Abstract
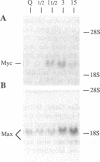
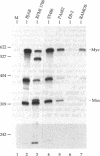
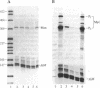
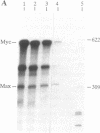
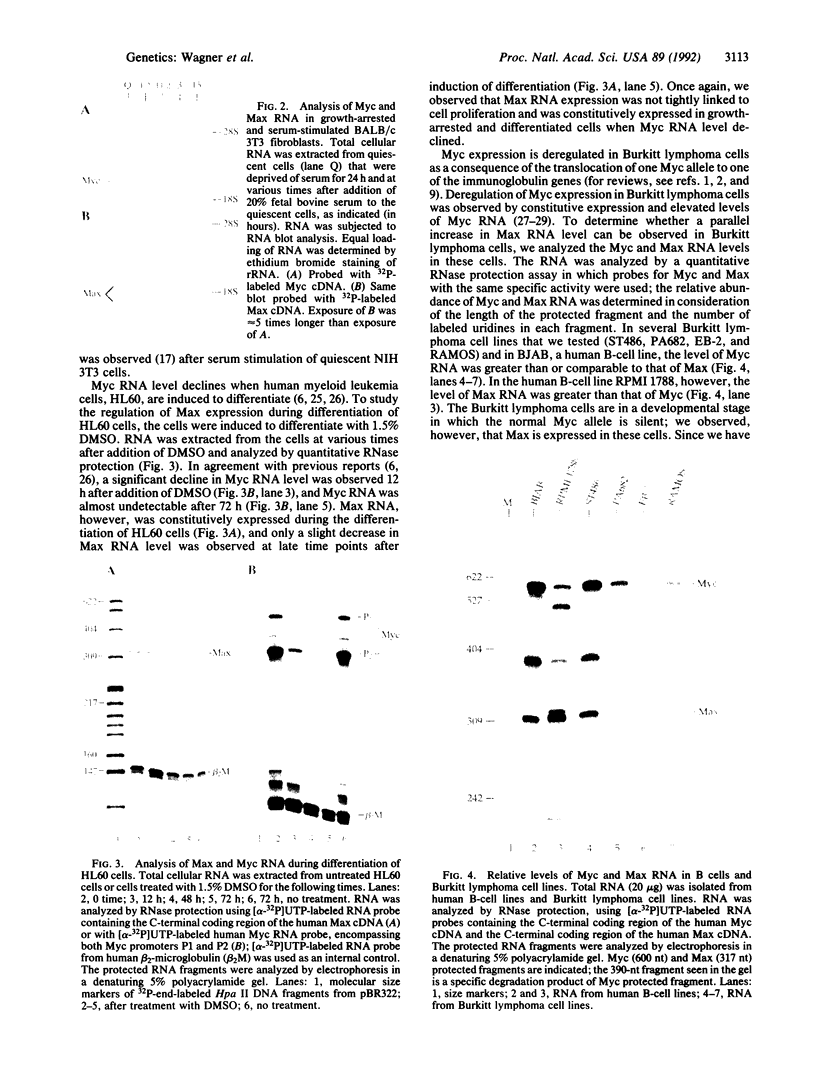
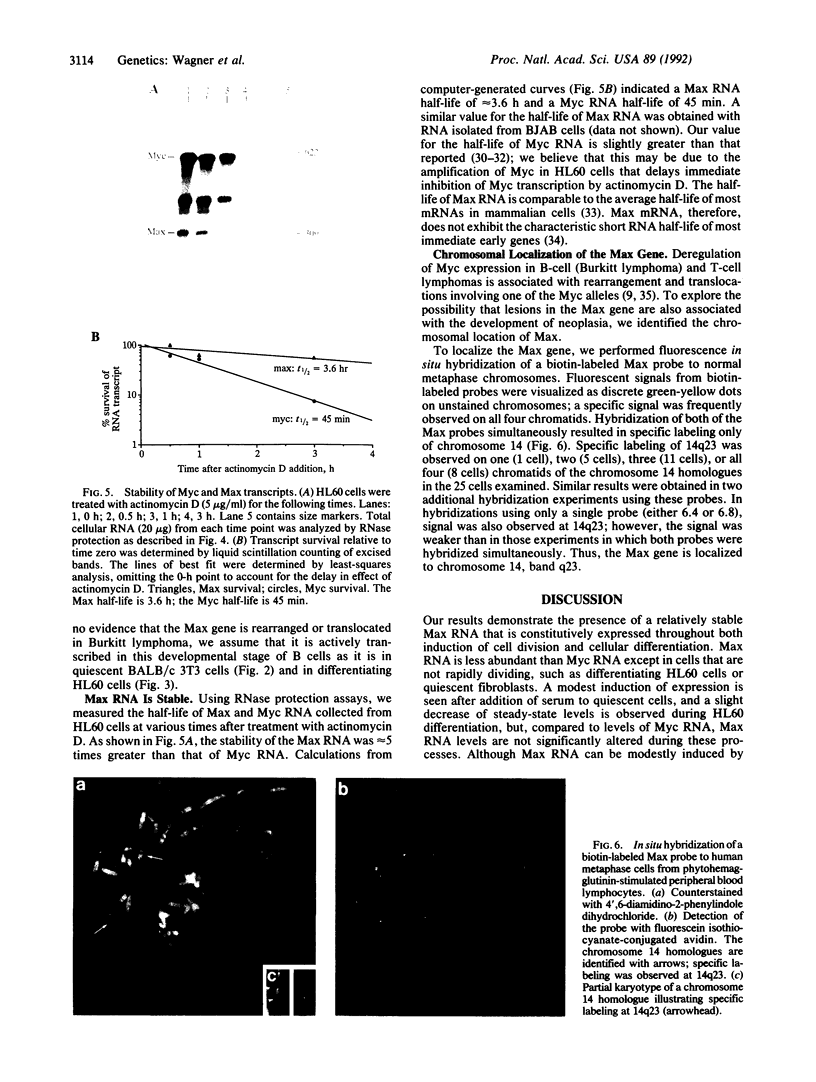
The Max gene encodes a protein that interacts specifically with the Myc protein to form a heterodimer with high affinity for the specific cognate DNA binding site of Myc. Here we examine the expression of Max RNA in comparison to Myc RNA during cell growth and differentiation. Two species of RNA, a major 2.0- and a minor 1.7-kilobase species, hybridized specifically to a Max cDNA probe in all human and murine cell lines that were tested. Unlike Myc, the steady-state level of Max RNA is not significantly modulated with respect to proliferation or differentiation. Max RNA is expressed in quiescent BALB/c 3T3 cells and is modestly increased 3 h after addition of serum to the quiescent cells. In contrast to Myc RNA, Max RNA does not decline immediately upon induction of differentiation of HL60 cells by dimethyl sulfoxide, and only a modest decrease of Max RNA was observed 72 h after induction of differentiation. Unlike Myc RNA, Max RNA is relatively stable with a half-life of greater than 3 h and, therefore, does not exhibit the characteristic short half-life of RNAs encoded by most immediate early genes. The human Max gene was localized to chromosome 14, band q23. With respect to the recurring abnormalities in human tumors, this region of chromosome 14 is involved in deletions in B-cell chronic lymphocytic leukemia and malignant lymphomas and in the 12;14 translocation in uterine leiomyomas.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bishop J. M. Molecular themes in oncogenesis. Cell. 1991 Jan 25;64(2):235–248. doi: 10.1016/0092-8674(91)90636-d. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Broome H. E., Reed J. C., Godillot E. P., Hoover R. G. Differential promoter utilization by the c-myc gene in mitogen- and interleukin-2-stimulated human lymphocytes. Mol Cell Biol. 1987 Aug;7(8):2988–2993. doi: 10.1128/mcb.7.8.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J., Gray H. E., Pardee A. B., Dean M., Sonenshein G. E. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984 Feb;36(2):241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- Cole M. D. Myc meets its Max. Cell. 1991 May 31;65(5):715–716. doi: 10.1016/0092-8674(91)90377-b. [DOI] [PubMed] [Google Scholar]
- Cole M. D. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Cory S. Activation of cellular oncogenes in hemopoietic cells by chromosome translocation. Adv Cancer Res. 1986;47:189–234. doi: 10.1016/s0065-230x(08)60200-6. [DOI] [PubMed] [Google Scholar]
- Cox D. W., Donlon T. A. Report of the committee on the genetic constitution of chromosomes 14 and 15. Cytogenet Cell Genet. 1989;51(1-4):280–298. doi: 10.1159/000132795. [DOI] [PubMed] [Google Scholar]
- Dani C., Blanchard J. M., Piechaczyk M., El Sabouty S., Marty L., Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Levine R. A., Ran W., Kindy M. S., Sonenshein G. E., Campisi J. Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J Biol Chem. 1986 Jul 15;261(20):9161–9166. [PubMed] [Google Scholar]
- Endo T., Nadal-Ginard B. Transcriptional and posttranscriptional control of c-myc during myogenesis: its mRNA remains inducible in differentiated cells and does not suppress the differentiated phenotype. Mol Cell Biol. 1986 May;6(5):1412–1421. doi: 10.1128/mcb.6.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Bishop J. M., Levens D. Regulatory elements that modulate expression of human c-myc. Genes Dev. 1987 Sep;1(7):659–671. doi: 10.1101/gad.1.7.659. [DOI] [PubMed] [Google Scholar]
- Hollis G. F., Gazdar A. F., Bertness V., Kirsch I. R. Complex translocation disrupts c-myc regulation in a human plasma cell myeloma. Mol Cell Biol. 1988 Jan;8(1):124–129. doi: 10.1128/mcb.8.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Rosenfeld P. J., Kelly T. J., Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987 Jan 16;48(1):79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lau L. F., Nathans D. Identification of a set of genes expressed during the G0/G1 transition of cultured mouse cells. EMBO J. 1985 Dec 1;4(12):3145–3151. doi: 10.1002/j.1460-2075.1985.tb04057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P., Cremer T., Borden J., Manuelidis L., Ward D. C. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum Genet. 1988 Nov;80(3):224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R. N. New light on Myc and Myb. Part I. Myc. Genes Dev. 1990 Dec;4(12A):2025–2035. doi: 10.1101/gad.4.12a.2025. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nishikura K., ar-Rushdi A., Erikson J., Watt R., Rovera G., Croce C. M. Differential expression of the normal and of the translocated human c-myc oncogenes in B cells. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4822–4826. doi: 10.1073/pnas.80.15.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechaczyk M., Yang J. Q., Blanchard J. M., Jeanteur P., Marcu K. B. Posttranscriptional mechanisms are responsible for accumulation of truncated c-myc RNAs in murine plasma cell tumors. Cell. 1985 Sep;42(2):589–597. doi: 10.1016/0092-8674(85)90116-3. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Ramsay R. G., Ikeda K., Rifkind R. A., Marks P. A. Changes in gene expression associated with induced differentiation of erythroleukemia: protooncogenes, globin genes, and cell division. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6849–6853. doi: 10.1073/pnas.83.18.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley J. D., Diaz M. O., Espinosa R., 3rd, Patel Y. D., van Melle E., Ziemin S., Taillon-Miller P., Lichter P., Evans G. A., Kersey J. H. Mapping chromosome band 11q23 in human acute leukemia with biotinylated probes: identification of 11q23 translocation breakpoints with a yeast artificial chromosome. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9358–9362. doi: 10.1073/pnas.87.23.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showe L. C., Ballantine M., Nishikura K., Erikson J., Kaji H., Croce C. M. Cloning and sequencing of a c-myc oncogene in a Burkitt's lymphoma cell line that is translocated to a germ line alpha switch region. Mol Cell Biol. 1985 Mar;5(3):501–509. doi: 10.1128/mcb.5.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Bressler P., Kelly K. Two distinct mechanisms of transcriptional control operate on c-myc during differentiation of HL60 cells. Mol Cell Biol. 1988 Feb;8(2):867–874. doi: 10.1128/mcb.8.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. U., Hsu T., Begley D. A., Mitchell B. S., Alizadeh B. N. Transcriptional regulation of the c-myc protooncogene by 1,25-dihydroxyvitamin D3 in HL-60 promyelocytic leukemia cells. J Biol Chem. 1987 Mar 25;262(9):4104–4108. [PubMed] [Google Scholar]
- Small M. B., Hay N., Schwab M., Bishop J. M. Neoplastic transformation by the human gene N-myc. Mol Cell Biol. 1987 May;7(5):1638–1645. doi: 10.1128/mcb.7.5.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- Suggs S. V., Wallace R. B., Hirose T., Kawashima E. H., Itakura K. Use of synthetic oligonucleotides as hybridization probes: isolation of cloned cDNA sequences for human beta 2-microglobulin. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6613–6617. doi: 10.1073/pnas.78.11.6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub R., Moulding C., Battey J., Murphy W., Vasicek T., Lenoir G. M., Leder P. Activation and somatic mutation of the translocated c-myc gene in burkitt lymphoma cells. Cell. 1984 Feb;36(2):339–348. doi: 10.1016/0092-8674(84)90227-7. [DOI] [PubMed] [Google Scholar]
- Trent J. M., Kaneko Y., Mitelman F. Report of the committee on structural chromosome changes in neoplasia. Cytogenet Cell Genet. 1989;51(1-4):533–562. doi: 10.1159/000132807. [DOI] [PubMed] [Google Scholar]
- Ullrich A., Shine J., Chirgwin J., Pictet R., Tischer E., Rutter W. J., Goodman H. M. Rat insulin genes: construction of plasmids containing the coding sequences. Science. 1977 Jun 17;196(4296):1313–1319. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- Welsh M., Nielsen D. A., MacKrell A. J., Steiner D. F. Control of insulin gene expression in pancreatic beta-cells and in an insulin-producing cell line, RIN-5F cells. II. Regulation of insulin mRNA stability. J Biol Chem. 1985 Nov 5;260(25):13590–13594. [PubMed] [Google Scholar]