Abstract
Many proteins of the outer membrane of Gram-negative bacteria and of the outer envelope of the endosymbiotically derived organelles mitochondria and plastids have a β-barrel fold. Their insertion is assisted by membrane proteins of the Omp85-TpsB superfamily. These proteins are composed of a C-terminal β-barrel and a different number of N-terminal POTRA domains, three in the case of cyanobacterial Omp85. Based on structural studies of Omp85 proteins, including the five POTRA-domain-containing BamA protein of Escherichia coli, it is predicted that anaP2 and anaP3 bear a fixed orientation, whereas anaP1 and anaP2 are connected via a flexible hinge. We challenged this proposal by investigating the conformational space of the N-terminal POTRA domains of Omp85 from the cyanobacterium Anabaena sp. PCC 7120 using pulsed electron-electron double resonance (PELDOR, or DEER) spectroscopy. The pronounced dipolar oscillations observed for most of the double spin-labeled positions indicate a rather rigid orientation of the POTRA domains in frozen liquid solution. Based on the PELDOR distance data, structure refinement of the POTRA domains was performed taking two different approaches: 1) treating the individual POTRA domains as rigid bodies; and 2) using an all-atom refinement of the structure. Both refinement approaches yielded ensembles of model structures that are more restricted compared to the conformational ensemble obtained by molecular dynamics simulations, with only a slightly different orientation of N-terminal POTRA domains anaP1 and anaP2 compared with the x-ray structure. The results are discussed in the context of the native environment of the POTRA domains in the periplasm.
Introduction
The central components of the protein complexes facilitating protein insertion into or translocation across the outer membrane (OM) of Gram-negative bacteria, mitochondria, and chloroplasts are members of the Omp85 superfamily (1). Omp85 proteins are composed of a transmembrane, pore-forming β-barrel at their C-terminus with two characteristic motifs (2) and a varying number of polypeptide-transport-associated (POTRA) domains in their N-terminal region (3, 4). Although the β-barrel domain follows the evolutionary path in its divergence, phylogenetic analysis of the POTRA domains revealed a functional classification (5). POTRA domains of Omp85 proteins have been implicated in complex assembly and recognition of substrate proteins (6).
In Escherichia coli, BamA is the central component of the β-barrel assembly machinery (BAM) complex that performs membrane insertion of OM proteins (OMPs) (7). It consists of the characteristic C-terminal β-barrel and five POTRA domains at the N-terminus. In addition to BamA, the BAM complex is composed of several lipoproteins (BamB/C/D/E (7, 8, 9, 10)). It is described that the third POTRA domain of BamA binds BamB (11) and thus is involved in complex formation. In turn, the first, and thus most N-terminal, POTRA domain is an essential interaction partner of SurA, which has been proposed to deliver β-barrel proteins from the plasma membrane to the BAM complex (12). In line with this, NMR titration experiments revealed that ecP1 (for POTRA 1 of the E. coli protein BamA) and ecP2 interact with peptides comprising transmembrane β-strands of PhoE, which itself is a substrate of the BAM complex (13).
Omp85 proteins in cyanobacteria were discovered first in Synechocystis sp. PCC 6803 (14) and in Anabaena sp. PCC 7120 (hereafter, Anabaena sp. (15)). They are considered to be ancestors of the translocation pore of the translocase of the outer envelope membrane of the chloroplast (TOC (16)). Although Anabaena sp. contains three open reading frames coding for distinct Omp85 proteins (17), the one encoded by alr2269 (hereafter referred to as anaOmp85) is considered the most abundant Omp85 protein in the outer membrane (18) and is the subject of this study. anaOmp85 contains three POTRA domains (anaP1, anaP2, and anaP3), which were found to regulate the pore gating of the β-barrel (5, 15, 19). In contrast to BamA, anaOmp85 contains an N-terminal proline-rich region of ∼200 amino acids that precedes the POTRA domains and for which structural information does not exist (19).
The importance of the POTRA domains in the eukaryotic systems remains under debate. A mutant of the Omp85 protein in the sorting and assembly machinery (SAM, also referred to as topogenesis of mitochondrial outer membrane β-barrel proteins (20)), Sam50, which lacks the POTRA domain, is viable (21), although it is said that this domain is involved in substrate release from the SAM complex (22). Similarly, the POTRA domain of Omp85 in the TOC complex, Toc75, recognizes the transit peptide of the chloroplast proteins that have to be translocated across the outer envelope and also interacts with another complex component, the receptor Toc34 (15). However, in Arabidopsis thaliana, two Omp85 homologs without the POTRA domain have been described experimentally (23, 24), which is in line with the notion that Omp85 with either alternative or without soluble domains has evolved to perform alternative functions (25). POTRA domains might function as a hub for interaction with other complex partners, which might be absent in the case of the Omp85 proteins without POTRA domains. In support of this suggestion, a POTRA domain is also found in the cell-division protein FtsQ, which is not involved in the translocation of other proteins, and the POTRA domain is essential for FtsQ positioning by interaction with a complex partner (26).
POTRA domains have a characteristic β-α-α-β-β fold, where the two helices are packed from the same side against the three-stranded β-sheet (19, 22, 27, 28, 29). The length of the linker and the interface between two adjacent POTRA domains determines their angular range of motion relative to each other. This flexibility is thought to be of functional relevance. The POTRA domains of BamA provide a scaffold for assembling the BAM complex, which implies that the orientation of the POTRA domains relative to each other is an important factor. This was also shown to be the case for FhaC, as a two-amino-acid insertion between its two POTRA domains reduced substrate recognition (30).
The published x-ray structures of the POTRA domains of BamA revealed a fishhook-like arrangement of the POTRA domains, with a kink between ecP2 and ecP3 (29), as well as an extended conformation of the domains (28). As a result, a conformational change upon substrate recognition was proposed for the hinge region in proteobacterial Omp85 proteins. However, the conformational exchange between ecP2 and ecP3 in full-length BamA in lipid bilayers, if it exists at all, is slow on the timescale of solid-state NMR (31). A conformational change similar to that in BamA x-ray structures was proposed for full-length FhaC by electron paramagnetic resonance (EPR) spectroscopy (32). Analysis of the orientation and conformational flexibility of the POTRA domains of BamA by solution and solid-state NMR, EPR, or small-angle x-ray scattering measurements revealed a rigid connection between ecP1 and ecP2 with a reorientation at timescales not detectable by these experimental methods, especially in lipid bilayers (13, 31, 33, 34). Similarly, ecP4-ecP5 has a stable/rigid conformation in lipid bilayers as well as in detergent (31, 35, 36), which is similar to the x-ray structure of those domains.
Sequence information in connection with existing results on BamA led to the proposal that at least the last two C-terminal POTRA domains of anaOmp85 (anaP2 and anaP3) form a stable hub, whereas the preceding POTRA domain (anaP1) might be more flexibly connected (1). To experimentally challenge this interpretation, we analyzed the anaOmp85-POTRA domains by pulsed electron-electron double resonance (PELDOR, also called DEER) spectroscopy (37, 38, 39). PELDOR is a method for measuring long-range distances and their distribution in spin-labeled macromolecules. Thus, PELDOR measurements can provide valuable information on conformational changes (40, 41) as well as wide-range distance restraints (1.8 nm to 6–10 nm in deuterated samples) for structural modeling (33, 42, 43, 44, 45, 46). The spin label (SL) utilized, MTSSL ((1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate)), possesses an intrinsic linker flexibility, which needs to be taken into account when a structure is refined or modeled based on PELDOR distance constraints. Several approaches to describe the intrinsic flexibility of the SL, such as rotamer libraries, as well as molecular dynamics (MD) simulations, have been introduced to enhance the reliability of structural modeling (46, 47, 48). We used site-directed spin labeling (SDSL) to investigate the relative orientation and conformational flexibility between POTRA domains of anaOmp85 by PELDOR spectroscopy using 27 double cysteine mutants including intra- and interdomain distance constraints. We compare our experimental results to the available SL rotamer libraries, as well as MD simulations, to account for the SL linker flexibility. The difficulties arising from ambiguities in rotamer libraries are evaluated and discussed in the context of structure refinement using our experimental PELDOR constraints. Despite these ambiguities in rotamer prediction, structure refinement using Rosetta revealed a rather restricted conformational ensemble of the POTRA domains in frozen liquid solution compared to the same orientation between anaP2 and anaP3, and only a slightly different orientation of anaOmp85 POTRA domains anaP1 and anaP2, as observed in the x-ray structure.
Materials and Methods
Sample preparation and purification
Surface-exposed residues were substituted by cysteine via Quick-change polymerase chain reaction on a construct containing amino acids 161–467 of anaOmp85, as described in (19) (for oligonucleotides, see Table S1 in the Supporting Material). Transformed E. coli BL21 DE3 (Invitrogen, Carlsbad, CA) were inoculated in lysogeny broth at 37°C and expression was induced by addition of 1 mM isopropyl 1-thio-β-d-galacto-pyranoside at A600 = 0.8. Cells were harvested after 4 h incubation, resuspended in lysis buffer (25 mM HEPES/KOH, pH 7.0, 250 mM NaCl, and 5 mM MgCl2), and lysed via sonication. After 30 min centrifugation at 25,000 × g, the proteins were immobilized on Ni-NTA (Qiagen, Hilden, Germany) and washed with wash buffer (25 mM HEPES/KOH, pH 7.0, 250 mM NaCl, 5 mM MgCl2, and 10 mM imidazole). The protein was labeled with MTSSL (Enzo Biochem, New York, NY) in wash buffer at 4°C overnight. After extensively washing, the labeled proteins were eluted with elution buffer (25 mM HEPES/KOH, pH 7.0, 250 mM NaCl, and 500 mM imidazole). Protein concentration was determined using Bradford reagent (Bio-Rad, Hercules, CA) and adjusted to 100 μM in PELDOR buffer (25 mM HEPES/KOH, pH 7.0, 250 mM NaCl, 500 mM imidazole, and 30% glycerol). X-band samples (180 μL or 40 μL with 100 μM protein concentration) were transferred to standard 4-mm or 2.8-mm-diameter quartz EPR tubes (Wilmad-Labglass, Vineland, NJ). The samples were shock-frozen in liquid nitrogen. The Q-band samples (10 μL with 100 μM protein concentration) were transferred to a 1.6-mm-diameter suprasil EPR tube (Wilmad).
PELDOR data collection and data analysis
Pulsed EPR data were measured on an Elexsys E580 EPR spectrometer (Bruker, Billerica, MA) equipped with a Bruker PELDOR unit (E580-400U). Temperature was kept at 50 K with a continuous-flow helium cryostat (CF935) and temperature control system (ITC 502), both from Oxford Instruments (Abingdon, UK). For PELDOR experiments, the dead-time free four-pulse sequence was used (49). For measurements at X-Band frequencies (9.6 GHz), a 1 kW TWT amplifier (ASE 117x) and a dielectric ring resonator or split ring resonator (MD5 W1 or MS3) were used. The pulse lengths were 32 ns (π/2 and π) for the probe pulses and 12 ns for the pump pulse. The delay between the first and second probe pulses was varied between 136 and 192 or 200 and 256 ns in 8 ns steps (protonated samples), and between 456 and 848 ns in 56 ns steps (for buffer-deuterated sample I292C-V460C) to reduce contributions from nuclear modulations (50). The pulse separation between the second and third probe pulses was between 1.2 and 7.3 μs, depending on the probed distances and transversal relaxation time (T2) of the samples. The shot repetition time was 4–5 ms. The frequency of the pump pulse was set to the resonance frequency of the over-coupled resonator (Q ∼50) and the magnetic field was such that the excitation coincided with the maximum of the nitroxide powder spectrum to obtain optimum pumping efficiency. The probe frequency was chosen to be 70 MHz higher (80 MHz for the A319C-E344C sample). Possible SL orientational effects on the distance determination were probed for I292C-E344C, I292C-V370C, N265C-A319C, Q429C-A319C, Q429C-E344C, and V460C-E344C by experiments using a 40 MHz offset between pump and probe frequency. No changes in the PELDOR time traces were observed, so SL orientational contributions could be excluded (data not shown). For measurements at Q-band frequencies (33 GHz) experiments were performed with the Elexsys SuperQ-FT accessory, with a 10 W AmpQ amplifier (Bruker) and an EN5107D2 resonator. The pulse lengths were 32 ns (π/2 and π) for the probe pulses and 20 ns for the pump pulse. The delay between the first and second probe pulses has not been varied due to the negligible effect of nuclear modulations for protons at this frequency band (51). The pulse separation between the second and third probe pulses was between 3 and 4 μs, depending on the probed distances and transversal relaxation time (T2) of the samples. The shot repetition time was 4.5 ms. The pump pulse frequency was set to the resonance frequency of the over-coupled resonator (Q ∼300) and the magnetic field was such that the excitation coincides with the maximum of the nitroxide powder spectrum to obtain optimum pumping efficiency. The probe frequency was chosen 70 MHz lower. Data analysis was performed using the DeerAnalysis software package (52) to obtain the distance distributions between the spin pairs of the double-labeled mutants. The PELDOR time traces were corrected for background decay using a homogeneous three-dimensional spin distribution.
Prediction of interspin distances and intramolecular dipolar evolution functions
In silico spin-labeling of the protein structure (PDB: 3MC8) was used to predict the interspin distances and intramolecular dipolar evolution functions for all SL pairs. Hereby, different rotamer libraries to model the conformational flexibility of the MTSSL were used for the predictions and compared to the experimental results. For rotamer libraries from multiscale modeling of macromolecular systems (MMM) (47) (175 K and 298 K), prediction of distance distributions and intramolecular dipolar evolution function were directly performed in MMM. Distance distributions from MD simulation trajectories were extracted using every 40th frame by calculating the distance between geometric midpoints of N1 and O1 atoms of SLs. Intramolecular dipolar evolution functions for distance distributions were derived from rotamer libraries of mtsslWizard (53), using thorough search and loose vdW restraints (cutoff 2.5 Å, five clashes allowed), and MD simulations were simulated in a home-written MATLAB script.
Structure refinement
Structural refinement of the x-ray structure was done using two approaches, a rigid-body-motion approach using a home-written MATLAB script, and a more structure-based approach. In the first approach, the N-O midpoint coordinates of each SL position derived from rotamer libraries described above were fixed to the respective POTRA domain (rigid-body assumption). Then a displacement (x, y, and z) and rotation (α, β, and γ) of domains anaP1 and anaP3 with respect to anaP2 were allowed to minimize the RMSD between the experimental and predicted distance distribution function. Typically, 20,000 minimizations (with different starting conditions) were performed, and the 100 best solutions were stored. The linker regions between the domains anaP1, anaP2, and anaP3 of the best solutions were rebuilt using Yasara. For the second approach, a YASARA (54) Python script was developed to screen the relative orientational space of the POTRA domains (with respect to the twist and swing angles) utilizing Rosetta (www.rosettacommons.org, release 2015.05.57576). The angular space, which was explored in the MD simulations (anaP1-anaP2: twist 0–160°, swing 0–120°; anaP2-anaP3: twist 10–105°, swing 0–75°), was screened in steps of 18°. For a total of 143 angle combinations, the corresponding structure was extracted from the trajectories of the MD simulations. In case a structure with the required angle combination was not available, two structures were extracted from the trajectories, i.e., one with the required anaP1-anaP2 angles and another that fulfilled the requirements for anaP2-anaP3. These two structures were then further processed and joined to form a single structure whose angles correspond to the requested angle combination for anaP1-anaP2, as well as anaP2-anaP3. Any missing or superfluous SLs in these structures were introduced or removed. The above-selected structures were subjected to Rosetta relax protocol (55), creating 500 models for each input structure (143 × 500 ≈ 72,000 Rosetta models) and enforcing distance constraints to ensure that the pairwise SL distances lie within the range of the experimentally determined distance distributions. Briefly, the better SL pairs match the distances associated with the highest normalized frequencies, the larger their contribution to the total Rosetta score of a model. A virtual atom was introduced at the geometric center of the N1 and O1 atoms of each SL by extending the R1A parameter file included in Rosetta. Distance constraints of the type “AtomPair” were defined between these virtual atoms of SL pairs, and the experimentally determined SL-pair distance distributions were used as SPLINE functions for constraint scoring. The frequencies of each distance distribution were normalized to (0, 1); they were multiplied by −1 to fit into Rosetta’s scoring scheme. The constraint weight was set to 4, which leads to optimal results with RosettaEPR (45). It employs a related approach for refining structures with EPR/PELDOR data.
MD simulations: wild-type
The rotational freedom of the POTRA domains of anaOmp85 was explored by replica-exchange MD (REMD) across 128 replicas in the temperature range from 300 to 405 K. A webserver (http://folding.bmc.uu.se/remd/ (56)) was used to calculate the exponential temperature distribution of the replicas with an exchange probability of 0.2. The 20 ns REMD simulation (20 ns × 128 replicas = 2560 ns) using the AMBER03 force field (57) was performed with Gromacs 5.0.4. A rhombic dodecahedron was selected as the shape of the simulation box, with the protein being at least 18 Å away from the periodic boundaries. A 5 fs time step was enabled by converting hydrogen atoms to virtual sites (pdb2gmx option –vsite hydrogens). The Verlet cutoff scheme was used for neighbor searching (58). The short-range cutoff was 1.4 nm. Long-range electrostatics were calculated with particle-mesh Ewald (PME) (59). The PME order was 4 and the Fourier spacing 0.16 nm. Two groups (protein and water/ions) were used for temperature coupling with a time constant of 0.1 ps using the V-rescale thermostat (60). Isotropic pressure coupling was performed with a Parrinello-Rahman barostat (61, 62) using a time constant of 2.0 ps to keep the pressure at 1.0 bar. The 1 μs wild-type MD simulation of anaOmp85 POTRA domains used the same parameters as described for the REMD setup, but the temperature was set to 300 K.
MD simulations: spin-labeled mutants
For spin-labeled mutants, MTSSL was modeled, quantum-mechanically optimized with YASARA (63), and introduced into the crystal structure of the anaOmp85-POTRA domains (PDB: 3MC8) at appropriate sites. In total, five sets of mutants were constructed, which carry MTSSLs at the following positions in anaP1 and anaP3: 1) Q259, I292, Q429, and V460; 2) N265 and Q429; 3) Q259, I292, and L448; 4) Q259, I292, and V457; and 5) G233 and V460. For each set of mutants, three MD simulations were performed, where anaP2 carries MTSSL at residue A319, E344, or V370, respectively. The SL combinations of these mutants minimize the number of required simulations as well as the putative contact between the SLs. MD simulations were performed with YASARA using the AMBER03 force field (57). The structures were put in cubic simulation boxes of 128 Å side length with periodic boundaries. The three POTRA domains of the initial (x-ray) conformation of anaOmp85 span ∼9 nm; also, a fully elongated (∼10.5 nm) structure can be accommodated in the box. Placement of water and ions, as well as pKa prediction of protonatable groups of the protein, were performed with YASARA’s neutralization experiment (64, 65). The van der Waals interactions were cut off at 10.48 Å and Coulomb interactions were calculated with PME (59). Proteins were simulated for 100 ns at 298 K.
Results
The N-terminal anaOmp85-POTRA-domain-containing region is monomeric in the absence of the β-barrel and other interaction partners
To verify the monomeric state of the recombinantly expressed anaOmp85-POTRA domains, we probed for the potential presence of POTRA domain dimers using a single-Cys mutant in domain anaP2 (V370C). A monotonously decaying background due to random intermolecular interactions was detected (Fig. S1 (39)). Calculation of the spin concentration from this decay function, assuming an intermolecular interaction between randomly distributed proteins, resulted in a value of 86 μmol/L, very similar to the spin concentration of 103 μmol/L determined by continuous wave (CW) EPR. This proves the monomeric behavior of the domains under the experimental conditions. In addition, different cryoprotectants and freezing procedures had no impact on PELDOR results probed by the I292C-V370C double mutant (Fig. S2).
Structure of individual anaOmp85-POTRA domains in frozen solution compares well to the x-ray structure
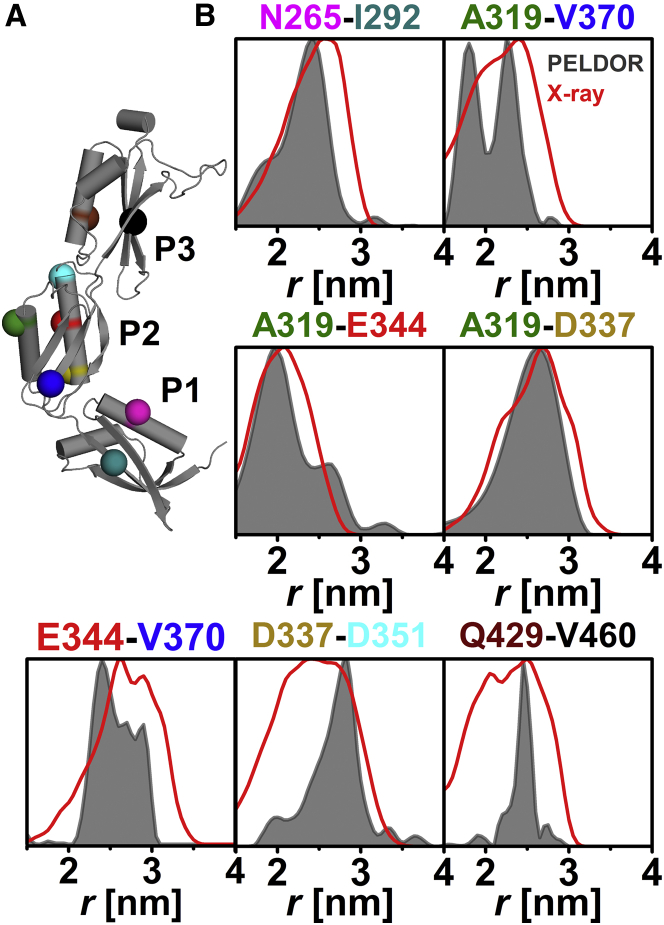
Seven intradomain PELDOR constraints (Fig. 1 A) located within regular α-helix or β-sheet elements of the POTRA motif (β-α-α-β-β) were used to probe the structural integrity of individual POTRA domains in the absence of substrate and possible interaction partners. We observed well-defined oscillations that allow a clear differentiation between intra- and intermolecular dipolar interactions (Fig. S3), allowing the extraction of well-resolved distance distributions by Tikhonov regularization (Fig. 1 B; Table S2). We applied in silico modeling of the internal conformational flexibility of the MTSSL using rotamer libraries obtained from different programs (47, 53) to simulate distance distributions and intramolecular dipolar evolution functions to compare our results to the available x-ray structure. Comparison of the experimental distance distributions with predicted distributions based on the x-ray structure (19), using MMM in 298 K mode, shows that experimental and predicted mean distances are in good agreement (Table S2). This is true for all other rotamer libraries compared in this study (see Materials and Methods, Figs. S4 and S5, and Tables S2 and S3 (48, 66)). Analysis of the root mean-square fluctuation between MD simulations and x-ray structure for the individual POTRA domains showed that large fluctuations within each domain are mostly restricted to loop regions (Fig. S6). Fluctuations in anaP3 are primarily caused by the presence of an elongated loop in this domain (Fig. S7 and Table S4 (1)).
Figure 1.
Analysis of the intradomain distances in anaOmp85-POTRA domains anaP1, anaP2, and anaP3. (A) The crystal structure of the three POTRA domains, indicating the spin-labeled residues. (B) The obtained intradomain experimental distance distributions (gray) compared with distance distributions generated on the crystal structure by MMM in 298 K mode (red).
Especially for SL pairs N265C-I292C, A319C-E344C, and A319C-D337C, the predicted distances and widths of the distributions are in good agreement with the experimental results (Fig. 1 B). This indicates that the rotamer libraries for these positions are able to reflect the true distance and flexibility of the SLs in these pairs with high precision (Figs. 1 B and S5). However, the anaP3 (Q429C-V460C), as well as the anaP2 (D337C-D351C), label pair (Fig. 1 B) exhibits experimental distance distributions much narrower than the predictions by any of the applied rotamer libraries (Fig. S5). In addition, we observe distance distributions of similar width but different shape compared with the predictions for SL pairs related to residue 370 in anaP2 (Fig. 1 B). The rigidity of the SL at this position was also observed in liquid solution via CW EPR (Fig. S1).
All these aforementioned deviations of experiment from prediction are most likely due to a more confined rotamer flexibility of at least one partner in an SL pair within the experiment. Thus, it can be concluded that the architecture of the individual domains in frozen solution is very similar to the x-ray structure, allowing the assumption that the individual domains can be treated as rigid bodies.
Interdomain distance constraints suggest a confined relative orientation of the anaOmp85-POTRA domains
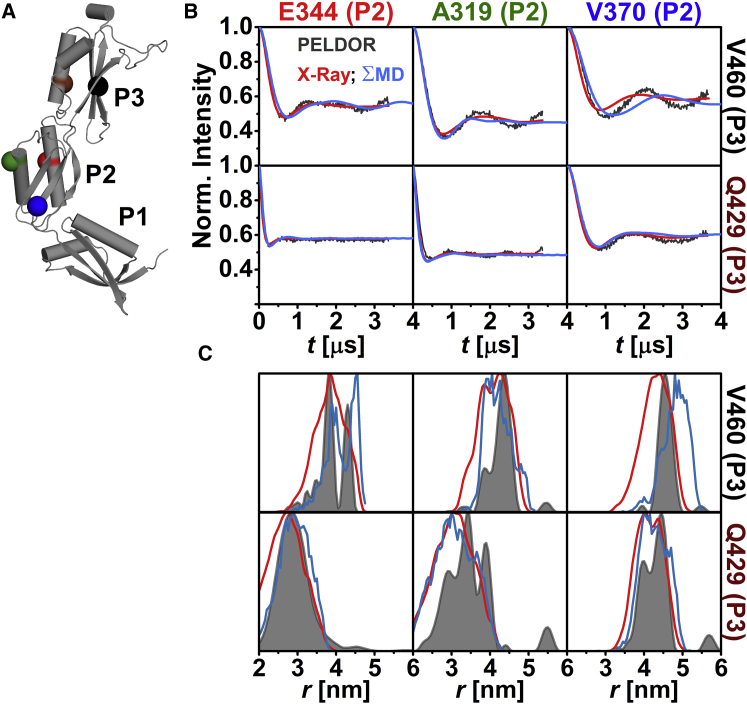
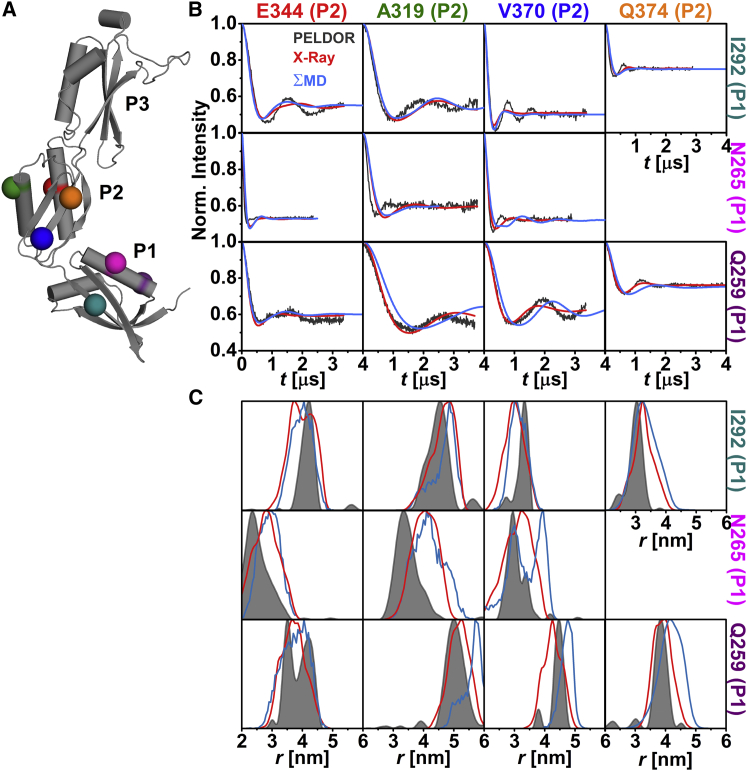
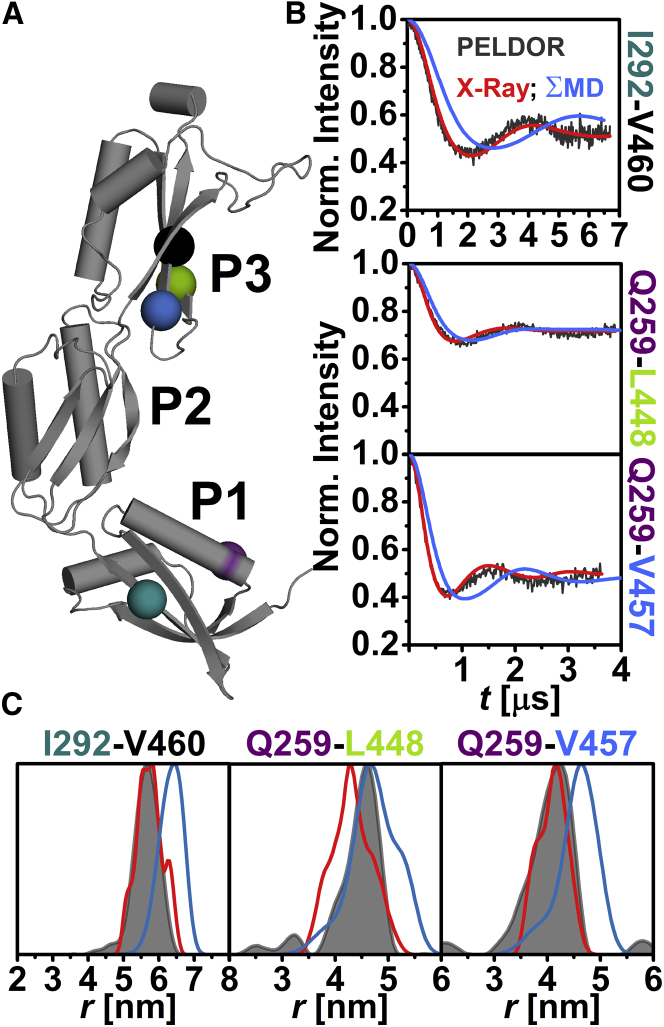
The relative orientation of the individual domains with respect to each other in frozen-solution samples was investigated by PELDOR spectroscopy using 20 interdomain distance constraints (Figs. 2 A, 3 A, and 4 A). These constraints were selected such that a triangulation of a position in P1 and P3 with a position in P2 was possible. The majority of the interdomain SL pairs exhibit pronounced dipolar oscillations (Figs. 2 B, 3 B, 4 B, and S8), leading to well-resolved interspin distances (Figs. 2 C, 3 C, and 4 C; Table S2) ranging from ∼2.4 nm (N265C-E344C (Fig. 3 C)) to ∼5.7 nm (I292C-V460C (Fig. 4 C)). Especially distance distributions related to residues V370 or Q374 are narrow, indicating a fairly defined domain-domain orientation for anaP1 and anaP2 as well as anaP2 and anaP3. Distance distributions obtained for different rotamer libraries and temperatures show comparable predictions (Figs. S9, S11, and S13); however, comparison to the experimental distances and distributions reveals discrepancies especially in the width of the predicted distributions, whereas the mean distances are close to those determined experimentally. The discrepancies in distribution width are most likely due to a smaller inherent flexibility of the SL in frozen liquid solution compared to the flexibility predicted by the different rotamer libraries.
Figure 2.
Analysis of the distances between anaOmp85-POTRA domains anaP2 and anaP3. (A) The crystal structure of the three POTRA domains, indicating the spin-labeled residues. (B) Comparison of background-corrected PELDOR time traces for measurements between domains anaP2 and anaP3 (gray) with predicted intramolecular dipolar evolution functions for MD (blue) and x-ray (red; MMM 298 K). (C) Comparison of distance distributions for interdomain SL pairs between anaP2 and anaP3 obtained by PELDOR (gray), MD (blue), and x-ray (red; MMM 298 K).
Figure 3.
Analysis of the distances between anaOmp85-POTRA domains anaP1 and anaP2. (A) The crystal structure of the three POTRA domains, indicating the spin-labeled residues. (B) Comparison of background-corrected PELDOR time traces for measurements between domains anaP1 and anaP2 (gray) with simulated intramolecular dipolar evolution functions for MD (blue) and x-ray (red; MMM 298 K). (C) Comparison of distance distributions for interdomain SL pairs between anaP1 and anaP2 obtained by PELDOR (gray), MD (blue), and x-ray (red; MMM 298 K).
Figure 4.
Analysis of the distances between anaOmp85-POTRA domains anaP1 and anaP3. (A) The crystal structure of the three POTRA domains, indicating the spin-labeled residues. (B) Comparison of background-corrected PELDOR time traces for measurements between domains anaP1 and anaP3 (gray) with simulated intramolecular dipolar evolution functions for MD (blue) and x-ray (red; MMM 298 K). (C) Comparison of distance distributions for interdomain SL pairs between anaP1 and anaP3 obtained by PELDOR (gray), MD (blue), and x-ray (red; MMM 298 K).
Comparison of the experimental intramolecular dipolar evolution functions observed between anaP2 and anaP3 with the predictions from x-ray (MMM 298 K) reveals that V370C-V460C exhibits stronger intramolecular dipolar oscillation and therefore a narrower experimental distance distribution than predicted by in silico labeling (Figs. 2 and S9–S14). E344C-V460C yields a bimodal experimental distance distribution. These observations are in line with our intradomain results, where distances involving residues 370 or 460 showed deviations between experiment and rotamer library prediction, emphasizing that the SL flexibility is strongly hindered at these positions. Apart from the rotamer-library-based predictions, distance distributions were extracted from MD simulations on spin-labeled POTRA domains of anaOmp85 by in silico labeling with MTSSL at the desired positions in the x-ray structure. These modified structures were simulated for a total of 1.9 μs with YASARA using the AMBER03 force field, which was validated by a 1 μs wild-type MD simulation (see Materials and Methods).
The extracted distance distributions are slightly broadened compared to the experimental result, leading to an increased dampening of the simulated intramolecular dipolar evolution functions of the SL MD simulations. This indicates a more constrained conformational flexibility of the domains in frozen solution than predicted by the MD simulations (Fig. 2). In the case of E344C-V460C, the MD simulations better reflect the shape of the experimental distance distributions than predictions based on the x-ray structure (Fig. 2 C). A reason for this might be that rotamer libraries are calculated based on the static crystal structure, whereas MD simulations allow us to account for backbone fluctuations and side-chain rearrangements of the protein. However, this improvement in rotamer prediction is not observed as a general trend (see, e.g., SL pair Q259C-E344C (Fig. 3 C)). Altogether our PELDOR results for SL pairs between anaP2 and anaP3 reveal a rigid conformation of these domains, which is comparable to the x-ray structure. At first glance, this seems to be the case for anaP1 and anaP2 as well (Fig. 3).
In-depth inspection revealed that the predicted time traces not only exhibit a stronger damping, but also some shift in dipolar coupling frequency (Fig. 3 B), leading to some deviations between experimental and predicted mean distances, especially for SL pairs N265C-E344C and N265C-A319C (Fig. 3 C), as well as width of the distribution (e.g., SL pair I292C-V370V (Fig. 3)). Since residue 265 was already probed in the intradomain experiments, where reasonable agreement was observed between experiment and prediction, we suggest that the alterations in mean distance might be due to a slightly different orientation of the domains, rather than rotamer inaccuracy (Fig. 1). Interestingly, this was not confirmed by the MD simulations on spin-labeled domains, which reveal distance distributions similar to those predicted on the basis of the x-ray structure (Fig. 3 C). This is most likely a freezing effect that results in a smaller selection of rotamer states of the SL. In the case of residue 370, limited flexibility of the SL side chain was observed by CW EPR as well as intradomain constraints. Thus, the width of the related distance distributions is much narrower than the rotamer library predictions (Figs. 1 and S5). Here, MD simulations report a bimodal distance distribution of the SL pair N265-V370. This might be a hint that also the flexibility of the SL at position 265 is influenced by backbone fluctuations and side-chain rearrangements of the protein. Taken together, a slightly different domain-domain orientation as well as a more confined rotamer ensemble compared to that predicted on the basis of the x-ray structure is observed in frozen solution. For the three constraints between anaP1 and anaP3 (Fig. 4 A), reasonable agreement between experimental results and simulations on the x-ray structure is observed (Fig. 4, B and C). Only the mean distance for SL pair Q259C-L448C is slightly longer than that predicted on the x-ray structure. However, this seems to be the cause of population differences of the rotamers at these positions. Interestingly, the distance distributions extracted from our MD simulations on spin-labeled domains show differences in mean distances for SL pairs Q259C-V457C and I292C-V460 (Fig. 4 C). These deviations might be due to a lower sampling of these distances within the MD simulations.
PELDOR in frozen solution yields a restricted conformational ensemble
To unravel the orientation of anaP1 and anaP3 with respect to anaP2 in frozen solution we made use of two structure refinement approaches considering the high-quality PELDOR distance constraints (see Materials and Methods). For the rigid-body approach, the orientation of the domains was varied such that the RMSD between the experimental and simulated distance distributions of pairs of rotamer bundles was minimized, whereas for the Rosetta refinement, the optimal orientation of the domains, as well as pairs of single rotamers, was optimized, using the best match to the experimentally most populated distances. Comparison of the refined distance distributions for the best rigid-body refinement reveals that especially distances for SL pairs N265-A319 and N265-E344 are shifted to match the experimental distribution (Figs. S15 and S16). The width of the simulated distributions is mainly unchanged, especially for distance distributions related to position V370, preserving the increased dampening of the dipolar oscillations of the time domain signal (Fig. S15). This overestimation of the SL flexibility by the rotamer library leads to several solutions giving similar good RMSDs between experimental and simulated distance distributions. Thus, this approach is strongly dependent on the quality of the rotamer library prediction.
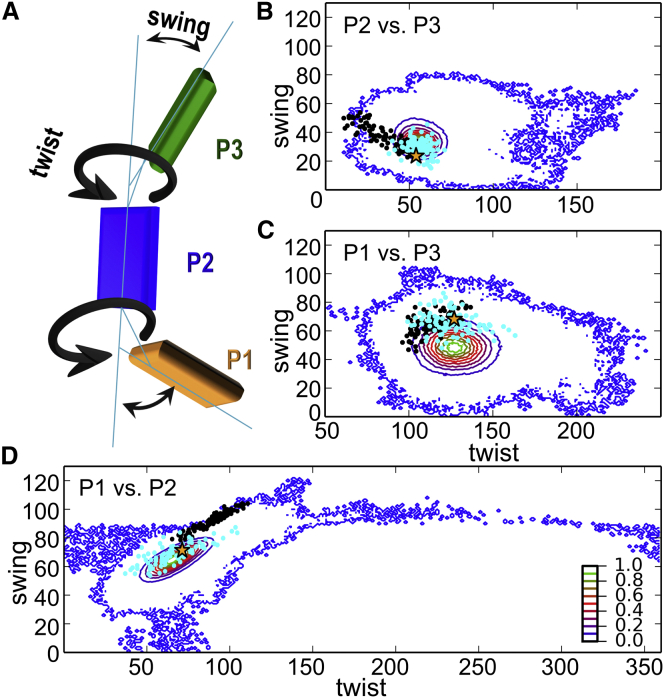
The best Rosetta refined model gives an almost complete agreement (∼98%) with the most probable distances of the experimental distributions by selecting a single rotamer for each position, which fulfills all distance constraints at once (Fig. S16). The precision of the underlying rotamer library, as well as the increased degrees of freedom due to full protein flexibility, allow several models with similar good agreement with the experimental results in frozen solution. Ensembles of the top 100 models of both refinement strategies were selected. In terms of rigid-body refinement, all models have an RMSD variation that is below the error of Tikhonov regularization. For Rosetta refinement, these models fall into a score range of ∼0.6% (Fig. 5; Table S5). To compare the models in each ensemble, the twist and swing angles between adjacent domains were used to describe the relative orientation of the domains with respect to each other (Fig. 5). The twist angle describes a rotational motion about the main axis of the respective domain anaP1 or anaP3 (Fig. 5 A). The swing angle describes a bending of either anaP1 or anaP3 from the main axis of anaP2 (Fig. 5 A). The small angular spread observed for the anaP2-anaP3 pair (40° and 30° for the rigid-body and Rosetta refinements, respectively) is in line with the well-defined oscillations of the dipolar evolution functions obtained for these constraints (Fig. 2), indicating that the observed inaccuracy in orientation prediction is mostly due to the inaccuracy in SL flexibility prediction by the rotamer libraries in both approaches (Fig. 5 B). Nonetheless, the orientation of this domain pair in the x-ray structure resembles the putative orientation in frozen solution (Fig. 5 B). In case of anaP1 and anaP2, we observed an elliptic distribution of angles with a small swing angle spread of 30° and 40° and a larger twist angle spread of 50° and 70° for the rigid-body and Rosetta refinements, respectively (Fig. 5 D). However, the large twist-angle spread, as well as the deviation of the centroids (rigid body: twist 90°, swing 92°; Rosetta: twist 66°, swing 69°) is in contradiction to the observed pronounced dipolar oscillations for constraints of this domain pair (Fig. 3 B). The relative orientations obtained for anaP1 and anaP3 show a comparable spread of twist/swing angles for both strategies, which is similar to the one observed for the domain pair anaP2 and anaP3. As concluded for anaP2 and anaP3, the relative orientation of anaP1 and anaP3 present in the x-ray structure bears a close resemblance to the model ensembles (Fig. 5).
Figure 5.
Comparison of the orientational space of spin-labeled anaOmp85-POTRA domains obtained by REMD simulations by PELDOR distance constraints. The density maps (10 contour levels) indicate the relative population of the orientational space between adjacent POTRA domains. (A) Cartoon representation of the POTRA domains, indicating the twist and swing angles describing the relative orientation of the domains. (B–D) Comparisons of anaP2 and anaP3 (B), anaP1 and anaP3 (C), and anaP1 and anaP2 (D) based on REMD simulations. In all plots, the asterisk indicates the conformation of the respective domains in the crystal structure. The refined structures obtained by rigid-body refinement (black dots) and Rosetta (cyan dots) are indicated.
To check whether the orientational space of the POTRA domains of anaOmp85 was sufficiently scanned during Rosetta refinement (Fig. S17), the total possible orientational space was estimated by REMD simulations (Fig. 5) in addition to the MD simulations on spin-labeled domains presented above. All MD simulations show a large overlap of the orientational space for all domain pairs (Figs. 5, B–D, and S17). This is also the case for the previously published MD simulations (19); however, the simulations presented in this study enable a more comprehensive view of the conformational space of anaOmp85 POTRA domains due to their multitude, time length, and an enhanced sampling method. The orientational ensemble of anaP3 with respect to anaP2 describes a circular space with a diameter of ∼30° for the most populated conformations and a larger area with a diameter of ∼90° in the case of low-populated structures (Fig. 5 B). The refined models of both strategies are located within the conformational space of the REMD simulations. Especially the models found by Rosetta refinement overlap with the most populated MD conformations. The conformational ensemble of anaP1 with respect to anaP2 describes an ellipse with a spread of ∼50° for the twist angle and ∼40° for the swing angle for the most populated conformations (Fig. 5 D), leading to a rather restricted conformational space of these anaOmp85 POTRA domains in frozen solution. Since the overall conformational space of the MD simulations including the area of lower-populated conformations is much larger for these domains, especially the full range of twist motion is explored during the REMD simulation in a small corridor bounded by the swing angle mostly between 80° and 100°.
Looking at the relative orientation of anaP1 and anaP3 reveals a circular orientational space; however, the diameter of the space is ∼20° larger than the one observed for anaP2-anaP3. The lower-populated conformations show a spread of ∼140° in the twist angle and ∼90° in the swing angle; this again covers the conformations observed upon structure refinement. The orientational space of the most populated REMD conformations is the same size as that for the model ensembles obtained by our refinement approaches. Furthermore, the Cα distance distributions of the investigated interdomain constraints in the REMD simulation reveal that the resulting distributions due to interdomain movement are much narrower than those of the experiment, which is due to the rather small size of POTRA domains and the short linkers between them (Fig. S16). Thus, the envelope introduced by the flexibility of the SL imposes a resolution limit to the presented structure refinements. Nonetheless, we conclude that the conformational ensemble of anaOmp85-POTRA domains in frozen liquid solution is restricted in comparison to the conformational space sampled by the REMD simulation.
Discussion
PELDOR spectroscopy has become a valuable tool to gain insight into conformational dynamics of biomolecules (67). This is achieved by introducing paramagnetic spin centers at selected sites in the biomolecule. In our study, 27 distance constraints within and between the POTRA domains of anaOmp85 were measured, which showed strong dipolar oscillations and narrow distance distributions, strongly indicative of a defined relative orientation of the domains. Nevertheless, the large intrinsic flexibility of the most commonly used SL, MTSSL, needs to be taken into account, especially when it comes to structure determination. The comparison of our experimental data to distance distributions from rotamer library predictions on the x-ray structure revealed some discrepancies, which can most likely be attributed to differences in rotamer populations. In addition, distance distributions extracted from MD simulations on spin-labeled POTRA domains are slightly broader than the experimental distributions. The deviations are observed especially for constraints between anaP1 and anaP2, suggesting a slightly different orientation from that observed in the x-ray structure, as well as a smaller flexibility of these anaOmp85-POTRA domains in frozen liquid solution compared to MD simulations. We used our PELDOR constraints in two structure refinement approaches. In the case of the rigid body refinement, rotamer libraries based on the static x-ray structure were used to account for SL flexibility at a specific site, disregarding side-chain rearrangement of surrounding residues or putative tertiary interactions. This leads to an overestimated flexibility of the SL, which is reflected by broader distance distributions, as previously reported as well (68). By considering the intraresidue Cα—H ∙∙∙Sδ interaction (48, 66, 69), the width of the distributions is slightly reduced, but the most probable distances differ as well (Figs. S5, S11, and S13). Thus, overall agreement of predicted and experimental distance distributions is not greatly increased. Nonetheless, the obtained models suggest a somewhat reduced angular space compared to our MD simulations (Fig. 5). To overcome the inherent inaccuracy of the rotamer libraries and to account for protein flexibility as well as interactions of the SL in structure refinement we used Rosetta. Here, a specific rotamer is selected to match the experimental constraints. By choosing the labeling sites such that each position is correlated to two or more different sites, the degrees of freedom introduced by the linker of the SL are drastically reduced. This approach yielded similar precision for the top 100 models as the rigid body refinement. However, a more stringent selection of allowed models significantly reduces the spread in angular space of the Rosetta models, which in the case of the rigid-body refinement is not true for the orientation of anaP2-anaP3 (Fig. S17). In addition, the Rosetta ensemble shows a larger overlap with the most populated orientations in the MD simulations, which therefore are a good representation of the ensemble in frozen solution. Yet the ensembles of both refinement approaches reveal a small intrinsic flexibility of the POTRA domain pairs anaP1-anaP2 and anaP2-anaP3 in frozen solution. In terms of anaP2-anaP3, this is in line with earlier findings on BamA, where the C-terminal POTRA domains ecP3–ecP5 were assumed to have a rigid architecture (36). However, recent crystal structures of assembled BAM complexes challenge this assumption (8, 9, 10). In addition, the orientation of anaP2-anaP3 in the x-ray structure is well within this structure bundle, and thus seems to be a good representative of this ensemble. Unfortunately, the resolution of structure refinement cannot be increased any further, due to the small size of the POTRA domains, which results in Cα-distance distributions narrower than the experimental PELDOR constraints (Fig. S16). Furthermore, due to the inherent flexibility of the SL, and thus the uncertainty in prediction of the mean label position with respect to the protein backbone, structural refinement cannot be improved higher than a limit (70).
Looking at the Cα interdomain distances for the pair anaP1 and anaP2, we observe slightly broader distributions for distances related to residue 374 than for the Cα atoms of other SL pairs. Thus, we suggest that anaP1 and anaP2 are connected in a slightly more flexible fashion than anaP2 and anaP3; likewise, a flexible connection of ecP2 and ecP3 was observed for a folded precipitate of BamA by solid-state NMR (34). However, a switch between different conformations of anaP1 and anaP2, as reported for ecP2 and ecP3 constructs of POTRA domains ecP1–ecP4 of BamA (28, 29), is not observed in frozen solution. This is in agreement with solid-state NMR studies on full-length BamA in lipid bilayers, where a reorientation of ecP2-ecP3 was found to be slow, if present at all (31).
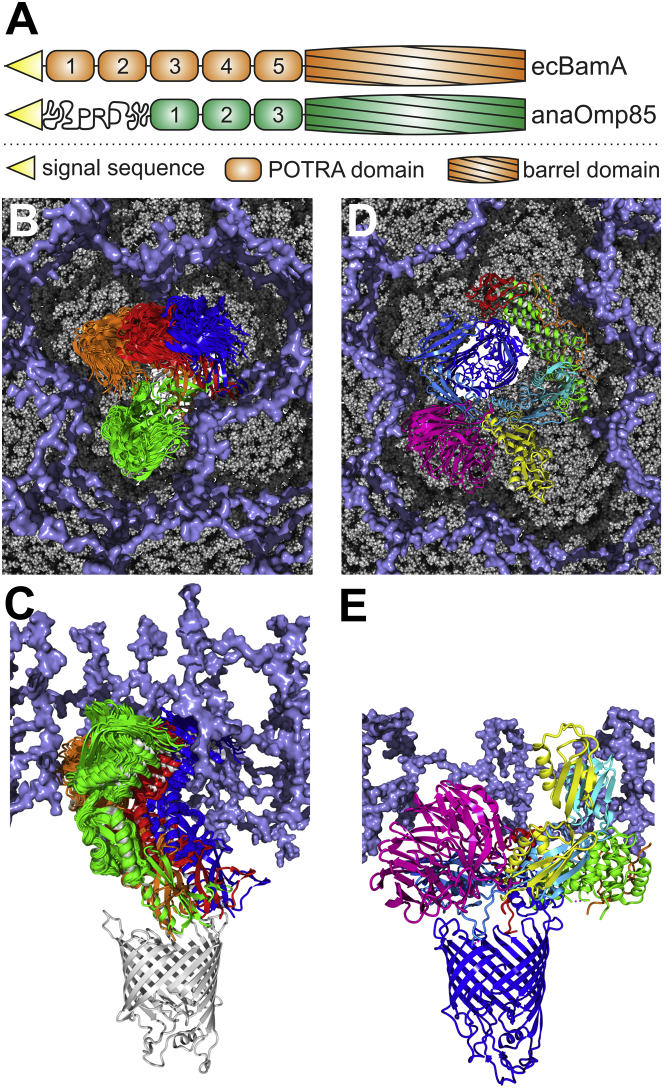
Our study considers the POTRA domains of anaOmp85, and thus, the orientation of anaP3 relative to the β-barrel, as well as its interaction with other complex components, remains unknown. However, extrapolating the position of ecP5 from known crystal structures of BamA ((8, 9, 28, 29)), we propose, to our knowledge, a first model for the relation between flexibility and the dimension of the peptidoglycan layer (PGL, Fig. 6). In general, the PGL is attached to the outer membrane and forms a porous network (71, 72), but its dimension appears to be species-specific. The PGL in Anabaena sp. is at 12 nm about twice as thick as that in E. coli ((73); Fig. 6, B and C). All current models suggest a close vicinity of the PGL to the outer membrane (74, 75) and thus, we selected a spacing between the outer membrane and the PGL of ∼3 nm. This positioning is in agreement with the observed BAM structure when considered in combination with the finding that the BAM components Pal in Caulobacter crescentus (76) and ComL/BamD in Neisseria gonorrheae (77) bind to PG and that PG binding motifs have been identified in Mlp/BamE in N. meningitidis (78).
Figure 6.
Model of anaOmp85-POTRA domains embedded in the PGL. (A) The domain compositions of anaOmp85 and ecBamA are shown. PRD denotes the proline-rich domain of anaOmp85. (B and C) An ensemble of MD structures on the outer contour line of the elliptic region in Fig. 5D, representing the most populated orientations of anaP1-anaP2 in MD simulations, and overlapping with the top-scoring Rosetta ensemble, was superimposed onto anaP2 in the crystal structure to indicate the space in which anaP1 rotates relative to anaP2 (Fig. S18). To estimate the rotation of anaP3 relative to the β-barrel, we selected BamA structures that exhibit different orientations of ecP5 relative to the β-barrel (PDB: 4K3B (green), 4K3C (blue), 5AWY (red), and 5EKQ (orange)) and aligned anaP3 onto ecP5. Another structure (PDB: 4C4V) was omitted, because it was a construct including ecP5 and the β-barrel only, which probably resulted in an artificial ecP5 orientation (35). A homology model of the β-barrel of anaOmp85 was built with YASARA based on an alignment with the template structure 4n75 constructed by the HHpred server (80). An ∼10-nm-thick model of the PGL (ice blue) (71) is shown in surface representation. (D and E) The BAM complex (PDB: 5AWY (9)) is shown with its components BamA (domains colored cyan to blue from N- to C-terminus), BamB (magenta), BamC (orange), BamD (green), and BamE (red). For ecP1 and ecP2, an additional conformation (PDB: 3EFC) is shown, which exhibits a different angle at the switch region between ecP2 and ecP3 (yellow). The PGL was cut to match the reported thickness of 5–6 nm in E. coli. The models in (B) and (D) are not shown on the same scale.
The three POTRA domains of anaOmp85 span ∼9.5 nm in the conformation known from the crystal structure (19), revealing that anaOmp85 might only require two POTRA domains to reach about as far as BamA ecP2–ecP5 into the PGL (∼5–6 nm). The positioning of the PGL would render ecP1 accessible for interaction with SurA and its substrates in the periplasm (Fig. 6, D and E), as recently proposed (8). anaP1 adds an additional 3–3.5 nm, and thus, all three POTRAS together would still not span the PGL. The remaining ∼6 nm of the PGL could be overcome by the proline-rich N-terminal domain, which is specific for cyanobacterial Omp85 proteins and potentially provides an initial interaction platform at the PGL boundary.
Thus, the observed conformational space would be consistent with constraints existing in vivo, although complex components might affect the conformational space of the POTRA domains as well. Based on our model, the conformational space of the POTRA domains determined for anaOmp85 would be consistent with thermal fluctuations within the confined environment of the PGL (Fig. 6, B and C), whereas the hinge between ecP2 and ecP3 found in BamA is likely absent in anaOmp85, as the anaPOTRA domains do not span the PGL, and only the proline-rich N-terminal domain is exposed to the periplasm.
Author Contributions
The project was designed by E.S. and T.F.P. Biochemical work was performed by E.-M.B. PELDOR measurements were performed by R.D. and D.S., and molecular modeling was performed by O.M. The manuscript was written through contributions from all authors. All authors have given approval to the final version of the manuscript.
Acknowledgments
We thank Dr. Alexey Cherepanov for help in freezing of a control sample by the freeze-quench technique and Dr. Sevdalina Lyubenova for initial EPR experiments. The model of the PGL was kindly provided by Shahriar Mobashery. The REMD simulation was performed on the LOEWE cluster of the Computer Sciences Corporation, Frankfurt, Germany.
The authors acknowledge the collaborative research center international symposium, CRC 807, “Transport and Communication across Membranes”, from the German Research Society (DFG) (P7 to T.F.P. and P17 to E.S.), and the Center of Biomolecular Magnetic Resonance (BMRZ) for financial support. E.S. received funding from DFG grant SCHL585/7-1. E.S. and T.F.P. are members of the DFG-funded Cluster of Excellence Frankfurt, a center for the investigation of macromolecular complexes.
Editor: David Cafiso.
Footnotes
Reza Dastvan and Eva-Maria Brouwer contributed equally to this work.
Reza Dastvan’s present address is Department of Molecular Physiology & Biophysics, Vanderbilt University, Nashville, Tennessee.
Eighteen figures and five tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30233-8.
Contributor Information
Enrico Schleiff, Email: schleiff@bio.uni-frankfurt.de.
Thomas F. Prisner, Email: prisner@chemie.uni-frankfurt.de.
Supporting Citations
Reference (79) appears in the Supporting Material.
Supporting Material
References
- 1.Schleiff E., Maier U.G., Becker T. Omp85 in eukaryotic systems: one protein family with distinct functions. Biol. Chem. 2011;392:21–27. doi: 10.1515/BC.2011.005. [DOI] [PubMed] [Google Scholar]
- 2.Moslavac S., Mirus O., Schleiff E. Conserved pore-forming regions in polypeptide-transporting proteins. FEBS J. 2005;272:1367–1378. doi: 10.1111/j.1742-4658.2005.04569.x. [DOI] [PubMed] [Google Scholar]
- 3.Bohnsack M.T., Schleiff E. The evolution of protein targeting and translocation systems. Biochim. Biophys. Acta. 2010;1803:1115–1130. doi: 10.1016/j.bbamcr.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Sánchez-Pulido L., Devos D., Valencia A. POTRA: a conserved domain in the FtsQ family and a class of β-barrel outer membrane proteins. Trends Biochem. Sci. 2003;28:523–526. doi: 10.1016/j.tibs.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Bredemeier R., Schlegel T., Schleiff E. Functional and phylogenetic properties of the pore-forming β-barrel transporters of the Omp85 family. J. Biol. Chem. 2007;282:1882–1890. doi: 10.1074/jbc.M609598200. [DOI] [PubMed] [Google Scholar]
- 6.Simmerman R.F., Dave A.M., Bruce B.D. Structure and function of POTRA domains of Omp85/TPS superfamily. Int. Rev. Cell Mol. Biol. 2014;308:1–34. doi: 10.1016/B978-0-12-800097-7.00001-4. [DOI] [PubMed] [Google Scholar]
- 7.Noinaj N., Rollauer S.E., Buchanan S.K. The β-barrel membrane protein insertase machinery from Gram-negative bacteria. Curr. Opin. Struct. Biol. 2015;31:35–42. doi: 10.1016/j.sbi.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakelar J., Buchanan S.K., Noinaj N. The structure of the β-barrel assembly machinery complex. Science. 2016;351:180–186. doi: 10.1126/science.aad3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han L., Zheng J., Huang Y. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat. Struct. Mol. Biol. 2016;23:192–196. doi: 10.1038/nsmb.3181. [DOI] [PubMed] [Google Scholar]
- 10.Gu Y., Li H., Dong C. Structural basis of outer membrane protein insertion by the BAM complex. Nature. 2016;531:64–69. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 11.Jansen K.B., Baker S.L., Sousa M.C. Crystal structure of BamB bound to a periplasmic domain fragment of BamA, the central component of the β-barrel assembly machine. J. Biol. Chem. 2015;290:2126–2136. doi: 10.1074/jbc.M114.584524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennion D., Charlson E.S., Misra R. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol. Microbiol. 2010;77:1153–1171. doi: 10.1111/j.1365-2958.2010.07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knowles T.J., Jeeves M., Henderson I.R. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol. Microbiol. 2008;68:1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- 14.Bölter B., Soll J., Wagner R. Origin of a chloroplast protein importer. Proc. Natl. Acad. Sci. USA. 1998;95:15831–15836. doi: 10.1073/pnas.95.26.15831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ertel F., Mirus O., Schleiff E. The evolutionarily related β-barrel polypeptide transporters from Pisum sativum and Nostoc PCC7120 contain two distinct functional domains. J. Biol. Chem. 2005;280:28281–28289. doi: 10.1074/jbc.M503035200. [DOI] [PubMed] [Google Scholar]
- 16.Schleiff E., Soll J. Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep. 2005;6:1023–1027. doi: 10.1038/sj.embor.7400563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicolaisen K., Mariscal V., Flores E. The outer membrane of a heterocyst-forming cyanobacterium is a permeability barrier for uptake of metabolites that are exchanged between cells. Mol. Microbiol. 2009;74:58–70. doi: 10.1111/j.1365-2958.2009.06850.x. [DOI] [PubMed] [Google Scholar]
- 18.Hahn A., Stevanovic M., Schleiff E. The TolC-like protein HgdD of the cyanobacterium Anabaena sp. PCC 7120 is involved in secondary metabolite export and antibiotic resistance. J. Biol. Chem. 2012;287:41126–41138. doi: 10.1074/jbc.M112.396010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koenig P., Mirus O., Tews I. Conserved properties of polypeptide transport-associated (POTRA) domains derived from cyanobacterial Omp85. J. Biol. Chem. 2010;285:18016–18024. doi: 10.1074/jbc.M110.112649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schleiff E., Becker T. Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol. 2011;12:48–59. doi: 10.1038/nrm3027. [DOI] [PubMed] [Google Scholar]
- 21.Habib S.J., Waizenegger T., Rapaport D. The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial β-barrel proteins. J. Cell Biol. 2007;176:77–88. doi: 10.1083/jcb.200602050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stroud D.A., Becker T., Wiedemann N. Biogenesis of mitochondrial β-barrel proteins: the POTRA domain is involved in precursor release from the SAM complex. Mol. Biol. Cell. 2011;22:2823–2833. doi: 10.1091/mbc.E11-02-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsueh Y.C., Flinner N., Schleiff E. The chloroplast outer envelope protein P39 in Arabidopsis thaliana belongs to the Omp85 protein family. Proteins. 2014 doi: 10.1002/prot.24725. Published online November 17, 2014. [DOI] [PubMed] [Google Scholar]
- 24.Nicolaisen K., Missbach S., Schleiff E. The Omp85-type outer membrane protein p36 of Arabidopsis thaliana evolved by recent gene duplication. J. Plant Res. 2015;128:317–325. doi: 10.1007/s10265-014-0693-4. [DOI] [PubMed] [Google Scholar]
- 25.Heinz E., Lithgow T. A comprehensive analysis of the Omp85/TpsB protein superfamily structural diversity, taxonomic occurrence, and evolution. Front. Microbiol. 2014;5:370. doi: 10.3389/fmicb.2014.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Ent F., Vinkenvleugel T.M., Löwe J. Structural and mutational analysis of the cell division protein FtsQ. Mol. Microbiol. 2008;68:110–123. doi: 10.1111/j.1365-2958.2008.06141.x. [DOI] [PubMed] [Google Scholar]
- 27.Clantin B., Delattre A.-S., Villeret V. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science. 2007;317:957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- 28.Gatzeva-Topalova P.Z., Walton T.A., Sousa M.C. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure. 2008;16:1873–1881. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S., Malinverni J.C., Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 30.Guédin S., Willery E., Jacob-Dubuisson F. Novel topological features of FhaC, the outer membrane transporter involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 2000;275:30202–30210. doi: 10.1074/jbc.M005515200. [DOI] [PubMed] [Google Scholar]
- 31.Sinnige T., Weingarth M., Baldus M. Solid-state NMR studies of full-length BamA in lipid bilayers suggest limited overall POTRA mobility. J. Mol. Biol. 2014;426:2009–2021. doi: 10.1016/j.jmb.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 32.Guérin J., Saint N., Jacob-Dubuisson F. Dynamic interplay of membrane-proximal POTRA domain and conserved loop L6 in Omp85 transporter FhaC. Mol. Microbiol. 2015;98:490–501. doi: 10.1111/mmi.13137. [DOI] [PubMed] [Google Scholar]
- 33.Ward R., Zoltner M., Norman D.G. The orientation of a tandem POTRA domain pair, of the β-barrel assembly protein BamA, determined by PELDOR spectroscopy. Structure. 2009;17:1187–1194. doi: 10.1016/j.str.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Renault M., Bos M.P., Baldus M. Solid-state NMR on a large multidomain integral membrane protein: the outer membrane protein assembly factor BamA. J. Am. Chem. Soc. 2011;133:4175–4177. doi: 10.1021/ja109469c. [DOI] [PubMed] [Google Scholar]
- 35.Sinnige T., Houben K., Baldus M. Insight into the conformational stability of membrane-embedded BamA using a combined solution and solid-state NMR approach. J. Biomol. NMR. 2015;61:321–332. doi: 10.1007/s10858-014-9891-6. [DOI] [PubMed] [Google Scholar]
- 36.Gatzeva-Topalova P.Z., Warner L.R., Sousa M.C. Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure. 2010;18:1492–1501. doi: 10.1016/j.str.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milov A.D., Salikhov K.M., Shirov M.D. Use of the double resonance in electron-spin echo method for the study of paramagnetic center spatial distribution in solids. Fizika Tverdogo Tela. 1981;23:975–982. [Google Scholar]
- 38.Schiemann O., Prisner T.F. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q. Rev. Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 39.Jeschke G. DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 40.Altenbach C., Kusnetzow A.K., Hubbell W.L. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc. Natl. Acad. Sci. USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grote M., Polyhach Y., Bordignon E. Transmembrane signaling in the maltose ABC transporter MalFGK2-E: periplasmic MalF-P2 loop communicates substrate availability to the ATP-bound MalK dimer. J. Biol. Chem. 2009;284:17521–17526. doi: 10.1074/jbc.M109.006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhatnagar J., Freed J.H., Crane B.R. Rigid body refinement of protein complexes with long-range distance restraints from pulsed dipolar ESR. Methods Enzymol. 2007;423:117–133. doi: 10.1016/S0076-6879(07)23004-6. [DOI] [PubMed] [Google Scholar]
- 43.Hilger D., Polyhach Y., Jeschke G. High-resolution structure of a Na+/H+ antiporter dimer obtained by pulsed electron paramagnetic resonance distance measurements. Biophys. J. 2007;93:3675–3683. doi: 10.1529/biophysj.107.109769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hilger D., Polyhach Y., Jeschke G. Backbone structure of transmembrane domain IX of the Na+/proline transporter PutP of Escherichia coli. Biophys. J. 2009;96:217–225. doi: 10.1016/j.bpj.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirst S.J., Alexander N., Meiler J. RosettaEPR: an integrated tool for protein structure determination from sparse EPR data. J. Struct. Biol. 2011;173:506–514. doi: 10.1016/j.jsb.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alexander N.S., Stein R.A., Meiler J. RosettaEPR: rotamer library for spin label structure and dynamics. PLoS One. 2013;8:e72851. doi: 10.1371/journal.pone.0072851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polyhach Y., Bordignon E., Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Phys. Chem. Chem. Phys. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- 48.Sezer D., Freed J.H., Roux B. Parametrization, molecular dynamics simulation, and calculation of electron spin resonance spectra of a nitroxide spin label on a polyalanine α-helix. J. Phys. Chem. B. 2008;112:5755–5767. doi: 10.1021/jp711375x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pannier M., Veit S., Spiess H.W. Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 50.Jeschke G., Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys. Chem. Chem. Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 51.Polyhach Y., Bordignon E., Jeschke G. High sensitivity and versatility of the DEER experiment on nitroxide radical pairs at Q-band frequencies. Phys. Chem. Chem. Phys. 2012;14:10762–10773. doi: 10.1039/c2cp41520h. [DOI] [PubMed] [Google Scholar]
- 52.Jeschke G., Chechik V., Jung H. DeerAnalysis2006: a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 2006;30:473–498. [Google Scholar]
- 53.Hagelueken G., Ward R., Schiemann O. MtsslWizard: in silico spin-labeling and generation of distance distributions in PyMOL. Appl. Magn. Reson. 2012;42:377–391. doi: 10.1007/s00723-012-0314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krieger E., Vriend G. YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics. 2014;30:2981–2982. doi: 10.1093/bioinformatics/btu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tyka M.D., Keedy D.A., Baker D. Alternate states of proteins revealed by detailed energy landscape mapping. J. Mol. Biol. 2011;405:607–618. doi: 10.1016/j.jmb.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patriksson A., van der Spoel D. A temperature predictor for parallel tempering simulations. Phys. Chem. Chem. Phys. 2008;10:2073–2077. doi: 10.1039/b716554d. [DOI] [PubMed] [Google Scholar]
- 57.Duan Y., Wu C., Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 58.Páll S., Hess B. A flexible algorithm for calculating pair interactions on SIMD architectures. Comput. Phys. Commun. 2013;184:2641–2650. [Google Scholar]
- 59.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 60.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 61.Nosé S., Klein M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 62.Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 63.Stewart J.J. MOPAC: a semiempirical molecular orbital program. J. Comput. Aided Mol. Des. 1990;4:1–105. doi: 10.1007/BF00128336. [DOI] [PubMed] [Google Scholar]
- 64.Krieger E., Nielsen J.E., Vriend G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 2006;25:481–486. doi: 10.1016/j.jmgm.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 65.Reference deleted in proof.
- 66.Warshaviak D.T., Serbulea L., Hubbell W.L. Conformational analysis of a nitroxide side chain in an α-helix with density functional theory. J. Phys. Chem. B. 2011;115:397–405. doi: 10.1021/jp108871m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McHaourab H.S., Steed P.R., Kazmier K. Toward the fourth dimension of membrane protein structure: insight into dynamics from spin-labeling EPR spectroscopy. Structure. 2011;19:1549–1561. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klose D., Klare J.P., Steinhoff H.-J. Simulation vs. reality: a comparison of in silico distance predictions with DEER and FRET measurements. PLoS One. 2012;7:e39492. doi: 10.1371/journal.pone.0039492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fleissner M.R., Cascio D., Hubbell W.L. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 2009;18:893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fischer A.W., Alexander N.S., Meiler J. BCL:MP-fold: Membrane protein structure prediction guided by EPR restraints. Proteins. 2015;83:1947–1962. doi: 10.1002/prot.24801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meroueh S.O., Bencze K.Z., Mobashery S. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc. Natl. Acad. Sci. USA. 2006;103:4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gan L., Chen S., Jensen G.J. Molecular organization of Gram-negative peptidoglycan. Proc. Natl. Acad. Sci. USA. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilk L., Strauss M., Schleiff E. Outer membrane continuity and septosome formation between vegetative cells in the filaments of Anabaena sp. PCC 7120. Cell. Microbiol. 2011;13:1744–1754. doi: 10.1111/j.1462-5822.2011.01655.x. [DOI] [PubMed] [Google Scholar]
- 74.Leduc M., Fréhel C., Van Heijenoort J. Multilayered distribution of peptidoglycan in the periplasmic space of Escherichia coli. J. Gen. Microbiol. 1989;135:1243–1254. doi: 10.1099/00221287-135-5-1243. [DOI] [PubMed] [Google Scholar]
- 75.Beveridge T.J. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anwari K., Poggio S., Lithgow T. A modular BAM complex in the outer membrane of the α-proteobacterium Caulobacter crescentus. PLoS One. 2010;5:e8619. doi: 10.1371/journal.pone.0008619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fussenegger M., Facius D., Meyer T.F. A novel peptidoglycan-linked lipoprotein (ComL) that functions in natural transformation competence of Neisseria gonorrhoeae. Mol. Microbiol. 1996;19:1095–1105. doi: 10.1046/j.1365-2958.1996.457984.x. [DOI] [PubMed] [Google Scholar]
- 78.Volokhina E.B., Beckers F., Bos M.P. The β-barrel outer membrane protein assembly complex of Neisseria meningitidis. J. Bacteriol. 2009;191:7074–7085. doi: 10.1128/JB.00737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waterhouse A.M., Procter J.B., Barton G.J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Söding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.