Abstract
We report the sequences of 1,244 human Y chromosomes randomly ascertained from 26 worldwide populations by the 1000 Genomes Project. We discovered more than 65,000 variants, including SNVs, MNVs, indels, STRs, and CNVs. Of these, CNVs contribute the greatest predicted functional impact. We constructed a calibrated phylogenetic tree based on binary SNVs and projected the more complex variants onto it, estimating the numbers of mutations for each class. Our phylogeny reveals bursts of extreme expansions in male numbers that have occurred independently among each of the five continental superpopulations examined, at times of known migrations and technological innovations.
Introduction
Due to its male-specific inheritance and the absence of crossover for most of its length, which together link it completely to male phenotype and behavior, the Y chromosome bears a unique record of human history1. Previous studies have demonstrated the value of full sequences for characterizing and calibrating the human Y-chromosome phylogeny2,3. This work has led to insights into male demography, but further work is needed: to more comprehensively describe the range of Y-chromosome variation, including non-SNV classes of variation; to investigate the mutational processes operating in the different classes; and to understand the relative roles of selection4 and demography5 in shaping Y-chromosome variation. The role of demography has risen to prominence with reports of male-specific bottlenecks in several geographical areas after 10 thousand years ago (kya)5–7, at times putatively associated with the spread of farming5 or Bronze Age culture6. With improved calibration of the Y-SNV mutation rate8–10 and, consequently, more secure dating of relevant features of the Y-chromosome phylogeny, it is now possible to hone such interpretations.
We have conducted a comprehensive analysis of Y-chromosome variation using the largest extant sequence-based survey of global genetic variation, phase 3 of the 1000 Genomes Project11. We have documented the extent of, and biological processes acting on, six types of genetic variation, and we have generated new insights into human male history.
Results
Dataset
Our dataset comprises 1,244 Y chromosomes sampled from 26 populations (Supplementary Table 1) and sequenced to a median haploid coverage of 4.3×. Reads were mapped to the GRCh37 human reference assembly used by phase 3 of the 1000 Genomes Project11 and to the GRCh38 reference for our analysis of short tandem repeats (STRs). We used multiple haploid-tailored methods to call variants and generate callsets containing more than 65,000 variants of six types, including single nucleotide variants (SNVs) (Supplementary Fig. 1 and Supplementary Tables 2 and 3), multiple nucleotide variants (MNVs), short insertions/deletions (indels), copy-number variants (CNVs) (Supplementary Figs. 2–12), and STRs (Supplementary Tables 4–6). We also identified karyotype variation that included one instance of 47,XXY and several mosaics of the karyotypes 46,XY and 45,X (Supplementary Table 7). We applied stringent quality control to meet the Project’s requirement of false discovery rate (FDR) < 5% for SNVs, indels and MNVs, and CNVs. In our validation analysis with independent datasets, genotype concordance was greater than 99% for SNVs and was 86%–97% for the more complex variants (Table 1).
Table 1.
Y-chromosome variants discovered in 1,244 males.
| Variant Type | Number | FDR (%) | Concordance (%) |
|---|---|---|---|
| SNVs | 60,555 | 3.9 | 99.6 |
| Indels & MNVs | 1,427 | 3.6 | 96.4 |
| CNVs | 110 | 2.7 | 86 |
| STRs | 3,253 | N/A | 89–97 |
FDR, false discovery rate; Concordance, with independent genotype calls. CNVs considered are those computationally inferred using Genome STRiP. N/A, not available.
To construct a set of putative SNVs, we generated six distinct callsets, which we input to a consensus genotype caller. In an iterative process, we leveraged the phylogeny to tune the final genotype calling strategy. We used similar methods for MNVs and indels, and we ran HipSTR to call STRs (Supplementary Note).
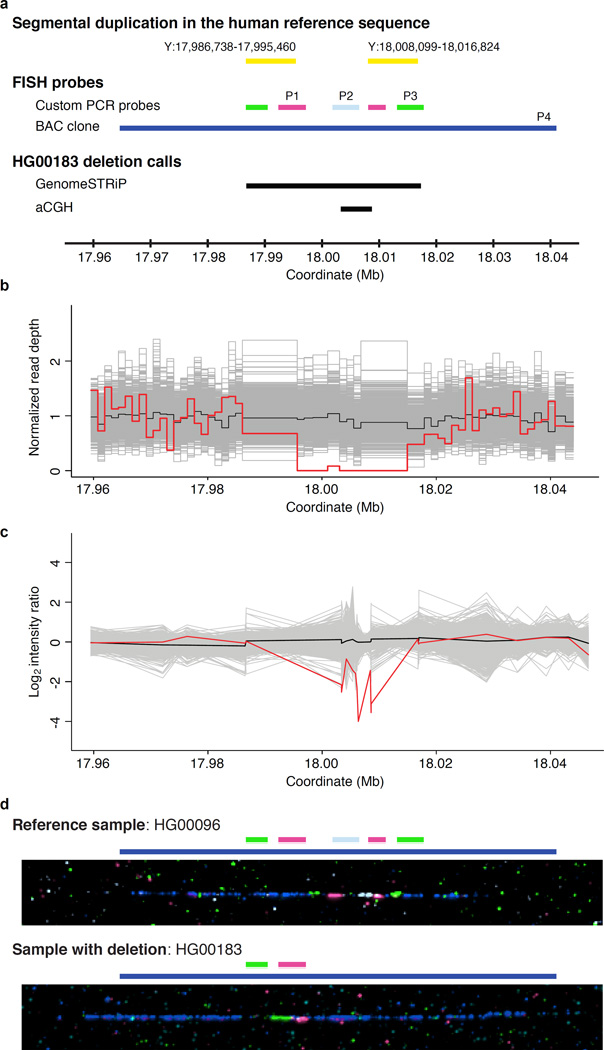
We discovered CNVs from the sequence data using two approaches, GenomeSTRiP12 and CnvHitSeq13 (Supplementary Note), and we validated calls using array comparative genomic hybridization (aCGH), supplemented by fluorescence in situ hybridization onto DNA fibres (fibre-FISH) in a few cases (Supplementary Figs. 8 and 9 and Supplementary Note). Figure 1 illustrates a representative large deletion we discovered in a single individual using GenomeSTRiP (Fig. 1b). We validated its presence by aCGH (Fig. 1c) and ascertained its structure with fibre-FISH (Fig. 1d). Notably, the event that gave rise to this variant was not a simple recombination between the segmental duplication elements it partially encompasses (Fig. 1a and Fig. 1d).
Figure 1.
Discovery and validation of a representative Y-chromosome CNV. (a) The GRCh37 reference sequence contains an inverted segmental duplication (yellow bars) within Y:17,986,738–18,016,824. We designed FISH probes to target the 3’ termini of the two segments (magenta and green bars labeled “P1” and “P3,” respectively) and the unique region between them (light blue, “P2”). A fourth probe used reference sequence BAC clone RP11-12J24 (dark blue, “P4”). Unlabeled green and magenta bars indicate expected cross-hybridization, and black bars indicate CNV events called by Genome STRiP and aCGH, respectively. GenomeSTRiP called a 30-kb deletion that includes the duplicated segments and the unique spacer region, whereas aCGH lacks probes in the duplicated regions. (b) Genome STRiP discovery plot. The red curve indicates the normalized read depth of HG00183, as compared to those for 1,232 other samples (grey) and the median (black). (c) Validation by aCGH. Intensity ratio for HG00183 (red), versus 1,233 other samples (grey) and the median (black). (d) Fibre-FISH validation using the probes illustrated in (a). The reference sample, HG00096, matches the human reference sequence, with green, magenta, light blue, magenta, and green hybridizations occurring in sequence. In contrast, we observed just one green and one magenta hybridization in HG00183, indicating the deletion of one copy of the segmental duplication and the central unique region. The coordinate scale that is consistent across (a–c) does not apply to (d), and BAC-clone hybridization lengths (dark blue) differ between the two samples solely due to the molecular combing process.
Phylogeny
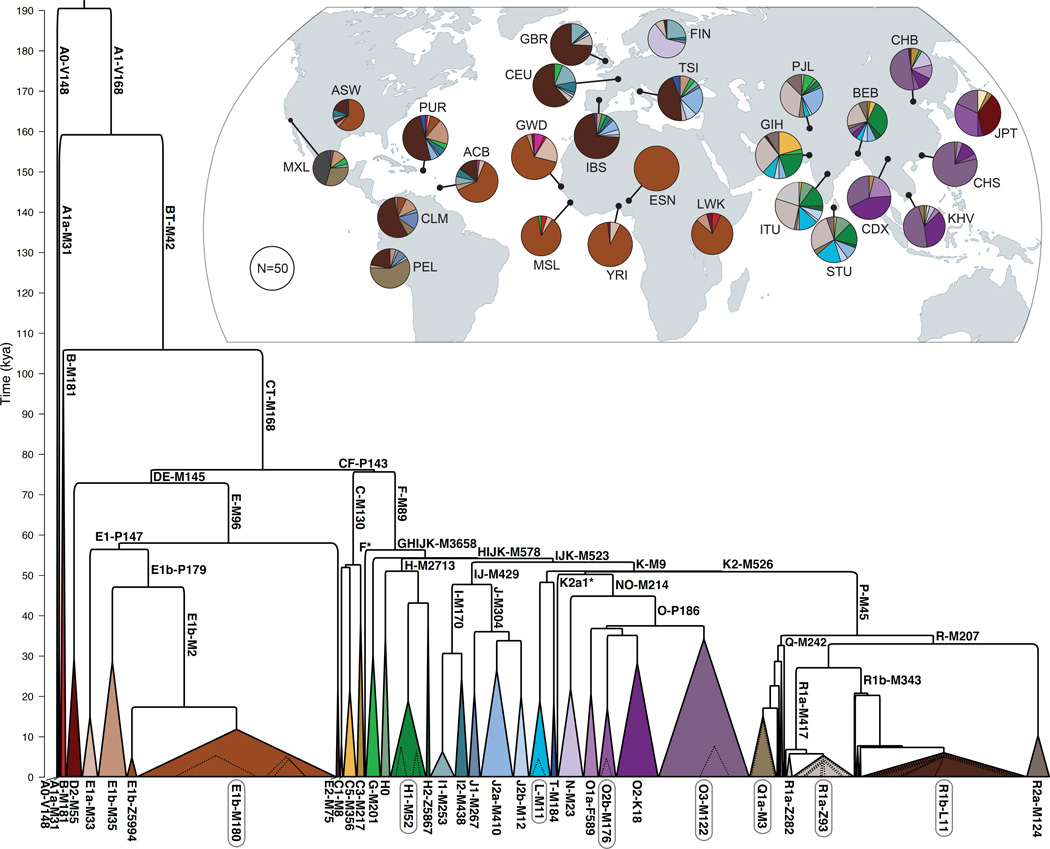
We identified each individual’s Y-chromosome haplogroup (Supplementary Tables 8 and 9 and Supplementary Data File) and constructed a maximum-likelihood phylogenetic tree using 60,555 biallelic SNVs derived from 10.3 megabases of accessible DNA (Fig. 2, Supplementary Figs. 13–17, Supplementary Note, and Supplementary Data File). Our tree recapitulates and refines the expected structure2,3,5, with all but two major haplogroups from A0 through T represented. The only haplogroups absent are M and S, both subgroups of K2b1 that are largely specific to New Guinea, which was not included in the 1000 Genomes Project. Notably, the branching patterns of several lineages suggest extreme expansions ~50–55 kya and also within the last few millennia. We investigated these later expansions in some detail and describe our findings in the “Haplogroup Expansions” section below.
Figure 2.
Y-chromosome phylogeny and haplogroup distribution. Branch lengths are drawn proportional to the estimated times between successive splits, with the most ancient division occurring ~190 kya. Colored triangles represent the major clades, and the width of each base is proportional to one less than the corresponding sample size. We modeled expansions within eight of the major haplogroups (circled) (Figure 4), and dotted triangles represent the ages and sample sizes of the expanding lineages. (Inset) World map indicating, for each of the 26 populations, the geographic source, sample size, and haplogroup distribution.
When calibrated with a mutation rate estimate of 0.76 × 10−9 mutations per base pair per year9, the time to the most recent common ancestor (TMRCA) of the tree is ~190 ky, but we consider the implications of alternative mutation rate estimates in the “Discussion” section. Of the clades resulting from the four deepest branching events, all but one are exclusive to Africa, and the TMRCA of all non-African lineages (i.e., the TMRCA of haplogroups DE and CF) is ~76 ky (Fig. 1, Supplementary Figs. 18 and 19, Supplementary Table 10, and Supplementary Note). We see a notable increase in the number of lineages outside Africa ~50–55 kya, perhaps reflecting the geographic expansion and differentiation of Eurasian populations as they settled the vast expanse of these continents. Consistent with previous proposals14, a parsimonious interpretation of the phylogeny is that the predominant African haplogroup, E, arose outside the continent. This model of geographic segregation within the CT clade requires just one continental haplogroup exchange (E to Africa), rather than three (D, C, and F out of Africa). Furthermore, the timing of this putative return to Africa—between the emergence of E and its differentiation within Africa by 58 kya—is consistent with proposals, based on non-Y data, of abundant gene flow between Africa and nearby regions of Asia 50–80 kya15.
Three novel features of the phylogeny underscore the importance of South and Southeast Asia as likely locations where lineages currently distributed throughout Eurasia first diversified (Supplementary Note). First, we observed in a Vietnamese individual a rare F lineage that is an outgroup for the rest of the megahaplogroup (Fig. 1 and Supplementary Fig. 14b). This sequence includes the derived allele for 147 SNVs shared by, and specific to, the 857 F chromosomes in our sample, but the lineage split off from rest of the group ~55 kya. This finding enabled us to define a new megagroup, GHIJK-M3658, whose subclades include the vast majority of the world’s non-African males1. Second, we identified in 12 South Asian individuals a new clade, here designated “H0,” that split with the rest of haplogroup H ~51 kya (Supplementary Fig. 14b). This new structure highlights the ancient diversity within the haplogroup and requires a more inclusive redefinition using, for example, the deeper SNV M2713, a G→A mutation at GRCh37 coordinate 6,855,809. Third, a lineage carried by a South Asian Telugu individual, HG03742, enabled us to refine early differentiation within the K2a clade ~50 kya (Fig. 1 and Supplementary Figs. 14d and 15). Using the high resolving power of the SNVs in our phylogeny, we determined that this lineage split off from the branch leading to haplogroups N and O (NO) not long after the ancestors of two individuals with well-known ancient DNA (aDNA) sequences did. Ust’-Ishim9 and Oase116 lived, respectively, in Western Siberia 43–47 kya and in Romania 37–42 kya. Their Y chromosomes join HG03742 in sharing with haplogroup NO the derived T allele at M2308 (GRCh37 Y:7,690,182), and the modern sample shares just four additional mutations with the NO clade.
Mutations
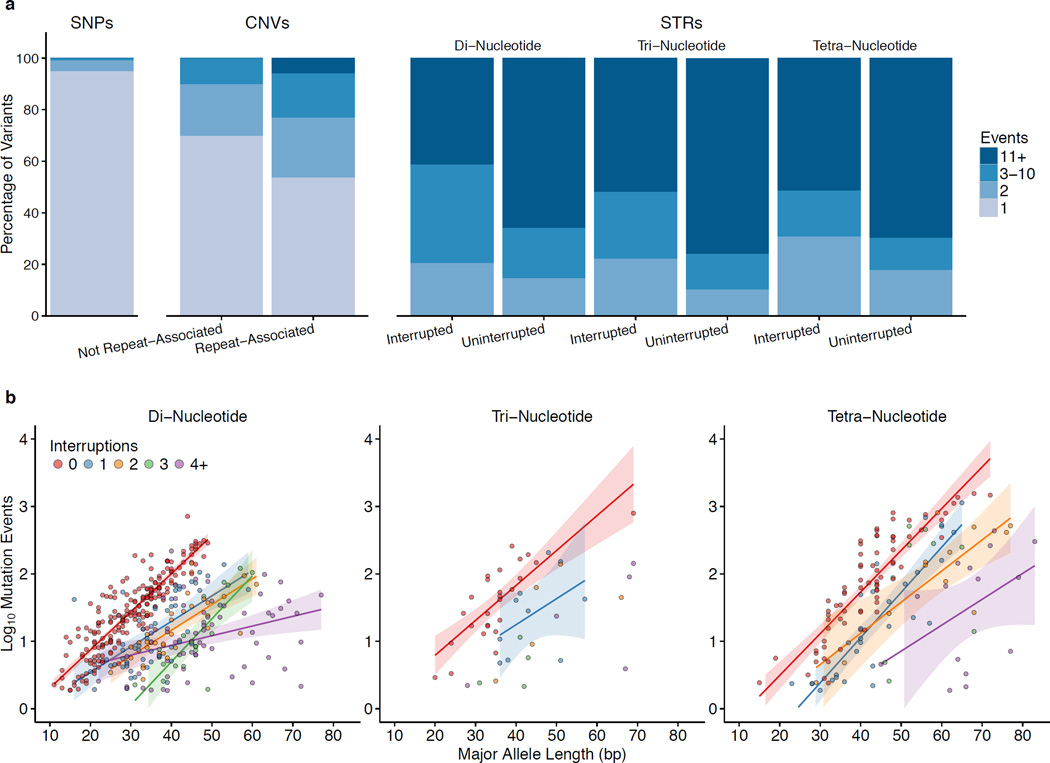
To map each SNV to a branch (or branches) of the phylogeny, we first partitioned the tree into eight overlapping subtrees (Supplementary Fig. 13). Within each subtree, we provisionally assigned each SNV to the internal branch constituting the minimum superset of carriers of one allele or the other, designating the derived state to the allele specific to this clade. When no member of the clade bore the ancestral allele, we deemed the site compatible with the subtree and assigned the SNV to the branch (Supplementary Note and Supplementary Data File). Most SNVs (94%) mapped to a single branch of the phylogeny, corresponding to a single mutation event during the Y-chromosome history captured by this tree. We projected the other variants onto the tree to infer the number of mutations associated with each (Fig. 3a).
Figure 3.
Mutation events. (a) Bar plots show the percentage of each variant type stratum associated with 1, 2, 3–10, or more mutations across the phylogeny. (b) For STRs, scatter plots show the logarithm of the number of mutation events versus major allele length, stratified by motif length and the number of interruptions to the repeat structure. We have plotted regression lines for categories with at least 10 data points, and we have omitted from the plots 44 STRs with motif lengths greater than four and 91 STRs whose mutation rate estimates were equal to the minimum threshold of 10−5 mutations per generation.
Supplementary Figure 10 summarizes our workflow to count the number of independent mutation events associated with each CNV (Supplementary Note). We found that 39% of CNVs have mutated multiple times, a much higher proportion than SNVs (Fig. 3a and Supplementary Data File). CNVs can arise by several different mutation mechanisms, one of which is homologous recombination between misaligned repeated sequences. This mechanism is particularly susceptible to recurrent mutations17 but, in comparing CNVs associated with repeated sequences to those that are not repeat-associated, we did not observe a significant difference in the proportion that have mutated multiple times (Mann-Whitney two-sided test). We did, however, observe that repeat-associated CNVs tend to be longer (p = 0.01).
We inferred more than six independent mutation events for each of three CNVs. One in particular stood out with 154 events. An apparent CNV hotspot spans a gene-free stretch of the chromosome’s long arm at GRCh37 Y:22,216,565–22,512,935. The region includes two arrays of long terminal repeat 12B (LTR12B) elements that together harbor 48 of the genome’s 211 copies (23%). In principle, our inference of numerous independent mutations could have been due to a “shadowing” effect from LTR12B elements elsewhere in the genome. That is, mismapping sequencing reads, and cross-hybridizing CGH probes, can lead to false inference of variation. But, in a phylogenetic analysis of all 211 LTR12B elements (Supplementary Figure 11), those within the putative CNV hotspot formed a pure monophyletic clade, demonstrating that the copy-number signal was genuine. The CNV has no predicted functional consequence.
Short tandem repeats (STRs) constituted the most mutable variant class, with a median of 16 mutations per locus and an average mutation rate of 3.9 × 10−4 mutations per generation. Assuming a generation time of 30 years, this equates to 1.3 × 10−5 mutations per year. Allele length explains more than half the variance of the log mutation rate for uninterrupted STRs. Longer STRs mutate more rapidly, and, conditional on allele length, mutability decreases when the repeat structure is interrupted, with a general trend toward slower mutations rates for STRs with more interruptions (Fig. 3b). Please see our Y-STR companion paper for more details18.
Functional Impact
A small proportion of SNVs have a predicted functional impact (Supplementary Figs. 20–23, Supplementary Tables 11–14, Supplementary Note, and Supplementary Data File). Among 60,555 SNVs, we observed two singleton premature stop-codons, one each in AMELY and USP9Y, and one splice-site SNV that affects all known transcripts of TBL1Y. Among 94 missense SNVs with SIFT19 scores, all 30 deleterious variants are singletons or doubletons, while 17/64 tolerated variants are present at higher frequency (p = 0.001), underscoring the impact of purifying selection on variation at protein-coding genes. No STRs overlapped protein-coding regions, but, in contrast to the SNVs, a high proportion of CNVs have a predicted functional impact.
Twenty of 100 CNVs in our final callset overlap with 27 protein-coding genes from 17 of the 33 Y-chromosome gene families. In our analysis of 1000 Genomes Project autosomal data, we observed that the ratio of the proportion of deletions overlapping protein-coding genes to the proportion of duplications overlapping protein-coding genes is 0.84. Whereas on the autosomes deletions are less likely to overlap protein-coding genes than duplications are, as others have also reported20, we found the reverse to be true for the Y chromosome. Despite its haploidy, we calculated its ratio of proportions to be 1.5, indicating a surprising increased tolerance of gene loss, as compared with the diploid genes on autosomes.
Diversity
Given observed diversity levels of the autosomes, the X chromosome, and the mitochondrial genome (mtDNA) (Supplementary Table 15, Supplementary Note, and Supplementary Data File), Y-chromosome diversity was reported to be lower than expected from simple population-genetic models that assume a Poisson-distributed number of offspring4, and the role of selection in this disparity is debated. We confirmed that Y-chromosome diversity in our sample is low (Supplementary Fig. 24) and found that positing extreme male-specific bottlenecks in the last few millennia can lead to a good fit between modeled and observed relative diversity levels of the autosomes, the X chromosome, the Y chromosome, and the mtDNA (Supplementary Figs. 25–28, Supplementary Table 16, and Supplementary Note). Therefore, we conclude that Y diversity may be shaped primarily by neutral demographic processes.
Haplogroup Expansions
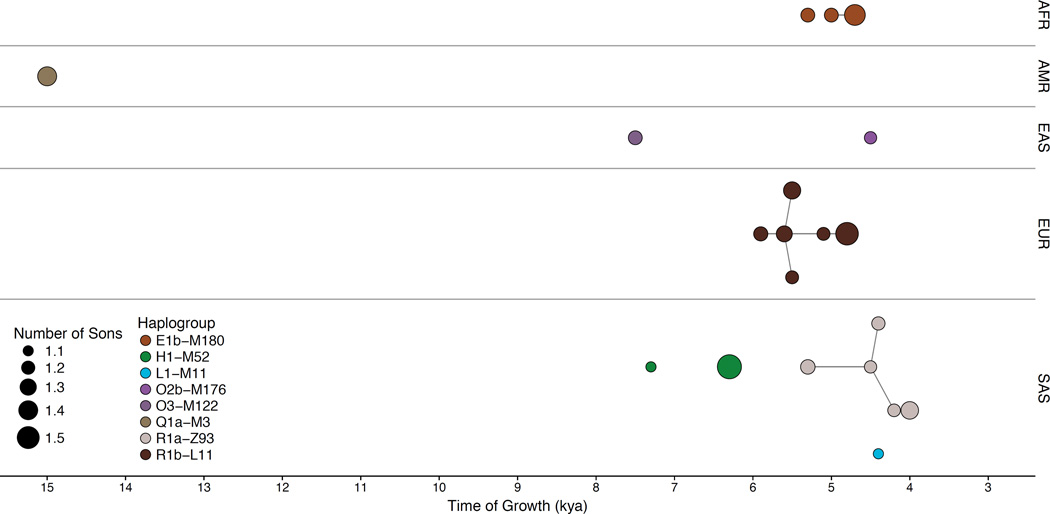
To investigate punctuated bursts within the phylogeny and estimate growth rates, we modeled haplogroup growth as a rapid phase followed by a moderate phase and applied this model to lineages showing rapid expansions (Supplementary Figs. 29–31, Supplementary Tables 17–19, Supplementary Note, and Supplementary Data File), noting that such extreme expansions are seldom seen in the mtDNA phylogeny here or in other studies5. We examined 20 nodes of the tree whose branching patterns were well-fit by this model. These nodes were drawn from eight haplogroups and included at least one lineage from each of the five continental regions surveyed (Fig. 4). As the haplogroup expansions we report are among the most extreme yet observed in humans, we think it more likely than not that such events correspond to historical processes that have also left archaeological footprints. Therefore, in what follows, we propose links between genetic and historical or archaeological data. We caution that, especially in light of as yet imperfect calibration, these connections remain unproven. But they are testable, for example using aDNA.
Figure 4.
Explosive male-lineage expansions of the last 15 thousand years. Each circle represents a phylogenetic node whose branching pattern suggests rapid expansion. The x-axis indicates the timings of the expansions, and circle radii reflect growth rates—the minimum number of sons per generation, as estimated by our two-phase growth model. Nodes are grouped by continental superpopulation (AFR, African; AMR, Admixed American; EAS, East Asian; EUR, European; SAS, South Asian) and colored by haplogroup. Line segments connect phylogenetically nested lineages.
First, in the Americas, we observed expansion of Q1a-M3 (Supplementary Figs. 14e and 17) at ~15 kya, the time of the initial colonization of the hemisphere21. This correspondence, based on one of the most thoroughly examined dates in human prehistory, attests to the suitability of the calibration we have chosen. Second, in sub-Saharan Africa, two independent E1b-M180 lineages expanded ~5 kya (Supplementary Figs. 14a), a period before the numerical and geographical expansions of Bantu speakers in whom E1b-M180 now predominates22. The presence of these lineages in non-Bantu speakers (e.g., Yoruba, Esan) indicates an expansion pre-dating the Bantu migrations, perhaps triggered by the development of ironworking23. Third, in Western Europe, related lineages within R1b-L11 expanded ~4.8–5.9 kya (Supplementary Figs. 14e), most markedly around 4.8 and 5.5 kya. The earlier of these times, 5.5 kya, is associated with the origin of the Bronze Age Yamnaya culture. The Yamnaya have been linked by aDNA evidence to a massive migration from the Steppe, which may have replaced much of the previous European population24,25, but the six Yamnaya with informative genotypes did not bear lineages descending from or ancestral to R1b-L11, so a Y-chromosome connection has not been established. The later time, 4.8 kya, coincides with the origins of the Corded Ware (Battle Axe) culture in Eastern Europe and the Bell-Beaker culture in Western Europe26.
Potential correspondences between genetics and archaeology in South and East Asia have received less investigation. In South Asia, we detect eight lineage expansions dating to ~4.0–7.3 kya and involving haplogroups H1-M52, L-M11, and R1a-Z93 (Supplementary Figs. 14b, 14d, and 14e). The most striking are expansions within R1a-Z93, ~4.0–4.5 kya. This time predates by a few centuries the collapse of the Indus Valley Civilization, associated by some with the historical migration of Indo-European speakers from the western steppes into the Indian sub-continent27. There is a notable parallel with events in Europe, and future aDNA evidence may prove to be as informative as it has been in Europe. Finally, East Asia stands out from the rest of the Old World for its paucity of sudden expansions, perhaps reflecting a larger starting population or the coexistence of multiple prehistoric cultures wherein one lineage could rarely dominate. We observed just one notable expansion within each of the O2b-M176 and O3-M122 clades (Supplementary Figs. 14d).
Discussion
The 1000 Genomes Project dataset provides a rich and unparalleled resource of Y-chromosome variation coupled with open access to DNA and cell lines that will facilitate diverse further investigations. By cataloging the phylogenetic position of ~60,000 SNVs, we have constructed a database of diagnostic variants with which one can assign Y-chromosome haplogroups to DNA samples (Supplementary Data File). This resource is particularly valuable for SNP-chip design and for aDNA studies, in which sequencing coverage is often quite low, as exemplified by our reanalysis of the Ust’-Ishim and Oase1 Y chromosomes.
The variants we report have well-calibrated FDRs. Nevertheless, due to the modest sequencing coverage, data missingness was a principal concern. Small CNVs and long STRs are largely undetected, and low frequency variants in general, including SNVs, are under-represented. We therefore took great care to minimize the impact of missing variants. In particular, we designed the relevant downstream analyses to only use information from higher frequency, shared, variation, corresponding to mutations on internal branches of the tree.
Since many DNA samples were extracted from lymphoblastoid cells, another potential concern was variation that has arisen during cell culture28. However, these false discoveries are inherently not shared. Therefore, the precautions we took to minimize the impact of missingness also precluded in vitro mutations from influencing our findings. We discuss additional caveats on the mapping of SNVs to branches in the Supplementary Note.
Our findings illustrate unique properties of the Y chromosome. Foremost, the abundance of extreme male-lineage expansions underscores differences between male and female demographic histories. A caveat to our expansion analysis is that our inference method assumes that population structure did not affect the branching patterns immediately downstream of the particular phylogenetic node under investigation. This is reasonable, because population structure is unlikely when a very rapid expansion is in progress, but to accommodate this strong assumption, we limited all analyses to pruned internal subtrees short enough for it to hold. A second caveat regards the choice of calibration metric, which is relevant to the links we have suggested between expansions and historical or archaeological events. Present-day geographical distributions provide strong support for the correspondences we proposed for the initial peopling of most of Eurasia by fully modern humans ~50–55 kya and for the first colonization of the Americas ~15 kya. For later male-specific expansions, we should consider the consequences of alternative mutation rate estimates, as pedigree-based methods relying on variation from the most recent several centuries8,10,28 may be more relevant. The pedigree-based estimate from the largest set of mutations8 would lead to a decrease in expansion times by ~15%, increasing the precision of the correspondences proposed for E1b and R1a. For R1b, a 15% decrease would suggest an expansion postdating the Yamnaya migration, perhaps better explaining the distinction between the Yamnaya R1b chromosomes and the expanding R1b-L11 lineage. Either way, the lineage expansions seem to have followed innovations that may have elicited increased variance in male reproductive success29, innovations such as metallurgy, wheeled transport, or social stratification and organized warfare. In each case, privileged male lineages could undergo preferential amplification for generations. We find that rapid expansions are not confined to unusual circumstances30,31. Rather, they can dominate on a continental scale and do so in some of the populations most studied by medical geneticists. Inferences incorporating demography may benefit from taking these male-female differences into account.
Online Methods
Study samples
The 1000 Genomes Project Consortium sequenced the genomes of 2,535 individuals from 26 populations representing five global super-populations (Supplementary Table 1). The Project’s phase 3 analysis included 2,504 of these11, and we used the Y-chromosome reads from the 1,244 males for this study.
SNVs, MNVs, and indels
To identify putative SNVs within the 10.3 Megabases of the Y chromosome that are amenable to short-read sequencing3, we generated six callsets using SAMtools33, FreeBayes34, Platypus35, Cortex_var36, and GATK Unified Genotyper37,38 in both haploid and diploid modes. We used FreeBayes to construct a preliminary consensus callset, imposed filters for the number of alleles, genotype quality, read depth, mapping quality, missingness, and called heterozygosity. Finally, we called each genotype as the maximum-likelihood allele whenever a two-log-unit difference in likelihoods existed between the two possible states. For MNVs and indels, we imposed additional filters to exclude repetitive regions of the genome.
We used 11 high-coverage PCR-free genome sequences to estimate the false discovery rate (FDR) and 143 high-coverage Complete Genomics (CG) sequences to estimate the false negative rate and genotype concordance. We also estimated the singleton false-positive rate by comparing the transition-transversion ratio among singletons to the corresponding ratio among shared SNVs.
CNVs
We discovered and genotyped CNVs using aCGH and two computational methods, Genome STRiP12 and cnvHitSeq13, across the entire euchromatic region. We ran Genome STRiP separately for uniquely alignable sequences and segmental duplications, using 5-kb and 10-kb windows and filtering calls based on call rate, density of alignable positions, cluster separation, and manual review to assess duplication of findings and strength of evidence. We excluded 10 samples with evidence for cell-line-specific clonal aneuploidy. To estimate FDR, we used the intensity rank-sum method12 and probe intensity data from Affymetrix 6.0 SNP arrays.
We generated a second callset using the cnvHitSeq algorithm, which we modified to model read-depth variation in a manner robust to the presence of repetitive regions and to estimate mosaicism. For the third callset, we used intensity ratios of 2,714 aCGH probes, with sample NA10851 as the reference. We segmented with the GADA algorithm39,40, called genotypes based on the distribution of mean log2 intensity ratios using the additive background model of Conrad et al.41, and imposed stringent criteria to minimise the FDR.
To validate the computational callsets, we used: aCGH; alkaline lysis fibre-FISH, following the protocol of Perry, et al.42; and molecular combing fibre-FISH, following Polley et al.43, Carpenter et al.44, and instructions from the manufacturer, Genomic Vision.
Karyotyping for sex-chromosome aneuploidies
Metaphase chromosome spreads were prepared from lymphoblastoid cell lines (Coriell Biorepository) according to a standard protocol45. Chromosome-specific paint probes for the human X and Y chromosomes were generated from 5,000 copies of flow-sorted chromosomes, using the GenomePlex Whole Genome Amplification kit (Sigma-Aldrich). Probes were labeled and FISH was performed following the strategy described in Gribble et al.46.
STRs
We called genotypes using HipSTR and assessed call quality by comparing genotypes across 3 father-son pairs and by measuring concordance with capillary electrophoresis for 15 loci in the PowerPlex Y23 panel. To estimate Y-STR mutation rates, we used an approach we have fully described in a companion manuscript18. We modeled mutations with a geometric step size distribution and a spring-like length constraint, and, to account for PCR stutter artifacts and alignment errors, we learned an error model for each locus. We then leveraged the Y-SNP phylogeny to compute each sample’s genotype posteriors, used a variant of Felsenstein’s tree-pruning algorithm47 to evaluate the likelihood of a given mutation model, and optimized the model until convergence. We validated our estimates with simulations and compared them to published estimates when available.
Phylogeny
We assigned haplogroups using the January 18, 2014 version of the SNP Compendium maintained by the International Society of Genetic Genealogy (ISOGG). To construct a total-evidence maximum-likelihood (ML) tree, we converted genotype calls for the 60,555 biallelic SNVs to nexus format and ran RAxML848 using the ASC_GTRGAMMA model. We then conducted 100 ML bootstraps and mapped these to the total-evidence tree. We partitioned the ML tree into eight overlapping subtrees, and for each subtree, we defined a set of SNVs that were variable within it and assigned each site to the internal branch constituting the minimum superset of carriers of one allele or the other. To estimate split times, we used two approaches to account for the modest coverage of our sequences. In the first, we pruned the sample to those sequences with 5× or greater coverage, and in the second, we traversed exclusively internal branches of tree, as internal branches have high effective sequencing coverage due to the superposition of descending lineages. We calibrated using two mutation rate estimates from the literature8,9.
Functional annotation
We used Ensembl’s Variant Effect Predictor49 to functionally annotate SNVs. To evaluate deleteriousness, we used Combined Annotation-Dependent Depletion scores50, SIFT19, and PolyPhen51.
MtDNA
We excluded deletions and those mutations proscribed by PhyloTree v.1652, generated a FASTA file using VCFtools53, and aligned mtDNA sequences to the revised Cambridge Reference Sequence (rCRS) using MEGA654. We assigned haplogroups to each sample using HaploGrep55, manually checked all variant calls, inferred the mtDNA phylogeny using RAxML48, and plotted the tree using FigTree.
Diversity
We used 141 high-coverage CG sequences to compare mtDNA diversity to that of the Y chromosome. Seeking to recapitulate this observed relative diversity, as well the observed diversity of the X chromosome and the autosomes, we used standard neutral coalescent simulations implemented in the program ms56 to simulate data for the four chromosome types under a series of demographic models. In all models, we held the autosomal effective population size fixed to values previously described for African and European demographic histories57,58, but we varied the ratio of male-to-female effective population sizes.
Haplogroup expansions
To estimate male-lineage growth rates, we developed a two-phase exponential growth model wherein the first phase coincides with an apparent rapid haplogroup expansion and the second phase links the first phase to the earliest time for which reasonable estimates exist for the size of the relevant population. Our primary objective was to estimate the duration of the first phase, T1, and the effective number of carriers of a haplogroup at its conclusion, N1, in order to estimate the growth rate during this period—the mean number of sons per man per generation. To do so, we conducted maximum-likelihood inference over a grid of (T1, N1) points for each of a sequence of “sampling” times, Ts, defined by pruning the subtree of a phylogenetic node of interest to a fixed root-to-tip height (number of SNPs) (Supplementary Fig. 29).
With N2 fixed, we needed one additional parameter, T2, to specify the full demographic model corresponding to each (T1, N1) in order to simulate two-phase growth. We estimated T2 using 10,000 ms coalescent simulations56 constrained by the TMRCA of the node of interest. With T2 and N2 in hand, we simulated two-phase growth to assemble a reference distribution of site frequency spectra (SFS) against which to compare the observed data. We did so for each point of a three-dimensional lattice of (T1, N1, Ts) values, allowing T1 to range from 1 to 48 generations and distributing 32 N1 values in a geometric progression between 13.6 and 200,000 individuals. With up to ten possible Ts values, the lattice contained up to 15,360 points, and for each, we conducted 16,384 ms simulations of two-phase growth, fixing the number of lineages equal to that of the pruned observed tree. For each Ts, we approximated the likelihood of a particular (T1, N1) point by comparing the SFS of the observed tree to those of the corresponding reference distribution, using an SFS distance measure we defined. Finally, we used the resulting likelihood contours to infer the magnitude of phase-1 growth.
Supplementary Material
Acknowledgments
We thank the 1000 Genomes Project sample donors for making this work possible and all Project members for their contributions. Figures were generated with FigTree and ggplot232. Thanks to A. Martin for ADMIXTURE results. G.D.P. was supported by the National Science Foundation (NSF) Graduate Research Fellowship under grant number DGE-1147470 and by the National Library of Medicine training grant LM-007033. Work at The Wellcome Trust Sanger Institute (Q.A., R.B., M.C., Y.C., S.L., A.M., S.A.M., C.T.-S., Y.X., and F.Y.) was supported by Wellcome Trust grant number 098051. F.L.M. was supported by the National Institutes of Health (NIH) grant number 1R01GM090087, by NSF grant number DMS-1201234, and by a postdoctoral fellowship from the Stanford Center for Computational, Evolutionary and Human Genomics (CEHG). T.W. was supported by an AWS Education Grant, and the work of T.W., M.G., and Y.E was supported in part by an NIJ Award 2014-DN-BX-K089. M.C. is supported by a Fundacion Barrie Fellowship. H.S. and L.Coin are supported by Australian Research Council Grants DP140103164 and FT110100972, respectively. M.G. was supported by a National Defense Science & Engineering Graduate Fellowship. G.R.S.R. was supported by the European Molecular Biology Laboratory and the Sanger Institute through an EBI-Sanger Postdoctoral Fellowship. X.Z.-B., P.F., D.R.Z., and L.Clarke were supported by Wellcome Trust grant numbers 085532, 095908, 104947 and by the European Molecular Biology Laboratory. P.A.U. was supported by SAP grant SP0#115016. C.L. was supported in part by NIH grant U41HG007497. Y.E. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. C.D.B. was supported by NIH grant number 5R01HG003229-09.
Footnotes
URLs
1000 Genomes Project, http://www.1000genomes.org/using-1000-genomes-data
ISOGG, http://www.isogg.org/
Author Contributions
G.D.P., Y.X., C.D.B, and C.T.-S. conceived and designed the project. R.B., S.L., and F.Y. generated FISH data. A.Malhorta, M.R., E.C., C.Z., and C.L. generated array-CGH data. G.D.P., Y.X., F.L.M., T.F.W., A.Massaia, M.A.W.S., Q.A., S.A.McC., A.N., S.K., Y.C., J.L.R.-F., M.C., H.S., M.G., R.D., G.R.S.R., T.W.F., E.G., A.Marcketta, D.M., X.Z.-B., G.R.S., S.A.McC., P.F., P.A.U., L.Coin, D.R.Z., L.Clarke, A.A., Y.E., R.E.H., C.D.B., and C.T.-S. analyzed the data. G.D.P., Y.X., F.L.M., T.F.W., A.Massaia, M.A.W.S., Q.A., and C.T.-S. wrote the manuscript. All authors reviewed, revised and provided feedback on the manuscript.
Competing Financial Interests
G.D.P. and A.A. are employees of 23andMe. P.F. is a member of the Scientific Advisory Board (SAB) for Omicia, Inc. P.A.U. has consulted for and owns stock options of 23andMe. Y.E. is an SAB member of Identify Genomics, BigDataBio, and Solve Inc. C.D.B. is on the SABs of AncestryDNA, BigDataBio, Etalon DX, Liberty Biosecurity, and Personalis. He is also a founder and SAB chair of IdentifyGenomics. None of these entities played a role in the design, execution, interpretation, or presentation of this study.
References
- 1.Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat. Rev. Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- 2.Wei W, et al. A calibrated human Y-chromosomal phylogeny based on resequencing. Genome Res. 2013;23:388–395. doi: 10.1101/gr.143198.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poznik GD, et al. Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females. Science. 2013;341:562–565. doi: 10.1126/science.1237619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson Sayres MA, Lohmueller KE, Nielsen R. Natural selection reduced diversity on human Y chromosomes. PLoS Genet. 2014;10:e1004064. doi: 10.1371/journal.pgen.1004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karmin M, et al. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 2015;25:459–466. doi: 10.1101/gr.186684.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batini C, et al. Large-scale recent expansion of European patrilineages shown by population resequencing. Nat. Commun. 2015;6:7152. doi: 10.1038/ncomms8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sikora MJ, Colonna V, Xue Y, Tyler-Smith C. Modeling the contrasting Neolithic male lineage expansions in Europe and Africa. Investig. Genet. 2013;4:25. doi: 10.1186/2041-2223-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helgason A, et al. The Y-chromosome point mutation rate in humans. Nat. Genet. 2015;47:453–457. doi: 10.1038/ng.3171. [DOI] [PubMed] [Google Scholar]
- 9.Fu Q, et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514:445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balanovsky O, et al. Deep phylogenetic analysis of haplogroup G1 provides estimates of SNP and STR mutation rates on the human Y-chromosome and reveals migrations of Iranic speakers. PLoS One. 2015;10:e0122968. doi: 10.1371/journal.pone.0122968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handsaker RE, et al. Large multiallelic copy number variations in humans. Nat. Genet. 2015;47:296–303. doi: 10.1038/ng.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellos E, Johnson MR, Coin LJM. cnvHiTSeq: integrative models for highresolution copy number variation detection and genotyping using population sequencing data. Genome Biol. 2012;13:R120. doi: 10.1186/gb-2012-13-12-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammer MF, et al. Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol. Biol. Evol. 1998;15:427–441. doi: 10.1093/oxfordjournals.molbev.a025939. [DOI] [PubMed] [Google Scholar]
- 15.Groucutt HS, et al. Rethinking the dispersal of Homo sapiens out of Africa. Evol. Anthropol. 2015;24:149–164. doi: 10.1002/evan.21455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Q, et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature. 2015;524:216–219. doi: 10.1038/nature14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willems T, et al. Population-Scale Sequencing Data Enables Precise Estimates of YSTR Mutation Rates. Am. J. Hum. Genet. 2016 doi: 10.1016/j.ajhg.2016.04.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 20.Sudmant PH, et al. Global diversity, population stratification, and selection of human copy-number variation. Science. 2015;349:aab3761. doi: 10.1126/science.aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raghavan M, et al. Genomic evidence for the Pleistocene and recent population history of Native Americans. Science. 2015;349:aab3884. doi: 10.1126/science.aab3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Filippo C, Bostoen K, Stoneking M, Pakendorf B. Bringing together linguistic and genetic evidence to test the Bantu expansion. Proc. R. Soc. B Biol. Sci. 2012;279:3256–3263. doi: 10.1098/rspb.2012.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jobling MA, Hollox E, Hurles M, Kivisild T, Tyler-Smith C. Human Evolutionary Genetics. 2nd. Garland Science; 2014. [Google Scholar]
- 24.Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 25.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harding AF. European Societies in the Bronze Age. Cambridge University Press; 2000. [Google Scholar]
- 27.Bryant EF, Patton LL. The Indo-Aryan Controversy: Evidence and Inference in Indian History. Routledge; 2005. [Google Scholar]
- 28.Xue Y, et al. Human Y chromosome base-substitution mutation rate measured by direct sequencing in a deep-rooting pedigree. Curr. Biol. 2009;19:1453–1457. doi: 10.1016/j.cub.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betzig L. Means, variances, and ranges in reproductive success: comparative evidence. Evol. Hum. Behav. 2012;33:309–317. [Google Scholar]
- 30.Zerjal T, et al. The genetic legacy of the Mongols. Am. J. Hum. Genet. 2003;72:717–721. doi: 10.1086/367774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balaresque P, et al. Y-chromosome descent clusters and male differential reproductive success: young lineage expansions dominate Asian pastoral nomadic populations. Eur. J. Hum. Genet. 2015;23:1413–1422. doi: 10.1038/ejhg.2014.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; 2009. [Google Scholar]
References for Online Methods
- 33.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv Prepr. 2012:1–9. [Google Scholar]
- 35.Rimmer A, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 2014;46:912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat. Genet. 2012;44:226–232. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DePristo MA, et al. A framework for variation discovery and genotyping using nextgeneration DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pique-Regi R, et al. Sparse representation and Bayesian detection of genome copy number alterations from microarray data. Bioinformatics. 2008;24:309–318. doi: 10.1093/bioinformatics/btm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pique-Regi R, Cáceres A, González JR. R-Gada: a fast and flexible pipeline for copy number analysis in association studies. BMC Bioinformatics. 2010;11:380. doi: 10.1186/1471-2105-11-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conrad DF et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perry GH, et al. Copy number variation and evolution in humans and chimpanzees. Genome Res. 2008;18:1698–1710. doi: 10.1101/gr.082016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polley S, et al. Evolution of the rapidly mutating human salivary agglutinin gene (DMBT1) and population subsistence strategy. Proc. Natl. Acad. Sci. 2015;112:5105–5110. doi: 10.1073/pnas.1416531112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carpenter D, et al. Obesity, starch digestion and amylase: association between copy number variants at human salivary (AMY1) and pancreatic (AMY2) amylase genes. Hum. Mol. Genet. 2015;24:3472–3480. doi: 10.1093/hmg/ddv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verma RS, Babu A. Human Chromosomes: Principles & Techniques. 2nd. McGraw-Hill, Inc.; 1995. [Google Scholar]
- 46.Gribble SM, et al. Massively Parallel Sequencing Reveals the Complex Structure of an Irradiated Human Chromosome on a Mouse Background in the Tc1 Model of Down Syndrome. PLoS One. 2013;8:e60482. doi: 10.1371/journal.pone.0060482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 48.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 53.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kloss-Brandstätter A, et al. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum. Mutat. 2011;32:25–32. doi: 10.1002/humu.21382. [DOI] [PubMed] [Google Scholar]
- 56.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 57.Lohmueller KE, Bustamante CD, Clark AG. Methods for human demographic inference using haplotype patterns from genomewide single-nucleotide polymorphism data. Genetics. 2009;182:217–231. doi: 10.1534/genetics.108.099275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lohmueller KE, Bustamante CD, Clark AG. The effect of recent admixture on inference of ancient human population history. Genetics. 2010;185:611–622. doi: 10.1534/genetics.109.113761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.