Abstract
Morphometric correlation networks of cortical thickness, surface area, and gray matter volume have statistically different structural topology. However, there is no report directly describing their correlation patterns in view of interregional covariance. Here, we examined the characteristics of the correlation patterns in three morphometric networks of cortical thickness, surface area, and gray matter volume using a Venn diagram concept across 314 normal subjects. We found that over 60% of all nonoverlapping correlation patterns emerged with divergent unique patterns, while there were 10% of all common edges in ipsilateral and homotopic regions among the three morphometric correlation networks. It was also found that the network parameters of the three networks were different. Our findings showed that correlation patterns of the network itself can provide complementary information when compared with network properties. We demonstrate that morphometric correlation networks of distinct structural phenotypes have different correlation patterns and different network properties. This finding implies that the topology of each morphometric correlation network may reflect different aspects of each morphometric descriptor.
Morphometric-based correlation networks (MCNs) have been commonly constructed using divergent structural phenotypes such as cortical thickness, cortical surface area, and gray matter (GM) volume to investigate the patterns of large-scale structural association between brain regions in healthy subjects and diverse patients such as those with Alzheimer’s disease, multiple sclerosis, schizophrenia, and epilepsy (see Supplementary Table S1). While most studies to date have employed MCNs without explicit distinction of structural phenotypes, some studies have begun to look at the difference in the network topology between the structural phenotypes. Sanabria-Diaz et al. found that the topologies of cortical thickness and surface area networks were significantly different, demonstrating that both capture distinct properties of the interaction or different aspects of the same interaction (e.g., mechanical, anatomical, or chemical) between brain structures1. Anand et al. also evaluated a comparison of MCN properties with surface area, GM volume, and thickness on the same population to show that the topology of cortical thickness, surface area, and volume networks was significantly different2.
While these two studies evaluated the network parameters that summarize an entire topology down to a single number such as clustering coefficient, characteristic path length, global network efficiency, and small-worldness, the correlation pattern of the network itself could provide a complementary insight to compare the topology of different structural phenotypes. The network parameters provide a global measure for an entire topology and local measures allowing the edge properties for a node, and are thus attractive because of their simplicity; however, they can only compare the networks statistically. By contrast, the correlation patterns of the networks reveal the spatial distribution of the edges and can detect the edge-based similarity of topological features inherent in each network. For example, Gong et al. found that approximately 35–40% of cortical thickness correlations showed convergent diffusion connectivity across the cerebral cortex3. Although MCNs have the same number of nodes with the same regions, they may have different interregional correlation patterns driven by distinct cellular mechanisms as the result of a distinct genetic origin. Different genetic factors influence patterns of structural covariance most strongly within different brain networks4,5,6,7,8; that is, there seem to be network-specific genetic influences9. Chen et al. showed that the differences in genetic influence on cortical surface area were along an anterior–posterior axis; the differences in genetic influences on cortical thickness were along a dorsal–ventral axis in the same cohort8,10. The distinct structural phenotypes could result in different interregional correlation patterns, which in turn have an effect on the network properties.
To our knowledge, the quantitative evaluation of the similarity of the correlation patterns between the network topology of distinct structural phenotypes including cortical thickness, surface area, and GM volume has not been considered in detail. The similarity of the correlation patterns can be evaluated by the overlap ratio (OR) of the spatially distributed edges and the types of edge. A Venn diagram concept dividing all possible cases of the edges is useful to investigate the OR of the spatially distributed edges in a detailed manner: common edges overlapping in all three networks, three parts of overlapping edges in two networks only, and three parts of nonoverlapping edges in each network exclusively (Fig. 1). Furthermore, the classification of edge types according to inter- and intrahemispheric connections may provide useful information regarding the edge patterns in terms of lateralization: (1) homotopic connection to indicate the same areas in opposite hemispheres; (2) the left and right ipsilateral connection to indicate different areas in the same hemispheres; and (3) heterotopic connection to indicate different areas in opposite hemispheres11. Therefore, the number of edges belonging to each connection type in seven partitions respectively yields quantitative information about the correlation patterns of MCNs.
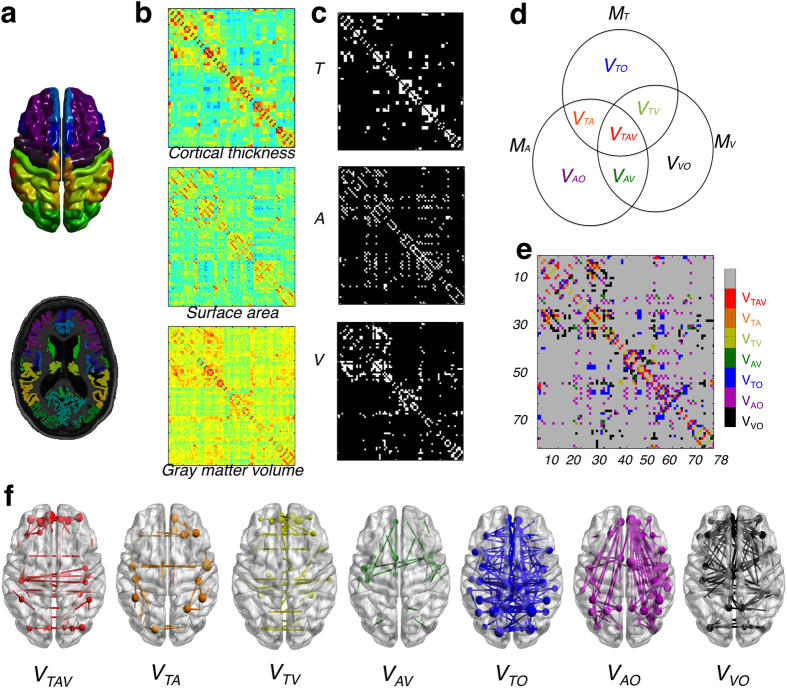
Figure 1. Schematic showing processing for constructing morphometric correlation networks (MCN) and the Venn diagram concept.
(a) The AAL template masks on the cortical surface and volume on the MNI 152 template. Each color represents 78 cortical regions. (b) Interregional correlation matrix in each cortical thickness, surface area, and gray matter (GM) volume was performed using Pearson correlation analysis from the principal data set, after removing the age, sex, and global measures. The colors of the matrix ranged from − 1 to 1. (c) The adjacency matrix was thresholded at 8% of sparsity. T, A, and V indicate the adjacency matrix of cortical thickness, surface area, and GM volume, respectively. (d) A Venn diagram concept was employed to identify edges of all possible cases in three networks. MT, MA, and MV are the set of the elements representing edges in each adjacency matrix. VTAV, VTA, VTV, VAV, VTO, VAO, and VVO were the set of the elements representing edges in each partition. (e) The colors of the element in the matrix indicate the edges in each partition. (f) The regional pairs in each partition for visualization (8% sparsity).
In this study, we partitioned the spatially distributed edges into seven parts and classified them into four different edge types to characterize the various correlation patterns of cortical volume, thickness, and surface area networks. We also calculated network parameters to explore whether the characteristics of correlation pattern have an effect on the network parameters.
Results
Similarity of the correlation pattern
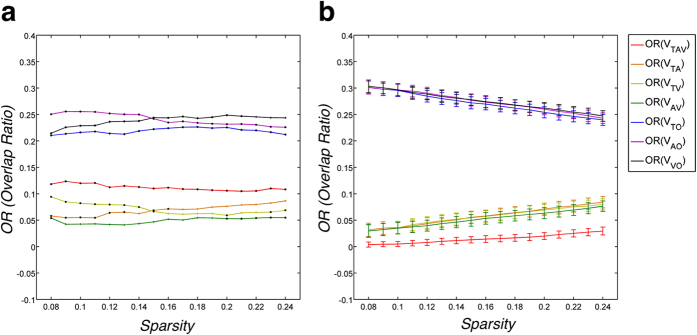
The overlap ratio (OR) is the number of pairs present in each partition divided by the number of all edges surviving the threshold in the selected network sparsity across 314 normal subjects from the principal data set. VTAV is the set of the common edges of the three networks; VTA is the set of the shared edges of thickness and surface area networks; VTV is the set of the shared edges of thickness and GM volume networks; VAV is the set of the shared edges of surface area and GM volume networks; VTO is the set of the exclusive edges in thickness networks; VAO is the set of the exclusive edges in surface area networks; and VVO is the set of the exclusive edges in GM volume networks. We calculated the OR in each partition and plotted the ORs according to the sparsity range of 8–24%, where OR(VTAV) was 10–12%, OR(VTA), OR(VTV) and OR(VAV) were under 10%, and OR(VTO), OR(VAO) and OR(VVO) were 20–25% (Fig. 2). The results indicate that each network had different correlation patterns that were distinct from each other. The observed OR(VTAV), OR(VTV), OR(VTV) and OR(VAV) were higher, and the observed OR(VTO), OR(VAO) and OR(VVO) were lower than the expected value from 1000 simulated pairs of random networks with statistical significance under P < 0.05.
Figure 2. The overlap ratio (OR) in seven partitions.
(a) The plot figure shows the observed OR of each partition as a function. Data points with black color indicate statistically significance (P < 0.05). The significance was reached if less than 5 percentile in the ORs distribution of the random networks. (b) The ORs of 1000 random simulated networks show the mean and standard deviation of each partition as a function.
Edge types in each partition
We examined the edge types and the signs in each partition to detect the characteristics of spatially distributed edges. The numbers of edges of four types were counted and divided by the total number of edges in each partition along the entire sparsity range (Fig. 3). The edges of VTAV were mostly shown ipsilateral with slightly more left-dominant lateralized connections than right connections and bilateral homotopic connections. The correlation patterns of VTA and VAV were stronger in order of ipsilateral (R > L) connections, and the correlation patterns of VTV showed a different order of high heterotopic connections. The correlation patterns in VTO, VAO, and VVO were considerably different from each other. The correlation patterns of VTO and VVO showed highly heterotopic connections, and the correlation patterns of VAO were in order of right-dominant ipsilateral connections. Furthermore, only positive associations were shown in VVO, whereas both positive and negative associations were revealed in VTO with heterotopic connections and VAO with mostly ipsilateral connections.
Figure 3. The percentages of the four types of edges, that is, the homotopic, the left and right ipsilateral, and heterotopic connections were plotted.
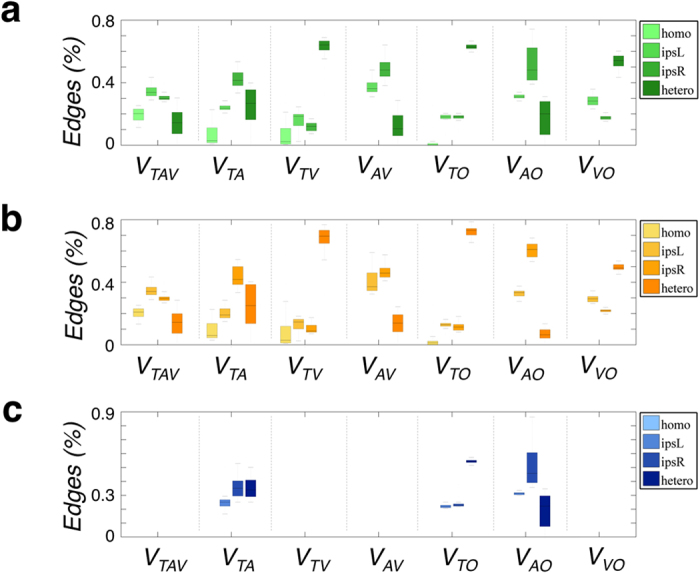
(a) The edges show both signs, with positive and negative correlation coefficients between regions. (b) The edges show only positive correlation. (c) The edges show only negative correlation. In each box, the central mark is the median, the edges of the box are the 25th and 75th percentiles, and the box color indicates the connection type.
Hub regions
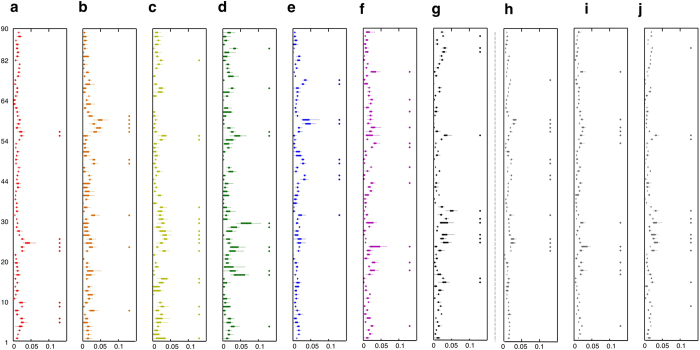
We examined hub regions of MT, MA, and MV and in each partition of VTAV, VTA, VTV, VAV, VTO, VAO, and VVO (Fig. 4). We found that the detected hub regions of MT had the maximum Jaccard index with VTO (Jaccard index: 0.53), and those of MV with VVO (Jaccard index: 0.6). Note that the detected hub regions of MA had a maximum Jaccard index in VAV (Jaccard index: 0.5) and the second in VAO (Jaccard index: 0.39) (Table 1).
Figure 4. The hub regions in seven partitions and MCNs.
Each partition (a–j) was marked as follows: (a) VTAV, (b) VTA, (c) VTV, (d) VAV, (e) VTO, (f) VAO, (g) VVO, and (h) MT, (i) MA, and (j) MV indicate the adjacency matrix of cortical thickness, surface area, and gray matter volume, respectively. In each box, the central mark is the median, the edges of the box are the 25th and 75th percentiles of the normalized degree of a node (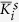 ) at each sparsity. Hi is the average of the normalized degree of a node (
) at each sparsity. Hi is the average of the normalized degree of a node (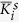 ) over the sparsity range. The hub regions were defined as the nodes with higher than mean (Hi) + standard deviation (Hi) and marked as circle with same color in each box plot. Abbreviations for the cortical regions and number of AAL were listed in Supplementary Table S3.
) over the sparsity range. The hub regions were defined as the nodes with higher than mean (Hi) + standard deviation (Hi) and marked as circle with same color in each box plot. Abbreviations for the cortical regions and number of AAL were listed in Supplementary Table S3.
Table 1. Jaccard index for the coincident hub nodes.
| VTAV | VTA | VTV | VAV | VTO | VAO | VVO | |
|---|---|---|---|---|---|---|---|
| MT | 0.263 | 0.500 | 0.111 | 0.136 | 0.533* | 0.083 | 0.083 |
| MA | 0.211 | 0.211 | 0.115 | 0.500* | 0.048 | 0.389 | 0.087 |
| MV | 0.294 | 0 | 0.556 | 0.211 | 0 | 0 | 0.600* |
The Jaccard index was calculated for the coincident hub regions between MCNs of MT, MA and MV and subnetworks of each partition such as VTAV, VTA, VTV, VAV, VTO, VAO, VVO. The hub regions of each network are marked in Fig. 4. Asterisk indicates the highest Jaccard index between hub regions of MCNs and subnetworks of each partition.
Topological properties of the MCNs
We compared the global network parameters including clustering coefficient, characteristic path length, small-worldness index, and global efficiency according to the sparsity range of 8–24% (see Supplementary Fig. S7). Three MCNs showed small-world network properties. Furthermore, three MCNs showed statistically different network parameters from ANOVA test with P < 0.05, and significantly reduced clustering coefficient, characteristic path length, and increased global efficiency in common. The thickness network showed higher clustering coefficient than those of the surface area and GM volume networks.
Subgroup analysis
Group differences were also investigated by comparing the ORs (1) between male and female, (2) young and old, (3) patients with dementia and old normal controls. The ORs of each group were similar to those of the principal data, for example, over 60% of all nonoverlapping edges with divergent patterns, approximately 10% of all common edges with the edges showing ipsilateral and homotopic pairs. There were little differences between both comparisons in the ORs (P < 0.05) (see Supplementary Fig. S1). The edge types of negative correlation in VVO were shown in patients with dementia and old group (see Supplementary Fig. S2). The hub regions of MCNs were highly overlapped to those of each partition in each group, with results shown in Supplementary Fig. S3 and Supplementary Table S41.
Replication of the study
In order to demonstrate the reproducibility of the results with larger sample size and with more young adult subjects, we calculated the ORs, edge types and hub regions. To investigate the group difference, we subdivided the total subjects of the replication data into males and females and analyzed them separately, with results shown in Supplementary Figs S4–S6. As shown in Supplementary Fig. S4, overall patterns of the ORs were similar to those of the principal data, although the ORs of VTAV were higher than those of the principal data. There were some different patterns showing the edge types of negative correlation in total and male group while the edge types of negative correlation in female group was similar to those of the principal data (see Supplementary Fig. S5). The hub regions of MCNs were similar to those of each partition in each group (see Supplementary Fig. S6, and Supplementary Table S5).
Discussion
In this study, we examined the characteristics of correlation patterns to determine the difference in structural topology between the three MCNs of cortical thickness, surface area, and GM volume. This may allow complementary accounting for spatial and topological information when combined with network parameters.
Our findings showed that over 60% of all nonoverlapping correlation patterns emerged from distinct structural phenotypes, that is, thickness, surface area, and GM volume-based networks. The findings support the hypothesis that the different structural phenotypes of cerebral cortex have different network properties1,2. Sanabria-Diaz et al. and Anand et al. demonstrated the hypothesis statistically comparing the network parameters such as clustering coefficient, characteristic path length, and global efficiency. The network parameters may help identify significant differences in the network topology. While the global network parameters gave us an abstraction of the complex human brain topology as a single value, we provided the complementary characteristics of network topology such as the percentage of similarities between MCNs and the types of edges. We can better understand and explain quantitatively the differences of brain network topology in terms of how they are similar or dissimilar between MCNs and which types of edge they have. In this study, we observed that the OR(VTO), OR(VAO) and OR(VVO) was respectively 20% with the different correlation patterns in case of the only existing edges in each network. The correlation patterns were considerably different than those of VTO and VVO, which had more heterotopic connections, while the correlation patterns of VAO showed right-dominant ipsilateral connections (R > L). We found only positive associations were shown in VVO, whereas both positive and negative associations were revealed in VTO and VAO. The negative correlation patterns of VTO were shown to be the pairs of regions between the frontal cortex (i.e. superior frontal gyrus) and the temporal cortex (i.e. parahippocampal gyrus), and those of VAO existed predominantly in the pairs of regions between the frontal cortex (i.e. superior frontal gyrus (dorsolateral, medial), supplementary motor area) and parieto-occipital cortex. The findings may be related to the distinct genetic mechanisms supporting the findings of previous studies8,10. Chen et al. reported the dorsal–ventral (D-V) regions, such as the motor/premotor, parietal, and occipital region (the dorsal cluster) from prefrontal and temporal regions (the ventral cluster) division as the most distinctive partition in the genetic patterns of cortical thickness. The anterior–posterior (A-P) regions, such as the motor and sensory cortices, like the basic frontal/nonfrontal division, were observed in the genetic patterning of the cortical surface area8,10. It is well known that cortical thickness and surface area are influenced by distinct cellular mechanisms because of their distinct genetic origins1,6,12,13,14,15. The measures of cortical surface area and cortical thickness represent respectively the number of cortical columns as an overall degree of folding and the amount of cell density in the cortex16, although the measure of cortical volume captures some combination of both cortical thickness and surface area such as structural phenotype and genetic factors, because the volume is calculated by multiplying cortical thickness and surface area. However, the exact biological nature of this relationship remains unknown and warrants further study.
The correlation patterns of two-by-two matrices such as VTA, VTV, and VAV showed quite different patterns between them. The differences could be related to intrinsic characteristics or different evolutionary development, or genetic effects of each morphometric descriptor. The interactions between the two-by-two networks may reflect a complex mechanism of structural covariance. Currently, it is difficult to compare our findings with previous work. It is important to investigate the interaction between the morphometric descriptors as to how those interactions are related to the information about the complex mechanical, genetic, and chemical relationships between different brain sites1. The exact biological nature of these relationships may be revealed by future study.
We found an OR of approximately 10% between the MCNs with all positive correlations. It is also important to note that the correlation patterns of VTAV showed left-dominant ipsilateral (L > R) and bilateral homotopic connections, which was consistent with previous findings by Gong et al. who reported that 30–40% of only positive thickness correlations shared white matter connectivity in ipsilateral or bilateral homotopic regions3. Lerch et al. were the first to show that cortical thickness correlations qualitatively match a diffusion tensor imaging traced track, implying that anatomical connectivity could be measured indirectly using information from the cortical surface17. Our findings are consistent with the hypothesis of anatomical connectivity induced by mutually tropic influences1,3,11,15,17,18,19,20 supporting the common morphometric changes of three structural phenotypes in the same direction leading to positive correlations between related regions.
We examined which hub regions of MCNs were similar to those of each partition. The hub regions of the cortical thickness network were closer to that of VTO. The hub regions of a surface area network were likely to be one of VAV and VAO. The hub regions of GM volume networks were similar to that of VVO. The similarity between hub regions of MCN networks and subnetworks of each partition showed that the hubs were strongly associated with their intrinsic correlation patterns. The hub has an important role in integrating and distributing information because of the number and position of its correlations in a network. We also examined the network parameters such as the clustering coefficient, characteristic path length, and global efficiency; these were significantly different between MCNs, which was consistent with previous studies1,2.
We investigated the group differences of the characteristics of correlation patterns between male and female, young and old, patients with dementia and old normal controls from the principal data set. We found that all subgroups had similar characteristics of correlation patterns in general, whereas patients with dementia and old normal subjects had the edges of the negative correlation in VVO. It might reflect altered inter-regional correlations in aging21. However, the underlying biological nature of the morphological correlations is still unknown.
We demonstrated that the reproducibility of the results were similar with the characteristics of correlation pattern showing over 60% of all nonoverlapping correlation patterns, 10–18% of all common edges with ipsilateral and homotopic pairs with only positive correlations. We examined some different types of negative correlation in total and male group, but overall trend of negative correlations were shown in bilateral heterotopic regions while those of positive correlations are mostly among ipsilateral or homotopic regions supporting with previous study3. It is still unknown the existence of different mechanisms of the positive and negative correlation of morphology.
This study has several limitations. The main limitation is that, we used the AAL atlas for a node definition, as can be done by alternative choices of different brain parcellations such as high-resolution parcellations or another anatomical atlas like Brodmman, but provides biological and anatomical meaningful cortical regions comparing to high-resolution parcellations23. Since previous studies have successfully employed this parcellation scheme, we used AAL atlas to keep our findings consistent with the previous studies1,3,21. It is also notable that inter-regional correlation measure was evaluated by Pearson correlation test3,15,21. Adoption of different methods such as partial correlation or mutual information may have some effect on the pattern of results2. The different connectivity measures need to be addressed in the future. Furthermore, although we tried to remove the confounding effect by using the regression model, it might not explicitly deal with still remain. Another consideration is that, because cortical thickness and surface area were defined on the cerebral cortex, we excluded the sub-cortical structures. And we constructed the binary adjacency matrix because of its simplicity for interpretation. It is needed to address in terms of weighted network.
Our results demonstrated that morphometric correlation networks of distinct structural phenotypes have different correlation patterns and different network properties.
Methods
Data acquisition
We conducted the experiments using two data sets in this study: (1) the set of cross-sectional data from the Open Access Series of Imaging Studies (OASIS) database (www.oasis-brains.org), referred to as the principal data set of the study and (2) Human Connectome Project (HCP) data (www.humanconnectome.org), referred to as the replication data set. Supplementary Data provides more information about study population and image acquisition.
Image preprocessing for a morphometric descriptor
We computed three cortical features such as cortical thickness, surface area and GM volume. CIVET software was used to extract two cortical surfaces, the inner and outer surfaces, which are composed of 3-D meshes. The procedures are well validated and have been extensively described23,24,25,26,27. Further image processing was described in Supplementary Methods section. Cortical thickness was calculated as the Euclidean distance between the corresponding vertices of inner and outer surfaces using a t-link method23. Surface area was measured by calculating the Voronoi area assigned to any vertices28. Cortical thickness and surface area were blurred using a 20 mm full-width half-maximum surface-based diffusion kernel29,30. GM volume was obtained from the segmented GM images in native space.
Morphometric network construction
We divided the cortical surface into 78 regions (39 for each hemisphere) to identify nodes of the MCN with the automated anatomical labeling (AAL) template excluding the cerebellum and subcortical regions (i.e. amygdala, caudate, hippocampus, pallidum, putamen, and thalamus). We applied the same procedures described in the previous work to cortical parcellation3,22 and network construction3. Supplementary Methods section provides more details of the network construction. Finally, three 78 × 78 symmetric correlation matrices were obtained for cortical thickness, cortical surface area, and GM volume. A binary adjacency matrix with 1 or 0 indicating the simple presence or absence of an edge was created by thresholding the correlation matrix in terms of sparsity3. Sparsity measures the percentage of the number of existing edges in all possible links31. Because there is no definitive criterion for a single threshold, we observed the results through a sparsity threshold of 8–24% satisfying a fully connected MCN ensuring the same number of suprathreshold regional pairs for all the networks3. In equations (1, 2, 3) below, T, A, and V indicate the adjacency matrix of cortical thickness, surface area, and GM volume, respectively, and MT, MA, and MV are the set of the elements representing edges in each adjacency matrix:
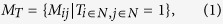 |
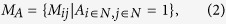 |
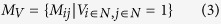 |
where index i and j indicates the node ith row and jth column in an adjacency matrix and N is the set of all nodes in the networks. Note that only the elements of upper triangles in the adjacency matrix were considered because the adjacency matrix is symmetric.
Partitioning the edges
All the edges in the three MCNs were partitioned into seven parts: portions of their common edges overlapped in three networks, three parts of overlapped edges between two networks, and three parts of exclusive edges in each network (Fig. 1). Each partition was defined as follows.
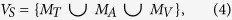 |
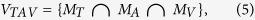 |
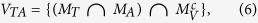 |
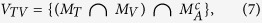 |
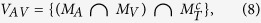 |
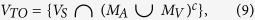 |
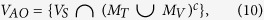 |
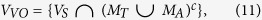 |
VS is the set of the union edges of the three networks. VTAV, VTA, VTV, VAV, VTO, VAO and VVO were the set of the elements representing edges in each partition.
Identifying characteristics of correlation patterns
We defined the OR of the spatially distributed edges as follows.
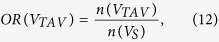 |
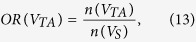 |
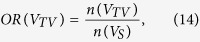 |
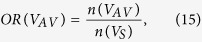 |
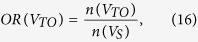 |
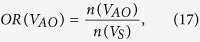 |
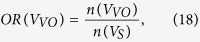 |
Note that the total sum of ORs is 1. We classified the edges in each partition into the four types to investigate the connection patterns in terms of lateralization: (1) homotopic connection between the same areas in opposite hemispheres; (2) the left and (3) right ipsilateral connection between different areas in the same hemispheres; and (4) heterotopic connection between different areas in opposite hemispheres11. Note that we have separated the ipsilateral connections into left and right hemispheres, thereby expanding the three types described by Mechelli et al.11. The edges of each connection type in each partition at each sparsity level were counted, and the number of edges of each case was divided by the total number of edges in the corresponding partition to normalize the value. We also examined the positive and negative correlation patterns between brain regions.
Identifying hub nodes
We calculated the network parameters such as nodal degree, clustering coefficient, characteristic path length, and global efficiency, which represent the topological properties of the network, to elucidate the associations between the correlation patterns and the network parameters. The definitions and formulations of the network parameters were described in Supplementary Table S2.
Hubs can be defined as the nodes with either high degree or high centrality31,32. The degree of a node (ki) indicates the number of links connected to a node. In this study, the degree of a node was divided by degree sum of all nodes at each level of sparsity to normalize the value as follows.
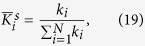 |
The average of the normalized degree of a node over the sparsity range is defined as:
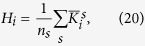 |
where ns is the number of the sparsity range from 8–24% with 1% increment, s = {0.08:0.01:0.24}.
Nodes with the average of the normalized degree of a node (Hi) larger than the mean plus standard deviation are considered as hubs of a network. We calculated the network parameters using the Brain Connectivity Toolbox (www.brain-connectivity-toolbox.net) and used BrainNet Viewer for visualization of networks on brain templates (www.nitrc.org/projects/bnv).
Statistical analysis
A non-parametric test was employed to assess the statistical differences of ORs between observed and simulated pairs of random networks. First, matched random networks with all morphological measures were constructed with the same node degree of distribution18. Then, we recomputed the partitioning edges and ORs. This procedure was repeated 1000 times for each sparsity threshold and significance was reached if less than 5 percentile in the ORs distribution of the random networks.
We measured the Jaccard index of the hub regions of MT, MA, and MV and in each partition of VTAV, VTA, VTV, VAV, VTO, VAO, and VVO to see the overlap of the hub regions quantitatively. If A and B are the hub node sets from two networks, then the Jaccard index of the hub regions is defined as:
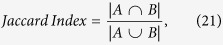 |
where 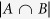 is the number of hub nodes that overlap in network A and B, and
is the number of hub nodes that overlap in network A and B, and 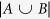 is the total number of hub nodes in the two networks. Note that the Jaccard index, ranging from 0 (no overlap) to 1 (perfect overlap), indicates how much overlap there is between the hub sets A and B.
is the total number of hub nodes in the two networks. Note that the Jaccard index, ranging from 0 (no overlap) to 1 (perfect overlap), indicates how much overlap there is between the hub sets A and B.
We also obtained bootstrap samples by selecting random samples with replacement from principal data (n = 251, 80% of 314 samples)2,3. MCNs were constructed and network parameters were computed from the bootstrap samples. This procedure was repeated 1000 times and analysis of variance (ANOVA) test was performed to compare the network parameters among the three MCNs.
To evaluate the difference in ORs between groups; (1) male and female, (2) young and old, (3) patients with dementia and old, we used a permutation test. Subgroups were randomly reassigned to one of two groups comprising the same number of subjects. MCNs were then reconstructed, the ORs were calculated at each sparsity range, and the between group difference in ORs was calculated for each permutation. This procedure was repeated 1000 times. The p value was then calculated as the proportion of entries in the corresponding permutation distribution that were greater than the observed group difference.
Replication analysis
To test the reproducibility of the results, we performed the same analyzing procedures aforementioned to an independent dataset of HCP data, which is composed of young adult subjects (age ranged from 20–35 years old).
Additional Information
How to cite this article: Yang, J.-J. et al. Complementary Characteristics of Correlation Patterns in Morphometric Correlation Networks of Cortical Thickness, Surface Area, and Gray Matter Volume. Sci. Rep. 6, 26682; doi: 10.1038/srep26682 (2016).
Supplementary Material
Acknowledgments
This research was supported by the Brain Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning (NRF-2014M3C7A1046050).
Footnotes
Author Contributions Conceived and designed the experiments: J.-J.Y., H.K. and J.-M.L. Performed the experiments: J.-J.Y. Analyzed the data: J.-J.Y. Contributed reagents/materials/analysis tools: J.-J.Y. and H.K. Wrote the paper: J.-J.Y. and J.-M.L. All authors reviewed the manuscript.
References
- Sanabria-Diaz G. et al. Surface area and cortical thickness descriptors reveal different attributes of the structural human brain networks. Neuroimage 50, 1497–1510, 10.1016/j.neuroimage.2010.01.028 (2010). [DOI] [PubMed] [Google Scholar]
- Anand A. Joshi, S. H. J. Moriah E. Thomason, Ivo Dinov & Arthur W. Toga Evaluation of connectivity measures and anatomical features for statistical brain networks. Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium on. 836–840, 10.1109/ISBI.2011.5872534 (2011). [DOI] [Google Scholar]
- Gong G., He Y., Chen Z. J. & Evans A. C. Convergence and divergence of thickness correlations with diffusion connections across the human cerebral cortex. Neuroimage 59, 1239–1248, 10.1016/j.neuroimage.2011.08.017 (2012). [DOI] [PubMed] [Google Scholar]
- Schmitt J. E. et al. A twin study of intracerebral volumetric relationships. Behavior genetics 40, 114–124, 10.1007/s10519-010-9332-6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J. E. et al. Identification of genetically mediated cortical networks: a multivariate study of pediatric twins and siblings. Cereb Cortex 18, 1737–1747, 10.1093/cercor/bhm211 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler L. T. et al. Genetic and environmental contributions to regional cortical surface area in humans: a magnetic resonance imaging twin study. Cereb Cortex 21, 2313–2321, 10.1093/cercor/bhr013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright I. C., Sham P., Murray R. M., Weinberger D. R. & Bullmore E. T. Genetic contributions to regional variability in human brain structure: methods and preliminary results. Neuroimage 17, 256–271 (2002). [DOI] [PubMed] [Google Scholar]
- Chen C. H. et al. Hierarchical genetic organization of human cortical surface area. Science 335, 1634–1636, 10.1126/science.1215330 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A., Giedd J. N. & Bullmore E. T. Imaging structural co-variance between human brain regions. Nat Rev Neurosci 14, 322–336, 10.1038/Nrn3465 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H. et al. Genetic topography of brain morphology. Proc Natl Acad Sci USA 110, 17089–17094, 10.1073/pnas.1308091110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechelli A., Friston K. J., Frackowiak R. S. & Price C. J. Structural covariance in the human cortex. Journal of Neuroscience 25, 8303–8310, 10.1523/Jneurosci.0357-05.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler A. M. et al. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage 53, 1135–1146, 10.1016/j.neuroimage.2009.12.028 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapellou O. et al. Abnormal cortical development after premature birth shown by altered allometric scaling of brain growth. Plos Med 3, 1382–1390, doi: ARTN e265 10.1371/journal.pmed.0030265 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimol L. M. et al. Sex-dependent association of common variants of microcephaly genes with brain structure. Proc Natl Acad Sci USA 107, 384–388, 10.1073/pnas.0908454107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini S. M. et al. Topological properties of large-scale structural brain networks in children with familial risk for reading difficulties. Neuroimage 71, 260–274, 10.1016/j.neuroimage.2013.01.013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Specification of Cerebral Cortical Areas. Science 241, 170–176, 10.1126/science.3291116 (1988). [DOI] [PubMed] [Google Scholar]
- Lerch J. P. et al. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. Neuroimage 31, 993–1003, 10.1016/j.neuroimage.2006.01.042 (2006). [DOI] [PubMed] [Google Scholar]
- He Y., Chen Z. J. & Evans A. C. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex 17, 2407–2419, 10.1093/cercor/bhl149 (2007). [DOI] [PubMed] [Google Scholar]
- Bernhardt B. C. et al. Mapping limbic network organization in temporal lobe epilepsy using morphometric correlations: Insights on the relation between mesiotemporal connectivity and cortical atrophy. Neuroimage 42, 515–524, 10.1016/j.neuroimage.2008.04.261 (2008). [DOI] [PubMed] [Google Scholar]
- Pezawas L. et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. Journal of Neuroscience 24, 10099–10102, 10.1523/Jneurosci.2680-04.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J., He Y., Rosa-Neto P., Gong G. & Evans A. C. Age-related alterations in the modular organization of structural cortical network by using cortical thickness from MRI. Neuroimage 56, 235–245, 10.1016/j.neuroimage.2011.01.010 (2011). [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N. et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15, 273–289, 10.1006/nimg.2001.0978 (2002). [DOI] [PubMed] [Google Scholar]
- Lerch J. P. & Evans A. C. Cortical thickness analysis examined through power analysis and a population simulation. Neuroimage 24, 163–173, 10.1016/j.neuroimage.2004.07.045 (2005). [DOI] [PubMed] [Google Scholar]
- Kabani N., Le Goualher G., MacDonald D. & Evans A. C. Measurement of cortical thickness using an automated 3-D algorithm: A validation study. Neuroimage 13, 375–380, 10.1006/nimg.2000.0652 (2001). [DOI] [PubMed] [Google Scholar]
- Lerch J. P. et al. Focal decline of cortical thickness in Alzheimer’s disease identified by computational neuroanatomy. Cereb Cortex 15, 995–1001, 10.1093/cercor/bhh200 (2005). [DOI] [PubMed] [Google Scholar]
- Lee J. K. et al. A novel quantitative cross-validation of different cortical surface reconstruction algorithms using MRI phantom. Neuroimage 31, 572–584, 10.1016/j.neuroimage.2005.12.044 (2006). [DOI] [PubMed] [Google Scholar]
- Singh V. et al. Spatial patterns of cortical thinning in mild cognitive impairment and Alzheimer’s disease. Brain 129, 2885–2893, 10.1093/Brain/Awl256 (2006). [DOI] [PubMed] [Google Scholar]
- Im K. et al. Brain size and cortical structure in the adult human brain. Cereb Cortex 18, 2181–2191, 10.1093/cercor/bhm244 (2008). [DOI] [PubMed] [Google Scholar]
- Robbins S., Evans A. C., Collins D. L. & Whitesides S. Tuning and comparing spatial normalization methods. Medical image analysis 8, 311–323, 10.1016/j.media.2004.06.009 (2004). [DOI] [PubMed] [Google Scholar]
- Lyttelton O., Boucher M., Robbins S. & Evans A. An unbiased iterative group registration template for cortical surface analysis. Neuroimage 34, 1535–1544, 10.1016/j.neuroimage.2006.10.041 (2007). [DOI] [PubMed] [Google Scholar]
- Rubinov M. & Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069, 10.1016/j.neuroimage.2009.10.003 (2010). [DOI] [PubMed] [Google Scholar]
- Bullmore E. & Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci 10, 186–198, 10.1038/nrn2575 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.