Abstract
The recent availability of genomic data has spurred many genome-wide studies of human adaptation in different populations worldwide. Such studies have provided insights into novel candidate genes and pathways that are putatively involved in adaptation to different environments, diets and disease prevalence. However, much work is needed to translate these results into candidate adaptive variants that are biologically interpretable. In this Review, we discuss methods that may help to identify true biological signals of selection and studies that incorporate complementary phenotypic and functional data. We conclude with recommendations for future studies that focus on opportunities to use integrative genomics methodologies in human adaptation studies.
Evidence from the archaeological record as well as from genetic and genomic studies demonstrates that anatomically modern humans emerged in Africa ~200 thousand years ago (kya) and rapidly migrated across the globe into new environments ~80–50 kya (reviewed in REFS 1,2). One of the implications of this history is that our Late Pleistocene (~125–12 kya) ancestors were subjected to a diverse range of novel selective pressures. Therefore, phenotypes such as thermoregulation in cold environments, tolerance to hypoxia at high altitude and light skin pigmentation in regions with low amounts of sunlight are likely to have increased reproductive fitness and thus to have been affected by adaptive pressures. In addition, during the Neolithic period (~12–4 kya), many of our ancestors began to adopt more sedentary lifestyles, resulting in increased population densities, as well as distinct diets that were associated with pastoral and agricultural technologies. These changes are also likely to have resulted in distinct adaptive pressures. For example, increased population densities are correlated with increased infectious-disease loads, and phenotypes related to the immune response are thought to have been affected by this environmental shift3. Thus, the identification of genetic signatures of human adaptation to such pressures is informative not only for understanding the ways in which adaptation has shaped genetic variation in contemporary populations, but also because the phenotypic consequences of adaptive genetic variants may have a role in the biological variation and health of contemporary humans.
Recent advances in genomic technology have led to the availability of genome-wide single nucleotide polymorphism (SNP) and whole-genome sequencing (WGS) data from many different populations, giving us the ability to take a ‘top-down’ approach to the study of human adaptation in globally diverse populations. Thus, studies of human adaptation have moved from focusing on specific genes that are known to be involved in certain traits of interest to looking across the genome for signatures of selection. There are several advantages to genome-wide approaches, the most important of which is that there is no need for a priori candidate gene hypotheses. Recent work using these genomic approaches has identified novel candidate adaptive genes as well as gene pathways that are potentially involved in human adaptation, several examples of which are discussed below4–6.
However, the genomic strategies to identify candidate adaptive genes have their limitations, and it is therefore crucial to use appropriate methodologies and to carry out in-depth follow-up studies of putative functional variation to separate false positives from biological signals. False-positive signals (also known as type I errors) occur when negative results are not rejected. In the case of genome-wide scans for selection, false positives generally refer to candidate adaptive loci which are actually neutrally evolving or deleterious. The primary difficulty in identifying candidate adaptive loci is that the evolutionary processes generating genomic variation are variable across loci, resulting in a low signal-to-noise ratio for tests of selection. Furthermore, most strong candidate adaptive loci that have been identified in global human populations are located not in protein-coding regions but in intergenic regions instead, and these loci are more likely to be involved in complex mechanisms such as gene regulation, which generate phenotypic variation. Indeed, regulatory elements can be located hundreds of thousands of base pairs away from the genes that they regulate7. These combined characteristics make it particularly challenging to identify genetic loci that are involved in recent human adaptation.
In this Review, we describe the current status of genome-wide tests of neutrality, regions of the genome that contain the strongest adaptive signatures and integrative methods to investigate the functional consequences of variation at these candidate adaptive loci. In particular, we highlight examples of recently characterized candidate adaptive genes and pathways, some of which have been studied in geographically diverse populations. In addition, we discuss some of the recent opportunities to take advantage of phenotypic, regulatory and functional data in the interpretation of results that have been generated with genome-wide scans for adaptation, as well as some of the challenges that this field of research has faced.
Identifying signatures of selection
Genomic technologies have made the collection and analysis of large amounts of population-level data possible. In turn, these data have been used to study the ways in which adaptation has shaped genomic variation across human populations. There are several genome-wide strategies to identify signatures of selection; however, the general principle involves identifying regions of the genome with patterns of variation that are unlikely to have been shaped by neutral processes (such as demographic history) alone. Here, we consider the data that are currently available, the various methods that have been used to collect these data and the limitations of these methods.
Genomic data sets
Over the past decade or so, several genome-wide sets of common SNPs in global population samples have been generated and interrogated for adaptive signatures8–12. Most of the earliest genomic scans for adaptation5,13–19 focused on population samples from phases I and II of the International HapMap Project, including Yoruba individuals living in Ibadan, Nigeria; Han Chinese individuals living in Beijing, China; Japanese individuals living in Tokyo, Japan; individuals with European ancestry living in Utah, United States; and/or Perlegen population samples (African American, Asian American and European American). This work has been complemented by more recent studies that have included genomic scans for adaptation in the Human Genome Diversity Project (HGDP), which includes 51 worldwide population samples11,12,20–22.
SNP arrays are reasonably inexpensive and constitute the largest worldwide data sets that are currently available. However, they are not an ideal platform for identifying functional targets of adaptation, as the majority of functional variants are not captured on these arrays, and because both the allele frequency spectrum and patterns of linkage disequilibrium (LD) are influenced by ascertainment biases. Therefore, most studies of genetic signatures of positive selection have focused on candidate genes or genomic regions that are tagged or represented by common SNPs8–12. The variants on SNP arrays were chosen according to various criteria23, most often based on variant identification, allele frequencies and patterns of LD in a limited number of populations, mostly in Eurasians9,24; however, the ascertainment strategy is often unclear. This design was optimized for genome-wide association studies (GWASs), but the same data have also been used for genome-wide scans for adaptation. Thus, the variants captured were mainly those common to a particular population, and many common variants in other continental regions were probably missed. Another problem that has arisen from this strategy involves allele frequency bias that can affect inferences of demographic history23, although the Human Origins SNP array uses a transparent ascertainment scheme that is particularly designed for demographic inferences25,26. Thus, genome-wide tests of neutrality that incorporate demographic parameters using genome-wide SNP data may be limited.
More recently, WGS data from global populations (for example, 1000 Genomes and Complete Genomics 69 Genomes) have been made available for interrogation. These types of data have several advantages, including the removal of the bias introduced by variant ascertainment strategies, as well as the ability to directly capture rare and functional variants.
Outlier approaches
The most common approach to identify signatures of positive selection in genome-wide data is to apply a statistical test that is sensitive to genetic signatures of adaptation and to identify the ‘outlier’ variants or the variants with values that are in the extreme tails of the empirical distribution27. The statistical tests that have been developed for this strategy are primarily designed to identify classic selective sweeps (FIG. 1a), in which a novel adaptive variant arises de novo in a population and rapidly increases in frequency to (or near) fixation. Methodologies that have been designed to identify classic sweeps include tests of neutrality based on the allele frequency spectrum, such as Tajima’s D test28, Fay and Wu’s H test29, Fu and Li’s D and F tests30 and the composite likelihood ratio (CLR) test16. They also include tests that are based on unusual patterns of LD, such as the extended haplotype homozygosity (EHH)5 and the integrated haplotype score (iHS)14 methods, and tests that are based on population structure (as measured by allele frequency differences among populations), such as FST (REF. 31), the locus-specific branch length (LSBL)32, the population branch statistic33 and the cross-population CLR test34, or some combination of these, such as the cross-population EHH test13 and the composite of multiple signals (CMS) test35. Another recent method has extended the long-range haplotype approach to distinguish between convergent evolution and single adaptive founder mutations36. These tests are most sensitive to recent signals of adaptation (over the past 80 kya37,38). However, one of the limitations to the genome-wide approaches for detecting adaptation lies in the methods that are available for data phasing and imputation. LD-based methods require phased data as well as a known recombination map. Therefore, any uncertainty or error in these input data will bias the results and inferences made from such analyses. Moreover, genomic heterogeneity in background levels of purifying selection39, mutation rates, structural variation and recombination rates can result in spurious candidates for natural selection40,41.
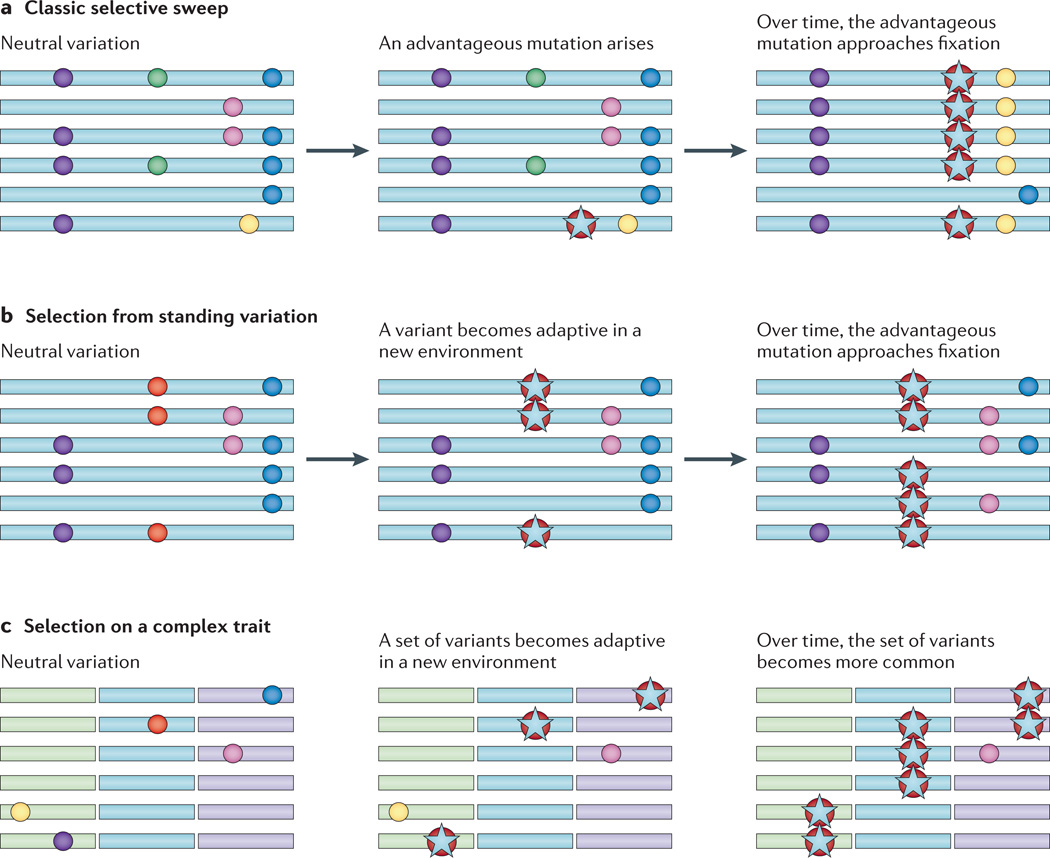
Figure 1. Genetic signatures of positive selection.
Each panel depicts changes in variant frequencies over time. Variants are shown as circles on the oblong chromosomes, and advantageous variants are represented with a star. a | A classic selective sweep, in which a novel adaptive variant arises in a population and increases in frequency over time until it approaches fixation, leaving an excess of linkage disequilibrium with surrounding variants and a decrease in levels of genetic variation. b | Selection from standing variation, in which a variant that is already present in the population becomes advantageous in a new environment and increases in frequency over time until it approaches fixation. Because the variant exists on different haplotype backgrounds, it cannot be easily detected using tests of extended haplotype homozygosity. However, when this happens in a regionally restricted manner, there is a resultant excess of allele frequency differentiation in the population being subjected to adaptive pressures, relative to a population for which the variant does not confer a selective advantage. c | Selection on a complex trait involving multiple loci on different chromosomes (represented by oblongs in different colours); when this trait becomes advantageous, it increases in frequency as a set, leaving a more subtle signature of adaptation which may include subtle shifts in allele frequencies at multiple loci.
In addition, much of the variation that would have already been present at the onset of rapid transitions in recent human history (for example, migrations to new environments and shifts in subsistence strategies) would have been pre-existing, and selection from standing variation (for example, a soft sweep) leaves a signature of selection that is more difficult to detect than a classic sweep6,42–45 (FIG. 1b). Such signals are even more difficult to detect when an adaptive trait is influenced by multiple loci44,46 (FIG. 1c); in these polygenic cases, it is also likely that selection from standing variation is the more relevant model of adaptation, rather than a classic sweep47. Indeed, a study of nucleotide diversity in the 1000 Genomes data set suggests that classic sweeps have not been a common mechanism of adaptation in recent human history48. Theoretical work in the past few years has demonstrated that measures of population structure are sensitive to selection from standing variation43 (FIG. 1b); therefore, statistical tests that use population structure may be particularly useful in studies of recent human adaptation. However, new statistical approaches that are able to take advantage of the allele frequency spectrum and patterns of haplotype diversity in WGS data will complement neutrality tests that are based on population structure.
The traditional outlier approach evaluates one variant at a time. However, several studies have also focused on identifying signatures of polygenic selection44 and of selection on gene pathways4 that are involved in a given phenotype, as the biological phenotype is what contributes to the overall reproductive fitness. This approach has been used in cases in which the pathway of interest has been known a priori or is reasonably well annotated21,47,49–53, such as the hypoxia-inducible factor 1 (HIF1) pathway, which is involved in the physiological response to hypoxic conditions at high altitude. However, in cases in which the biological pathway is not well understood or well annotated, this is a more difficult and complex problem that will require better annotation of gene function, as well as more sophisticated statistical approaches. For example, one study47 tested more than 1,000 sets of gene pathways for an enrichment of signals of positive selection (as defined by a genome-wide neutrality test using FST) and for epistatic interactions among the candidate adaptive loci using a statistical test to detect unusual levels of long-range LD. The results from this study highlight pathways that are involved in the immune response as having particularly strong signatures of polygenic adaptation.
Incorporating demography
One of the advantages of using genome-wide data sets for the study of adaptation is that, theoretically, the genome-wide patterns of variation can be used to model past demographic processes, and the outliers in the tails of the genome-wide distribution of a given summary statistic can be investigated for candidate adaptive variants. Indeed, the primary justification for outlier approaches is that demography generally has a uniform effect on the genome, whereas selection affects particular genes54. This strategy, however, poses several limitations. Most notably, a true demographic model (that is, a null model for adaptive scans) to predict patterns of neutral variation is lacking, and several studies have demonstrated high false-positive rates using outlier approaches, which is compounded by the sheer volume of genome-wide scans for human adaptation that have been reported27,54–56. One way to improve the resolution of genomic scans for adaptation is to explicitly model demographic parameters using genome-wide patterns of neutral or nearly neutral variation, thereby increasing the ability to identify variants that cannot be explained by demography alone57–61, although this is a non-trivial undertaking and generally requires genome-wide sequencing data (reviewed in REF. 55).
Insights into diverse traits
Given what is already known about recent human history — that anatomically modern humans migrated within and outside Africa at least 60 kya into many diverse and new environments with distinct selective pressures — we can infer many of the phenotypes that would have been involved in recent human adaptation, as discussed above. Here, we provide examples of putatively adaptive traits that have been studied in geographically diverse populations.
Low-hanging fruit
The earliest genome-wide searches for adaptation identified several strong signatures of adaptation in genes of major effect that also have obvious connections to the phenotypes involved in recent human adaptation. These early success stories involve situations in which there is a reasonably clear relationship between genotypic and phenotypic variation, as well as between phenotypic variation and reproductive fitness, hence suggesting a role for the genes in adaptation. For example, an extended region of haplotype homozygosity surrounds the lactase gene (LCT) in the genomes of Europeans and Africans14,62,63. This extended haplotype contains regulatory mutations that are associated with the lactase persistence phenotype in adults, which is thought to have been important in the past because it allows an individual to metabolize lactose (the complex sugar present in milk) without gastric distress. The ingestion of milk is not only an important source of nutrition but also a source of liquid in arid environments, thus providing a potential advantage to the ancestors who practised pastoralism and dairying, particularly those living in the desert63.
Similarly, solute carrier family 24, member 5 (SLC24A5) is surrounded by a region of extended LD13,14 and contains a non-synonymous SNP (rs1426654) that is associated with light skin pigmentation in Europeans64, whose ancestors are thought to have moved into environments with lower ultraviolet radiation levels 60–50 kya. There are several adaptive hypotheses to explain light skin pigmentation at high latitudes, including the possibility that it relates to vitamin D synthesis65. Although it is not entirely clear how SLC24A5 affects skin pigmentation, work in zebrafish suggests that the encoded protein regulates calcium levels in melanosomes64; moreover, the SLC24A5 candidate adaptive variants in Europeans are also found to be associated with skin pigmentation in African American, African Caribbean and Cape Verdean populations, members of which have high levels of recent European admixture66. These examples represent some of the most striking signatures of recent human adaptation. It is important to note that the ‘low-hanging fruit’ in these scenarios comprise phenotypic traits with strong links to reproductive fitness and with genetic architectures that include few genetic variants but large effect sizes per variant.
Complex traits
Traits with complex genetic architectures and/or a less clear influence on survival and reproduction have been difficult to study with the traditional outlier approach that looks at one locus at a time, particularly when each locus contributes a small effect to the trait. One example is stature, a highly heritable trait, that has been shown to involve hundreds of genetic variants, each with a small effect size (together, these variants account for only ~10% of the variance in phenotype) in studies of European populations67. However, the genetic architecture underlying the heritable component of stature might not be consistent across populations68,69. For example, so-called ‘Pygmy’ populations exhibit shorter average adult height relative to other populations70,71 (BOX 1), and the genetic architecture of stature in Baka, Bakola and Bedzan Pygmy individuals living in Cameroon seems to be distinct from what has been documented in Europeans69.
Box 1 | An example of integrative analyses of positive selection.
Our ability to identify biologically meaningful results from analyses of adaptive traits can be enhanced by the integration of genome-wide single nucleotide polymorphism or whole-genome sequencing (WGS) data with phenotypic data that are related to an adaptive hypothesis. For example, two recent studies looked at signatures of adaptation and stature in Baka, Bakola and Bedzan Pygmy individuals69,109. The short-stature phenotype is found in populations around the world, many of whom practise a hunting and gathering lifestyle in tropical rainforest environments. This phenotype is thought to have arisen independently in continental populations and either to have been adaptive in the past owing to thermoregulation, diet or locomotion71, or to be a by-product of some other adaptive trait such as reproduction, immune function or metabolism69–71. In a study69 that integrated genome-wide signatures of adaptation with data from a genotype-phenotype association study involving stature, the authors identified a strong candidate region on chromosome 3 (45–60 Mb) which contained overlapping signals of adaptation and association with stature. This region contains a cluster of candidate genes, including dedicator of cytokinesis 3 (DOCK3), previously shown to be associated with height in Europeans110, and the cytokine-inducible SH2-containing protein gene (CISH), which has an important role in the immune response and also downregulates growth hormone receptor action69. A more recent study109, which included a complimentary set of WGS data together with stature phenotypes from a subset of the same individuals, identified two additional candidate functional regions: a region containing homeobox 1 (HESX1), which is located within the chromosome 3 region identified in the earlier study, and a region containing POU class 1 homeobox 1 (POU1F1), which is located ~27 Mb upstream. Interestingly, both genes are thought to play a part in pituitary development and growth hormone regulation109. Thus, we can highlight biological mechanisms that are involved in an adaptive trait by taking advantage of phenotypic variation. However, future in vitro studies of gene expression and in vivo studies of the influence of these candidate adaptive loci on pituitary development and stature will be necessary to verify their relationship to the short-stature phenotype in humans.
One general approach that has been developed and used for identifying genetic variants that are associated with stature integrates genome-wide SNP data with indirect phenotypic measurements72. More specifically, the authors used genome-wide African population data from the HGDP and HapMap phase III data sets together with the reported mean and standard deviation measures of stature in five African population samples (including Mbuti and Biaka individuals) and identified several candidate adaptive loci containing SNPs that co-vary with stature (for example, protein phosphatase 3, catalytic subunit, β-isozyme (PPP3CB)). An alternative approach73 used a set of SNPs that has previously been shown to be associated with height in Europeans. Under a model of polygenic adaptation, the authors applied a method discussed below, and from this analysis, they predicted that alleles which are associated with increased stature would be present at higher frequencies (relative to the frequency expected by chance under a neutral model) in populations with a taller stature on average66. They demonstrated that height-increasing SNPs exhibit significantly higher frequencies in a population sample of individuals with Northern European ancestry (who generally have taller stature) than in a sample of individuals with Southern European ancestry (who generally have shorter stature). This result suggests that stature was adaptive in the ancestors of contemporary Europeans, but its relationship to reproductive fitness remains unclear.
More generally, when SNPs associated with phenotypic traits have been identified in one population, caution must be used when applying these results to other populations because patterns of LD may differ and the genetic architecture of the trait may be distinct. For example, the genetic architecture of stature in Africans may be distinct from that in Europeans69, in which case it will not be possible to make inferences about stature in Africans based on results from European GWASs. Furthermore, it is important to take pleiotropic effects into account (BOX 2). Taken together, these studies illustrate the difficulties in studying the genetic contribution to complex traits, and how adaptation may have shaped the heritable component of complex traits in particular.
Box 2 | Pleiotropy.
The interpretation of putatively adaptive traits, genetic variants and gene pathways is limited by what is already known about the phenotype, the gene function and the interactions among genes that are related to the phenotype. Moreover, when candidate adaptive genes and/or pathways have pleiotropic effects, biological interpretations are even more difficult. Several examples discussed in this Review (high-altitude physiology, lactase persistence, skin colour, insulin resistance and stature) involve phenotypes that are a priori thought to have been involved in reproductive fitness in human history; however, many of the putative adaptive loci identified in genome-wide association studies are thought to have pleiotropic effects6,69,90,97. For example, Alström syndrome 1 (ALMS1)6,90 is involved in fertility, insulin resistance, sensory perception and leprosy; ectodysplasin A receptor (EDAR)97 affects dentition, thermoregulation and mammary gland density; solute carrier family 24, member 5 (SLC24A5)87,90 is involved in skin pigmentation and leprosy; and the cytokine-inducible SH2-containing protein gene (CISH)69 affects immune function and growth. Therefore, selective pressure could be acting on one or more traits, and we must use caution when relying on simplistic interpretations and conclusions related to how past selective pressures have affected contemporary biological variation, particularly when there is a lack of functional understanding of the genes and the putatively functional gene variants involved.
Insights from studying diverse populations
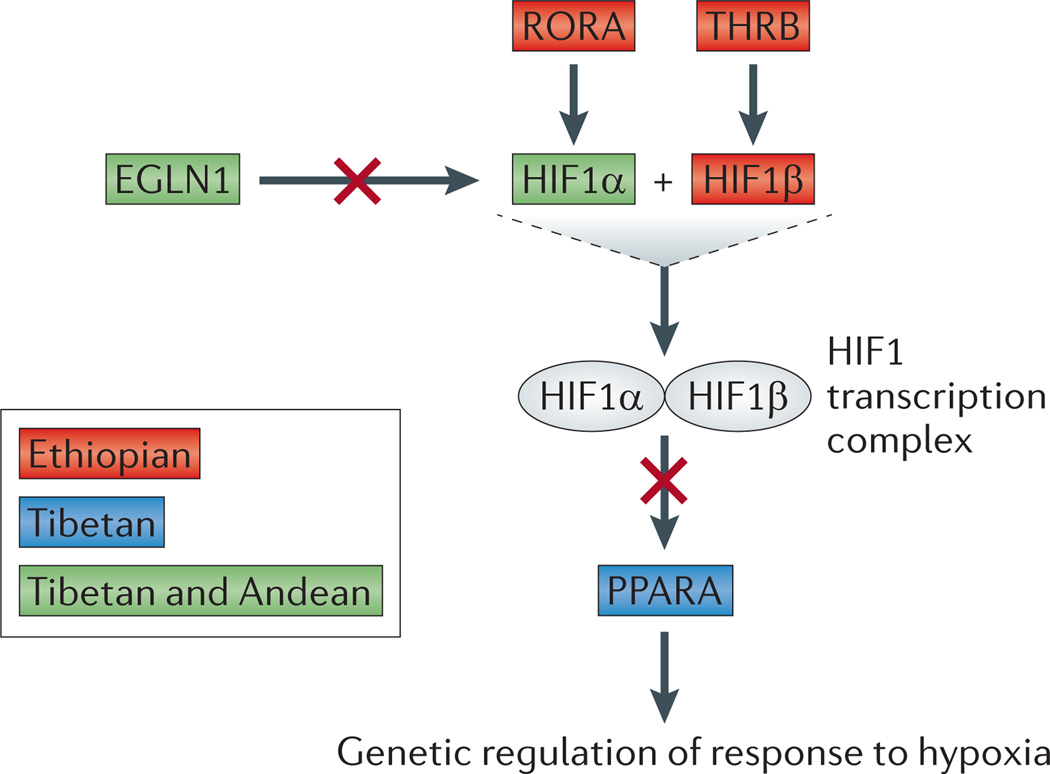
Although the inclusion of a wide range of populations, which often requires extensive field studies, has been challenging, there has been some success with this approach. For example, genome-wide studies of adaptation to high altitude have been reported for highland Andean74,75, Tibetan33,49,76,77 and Ethiopian populations78–80. One of the important findings that has emerged from this combined work is that although the biological pathway (HIF1) implicated in each continental region is consistent, the specific loci that have been identified in high-altitude populations (living >2,500 metres above sea level) are often not consistent74,79,81 (FIG. 2), which indicates convergent adaptation among different geographical regions. Additionally, results from studies of the same ethnic group (for example, Amhara individuals living at high altitude (>3,200 metres above sea level)78,79 and those living at moderate altitude (~1,800 metres above sea level)80 in Ethiopia) may differ owing to different environmental conditions and to the complex genetic architecture of adaptation to hypoxic conditions.
Figure 2. An abridged hypoxia-inducible factor 1 pathway.
Distinct genes in the hypoxia-inducible factor 1 (HIF1) pathway are implicated in adaptation in different high-altitude populations (living at altitudes >2,500 metres above sea level) owing to convergent adaptation. Each gene interaction involving a candidate adaptive gene for high altitude is shown in relation to the HIF1 pathway. Genes that have been implicated as candidate adaptive genes in high-altitude Ethiopian populations78,79, high-altitude Tibetan populations33,49,76,77, and high-altitude Tibetan and Andean populations74 are indicated. HIF1α (also known as EPAS1) and HIF1β (also known as ARNT2) form a heterodimer known as the HIF1 transcription complex. Thyroid hormone receptor-β (THRB) is required for the expression of the HIF1 transcription complex, RAR-related orphan receptor A (RORA) induces transcription of HIF1α, egl nine homologue 1(EGLN1) is involved in the degradation of the HIF1 transcription complex, and the HIF1 transcription complex inhibits peroxisome proliferator-activated receptor-α (PPARA) expression.
Another example of convergent evolution is the genetic basis of lactase persistence. Several variants that regulate lactase gene expression and are associated with the lactase persistence trait have been identified in Middle Eastern and East African populations, and these variants seem to have arisen independently from the European lactase persistence-associated variant63,82–86. Similarly, a study of skin pigmentation in Asian populations suggests that light skin pigmentation is not affected by SLC24A5 but is affected by other genes instead, such as ADAM metallopeptidase domain 17 (ADAM17) and attractin (ATRN)87. These studies demonstrate that functionally important genetic variants which have been targets of selection can be found at high frequency in geographically restricted regions owing to local adaptation and convergent adaptation.
A more systematic search for examples of convergent adaptation on the basis of EHH suggests that convergent evolution is rare on a genome-wide scale36. However, this method is sensitive to classic sweeps, and a more comprehensive estimate of the prevalence of convergent evolution will be facilitated by the development of new approaches that detect other types of adaptation as well, including polygenic adaptation and selection from standing variation.
Recent work has also evaluated the degree to which signatures of adaptation are present within populations versus across diverse populations. An investigation of the distribution of adaptive signals across the HGDP population samples found extensive sharing (44%) of the top 5% of candidate adaptive loci among population samples from Europe, the Middle East and Central Asia; however, sharing among more distantly related groups such as Europeans and East Asians was noticeably lower (12%). Similarly, another study88 found little overlap among the top 1% of iHS candidate loci that have been identified in five African population samples. These results make intuitive sense, given that the chosen statistical test (iHS) is most sensitive to recent phenomena (over the past 25 kya37) and would therefore be identifying signals of adaptation that arose after the population split of the ancestors of many of the populations included in these studies.
Taken together, these examples demonstrate the importance of including diverse human populations in genomic studies of adaptation, particularly when the results have implications for human health and disease. It is also of note that the ‘replication’ or reproducibility of particular candidate adaptive genes across diverse populations is not necessarily a reasonable requirement for the identification of candidate adaptive loci owing to the possibility of local adaptation and/or convergent evolution. Nevertheless, the inclusion of diverse populations may be one way to identify pathways of interest, even when the adaptive variants differ, and might thereby elucidate the underlying biology of the adaptive phenotype.
Using phenotypic and functional data
One of the ways that studies in the past few years have been able to recover biologically interpretable signals of recent human adaptation has been to integrate genome-wide scans of positive selection with genome-wide genotype–phenotype association studies (BOX 1) and/or with genome-wide regulatory data and functional data using in vitro and in vivo methodologies. Given the accessibility of next-generation technologies, the opportunities for generating integrative, genome-wide and functional data sets (including epigenetic, proteomic, metabolomic and transcriptomic data) are growing, and such work can improve the identification of candidate adaptive loci for in-depth follow-up functional studies.
When the identification of adaptive variation is the ultimate goal of a study, there are at least four required stages: identifying candidate adaptive loci; identifying the underlying functional variants; quantifying the phenotypic consequences of the candidate adaptive allele (or alleles), with in vitro experiments, model organisms and/ or genotype–phenotype association studies in humans; and finally, clarifying the relationship between the functional allele (or alleles) and reproductive fitness in the environment in which the allele (or alleles) rose to a high frequency in the ancestors of the study population. There are, however, only a few examples in which all of these stages have been achieved (such as the discussions of lactose tolerance above and below). More commonly, these success stories involve the identification of candidate adaptive alleles (such as ectodysplasin A receptor (EDAR)) that functionally contribute to biological variation in contemporary populations. In these cases, it is not always possible to decipher an obvious relationship between contemporary phenotypic variation and past reproductive success. Therefore, the inability to determine past selective pressures or to directly demonstrate phenotypic effects on reproductive fitness should not rule out the possibility that a locus has been adaptive.
Integrating data sets
Some studies have integrated genome-wide scans of adaptation with phenotypic data that are related to a particular adaptive hypothesis. One of the potential limitations of using putatively adaptive phenotype data is that many of the populations living in diverse environmental conditions that are relevant to adaptive hypotheses are difficult to access for genomic research studies. Therefore, it may be challenging to obtain samples sizes that are large enough to detect significant associations between phenotypic traits and variants with modest or small effect sizes. However, studies including geographically and ethnically diverse populations can be used to determine whether GWAS hits that have been identified in urban populations are replicated in other populations, which is also informative for distinguishing between environmental and genetic factors that influence the trait of interest. In addition, some populations living in extreme environments may have mean phenotypes in the tails of the phenotypic distribution (for example, stature in Pygmy populations) and may be particularly informative for identifying loci with larger effects. In particular, when strong selective pressures have acted on these populations, their genetic architecture may be distinct from that of other populations, and smaller samples sizes may still be informative.
For example, several studies of high-altitude adaptation have tested candidate adaptive regions for an association with haemoglobin levels33,49,74,77–79. This approach, in which candidate adaptive genes are used in tests of genotype–phenotype association in place of a GWAS, is especially useful when sample sizes are modest and may not permit genome-wide levels of significance after correcting for multiple testing51. Moreover, the strongest candidate adaptive genes that also contain variants associated with haemoglobin tend to have an obvious biological role in the physiological response to hypoxic conditions (that is, these genes are involved in the HIF1 pathway) (FIG. 2). Thus, this integrated approach can narrow down large lists of candidate adaptive genes to a manageable subset of strong candidates for follow-up functional studies.
Other studies have focused more generally on the integration of candidate adaptive regions with genome-wide regulatory variation. For example, one study89 used a genome-wide set of expression quantitative trait loci (eQTLs) to test the hypothesis that genetic variants involved in gene expression have been important in recent human adaptation. The authors integrated results from an LD-based neutrality test (iHS) with eQTLs that have been identified in the HapMap European, Asian and African population samples, and identified several overlapping signals, for example, genes involved in the immune response, including the human leukocyte antigen C (HLA-C) in Europeans and Asians, and HLA-DQA1, HLA-DPB2 and HLA-DRB5 in Asians. The authors more formally tested whether these regions containing signatures of adaptation are correlated with regions containing eQTLs, and found a significant association in the African sample and a suggestive association in the European and Asian samples. Given that their search for adaptive signals was restricted to recent, classic sweeps and that their sample sizes were modest, their power to identify eQTLs was limited. This result further supports the claim that in addition to protein-altering genetic variants, the variants that are involved in the regulation of gene expression have also had an important role in recent human adaptation.
The importance of gene expression variation in adaptation is further supported by an analysis that looked at the overlap between candidate adaptive variants which were identified in the 1000 Genomes data and an eQTL study90. This study also integrated other types of gene regulatory variation, including the variation in long intergenic non-coding RNAs (lincRNAs), and in predicted enhancers and promoters. Consistent with the eQTL study89, the authors documented several candidate adaptive regulatory variants that are involved in immune function. Although these putatively adaptive eQTLs and functional elements have not yet been interrogated for their phenotypic consequences or relationships to reproductive fitness, these studies suggest why so many of the candidate adaptive loci identified in genome-wide scans for selection have been intergenic. Furthermore, the broad biological classifications that have been implicated by this work (such as immune function) recapitulate phenotypes that have been implicated in functional studies of single candidate genes, such as genes involved in malarial resistance (those encoding haemoglobin and duffy91,92). Thus, functional follow-up studies are likely to include a wide range of regulatory variants.
Other functional data such as DNAase I-hypersensitive sites may also be integrated with adaptation data93. For example, one study integrated DNase I-hypersensitive sites with signatures of adaptation that were discovered in the Complete Genomics 69 Genomes data set using the LSBL method. DNase I-hypersensitive sites are of particular interest because they are located in regions where gene-regulatory proteins bind93. The authors found that genes involved in melanogenesis (which is involved in skin pigmentation) were significantly over-represented in regions containing both DNase I-hypersensitive sites and strong signals of adaptation in the European population sample, and that genes involved in chemokine and adipocytokine signalling (which is involved in insulin resistance) were significantly over-represented in regions containing both DNase I-hypersensitive sites and strong signals of adaptation in the African population sample93. Both of these analyses identified gene-regulatory variants that may have a role in adaptive phenotypic variation such as skin pigmentation and insulin resistance.
A recent integration of local candidate adaptive loci with putative regulatory variants extended the approaches that look at one locus at a time (as mentioned above) to incorporate sets of genes with coordinated upregulation or downregulation of a given class of genes4. With this method, previously reported candidate loci50 that have a role in polygenic local adaptation were used to identify positive selection on the expression levels of genes, including those that play a part in response to ultraviolet radiation, those that are involved in diabetes pathways and those that are involved in immune cell proliferation.
There are, however, several limitations to using genome-wide sets of regulatory variation in this way. Of note is the unknown generality of the phenotypic impact of regulatory variants across developmental time and tissue types. Thus, it will be a non-trivial undertaking to conduct follow-up functional studies of particular candidate adaptive regulatory variants, and this must take place before any clarification of how these variants have contributed to phenotypic variation and, ultimately, to reproductive fitness in the past. Moreover, studies that analyse two or more large genomic data sets are compounding false-positive signals and must therefore be corrected for multiple testing within and between data sets.
Follow-up functional studies
When candidate adaptive genes have been identified, there are several ways that functional follow-up studies of particular variants can be conducted in more detail using in vitro and in vivo techniques.
In the case of regulators of LCT expression, several putative adaptive functional variants were identified using genotype–phenotype association studies in human populations63,94. These candidate variants were then studied in vitro. For example, the expression of LCT in vitro was shown to be increased by several cis-regulatory variants located upstream of LCT that are commonly found in European populations (T-13910) and African populations (C-14010, G-13907 and G-13915). These variants are located ~14 kb upstream of the LCT gene within a non-coding region of the minichromosome maintenance complex component 6 (MCM6) gene63,94; thus, functionally important adaptive variants may be located at long distances from the genes that they regulate. Another in vitro follow-up study investigated functional differences between a set of three derived and ancestral amino acid substitutions located in the transient receptor potential cation channel, subfamily V, member 6 (TRPV6) locus95. In this case, no significant differences in intracellular TRPV6 expression between the derived and ancestral constructs were found95, despite a previously identified dramatic signature of selection at the TRPV6 locus96. One explanation for this discrepancy is that there is a neutral reason for the patterns of variation at TRPV6; however, it is also possible that the phenotypic consequences of adaptive TRPV6 variants do not involve intracellular expression, and the putative adaptive functional impact of variation at TRPV6 remains elusive.
Other studies have used in vivo methodologies to explore the functional consequences of putatively adaptive variation. For example, one study97 generated a knock-in mouse model to characterize the impact of a derived Edar non-synonymous SNP (370A) that is located within a haplotype background which is consistent with recent positive selection in Asian populations13. The authors found that the 370A mouse model had several potentially adaptive phenotypes, such as increased hair thickness, increased mammary gland density, smaller fat pads and altered sweat glands relative to the wild-type mice. Furthermore, the authors demonstrated that the 370A variant is associated with sweat gland activity in human populations.
These examples illustrate the ways in which putatively adaptive genetic variants can be interrogated for phenotypic consequences in vitro and in vivo in ways that are complementary to studies of genotype–phenotype association in human populations. One of the limitations of these approaches is that the variants may behave differently in vitro and in model systems compared with in humans, especially when the impact of a given variant is restricted to particular tissues and/or developmental times. For example, the stickleback fish98 was used as a model system to identify a candidate locus, a regulatory kit ligand (KITLG), as having a contribution to skin pigmentation and demonstrated its phenotypic impact on stickleback skin pigmentation. KITLG has been implicated as a candidate adaptive gene in genome-wide human studies as well99; however, a more recent human GWAS of skin pigmentation in a population sample of individuals living in Cape Verde with mixed European and African ancestry failed to identify a significant association between variation at the KITLG locus and skin pigmentation66, even though the authors validated other candidate adaptive skin pigmentation genes (for example, SLC24A5). Therefore, it remains unclear whether the variation at KITLG has a role in human pigmentation, and this example highlights the challenges in identifying genetic variants that contribute to complex traits.
In summary, in vitro and in vivo studies provide an opportunity to explore functional relationships that may not always be possible to measure directly in human populations. However, the results of these experiments may be inconsistent with those of human genotype– phenotype association studies or inconclusive in the context of human adaptive variation.
Perspectives and future directions
Next-generation technologies are being used to characterize a wide range of functional data, including epigenomic, metabolomic, metagenomic and transcriptomic data. Moreover, the Encyclopedia of DNA Elements (ENCODE) project has identified functional DNA elements in both coding and non-coding regions of the genome across a wide range of tissues. Furthermore, induced pluripotent stem cell (iPSC) technologies have revolutionized the ability to construct a range of cell types in vitro. Thus, the emerging functional data can and, in our opinions, should be used in conjunction with genomic adaptation studies to identify the underlying functional variation responsible for adaptive genetic signatures (FIG. 3A). For example, these functional data can be used to study differences in gene expression that are associated with adaptive variants (as previously carried out for lactase regulatory variation63,82–86), even in cases in which the gene expression variation is specific to certain tissues.
Figure 3. Data integration approaches.
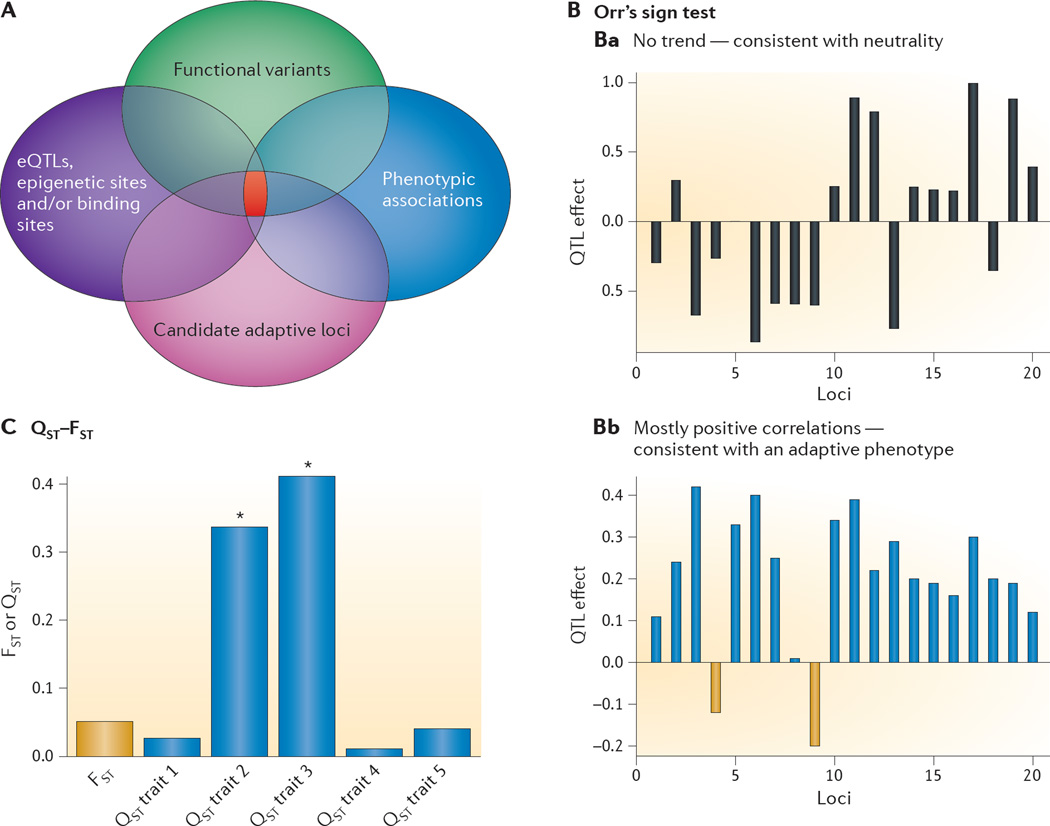
A | An integrative genomics approach. Taking advantage of multiple types of data can direct us to functionally important regions of the genome, which could in turn narrow down the candidate regions at which selective pressures may have acted. Each circle represents a distinct type of data that can be overlaid to identify the subset of loci (shown in red) that contains candidate adaptive variants, functional variants and associations with the phenotypes of interest. B | Orr’s sign test of quantitative trait locus (QTL) effects106. The x axis shows multiple loci, each with an effect on the trait of interest, which is shown on the y axis. In the neutral scenario, under the null hypothesis of neutrality, the expectation is that genetic drift will result in a situation in which positive and negative effects are randomly distributed among the loci involved in the trait. Under the alternative hypothesis of positive selection, however, the expectation is that the majority of loci involved in the trait will have either a positive or negative effect (part Ba). In the putatively adaptive scenario, the majority of loci have a positive effect on the trait of interest (shown in blue), consistent with adaptation (part Bb). C | The QST–FST test of neutrality. FST is a statistical measure of population structure or differences in variant frequencies between populations using genotypic data; QST is a statistical measure of population differentiation based on quantitative traits. The assumption is that when levels of QST exceed average levels of FST, the trait is exhibiting more inter-population divergence than expected under neutrality. The x axis includes genetic and multiple phenotypic traits, each with a measure of population structure that is displayed on the y axis, either by FST (measured with genotypic data) or QST (measured with phenotypic data). Traits 2 and 3 both have QST values that far exceed the average FST (shown by the asterisks), consistent with adaptation. eQTLs, expression quantitative trait loci.
Moreover, integrative genomics methods have been used to identify potentially functional variants that affect the traits of interest. For example, data integration methods have been successfully developed and used by researchers to more formally integrate large ‘omics’ data in the study of gene expression, as well as the investigation of particular complex traits related to bone disorders and cancer100–105. However, these types of studies have not yet incorporated candidate adaptive loci. Incorporating genome-wide neutrality tests into data integration approaches generally requires the assignment of P values to each genetic variant. Outlier approaches, however, do not provide formal P values. Therefore, future methods that can integrate data without the required P values will be of great use.
Several methods to integrate genomic data have also been developed to distinguish between adaptive traits and neutrally evolving traits. One of the earlier methods, the QTL sign test, which was first proposed in 1998 by Orr106, was designed to address whether a phenotype of interest is adaptive when it has a complex genetic architecture involving several loci (FIG. 3B). This approach was originally developed and applied to morphological variation73, as discussed above; however, it has been extended to gene expression data to detect non-neutral cis-regulatory elements107 and could also be applied to any intermediate molecular phenotypes (for example, epigenomic, transcriptomic and metabolomic data). Similarly, QST–FST comparisons are designed to differentiate between neutral evolutionary processes and positive selection by combining information across quantitative traits and genetic variation data (reviewed in REF. 108) (FIG. 3C). Future studies of adaptation involving complex traits will benefit from these types of approaches, as well as those that use sets of interacting genes or gene pathways as the functional unit, as demonstrated by a study that searched for signatures of ‘local adaptations’ (REF. 4).
More generally, we would like to see an expansion of the existing theoretical models, simulation studies and neutrality tests that can be used to further investigate the more complex adaptive scenarios such as polygenic adaptation and selection from standing variation. We predict that these developments, coupled with emerging large-scale functional technologies, will expedite the ability to identify candidate functional variants that have had a role in adaptation. This, in turn, will facilitate the in-depth interrogation of the phenotypic consequences of putatively adaptive functional variants. Genomic data will also improve our ability to study interactions among loci and will therefore permit the continued study of adaptive phenotypes with complex, polygenic architectures.
Ultimately, this field of research will benefit from more collaboration between researchers who are conducting in vivo and in vitro studies of genetic variants so that candidate adaptive variants can be interrogated for functional consequences on phenotype, and the biological impact of adaptation can be better understood. This would enable the field to move past simple ‘just-so stories’ to the more complex relationship between reproductive fitness, genetic variation and phenotypic variation over evolutionary time.
Acknowledgments
The authors thank S. Soi and J. Lachance for their discussions and suggestions. This work was funded by the US National Institutes of Health (NIH) Pioneer Award (DP1OD0644S), the NIH (R0 1GM076637) and the US National Science Foundation Hominid grant (BCS0827436) to S.A.T.
Glossary
- Reproductive fitness
A measure of how well an organism survives and reproduces it is generally determined by the genetic contribution of an individual to the gene pool of the next generation.
- Gene pathways
Sets of interacting gene products that are related to particular functions, including signalling and metabolic pathways
- Demographic history
The study of population-level changes over time, including changes in population size, migration, and gene flow between populations
- International HapMap Project
A publically available genome-wide data set of common single nucleotide polymorphisms generated from global populations
- Linkage disequilibrium
The non-random association of two or more genetic variants
- Positive selection
A type of natural selection in which a trait that increases reproductive fitness becomes more common over time in a population
- FST
A statistical measure of population structure based on differences in variant frequencies between populations using genotypic data
- Locus-specific branch length
A measure of population structure in one population sample relative to two other population samples based on FST values it is useful for identifying population-specific adaptation
- Haplotype
A set of genetic variants that are inherited together on a single chromosome
- Convergent evolution
The independent evolution of similar phenotypic traits
- Locus
A region of the genome that contains a particular gene or genetic sequence
- Pygmy
Typically used in the genetic literature to refer to an individual member of a population in which the average male and female stature is <150 cm and <140 cm, respectively. Historically, the term has been used in a pejorative manner however some recent populations that culturally identify themselves as Pygmies have revived the term themselves
- Pleiotropic
Pertaining to a gene: affecting multiple phenotypes
- Epigenetic
Pertaining to an inherited phenotypic change: caused by mechanisms other than changes in the underlying DNA sequence, such as DNA methylation or histone modification
- Metabolomic
Pertaining to the metabolome, which is the combined set of metabolites that are present in a given tissue at a given time
- Transcriptomic
Pertaining to the transcriptome, which is the combined set of RNA transcripts that are present in a given tissue at a given time
- Expression quantitative trait loci
Regions of the genome containing genetic polymorphisms that either alter or are in linkage disequilibrium with variants which affect gene regulation to influence the levels of RNA or protein produced
- Enhancers
Regulatory DNA elements that usually bind several transcription factors they can activate transcription at long distances from the promoter and in an orientation-independent manner
- DNase I-hypersensitive sites
Genetic regions that are sensitive to the DNase I enzyme. These are important regulatory regions because they are available for binding by gene-regulatory proteins
- Induced pluripotent stem cells
Somatically derived cells that have been synthetically induced to behave as pluripotent stem cells
- Integrative genomics
The integration of data from multiple ‘omics’ platforms
- QST
A statistical measure of population structure or differences in quantitative trait measurements between populations using phenotypic data.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
1000 Genomes Project: http://www.1000genomes.org/
Complete Genomics 69 Genomes: http://www.completegenomics.com/public-data/69Genomes/
Encyclopedia of DNA Elements: http://genome.ucsc.edu/ENCODE/
Human Genome Diversity Project: http://www.hagsc.org/hgdp/
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Contributor Information
Laura B. Scheinfeldt, Email: lscheinfeldt@coriell.org.
Sarah A. Tishkoff, Email: tishkoff@mail.med.upenn.edu.
References
- 1.Scheinfeldt LB, Soi S, Tishkoff SA. Colloquium paper: working toward a synthesis of archaeological, linguistic, and genetic data for inferring African population history. Proc. Natl Acad. Sci. USA. 2010;107(Suppl. 2):8931–8938. doi: 10.1073/pnas.1002563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henn BM, Cavalli-Sforza LL, Feldman MW. The great human expansion. Proc. Natl Acad. Sci. USA. 2012;109:17758–17764. doi: 10.1073/pnas.1212380109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jobling MA, Hurles M, Tyler-Smith C. Human Evolutionary Genetics: Origins, Peoples and Disease. Garland Publishing; 2004. [Google Scholar]
- 4. Fraser HB. Gene expression drives local adaptation in humans. Genome Res. 2013;23:1089–1096. doi: 10.1101/gr.152710.112. This study systematically evaluates the relative abundances of regulatory variation and coding variation in candidate adaptive regions and includes a novel method for identifying polygenic adaptation.
- 5.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 6.Scheinfeldt LB, et al. Population genomic analysis of ALMS1 in humans reveals a surprisingly complex evolutionary history. Mol. Biol. Evol. 2009;26:1357–1367. doi: 10.1093/molbev/msp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorisson GA, Stein LD. The SNP Consortium website: past, present and future. Nucleic Acids Res. 2003;31:124–127. doi: 10.1093/nar/gkg052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinds DA, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 11.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 12.Jakobsson M, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 13.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang ET, Kodama G, Baldi P, Moyzis RK. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc. Natl Acad. Sci. USA. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williamson SH, et al. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, et al. A whole genome long-range haplotype (WGLRH) test for detecting imprints of positive selection in human populations. Bioinformatics. 2006;22:2122–2128. doi: 10.1093/bioinformatics/btl365. [DOI] [PubMed] [Google Scholar]
- 18.Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 2006;16:980–989. doi: 10.1101/gr.5157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura R, Fujimoto A, Tokunaga K, Ohashi J. A practical genome scan for population-specific strong selective sweeps that have reached fixation. PLoS ONE. 2007;2:e286. doi: 10.1371/journal.pone.0000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fumagalli M, et al. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 2011;7:e1002355. doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brodwin P. “Bioethics in action” and human population genetics research. Cult. Med. Psychiatry. 2005;29:145–178. doi: 10.1007/s11013-005-7423-2. [DOI] [PubMed] [Google Scholar]
- 23.Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biswas S, Scheinfeldt LB, Akey JM. Genome-wide insights into the patterns and determinants of fine-scale population structure in humans. Am. J. Hum. Genet. 2009;84:641–650. doi: 10.1016/j.ajhg.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pickrell JK, et al. The genetic prehistory of southern Africa. Nature Commun. 2012;3:1143. doi: 10.1038/ncomms2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teshima KM, Coop G, Przeworski M. How reliable are empirical genomic scans for selective sweeps? Genome Res. 2006;16:702–712. doi: 10.1101/gr.5105206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting FST . Nature Rev. Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shriver MD, et al. The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum. Genom. 2004;1:274–286. doi: 10.1186/1479-7364-1-4-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grossman SR, et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010;327:883–886. doi: 10.1126/science.1183863. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, et al. Detecting and characterizing genomic signatures of positive selection in global populations. Am. J. Hum. Genet. 2013;92:866–881. doi: 10.1016/j.ajhg.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 38.Oleksyk TK, Smith MW, O’Brien SJ. Genome-wide scans for footprints of natural selection. Phil. Trans. R. Soc. B. 2010;365:185–205. doi: 10.1098/rstb.2009.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lohmueller KE, et al. Natural selection affects multiple aspects of genetic variation at putatively neutral sites across the human genome. PLoS Genet. 2011;7:e1002326. doi: 10.1371/journal.pgen.1002326. This paper demonstrates that deleterious mutations and hitch-hiking of neighbouring genetic variants are affecting patterns of nucleotide diversity across the human genome.
- 40.Price AL, et al. Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet. 2008;83:132–135. doi: 10.1016/j.ajhg.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang H, et al. Response to Price et al. Am. J. Hum. Genet. 2008;83:135–139. [Google Scholar]
- 42.Przeworski M, Coop G, Wall JD. The signature of positive selection on standing genetic variation. Evolution. 2005;59:2312–2323. [PubMed] [Google Scholar]
- 43.Innan H, Kim Y. Detecting local adaptation using the joint sampling of polymorphism data in the parental and derived populations. Genetics. 2008;179:1713–1720. doi: 10.1534/genetics.108.086835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cutter AD, Payseur BA. Genomic signatures of selection at linked sites: unifying the disparity among species. Nature Rev. Genet. 2013;14:262–274. doi: 10.1038/nrg3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coop G, et al. The role of geography in human adaptation. PLoS Genet. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daub JT, et al. Evidence for polygenic adaptation to pathogens in the human genome. Mol. Biol. Evol. 2013;30:1544–1558. doi: 10.1093/molbev/mst080. [DOI] [PubMed] [Google Scholar]
- 48. Hernandez RD, et al. Classic selective sweeps were rare in recent human evolution. Science. 2011;331:920–924. doi: 10.1126/science.1198878. This study demonstrates that amino acid substitutions and putative regulatory sites are not significantly enriched in alleles that are highly differentiated between populations, suggesting that classic sweeps were not a dominant mode of human adaptation over the past ~250 kya.
- 49.Simonson TS, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 50. Hancock AM, et al. Adaptations to climate-mediated selective pressures in humans. PLoS Genet. 2010;7:e1001375. doi: 10.1371/journal.pgen.1001375. This study looks at correlations between SNP allele frequencies and climate-related variables across global populations (a method that may be useful for detecting polygenic adaptation), demonstrating an enrichment for gene sets that are related to ultraviolet radiation, cancer, infection and immunity.
- 51.Moore LG, et al. Maternal adaptation to high-altitude pregnancy: an experiment of nature — a review. Placenta. 2004;25(Suppl. A):S60–S71. doi: 10.1016/j.placenta.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 52.Buzbas EO, Joyce P, Rosenberg NA. Inference on the strength of balancing selection for epistatically interacting loci. Theor. Popul. Biol. 2011;79:102–113. doi: 10.1016/j.tpb.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheinfeldt LB, Biswas S, Madeoy J, Connelly CF, Akey JM. Clusters of adaptive evolution in the human genome. Frontiers Genet. 2011;2:50. doi: 10.3389/fgene.2011.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Akey JM. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res. 2009;19:711–722. doi: 10.1101/gr.086652.108. This paper evaluates results across several genome-wide scans for selection and highlights the degree to which false positives are common in these studies.
- 55.Li J, et al. Joint analysis of demography and selection in population genetics: where do we stand and where could we go? Mol. Ecol. 2012;21:28–44. doi: 10.1111/j.1365-294X.2011.05308.x. [DOI] [PubMed] [Google Scholar]
- 56.Granka JM, et al. Limited evidence for classic selective sweeps in African populations. Genetics. 2012;192:1049–1064. doi: 10.1534/genetics.112.144071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quach H, et al. Signatures of purifying and local positive selection in human miRNAs. Am. J. Hum. Genet. 2009;84:316–327. doi: 10.1016/j.ajhg.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andres AM, et al. Targets of balancing selection in the human genome. Mol. Biol. Evol. 2009;26:2755–2764. doi: 10.1093/molbev/msp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nielsen R, et al. Darwinian and demographic forces affecting human protein coding genes. Genome Res. 2009;19:838–849. doi: 10.1101/gr.088336.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bazin E, Dawson KJ, Beaumont MA. Likelihood-free inference of population structure and local adaptation in a Bayesian hierarchical model. Genetics. 2010;185:587–602. doi: 10.1534/genetics.109.112391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sousa VM, Carneiro M, Ferrand N, Hey J. Identifying loci under selection against gene flow in isolation with migration models. Genetics. 2013;194:211–233. doi: 10.1534/genetics.113.149211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bersaglieri T, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nature Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lamason RL, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 65.Jablonski NG, Chaplin G. The evolution of human skin coloration. J. Hum. Evol. 2000;39:57–106. doi: 10.1006/jhev.2000.0403. [DOI] [PubMed] [Google Scholar]
- 66.Beleza S, et al. Genetic architecture of skin and eye color in an African-European admixed population. PLoS Genet. 2013;9:e1003372. doi: 10.1371/journal.pgen.1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang SJ, et al. Genome-wide association of anthropometric traits in African- and African-derived populations. Hum. Mol. Genet. 2010;19:2725–2738. doi: 10.1093/hmg/ddq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jarvis JP, et al. Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African Pygmies. PLoS Genet. 2012;8:e1002641. doi: 10.1371/journal.pgen.1002641. This study integrates signatures of natural selection together with GWASs to identify candidate loci that have a role in the short-stature trait in African pygmies.
- 70.Migliano AB, Vinicius L, Lahr MM. Life history trade-offs explain the evolution of human pygmies. Proc. Natl Acad. Sci. USA. 2007;104:20216–20219. doi: 10.1073/pnas.0708024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perry GH, Dominy NJ. Evolution of the human Pygmy phenotype. Trends Ecol. Evol. 2009;24:218–225. doi: 10.1016/j.tree.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 72.Mendizabal I, Marigorta UM, Lao O, Comas D. Adaptive evolution of loci covarying with the human African Pygmy phenotype. Hum. Genet. 2012;131:1305–1317. doi: 10.1007/s00439-012-1157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Turchin MC, et al. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nature Genet. 2012;44:1015–1019. doi: 10.1038/ng.2368. This paper demonstrates evidence for polygenic selection that acts on genetic variants influencing height in Northern Europeans compared to Southern Europeans.
- 74.Bigham AW, et al. Andean and Tibetan patterns of adaptation to high altitude. Am. J. Hum. Biol. 2013;25:190–197. doi: 10.1002/ajhb.22358. [DOI] [PubMed] [Google Scholar]
- 75.Bigham AW, et al. Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum. Genom. 2009;4:79–90. doi: 10.1186/1479-7364-4-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peng Y, et al. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol. Biol. Evol. 2011;28:1075–1081. doi: 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- 77.Beall CM, et al. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl Acad. Sci. USA. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scheinfeldt LB, et al. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 2012;13:R1. doi: 10.1186/gb-2012-13-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alkorta-Aranburu G, et al. The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet. 2012;8:e1003110. doi: 10.1371/journal.pgen.1003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huerta-Sanchez E, et al. Genetic signatures reveal high-altitude adaptation in a set of Ethiopian populations. Mol. Biol. Evol. 2013;30:1877–1888. doi: 10.1093/molbev/mst089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scheinfeldt LB, Tishkoff SA. Living the high life: high-altitude adaptation. Genome Biol. 2010;11:133. doi: 10.1186/gb-2010-11-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Enattah NS, et al. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am. J. Hum. Genet. 2008;82:57–72. doi: 10.1016/j.ajhg.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Imtiaz F, et al. The T/G 13915 variant upstream of the lactase gene (LCT) is the founder allele of lactase persistence in an urban Saudi population. J. Med. Genet. 2007;44:e89. doi: 10.1136/jmg.2007.051631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ingram CJ, et al. A novel polymorphism associated with lactose tolerance in Africa: multiple causes for lactase persistence? Hum. Genet. 2007;120:779–788. doi: 10.1007/s00439-006-0291-1. [DOI] [PubMed] [Google Scholar]
- 85.Ingram CJ, Mulcare CA, Itan Y, Thomas MG, Swallow DM. Lactose digestion and the evolutionary genetics of lactase persistence. Hum. Genet. 2009;124:579–591. doi: 10.1007/s00439-008-0593-6. [DOI] [PubMed] [Google Scholar]
- 86.Ingram CJ, et al. Multiple rare variants as a cause of a common phenotype: several different lactase persistence associated alleles in a single ethnic group. J. Mol. Evol. 2009;69:579–588. doi: 10.1007/s00239-009-9301-y. [DOI] [PubMed] [Google Scholar]
- 87.Norton HL, et al. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol. Biol. Evol. 2007;24:710–722. doi: 10.1093/molbev/msl203. [DOI] [PubMed] [Google Scholar]
- 88.Henn BM, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc. Natl Acad. Sci. USA. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kudaravalli S, Veyrieras JB, Stranger BE, Dermitzakis ET, Pritchard JK. Gene expression levels are a target of recent natural selection in the human genome. Mol. Biol. Evol. 2009;26:649–658. doi: 10.1093/molbev/msn289. One of the first studies to integrate eQTLs with candidate adaptive loci that have been identified using genome-wide SNP data, to explore the degree to which regulatory variation has been involved in human adaptation.
- 90. Grossman SR, et al. Identifying recent adaptations in large-scale genomic data. Cell. 2013;152:703–713. doi: 10.1016/j.cell.2013.01.035. This paper integrates candidate adaptive loci identified in WGS data with several different types of regulatory variation (eQTLs, lincRNAs, enhancers and promoters).
- 91.Lambert CA, Tishkoff SA. Genetic structure in African populations: implications for human demographic history. Cold Spring Harb. Symp. Quant. Biol. 2009;74:395–402. doi: 10.1101/sqb.2009.74.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Luzzatto L. Sickle cell anaemia and malaria. Mediterr. J. Hematol. Infect. Dis. 2012;4:e2012065. doi: 10.4084/MJHID.2012.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Vernot B, et al. Personal and population genomics of human regulatory variation. Genome Res. 2012;22:1689–1697. doi: 10.1101/gr.134890.111. This study integrates candidate adaptive loci with DNase I-hypersensitive sites that have been identified in the ENCODE project.
- 94.Olds LC, Sibley E. Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum. Mol. Genet. 2003;12:2333–2340. doi: 10.1093/hmg/ddg244. [DOI] [PubMed] [Google Scholar]
- 95.Hughes DA, et al. Parallel selection on TRPV6 in human populations. PLoS ONE. 2008;3:e1686. doi: 10.1371/journal.pone.0001686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Akey JM, Swanson WJ, Madeoy J, Eberle M, Shriver MD. TRPV6 exhibits unusual patterns of polymorphism and divergence in worldwide populations. Hum. Mol. Genet. 2006;15:2106–2113. doi: 10.1093/hmg/ddl134. [DOI] [PubMed] [Google Scholar]
- 97. Kamberov YG, et al. Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell. 2013;152:691–702. doi: 10.1016/j.cell.2013.01.016. This study is an example of how mouse models combined with human GWASs can be used to determine the functional significance of an adaptive variant.
- 98.Miller CT, et al. cis-regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lao O, de Gruijter JM, van Duijn K, Navarro A, Kayser M. Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann. Hum. Genet. 2007;71:354–369. doi: 10.1111/j.1469-1809.2006.00341.x. [DOI] [PubMed] [Google Scholar]
- 100.Varemo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grundberg E, et al. Population genomics in a disease targeted primary cell model. Genome Res. 2009;19:1942–1952. doi: 10.1101/gr.095224.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xiong Q, Ancona N, Hauser ER, Mukherjee S, Furey TS. Integrating genetic and gene expression evidence into genome-wide association analysis of gene sets. Genome Res. 2012;22:386–397. doi: 10.1101/gr.124370.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zaykin DV, Zhivotovsky LA, Czika W, Shao S, Wolfinger RD. Combining p-values in large-scale genomics experiments. Pharm. Statist. 2007;6:217–226. doi: 10.1002/pst.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mo Q, et al. Pattern discovery and cancer gene identification in integrated cancer genomic data. Proc. Natl Acad. Sci. USA. 2013;110:4245–4250. doi: 10.1073/pnas.1208949110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schadt EE, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nature Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Orr HA. Testing natural selection versus genetic drift in phenotypic evolution using quantitative trait locus data. Genetics. 1998;149:2099–2104. doi: 10.1093/genetics/149.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fraser HB, et al. Systematic detection of polygenic cis-regulatory evolution. PLoS Genet. 2011;7:e1002023. doi: 10.1371/journal.pgen.1002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Leinonen T, McCairns RJ, O’Hara RB, Merila J. QST-FST comparisons: evolutionary and ecological insights from genomic heterogeneity. Nature Rev. Genet. 2013;14:179–190. doi: 10.1038/nrg3395. [DOI] [PubMed] [Google Scholar]
- 109. Lachance J, et al. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell. 2012;150:457–469. doi: 10.1016/j.cell.2012.07.009. This is the first high-coverage WGS study across diverse African populations, demonstrating signatures of local adaptation, including candidate genes for the short-stature trait in pygmies.
- 110.Lanktree MB, et al. Meta-analysis of dense genecentric association studies reveals common and uncommon variants associated with height. Am. J. Hum. Genet. 2011;88:6–18. doi: 10.1016/j.ajhg.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]