Abstract
Background
Indigenous populations of animals have developed unique adaptations to their local environments, which may include factors such as response to thermal stress, drought, pathogens and suboptimal nutrition. The survival and subsequent evolution within these local environments can be the result of both natural and artificial selection driving the acquisition of favorable traits, which over time leave genomic signatures in a population. This study’s goals are to characterize genomic diversity and identify selection signatures in chickens from equatorial Africa to identify genomic regions that may confer adaptive advantages of these ecotypes to their environments.
Results
Indigenous chickens from Uganda (n = 72) and Rwanda (n = 100), plus Kuroilers (n = 24, an Indian breed imported to Africa), were genotyped using the Axiom® 600 k Chicken Genotyping Array. Indigenous ecotypes were defined based upon location of sampling within Africa. The results revealed the presence of admixture among the Ugandan, Rwandan, and Kuroiler populations. Genes within runs of homozygosity consensus regions are linked to gene ontology (GO) terms related to lipid metabolism, immune functions and stress-mediated responses (FDR < 0.15). The genes within regions of signatures of selection are enriched for GO terms related to health and oxidative stress processes. Key genes in these regions had anti-oxidant, apoptosis, and inflammation functions.
Conclusions
The study suggests that these populations have alleles under selective pressure from their environment, which may aid in adaptation to harsh environments. The correspondence in gene ontology terms connected to stress-mediated processes across the populations could be related to the similarity of environments or an artifact of the detected admixture.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2711-5) contains supplementary material, which is available to authorized users.
Keywords: Homozygosity, Selection signatures, Stress response
Background
In nature, environmental stressors can influence the phenotypic characteristics that individuals and populations develop over time. A challenging environment can also shape the genomic landscape that underlies a population’s adaption to weather, resources, and predators [1–3]. These variables can take many abiotic and biotic forms, all with varying levels of intensity leading to a complex balance of genetics and environment. Climate change, especially in the form of weather extremes, has the ability to disrupt this balance and place a population under increased environmental stress [4–7]. For livestock production, this shift in climate driven environmental stressors has been detrimental to commercial traits [8–10]. Climate change has led to higher temperatures and drought, contributing to losses in livestock production worldwide related to reduced reproduction, growth, and immune function [8–10]. For example, high ambient temperatures can operate as a primary environmental stressor. Environmentally stressed chickens can experience oxidative-stress, lipid peroxidation, disruption of internal energy balance, and immunosuppression [10–17]. A major cellular effect caused by multiple environmental stressors is the generation of reactive oxygen (ROS) species that leads to oxidative stress and lipid peroxidation. This is brought about by changes in intracellular oxidation, which results in a state of imbalance between ROS and antioxidants [12–15]. Oxidative stress can be detrimental to gene expression causing post-transcriptional changes to signaling genes [18, 19] disrupting the health of an animal at the genetic level. Oxidative stress in chickens can also cause endothelial dysfunction and vasoconstriction [20–22]. However, the differences in how chickens respond to stressors may depend upon their evolutionary course and how it was influenced by selective pressure to adapt for survival. Constant selection on these survival traits can lead to the presence of genomic signatures that indicate what genomic regions responded to selective pressure. Selection for survival traits can lead to reduction in the variability around genomic regions associated with that trait. This reduction in variability, referred to as a selection signature or selective sweep, can be detected and examined for its biological importance. Researchers have employed multiple methods for detecting the presence of these areas under selection by measuring the reductions in diversity of a genomic region [23, 24] both within and between populations. Studies examining selection signatures within populations have used methods based on the reduction of heterozygosity to detect selection and identify important domestication loci [25, 26] for traits related to reproduction, growth, feeding behavior, and skin color. Other studies have successfully used Fst measurements, pooled heterozygosity, and extended homozygosity to investigate the chicken genome for signatures of selection underlying key economic traits such as growth and egg production in commercial breeds [27–29]. Through comparisons of both wild and domesticated chickens these studies have been able to separate traits driven by natural and artificial selection. Studies have also been conducted on indigenous chicken breeds that have uncovered evidence of possible independent selection events towards pathways related to niche survival environments, such as low oxygen at high altitudes [30] that would normally be considered as stressful.
The current study examined indigenous chicken ecotypes from the countries of Uganda and Rwanda for the presence of genomic signatures that may indicate that selective pressure from their environment helped shape the genetics underlying their adaption to various stressors. The two countries have environments that present many challenges, such as weather and food availability and quality, which may have led to chickens adapted to pressure from their environment. In addition to the indigenous birds, we also examined Kuroiler chickens in Uganda. Kuroilers were bred in India to be tolerant of heat, dual-purpose for meat and egg production, and with the ability to scavenge when food is scarce [31]. Studying populations that have developed under natural and artificial selection for a challenging environment may reveal genomic signatures related to these populations’ mechanisms of tolerance, resistance, or resilience. This may lead to a greater understanding of the genomic control of response to environmental stressors and aid in breeding of animals that are better able to tolerate stressors related to harsh environments and shifting climate patterns.
Methods
Sample collection
Blood samples were collected from 196 African chickens: Ugandan (n = 72), Rwandan (n = 100), and Kuroilers (n = 24). Kuroilers, originally imported from India, were sampled from farms in Uganda. Five physically distinct farms were sampled in each sampling area to make up a single geographically defined ecotype, in an attempt to reduce stratification. There were six ecotypes (30 farms) for Rwanda from the areas of Huye, Kicukiro, Kirehe, Musanze, Nyagatare, and Rubavu. For Uganda there were three ecotypes (15 farms) from the areas of Kamuli, Luweero, and Masaka. Blood was collected using FTA cards (Additional file 1: Table S1).
Genotyping and quality control
Genotyping was conducted at GeneSeek (Lincoln, NE) using the Affymetrix Axiom ® 600 k Chicken Genotyping Array. SNPs were put through a quality control step in Plink [32, 33] based on the parameters of > 97 % call rate (-geno 0.03) and minor allele frequency (MAF) > 0.02. After filtering, 506,965 total SNPs remained. A total of 476,106 autosomal SNPs were available for downstream analyses.
Population stratification analysis
Hierarchical clustering analysis of the genotype data in JMP ® (www.jmp.com) was used to determine the relationships between the samples. This cluster analysis was run using the ward method [34]. PLINK [32, 33] was used to construct a multi-dimension scaling plot (MDS-plot) to examine population structure for stratification. MDS plots for this analysis were based on 196 × 196 matrix of genome-wide Identity-By-State (IBS) scores calculated based on pairwise comparisons of the genetic distances for all individuals. The APE [35] and PhanGorn [36] R packages were used to examine the relatedness of the birds by phylogenetic analysis using neighbor-joining (NJ) distance based tree construction methods (data not shown). Visualization of the corresponding trees was done using FigTree (http://tree.bio.ed.ac.uk/software/figtree) and Ninja (http://mesquiteproject.wikispaces.com/Additional+Mesquite+Packages). IBS values and inbreeding coefficients were calculated within Plink. Principal component analysis (PCA) was done using SNP and Variant Suite (SVS) [37] and JMP ® (www.jmp.com). Shared ancestry was also explored using the Admixture software [38] set at varying values of k, ranging from 1 to 9 to represent the number of ecotypes sampled, with a final optimal k = 3.
Runs of homozygosity analysis
Runs of homozygosity (ROH) analyses were carried out in Plink [32, 33] to examine genomic regions that harbor alleles driven to fixation using a SNP based sliding window approach. Runs of homozygosity were calculated for each individual and any ROH that overlapped between individuals within and between populations. For the individual and overlapping ROH analysis, a run was defined in Plink as ≥ 250 SNPs, density of 50 kb/SNP, allowed gap of 1000 kb, and 3 heterozygous calls allowed within a run. The analysis focused on the overlapping ROH that were also analyzed using the additional parameters of allelic match threshold of 0.95 identity and 20 or more informative SNPs. The overlapping ROH are regions that overlapped across all populations and contained 10 or more individuals. Consensus regions between populations were defined as the region that was common to every bird, irrespective of length of the ROH, that Plink assigned to a “pool” because of shared overlapping ROH. The analysis of the overlapping/consensus regions was based on all samples as a whole and a “pool” of individuals sharing overlapping/consensus ROH was made up of birds from all three populations. A gene ontology enrichment analysis was conducted on a gene list from the ROH consensus regions [39, 40]. The gene list is based only on genes mapped to within the consensus region of the ROH. Gene ontology enrichment analysis results were considered statistically significant at a FDR cut-off < 0.15.
Analysis of putative selection signatures
Sample haplotypes were phased using FastPhase [41] for downstream analysis of selection signatures. The R package REHH [42] was used to calculate integrated haplotype score (iHS) and standardized log-ratio of the integrated EHHS (iES) between pairs of populations (Rsb) values to examine the populations for SNPs that displayed signals of selection. Both iHS and Rsb values were log transformed to normalize the data and calculated as per the method established by Voight et al., 2006 [43]. Statistical significance of iHS values were determined by use of the-log p-values generated by the REHH software package. The -log p-values were not adjusted for multiple test correction because all multiple test correction procedures proved to be too conservative due to the number of tests exceeding 400,000. To address the issue of statistical significance a very stringent nominal p-value (α = 0.001, −log α = 4) was set and iHS data was then ranked from lowest to highest. After ranking, the p-value (α = 0.001, −log α = 4) was used as the cut-off for all samples and populations. iHS values were considered extreme at iHS > |3.29| because this was the lowest iHS value at the p-value cut-off. Pairwise comparisons between populations were examined using the Rsb statistic. P-values for the Rsb statistic were low enough to be subjected to multiple test correction using a FDR cut-off of < 0.05. The use of a lower FDR was also necessary to reduce the number of significant results for a more focused downstream analysis. To carry out gene ontology term enrichment, SNPs were annotated to genes of interest using the Affymetrix NetAffx™ Analysis Center which, when supplied with a list of markers from one genotyping platform, will give a list of genes within, upstream, or downstream of a gene in that region.
Gene ontology enrichment analysis
Gene ontology (GO) term enrichment was performed using (GO)TermFinder [39]. Visualization and reduction of redundant terms of the GO enrichment results were carried out using ReVigo [40]. The significant GO terms were filtered for redundant terms to produce a focused list of the functions and processes under selection. The FDR for the enrichment tests was set at < 0.15. This cut-off was set higher than the threshold for the iHS or Rsb tests since it was a separate independent test. Another point that prompted the use of a higher FDR was the lack of annotations for the chicken genome, which effectively reduced the overall number of genes that could be analyzed.
Results
Population structure analysis
The MDS plot (Fig. 1) showed overlap among Ugandan, Rwandan, and Kuroiler populations. The Ugandan and Rwandan ecotypes showed the highest degree of overlap between populations, with the Kuroilers showing the most discrete clustering of individuals. The amount of admixture, based upon identity by state, within each country (3 Ugandan ecotypes, 6 Rwandan ecotypes) showed crossover between sampling areas (Fig. 2). The clustering analysis based on the SNP genotyping calls indicated that the ecotypes (sampling location) assigned to the birds showed shared ancestry of genotypes between individuals across populations and ecotypes. These results were visualized as an admixture plot that also showed evidence of shared ancestry (Fig. 2).
Fig. 1.
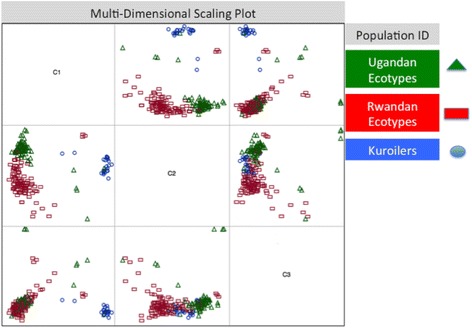
Multi-dimensional scaling plot showing distinct sampling populations. The multi-dimensional scaling plot was constructed using genomic distances to examine for the presence of population stratification
Fig. 2.
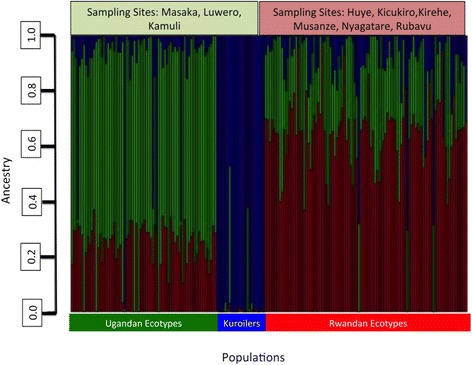
Admixture plot showing mixed ancestry between populations and individuals. Green denotes Ugandan ecotypes, red denotes Rwandan ecotypes, blue denotes Kuroilers. Based on optimal k = 3
Examination of runs of homozygosity (ROH)
The number and extent of individual ROH differed widely among the populations (unpublished data). Kuroilers had the fewest chromosomes that contained ROH, while the combined Ugandan ecotypes were the only population to have ROH on every chromosome except chromosome 16. Chromosome 16 showed no evidence of ROH based on the parameters in any of the populations. The median length of the ROH in a population was longest in Ugandan and shortest in Kuroiler. The amount of the genome covered by ROH per individual ranged from ~2 % to 40 % (unpublished data).
Examination of ROH overlapping consensus regions
The data were also analyzed for consensus overlapping ROH regions between all three populations that were identical by coordinates (Table 1). For the consensus ROH, the macro-chromosomes had the highest number of overlapping regions, span (kb), and number of SNPs comprising the runs on a chromosome. A gene list was created for the combined consensus regions and then analyzed using GO enrichment to determine over-enriched GO terms. There were 150 consensus overlapping ROH meeting criteria that harbored 343 genes that were used to conduct the GO enrichment analysis. The consensus ROH varied widely in the number of genes they contained, ranging from 0 to 24 genes within a given ROH. Statistically significant (FDR < 0.15) GO terms for biological processes and molecular functions related to both endogenous and external stressors (Table 2). Some of the most striking over-enriched gene ontology processes included regulation of cellular response to stress (GO:0080135), regulation of reactive oxygen species metabolic process (GO:2000377), regulation of apoptotic process (GO:0042981), and calcium ion transmembrane transport (GO:0070588) (FDR < 0.15). Over-enriched molecular functions that may be related to oxidative stress induced by the environment included calcium ion binding (GO:0005509), protein serine/threonine kinase activity (GO:0004674), and transforming growth factor beta receptor binding (GO:0005160) (Table 2).
Table 1.
Summary of ROH consensus regions present amongst all 3 populations, by chromosome
| Chr | N (Consensus) | Mean (# of birds) | Mean ROH length (Kb) | Sum ROH length (Kb) | Mean number Of SNPs | Sum Of SNPs |
|---|---|---|---|---|---|---|
| 1 | 46 | 13.83 | 298.88 | 13748.66 | 124.37 | 5721 |
| 2 | 20 | 16.70 | 572.33 | 11446.66 | 198.65 | 3973 |
| 3 | 17 | 12.65 | 326.66 | 5553.24 | 144.35 | 2454 |
| 4 | 21 | 17.19 | 153.77 | 3229.19 | 64.00 | 1344 |
| 5 | 9 | 14.33 | 482.82 | 4345.38 | 153.00 | 1377 |
| 6 | 4 | 10.75 | 256.77 | 1027.10 | 143.50 | 574 |
| 7 | 7 | 13.43 | 211.29 | 1479.02 | 102.14 | 715 |
| 8 | 6 | 21.33 | 218.13 | 1308.76 | 107.17 | 643 |
| 9 | 3 | 11.67 | 204.61 | 613.82 | 138.67 | 416 |
| 10 | 7 | 11.57 | 171.48 | 1200.37 | 158.29 | 1108 |
| 11 | 3 | 20.67 | 371.64 | 1114.93 | 207.00 | 621 |
| 12 | 1 | 12.00 | 1252.64 | 1252.64 | 778.00 | 778 |
| 13 | 1 | 10.00 | 984.15 | 984.15 | 408.00 | 408 |
| 14 | 1 | 18.00 | 1034.33 | 1034.33 | 645.00 | 645 |
| 15 | 2 | 10.50 | 239.67 | 479.33 | 182.50 | 365 |
| 19 | 2 | 13.00 | 511.86 | 1023.72 | 276.50 | 553 |
Each population is represented in each consensus group by at least one bird. Only consensus groups with 15 or more individuals shown. Consensus regions in between flanking markers were annotated for genes and coding non-synonymous and splice site category SNPs
Table 2.
Gene ontology (GO) enrichment of consensus ROH analysisa
| GO: ID | Go: term |
|---|---|
| GO:0006915 | Apoptotic process |
| GO:0002209 | Behavioral defense response |
| GO:0001662 | Behavioral fear response |
| GO:0070588 | Calcium ion transmembrane transport |
| GO:0071345 | Cellular response to cytokine stimulus |
| GO:0071495 | Cellular response to endogenous stimulus |
| GO:0071396 | Cellular response to lipid |
| GO:0033554 | Cellular response to stress |
| GO:0019221 | Cytokine-mediated signaling pathway |
| GO:0006281 | DNA repair |
| GO:0007631 | Feeding behavior |
| GO:0007599 | Hemostasis |
| GO:0031663 | Lipopolysaccharide-mediated signaling pathway |
| GO:0032873 | Negative regulation of stress-activated MAPK cascade |
| GO:0070303 | Negative regulation of stress-activated protein kinase signaling cascade |
| GO:0016310 | Phosphorylation |
| GO:0042981 | Regulation of apoptotic process |
| GO:0080135 | Regulation of cellular response to stress |
| GO:0001959 | Regulation of cytokine-mediated signaling pathway |
| GO:0043408 | Regulation of MAPK cascade |
| GO:2000377 | Regulation of reactive oxygen species metabolic process |
| GO:0080134 | Regulation of response to stress |
| GO:0032319 | Regulation of rho gtpase activity |
| GO:1901700 | Response to oxygen-containing compound |
| GO:0009314 | Response to radiation |
| GO:0006950 | Response to stress |
| GO:0023014 | Signal transduction by phosphorylation |
| GO:0033209 | Tumor necrosis factor-mediated signaling pathway |
| GO:0042060 | Wound healing |
| GO:0003684 | Damaged DNA binding |
| GO:0005246 | Calcium channel regulator activity |
| GO:0005488 | Binding |
| GO:0008289 | Lipid binding |
| GO:0030234 | Enzyme regulator activity |
| GO:0005509 | Calcium ion binding |
| GO:1901363 | Heterocyclic compound binding |
| GO:0005160 | Transforming growth factor beta receptor binding |
| GO:0005543 | Phospholipid binding |
| GO:0004674 | Protein serine/threonine kinase activity |
| GO:0035258 | Steroid hormone receptor binding |
| GO:0008083 | Growth factor activity |
Subset of GO terms that could putatively affect responses to environmental stressors. aAll terms were statistically significant at FDR < 0.15. This analysis lends evidence to support that all three populations experienced selective pressures for variants effecting stress response, immune response, and behavior
Genes under putative selection within populations
The strongest iHS signal (p< 0.001; iHS > |4|) across all three populations was on chromosome 18 in variants annotated to intronic regions of the protein kinase C alpha (PRKCA) gene (Figs. 3 and 4) (Additional file 2: Table S2). It was the strongest overall signal in the Ugandan birds, and second strongest signal in the Rwandan and Kuroilers (p< 0.001; iHS > |4|). There were also strong selection signals that were unique to each of the three populations. In the Uganda ecotypes the gene cyclin-dependent kinase inhibitor 3 (CDKN3) on chromosome 5 had the next highest iHS value (iHS = |4.82|) with 41 statistically significant variants (p < 0.001). The gene deleted in liver cancer 1 (DLC1) on chromosome 4 had the highest iHS value in the Rwandan birds (iHS = |4.29|) supported by 44 statistically significant variants (p < 0.001). In Kuroilers, variants annotated to adrenomedullin (ADM) (iHS = |5.79|) on chromosome 5 and glutamate-cysteine ligase, modifier subunit (GCLM) on chromosome 8 (iHS = |4.40|) had the highest iHS values (Additional file 3: Table S3).
Fig. 3.
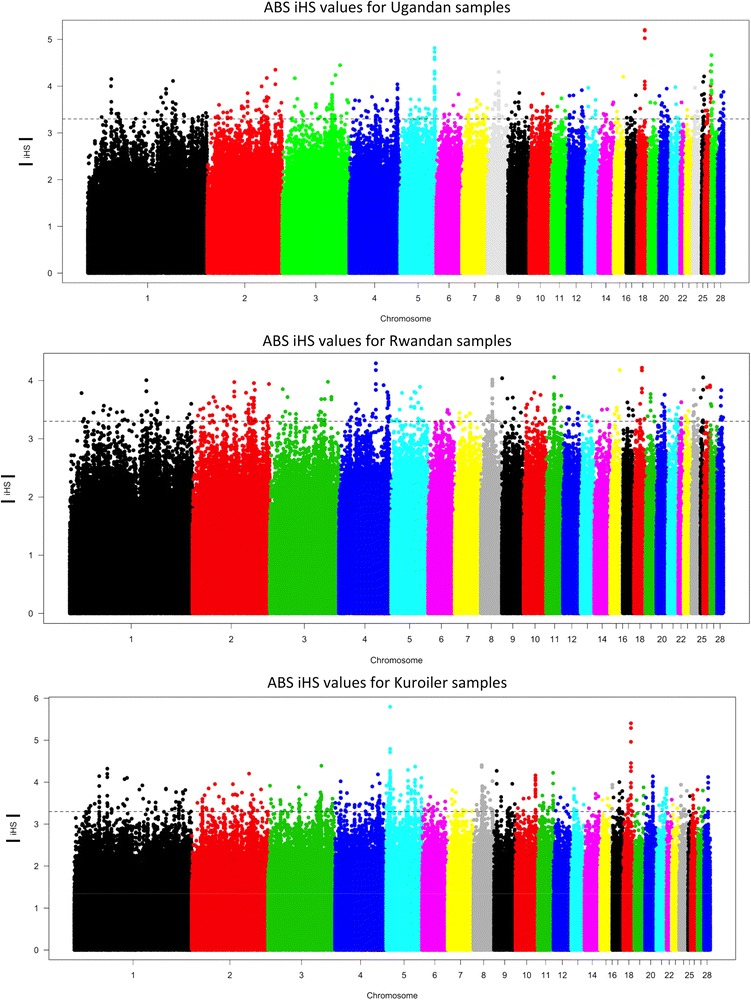
Manhattan plots of iHS values for each population. The horizontal lines represent the p-value cut-off for significance (< 0.001). iHS scores were recorded as absolute values. iHS cut-off for significance > |4| for variants to be considered as signal of selection
Fig. 4.
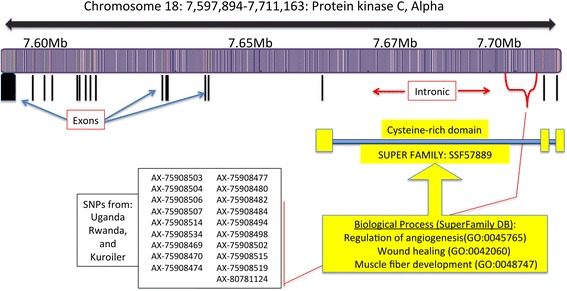
Statistically significant SNPs annotated to PRKCA fall within an intronic region of the gene near a cysteine-rich domain superfamily [86]. Variants are located near the protein kinase cysteine-rich domain, a group of small proteins that have functions possibly involved in the response to environmental stressors
Putative genes under selection between populations
The populations were also compared pairwise by means of the Rsb statistic to identify additional genomic regions under selection. Statistical significance (FDR < 0.05) and the highest Rsb value (>|7|) were used to determine genes under possible selection. The Ugandan ecotypes vs. the Rwandan ecotypes showed selective pressure on chromosome 13 for Gamma-aminobutyric acid receptor, gamma-2 (GABRG2). Also annotated to the highest Rsb value on chromosome 13 was the gene teneurin-2 isoform 1 (TENM2). On chromosome 11, Beta, beta-carotene 15,15'-monooxygenase (BCMO1) reached statistical significance along with having a Rsb above |7|. The Ugandan and the Rwandan ecotypes both yielded similar results when compared to the Kuroilers. The strongest selection occurred on chromosome 1 with the gene olfactomedin 4 (OLFM4).
Gene set enrichment analysis
ROH
Gene ontology (GO) enrichment was conducted using a list composed of genes located between the flanking markers of the ROH consensus regions (Table 2). The GO enrichment was subjected to a FDR < 0.15 then filtered for redundancy of terms using the software Revigo [40], which produced a list of significant GO terms related to selective pressures for variants effecting oxidative and cellular stress, immune response, and behavior. The GO enrichment also provided information on possible molecular functions and biological processes under selection related to calcium, lipid, and kinase activity and binding. Significance was also reached for processes related to UV radiation and DNA repair, possibly as a result of the birds living at the equator.
iHS
A GO enrichment was conducted using a list of genes annotated to statistically significant (p < 0.001) variants on the chip. Statistically significant (FDR ≤ 0.15) enriched GO terms relating to a host of environmental stressors emerged from the analysis. Regions of the genome in each population showed evidence of selective pressure on biological processes related to multiple stress-related signaling pathways. There was also evidence of selective pressure on molecular functions involved in kinase and lipid activity. In the Ugandan ecotypes and Kuroilers GO enrichment showed selective pressure on protein kinase activity (Additional file 4: Figure S4). The Rwandan and Kuroiler populations were both associated with PRKCA through the GO term protein kinase, this was not the case in the Ugandan ecotypes (Tables 3, 4 and 5).
Table 3.
Gene ontology (GO) enrichment of statistically significant (FDR < 0.15) GO terms for the Ugandan ecotypes
| Population | GO ID | Gene ontology term | Genes under GO term | Related environmental stressor(s) | Reference |
|---|---|---|---|---|---|
| Ugandan Ecotypes | GO:0065008 | Regulation of biological quality | PDIA5, CADPS2, TNIP2, CNOT6L, DISP3, ABCA4, PRKCA, SIN3A, PEX11A, DLG1, MC4R, RC3H1, NPY, SMAD1, CNGA2, RCOR1, RAP1GDS1, NAT8L, BICD2, SYNE1, ATP9A, STX12 | Growth response to Environmental Stress | UniProt [67], Jenuth 2000 [69, 77] |
| GO:0001816 | Cytokine production | DLG1, TRIB2, MAP3K7, EIF2AK2, SNAI2 | Pathogen/Predator | Welc et al. 2013 [56] | |
| GO:0043122 | Regulation of I- kappaB kinase/NF-kappaB signaling | TNIP2, ZFAND6, ZDHHC17, MAP3K7 | Inflammation/Oxidative Stress | Evans et al. 2002 [46], Perkins et al. 2007 [55] Salminen et al. 2008 [87], Tam et al. 2012 [88] | |
| GO:0016627 | Oxidoreductase activity, acting on the CH-CH group of donors | TECRL, DYPD | Oxidative stress | UniProt [67], Jenuth 2000 [69, 77], Lee et al. 2007 [89] | |
| GO:0005215 | Phospholipid transporter activity | ABCA4, ATP9A | Lipid Metabolism, phospholipid transfer to membrane | UniProt [67], Jenuth 2000 [69, 77] | |
| GO:0080134 | Regulation of response to stress | TNIP2, PSMB4, MAP3K7, SIN3A, MID1, EIF2AK2, SNAI2 | Oxidative/Metabolic/Environmental Stress | UniProt [67], Puvadolpirod et al. 2000 [48], Li et al. 2011 [70], Yen et al. 2014 [90] | |
| GO:1901224 | Positive regulation of NIK/NF-kappaB signaling | RC3H1, MAP3K7, EIF2AK2 | Inflammation/Oxidative Stress | Evans et al. 2002 [46], Perkins et al. 2007 [55], Salminen et al. 2008 [87], Tam et al. 2012 [88] | |
| GO:0031098 | Stress-activated protein kinase signaling cascade | MAP3K7, MID1, EIF2AK2 | Oxidative/Environmental Stress | Li et al. 2011 [70], Yen et al. 2014 [90] | |
| GO:0006915 | Apoptotic process | TNIP2, SUDS3, ROBO1, ZFAND6, DNAJC5, MAP3K7, LCMT1, SIN3A, GRK5, EIF2AK2, SHQ1, CHL1, COMP, LFABP, SNAI2 | Oxidative/Environmental Stress | Galvez et al. 2001 [47] | |
| GO:0032874 | Positive regulation of stress-activated MAPK cascade | MAP3K7, MID1, EIF2AK2 | Oxidative/Environmental Stress | UniProt [67], Jenuth 2000 [69, 77] | |
| GO:0006950 | Response to stress | PDIA5, CADPS2, TNIP2, PSMB4, MAP3K7, UBE2T, SIN3A, EIF2AK2, MMP7, CXCR4, MID1, PAX-7, LFABP, SMAD1, SNAI2 | Oxidative/Environmental Stress | UniProt [67], Puvadolpirod et al. 2000 [48], Li et al. 2011 [70], Yen et al. 2014 [90] | |
| GO:0009719 | Response to endogenous stimulus | MC4R, GRB10, DISP3, MAP3K7, SMAD1, STMN2, SNAI2 | Oxidative/Environmental Stress | Evans et al. 2002 [46] | |
| GO:0034976 | Response to endoplasmic reticulum stress | PDIA5, EIF2AK2 | Oxidative/Environmental Stress | Evans et al. 2002 [46], Zhang et al. 2008 [54] |
Table 4.
Gene ontology (GO) enrichment of statistically significant (FDR < 0.15) GO terms for the Rwandan ecotypes
| Population | GO ID | Gene ontology term | Genes under GO term | Related environmental stressor(s) | Reference |
|---|---|---|---|---|---|
| Rwandan Ecotypes | GO:0004697 | Protein kinase C activity | PRKCA, PRKD1 | Oxidative stress | UniProt [67], Jenuth 2000 [69, 77] |
| GO:0046649 | Lymphocyte activation | DLG1, GPAM, ZBTB16, PREX1, CD151 | Pathogen/Predator | Flint et al. 2011 [91] | |
| GO:0006954 | Inflammatory response | PSMB4, UACA | Pathogen/Predator | Evans et al. 2002 [46], Zhang et al. 2008 [54] | |
| GO:0006950 | Response to stress | SEL1L, PSMB4, SIRT6, NEK6, BCAS3, GPAM, LIG3, UACA, PRKD1, SNAI2, SMAD1 | Growth response to Environmental Stress | UniProt [67], Puvadolpirod et al. 2000 [48], Li et al. 2011 [70], Yen et al. 2014 [90] | |
| GO:0007249 | I-kappaB kinase/NF-kappaB signaling | UACA, ZDHHC17, NEK6, PRKD1 | Inflammation/Oxidative Stress | Evans et al. 2002 [46], Perkins et al. 2007 [55], Salminen et al. 2008 [87], Tam et al. 2012 [88] | |
| GO:0034599 | Cellular response to oxidative stress | UACA, PRKD1 | Oxidative/Environmental Stress | UniProt [67], Jenuth 2000 [69, 77] | |
| GO:0009719 | Response to endogenous stimulus | HPGD, GRB10, CITED3, GABRB3, BCAS3, SMAD1, STMN2, SNAI2 | Oxidative/Metabolic stress | Evans et al. 2002 [46] | |
| GO:0048010 | Vascular endothelial growth factor receptor signaling pathway | GRB10, PRKD1 | Oxidative/Metabolic Stress | Koch et al. 2012 [57] | |
| GO:0043281 | Regulation of cysteine-type endopeptidase activity involved in apoptotic process | DLC1, RFFL, UACA | Metabolic stress | Galvez et al. 2001 [47] | |
| GO:0065008 | Regulation of biological quality | DLC1, PRKCA, RAP1GDS1, DLG1, GPAM, FRMPD4, GRM8, CRTC1, NPY, PTPN3, STX12, SMAD1 | Growth response to Environmental Stress | UniProt [67], Jenuth 2000 [69, 77] | |
| GO:0006915 | Apoptotic process | ZBTB16, DLC1, RFFL, SUDS3, DNAJC5, SHQ1, GPAM, UACA, GABRB3, UTP11L, SNAI2 | Metabolic stress | Galvez et al. 2001 [47] |
Table 5.
Gene ontology (GO) enrichment of statistically significant (FDR < 0.15) GO terms for the Kuroilers
| Population | GO ID | Gene ontology term | Genes under GO term | Related environmental stressor(s) | Reference |
|---|---|---|---|---|---|
| Kuroilers | GO:0060584 | Regulation of prostaglandin-endoperoxide synthase activity | EDN2, EDN1 | Oxidative stress/Pathogen | Lee et al. 2007 [89] |
| GO:0007631 | Feeding behavior | HCRTR2, HTR1B, GRIN1, NPY1R, ASIP | Nutrient stress | UniProt [67], Jenuth 2000 [69, 77] | |
| GO:0032693 | Negative regulation of interleukin-10 production | TRIB2, IL-12B | Pathogen | UniProt [67], Jenuth 2000 [69, 77] | |
| GO:0051341 | Regulation of oxidoreductase activity | ACVR2A, EDN2, EDN1, SNCA | Oxidative stress/Pathogen | UniProt [67], Jenuth 2000 [69, 77], Lee et al. 2007 [89] | |
| GO:0007618 | Mating | ACVR2A, SERPINE2, GRIN1 | Survival Behavior | UniProt [67], Jenuth 2000 [69, 77] | |
| GO:0016627 | Oxidoreductase activity, acting on the CH-CH group of donors | IVD, CPOX, ACAD9, DHCR24 | Oxidative/Environmental Stress | UniProt [67], Jenuth 2000 [69, 77], Lee et al. 2007 [89] | |
| GO:0006950 | Response to stress | ENTPD1, ETFDH, WNT5A CHID1, RBBP5, NEDD4, NSMCE2, GLP1R, STX2, LEAP2, RAB23, DEAF1, TNFSF11, PPP2R5C, DRD4, ZC3HAV1, CDH8, PPAP2B, GTF2H5, MCPH1, EDN1, FAM175B, GCLM, JAK1, UBXN2A, MID1, BATF, CPEB2, IL12B, PIK3AP1, SOX2, SNCA, CITED2, CHAC1, DTL, HIPK3, NOS1, STT3B, PDIA4, SBNO2, NUAK1, CDC7, SERPINE2, ULK1, MASP2, WFS1, DHCR24, PRKD1 | Oxidative/Metabolic/Environmental Stress | UniProt [67], Puvadolpirod et al. 2000 [48], Li et al. 2011 [70], Yen et al. 2014 [90] | |
| GO:0080134 | Regulation of response to stress | WNT5A, CHID1, PIK3AP1, IL12B, STX2, SNCA, HIPK3, TNFSF11, ZC3HAV1, PPAP2B, SBNO2, NUAK1, MCPH1, EDN1, SERPINE2, ULK1, WFS1, MID1 | Oxidative/Metabolic/Environmental Stress | UniProt [67], Puvadolpirod et al. 2000 [48], Li et al., 2011, Yen et al., 2014 | |
| GO:0033554 | Cellular response to stress | WNT5A, RBBP5, BATF, CPEB2, NEDD4, NSMCE2, SNCA, CHAC1, CITED2, DTL, HIPK3, RAB23, TNFSF11, PPP2R5C, PDIA4, GTF2H5, NUAK1, FAM175B, EDN1, CDC7, ULK1, WFS1, UBXN2A, MID1, PRKD1 | Oxidative/Metabolic/Environmental Stress | UniProt [67], Puvadolpirod et al. 2000 [48] | |
| GO:0045859 | Regulation of protein kinase activity | TNFSF11, DRD4, WNT5A, TGFBR2, RAPGEF2, PDGF, PIK3CA, KIAA1199, LATS2, IL12B, EDN1, PAQR3, SNCA, GTPBP4, HIPK3 | Oxidative/Metabolic/Environmental Stress | UniProt [67], Jenuth 2000 [69, 77] | |
| GO:0003100 | Regulation of systemic arterial blood pressure by endothelin | EDN2, EDN1 | Oxidative/Metabolic Stress | Endemann et al. 2004 [20], Gomez et al. 2007 [21] | |
| GO:0048010 | Vascular endothelial growth factor receptor signaling pathway | FIGF, NEDD4, GRB10, PRKD1 | Oxidative/Metabolic Stress | Koch et al., 2012 [57] | |
| GO:0004674 | Protein serine/threonine kinase activity | CDC42BPB, DCLK1, TGFBR3, TGFBR2, PRKCA, LATS2, ACVR2A, NUAK1, CDC7, ULK1, CDC2L1, HIPK3, VRK1, PRKD1 | Oxidative/Metabolic/Environmental Stress | UniProt [67], Evans et al. 2002 [46], Perkins et al. 2007 [55], Salminen et al. 2008 [87], Tam et al., 2012 [88] | |
| GO:0009628 | Response to abiotic stimulus | GPR65, GRIN1, CASP8, IL12B, CPEB2, SOX2, NEDD4, TNFRSF8, CITED2, DTL, ER81, NOS1, DEAF1, CNGA2, CDH8, MME, GTF2H5, HR1B, SERPINE2, EDN1, FAM175B | Oxidative/Metabolic/Environmental Stress | Mashaly et al. 2004 [92], Mujahid et al. 2005 [16] | |
| GO:0004672 | Protein kinase activity | CDC42BPB, DCLK1, PRKCA, HIPK3, VRK1, ROR1, TRIB2, TGFBR3, TGFBR2, LATS2, ACVR2A, NUAK1, CDC7, ULK1, CDC2L1, PEAK1, JAK1, RPS6KC1, PRKD1 | Oxidative/Metabolic/Environmental Stress | UniProt [67], Jenuth 2000 [69, 77] |
Discussion
Population structure and admixture of populations indicate mixed genetic backgrounds
The observed population structure could partially be a result of using IBS values, which are based on the likelihood that identical alleles inherited by two individuals came from the same parent. This can lead to some uncertainty due to not having pedigree information, but it still allowed us to capture information on the population structure [44]. The amount of overlap seen across ecotype and populations may stem from flock management that allows unrestricted inter-mating of chickens from different genetic backgrounds. The admixture may be due to individuals having ancestors in multiple source groups that contribute to the shared ancestry. Another contributor to this admixture may be related to the movement of trade and selection parameters used by farmers to purchase new birds at market.
Possible factors contributing to signatures of selection
There are multiple environmental challenges that could lead to the occurrence of the discovered selection signals in the three populations. The geographic location of Uganda and Rwanda is near the equator and in tropical climates (Additional file 5: Figure S5). This, coupled with smallholder farm practices, places the studied birds in situations in which they would likely contact multiple environmental stressors that may affect their fitness. This would include challenges to their immune system from pathogens in the absence of vaccination. Also the equatorial locations of the samplings were exposed to high ultraviolet rays. The birds may also have experienced nutritive stress brought on by sub-optimal food in an environment in which birds must forage for food. The birds used in this study would have adapted to the environment over the years, likely through changes in allele frequency of a beneficial or detrimental allele.
Evidence of common signatures of selection across ecotypes for stress response
In livestock species, as well as humans and mice, environmental stressors such as pathogens or non-optimal temperatures can create cellular oxidative stress by generating reactive oxygen and nitrogen species effecting calcium signaling, apoptosis, vascular plasticity, growth and immune functions [16, 22, 45–50]. The chickens in our study displayed evidence of population similarities in how selection may have affected their response to environmental stress. Signatures are related to genes and signaling pathways involved in the reduction of ROS through utilization of Ca2+, lipids, and phosphorylated kinases. If the mobilization of Ca2+ is part of a prolonged response to chronic stressors it is possible that birds that scavenge for food have to make trade-offs in Ca2+ usage. An eggshell can contain a concentration 1.99 M Ca2+ [51]. Signal transduction of Ca2+ stored in the endoplasmic reticulum can be identified by increased cytoplasmic Ca2+ concentrations of 300–500nM [52]. The Uganda population showed GO terms related to endoplasmic reticulum (ER) stress response where Ca2+ is stored. Despite being such a small amount (300–500 nM) this diversion of resources may hinder other functions dependent on calcium. This mobilization of calcium for stress tolerance could also reduce the ability of birds to produce structurally competent shells and limit egg production. Pathways affected by calcium were also upregulated in heat stressed broilers [53]. We also uncovered selective pressure on negative regulation of multiple kinases involved in stress responses that may function to reduce pro-inflammatory cytokines. This reduction of pro-inflammatory cytokines had a lowering effect on metabolism, reducing stress during an immune response [10, 46, 54–56]. The most marked effect of the selection was the enrichment of terms related to response to stress, especially those indicating selection for or against cellular, endoplasmic reticulum (ER), and oxidative stress [46, 54, 57]. Other processes that reached significance include cell-cell signaling (all populations), VEGF signaling, cytokine binding/production (Uganda and Rwanda), and inflammatory response (Rwanda) (Tables 3, 4 and 5). Selection on the genes and processes reported may be related to cyto-protective effects being used by chickens to tolerate heat as a general stress response, because it is a constant in their environment [58, 59].
Selection pressure identified by ROH analysis indicate similar biological processes between populations
The results from the ROH analysis indicate that the populations have experienced selective pressure on their genomes from the environment. Runs of homozygosity (ROH) are defined as long stretches of homozygous genotypes within a genome, thought to be the result of consanguinity and identity by descent (IBD) inheritance [44, 60–62]. It is possible that ROH carry genes or variants under positive selection or that smaller ROH are related to the hitchhiking effects of selective sweeps [60, 63]. Analysis of ROH has been used to uncover alleles detrimental to health in humans and livestock [61, 63–65]. However, it may be possible that the ROH shared between the three populations point to conservation of alleles necessary for survival. These may be alleles that provide protection from oxidative and other stressors due to the environment. The existence of statistically significant hits for calcium ion activity and apoptotic regulation may, respectively, be related to kinase activation and amino acid recycling in the birds as a means of maintaining energy to put toward redox functions. It is possible that the birds are diverting resources to autophagy to conserve resources to put into increasing calcium activity to lower metabolic temperatures. This may be possible since it has been shown that blood Ca2+ levels are inversely related to body temperature [66].
Genes putatively under selection from environmental challenges among and within ecotypes
Protein kinase C alpha (PRKCA) found on chromosome 18 appears as a strong signal in all three populations. The gene PRKCA is a calcium activated, phospholipid dependent kinase that is involved in gene expression, inflammation, prolactin secretion, as well as, the regulation of multiple cell processes including inflammation and wounding [67, 68] (Fig. 4). It is also upregulated in response to endothelial injury. In chicken, PRKCA carries out functions involved in gene expression and kinase activity, and is involved in prolactin secretion [67, 69]. The selective pressure on PRKCA may be related to the negative effect that a challenging environment has on overall body weight and feed efficiency [22, 70], which could indicate nutritive stress. In addition to its functions in livestock, PRKCA acts as a messenger to stimulate prolactin secretion [71], which in humans has been linked to psychosocial stress responses [72].
Protein kinase C alpha is also involved in the biological process of prolactin secretion (GO:0070459) possibly through protein-protein interactions with the prolactin receptor (PRLR). This is interesting due to recent studies showing evidence of PRLR’s connection to heat tolerance in livestock [73, 74]. The interaction between PRKCA and PRLR indicates that both genes may contribute to the birds’ response to environmental stressors. In humans, PRKCA regulates induction of NF-kappa-B inhibitor alpha (NFKBIA/IKBA), which mediates host defense and inflammation responses, as well as plays a role in the activation of MAPK signaling and cyclin-dependent kinase (CDK) complex formation. Previous studies have indicated a role of MAPK and NF-κB pathways in the regulation of heat [70]. Regulation of NF-κB may be part of the response to inflammation triggered by ROS. The possible reduction in metabolic heat by PRKCA may be related the protein-protein interactions it has with suppressor of cytokine signaling 1 (SOCS1) and suppressor of cytokine signaling 3 (SOCS3) (Fig. 5). Both SOCS1 and SOCS3 help to battle ROS and inflammation and may play a role in the cascade needed to restore lost homeostasis [53, 75, 76]. Other SOCS genes have also been shown to respond under heat stress, which is a possible indication of a link between heat stress tolerance and cytokines [53].
Fig. 5.
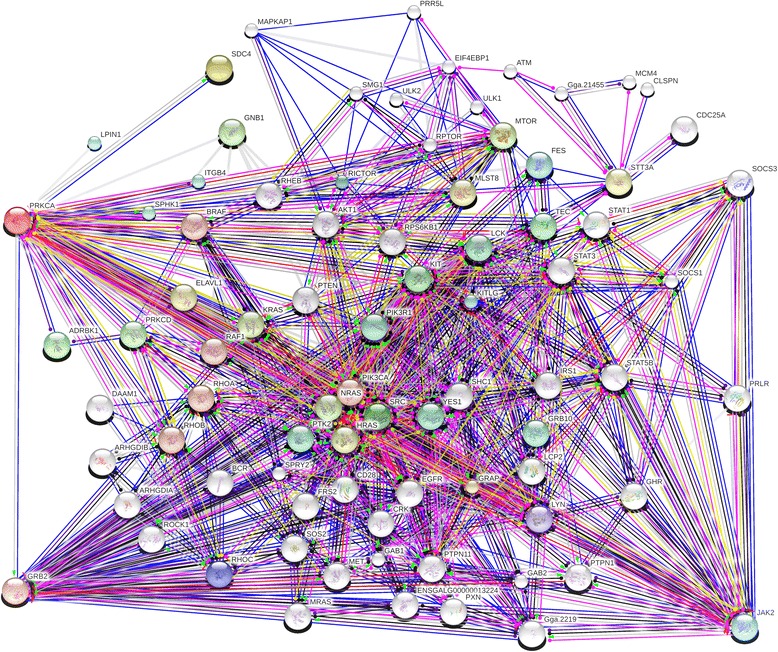
Network of predicted functional proteins associated with PRKCA created using STRING. Lines represent interactions that PRKCA has with predicted proteins in its network. Figure shows that PRKCA has protein-protein interactions with SOCS1 and SOCS3 that may be related to regulation of inflammation, while its interaction with PRLR may be related to stress pathways
The other kinase, cyclin-dependent kinase inhibitor 3 (CDKN3) found on chromosome 5 only appears as a strong signal in the Ugandan ecotypes. As a dual specific protein phosphatase, CDKN3 functions to inhibit cell cycle processes by blocking kinases targeting cyclin-dependent kinase 2 (CDK2), which causes cell cycle arrest and apoptosis in response to stress signals. The CDK2 gene is also part of the FoxO signaling pathway, which regulates transcription factors involved in gene activation of physiological cellular processes such as apoptosis, glucose, oxidative and nutrient stimuli stress resistance. Another function of CDKN3 is its ability to associate with cyclin C for activation and regulate different cell cycle events [67, 69, 77]. The selection pressure on CDKN3 in the Ugandan ecotypes may be related to its part in cell cycle regulation and ability to interact with phosphotyrosine or phosphoserine residues [67]. It functions to inactivate CDK2 which is part of a pathway engaged in DNA damage/telomere stress induced senescence, cell cycle arrest, and apoptosis in response to stress signals. To prevent senescence caused by oxidative stress, CDKN3 may function to inhibit CDK2. Studies have shown that oxidative stress can trigger activation of Protein Tyrosine Kinases (PTKs), which leads to the release of intracellular calcium in in vitro chicken cells and rat macrophages [49, 50]. The gene CDKN3 has also been shown to be considerably down regulated in mice undergoing oxidative stress from simulated environmental exposures [78].
In addition to the strong signal on chromosome 18, the Rwandan ecotypes also had a strong signal on chromosome 4 variants connected to the Rho GTPase activating protein, DLC1. This gene has functions involved in lipid binding and Rho GTPase activator activity and is also involved in negative regulation of stress fiber assembly. The gene DLC1 is also involved in the activation of cysteine-type endopeptidase activity involved in the apoptotic process, as well as positive regulation of the execution phase of apoptosis [67, 69, 77, 79]. For the Rwandan ecotypes, DLC1 may be involved in a population-specific mechanism of increasing cell migration and expanding cellular redox and lipid binding capabilities [67, 79, 80].
The gene with the strongest signal in the Kuroilers, ADM, is a hypotensive peptide that functions in hormone activity and in chicken is part of the biological processes of vasculogenesis (GO:0001570), positive regulation of angiogenesis (GO:0045766), and negative regulation of vasoconstriction (GO:0045906) [67, 69, 77, 81]. It also protects endothelial cells from cardiac stress [21]. In humans it is a vasodilator with functions that regulate fluid and electrolyte homeostasis and also has a role in the biological processes of response to cold (GO:0009409), hypoxia (GO:0001666), starvation (GO:0042594), LPS (GO:0032496), and wounding (GO:0009611) [45, 67, 82]. Other studies have shown ADM to also have anti-microbial, anti-apoptosis, and antioxidant functions [45, 67, 82, 83]. In the chicken, ADM has strong vascular modulation and functions as an antioxidant. Oxidative stress increases expression of inflammatory responses by endothelial cells [45, 68, 82] and ADM is shown to protect cardiovascular cells from oxidative damage [82, 83]. The selective pressure on this gene seen in the Kuroilers does not occur in the African ecotypes and may represent a result of artificial selection for stress tolerance in the Kuroilers during their development in India. The gene GCLM in chickens is part of the response to oxidative stress (GO:0006979), apoptotic mitochondrial changes (GO:0008637), and negative regulation of extrinsic apoptotic signaling pathway (GO:2001237). In humans, GCLM is also part of the response to nitrosative stress (GO:0051409) [67, 69, 77]. Nitrosative stress is caused by the formation of reactive nitrogen species (RNS) from cellular nitric oxide (NO) or it reacting with oxidative stress molecules to inflict cellular and vascular damage [84]. Other genes of interest that were in statistically significant regions of the genome based on the iHS values included DnaJ (Hsp40) homolog, subfamily C, member 5 (DNAJC5) and Collagen alpha-2(VI) chain (COL6A2). The heat shock protein DNAJC5 appeared in all 3 populations and functions as a molecular chaperone and a negative regulator of neuron apoptosis. It is also part of the GABA synthesis, release, re-uptake and degradation pathway and the protein processing in endoplasmic reticulum pathway. Collagen alpha-2(VI) chain (COL6A2) only appeared in the Kuroiler samples and was the only stop-gain (high impact) variant to reach statistical significance based on the iHS analysis. It is involved in focal adhesion and is part of the collagen biosynthesis and modifying enzymes, collagen degradation, and integrin cell surface interaction pathways [67, 69].
Pairwise comparisons of populations reinforced selection toward genes and functions related to oxidative stress
The comparison between the Ugandan and Rwandan ecotypes pointed selective pressure on gamma-aminobutyric acid A receptor, gamma 2 (GABRG2), an inhibitory neurotransmitter in vertebrates that mediates neuronal inhibition by binding to GABA receptors and opening an integral chloride channel. This is consistent with the results we observed in DNAJC5. Gamma-aminobutyric acid A receptor, gamma 2 (GABRG2) is also involved in the response to hypoxia in chickens [67, 69, 77]. The gene teneurin transmembrane protein 2 (TENM2) also plays a part in neural development and regulation of proper nervous system connections and carries out calcium-mediated signaling using intracellular calcium sources, hemophilic and heterophilic cell-cell adhesion [67, 69, 77]. The calcium functions of TENM2 may help to stimulate the necessary calcium release to activate PRKCA into activation when stressors appear. On chromosome 11, beta, beta-carotene 15,15’-monooxygenase (BCMO1), functions as a mono-oxygenase activator involved in cellular redox reactions as scavengers of oxygen radicals for photo-protection [67, 69, 77]. BCMO1 might experience selection related to protection from uv-induced DNA damage related to the generation of oxidative stress. When Ugandan and Rwandan ecotypes were compared to Kuroilers, the genes with the strongest signals for selection were the same but had different markers for the same gene regions in each comparison. Olfactomedin 4 (OLFM4) functions as a negative apoptotic factor and an extracellular matrix glycoprotein involved in cell adhesion. In chickens, it negatively regulates I-kappaB kinase/NF-kappaB signaling and the immune response. The between-population results for GO enrichment also reinforce the GO term results seen within population with slightly more emphasis on signaling, activation, and transport functions.
Unique and shared features under selection in Kuroilers compared to native African ecotypes
Kuroilers showed a great deal of overlap with the African ecotypes in biological processes that reduce the effects of oxidative and metabolic stress. Unique to the Kuroilers were selection on prostaglandin-endoperoxide synthase activity, a target of NF-κB and negative regulation of interleukin-10 (IL-10) production. Both are a part of reduction of oxidative stress and have anti-inflammatory effects. The differences in the guided selection of the Kuroilers can be seen in the many growth and behavior related GO terms that are enriched within the Kuroiler population and not observed in the African ecotypes. In Kuroilers, the GO enrichment results from the iHS analysis have larger gene lists, which may signify that under artificial selective breeding for stress tolerance, larger genomic areas or QTL regions may have been selected upon. This may be the opposite of natural selection in the African ecotypes, which may be focused on smaller regions of the genome. This is reflected in the genes annotated to similar terms seen in the Ugandan and Rwandan populations related to stress, inflammatory responses, and apoptosis. The length of the natural selection experienced by the African populations may have affected this also. This would have led to more recombination events, leading to smaller genomic regions under selection than what was identified in the Kuroilers, which is a recently developed breed. It appears that all three populations have found ways through natural and artificial selection to tolerate environmental stressors. What this overlap in GO terms also shows is that both the artificial selection for stress tolerance (Kuroilers) and natural selection (Ugandan and Rwandan ecotypes) share a biological link. Previous studies showed evidence that exposure to one stressor, such as heat, can lead to protection from other stressors through cyto-protective memory present in the activation of protective signaling [58, 59, 85]. It is possible that the underlying purpose of the selection seen in the populations are involved in histone and transcriptional modifications leading to cross-tolerance brought about by numerous small adaptions to a challenging environment. The regulation of these types of signaling processes (i.e. MAPK, NF-κB) and the initiation of regulatory kinases and genes (PRKCA, CDKN3, ADM, OFLM4) that assist in activating processes that protect cells from oxidative damage. Selective pressure on PRKCA also indicates the presence of prolactin signaling, shown to be a heat stress related signaling system in cattle. There is also evidence of selective pressure for protection of cardiovascular health under metabolic stress. In the sampling populations of Uganda and Rwanda there was some indication of adaption to environmental challenges from heat present in the some of the observable phenotypes of the birds. Heat stress related phenotypes such as frizzled and Naked Neck were observed within the sampling populations.
Conclusion
The three populations displayed evidence of stratification, which indicated admixture among the populations and especially among the ecotypes within a country. The strong selection signal in all populations at PRKCA, as well as, the population-specific selection signals for CDKN3, DLC1, and ADM, strongly indicates that these populations have developed means to maintain cellular homeostasis despite the presence of oxidative and metabolic stress. Our results indicate that the birds may use calcium-mediated responses to counteract environmentally generated oxidative stress. Results from the pairwise comparison, along with the over-representation of GO terms related to stress responses, also support this notion. The populations shared multiple statistically significant GO terms and genes related to selection pressure on kinases and calcium activity. Overall, this evidence of selective pressure on genes related to kinases, calcium activity, and oxidative stress responses provides a window through which to discern mechanisms used by chickens to tolerate the effects of a challenging environment.
Acknowledgements
Funding for project provided by USDA-NIFA-AFRI Climate Change Award #2011-67003-30228. DSF received a USDA National Needs Fellow Award #2011-38420-2005. The authors thank Donald Kugonza, Makerere University, and Dr. Gideon Nadiope, Uganda Program, Iowa State University, and their groups for assistance in collecting the samples; and Antonio Bagnato and lab group, University of Milan; Dave Burt and lab group, Roslin Institute; Eui-SooKim Iowa State University; Nick Serão, North Carolina State University; Henner Simianer and lab group, Georg-August-University Goettingen; Steffen Wiegend and Annette Wiegend, Friedrich Loeffler Institute, for helpful input into analyses, interpretation, and presentation of data.
Authors’ contributions
Design of experiment: MFR, CJS, CMA, MEP, JMR, SJL. Analysis tools: DSF, JEK. Data analysis: DSF, ADM. Drafting of manuscript: DSF, JMR, SJL. All authors read and approved the final manuscript.
Competing interests
The authors declare that there are no competing interests.
Ethics statement
Animal handling and sample collection was carried out following national guidelines and under the supervision of an qualified veterinarian and animal care specialist, Dr Gideon Nadiope.
Additional files
Table showing family, individual, and sex identification for study samples. (XLSX 37 kb)
Locations and relations of shared genes and statistically significant SNPs for iHS analysis. (DOCX 15 kb)
Locations and relations of unique genes and statistically significant SNPs for iHS analysis. (DOCX 16 kb)
Figure of Fst sliding window analysis showing selection pressure around PRKCA. (PDF 10746 kb)
Figure showing regions where Ugandan and Rwandan chickens were sampled. (PDF 4238 kb)
References
- 1.Badyaev AV. Stress-induced variation in evolution: from behavioural plasticity to genetic assimilation. Proc Biol Sci. 2005;272(1566):877–86. doi: 10.1098/rspb.2004.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elena SF, de Visser JA. Environmental stress and the effects of mutation. J Biol. 2003;2(2):12. doi: 10.1186/1475-4924-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann AA, Hercus MJ. Environmental stress as an evolutionary force. Bioscience. 2000;50(3):217–26. doi: 10.1641/0006-3568(2000)050[0217:ESAAEF]2.3.CO;2. [DOI] [Google Scholar]
- 4.Ahuja I, de Vos RC, Bones AM, Hall RD. Plant molecular stress responses face climate change. Trends Plant Sci. 2010;15(12):664–74. doi: 10.1016/j.tplants.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Nardone A, Ronchi B, Lacetera N, Ranieri MS, Bernabucci U. Effects of climate changes on animal production and sustainability of livestock systems. Livest Sci. 2010;130(1–3):57–69. doi: 10.1016/j.livsci.2010.02.011. [DOI] [Google Scholar]
- 6.Thornton PK, Ericksen PJ, Herrero M, Challinor AJ. Climate variability and vulnerability to climate change: a review. Glob Chang Biol. 2014;20(11):3313–28. doi: 10.1111/gcb.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koenraadt CJ, Githeko AK, Takken W. The effects of rainfall and evapotranspiration on the temporal dynamics of Anopheles gambiae s.s. and Anopheles arabiensis in a Kenyan village. Acta Trop. 2004;90(2):141–53. doi: 10.1016/j.actatropica.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 8.St-Pierre NR, Cobanov B, Schnitkey G. Economic losses from heat stress by US livestock industries. J Dairy Sci. 2003;86:E52–77. doi: 10.3168/jds.S0022-0302(03)74040-5. [DOI] [Google Scholar]
- 9.Renaudeau D, Collin A, Yahav S, de Basilio V, Gourdine JL, Collier RJ. Adaptation to hot climate and strategies to alleviate heat stress in livestock production. Animal. 2012;6(5):707–28. doi: 10.1017/S1751731111002448. [DOI] [PubMed] [Google Scholar]
- 10.Quinteiro-Filho WM, Ribeiro A, Ferraz-de-Paula V, Pinheiro ML, Sakai M, Sa LR, Ferreira AJ, Palermo-Neto J. Heat stress impairs performance parameters, induces intestinal injury, and decreases macrophage activity in broiler chickens. Poult Sci. 2010;89(9):1905–14. doi: 10.3382/ps.2010-00812. [DOI] [PubMed] [Google Scholar]
- 11.Ajakaiye JJ, Ayo JO, Ojo SA. Effects of heat stress on some blood parameters and egg production of Shika Brown layer chickens transported by road. Biol Res. 2010;43(2):183–9. doi: 10.4067/S0716-97602010000200006. [DOI] [PubMed] [Google Scholar]
- 12.Altan O, Pabuccuoglu A, Altan A, Konyalioglu S, Bayraktar H. Effect of heat stress on oxidative stress, lipid peroxidation and some stress parameters in broilers. Brit Poultry Sci. 2003;44(4):545–50. doi: 10.1080/00071660310001618334. [DOI] [PubMed] [Google Scholar]
- 13.Azad MAK, Kikusato M, Sudo S, Amo T, Toyomizu M. Time course of ROS production in skeletal muscle mitochondria from chronic heat-exposed broiler chicken. Comp Biochem Phys A. 2010;157(3):266–71. doi: 10.1016/j.cbpa.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Lin H, Decuypere E, Buyse J. Acute heat stress induces oxidative stress in broiler chickens. Comp Biochem Physiol A Mol Integr Physiol. 2006;144(1):11–7. doi: 10.1016/j.cbpa.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 15.Mujahid A, Akiba Y, Toyomizu M. Acute heat stress induces oxidative stress and decreases adaptation in young white leghorn cockerels by downregulation of avian uncoupling protein. Poult Sci. 2007;86(2):364–71. doi: 10.1093/ps/86.2.364. [DOI] [PubMed] [Google Scholar]
- 16.Mujahid A, Yoshiki Y, Akiba Y, Toyomizu M. Superoxide radical production in chicken skeletal muscle induced by acute heat stress. Poult Sci. 2005;84(2):307–14. doi: 10.1093/ps/84.2.307. [DOI] [PubMed] [Google Scholar]
- 17.Pamok S, Aengwanich W, Komutrin T. Adaptation to oxidative stress and impact of chronic oxidative stress on immunity in heat-stressed broilers. J Therm Biol. 2009;34(7):353–7. doi: 10.1016/j.jtherbio.2009.06.003. [DOI] [Google Scholar]
- 18.Allen RG, Tresini M. Oxidative stress and gene regulation. Free Radic Biol Med. 2000;28(3):463–99. doi: 10.1016/S0891-5849(99)00242-7. [DOI] [PubMed] [Google Scholar]
- 19.Covarrubias L, Hernandez-Garcia D, Schnabel D, Salas-Vidal E, Castro-Obregon S. Function of reactive oxygen species during animal development: passive or active? Dev Biol. 2008;320(1):1–11. doi: 10.1016/j.ydbio.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 20.Endemann DH, Schiffrin EL. Endothelial dysfunction. J Am Soc Nephrol. 2004;15(8):1983–92. doi: 10.1097/01.ASN.0000132474.50966.DA. [DOI] [PubMed] [Google Scholar]
- 21.Gomez AP, Moreno MJ, Iglesias A, Coral PX, Hernandez A. Endothelin 1, its endothelin type A receptor, connective tissue growth factor, platelet-derived growth factor, and adrenomedullin expression in lungs of pulmonary hypertensive and nonhypertensive chickens. Poult Sci. 2007;86(5):909–16. doi: 10.1093/ps/86.5.909. [DOI] [PubMed] [Google Scholar]
- 22.Pan JQ, Tan X, Li JC, Sun WD, Wang XL. Effects of early feed restriction and cold temperature on lipid peroxidation, pulmonary vascular remodelling and ascites morbidity in broilers under normal and cold temperature. Br Poult Sci. 2005;46(3):374–81. doi: 10.1080/00071660500098152. [DOI] [PubMed] [Google Scholar]
- 23.Qanbari S, Seidel M, Strom TM, Mayer KF, Preisinger R, Simianer H. Parallel selection revealed by population sequencing in chicken. Genome Biol Evol. 2015;7(12):3299–306. doi: 10.1093/gbe/evv222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qanbari S, Strom TM, Haberer G, Weigend S, Gheyas AA, Turner F, Burt DW, Preisinger R, Gianola D, Simianer H. A high resolution genome-wide scan for significant selective sweeps: an application to pooled sequence data in laying chickens. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0049525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubin CJ, Zody MC, Eriksson J, Meadows JR, Sherwood E, Webster MT, Jiang L, Ingman M, Sharpe T, Ka S, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464(7288):587–91. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- 26.Fan WL, Ng CS, Chen CF, Lu MY, Chen YH, Liu CJ, Wu SM, Chen CK, Chen JJ, Mao CT, et al. Genome-wide patterns of genetic variation in two domestic chickens. Genome Biol Evol. 2013;5(7):1376–92. doi: 10.1093/gbe/evt097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elferink MG, Megens HJ, Vereijken A, Hu X, Crooijmans RP, Groenen MA. Signatures of selection in the genomes of commercial and non-commercial chicken breeds. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0032720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stainton JJ, Haley CS, Charlesworth B, Kranis A, Watson K, Wiener P. Detecting signatures of selection in nine distinct lines of broiler chickens. Anim Genet. 2015;46(1):37–49. doi: 10.1111/age.12252. [DOI] [PubMed] [Google Scholar]
- 29.Gheyas AA, Boschiero C, Eory L, Ralph H, Kuo R, Woolliams JA, Burt DW. Functional classification of 15 million SNPs detected from diverse chicken populations. DNA Res. 2015;22(3):205–17. doi: 10.1093/dnares/dsv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang MS, Li Y, Peng MS, Zhong L, Wang ZJ, Li QY, Tu XL, Dong Y, Zhu CL, Wang L, et al. Genomic analyses reveal potential independent adaptation to high altitude in Tibetan chickens. Mol Biol Evol. 2015;32(7):1880–9. doi: 10.1093/molbev/msv071. [DOI] [PubMed] [Google Scholar]
- 31.Sharma J, Semambo D, Galukande E. Pan-african conference on the launch of the Kuroiler in Uganda. Uganda: National Animal Genetic Resources Centre And Data B; 2011. Kuroiler project overview. [Google Scholar]
- 32.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye C, Liu J, Ren F, Okafo N. Design of experiment and data analysis by JMP (SAS institute) in analytical method validation. J Pharm Biomed Anal. 2000;23(2–3):581–9. doi: 10.1016/S0731-7085(00)00335-6. [DOI] [PubMed] [Google Scholar]
- 35.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–90. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 36.Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27(4):592–3. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.SNP & Variation Suite (Version 8.x) [Software]. Bozeman, MT: Golden Helix, Inc. Available from http://www.goldenhelix.com. Accessed on 24 May 2015, and downloaded to local server for consistent use of this version.
- 38.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20(18):3710–5. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78(4):629–44. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gautier M, Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28(8):1176–7. doi: 10.1093/bioinformatics/bts115. [DOI] [PubMed] [Google Scholar]
- 43.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3) doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luan T, Yu X, Dolezal M, Bagnato A, Meuwissen TH. Genomic prediction based on runs of homozygosity. Genet Sel Evol. 2014;46:64. doi: 10.1186/s12711-014-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawai J, Ando K, Tojo A, Shimosawa T, Takahashi K, Onozato ML, Yamasaki M, Ogita T, Nakaoka T, Fujita T. Endogenous adrenomedullin protects against vascular response to injury in mice. Circulation. 2004;109(9):1147–53. doi: 10.1161/01.CIR.0000117231.40057.6D. [DOI] [PubMed] [Google Scholar]
- 46.Evans JL, Goldfine ID, Maddux BA, Grodsky GM. Oxidative stress and stress-activated signaling pathways: a unifying hypothesis of type 2 diabetes. Endocr Rev. 2002;23(5):599–622. doi: 10.1210/er.2001-0039. [DOI] [PubMed] [Google Scholar]
- 47.Galvez A, Morales MP, Eltit JM, Ocaranza P, Carrasco L, Campos X, Sapag-Hagar M, Diaz-Araya G, Lavandero S. A rapid and strong apoptotic process is triggered by hyperosmotic stress in cultured rat cardiac myocytes. Cell Tissue Res. 2001;304(2):279–85. doi: 10.1007/s004410100358. [DOI] [PubMed] [Google Scholar]
- 48.Puvadolpirod S, Thaxton JP. Model of physiological stress in chickens 1. Response parameters. Poult Sci. 2000;79(3):363–9. doi: 10.1093/ps/79.3.363. [DOI] [PubMed] [Google Scholar]
- 49.Kubista H, Hawkins T, Moss SE. Characterisation of calcium signalling in DT40 chicken B-cells. Bba Mol Cell Res. 1998;1448(2):299–310. doi: 10.1016/s0167-4889(98)00132-3. [DOI] [PubMed] [Google Scholar]
- 50.Murphy JK, Hoyal CR, Livingston FR, Forman HJ. Modulation of the Alveolar Macrophage Respiratory Burst by Hydroperoxides. Free Radical Bio Med. 1995;18(1):37–45. doi: 10.1016/0891-5849(94)00101-O. [DOI] [PubMed] [Google Scholar]
- 51.Rajan Choudhary SK, Swamiappan S. Bioactivity studies of calcium magnesium silicate prepared from eggshell waste by sol–gel combustion synthesis. J Asian Ceram Soc. 2015;3(2):173–7. doi: 10.1016/j.jascer.2015.01.002. [DOI] [Google Scholar]
- 52.Deng XX, Wang YJ, Zhou YD, Soboloff J, Gill DL. STIM and Orai: Dynamic intermembrane coupling to control cellular calcium signals. J Biol Chem. 2009;284(34):22501–5. doi: 10.1074/jbc.R109.018655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coble DJ, Fleming D, Persia ME, Ashwell CM, Rothschild MF, Schmidt CJ, Lamont SJ. RNA-seq analysis of broiler liver transcriptome reveals novel responses to high ambient temperature. BMC Genomics. 2014;15:1084. doi: 10.1186/1471-2164-15-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang KZ, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454(7203):455–62. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perkins ND. Integrating cell-signalling pathways with NF-[kappa]B and IKK function. Nat Rev Mol Cell Biol. 2007;8(1):49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 56.Welc SS, Clanton TL, Dineen SM, Leon LR. Heat stroke activates a stress-induced cytokine response in skeletal muscle. J Appl Physiol. 2013;115(8):1126–37. doi: 10.1152/japplphysiol.00636.2013. [DOI] [PubMed] [Google Scholar]
- 57.Koch S, Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb Perspect Med. 2012;2(7):a006502. doi: 10.1101/cshperspect.a006502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horowitz M. Heat acclimation and cross-tolerance against novel stressors: genomic-physiological linkage. Prog Brain Res. 2007;162:373–92. doi: 10.1016/S0079-6123(06)62018-9. [DOI] [PubMed] [Google Scholar]
- 59.Horowitz M, Eli-Berchoer L, Wapinski I, Friedman N, Kodesh E. Stress-related genomic responses during the course of heat acclimation and its association with ischemic-reperfusion cross-tolerance. J Appl Physiol. 2004;97(4):1496–507. doi: 10.1152/japplphysiol.00306.2004. [DOI] [PubMed] [Google Scholar]
- 60.Liu HM, Sorensen AC, Meuwissen THE, Berg P. Allele frequency changes due to hitch-hiking in genomic selection programs. Genet Sel Evol. 2014;46. [DOI] [PMC free article] [PubMed]
- 61.MacLeod IM, Larkin DM, Lewin HA, Hayes BJ, Goddard ME. Inferring demography from runs of homozygosity in whole-genome sequence, with correction for sequence errors. Mol Biol Evol. 2013;30(9):2209–23. doi: 10.1093/molbev/mst125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Purfield DC, Berry DP, McParland S, Bradley DG. Runs of homozygosity and population history in cattle. BMC Genet. 2012;13:70. doi: 10.1186/1471-2156-13-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bosse M, Megens HJ, Madsen O, Paudel Y, Frantz LA, Schook LB, Crooijmans RP, Groenen MA. Regions of homozygosity in the porcine genome: consequence of demography and the recombination landscape. PLoS Genet. 2012;8(11) doi: 10.1371/journal.pgen.1003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lencz T, Lambert C, DeRosse P, Burdick KE, Morgan TV, Kane JM, Kucherlapati R, Malhotra AK. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci U S A. 2007;104(50):19942–7. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim ES, Sonstegard T, Van Tassell CP, Wiggans G, Rothschild MF. The relationship between runs of homozygosity and inbreeding in Jersey cattle under selection. PLoS One. 2015;10(7):e0129967. doi: 10.1371/journal.pone.0129967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aitboulahsen A, Garlich JD, Edens FW. Calcium deficiency and food-deprivation improve the response of chickens to acute heat-stress. J Nutr. 1993;123(1):98–105. doi: 10.1093/jn/123.1.98. [DOI] [PubMed] [Google Scholar]
- 67.UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(Database issue):D204–12. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mei N, Guo L, Liu R, Fuscoe JC, Chen T. Gene expression changes induced by the tumorigenic pyrrolizidine alkaloid riddelliine in liver of Big Blue rats. BMC Bioinformatics. 2007;8(Suppl 7):S4. doi: 10.1186/1471-2105-8-S7-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jenuth JP. The NCBI. Publicly available tools and resources on the Web. Methods Mol Biol. 2000;132:301–12. doi: 10.1385/1-59259-192-2:301. [DOI] [PubMed] [Google Scholar]
- 70.Li C, Wang X, Wang G, Li N, Wu C. Expression analysis of global gene response to chronic heat exposure in broiler chickens (Gallus gallus) reveals new reactive genes. Poult Sci. 2011;90(5):1028–36. doi: 10.3382/ps.2010-01144. [DOI] [PubMed] [Google Scholar]
- 71.Sun SS, Elhalawani ME. Protein-Kinase-C mediates chicken vasoactive-intestinal-peptide stimulated prolactin secretion and gene-expression in turkey primary pituitary-cells. Gen Comp Endocrinol. 1995;99(3):289–97. doi: 10.1006/gcen.1995.1112. [DOI] [PubMed] [Google Scholar]
- 72.Lennartsson AK, Jonsdottir IH. Prolactin in response to acute psychosocial stress in healthy men and women. Psychoneuroendocrinology. 2011;36(10):1530–9. doi: 10.1016/j.psyneuen.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 73.Littlejohn MD, Henty KM, Tiplady K, Johnson T, Harland C, Lopdell T, Sherlock RG, Li W, Lukefahr SD, Shanks BC, et al. Functionally reciprocal mutations of the prolactin signalling pathway define hairy and slick cattle. Nat Commun. 2014;5:5861. doi: 10.1038/ncomms6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mariasegaram M, Chase CC, Jr, Chaparro JX, Olson TA, Brenneman RA, Niedz RP. The slick hair coat locus maps to chromosome 20 in Senepol-derived cattle. Anim Genet. 2007;38(1):54–9. doi: 10.1111/j.1365-2052.2007.01560.x. [DOI] [PubMed] [Google Scholar]
- 75.Ye S, Lowther S, Stambas J. Inhibition of reactive oxygen species production ameliorates inflammation induced by influenza A viruses via upregulation of SOCS1 and SOCS3. J Virol. 2015;89(5):2672–83. doi: 10.1128/JVI.03529-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoshimura A, Suzuki M, Sakaguchi R, Hanada T, Yasukawa H. SOCS, Inflammation, and Autoimmunity. Front Immunol. 2012;3:20. doi: 10.3389/fimmu.2012.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33(Database issue):D54–8. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu XZ, Robinson JF, Sidhu JS, Hong S, Faustman EM. A system-based comparison of gene expression reveals alterations in oxidative stress, disruption of ubiquitin--proteasome system and altered cell cycle regulation after exposure to cadmium and methylmercury in mouse embryonic fibroblast. Toxicol Sci. 2010;114(2):356–77. doi: 10.1093/toxsci/kfq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shih YP, Takada Y, Lo SH. Silencing of DLC1 upregulates PAI-1 expression and reduces migration in normal prostate cells. Mol Cancer Res. 2012;10(1):34–9. doi: 10.1158/1541-7786.MCR-11-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hobbs GA, Zhou B, Cox AD, Campbell SL. Rho GTPases, oxidation, and cell redox control. Small GTPases. 2014;5(2) doi: 10.4161/sgtp.28579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zudaire E, Cuesta N, Martinez A, Cuttitta F. Characterization of adrenomedullin in birds. Gen Comp Endocrinol. 2005;143(1):10–20. doi: 10.1016/j.ygcen.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 82.Yoshimoto T, Gochou N, Fukai N, Sugiyama T, Shichiri M, Hirata Y. Adrenomedullin inhibits angiotensin II-induced oxidative stress and gene expression in rat endothelial cells. Hypertens Res. 2005;28(2):165–72. doi: 10.1291/hypres.28.165. [DOI] [PubMed] [Google Scholar]
- 83.Rahman M, Nishiyama A, Guo P, Nagai Y, Zhang GX, Fujisawa Y, Fan YY, Kimura S, Hosomi N, Omori K, et al. Effects of adrenomedullin on cardiac oxidative stress and collagen accumulation in aldosterone-dependent malignant hypertensive rats. J Pharmacol Exp Ther. 2006;318(3):1323–9. doi: 10.1124/jpet.106.105106. [DOI] [PubMed] [Google Scholar]
- 84.Ridnour LA, Thomas DD, Mancardi D, Espey MG, Miranda KM, Paolocci N, Feelisch M, Fukuto J, Wink DA. The chemistry of nitrosative stress induced by nitric oxide and reactive nitrogen oxide species. Putting perspective on stressful biological situations. Biol Chem. 2004;385(1):1–10. doi: 10.1515/BC.2004.001. [DOI] [PubMed] [Google Scholar]
- 85.Tetievsky A, Horowitz M. Posttranslational modifications in histones underlie heat acclimation-mediated cytoprotective memory. J Appl Physiol. 2010;109(5):1552–61. doi: 10.1152/japplphysiol.00469.2010. [DOI] [PubMed] [Google Scholar]
- 86.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001;313(4):903–19. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 87.Salminen A, Paimela T, Suuronen T, Kaamiranta K. Innate immunity meets with cellular stress at the IKK complex: Regulation of the IKK complex by HSP70 and HSP90. Immunol Lett. 2008;117(1):9–15. doi: 10.1016/j.imlet.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 88.Tam AB, Mercado EL, Hoffmann A, Niwa M. ER stress activates NF-kappaB by integrating functions of basal IKK activity, IRE1 and PERK. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0045078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee BR, Kim H, Park TS, Moon S, Cho S, Park T, Lim JM, Han JY. A set of stage-specific gene transcripts identified in EK stage X and HH stage 3 chick embryos. BMC Dev Biol. 2007;7:60. doi: 10.1186/1471-213X-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yen SY, Tseng JK, Chuang SM, Chen SE, Ju JC. Expression and activation of mitogen-activated protein kinases in matured porcine oocytes under thermal stress. J Reprod Dev. 2014;60(5):388–394. doi: 10.1262/jrd.2014-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Flint MS, Budiu RA, Teng PN, Sun M, Stolz DB, Lang MG, Hood BL, Vlad AM, Conrads TP. Restraint stress and stress hormones significantly impact T lymphocyte migration and function through specific alterations of the actin cytoskeleton. Brain Behav Immun. 2011;25(6):1187–1196. doi: 10.1016/j.bbi.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 92.Mashaly MM, Hendricks GL, 3rd, Kalama MA, Gehad AE, Abbas AO, Patterson PH. Effect of heat stress on production parameters and immune responses of commercial laying hens. Poult Sci. 2004;83(6):889–894. doi: 10.1093/ps/83.6.889. [DOI] [PubMed] [Google Scholar]


