Abstract
Pectin acetylation influences the gelling ability of this important plant polysaccharide for the food industry. Plant apoplastic pectinacetylesterases (PAEs) play a key role in regulating the degree of pectin acetylation and modifying their expression thus represents one way to engineer plant polysaccharides for food applications. Identifying the major active enzymes within the PAE gene family will aid in our understanding of this biological phenomena as well as provide the tools for direct trait manipulation. Using comparative genomics we propose that there is a minimal set of 4 distinct PAEs in plants. Possible functional diversification of the PAE family in the grasses is also explored with the identification of 3 groups of PAE genes specific to grasses.
Keywords: acetylation, cell walls, comparative genomics, pectin, pectinacetylesterases, phylogeny
Abbreviations
- PAE
pectinacetylesterase.
Pectins are a structurally diverse set of plant cell wall polysaccharides that harbor the most complex glycan structures in nature.1 All pectins contain the charged sugar galacturonic acid and can contain many substitutents, one of which is an O-acetate. Pectins are known to be involved in many aspects of the plant cell biology including but not limited to coordinating wall architecture, stress signaling, cell to cell adhesion, wall porosity and cohesion.1-3 Pectic polymers are synthesized in the Golgi apparatus and secreted into the wall, where they can be further modified by apoplastic enzymes. Among these plant enzymes are pectinacetylesterases (PAEs), which can remove acetyl-substituents. While the biological role of these enzymes is not fully established, mutants lacking specific PAEs exhibit changes in cell wall rigidity and display plant developmental aberrations.4-6 Complete elucidation of PAE gene function and biochemistry has not yet been achieved, mostly due to genetic redundancy and difficulties in determining the exact substrate specificity. Due to its gelling and thickening properties pectins are extensively used by the food industry. The acetyl-esters on the pectic polymers can directly influence its gelling ability.7 Thus, acetyltransferases and esterases represent targets to engineer pectin structures for commercial applications.
One way to identify genes that influence wall structures is through comparative genomics and phylogenetic approaches. In a recent comparative genomics and phylogeny study uncharacterized wall hemicellulosic glycosyltransferases were identified.8 Using a similar phylogenetic approach Gille et al.9 were able to propose candidate genes likely to be involved in hemicellulose acetylation. These studies took advantage of a detailed hemicellulosic characterization in several plant species10 as well as the recent available genomic data.11
The pectinacetylesterase gene family in Arabidopsis includes 12 genes and is part of the carbohydrate esterase family 13.12 Due to the available resources for this model species, the characterization of the Arabidopsis PAEs will aid in uncovering specific function for each member of this gene family. Utilizing a reverse genetic approach recently 2 of these genes were characterized biochemically in Arabidospsis [PAE8 and PAE9;4] and shown to de-acetylate pectic fractions enriched for rhamnogalacturonan I in vitro. Null mutants of PAE8 and PAE9 produced rosette leaf walls with increased acetate content (+20%) and the double mutant showed not only an additive cell wall acetate phenotype (+37%) but also reduced inflorescence height.4 An ortholog of the Arabidopsis PAE8 from mung bean (Vigna unguiculata) was overexpressed in potato, resulting in decreased pectin acetylation, stiffer tubers and a stronger wall matrix.6 A poplar ortholog of the Arabidopsis PAE7, when overexpressed in tobacco, produced profound developmental alterations in pollen grains and floral structures.5 These results demonstrate the significance of the PAE genes for plant developmental processes. In an attempt to gain further insights into the structure and function of the PAE gene family, a phylogenetic and comparative genomic approach was taken here using Arabidopsis as a model system.
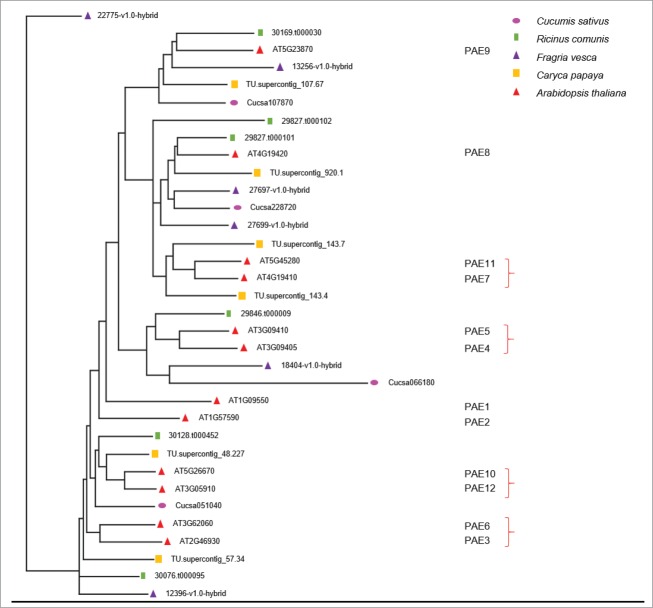
According to the Phytozome data base, a repository of plant genomes,11 the number of PAE genes currently varies from 1 in Physcomitrella patens to 22 in Panicum virgatum. On average plants have ˜10 PAE genes.11 To date 7 distinct acetyl substitutions have been decribed for pectic polymers [O-2 and O-3 of galacturonic acid in homogalacturonan and rhamnogalacturonan I13, 14; O-3 of rhamnose in rhamnogalacturonan I15; O-3 of Aceric acid and fucosyl residues of rhamnogalacturon II side chain16] suggesting that a set of at least 7 different genes would be required for the complete de-acetylesterification of pectins in plants, if PAEs are non-promiscuous and harbor specific, defined substrate specificities. However, comparative analysis show that at least 4 species (Ricinus comunis, Caryca papaya, Cucumis sativus and Fragria vesca) have 6 or less PAE genes in their genomes suggesting that some of these genes might be promiscuous and can remove O-acetyl substituents at multiple locations. To investigate what would be a sufficient set of PAEs in eudicots and identify these genes in Arabidopsis, a phylogenetic tree was constructed (Fig. 1).17 The tree shows that the Arabidopsis AtPAE4, 5, 8, 9, 10 and 12 genes are candidates for a minimal PAE set in eudicots based on the similarity to the C. sativus genes (Fig. 1). A larger tree containing 35 species from the Phytozome database (Fig. S1) also indicates AtPAE8 and AtPAE9 as being the closest orthologs to 2 of the 4 C. sativus genes. Supporting this finding is our recent description of distinct mutant phenotypes in knockouts of PAE8 and 9,4 which proved that these genes are functional and likely to part of a sufficient set of PAEs.
Figure 1.
Phylogenetic tree of Arabidopsis thaliana and species containing reduced numbers of PAE genes in their genomes. Arabidopsis PAEs numbers are indicated to the right. Red brackets indicate paralogous genes. Protein sequence alignments were performed using MUSCLE and Tree calculated by neighborjoining using BLOSUM62 by the Jalview 2.8.2 software package.17
Another observation is that species with reduced amounts of PAEs in their genome harbor orthologs of the Arabidopsis PAE8 and PAE9 genes (Fig. 1), however when analyzing a larger set of species from the Phytozome data base (Fig. S1), some plants have lost PAE8 (Capsella rubella, Zea mayz and Setaria italica) or PAE9 (Arabidopsis lyrata, Solanum tuberosum, Sroghum bicolor and Eucalyptus grandis), but not both genes simultaneously. This suggests that retaining at least one of these genes is important for plant fitness. This idea is consistent with the finding that in a pae8/pae9 double mutant plants exhibited shorter inflorescences,4 a trait that could significantly impact fitness in natural conditions.
The identification of a minimal set of PAEs using this phylogenetic approach enables the selection of reverse genetics strategies to uncover wall acetylation phenotypes by knocking out PAEs likely to be functional or producing double mutants from redundant genes. Based on this analysis we propose that a double knockout mutants of PAE10/12 and PAE4/5 is likely to result in a cell wall acetylation phenotype, due to its similarity to a C. sativus gene (Fig. 1). The remaining 2 pairs of paralogs in Arabidopsis [(PAE3/6 and PAE7/11); Fig. 1] might have undergone some form of neo functionalization and would possibly present phenotypes if double mutants were constructed.
When overexpressing all of the Arabidopsis PAE genes (for method see ref.4) in the Arabidopsis wild type background no alteration in wall O-acetylation level was observed with the exception of PAE2, whose constitutive expression leads to a ˜10% decrease in rosette leaf cell wall acetate content (Fig. 2A). This gene is closely related to the PAE10/12 paralogs (Fig. 1), which further supports the idea that these proteins could be active. When pectin extracts from PAE2 overexpression lines were measured for acetate content, 2 of the 3 lines tested showed ˜10% less acetate when compared to WT (Fig. 2B).
Figure 2.
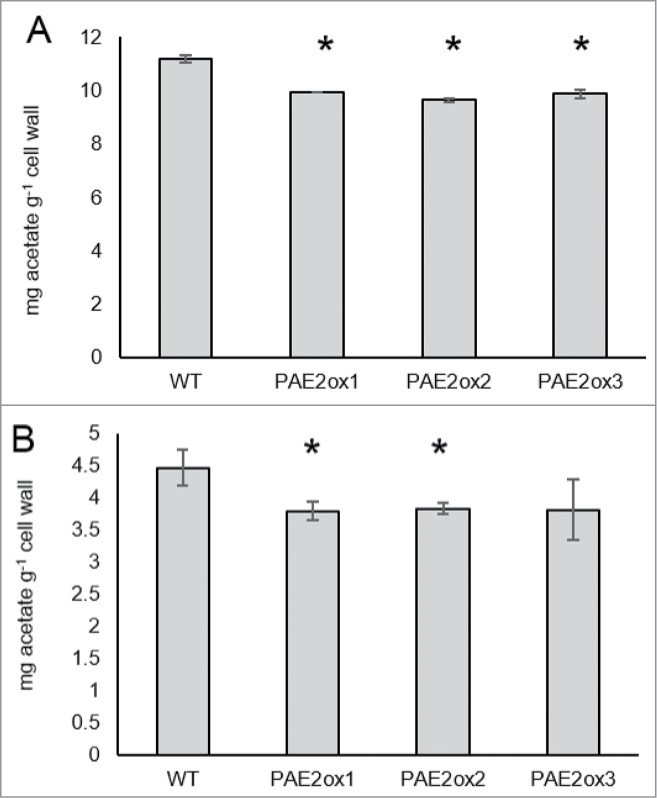
Acetate content of PAE2 overexpression lines cell walls (A) and pectin extracts (B). Measurements made on 35 day old rosette leaf material (for method detail see ref. 4). Asterisk indicates significant differences based on t test (P < 0.05; n ≥ 2).
Although there is mounting evidence suggesting that the PAE genes are acting primarily on pectins, the possibility that they may target other cell wall polymers cannot be discarded. In fact, an Arabidospsis PAE8 ortholog identified in Voodoo lily (Amorphophalus konjac) corm, which contains large quantities of acetylated glucomannans, is a likely candidate for a mannan acetylesterase.18 Most hemicellulosic polysaccharides are known to be acetylated 10 and no plant carbohydrate acetylesterase, aside from the PAEs, have been identified to date. The PAE family remains therefore a potential source for such activities against hemicelluloses thought to exist in plants.
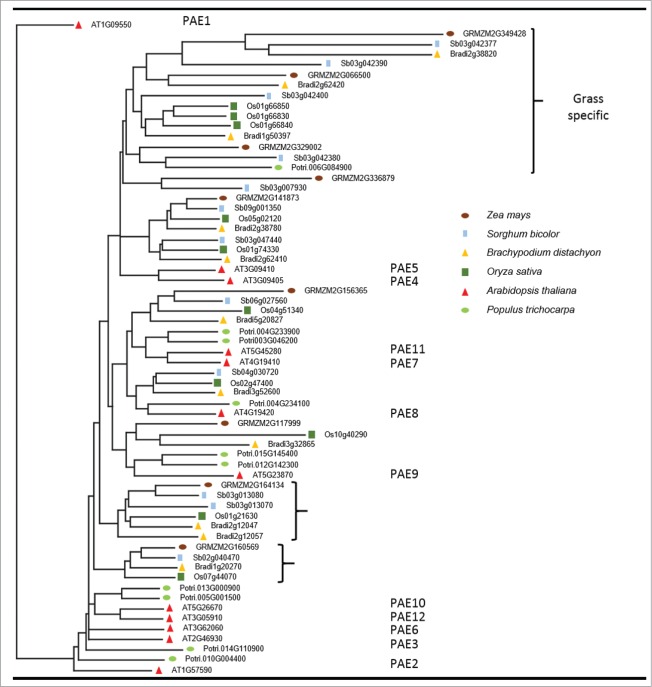
Since grasses contain very little pectin in their walls,19 a phylogenetic tree including Arabidopsis and poplar as well as the grasses Z. mays, S. bicolor, Brachypodium distachyon and Oryza sativa was constructed with the objective of investigating possible diverging branches of the PAE family (Fig. 3). Novel PAE branches might have acquired new functions and act on other polymers instead of pectins. At least 3 clades of genes were identified as grass specific (Fig. 3, top). The clade containing the most genes have long branches which suggest low levels of conservation consistent with the idea of neo functionalization of the PAE family in grasses. These grass specific PAEs could possibly be acting on hemicelluloses, which are highly abundant and highly O-acetylated in the grass walls.10
Figure 3.
Phylogenetic tree of Grass PAEs along with other reference species. Arabidopsis PAEs numbers are indicated to the right. Black brackets indicates clades exclusive to grasses. Protein sequence alignments were performed using MUSCLE and Tree calculated by neighborjoining using BLOSUM62 by the Jalview 2.8.2 software package.17
In conclusion, we have shown that by using comparative genomics new insights into the structure of the PAE gene family were obtained. A putative minimum set of PAEs in eudicots was proposed as well as the identification of gene expansion within the grasses, which could suggest neo functionalization within the PAE family.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was funded by the Energy Biosciences Institute at UC Berkeley.
References
- 1.Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A. Plant Cell Walls: From Chemistry to Biology. New York: Garland Science, 2011. [Google Scholar]
- 2.Atmodjo MA, Hao ZY, Mohnen D. Evolving views of pectin biosynthesis. Annu Rev Plant Biol 2013; 64:747-79; PMID:23451775; http://dx.doi.org/ 10.1146/annurev-arplant-042811-105534 [DOI] [PubMed] [Google Scholar]
- 3.Harholt J, Suttangkakul A, Scheller HV. Biosynthesis of pectin. Plant Physiol 2010; 153:384-95; PMID:20427466; http://dx.doi.org/ 10.1104/pp.110.156588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Souza A, Hull PA, Gille S, Pauly M. Identification and functional characterization of the distinct plant pectin esterases PAE8 and PAE9 and their deletion mutants. Planta 2014; 240:1123-38; PMID:25115560; http://dx.doi.org/ 10.1007/s00425-014-2139-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gou JY, Miller LM, Hou G, Yu XH, Chen XY, Liu CJ. Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. The Plant Cell 2012; 24:50-65; PMID:22247250; http://dx.doi.org/ 10.1105/tpc.111.092411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orfila C, Dal Degan F, Jorgensen B, Scheller HV, Ray PM, Ulvskov P. Expression of mung bean pectin acetyl esterase in potato tubers: effect on acetylation of cell wall polymers and tuber mechanical properties. Planta 2012; 236:185-96; PMID:22293853; http://dx.doi.org/ 10.1007/s00425-012-1596-z [DOI] [PubMed] [Google Scholar]
- 7.Ralet MC, Crepeau MJ, Buchholt HC, Thibault JF. Polyelectrolyte behaviour and calcium binding properties of sugar beet pectins differing in their degrees of methylation and acetylation. Biochem Eng J 2003; 16:191-201; http://dx.doi.org/ 10.1016/S1369-703X(03)00037-8 [DOI] [Google Scholar]
- 8.Schultink A, Cheng K, Park YB, Cosgrove DJ, Pauly M. The identification of two arabinosyltransferases from tomato reveals functional equivalency of xyloglucan side chain substituents. Plant Physiol 2013; 163:86-94; PMID:23893172; http://dx.doi.org/ 10.1104/pp.113.221788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gille S, de Souza A, Xiong G, Benz M, Cheng K, Schultink A, Reca IB, Pauly M. O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. The Plant Cell 2011; 23:4041-53; PMID:22086088; http://dx.doi.org/ 10.1105/tpc.111.091728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pauly M, Gille S, Liu L, Mansoori N, de Souza A, Schultink A, Xiong G. Hemicellulose biosynthesis. Planta 2013; 238:627-42; PMID:23801299; http://dx.doi.org/ 10.1007/s00425-013-1921-1 [DOI] [PubMed] [Google Scholar]
- 11.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al.. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 2012; 40:D1178-86; PMID:22110026; http://dx.doi.org/ 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 2014; 42:D490-D5; PMID:24270786; http://dx.doi.org/ 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishii T. O-acetylated oligosaccharides from pectins of potato tuber cell walls. Plant Physiol 1997; 113:1265-72; PMID:9112775; http://dx.doi.org/ 10.1104/pp.113.4.1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keenan MHJ, Belton PS, Matthew JA, Howson SJ. A C-13Nmr study of sugar-beet pectin. Carbohydr Res 1985; 138:168-70; http://dx.doi.org/ 10.1016/0008-6215(85)85236-8 [DOI] [Google Scholar]
- 15.Sengkhamparn N, Bakx EJ, Verhoef R, Schols HA, Sajjaanantakul T, Voragen AG. Okra pectin contains an unusual substitution of its rhamnosyl residues with acetyl and alpha-linked galactosyl groups. Carbohydr Res 2009; 344:1842-51; PMID:19195648; http://dx.doi.org/ 10.1016/j.carres.2008.11.022 [DOI] [PubMed] [Google Scholar]
- 16.Buffetto F, Ropartz D, Zhang XJ, Gilbert HJ, Guillon F, Ralet MC. Recovery and fine structure variability of RGII sub-domains in wine (Vitis vinifera Merlot). Ann Bot 2014; 114:1327-37; PMID:24908680; http://dx.doi.org/ 10.1093/aob/mcu097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009; 25:1189-91; PMID:19151095; http://dx.doi.org/ 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gille S, Cheng K, Skinner ME, Liepman AH, Wilkerson CG, Pauly M. Deep sequencing of voodoo lily (Amorphophallus konjac): an approach to identify relevant genes involved in the synthesis of the hemicellulose glucomannan. Planta 2011; 234:515-26; PMID:21538106; http://dx.doi.org/ 10.1007/s00425-011-1422-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J: Cell Molecular Biol 2008; 54:559-68; PMID:18476863; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03463.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.