Abstract
The molecular basis of epileptogenesis is poorly characterized. Studies in humans and animal models have identified an electrophysiological signature that precedes the onset of epilepsy, which has been termed fast ripples (FRs) based on its frequency. Multiple lines of evidence implicate regions generating FRs in epileptogenesis, and FRs appear to demarcate the seizure onset zone, suggesting a role in ictogenesis as well. We performed gene expression analysis comparing areas of the dentate gyrus that generate FRs to those that do not generate FRs in a well-characterized rat model of epilepsy. We identified a small cohort of genes that are differentially expressed in FR versus non-FR brain tissue and used quantitative PCR to validate some of those that modulate neuronal excitability. Gene expression network analysis demonstrated conservation of gene co-expression between non-FR and FR samples, but examination of gene connectivity revealed changes that were most pronounced in the cm-40 module, which contains several genes associated with synaptic function and the differentially expressed genes Kcna4, Kcnv1, and Npy1r that are down-regulated in FRs. We then demonstrate that the genes within the cm-40 module are regulated by seizure activity and enriched for the targets of the RNA binding protein Elavl4. Our data suggest that seizure activity induces co-expression of genes associated with synaptic transmission and that this pattern is attenuated in areas displaying FRs, implicating the failure of this mechanism in the generation of FRs.
Keywords: Epilepsy, Fast ripples, Microarray, Coexpression network analysis
Introduction
The epileptogenic regions of both patients with temporal lobe epilepsy (TLE) and animal models of TLE display abnormal electrophysiological oscillations between 250 and 600 Hz, which have been termed fast ripples (FRs) (Bragin et al., 1999; Staba et al., 2002). Within the dentate gyrus of animals with TLE, FRs can appear before the onset of epilepsy and have not been observed in non-epileptic animals, suggesting that they may reflect pathological mechanisms involved in epileptogenesis (Bragin et al., 2000). In patients with epilepsy, FRs have been shown to reliably demarcate the seizure onset zone (Jacobs et al., 2008). In addition, surgical resection of regions generating FRs is a better predictor of seizure freedom than resection of the seizure onset zone itself (Jacobs et al., 2010), demonstrating that FRs may play a role in ictogenesis. Therefore, understanding the cellular and molecular alterations that occur in areas generating FRs may provide insight into the key processes of epileptogenesis and ictogenesis in TLE.
In the hippocampus of non-epileptic animals, there are physiological oscillations between 120 and 200 Hz that occur during immobility or slow wave sleep (Buzsaki et al., 1992). Therefore, it has been suggested that FRs are related to these sharp-wave ripple oscillations. However, physiological sharp-wave ripples have been shown to be dependent on inhibitory post-synaptic potentials (Buzsaki et al., 1992; Ylinen et al., 1995), whereas areas generating FRs are enlarged when inhibitory transmission is blocked (Bragin et al., 2002). Recent evidence suggests that FRs represent abnormal population spikes within the dentate gyrus (Bragin et al., 2011; Ibarz et al., 2010). However, the cellular dysregulation and network abnormalities that cause this behavior are unknown.
Studies examining the molecular mechanisms underlying FRs have been difficult because FRs are not diffusely distributed throughout the epileptic hippocampus, but are generated by small clusters of neurons surrounded by large areas of tissue that do not generate FRs. However, advances in applying functional genomics techniques on limited biological samples provide the ability to study molecular processes within electrophysiologically identified neuronal clusters (Bragin et al., 2004). Therefore, we designed our study to compare gene expression in areas of the dentate gyrus that generated FRs to neighboring regions that did not display abnormal activity. After isolating these regions, we examined the gene expression changes between FR and non-FR generating areas, and we found that there were differences in the molecular networks between these regions, indicating changes in gene expression associated with synaptic transmission that are regulated by seizure activity.
Materials and methods
Animals
All procedures described in this study were approved by the University of California, Los Angeles, Institutional Animal Care and Use Committee. Wistar rats weighing 250–300 g were used for experiments. Twenty hours prior to the pilocarpine injection, each animal was injected intraperitoneally with 3 mg/kg of LiCl. Thirty minutes before pilocarpine injection, methylscopolamine (1 mg/kg) was injected subcutaneously, followed by 25 mg/kg subcutaneous injection of pilocarpine. Two hours after status began, it was terminated by intraperitoneal injection of pentobarbital (30 mg/kg). During the post status period, 2 cm3 of Lactated Ringers solution was injected subcutaneously and repeated as necessary to maintain hydration. The following day, Lactated Ringers solution with 5% Dextrose was injected every 4–8 h until fluids were accepted by mouth. Beginning one week after status, animals were subjected to 24 hour video monitoring for identification of epileptic and non-epileptic rats. Seven rats with recurrent spontaneous seizures within 2–4 months after status epilepticus were used for electro-physiological experiments.
Electrophysiology
Rats were deeply anesthetized with 5% isoflurane, the brains were removed, and horizontal 400-μm-thick slices containing the EC, subiculum, and hippocampal formation in the horizontal plane were prepared using a Leica series VT1000S vibratome. The brains were cooled for 1 min in 4 °C artificial CSF (ACSF) containing (in mM): 126 NaCl, 1.25 NaH2PO4, 26 NaHCO3, 2.5 KCl, 2 CaCl2, 2 MgCl2, and 10 D-glucose, pH 7.4. Before recordings, the slices were incubated at 32 °C for 1–6 h in a 300 ml storage chamber filled with ACSF and continuously bubbled with a mixture of 95% O2–5% CO2.
Eleven slices from six animals were used for electrophysiological characterization. Before recording from each slice, a picture was taken of each slice and a 200 μm mesh was superimposed on each picture. This map of each slice was used to track the electrical response of each area to perforant path stimulation. Electrical activity was recorded using standard, extracellular, ACSF-filled glass microelectrodes (~10 MΩ impedance) in the DG granular cell layer at a depth of 200 μm in the middle of the slice. The stimulating electrodes (tungsten wires OD 50 μm) were placed in the perforant path. Rectangular pulses with 0.2 ms duration and currents ranging from 0.1 to 0.7 mA were delivered every 10 s for stimulation. Under visual control, the recording microelectrode was moved with 200 μm steps along the granular layer and evoked potentials were recorded after low-pass filtering at 3 kHz, with 10 kHz sampling rate using DataPac data acquisition software. Four different current amplitudes (0.1; 0.3; 0.5; 0.7 mA) were used to stimulate each point within the slice 10–15 times, and it was determined on-line whether responses contained single or multiple population spikes. FR generating areas were dissected based on the map of multiple population spikes elicited (number of population spikes ≥ 3), and dissection included an approximately 200 μm buffer area around the FR generating area. The area of dissection included the dentate gyrus granule cell layer and their dendrites. These pieces of tissue were all approximately 1 mm2, which is consistent with our prior data demonstrating the size of FR generating areas (Bragin et al., 2002). An equivalently sized tissue area encompassing the same cellular regions in an area that did not generate multiple population spikes was also dissected from the same slice. After dissection, the pieces of tissue were immediately placed in microcentrifuge tubes on dry ice.
RNA extraction and microarray hybridization
Areas generating FRs and those that did not were isolated as above. Total RNA was extracted from each sample using the Qiagen miRNeasy kit, and its quality was checked using the Agilent 2100 Bioanalyzer (Agilent). RNA was amplified using a two-step protocol, TargetAmp 2-Round Aminoallyl-aRNA amplification kit (Epicentre). Amplified RNA was directly labeled with biotin using the Biotin-X-X-NHS reagent (Epicentre). This labeled RNA was hybridized to Illumina RatRef12 microarrays, and a total of 22 microarrays were used for this experiment (11 FR and 11 non-FR samples). Array hybridization and scanning were performed following the manufacturer’s protocol at UCLA microarray core facility (http://microarray.genetics.ucla.edu/xowiki/).
Data normalization
Microarray data was imported into R (http://www.r-project.org/) and normalized using the Variance Stabilizing Transform and Robust Spline Normalization, which are a part of the lumi package in Bioconductor (http://www.bioconductor.org/). The Variance Stabilizing Transform was specifically designed to take advantage of the multiple technical replicates present on Illumina arrays, and Robust Spline Normalization is a simple scaling algorithm designed for use with Illumina data (Du et al., 2008). We then log transformed the data using Log2. We limited subsequent analyses to genes that either met thresholds for mean expression in either the FR or non-FR samples (Expression > 5.6), or overall variance (variance > 0.02), which leads to the selection of 12,581 genes for further analysis.
Data analysis
Among the 12,581 genes that met the criteria for mean expression and variance, we limited this analysis to those with at least ten values with a detection p-value < 0.01 (10 out of 22 samples, >45%), which resulted in the analysis of 10,083 genes for differential expression. We used a Bayesian ANOVA to evaluate differential expression because it provides estimates of a gene’s variance based on the variance of other genes with similar expression (Baldi and Long, 2001) and has been shown to be a robust method for determining differential expression in microarray experiments (Choe et al., 2005). Because the algorithm utilizes a composite of similarly expressed genes to estimate variance, we confirmed normality of all groups of genes with similar expression in the dataset using the Shapiro–Wilk test (p < 2e–16). Each FR sample was paired with an accompanying non-FR sample from the same slice, and these data were used in a paired Bayesian ANOVA to evaluate the differential expression. The threshold for significance was set at p < 0.05 and Bonferroni correction was used for multiple testing (p < 5e–6).
Weighted gene co-expression network analysis was performed as described previously (Oldham et al., 2006; Winden et al., 2009). Briefly, the top 15% of genes (among the 12,581 previously selected) with the highest variance in either the FR or non-FR datasets were selected, representing 2013 genes. Pearson correlations between all these genes were calculated, and this correlation matrix was scaled using an exponent (beta = 8) to best approximate scale free topology. This scaled matrix was then used to calculate the topological overlap matrix (Ravasz et al., 2002), which was used an input into the hierarchical clustering algorithm. From these clusters, modules were identified using a dynamic tree-cutting algorithm (Langfelder et al., 2008). Module eigengenes were calculated using singular value decomposition. The functions used here can be found in the WGCNA R package (http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/).
Quantitative PCR
To validate the differential expression of specific genes, we used quantitative PCR as previously described (Konopka et al., 2009). Briefly, RNA isolated and amplified from 22 samples (11 FR and 11 non-FR) as described previously was converted to cDNA using the First Strand Synthesis Kit (Invitrogen). Gene specific primers were used to amplify ~100 bp regions of target genes. Amplification was detected using Sybr Green (Bioline) using the LightCycler 480 Real Time PCR System (Roche). Each sample was evaluated with four replicates. The delta delta Ct method was used to calculate the fold change of each gene between the non-FR and FR samples, using Gapdh as a loading control. Significance was determined using paired t-test.
Functional annotation
To understand the functions of different modules identified by WGCNA, we used the gene ontology tool DAVID (http://david.abcc.ncifcrf.gov/). This tool associates genes with functional categories, including biological process, cellular compartment, and molecular function. The method examines a set of genes of interest (genes within a module) for over-representation within these functional categories. We used only gene ontology terms associated with the GO_FAT categories, which filters out broad terms.
Results
We used the pilocarpine model of epilepsy to investigate the molecular alterations associated with areas of the dentate gyrus that generate FRs in epileptic animals. Animals were injected with pilocarpine to induce status epilepticus, and animals that developed epilepsy were sacrificed two to four months after injection. In vitro electrophysiological recordings were performed on hippocampal slices from these animals to identify specific areas that generated FRs upon electrical stimulation (Fig. 1a). Regions that generated FRs displayed multiple population spikes (Fig. 1b), and these areas were manually dissected with an approximately 200 μm border around the defined FR area. These samples included the granule cell layer of the dentate gyrus and their dendrites and were about 1 mm2, which is similar to previous descriptions of FR generating areas (Bragin et al., 2002). An equivalently sized region from the same slice that did not show FRs was also obtained. We estimate that hundreds of neurons were recorded based on the characteristic population spikes elicited (Andersen et al., 1971; Lomo, 1971), but the total number of cells within these samples was likely much greater. Total RNA was extracted from these microbiopsies and gene expression was analyzed on Illumina microarrays. We used a principal component analysis to evaluate major drivers of gene expression, and none of the principal components were significantly associated with changes due to slice identity, animal identity, or presence of FRs (ANOVA p > 0.0025; Bonferroni correction). We then calculated the Pearson correlation between all of the arrays and used the average hierarchical clustering algorithm to cluster the samples (Supp Fig. 1). Visual inspection of these data corroborated the principal component analysis, demonstrating that there were no batch effects due to the animal from which the slice was taken, which could confound subsequent analyses.
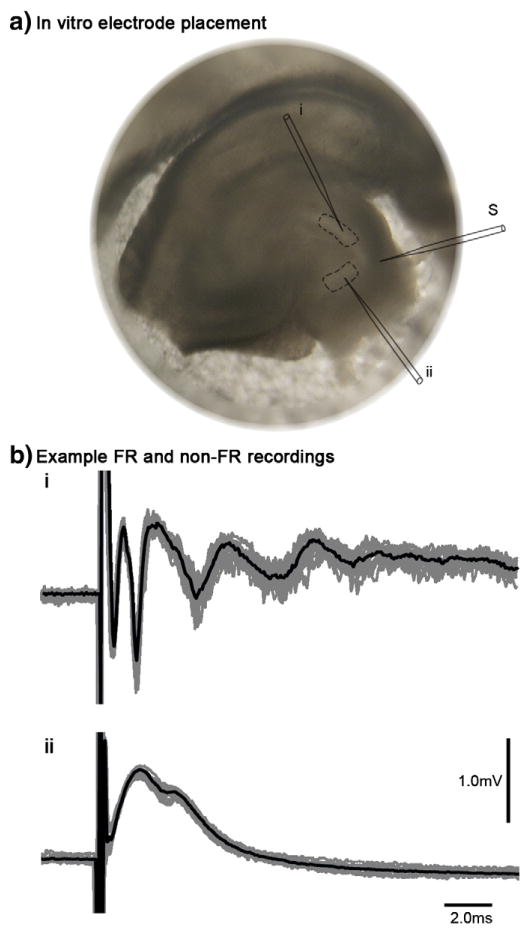
Fig. 1.
In vitro electrophysiological recordings. a) This is an example of a hippocampal slice from an animal with epilepsy. The stimulating electrode (S) was placed in the perforant path. Recording electrodes were used to systematically evaluate population spikes throughout the entire dentate gyrus. Areas that were found to display FRs (i) were subsequently dissected and used for gene expression analysis (dashed lines), as well as a neighboring region from each slice that did not demonstrate FRs (ii). b) Example of electrophysiological recordings within FR and non-FR regions are shown with time (ms) on the x-axis and voltage (mV) on the y-axis. The gray lines demonstrate eleven superimposed responses to perforant path stimuli, and the black line represents the average across all responses. FR areas display multiple population spikes (i), whereas non-FR areas display a single population spike (ii). Scale bars represent time and voltage for both plots.
Differential expression analysis
To identify gene expression changes between non-FR and FR generating areas, we initially performed a differential expression analysis. We used the paired Bayesian ANOVA algorithm to compare gene expression between these hippocampal regions (Baldi and Long, 2001). To limit false positives, we restricted this analysis to genes that were present within at least half of the samples (10,083 genes). We identified a relatively small group of 35 genes that were differentially expressed (p < 0.05; uncorrected), but there were no genes that were significantly differentially expressed after correction for multiple comparisons (p > 5e–6). Within the group of differentially expressed genes that were not significant after multiple comparisons, there were 22 more highly expressed in non-FR areas (Table 1) and 13 more highly expressed in FR areas (Table 2).
Table 1.
Down-regulated genes within areas generating FRs. This table displays the genes that are expressed at a significantly lower level in the FR than non-FR areas (P < 0.05, uncorrected). The fold change represents the expression in the FR areas relative to the non-FR areas.
| Refseq | Fold change | p-Value | Detection p-value < 0.01 | Symbol | Description |
|---|---|---|---|---|---|
| XM_346935.2 | 0.885 | 0.013 | 12 | NA | NA |
| XM_236187.3 | 0.816 | 0.017 | 15 | Mizf | MBD2-interacting zinc finger |
| XM_344018.2 | 0.824 | 0.020 | 15 | Plcxd2 | Phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| NM_012803.1 | 0.882 | 0.028 | 16 | Proc | Protein C |
| XM_576272.1 | 0.906 | 0.028 | 12 | BC030396 | cDNA sequence BC030396 |
| NM_173290.1 | 0.876 | 0.033 | 10 | Yy1 | YY1 transcription factor |
| XM_216497.3 | 0.896 | 0.034 | 11 | Rad54l | RAD54 like (S. cerevisiae) |
| XM_215196.2 | 0.859 | 0.036 | 16 | Fkbp2 | FK506 binding protein 2 |
| XM_576127.1 | 0.876 | 0.037 | 11 | Rapgef5 | Rap guanine nucleotide exchange factor (GEF) 5 |
| XM_344009.2 | 0.586 | 0.040 | 21 | Arl6 | ADP-ribosylation factor-like 6 |
| XM_213963.3 | 0.858 | 0.040 | 15 | Nvl | Nuclear VCP-like |
| XM_221424.3 | 0.847 | 0.042 | 16 | Stxbp5l | Syntaxin binding protein 5-like |
| XM_344502.2 | 0.775 | 0.044 | 17 | Npy1r | Neuropeptide Y receptor Y1 |
| NM_031838.2 | 0.881 | 0.044 | 15 | LOC100043752 | Ribosomal protein S2 pseudogene |
| NM_021697.1 | 0.742 | 0.044 | 20 | Kcnv1 | Potassium channel, subfamily V, member 1 |
| XM_230560.2 | 0.889 | 0.046 | 16 | LOC100043752 | Ribosomal protein S2 pseudogene |
| XM_229086.3 | 0.914 | 0.046 | 11 | 4932411N23Rik | RIKEN cDNA 4932411 N23 gene |
| XM_228611.3 | 0.917 | 0.047 | 13 | 4932429P05Rik | RIKEN cDNA 4932429P05 gene |
| XM_346261.1 | 0.885 | 0.049 | 17 | Ankrd58 | Ankyrin repeat domain 58 |
| XM_226486.3 | 0.911 | 0.049 | 16 | Terf2ip | Telomeric repeat binding factor 2, interacting protein |
| NM_012971.1 | 0.720 | 0.050 | 16 | Kcna4 | Potassium voltage-gated channel, shaker-related subfamily, member 4 |
| NM_017263.1 | 0.869 | 0.050 | 10 | Gria4 | Glutamate receptor, ionotropic, 4 |
Table 2.
Up-regulated genes within areas generating FRs. This table displays the genes that are expressed at a significantly higher level in the FR than non-FR areas (P < 0.05, uncorrected). The fold change represents the expression in the FR areas relative to the non-FR areas.
| Refseq | Fold change | p-Value | Detection p-value < 0.01 | Symbol | Description |
|---|---|---|---|---|---|
| NM_019312.1 | 1.171 | 0.003 | 11 | Itpkb | Inositol 1,4,5-trisphosphate 3-kinase B |
| NM_001005882.1 | 1.160 | 0.009 | 13 | Rbm47 | RNA binding motif protein 47 |
| XM_341327.2 | 1.125 | 0.017 | 15 | F630043A04Rik | RIKEN cDNA F630043A04 gene |
| XM_576628.1 | 1.121 | 0.024 | 13 | EG622645 | Predicted gene, EG622645 |
| XM_345345.2 | 1.117 | 0.025 | 11 | Zbtb6 | Zinc finger and BTB domain containing 6 |
| XM_225699.3 | 1.103 | 0.028 | 10 | 8030462N17Rik | RIKEN cDNA 8030462 N17 gene |
| XM_343667.2 | 1.127 | 0.030 | 14 | Ticam1 | Toll-like receptor adaptor molecule 1 |
| XM_237359.2 | 1.095 | 0.033 | 10 | Slc16a14 | Solute carrier family 16 (monocarboxylic acid transporters), member 14 |
| XM_573886.1 | 1.745 | 0.037 | 21 | Yjefn3 | YjeF N-terminal domain containing 3 |
| NM_175762.2 | 1.097 | 0.037 | 15 | Ldlr | Low density lipoprotein receptor |
| XM_214338.3 | 1.590 | 0.044 | 15 | Palld | Palladin, cytoskeletal associated protein |
| XM_217689.3 | 1.380 | 0.048 | 15 | Clca3 | Chloride channel calcium activated 3 |
| XM_577560.1 | 1.120 | 0.049 | 18 | LOC667488 | Hypothetical LOC667488 |
Among the genes that were more highly expressed within non-FR areas, there were four genes that are known to impact neuronal excitability, including Kcna4, Kcnv1, Gria4, and Npy1r1. Kcna4 encodes a rapidly inactivating potassium channel (Wymore et al., 1994), while Kcnv1 encodes a non-functional potassium channel that alters the properties of Shab and Shaw type channels (Hugnot et al., 1996). Gria4 and Npy1r are neurotransmitter receptors for glutamate and neuropeptide Y, respectively. Given the small number of genes identified and the lenient significance threshold, we examined these expression changes using quantitative PCR in the same samples used for microarray analysis and confirmed that Gria4, Npy1r, Kcna4, and Kcnv1 were all expressed at significantly lower levels within the FR samples (paired t-test, p < 0.05). Although some samples did not meet the detection threshold (p-value < 0.01) on the microarray, these genes were reliably detected using quantitative PCR in all samples, demonstrating that our detection threshold was relatively conservative. We also found that the quantitative PCR and microarray results were significantly correlated (p < 0.01; Pearson correlation) and that the fold changes obtained using quantitative PCR were in the same direction but greater than on the microarray (Fig. 2), which has previously been observed with Illumina microarrays (Konopka et al., 2009). These data demonstrate the differential expression of a small number of genes involved in neuronal excitability that display decreased expression levels within areas that generate FRs.
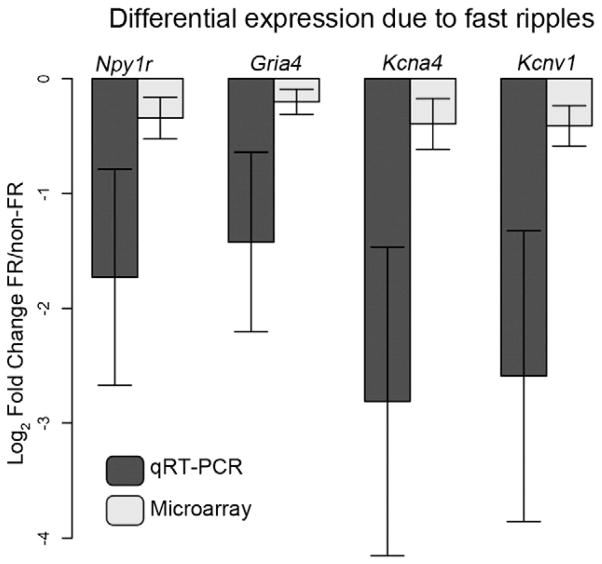
Fig. 2.
Differential expression between FR and non-FR samples. To confirm gene expression changes observed in the microarray experiment, we performed quantitative PCR on four genes, Gria4, Npy1r, Kcna4, and Kcnv1. The barplot demonstrates the average fold change between the FR and non-FR samples from the same slice across all eleven slices. Fold changes were also calculated from the microarray data, and these values are shown as a comparison. Each of these genes demonstrates significantly decreased expression within the FR samples as compared to the non-FR samples (P < 0.05). Each sample was evaluated with four replicates, and Gapdh was used as the loading control for quantitative PCR. Significance was determined using a paired t-test (n = 11), and error bars represent the standard error.
We hypothesized that the paucity of differentially expressed genes was due to biological heterogeneity between samples, reflected in high variability. To investigate this hypothesis, we compared the variance of each gene within the FR samples or non-FR samples separately to the variance of each gene across all the samples (Supp Fig. 2). We expected that the variance within either the FR or non-FR specimens would be less than the variance across all the samples, but the distributions of variance were not significantly different (p > 0.05; Mann–Whitney test). Therefore, these data demonstrate that factors other than the presence of FRs strongly contributed to gene expression variability and impeded our ability to identify differentially expressed genes between FR and non-FR areas.
Network analysis
To compensate for the biological heterogeneity within this dataset, we performed Weighted Gene Co-expression Network Analysis (WGCNA), which utilizes variance between the samples to identify patterns of gene expression (Oldham et al., 2006; Oldham et al., 2008; Zhang and Horvath, 2005). WGCNA is an unbiased and unsupervised method to identify co-expressed groups of genes or modules that correspond to major functional elements within a dataset. In order to identify differential molecular processes between FR and non-FR areas, we utilized a differential network analysis, which can be used to identify groups of genes whose co-expression becomes stronger or weaker under different conditions (Miller et al., 2008, 2010; Oldham et al., 2006). Rearrangement of co-expression within a tissue sample could suggest alterations such as loss of normal molecular processes or migration of abnormal cell types, which could point to biological processes underlying FR.
We first created separate networks for non-FR and FR samples. Probes within the top 15% of variance in either the non-FR or FR generating samples were used for network analysis, which led to the selection of 2013 probes. Networks for the non-FR and FR samples were created separately using previously published methods (Langfelder et al., 2008; Zhang and Horvath, 2005), and we identified 36 modules within the non-FR network (nf1–36) and 38 modules within the FR network (fr1–38). To ensure that the initial networks were representative of the dataset, we created 20 additional networks from random sets of 2013 probes in both the FR and non-FR datasets. Although no single network contained all modules from the original network, each module from these random networks was similar to an original module (mean correlation 0.92), demonstrating that these expression patterns were representative of both datasets. We then compared all the modules in the FR and non-FR networks and found that each module overlapped significantly with at least one module from the other network (Fig. 3; Hypergeometric probability, p < 1.0e–4). These data demonstrate that basic co-expression relationships are conserved between the non-FR and FR areas, suggesting that there is no large-scale reorganization of co-expression between the two areas that might account for the presence of FR.
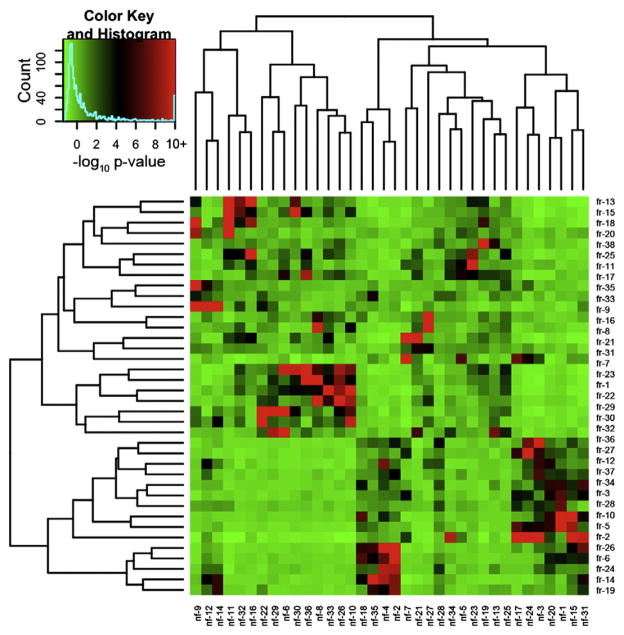
Fig. 3.
Separate networks in the FR and non-FR datasets contain overlapping modules. To compare network organization of gene expression within FR and non-FR samples, we identified overlapping genes between all modules in the two networks. This heatmap demonstrates the significance of the overlap between FR (rows) and non-FR (columns) modules calculated using the hypergeometric probability. The colors within the heatmap represent the log10 transformed p-value of the overlap between two corresponding modules, and the key on the top left shows the correspondence between colors and log10 p-values with red representing more significant p-values. The dendrograms on either side of the heatmap demonstrate the clustering of the modules based on Euclidean distance between their MEs. Each module in the FR network is significantly related to a module in the non-FR network and the converse is also true, demonstrating that gene expression patterns are conserved between FR and non-FR areas.
Changes in connectivity between FR and non-FR areas
In the absence of gross changes in co-expression, we performed analyses to determine whether connections between co-expressed genes were altered between non-FR and FR areas. We first developed consensus modules between the non-FR and FR networks by assigning overlapping genes from two significantly related modules to a consensus module, which led to the creation of 44 consensus modules (cm1–44). We summarized gene expression within each module using the first principal component, which we term the module eigengene (ME). The ME provides a summary of gene expression across all genes within a module, and we confirmed that each ME explained a substantial proportion of variance of gene expression (>50%) within each module in both datasets. Within each module, we calculated the Pearson correlation between the expression of each gene within a module and the ME to determine ME-based connectivity (kME), which is a measure of a gene’s connectivity within a module (Horvath and Dong, 2008). Connectivity has been shown to be an effective measure to compare different modules and determine module conservation between datasets (Oldham et al., 2006). We then compared kME values for each module between the non-FR and FR datasets and found that kME values between the datasets were correlated for all consensus modules (p < 1.0e–16), confirming that these modules were conserved across both datasets.
We used these consensus modules as a starting point for determining whether there might be connectivity changes within specific modules that might explain the presence of FRs. We examined each module to determine whether there was a tendency to display higher connectivity in one dataset relative to the other. We used topological overlap (TO) to determine connectivity between genes because it has been shown to be a robust measure in biological systems (Ravasz et al., 2002). For each consensus module, we identified all of the genes that were highly connected in either the non-FR or FR dataset (kME > 0.7), and then calculated the TO between this group of genes in the non-FR and FR datasets separately. We then examined the difference in average TO between the non-FR and FR datasets for each module. We found that many modules demonstrated higher TO in the non-FR dataset compared to the FR dataset, but the cm-40 module demonstrated the greatest difference in TO between the datasets (Fig. 4a). We examined the distribution of the TO difference across all genes within the cm-40 module, and we found that 98% of the genes demonstrated higher TO in the non-FR dataset (Fig. 4b). Interestingly, Kcna4, Kcnv1, and Npy1r were each previously found to be differentially expressed between the FR and non-FR samples, and they are all present within the cm-40 module and display higher TO within the non-FR dataset compared to the FR dataset, which further implicates this module in differences between the non-FR and FR areas.
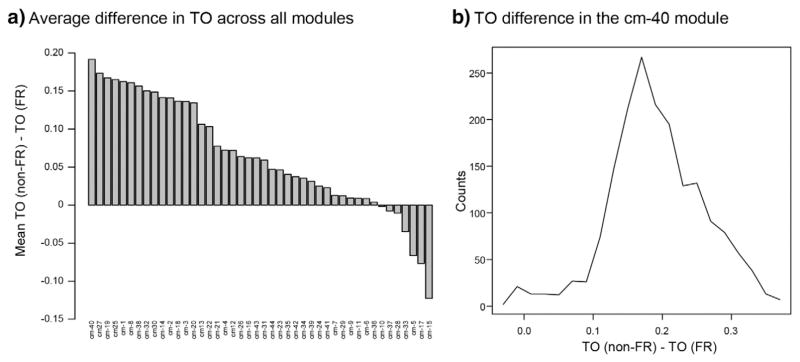
Fig. 4.
Comparison of co-expression measured by TO between the FR and non-FR datasets for each module in the consensus network. a) The barplot demonstrates the average difference in TO between non-FR and FR datasets within each consensus module. Genes with a kME > 0.7 in either the non-FR or FR network were selected for each module, and TO between all genes within a module was calculated separately between the non-FR and FR datasets. The y-axis displays the average difference in TO between the non-FR and FR datasets across all genes in a module. There is a notable skew towards higher connectivity in the non-FR dataset in multiple consensus modules, but the cm-40 module displays the largest average difference. b) The histogram shows the TO difference between the non-FR and FR datasets across all genes within the cm-40 module. The x-axis represents the difference in TO between the non-FR and FR datasets for each gene, and the y-axis represents the number of genes with the specific value of difference in TO. There is a dramatic shift towards higher TO values in the non-FR dataset, demonstrating stronger co-regulation of the genes within the cm-40 module in non-FR areas.
Functional annotation of the cm-40 module
We attempted to understand the biological significance of the cm-40 module by first examining molecular functions and characteristics associated with the genes within the module using the gene ontology tool DAVID. We limited this analysis to the top 750 genes with the greatest difference in TO between the non-FR and FR datasets. We found that there was a significant enrichment of genes related to protein transport (p = 2.6e–8), especially vesicle-mediated transport (p = 2.2e–3) (Table 2). There were also many genes associated with RNA splicing (p = 1.7e–7) and RNA binding (p = 1.2e–6). Finally, there were many categories associated with synapses, including localization to synapses (p = 0.018) or the post-synaptic density (p = 0.022). These data demonstrate enrichment in various functional categories, but many of these categories have important functions in neuronal processes.
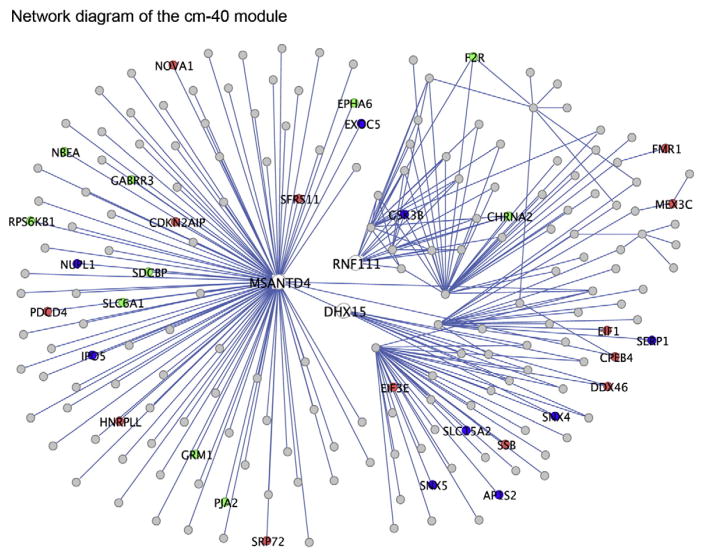
To further examine the functions of this group of genes, we examined the genes that were highly connected within this module because hub genes have been shown to be key drivers of module organization and function (Carlson et al., 2006; Winden et al., 2009). The most highly connected gene within this module is Msantd4, which is a poorly characterized gene that contains a MYB DNA binding domain (Fig. 5). Interestingly, the region outside of the DNA binding domain is similar to the synaptic vesicle protein Eps15 (E value = 6e–10, blastp), which suggests that this gene could also be associated with synaptic vesicles. Dhx15 is another highly connected gene within this module, and it is a known RNA helicase and part of the spliceosome complex (Fouraux et al., 2002; Niu et al., 2012), which is consistent with the GO analysis demonstrating enrichment of genes involved in RNA processing. Another hub gene within this module, Rnf111, is an E3 ubiquitin ligase (Koinuma et al., 2003), which is consistent with enrichment of genes associated with ubiquitination. Interestingly, two key enzymes involved in GABA synthesis and transport, Gad1 and Slc6a1, are highly connected within this module, as well as several neurotransmitter receptors, including Chrna7 and Grik1 (Frerking et al., 1998; Son and Winzer-Serhan, 2008). Therefore, these data highlight multiple potential regulators of neuronal processes, and they suggest that alterations could impact synaptic transmission.
Fig. 5.
Network plot of the genes with the greatest difference in TO in the cm-40 module. This plot displays the strongest connections based on TO between the top 750 genes within the cm-40 module that had the greatest TO difference between the non-FR and FR datasets. The TO values between these genes were calculated using data from the non-FR dataset, and the plot is limited to the 250 connections with the greatest TO. The genes that clearly display the most connections include Msantd4, Acsl1, and Rnf111. The genes associated with enriched GO categories such as synaptic functions (green), RNA binding (red), and protein transport (blue) are also highlighted.
Mechanisms underlying co-expression in the cm-40 module
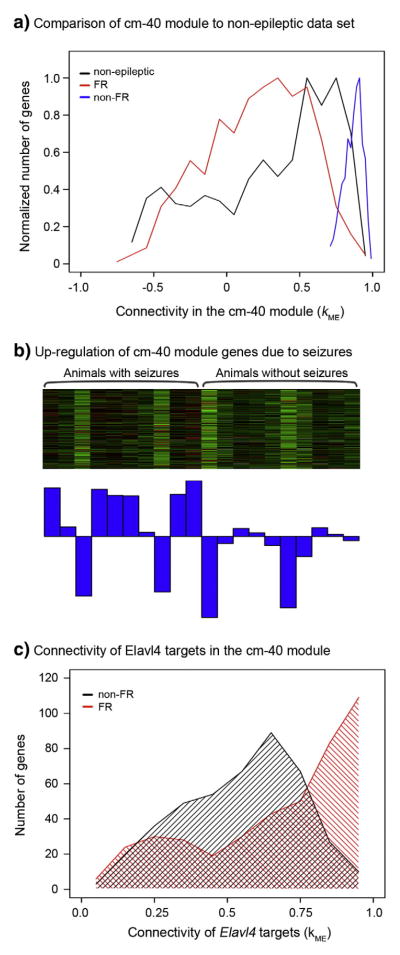
Our data indicate that there is a difference between the regulation of a large number of genes between areas that generate FRs and those that do not generate FRs. However, this could either reflect a loss of normal neuronal functions in areas generating FR or a gain of function in non-FR areas. To address this question, we compared connectivity within the cm-40 module to connectivity in a dataset consisting of 80 microarrays examining gene expression in the non-epileptic rat dentate gyrus (Burger et al., 2008). We used the genes composing the cm-40 module to define a module within the non-epileptic dataset and calculated kME for all genes within the non-epileptic dataset. We used the top 750 genes in the cm-40 module with the greatest TO difference between the non-FR and FR datasets, and we compared their connectivity in the non-FR and FR datasets to the non-epileptic dataset (Fig. 6a). We found that the connectivity in the non-FR dataset was significantly greater than connectivity within the non-epileptic dataset for this group of genes (p < 2.2e–16; t-test). However, we also found that connectivity within the non-epileptic dataset was significantly greater than connectivity in the FR dataset (p = 2e–5; t-test). These data demonstrate that connectivity within the cm-40 module is stronger within non-FR areas when compared to the non-epileptic dentate gyrus, whereas connectivity of these genes is weaker within FR areas. Therefore, these data suggest that there is a gene expression pattern that is induced by epileptic processes in non-FR regions, and this pattern is attenuated in areas that generate FRs compared to the non-epileptic brain.
Fig. 6.
Comparison of connectivity in the cm-40 module to a non-epileptic dataset, and identification of seizure activity and the RNA binding protein Elavl4 as regulators of expression. a) This histogram demonstrates the comparison of connectivity within non-FR and FR datasets to a dataset non-epileptic rat hippocampus (Burger et al., 2008). The top 750 genes with the greatest TO difference between the non-FR and FR datasets were used to create all three distributions. Connectivity in the non-FR dataset is significantly greater than connectivity in the non-epileptic dataset, but interestingly, connectivity in the non-epileptic dataset is significantly greater than connectivity within the FR dataset. b) This heatmap demonstrates the expression of the genes within cm-40 module in previously published data that analyzed gene expression within animals with and without epilepsy (Winden et al., 2011). In this heatmap, samples are in columns and genes are in the rows. Green denotes low expression, while red represents high expression of a specific gene. Below the heatmap, the barplot depicts the ME, which is a summary of gene expression across the genes within the module. Gene expression within this module demonstrates significant up-regulation within animals with seizures (P < 0.05, Kruskal–Wallis test). c) This histogram demonstrates the connectivity of known targets of the RNA binding protein Elavl4 in the non-FR and FR datasets within the cm-40 module. The red area shows the distribution of connectivity of the group of Elavl4 targets in the non-FR dataset, and the black area shows the distribution of the same genes in the FR dataset. This group of genes demonstrates significantly higher connectivity within the non-FR dataset than in the FR dataset.
We hypothesized that this epilepsy specific pattern of gene expression could represent a previously identified expression pattern that was up-regulated due to seizures (Winden et al., 2011). The expression of the same group of 750 genes that had the greatest difference in TO between the non-FR and FR datasets in the cm-40 module was examined in a previously characterized microarray dataset from epileptic and non-epileptic animals. The heatmap and first principal component of expression of these genes demonstrate that this group of genes is significantly up-regulated within animals with seizures compared to animals without seizures (Fig. 6b; p < 0.05, Kruskal–Wallis test). These data demonstrate that seizure activity itself is an important regulator of expression within the cm-40 module.
We had also previously described that there was an enrichment of targets of the RNA binding protein, Elavl4, within the set of genes that were up-regulated by seizures (Winden et al., 2011). Therefore, we tested whether the set of genes that were highly connected within the non-FR dataset were also enriched for Elavl4 targets. We used a list of Elavl4 targets generated from immunoprecipitation and subsequent microarray analysis (Bolognani et al., 2010), and we found that there was a significant enrichment of Elavl4 targets within the group of 750 genes that had the greatest TO difference between the non-FR and FR datasets (p = 1.3e–29; hypergeometric probability). We then compared connectivity of known Elavl4 targets within the cm-40 module between the non-FR and FR datasets, and we found that there is a dramatic shift towards higher connectivity of these genes within the non-FR dataset (Fig. 6c). Therefore, these data suggest that gene expression changes induced by seizures and Elavl4 are not present in areas that generate FRs, which may explain the epileptogenic and ictogenic properties of these areas.
Discussion
This study focused on the molecular changes associated with areas of the dentate gyrus that generate pathological FRs in a model of TLE. We recorded evoked field potentials from the dentate gyrus in vitro to identify FRs and analyzed gene expression within those regions. Using the differential expression analysis, we identified only a small group of genes and confirmed the differential expression of four genes that could directly affect neuronal activity. We hypothesized that the paucity of differentially expressed genes was due to sample heterogeneity, and therefore, we used WGCNA to circumvent biological variability and understand gene co-expression relationships in both non-FR and FR areas. We compared co-expression patterns between non-FR and FR regions and found that there was a conserved co-expression structure between these regions. However, we found that the cm-40 module contained many genes with stronger co-expression in the non-FR dataset compared to the FR dataset. Function annotation demonstrated that these genes were associated with several processes integral to both pre- and post-synaptic functions. By comparing connectivity within the cm-40 module to a non-epileptic dataset, we found that the non-FR dataset demonstrated significantly higher connectivity than the non-epileptic dataset, whereas the FR dataset demonstrated significantly lower connectivity. We also found that seizure activity could regulate expression within the cm-40 module and that it was enriched for targets of the RNA binding protein Elavl4. These data lead to the hypothesis that a gene expression program associated with synaptic transmission is induced by seizures and is not up-regulated in areas that demonstrate FR, implicating the absence of these processes in the generation of FRs.
Using the differential expression analysis, we were able to identify a small number of significant expression changes, and we were able to confirm four differentially expressed genes that affect neuronal activity (Kcnv1, Kcna4, Gria4, Npy1r). Expression of Gria4 has been shown to be limited to basket cells within the dentate gyrus (Geiger et al., 1995), and Gria4 knockout animals demonstrate spike-wave discharges consistent with absence epilepsy (Beyer et al., 2008). Npy1r is expressed in both granule cells and interneurons within the dentate gyrus and localized post-synaptically where it interacts with calcium channels (Larsen et al., 1993; McQuiston et al., 1996), but knockout studies have shown that Npy1r may have pro-convulsant properties (Lin et al., 2006). Although these genes may modulate epileptic processes, we cannot identify possible mechanisms underlying FRs because of the limited gene expression changes. Therefore, we focused on the network analysis to circumvent the lack of expression changes due to sample heterogeneity.
Through comparing the networks between the non-FR and FR datasets, we were able to identify a number of modules demonstrating stronger co-expression within the non-FR dataset with the largest changes found in the cm-40 module. Functional annotation demonstrated that there were several genes involved in protein transport and vesicles associated with the pre-synaptic processes of synaptic vesicle release and recycling, including Atp6v1g2 that is involved in synaptic vesicle acidification (Murata et al., 2002). Neurotransmitter receptors such as Grik1 and Chrna7 were also present within this module, and these receptors have been shown to be localized to the pre-synaptic membrane and modulate inhibitory and excitatory transmission within the hippocampus (Frerking et al., 1998; Jones and Yakel, 1997; Vignes et al., 1998). In addition, Gad1 and Slc6a1 were present within this group of genes and are key enzymes involved in GABA synthesis and transport (Asada et al., 1997; Jensen et al., 2003). There were also several neurotransmitter receptors localized to the post-synaptic membrane, such as Gria2 and Drd1a, and both receptors have been shown to be expressed in dentate gyrus granule cells and interneurons (Gangarossa et al., 2012; Geiger et al., 1995). In addition, RNA binding proteins such as Nova1 were enriched within this module, and Nova1 has been shown to regulate splicing of multiple genes involved in inhibitory transmission (Ule et al., 2005). These data demonstrate that this module represents processes associated with both pre- and post-synaptic functions within multiple cell types in the dentate gyrus.
By comparing the connectivity of genes in the cm-40 module to a non-epileptic dataset, we found stronger co-expression in non-FR areas and weaker co-expression in FR areas compared to non-epileptic hippocampus. Therefore, we hypothesized that this represents increased predominance of the synaptic functions associated with the cm-40 module in non-FR areas, and weakening of these same processes in FR areas. Prior studies have demonstrated that inhibition and inhibitory synapses are increased in epileptic animals (Engel and Wilson, 1986; Haas et al., 1996; Milgram et al., 1991; Thind et al., 2010; Wilson et al., 1998). However, recordings of epileptic animals in vitro and in vivo suggest local decreased inhibition in areas generating FRs (Bragin et al., 2002; Bragin et al., 2005), suggesting that ictal events could arise from breakdown of inhibitory mechanisms leading to coalescence and synchronization of FR areas. Studies using juxtacellular recordings of dentate gyrus in vivo have demonstrated that FRs can arise from bursting granule cells and that interneurons showed decreased activity during episodes of FRs (Bragin et al., 2011; Ibarz et al., 2010), demonstrating changes in the temporal profile of inhibition in areas that generate FRs. Alternatively, these data could reflect the various synaptic alterations of dentate granule cells in epilepsy, such as mossy fiber sprouting or phenotype modifications by seizures (Dudek and Sutula, 2007; Schwarzer and Sperk, 1995). However, one study that examined the relationship between mossy fiber sprouting and FR found no correlation (Foffani et al., 2007). Therefore, our data suggesting changes associated with the synaptic function is consistent with prior studies demonstrating changes in synaptic growth and transmission caused by epilepsy.
Our data also demonstrate that seizure activity itself is capable of up-regulating activity within this module. Previously, we demonstrated that the RNA binding protein Elavl4 is up-regulated with seizure activity (Winden et al., 2011), and here we show that targets of this RNA binding protein show increased connectivity in the non-FR dataset compared to the FR dataset. Therefore, we hypothesize that seizures induce gene expression through alteration of Elavl4 expression, but this mechanism apparently fails in small regions, underlying the generation of FRs in those areas. Elavl4 has been shown to be expressed in interneurons in the hippocampus (Bolognani et al., 2004), and it has been shown to be involved in transporting mRNA in neuronal processes, leading to neurite outgrowth and changes in neuroplasticity (Aronov et al., 2002; Bolognani et al., 2007; Tanner et al., 2008). These processes could underlie several alterations in inhibitory interneurons or dentate granule neurons that have been observed in the epileptic brain, including increased axonal growth and synaptogenesis (Dudek and Sutula, 2007; Thind et al., 2010). In addition, properties of synaptic vesicle release probabilities have been shown to be altered in interneurons and mossy fibers in epileptic animals (Goussakov et al., 2000; Kobayashi and Buckmaster, 2003). Therefore, failure of these compensatory mechanisms could result in dysfunctional synaptic transmission and the generation of FR.
The aim of this work was to examine the molecular composition of areas that generate FR to attempt to understand the basic mechanisms that generate FRs. We have demonstrated that the changes are subtle and involve reorganization of co-expression relationships in genes related to synaptic function in both inhibitory and excitatory cells. These data lead to the hypothesis that epileptic processes induce a co-expression program associated with synaptic transmission and that this gene expression program is absent within areas that generate FRs, which may alter synaptic transmission and lead to the generation of FRs.
Supplementary Material
Acknowledgments
We acknowledge the support from NIH grants 5R37NS033310-12 (KDW, AB, DHG, JE), NIGMS GM08042 (KDW), Neurobehavioral Genetics Training Grant T32MH073526-01A1 (KDW), Medical Scientist Training Program (KDW), and Aesculapians Fund of the UCLA School of Medicine (KDW). These funders had no role in the study design, data collection and analysis, decision to publish, or in the preparation of the manuscript. The authors declare no competing interests.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nbd.2015.02.011.
Footnotes
Available online on ScienceDirect (www.sciencedirect.com).
References
- Andersen P, Bliss TV, Skrede KK. Unit analysis of hippocampal polulation spikes. Exp Brain Res. 1971;13:208–221. doi: 10.1007/BF00234086. [DOI] [PubMed] [Google Scholar]
- Aronov S, Aranda G, Behar L, Ginzburg I. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J Cell Sci. 2002;115:3817–3827. doi: 10.1242/jcs.00058. [DOI] [PubMed] [Google Scholar]
- Asada H, Kawamura Y, Maruyama K, Kume H, Ding RG, Kanbara N, Kuzume H, Sanbo M, Yagi T, Obata K. Cleft palate and decreased brain gamma-aminobutyric acid in mice lacking the 67-kDa isoform of glutamic acid decarboxylase. Proc Natl Acad Sci U S A. 1997;94:6496–6499. doi: 10.1073/pnas.94.12.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Beyer B, Deleuze C, Letts VA, Mahaffey CL, Boumil RM, Lew TA, Huguenard JR, Frankel WN. Absence seizures in C3H/HeJ and knockout mice caused by mutation of the AMPA receptor subunit Gria4. Hum Mol Genet. 2008;17:1738–1749. doi: 10.1093/hmg/ddn064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognani F, Merhege MA, Twiss J, Perrone-Bizzozero NI. Dendritic localization of the RNA-binding protein HuD in hippocampal neurons: association with polysomes and upregulation during contextual learning. Neurosci Lett. 2004;371:152–157. doi: 10.1016/j.neulet.2004.08.074. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Qiu S, Tanner DC, Paik J, Perrone-Bizzozero NI, Weeber EJ. Associative and spatial learning and memory deficits in transgenic mice overexpressing the RNA-binding protein HuD. Neurobiol Learn Mem. 2007;87:635–643. doi: 10.1016/j.nlm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Contente-Cuomo T, Perrone-Bizzozero NI. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 2010;38:117–130. doi: 10.1093/nar/gkp863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragin A, Engel J, Jr, Wilson CL, Fried I, Mathern GW. Hippocampal and entorhinal cortex high-frequency oscillations (100–500 Hz) in human epileptic brain and in kainic acid-treated rats with chronic seizures. Epilepsia. 1999;40:127–137. doi: 10.1111/j.1528-1157.1999.tb02065.x. [DOI] [PubMed] [Google Scholar]
- Bragin A, Wilson CL, Engel J., Jr Chronic epileptogenesis requires development of a network of pathologically interconnected neuron clusters: a hypothesis. Epilepsia. 2000;41(Suppl 6):S144–S152. doi: 10.1111/j.1528-1157.2000.tb01573.x. [DOI] [PubMed] [Google Scholar]
- Bragin A, Mody I, Wilson CL, Engel J., Jr Local generation of fast ripples in epileptic brain. J Neurosci. 2002;22:2012–2021. doi: 10.1523/JNEUROSCI.22-05-02012.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragin A, Karsten SL, Almajano J, Wilson CL, Geschwind DH, Engel J., Jr Large-scale microarray gene expression analysis in discrete electrophysiologically identified neuronal clusters. J Neurosci Methods. 2004;133:49–55. doi: 10.1016/j.jneumeth.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Bragin A, Azizyan A, Almajano J, Wilson CL, Engel J., Jr Analysis of chronic seizure onsets after intrahippocampal kainic acid injection in freely moving rats. Epilepsia. 2005;46:1592–1598. doi: 10.1111/j.1528-1167.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Bragin A, Benassi SK, Kheiri F, Engel J., Jr Further evidence that pathologic high-frequency oscillations are bursts of population spikes derived from recordings of identified cells in dentate gyrus. Epilepsia. 2011;52:45–52. doi: 10.1111/j.1528-1167.2010.02896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger C, Lopez MC, Baker HV, Mandel RJ, Muzyczka N. Genome-wide analysis of aging and learning-related genes in the hippocampal dentate gyrus. Neurobiol Learn Mem. 2008;89:379–396. doi: 10.1016/j.nlm.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G, Horvath Z, Urioste R, Hetke J, Wise K. High-frequency network oscillation in the hippocampus. Science. 1992;256:1025–1027. doi: 10.1126/science.1589772. [DOI] [PubMed] [Google Scholar]
- Carlson MR, Zhang B, Fang Z, Mischel PS, Horvath S, Nelson SF. Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics. 2006;7:40. doi: 10.1186/1471-2164-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe SE, Boutros M, Michelson AM, Church GM, Halfon MS. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 2005;6:R16. doi: 10.1186/gb-2005-6-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Dudek FE, Sutula TP. Epileptogenesis in the dentate gyrus: a critical perspective. Prog Brain Res. 2007;163:755–773. doi: 10.1016/S0079-6123(07)63041-6. [DOI] [PubMed] [Google Scholar]
- Engel J, Jr, Wilson C. Evidence for enhanced synaptic inhibition in human epilepsy. In: Nistico G, Morselli PL, Lloyd KG, Fariello RG, Engel J Jr, editors. Neurotransmitters, Seizures, and Epilepsy, III. Raven Press; New York: 1986. pp. 1–10. [Google Scholar]
- Foffani G, Uzcategui YG, Gal B, Menendez de la Prida L. Reduced spike-timing reliability correlates with the emergence of fast ripples in the rat epileptic hippocampus. Neuron. 2007;55:930–941. doi: 10.1016/j.neuron.2007.07.040. [DOI] [PubMed] [Google Scholar]
- Fouraux MA, Kolkman MJ, Van der Heijden A, De Jong AS, Van Venrooij WJ, Pruijn GJ. The human La (SS-B) autoantigen interacts with DDX15/hPrp43, a putative DEAH-box RNA helicase. RNA. 2002;8:1428–1443. doi: 10.1017/s1355838202021076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerking M, Malenka RC, Nicoll RA. Synaptic activation of kainate receptors on hippocampal interneurons. Nat Neurosci. 1998;1:479–486. doi: 10.1038/2194. [DOI] [PubMed] [Google Scholar]
- Gangarossa G, Longueville S, De Bundel D, Perroy J, Herve D, Girault JA, Valjent E. Characterization of dopamine D1 and D2 receptor-expressing neurons in the mouse hippocampus. Hippocampus. 2012;22:2199–2207. doi: 10.1002/hipo.22044. [DOI] [PubMed] [Google Scholar]
- Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Goussakov IV, Fink K, Elger CE, Beck H. Metaplasticity of mossy fiber synaptic transmission involves altered release probability. J Neurosci. 2000;20:3434–3441. doi: 10.1523/JNEUROSCI.20-09-03434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas KZ, Sperber EF, Moshe SL, Stanton PK. Kainic acid-induced seizures enhance dentate gyrus inhibition by downregulation of GABA(B) receptors. J Neurosci. 1996;16:4250–4260. doi: 10.1523/JNEUROSCI.16-13-04250.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 2008;4:e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugnot JP, Salinas M, Lesage F, Guillemare E, de Weille J, Heurteaux C, Mattei MG, Lazdunski M. Kv8.1, a new neuronal potassium channel subunit with specific inhibitory properties towards Shab and Shaw channels. EMBO J. 1996;15:3322–3331. [PMC free article] [PubMed] [Google Scholar]
- Ibarz JM, Foffani G, Cid E, Inostroza M, Menendez de la Prida L. Emergent dynamics of fast ripples in the epileptic hippocampus. J Neurosci. 2010;30:16249–16261. doi: 10.1523/JNEUROSCI.3357-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J, LeVan P, Chander R, Hall J, Dubeau F, Gotman J. Interictal high-frequency oscillations (80–500 Hz) are an indicator of seizure onset areas independent of spikes in the human epileptic brain. Epilepsia. 2008;49:1893–1907. doi: 10.1111/j.1528-1167.2008.01656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J, Zijlmans M, Zelmann R, Chatillon CE, Hall J, Olivier A, Dubeau F, Gotman J. High-frequency electroencephalographic oscillations correlate with outcome of epilepsy surgery. Ann Neurol. 2010;67:209–220. doi: 10.1002/ana.21847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K, Chiu CS, Sokolova I, Lester HA, Mody I. GABA transporter-1 (GAT1)-deficient mice: differential tonic activation of GABAA versus GABAB receptors in the hippocampus. J Neurophysiol. 2003;90:2690–2701. doi: 10.1152/jn.00240.2003. [DOI] [PubMed] [Google Scholar]
- Jones S, Yakel JL. Functional nicotinic ACh receptors on interneurones in the rat hippocampus. J Physiol. 1997;504(Pt 3):603–610. doi: 10.1111/j.1469-7793.1997.603bd.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Buckmaster PS. Reduced inhibition of dentate granule cells in a model of temporal lobe epilepsy. J Neurosci. 2003;23:2440–2452. doi: 10.1523/JNEUROSCI.23-06-02440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinuma D, Shinozaki M, Komuro A, Goto K, Saitoh M, Hanyu A, Ebina M, Nukiwa T, Miyazawa K, Imamura T, Miyazono K. Arkadia amplifies TGF-beta superfamily signalling through degradation of Smad7. EMBO J. 2003;22:6458–6470. doi: 10.1093/emboj/cdg632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, Peng S, Preuss TM, Wohlschlegel JA, Geschwind DH. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- Larsen PJ, Sheikh SP, Jakobsen CR, Schwartz TW, Mikkelsen JD. Regional distribution of putative NPY Y1 receptors and neurons expressing Y1 mRNA in forebrain areas of the rat central nervous system. Eur J Neurosci. 1993;5:1622–1637. doi: 10.1111/j.1460-9568.1993.tb00231.x. [DOI] [PubMed] [Google Scholar]
- Lin EJ, Young D, Baer K, Herzog H, During MJ. Differential actions of NPY on seizure modulation via Y1 and Y2 receptors: evidence from receptor knockout mice. Epilepsia. 2006;47:773–780. doi: 10.1111/j.1528-1167.2006.00500.x. [DOI] [PubMed] [Google Scholar]
- Lomo T. Patterns of activation in a monosynaptic cortical pathway: the perforant path input to the dentate area of the hippocampal formation. Exp Brain Res. 1971;12:18–45. [PubMed] [Google Scholar]
- McQuiston AR, Petrozzino JJ, Connor JA, Colmers WF. Neuropeptide Y1 receptors inhibit N-type calcium currents and reduce transient calcium increases in rat dentate granule cells. J Neurosci. 1996;16:1422–1429. doi: 10.1523/JNEUROSCI.16-04-01422.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milgram NW, Yearwood T, Khurgel M, Ivy GO, Racine R. Changes in inhibitory processes in the hippocampus following recurrent seizures induced by systemic administration of kainic acid. Brain Res. 1991;551:236–246. doi: 10.1016/0006-8993(91)90938-r. [DOI] [PubMed] [Google Scholar]
- Miller JA, Oldham MC, Geschwind DH. A systems level analysis of transcriptional changes in Alzheimer’s disease and normal aging. J Neurosci. 2008;28:1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci U S A. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Sun-Wada GH, Yoshimizu T, Yamamoto A, Wada Y, Futai M. Differential localization of the vacuolar H+ pump with G subunit isoforms (G1 and G2) in mouse neurons. J Biol Chem. 2002;277:36296–36303. doi: 10.1074/jbc.M200586200. [DOI] [PubMed] [Google Scholar]
- Niu Z, Jin W, Zhang L, Li X. Tumor suppressor RBM5 directly interacts with the DExD/H-box protein DHX15 and stimulates its helicase activity. FEBS Lett. 2012;586:977–983. doi: 10.1016/j.febslet.2012.02.052. [DOI] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci U S A. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- Schwarzer C, Sperk G. Hippocampal granule cells express glutamic acid decarboxylase-67 after limbic seizures in the rat. Neuroscience. 1995;69:705–709. doi: 10.1016/0306-4522(95)00348-m. [DOI] [PubMed] [Google Scholar]
- Son JH, Winzer-Serhan UH. Expression of neuronal nicotinic acetylcholine receptor subunit mRNAs in rat hippocampal GABAergic interneurons. J Comp Neurol. 2008;511:286–299. doi: 10.1002/cne.21828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staba RJ, Wilson CL, Bragin A, Fried I, Engel J., Jr Quantitative analysis of high-frequency oscillations (80–500 Hz) recorded in human epileptic hippocampus and entorhinal cortex. J Neurophysiol. 2002;88:1743–1752. doi: 10.1152/jn.2002.88.4.1743. [DOI] [PubMed] [Google Scholar]
- Tanner DC, Qiu S, Bolognani F, Partridge LD, Weeber EJ, Perrone-Bizzozero NI. Alterations in mossy fiber physiology and GAP-43 expression and function in transgenic mice overexpressing HuD. Hippocampus. 2008;18:814–823. doi: 10.1002/hipo.20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thind KK, Yamawaki R, Phanwar I, Zhang G, Wen X, Buckmaster PS. Initial loss but later excess of GABAergic synapses with dentate granule cells in a rat model of temporal lobe epilepsy. J Comp Neurol. 2010;518:647–667. doi: 10.1002/cne.22235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, Zeeberg BR, Kane D, Weinstein JN, Blume J, Darnell RB. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Vignes M, Clarke VR, Parry MJ, Bleakman D, Lodge D, Ornstein PL, Collingridge GL. The GluR5 subtype of kainate receptor regulates excitatory synaptic transmission in areas CA1 and CA3 of the rat hippocampus. Neuropharmacology. 1998;37:1269–1277. doi: 10.1016/s0028-3908(98)00148-8. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Khan SU, Engel J, Jr, Isokawa M, Babb TL, Behnke EJ. Paired pulse suppression and facilitation in human epileptogenic hippocampal formation. Epilepsy Res. 1998;31:211–230. doi: 10.1016/s0920-1211(98)00063-1. [DOI] [PubMed] [Google Scholar]
- Winden KD, Oldham MC, Mirnics K, Ebert PJ, Swan CH, Levitt P, Rubenstein JL, Horvath S, Geschwind DH. The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol. 2009;5:291. doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winden KD, Karsten SL, Bragin A, Kudo LC, Gehman L, Ruidera J, Geschwind DH, Engel J., Jr A systems level, functional genomics analysis of chronic epilepsy. PLoS One. 2011;6:e20763. doi: 10.1371/journal.pone.0020763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymore RS, Korenberg JR, Kinoshita KD, Aiyar J, Coyne C, Chen XN, Hustad CM, Copeland NG, Gutman GA, Jenkins NA, et al. Genomic organization, nucleotide sequence, biophysical properties, and localization of the voltage-gated K+ channel gene KCNA4/Kv1.4 to mouse chromosome 2/human 11p14 and mapping of KCNC1/Kv3.1 to mouse 7/human 11p14.3–p15.2 and KCNA1/Kv1.1 to human 12p13. Genomics. 1994;20:191–202. doi: 10.1006/geno.1994.1153. [DOI] [PubMed] [Google Scholar]
- Ylinen A, Bragin A, Nadasdy Z, Jando G, Szabo I, Sik A, Buzsaki G. Sharp wave-associated high-frequency oscillation (200 Hz) in the intact hippocampus: network and intracellular mechanisms. J Neurosci. 1995;15:30–46. doi: 10.1523/JNEUROSCI.15-01-00030.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article 17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.