Abstract
The inter-follicular epidermis regenerates from heterogeneous basal skin cell populations that divide at different rates. It has previously been presumed that infrequently dividing basal cells, label retaining cells (LRCs), are stem cells, while non-LRCs are short-lived progenitors. Here we employ the H2B-GFP pulse-chase system in adult mouse skin and find that epidermal LRCs and non-LRCs are molecularly distinct and can be differentiated by Dlx1CreER and Slc1a3CreER genetic marking, respectively. Long-term lineage tracing and mathematical modelling of H2B-GFP dilution data show that LRCs and non-LRCs constitute two distinct stem cell populations with different patterns of proliferation, differentiation, and upward cellular transport. During homeostasis, these populations are enriched in spatially distinct skin territories and can preferentially produce unique differentiated lineages. Upon wounding or selective killing, they can temporarily replenish each other’s territory. These two discrete inter-follicular stem cell populations are functionally interchangeable and intrinsically well adapted to thrive in distinct skin environments.
Introduction
A classical hierarchical model for adult tissue homeostasis suggests that infrequently dividing cells are long-lived stem cells (SCs) that generate rapidly dividing, short-lived progenitor cells1. Examples include the hair follicle (HF) and blood, where infrequently dividing cells, identified as H2B-GFP label retaining cells (LRCs), have unique long-term SC potential2–5. Mouse intestine studies suggest other LRC functions; e.g., committed secretory precursors6, independent SCs co-existing with frequently dividing SCs7–13, or reserve SCs specialized in injury repair14. Heterogeneity in SC potential can result from spatial positioning within niche environments or from cell-intrinsic differences15.
Although the inter-follicular epidermis (IFE) is an essential body barrier, its SCs are poorly understood16,17. Proliferative cells are located in the basal layer (BL) and express high levels of α6-integrin and keratin (K) 14/K518 (Fig. 1a). When differentiating, the basal cells move upwards to form the spinous layer (SL), granular layer (GL), and the outermost cornified layer (CL). The mouse epidermis turns over every 7–10 days19 and is one of the most rapidly regenerative tissues in the body.
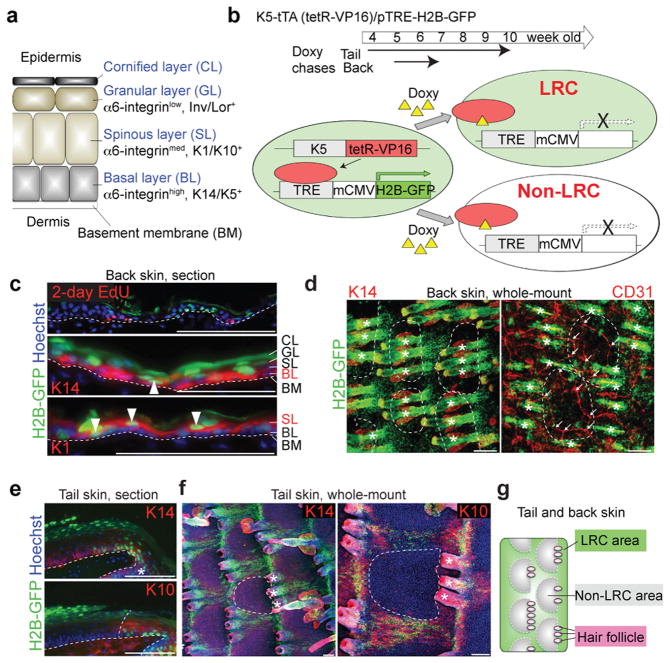
Figure 1. H2B-GFP LRCs and non-LRCs reside in distinct territories of the epidermis.
a, Structure of mouse inter-follicular epidermis and associated markers. b, Scheme of the strategy to detect infrequently dividing cells as H2B-GFP LRCs in adult skin of transgenic mice. c-i, Immunostaining of back and tail skin after H2B-GFP pulse-chase is shown in 10 μm tissue sections or whole mount using antibodies to differentiated markers, as indicated. CD31 is a vascular marker. Hoechst is a DNA-specific stain. The dashed line surrounds non-LRC areas (d,e,h,i) or represents epidermal-dermal junctions (c,f,g). Arrowheads indicate LRCs in the K14+ BL (middle) or K1+ SL (bottom) (c). (e) Arrows indicate branch points of blood vessels. Asterisks indicate HFs. Scale bars, 100 μm (c-i). j, Schematic view of the tail and back epidermis indicate LRC and non-LRC areas (territories) organization relative to hair follicles. Experiments are repeated twice with 2 mice for all representative images (b-i).
Early studies using pulse-chase experiments with labeled nucleotides20 or H2B-GFP21 and reconstitution assays in human22 revealed that BL cells divide at different rates and suggested a hierarchical stem/progenitor model in the epidermis. But this model was challenged when the behaviors of clones derived from AhCreER+ or Axin2CreER+ cells were analyzed by mathematical modelling23–25. This was taken to suggest that all basal cells comprise a single population of functionally identical progenitors. However, the stem/progenitor model was subsequently reinstated when K14CreER+ cells marked with low tamoxifen (K14CreER with low TM) showed distinct clonal behavior from InvCreER+ cells26; in this model the two populations were interdependent and the K14CreER with low TM SCs were the source of the InvCreER+ progenitors. Finally, a recent lineage analysis of K14CreER+ cells suggested a third model, which had two independent SC populations, although specific markers to confirm this were lacking27 (Supplementary Fig. 1a).
To distinguish among these models and to define the long-sought epidermal SCs, we identified genetic tools and characterized epidermal LRCs or non-LRCs behavior and lineages. We discovered two independent SC populations that comprise the bulk of the BL, that are spatially segregated and can preferentially differentiate to distinct lineages, and are able to partially interchange their functions in injury conditions.
Results
LRCs and non-LRCs reside in distinct territories
To visualize epidermal LRCs and non-LRCs in vivo, we used K5-tTA (tet-off)/pTRE-H2B-GFP mice21 in which doxycycline (doxy) turns off H2B-GFP mRNA expression (chase). Fluorescence is diluted 2-fold at each division, and so it is retained preferentially in infrequently dividing cells (LRCs) (Fig 1b). Skin sections from doxy-treated mice showed that LRCs cluster together, exist in both basal and suprabasal layers, and are inversely marked by short BrdU pulses (Fig. 1c, e; Supplementary Fig. 1c, d, f).
To view large epidermal areas and spatially relate LRC clusters to other skin structures, we performed whole-mount immunostaining28,29. This revealed spatially distinct distributions of epidermal LRCs and non-LRCs in both tail and back skin (Fig. 1d, f; Supplementary Fig. 1b, e, f). The LRC-dense regions surrounded circular regions predominantly made of non-LRCs. On average, three HFs flank each non-LRC region; this number varies somewhat regionally in the back, while it is exactly three in the tail (Fig. 1d, f; Supplementary Fig. 1g). The LRCs and non-LRC arrangements in both back and tail skin resembled the “interscale” and “scale” structures reported for tail skin27. Scales are circular inter-follicular regions with unique differentiation features: they retain nuclei in the CL, uniquely express K31, and lack K10 expression27. Therefore, the observation of strong H2B-GFP fluorescence in K10+ regions confirmed the presence of LRCs in interscale (Fig. 1e, f). Back skin does not display the scale differentiation27 but, importantly, the segregation of the LRC and non-LRC areas suggests previously unrecognized structural similarities between back and tail skin (Fig. 1g). Finally, analysis of back skin whole mounts showed that that blood vessel branch points predominate in the vicinity of the non-LRC areas (Fig. 1d; Supplementary Fig. 1h). This relationship between the 3D organization of blood vessel branch points and the non-LRC territories suggests that the LRCs and non-LRCs are non-randomly segregated and that the distribution of blood vessels and/or their associated cells may influence non-LRC behaviour (see Discussion). We conclude that epidermal LRCs and non-LRCs cluster in both basal and suprabasal layers and segregate in spatially distinct territories that are organized in structured arrays relative to skin landmarks such as HFs and blood vessels.
LRCs and non-LRCs are molecularly distinct
The distinct territorial localization of LRC and non-LRC populations suggests they are functionally and molecularly distinct. To determine their molecular profiles, we isolated LRCs and non-LRCs from quadruple-transgenic K5-tTA/pTRE-H2B-GFP/K14CreER/Rosa-tdTomato mouse back skin by fluorescence-activated cell sorting (FACS; Fig. 2a), and then assayed mRNA expression using Affymetrix microarrays. We used reduced TM doses that predominantly mark epidermis away from HFs30 (Fig. 2b, d), minimized residual HF contamination by using postnatal day (PD) 49-53 mice when HFs lack differentiated lineages31, and excluded CD34+ (a HFSC marker) cells32,33 (Fig. 2d; Supplementary Fig. 2a). TdTomato+/CD34− cells were separated on high, medium and low α6-integrin levels into fractions enriched in BL, SL, and GL epidermal layers, respectively (Fig. 2a–d). This fractionation was verified by quantitative reverse transcriptase PCR (qRT-PCR) for known epidermal layer markers (Supplementary Fig. 2b). The BL and SL fractions were further divided in H2B-GFP LRCs and non-LRCs (Fig. 2c, d).
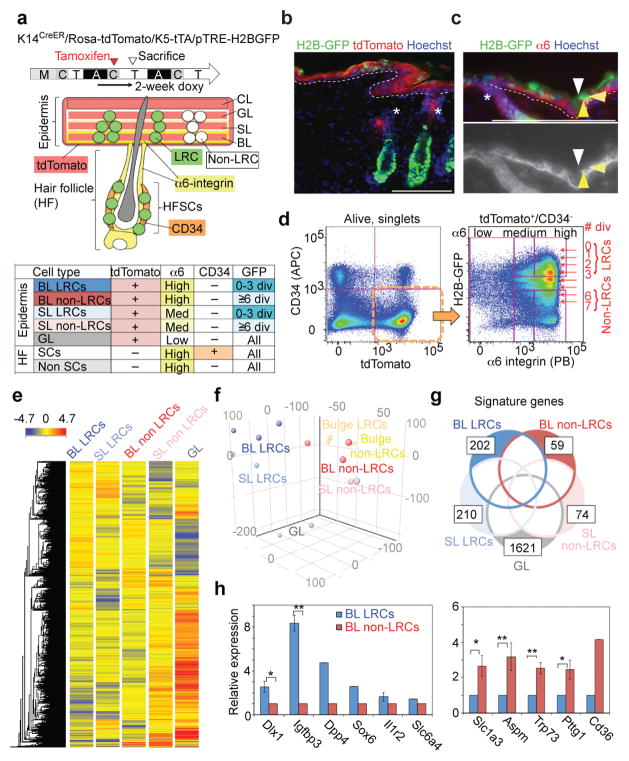
Figure 2. H2B-GFP LRCs and non-LRCs are molecularly distinct.
a, Experimental time-line and scheme for FACS-cell sorting. M, morphogenesis; A, anagen; T. telogen; C, catagen stages of hair cycle. b, c, Immunostaining of back skin from K5-tTA/pTRE-H2B-GFP/K14CreER/Rosa26-tdTomato quadruple-transgenic mice. Arrowheads in c and c′ indicate LRCs in the BL (yellow), SL (light yellow) and GL (white) with different levels of α6-integrin expression. The dashed line represents epidermal-dermal junctions. Asterisks indicate the upper hair follicle region (infundibulum). Scale bars, 100 μm. d, FACS scatter plot and sorting gates for isolating skin sub-populations subsequently analysed by microarray. e, Heat map and associated hierarchical clustering of microarray signals detected in all sorted populations indicated at the top. f, Principal component analysis of microarray signals from all the sorted populations. Each dot represents one sample derived from a different mouse (triplicate for BL LRCs and BL non-LRCs and duplicate for SL LRCs, SL non-LRCs, GL, bulge LRCs and bulge non-LRCs). g, Number of signature genes in each population and extent of overlap between the indicated populations. h, QRT-PCR for select BL LRC and non-LRC up-regulated genes. Error bars show s.e.m.; **: P<0.01; *: P<0.05. (Dlx1, n = 5 mice, P=0.01; Igfbp3, n = 3, P=0.0005; Il1r2, n = 4; Slc1a3, n = 5, P=0.04; Aspm, n = 4, P=0.001; Trp73, n = 3, P=0.008; Pttg1, n = 3, P=0.03). For Dpp4, Sox6, Slc6a4 and Cd36, the average of two mice is shown. Statistical analyses were performed using the two-tailed Student’s t-test wherever n ≥ 3. Experiments are repeated twice with 2 different mice for representative images (b, c) and 5 times with 10 mice for FACS (d). Microarray are repeated triplicate for BL LRCs and BL non-LRCs and duplicate for SL LRCs, SL non-LRCs, GL, bulge LRCs and bulge non-LRCs (e-g).
Microarray analysis generated molecular profiles and revealed putative LRC and non-LRC markers. Hierarchical clustering and principal component analysis showed that BL LRCs and non-LRCs were closely related but molecularly distinct (Fig. 2e, f). We identified 2,450 and 1,452 genes that were differentially expressed (≥2-fold) between BL LRCs vs non-LRCs (Supplementary Fig. 2c; Supplementary Table 1). After excluding genes that were also expressed in SL or GL fractions, 202 and 59 were identified as BL LRC or non-LRC signature genes, respectively (Fig. 2g; Supplementary Table 2). Several LRC- or non-LRC-enriched genes were confirmed by qRT-PCR (Fig. 2h).
We compared genes changed in BL LRCs vs non-LRCs with those differentially expressed in related quiescent vs proliferative populations, specifically, telogen vs anagen HFSCs34. This comparison showed that only 20–30% of the BL LRC/non-LRC gene expression differences could be related to differences in proliferation status (Supplementary Fig. 2d, e, top). Furthermore, only ~10–25% of these genes changed expression levels in a manner similar to the changes between the K14CreER with low TM vs InvCreER+ populations26 (Supplementary Fig. 2d, e, bottom). We conclude that the molecular characteristics of our LRC and non-LRC populations are different from those previously reported for epidermal populations.
Gene Ontology (GO) analysis showed that the BL LRC/non-LRC genes function in metabolism, development, signalling, mitosis, and cell migration (Supplementary Table 3–5). GO categories of apoptosis inhibition and Ras-GTPase and insulin signalling pathways35–37 were overrepresented specifically in BL LRCs, while chromatin regulation, blood vessel development, and MAPK pathway genes were overrepresented in BL non-LRCs. In summary, the microarray analysis revealed distinct gene expression patterns and putative markers for BL LRCs and non-LRCs.
Modelling implies two SC populations
To determine the kinetics of epidermal LRC and non-LRC division and differentiation, we isolated back skin cells from K5-tTA/pTRE-H2B-GFP/K14CreER/Rosa-tdTomato quadruple-transgenic mice after different doxy chases and measured the H2B-GFP dilution patterns by FACS (Fig. 3a, b). Beyond analyzing the BL, which informs on cell division, we quantitatively analyzed H2B-GFP dilution data in the SL and GL. This provided information describing the kinetics of differentiation and upward transport from the BL into the suprabasal layers that was not available in a previous similar study26. We then quantitatively tested multiple mathematical models of population dynamics (Fig. 3c; Supplementary Fig. 3a), including two models previously described23,26, against the data.
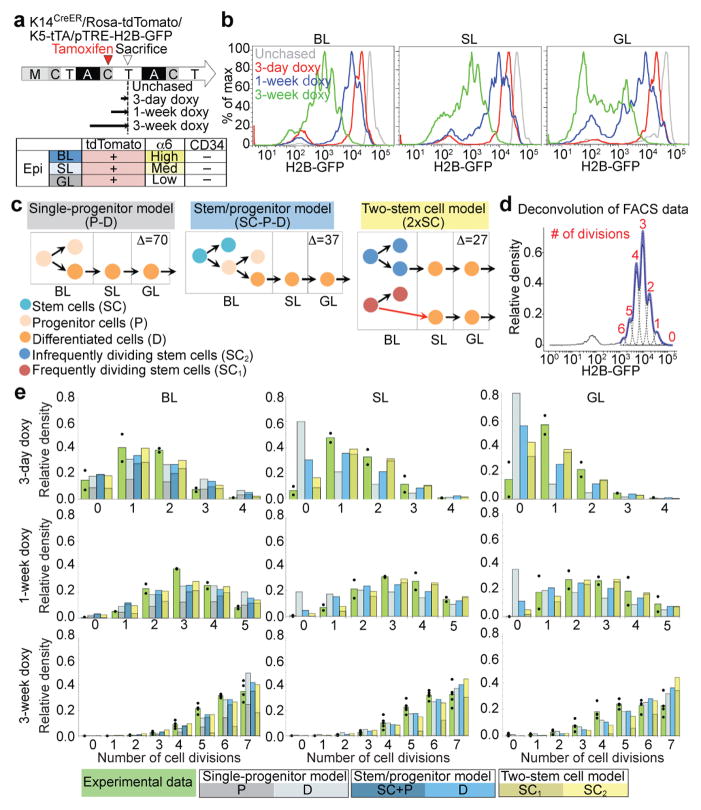
Figure 3. H2B-GFP dilution implies two independent cell populations with different proliferation and transport rates.
a, Doxy-chase scheme and epidermal compartment markers. b, FACS histograms showing H2B-GFP dilution patterns for different chases in basal layer (BL), spinous layer (SL) and granular layer (GL). c, Models. Δ assesses the best-fit; smaller is better. Red arrows denote division coupled to transport. d, Example of deconvolution of H2B-GFP fluorescence histogram and determination of cell division number (d)-distribution. e, Normalized compartment d-distributions after different chase periods. The data from individual mice are shown as dots with means given by the green bars. The other bars are the predictions from the indicated models using the best-fit parameters; they are subdivided to represent individual model component contributions (see Supplementary Note). The FACS experiments are repeated twice with 2 mice for day 3 and 7; twice with 5 mice for day 21 (b, d, e).
H2B-GFP histograms were deconvolved38 to determine the relative proportions of cells that had divided 0, 1, 2… times (Fig. 3d; Supplementary Note). This showed that a majority of the BL cells divide rapidly---more than half had divided at least once by 3 days and essentially all had divided by 1 week (Fig. 3e, green). Strikingly, essentially all the original SL and GL cells were replaced at similar fast rates by divided BL descendants: at day 3 only 7% of the SL cells and 15% of the GL cells had not divided, implying that at least 93% (SL) and 85% (GL) were BL cells that had divided and been transported during the 3-day chase. Essentially all the original SL and GL cells were replaced by 1 week. This extremely rapid replacement with newly divided BL cells suggested that the majority of BL divisions are tightly coupled to upward transport and differentiation into SL (Supplementary Note). Mathematical modelling showed that this cannot be explained by previous models, such as single-progenitor23 (Fig. 3c, e, grey) or stem/progenitor26 models (Fig. 3c, e, blue), that do not couple division to upward transport and instead posit substantial terminal differentiation within the BL23,26 (Fig. 3c, orange cells within BL). Moreover, a model of a single SC population undergoing division coupled to transport (Supplementary Fig. 3a, dark blue, red BL→SL arrow) could provide a good fit to the 3-day, but not the 1-week, data (Supplementary Fig. 3b, dark blue). The combined data could only be well-modelled by adding a second, independent SC population that divided at a slower rate (Fig. 3c, e; Supplementary Fig. 3a, b, yellow). The best-fit was obtained with two independent SC populations comprised of: (1) ~65% fast SCs (corresponding to our non-LRCs), which divide every ~2 days with tight coupling of most division to transport; and (2) ~35% slow SCs (corresponding to our LRCs), which divide every ~5 days.
Analysis with this model indicated that the BL LRC and non-LRC populations used for microarray analysis (Fig. 2) were separated to >80% purity (Supplementary Note).
The computed division rate of the LRC population is comparable with that of the previously described AhCreER, AxinCreER, and InvCreER populations23,25,26. However, by using SL and GL, in addition to BL, H2B-GFP dilution data, we were able to identify a distinct population (our non-LRCs) that divides faster than any previously described. Moreover, since it was identified with highly efficient H2B-GFP labeling, we were able to determine that this fast population comprises two-thirds of the entire BL, and so must contribute substantially to tissue homeostasis.
Because the tail scale (enriched in non-LRCs) and interscale (enriched in LRCs) regions are morphologically distinct, we could use pulse-chase experiments to determine the H2B-GFP fluorescence turnover rate and the corresponding overall BL division rate within each region separately (Supplementary Fig. 3c–e; Supplementary Note). The 3:1 division rate ratio of BL cells between these regions closely matches the ~2.7 ratio in the back computed from the H2B-GFP dilution experiments, although division in the tail was 20—25% slower overall. We also were able to compute the overall epidermal turnover half-lives in the LRC vs the non-LRC tail regions from the H2B-GFP decay rates in their BL and BL+SL+GL layers (CL was excluded). We found that the LRC and non-LRC skin region turnover half-lives were in ~2:1 ratio (i.e. 6.2 days vs 3.5 days). This highlights how the differences in BL divisions may drive different overall epidermal regeneration speeds in each skin region.
In summary, quantitative analysis of the H2B-GFP dilution data implies that the epidermal BL contains at least two independent SC populations with different rates of proliferation and upward differentiation/transport and that epidermis in the LRC and non-LRC territories regenerates overall at different rates.
Lineage tracing implies two SC populations
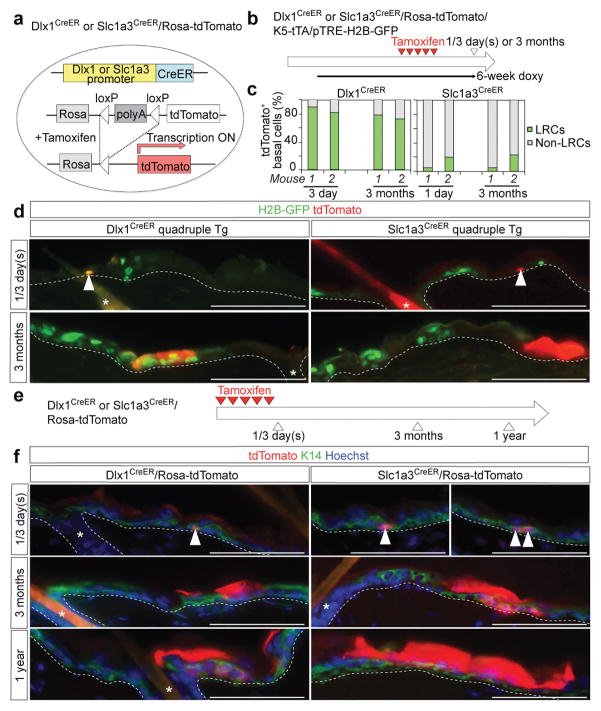
To directly address the long-term SC potential and the lineage relationship between LRCs and non-LRCs, we examined their behavior by in vivo lineage tracing. We used transgenic mice39,40 carrying the CreER driven by Dlx1 and Slc1a3 genomic loci, two genes selected from our microarray analysis as up-regulated in LRCs and non-LRCs, respectively (Fig. 2h; Fig. 4a). TdTomato expression was strictly induced by TM and was dose-dependent (Supplementary Fig. 4). A few days after the last high-dose TM injection, both Dlx1CreER and Slc1a3CreER mice showed tdTomato expression in the BL (Fig. 4d, f). The Dlx1CreER labelled back skin inefficiently, and the clones were mostly single cells. In contrast, the Slc1a3CreER induced multiple labeling events interspersed throughout the epidermis within each section of back skin tissue analyzed. Back skin from Dlx1- or Slc1a3- CreER/Rosa-tdTomato/pTRE-H2B-GFP/K5-tTA transgenic mice subjected to co-temporal TM injections and doxy chases showed preferential co-localization of tdTomato cells with BL LRCs (Dlx1CreER mice) or non-LRCs (Slc1a3CreER mice) a few days after TM induction (Fig. 4b–d). This demonstrated that Dlx1CreER and Slc1a3CreER can be used as genetic tools to distinguish LRCs and non-LRCs, respectively, and to study their behaviour as SCs.
Figure 4. Slc1a3CreER and Dlx1CreER mark long-lived stem cells in the back epidermis.
a, Schematic representation of the tamoxifen (TM)-inducible CreER system. b, Tamoxifen and doxycycline pulse-chase scheme to examine the relationship of Dlx1CreER- and Slc1a3CreER-marked cells with H2B-GFP LRCs in quadruple transgenic mice. c, Fraction of LRCs and non-LRCs in tdTomato+ basal cells in back skin from Dlx1CreER or Slc1a3CreER at 1/3 days or 3 months post-TM. Data have been obtained from 2 mice per condition; data from each mouse are shown as an individual bar. d, Fluorescence images of back skin sections of quadruple-transgenic mice show co-localization of Dlx1CreER, but not Slc1a3CreER, marked cells and generated clones with LRCs. e, Scheme for long-term lineage tracing. f, Lineage tracing in back skin. Arrowheads indicate tdTomato+ basal cells. The dashed line represents epidermal-dermal junctions. The tdTomato signal is extremely intense in the cornified layer (CL), the outermost skin layer, when compared with the basal and suprabasal layers. This is likely because CL contains multiple layers of stacked cells or because the tdTomato reporter is more active there. Interestingly, Slc1a3CreER clones spread away in CL from their BL-marked areas, suggesting that Slc1a3CreER BL cells contribute broadly to the outer skin layer. Asterisks indicate HFs. Scale bars, 100 μm (d, f). Experiments are repeated twice with 2 mice for representative images in (d); and twice with 4 mice (Slc1a3 1 day), 3 mice (Dlx1 3 days and Dlx1 3 months) and 2 mice (Dlx1 1 year, Slc1a3 3 months and Slc1a3 1 year) for images in (f).
By 3-months we still found Dlx1CreER- and Slc1a3CreER-marked clones that co-localized with LRC- and non-LRC-clusters in the basal and suprabasal layers (Fig. 4c, d). This demonstrated differential rates of proliferation and upward differentiation/transport for the two populations, consistent with the H2B-GFP analyses. Importantly, both Dlx1CreER- and Slc1a3CreER-marked cells clones were maintained for 1-year, as expected of long-lived SCs (Fig. 4e, f).
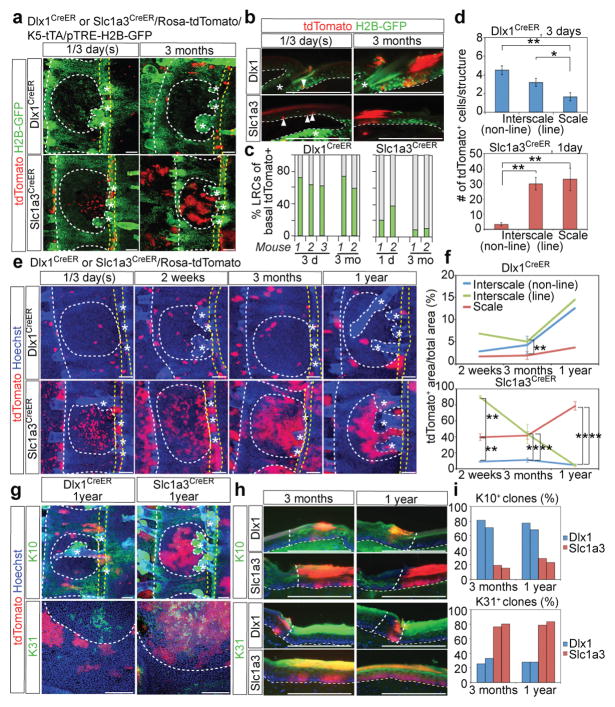
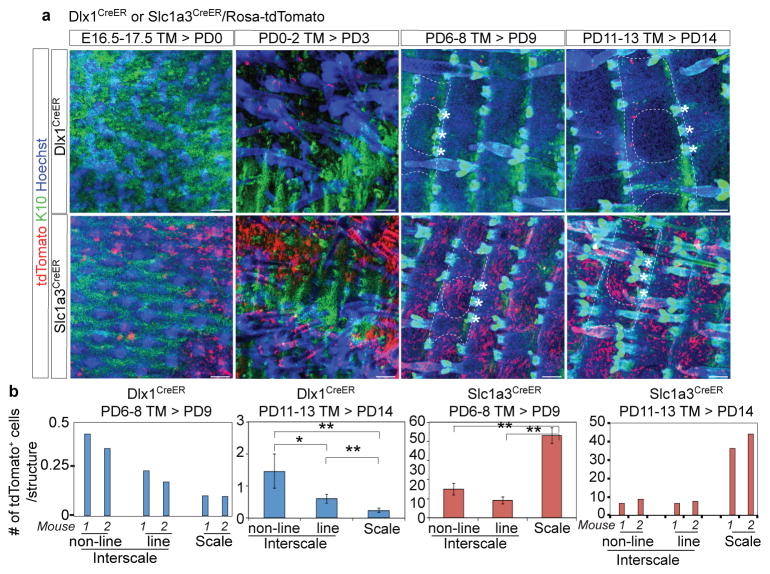
Slc1a3CreER- and Dlx1CreER-induced cells were more frequent in tail than in back skin, and clones were readily detectable at all time points after TM induction. As in the back skin, in tail skin after chases of a few days and 3 months, Dlx1CreER- and Slc1a3CreER-marked basal cells were enriched in LRCs and non-LRCs, respectively (Fig. 5a–c). Strikingly, in whole-mount microscopy, Slc1a3CreER-marked cells were preferentially located in the scale (non-LRC) areas and also showed a remarkable linear clustering within an interscale substructure that spanned the HF-neighbouring region (Fig. 5a, d; Supplementary Fig. 5a–c). We call this substructure the “line” and the remaining interscale the “non-line” areas. In contrast, Dlx1CreER-marked cells were biased towards the interscale for both line and non-line substructures. We examined Dlx1CreER- and Slc1a3CreER-marked populations throughout tail skin development and found that this distinct spatial localization occurs around PD9 (Fig. 6), which is at the onset of scale differentiation27.
Figure 5. Dlx1CreER- and Slc1a3CreER-marked cells replenish distinct tail skin territories with distinct differentiated progeny.
a, Tail epidermis whole-mounts from quadruple-transgenic mice show preferential marking (red signal) of Slc1a3CreER in the scale area (white dotted circle) and in the interscale “line” area (yellow dotted lines). The scale area is enriched in non-LRCs. Dlx1CreER marking is enriched in the interscale (LRC area). b, Tail skin sections from Dlx1CreER or Slc1a3CreER quadruple-transgenic mice show co-localization of these markers with LRCs and non-LRCs, respectively. Scale bars, 100 μm (a, b). c, Fraction of LRCs and non-LRCs in tdTomato+ basal cells in tail skin. Data from each mouse is shown as an individual bar. d, Number of tdTomato+ cells within each structure. Error bars show s.e.m (n = 5 mice); **: P<0.01; *: P<0.05. [Dlx1CreER, interscale (non-line) vs scale, P=0.002; Dlx1CreER, interscale (line) vs scale, P=0.03; Slc1a3CreER, interscale (non-line) vs scale, P=1×10−5; Slc1a3CreER, interscale (non-line) vs interscale (line), P=1×10−5.] e, Lineage tracing in tail skin. Scale bars, 100 μm. f, Quantification of tdTomato+ area per total area. [Dlx1CreER, 3 months, n = 4 mice, interscale (line) vs scale, P=1×10−4; Slc1a3CreER, 2 weeks, n = 3, interscale (line) vs scale, P=1×10−4; interscale (non-line) vs scale, P=0.008; Slc1a3CreER, 3 months, n = 4, interscale (non-line) vs interscale (line), P=0.007, interscale (non-line) vs scale, P=1×10−5; Slc1a3CreER, 1 year, n = 3, interscale (line) vs scale, P=1×10−12, interscale (non-line) vs scale, P=1×10−12. For Dlx1CreER, 2 weeks and 1 year, the average of two mice is shown.] g, h, Immunostaining of tail skin. Dotted lines delineate scale-interscale boundaries (white) and “line” structures (yellow) or epidermal-dermal junctions (white). Asterisks indicate HFs. Scale bars, 100 μm. i, Fraction of K10+ or K31+ clones. Data from each of two assessed mice are shown as individual bars. All P-values were calculated using the two-tailed Student’s t-test wherever n ≥ 3. Experiments are repeated twice with 2 mice for representative images in (a, b, g, h); and twice with 4 mice (Slc1a3 1 day, Slc1a3 2 weeks), 3 mice (Dlx1 3 days and Dlx1 3 months) and 2 mice (Dlx1 2 weeks, Dlx1 1 year, Slc1a3 3 months and Slc1a3 1 year) for images in (e).
Figure 6. The emergence of Dlx1CreER- and Slc1a3CreER-marked cells during skin development.
a, Whole-mount immunostaining of the tail skin in double-transgenic Dlx1CreER/or Slc1a3CreER/Rosa-tdTomato mice. Mice were injected with tamoxifen (TM) and sacrificed at the indicated ages. Dlx1CreER- or Slc1a3CreER-marked cells were first detected at PD3 or PD0, respectively, in the K10− region. By PD9, Dlx1CreER- or Slc1a3CreER-marked cells were preferentially observed in interscale (K10+) or scale (K10–), respectively; this pattern was maintained in adults. The dashed line represents the boundary of tail epidermis structure (circle is scale, while the remaining marked area is interscale). Asterisks indicate HFs. Scale bars, 100 μm. b, Number of tdTomato+ cells within each structure. Error bars show s.e.m.; **: P<0.01; *: P<0.05. Statistical analyses were performed using the two-tailed Student’s t-test when n ≥ 3; an individual bar is shown for each mouse otherwise. [Dlx1CreER PD14, n = 4 mice, interscale (non-line) vs interscale (line), P=0.05, interscale (line) vs scale, P=0.006, interscale (line) vs scale, P=0.005; Slc1a3CreER PD9, n = 5, interscale (non-line) vs scale, P=1×10−13, interscale (line) vs scale, P=1×10−18.] Experiments are repeated twice with number of mice indicated above for images in (a).
Next, we used the Dlx1CreER and Slc1a3CreER systems for long-term lineage tracing in the adult tail epidermis. By 1-year chase, Slc1a3CreER-marked cells remained scarce in the interscale non-line, were dramatically lost within the interscale line, and expanded to form large colonies within scales. Conversely, Dlx1CreER showed significant preference for the interscale structures (Fig. 5e, f; Supplementary Fig. 5c, d). The inefficient Dlx1CreER marking allowed single cell labelling and clonal analysis over the extended chase period. (This was not possible for Slc1a3CreER since it marked a substantial fraction of cells, which generated overlapping clones, in the scale areas.) About 20% of the Dlx1CreER-derived clones survived for 1 year in all structures (Supplementary Fig. 5f). The areas of the surviving clones at 1-year were nearly twice as large in interscale non-line as in scale (Supplementary Fig. 5e). This was especially interesting since the interscale is an LRC area, which we found regenerates 2× more slowly, yet Dlx1CreER-marked cells showed enhanced regenerative ability in this environment. Thus, these data suggest that the Slc1a3CreER and Dlx1CreER lineages generally show biased localization within the scale vs interscale (especially pronounced in the non-line) areas, and each of them thrives more in their respective regions in long-term homeostasis.
The gradual decrease in the number of Dlx1CreER+ clones over 1 year was consistent with neutral drift41 of this SC population (Supplementary Fig. 5g; Supplementary Note). Moreover, combining this analysis with that of the H2B-GFP data suggested that division of these cells, like that of the non-LRC cells, may be partially coupled to transport and differentiation.
Next we explored the contribution of the Dlx1CreER- and Slc1a3CreER-marked cells to differentiation lineages specific to interscales and scales. As expected from their biased localization within these regions, we found that overall Dlx1CreER and Slc1a3CreER lineages preferentially produced K10+ (interscale) vs K31+ (scale) differentiated cells, respectively (Fig. 5g–i). Clones showing the non-preferred differentiation pattern also localized in the territory atypical for that lineage. That is, Dlx1CreER clones are preferentially located in the interscale where they make the interscale-specific lineages, but the rare Dlx1CreER clones found in scales differentiate to scale-type lineages; the converse is true for Slc1a3CreER.
Together, these data showed that Dlx1CreER- and Slc1a3CreER-marked cells comprise two distinct SC populations with distinct proliferation and preferred differentiation patterns. These two populations are enriched in spatially segregated epidermal territories, where they thrive preferentially and function to replenish specialized differentiated lineages during steady-state homeostasis.
The two populations are interchangeable in injury
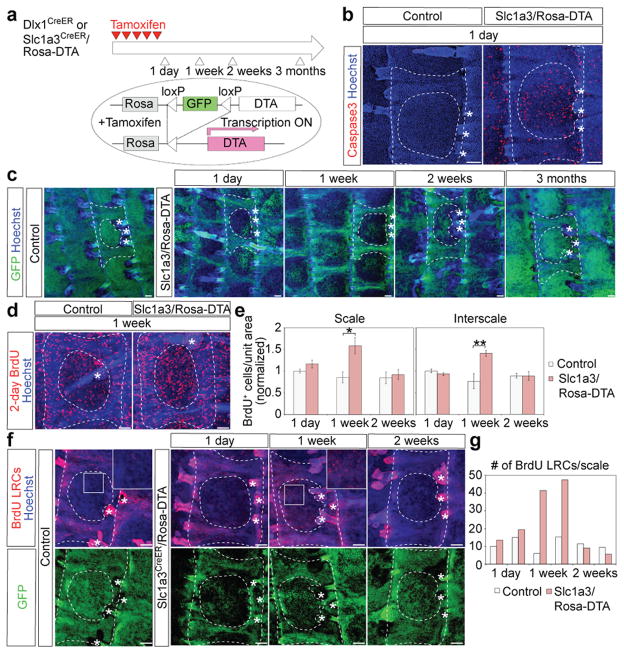
Next, we asked whether Dlx1CreER- and Slc1a3CreER-marked SCs have the ability to interchange their functions upon injury or selective cell loss. First, we used transgenic mice that carry the Rosa-Diphtheria toxin fragment A (DTA)42 crossed with our CreER mice for selective cell killing (Fig. 7a). Upon CreER activation, GFP is suppressed and DTA is expressed causing death of the CreER-marked cells. Depleting Dlx1CreER-marked cells had no effect, probably due to low marking efficiency (Supplementary Fig. 6).
Figure 7. Dlx1CreER- and Slc1a3CreER-marked cells replenish each other’s territories following selective killing or injury.
a, Scheme to ablate Dlx1CrER+ or Slc1a3CreER+ cells using DTA induction via tamoxifen (TM) injection. b-d, Whole-mount immunostaining of the tail epidermis. (b) Note frequent Caspase3+ cells, a marker of apoptosis, within the scale area one day after the last TM injection in Slc1aCreER/Rosa-DTA (right) but not control (left) skin. (c) Note the striking loss of GFP (indicative of cell death) in scale regions relative to control in skin collected at 1 day, 1 week, and 2 weeks after the last TM injection, and the recovery of signal by 3 months post-TM. (d) Note the increased frequency of BrdU+ cells (indicative of proliferation) relative to control at 1-week post-TM. e, Quantification of BrdU+ cells per unit area at the indicated post-TM time points Error bars show s.e.m. (n = 3 mice).; **P<0.01; *P<0.05*. Statistical analyses were performed using the two-tailed Student’s t-test. (Scale 1 week, control vs Slc1a3CreER/DTA, P=0.02; interscale 1 week, control vs Slc1a3CreER/DTA, P=0.01.) f, Tail epidermis whole-mounts from mice subjected to BrdU pulse-chase, which marks the interscale area as BrdU-LRCs, followed by selective killing within the scale area induced by TM injection in Slc1a3CreER/Rosa-DTA mice. Insets are higher magnifications of the boxed area and illustrate massive migration of LRCs into the scale area by 1-week post-TM. g, Quantification of BrdU LRCs in scales in mice from (f) (data from each mouse shown as an individual bar). Dashed lines delineate boundaries of the tail epidermis structures. Asterisks indicate HFs. Scale bars, 100 μm (b, c, d, f). Experiments are repeated twice with 2 different mice for all representative images (b, c, d, f).
One day post-TM, targeting the Slc1a3CreER+ cells induced large numbers of apoptotic cells and a dramatic reduction of GFP fluorescence within scales (Fig. 7b, c; Supplementary Fig. 6b). The GFP signal began to recover after 1 week and completely filled the scales by 3 months, and BrdU incorporation transiently increased in both scale and interscale, indicating the production of new cells (Fig. 7c–e). By 2-weeks, the few surviving tdTomato+ cells in Slc1a3CreER/Rosa-DTA/Rosa-tdTomato mice expanded only modestly (Supplementary Fig. 7a–d), indicating that they cannot account for the rapid GFP recovery and the observed repopulation of the scale region after cell killing. Instead the scale was likely repopulated from the interscale, as LRCs marked by BrdU pulse-chase massively migrated into the scale and eventually diluted their BrdU label, probably through activated cell divisions (Fig. 7f, g; Supplementary Fig. 7e–i). Similarly to tail skin, LRCs in the back skin became more proliferative upon the loss of Slc1a3CreER+ cells (Supplementary Fig. 8). We also detected up-regulation of “wounding” markers, such as Tenascin and K6 in the back skin, demonstrating that the Slc1a3CreER-induced population is needed for the integrity of back skin epidermis 43, 44. We conclude that LRCs are activated and recruited into neighboring regions enriched in non-LRCs to replenish the skin upon massive cell loss in those areas.
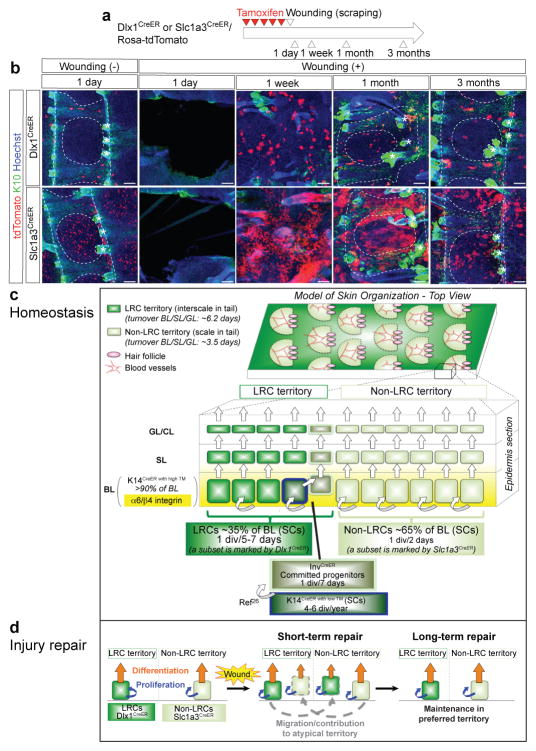
Next, we examined the behavior of Dlx1CreER- and Slc1a3CreER-marked cells after mechanical epidermal wounding in tail skin (Fig. 8a). TdTomato+ cells migrated and expanded into the wounded area by 1 week and massively contributed to both K10+ interscale and K10− scale lineages by 1 month in both Dlx1CreER- and Slc1a3CreER-mice (Fig. 8b). Strikingly, by 3 months, Dlx1CreER- and Slc1a3CreER-marked cells were mostly lost from their atypical regions and again localized preferentially within interscales and scales, respectively, where they normally primarily reside.
Figure 8. Model of epidermal lineage organization and SC dynamics.
a, Experimental scheme of wounding experiments. b, Whole-mount immunostaining of the tail epidermis at different time points post-wounding. Dashed lines delineate boundaries of the tail epidermis structures. Asterisks indicate HFs. Scale bars, 100 μm. Experiments are repeated twice with 2 different mice for all representative images. c, Two distinct territory types (LRCs and non-LRCs) with different turnover rates pattern the skin by forming structured arrays that are spatially coordinated with tissue structures such as blood vessels (back) and hair follicles (back and tail). In tail, the LRC territories correspond to interscales and the non-LRC territories correspond to scales. Turnover rates for the nucleated epidermis (BL/SL/GL) computed from H2B-GFP dilution data in tail skin are shown on the left. d, Independent SC populations fuel homeostasis in the LRC and non-LRC territories. The H2B-GFP LRC and non-LRC populations are preferentially located within their respective territories; their SCs can be distinguished by marking with Dlx1CreER and Slc1a3CreER, which identify LRC and non-LRC subsets, respectively. The two SC populations are largely independent and are not hierarchically related. The data do not exclude the possibility that each SC population may generate its own short-lived progenitor cells prior to differentiation. Populations previously defined by Mascre et al26 as K14CreER with low TM and InvCreER are represented as subsets within our LRC territories and are distinct from our Dlx1CreER population as indicated by comparison of microarray data from this study with that of Mascre et al26. The InvCreER population is designated as a special population since it shows lower levels of α6/β4 integrin (yellow gradient) relative to the rest of the BL26. High TM doses activate the K14CreER in >90% of the BL. b, Behavior of two SC populations and their descendants, represented by Dlx1CreER- (LRCs) and Slc1a3CreER- (non-LRCs) marked cells, during injury repair. Both SC populations can temporarily exchange function in response to injury by migrating and repopulating their neighboring atypical territories in short-term. However, they are largely lost from their atypical territory and remain present only in their typical territory where they thrive better post-injury and during homeostasis in long-term.
In conclusion, skin injury can temporarily stimulate migration and increased regenerative activity of Dlx1CreER- and Slc1a3CreER-marked SCs in both their typical and atypical territories for rapid and robust skin regeneration. However, these two lineages do not fully inter-convert to each other, as indicated by inefficient long-term maintenance in their atypical territory.
Discussion
Here we identify two distinct IFE subpopulations, defined as H2B-GFP LRCs and non-LRCs, that comprise ~35% and ~65% of the BL, are regionally clustered, and are molecularly distinct. Furthermore, we provide Dlx1CreER and Slc1a3CreER as mouse genetic marking tools to distinguish IFE LRC and non-LRC behavior by lineage tracing. The current hierarchical stem-progenitor model endows SC properties exclusively to infrequently dividing cells (i.e., that are label-retaining) while positing that frequently dividing cells are short-lived progenitors incapable of injury repair26. In contrast, here we demonstrate that our LRCs and non-LRCs act as two independent SC populations that contribute to homeostasis within distinct skin territories that regenerate at different rates, and that they both substantially participate in injury repair of both territories. Based on these findings, we re-define the model of IFE lineage organization and SC dynamics (Fig. 8c, d).
Our H2B-GFP pulse-chase data revealed the existence of spatially segregated LRC and non-LRC territories relative to HFs in both back and tail skin, suggesting a potential parallel between the organization of these two tissue types (Fig. 8c, top). Slc1a3CreER and Dlx1CreER, which surfaced as potential genetic marking tools from our microarray of back skin cells, mark cells enriched in non-LRCs and LRCs, respectively, and both marked cell types act as long-term SCs in lineage tracing. Furthermore, the Slc1a3CreER and Dlx1CreER clones by 3 months post-induction are enriched in LRCs in non-LRCs in both basal and suprabasal layers, demonstrating that the two populations proliferate, differentiate and transport upwards at different rates. This occurs in both back and tail skin, suggesting further parallels in the organization of these two tissue types. The defined structure, ease of manipulation, and superior imaging amenability of tail skin allowed us to demonstrate that these two SC populations reside within and fuel the LRC and non-LRC tail territories that turn over at different rates (Fig. 8c). These territories display previously described differentiation characteristics of tail skin scales and interscales27. After injury, both SC types migrate, proliferate, and temporarily inter-convert their functions to regenerate both territory types. Analogously, ablation of Slc1a3CreER+ non-LRCs in both tail and back skin stimulated migration and proliferation of the LRCs. The rapid expansion of these SCs upon injury might reflect the switching from “balanced” to “expanding” mode of division proposed in human keratinocytes45. Importantly, long-term after injury, each SC is lost from its atypical territory within the tail (Fig. 8d). This echoes our observation that during normal homeostasis clones generated by Dlx1CreER in their typical territory (interscale) have larger area than the occasional clones generated by Dlx1CreER in the atypical territory, despite the fact that the interscales regenerate at overall rates half those of scales. Together these findings suggest that there are cell-intrinsic differences that enhance the adaptation of each type of SC to its normal environment (territory). The non-LRC territory within the back skin coincides with areas containing organized blood vessel arrays (Fig 8c, top), which are known to associate with pericytes that promote human epidermal SC proliferation46. This is consistent with the more rapid divisions of non-LRCs. In addition, the non-LRCs overexpress genes known to participate in blood vessel development, suggesting that they may in turn actively recruit the vessels and that there is crosstalk between epidermis and blood vessels.
Multiple IFE SC populations have previously been characterized in tail, plantar, and ear epidermis23,25,26; they all divide at rates comparable to, or slower than, the overall division rate determined for the bulk of our LRC population (Fig. 8c, bottom). We propose that the LRC population is heterogeneous and represents a collection of basal cell cohorts with different gene signatures, all located within the LRC territory and dividing at slow, but somewhat different, rates. In particular, we suggest that the rare K14CreER with low TM and rare InvCreER populations identified by Mascre et al26 in tail skin localize within LRC (interscale) regions, along side the LRC subset we found here marked by Dlx1CreER (Fig. 8c, bottom). Consistent with this, the long-term clones marked via K14CreER with low TM survived in skin regions referred to there26 as “undulations” that were reminiscent of the rete ridges in human skin. These regions, which morphologically overlap with the interscale, were enriched in H2B-GFP LRCs. Furthermore, previous analysis of Inv promoter activity in tail skin suggested that the InvCreER might mark BL cells confined to the scale/interscale junction27, 47, that is, neighboring the LRC territories.
The heterogeneity in gene expression and division rates of at least some of these LRC subpopulations could be mere transient fluctuations of the same SC state in response to micro-local needs within their territory. This would fit with the implication from our selective killing of BL SC subsets and wounding experiments that epidermis maintains flexibility in utilizing its SC populations, a likely explanation for the known robustness of skin regeneration. However, other molecular profile differences may be static and related to different physiological behaviors; for instance, the InvCreER population does not contribute to injury repair26.
On-the-other-hand, the non-LRC population discovered here is the most rapidly dividing epidermal population ever reported (1×/2 days) and stands out as the only population to date located preferentially within areas of rapid regeneration (non-LRC territories in back and tail, known as scales in tail)23, 25, 26. Our H2B-GFP dilution experiments demonstrated that its division is tightly coupled with upward transport, which uniquely explains the previously described19 rapid BL→GL transit of incorporated thymidine. The rapid turnover of H2B-GFP in the non-LRC territory, which comprises the majority of the BL, cannot be explained by relatively slow division rates of populations previously defined by Mascre et al26. Marking with Slc1a3CreER efficiently targeted the non-LRC SC population and allowed us to demonstrate that non-LRCs represent a distinct lineage that stands alone, independent of the LRCs, maintaining their own territories (scales in tail) during normal homeostasis.
In the future, it will be important to examine in more detail the potential heterogeneity of skin and SC organization in the mouse back skin and in human skin. The Slc1a3CreER transgene marks a substantial subset of the mouse back epidermis, and thus provides a unique means for studying this rapidly regenerating type of epithelium. Better markers will be required for the back skin LRCs, since Dlx1CreER (this work), K14CreER with low TM 26, and InvCreER 26 only target rare subsets of this population, which according to our H2B-GFP pulse-chase data and modelling data, represent ~35% of the BL. Furthermore, the fact that different doses of TM (e.g., the low doses used to identify K14CreER with low TM 26 versus the medium doses used in this work) can enrich different populations with distinct molecular profiles and behavior must be considered carefully when interpreting clonal data. Our microarray data provides a number of potential candidates for additional LRC and non-LRC markers that, if substantiated at the protein level in mouse and human skin, will be valuable reagents for investigating the physiological significance of the heterogeneity in IFE SC and skin organization revealed here. Suggestively, some of our LRC and non-LRC signature genes are known to be heterogeneously expressed in the human epidermis48, 49, and thus may be useful for understanding the heterogeneity of human skin and provide a platform for examining heterogeneous SC populations in aging, cancer and other diseases.
Methods
Mice
All mouse experiments were carried out according to Cornell University Institutional Animal Care and Use Committee guidelines (protocol number #2007-0125). To employ the H2B-GFP tet-off system, double-transgenic K5-tTA (FVB)50/pTRE-H2B-GFP (CD1)21 mice were used. Sequential crosses were employed to obtain quadruple-transgenic mice [K14CreER (CD1)51 (a gift from Dr. Elaine Fuchs, Rockefeller University, New York, USA)/Rosa-tdTomato (C57BL6)52 (The Jackson Laboratory, #007905)/K5-tTA/pTRE-H2B-GFP].
For lineage tracing, Dlx1CreER (C57BL6)39 (The Jackson Laboratory, #014551) or Slc1a3CreER (C57BL6)40 (The Jackson Laboratory, #012586) mice were crossed with Rosa-tdTomato reporter mice. CreER/Rosa-tdTomato mice without TM injections were used to examine the leakiness of Cre. Dlx1CreER or Slc1a3CreER quadruple transgenic mice [CreER/Rosa-tdTomato/K5-tTA/pTRE-H2B-GFP] were obtained after several steps of intercrossing the above lines.
For depleting Cre+ cells, Dlx1CreER or Slc1a3CreER mice were crossed with the Rosa-DTA line (C57BL6)42 (The Jackson Laboratory, #006331). The Rosa-tdTomato reporter was additionally introduced in Slc1a3CreER/Rosa-DTA mice to trace the lineage of surviving Slc1a3+ cells. Controls were either single-transgenic CreER or Rosa-DTA mice. Both male and female mice were used for all experiments.
The sample size was dictated by experimental considerations and not by a statistical method. The experiments were not randomized. The investigators were not blinded to allocation and outcome assessment during experiments.
H2B-GFP pulse-chase
Mice were fed with doxy chow (1g doxy/1kg, Bio-serv) for the indicated chase periods, starting at 1–3 months of age. GFP antibody was used to detect the H2B-GFP signal for 6 weeks of doxy chase.
Tamoxifen injection
In K14CreER quadruple transgenic mice were injected intraperitoneally with a single dose of TM (75 μg/g body weight; Sigma) and sacrificed after 1 week at second telogen. For lineage tracing, DTA, and wounding experiments, Dlx1CreER and Slc1a3CreER lines, mice were injected with TM (100 μg/g body weight) for 5 consecutive days at 4–7 weeks of age, unless noted elsewhere. Mice were sacrificed at the indicated times after the last injection.
BrdU labeling
To label LRCs, 5-bromo-2-deoxyuridine (BrdU; Calbiochem) was administrated in drinking water (0.8 mg/ml) for 1 week, followed by 5 weeks of chase without BrdU. To label proliferative cells, BrdU was administrated in drinking water (0.8 mg/ml) for 2 days or EdU (50 μg/g body weight, Invitrogen) is injected intraperitoneally twice a day for 2 days before sacrifice.
Whole-mount immunostaining in the tail epidermis
Tail skin pieces (5 mm × 5 mm) were incubated in EDTA (20 mM)/PBS on a shaker at 37°C for 2 h to separate the epidermis from the dermis as an intact sheet. Epidermal sheets were fixed in 4% paraformaldehyde (PFA) overnight at 4°C. The skin pieces were washed, incubated in blocking buffer (1% BSA, 2.5% donkey serum, 2.5% goat serum, 0.8% Triton in PBS) for 3 hours at room temperature (RT), and incubated with primary antibodies/blocking buffer overnight at RT. Samples were washed 4× in PBS with 0.2% Tween (PBS-T) for 1 h at RT, and were incubated overnight with secondary antibodies at 4°C. After washing, samples were counterstained with Hoechst for 1 h and mounted. For BrdU staining, the epidermal sheets were incubated for 30 mins in 2M HCl at 37°C after blocking and analyzed as indicated above.
Primary antibody dilutions: rabbit anti-K14 (1:1000, BioLegend #905301), chick anti-GFP (1:500, Abcam #ab13970), mouse anti-K10 (1:100, BioLegend #904301), rat anti-BrdU (1:300, Abcam #ab6326, clone # BU1/75 (ICR1)), guinea pig anti-K31 (1:100, PROGEN Biotechnik #GP-hHa1), rabbit anti-Caspase3 (1:300, R&D Systems #AF835) and rabbit anti-Ki67 (1:100, Leica Biosystems #NCL-Ki67p). All secondary antibodies (TxR, FITC, Cy5 or Alexa-594, Jackson ImmunoResearch) were used at a 1:500 dilution. The MOM kit (Vector Laboratories) was used for blocking with primary antibodies.
Preparations were analyzed by confocal microscopy (Zeiss LSM710 or Zeiss LSM880) with Zen 2012 software. All confocal data are shown as projected Z-stack images viewed from the basal surface.
Whole-mount immunostaining in back skin
Telogen back skin was fixed in 4% PFA overnight at 4°C. After washing 10× with PBS containing 0.3% Triton X-100 (0.3% PBST) every 30 min, the skin pieces (5 mm × 1 cm) were incubated with primary antibodies in blocking buffer (0.3% PBST containing 5% goat serum and 20% DMSO) for 5 d at RT. Samples were washed with 0.3% PBST every 30 min for 10× and were incubated overnight with secondary antibodies for 3 days at RT. After washing, samples were dehydrated sequentially in 25%, 50%, and 75% methanol for 5 min and 100% methanol for 20 min, 3 times. After incubating with benzyl benzoate and benzyl alcohol (BBBA) at 2:1 (v:v) overnight at RT, samples became transparent and could be observed under the confocal microscope.
Primary antibodies were used at the following dilutions: rabbit anti-K14 (1:1000, BioLegend #905301), chick anti-GFP (1:500, Abcam #ab13970) and rat anti-CD31 (1:100, BD Biosciences #550274, clone #MEC 13.3). All secondary antibodies (TxR, FITC or Alexa-594, Jackson ImmunoResearch) were used at 1:500 dilution.
Immunostaining of skin sections
Back and tail skin was directly embedded in Optimal Cutting Temperature (OCT) compound (Tissue Tek, Sakura). The frozen sections (10 μm) were fixed with 4% PFA for 10 min at RT. After blocking in normal serum, sections were incubated with primary antibodies overnight at 4°C. The following day, the sections were washed and incubated for 1 h with secondary antibodies at RT. After washing, the sections were counterstained with Hoechst and mounted. For staining with anti-BrdU antibody, the sections were treated with 2M HCl for 55 min at 37°C after blocking and stained as described above.
Primary antibodies dilutions were: rabbit anti-K14 (1:1000, BioLegend #905301), chick anti-GFP (1:500, Abcam #ab13970), rabbit anti-K1 (1:500, BioLegend #905601), rat anti-α6integrin (CD49f) (1:150, BD Biosciences #555734, Clone #GoH3), mouse anti-K10 (1:100, BioLegend #904301), guinea pig anti-K31 (1:100, PROGEN Biotechnik #GP-hHa1), rat anti-CD34 (1:150, BD Biosciences #553731), rat anti-BrdU (1:300, Abcam #ab6326, clone # BU1/75 (ICR1)), rabbit anti-Ki67 (1:100, Leica Biosystems #NCL-Ki67p), rabbit anti-Caspase3 (1:300, R&D Systems #AF835), rabbit Tenascin (1:300, EMD Millipore #AB19013) and anti-K6 (1:500, Abcam #ab24646). All secondary antibodies (TxR, FITC, Cy5 or Alexa-594, Jackson ImmunoResearch) were used at 1:500 dilution. For mouse primary antibodies, the MOM kit (Vector Laboratories) was used for blocking.
EdU staining was performed according to the manufacturer’s protocol (Click-iT EdU Imaging Kits, Invitrogen). To measure GFP fluorescence, slides were washed 3× for 20 min in 0.1M EDTA, followed by 5 min in PBS.
Preparations were examined using a fluorescent microscope (Nikon) and digitally imaged using a CCD 12-bit digital camera (Retiga EXi; QImaging) and IP-Lab software (MVI).
FACS
Mouse back skin was incubated in 0.25% trypsin/versene overnight at 4°C and for 30 min at 37°C. Single cell suspensions were prepared by scraping off the fat and subcutaneous tissue from the dermal side of the skin followed by enzymatic digestions and subsequent filtering with strainers (70 mm, followed by 40 mm). Cells were stained with the following antibodies for 30 min on ice: CD34-biotin (1:50, eBioscience), Streptavidin-APC (1:100, BD Biosciences) and α6-integrin-Pacific blue (1:100, BD Biosciences, custom order). Dead cells were excluded by propidium iodide (PI) (Sigma) staining. FACS (FACS Aria, BD Biosciences) were performed in the Cornell Flow Cytometry facility. FACS data were analyzed with the FlowJo software.
RNA isolation and RT-PCR
Total RNAs were isolated from sorted skin cells prepared by using RNeasy (Qiagen) and used for reverse-transcription by Super script III (Invitrogen). The primers used were as follows (5′ to 3′): Gapdh, ACTGCCACCCAGAAGACTGT and GATGCAGGGATGATGTTCT; Dlx1, ATGCCAGAAAGTCTCAACAGC and AACAGTGCATGGAGTAGTGCC; Igfbp3, TCTAAGCGGGAGACAGAATACG and CTCTGGGACTCAGCACATTGA; Dpp4; CCGTGGAAGGTTCTTCTGGG and GCTGCCGCTTCATCTTTGC; Sox6 GGTCATGTTTCCCACCCACAA and TTCAGAGGGGTCCAAATTCCT; Il1r2, CCCCTGGAGACAATACCAGC and TTAGCCAACCACCACACAATG; Slc6a4, TATCCAATGGGTACTCCGCAG and CCGTTCCCCTTGGTGAATCT; Slc1a3, ACCAAAAGCAACGGAGAAGAG and GGCATTCCGAAACAGGTAACTC; Aspm, TGGCTATGAGTGAATGCTCTTCC and TCGCGTAAAAACAGTGGCAAG; Trp73, CCCACCACTTCGAGGTCAC and GGCATGTCTTAGCAATCTGACAG; Pttg1, AACCCCTCCAACCAAAACAG and TCTGGGTAGGCATCATCAGGA; Cd36, AGATGACGTGGCAAAGAACAG and CCTTGGCTAGATAACGAACTCTG; tdTomato, CACCATCGTGGAACAGTACG and TGAAGCGCATGAACTCTTTG; Cd34, AAGGCTGGGTGAAGACCCTTA and TGAATGGCCGTTTCTGGAAGT; Ctgf, GGGCCTCTTCTGCGATTTC and ATCCAGGCAAGTGCATTGGTA; Ltbp2, AACAGCACCAACCACTGTATC and CCTGGCATTCTGAGGGTCAAA; α6-integrin, TGCAGAGGGCGAACAGAAC and GCACACGTCACCACTTTGC; β4-integrin, GCAGACGAAGTTCCGACAG and GGCCACCTTCAGTTCATGGA; K5, CCTGCAGAAGGCCAAGCA and TGGTGTTCATGAGCTCCTGGTA; K14, AAGGTCATGGATGTGCACGAT and CAGCATGTAGCAGCTTTAGTTCTTG; K1, AACCCGGACCCAAAACTTAG and CCGTGACTGGTCACTCTTCA; K10, GGAGGGTAAAATCAAGGAGTGGTA and TCAATCTGCAGCAGCACGTT; Involucrin, CACAATGCCAGGTCTTCACTGA and AGGGTTTGGCCGCTTCTC; Loricrin, TCACTCATCTTCCCTGGTGCTT and GTCTTTCCACAACCCACAGGA. QRT-PCR for each gene is normalized to GAPDH. The relative level for each gene is set to 1 in the control population.
Microarray
5 ng total RNA isolated from sorted cell populations was used with the “Ovation Pico WTA System (NuGEN)” for cDNA amplification, followed by “Encore Biotin Module (NuGEN)” for labeling and fragmentation of cDNA. The labeled cDNA was hybridized to Mouse Genome 430 2.0 genechips (Affymetrix) at Cornell Core Facilities. Microarrays were performed in triplicate (BL LRCs and BL non-LRCs) or duplicate (SL LRCs, SL non-LRCs and GL) isolated from two or three different mice.
Microarray analysis was carried out with GeneSpring GX13 software (Agilent Technologies). After normalization, the probes with a signal value <100 or reported “absent” were excluded. The remaining probes that are ≥2-fold up- or down-regulated in BL LRCs relative to BL non-LRCs were selected. Signature genes were defined as probes that were ≥2-fold up-regulated in one fraction over all the others. Gene Ontology analyses were performed on differentially expressed genes (Supplementary Fig. 2c) or signature genes (Fig. 2g). Principal component analysis was performed on the entire set of probes (45,101 probes) with 5 epidermal populations and infrequently or frequently dividing HFSC (bulge) populations, and the scores were represented in a 3D scatter plot. Hierarchical clustering was performed on 4,221 probes that were ≥5-fold up- or down-regulated in any of two comparisons between 5 epidermal populations.
To compare the telogen vs anagen bulge populations with our populations, lists of “≥2-fold increased in telogen bulge over anagen bulge” (4,961 probes) and “≥2-fold increased in anagen bulge over telogen bulge” (3,105 probes) were used from the previously published microarray34. For the comparison with K14+ and Inv+ populations, lists of “≥2-fold increased in K14+ over Inv+ population” (2,456 probes) and “≥2-fold increased in Inv+ over K14+ population” (2,671 probes) were used26. The genes enriched in our BL LRCs or non-LRCs were then compared with the above populations; the numbers of overlapping probes are shown in the pie charts (Fig. 2g).
Wounding
Mice were injected with ketoprofen (2 μg/g body weight) and amoxicillin (100 μg/g body weight) to prevent infections. Wounding was induced by removing the surface of the tail epidermis using #11 scalpel (Miltex). Isoflurane was used for the mouse anesthesia.
Quantification of microscope images
Area or cell numbers were quantified by using ImageJ software. For calculating turnover times and division rates in the tail epidermis, the average intensities of H2B-GFP fluorescent signals on BL, SL, GL and CL on tissue sections were measured by using AutoQuant X software. The scale/interscale regions are defined based on the retention of nuclei in the CL. The number of tdTomato+ clones within interscale or scale structure of the tail epidermis were counted on projected Z-stack confocal images. Clones are defined as single or cluster of cell(s) that contain at least one basal or suprabasal cell.
Quantifications were independently performed on ≥2 mice, and ≥50 cells/structures were counted per mouse.
Statistics & reproducibility
All experiments with or without quantification were independently performed at least twice with different mice and the representative data are shown. All statistical analyses were performed using the two-tailed Student’s t-test; statistical significance was defined as P<0.05.
Mathematical modelling
All details on mathematical modelling can be found in the Supplementary Note.
Data availability
Microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE70523. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
We thank L.G. Sayam for FACS; C.J. Bayles, R.M. Williams and J.M. DelaCruz for imaging; B. Hover and J. Mosher for microarray; the Cornell Center for Animal Resources and Education facility for mouse care; D.J. McDermitt for help with mouse crossings; and K.L. Goodyear for help with mathematical modelling. Support was from: Empire State Stem Cell Foundation, New York State-Department of Health (NYS-DOH), Contract #C026718 (in part) for Cornell Cytometry core; NIH Grant 1S10RR025502-01 for Cornell Biotechnology Research Center and Imaging Facility. Research funding was from NYSTEM Grant C024354, NIH Grant R21AR063278 to T.T and a Long-Term Fellowship from Human Frontier Science Program to A.S. and a postdoctoral fellowship for research abroad from the Japan Society for the Promotion of Science to A.S.
Footnotes
Authors Contributions
A.S. and T.T. designed the experiments. A.S., D.S. and T.T. wrote the manuscript. A.S., F.J., E.L. and S.W. performed experiments. D.S. and B.S.W. performed mathematical modelling.
References
- 1.Sada A, Tumbar T. New insights into mechanisms of stem cell daughter fate determination in regenerative tissues. International review of cell and molecular biology. 2013;300:1–50. doi: 10.1016/B978-0-12-405210-9.00001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foudi A, et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson A, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 4.Zhang YV, Cheong J, Ciapurin N, McDermitt DJ, Tumbar T. Distinct self-renewal and differentiation phases in the niche of infrequently dividing hair follicle stem cells. Cell Stem Cell. 2009;5:267–278. doi: 10.1016/j.stem.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waghmare SK, et al. Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. EMBO J. 2008;27:1309–1320. doi: 10.1038/emboj.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buczacki SJ, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 7.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeda N, et al. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 11.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montgomery RK, et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A. 2010;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metcalfe C, Kljavin NM, Ybarra R, de Sauvage FJ. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149–159. doi: 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Tian H, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritsma L, et al. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature. 2014;507:362–365. doi: 10.1038/nature12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaks V, Kasper M, Toftgard R. The hair follicle—a stem cell zoo. Exp Cell Res. 2010;316:1422–1428. doi: 10.1016/j.yexcr.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Plikus MV, et al. Epithelial stem cells and implications for wound repair. Seminars in cell & developmental biology. 2012;23:946–953. doi: 10.1016/j.semcdb.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai X, Segre JA. Transcriptional control of epidermal specification and differentiation. Curr Opin Genet Dev. 2004;14:485–491. doi: 10.1016/j.gde.2004.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potten CS, Saffhill R, Maibach HI. Measurement of the transit time for cells through the epidermis and stratum corneum of the mouse and guinea-pig. Cell Tissue Kinet. 1987;20:461–472. doi: 10.1111/j.1365-2184.1987.tb01355.x. [DOI] [PubMed] [Google Scholar]
- 20.Bickenbach JR. Identification and behavior of label-retaining cells in oral mucosa and skin. J Dent Res. 1981;60(Spec No C):1611–1620. doi: 10.1177/002203458106000311011. [DOI] [PubMed] [Google Scholar]
- 21.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schluter H, Paquet-Fifield S, Gangatirkar P, Li J, Kaur P. Functional characterization of quiescent keratinocyte stem cells and their progeny reveals a hierarchical organization in human skin epidermis. Stem Cells. 2011;29:1256–1268. doi: 10.1002/stem.675. [DOI] [PubMed] [Google Scholar]
- 23.Clayton E, et al. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 24.Doupe DP, Klein AM, Simons BD, Jones PH. The ordered architecture of murine ear epidermis is maintained by progenitor cells with random fate. Dev Cell. 2010;18:317–323. doi: 10.1016/j.devcel.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 25.Lim X, et al. Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science. 2013;342:1226–1230. doi: 10.1126/science.1239730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mascre G, et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 2012;489:257–262. doi: 10.1038/nature11393. [DOI] [PubMed] [Google Scholar]
- 27.Gomez C, et al. The interfollicular epidermis of adult mouse tail comprises two distinct cell lineages that are differentially regulated by Wnt, Edaradd, and Lrig1. Stem cell reports. 2013;1:19–27. doi: 10.1016/j.stemcr.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braun KM, et al. Manipulation of stem cell proliferation and lineage commitment: visualisation of label-retaining cells in wholemounts of mouse epidermis. Development. 2003;130:5241–5255. doi: 10.1242/dev.00703. [DOI] [PubMed] [Google Scholar]
- 29.Chang H, Nathans J. Responses of hair follicle-associated structures to loss of planar cell polarity signaling. Proc Natl Acad Sci U S A. 2013;110:E908–917. doi: 10.1073/pnas.1301430110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang YV, White BS, Shalloway DI, Tumbar T. Stem cell dynamics in mouse hair follicles: a story from cell division counting and single cell lineage tracing. Cell Cycle. 2010;9:1504–1510. doi: 10.4161/cc.9.8.11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller-Rover S, et al. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117:3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- 32.Trempus CS, et al. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J Invest Dermatol. 2003;120:501–511. doi: 10.1046/j.1523-1747.2003.12088.x. [DOI] [PubMed] [Google Scholar]
- 33.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Lien WH, et al. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edmondson SR, et al. Insulin-like growth factor binding protein-3 (IGFBP-3) localizes to and modulates proliferative epidermal keratinocytes in vivo. The British journal of dermatology. 2005;152:225–230. doi: 10.1111/j.1365-2133.2004.06350.x. [DOI] [PubMed] [Google Scholar]
- 36.Weger N, Schlake T. Igf-I signalling controls the hair growth cycle and the differentiation of hair shafts. J Invest Dermatol. 2005;125:873–882. doi: 10.1111/j.0022-202X.2005.23946.x. [DOI] [PubMed] [Google Scholar]
- 37.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocrine reviews. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 38.Lee J, et al. Runx1 and p21 synergistically limit the extent of hair follicle stem cell quiescence in vivo. Proc Natl Acad Sci U S A. 2013;110:4634–4639. doi: 10.1073/pnas.1213015110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taniguchi H, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nathans J. Generation of an inducible Slc1a3-cre/ERT transgenic allele. MGI Direct Data Submission. 2010 MGI Ref ID J:157151. [Google Scholar]
- 41.Klein AM, Simons BD. Universal patterns of stem cell fate in cycling adult tissues. Development. 2011;138:3103–3111. doi: 10.1242/dev.060103. [DOI] [PubMed] [Google Scholar]
- 42.Ivanova A, et al. In vivo genetic ablation by Cre-mediated expression of diphtheria toxin fragment A. Genesis. 2005;43:129–135. doi: 10.1002/gene.20162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coulombe PA. Wound epithelialization: accelerating the pace of discovery. J Invest Dermatol. 2003;121:219–230. doi: 10.1046/j.1523-1747.2003.12387.x. [DOI] [PubMed] [Google Scholar]
- 44.Mackie EJ, Halfter W, Liverani D. Induction of tenascin in healing wounds. J Cell Biol. 1988;107:2757–2767. doi: 10.1083/jcb.107.6.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roshan A, et al. Human keratinocytes have two interconvertible modes of proliferation. Nat Cell Biol. 2016;18:145–156. doi: 10.1038/ncb3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paquet-Fifield S, et al. A role for pericytes as microenvironmental regulators of human skin tissue regeneration. The Journal of clinical investigation. 2009;119:2795–2806. doi: 10.1172/JCI38535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez-Rovira T, Silva-Vargas V, Watt FM. Different consequences of beta1 integrin deletion in neonatal and adult mouse epidermis reveal a context-dependent role of integrins in regulating proliferation, differentiation, and intercellular communication. J Invest Dermatol. 2005;125:1215–1227. doi: 10.1111/j.0022-202X.2005.23956.x. [DOI] [PubMed] [Google Scholar]
- 48.Ishitsuka Y, et al. Pituitary tumor-transforming gene 1 enhances proliferation and suppresses early differentiation of keratinocytes. J Invest Dermatol. 2012;132:1775–1784. doi: 10.1038/jid.2012.74. [DOI] [PubMed] [Google Scholar]
- 49.Sugiyama-Nakagiri Y, Ohuchi A, Hachiya A, Kitahara T. Involvement of IGF-1/IGFBP-3 signaling on the conspicuousness of facial pores. Arch Dermatol Res. 2010;302:661–667. doi: 10.1007/s00403-010-1062-3. [DOI] [PubMed] [Google Scholar]
- 50.Diamond I, Owolabi T, Marco M, Lam C, Glick A. Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J Invest Dermatol. 2000;115:788–794. doi: 10.1046/j.1523-1747.2000.00144.x. [DOI] [PubMed] [Google Scholar]
- 51.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci U S A. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE70523. All other data supporting the findings of this study are available from the corresponding author on reasonable request.