Abstract
αvβ8 is an integrin that recognizes an Arg-Gly-Asp (RGD) motif and interacts with fibronectin, vitronectin, and latent TGF-β1. We comprehensively determined the binding activity of the αvβ8 integrin toward 25 secreted proteins having an RGD motif. The αvβ8 integrin strongly bound to latent TGF-β1 but showed marginal activity for other RGD-containing proteins, including fibronectin and vitronectin. Site-directed mutagenesis of latent TGF-β1 demonstrated that the high affinity binding of αvβ8 integrin to latent TGF-β1 was defined by Leu-218 immediately following the RGD motif within the latency-associated peptide of TGF-β1. Consistent with the critical role of Leu-218 in latent TGF-β1 recognition by αvβ8 integrin, a 9-mer synthetic peptide containing an RGDL sequence strongly inhibited interactions of latent TGF-β1 with αvβ8 integrin, whereas a 9-mer peptide with an RGDA sequence was ∼60-fold less inhibitory. Because αvβ3 integrin did not exhibit strong binding to latent TGF-β1 or distinguish between RGDL- and RGDA-containing peptides, we explored the mechanism by which the integrin β8 subunit defines the high affinity binding of latent TGF-β1 by αvβ8 integrin. Production of a series of swap mutants of integrin β8 and β3 subunits indicated that the high affinity binding of αvβ8 integrin with latent TGF-β1 was ensured by interactions between the Leu-218 residue and the β8 I-like domain, with the former serving as an auxiliary recognition residue defining the restricted ligand specificity of αvβ8 integrin toward latent TGF-β1. In support of this conclusion, high affinity binding toward the αvβ8 integrin was conferred on fibronectin by substitution of its RGDS motif with an RGDL sequence.
Keywords: cell adhesion, extracellular matrix, fibronectin, integrin, transforming growth factor beta (TGF-β), RGD motif
Introduction
Integrins are a family of adhesion receptors that bind to a variety of extracellular ligands, typically cell adhesion proteins in the extracellular matrix (ECM).2 Integrins play mandatory roles in embryonic development and the maintenance of tissue architecture by providing essential links between cells and the ECM (1). Integrins are composed of two non-covalently associated subunits, termed α and β. In mammals, 18 α and 8 β subunits have been identified, and combinations of these subunits give rise to at least 24 distinct integrin heterodimers, among which 18 isoforms function as ECM receptors. Based on their ligand binding specificities, ECM-binding integrins are classified into three major groups as follows: laminin-, collagen-, and Arg-Gly-Asp (RGD)-binding integrins (1, 2), of which the RGD-binding integrins have been most extensively investigated. The RGD-binding integrins include α5β1, α8β1, αIIbβ3, and αv-containing integrins, which interact with a variety of ECM ligands containing RGD motifs with distinct binding specificities.
The integrin αv subunit was originally identified as a receptor for vitronectin (3). The αv-containing integrins are widely expressed on many cell types, including neural crest cells, glial cells, muscle cells, osteoclasts, epithelial cells, and vascular endothelial cells during embryonic development (4–8) and during angiogenesis in response to tumors (9). Mouse embryos containing a null mutation in the αv gene exhibit placental defects and intracerebral hemorrhage, indicative of its important role in placentation and vasculogenesis (10, 11). To date, the αv subunit has been shown to combine with five β subunits, β1, β3, β5, β6, and β8. Among the five αv-containing integrins, αvβ8 is the major αv integrin and has a critical role in placentation and vasculogenesis, because the phenotypes of mice lacking integrin β8 expression largely overlap with those lacking the αv subunit (12).
The amino acid sequence of the β8 subunit is highly conserved among vertebrates but is divergent from other integrin β subunits (13), suggesting the αvβ8 integrin may have unique functions among the αv-containing integrins. The exogenous expression of integrin β8 inhibited cell growth, spreading, and focal contact formation (14, 15), in contrast to the exogenous expression of other αv-associated integrin β subunits. The β8 subunit is found in the brain, kidneys, airways, and placenta and is localized at brain vessels, synapses, glial cells, and dendritic spines, implying a specific function in the brain (13, 16). To date, the αvβ8 integrin has been shown to bind vitronectin (14), fibronectin (17), and a latency-associated peptide of TGF-β1/3 (18, 19), among which TGF-β1 is the most characterized ligand for αvβ8 integrin. The αvβ8 integrin binds to the latent form of TGF-β1 (designated latent TGF-β1) and activates it by releasing the mature TGF-β1 from the latency-associated peptide (18). However, the binding capability of the αvβ8 integrin to other RGD-containing proteins has not been comprehensively analyzed at the molecular level, suggesting unknown proteins containing the RGD motif might also serve as ligands that specifically interact with the αvβ8 integrin.
Although ligand recognition by RGD-binding integrins is primarily determined by the RGD motif in the ligands, several studies have demonstrated that residues outside the RGD motif also define binding specificities and affinities toward individual RGD-binding integrins (2, 20). For example, α5β1 integrin specifically binds to fibronectin through the bipartite recognition of an RGD motif in the 10th type III repeat, together with the PHSRN sequence and several basic residues within the 9th type III repeat, the latter serving as a “synergy site” (22, 23). α8β1 integrin selectively binds to nephronectin via a bipartite interaction with the RGD motif and LFEIFEIER sequence, the latter located at the C-terminal ∼10 amino acids from the RGD motif (24). The high affinity binding of αvβ6 integrin to its ligands, foot-and-mouth disease virus and latent TGF-β1, requires the RGD motif and an LXX(L/I) sequence, of which the latter forms an α-helix to align the two conserved hydrophobic residues along the length of the helix (25, 26). Thus, the ligand-binding specificity of RGD-binding integrins might be defined by the bipartite recognition site comprising an RGD motif and residues flanking the RGD motif or those in neighboring domains that come into close proximity with the RGD motif in an intact ligand protein. However, the ligand-binding specificity of the αvβ8 integrin toward a broad range of RGD-containing proteins as well as the molecular mechanisms defining its ligand specificity remain poorly understood.
In this study, we comprehensively investigated the binding activities of the αvβ8 integrin toward 25 RGD-containing proteins selected by in silico screening for putative secreted proteins containing a conserved RGD motif. Our results showed that the αvβ8 integrin has a restricted binding specificity toward latent TGF-β1, which is primarily defined by the Leu-218 residue located immediately after the RGD motif. The mechanism by which the αvβ8 integrin recognizes the Leu-218 residue was investigated by constructing a series of swap mutants between integrin β8 and β3 subunits.
Experimental Procedures
Cells, Antibodies, and Reagents
FreeStyleTM 293-F cells were obtained from Life Technologies, Inc., and cultured in FreeStyle 293-F expression medium. Human plasma fibronectin was purified from outdated human plasma by gelatin affinity chromatography as described previously (27). HRP-conjugated mAbs against FLAG and penta-His tags were purchased from Sigma and Qiagen (Valencia, CA), respectively. An anti-“Velcro” (ACID/BASE coiled-coil) antibody was raised in rabbits by immunization with coiled-coil ACID and BASE peptides, as described previously (28), and biotinylated by an EZ-link NHS-Sulfo-LC-biotin kit (Pierce) to detect recombinant integrins. HRP-conjugated streptavidin was purchased from Pierce. Synthetic peptides were purchased from Thermo Fisher Scientific (Dreieich, Germany) and dissolved in 100% DMSO.
In Silico Screening of RGD Motif-containing Proteins
The Protein Information Resource (PIR) Perfect Peptide Match program was used to screen proteins registered in the UniProt Knowledgebase (UniprotKB) (29, 30). Non-redundant proteins possessing at least one RGD motif were selected and then further screened for their ability to be secreted into the extracellular space, based on their annotation in the Uniprot database or the presence or absence of signal peptides and transmembrane regions, respectively, which were predicted by using PSORT II (31) and SOSUI (32). Conservation of RGD motifs in vertebrates was assessed using the Ensembl genome database (33).
cDNA Cloning and Construction of Expression Vectors
cDNA encoding human latent TGF-β1 was amplified by PCR using a latent TGF-β1 cDNA clone purchased from Life Technologies, Inc. (IMAGE clone 3356605), as a template. The amplified cDNA was subcloned into pBluescript II KS+ vector (Stratagene, La Jolla, CA). After verification by DNA sequencing, the amplified cDNA was digested with HindIII/EcoRI and inserted into the corresponding restriction sites of the pSecTag2B vector (Invitrogen), yielding the latent TGF-β1 expression vector pSecTag-TGF-β1. cDNAs encoding human angiopoietin-related protein 7 (ANGPTL7, IMAGE clone 3544149), human EGF-like repeat and discoidin-I-like domain-containing protein 3 (EDIL3, IMAGE clone 4791845), human osteopontin (SPP1, IMAGE clone 4284921), and human vitronectin (IMAGE clone 4040317) were obtained from the Mammalian Gene Collection and amplified by PCR using individual cDNA clones as templates. cDNAs encoding insulin-like growth factor-binding protein 2 (IGFBP2), lactadherin (MFGE8, deleted for its second discoidin-like domain), thrombospondin-1 (THBS1), thrombospondin-2 (THBS2), bone sialoprotein (IBSP), EGF-like, fibronectin type III and laminin G domains (EGFLAM), prothrombin (F2), TGF-β1-induced protein ig-h3 (BIGH3), proprotein convertase subtilisin/kexin type 6 (PCSK6), wingless-type murine mammary tumor virus integration site family, member 10A (WNT10A), fibrillin-1 (FBN1; a truncated form consisting of 23rd to 28th EGF-like repeats and 6th to 7th TGF-β-binding (TB) domains), fibrillin-2 (FBN2; a truncated form consisting of 5th TB, 21st to 26th EGF-like repeats and 6th TB domains), fibulin-5 (FBLN5), netrin-1 (NTN1), and hemicentin-2 (HMCN2, a truncated form consisting of 4th and 5th immunoglobulin-like domains) were amplified by reverse transcription-PCR. Template RNAs used for PCR amplification were obtained from A549 cells (for IGFBP2 and MFGE8), HeLa-S3 cells (for THBS1 and THBS2), human fetal brain (Clontech; for IBSP and EGFLAM), human fetal liver (Clontech; for F2, BIGH3, PCSK6, and WNT10A), and human fetal heart (Clontech; for FBN1, FBN2, FBLN5, NTN1, and HMCN2). PCR-amplified cDNAs except for latent TGF-β1 were subcloned into pSecTag2B vector (Invitrogen) in which a FLAG tag was inserted in-frame to the Igκ leader sequence at the 5′ end and verified by DNA sequencing. A list of the primer sequences used for PCR is available upon request. A cDNA encoding human fibronectin III7–10 (FNIII7–10; a truncated form consisting of the 7th to 10th type III domains) was amplified by PCR using a fibronectin cDNA clone (34) as a template. The amplified cDNA was subcloned into the pBluescript II KS+ vector. After verification by DNA sequencing, the amplified cDNA was digested with HindIII/PstI and inserted into the corresponding restriction sites of the pSecTag2B vector, yielding an expression vector for FNIII7–10 designated pSecTag-FNIII7–10.
A cDNA encoding the extracellular region of human integrin β8 was amplified by reverse transcription-PCR using total RNA extracted from WiDr human colon carcinoma cells as a template. The PCR-amplified cDNA was digested with BamHI/PmeI and inserted into the corresponding restriction sites of the pEF expression vector in-frame to the sequence encoding the “BASE” peptide and a His6 tag at the 3′ end of the integrin β8 cDNA as described previously (35). Another pEF vector encoding the extracellular region of integrin β8 lacking a C-terminal His6 tag was also constructed by overlap extension PCR for expression of recombinant αvβ8 integrin lacking the His6 tag (designated as αvβ8(ΔHis)). The expression vectors for the extracellular regions of integrin αv and β3 were described previously (28, 35). cDNAs encoding a series of swap mutants of the extracellular region of integrin β8 and β3 were amplified by PCR using cDNAs encoding the integrin β8 and β3 as a template, respectively. The primer sequences for PCR are available upon request. After verification by DNA sequencing, PCR-amplified cDNA fragments were digested with BamHI and NheI and inserted into the corresponding restriction sites of pEF-integrin β8-BASE-His6 or pEF-integrin β3-BASE-His6 and verified by DNA sequencing, respectively.
Site-directed Mutagenesis
Site-directed mutagenesis of latent TGF-β1 and a truncated form of fibronectin (FNIII7–10) was accomplished by overlap extension PCR with KOD polymerase using pSecTag-TGF-β1 and pSecTag-FNIII7–10 as templates, respectively. The primer sequences for the site-directed mutagenesis are available upon request. After verification by DNA sequencing, the PCR products containing the mutations were subcloned into the pSecTag2B vector.
Expression and Purification of Recombinant Proteins and Integrins
GST-fused EGF-like protein 6 (EGFL6), GST-fused FRAS-1-related extracellular matrix protein 1 (FREM1), laminin-α5 (LAMA5, as laminin-511), and nephronectin were purified as described previously (24, 36–38). For purification of other RGD-containing proteins, FreeStyle 293-F cells (Life Technologies, Inc.) were transiently transfected with individual expression vectors according to the manufacturer's instructions. The conditioned media were collected at 72 h after transfection and centrifuged to remove cells and debris, followed by addition of Pefabloc SC (Roche Diagnostics, Basel, Switzerland; 0.4 mm), imidazole (10 mm), and sodium azide (0.02%). The conditioned media were incubated with nickel-nitrilotriacetic acid (Ni-NTA)-agarose beads (Qiagen), followed by washing with TBS. Bound proteins were eluted with TBS containing 200 mm imidazole. The eluted fractions except for latent TGF-β1, vitronectin, thrombospondin-1, and PCSK6 were applied to columns of anti-FLAG M2-agarose beads (Sigma), and the bound proteins were eluted with 100 μg/ml FLAG peptide (Sigma). Recombinant latent TGF-β1, vitronectin, thrombospondin-1, and PCSK6 were purified by one-step Ni-NTA affinity chromatography. The purified proteins were dialyzed against TBS and quantified by a protein assay kit (Bio-Rad) using BSA as a standard. Recombinant integrins were expressed in FreeStyle 293-F cells by cotransfection with expression vectors encoding integrin α and β subunits and purified as described above by two-step affinity chromatography, except for αvβ8(ΔHis), which was purified by one-step chromatography using anti-FLAG-M2-agarose (28, 35).
SDS-PAGE and Western Blotting
SDS-PAGE was carried out according to Laemmli (39) using 8, 12, or 5–20% gradient gels. Separated proteins were visualized by Coomassie Brilliant Blue staining or transferred onto PVDF membranes (Millipore, Billerica, MA) for immunoblotting. The membranes were treated with TBS containing 5% skim milk and 0.05% Tween 20 for detection of FLAG tags or anti-His blocking reagent (Qiagen) containing 0.05% Tween 20 for detection of His6 tags. The membranes were then probed with HRP-conjugated antibodies against FLAG or penta-His tags, followed by visualization with the ECL Western blotting substrate (GE Healthcare).
Integrin Binding Assay
Integrin binding assays were performed as described previously (40). Briefly, microtiter plates (Nunc-ImmunoTM MicroWellTM 96-well plates; Thermo Fisher Scientific) were coated with various RGD-containing proteins (10 nm) overnight at 4 °C and then blocked with TBS containing 10 mg/ml BSA. The plates were incubated with integrins in the presence of 1 mm MnCl2 with or without 10 mm EDTA. In the inhibition assays, integrins were incubated on the plates in the presence of synthetic peptides at various concentrations to evaluate their inhibitory activities. The plates were washed with TBS containing 1 mm MnCl2, 0.1% BSA, and 0.02% Tween 20 with or without 10 mm EDTA, followed by quantification of bound integrins by an ELISA using a biotinylated rabbit anti-Velcro antibody and HRP-conjugated streptavidin. Integrin binding assays were also performed in a reverse manner. Briefly, microtiter plates were coated with 10 nm αvβ8(ΔHis) integrin and then incubated with various RGD-containing proteins having a His6 tag. Bound proteins were quantified by ELISA using an HRP-conjugated anti-penta-His antibody. The results represent the means of triplicate determinations. Apparent dissociation constants were calculated by saturation binding assays as described previously (40).
Results
In Silico Screening of RGD-containing Proteins as Candidates for αvβ8 Integrin Ligands
Among the currently available databases for protein sequences, UniProtKB is a suitable resource for the screening of putative αvβ8 integrin ligands because it contains ∼91,800 human protein sequences, some of which are manually annotated with information extracted from the literature. We performed protein sequence-based screening for proteins containing an RGD motif(s) using the Protein Information Resource Perfect Peptide Match program and extracted 5,083 proteins containing at least one RGD sequence. Given that this program often assigns different ID numbers for alternatively spliced variants and/or fragments, we reassigned the same ID numbers for such variants, thus yielding a non-redundant protein list comprising 1,909 proteins possessing at least one RGD motif. Because integrins are cell-surface receptors that recognize extracellular proteins, we next screened for putative secreted proteins based on the annotation of UniProtKB or the presence of a signal peptide and the absence of transmembrane region(s), respectively, as predicted by PSORTII and SOSUI. This yielded 190 putative secreted proteins with an RGD motif. Because integrin β8 is evolutionarily conserved among vertebrates, we assumed that the ligands for the αvβ8 integrin should have an RGD motif conserved among vertebrates. Screening of 190 putative secreted proteins among vertebrate orthologs that harbored the conserved RGD motif yielded 29 candidates for the αvβ8 integrin ligands (Table 1).
TABLE 1.
Candidate proteins for αvβ8 integrin ligands
| Gene symbols | Protein names | N-terminal tag | C-terminal tag | Purification methods |
|---|---|---|---|---|
| ANGPTL7 | Angiopoietin-related protein 7 | FLAG | His | Ni-NTA and anti-FLAG |
| BIGH3 | Transforming growth factor-β induced protein ig-h3 | FLAG | His | Ni-NTA and anti-FLAG |
| ECM2a | Extracellular matrix protein 2 | FLAG | His | Ni-NTA and anti-FLAG |
| EDIL3 | EGF-like repeat and discoidin l-like domain containing protein 3 | FLAG | His | Ni-NTA and anti-FLAG |
| EGFL6 | EGF-like protein 6 | GST | 38 | |
| EGFLAM | EGF-like, fibronectin type III, and laminin G domains (pikachurin) | FLAG | His | Ni-NTA and anti-FLAG |
| F2 | Prothrombin | FLAG | His | Ni-NTA and anti-FLAG |
| FBLN5 | Fibulin-5 | FLAG | His | Ni-NTA and anti-FLAG |
| FBN1b | Fibrillin-1 | FLAG | His | Ni-NTA and anti-FLAG |
| FBN2b | Fibrillin-2 | FLAG | His | Ni-NTA and anti-FLAG |
| FINCc | Fibronectinc | 27 | ||
| FREM1 | FRAS1-related extracellular matrix protein 1 | GST | 37 | |
| HMCN2b | Hemicentin-2 | FLAG | His | Ni-NTA and anti-FLAG |
| IBSP | Bone sialoprotein 2 | FLAG | His | Ni-NTA and anti-FLAG |
| IGFBP2 | Insulin-like growth factor-binding protein 2 | FLAG | His | Ni-NTA and anti-FLAG |
| KARSa | Lysine-tRNA ligase | FLAG | His | Ni-NTA and anti-FLAG |
| LAMA5 | Laminin α5 subunit | 36 | ||
| MFGE8b | Lactadherin | FLAG | His | Ni-NTA and anti-FLAG |
| NPNT | Nephronectin | FLAG | 24 | |
| NTN1 | Netrin-1 | FLAG | His | Ni-NTA and anti-FLAG |
| PCSK5a | Proprotein convertase subtilisin/kexin type 5 | FLAG | His | Ni-NTA and anti-FLAG |
| PCSK6b | Proprotein convertase subtilisin/kexin type 6 | FLAG | His | Ni-NTA |
| SEMA3Ca | Semaphorin-3C | FLAG | His | Ni-NTA and anti-FLAG |
| SPP1 | Osteopontin | FLAG | His | Ni-NTA and anti-FLAG |
| TGFB1c | Transforming growth factor-β1c | His | Ni-NTA | |
| THBS1b | Thrombospondin-1 | FLAG | His | Ni-NTA |
| THBS2 | Thrombospondin-2 | FLAG | His | Ni-NTA and anti-FLAG |
| VTNc | Vitronectinc | FLAG | His | Ni-NTA |
| WNT10A | Wingless-type MMTVd integration site family, member 10A | FLAG | His | Ni-NTA and anti-FLAG |
a The candidate proteins that could not be purified because of low expression are shown.
b These are known αvβ8 ligands.
c The candidate proteins that were purified as truncated forms are shown.
d MMTV is murine mammary tumor virus.
Recombinant Expression and Purification of the Candidates for αvβ8 Integrin Ligands
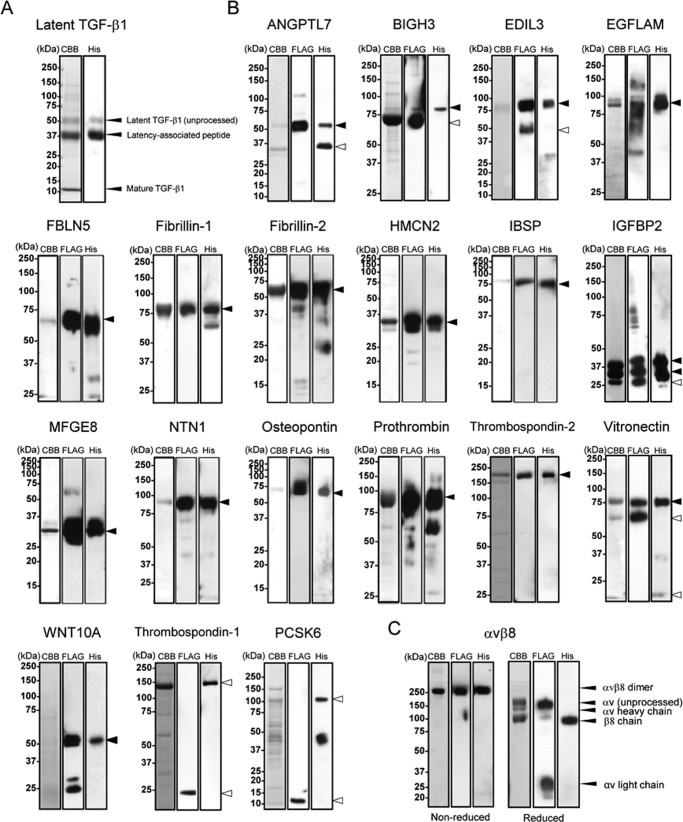
We expressed and purified the 29 RGD-containing proteins listed in Table 1. Fibronectin, EGFL6, FREM1, laminin-α5 (LAMA5, as laminin-511), and nephronectin were purified as described previously (24, 27, 36–38). Latent TGF-β1 was expressed as a recombinant protein using a mammalian expression system with a His6 tag at the N terminus and purified from conditioned medium by Ni-NTA affinity chromatography. Other proteins were expressed with N-terminal FLAG and C-terminal His6 tags to ensure the secretion of full-length proteins. Secreted recombinant proteins were purified from the conditioned medium by Ni-NTA affinity chromatography. When protein purity was not sufficient, they were further purified by anti-FLAG mAb affinity chromatography. The authenticity of the purified proteins was verified by SDS-PAGE under reducing conditions, followed by Coomassie Brilliant Blue staining and immunoblotting against the FLAG and His6 tags. Purified latent TGF-β1 gave three bands migrating at 50 kDa (unprocessed latent TGF-β1), 37 kDa (the latency-associated peptide), and 10 kDa (mature TGF-β1) (Fig. 1A), confirming the authenticity of the purified protein (41). Except for thrombospondin-1, proprotein convertase subtilisin/kexin-type 6 (PCSK6), semaphorin-3C (SEMA3C), extracellular matrix protein 2 (ECM2), lysine-tRNA ligase (KAR2), and PCSK5, the other proteins were also purified at their predicted molecular mass regions (Fig. 1B). Thrombospondin-1 and PCSK6 were obtained as proteolytically processed forms comprising N- and C-terminal fragments by Ni-NTA affinity chromatography. It was difficult to obtain sufficient amounts of recombinant SEMA3C, ECM2, KAR2, and PCSK5 for subsequent integrin binding assays because of their low levels of expression and/or proteolytic degradation.
FIGURE 1.
Purification of recombinant RGD proteins and αvβ8 integrin. Purified latent TGF-β1 (A), other recombinant RGD proteins (B), and αvβ8 integrin (C) were subjected to SDS-PAGE on 5–20% gradient gels (ANGPTL7, EDIL3, PCSK6, and latent TGF-β1), 8% gels (EGFLAM, BIGH3, FBLN5, IGFBP2, NTN1, prothrombin, thrombospondin-2, vitronectin, WNT10A, thrombospondin-1, and αvβ8 integrin), or 12% gels (fibrillin-1, fibrillin-2, HMCN2, IBSP, MFGE8, and osteopontin) under reducing conditions, followed by Coomassie Brilliant Blue staining (left), immunoblotting with an anti-FLAG monoclonal antibody (middle), or with an anti-penta-His monoclonal antibody (right), except for αvβ8 integrin that was analyzed under both reducing and non-reducing conditions. Molecular masses are indicated on the left of panels. Arrowheads indicate predicted molecular size of full-length (close) or processed form (open) of each recombinant protein.
αvβ8 Integrin Preferentially Binds to Latent TGF-β1
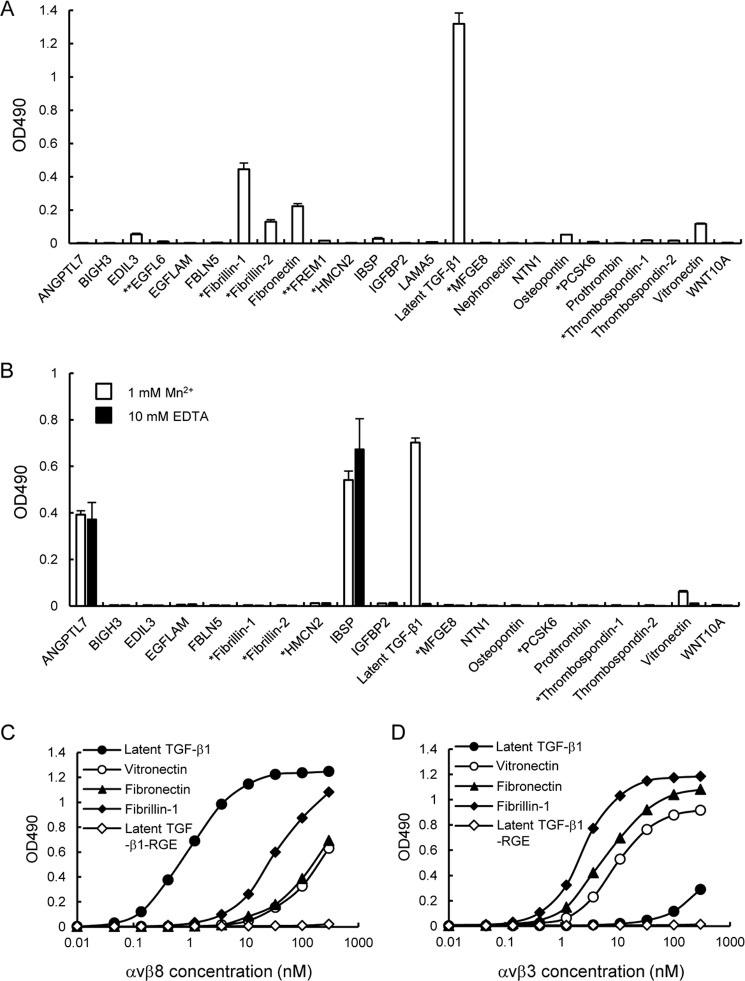
A total of 25 RGD-containing proteins (hereafter designated as “RGD proteins”) were purified and subjected to integrin binding assays using recombinant αvβ8 integrin expressed and purified as a disulfide-linked heterodimer of the extracellular domains of αv and β8 chains. The purified αvβ8 integrin gave a single band migrating at ∼250 kDa upon SDS-PAGE under non-reducing conditions and was resolved into four bands, i.e. ∼150 kDa (unprocessed αv chain), ∼120 kDa (αv heavy chain), ∼100 kDa (β8 chain), and ∼30 kDa (αv light chain), under reducing conditions (Fig. 1C). The integrin binding assays were performed in the presence of 1 mm Mn2+ to fully activate αvβ8 integrin. Among the 25 RGD proteins examined, EDIL3, fibrillin-1, fibrillin-2, fibronectin, osteopontin, latent TGF-β1, and vitronectin bound to the αvβ8 integrin with different binding affinities, but other RGD proteins did not show any significant binding to the αvβ8 integrin (Fig. 2A). Because the coating efficiency varies among the 25 RGD proteins, we also performed reverse binding assays in which microtiter plates were coated with αvβ8 integrin without a His6 tag and then incubated with a panel of RGD proteins added in the solution phase. Bound RGD proteins were detected with an HRP-conjugated anti-penta-His antibody. EGFL6, fibronectin, FREM1, laminin-α5, and nephronectin without a His6 tag were not included in this assay. Among the 20 RGD proteins tested, only latent TGF-β1 bound strongly to the αvβ8 integrin in a divalent cation-dependent manner (Fig. 2B). ANGPTL7 and IBSP also gave positive signals in the assay, but the signals still persisted in the presence of 10 mm EDTA, suggesting that they did not represent authentic interactions of the αvβ8 integrin with its ligand through the divalent cation in the β8 subunit. The results obtained in two separate assays of the reverse format indicated that the αvβ8 integrin preferentially bound to latent TGF-β1 with an affinity far exceeding those of fibronectin and vitronectin, the known ligands for the αvβ8 integrin. Saturation binding assays revealed that αvβ8 integrin bound to latent TGF-β1 with an apparent dissociation constant of 2.3 ± 0.2 nm, which was approximately 1 or 2 orders of magnitude lower than that of fibrillin-1, fibronectin, and vitronectin (Fig. 2C). Substitution of the RGD motif of latent TGF-β1 with an inactive RGE sequence completely abrogated the ability of latent TGF-β1 to bind to the αvβ8 integrin, confirming the RGD-dependent interaction of latent TGF-β1 with αvβ8 integrin. The low binding affinities of the αvβ8 integrin toward vitronectin, fibronectin, and fibrillin-1 were not caused by inactivation of their RGD ligand activity because they retained the ability to bind to αvβ3 integrin, an integrin that binds to these RGD proteins (Fig. 2D) (42–44). It should be noted that the αvβ3 integrin showed only marginal binding activity to latent TGF-β1. Its activity was nullified by substitution of the RGD motif with an inactive RGE sequence (Fig. 2D). These results indicated that the ligand specificity and binding affinity of αvβ8 integrin toward latent TGF-β1 differ significantly from the αvβ3 integrin.
FIGURE 2.
Binding activities of αvβ8 integrin toward 25 RGD proteins. A, microtiter plates were coated with RGD proteins (10 nm) and then incubated with αvβ8 integrin (10 nm) in the presence of 1 mm Mn2+. The bound integrins were quantified using a biotinylated anti-Velcro polyclonal antibody and HRP-conjugated streptavidin as described under “Experimental Procedures.” The amounts of integrin bound in the presence of 10 mm EDTA were used as negative controls and subtracted as background. The results represent S.E. of triplicate determinations. *, candidate proteins expressed as fragments containing an RGD motif. **, candidate proteins expressed as recombinant fragments fused to GST at their N termini. B, microtiter plates were coated with αvβ8(ΔHis) integrin (10 nm) and then incubated with RGD proteins (10 nm) in the presence of 1 mm Mn2+ or 10 mm EDTA. The bound RGD proteins were quantified using an HRP-conjugated anti-His6 antibody as described under “Experimental Procedures.” The results represent the means ± S.E. of triplicate determinations. C and D, titration curves of αvβ8 (left) and αvβ3 (right) integrins bound to latent TGF-β1 (closed circles), vitronectin (open circles), fibronectin (closed triangles), fibrillin-1 (closed diamonds), and the RGD → RGE substitution mutant of latent TGF-β1 (open diamonds). The results represent the means of three independent determinations. Bound integrins were quantified as described under “Experimental Procedures.”
Leu-218 Residue Immediately Following the RGD Motif Is Required for High Affinity Binding to αvβ8 Integrin
To explore the molecular basis of the restricted ligand specificity of αvβ8 integrin, we focused on the LXXI sequence immediately following the RGD motif, because the LXXI sequence was required for the high affinity binding of latent TGF-β1 to αvβ6 integrin in concert with the RGD motif (25, 26). We constructed latent TGF-β1 mutants, in which the Leu-218 and/or Ile-221 residues of the LXXI sequence were substituted with Ala, and we assessed their ability to bind to the αvβ8 integrin (Fig. 3). Although the I221A substitution did not affect the binding of latent TGF-β1 to αvβ8 integrin, the L218A substitution almost completely abrogated the binding, similar to an RGD → RGE substitution. L218A/I221A double substitution also inactivated binding to αvβ8 integrin. These results indicated that both the RGD motif and the Leu-218 residue were strictly required for the high affinity binding of latent TGF-β1 to αvβ8 integrin. To confirm the importance of Leu-218 immediately following the RGD motif in latent TGF-β1 binding by the αvβ8 integrin, we examined whether synthetic peptides modeled after the RGDL-containing sequence in latent TGF-β1 could inhibit the binding of latent TGF-β1 to αvβ8 integrin. We synthesized a 9-mer peptide containing the RGDL sequence (RRGDLATIH, designated as RGDL) and its mutant forms with RGDL → RGDA, RGDL → RGEL, and RGDL → RGEA substitutions (Fig. 4A), and we examined their inhibitory effects on the binding of the αvβ8 integrin to latent TGF-β1. The RGDL peptide strongly inhibited the binding of αvβ8 integrin to latent TGF-β1 with an IC50 of ∼0.3 μm (Fig. 4B and Table 2). Substitution of the Leu residue with Ala resulted in an ∼60-fold decrease in the potency of the peptide to inhibit the αvβ8 integrin-latent TGF-β1 interaction, although the RGEL peptides, in which the Asp residue of the RGD motif was substituted with Glu, resulted in an ∼500-fold decrease. The RGEA peptide showed little inhibitory effect at the highest peptide concentration used. These results indicated that although the RGD motif is the primary determinant, the Leu residue immediately after the RGD motif is critically required for the binding of the αvβ8 integrin to latent TGF-β1. We also examined the potency of the peptides to inhibit the binding of αvβ3 integrin to fibronectin (Fig. 4C). Both the RGDL and RGDA peptides were equally inhibitory with an IC50 of 0.3–0.4 μm, regardless of the presence or absence of the Leu residue following the RGD motif, although RGEL and RGEA peptides were only weakly inhibitory. These results indicated that the Leu residue immediately after the RGD motif is not involved in ligand recognition by the αvβ3 integrin, although it is indispensable for latent TGF-β1 recognition by the αvβ8 integrin.
FIGURE 3.
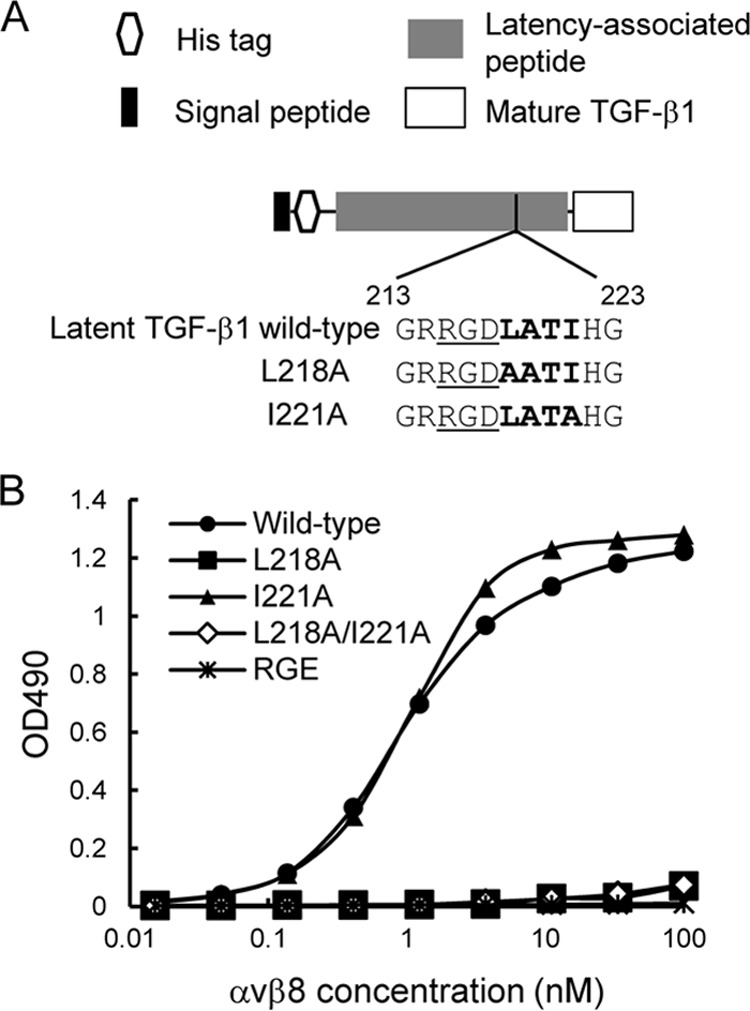
Effect of alanine substitutions within the LATI sequence on αvβ8 integrin binding activity to latent TGF-β1. A, schematic of full-length TGF-β1 and the amino acid sequences of wild-type and alanine substitution mutants of latent TGF-β1. RGD motifs are underlined, and the following LATI sequences are shown in bold. B, titration curves of αvβ8 integrin bound to full-length TGF-β1 (wild-type, circles), L218A substitution mutant (L218A, squares), I221A substitution mutant (I221A, triangles), L218A/I221A double substitution mutant (L218A/I221A, open diamonds), and RGD → RGE mutant (RGE, asterisks). The assays were performed as described in the Fig. 2 legend. The results represent the means of three independent determinations.
FIGURE 4.
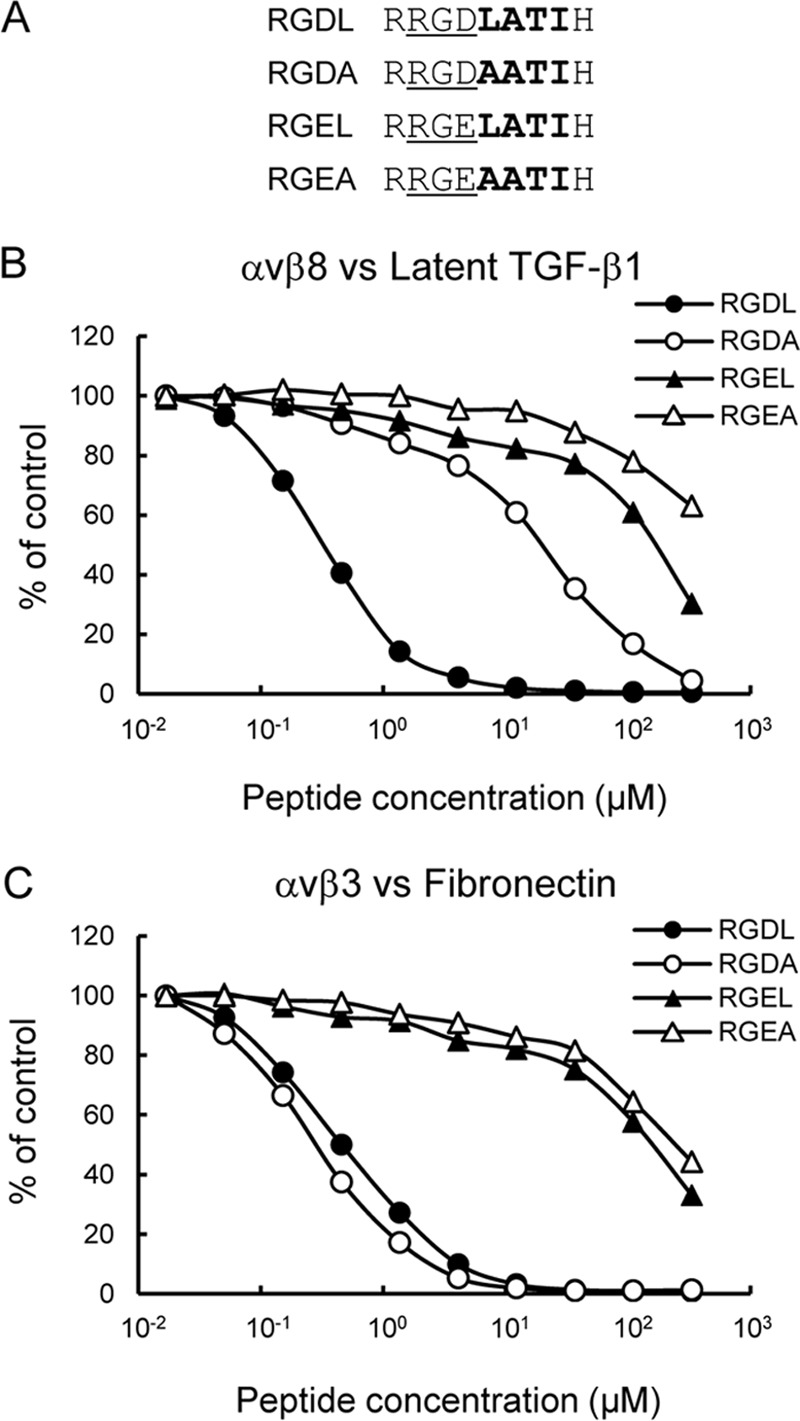
Inhibition of αvβ8 integrin binding to latent TGF-β1 by synthetic peptides. A, amino acid sequences of the synthetic peptides tested. RGD motifs are underlined, and the following LATI sequences are shown in bold. B and C, integrins (10 nm) were incubated on microtiter plates coated with latent TGF-β1 (10 nm; B) or fibronectin (10 nm; C) in the presence of increasing concentrations of synthetic peptides. To prevent precipitation of the peptides, the integrin binding assays were performed in the presence of 10% DMSO. The amounts of bound integrins are shown as percentages relative to the control, in which integrins were incubated on latent TGF-β1- or fibronectin-coated plates in the presence of 10% DMSO. The results represent the means of three independent determinations. Closed circles, RGDL (9-mer containing both RGD motif and Leu residue); open circles, RGDA (9-mer with the Leu → Ala substitution); closed triangles, RGEL (9-mer with RGD → RGE substitution); open triangles, RGEA (9-mer with RGDL → RGEA double substitution).
TABLE 2.
Inhibition of αvβ integrin and swap mutant binding to ligands by synthetic peptides
| Integrin vs. ligand | Peptides | IC50a |
|---|---|---|
| μm | ||
| αvβ8 vs. latent TGF-β1 | RGDL | 0.31 ± 0.03 |
| RGDA | 19 ± 1 | |
| RGEL | 170 ± 60 | |
| RGEA | NDb | |
| αvβ3 vs. fibronectin | RGDL | 0.40 ± 0.10 |
| RGDA | 0.29 ± 0.04 | |
| RGEL | 170 ± 30 | |
| RGEA | 220 ± 30 | |
| αvβ3–8BI vs. latent TGF-β1 | RGDL | 0.07 ± 0.01 |
| RGDA | 3.9 ± 0.9 | |
| RGEL | 32 ± 3 | |
| RGEA | 63 ± 29 | |
| αvβ8–3DLL vs. latent TGF-β1 | RGDL | 0.06 ± 0.02 |
| RGDA | 4.7 ± 0.4 | |
| RGEL | 120 ± 50 | |
| αvβ3–8DLL vs. latent TGF-β1 | RGDL | 0.17 ± 0.03 |
| RGDA | 0.13 ± 0.03 | |
| RGEL | 46 ± 7 |
To further corroborate the critical role of Leu-218 immediately after the RGD motif in latent TGF-β1 recognition by αvβ8 integrin, we examined whether the high affinity binding of latent TGF-β1 toward the αvβ8 integrin could be conferred on fibronectin by substituting the RGDL sequence for the RGDS cell-adhesive motif in the 10th FNIII domain. We produced a truncated form of fibronectin consisting of the 7th to 10th type III domains (FNIII7–10) and corresponding mutants with substitution of its RGDS sequence for RGDL or RGEL, and we assessed their abilities to bind to the αvβ8 integrin (Fig. 5A). Although control FNIII7–10 bound to the αvβ8 integrin with only moderate affinity, the RGDL mutant bound strongly to the αvβ8 integrin with an apparent Kd of 2.2 nm, which was comparable with that of latent TGF-β1 (Fig. 5B). Substitution with the RGEL sequence completely abrogated the ability of FNIII7–10 to bind to αvβ8 integrin, underscoring the prerequisite role of the RGD motif in ligand recognition by αvβ8 integrin. These results provide further support for a critical role of the Leu residue immediately after the RGD motif in high affinity binding of the αvβ8 integrin to its ligands.
FIGURE 5.
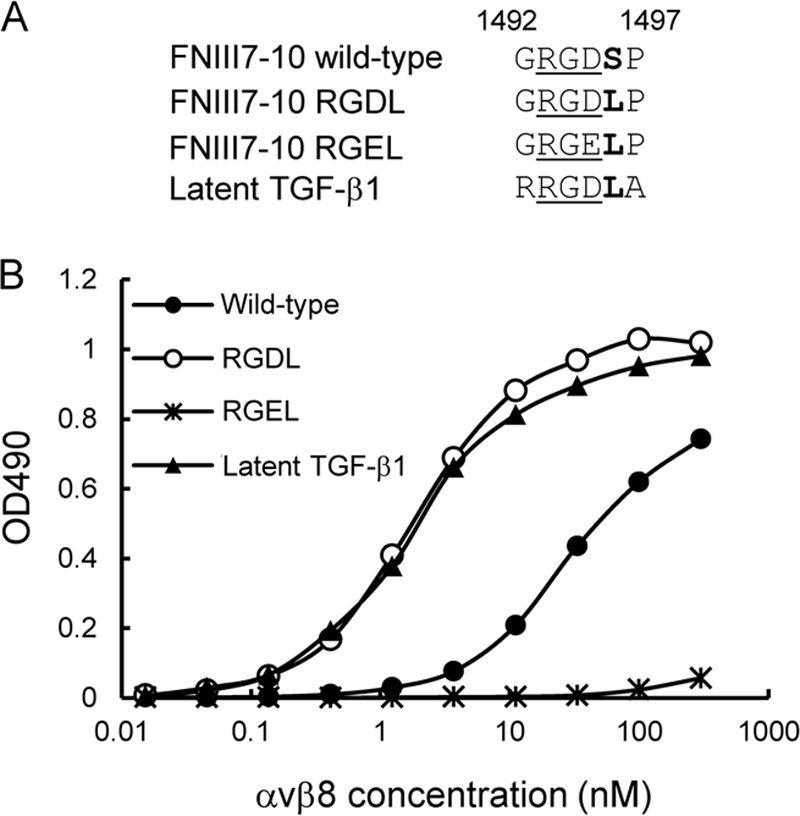
Effect of leucine substitution for the serine residue immediately after the RGD motif in the 10th FNIII domain of fibronectin on its αvβ8 integrin binding activity. A, amino acid sequences of wild-type and leucine-substituted mutants of FNIII7–10. RGD motifs are underlined, and the subsequent residues are shown in bold. B, titration curves of αvβ8 integrin bound to wild-type FNIII7–10 (closed circles), RGDL mutant (open circles), RGEL mutant (asterisks), and latent TGF-β1 (triangles). The assays were performed as described in the legend for Fig. 2. The results represent the means of three independent determinations.
β8 I-like Domain Defines the Binding Specificity of αvβ8 Integrin to Latent TGF-β1
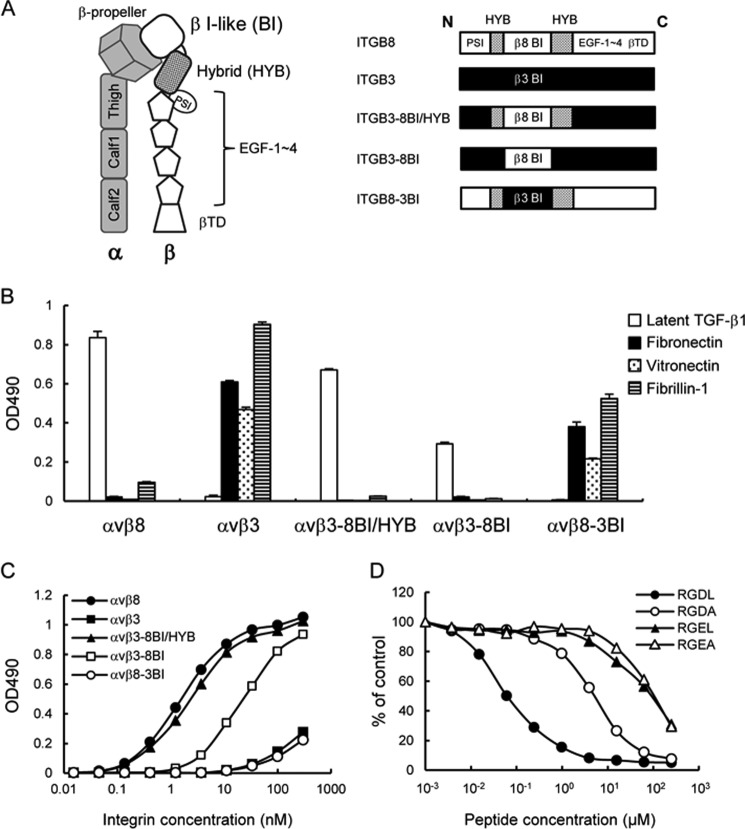
Given that the αvβ8 and αvβ3 integrins share the same αv subunit, the β8 subunit should be responsible for latent TGF-β1 recognition by the αvβ8 integrin. To explore the region within the β8 subunit that defines the ligand-binding specificity of αvβ8 integrin, we focused on the β I-like and hybrid domains of the β8 subunit because accumulating evidence suggests that these domains are directly involved in ligand recognition by αvβ3 and αIIbβ3 integrins (45–47). Given that αvβ3 integrin lacks the ability to bind to latent TGF-β1, we produced a mutant of the αvβ3 integrin, designated αvβ3–8BI/HYB, in which the β3 I-like and hybrid domains were swapped with those of the β8 subunit (Fig. 6A), and we assessed its binding activities toward latent TGF-β1, fibronectin, vitronectin, and fibrillin-1. Although wild-type αvβ3 integrin showed weak binding to latent TGF-β1, the αvβ3–8BI/HYB mutant exhibited strong binding, recapitulating the high affinity binding of αvβ8 integrin to latent TGF-β1 (Fig. 6B). In contrast, the binding activities toward fibronectin, vitronectin, and fibrillin-1 were abrogated in αvβ3–8BI/HYB, demonstrating that ligand-binding specificity of the αvβ8 integrin was conferred to αvβ3 integrin by swapping the β I-like and hybrid domains. Saturation binding assays revealed that the binding affinity of the αvβ3–8BI/HYB mutant toward latent TGF-β1 was similar to that of wild-type αvβ8 integrin, yielding an apparent dissociation constant of 3.3 ± 0.5 nm (Fig. 6C and Table 3). These results indicated that the ligand specificity and binding affinity of αvβ8 integrin to latent TGF-β1 is defined by the β8 I-like and hybrid domains.
FIGURE 6.
Ligand-binding specificities of domain swap mutants of αvβ8 and αvβ3 integrins. A, schematic of the ectodomain of integrin (left) and representations of the β8/β3 swap mutants (right). The β8- and β3-derived domains are represented by open boxes and closed boxes, respectively. The β8 hybrid domain is represented by dotted boxes. B, binding activities of domain swap mutants of αvβ8 and αvβ3 integrins toward latent TGF-β1, fibronectin, vitronectin, and fibrillin-1. C, titration curves of swap mutants bound to latent TGF-β1. Increasing concentrations of αvβ8 integrin (closed circles), αvβ3 integrin (closed squares), αvβ3–8BI/HYB (closed triangles), αvβ3–8BI (open squares), and αvβ8–3BI (open circles) were allowed to bind to microtiter plates coated with latent TGF-β1 in the presence of 1 mm MnCl2. Bound integrins were quantified as described under “Experimental Procedures.” The results represent the means of three independent determinations. Apparent dissociation constants of recombinant integrins are summarized in Table 3. D, inhibition of αvβ3–8BI binding to latent TGF-β1 by synthetic peptides. The assays were performed as described in the Fig. 4 legend. The results represent the means of three independent determinations. Closed circles, RGDL (9-mer containing both RGD motif and Leu residue); open circles, RGDA (9-mer with the Leu → Ala substitution); closed triangles, RGEL (9-mer with RGD → RGE substitution); open triangles, RGEA (9-mer with RGDL → RGEA double substitution).
TABLE 3.
Dissociation constants of αvβ8 integrin and its swap mutants towards latent TGF-β1
| β-Chains | Kda |
|---|---|
| nm | |
| β8 | 2.3 ± 0.2 |
| β3 | NDb |
| β3–8BI/HYB | 3.3 ± 0.5 |
| β3–8BI | 18 ± 1 |
| β8–3BI | ND |
| β3–8DLL | ND |
| β8–3DLL | 4.7 ± 1.2 |
a Values represent the means ± S.D. of three independent experiments.
b ND means not determined. The dissociation constant was not determined because of the partial saturation only being evaluated at the highest integrin concentration.
To identify further the region responsible for latent TGF-β1 binding by αvβ8 integrin, we constructed another swap mutant of αvβ3 integrin, αvβ3–8ΒΙ, in which only the β3 I-like domain was swapped with the β8 I-like domain (Fig. 6A). The binding specificity toward latent TGF-β1 was retained by the αvβ3–8BI mutant, although its binding activity was lower than for the αvβ8 integrin and αvβ3–8BI/HYB mutants (Fig. 6B). The apparent dissociation constant of the αvβ3–8BI mutant for latent TGF-β1 was 18 ± 1 nm (Table 3), demonstrating that the binding affinity toward latent TGF-β1 was ∼6-fold lower than those of αvβ8 integrin and αvβ3–8BI/HYB. These results indicated that the β8 I-like domain primarily defines the ligand specificity of the αvβ8 integrin, although the β8 hybrid domain potentiates the binding affinity toward latent TGF-β1. Consistent with these results, αvβ8–3BI, a mutant of the αvβ8 integrin whose β I-like domain was swapped with the β3 I-like domain, lost the ability to bind to latent TGF-β1 but bound avidly to fibronectin, vitronectin, and fibrillin-1 (Fig. 6B), recapitulating the ligand specificity of wild-type αvβ3 integrin. The binding of αvβ3–8BI to latent TGF-β1 was strongly inhibited by the RGDL peptide with an IC50 of ∼0.07 μm, although substitution of the Leu residue with Ala resulted in an ∼60-fold decrease in the inhibitory potency of the peptide (Fig. 6D and Table 2). These results indicated that the β I-like domains primarily determine the binding specificity of αv-containing integrins and that the β8 I-like domain recognizes the Leu residue following the RGD motif and is necessary for the high affinity binding of αvβ8 integrin to latent TGF-β1.
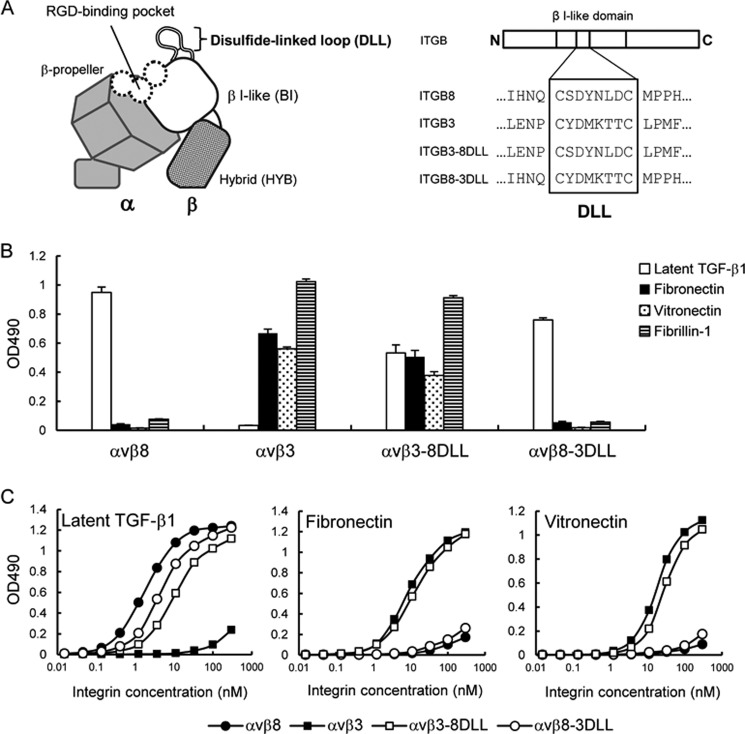
Role of the Disulfide-linked Loop in Latent TGF-β1 Recognition by αv Integrins
To identify the integrin β8 subunit region involved in latent TGF-β1 binding further, we focused on a small disulfide-linked loop consisting of 6–8 amino acid residues that resides on the top of the β I-like domain, designated as the “disulfide-linked loop (DLL)” (Fig. 7A). The DLL was reported to determine the ligand-binding specificities of αvβ1 and αvβ3 integrins (48). To address the role of the β8 subunit DLL (β8-DLL) in latent TGF-β1 recognition by the αvβ8 integrin, we produced a swap mutant of the αvβ3 integrin termed αvβ3–8DLL, in which the β3-DLL was swapped with the corresponding residues of the β8 subunit. The αvβ3–8DLL mutant exhibited binding to latent TGF-β1 while retaining the ability of the αvβ3 integrin to bind to fibronectin, vitronectin, and fibrillin-1 (Fig. 7B). A saturation binding assay demonstrated that the binding affinity of αvβ3–8DLL to latent TGF-β1 was significantly lower than that of wild-type αvβ8 integrin, although the affinity toward fibronectin and vitronectin was only slightly affected (Fig. 7C). These results indicated that β8-DLL confers latent TGF-β1 binding activity to the αvβ3 integrin without compromising the ability of the αvβ3 integrin to bind to its cognate ligands. We also produced another swap mutant, αvβ8–3DLL, in which the β8-DLL was swapped with the β3-DLL. Unexpectedly, the overall binding specificity of the αvβ8 integrin remained unaltered after DLL swapping; αvβ8–3DLL selectively bound to latent TGF-β1 without acquiring the ability to bind to fibronectin, vitronectin, and fibrillin-1 (Fig. 7B). Saturation binding assays indicated only a small decrease in the binding affinity toward latent TGF-β1 with αvβ8–3DLL (Fig. 7C). Consistent with these results, the binding affinities of αvβ8–3DLL toward fibronectin and vitronectin were also unchanged after DLL swapping. These results indicated that DLL is not the primary determinant for ligand specificity of the αvβ8 integrin, although it conferred latent TGF-β1 binding activity to the αvβ3 integrin upon DLL swapping. Therefore, it is puzzling why β8-DLL, which is dispensable for defining the ligand specificity of αvβ8 integrin, confers latent TGF-β1 binding activity to αvβ3 integrin without compromising its ability to bind to fibronectin, vitronectin, and fibrillin-1.
FIGURE 7.
Ligand-binding specificities of DLL swap mutants. A, schematic of the head region of integrin (left) and amino acid sequences of the DLL regions of the β8 and β3 subunits and their swap mutants (right). Swapped amino acids between the β8 and β3 subunits are indicated in the boxed area. B, binding activities of DLL swap mutants of αvβ8 and αvβ3 integrins toward latent TGF-β1, fibronectin, vitronectin, and fibrillin-1. C, titration curves of DLL swap mutants bound to latent TGF-β1 (left), fibronectin (middle), and vitronectin (right). Increasing concentrations of αvβ8 integrin (closed circles), αvβ3 integrin (closed squares), αvβ3–8DLL (open squares), and αvβ8–3DLL (open circles) were allowed to bind to microtiter plates coated with latent TGF-β1, fibronectin, or vitronectin in the presence of 1 mm MnCl2. Bound integrins were quantified as described under “Experimental Procedures.” The results represent the means of three independent determinations. Apparent dissociation constants of recombinant integrins are summarized in Table 3.
β8-DLL Is Not Involved in the Specific Recognition of Leu-218 Immediately after the RGD Motif
To address the apparent discrepancy of the role of β8-DLL in defining the ligand specificity of the αvβ8 integrin, we examined whether β8-DLL is involved in the recognition of Leu-218 required for high affinity binding of latent TGF-β1 to αvβ8 integrin. We examined the inhibitory effects of the RGDL and RGDA peptides on the binding of latent TGF-β1 to αvβ8–3DLL and αvβ3–8DLL, both of which were capable of binding to latent TGF-β1 (Fig. 8). The binding of αvβ8–3DLL to latent TGF-β1 was strongly inhibited by the RGDL peptide with an IC50 of 0.06 μm, although substitution of the Leu residue with Ala resulted in an ∼80-fold decrease in the inhibitory potency of the peptide (Fig. 8A and Table 2). This indicated that Leu-218 is recognized by αvβ8–3DLL to sustain its specific binding to latent TGF-β1. In contrast, RGDL and RGDA peptides equally inhibited the binding of αvβ3–8DLL to latent TGF-β1 with an IC50 of 0.17 and 0.13 μm, respectively, irrespective of the presence or absence of a Leu residue immediately after the RGD motif (Fig. 8B and Table 2). These results demonstrated that β8-DLL in αvβ3–8DLL does not recognize the Leu residue immediately after the RGD motif, consistent with the conclusion that β8-DLL is not involved in the Leu-218-dependent high affinity binding of αvβ8 integrin to latent TGF-β1. The RGEL peptide did not inhibit the interaction of αvβ8–3DLL and αvβ3–8DLL mutants with latent TGF-β1, highlighting the critical importance of the RGD motif in latent TGF-β1 recognition by the αvβ8 integrin. Taken together, these results indicate that β8-DLL is dispensable for the specific binding of the αvβ8 integrin to latent TGF-β1, although the β8 I-like domain is necessary and sufficient for recognition of the Leu-218 residue by the αvβ8 integrin.
FIGURE 8.
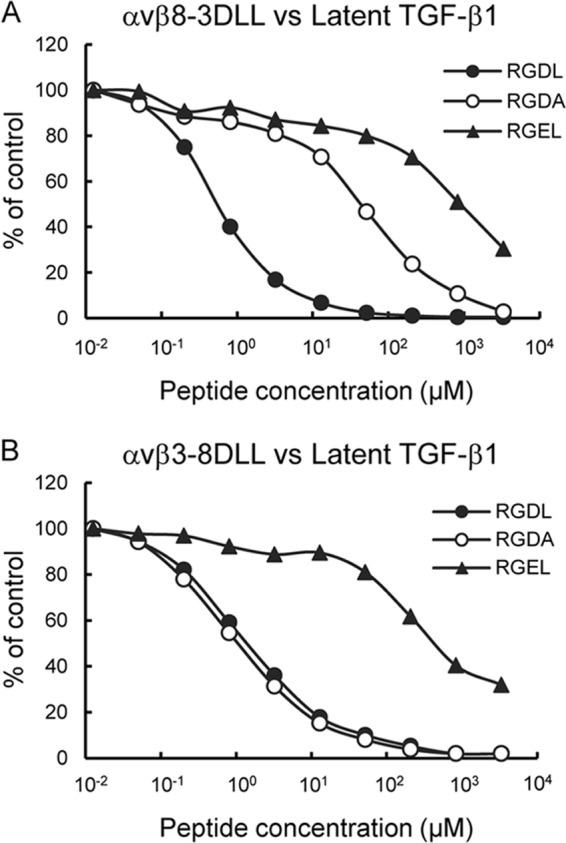
Inhibition of DLL swap mutant binding to latent TGF-β1 by synthetic peptides. αvβ8–3DLL (A) and αvβ3–8DLL (B) mutants (10 nm) were incubated on microtiter plates coated with latent TGF-β1 (10 nm) in the presence of increasing concentrations of synthetic peptides. To prevent precipitation of the peptides, the integrin binding assays were performed in the presence of 10% DMSO. The amounts of bound integrins are shown as percentages relative to the control, in which integrins were incubated on latent TGF-β1-coated plates in the presence of 10% DMSO. The results represent the means of three independent determinations. Closed circles, RGDL (9-mer containing both RGD motif and Leu residue); open circles, RGDA (9-mer with the Leu → Ala substitution); closed triangles, RGEL (9-mer with RGD → RGE substitution).
Discussion
This study demonstrated that αvβ8 integrin binds strongly and preferentially to latent TGF-β1 with an affinity of ∼100-fold higher than for other RGD proteins, including fibronectin, vitronectin, and fibrillin-1. The high affinity interaction of the αvβ8 integrin with latent TGF-β1 is determined by the Leu-218 residue immediately following the RGD motif within the latency-associated peptide of latent TGF-β1. Substitution of the Leu-218 residue with Ala resulted in a dramatic reduction of the latent TGF-β1 binding affinity of αvβ8 integrin, even though the RGD motif remained unperturbed. Accumulating evidence indicates that the binding affinities of RGD-containing ligands toward integrins are potentiated by sequences residing outside the RGD motif. The occurrence of such an auxiliary binding sequence was originally proposed in the central cell-binding domain of fibronectin, where a set of residues within the 9th type III repeat (designated synergy site) potentiates the α5β1 integrin-mediated cell adhesive activity of the RGD motif within the 10th type III repeat, although electron microscopic analyses failed to confirm a direct interaction of the 9th type III module harboring the synergy site with α5β1 integrin (49). Nephronectin also contains an auxiliary sequence LFEIFEIER required for the high affinity binding of nephronectin to the α8β1 integrin, which functions in concert with an RGD motif (24). DiCara et al. (25) reported that the interaction of latent TGF-β1 with αvβ6 integrin is determined by an LXXI sequence immediately C-terminal to the RGD motif, in which two hydrophobic residues Leu-218 and Ile-221 are required for the high affinity binding of latent TGF-β1 to the αvβ6 integrin. Our results show that Leu-218 is critically required for latent TGF-β1 recognition by αvβ8 integrin, but Ile-221 is dispensable for recognition, because the substitution of Ile-221 with Ala did not affect the high affinity binding of latent TGF-β1 to αvβ8 integrin. The substitution of Ile-221 with Pro had no impact on the latent TGF-β1 recognition by αvβ8 integrin either (data not shown). In support of this conclusion, a 9-mer peptide containing an RGDL sequence strongly inhibited the interaction of latent TGF-β1 with αvβ8 integrin, whereas a 9-mer peptide with an RGDA sequence had an ∼60-fold lower inhibitory effect on the interaction. The critical role of Leu-218 in latent TGF-β1 recognition by αvβ8 integrin was further corroborated by site-directed mutagenesis of fibronectin, in which the high affinity binding toward αvβ8 integrin was conferred on fibronectin by substitution of its RGDS motif with an RGDL sequence. The Leu-218 residue immediately following the RGD motif is conserved in latent TGF-β1 among vertebrates, underscoring the importance of Leu-218 as an auxiliary recognition residue defining the high affinity interaction of latent TGF-β1 with αvβ8 integrin.
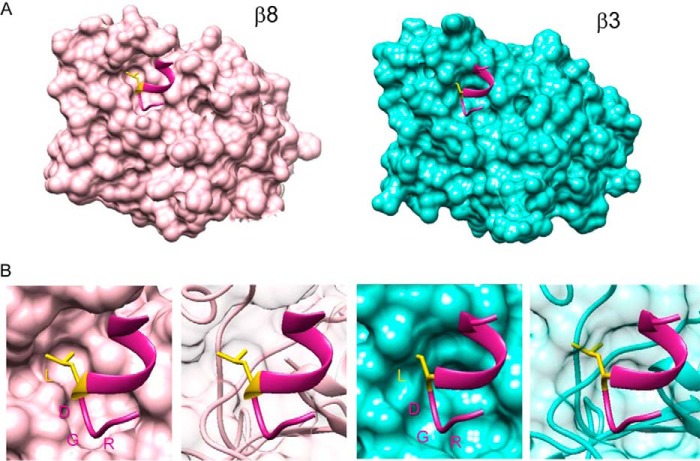
The high affinity binding of αvβ8 integrin with latent TGF-β1 was conferred upon αvβ3 integrin (which normally exhibits marginal latent TGF-β1 binding) by swapping the I-like domain of the β3 subunit with the β8 subunit. This demonstrated the β I-like domain primarily defines the high affinity interaction of αvβ8 integrin with latent TGF-β1. Recently, Dong et al. (26) reported the crystal structure of αvβ6 integrin complexed with an 11-mer peptide HGRGDLGRLKK derived from latent TGF-β3. They demonstrated that the β6 I-like domain interacted with the LGRLK sequence immediately following the RGD motif, which forms an amphipathic α-helix and confers high affinity binding of αvβ6 integrin to latent TGF-β3. The binding to latent TGF-β1 by αvβ3–8BI, in which the I-like domain of β3 subunit was swapped with the β8 subunit, was strongly inhibited by the RGDL peptide but less so by the RGDA peptide, indicating the β I-like domain of the β8 subunit might harbor the region(s) responsible for Leu-218 recognition. To explore this possibility further, we predicted the structures of the β I-like domain of αvβ8 and αvβ3 integrins complexed with an RGDL-containing peptide using Swiss-Model with the crystal structure of αvβ6 integrin complexed with the latent TGF-β3 peptide (26) and that of the αvβ3 integrin (50) as templates (Fig. 9). The β8 I-like domain was predicted to assume a structure of open conformation that allows the side chain of the Leu residue to fit into the β8 I-like domain, thus enabling the high affinity binding of latent TGF-β1 with αvβ8 integrin. In contrast, the β3 I-like domain was predicted to assume a closed structure, failing to accommodate the side chain of the Leu residue, and therefore resulting in a low affinity binding to latent TGF-β1.
FIGURE 9.
Predicted structures of the head region of αvβ8 integrin with an 11-mer peptide containing RGD motif and Leu residue. A, ribbon models of β I-like domain of αvβ8 integrin (left) and αvβ3 integrin (right) with 11-mer peptide HGRGDLGRLKK derived from latent TGF-β3 were created using the crystal structure of αvβ6 integrin with this peptide (Protein Data Bank code 4UM9) as the template. The models were predicted with the Swiss-Model and fine-tuned by energy minimization with UCSF Chimera. β I-like domains of β8 and β3 are shown in pink and cyan, respectively. 11-mer peptides are colored in violet red, and the Leu residue immediately following the RGD motif is shown with side chain in yellow. B, molecular surfaces of the β I-like domain of integrin β8 and β3 subunits with the 11-mer peptide containing RGD motif and Leu residue were generated with the Chimera. The β8 I-like domain is predicted to assume a structure of open conformation that allows the side chain of the Leu residue to fit into the β8 I-like domain, although the β3 I-like domain is predicted to assume a closed structure, failing to accommodate the side chain of the Leu residue.
The DLL of β I-like domain has been shown to modulate the ligand specificity of αv-containing integrins (48). Although cells expressing αvβ1 integrin did not bind to the substrate coated with von Willebrand factor and fibrinogen, which are high affinity ligands for the αvβ3 integrin, cells expressing the mutant αvβ1 integrin, where β1-DLL was swapped with β3-DLL, bound to these ligands, demonstrating that the ligand-binding specificity of αvβ3 integrin can be conferred upon αvβ1 integrin by swapping the DLL. In contrast to this report, our results showed that β3-DLL had little involvement in the recognition of fibronectin, vitronectin, and fibrillin-1 by αvβ3 integrin, because ligand-binding specificities of αvβ3 integrin toward these proteins remained unchanged after DLL swapping. Furthermore, the ability of αvβ8 integrin to bind to latent TGF-β1 was retained after swapping β8-DLL with β3-DLL, indicating that β8-DLL is not the primary determinant of the specific binding of the αvβ8 integrin to latent TGF-β1. Because Leu-218 is still recognized by αvβ8–3DLL, which retains specific binding to latent TGF-β1, it seems likely that the Leu-218 binding pocket is maintained in the β8 I-like domain even after replacement of β8-DLL with β3-DLL. However, why β8-DLL confers latent TGF-β1 binding activity to αvβ3 integrin, despite β8-DLL being dispensable for the recognition of latent TGF-β1 by αvβ8 integrin remains to be elucidated. The replacement of β3-DLL with β8-DLL might alter the conformation of the β3 I-like domain so that the resulting β I-like domain adopts a structure reminiscent of the open conformation that is competent for latent TGF-β1 binding, whereas the normal β3 I-like domain assumes a closely packed structure that constrains accommodation of Leu-218.
Several lines of evidence indicate that the β hybrid domain acts as a mechanical device that regulates the affinity state of integrins by rearrangement at the interface between the β Ι-like domain and the β-propeller domain of the α subunit, which together form the ligand-binding site of integrins. Thus, binding of α5β1 integrin to its ligand causes a dramatic change in the position of the β hybrid domain relative to the β Ι-like domain to induce an open conformation of the integrin headpiece (49). Xiao et al. (46) demonstrated that the hybrid domain of the β3 subunit extends laterally away from the ligand-binding site to stabilize the open headpiece conformation. Consistent with the role of the β hybrid domain in the affinity state modulation of integrins, the limiting swing-out of the β3 hybrid domain prevents αIIbβ3 integrin from binding to its high affinity ligand fibrinogen, indicating that the swing-out of the β3 hybrid domain is required for activation of the αIIbβ3 integrin (47). Our results showed that αvβ3–8BI/HYB and αvβ8 integrin had a similar binding affinity to latent TGF-β1, although αvβ3–8BI had a lower latent TGF-β1 binding affinity. These results are consistent with previous studies and support the consensus that the β hybrid domain regulates, in collaboration with the β I-like domain, the conformational change of the integrin headpiece from a low to a high affinity state for ligand binding.
There is compelling evidence that the αvβ8 integrin plays a dominant role in activating latent TGF-β1 in the developing brain. Mice deficient in the expression of the integrin β8 subunit exhibit variable embryonic lethality because of vasculogenesis failure and severe brain hemorrhage (12), as is the case with mice deficient for integrin αv subunit expression (11). Notably, when mice with a TGF-β1 gene knock-in mutation that causes an RGE substitution of the RGD motif are crossed with TGF-β3-deficient mice they die as a result of severe brain hemorrhage (51), recapitulating the phenotype of integrin β8-deficent mice. Given the similarities in phenotypes between αvβ8 integrin-deficient mice and TGF-β1(RGE)/TGF-β3 double mutant mice, αvβ8 integrin has been proposed to act as an “angiogenic switch” in the brain through TGF-β activation (52, 53). Consistent with the role of αvβ8 integrin in TGF-β1 activation, Yamazaki et al. (54) reported that β8 integrin was specifically expressed by Schwann cells and involved in latent TGF-β activation in the bone marrow, thereby regulating hematopoietic stem cell hibernation. Furthermore, the activation of latent TGF-β1 by αvβ8 integrin may occur via mechanisms distinct from those for TGF-β1 activation by αvβ6 integrin, which requires the β6 subunit cytoplasmic domain and a functional actin cytoskeleton. However, TGF-β activation by αvβ8 integrin is considered independent of the β8 subunit cytoplasmic domain and actin cytoskeleton (18, 21). Indeed, the amino acid sequence of the cytoplasmic domain of integrin β8 subunit differs significantly from those of other β subunits (13), making it unlikely that the β8 cytoplasmic domain interacts with the actin cytoskeleton and generates traction force for cell spreading and migration as well as the conformational activation of latent TGF-βs (15). Given its restricted binding specificity and prominent expression in astrocytes surrounding cerebral blood vessels (52), αvβ8 integrin may function as an anchor specialized for latent TGF-β1, thereby ensuring the localized action of active-TGF-β1 at the neurovascular unit where astrocytes cross-talk with endothelial cells to facilitate brain vascular development.
In conclusion, we have shown that the αvβ8 integrin binds strongly and preferentially to latent TGF-β1. Its high affinity binding is primarily defined by the Leu-218 residue located immediately after the RGD motif within the latency-associated peptide of latent TGF-β1. Although it remains to be determined how the β8 I-like domain recognizes the Leu-218 residue at the atomic level by x-ray crystallography of the αvβ8 integrin complexed with a latent TGF-β1, our comprehensive study on the binding activities of αvβ8 integrin and its mutant proteins toward a wide range of RGD proteins has identified molecular mechanisms involved in the specific recognition of latent TGF-β1 by the αvβ8 integrin that might help our understanding of its physiological and pathological roles.
Author Contributions
A. O. designed and performed the experiments, analyzed the results shown in all figures, and wrote the paper. K. S. conceived and coordinated the study, analyzed the results, and wrote the paper. Y. S. coordinated the study, performed and analyzed the experiments shown in Fig. 1, and wrote the paper. T. I. performed and analyzed the experiments shown in Fig. 1. T. U. constructed vectors for expression of integrins and performed preliminary experiments exploring their ligand specificity. J. T. provided expression vectors for the integrin αv and β3 subunits and contributed to interpretation of the results. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank Dr. Daiji Kiyozumi (Immunology Frontier Research Center, Osaka University) for the gift of the GST-FREM1 protein.
This work was supported by a grant-in-aid for scientific research on innovative areas from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. The authors declare that they have no conflicts of interest with the contents of this article.
- ECM
- extracellular matrix
- BI
- βI-like domain
- DLL
- disulfide-linked loop
- HYB
- β hybrid domain
- Ni-NTA
- nickel-nitrilotriacetic acid-agarose
- RGD
- Arg-Gly-Asp
- TB
- TGF-β-binding
- IBSP
- integrin binding sialoprotein/bone sialoprotein.
References
- 1. Hynes R. O. (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 2. Takagi J. (2007) Structural basis for ligand recognition by integrins. Curr. Opin. Cell Biol. 19, 557–564 [DOI] [PubMed] [Google Scholar]
- 3. Pytela R., Pierschbacher M. D., and Ruoslahti E. (1985) A 125/115-kDa cell surface receptor specific for vitronectin interacts with the arginine-glycine-aspartic acid adhesion sequence derived from fibronectin. Proc. Natl. Acad. Sci. U.S.A. 82, 5766–5770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brooks P. C., Clark R. A., and Cheresh D. A. (1994) Requirement of vascular integrin αvβ3 for angiogenesis. Science 264, 569–571 [DOI] [PubMed] [Google Scholar]
- 5. Delannet M., Martin F., Bossy B., Cheresh D. A., Reichardt L. F., and Duband J. L. (1994) Specific roles of the αVβ1, αVβ3 and αVβ5 integrins in avian neural crest cell adhesion and migration on vitronectin. Development 120, 2687–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hirsch E., Gullberg D., Balzac F., Altruda F., Silengo L., and Tarone G. (1994) αv integrin subunit is predominantly located in nervous tissue and skeletal muscle during mouse development. Dev. Dyn. 201, 108–120 [DOI] [PubMed] [Google Scholar]
- 7. Drake C. J., Cheresh D. A., and Little C. D. (1995) An antagonist of integrin αvβ3 prevents maturation of blood vessels during embryonic neovascularization. J. Cell Sci. 108, 2655–2661 [DOI] [PubMed] [Google Scholar]
- 8. Friedlander M., Theesfeld C. L., Sugita M., Fruttiger M., Thomas M. A., Chang S., and Cheresh D. A. (1996) Involvement of integrins αvβ3 and αvβ5 in ocular neovascular diseases. Proc. Natl. Acad. Sci. U.S.A. 93, 9764–9769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weis S. M., and Cheresh D. A. (2011) αV integrins in angiogenesis and cancer. Cold Spring Harb. Perspect. Med. 1, a006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Damsky C., Sutherland A., and Fisher S. (1993) Extracellular matrix 5: adhesive interactions in early mammalian embryogenesis, implantation, and placentation. FASEB J. 7, 1320–1329 [DOI] [PubMed] [Google Scholar]
- 11. Bader B. L., Rayburn H., Crowley D., and Hynes R. O. (1998) Extensive vasculogenesis, angiogenesis, and organogenesis precede lethality in mice lacking all α v integrins. Cell 95, 507–519 [DOI] [PubMed] [Google Scholar]
- 12. Zhu J., Motejlek K., Wang D., Zang K., Schmidt A., and Reichardt L. F. (2002) β8 integrins are required for vascular morphogenesis in mouse embryos. Development 129, 2891–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moyle M., Napier M. A., and McLean J. W. (1991) Cloning and expression of a divergent integrin subunit β8. J. Biol. Chem. 266, 19650–19658 [PubMed] [Google Scholar]
- 14. Nishimura S. L., Sheppard D., and Pytela R. (1994) Integrin αvβ8. Interaction with vitronectin and functional divergence of the β8 cytoplasmic domain. J. Biol. Chem. 269, 28708–28715 [PubMed] [Google Scholar]
- 15. Cambier S., Mu D. Z., O'Connell D., Boylen K., Travis W., Liu W. H., Broaddus V. C., and Nishimura S. L. (2000) A role for the integrin αvβ8 in the negative regulation of epithelial cell growth. Cancer Res. 60, 7084–7093 [PubMed] [Google Scholar]
- 16. Nishimura S. L., Boylen K. P., Einheber S., Milner T. A., Ramos D. M., and Pytela R. (1998) Synaptic and glial localization of the integrin αvβ8 in mouse and rat brain. Brain Res. 791, 271–282 [DOI] [PubMed] [Google Scholar]
- 17. Venstrom K., and Reichardt L. (1995) β8 integrins mediate interactions of chick sensory neurons with laminin-1, collagen IV, and fibronectin. Mol. Biol. Cell 6, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mu D., Cambier S., Fjellbirkeland L., Baron J. L., Munger J. S., Kawakatsu H., Sheppard D., Broaddus V. C., and Nishimura S. L. (2002) The integrin α(v)β8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-β1. J. Cell Biol. 157, 493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Worthington J. J., Klementowicz J. E., and Travis M. A. (2011) TGFβ: a sleeping giant awoken by integrins. Trends Biochem. Sci. 36, 47–54 [DOI] [PubMed] [Google Scholar]
- 20. Ruoslahti E. (1996) RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 12, 697–715 [DOI] [PubMed] [Google Scholar]
- 21. Munger J. S., Huang X., Kawakatsu H., Griffiths M. J., Dalton S. L., Wu J., Pittet J. F., Kaminski N., Garat C., Matthay M. A., Rifkin D. B., and Sheppard D. (1999) The integrin αvβ6 binds and activates latent TGFβ1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell 96, 319–328 [DOI] [PubMed] [Google Scholar]
- 22. Aota S., Nomizu M., and Yamada K. M. (1994) The short amino acid sequence Pro-His-Ser-Arg-Asn in human fibronectin enhances cell-adhesive function. J. Biol. Chem. 269, 24756–24761 [PubMed] [Google Scholar]
- 23. Redick S. D., Settles D. L., Briscoe G., and Erickson H. P. (2000) Defining fibronectin's cell adhesion synergy site by site-directed mutagenesis. J. Cell Biol. 149, 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sato Y., Uemura T., Morimitsu K., Sato-Nishiuchi R., Manabe R., Takagi J., Yamada M., and Sekiguchi K. (2009) Molecular basis of the recognition of nephronectin by integrin α8β1. J. Biol. Chem. 284, 14524–14536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. DiCara D., Rapisarda C., Sutcliffe J. L., Violette S. M., Weinreb P. H., Hart I. R., Howard M. J., and Marshall J. F. (2007) Structure-function analysis of Arg-Gly-Asp helix motifs in αvβ6 integrin ligands. J. Biol. Chem. 282, 9657–9665 [DOI] [PubMed] [Google Scholar]
- 26. Dong X., Hudson N. E., Lu C., and Springer T. A. (2014) Structural determinants of integrin β-subunit specificity for latent TGF-β. Nat. Struct. Mol. Biol. 21, 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sekiguchi K., and Hakomori S. (1983) Domain structure of human plasma fibronectin. Differences and similarities between human and hamster fibronectins. J. Biol. Chem. 258, 3967–3973 [PubMed] [Google Scholar]
- 28. Takagi J., Erickson H. P., and Springer T. A. (2001) C-terminal opening mimics 'inside-out' activation of integrin α5β1. Nat. Struct. Biol. 8, 412–416 [DOI] [PubMed] [Google Scholar]
- 29. Chen C., Li Z., Huang H., Suzek B. E., Wu C. H., and UniProt Consortium (2013) A fast peptide match service for UniProt knowledgebase. Bioinformatics 29, 2808–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Apweiler R., Bairoch A., Wu C. H., Barker W. C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., Martin M. J., Natale D. A., O'Donovan C., Redaschi N., and Yeh L. S. (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32, D115–D119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakai K., and Horton P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–36 [DOI] [PubMed] [Google Scholar]
- 32. Hirokawa T., Boon-Chieng S., and Mitaku S. (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379 [DOI] [PubMed] [Google Scholar]
- 33. Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T., Durbin R., Eyras E., Gilbert J., Hammond M., Huminiecki L., et al. (2002) The Ensembl genome database project. Nucleic Acids Res. 30, 38–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Manabe R., Ohe N., Maeda T., Fukuda T., and Sekiguchi K. (1997) Modulation of cell adhesive activity of fibronectin by the alternatively spliced EDA segment. J. Cell Biol. 139, 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ido H., Nakamura A., Kobayashi R., Ito S., Li S., Futaki S., and Sekiguchi K. (2007) The requirement of the glutamic acid residue at the third position from the carboxyl termini of the laminin γ chains in integrin binding by laminins. J. Biol. Chem. 282, 11144–11154 [DOI] [PubMed] [Google Scholar]
- 36. Ido H., Harada K., Futaki S., Hayashi Y., Nishiuchi R., Natsuka Y., Li S., Wada Y., Combs A. C., Ervasti J. M., and Sekiguchi K. (2004) Molecular dissection of the α-dystroglycan- and integrin-binding sites within the globular domain of human laminin-10. J. Biol. Chem. 279, 10946–10954 [DOI] [PubMed] [Google Scholar]
- 37. Kiyozumi D., Osada A., Sugimoto N., Weber C. N., Ono Y., Imai T., Okada A., and Sekiguchi K. (2005) Identification of a novel cell-adhesive protein spatiotemporally expressed in the basement membrane of mouse developing hair follicle. Exp. Cell Res. 306, 9–23 [DOI] [PubMed] [Google Scholar]
- 38. Osada A., Kiyozumi D., Tsutsui K., Ono Y., Weber C. N., Sugimoto N., Imai T., Okada A., and Sekiguchi K. (2005) Expression of MAEG, a novel basement membrane protein, in mouse hair follicle morphogenesis. Exp. Cell Res. 303, 148–159 [DOI] [PubMed] [Google Scholar]
- 39. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 40. Nishiuchi R., Takagi J., Hayashi M., Ido H., Yagi Y., Sanzen N., Tsuji T., Yamada M., and Sekiguchi K. (2006) Ligand-binding specificities of laminin-binding integrins: a comprehensive survey of laminin-integrin interactions using recombinant α3β1, α6β1, α7β1 and α6β4 integrins. Matrix Biol. 25, 189–197 [DOI] [PubMed] [Google Scholar]
- 41. Dubois C. M., Laprise M. H., Blanchette F., Gentry L. E., and Leduc R. (1995) Processing of transforming growth factor β1 precursor by human furin convertase. J. Biol. Chem. 270, 10618–10624 [DOI] [PubMed] [Google Scholar]
- 42. van der Flier A., and Sonnenberg A. (2001) Function and interactions of integrins. Cell Tissue Res. 305, 285–298 [DOI] [PubMed] [Google Scholar]
- 43. Humphries J. D., Byron A., and Humphries M. J. (2006) Integrin ligands at a glance. J. Cell Sci. 119, 3901–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jovanović J., Iqbal S., Jensen S., Mardon H., and Handford P. (2008) Fibrillin-integrin interactions in health and disease. Biochem. Soc. Trans. 36, 257–262 [DOI] [PubMed] [Google Scholar]
- 45. Xiong J. P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S. L., and Arnaout M. A. (2002) Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science 296, 151–155 [DOI] [PubMed] [Google Scholar]
- 46. Xiao T., Takagi J., Coller B. S., Wang J. H., and Springer T. A. (2004) Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature 432, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cheng M., Li J., Negri A., and Coller B. S. (2013) Swing-out of the β3 hybrid domain is required for αIIbβ3 priming and normal cytoskeletal reorganization, but not adhesion to immobilized fibrinogen. PLoS ONE 8, e81609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takagi J., Kamata T., Meredith J., Puzon-McLaughlin W., and Takada Y. (1997) Changing ligand specificities of αvβ1 and αvβ3 integrins by swapping a short diverse sequence of the β subunit. J. Biol. Chem. 272, 19794–19800 [DOI] [PubMed] [Google Scholar]
- 49. Takagi J., Strokovich K., Springer T. A., and Walz T. (2003) Structure of integrin α5β1 in complex with fibronectin. EMBO J. 22, 4607–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dong X., Mi L. Z., Zhu J., Wang W., Hu P., Luo B. H., and Springer T. A. (2012) α(V)β(3) integrin crystal structures and their functional implications. Biochemistry 51, 8814–8828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mu Z., Yang Z., Yu D., Zhao Z., and Munger J. S. (2008) TGFβ1 and TGFβ3 are partially redundant effectors in brain vascular morphogenesis. Mech. Dev. 125, 508–516 [DOI] [PubMed] [Google Scholar]
- 52. Cambier S., Gline S., Mu D., Collins R., Araya J., Dolganov G., Einheber S., Boudreau N., and Nishimura S. L. (2005) Integrin α(v)β8-mediated activation of transforming growth factor-β by perivascular astrocytes: an angiogenic control switch. Am. J. Pathol. 166, 1883–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Arnold T. D., Niaudet C., Pang M. F., Siegenthaler J., Gaengel K., Jung B., Ferrero G. M., Mukouyama Y. S., Fuxe J., Akhurst R., Betsholtz C., Sheppard D., and Reichardt L. F. (2014) Excessive vascular sprouting underlies cerebral hemorrhage in mice lacking αVβ8-TGFβ signaling in the brain. Development 141, 4489–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamazaki S., Ema H., Karlsson G., Yamaguchi T., Miyoshi H., Shioda S., Taketo M. M., Karlsson S., Iwama A., and Nakauchi H. (2011) Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell 147, 1146–1158 [DOI] [PubMed] [Google Scholar]