Abstract
Fanconi anemia (FA) pathway genes are important tumor suppressors whose best-characterized function is repair of damaged nuclear DNA. Herein, we describe an essential role for FA genes in two forms of selective autophagy. Genetic deletion of Fancc blocks the autophagic clearance of viruses (virophagy) and increases susceptibility to lethal viral encephalitis. FANCC interacts with Parkin; is required in vitro and in vivo for clearance of damaged mitochondria; and decreases mitochondrial ROS production and inflammasome activation. The mitophagy function of FANCC is genetically distinct from its role in genomic DNA damage repair. Moreover, additional genes in the FA pathway, including FANCA, FANCF, FANCL, FANCD2, BRCA1 and BRCA2, are required for mitophagy. Thus, members of the FA pathway represent a previously undescribed class of selective autophagy genes that function in immunity and organellar homeostasis. These findings have implications for understanding the pathogenesis of FA and cancers associated with mutations in FA genes.
INTRODUCTION
Homozygous or compound heterozygous loss-of-function mutations in any of ~19 genes of the Fanconi Anemia (FA) pathway lead to a spectrum of clinical disorders, including birth defects, cognitive impairment, bone marrow failure, cancer and premature aging (Bogliolo and Surralles, 2015; Neveling et al., 2009). Although FA is rare, somatic mutations or epigenetic silencing of FA genes are commonly found in sporadic cancers, particularly of the breast, ovaries, and pancreas (D'Andrea, 2010). Moreover, heterozygous germline mutations in at least five genes in the FA pathway, BRCA2/FANCD1, BRIP1/FANCJ, PALB2/FANCN, RAD51C/FANCO, and BRCA1/FANCS, are important cancer risk alleles (Bogliolo and Surralles, 2015).
The best characterized function of FA genes is genomic DNA damage repair – specifically the repair of DNA interstrand crosslinks formed in the presence of endogenous ROS or aldehyde substrates (Garaycoechea and Patel, 2014). In response to DNA crosslinker damage, eight FA proteins assemble to form the FA core complex containing the E3 ubiquitin ligase FANCL, which mono-ubiquitinates FANCD2 and FANCI in the nucleus and thereby orchestrates downstream DNA repair (Garaycoechea and Patel, 2014). Some FA proteins also have cytoplasmic functions, such as protection from proinflammatory cytokine-induced cell death (Haneline et al., 1998), maintenance of mitochondrial respiratory function, suppression of intracellular ROS levels (Pagano et al., 2013), and reduction of inflammasome activity (Garbati et al., 2013).
The mechanism(s) by which FA proteins function in cytoplasmic processes and the extent to which such processes can be separated from genomic DNA repair activity are poorly understood. However, a naturally occurring hypomorphic mutant of FANCC (c.67delG) encodes a shorter protein lacking the FANCC N-terminal 55 amino acids and fully complements the cytoprotective but not the genomic DNA repair functions in FANCC null mutant cells (Bagby and Alter, 2006). FANCC-deficient patients with at least one FANCC c.67delG allele have fewer congenital abnormalities and a milder clinical phenotype when compared to FA patients without any detectable FANCC protein (Yamashita et al., 1996). This suggests that DNA repair-independent functions of FANCC (and perhaps other FA genes) may play a crucial role in the pathophysiology of FA and/or cancers that occur in the setting of mutations in FA genes.
Three FA genes, FANC, FANCF and FANCL, were identified in a genome-wide screen as candidate selective autophagy factors (Orvedahl et al., 2011). Selective autophagy is a homeostatic process that targets unwanted cellular cargo to autophagosomes with subsequent degradation in autolysosomes (Rogov et al., 2014). Diverse cargos have been recognized as substrates for selective autophagy, including damaged mitochondria (mitophagy) and viruses (virophagy). Selectivity is achieved through autophagy receptors (such as p62, NBR1, NDP52, and optineurin), which recognize cargos tagged with degradation signals as well as the autophagosomal membrane protein LC3. Although multiple autophagy receptors, as well as ubiquitin ligases such as Parkin (which mediates the ubiquitination of damaged mitochondria and intracellular bacteria) have been identified (Rogov et al., 2014), it is likely that other factors are involved in selective autophagy.
Defects in selective autophagy have profound effects on organismal biology, including several that overlap with effects of mutations in FA pathway genes. Both mutations in core autophagy genes that function in autophagosome membrane formation and mutations in mitophagy-specific genes promote tumorigenesis through a mechanism thought to involve failure to remove damaged mitochondria and subsequent increases in mitochondrial ROS (mtROS) and genotoxic stress (Chourasia et al., 2015). Deletion of essential autophagy genes in hematopoietic stem cells (HSCs) results in bone marrow failure characterized by accumulation of damaged mitochondria and mtROS (Mortensen et al., 2011) and defective mitophagy results in enhanced inflammasome-mediated IL-1 β secretion (Nakahira et al., 2011; Zhou et al., 2011).
We therefore hypothesized that FA genes may function in selective autophagy, and that such a role may represent a previously undescribed function that could contribute to the pathogenesis of congenital FA and of cancers that occur in patients with germline or somatic FA gene mutations. Our data demonstrate that FA genes function in the selective autophagy of genetically distinct viruses, in mitochondrial quality control, and in preventing inflammasome activation due to mtROS. Moreover, we show that the activity of FANCC in mitophagy is independent of its role in nuclear DNA damage repair. These findings have important implications for understanding the mechanisms by which mutations in FA genes contribute to human disease.
RESULTS
FANCC is required for virophagy but not for starvation-induced autophagy
FANCC, FANCF and FANCL were identified in a genome-wide siRNA screen as potential selective autophagy factors (Orvedahl et al., 2011). We focused on FANCC, as its role in cytokine hypersensitivity is independent of its function in nuclear DNA damage repair (Pang et al., 2001) and as Fancc knockout mice exhibit increased susceptibility to bone marrow failure when exposed to proinflammatory cytokines or endotoxin (Haneline et al., 1998; Parmar et al., 2009). To determine whether FANCC is a selective autophagy factor, we used siRNAs targeting FANCC and murine embryonic fibroblasts (MEFs) derived from Fancc−/− mice and Fancc+/+ littermate controls.
First, we evaluated whether Fancc is required for starvation-induced autophagy, a form of nonselective autophagy (Mizushima and Komatsu, 2011). Fancc−/− and Fancc+/+ MEFs did not differ in starvation-induced autophagic flux, as measured by the conversion of the non-lipidated form of LC3 (LC3-I) to the lipidated, autophagosome-associated form, LC3-II, in the presence and absence of the lysosomal inhibitor bafilomycin A1 (Baf A1) (Figure 1A) or by the degradation of the autophagy substrate p62 (Figure 1B). The numbers of GFP-LC3 puncta (a marker of autophagosomes) (Mizushima et al., 2010) were not decreased during basal or starvation conditions in MEFs derived from Fancc−/− mice crossed to GFP-LC3 transgenic mice (Mizushima et al., 2004) (Fancc−/−/GFP-LC3) compared to wild-type (WT) (Fancc+/+/GFP-LC3) controls (Figure 1C). There was also no difference in GFP-LC3 puncta in the presence of Baf A1, confirming that Fancc is not required for starvation-induced autophagic flux. Upon ultrastructural analysis, Fancc−/− and Fancc+/+ MEFs had a similar robust increase in autophagosomes and autolysosomes in response to starvation (Figure 1D). Thus, Fancc deficiency in MEFs does not impair starvation-induced autophagy.
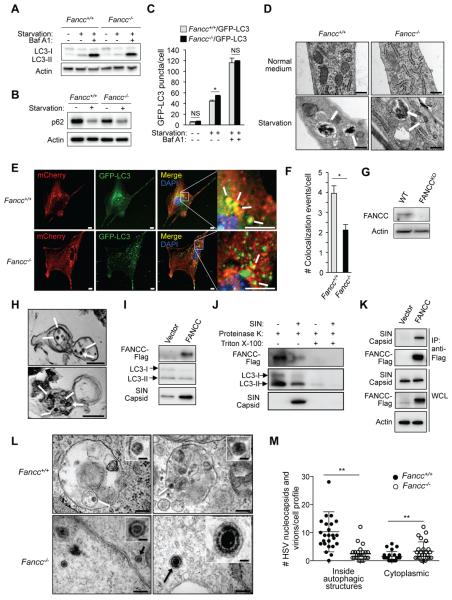
Figure 1. Fancc is Required for Autophagic Targeting of Genetically Distinct Viruses, but Not for Starvation-Induced Autophagy.
(A) Western blot of LC3-I/LC3-II in primary murine embryonic fibroblasts (MEFs) during growth in normal (starvation −) or EBSS (starvation +) medium for 4 hr +/− 100 nM bafilomycin A1 (Baf A1).
(B) Western blot of p62 in MEFs in culture conditions as in (A).
(C) Quantification of GFP-LC3 puncta (autophagosomes) in Fancc+/+/GFP-LC3 and Fancc−/−/GFP-LC3 MEFs during growth in normal or starvation medium for 4 hr +/− 100 nM Baf A1. Bars are mean ± SEM for triplicate samples (>100 cells analyzed per sample by an observer blinded to experimental condition). NS, not significant; *, P<0.05; t-test.
(D) EM of autophagic structures in MEFs during growth in normal or starvation medium (EBSS, 4 hr). White arrows indicate autolysosomes. Scale bars, 1 μm.
(E and F) Representative light micrographs of images (E) used for quantification (F) of colocalization of SIN/mCherry.capsid and GFP-LC3 in MEFs infected with SIN mCherry.capsid/GFP-LC3 (multiplicity of infection [MOI] = 2.5 plaque-forming units [PFUs], 18 hr). In (E), white arrows denote representative colocalized mCherry.capsid and GFP-LC3 signal. Scale bars, 10 μm. In (F), bars are mean ± SEM for triplicate samples (> 50 cells analyzed per sample by an observer blinded to experimental condition). *, P<0.05; t-test.
(G) Western blot of FANCC in HeLa WT and HeLa FANCCKO cells.
(H) EM of immunoprecipitated autophagic structures using an anti-LC3 antibody from HeLa FANCCKO cells transduced with a lentivirus expressing FANCC-Flag and infected with SIN strain SVIA (MOI = 5, 6 hr). White arrows indicate SIN nucleocapsids inside autolysosomes. Asterisks indicate dynabeads used for IP. Scale bars, 200 nm.
(I) Western blot of indicated proteins in anti-LC3 immunoprecipitates (representative electron microscopes shown in (H)) from HeLa FANCCKO cells transduced with lentivirus vector (HeLa FANCCKO/vector) or lentivirus expressing FANCC-Flag (HeLa FANCCKO/FANCC) and infected with SIN strain SVIA (MOI = 5, 6 hr).
(J) Western blot of indicated proteins in control and SIN-infected HeLa FANCCKO/FANCC cells subjected to anti-LC3 IP as in (H-I) and then 30 min digestion on ice with 2.5 ng/ml proteinase K +/− 0.5% triton X-100.
(K) Co-IP of SIN capsid protein with FANCC-Flag in HeLa FANCCKO/vector (Vector) or HeLa FANCCKO/FANCC (FANCC) cells infected with SIN strain dsTE12Q (MOI = 5, 6 hr).
(L) Representative EMs of MEFs infected with HSV-1 ICP34.5Δ68-87 (MOI = 5, 6 hr). White arrows denote representative HSV-1 nucleocapsids being degraded inside autolysosomes. Black arrows denote representative HSV-1 nucleocapsid (bottom left) or assembled HSV-1 virion (bottom right) inside the cytoplasm. Scale bars, 200 nm for large panels; 50 nm for insets.
(M) Quantification of HSV-1 nucleocapsids and virions within autophagosomes or autolysosomes (inside autophagic structures) or free within the cytoplasm (cytoplasmic) in experiment shown in (L). Twenty-five HSV-1-infected cell profiles were counted per genotype by an observer blinded to experimental conditions. **, P<0.01; Mann-Whitney U test.
In A-C, E and F, and I-K, similar results were observed in three independent experiments.
See also Figure S1.
We then evaluated whether Fancc is required for virophagy (Sumpter and Levine, 2011). We used two neuronotropic viruses, Sindbis virus (SIN), a positive-strand RNA virus in the Togaviridae family, and a herpes simplex virus type 1 mutant strain HSV-1 ICP34.5Δ68-87 (Orvedahl et al., 2007), a DNA virus genetically engineered to lack autophagy inhibitory activity. This mutant virus contains a 20 amino acid deletion in the neurovirulence protein ICP34.5 which abrogates its inhibition of the essential autophagy protein, Beclin 1 and blocks HSV-1 neurovirulence in WT mice (Orvedahl et al., 2007). We chose these viruses because they are targeted for autophagic degradation (Sumpter and Levine, 2011) and because autophagic degradation of viral components in neurons is important in antiviral defense (Yordy and Iwasaki, 2013).
We confirmed that three siRNAs targeting FANCC decreased FANCC gene expression and decreased the colocalization of an mCherry SIN capsid fusion protein with GFP-LC3 in HeLa/GFP-LC3 cells infected with the recombinant SIN virus, SIN/mCherry.capsid, as efficiently as knockdown of an essential autophagy gene, ATG7 (Mizushima et al., 2010) (Figures S1A-S1C). To rule out off-target effects of siRNA, we evaluated SIN capsid and GFP-LC3 colocalization in Fancc−/− and Fancc+/+ MEFs infected with SIN-mCherry.capsid/GFP-LC3 (Orvedahl et al., 2010), a virus genetically engineered to express both an mCherry capsid fusion protein and, from a duplicated internal subgenomic promoter, GFP-LC3. Fewer colocalized mCherry.capsid and GFP-LC3 puncta were observed in SIN-infected Fancc−/− as compared to Fancc+/+ MEFs (Figure 1E and 1F). No differences were observed in numbers of mCherry.capsid or GFP-LC3 puncta between the two genotypes (data not shown), indicating that this decrease is not due to differences in viral replication or autophagosome formation but rather represents a specific defect in targeting SIN nucleocapsids to autophagic structures.
We next examined whether FANCC localizes to SIN capsid-containing autophagic structures. We generated HeLa FANCCKO cells using the CRISPR/Cas9 system (Figure 1G), and performed immunoprecipitation (IP) using an anti-LC3 antibody of the microsomal + mitochondrial fractions (Figure S1D) from mechanically lysed SIN-infected HeLa FANCCKO cells transduced with a lentivirus expressing empty vector or FANCC-Flag. Electron microscopic (EM) analysis of structures IP’d with anti-LC3 revealed numerous autophagosomal and autolysosomal vesicles containing SIN nucleocapsids (Figure 1H). Western blot analysis of these structures revealed the presence of both FANCC-Flag and SIN capsid protein (Figure 1I). FANCC-Flag was contained within these autophagolysosomal membrane structures as it was protected from proteinase K-mediated proteolysis and this protection was lost upon membrane disruption with detergent (Figure 1J). Moreover, similar to previously identified SIN virophagy factors such as p62 and SMURF1 (Orvedahl et al., 2010; Orvedahl et al., 2011), we found that FANCC-Flag and SIN capsid interact by performing co-immunoprecipitation (co-IP) of FANCC-Flag and SIN capsid in SIN-infected HeLa FANCCKO/vector and HeLa FANCCKO/FANCC cells subjected to detergent lysis (Figure 1K).
Thus, FANCC-Flag is not only required for SIN viral capsid targeting to GFP-LC3 positive structures; it is also present inside SIN capsid-containing autophagic vesicles and interacts with SIN capsid protein, consistent with a direct role in targeting SIN capsid for autophagy. However, FANCC-Flag is not a classical autophagy adaptor that binds both cargo and LC3 and/or other Atg8 family members (e.g. GABARAP, GATE-16) as these proteins did not co-IP with FANCC-Flag in uninfected or SIN-infected cells (Figure S1E). As there are no suitable antibodies for IP of endogenous FANCC, we performed IP of another FA core complex protein, FANCA (the most commonly mutated protein in FA patients (Bagby and Alter, 2006)), and found that endogenous FANCA co-IPs with SIN capsid (Figure S1F).
To determine whether FANCC may have a more generalized role in virophagy, we performed EM of Fancc+/+ and Fancc−/− MEFs infected with HSV-1 ICP34.5Δ68-87 (Figure 1L and 1M). HSV-1 replicates and assembles its nucleocapsids in the nucleus but can be captured by autophagosomes during its transit through the cytoplasm (Talloczy et al., 2006). In Fancc+/+ MEFs, most HSV-1 nucleocapsids and viral particles were detected (often in a partially degraded form) inside autolysosomal structures. In contrast, in Fancc−/− MEFs (similar to virophagy-deficient Pkr−/− and Smurf1−/− MEFs (Orvedahl et al., 2011; Talloczy et al., 2006)), there were fewer HSV-1-containing autophagosomes/autolysosomes and more free nucleocapsids and assembled virions inside in the cytoplasm.
Taken together, these data demonstrate that FANCC is required for virophagy of two genetically distinct viruses, SIN (ssRNA genome) and HSV-1 (dsDNA genome), but not for starvation-induced autophagy.
FANCC is required for host defense against neuronotropic viruses
To investigate whether Fancc-mediated virophagy is associated with antiviral defense in vivo, we studied the pathogenesis of CNS infection in Fancc+/+ and Fancc−/− mice in well-established animal models of SIN and HSV-1 ICP34.5Δ68-87 infection (Orvedahl et al., 2007; Orvedahl et al., 2010). Following intracerebral inoculation of one week-old animals with SIN (strain dsTE12Q), Fancc−/− mice had increased mortality compared to Fancc+/+ littermate controls (Figure 2A). This was associated with an increase in neuronal cell death by TUNEL staining (Figure 2B and 2C). Similar to findings in mice lacking neuronal expression of the Atg5 autophagy protein (Orvedahl et al., 2010), this increase in cell death was accompanied by impaired SIN capsid antigen clearance but not by increases in SIN viral titers in the brain (Figure S2A and S2C). Six-eight week-old Fancc−/− mice were also more susceptible to lethal CNS infection (following intracerebral inoculation) with HSV-1 ICP34.5Δ68-87 than were Fancc+/+ or Fancc+/− mice (Figure 2D). The brains of Fancc−/− mice had more neuronal death at day 3 after infection and increased HSV-1 antigen at days 3 and 5 without an increase in CNS viral titers (Figures 2E and 2F, Figures S2B and S2D). Fancc−/− mice had no increase in susceptibility to another HSV-1 strain, HSV-1 TK− (Pyles and Thompson, 1994), which is neuroattentuated because of a deletion in the viral thymidine kinase gene rather than a mutation that affects autophagy (Figure S2E), providing genetic evidence that Fancc-mediated antiviral host defense against lethal CNS HSV-1 infection involves the autophagy pathway. Thus, taken together, our findings demonstrate that Fancc is required for antiviral host defense against two unrelated viruses in vivo and its antiviral effects are likely mediated through autophagy.
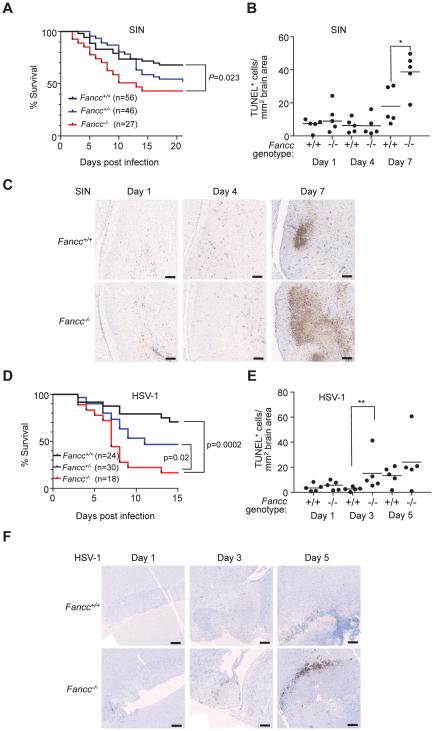
Figure 2. Fancc is Required for Host Defense against SIN and HSV-1 CNS Viral infections.
(A) Survival of 7 day-old mice infected intracerebrally (i.c.) with 1000 PFUs SIN strain dsTE12Q. Results shown represent combined survival for independent infections of more than 5 litters (similar results obtained for each infection). P-value (log-rank test) and number of mice per genotype indicated in graph.
(B and C) Quantitation of TUNEL-positive area per brain section (B) and representative light micrographs (C) after infection of 7 day-old mice of indicated genotype infected as in (A). Bars represent median value for each group. *, P<0.05; Mann-Whitney U test. Scale bars, 500 μm.
(D) Survival of 6-8 week-old mice infected i.c. with 5x104 PFUs HSV-1 ICP34.5Δ68-87. Results shown represent combined survival data combined from three independent infections (similar results obtained for each infection). P-value (log-rank test) and number of mice per genotype indicated in graph.
(E and F) Quantitation of TUNEL-positive area per brain section (E) and representative light micrographs (F) at indicated time points of 2 month-old mice infected as in (D). Bars represent median value for each group. **, P<0.01; Mann-Whitney U test. Scale bars, 500 μm.
See also Figure S2.
FANCC is required for mitophagy in vitro and mitochondrial quality control in vivo
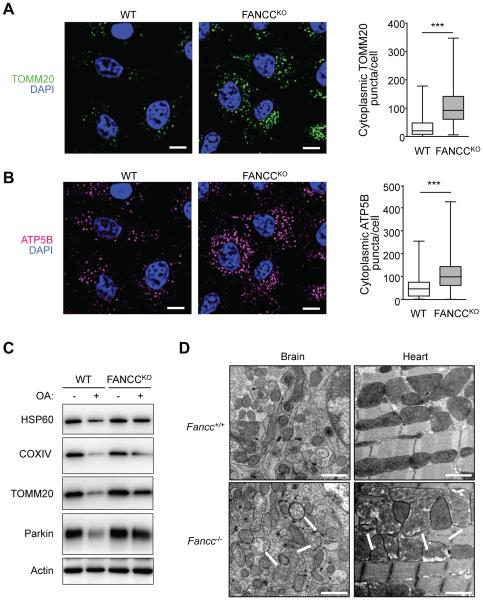
In our previous genome-wide screen, FANCC was required not only for SIN virophagy, but also for mitophagy mediated by the E3 ligase Parkin upon treatment with the mitochondrial uncoupling agent, CCCP (Orvedahl et al., 2011). We confirmed that siRNA knockdown of FANCC blocked CCCP-induced, Parkin-mediated mitophagy as efficiently as knockdown of the essential autophagy gene, ATG7 (Figure S3A-S3D). We verified an essential role of FANCC in Parkin-mediated mitophagy using parental HeLa cells or HeLa FANCCKO cells transduced with a retrovirus expressing HA-Parkin. HeLa FANCCKO/Parkin cells were deficient in mitophagy after treatment with a combination of two specific inhibitors of mitochondrial respiration, Oligomycin + Anitmycin A, (OA), as measured by quantitation of TOMM20 (outer mitochondrial membrane protein) and ATP5B (mitochondrial matrix protein) puncta (Figures 3A and 3B) or by western blot detection of TOMM20, HSP60 (mitochondrial matrix protein) and COXIV (inner mitochondrial membrane protein) (Figure 3C). To determine whether Fancc deficiency results in a defect in mitochondrial quality control in vivo, we performed EM analysis of brains and hearts from one year-old Fancc+/+ or Fancc−/− mice (Figure 3D). In Fancc−/− but not Fancc+/+ mice, we observed a marked accumulation of abnormal mitochondria. Thus, Fancc is essential for Parkin-dependent mitophagy in vitro and for mitochondrial homeostasis in vivo.
Figure 3. FANCC is Required for the Clearance of Damaged Mitochondria in vitro and in vivo.
(A and B) Mitophagy analysis in FANCCKO/FANCC (WT) or FANCCKO/vector (FANCCKO) cells stably expressing HA-Parkin assessed by clearance of TOMM20 (A) or ATP5B (B). Cells were treated with OA (Oligomycin, 2.5 μM; Anitmycin A, 250 nM) for 8 hr prior to imaging and automated image analysis. Left, representative images of immunoflourescence staining. Right, quantitative image analysis. Shown are box plots of at least 150 cells analyzed per condition. Similar results were observed in three independent experiments. ***, P<0.001, Mann-Whitney U-test. Scale bars, 20 μm.
(C) Mitophagy analysis by western blot detection of HSP60, COXIV or TOMM20 in WT or FANCCKO cells stably expressing HA-Parkin and treated +/− OA for 8 hr.
(D) EM analysis of brain (cerebellum) or heart tissue (left ventricle) from 1 year-old mice. Shown are images from one representative mouse per genotype. Similar results were observed in 6 mice per genotype. White arrows indicate damaged mitochondria. Scale bars, 1 μm.
See also Figure S3.
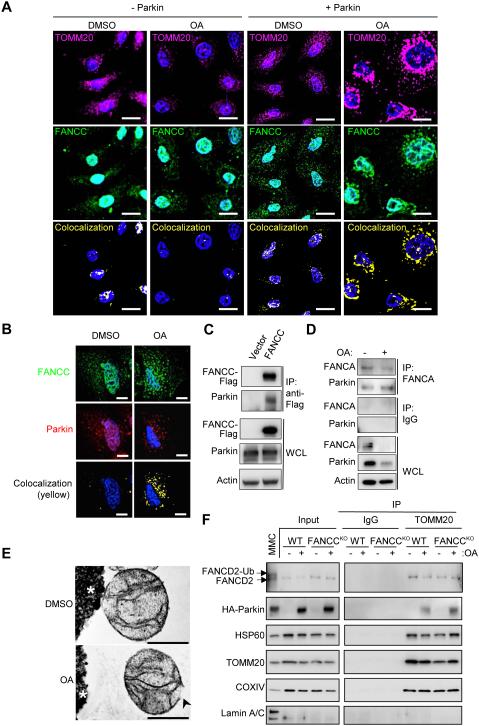
We next evaluated whether Parkin expression affects the subcellular localization of FANCC and mitochondria using HeLa FANCCKO/FANCC cells transduced with a retrovirus expressing HA-Parkin (HeLa FANCCKO/FANCC/Parkin). In the absence of Parkin expression, little colocalization was observed between FANCC-Flag and the mitochondrial marker, TOMM20, even after treatment with OA (Figure 4A, left panels). In the presence of Parkin expression and OA, there was a marked increase in FANCC and TOMM20 colocalization (Figure 4A, right panels). This increase in FANCC and TOMM20 colocalization was accompanied by an increase in FANCC and Parkin colocalization (Figure 4B). Furthermore, Parkin co-IP’d with FANCC-Flag in HeLa FANCCKO/FANCC/Parkin cells (Figure 4C). Thus, Parkin increases the mitochondrial localization of FANCC during OA treatment, colocalizes with FANCC at damaged mitochondria and biochemically interacts with FANCC. Although FANCC interacts with Parkin, it does not function as a classical mitophagy receptor that binds cargo and LC3, as no interaction was detected between FANCC-Flag and Atg8 family proteins in the presence or absence of OA (Figure S4A). The interaction between Parkin and FA proteins was not an artifact of ectopic FA protein expression, as Parkin interacted with endogenous FANCA in HeLa/Parkin cells, and this increased with OA (Figure 4D).
Figure 4. FANCC Interacts with Parkin and Increases its Localization to Mitochondria in a Parkin-Dependent Fashion.
(A) Mitochondrial colocalization of FANCC-Flag in HeLa FANCCKO/FANCC/Parkin cells treated with DMSO or OA for 8 hr, and subjected to immunofluorescence staining to detect TOMM20 and FANCC-Flag. Similar results were observed in three independent experiments. Scale bars, 20 μm.
(B) Representative light micrographs of immunofluorescence staining of HeLa FANCCKO cells transiently transfected with FANCC-Flag and mCherry-Parkin for 24 hr and then treated with DMSO or OA for 4 hr. Scale bars, 10 μm.
(C) Co-IP of Parkin with FANCC-Flag in HeLa FANCCKO cells transiently transfected with mCherry-Parkin and vector or FANCC-Flag for 24 hr.
(D) Co-IP of Parkin with endogenous FANCA in HeLa WT/Parkin cells +/− OA for 4 hr.
(E) EMs of immunoprecipitated mitochondria using an anti-TOMM20 antibody from HeLa WT/Parkin cells +/− OA for 4 hr. Black arrowhead, ruptured mitochondrial membrane.
(F) Western blot of indicated proteins in TOMM20 immunoprecipitates from HeLa WT/Parkin or HeLa FANCCKO/Parkin cells +/− OA for 4 hr. Input, crude mitochondrial fractions. MMC, whole cell lysate from HeLa FANCCKO/FANCC/Parkin cells treated with MMC for 24 hr is included as a loading control to show FANCD2 mono-ubiquitination. For mitochondrial samples, protein loading was normalized by densitometry for HSP60 in crude mitochondrial fractions (input). Left and right gel panels for each protein are cropped from the same gel image.
See also Figure S4.
To confirm that FA proteins are present in mitochondria, we isolated highly purified mitochondria from HeLa WT/Parkin cells using differential centrifugation followed by anti-TOMM20 IP of crude mitochondrial fractions (Fig. S4B). We confirmed this approach yielded mitochondria by EM evaluation of the anti-TOMM20 immunoprecipitates; the mitochondria were intact in the absence of OA but were ruptured in the presence of OA (Figure 4E). Western blot analyses of these anti-TOMM20 immunoprecipitates (mitochondria) revealed the presence of other mitochondrial proteins such as HSP60 and COXIV and no evidence of nuclear lamin A/C contamination (Figure 4F). With (but not without) OA, HA-Parkin was also present in mitochondria isolated from both HeLa WT and HeLa FANCCKO cells, suggesting that FANCC is not required for mitochondrial localization of Parkin. Moreover, endogenous FANCD2 was present in mitochondrial fractions in the presence or absence of FANCC or OA. Unlike nuclear FANCD2 which is mono-ubiquitinated in response to treatment with the DNA cross-linking agent, MMC, mitochondrial FANCD2 was not mono-ubiquitinated during mitophagy-inducing conditions.
FANCC inhibits mtROS production and inflammasome activation
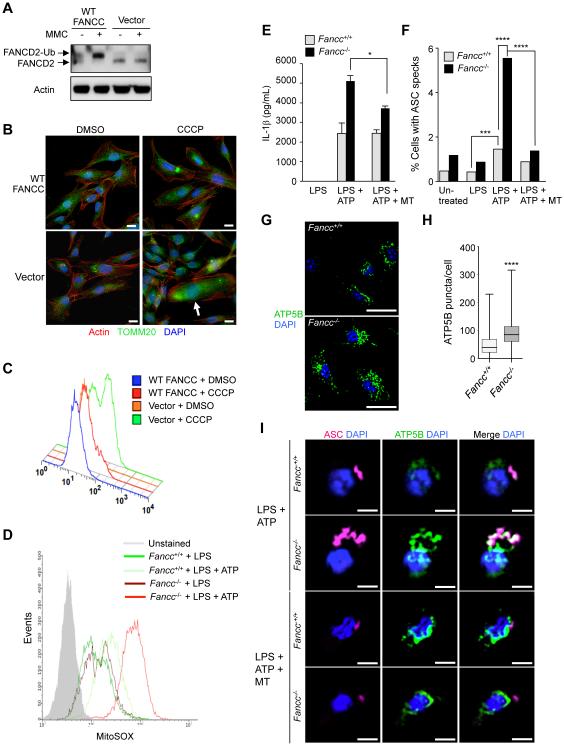
We evaluated whether fibroblasts from patients with FA exhibit abnormalities in the clearance of damaged mitochondria. FANCC mutant patient fibroblasts are deficient in FANCD2 mono-ubiquitination when treated with MMC, and this is rescued by retroviral transduction of WT FANCC (Figure 5A). In response to CCCP, FANCC patient mutant fibroblasts accumulate diffuse fragmented mitochondria as detected by TOMM20 immunostaining (Figure 5B). This accumulation of damaged mitochondria was associated with increased mtROS levels which were rescued by expression of WT FANCC (Figure 5C). Thus, FANCC removes damaged mitochondria and decreases levels of mtROS in human fibroblasts.
Figure 5. FANCC Deficiency in Patient Fibroblasts and Murine BMDMs Results in Abnormal mtROS Production.
(A) FANCC-deficient patient fibroblasts are defective in FA core complex activity assessed by western blot of MMC-induced FANCD2 mono-ubiquitination. FANCC-deficient patient fibroblasts were transduced with a FANCC-expressing retrovirus (WT FANCC) or a vector control retrovirus (Vector) and then treated with 1 μM MMC for 24 hr.
(B) Mitochondrial morphology in WT FANCC or vector transduced immortalized patient fibroblasts. Cells were treated with 10 μM CCCP for 24 hr prior to immunofluorescence staining with anti-TOMM20. White arrow, representative cell with diffuse accumulation of fragmented mitochondria. Scale bars, 20 μm.
(C) MtROS production assessed by flow cytometric analysis of MitoSOX fluorescence levels in WT FANCC or vector transduced immortalized patient fibroblasts treated with DMSO or 10 μM CCCP for 24 hr.
(D) MtROS production assessed by flow cytometric analysis of MitoSOX fluorescence levels in primary BMDMs from Fancc+/+ and Fancc−/− mice. BMDMs were treated with LPS (100 ng/mL, 4 hr) +/− ATP (5 mM) during the final 30 min prior to flow cytometric analysis.
(E) Inflammasome activation measured by IL-1β levels in supernatants of BMDMs of treated with LPS (100 ng/mL, 4 hr) +/− ATP (5 mM) and MitoTEMPO (500 μM) for the final 30 min of LPS treatment. Bars are mean ± SEM for triplicate samples. *P<0.05; t-test.
(F) Inflammasome activation measured by ASC speck formation in BMDMs treated with LPS (100 ng/mL, 4 hr), LPS and ATP (5 mM for final 15 min of LPS), or LPS, ATP and MitoTEMPO (500 μM for the final hr of LPS) prior to immunostaining and automate image analysis. At least 500 cells were analyzed per condition. **, P<0.01, ****, P<0.0001, chi-square test.
(G-H) Representative images of ATP5B immunoflourescence staining (G) and quantitation of cytoplasmic ATP5B puncta (H) in LPS-treated (100 ng/mL, 4 hr) BMDMs. Shown in (H) are box plots of at least 450 cells analyzed per condition. ****, P<0.0001, Mann-Whitney U-test. Scale bars, 10 μm.
(I) Representative light micrographs of immunofluorescence staining of ASC and ATP5B in Fancc+/+ and Fancc−/− BMDMs treated with LPS + ATP or LPS + ATP + MT as described in (F). Scale bars, 5 μm.
For (A-I), similar results were observed in three independent experiments. MT, mitoTEMPO.
See also Figure S5.
Given the established role of mtROS in inflammasome activation (Tschopp, 2011), we hypothesized that increased mtROS generation in FANCC mutant cells leads to enhanced inflammasome-dependent cytokine secretion. When we stimulated bone marrow-derived macrophages (BMDMs) from Fancc+/+ and Fancc−/− mice with inflammasome activators, bacterial lipopolysaccharides (LPS) and ATP (Figure 5D), BMDMs from Fancc−/− mice produced more mtROS than BMDMs from WT littermates. BMDMs from Fancc−/− mice also secreted more IL-1β than BMDMs from Fancc+/+ mice after LPS + ATP (Figure 5E) indicating increased inflammasome activation. This effect was partially reversed by MitoTEMPO, a mitochondria-localized oxygen scavenger (Trnka et al., 2009), indicating a role for increased mtROS generation in enhanced inflammasome activation in Fancc-deficient BMDMs.
To confirm that mtROS causes enhanced inflammasome activation in Fancc−/− BMDMs, we analyzed an additional readout of inflammasome activation, the ASC speck (Stutz et al., 2013). The ASC speck is a macromolecular structure consisting of cytoplasmic sensors that interact with the adaptor protein ASC, which serves as a scaffold to link sensor(s) with caspase-1; inside these structures, caspase-1 processes pro-IL-1β to its mature, secreted form (Man et al., 2014). Consistent with our data on IL-1β secretion, cells containing an ASC speck were more frequent in Fancc−/− than in Fancc+/+ BMDMs and this was reduced by MitoTEMPO (Figure 5F).
To determine whether the elevated levels of mtROS and inflammasome activation in Fancc-deficient BMDMs are associated with a defect in mitochondrial clearance, we stained the mitochondrial matrix protein ATP5B in untreated and LPS-treated Fancc+/+ and Fancc−/− BMDMs (LPS + ATP treatment induced cell death too rapidly to allow measurement of mitochondrial clearance [data not shown]). At baseline, fragmented mitochondria accumulated throughout the cytoplasm in Fancc−/− but not Fancc+/+ BMDMs (Figure S5). Following LPS, Fancc+/+ BMDMs had significantly more clearance of mitochondria than Fancc−/− BMDMs (Figure 5G and 5H). These observations suggest that Fancc−/− BMDMs have a basal defect in mitochondrial quality control and an impaired ability to clear mitochondria in response to LPS.
In parallel with the effects of MitoTEMPO on decreased inflammasome activation, Fancc−/− BMDMs treated with LPS + ATP had a striking accumulation of large ASC specks which colocalized with mitochondria and which decreased in size and number with MitoTEMPO treatment (Figure 5F and 5I). Taken together, these findings suggest that the failure to clear damaged mitochondria leads to elevated mtROS levels and inflammasome activation in Fancc-deficient BMDMs.
The role of FANCC in mitophagy is genetically distinct from its role in nuclear DNA damage repair
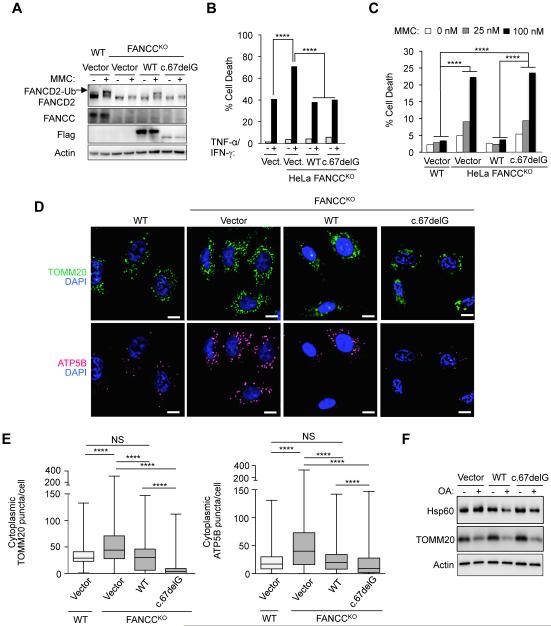
FANCC-mutated cells carrying a hypomorphic allele of a naturally occurring mutation in FANCC, FANCC c.67delG, are not hypersensitive to inflammatory cytokine-induced cell death (Pang et al., 2001). However, these cells are still defective in the DNA damage repair function of the FA core complex, which is commonly assessed by sensitivity to cell death induced by MMC and immunoblot analysis of MMC-induced FANCD2 mono-ubiquitination. Although FANCC c.67delG partially rescued MMC-induced cell death in a previous study, this was an artifact of overexpression as patient cells harboring the FANCC c.67delG allele were indistinguishable from patient cells with null alleles for FANCC in the MMC-induced cell death assay (Yamashita et al., 1996). Therefore, we used FANCC c.67delG as a naturally occurring mutant form of FANCC to investigate whether the mitophagy function of FANCC is linked to its role in genomic DNA damage repair.
We expressed Flag epitope-tagged WT FANCC or FANCC c.67delG in HeLa FANCCKO cells (Figure 6A) and examined cytokine hypersensitivity (Figure 6B), MMC-induced FANCD2 mono-ubiquitination (Figure 6A) and cell death (Figure 6C), and Parkin-mediated mitophagy (Figure 6D-F). Although FANCC protein levels were lower in FANCCKO cells transduced with a vector expressing FANCC c.67delG than WT FANCC (and the protein levels of both WT FANCC and FANCC c.67delG were lower than endogenous FANCC protein levels) (Figure 6A), FANCC c.67delG rescued cytokine hypersensitivity (as measured by cell death in response to TNF-α and IFN-γ) as efficiently as WT FANCC (Figure 6B). Although FANCC c.67delG rescued cytokine hypersensitivity, it failed to rescue nuclear DNA damage repair functions of FANCC. WT FANCC but not FANCC c.67delG rescued both MMC-induced FANCD2 mono-ubiquitination (Figure 6A) and cell death (Figure 6C), confirming that the FANCC c.67delG mutant is a null allele for the genomic DNA repair function of the FA pathway.
Figure 6. The Mitophagy Function of FANCC is Genetically Dissociated from its Role in Nuclear DNA Damage Repair.
(A) FA core complex activity assessed by western blot of MMC (1 μM, 24 hr)-induced FANCD2 mono-ubiquitination in parental HeLa cells (WT) or HeLa FANCCKO cells transduced with indicated FANCC vector. A longer exposure of FANCC reveals FANCC-Flag protein expression in lanes 4-8 (data not shown).
(B) Cytokine-induced cell death in HeLa FANCCKO cells transduced with indicated FANCC vector. Cells were treated with TNF-α and IFN-γ (10 ng/mL each, 48 hr) and cell death was measured by flow cytometric analysis of 7-AAD (a vital dye) staining. ****, P<0.0001, chi-square test.
(C) MMC-induced cell death in HeLa FANCCKO cells transduced with indicated FANCC vector. Cells were treated with MMC (1 μM) for 48 hr and cell death was measured by flow cytometric analysis of 7-AAD (a vital dye) staining. ****, P<0.0001, chi-square test.
(D) Representative images of TOMM20 (upper panels) or ATP5B (lower panels) immunoflourescence staining of parental HeLa/Parkin cells (WT) or HeLa FANCCKO/Parkin cells transduced with indicated FANCC vector treated with OA for 8 hr prior to imaging and automated image analysis (D). Scale bars, 10 μm.
(E) Quantitation of cytoplasmic TOMM20 puncta or ATP puncta in cells treated as shown in representative images in (D). Shown are box plots of at least 150 cells analyzed per condition. ****, P<0.0001, Mann-Whitney U-test.
(G) Mitophagy analysis in HeLa FANCCKO/Parkin cells transduced with indicated FANCC vector assessed by western blot of HSP60 or TOMM20 proteins in cells +/− OA for 8 hr.
For (A-F), similar results were observed in three independent experiments.
In FANCCKO cells expressing FANCC c.67delG, Parkin-mediated mitochondrial clearance after OA treatment (measured by quantitative image analysis of TOMM20 and ATP5B puncta and western blot analysis of HSP60 and TOMM20) was equal to or greater than in FANCCKO cells expressing WT FANCC (Figures 6D-G, Figure S6). Thus, the mitophagy function of FANCC segregates with its role in cytokine hypersensitivity rather than its role in genomic DNA damage repair.
Multiple Fanconi anemia genes are required for mitophagy
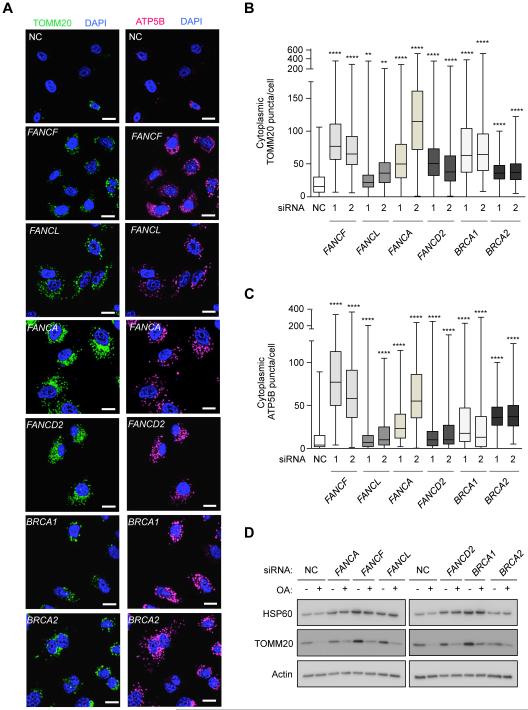
To evaluate whether selective autophagy represents a function of FA pathway genes other than FANCC, we evaluated Parkin-mediated mitophagy in HeLa cells with knockdown of additional FA core complex genes, including FANCA, FANCF and FANCL, as well as FANCD2, BRCA1/FANCS, and BRCA2/FANCD1, which coordinate the repair of genomic DNA (Bagby and Alter, 2006). We confirmed that all four siRNAs targeting FANCF and FANCL used in our previous screen (Orvedahl et al. 2011) decreased CCCP-induced Parkin-dependent clearance of TOMM20-positive mitochondria as well as knockdown of the essential autophagy gene, ATG7 (Figure S7A). By quantitative image analysis of two mitochondrial markers, TOMM20 and ATP5B, independent siRNAs targeting each of the following FA genes – FANCA, FANCF, FANCL, FANCD2, BRCA1 and BRCA2 – impaired the clearance of mitochondria after OA (Figure 7A-C, Figure S7B). Western blot analysis of mitochondrial proteins, HSP60 and TOMM20, confirmed a mitophagy defect in cells with knockdown of each of these FA genes (Figure 7D). Thus, multiple FA genes, including those lacking known cytoplasmic functions, are essential for mitophagy.
Figure 7. Multiple FA Genes are Required for Mitophagy.
(A) Representative light micrographs of images used for quantification of TOMM20 (left panels) in (B) and ATP5B (right panels) clearance in (C) in HeLa/Parkin cells transfected with indicated siRNAs and 48 hr later treated with OA for 8 hr. Scale bars, 20 μm.
(B-C) Quantitation of cytoplasmic TOMM20 (B) or ATP5B (C) puncta in cells transfected with indicated siRNAs and treated as in (A). Shown are box plots of at least 150 cells analyzed per condition. Similar results were observed in three independent experiments. **, P<0.01, ****, P<0.0001; Mann-Whitney U-test.
(D) Mitophagy analysis in HeLa FANCCKO/Parkin cells transfected with indicated siRNA assessed by western blot of indicated proteins +/− OA for 8 hr. See also Figure S7.
DISCUSSION
Our data demonstrate a previously unappreciated role for FA genes in the selective autophagy of viruses and damaged mitochondria. The FA pathway possesses not only a nuclear function in DNA damage repair but also a cytoplasmic function in selective (but not general) autophagy, and thereby, organellar quality control. Both of these functions may act in concert to protect the host genome in embryonic development, cancer and aging; specifically, by restraining mtROS production via the proper removal of damaged mitochondria, the FA pathway can limit genotoxic stress in parallel with its ability to repair damaged nuclear DNA, thus serving a “double-duty” role in protection of the host genome. Moreover, the role of FA genes in selective virophagy suggests yet another level of host cellular protection mediated by the FA pathway that may interplay with mitophagy, mtROS-dependent inflammasome activation and nuclear DNA damage repair to protect organisms against bone marrow failure and/or cancers in the setting of infection and/or proinflammatory signals. Thus, the cytoplasmic functions of FA pathway genes in selective autophagy may coordinate with their nuclear functions to maintain cellular homeostasis.
Role of FA genes in the control of viral infection
We found that Fancc was required for virophagy of two genetically diverse viruses, including Sindbis virus (SIN), a single-stranded positive-sense RNA virus, and herpes simplex virus type 1 (HSV-1), a double-stranded DNA virus. Furthermore, Fancc−/− mice were more susceptible to lethal CNS infection with these two pathogens, thus identifying Fancc as a host innate antiviral immunity factor.
Similar to previously identified SIN virophagy factors, p62 and SMURF1 (Orvedahl et al., 2010; Orvedahl et al., 2011), FANCC also interacts with SIN capsid protein (which is not known to be ubiquitinated (Sumpter and Levine, 2011)). In addition, we found that FANCC was present in immunoprecipitated autophagic structures containing SIN nucleocapsids. Thus, FANCC may act directly in targeting SIN nucleocapsids for delivery to autophagosomes, although the precise molecular mechanism by which FANCC functions in SIN virophagy is unknown. Nonetheless, histopathological analyses of SIN-infected mouse brains suggest that Fancc functions similarly to core autophagy genes in protecting neurons against SIN infection by clearing viral antigen, thereby decreasing virus-induced neuronal death (Orvedahl et al., 2010).
The increased susceptibility to lethal CNS HSV-1 infection is consistent with previous reports indicating that the autophagy pathway, rather than interferon responses, protects against HSV-1 neuronal disease (Yordy and Iwasaki, 2013). The restored neurovirulence of an HSV-1 mutant strain lacking the ability to block autophagy, but not of another HSV-1 strain neuroattenuated due to a mutation in a gene unrelated to autophagy, provides genetic evidence (Leib et al., 2000) that the protective function of Fancc in CNS neurons is mediated by autophagy and not other host immunity pathways.
These findings raise intriguing questions about the interrelationship between FA gene deficiency and viral infection, both in the context of antiviral defense and in FA pathogenesis. A role for FANCC in virophagy may mechanistically explain either the increased prevalence of certain viral infections (such as human papilloma virus [HPV] or SV40) in FA patients (Comar et al., 2013; Sauter et al., 2015) or the role of FA pathway components (such as FANCA and FANCD2) in restricting the HPV life cycle (Hoskins et al., 2012). Furthermore, the failure to remove aggregates of viral proteins in the setting of Fancc deficiency may represent an environmental trigger that functions in the pathophysiology of FA (Parmar et al., 2009). Defective general autophagy during single-stranded RNA virus infection has been linked to increased mtROS production and proinflammatory signaling (Tal et al., 2009). Thus, FA gene deficiency could lead to increased generation of known and potential cytokine triggers of bone marrow failure in FA patients (Dufour et al., 2003; Garbati et al., 2013) through the dual effects of defective viral autophagic targeting (and increased accumulation of pathogen-associated molecular patterns) and defective removal of damaged mitochondria.
Role of FA genes in mitophagy, mtROS generation, and inflammasome activation
Our data indicate that FANCC is required for mitophagy, FANCC interacts with Parkin, and FANCC accumulates in damaged mitochondria in a Parkin-dependent manner. In addition, FANCC likely plays a role in removing damaged mitochondria independently of Parkin, as FANCC patient mutant fibroblasts (that lack Parkin expression) accumulated damaged mitochondria when treated with a mitochondrial uncoupling agent, and the brains and hearts of Fancc−/− mice accumulated damaged mitochondria with aging. The role of FANCC in Parkin-dependent mitophagy is independent of its role in genomic DNA repair, as the FANCC c.67delG mutant (which is as effective as WT FANCC in decreasing cytokine hypersensitivity but cannot rescue MMC-induced cell death or FANCD2 mono-ubiquitination) fully restored mitophagy in FANCC-deficient cells. The mechanism by which FANCC functions in the targeting of diverse substrates (two unrelated viruses and mitochondria), without interacting with LC3 family members, is an important area of future study.
We found that multiple other FA genes including FANCA, FANCF, FANCL, FANCD2, and BRCA1 and BRCA2 are required for Parkin-mediated mitophagy, suggesting that the FA pathway (rather than FANCC alone) participates in organellar homeostasis. While some FA proteins may regulate specific cytoplasmic functions through transcriptional effects and/or function directly in the cytoplasm (Du et al., 2012; Mullan et al., 2006; Preobrazhenska et al., 2002), some FA proteins, such as FANCD2, BRCA1 and BRCA2, have not been described to function directly in the cytoplasm. Besides observing co-localization of FANCC with damaged mitochondria in Parkin-expressing cells, we also biochemically detected FANCD2 in purified mitochondrial fractions. Taken together, our data suggest that the FA pathway, including those components downstream of the FA core complex which are part of nuclear DNA damage foci, functions in the cytoplasm (and potentially at the mitochondrion itself) to mediate the selective removal of damaged mitochondria. The precise mechanism by which FA pathway proteins mediate mitophagy remains to be determined. For FA proteins other than those in the core FA complex, we cannot rule out that defects in mitophagy with knockdown of protein expression are an indirect consequence of defects in genomic DNA damage repair.
Previous studies have indicated that FA gene-deficient cells (from humans and from knockout mouse models) have increased ROS, increased sensitivity to cytokine-induced cell death (Garaycoechea and Patel, 2014), and increased TLR-induced inflammasome activation (Garbati et al., 2013). Our data indicate that damaged mitochondria are a major source of ROS in FA-deficient cells and directly link increased mtROS generation with increased inflammasome activation. Furthermore, our findings are consistent with a model in which damaged, ROS-producing mitochondria in LPS-treated Fancc-deficient BMDMs act as “scaffolds” for the assembly of large ASC specks to drive pro-IL-1β processing. These observations provide a mechanism by which defective mitophagy (which leads to increased mtROS) in the setting of FA gene deficiency may lead to aberrant inflammasome activation and other adverse consequences of increased mtROS in patients with FA mutations. Although low levels of mtROS are required for many homeostatic functions (including control of cell growth, differentiation, death, senescence and inflammation (Hamanaka and Chandel, 2010)), alterations in cellular signaling pathways and genotoxic stress arising from dysregulated mtROS production are sufficient to drive cellular transformation and tumor progression in multiple cancer models (Costa et al., 2014). We speculate that the failure to clear damaged mitochondria may be an important driver of bone marrow failure and tumorigenesis in the setting of FA gene defects/mutations, either as a result of mtROS-amplified proinflammatory cytokine production, increased cell death, and/or accumulation of protumorigenic mutations.
Defective mitophagy may contribute to cancers that occur in patients with mutations in FA pathway genes
Hematological malignancies in FA patients are thought to result from clonal expansions of HSCs that survive after generalized bone marrow failure (Bagby and Alter, 2006). The pathogenesis of solid malignancies in these patients is not understood, but may be related to increased susceptibility to HPV infection (Liu et al., 2015). Consequently, as noted above, defects in virophagy and/or mitophagy (and downstream consequences of such defects) may contribute not only to bone marrow failure, but also to the increased risk of malignancy in FA patients. Given the essential role of autophagy in aging (Rubinzstein et al., 2011) and development (Mizushima and Levine, 2010), defects in selective autophagy may also contribute to the congenital abnormalities and the aging phenotype in FA patients.
Defective mitophagy may also play a role in tumorigenesis that occurs in the setting of non-FA patients with driver mutations in FA genes. The mild phenotype of FA patients with the hereditary FANCC c.67delG mutation, which is functional in mitophagy but not in genomic DNA repair, argues for an important role of cytoplasmic functions of FA genes in protection against cancer. Of note, other studies suggest that defective mitophagy (as a result of mutations in genes that function in selective but not general autophagy) contributes to tumorigenesis (Chourasia et al. 2015). PARK2 encodes Parkin, an E3 ligase that is essential in mitophagy but not general autophagy (Rogov et al., 2014), and it is likely that Parkin is a tumor suppressor protein (Xu et al., 2014). PARK2 is deleted or mutated in many human cancers (including bladder, lung, breast, ovarian, and colon), and Parkin null mice display spontaneous liver carcinomas, increased colorectal neoplasia when crossed with Apc mutant mice, and increased susceptibility to radiation-induced lymphogenesis (Chourasia et al., 2015; Poulogiannis et al., 2010). These observations, coupled with our findings that multiple FA pathway genes (including the well-characterized tumor suppressors commonly mutated in familial breast and ovarian cancer, BRCA1 and BRCA2) function in mitophagy, support the hypothesis that FA gene mutations may promote cancer through their essential roles in mitophagy in addition to through their roles in genomic DNA damage repair.
Conclusion
Our results reveal a previously undescribed cytoplasmic role for FA pathway genes, specifically in the cellular homeostasis pathway involving selective autophagy of unwanted cargo. These findings provide new directions for investigation into the pathophysiology of not only FA and cancers that arise from driver mutations of FA genes, but also of congenital abnormalities, infectious diseases, aging and other processes in which the autophagy pathway plays an important role (Levine and Kroemer, 2008; Mizushima and Komatsu, 2011; Rubinsztein et al., 2011).
EXPERIMENTAL PROCEDURES
Primary Murine Cells and HeLa FANCCKO Cells
Primary murine embryonic fibroblasts (MEFs) were established from Fancc+/+ and Fancc−/− mice at day e13.5. Primary murine bone marrow-derived macrophages (BMDMs) were generated from Fancc+/+ and Fancc−/− mice by culturing cells from mouse femurs and tibia in DMEM supplemented with 20% FBS and 30% L929 cell-conditioned media. HeLa FANCCKO cells were generated by CRISPR/Cas9 genome editing at the Washington University Genome Engineering and iPSC center (St. Louis, MO). See Supplemental Experimental procedures for details.
LC3 and TOMM20 IP’s
HeLa cells were mechanically lysed and fractionated using the QProteome mitochondria isolation kit (Qiagen) according to the manufacturer’s instructions. Microsomal + mitochondrial fractions were used to isolate LC3-positive membranous structures in cells infected with SIN strain SVIA by performing IP with an anti-LC3 antibody. Mitochondrial fractions were used to isolated purified mitochondria from cells treated DMSO- or OA-treated (4 hr) by performing IP with an anti-TOMM20 antibody. See Supplemental Experimental procedures for details.
Mouse Strains and Animal Studies
See Supplemental Experimental procedures for details.
siRNAs and qRT-PCR
See Supplemental Experimental procedures and Supplemental Table 1 for details.
Light and Electron microscopy, Quantitative Image Analyses, and Flow cytometry
See Supplemental Experimental procedures for details.
Mammalian Cells, Viruses, Plasmids, Antibodies and Chemicals
See Supplemental Experimental procedures for details.
Western Blot Analyses, Co-Immunoprecipitations and IL-1β ELISA
See Supplemental Experimental procedures for details.
Statistical Analyses
Log-rank tests were used to analyze all mortality studies. t-tests were used for comparisons of means of normally distributed data and Mann-Whitney U tests were used for comparison of non-normally distributed data. Chi-square analysis was used to determine difference between groups in ASC speck and flow cytometry experiments.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. MacDonald, N. Mizushima, R. Thompson, and R. Youle for providing critical reagents; D. Chan, A. Orvedahl, and T. Ross for helpful discussions; L. Nguyen and S.L. Kelich for technical assistance; A. Bugde, P. Doss, R. Leidel, and A. Darehshouri for assistance with light and electron microscopy; and H. Smith for assistance with manuscript preparation. This work was supported by NIH grants K08AI099150 (R.S.), CA155294 (H.H.), U19 AI109725 (B.L.) and RO1 109618 (B.L), CPRIT grant RP120718 (B.L.), a UTSW PRC Award (R.S.), a Burroughs Wellcome CAMS (R.S.), a fellowship from Ministerio de Educación of Spain (A.F.), the Lilly Foundation Physician/Scientist initiative (H.H.) and the Walther Oncology Foundation (H.H).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, one table, and seven figures.
AUTHOR CONTRIBUTIONS
R.S., D.C., H.H. and B.L. designed the study; R.S., S.S., A.F., Y.W., X.D., L.F., Z.Z., C.M. and M.L. performed experiments; R.S., H.H. and B.L. analyzed the data; and R.S. and B.L. wrote the manuscript.
REFERENCES
- Bagby GC, Alter BP. Fanconi anemia. Semin Hematol. 2006;43:147–156. doi: 10.1053/j.seminhematol.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Bogliolo M, Surralles J. Fanconi anemia: a model disease for studies on human genetics and advanced therapeutics. Curr Opin Genet Dev. 2015;33:32–40. doi: 10.1016/j.gde.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Chourasia AH, Boland ML, Macleod KF. Mitophagy and cancer. Cancer Metabol. 2015;3:4. doi: 10.1186/s40170-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comar M, De Rocco D, Cappelli E, Zanotta N, Bottega R, Svahn J, Farruggia P, Misuraca A, Corsolini F, Dufour C, et al. Fanconi anemia patients are more susceptible to infection with tumor virus SV40. PloS one. 2013;8:e79683. doi: 10.1371/journal.pone.0079683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Scholer-Dahirel A, Mechta-Grigoriou F. The role of reactive oxygen species and metabolism on cancer cells and their microenvironment. Semin Cancer Biol. 2014;25:23–32. doi: 10.1016/j.semcancer.2013.12.007. [DOI] [PubMed] [Google Scholar]
- D'Andrea AD. Susceptibility pathways in Fanconi's anemia and breast cancer. N Engl J Med. 2010;362:1909–1919. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Rani R, Sipple J, Schick J, Myers KC, Mehta P, Andreassen PR, Davies SM, Pang Q. The FA pathway counteracts oxidative stress through selective protection of antioxidant defense gene promoters. Blood. 2012;119:4142–4151. doi: 10.1182/blood-2011-09-381970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour C, Corcione A, Svahn J, Haupt R, Poggi V, Béka'ssy AN, Scime R, Pistorio A, Pistoia V. TNF-α and IFN-γ are overexpressed in the bone marrow of Fanconi anemia patients and TNF-α suppresses erythropoiesis in vitro. Blood. 2003;102:2053–2059. doi: 10.1182/blood-2003-01-0114. [DOI] [PubMed] [Google Scholar]
- Garaycoechea JI, Patel KJ. Why does the bone marrow fail in Fanconi anemia? Blood. 2014;123:26–34. doi: 10.1182/blood-2013-09-427740. [DOI] [PubMed] [Google Scholar]
- Garbati MR, Hays LE, Keeble W, Yates JE, Rathbun RK, Bagby GC. FANCA and FANCC modulate TLR and p38 MAPK-dependent expression of IL-1beta in macrophages. Blood. 2013;122:3197–3205. doi: 10.1182/blood-2013-02-484816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci. 2010;35:505–513. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneline LS, Broxmeyer HE, Cooper S, Hangoc G, Carreau M, Buchwald M, Clapp DW. Multiple inhibitory cytokines induce deregulated progenitor growth and apoptosis in hematopoietic cells from Fac−/− mice. Blood. 1998;91:4092–4098. [PubMed] [Google Scholar]
- Hoskins EE, Morreale RJ, Werner SP, Higginbotham JM, Laimins LA, Lambert PF, Brown DR, Gillison ML, Nuovo GJ, Witte DP, et al. The fanconi anemia pathway limits human papillomavirus replication. J Virol. 2012;86:8131–8138. doi: 10.1128/JVI.00408-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leib DA, Machalek MA, Williams BR, Silverman RH, Virgin HW. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc Natl Acad Sci U S A. 2000;97:6097–6101. doi: 10.1073/pnas.100415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GB, Chen J, Wu ZH, Zhao KN. Association of human papillomavirus with Fanconi anemia promotes carcinogenesis in Fanconi anemia patients. Rev Med Virol. 2015;6:345–53. doi: 10.1002/rmv.1834. [DOI] [PubMed] [Google Scholar]
- Man SM, Hopkins LJ, Nugent E, Cox S, Gluck IM, Tourlomousis P, Wright JA, Cicuta P, Monie TP, Bryant CE. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc Natl Acad Sci U S A. 2014;111:7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, Stranks AJ, Glanville J, Knight S, Jacobsen SE, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–5863. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K, Endt D, Hoehn H, Schindler D. Genotype-phenotype correlations in Fanconi anemia. Mutat Res. 2009;668:73–91. doi: 10.1016/j.mrfmmm.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host & Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, MacPherson S, Sumpter R, Jr., Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host & Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Sumpter R, Jr., Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Q, Christianson TA, Keeble W, Diaz J, Faulkner GR, Reifsteck C, Olson S, Bagby GC. The Fanconi anemia complementation group C gene product: structural evidence of multifunctionality. Blood. 2001;98:1392–1401. doi: 10.1182/blood.v98.5.1392. [DOI] [PubMed] [Google Scholar]
- Poulogiannis G, McIntyre RE, Dimitriadi M, Apps JR, Wilson CH, Ichimura K, Luo F, Cantley LC, Wyllie AH, Adams DJ, et al. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc Natl Acad Sci U S A. 2010;107:15145–15150. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preobrazhenska O, Yakymovych M, Kanamoto T, Yakymovych I, Stoika R, Heldin CH, Souchelnytskyi S. BRCA2 and Smad3 synergize in regulation of gene transcription. Oncogene. 2002;21:5660–5664. doi: 10.1038/sj.onc.1205732. [DOI] [PubMed] [Google Scholar]
- Pyles RB, Thompson RL. Mutations in accessory DNA replicating functions alter the relative mutation frequency of herpes simplex virus type 1 strains in cultured murine cells. J Virol. 1994;68:4514–4524. doi: 10.1128/jvi.68.7.4514-4524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Sauter SL, Wells SI, Zhang X, Hoskins EE, Davies SM, Myers KC, Mueller R, Panicker G, Unger ER, Sivaprasad U, et al. Oral human papillomavirus is common in individuals with Fanconi anemia. Cancer Epidemiol Biomarkers Prev. 2015;24:864–872. doi: 10.1158/1055-9965.EPI-15-0097-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz A, Horvath GL, Monks BG, Latz E. ASC speck formation as a readout for inflammasome activation. Methods Mol Biol. 2013;1040:91–101. doi: 10.1007/978-1-62703-523-1_8. [DOI] [PubMed] [Google Scholar]
- Sumpter R, Jr., Levine B. Selective autophagy and viruses. Autophagy. 2011;7:260–265. doi: 10.4161/auto.7.3.14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talloczy Z, Virgin HWIV, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- Trnka J, Blaikie FH, Logan A, Smith RA, Murphy MP. Antioxidant properties of MitoTEMPOL and its hydroxylamine. Free Radic Res. 2009;43:4–12. doi: 10.1080/10715760802582183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp J. Mitochondria: Sovereign of inflammation? Eur J Immunol. 2011;41:1196–1202. doi: 10.1002/eji.201141436. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Wu N, Kupfer G, Corless C, Joenje H, Grompe M, D'Andrea AD. Clinical variability of Fanconi anemia (type C) results from expression of an amino terminal truncated Fanconi anemia complementation group C polypeptide with partial activity. Blood. 1996;87:4424–4432. [PubMed] [Google Scholar]
- Yordy B, Iwasaki A. Cell type-dependent requirement of autophagy in HSV-1 antiviral defense. Autophagy. 2013;9:236–238. doi: 10.4161/auto.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.