 260 unrelated and nondiabetic AAs. SI measured by the frequently sampled iv glucose tolerance test (FSIGT) was the primary physiological outcome. Correlated secondary phenotypes were also assessed. These data were used in an integrative multiomics approach (Supplemental Figure 1) to test two hypotheses: 1) transcription profiles in adipose and muscle would correlate with SI and might reveal tissue-specific modulation of physiologic pathways, and 2) a subset of SI-associated transcripts would be controlled by DNA sequence variants as expression quantitative traits, and these variants in turn would be associated with SI.
Materials and Methods
Human subjects
Studies were performed at the Wake Forest School of Medicine (WFSM) Clinical Research Unit. The study was approved by the WFSM Institutional Review Board and all participants provided written informed consent. Participants were healthy, self-described AAs age 18–60 years, with a BMI between 18 and 42 kg/m2. Individuals taking medications that increased bleeding risk or altered SI and/or gene expression in insulin-responsive tissues (eg, steroids or thyroid hormone replacement) were excluded. Methods for recruitment, physical examination, physiological experiments, and performance of adipose and muscle biopsies have been reported (8–11).
Height and weight and waist and hip circumference were measured, body fat was determined by a bioelectrical impedance analyzer (RJL analyzer Quantum Desktop), and fasting blood was drawn for DNA isolation and biochemical analyses (complete metabolic panel, hemoglobin A1c, and lipid profile) at the screening visit. A standard 75-g oral glucose tolerance test (OGTT) was completed during the screening visit to exclude diabetes, and glucose and insulin were measured at baseline and 30-minute intervals for 2 hours. Of 319 screened participants, 31 were excluded based on abnormal laboratory results (13 had T2D) and 28 were lost to followup. Nondiabetic individuals (fasting plasma glucose < 126 mg/dL and 2-h OGTT glucose < 199 mg/dL) were invited to a second visit for adipose and muscle tissue biopsies under fasting conditions and an insulin-modified (0.03 U/kg) FSIGT. Abdominal sc adipose near the umbilicus and vastus lateralis skeletal muscle biopsies were obtained under local anesthesia (lidocaine). Biopsies was performed by opening a Bergstrom needle to cut 100–200-mg pieces of tissue, then applying suction using a 50-mL syringe. Tissues were immediately rinsed in sterile saline, quick-frozen in liquid nitrogen, and stored at −80°C. A total of 260 unrelated individuals completed all study visits and are referred to as the “African American Genetics of Metabolism and Expression” (AAGMEx) cohort. Adipose and muscle biopsies were collected from 256 individuals and high-quality FSIGT data were available from 235 participants.
Laboratory measurements and physiological phenotyping
Details of clinical/biochemical methods are presented in Supplemental Methods. In brief, OGTT data were analyzed by homeostatic model assessment (HOMA; http://mmatsuda.diabetes-smc.jp/MIndex.html) (12, 13). The MINMOD Millennium program was used to analyze FSIGT data to determine SI and AIRG (14).
Gene expression analysis and genotyping methods
Details of gene expression analysis, genotyping, and data quality control methods are presented in Supplemental Methods. In brief, genome-wide expression data were generated using HumanHT-12 v4 Expression BeadChip (Illumina) whole-genome gene expression arrays for quantitative analyses of transcript expression in adipose and muscle samples. Infinium HumanOmni5Exome-4 v1.1 DNA Analysis BeadChips (Illumina) were used to genotype DNA samples based on manufacturer instructions.
Statistical and bioinformatic methods
To test for an association between glucose homeostasis or obesity traits and expression level, a linear regression model was computed with the trait (transformed, if necessary; see Supplemental Methods) as the outcome and expression level (log2) as the predictor. Models included age, sex, and ancestry proportion (admixture) as covariates. Benjamini-Hochberg false-discovery rate (FDR) adjusted P-values were computed by trait (15). Expression of a transcript associated with a trait at an FDR-adjusted P < .01 (1%) was considered significant. In a secondary analysis, comparison of transcript expression between participants in the top and bottom quartiles of the SI distribution (adjusted for age, sex, and African ancestry proportion) was performed to identify transcripts with significantly different expression between groups. Standard two-class unpaired analysis as implemented in Statistical Analysis for Microarray (SAM) software was used (16). To identify expression quantitative trait loci (eQTLs), linear regression was computed with the log2-transformed expression values as the outcome and an additive genetic model for the SNP as implemented in the R-package MatrixEQTL (17); age, sex, and African ancestry proportion were covariates. Detailed statistical and bioinformatic data analysis methods are presented in Supplemental Methods. Sample sizes in each analysis varied based on available data after quality control (Supplemental Table 1).
In vitro studies to identify functional significance of a cis-eGene
The effect of experimental manipulation of the expression of a selected (see Results) cis-eGene on cellular pathways was tested in THP1 cells, a widely used human monocytic leukemia cell line model for monocyte-macrophage differentiation (18). Gene-specific lentiviral short hairpin RNA (shRNA) was used for transduction and to silence the expression of the gene by stable RNA interference. Detailed descriptions of cell culture experiments, RNA sequencing (RNA-Seq), qRT-PCR and analytic methods are presented in Supplemental Methods.
Results
Phenotypic characteristics
Anthropometric and physiological characteristics are summarized in Table 1. Participants had a broad range of BMIs (28.9 ± 5.6 kg/m2), adiposity (waist:hip ratio = 0.91 ± 0.08; fat mass = 32.9 ± 9.7%), and glucose homeostasis-related phenotypes, including FSIGT-derived insulin sensitivity (SI = 4.0 ± 3.5). Hemoglobin A1c levels were at least 5.7% in 113 participants, fasting plasma glucose concentrations were at least 100 mg/dL in 47 participants, and 2-h glucose values during the 75-g OGTT were at least 140 mg/dL in 28 participants. In addition, high-density lipoprotein cholesterol concentrations were less than or equal to 40 mg/dL in 32 participants, and triglycerides were at least 150 mg/dL in 21 individuals. Consistent with other studies, age- and sex-adjusted SI showed significant negative correlation with BMI (r = −0.48; P = 4.7 × 10−15) and insulin resistance (HOMA-IR, r = −0.47; P = 5.5 × 10−14). SI had a negative correlation with AIRG (r = −0.58; P = 5.3 × 10−22) and a positive correlation with OGTT-derived insulin sensitivity index (Matsuda insulin sensitivity index, r = 0.57; P = 1.1 × 10−20).
Table 1.
Demographics of the Study Population: AAGMEx Cohort
| Trait | N | Mean | Median | SD |
|---|---|---|---|---|
| Sex, M/F | 139/121 | |||
| Age, y | 260 | 40.6 | 41.4 | 11.5 |
| BMI, kg/m2 | 260 | 28.9 | 28.5 | 5.6 |
| Waist-to-hip ratio | 256 | 0.91 | 0.91 | 0.08 |
| Fat mass, % | 236 | 32.9 | 32.3 | 9.7 |
| Systolic BP, mm Hg | 260 | 122.1 | 119.8 | 15.8 |
| Diastolic BP, mm Hg | 260 | 75.3 | 74.3 | 10.6 |
| Cholesterol, total, mg/dL | 259 | 176.5 | 173.0 | 36.3 |
| Triglycerides, mg/dL | 259 | 84.0 | 73.0 | 43.3 |
| HDL cholesterol, mg/dL | 259 | 56.2 | 54.0 | 15.6 |
| VLDL cholesterol, mg/dL | 259 | 16.8 | 15.0 | 8.6 |
| LDL cholesterol, mg/dL | 259 | 103.5 | 99.0 | 32.3 |
| Hemoglobin A1c, % | 260 | 5.6 | 5.6 | 0.3 |
| Fasting insulin, mU/L | 260 | 10.0 | 8.0 | 8.4 |
| 2-h insulin, mU/La | 260 | 62.2 | 40.1 | 77.5 |
| Fasting glucose, mg/dL | 260 | 91.3 | 90.0 | 9.4 |
| 2-h glucose, mg/dLa | 259 | 100.7 | 97.0 | 29.6 |
| Matsuda indexa | 252 | 6.2 | 4.6 | 6.6 |
| HOMA-IR | 260 | 2.3 | 1.8 | 2.2 |
| SI [×10−4 (mU/L)−1 · min−1]b | 235 | 4.0 | 3.1 | 3.5 |
| AIRG, mU.L−1 · minb | 235 | 779.4 | 581.8 | 645.8 |
| DIb | 235 | 2287.5 | 1886.6 | 1519.5 |
| SG, min−1b | 235 | 0.019 | 0.017 | 0.010 |
Abbreviations: BP, blood pressure; HDL, high-density lipoprotein; LDL, low-density lipoprotein; VLDL, very low-density lipoprotein.
SG, measure of the ability of glucose to promote its own uptake (glucose effectiveness). Units are taken from MINMOD Millennium program.
Metabolic measurements from OGTT.
Metabolic traits from FSIGT evaluation of nondiabetic individuals.
SI-associated transcripts in adipose tissue
SI was positively and negatively associated, respectively, with expression of 1169 and 1043 adipose transcripts (FDR < 1%) (Supplemental Table 2A). Expression levels of adipose transcripts were significantly correlated with glucose homeostasis and obesity phenotypes. The clustering of Matsuda index and SI indicated significant overlap in direction of association of genes correlated with these insulin sensitivity traits (Supplemental Figure 2). SI was strongly correlated with BMI; after adjustment for BMI, only 17 adipose tissue transcripts remained significantly associated (FDR < 5%) with SI (Supplemental Table 2B).
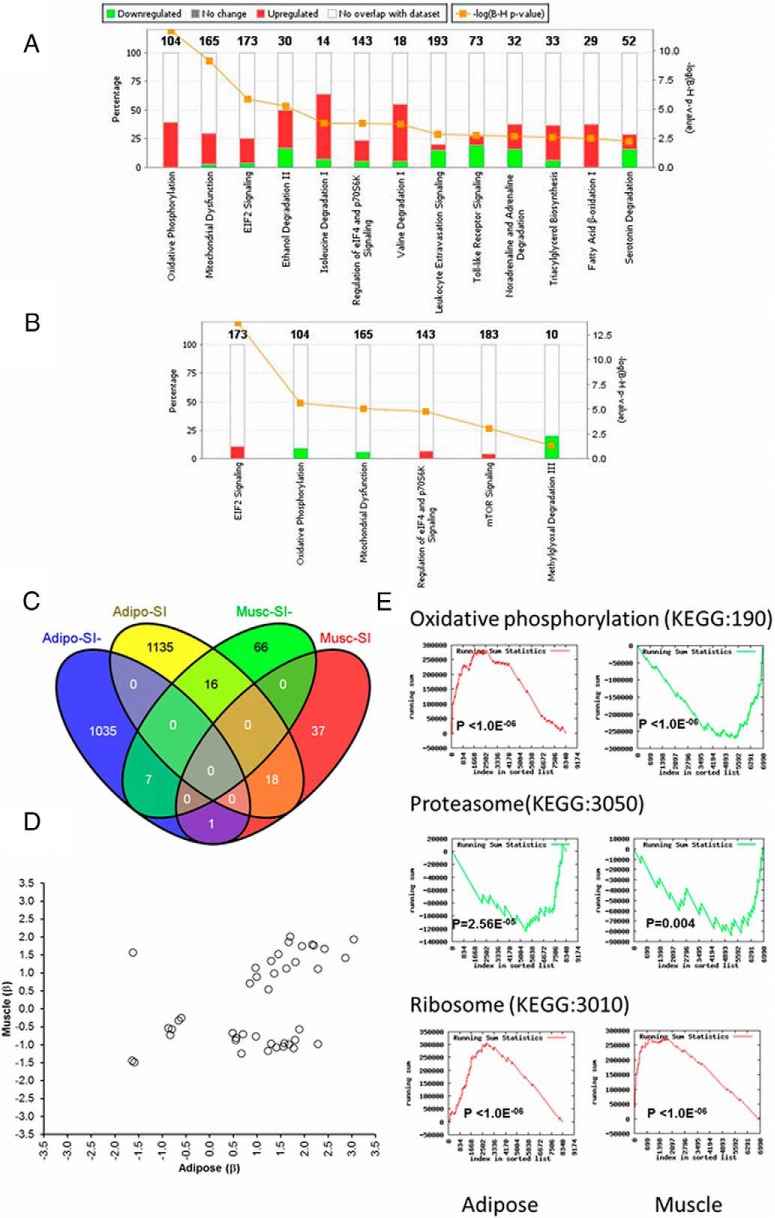
Ingenuity pathway analysis (IPA) identified significant enrichment of biological pathways among the 2212 adipose transcript probes associated with SI (FDR < 1%; Supplemental Table 3). Genes involved in metabolic pathways including oxidative phosphorylation (P = 2.0 × 10−12), EIF2 signaling (P = 1.4 × 10−6), regulation of eIF4 and p70S6K signaling (P = 1.8 × 10−4), and valine and isoleucine degradation (P ≤ .0002) were positively associated with SI. Genes involved in leukocyte extravasation and toll-like receptor signaling pathways (P ≤ .002) were negatively associated with SI (Figure 1A; Supplemental Figures 3 and 4). Furthermore, gene set enrichment analysis (GSEA), using the ranked list of all transcripts expressed in adipose tissue, also identified enrichment of SI-associated transcripts (Supplemental Table 4). Genes involved in ribosome function (Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway:3010; P < 1.0 × 10−6) were positively associated with SI, and genes in lysosome (KEGG: 4142) and proteasome (KEGG: 3050) pathways were negatively associated with SI (P < .001).
Figure 1.
Common and tissue-specific characteristics of adipose and muscle tissue transcripts associated with SI in AAs. A stacked bar graph shows enrichment of adipose (A) and muscle (B) tissue transcripts associated with SI in IPA biological pathway analysis. A Venn diagram (C) shows overlap of adipose and muscle tissue transcripts positively or negatively associated with SI. Scatter plot (D) shows the concordance and discordance in direction of association (effect, β from regression analysis) of transcripts associated with SI in both adipose and muscle tissue. Visualization of running sum statistics from advanced gene set enrichment analysis (GSEA, GeneTrail) shows (E) common and tissue-specific modulation of transcripts with SI and enrichment in biological pathways.
In a secondary analysis, comparison of adipose transcript expression between individuals in the top and bottom quartiles of the SI distribution (adjusted for age, sex, and ancestry proportion) identified 247 differentially expressed transcripts (≥1.2-fold; FDR ≤ 5%). Those in the bottom quartile were insulin resistant (IR; SI = 1.37 ± 0.47; N = 57) and those in the top quartile were insulin sensitive (IS; SI = 8.17 ± 4.09, N = 58). Differentially expressed probes included 156 up-regulated and 91 down-regulated mRNA transcripts of RefSeq genes with known function (Supplemental Table 5). Genes with higher expression in IR participants included dermatopontin (DPT), NADPH dehydrogenase quinone 1 (NQO1), ubiquitin carboxyl-terminal esterase L1 (UCHL1), leptin (LEP), Fc fragment of IgG binding protein (FCGBP), integrin beta 2 (ITGB2), secreted frizzled-related protein 4 (SFRP4), lymphocyte cytosolic protein 1 (LCP1), and monocyte/macrophage serine esterase 1 and carboxylesterase 1(CES1). Genes with lower expression in IR individuals included α-2-glycoprotein 1 zinc-binding (AZGP1), lipin 1 (LPIN1), 1-acylglycerol-3-phosphate O-acyltransferase 9 (AGPAT9), and NDRG family member 4 (NDRG4). However, differential expression between IR and IS subjects for all of these transcripts were less than 2-fold.
Fewer muscle tissue transcripts were associated with SI and showed distinct tissue-specific characteristics
Compared with adipose, fewer transcripts in muscle were positively (n = 56) or negatively (n = 89) associated with SI (Supplemental Table 6A). Only three muscle tissue transcripts were significantly associated (FDR < 5%) with SI after adjustment for BMI (Supplemental Table 6B).
As in adipose, muscle tissue transcripts positively associated with SI were enriched for eIF2, eIF4-p70S6K, and mTOR signaling (P = .001–2.0 × 10−14; Supplemental Tables 7 and 8; Figure 1B). SI was associated with expression levels of 42 transcripts in both adipose and muscle (FDR < 1%). However, 17 of these transcripts show discordant directions of association, indicating tissue-specific transcript modulation (Figure 1, C and D). SI-associated transcripts in muscle were enriched for oxidative phosphorylation (P = 2.5 × 10−6). In contrast with the predominantly positive association in adipose (41 genes), the expression of transcripts in this pathway (9 genes) were negatively (inversely) associated in muscle (Supplemental Table 9A; Supplemental Figure 3). GSEA validated discordant and tissue-specific modulation of genes in the oxidative phosphorylation pathways (Figure 1E; Supplemental Table 9B).
In a secondary analysis, comparison of muscle transcript expression between those in the top and bottom quartiles of the SI distribution identified only 57 differentially expressed transcripts using less stringent significance thresholds (≥ 1.2-fold; FDR ≤ 5%; Supplemental Table 10). Consistent with animal models and human studies in insulin resistance and T2D, genes with higher expression in IR participants included G0/G1switch 2 (G0S2) (19), dysbindin domain containing 1 (DBNDD1) (20), RAP2A member of RAS oncogene family (RAP2A) (21), and aldo-keto reductase family 1 member C3 (AKR1C3) (22). Genes down-regulated in IR individuals included lipoprotein lipase (LPL) (22) and insulin receptor substrate 1 (IRS1). Expression of G0S2 was 1.5-fold higher in IR participants in this cohort. Interestingly, Parikh et al (23) reported that the expression of G0S2 in skeletal muscle was strongly induced by insulin. The higher expression of the G0S2 gene observed in IR individuals may reflect elevated fasting concentrations of insulin compared with IS individuals. The function of this gene in muscle is poorly understood; however, a recent study showed that whole-body knockout of the G0S2 gene enhanced adipose lipolysis, attenuated gain of adiposity, and improved insulin sensitivity (24).
Replication of SI-transcript associations
Strong concordance was observed between correlations of adipose tissue transcripts and SI in nondiabetic AAGMEx AAs (N = 230) in this cohort and 99 European Americans from a previously published data set (see Supplemental Methods and Results; Supplemental Figure 5A) (8). The correlation between four selected muscle transcript expression and SI identified in AAs was also validated by qRT-PCR in European American and AA nondiabetic individuals from Arkansas (N = 127) (11) (see Supplemental Methods and Results; Supplemental Figure 5, C–F).
Coexpressed adipose and muscle transcripts support novel regulatory interactions
Weighted gene coexpression network analysis (25, 26) in the AAGMEx cohort identified 28 coexpressed transcript modules in adipose and 22 modules in muscle (Supplemental Figure 6; Supplemental Tables 11 and 12). Modules are denoted by unique colors. A subset of these “modules” was correlated with SI or BMI and enriched for genes in biological pathways (Table 2; Supplemental Tables 13 and 14; Supplemental Results).
Table 2.
SI-associated Coexpression Modules in Adipose and Muscle Tissue Are Enriched for Biological Pathways and Processes
| Module | No. of Probe in Module | Correlation (P) of Module Eigengene |
Percentage of Gene Associated With SI (P < .003) |
Enrichment (n, p, %FDR) | Hub Genes | |||
|---|---|---|---|---|---|---|---|---|
| BMI | PFAT | SI | % β+ | % β− | ||||
| Adipose | ||||||||
| Yellow | 538 | −0.44 (2e-13) | −0.43 (8e-13) | 0.23 (3e-04) | 27.5 | 8.9 | hsa00230:Purine metabolism (16, 1.61e-05, 0.018) | RNASE4, PSMD14, CIDEA, CORO1C, ANG, SLBP, DNAJC27, LETMD1, ZCCHC8, TUBB4Q, PPWD1, PKM2, PABPC1, EIF3H, ADH1B, HAT1, TM7SF2, MLLT10, PHF13, CSNK2A2, PGAM4, SIN3A |
| Blue | 704 | 0.6 (9e-26) | 0.53 (1e-19) | −0.31 (3e-07) | 29.5 | 31.1 | hsa04142:Lysosome (16, 2.95e-05, 0.035) | CD163, ITGB5, GLB1, HTRA1, AZGP1, GRN, UCHL1, VSIG4, GPR116, NPC2, PFKFB3, EFEMP1, CRTAP, FOLR2, KIAA1598, TTYH3, IRAK1, MYO9B, TIMP1, TXN, M6PRBP1, AADACL1, STOX1, CYYR1 |
| Red | 416 | 0.37 (8e-10) | 0.27 (1e-05) | −0.16 (0.01) | 3.4 | 34.9 | hsa04670:Leukocyte transendothelial migration (16, 1.34e-06, 0.0015) | ARHGAP30, DOCK2, LCP1, PRKCB1, WAS, RASSF2, EVI2B, FCGR2A, CSF3R, NCKAP1 L, CD3D, CD86, LTB |
| Brown | 686 | 0.31 (5e-07) | 0.32 (2e-07) | −0.13 (0.04) | 10.5 | 22.6 | hsa03050:Proteasome (10, 1.98e-04, 0.030) | MYL6, AKAP1, HSPA8, PSME3, SIL1, ATP6AP1, ATMIN, TAX1BP3, DPP3, NCLN, AASS, PECI, SIRT1, MAGED1, ANXA5, IFP38, CROP, AKR1C3, KIAA0556, CCM2, UBE2E2, CAV2, CD33, BOP1, TMEM138, B4GALT7, DSG2, SPARC, ARID4A, TAGLN2, LIX1 L, GMPPA, COPB2, PSMC2, TUBA1A, GBA, TUBA1C, RPRC1, TNIP1, PSMB10 |
| Pink | 372 | −0.54 (2e-20) | −0.44 (1e-13) | 0.32 (2e-07) | 46.8 | 3.8 | hsa00190:Oxidative phosphorylation (21, 1.40e-10, 1.60e-07) | PCCB, CS, PRDX3, ATP5A1, UQCRFS1, COX5B, CYC1, MOSC1, PRDX2, GPAM, BRP44, ETS1, DCI, SDHB, UQCRH, AGPAT2, AKAP11, RPS15, PPP1R16A |
| Muscle | ||||||||
| Darkgreen | 26 | 0.2 (0.001) | 0.2 (0.001) | −0.26 (4e-05) | 0.00 | 15.38 | GO:0046320∼regulation of fatty acid oxidation (4, 4.37e-06, 0.0059) | SLC25A20, SLC25A34, MLYCD, ACSL1, PREB, ACACB, CD36 |
| Cyan | 131 | −0.36 (3e-09) | −0.29 (3e-06) | 0.24 (1e-04) | 11.45 | 7.63 | hsa03010:Ribosome (10, 2.76e-07, 3.5e-04) | RBM3, NT5C, RPL18A, SDHC, ENY2, UBXN1, PKIA, EIF3 L, SYF2 |
| Red | 324 | 0.2 (0.002) | 0.18 (0.003) | −0.16 (0.01) | 0.62 | 6.79 | hsa00190:Oxidative phosphorylation (47, 4.35e-43, 4.90e-40) | ATP5A1, CKMT2, BOLA3, ATP5J, NDUFS8, UQCRC1, DECR1, ETFB, UQCRH, BRP44 L |
Abbreviations: PFAT, percent fat mass; %β+ and %β−, percentages of transcripts positively and negatively associated with SI.
Algorithm for the Reconstruction of Accurate Cellular Networks (ARACNE) analysis (27) identified significant interactions between genes in each of these SI-associated modules and identified “hub genes” in the regulatory network (Table 2 and Supplemental Results).
Cis-eSNPs for SI-associated transcripts
We hypothesized that the expression level of a subset of SI-associated transcripts would be associated with genotype and manifest as eQTLs. We identified 1971 and 2078 transcripts with at least one significant cis-eQTL (top eQTL single nucleotide polymorphism [SNP] within ± 500 kb around the expressed transcript at FDR < 1%) in adipose (N = 250) and muscle (N = 247), respectively. Intersecting significant cis-eQTL results with data from transcript-SI association analysis to test our hypothesis, identified cis-regulatory single nucleotide polymorphisms (SNPs) (FDR < 1%) for 363 and 42 SI-associated transcripts in adipose and muscle, respectively (Supplemental Tables 15 and 16).
Cis-expression regulatory SNPs (cis-eSNPs) identified in adipose and muscle of AAs represented a prioritized set of SNPs providing statistical evidence for genotype-dependent variations in transcript abundance. To identify roles of these putatively functional SNPs in modulating insulin sensitivity, tissue eQTL data from this cohort was examined to identify whether a subset of cis-eSNPs was also associated with SI. The association of 2 296 925 autosomal SNPs (representing 2 210 735 unique high-quality genotyped SNPs with minor allele frequency (MAF) > 0.01 and Hardy-Weinberg equilibrium P-value > 1 × 10−6) was assessed with SI in 230 AAGMEx AAs. Of these, SI was associated with 2325 SNPs at P < .001 (additive genetic model), including 29 associated with SI at P ≤ 1 × 10−5. Intersecting cis-eQTLs with SNP-SI association results revealed that among SI-associated SNPs, 10 and 8 were cis-regulatory SNPs (at FDR < 0.01) for seven and five transcripts in adipose and muscle, respectively (Supplemental Table 17).
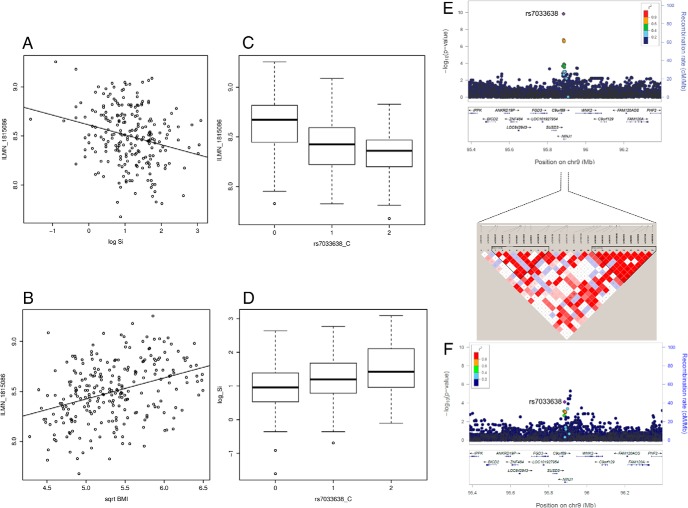
Finally, intersecting transcript-SI, SNP-transcript, and SNP-SI association data identified genetic polymorphisms in three loci modulating expression of SI-associated transcripts in adipose tissue as cis-eSNPs and also associated with SI in the AAGMEx cohort (Table 3). Expression of Ninjurin1 gene (NINJ1) transcript in adipose was significantly associated with SI (P = 7.97 × 10−6; β = −1.24). A 3′-UTR SNP (rs7033638) in NINJ1 was a significant cis-eSNP in adipose and muscle (P = 1.82 × 10−11 and 5.87 × 10−16). The rs7033638-C allele (MAF = 0.36) was associated with lower expression (β = −0.17 and −0.14) of NINJ1 transcript and significantly associated with increased SI (P = 6.24 × 10−5; β = 0.26; Figure 2). Cis-eSNPs for two other SI-associated adipose transcripts, aspartylglucosaminidase (AGA; Supplemental Figure 9) and C-type lectin domain family 10 member A (CLEC10A; Supplemental Figure 10) were also associated with SI in AAGMEx. Formal analysis by the causal inference test (CIT) (28) identified no genome-wide significant loci. However, SI-associated cis-eSNPs were nominally significant in CIT (PCIT < .05) for these three genes (Table 3), indicating putative causal association between the SNPs and SI mediated by transcript expression of these genes (SNP → mRNA → SI). Thus, a subset of regulatory SNPs may modulate transcript expression in tissues important for glucose homeostasis and could affect insulin sensitivity.
Table 3.
Genetic Polymorphisms Modulate Expression of SI-associated Transcripts in Adipose as cis-eSNPs and Associated With SI
| Gene | Association of Adipose Tissue Expression With SI |
Association of SNP with Expression and SI |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-Valuea (Q-value) | βb | Best cis-eSNP | SNP Chr | SNP Position (bp) | Distance to TSS | A1 | A2 | MAF | βc | P-Valuee | Q-Value/FDR | ||
| NINJ1 (ILMN_1815086) | 7.97e-06 | −1.24 | In adipose | ||||||||||
| (1.27e-04) | rs7033638 | 9 | 95 883 785 | 12 785 | C | T | 0.359 | −0.17 | 1.82e-11 | 3.07e-08 | |||
| In muscle | |||||||||||||
| rs7033638 | −0.14 | 5.87e-16 | 1.56e-12 | ||||||||||
| SI-associated SNP | βd | P-valuef | Model | PCIT | |||||||||
| rs7033638 | 0.261 | 6.24e-05 | add | 0.048 | |||||||||
| AGA (ILMN_1802631) | 7.56e-06 | −1.73 | Best cis-eSNP | βc | P-valuee | FDR | |||||||
| (1.23e-04) | In adipose | ||||||||||||
| rs77842019 | 4 | 178 625 097 | −261 440 | A | G | 0.145 | −0.11 | 6.77e-06 | 0.0029 | ||||
| SI-associated SNP | βd | P-valuef | Model | PCIT | |||||||||
| rs77842019 | 0.34 | 1.02e-03 | dom | 4.28e-03 | |||||||||
| 0.31 | 8.36e-04 | add | |||||||||||
| CLEC10A (ILMN_2415303) | 3.11e-04 | −1.38 | Best cis-eSNP | βc | P-valuee | FDR | |||||||
| (2.26e-03) | In adipose | ||||||||||||
| rs35318160 | 17 | 6 980 105 | 3495 | A | G | 0.026 | 0.28 | 8.11e-08 | 6.25e-05 | ||||
| Other cis-eSNP | |||||||||||||
| rs75437894 | 17 | 6 966 496 | 17 104 | G | A | 0.016 | 0.27 | 2.92e-05 | 9.87e-03 | ||||
| SI-associated SNP | βd | P-valuef | Model | PCIT | |||||||||
| rs75437894 | −0.93 | 2.56e-04 | dom | 0.025 | |||||||||
| −0.93 | 2.56e-04 | add | |||||||||||
Abbreviations: A1, minor allele; A2, major allele of cis-eSNP; add, addative genetic model; βc, effect size of minor allele (A1) on expression; βd, effect of minor allele on SI; dom, dominant genetic model. FDR, false discovery rate; MAF, minor allele frequency; PCIT, omnibus P-value from CIT for a SNP considering SI as the clinical trait. P-valuea and βb, significance of association and effect direction between SI and transcript expression. P-valuee, significance in additive model (MatrixEQTL analysis); P-valuef, significance of association between genetic marker and SI; TSS, transcription start site.
Figure 2.
Ninjurin1 (NINJ1) transcript is associated with SI and cis-eSNPs for this transcript also associate with SI in AAs. The scatter plot shows association of NINJ1 transcript expression (ILMN_1815086) in adipose tissue with SI and BMI (A, B). The box plot shows association of NINJ1 transcript expression (ILMN_1815086) in adipose with genotype of the cis-eSNP rs7033638 (C) and association of SI and with genotype of the SNP rs7033638 (D). LocusZoom plots show regional association of NINJ1 cis-eQTL region SNPs (genotyped and imputed) with transcript expression and SI (E, F). Significance level (-log10 P-values) indicated in LocusZoom plots are based on score test implemented in the program SNPTEST. LD plot shows LD relationship (r2) between genotyped SNPs in the marked region.
NINJ1 modulates expression of genes in cellular pathways involved in insulin sensitivity
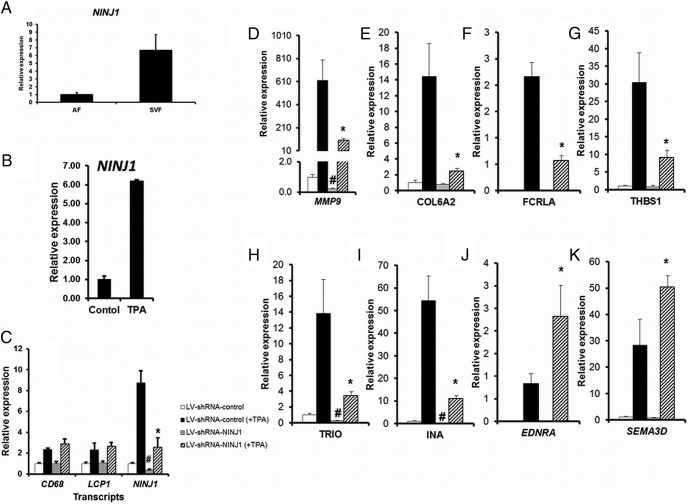
Among the cis-eGenes identified in this study, expression of Ninjurin 1 (NINJ1) in adipose tissue was negatively associated with SI, and the genotype of the strongest cis-eSNP for this gene also showed association with SI in the AAGMEx cohort. NINJ1 was further associated with BMI. Although mature adipocytes are the predominant cells in adipose tissue, preadipocytes, macrophages, and other cell types are present. To define the contribution of adipose tissue cells to the observed relationship between NINJ1 expression and SI and BMI, expression in RNA from adipocytes and adipose stromal vascular cells was compared. NINJ1 expression was approximately six times higher in RNA isolated from the stromal vascular fraction compared with the adipocyte fraction of adipose tissue (Figure 3A). NINJ1 is a genetically regulated transcript, and may play an important role in controlling SI and BMI by modulating expression of genes in macrophages. Thus, we tested the effect of experimental manipulation of NINJ1 expression on cellular pathways in THP1 cells, a human monocytic leukemia cell line widely used as a model for monocyte-macrophage differentiation (18). Exposure to phorbol esters ([phorbol 12-myristate 13-acetate (PMA/TPA)] ) induces differentiation of THP1 monocytes into macrophages (18). Differentiated THP1 macrophages showed a strong induction of NINJ1 expression (Figure 3B) compared with undifferentiated cells. Transduction of THP1 cells by NINJ1-specific lentiviral shRNA stably knocked down its expression at baseline and in the PMA/TPA-induced state (Figure 3C). Comparison of NINJ1 knockdown cells with control shRNA treated THP1 macrophage cells by RNA-Seq analysis identified genes modulated by NINJ1 (Supplemental Table 18). ShRNA mediated knockdown of NINJ1 induction in PMA/TPA-treated THP1 macrophages strongly abrogated induction of several genes including MMP9, COL6A2, FCRLA, and THBS1. Induction of genes including EDNRA and SEMA3D was enhanced in NINJ1 knockdown THP1 macrophages. These results were validated by independent experiments and by qRT-PCR (Figure 3, D–K). Bioinformatic analysis identified a significant enrichment of genes in chemokine signaling pathway, cytokine-cytokine receptor interaction, cell adhesion, and angiogenesis among genes modulated by NINJ1 (Supplemental Table 19). Thus, our in vitro studies identified target genes and pathways through which NINJ1 may modulate obesity and/or insulin sensitivity.
Figure 3.
Knockdown of NINJ1 expression in vitro in THP1 cells modulated expression of target genes. A, Relative expression of NINJ1 in pooled RNA samples from stromal vascular fractions compared with the adipocyte fraction of sc adipose tissue in AAs. B, Phorbol ester exposed (TPA/PMA, 10 ng/mL for 48 h) differentiated THP1 macrophages showed strong induction of NINJ1 expression compared with undifferentiated (control) cells. C, Transduction of THP1 cells by NINJ1 specific lentiviral shRNA stably knocked down its expression at baseline and in the TPA-induced differentiated state compared with control-shRNA lentiviral particle–treated cells. Stable knockdown efficiency of NINJ1 expression determined by qRT-PCR from two independent experiments (each with 2–3 biological replicates for each condition are shown. Expression of two other macrophage genes (CD68 and LCP1) was unchanged. D–K, ShRNA-mediated abrogation of NINJ1 induction in PMA/TPA treated THP1 macrophages significantly abrogated induction of several genes, while enhancing the expression of other genes. Validation of selected RNA-seq identified genes by qRT-PCR are shown. #, P < .01 lv-shRNA-control vs lv-shRNA-NINJ1 in basal or undifferentiated condition; *, P < .01 lv-shRNA-control vs lv-shRNA-NINJ1 in TPA treated or differentiated THP1 cells.
Discussion
T2D-related phenotypes are complex and heterogeneous; the pathophysiology of T2D likely involves altered gene expression in multiple tissues and pathways. The molecular processes and/or genetic variation that may contribute to population ancestry-specific pathophysiology of altered glucose homeostasis in AAs are poorly understood. The present study addresses this gap by combining quantitative intermediate traits related to glucose homeostasis, gene expression in tissues important to insulin action, and genome-wide genotype data in AAs. It successfully identified: 1) transcripts, networks of coexpressed transcripts, and biological pathways in adipose and muscle associated with SI; and 2) genetic controls on the expression of transcripts involved in modulating insulin sensitivity. Discordant modulation of transcripts in some pathways suggests tissue-specific mechanisms of cellular glucose homeostasis. Thus, this study provides novel insights into the molecular mechanisms of insulin sensitivity in AAs. Adipose and muscle expression datasets from cohorts of similar phenotypes and comparable statistical power, regardless of ethnicity, are not available for direct comparison. Thus, ethnic-specific characteristics of SI-modulating genes and pathways cannot be conclusively defined from this study.
SI, a quantitative estimate of insulin sensitivity, reflects the net capacity for insulin to promote glucose disposal in insulin-responsive tissues and to inhibit endogenous glucose production. The insulin-modified FSIGT followed by MINMOD analysis (a dynamic test) as implemented in our study to evaluate SI is comparable to steady-state measurement of insulin sensitivity by the euglycemic-hyperinsulinemic clamp (29, 30). However, MINMOD analysis merges the effects of insulin in promoting peripheral glucose utilization and suppressing hepatic (or endogenous) glucose production (31, 32). Thus, insulin sensitivity measured by this method is comparatively less reliable in individuals with impaired insulin secretion/and or significant insulin resistance (32), and some of the observations in this study may have been influenced by these inherent limitations of the FSIGT. Given that most insulin-mediated glucose disposal occurs in muscle, deranged expression of transcripts in skeletal muscle likely plays an important role in insulin resistance. However, a greater number of adipose tissue transcripts (relative to muscle) were associated with SI in this AA cohort (2212 vs 145; FDR < 1%), despite identical statistical power. Limitations of assessing insulin sensitivity by FSIGT may have driven this observation. Nevertheless, this result is similar to a study by Sears et al (22), which used the hyperinsulinemic-euglycemic clamp to measure insulin sensitivity. Comparison of basal adipose and muscle tissue gene expression between IR (N = 53) and IS (N = 19) individuals in that study identified 524 differentially expressed transcripts (at FDR 5%) in adipose and only 81 differentially expressed transcripts in muscle (22). Thus, distinct tissue-specific molecular mechanisms in modulating insulin sensitivity seem to be a plausible explanation for this observation. Stronger associations of SI were observed with sc adipose transcripts (maximum adjusted r2 for a transcript = 0.21) compared with skeletal muscle (r2 = 0.15), likely reflecting an important role of the adipose transcriptome in modulating insulin sensitivity (potentially via influence on glucose homeostasis in other tissues, including muscle). Recent studies revealed that adipose tissue has an important role (as a dominant regulator) in controlling whole-body glucose homeostasis in health and disease state (33). In adipocytes, insulin resistance and inflammation lead to production and release of free fatty acids and insulin resistance–provoking proinflammatory cytokines. These factors contribute to the accumulation of toxic lipid metabolites in muscle and liver, which impairs insulin signaling and activates pathways in those tissues, thereby modulating insulin sensitivity (34, 35). Many of the insulin resistance–associated molecular changes in these tissues are likely post-transcriptional and cannot be detected in this study that focused on transcriptional modulation.
Glucose homeostasis phenotypes are correlated with obesity; and we observed a significant overlap among transcripts associated with these traits. The magnitudes of associations between SI and adipose and muscle transcript levels were reduced after adjusting for BMI. Thus, the associations between expression levels of many transcripts and SI may reflect effects of BMI and are likely important in obesity-mediated insulin resistance.
It is thought that reduction in mitochondrial oxidative phosphorylation (OXPHOS) gene expression may be causally linked to impaired metabolic flexibility associated with insulin resistance and T2D. However, tissue-specific mechanisms and whether OXPHOS has a causal role in insulin resistance remain under debate (1, 36, 37). In this study, expression of PGC1α (PPARGC1A gene; probe ILMN_1750062), a key OXPHOS gene, was positively correlated with SI in adipose (β = 2.23; P = 2.12 × 10−9; q-value = 2.61 × 10−7), but its expression in muscle showed no significant correlation (β = −0.059; P = .75) with SI. Despite a relatively low mitochondrial mass compared with overall volume, the adipocyte interprets nutritional and hormonal cues in its microenvironment and determines appropriate mitochondrial responses (38). Expressions of many OXPHOS pathway genes are positively associated with SI in adipose under fasting conditions (9, 22, 39); this, supports down-regulation of this pathway in insulin resistant AA AAGMEx participants. Several nuclear encoded mitochondrial genes appeared as hub genes in a network of 372 coexpressed transcripts (the “pink” module) in adipose tissue. This network module was enriched for OXPHOS pathway genes (KEGG: hsa00190), and was positively and negatively correlated with SI and BMI, respectively. A recent study in mice showed that adipose-specific impairment of mitochondrial function may alter whole-body insulin sensitivity and produce insulin resistance and the metabolic syndrome (40). Thus, a compromised mitochondrial OXPHOS pathway in adipose may single-handedly, or collectively with other molecular defects, modulate insulin sensitivity.
Studies analyzing transcript expression in muscle have demonstrated reductions in expression of OXPHOS pathway genes in T2D (41, 42). However, this finding was not present in all studies, potentially reflecting differences in methodology, sample size, degree of insulin resistance, age, obesity, and ethnicity (1). No change in expression of OXPHOS pathway transcripts was seen in muscle of IR Europeans (43), whereas it was increased in insulin resistant Asian Indians (44). Furthermore, recent metabolomics and physiological studies in humans and animal models challenge the concept of muscle mitochondrial deficiency (including reduced fatty acid oxidation) as a common cause of insulin resistance (45, 46). Despite mild reductions in mitochondrial content in IR individuals, skeletal muscles have sufficient mitochondrial reserve to meet the excess demand of fatty acid oxidation (47, 48). Targeted metabolomic analysis reported excessive, not reduced, mitochondrial β-oxidation in initial stages of overnutrition and insulin resistance (45). Overload of the OXPHOS pathway may lead to incomplete oxidation of metabolic substrates, generation of reactive oxygen species, and trigger insulin resistance. Concordant with this, significant up-regulation of OXPHOS pathway genes was observed in muscle of IR AAs in AAGMEx. However, individual transcripts were only mildly modulated. The status of mitochondrial OXPHOS pathway genes identified in this cross-sectional study in resting individuals after an overnight fast suggests a unique tissue-specific mechanism of cellular energy homeostasis.
Most of the enriched biological pathways in SI-associated adipose transcripts related to cellular metabolic activities were positively associated with SI. Ingenuity pathway and gene set enrichment analyses also identified significant enrichment for canonical pathways not directly associated with metabolism, which may indirectly modulate SI by altering the adipose microenvironment and adipocyte signaling. These pathways included leukocyte extravasation signaling, Tec kinase signaling, integrin signaling, Fcγ receptor-mediated phagocytosis in macrophages and monocytes, IL-8 signaling, remodeling of epithelial adherens junctions, and toll-like receptor signaling. Thirty genes in the integrin-signaling pathway were significantly negatively associated with SI (Activation z score, −4.24), indicating up-regulation of this pathway in insulin resistance. A network of 416 coregulated transcripts (the “red” module) was enriched for leukocyte transendothelial migration pathway (KEGG: hsa04670); while a network of 704 transcripts (the “blue” module) was enriched for lysosomal genes (KEGG: hsa04142) in adipose. More than 30% of transcripts in these two modules were negatively correlated with SI. Interaction of the hub genes in these modules, namely ARHGAP30 in the “red” module and CD163 in the “blue” module, were identified with other genes in these modules. Some intra-modular interactions are supported by the literature (IPA interaction network database), whereas others support the existence of novel transcriptional regulatory mechanisms that may be relevant in modulating SI in AAs. The CD163 gene in macrophages is involved in endocytosis and lysosomal degradation of hemoglobin/haptoglobin complexes and may be involved in local and systemic insulin sensitivity by protecting tissues from free hemoglobin-mediated oxidative damage (49, 50). The ARHGAP30 gene was identified as a hub gene in an adipose coexpression module associated with circulating triglycerides in Finns and Mexican Americans (51). ARHGAP30 is near the USF1 gene in chromsome1q21–24, a region implicated in T2D susceptibility (52). Further statistical and functional studies are required to identify “key drivers” in these gene networks to achieve a mechanistic understanding of the processes modulating SI.
Considering significant cis-eQTL results with transcript-SI association data identified cis-eSNPs for 363 and 42 SI-associated transcripts, respectively, in adipose and muscle. Further integrative analysis successfully identified genetic polymorphisms in NINJ1, AGA, and CLEC10A, three novel loci associated with SI and expression of SI-associated transcripts in adipose from AAs. Expression of NINJ1 is negatively (inversely) associated with SI. A 3′UTR NINJ1 SNP (rs7033638) had the strongest association. The “C”-allele (freq = 0.36) was associated with higher SI and lower adipose NINJ1 transcript expression. This variant was also a cis-eSNP in muscle eQTL data in our AAGMEx cohort. A coding missense variant rs2275848 (in linkage disequilibrium [LD] with rs7033638; r2 = 0.81) was a cis-eSNP (P = 1.07 × 10−7) for NINJ1 in this study and, in the Multi Tissue Human Expression Resource adipose eQTL data (53) was associated with early-onset extreme obesity (54). The role of NINJ1 gene in human adipose is unknown; however, its expression is increased in adipose tissue of db/db mice (vs controls) (55) and adipose expression is induced by TNFα (56). NINJ1 is expressed by myeloid lineage cells in human blood, particularly CD14+ monocytes and CD68+ macrophages. NINJ1 is a cell-adhesion molecule; it mediates cell communication and enhances entry, migration, and activity of monocytes and macrophages in inflammation (57). In addition, NINJ1 regulates interactions between leukocytes and vascular endothelial cells and may play a role in vascular remodeling (57, 58). NINJ1 is a p53 target and may be involved in p53-dependent premature senescence, cell proliferation, and apoptosis (59). NINJ1 knockout (whole body) leads to postnatal lethality in mice (59). Analysis of transcripts in adipose tissue fractions in this study showed higher expression of NINJ1 in the stromal vascular fraction (including macrophages and vascular endothelial cells) compared with the adipocyte fraction. Its expression is increased during monocyte-to-macrophage differentiation. ShRNA-mediated abrogation of NINJ1 induction in phorbol ester–differentiated THP1 macrophages modulated expression of genes in chemokine signaling, cytokine-cytokine receptor interaction, cell adhesion, and angiogenesis pathways. Published genome-wide and gene-specific studies showed the association of these NINJ1-modulated pathways and genes, including matrix metalloproteinase, collagen VI, and thrombospondin with insulin sensitivity and obesity (22, 60–62). Thus, NINJ1 is a strong candidate gene that modulates inflammation/immune responses and adipose tissue angiogenesis; it may further modulate insulin sensitivity in adipose tissue.
In conclusion, this study presents a comprehensive characterization of adipose and muscle tissue transcripts, biological pathways, and gene-expression networks involved in the modulation of SI in AAs. Despite limited overlap of SI-associated transcripts between adipose and muscle at the individual transcript level, pathway-based, gene set enrichment analysis suggested modulation of the same pathways in both tissues. Transcript levels of genes involved in eIF2, eIF4-p70S6K, mTOR signaling pathways, and ribosome function were positively correlated with SI, whereas the transcript level of genes in lysosome and proteasome pathways were negatively (inversely) correlated with SI in both tissues. However, genes involved in leukocyte extravasation signaling showed adipose-specific regulation, and genes involved in oxidative phosphorylation had discordant regulation between tissues. Although multiple pathways are involved in the complex process of insulin resistance, discordant adipose- and muscle-specific regulation of oxidative phosphorylation seems to be important. Identification of genetic polymorphisms in NINJ1, AGA, and CLEC10A, three novel loci that modulate the expression of SI-associated transcripts in adipose tissue as cis-eSNPs and are also associated with SI, demonstrates the success of integrative multiomics approaches in mapping eQTLs of physiologic importance. Identification of SNPs associated with expression of a subset of SI-associated transcripts suggests a genetic regulatory mechanism of insulin resistance and may contribute to the distinctive pathophysiology of T2D in AAs.
Acknowledgments
We thank the dedicated staff of the Clinical Research Unit at WFSM and Kurt A. Langberg (WFSM, Department of Endocrinology) for support of the clinical studies and assistance with data management. We thank Mrs Joyce Byers for support in participant recruitment. We thank the staff in the genomics core laboratory at Center for Genomics and Personalized Medicine Research, and Center for Cancer Genomics, WFSM, especially Dr Siqun Zheng, Shelly Smith, Tracey Young, Dr Ge Li, and Lou Craddock for their extensive support in genotyping, gene expression analysis using the Illumina microarray platform, and RNA-seq analysis. We also thank Karen Klein (Biomedical Research Services Administration, Wake Forest University Health Sciences) for critical reading and editing of our manuscript. We acknowledge the support of the Center for Public Health Genomics, WFSM for computational resources.
Current address for J.C.-E.: Division of Endocrinology, Metro Health System, and School of Medicine, Case Western Reserve University, Cleveland, Ohio 44105.
S.K.D. and C.D.L. are the guarantors of this work, and as such, had full access to all study data and take responsibility for the integrity of the data and accuracy of data analysis.
We dedicate this manuscript to the memory of our colleague, the late Dr Steven C. Elbein.
This work was supported by the National Institutes of Health Grant R01 DK090111 (to S.K.D.) and the Section on Nephrology at WFSM.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AA
- African American
- AAGMEx
- African American Genetics of Metabolism and Expression
- AGA
- aspartylglucosaminidase
- AIRG
- acute insulin response to iv glucose
- ARACNE
- Algorithm for the Reconstruction of Accurate Cellular Networks
- BMI
- body mass index
- CIT
- causal inference test
- DI
- disposition index
- eQTL
- expression quantitative trait locus
- FDR
- false-discovery rate
- FSIGT
- frequently sampled iv glucose tolerance test
- GSEA
- gene set enrichment analysis
- HOMA
- homeostatic model assessment
- IPA
- ingenuity pathway analysis
- IR
- insulin resistant
- IS
- insulin sensitive
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- LD
- linkage disequilibrium
- MAF
- minor allele frequency
- OGTT
- oral glucose tolerance test
- OXPHOS
- oxidative phosphorylation
- SAM
- Statistical Analysis for Microarray
- shRNA
- short hairpin RNA
- SI
- insulin sensitivity index
- SNP
- single nucleotide polymorphism
- T2D
- type 2 diabetes
- TPA/PMA
- phorbol 12-myristate 13-acetate
- WFSM
- Wake Forest School of Medicine.
References
- 1. Sales V, Patti ME. The ups and downs of insulin resistance and type 2 diabetes: Lessons from genomic analyses in humans. Curr Cardiovasc Risk Rep. 2013;7(1):46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bergman RN, Stefanovski D, Kim SP. Systems analysis and the prediction and prevention of type 2 diabetes mellitus. Curr Opin Biotechnol. 2014;28:165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kahn SE, Cooper ME, Del Prato S. Pathophysiology and treatment of type 2 diabetes: Perspectives on the past, present, and future. Lancet. 2014;383(9922):1068–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson AM, Olefsky JM. The origins and drivers of insulin resistance. Cell. 2013;152(4):673–684. [DOI] [PubMed] [Google Scholar]
- 5. Tabák AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimaki M, Witte DR. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: An analysis from the Whitehall II study. Lancet. 2009;373(9682):2215–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haffner SM, D'Agostino R, Saad MF, et al. Increased insulin resistance and insulin secretion in nondiabetic African-Americans and Hispanics compared with non-Hispanic whites. The Insulin Resistance Atherosclerosis Study. Diabetes. 1996;45(6):742–748. [DOI] [PubMed] [Google Scholar]
- 7. Kodama K, Tojjar D, Yamada S, Toda K, Patel CJ, Butte AJ. Ethnic differences in the relationship between insulin sensitivity and insulin response: A systematic review and meta-analysis. Diabetes Care. 2013;36(6):1789–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Das SK, Sharma NK, Hasstedt SJ, et al. An integrative genomics approach identifies activation of thioredoxin/thioredoxin reductase-1-mediated oxidative stress defense pathway and inhibition of angiogenesis in obese nondiabetic human subjects. J Clin Endocrinol Metab. 2011;96(8):E1308–E1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elbein SC, Kern PA, Rasouli N, Yao-Borengasser A, Sharma NK, Das SK. Global gene expression profiles of subcutaneous adipose and muscle from glucose-tolerant, insulin-sensitive, and insulin-resistant individuals matched for BMI. Diabetes. 2011;60(3):1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sharma NK, Das SK, Mondal AK, et al. Endoplasmic reticulum stress markers are associated with obesity in nondiabetic subjects. J Clin Endocrinol Metab. 2008;93(11):4532–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sharma NK, Langberg KA, Mondal AK, Elbein SC, Das SK. Type 2 diabetes (T2D) associated polymorphisms regulate expression of adjacent transcripts in transformed lymphocytes, adipose, and muscle from Caucasian and African-American subjects. J Clin Endocrinol Metab. 2011;96(2):E394–E403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: Comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22(9):1462–1470. [DOI] [PubMed] [Google Scholar]
- 13. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: Insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. [DOI] [PubMed] [Google Scholar]
- 14. Boston RC, Stefanovski D, Moate PJ, Sumner AE, Watanabe RM, Bergman RN. MINMOD Millennium: A computer program to calculate glucose effectiveness and insulin sensitivity from the frequently sampled intravenous glucose tolerance test. Diabetes Technol Ther. 2003;5(6):1003–1015. [DOI] [PubMed] [Google Scholar]
- 15. Wright SP. Adjusted P-values for simultaneous inference. Biometrics. 1992;48:1005–1013. [Google Scholar]
- 16. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shabalin AA. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28(10):1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Auwerx J. The human leukemia cell line, THP-1: A multifacetted model for the study of monocyte-macrophage differentiation. Experientia. 1991;47(1):22–31. [DOI] [PubMed] [Google Scholar]
- 19. Heckmann BL, Zhang X, Xie X, Liu J. The G0/G1 switch gene 2 (G0S2): Regulating metabolism and beyond. Biochim Biophys Acta. 2013;1831(2):276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keildson S, Fadista J, Ladenvall C, et al. Expression of phosphofructokinase in skeletal muscle is influenced by genetic variation and associated with insulin sensitivity. Diabetes. 2014;63(3):1154–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Larrouy D, Barbe P, Valle C, et al. Gene expression profiling of human skeletal muscle in response to stabilized weight loss. Am J Clin Nutr. 2008;88(1):125–132. [DOI] [PubMed] [Google Scholar]
- 22. Sears DD, Hsiao G, Hsiao A, et al. Mechanisms of human insulin resistance and thiazolidinedione-mediated insulin sensitization. Proc Natl Acad Sci U S A. 2009;106(44):18745–18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parikh H, Carlsson E, Chutkow WA, et al. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 2007;4(5):e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang X, Xie X, Heckmann BL, Saarinen AM, Czyzyk TA, Liu J. Targeted disruption of G0/G1 switch gene 2 enhances adipose lipolysis, alters hepatic energy balance, and alleviates high-fat diet-induced liver steatosis. Diabetes. 2014;63(3):934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article 17. [DOI] [PubMed] [Google Scholar]
- 27. Margolin AA, Nemenman I, Basso K, et al. An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7 Suppl 1:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Millstein J, Zhang B, Zhu J, Schadt EE. Disentangling molecular relationships with a causal inference test. BMC Genet. 2009;10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bergman RN. Minimal model: Perspective from 2005. Horm Res. 2005; 64 Suppl 3:8–15. [DOI] [PubMed] [Google Scholar]
- 30. Saad MF, Anderson RL, Laws A, et al. A comparison between the minimal model and the glucose clamp in the assessment of insulin sensitivity across the spectrum of glucose tolerance. Insulin Resistance Atherosclerosis Study. Diabetes. 1994;43(9):1114–1121. [DOI] [PubMed] [Google Scholar]
- 31. Cobelli C, Dalla Man C, Toffolo G, Basu R, Vella A, Rizza R. The oral minimal model method. Diabetes. 2014;63(4):1203–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muniyappa R, Lee S, Chen H, Quon MJ. Current approaches for assessing insulin sensitivity and resistance in vivo: Advantages, limitations, and appropriate usage. Am J Physiol Endocrinol Metab. 2008;294(1):E15–E26. [DOI] [PubMed] [Google Scholar]
- 33. Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9(5):367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DeFronzo RA, Ferrannini E, Groop L, et al. Type 2 diabetes mellitus. Nat Rev Dis Prim. 2015;article 15019. [DOI] [PubMed] [Google Scholar]
- 35. Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156(1–2):20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Goodpaster BH. Mitochondrial deficiency is associated with insulin resistance. Diabetes. 2013;62(4):1032–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Holloszy JO. “Deficiency” of mitochondria in muscle does not cause insulin resistance. Diabetes. 2013;62(4):1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kusminski CM, Scherer PE. Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol Metab. 2012;23(9):435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qatanani M, Tan Y, Dobrin R, et al. Inverse regulation of inflammation and mitochondrial function in adipose tissue defines extreme insulin sensitivity in morbidly obese patients. Diabetes. 2013;62(3):855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vernochet C, Damilano F, Mourier A, et al. Adipose tissue mitochondrial dysfunction triggers a lipodystrophic syndrome with insulin resistance, hepatosteatosis, and cardiovascular complications. FASEB J. 2014;28(10):4408–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patti ME, Butte AJ, Crunkhorn S, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100(14):8466–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. [DOI] [PubMed] [Google Scholar]
- 43. Jin W, Goldfine AB, Boes T, et al. Increased SRF transcriptional activity in human and mouse skeletal muscle is a signature of insulin resistance. J Clin Invest. 2011;121(3):918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nair KS, Bigelow ML, Asmann YW, et al. Asian Indians have enhanced skeletal muscle mitochondrial capacity to produce ATP in association with severe insulin resistance. Diabetes. 2008;57(5):1166–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koves TR, Ussher JR, Noland RC, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7(1):45–56. [DOI] [PubMed] [Google Scholar]
- 46. Muoio DM, Neufer PD. Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab. 2012;15(5):595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fisher-Wellman KH, Weber TM, Cathey BL, et al. Mitochondrial respiratory capacity and content are normal in young insulin-resistant obese humans. Diabetes. 2014;63(1):132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hoeks J, Schrauwen P. Muscle mitochondria and insulin resistance: A human perspective. Trends Endocrinol Metab. 2012;23(9):444–450. [DOI] [PubMed] [Google Scholar]
- 49. Fjeldborg K, Christiansen T, Bennetzen M, Moller J, Pedersen SB, Richelsen B. The macrophage-specific serum marker, soluble CD163, is increased in obesity and reduced after dietary-induced weight loss. Obesity (Silver Spring). 2013;21(12):2437–2443. [DOI] [PubMed] [Google Scholar]
- 50. Hubler MJ, Peterson KR, Hasty AH. Iron homeostasis: A new job for macrophages in adipose tissue? Trends Endocrinol Metab. 2015;26(2):101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Haas BE, Horvath S, Pietiläinen KH, et al. Adipose co-expression networks across Finns and Mexicans identify novel triglyceride-associated genes. BMC Med Genomics. 2012;5:61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Das SK, Elbein SC. The search for type 2 diabetes susceptibility loci: The chromosome 1q story. Curr Diab Rep. 2007;7(2):154–164. [DOI] [PubMed] [Google Scholar]
- 53. Grundberg E, Small KS, Hedman AK, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44(10):1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wheeler E, Huang N, Bochukova EG, et al. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat Genet. 2013;45(5):513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sato C, Shikata K, Hirota D, et al. P-selectin glycoprotein ligand-1 deficiency is protective against obesity-related insulin resistance. Diabetes. 2011;60(1):189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruan H, Miles PD, Ladd CM, et al. Profiling gene transcription in vivo reveals adipose tissue as an immediate target of tumor necrosis factor-alpha: Implications for insulin resistance. Diabetes. 2002;51(11):3176–3188. [DOI] [PubMed] [Google Scholar]
- 57. Lee HJ, Ahn BJ, Shin MW, Choi JH, Kim KW. Ninjurin1: A potential adhesion molecule and its role in inflammation and tissue remodeling. Mol Cells. 2010;29(3):223–227. [DOI] [PubMed] [Google Scholar]
- 58. Matsuki M, Kabara M, Saito Y, et al. Ninjurin1 is a novel factor to regulate angiogenesis through the function of pericytes. Circ J. 2015;79(6):1363–1371. [DOI] [PubMed] [Google Scholar]
- 59. Cho SJ, Rossi A, Jung YS, et al. Ninjurin1, a target of p53, regulates p53 expression and p53-dependent cell survival, senescence, and radiation-induced mortality. Proc Natl Acad Sci U S A. 2013;110(23):9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Spencer M, Yao-Borengasser A, Unal R, et al. Adipose tissue macrophages in insulin-resistant subjects are associated with collagen VI and fibrosis and demonstrate alternative activation. Am J Physiol Endocrinol Metab. 2010;299(6):E1016–E1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Unal R, Yao-Borengasser A, Varma V, et al. Matrix metalloproteinase-9 is increased in obese subjects and decreases in response to pioglitazone. J Clin Endocrinol Metab. 2010;95(6):2993–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Varma V, Yao-Borengasser A, Bodles AM, et al. Thrombospondin-1 is an adipokine associated with obesity, adipose inflammation, and insulin resistance. Diabetes. 2008;57(2):432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]