Abstract
The PCR amplification of oligonucleotides enables the evolution of sequences called aptamers that bind specific targets with antibody-like affinity. However, the use of these aptamers is limited in many applications by nuclease-mediated degradation. In contrast, oligonucleotides that are modified at their sugar C2' positions with methoxy or fluorine substituents are stable to nucleases but cannot be synthesized by natural polymerases. Here, we report the development of a polymerase evolution system and its use to evolve thermostable polymerases that efficiently interconvert C2'-OMe modified oligonucleotides and their DNA counterparts via “transcription” and “reverse transcription,” or more importantly, PCR amplify partially C2'-OMe or C2'-F modified oligonucleotides. A mechanistic analysis demonstrates that the ability to amplify the modified oligonucleotides was evolved by optimizing interdomain interactions that stabilize the catalytically competent closed conformation of the polymerase. The evolved polymerases should find practical applications and the developed evolution system should be a powerful tool for the tailoring of polymerases to have other types of novel function.
Summary for Graphical Abstract
DNA polymerase can PCR amplify natural DNA efficiently, but cannot utilize C2’-modified substrates. Via a combination of selection and screening, Stoffel fragment DNA polymerase was evolved to “transcribe” C2’-modified-DNA from a DNA template, “reverse transcribe” C2’-modified-DNA back into DNA, and PCR amplify C2’-modified DNA.
Introduction
DNA is unique amongst all materials because it can be enzymatically replicated. This replication underlies not only the storage of genetic information, but also PCR amplification, which itself enables innumerable biotechnology applications such as cloning, sequencing, and the systematic evolution of ligands by exponential enrichment (SELEX). SELEX is a powerful method for aptamer evolution1–3, but the practical utility of the aptamers is often limited by their instability in biological solutions due to nuclease-mediated degradation. To circumvent this liability, the addition of methoxy or fluorine substituents to the C2’ of the sugar ring (C2’-OMe and C2’-F) has received much attention, as the corresponding oligonucleotides are resistant to nucleases. For example, only two nucleotides of the FDA approved therapeutic aptamer Macugen are unmodified ribonucleotides, the remaining twenty-five are modified with a C2’-OMe or C2’-F moiety4. These modifications have also been shown to underlie or contribute to optimized activities or properties of other therapeutic aptamers5–10. Their inclusion typically requires the modification of an evolved natural aptamer, because the modified oligonucleotides themselves are not recognized by DNA polymerases and thus are not amenable to the amplification step of the SELEX protocol. Unfortunately, post-selection modification commonly results in loss of activity, and typically the development of active, stable aptamers, if even possible, is a time-consuming process that requires the iterative examination of different substituents at different positions to determine which combination is tolerated.
To circumvent the need for post-selection modification, much effort has been directed toward the discovery of polymerases capable of recognizing nucleotides with modified sugars so that the modified oligonucleotides themselves may be subject to the SELEX process. While 4’-thio-2’-deoxyoligonucleotides may be PCR amplified with some natural polymerases11, and in some cases may also be more stable to nuclease digestion12, more invasive modifications, such as the C2’-OMe or C2’-F substituents, are not tolerated by natural polymerases. Thus, even recent efforts focused on evolving sugar modified aptamers13 or catalysts14 have relied on converting the libraries after selection into natural DNA for amplification, a strategy that appears to have been first employed by Lin, et al.15. Thus the amplification of C2’-OMe or C2’-F modified oligonucleotides will likely require polymerase optimization via directed evolution16–20.
Focusing on the evolution of thermostable DNA polymerases that should be suitable for PCR applications, in 2004 we reported the activity-based selection of several variants of the Stoffel fragment of Taq polymerase (Sf). The most notable of these variants is SFM19, which combines thermal stability with a ~10,000-fold increased ability to extend a primer with C2’-OMe modified nucleotides16. However, SFM19 synthesizes only short stretches of C2’-OMe modified oligonucleotides, and is incapable of the PCR amplification of C2’-modified oligonucleotides, which requires the more challenging synthesis of the modified polymer using a modified template.
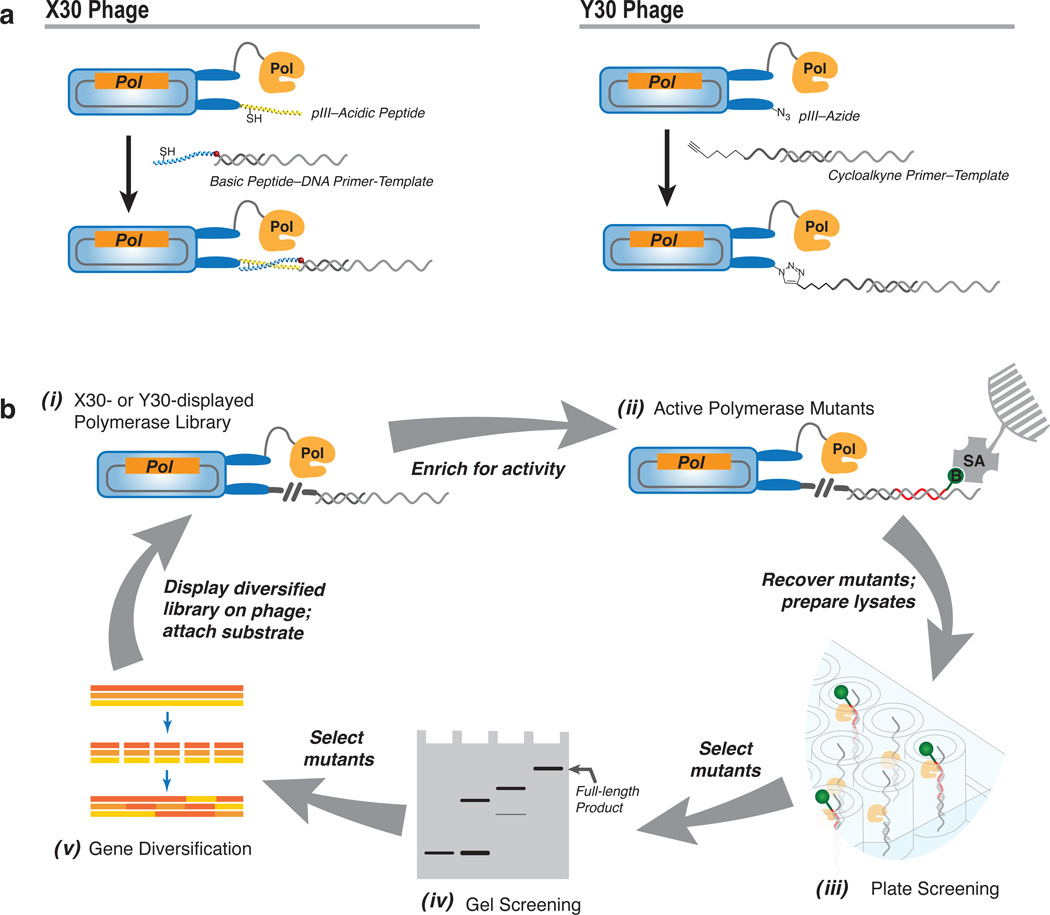
The activity-based selection system that yielded SFM19 (see Fig. 1a and b and Xia, et al.21) was based on the co-display of a polymerase variant and its modified substrate on phage particles. It employed the pFAB-Sf phagemid, which encodes a pIII-Sf fusion protein, and X30 helper phage, which encodes pIII proteins as N-terminal fusions with an acidic peptide, and production is optimized such that each phage displays ~1 polymerase. An oligonucleotide, which during selection will be used to prime DNA synthesis, is hybridized to an oligonucleotide template and covalently attached to a basic peptide. When combined with phage particles, the basic peptide forms a coiled-coil and disulphide bond with the displayed acidic peptide, and thereby attaches the primer/template oligonucleotide substrate. In this manner, the polymerase and its substrate are covalently linked to the same phage such that intramolecular incorporation of a biotinylated nucleoside triphosphate permits the recovery of phagemids encoding active polymerase mutants. Importantly, the primer and/or template may be designed such that the recognition of a modified nucleotide (in the primer, template, or provided triphosphate) is required prior to biotinylation, and thus the selection system allows for the application of selection pressure for the recognition of the modified substrates. Although we have used this system to evolve Sf variants with a wide variety of unnatural activities16,21,22 its use is labour intensive due to the demands of peptide-DNA conjugate synthesis and purification.
Figure 1. Preparation of phage particles and their use in the polymerase evolution system.
a, Attachment of substrates. X30 Phage: Substrates are attached via a fusion between phage pIII and an acidic peptide, which forms a coiled-coil and disulphide bond with a basic peptide-DNA primer/template conjugate; maleimidocaproic acid (2-nitro-4-sulfo) phenyl ester (red sphere) is used to couple the DNA to the basic peptide. Y30 Phage: Phage pIII protein is expressed with an N-terminal pAzF residue (indicated by an azide), which is then used to attach cycloalkyne–primer/template substrates (indicated by an alkyne) via a strain-promoted click reaction. b, Overview of the evolution process: i, Libraries displayed on X30 or Y30 phage with primer/template complexes attached as described in a are subjected to enrichment for active members via phage selection, in which phage are mixed with natural or modified dATP, dCTP, dGTP, and biotin (B)-labelled UTP to extend the primer, and ii, phage displaying active Sf mutants are isolated with streptavidin (SA) beads. After washing to remove non-specific binders, the phage are cleaved from the beads using DNase I and used to re-infect E. coli XL1-Blue to recover phagemids. iii, Heat-treated lysates of E. coli expressing the recovered Sf mutants are next subjected to plate-based screening using amine-binding 96-well plates (Corning) coated with amine-primer/template complex and extension buffer containing natural or modified dATP, dCTP, dGTP, and biotin-UTP. After incubation at 50 °C, the incorporated biotin-UTP tags are detected chromogenically using horseradish peroxidase (HRP)-streptavidin and o-phenylenediamine/H2O2. iv, Mutants giving rise to the most activity are selected for individual gel-based analysis, from which v, promising candidates are selected for further diversification (for example by gene shuffling, as shown), and then subjected to additional rounds of evolution.
Here, we develop an improved evolution system by optimizing substrate attachment to the phage via the use of an unnatural amino acid and strain-promoted click chemistry, which eliminates the use of the peptide-DNA conjugate, and by augmenting the selection system with a plate based screen, which facilitates the identification of active mutants. The improved system is then used to evolve polymerases that “transcribe” or “reverse transcribe” fully C2’-OMe modified oligonucleotides or which PCR amplify partially C2’-OMe or C2’-F modified oligonucleotides. An analysis of an evolved mutant suggests that the ability to amplify the C2’ modified oligonucleotides resulted from stabilizing an interaction between the fingers and thumb domains that favours the formation of the catalytically active closed complex.
Results and Discussion
Development of an improved evolution system
We first constructed a helper phage (Y30) in which the 5’ terminus of a truncated gIII gene encodes a pIII protein with an amber (TAG) stop codon. When E. coli SS320 cells are transformed with plasmid pEVOL-pAzF, which encodes a mutant Methanocaldococcus jannaschii aminoacyl-tRNA synthetase, the stop codon directs the incorporation of the unnatural amino acid p-azidophenylalanine (pAzF) when it is supplied to the growth media23. Superinfection of SS320/pEVOL-pAzF harbouring the pFAB-Sf phagemid with Y30 helper phage and growth in media supplemented with pAzF was optimized to produce phage particles containing phagemid DNA, and displaying zero to one copy of the encoded pIII-Sf fusion, with the remainder of the pIII proteins containing the N-terminal pAzF, which was used to attach a primer/template via strain-promoted click chemistry24 (Fig. 1a and b, see also Supplementary Fig. 1, Supplementary Tables 1 and 2, and Supplementary Methods).
To supplement the phage-based selection system and to increase the efficiency of single clone identification, we also developed a plate-based method to screen members of the enriched libraries for activity (Fig. 1a and b, Supplementary Fig. 2). The screen employs DNA binding 96-well microplates with an N-oxysuccinimide (NOS) decorated surface, which can be used to immobilize 5’-amine modified primer/templates. Lysates obtained from heat-treating cells transformed with library genes subcloned into pET-23b(+) are added to the wells, and activity is detected after incorporation of biotin-UTP using horseradish peroxidase (HRP)-streptavidin conjugate.
Evolution of SFM19 for increased recognition of C2’-modified substrates
SFM19 was selected from a library in which only the region proximal to the incoming triphosphate was diversified, and selection pressure was only applied for the incorporation of three modified nucleotides16, thus accounting for its ability to synthesize only short stretches of C2’-OMe modified oligonucleotides. For example, SFM19 was only able to extend a natural DNA primer by five to six C2’-OMe nucleotides and was unable to extend a fully C2’-OMe modified primer. To further optimize SFM19, we subjected it to iterative rounds of more general diversification and selection for more processive synthesis, including selection with both modified triphosphates and templates. These selections employed C2’-OMe substrates, as they are generally more challenging to recognize than their C2’-F counterparts25, and we expected that any evolved mutants might recognize both C2’-OMe and C2’-F modifications.
For the first two rounds of evolution, PCR mutagenesis was used to create polymerase genes with 1 to 5 mutations, and display on X30 phage resulted in libraries with 1011–1012 members. In the first round, selection pressure was applied for the efficient incorporation of a single C2’-OMe-CTP, and 800 members of the enriched library were subjected to the screen. After confirmation that fidelity had not been compromised (Supplementary Fig. 3), two mutants, SFM1-5 and SFM1-33 (Table 1, Fig. 2,Supplementary Table 3), were progressed to the second round, in which the incorporation of one C2’-OMe-CTP, one C2’-OMe-ATP, and two C2’-OMe-GTPs were required for biotinylation. One thousand members of this enriched library were then screened for activity with the most promising candidates assayed individually for fidelity (Supplementary Fig. 4), resulting in four mutants for further progression: SFM2-47, SFM2-51, SFM2-56, and SFM2-74 (Table 1, Fig. 2).
Table 1.
Polymerase mutants.
| Polymerase | Mutations |
|---|---|
| SFM19 | SF WT: I614E, E615G |
| SFM1-5 | SFM19: E295G, E681V |
| SFM1-33 | SFM19: V607A |
| SFM2-47 | SFM19: N415Y, N583S, E681V |
| SFM2-51 | SFM19: D655N, L657M, E681V |
| SFM2-56 | SFM19: R651W, E681V |
| SFM2-74 | SFM19: E295G, V518A, E681V |
| SFM3-2 | SFM19: N415Y, N583S, D655N, L657M, E681V, E742A, M747Q, E774K |
| SFM3-5 | SFM19: V518A, N583S, D655N, E681V, E742Q, M747R |
| SFM3-6 | SFM19: E295G, V518A, Y545H, D655N, L657M, E681V, E742Y, M747A |
| SFM3-14 | SFM19: N415Y, N583S, D655N, L657M, E681V, E742M, M747R |
| SFM3-16 | SFM19: N415Y, N583S, D655N, L657M, E681V, E742N, M747R, K793N |
| SFM4-3 | SFM19: V518A, N583S, D655N, E681K, E742Q, M747R |
| SFM4-6 | SFM19: D655N, L657M, E681K, E742N, M747R |
| SFM4-9 | SFM19: N415Y, V518A, D655N, L657M, E681V, E742N, M747R |
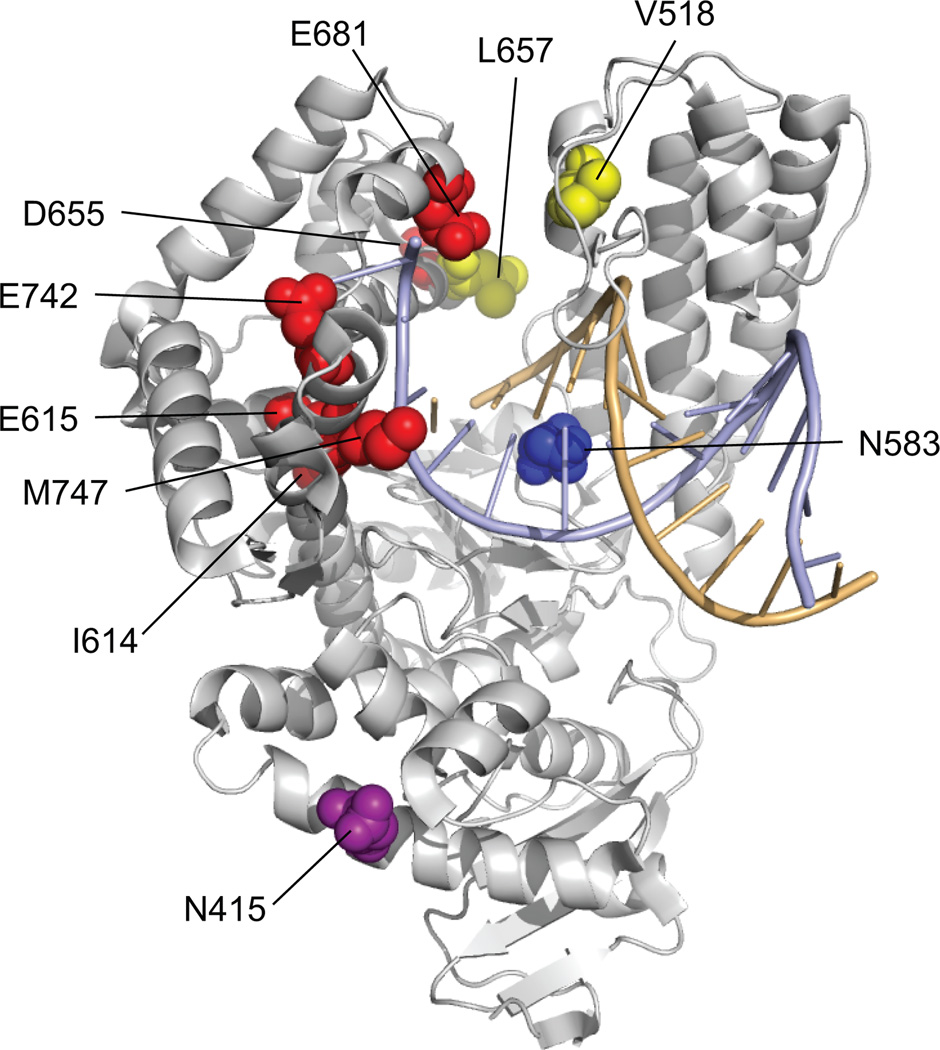
Figure 2. Distribution of Mutations Selected in Evolved Polymerases.
Mutation sites have been coloured by their representation among evolved polymerases. Mutation sites shared by SFM4-3, SFM4-6, and SFM4-9 (red); sites shared by SFM4-3 and SFM4-9 (yellow); and sites unique to SFM4-3 (blue) or SFM4-9 (purple). Also highlighted are the primer strand (orange) and the template strand (pale blue). Residue labels correspond to the wild type Sf protein. (PDB ID: 3KTQ)
The library for the third round of evolution was created by a combination of gene shuffling the second round progeny and saturation mutagenesis at E742 and M747, sites at which mutations have been shown to bestow Taq with reverse transcriptase activity26. Selection for the third round of evolution was performed via display on X30 phage and selection for primer extension via the incorporation of one C2’-OMe-ATP and two C2’-OMe-GTPs after incorporation of dCTP opposite a C2’-OMe-G in the template, followed by display of the enriched library on Y30 phage and repeating the selection. Finally, one thousand members of the twice-enriched library were subjected to the screening assay, and after gel-based analysis of transcription, reverse transcription, and fidelity (Supplementary Fig. 5), five mutants were identified: SFM3-2, SFM3-5, SFM3-6, SFM3-14, and SFM3-16 (Table 1, Fig. 2).
The library for the fourth and final round of evolution was created by gene shuffling the third round progeny and saturation mutagenesis at residue Y545, which structural data suggested may interact with the primer terminus27, followed by screening 500 members for both transcription and reverse transcription activity. An examination of activity and fidelity (Supplementary Fig. 5 and Supplementary Fig. 6) identified three promising mutants: SFM4-3, SFM4-6, and SFM4-9 (Table 1, Fig. 2).
Characterization of evolved polymerases
The most promising reverse transcription activity was observed with SFM4-9, which can reverse-transcribe a full-length, fully C2’-OMe-modified template into a DNA product (Supplementary Fig. 7). To more rigorously gauge the evolved activity, we characterized the ability of SFM4-9 and the parental polymerase SFM19 to extend a DNA primer by incorporation of a single dCTP opposite a C2’-OMe-G in a template (Supplementary Table 4). Under the steady-state conditions employed, SFM19 incorporated the nucleotide with a second order rate constant (kcat/KM) of 2.4 × 104 M−1min−1 (kcat= 3.3 min−1 and KM = 137 µM). SFM4-9 catalysed the same insertion with a second order rate constant of 3.5 × 106 M−1min-1 (kcat = 3.1 min−1 and KM = 0.9 µM). Thus, the efficiency of the evolved enzyme is greater than 100-fold that of its parent, and this improvement is entirely due to an increase in triphosphate binding. To characterize the fidelity of this reverse transcription, SFM4-9 was used to reverse transcribe a chemically synthesized C2’-OMe 60-mer template into DNA, which was then PCR amplified by Q5 DNA polymerase. Cloning and sequencing of the product revealed that the error rate of SFM4-9-mediated reverse transcription is less than 1.7 × 10−3 (Supplementary Fig. 7). This error rate compares favourably with those previously reported for other polymerases selected or evolved to reverse transcribe sugar modified oligonucleotides20.
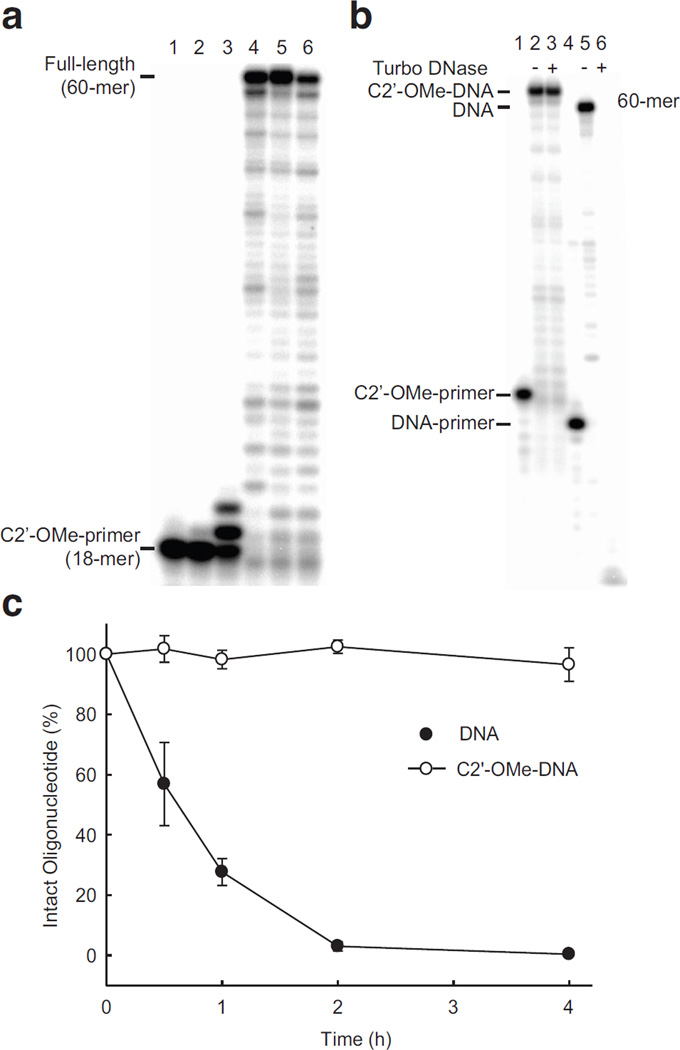
While each enzyme transcribes full-length, fully C2’-OMe-modified oligonucleotides (Fig. 3a), the greatest activity was observed with SFM4-6. To more rigorously gauge the extent of this evolved activity, we characterized the ability of SFM4-6 to extend a fully C2’-OMe modified primer by insertion of C2’-OMe-CTP opposite dG in a template under steady state conditions (Supplementary Table 4). SFM19 was again characterized for comparison. As expected due the use of a fully C2’-OMe modified primer, no activity was observed with SFM19. However, SFM4-6 catalysed extension of the modified primer with a second order rate constant of 7.3 × 103 M−1min-1 (kcat = 0.8 min−1 and KM = 103 µM). To characterize the fidelity of this transcription, SFM4-6 was used to transcribe a DNA template into its fully C2’-OMe modified complementary strand, which was then incubated with Turbo DNase, to remove template, and then reverse transcribed back into DNA using SFM4-9. The resulting DNA product was amplified by Q5 DNA polymerase, cloned, and sequenced, which revealed that the combined fidelity of SFM4-6-mediated transcription and SFM4-9-mediated reverse transcription is ~ 3.8 × 10−2, which allows us to estimate an error rate of SFM4-6 transcription of ~3.6 × 10−2 (Supplementary Fig. 7). While somewhat less than the fidelity of SFM4-9-mediated reverse transcription, this fidelity is still comparable to those previously reported for other polymerases selected or evolved to transcribe sugar modified oligonucleotides20, and is likely sufficient for practical applications. Incubation with Turbo DNase or 10% fetal bovine serum (FBS) confirmed the stability of the transcribed oligonucleotides (Fig. 3b,c and Supplementary Fig. 8).
Figure 3. Characterization of evolved polymerases and the nuclease stability of their transcribed products.
a, Transcription of fully C2’-OMe-modified oligonucleotides in the presence of all four C2’-OMe NTPs and different polymerases. No transcription is observed in the no polymerase control (lane 1) or when wild-type Sf is used (lane 2), and the parent SFM19 shows only slight extension (lane 3). In contrast, each of the mutants SFM4-3 (lane 4), SFM4-6 (lane 5), and SFM4-9 (lane 6) synthesizes the fully modified, full-length 60-mer C2’-OMe-DNA product. b, C2’-OMe-DNA resistance to Turbo DNase. A C2’-OMe DNA product synthesized by SFM4-6 using a C2’-OMe primer (lane 1) showed no degradation following incubation with Turbo DNase (compare lane 2, C2’-OMe-DNA product incubated in the absence of DNase, with lane 3 showing the same C2’-OMe-DNA treated with DNase). In contrast, a DNA product synthesized by Taq DNA polymerase using a DNA primer (lane 4) and treated analogously with DNase was completely degraded (compare lane 5, DNA product incubated in the absence of DNase, with lane 6 showing the same DNA treated with DNase) c, Stability of C2’-OMe oligonucleotide transcribed by SFM4-6 in 10% FBS. Natural DNA was rapidly degraded by serum enzymes, while a C2’-OMe-DNA product persisted with no degradation over 4 h. Data shown is the average ± s.d. of three, independent determinations.
PCR amplification of C2’-modified oligonucleotides
We next explored the ability of SFM4-3, SFM4-6 and SFM4-9 to PCR amplify C2’-OMe oligonucleotides. While each polymerase can interconvert natural and fully C2’-OMe oligonucleotides, further characterization demonstrated that under the conditions required for PCR amplification, sequential replacement of each modified nucleotide with its natural counterpart resulted in increasing efficiency, and we found that SFM4-3 with C2’-OMe-ATP was the most efficient (see Supplementary Figs. 9–11). To further characterize the SFM4-3 amplification of oligonucleotides containing C2’-OMe-A, a biotinylated DNA template was amplified, and streptavidin gel shift revealed a significant amount of amplified (unshifted) product (see Supplementary Fig. 11). To determine the fidelity of amplification, the reaction was repeated, the biotinylated template was removed with streptavidin-coated beads, and an aliquot of the amplified product was first converted back into DNA via PCR amplification with SFM4-3 and natural triphosphates, and then cloned and sequenced. When combined with the level of amplification, this allowed us to determine that the error rate is less than 1.7 × 10−2 nucleotide−1 cycle−1 (see Supplementary Methods for details). While reasonable for many practical applications, this error rate is somewhat increased relative to that for the wild type protein with fully natural substrates, which is ~2.8 × 10−4 nucleotide−1 cycle−1 28.
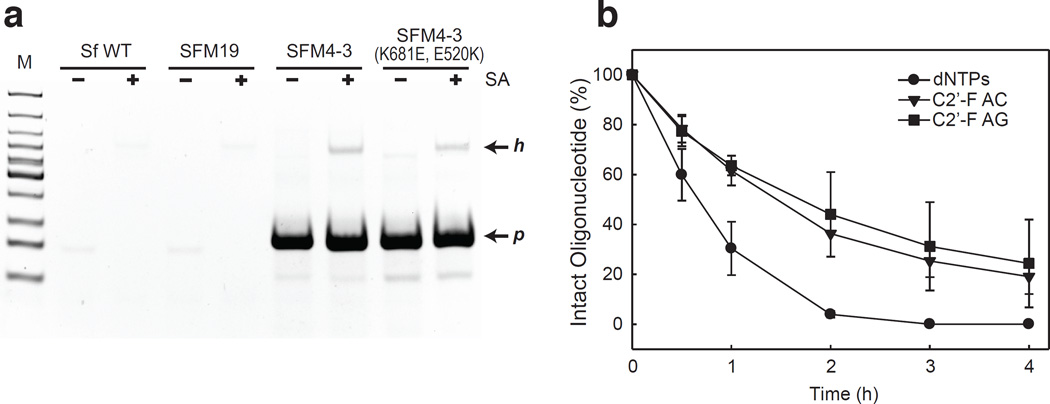
We next explored the PCR amplification of C2’-F-modified oligonucleotides. Initial experiments suggested that PCR amplification was very efficient when any pair of the four natural triphosphates was replaced with the corresponding C2’-F analogues (Supplementary Fig. 12). A biotinylated template was amplified with SFM4-3 and C2’-F-dATP, C2’-F-dGTP, dTTP, and dCTP, and streptavidin gel shift revealed that virtually all of the product was unshifted and thus resulted from very efficient amplification (Fig. 4a). Fidelity analysis, as described above, allowed us to estimate that the error rate is less than 2.8 × 10−3 nucleotide−1 cycle−1 (see Supplementary Methods for details), which is only 10-fold less than that reported for Taq polymerase with natural substrates28. Importantly, the conditions used to amplify both the C2’-OMe and C2’-F oligonucleotides are standard and not forcing (see Supplementary Methods for details), and thus the data reveal that the amplifications, especially of the C2’-F oligonucleotides, are both relatively efficient and high fidelity. Incubation in 5% FBS confirmed the increased stability of the partially modified amplification products (Fig. 4b).
Figure 4. Characterization of SFM4-3-mediated amplification of C2’-F oligonucleotides and analysis of the stability of the resulting PCR products.
a, Native PAGE analysis of PCR products obtained from a biotinylated DNA template in the presence of C2’-F-dATP, C2’-F-dGTP, dCTP, and dTTP, by different polymerases, without and with incubation with streptavidin (SA) (Lane M: low molecular weight marker (NEB)). Neither wild type Sf nor the parent mutant SFM19 produces any observable product. In contrast, the evolved mutant SFM4-3 and its derivative SFM4-3 (K681E, E520K) each produce a substantial amount of C2’-F-modified product (amplification product band, p), and only a small amount of the heteroduplex of DNA template and synthesized product strand (heteroduplex band, h) is observed. b, Stability of C2’-F-modified DNA product from SFM4-3 amplification in 5% FBS. Natural DNA (dNTPs) was rapidly degraded in the presence of FBS, while C2’-F-modified DNA products containing C2’-F-dA and C2’-F-dC (C2’-F AC), or C2’-F-dA and C2’-F-dG (C2’-F AG) exhibited significantly higher stability. Data shown is the average ± s.d. of three, independent determinations.
Finally, we explored the mechanism by which SFM4-3 acquired the ability to amplify the modified oligonucleotides. When considering the potentially adaptive mutations, position 681 was immediately interesting (Fig. 2). A Glu to Val mutation was selected in the first round of evolution (SFM1-5) in response to transcription pressure, and similar mutations at this site have been observed in several Taq polymerase mutants that have improved salt tolerance or modified triphosphate recognition29–31. The E681V mutation was retained in all selected mutants until the third or fourth round where pressure was applied for both increased transcription and reverse transcription activity, and SFM4-3 and SFM4-6 both acquired the Val to Lys mutation (Supplementary Table 3). Interestingly, residue 681 is located in the fingers domain, and based on structural data32, it is ~8 Å from residue E520 of the thumb domain (Fig. 2), and mutations of E520 to a neutral residue have also been found to confer Taq polymerase with reverse transcriptase activity or an altered substrate tolerance26,33. Thus, we hypothesized that the increased activity results from reducing electrostatic repulsion between the finger and thumb domains, and eventually stabilizing their interaction via a salt bridge. To test this hypothesis, we constructed the SFM4-3 (K681E, E520K) double mutant in which the potentially interacting residues are exchanged, as this mutant should only retain activity if the mutations act by forming a salt bridge. The efficient PCR amplification of a C2’-F-A/G oligonucleotide by the double mutant clearly reveals the retention of activity (Fig. 4a). A stabilized interaction between the fingers and thumb domains likely facilitates formation of the catalytically competent closed complex. Interestingly, antiterminators, which function to increase the processivity of RNA polymerases appear to act by a similar mechanism34.
Conclusion
The ability of SFM4-6 and SFM4-9, respectively, to transcribe and reverse transcribe fully C2’-OMe modified oligonucleotides with reasonable efficiency and fidelity should find immediate use, especially because the secondary structures of these modified oligonucleotides have higher stability, and the ability to run reactions at higher temperatures should preclude this from limiting product formation or from introducing sequence biases. Most importantly, SFM4-3 is the first polymerase with the demonstrated ability to PCR amplify any C2’-modified oligonucleotides, and the mechanism by which the activity was evolved, the stabilization of an interaction between the thumb and fingers domain, may be general and thus useful for the optimization of other polymerases. Regardless, based on the increased stability in biological solutions of these modified oligonucleotides, SFM4-3 should find immediate application as part of efforts to develop modified aptamers for diagnostic or therapeutic applications.
Methods
Phage construction
The X30 helper phage was constructed in a previous work21. The Y30 helper phage was constructed by fusing the 5’ terminus of the truncated gIII gene with DNA encoding the pIII extension GAXGGSGGSGGSGGS, where X is pAzF.
SF library construction
Libraries for the first and second rounds of evolution were constructed by standard error-prone PCR methodology using MnCl2 (0 mM or 0.05 mM). The resulting two libraries had 0~3 or 3~5 amino acid mutations per gene, and were mixed and subjected to the polymerase evolution system described in Fig. 1. For the third and fourth rounds of evolution, DNA shuffling conducted via staggered extension process (StEP) and site saturation mutagenesis using codon-degenerated oligonucleotides were combined to generate the libraries. All PCR products were digested with SfiI and NotI, and inserted into the similarly digested phagemid pFAB-Sf to replace the wild type Sf gene.
Phage selection
The ligation products from construction of the pFAB-Sf library were directly electroporated into E. coli SS320 or SS320/pEVOL-pAzF, and the library was displayed on X30 or Y30 phage as described in the Supplementary Information. After attaching the primer/template substrate onto X30 or Y30 phage and purifying the phage via PEG/NaCl precipitation, substrate-attached phage particles (~1×1011 CFU) were mixed with C2’-OMe-modified or natural dNTPs (50 µM; TriLink BioTechnologies or New England Biolabs), and biotin-UTP or biotin-dUTP (2 µM; TriLink BioTechnologies) in 1× Sf reaction buffer, and incubated at 50 °C for 10 min. After extension, the reaction was quenched by addition of 1/5 volume 0.5 M EDTA (pH 8.5). The phage particles with primer extended and biotin-UTP incorporated were then captured with streptavidin beads, washed, and cleaved off the beads by DNase I treatment. The enriched phage particles were directly used to infect E coli XL-Blue/MRF’ cells (log phase), and then plated onto LB/agar supplemented with spectinomycin to recover the pFAB-Sf phagemid library. After an overnight growth at 37 °C, colonies were scraped from the agar surface, and the phagemid pFAB-Sf library was recovered by spin column (ZR Plasmid Miniprep, Zymo Research).
96-well plate screening
The post-selection Sf libraries were excised from the phagemids extracted after phage selection with SfiI and NotI, subcloned into a modified pET-23b(+) plasmid21 (for details, see Supplementary Methods), and transformed into BL21(DE3)/ pLysS cells. Single colonies were grown in 96-well deep-well plates, and induced with 0.4 mM ITPG at log phase. After 6 h, the cells were pelleted, lysed with 1× Bugbuster (Novagen), and heat-treated to remove most of the cellular proteins. The heat-purified polymerases were then directly used for activity screening. Amine-binding 96-well plates (Corning) were coated with amine-primer/template complex, washed and blocked with BSA. The extension buffer containing the desired C2’-OMe-modified or natural dNTPs, as well as biotin-UTP, was then added into the wells and mixed with the heat-purified polymerases. The plates were then incubated at 50 °C for varying amounts of time (1–60 min). After primer extension, the incorporated biotin-UTP was detected with horseradish peroxidase (HRP)-streptavidin binding and chromogenic reaction catalysed by HRP.
General primer extension and gel characterization of the mutants
C2’-OMe-DNA or natural DNA primers were radiolabeled with [γ-32P] ATP (PerkinElmer) using T4 polynucleotide kinase (New England Biolabs), and purified with the Qiaquick nucleotide removal kit (Qiagen). Radiolabeled primer (100 nM) was mixed with template (200 nM) in 2 × Sf reaction buffer (or other defined buffers), and annealed by heating to 95 °C and slowly cooling to room temperature. The annealing product was then mixed with ½ volume of C2’-OMe-NTPs and/or dNTPs mixture of defined concentrations, as well as other defined components (see Supplementary Information for details), and ½ volume diluted heat-purified cell lysate or column-purified enzymes. The extension reaction was incubated at defined temperatures and times, and quenched by the addition of 2 volumes of quenching/gel loading buffer (95% formamide, 18 mM EDTA, SDS, xylene cyanol and bromophenol blue). The quenched reactions were heated to 95 °C for 5 min, and then analysed using denaturing PAGE (15% acrylamide, 8 M urea).
PCR with C2’-OMe-NTPs or C2’-F-NTPs
PCR with C2’-OMe-NTPs or C2’-F-dNTPs was performed using modified standard PCR conditions with an enhanced extension time and lower extension temperature. For a typical PCR with C2’-modified-dNTPs, the following program was applied: 94 °C for 2 min; 15–40 cycles of 94 °C for 30 s, 49 °C for 1 min, 50 °C for 20 min; 50 °C for 2 h. Reactions contained 0.2–3 µM polymerase, 0.5–1.25 mM C2’-modified or natural dNTPs, 1.5–3.5 mM MgCl2, and 0–1 mM MnCl2. The product fraction containing original template was followed using a biotinylated template. After PCR, purified products were mixed with excess streptavidin and incubated at 37 °C for 30 min. All PCR products were analysed using 6% or 8% PAGE.
Serum stability test of C2’-modified products
The C2’-OMe-transcription product or the C2’-F-PCR product were incubated with 10% or 5% fetal bovine serum (FBS) at 37 °C. Aliquots were removed and quenched with 20 mM EDTA at defined time points and then frozen. All samples were analysed using 15% denaturing PAGE containing 8 M urea or 8% native PAGE.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (GM097489) and the Defense Advanced Research Projects Agency (DARPA; N66001-14-2-4052) for supporting this work. We thank Peter Schultz for the plasmid pEVOL-pAzF.
Footnotes
Author contributions
T.C. and F.E.R. conceived and designed the experiments. T.C., N.H., Z.L, R.A., and S.S.T performed experiments. T.C., N.H., Z.L. and F.E.R. analysed the data. T.C. and F.E.R. co-wrote the paper. F.E.R. supervised the project.
Supplementary information is available in the online version of the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Joyce GF. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 3.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 4.Ng EW, et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 5.Metelev V, Lisziewicz J, Agrawal S. Study of antisense oligonucleotide phosphorothioates containing segments of oligodeoxynucleotides and 2′-o-methyloligoribonucleotides. Bioorg. Med. Chem. Lett. 1994;4:2929–2934. [Google Scholar]
- 6.Agrawal S, et al. Mixed-backbone oligonucleotides as second generation antisense oligonucleotides: in vitro and in vivo studies. Proc. Natl. Acad. Sci. USA. 1997;94:2620–2625. doi: 10.1073/pnas.94.6.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heemskerk H, et al. Preclinical PK and PD studies on 2'-O-methyl-phosphorothioate RNA antisense oligonucleotides in the mdx mouse model. Mol. Ther. 2010;18:1210–1217. doi: 10.1038/mt.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goemans NM, et al. Systemic administration of PRO051 in Duchenne's muscular dystrophy. N. Engl. J. Med. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 9.Manoharan M, et al. Unique gene-silencing and structural properties of 2'-fluoro-modified siRNAs. Angew. Chem. Int. Ed. 2011;50:2284–2288. doi: 10.1002/anie.201006519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rigo F, et al. Synthetic oligonucleotides recruit ILF2/3 to RNA transcripts to modulate splicing. Nat. Chem. Biol. 2012;8:555–561. doi: 10.1038/nchembio.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kojima T, et al. PCR amplification of 4'-thioDNA using 2'-deoxy-4'-thionucleoside 5'-triphosphates. ACS Synth. Biol. 2013;2:529–536. doi: 10.1021/sb400074w. [DOI] [PubMed] [Google Scholar]
- 12.Jones GD, Lesnik EA, Owens SR, Risen LM, Walker RT. Investigation of some properties of oligodeoxynucleotides containing 4'-thio-2'-deoxynucleotides: duplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1996;24:4117–4122. doi: 10.1093/nar/24.21.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J, et al. Cell-specific RNA aptamer against human CCR5 specifically targets HIV-1 susceptible cells and inhibits HIV-1 infectivity. Chem. Biol. 2015;22:379–390. doi: 10.1016/j.chembiol.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor AI, et al. Catalysts from synthetic genetic polymers. Nature. 2015;518:427–430. doi: 10.1038/nature13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, Qiu Q, Gill SC, Jayasena SD. Modified RNA sequence pools for in vitro selection. Nucleic Acids. Res. 1994;22:5229–5234. doi: 10.1093/nar/22.24.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fa M, Radeghieri A, Henry AA, Romesberg FE. Expanding the substrate repertoire of a DNA polymerase by directed evolution. J. Am. Chem. Soc. 2004;126:1748–1754. doi: 10.1021/ja038525p. [DOI] [PubMed] [Google Scholar]
- 17.Chen T, Romesberg FE. Directed polymerase evolution. FEBS Lett. 2014;588:219–229. doi: 10.1016/j.febslet.2013.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chelliserrykattil J, Ellington AD. Evolution of a T7 RNA polymerase variant that transcribes 2'-O-methyl RNA. Nat. Biotechnol. 2004;22:1155–1160. doi: 10.1038/nbt1001. [DOI] [PubMed] [Google Scholar]
- 19.Ibach J, et al. Identification of a T7 RNA polymerase variant that permits the enzymatic synthesis of fully 2'-O-methyl-modified RNA. J. Biotechnol. 2013;167:287–295. doi: 10.1016/j.jbiotec.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Pinheiro VB, et al. Synthetic genetic polymers capable of heredity and evolution. Science. 2012;336:341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia G, et al. Directed evolution of novel polymerase activities: Mutation of a DNA polymerase into an efficient RNA polymerase. Proc. Natl. Acad. Sci. USA. 2002;99:6597–6602. doi: 10.1073/pnas.102577799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leconte AM, et al. Directed evolution of DNA polymerases for next-generation sequencing. Angew. Chem. Int. Ed. 2010;49:5921–5924. doi: 10.1002/anie.201001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 24.Marks IS, et al. Strain-promoted "click" chemistry for terminal labeling of DNA. Bioconjug. Chem. 2011;22:1259–1263. doi: 10.1021/bc1003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong JL, Loakes D, Jaroslawski S, Too K, Holliger P. Directed evolution of DNA polymerase, RNA polymerase and reverse transcriptase activity in a single polypeptide. J. Mol. Biol. 2006;361:537–550. doi: 10.1016/j.jmb.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 26.Vichier-Guerre S, Ferris S, Auberger N, Mahiddine K, Jestin J-L. A population of thermostable reverse transcriptases evolved from Thermus aquaticus DNA polymerase I by phage display. Angew. Chem. Int. Ed. 2006;45:6133–6137. doi: 10.1002/anie.200601217. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.New England Biolabs Inc. Taq DNA Polymerase with Standard Taq Buffer. [Accessed Aug. 2015]; https://www.neb.com/products/m0273-taq-dna-polymerase-with-standard-taq-buffer. [Google Scholar]
- 29.Davis M, et al. Taq DNA polymerases having an amino acid substitution at E681 and homologs thereof exhibiting improved salt tolerance. 2,381,206. CA Patent. filed 10 Aug. 2000.
- 30.Brandis J, Bloom C, Richards J. DNA polymerases having improved labeled nucleotide incorporation properties. 6,265,193. US Patent. filed 12 Mar. 1998, issued 24 Jul. 2001.
- 31.Brandis J, Bloom C, Richards J. DNA polymerases having improved labeled nucleotide incorporation properties. 7,897,738. US Patent. filed 9 Dec. 2005, issued 1 Mar. 2011.
- 32.Holzberger B, Obeid S, Welte W, Diederichs K, Marx A. Structural insights into the potential of 4-fluoroproline to modulate biophysical properties of proteins. Chem. Sci. 2012;3:2924–2931. [Google Scholar]
- 33.Ghadessy FJ, et al. Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nat. Biotechnol. 2004;22:755–759. doi: 10.1038/nbt974. [DOI] [PubMed] [Google Scholar]
- 34.Santangelo TJ, Artsimovitch I. Termination and antitermination: RNA polymerase runs a stop sign. Nat. Rev. Microbiol. 2011;9:319–329. doi: 10.1038/nrmicro2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.