Abstract
The senses provide a means by which data on the physical and chemical properties of the environment may be collected and meaningfully interpreted. Sensation begins at the periphery, where a multitude of different sensory cell types are activated by environmental stimuli as different as photons and odorant molecules. Stimulus sensitivity is due to expression of different cell surface sensory receptors, and therefore the receptive field of each sense is defined by the aggregate of expressed receptors in each sensory tissue. Here, we review current understanding on patterns of expression and modes of regulation of sensory receptors.
INTRODUCTION
An animal’s most basic goals are to identify food and to avoid becoming it. The most powerful set of tools to attain these goals are the senses, which allow a multitude of fundamentally different types of environmental data to be extracted and converted to neural signals that control and modify behavior. The multimodal nature of the senses allows animals to integrate and become sensitive to the most important of cues, those that enable them to identify nutrients, mates, and threats. Because the types of environmental data that animals extract have distinct properties, their modes of extraction are distinct, beginning with different peripheral sensory organs and continuing with different neural circuits. Herein, we limit our discussion to the well-studied senses of olfaction and taste in rodents.
Sensation begins with activation of a cell surface sensory receptor. Each class of sensory cells expresses a subset of receptors, presumably specialized for the ligands to which each sensory tissue is exposed. Of the known sensory receptors, the vast majority are G protein–coupled receptors (GPCRs), seven-transmembrane-pass receptors whose activation stimulates G protein activity. Sensory GPCRs often interact with specific G proteins, allowing for signaling segregation between sensory and nonsensory GPCRs ( Jones & Reed 1989, Jones et al. 1990). The primary outcome of ligand-induced G protein signaling is the opening or closing of ion channels (Levy et al. 1991, Stryer 1991, Imai & Sakano 2008). However, G protein signaling can also influence transcription and epigenetic gene regulation or modify GPCR signaling (Sorkin & Von Zastrow 2009).
Sensory neurons may express one or several receptors, reflecting strategies of discriminatory power versus coordination of broad stimuli with uniform behavioral responses. For example, each olfactory sensory neuron (OSN) expresses only a single receptor gene, enabling high specificity but narrow tuning (Araneda et al. 2000). In contrast, bitter taste receptor cells (TRCs) express several receptors, resulting in broad tuning and suggesting that individuals do not need to discriminate between bitter stimuli, but instead need only to distinguish them from, for example, sweet stimuli (Chandrashekar et al. 2000).
Sensory receptor expression defines the functional identity of each sensory cell, and therefore the molecular mechanisms of receptor gene choice are paramount to understanding sensory neuronal development. It is not surprising that these gene regulatory events share features across sensory tissues, and thus we expect that determining gene regulatory strategies in one tissue will yield valuable insights into strategies employed by other tissues, generalizing findings and insights to advance the studies of these tissues simultaneously. Below we outline current understanding of sensory receptor gene regulation and propose feedback models based on those employed by olfactory receptors to explain the expression patterns of each class of sensory receptors.
THE MAIN OLFACTORY EPITHELIUM: OLFACTORY RECEPTORS
Olfaction begins in the main olfactory epithelium (MOE), which houses OSNs and their progenitors in a neurogenic pseudostratified epithelium. Following commitment to the neuronal lineage, OSNs express either olfactory receptors (ORs) (Buck & Axel 1991) or trace amine-associated receptors (TAARs) (Liberles & Buck 2006), both of which are GPCRs. The mouse genome encodes ~1,075 intact (and ~1,430 total) OR genes, making this the largest known gene family. ORs are found in clusters on most chromosomes and can be divided into two groups: the 160 fish-like type I ORs, and the 1,270 mammal-specific type II ORs (Sullivan et al. 1996, Zhang & Firestein 2002). ORs expressed heterologously fail to traffic to the plasma membrane, requiring specific chaperones such as receptor transporting proteins 1 and 2 (Rtp1 and Rtp2) for endoplasmic reticulum (ER) export (Saito et al. 2004).
Extensive evidence suggests that OR expression is monogenic and monoallelic (Chess et al. 1994, Ebrahimi & Chess 2000, Serizawa et al. 2003, Vassalli et al. 2002, Shykind et al. 2004, Clowney et al. 2011); however, only after comprehensive and careful single-cell RNAseq analysis is performed for multiple OSNs can this assertion be stated with conviction. Moreover, OR expression is generally viewed as stochastic. However, this stochasticity, highlighted by the fact that OSNs have the potential to express multiple ORs (Shykind et al. 2004), occurs within uncharacterized deterministic spatiotemporal restrictions (Ressler et al. 1994, Vassar et al. 1993, Rodriguez-Gil et al. 2010) that make OR expression frequencies highly reproducible within identical mouse strains (Ibarra-Soria et al. 2014). All OSNs expressing the same receptor allele send their axons to a common target in the olfactory bulb (Ressler et al. 1994, Vassar et al. 1994, Mombaerts et al. 1996). This extremely sensitive process depends on the identity of the expressed OR (Wang et al. 1998), which is also detected in the axon (Barnea et al. 2004); even single amino acid substitutions in ORs often result in targeting to new glomeruli (Feinstein & Mombaerts 2004, Zhang et al. 2012). Thus, the single chosen OR informs OSN connectivity to the brain and defines its receptive field, making OR choice the central event in establishing OSN identity. Below, we discuss the current understanding of the mechanisms of stochastic and monogenic OR expression.
Olfactory Receptor Choice: Transcriptional Activation
RNA in situ hybridization studies demonstrated that each OR gene is expressed in a distinct zone of the epithelium, indicating initial constraints on OR choice (Ressler et al. 1994, Vassar et al. 1993, Miyamichi et al. 2005). Experiments in which coding sequences for receptors expressed in different zones were swapped suggested that regions surrounding the OR coding sequence impose these zonal constraints (Wang et al. 1998), although it remains to be seen what these features are.
Numerous studies have assayed the structure of OR promoters and proximal regions to identify factors required for OR expression (Michaloski et al. 2006, Clowney et al. 2011, Plessy et al. 2012). A high-throughput analysis of OR promoters revealed a number of striking features. First, the majority share the same transcription factor binding sites, irrespective of their zone of expression, suggesting that OR promoters may be indistinguishable and supporting a mode of stochastic regulation. Second, OR promoters are extremely adenine/thymine (AT)-rich (Clowney et al. 2011). The importance of this finding has not been tested, but given that this feature is shared only with promoters from other rapidly-evolving gene families, including the rest of the chemoreceptors considered herein, we consider it to be of exceptional interest.
OR promoters are enriched for homeodomain sites as well as for O/E (Olf1/early B cell factor)-like sites (Michaloski et al. 2006, Clowney et al. 2011, Plessy et al. 2012). Two homeodomain proteins, LHX2 and EMX2, regulate OR gene expression. Electrophoretic mobility shift assays demonstrated that LHX2 can bind the promoter of the OR gene M71 (Hirota & Mombaerts 2004). Lhx2 mutant mice fail to express the majority of type II ORs, whereas expression of all except two type I ORs is unaffected (Hirota et al. 2007). In contrast, Emx2 mutants have reduced the expression of ~75% of type I and type II OR genes and increased expression of a handful of others (Levi et al. 2003, Hirota & Mombaerts 2004, Hirota et al. 2007, McIntyre et al. 2008). How Emx2 and Lhx2 cooperate to regulate OR transcription is not yet known, though it is tempting to speculate that OR gene choice involves the concomitant activity of numerous transcriptional activators.
Finally, OR enhancer elements appear to regulate OR expression. The first of these enhancers to be discovered, “H” (Serizawa et al. 2003), was associated in trans and in cis with the promoters of several OR genes. Furthermore, one copy of H per cell is methylated, suggesting that a single functional H could provide the singularity required for singular OR choice (Lomvardas et al. 2006). However, deletion of H resulted in loss of OR expression of only proximal OR genes (Fuss et al. 2007). Subsequent studies identified an additional element, “P,” with a similar function (Bozza et al. 2009, Khan et al. 2011). This work has been greatly extended by our lab, with the recent identification of dozens of new enhancer elements required for OR choice (Markenscoff-Papadimitriou et al. 2014).
An Epigenetic Platform for Monogenic Olfactory Receptor Choice
OR gene regulation has a surprising layer of epigenetic control. Native chromatin immunoprecipitation experiments revealed that the entire OR gene family displays trimethylation of histone 3, lysine 9 (H3K9me3) and trimethylation of histone 4, lysine 20 (H3K20me3), hallmarks of constitutive heterochromatin (Magklara et al. 2011). These marks are more prevalent on type II than on type I ORs and are deposited prior to OR choice, which indicates that ORs are silenced before they are expressed. By using fluorescence-activated cell sorting (FACS) to isolate a population of cells expressing the same receptor gene, it was also shown that these repressive marks are removed from the active OR allele. Thus, OR choice involves derepression of a silenced allele. Broad epigenetic silencing could provide a platform for OR choice by marking ORs with an epigenetic signature specifically recognized by the OR choice machinery. Moreover, it could allow deployment of a specialized feedback signal that stabilizes OR expression without affecting global OSN transcription. The only two type I ORs with H3K9me3 marks at levels comparable to type II ORs are also the only type I ORs whose expression is lost in Lhx2 mutants (our unpublished observations; Hirota et al. 2007).
OR gene regulation also has a spatial component. Following OR choice, the entire OR gene family condenses into a small number of densely compacted foci; reversing the formation of these foci by overexpression of lamin B receptor results in coexpression of many ORs in each OSN (Clowney et al. 2012). Thus, broad silencing and compaction appear to form the basis for monogenic OR choice.
Olfactory Receptor Feedback
OR choice involves the removal of silencing chromatin modifications by the demethylase Lsd1, which is expressed in immature OSNs during OR choice and is downregulated in mature OSNs. Lsd1 deletion results in a dramatic loss of OR expression. This causes loss of OSN maturation, as assayed by expression of the OR signaling molecule adenylyl cyclase III (Adcy3), and maturation can, in turn, be rescued in Lsd1 mutants by transgenic OR expression. Thus, OR expression is both required and sufficient for OSN maturation. Finally, loss of Adcy3 prolongs LSD1 expression, suggesting that ADCY3 signals for the termination of OR choice (Lyons et al. 2013).
The requirement and sufficiency of OR expression for Adcy3 expression suggest the presence of an OR-elicited feedback pathway that signals for the termination of OR choice. Evidence for this pathway was first provided by experiments showing that transgenic overexpression of a single OR precludes further OR choice (Serizawa et al. 2003). The expressed OR requires a start codon, indicating that OR protein is required to elicit feedback (Lewcock & Reed 2004). In addition, OR pseudogenes are not expressed stably, and thus OR feedback also acts to stabilize expression of OR alleles that pass some measure of quality control (Shykind et al. 2004). This property likely arises from the ability of LSD1 to both activate and then subsequently repress genes (Shi et al. 2004) (Figure 1). Together, these findings suggest that the OR feedback pathway serves to coordinate the appearance of intact ORs with OSN maturation to prevent further OR choice, to stabilize OR transcription, and to select against pseudogene expression.
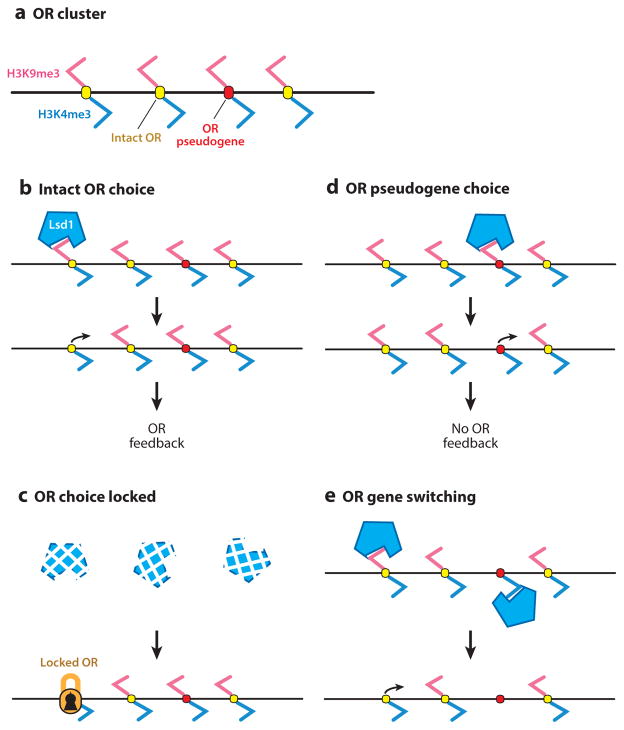
Figure 1.
A model for gene switching and stabilization of olfactory receptor (OR) gene choice. (a) Prior to OR choice, ORs are silenced by H3K9me3. They are also marked with H3K4me3. (b) LSD1 removes H3K9me3 from an intact OR gene, allowing for its transcription and the activation of unfolded protein response (UPR)-based feedback. (c) LSD1 is downregulated, locking the choice of the OR. (d ) Alternatively, LSD1 removes H3K9me3 from an OR pseudogene, allowing for its transcription. The pseudogene product fails to activate the UPR. (e) Prolonged LSD1 activity results in removal of H3K4me3, silencing the initially chosen allele, and the removal of H3K9me3 from another OR gene to allow for gene switching.
We recently discovered that OR feedback is elicited through the unfolded protein response (UPR), a ubiquitous signaling pathway that homeostatically adjusts the protein folding capacity and load of the ER in response to the detection of unfolded proteins in the ER lumen (Ron & Walter 2007). Unfolded proteins attenuate protein translation initiation by activating the ER-resident kinase PERK, which in turn phosphorylates the initiation factor EIF2A, in effect slowing assembly of translating ribosomes (Ron & Walter 2007). EIF2A phosphorylation, paradoxically, also drives a selective increase in translation of a small number of mRNAs, among them activating transcription factor 5 (Atf5) (Watatani et al. 2008), one of the most highly transcribed genes in OSNs. OR expression activates PERK, leading to EIF2A phosphorylation and Atf5 translation. ATF5 in turn directs Adcy3 transcription, stabilizes OR choice, and terminates LSD1 activity. Thus, ORs activate the UPR to elicit feedback (Dalton et al. 2013) (Figure 2).
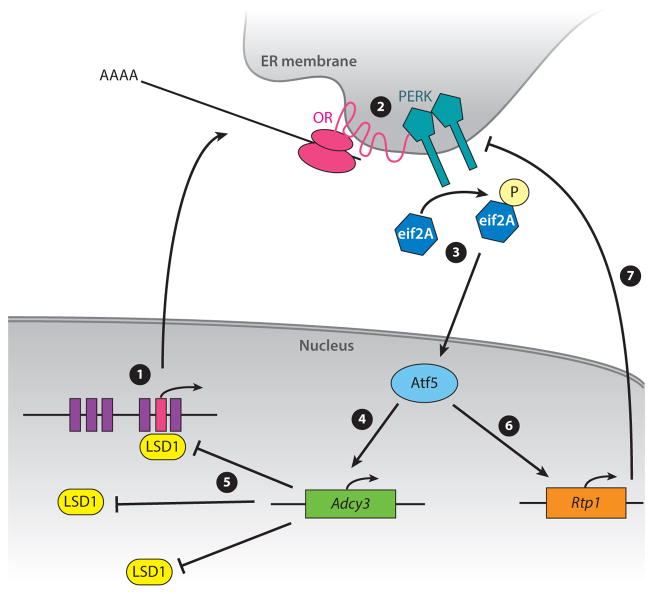
Figure 2.
Unfolded protein response (UPR)-mediated olfactory receptor (OR) feedback. ❶ Histone demethylation by LSD1 activates an OR gene. ❷ As the OR is being translated, it activates the PERK branch of the UPR. ❸ This activation results in EIF2A phosphorylation and Atf5 translation. ❹ ATF5 activates Adcy3 transcription, and ❺ ADCY3 then prevents further LSD1 activity. ❻ ATF5 also activates Rtp1 transcription. ❼ RTP1 suppresses further UPR activation. ER, endoplasmic reticulum.
The precise mechanism by which ORs activate Perk is a matter of intense interest for our group. OR-like structures may specifically activate Perk; alternatively, at the onset of OR expression, RTP1 may be absent, preventing OR trafficking and subsequently clogging the ER. Supporting this model, Rtp1 is indeed a transcriptional target of ATF5 (Wang et al. 2012). Thus, UPR activation by ORs could restore ER homeostasis by changing which proteins reside in the ER.
As described above, Adcy3 expression is required to terminate OR choice. In addition, in zebrafish, G protein beta/gamma signaling acts to prevent further OR choice (Ferreira et al. 2014). Thus, OR feedback may have three distinct steps: activation of the UPR, a structural test of the ORs’ ability to interact with specific chaperones, and an ultimate test of their ability to activate G protein signaling.
A Model for Stochastic Olfactory Receptor Choice
Finally, although findings over the past several years provide valuable clues into the processes of OR choice and demonstrate how, once a single OR appears, further OR choice is terminated, the means by which a single OR gene is chosen in the first place is still to be discovered. A recent modeling study suggested that monogenic choice may simply be the outcome of a very slow process of gene choice, followed by very rapid OR feedback (Tan et al. 2013). An additional (and not mutually exclusive) possibility is that singular OR gene choice requires the activity of a singular OR transcription-activating apparatus. Mechanistically, this would most likely involve a complex and extremely low-probability assembly of many elements required for transcriptional activation.
Recent data from our lab support a model whereby the transcriptional singularity is encoded by the infrequent coincidence of multiple enhancer elements over a transcriptionally active OR allele (Markenscoff-Papadimitriou et al. 2014). Using epigenetic analyses to identify enhancer regions in the MOE, we have identified dozens of potential OR enhancers in addition to the H and P elements. DNA FISH and 4C-seq experiments reveal that many of these elements, despite being located on different chromosomes, frequently congregate at the active OR gene locus (Markenscoff-Papadimitriou et al. 2014). These data suggest that an assembly of enhancers, or “enhanceosome” (Thanos & Maniatis 1995), may form the basis of singular OR choice. Because enhancer elements are limiting, if many are required to activate a single OR, their assembly at one OR would preclude their assembly at any other OR. However, this hypothesis has yet to be properly tested. Notably, the stochasticity imposed by the probabilistic association of multiple enhancers from various chromosomes interfaces with the epigenetic state of OR loci. Recent data from our lab demonstrated that loss of heterochromatic silencing from the OR cluster by the simultaneous deletion of H3K9 methyltransferases G9a and G9a-like (Glp) results in expression of only a few OR genes instead of the full repertoire. Thus, without chromatin-mediated silencing, it appears that the random selection of an OR allele is substituted by the selective choice of a few alleles that dominate the olfactory epithelium and are expressed in a nonmutually exclusive fashion (Lyons et al. 2014).
Trace Amine-Associated Receptors
A subset of OSNs do not express ORs, instead monogenically expressing TAARs, and these cells likely constitute an olfactory subsystem ( Johnson et al. 2012). TAARs are GPCRs found in a single genomic cluster of 15 genes, 14 of which are intact (Liberles & Buck 2006). TAARs are thought to be activated by aversive or attractive volatile odorants to elicit hardwired behavioral responses (Liberles & Buck 2006, Ferrero et al. 2011, Ferrero et al. 2012).
Following TAAR pseudogene choice, a cell is far more likely to choose another TAAR, indicating that some elements of TAAR versus OR identity may be imprinted prior to or coincident with receptor choice ( Johnson et al. 2012) or that TAAR identity may be, in some way, dominant to OR identity. This finding also indicates that TAAR-expressing cells use a receptor-elicited feedback system. However, unlike what is seen with OR pseudogene choice, following TAAR pseudogene choice, expression of the pseudogene continues. Thus, unlike ORs, TAARs do not require feedback to stabilize their expression; feedback is required only to prevent expression of another receptor. The second-chosen TAAR gene can be chosen in trans, although it is not yet known whether, as has been observed for vomeronasal receptors (VRs), the second allele must be chosen in trans.
TAARs are expressed zonally and in a salt-and-pepper fashion. As do ORs, TAARs inform axonal connectivity, although the location of TAAR glomeruli appears to be less stereotyped than for ORs. However, despite the apparent centrality of the chosen TAAR in cell identity, the basic principles of TAAR gene choice are yet to be elucidated. TAARs do not share an epigenetic signature with ORs ( Johnson et al. 2012), which may indicate that TAARs have a mechanism of choice that is both stochastic and distinct from that used for OR choice. This apparent lack of repression prior to choice could indicate that the scale of the OR choice problem mandates repression, that repressive elements are yet to be discovered, or that TAAR choice and feedback simply involve a single enhancer element near each TAAR cluster, a model that is discussed below in more detail with regard to VRs.
THE VOMERONASAL ORGAN: VOMERONASAL RECEPTORS
The vomeronasal organ (VNO), a second rodent olfactory apparatus, is thought to be responsible for detecting pheromones (Halpern 1987). Pheromones are detected by activating VRs (Dulac & Axel 1995) expressed by vomeronasal sensory neurons (VSNs). The VNO is neurogenic, with progenitors and immature VSNs confined to the tissue margins.
VSNs can be divided into two subclasses. Apically located type I VSNs express G protein subunit Gnai2 and type I VRs, whereas basally located type II VSNs express Gnao and type II VRs (Berghard & Buck 1996, Ryba & Tirindelli 1997). A subset of type II VSNs also express a family of nonclassical MHC 1b H2-Mv genes, which contribute to VR regulation, signaling, or both (Ishii et al. 2003, Loconto et al. 2003, Leinders-Zufall et al. 2014). Enomoto et al. (2011) recently showed that loss of the transcription factor Bcl11b results in an increase in the number of type I VSNs and a concomitant decrease in type II VSNs. This finding echoes what was observed for type I versus type II ORs in Lhx2 mutants and suggests that an early step in fate specification for sensory neurons may be a restriction of the types of receptors they can express.
A small number of type I and type II VSNs do not express VRs, instead expressing formyl peptide receptors (FPRs), a seven-gene family whose members detect ligands related to disease or inflammation (Liberles et al. 2009, Riviere et al. 2009). VNO-expressed receptors are instructive in VSN connectivity to the accessory olfactory bulb (AOB) (Rodriguez et al. 1999, Dietschi et al. 2013), and we therefore consider receptor choice to be central to VSN identity. Below, we review current understanding of VR expression and gene regulation. We also propose receptor-elicited feedback models to account for their unique patterns of expression.
Type I Vomeronasal Receptors and Their Regulation
Type I VRs, encoded by a family of 300 genes, ~150 of which are intact, are thought to be activated by volatile pheromones (Del Punta et al. 2002). Gene-targeting experiments have revealed that V1Rs are monogenically and monoallelically expressed (Rodriguez et al. 1999). Like OR promoters, V1R promoters contain O/E sites, indicating that V1Rs and ORs may have common transcriptional activators (Lane et al. 2002, Michaloski et al. 2011). However, whether V1Rs, like ORs, require Lhx2 or Emx2 for transcription has not been reported.
V1R promoters also share highly homologous regions containing potential binding motifs for transcriptional repressors. One such motif, MV12, binds nuclear protein in electrophoretic mobility shift assays (EMSA), although the identity and role of these binding partners have not been demonstrated (Michaloski et al. 2011). Their presence may suggest that V1Rs, like ORs, are widely repressed as part of their regulation. It will be interesting to determine whether V1Rs also display repressive chromatin modifications in the VNO. Finally, although it has yet to be tested whether enhancer elements regulate V1Rs, indirect evidence to be discussed below supports this possibility.
Type II Vomeronasal Receptors and Their Regulation
The V2R family contains 122 intact and 280 total members. The intact genes can be divided into families A, B, and D, which together total 115 genes, and the 7-member family C. Each type II VSN expresses an A, B, or D family member, as well as a single gene from family C, in nonrandom combinations (Martini et al. 2001, Ishii & Mombaerts 2011). Type II VSNs can be further divided into basal and apical cells, with basal cells also expressing at least one H2-Mv gene (Ishii & Mombaerts 2008).
The significance of V2R coexpression has not been studied. Type C V2Rs may act as chaperones or coreceptors, as has been observed for other GPCRs (George et al. 2002, Nelson et al. 2002). Even more interesting is coexpression with H2-Mvs. Although an initial study suggested that H2-Mvs act as chaperones or export molecules for V2Rs (Loconto et al. 2003), loss of a required cofactor for H2-Mv plasma membrane expression does not alter V2R protein distribution in VSNs (Ishii & Mombaerts 2008). A second model proposes that H2-Mvs increase the sensitivity of V2Rs for their ligands without affecting ligand specificity (Leinders-Zufall et al. 2014). Future studies will be needed to resolve these models.
Vomeronasal Receptor Feedback
Evidence for VR-elicited feedback first came from studies showing that when a V1R coding sequence is deleted, VSNs choosing this allele target broadly across the AOB, indicating that these VSNs eventually express other VRs (Rodriguez et al. 1999). In contrast with OR pseudogene choice, the mutant V1R continues to be expressed, and the second VR cannot be chosen from within the same cluster as the mutant VR, a phenomenon referred to as “cluster lock” (Roppolo et al. 2007).
Cluster lock suggests a simple model to explain monogenic V1R choice (see Figure 3): Each V1R cluster has a single nearby enhancer element that stochastically and permanently activates a V1R gene. Intact V1R expression then prevents further VR choice, most likely by preventing further V1R enhancer activity. Whether V1Rs activate the UPR to elicit feedback has not been tested, but this model seems likely, given that V1Rs fail to traffic from the ER when expressed heterologously (Dey & Matsunami 2011). In this model, the UPR would act to enhance V1R trafficking from the ER, possibly via transcriptional activation of V1R-specific chaperones such as Calreticulin 4 (Dey & Matsunami 2011). In addition to explicitly testing this model, determining the causes and functional importance of stable (i.e., V1R-like) versus unstable (i.e., OR-like) receptor choice will be interesting.
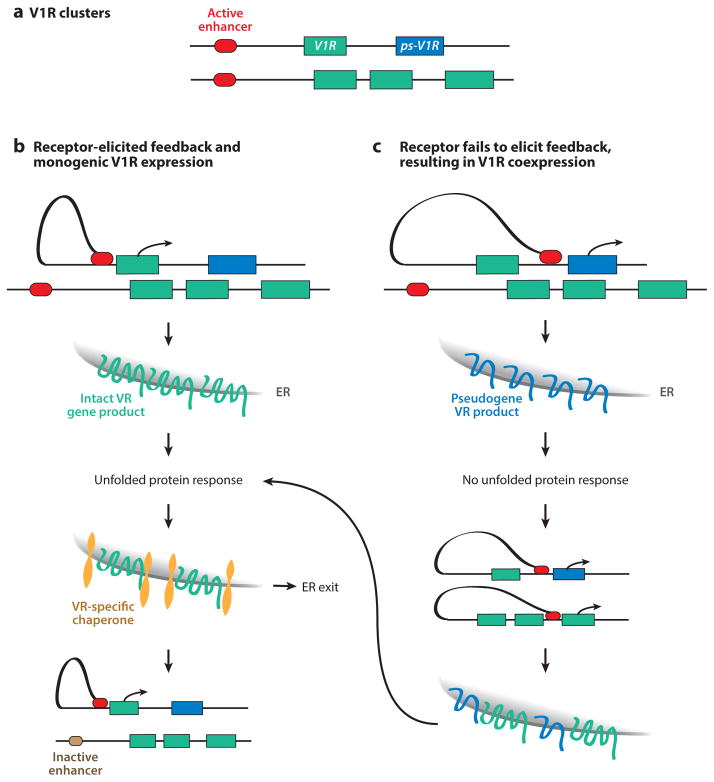
Figure 3.
An unfolded protein response (UPR)-based model for V1R choice and feedback. (a) Pictured are two V1R clusters, one of which contains a pseudogene (ps-V1R). (b) A cis enhancer stably binds a V1R promoter and activates transcription of an intact V1R, leading to production of the V1R protein and activation of the UPR. UPR activity results in transcription of V1R chaperones, allowing for endoplasmic reticulum (ER) exit of the V1R, completing the feedback. Poised but unbound V1R enhancers on other V1R clusters will also be targeted by feedback, preventing further V1R activation. (c) In the case of pseudogene V1R choice, the pseudogene product may fail to activate the UPR, which would prevent inactivation of other poised V1R enhancers, allowing for coexpression of multiple V1Rs. VR, vomeronasal receptor.
The coordination of feedback for V2Rs is more difficult to envision, given their multiple receptor expression. VSNs choose a class A, B, or D receptor, then a class C receptor, and then the optional H2-Mv. Deletion studies have elaborated on this pathway. VSNs choosing an intact type A allele subsequently express a specific type C receptor. This type C receptor is only infrequently expressed by VSNs choosing a coding sequence–deleted form of the type A allele, and axons from these cells broadly innervate the AOB, indicating that they have chosen other type A/B/D receptors and possibly other H2-Mvs (Ishii & Mombaerts 2008, 2011).
Together, these findings indicate that type C receptor choice is mechanistically linked to and dependent on type A/B/D choice. Given that only a subset of type A/B/D receptors are coexpressed with H2-Mv molecules (Ishii & Mombaerts 2008), an initial step in this process may be to limit the available A/B/D receptors for expression, analogous to zonality in the MOE. In this model, apical type II VSNs can choose only V2Rs that do not functionally require an H2-Mv. Thus, type A/B/D receptor choice could initiate feedback, for example through the UPR, driving expression of type C receptors. If type C receptors act as molecular chaperones, a VSN could choose one type C receptor at a time until selecting one that can functionally couple to the chosen A/B/D receptor. This receptor pair could then signal to terminate VR choice, with signaling in turn activating H2-Mv expression. In an alternative and much simpler model, each type C receptor has promoter features shared only with a subset of class A/B/D receptors, and each VSN has a unique set of transcription factors, such that each cell can choose only compatible receptor pairs. This model seems unlikely, however, given that type C receptor expression appears to depend on A/B/D receptor choice (Ishii & Mombaerts 2011).
LINGUAL EPITHELIUM: TASTE RECEPTORS
Taste begins with the activation of TRCs, which arise throughout the life of the animal from the lingual epithelium. TRCs are organized into onion-shaped aggregates of 50–150 TRCs known as taste buds, which are found in one of three types of sensory papillae with characteristic structure and location on the tongue. At the apical surface of each taste bud is a taste pore, into which TRCs extend microvilli. Taste information is carried by the VII and IX cranial nerves, which receive input from the TRCs. TRCs detect five basic taste modalities (sweet, sour, bitter, salty, and umami), although the peripheral taste-coding strategy has been a matter of intense debate, as studies have variously indicated that TRCs are sensitive to single or multiple stimulus modalities. However, the recent elucidation of the receptor genes mediating the five modalities has provided clear evidence that each TRC is tuned to only a single modality and has provided the tools to explore taste coding in the brain. Although for the most part the regulation of taste receptor genes is unstudied, evidence has shown that the specification of the TRC lineage, as with the OSN lineage, requires homeodomain transcription factors. In the case of TRCs, Skn-1a specified lineage for sweet, umami, and bitter TRCs (Matsumoto et al. 2011). Other transcription factors known to specify neuronal lineages, such as Ascl1 and Prox1, have been hypothesized to promote sour TRC differentiation (Matsumoto et al. 2013). Thus, as described for OSNs and VSNs, these transcription factors may act early to define the available fates for each developing TRC.
Below we propose gene-regulatory strategies that could account for the patterns of receptor expression and other features observed in TRCs.
Sweet and Umami Taste Receptors
Sweet and umami taste are mediated by the tas1R family of GPCRs, a group of 3 genes found on chromosome 4 in the mouse. A surprising link between sweet and umami taste came with the discovery that both of these modalities employ the tas1R3 gene, in combination with either tas1R1 (umami) or tas1R2 (sweet) (Nelson et al. 2001, 2002; Zhao et al. 2003). Tas1R3 deletion studies have demonstrated that this gene is required for both sweet and umami taste (Zhao et al. 2003).
This study as well as additional studies have found residual sensitivity to sweet tastants in tas1R3 null animals, and one study has found residual sensitivity to umami tastants in tas1R3 nulls (Nelson et al. 2002). However, this result probably does not indicate the presence of additional receptors and may instead hint at some of the more fascinating aspects of these receptors, specifically the manner in which they cooperate. Tas1R3 deletion abolishes behavioral responses to some sweet tastants while only attenuating responses to others, mirroring what is observed in tas1R2 nulls. Similarly, tas1R1 deletion abolishes behavioral responses to some umami tastants while attenuating responses to others (Zhao et al. 2003). Together, these data suggest that TAS1R1 or TAS1R2 functionally interact with TAS1R3 and that this interaction not only increases sensitivity but also modulates stimulus tuning. Indeed, TAS1R3 directly interacts with both TAS1R1 and TAS1R2 (Nelson et al. 2002), supporting a model in which tas1R family heterodimers constitute sweet and umami taste receptors.
The precise nature of this interaction has yet to be explored in detail, but the difficulty in expressing these receptors heterologously (Nelson et al. 2001) suggests an intriguing model that could account for the as-yet-unstudied topic of taste receptor gene regulation. TAS1R3 could act as a molecular chaperone for both TAS1R1 and TAS1R2, allowing them to exit the ER. In this model, a developing TRC stochastically chooses tas1R1 or tas1R2 expression, resulting in activation of the UPR, which would then promote expression of tas1R3 to allow cell surface expression of the receptor, to relieve the UPR, and to lock in the functional identity of the TRC. In this sense, TAS1R3 would act similarly to RTP1 in the MOE. Given that in tas1R1 mutants tas1R2 expression expands (and vice versa), we further suggest that in this receptor-elicited feedback model, deletion of tas1R1 should result in tas1R2 choice and subsequent tas1R3 choice. Given this dependence, tas1R1/tas1R2 double mutants should not exhibit tas1R3 expression (Figure 4).
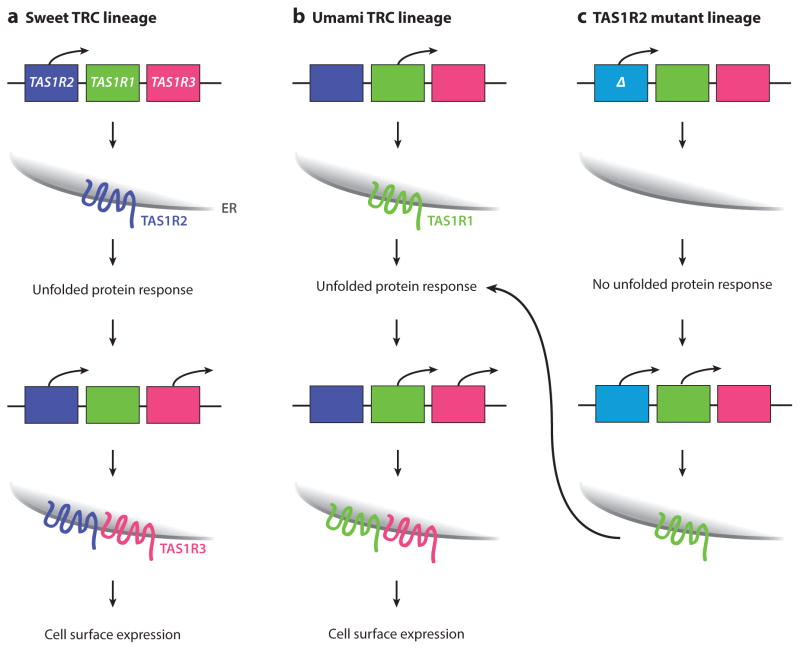
Figure 4.
An unfolded protein response (UPR)–based model for tas1R feedback. (a) Some cells choose tas1R2 for expression, leading to production of TAS1R2 protein. This protein fails to exit the endoplasmic reticulum (ER), activating the UPR. The UPR in turn promotes transcription of tas1R3, which couples with TAS1R2 to allow exit from the ER, similar to the role of RTP1 with olfactory receptors (ORs). Feedback could also act to prevent transcription of tas1R1. (b) Likewise, tas1R1 choice promotes UPR activation, driving transcription of tas1R3, ER exit of the coreceptor, and cell surface expression. (c) In contrast, choice of a deleted tas1R2 copy would fail to activate the UPR, allowing for choice of a second tas1R gene and explaining how, in tas1R1 mutants, tas1R2 expression expands.
Bitter Taste Receptors
Bitter taste is mediated by the tas2R family of GPCRs, which is composed of ~30 genes in 2 genomic clusters (Adler et al. 2000, Chandrashekar et al. 2000, Matsunami et al. 2000). Each bitter TRC expresses multiple tas2Rs, the likely functional consequence of which is broad tuning to bitter tastants at the expense of discriminatory power (Adler et al. 2000, though see Caicedo & Roper 2001). How many tas2Rs and which combinations of receptors are coexpressed are yet to be firmly demonstrated. Recordings from bitter TRCs suggest that these cells recognize only small numbers of bitter tastants, whereas comprehensive RNA ISH for the human tas2R family suggests that each bitter TRC probably expresses between 4 and 11 tas2Rs (Behrens et al. 2007). The number of coexpressed tas2Rs may be a compromise between sensitivity and tuning because expression of many tas2Rs has reduced bitter taste sensitivity (Behrens et al. 2007). How tas2Rs are regulated has yet to be addressed; however, like ORs, VRs, and tas1Rs, tas2Rs fail to traffic to the plasma membrane when expressed heterologously (Behrens et al. 2006), suggesting that a UPR-based feedback pathway may be in place in bitter TRCs to limit or otherwise control their receptor complements.
Sour and Salty Taste Receptors
Whereas sweet, umami, and bitter taste are mediated by GPCRs, both sour and salty taste are not. Sour taste is mediated by an ion channel, polycystic kidney disease 2-like 1 (PKD2L1) (Huang et al. 2006). TRCs expressing Pkd2l1 do not express receptors for other taste modalities (though see below), and animals lacking this gene have severely reduced (Horio et al. 2011) or abolished (Huang et al. 2006) responses to sour tastants. PKD2L1 interacts with the related PKD1L3, possibly forming a coreceptor or a receptor–chaperone pair (Ishimaru et al. 2010). However, deletion of Pkd1l3 does not abolish sour taste (Horio et al. 2011), and thus the role of Pkd1l3 has yet to be defined.
Salty taste is mediated by the epithelial sodium channel (ENAC), which is composed of alpha, beta, and gamma subunits and which is typically thought to be involved in the maintenance of salt homeostasis. ENAC channels are expressed by at least two populations of TRCs, one of which also expresses the sour TRC marker Car4. Deletion of ENaC using Car4-Cre does not affect salt responses, whereas deletion of ENaC from all TRCs abolishes salt responses, indicating that only the ENaC(+), Car4(−) cells are required for salt taste (Chandrashekar et al. 2010). Regulation of salty and sour taste receptor genes has yet to be studied; however, given that these lineages develop independently of Skn-1a (Matsumoto et al. 2011), it seems likely that either developing TRCs choose a fate from among the five modalities, with sour/salty cells not requiring subsequent Skn-1a expression, or that sour/salty versus sweet/umami/bitter lineages are independent of each other (see Figure 5). Uncovering the precise lineage relationships among the TRCs encoding the five taste modalities will be of great interest.
Figure 5.
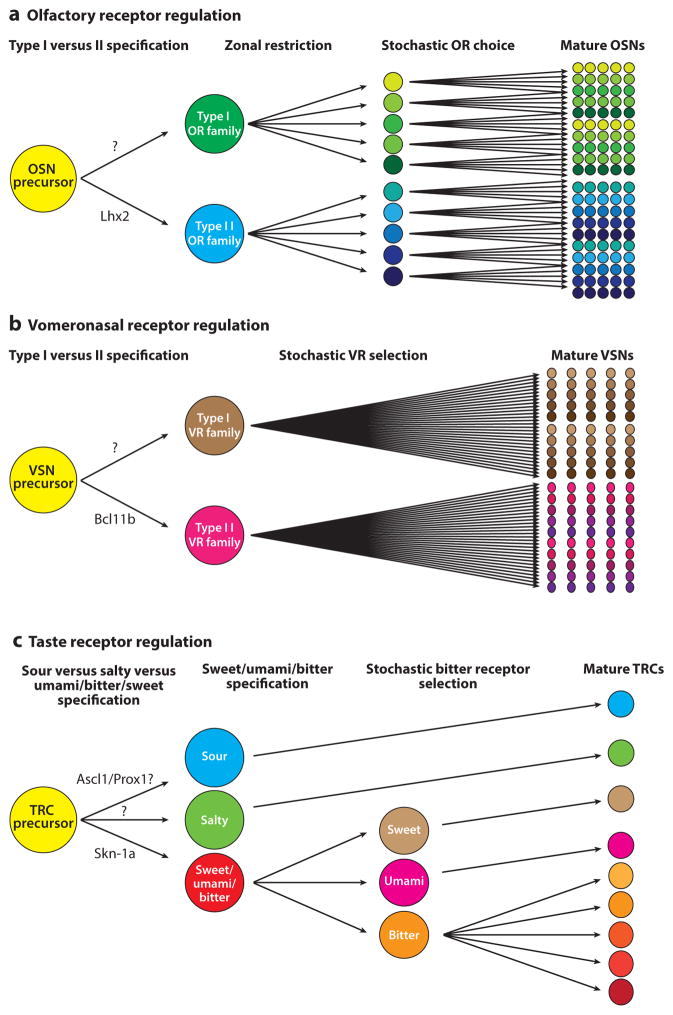
Current model of olfactory, vomeronasal, and taste receptor gene regulation and effects on cell lineages. (a) Olfactory sensory neuron (OSN) precursor cells are committed to type I olfactory receptor (OR) expression or type-II OR expression by Lhx2. The zonal location in the main olfactory epithelium (MOE) of each OSN then restricts the pool of ORs among which it can choose for expression. Each OSN then stochastically selects an OR gene, giving rise to a field of mature OSNs expressing a single OR. (b) Vomeronasal precursor cells are committed to V1R or V2R expression by Bcl11b. Type I and type II vomeronasal sensory neurons (VSNs) then select a vomeronasal receptor (VR) for monogenic expression, with V2R-choosing cells also selecting a second V2R and in some cases an H2-Mv (see text). (c) Taste receptor cell (TRC) precursors are committed to sour lineage by Ascl1 and/or Prox1, to salt lineage by unknown factors, or to sweet/umami/bitter lineages by Skn-1a. Sweet/umami/bitter cells select one modality for their mature identity by unknown mechanisms. Bitter cells then further diversify, expressing heterogeneous sets of bitter taste receptors, which gives rise to the five TRC modalities as well as to subclasses of sour and bitter cells (see text).
CONCLUDING REMARKS
The past 25 years have seen the elucidation of a great assembly of receptors mediating sensory reception at the periphery. But because sensory-coding strategies rely on expression of limited or defined subsets of receptors in each sensory cell, an equally important challenge is to understand the regulation of these genes. Recent breakthroughs, primarily in the regulation of olfactory receptor genes, suggest models that could be common to each sensory modality. In particular, we propose that receptor gene family repression and UPR-mediated receptor feedback will prove to be widespread features in receptor gene regulation. Despite the fact that these chemoreceptors share common, AT-rich promoter signatures (Clowney et al. 2011), differences in the specific molecular strategies followed by each regulatory system undoubtedly exist and are probably responsible for restricting their expression to the proper sensory organ. However, final refinement of their expression patterns may be mediated not only by their cis-regulatory elements and the transcription factors that stabilize them, but also by cofactors that govern their proper processing and targeting to the membrane. In this scenario, infrequent expression of the wrong chemoreceptor in a sensory organ may not lead to stable and productive expression because specialized chaperones for this receptor may be absent. Thus, deploying the UPR pathway in receptor regulation may provide an elegant solution for controlling gene expression of chemoreceptors with an ever-expanding number of family members. The exact regulatory solution will likely be determined by the exact number of genes, by the requirements for absolute singularity versus tolerance for receptor coexpression, by whether ligands have innate valence versus a value assigned by experience, and by a need for rapid evolvability versus stability. Further identification of the convergences and divergences between the gene-regulatory programs of each sensory modality will undoubtedly shed light not only on other sensory modalities, but on gene regulation in general.
Acknowledgments
We thank members of the Lomvardas lab for critical reading of this manuscript. This work was supported by the following R01 grants: R01DC014144 and R01DA036894.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- Araneda RC, Kini AD, Firestein S. The molecular receptive range of an odorant receptor. Nat Neurosci. 2000;12:1248–55. doi: 10.1038/81774. [DOI] [PubMed] [Google Scholar]
- Barnea G, O’Donnell S, Mancia F, Sun X, Nemes A, et al. Odorant receptors on axon termini in the brain. Science. 2004;304:1468. doi: 10.1126/science.1096146. [DOI] [PubMed] [Google Scholar]
- Behrens M, Bartelt J, Reichling C, Winnig M, Kuhn C, Meyerhof W. Members of RTP and REEP gene families influence functional bitter taste receptor expression. J Biol Chem. 2006;281:20650–59. doi: 10.1074/jbc.M513637200. [DOI] [PubMed] [Google Scholar]
- Behrens M, Foerster S, Staehler F, Raguse JD, Meyerhof W. Gustatory expression pattern of the human TAS2R bitter receptor gene family reveals a heterogeneous population of bitter responsive taste receptor cells. J Neurosci. 2007;27:12630–40. doi: 10.1523/JNEUROSCI.1168-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghard A, Buck LB. Sensory transduction in vomeronasal neurons: evidence for G alpha o, G alpha i2, and adenylyl cyclase II as major components of a pheromone signaling cascade. J Neurosci. 1996;16:909–18. doi: 10.1523/JNEUROSCI.16-03-00909.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza T, Vassalli A, Fuss S, Zhang JJ, Weiland B, et al. Mapping of class I and class II odorant receptors to glomerular domains by two distinct types of olfactory sensory neurons in the mouse. Neuron. 2009;61:220–33. doi: 10.1016/j.neuron.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–87. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Caicedo A, Roper SD. Taste receptor cells that discriminate between bitter stimuli. Science. 2001;291:1557–60. doi: 10.1126/science.291.5508.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Kuhn C, Oka Y, Yarmolinksy DA, Hummler E, et al. The cells and peripheral representation of sodium taste in mice. Nature. 2010;464:297–301. doi: 10.1038/nature08783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, et al. T2Rs function as bitter taste receptors. Cell. 2000;100:703–11. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–34. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenscoff-Papadimitriou EC, et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–37. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, Magklara A, Colquitt BM, Pathak N, Lane RP, Lomvardas S. High-throughput mapping of the promoters of mouse olfactory receptor genes reveals a new type of mammalian promoter and provides insight into olfactory receptor gene regulation. Genome Res. 2011;21:1249–59. doi: 10.1101/gr.120162.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155:321–32. doi: 10.1016/j.cell.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Punta K, Leinders-Zufall T, Rodriguez I, Jukam D, Wysockl C, et al. Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes. Nature. 2002;419:70–74. doi: 10.1038/nature00955. [DOI] [PubMed] [Google Scholar]
- Dey S, Matsunami H. Calreticulin chaperones regulate functional expression of vomeronasal type 2 pheromone receptors. PNAS. 2011;108:16651–56. doi: 10.1073/pnas.1018140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietschi Q, Assens A, Challet L, Carleton A, Rodriguez I. Convergence of FPR-rs3-expressing neurons in the mouse accessory olfactory bulb. Mol Cell Neurosci. 2013;56:140–47. doi: 10.1016/j.mcn.2013.04.008. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Ebrahimi F, Chess A. Olfactory neurons are interdependent in maintaining axonal projections. Curr Biol. 2000;10:219–22. doi: 10.1016/s0960-9822(00)00342-0. [DOI] [PubMed] [Google Scholar]
- Enomoto T, Ohmoto M, Iwata T, Uno A, Saitou M, et al. Bcl11b/Ctip2 controls the differentiation of vomeronasal sensory neurons in mice. J Neurosci. 2011;31:10159–73. doi: 10.1523/JNEUROSCI.1245-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein P, Mombaerts P. A contextual model for axon sorting into glomeruli in the mouse olfactory system. Cell. 2004;117:817–31. doi: 10.1016/j.cell.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Ferreira T, Wilson SR, Choi YG, Risso D, Dudoit S, et al. Silencing of odorant receptor genes by G protein βγ signaling ensures the expression of one odorant receptor per olfactory sensory neuron. Neuron. 2014;81:847–59. doi: 10.1016/j.neuron.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero DM, Lemon JK, Fluegge D, Pashkovski S, Korzan WJ, et al. Detection and avoidance of a carnivore odor by prey. PNAS. 2011;108:11235–40. doi: 10.1073/pnas.1103317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero DM, Wacker D, Roque M, Baldwin MW, Stevens C, Liberles SD. Agonists for 13 trace amine-associated receptors provide insight into the molecular basis of odor selectivity. ACS Chem Biol. 2012;7:1184–89. doi: 10.1021/cb300111e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130:373–84. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- George SR, O’Dowd BF, Lee SP. G-protein-coupled receptor oligomerization and its potential for drug discovery. Nat Rev Drug Discov. 2002;1:808–20. doi: 10.1038/nrd913. [DOI] [PubMed] [Google Scholar]
- Halpern M. The organization and function of the vomeronasal system. Annu Rev Neurosci. 1987;10:325–62. doi: 10.1146/annurev.ne.10.030187.001545. [DOI] [PubMed] [Google Scholar]
- Hirota J, Mombaerts P. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. PNAS. 2004;101:8751–55. doi: 10.1073/pnas.0400940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota J, Omura M, Mombaerts P. Differential impact of Lhx2 deficiency on expression of class I and class II odorant receptor genes in mouse. Mol Cell Neurosci. 2007;34:679–88. doi: 10.1016/j.mcn.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Horio N, Yoshida R, Yasumatsu K, Yanagawa Y, Ishimaru Y, et al. Sour taste responses in mice lacking PKD channels. PLOS ONE. 2011;6(5):e20007. doi: 10.1371/journal.pone.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AL, Chen X, Hoon MA, Chandrashekar J, Guo W, et al. The cells and logic for mammalian sour taste detection. Nature. 2006;442:934–38. doi: 10.1038/nature05084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Soria X, Levitin MO, Saraiva LR, Logan DW. The olfactory transcriptomes of mice. PLOS Genet. 2014;10:e1004593. doi: 10.1371/journal.pgen.1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Sakano H. Odorant receptor-mediated signaling in the mouse. Curr Opin Neurobiol. 2008;18:251–60. doi: 10.1016/j.conb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Ishii T, Hirota J, Mombaerts P. Combinatorial coexpression of neural and immune multigene families in mouse vomeronasal sensory neurons. Curr Biol. 2003;13:394–400. doi: 10.1016/s0960-9822(03)00092-7. [DOI] [PubMed] [Google Scholar]
- Ishii T, Mombaerts P. Expression of nonclassical class 1 major histocompatability genes defines a tripartite organization of the mouse vomeronasal system. J Neurosci. 2008;28:2332–41. doi: 10.1523/JNEUROSCI.4807-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Mombaerts P. Coordinated coexpression of two vomeronasal receptor V2R genes per neuron in the mouse. Mol Cell Neurosci. 2011;46:397–408. doi: 10.1016/j.mcn.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Katano Y, Yamamoto K, Akiba M, Misaka T, et al. Interaction between PKD1L3 and PKD2L1 through their transmembrane domains is required for localization of PKD2L1 at taste pores in taste cells of circumvallate and foliate papillae. FASEB J. 2010;24:4058–67. doi: 10.1096/fj.10-162925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Tsai L, Roy DS, Valenzuela DH, Mosley C, et al. Neurons expressing trace amine-associated receptors project to discrete glomeruli and constitute an olfactory subsystem. PNAS. 2012;109:13410–15. doi: 10.1073/pnas.1206724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Masters SB, Bourne HR, Reed RR. Biochemical characterization of three stimulatory GTP-binding proteins: the large and small forms of Gs and the olfactory-specific G-protein, Golf. J Biol Chem. 1990;265:2671–76. [PubMed] [Google Scholar]
- Jones DT, Reed RR. Golf: an olfactory neuron specific-G protein involved in odorant signal transduction. Science. 1989;244:790–95. doi: 10.1126/science.2499043. [DOI] [PubMed] [Google Scholar]
- Khan M, Vaes E, Mombaerts P. Regulation of the probability of mouse odorant receptor gene choice. Cell. 2011;147:907–21. doi: 10.1016/j.cell.2011.09.049. [DOI] [PubMed] [Google Scholar]
- Lane RP, Cutforth T, Axel R, Hood L, Trask BJ. Sequence analysis of mouse vomeronasal receptor gene clusters reveals common promoter motifs and a history of recent expansion. PNAS. 2002;99:291–96. doi: 10.1073/pnas.012608399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinders-Zufall T, Ishii T, Chamero P, Hendrix P, Oboti L, et al. A family of nonclassical class I MHC genes contributes to ultrasensitive chemodetection by mouse vomeronasal sensory neurons. J Neurosci. 2014;34:5121–33. doi: 10.1523/JNEUROSCI.0186-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi G, Puche A, Mantero S, Barbien O, Trombino S, et al. The Dlx5 homeodomain gene is essential for olfactory development and connectivity in the mouse. Mol Cell Neurosci. 2003;22:530–43. doi: 10.1016/s1044-7431(02)00041-6. [DOI] [PubMed] [Google Scholar]
- Levy NS, Bakalyar HA, Reed RR. Signal transduction in olfactory neurons. J Steroid Biochem Mol Biol. 1991;39:633–37. doi: 10.1016/0960-0760(91)90262-4. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic olfactory receptor expression. PNAS. 2004;101:1069–74. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. PNAS. 2006;442:645–50. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- Liberles SD, Horowitz LF, Kuang D, Contos JJ, Wilson KL, et al. Formyl peptide receptors are candidate chemosensory receptors in the vomeronasal organ. PNAS. 2009;106:9842–47. doi: 10.1073/pnas.0904464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loconto J, Papes F, Chang E, Stowers L, Jones EP, et al. Functional expression of murine V2R pheromone receptors involves selective association with the M10 and M1 families of MHC class 1b molecules. Cell. 2003;112:607–18. doi: 10.1016/s0092-8674(03)00153-3. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–13. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154:325–36. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DB, Magklara A, Goh T, Sampath SC, Schaefer A, et al. Heterochromatin-mediated gene silencing facilitates the diversification of olfactory neurons. Cell Rep. 2014;9:884–92. doi: 10.1016/j.celrep.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–70. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markenscoff-Papadimitriou E, Allen WE, Colquitt BM, Goh T, Murphy KK, et al. Enhancer interaction networks as a means for singular olfactory receptor expression. Cell. 2014;159:547–57. doi: 10.1016/j.cell.2014.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini S, Silvotti L, Shirazi A, Ryba NJ, Tirindelli R. Co-expression of putative pheromone receptors in the sensory neurons of the vomeronasal organ. J Neurosci. 2001;21:843–48. doi: 10.1523/JNEUROSCI.21-03-00843.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto I, Ohmoto M, Abe K. Functional diversification of taste cells in vertebrates. Sem Cell Dev Biol. 2013;24:210–14. doi: 10.1016/j.semcdb.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto I, Ohmoto M, Narukawa M, Yoshihara Y, Abe K. Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat Neurosci. 2011;14:685–87. doi: 10.1038/nn.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H, Montmayeur JP, Buck LB. A family of candidate taste receptors in human and mouse. Nature. 2000;404:601–4. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- McIntyre JC, Bose SC, Stromberg AJ, McClintock TS. Emx2 stimulates odorant receptor gene expression. Chem Senses. 2008;33:825–37. doi: 10.1093/chemse/bjn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloski JS, Galante PA, Malnic B. Identification of potential regulatory motifs in odorant receptor genes by analysis of promoter sequences. Genome Res. 2006;16:1091–98. doi: 10.1101/gr.5185406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloski JS, Galante PA, Nagai MH, Armelin-Correa L, Chien MS, et al. Common promoter elements in odorant and vomeronasal receptor genes. PLOS ONE. 2011;6(12):e29065. doi: 10.1371/journal.pone.0029065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K, Serizawa S, Kimura HM, Sakano H. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J Neurosci. 2005;25:3586–92. doi: 10.1523/JNEUROSCI.0324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao S, Nemes M, et al. Visualizing an olfactory sensory map. Cell. 1996;87:675–86. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, et al. An amino-acid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–90. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- Plessy C, Pascarella G, Bertin N, Akalin A, Carrieri C, et al. Promoter architecture of mouse olfactory receptor genes. Genome Res. 2012;22:486–97. doi: 10.1101/gr.126201.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler HJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79:1245–55. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- Riviere S, Challet L, Fluegge D, Spehr M, Rodriguez I. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemoreceptors. Nature. 2009;459:574–77. doi: 10.1038/nature08029. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Feinstein P, Mombaerts P. Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell. 1999;97:199–208. doi: 10.1016/s0092-8674(00)80730-8. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gil D, Treolar HB, Zhang H, Miller AM, Two A, et al. Chromosomal location-dependent nonstochastic onset of odorant receptor expression. J Neurosci. 2010;30:10067–75. doi: 10.1523/JNEUROSCI.1776-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Roppolo D, Vollery S, Kan CD, Lüscher C, Broillet MC, Rodriguez I. Gene cluster lock after pheromone receptor gene choice. EMBO J. 2007;26:3423–30. doi: 10.1038/sj.emboj.7601782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba NJ, Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19:371–79. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–91. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, et al. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–94. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, et al. Histone demethylation by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O’Donnell S, Nemes S, Mendelsohn M, et al. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–15. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Sorkin A, Von Zastrow M. Endocytosis and signaling: intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10:609–22. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryer L. Visual excitation and recovery. J Biol Chem. 1991;266:10711–14. [PubMed] [Google Scholar]
- Sullivan SL, Adamson MC, Ressler KJ, Kozak CA, Buck LB. The chromosomal distribution of mouse odorant receptor genes. PNAS. 1996;93:884–88. doi: 10.1073/pnas.93.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Zong C, Xie S. Rare event of histone demethylation can initiate singular gene expression of olfactory receptors. PNAS. 2013;110:21148–52. doi: 10.1073/pnas.1321511111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35:681–96. doi: 10.1016/s0896-6273(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, Nuñez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–91. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74:309–18. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. Odorant receptors govern the formation of a precise topographic map. Cell. 1998;93:47–60. doi: 10.1016/s0092-8674(00)81145-9. [DOI] [PubMed] [Google Scholar]
- Wang SZ, Ou J, Zhu LJ, Green MR. Transcription factor ATF5 is required for terminal differentiation and survival of olfactory sensory neurons. PNAS. 2012;109:18589–94. doi: 10.1073/pnas.1210479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watatani Y, Ichikawa K, Nakanishi N, Fujimoto M, Takeda H, et al. Stress-induced translation of ATF5 mRNA is regulated by the 5′-untranslated region. J Biol Chem. 2008;283:2543–53. doi: 10.1074/jbc.M707781200. [DOI] [PubMed] [Google Scholar]
- Zhang J, Huang G, Dewan A, Feinstein P, Bozza T. Uncoupling stimulus specificity and glomerular position in the mouse olfactory system. Mol Cell Neurosci. 2012;51:79–88. doi: 10.1016/j.mcn.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–33. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, et al. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–66. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]